- 1Natural Resources Institute Finland (Luke), Production Systems, Jokioinen, Finland
- 2Natural Resources Institute Finland (Luke), Production Systems, Maaninka, Finland
Identification of bacterial communities in both the raw material and the subsequent silages provides new insights into understanding the silage fermentation process. The objective was to evaluate how different silage management factors affect silage preservation characteristics, the microbiome, and their correlations. A red clover dominated sward was used as the raw material and ensiled in pilot scale using a 3 × 4 design, with three management conditions including the level of compaction (loose and tight), and further, for the tightly compacted silages contamination with soil and faeces was conducted; and four different additive treatments with different modes of action: Control without additive (CONT), formic and propionic acid-based additive (FPA), homofermentative lactic acid bacteria inoculant (LAB) and salt-based additive (SALT). Samples of the raw material and subsequent silages were taken and routinely analysed, including DNA extraction and PCR amplification using universal primers. Tight compaction reduced slightly the extent of silage fermentation, but contamination with soil and faeces stimulated a non-desired type of fermentation with higher concentrations of ethanol, acetic acid and propionic acid and a higher pH. Use of LAB and SALT had only minimal effects on silage fermentation, but FPA clearly restricted fermentation and resulted in a better fermentation quality (lower pH, ammonia N and acetic acid concentration) of the silages compared to CONT. The FPA silages presented greater diversity of bacterial communities compared to the other silages. Proteobacteria were the most abundant in raw material, followed by Firmicutes, and major shifts happened in these communities during the silage fermentation process. Weissella was found in small amounts in the raw material but it dominated in the silages. The most abundant communities in the silages, such as Weissella, Lactobacillus and Pseudomonas, were correlated with several silage fermentation characteristics. Use of FPA improved fermentation quality of silages, but SALT and LAB differed from CONT to a smaller extent. All additives modified the bacterial profiles of grasses ensiled under different management conditions. The combination of parameters related to silage quality and bacterial communities provided a deeper understanding of the silage fermentation process and how they can be manipulated to obtain better feed quality.
1. Introduction
Under Northern European conditions, red clover (Trifolium pratense) is commonly used as a silage crop, mainly because of its ability to withstand winter and good nutritional characteristics (Järvenranta et al., 2016), such as high intake potential (Huhtanen et al., 2007). Additionally, red clover, being a leguminous plant, increases agroecological resilience and productivity via nitrogen fixation through symbiotic interactions between rhizobia bacteria and subterranean nodules (Wyngaarden et al., 2015). The benefits in reduced need of N fertilization and increased biodiversity can be widely exploited in the livestock sector. Furthermore, forage legumes such as red clover are an essential part of low-input and organic milk and beef production chains in Northern Europe.
Ensiling is the mainstream technology to preserve forage for long periods of time to feed ruminants (Wilkinson and Rinne, 2018). Silage making helps to reduce seasonal imbalance between feed demand for livestock and the high-quality forage available, extending forage storage life (Wright et al., 2000). The ensiling process is a method of rapid anaerobic fermentation of fresh plant material, for example red clover, by epiphytic lactic acid bacteria (LAB), which are naturally found on the plant surface (Ren et al., 2020). Exogenously applied selected strains of LAB are also commercially available to be used as silage additives (Muck et al., 2018). The presence and proliferation of LAB produce organic acids under an environment absent of free oxygen, which lower the pH of the green biomass, suppressing the activities of potentially harmful bacteria (McDonald et al., 1991). The fermentation process involves a succession of bacterial communities in which various kinds of fermenting microorganisms produce a variety of metabolites that affect forage storage, utilization, and animal production (Kung et al., 2018; Wang et al., 2022). Various investigations have used integrated 16S rRNA sequencing to analyse the microbiome of fermented feeds to better comprehend biological mechanisms that underpin silage production systems (Guo et al., 2018; Xu et al., 2019). The identification of bacterial communities in the fresh biomass used as the raw material and subsequent silages provides new insights into the understanding of the silage fermentation process (McAllister et al., 2018; Bai et al., 2020; Li et al., 2021; Wang et al., 2021), as each and every particular bacterial community can modulate nutritional composition, hygienic quality and overall ensiling process of the feed and influence ensiling losses (Franco et al., 2022).
Contamination with soil and poor compaction of silages are the main reasons that cause spoilage and aerobic deterioration of silage before feeding (Holmes and Bolsen, 2009). Among good silage production practices, some of the most important factors include avoiding soil contamination to prevent inoculation with spoilage microorganisms. In addition, tight compaction of the green biomass is necessary to guarantee conditions of absence of free oxygen and accumulation of lactic acid which consequently causes a reduction in pH, thus inhibiting microbial metabolism and preserving the nutrients of the fresh biomass. In addition to silo management, use of different kinds of silage additives can be used to ameliorate fermentation quality of silages (Muck et al., 2018).
To the best of our knowledge, the effect of different ensiling management factors on bacterial ecology of red clover silage have not yet been investigated. The objective of this experiment was to assess the effect of different kinds of silage additives on the ensiling process and bacterial community composition of a red clover dominated silage under different management conditions, such as two levels of compaction and soil contamination. Moreover, we hypothesized that the different silage additive, compaction and contamination treatments shift the silage fermentation and the bacterial community towards different directions.
2. Materials and methods
2.1. Raw material for silage making
Organically grown mixed red clover and timothy (Phleum pratense) grass was mown from a first regrowth of the sward on August 1st 2018 at Häme University of Applied Sciences in Mustiala, Finland (60°83’N, 23°77’E). The botanical composition of the raw material was analysed from a representative sample of 1 kg fresh matter, which was manually divided into red clover and timothy. After a 24-hour wilting period, the green biomass was cut using a farm scale precision chopper (JF FCT 1350, JF-Fabriken-J Freudendahl A/S, Sonderborg, Denmark) without additive application. Green biomass samples were immediately collected for evaluation of chemical composition, microbial counts and bacterial communities before ensiling.
2.2. Experimental treatments and procedures
The experiment was conducted according to a 3 × 4 design, with three management conditions and four additive treatments. One of the management factors to manipulate the silage quality was the different levels of compaction. The two compactions were obtained by dropping (approximately 80 cm height) an 8-kg lead plummet ten or twice into a handful of raw material in a cylindrical silo for the tight and loose compactions, respectively. In order to challenge the potential effect of additives on both fermentation quality and bacterial ecology of a low-hygienic quality green biomass, contamination with soil from slurry-treated area and fresh cow faeces was performed for the tightly compacted silos. Soil, faeces and tap water were mixed 24 hours before silage preparation in a proportion of 1:1:7, vigorously shaken and kept for sedimentation in room temperature. The supernatant was taken and used to prepare the additive solutions. This schematic design resulted in three different types of ensiling management: loose compaction, tight compaction, and tight compaction with soil + faeces contamination.
For each of the management conditions, four additive treatments were used according to commercial instructions:
1. Control (CONT), as a negative treatment without additive
2. Formic and propionic acid-based additive (FPA) at 5 l/t of fresh matter (formic acid, propionic acid, sodium formate, and potassium sorbate; AIV Ässä Na, Eastman, Oulu, Finland)
3. Homofermentative strains of lactic acid bacteria (LAB) at 1.0 × 105 cfu/g of fresh matter (Lactobacillus plantarum; Kofasil® Lac, Addcon, Bitterfeld-Wolfen, Germany)
4. Salt-based additive (SALT) at 2 l/t of fresh matter (sodium nitrite, sodium benzoate and potassium sorbate; Safesil Challenge, Salinity AB, Göteborg, Sweden)
After careful homogenization of the raw material, it was divided into 36 batches of 15 kg each in which the treatments were applied. For the silage additives to be uniformly applied, they were all previously diluted with tap water so that the final amount of liquid applied was 17 L per ton for all treatments including CONT. In the case of silages contaminated with soil + faeces, the additive solution was prepared with the supernatant of the contaminating solution previously described. After applying the treatments, the raw material was ensiled in laboratory-scale cylindrical silos with 12-L capacity. Silages were produced with three replications per treatment. After preparing the silos, they were covered with a plastic cover, plastic lid, an 8-kg lead plummet, and a water lock. Silos were then stored at room temperature, protected from the incidence of light and opened after an ensiling period of 105 days. The deteriorated parts on the surface of the silos were removed and discarded. Silage was then carefully mixed, and representative samples collected and analysed for chemical composition, fermentation quality, aerobic stability, microbial counts and bacterial community diversity.
Height and weight of silage were measured before opening the silos in order to calculate silage density. Silos were weighed shortly after filling and immediately before opening to calculate ensiling losses according to Knicky and Spörndly (2015), assuming that the weight loss was CO2 leaving the silo during fermentation.
2.3. Laboratory analyses
The standard analytical methods used for dry matter (DM), ash, crude protein, water soluble carbohydrates (WSC), neutral detergent fibre, buffering capacity, organic matter digestibility, pH, ammonia-N, lactic acid, volatile fatty acids (VFA), formic acid, ethanol and microbial counts as well as the conduction of the aerobic stability test based on heat accumulation are described in detail by Franco et al. (2022).
Samples for DNA extraction and sequencing were kept in -80°C prior to analyses. The DNA extraction was performed from 0.25 g of freeze dried and ground raw material and silage samples following the protocol by Yu and Morrison (2004). For bacterial amplicon sequencing, universal primers 515F and 806R targeting the 16S rRNA gene V4 region were used (Caporaso et al., 2011). Libraries were prepared following the “16S metagenomics sequencing library preparation” protocol (Illumina) and sequenced on an Illumina MiSeq platform using 2 × 250 bp chemistry at the Finnish Functional Genomics Centre (Turku, Finland). Demultiplexing of sequences, adapter removal and sorting sequences by barcode were performed by the sequencing data provider. Sequencing data was further processed using QIIME v 2 (Bolyen et al., 2019). Briefly, quality control, filtering of chimeric reads, and clustering of bacterial sequences into amplicon sequence variants (ASV) were performed using DADA2 (Callahan et al., 2016). ASVs with less than 10 reads in total were removed. Bacterial ASV taxonomy was assigned using the Silva 138 database (Quast et al., 2012).
2.4. Data processing and statistical analyses
Data was analysed using a MIXED procedure (SAS Inc. 2002-2012, Release 9.4; SAS Inst. Inc., Cary, NC, USA) of SAS with treatment (three management factors × four additives resulting in 12 treatments) as fixed effect and replicates as a random effect. The data was tested regarding the normal distribution through Shapiro-Wilk test of the Univariate procedure. Least squares means and standard error of the means were reported per treatment and differences among treatment means were declared significant at 5% of probability. A pairwise comparison among treatment means was performed using a Tukey’s test and the effects of the management factors (compaction and soil contamination) were evaluated using contrasts.
Silage bacterial community alpha diversity was evaluated using Shannon and Simpson diversity indexes as well as observed number of ASVs. To evaluate treatment effect on the changes in silage bacterial community structure, between sample diversity was calculated as Bray-Curtis dissimilarities following Hellinger transformation and visualized using principal co-ordinate analysis (PCoA) as implemented in MicrobiotaProcess R package (Xu and Yu, 2021). The significance of groups was evaluated by distance-based permutational multivariate analysis of variance (adonis) and defined at P<0.05 level after 999 permutations, as implemented in vegan R package (Oksanen et al., 2019). To determine which bacterial taxa were affected by the silage preservation and management methods, a linear discriminant analysis was performed as implemented in MicrobiotaProcess. Significance was defined at P<0.05 with false discovery rate (FDR) correction (q<0.05).
To explore the magnitude of associations between bacterial communities and silage fermentation characteristics, the variables were ordered based on an analysis of a Spearman correlation plot (CORR procedure of SAS) and a heat map originated from two-dimensional display was created to characterize the effects of bacteria species on fermentation quality. Correlation data was filtered so that all genera below 0.01% were left out. This filtering reduced the number of genera from 88 to 28.
3. Results and discussion
3.1. Raw material characteristics
Botanical composition of the sward was dominated by red clover with proportions of red clover and timothy being 0.77 and 0.23, respectively, on DM basis. A high proportion of red clover is often found in organic swards due to the high competitiveness of red clover compared to grasses particularly in regrowth (Rinne and Nykänen, 2000). Chemical composition of the ensiled material was typical in terms of relatively high ash and crude protein concentrations and low neutral detergent fibre concentration (Huhtanen et al., 2006; Kuoppala et al., 2009), but the WSC concentration was particularly low, as often seen in red clover when compared to forage grasses (Kuoppala et al., 2009; King et al., 2012; Rinne et al., 2018). According to Li et al. (2019), WSC concentration in the raw material prior to ensiling is important for lactic acid fermentation, and the ideal is at least 50 g/kg DM. The low WSC concentration of the raw material accompanied with a relatively low DM content and high buffering capacity led to a low fermentation coefficient (Table 1), which reveals that this was a difficult material to ensile (fermentation coefficient <35; Weissbach et al., 1974; Weissbach, 1996). Silage raw material DM content is highly dependent on the extent of wilting, but is in general somewhat higher under Finnish practical conditions (321 g/kg; Salo et al., 2014) than in the current experiment (261 g/kg). This may partly be due to the inherently lower DM content of red clover compared to timothy (Rinne and Nykänen, 2000) as well as the wilting conditions. The DM fits well within the range of previous experiments using red clover, i.e., 170 and 430 g/kg in the 1st and 2nd cuts, respectively in the material of Kaldmäe et al. (2009) and 199 and 314 g/kg of red clover harvested fresh and after wilting, respectively by König et al. (2019). Microbial counts can be considered normal for raw material before ensiling.
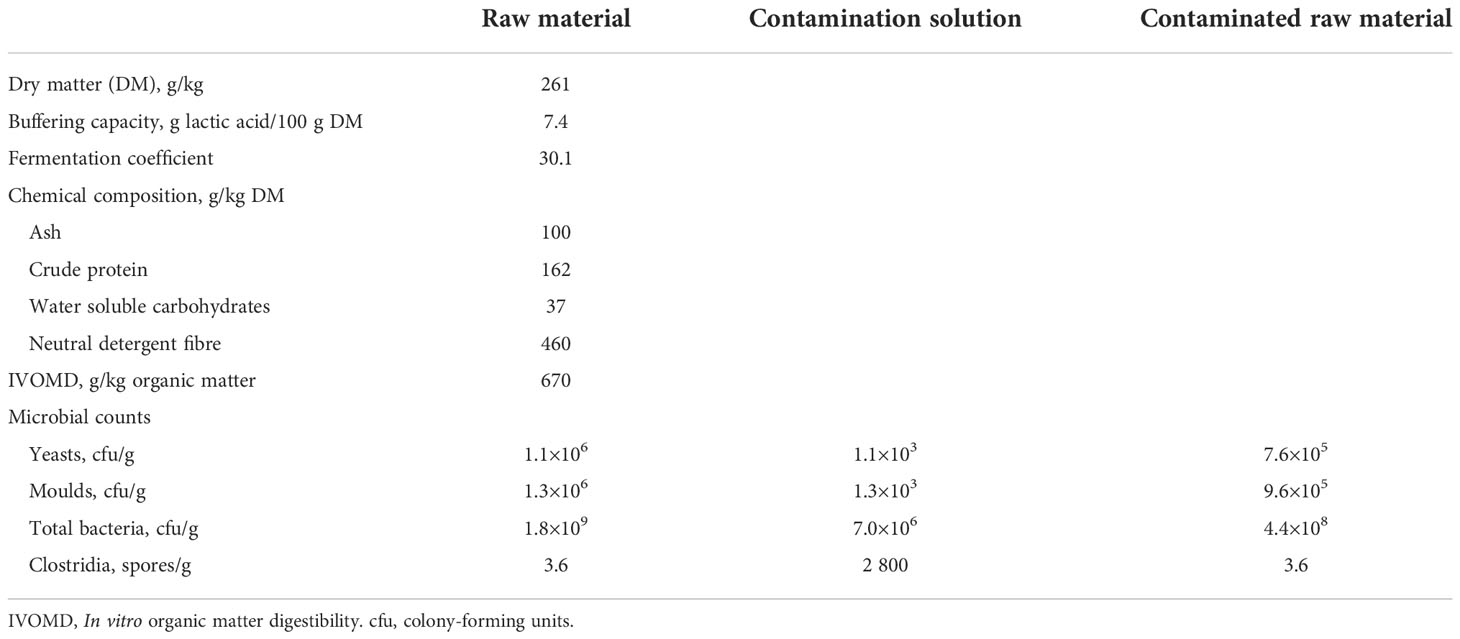
Table 1 Composition and microbial counts of red clover dominated raw material and the contamination solution.
3.2. Preservation characteristics of the experimental silages
Efficient compaction is considered an important management factor at farm level (Holmes and Bolsen, 2009) as it is required to achieve lactic acid fermentation as well as to prevent thriving of aerobic microbes such as yeasts and moulds, which have an important role in initiating the aerobic deterioration after opening the silo. The two compaction levels used in this experiment resulted in silage densities of 500 kg/m3 (136 kg DM/m3) and 665 kg/m3 (179 kg DM/m3) for loose and tight compactions, respectively. There were however hardly any significant effects on the analysed silage characteristics when loosely and tightly compacted silages were compared (Table 2) including the aerobic stability (194 and 234 h for loose and tight compaction, respectively, P>0.05). The only fermentation characteristic that was significantly (P<0.03) affected by compaction was the sum of all fermentation acids, which was higher for loosely rather than tightly compacted silages (95.8 vs. 91.9 g/kg DM). Limited responses to intensity of compaction have also been obtained earlier in pilot scale silos (McEniry et al., 2007; Franco et al., 2022). Use of airtight laboratory silos may not mimic sufficiently farm scale silos regarding the effects of compaction, which could explain the limited responses to compaction in the current experiment.
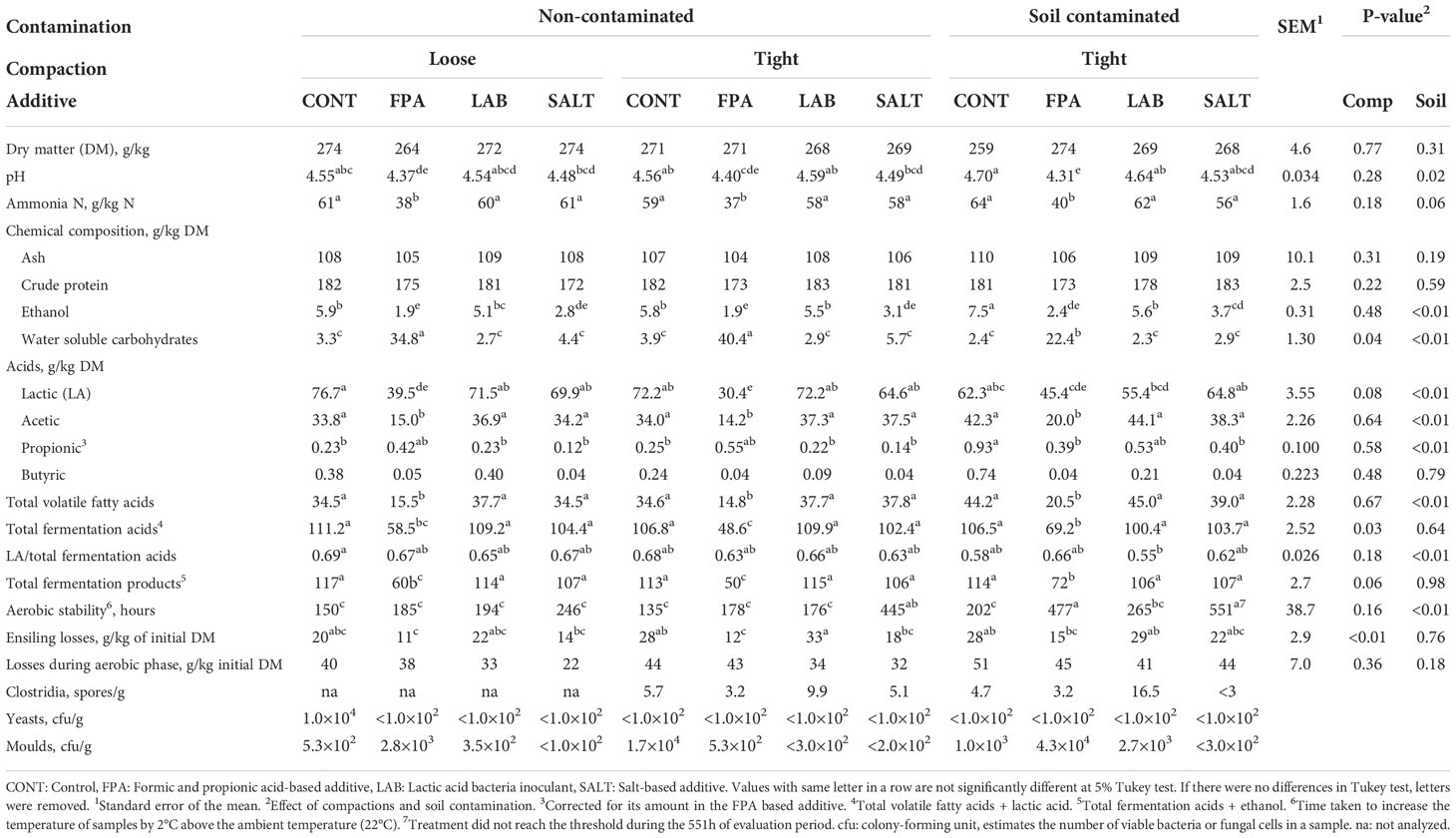
Table 2 Chemical composition, fermentation quality, aerobic stability, ensiling losses and microbial counts of red clover dominated silages treated with additives under different compaction (Comp) and soil contamination (Soil) levels.
Contaminating red clover with soil and faeces prior to ensiling negatively affected silage fermentation quality (Table 2). The effects included significant differences in pH (4.51 vs 4.55), lactic acid (59.9 vs 57.0 g/kg DM) and acetic acid (30.8 vs 36.2 g/kg DM), and proportion of ammonia N in total N tended (P<0.06) to increase (53 vs 56 g/kg N). The responses to contamination were not as clear as in Franco et al. (2022) indicating that the raw material characteristics in the current experiment were more resistant to contamination, or that the contaminants were less competitive. However, in both cases, increased acetic acid production was noted, and it probably contributed to the increased aerobic stability of the silages (234 vs. 374 h for non-contaminated and contaminated silages, respectively; P<0.01). As stated by Woolford (1975), acetic acid is a good antifungal agent. Although increased aerobic stability is desirable, achieving it through “wild-type” fermentation cannot be considered positive, as it may be linked with decreased hygienic quality of silage and high losses (Kung Jr, 2010).
Additives are commonly used at commercial farms to ensure high silage quality (Wilkinson and Rinne, 2018; Muck et al., 2018). In the current study, the three additives used present different modes of action. Organic acids included in FPA directly decrease grass material pH and they also have antimicrobial effects, LAB inoculants boost and direct natural lactic acid fermentation, while salt-based additives are particularly effective in preventing the growth of clostridia as well as in prolonging the aerobic stability of silages (Muck et al., 2018). The amount of formic acid recovered in the silages that were treated with FPA was on average 13.5 g/kg DM, corresponding to 4.97 litres of FPA additive per ton, which reflects well the targeted level of application (5 l/ton).
The challenging characteristics of the current red clover ensiled was reflected in slightly elevated final pH of all silages, but FPA was able to reduce it compared to the other additive treatments (4.60, 4.36, 4.59 and 4.50 for CONT, FPA, LAB and SALT, respectively; P<0.05). The higher pH in silages relying on production of fermentation acids has probably been due to the exhaustion of WSC in those silages, which has limited the acid formation. FPA resulted in the lowest proportion of ammonia N in total N (61, 38, 60 and 58 for CONT, FPA, LAB and SALT, respectively; P<0.05), although values for the other treatments were not particularly high either. König et al. (2019) also found lower ammonia N concentrations in red clover silages treated with FPA, but there was no evidence that the pH of the silages was lower. Proportion of ammonia N in total N is generally considered as a good indicator of silage fermentation quality (McDonald et al., 1991). The biological mechanism for the effect of FPA on lower ammonia N is due to restricted fermentation with less presence of fermenting microorganisms, which are less available to degrade plant cells and cause protein breakdown, concomitantly with rapid pH drop (McDonald et al., 1991). According to Wilkinson (1990), grass silage with ammonia N under 50 g/kg total N is considered very well fermented but silage with ammonia N ranging from 50 – 100 g/kg total N is still considered adequately fermented. In terms of extent of fermentation (concentrations of lactic acid and total fermentation acids in DM), CONT, LAB and SALT behaved in a very similar way, but FPA restricted it. Further, FPA clearly limited the acetic acid production (36.7, 16.4, 39.4 and 36.7 g/kg DM for CONT, FPA, LAB and SALT, respectively). The restricted fermentation caused by FPA application is the main driver of the lower acetic acid production in silages, which is also in agreement with Franco et al. (2022).
The efficiency of formic acid-based additives in restricting silage fermentation has been well established in grass (Jaakkola et al., 2006; Seppälä et al., 2016; Franco et al., 2018; Rinne et al., 2018; Franco et al., 2022) and red clover-based silages (Seppälä et al., 2013; Franco et al., 2018; Rinne et al., 2018). Restriction of silage fermentation positively affects voluntary silage intake of dairy cows (Huhtanen et al., 2007), which is a major factor affecting the nutrient supply and subsequent milk yield of dairy cows. Lower concentrations (P<0.05) of ethanol were found in FPA treated silages, unlike König et al. (2019), who found higher concentrations of ethanol for silages treated with formic acid at low DM, and no differences for silages prepared with higher DM raw material. The CONT and LAB silages were almost identical, and the only statistically significant difference being the slightly lower ethanol concentration in LAB compared to CONT in contaminated silages, so that under the conditions of the current experiment, benefits of using LAB could not be demonstrated, although in grass material described by Franco et al. (2022), the same inoculant was able to remarkably boost lactic acid production compared to CONT. The effects of SALT on silage fermentation were also minor as statistically significant differences (P<0.05) compared to CONT were limited to a small decrease of ethanol concentration in all silage groups, and a decrease in propionic acid concentration in contaminated silages.
Fermentation during ensiling decreased counts of yeasts and moulds in relation to raw material for all treatments. As reported by Pahlow et al. (2003), yeasts are usually considered as starters of aerobic spoilage, because they consume WSC and fermentation acids, and raise silage temperature and pH. Within loose compaction, the higher yeast count and numerically lower aerobic stability of CONT is in line with this, but for all other silages, the yeast counts were equally low.
Due to high dispersion of data, there were relatively few statistically significant differences in aerobic stability of silages (Table 2), but in tight, non-contaminated silages, SALT resulted in the longest aerobic stability (P<0.05), and in tight, contaminated silages, both FPA and SALT increased aerobic stability compared to CONT and LAB (P<0.05). The overall aerobic stabilities for CONT, FPA, LAB and SALT were 162, 280, 212 and 414 h, respectively. The positive effects of SALT and FPA on aerobic stability have been demonstrated earlier (e.g. Franco et al., 2022). The mechanisms that explain the improvement of aerobic stability in silages treated with SALT and FPA might be the antimicrobial actions on yeast and mould communities for SALT, and similarly for FPA this is due to the intentional application of propionic acid via additive, which also has antimicrobial activity.
3.3. Bacterial communities in the experimental silages
Alpha diversity estimates of raw material and experimental silages are shown in Table 3. Silages showed lower diversity of bacterial communities than the raw material for all indexes evaluated. This is in line with Fu et al. (2022), who investigating ryegrass silages found greater bacterial diversity in the raw material. There was no significant effect of compaction and contamination on alpha diversity, but some differences were observed between the additive treatments. Shannon’s and Simpson’s diversity indexes were significantly higher in the FPA treated silages, followed by CONT, while the smallest diversities were identified in the silages treated with LAB and SALT.
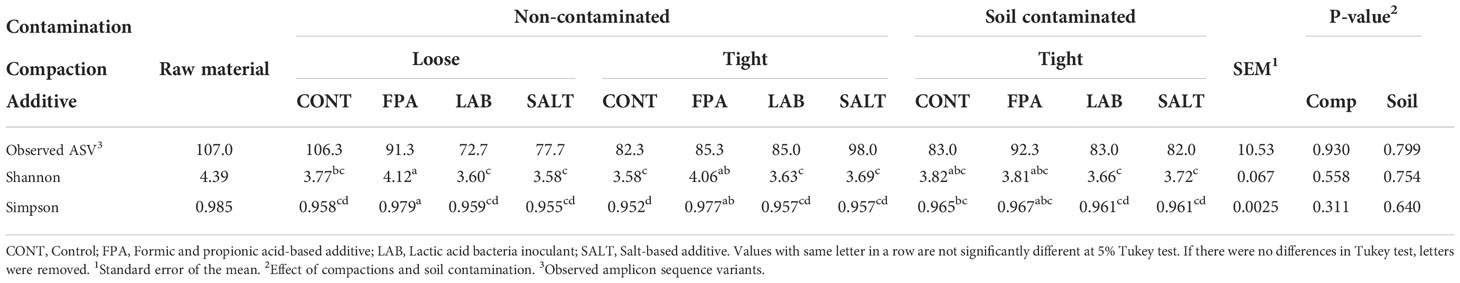
Table 3 Alpha diversity estimates of red clover dominated silages treated with additives under different compaction (Comp) and soil contamination (Soil) levels.
The bacterial community structure in raw material prior to ensiling was distinct compared to the silages. Among silages, axis 1 in the ordination plot discriminated silages treated with FPA from the remaining additive groups (adonis test P<0.001; Figure 1). There was no evidence that compaction or contamination significantly affected the community structure. In general terms, anaerobic fermentation is a metabolic process that starts with the activity of undesirable microorganisms from the raw materials, but these are suppressed and gradually replaced by lactic acid bacteria, which mostly dominate the fermentation process (Ni et al., 2017). Consequently, the raw material remained apart and distant from the clusters of silages, which was the same pattern found by Fu et al. (2022). This can potentially be explained by the inability of some epiphytic microorganisms on the foliar surface of the plant to survive under the anaerobic and acidic silage environments.
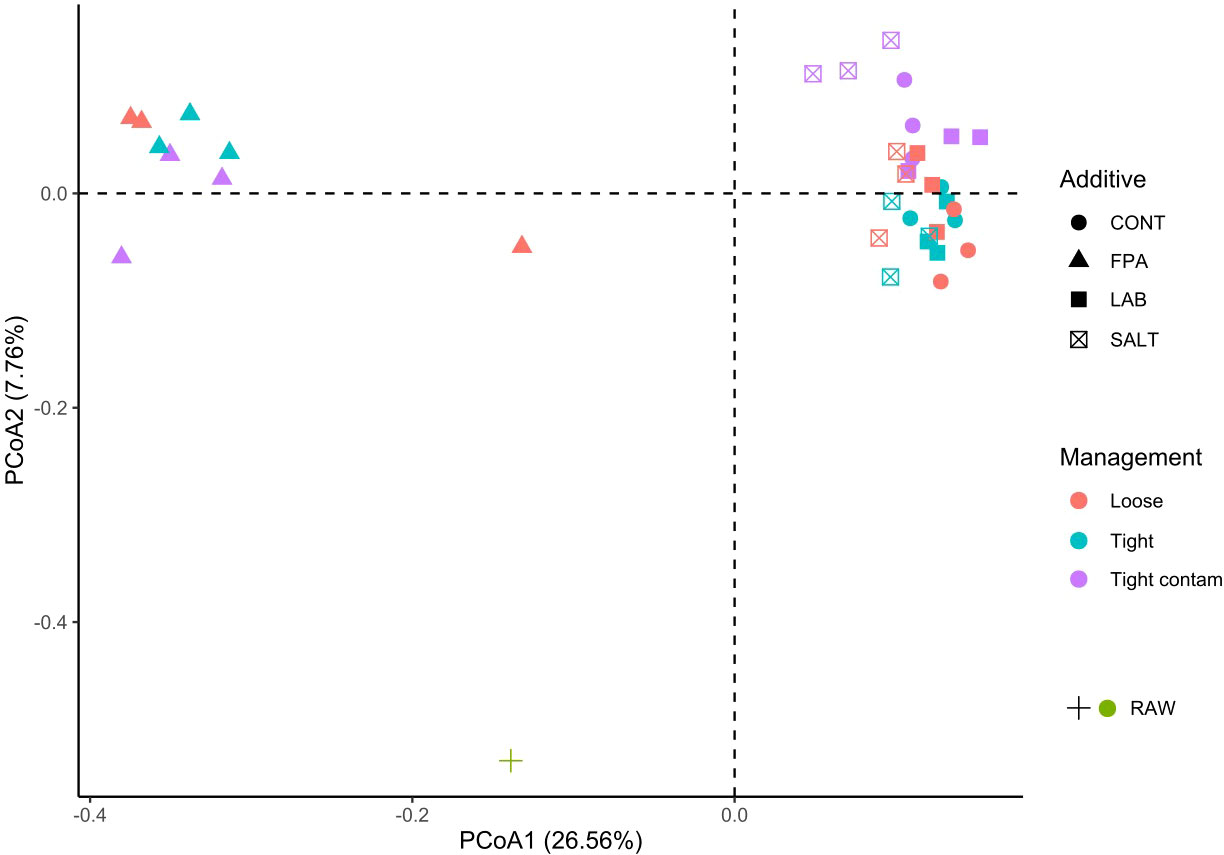
Figure 1 Principal coordinate analysis (PCoA) of the beta diversity analysis of red clover dominated silages treated with additives under different compaction and soil contamination levels. CONT, Control; FPA, Formic and propionic acid-based additive; LAB, Lactic acid bacteria inoculant; SALT, Salt-based additive; Loose, loosely compacted silages; Tight, tightly compacted silages; Tight contam, tightly compacted and contaminated silages; RAW, raw material prior ensiling.
The relative abundance of bacterial communities in red clover silages was affected by the additives (Figure 2). Abundance changes of several bacterial genera in all silage treatment groups were observed because of silage compaction or contamination, however, none of the changes remained significant after FDR adjustment and are not further discussed. The ensiling process without the use of inoculants, chemical additives, salts, enzymes, or any other initiator is a spontaneous fermentation process where fermentation relies on composition of epiphytic microorganisms on plant surface (Guo et al., 2018). Six phyla were identified in the current samples (Figure 2A). Raw material differed clearly from the silages. Although Firmicutes was a phylum of small contribution from epiphytic communities in the raw material, they dominated the fermentation in most silages. Proteobacteria have been observed to be the most abundant phylum in various silage raw materials such as ryegrass (Fu et al., 2022), maize (Keshri et al., 2018) and timothy (Franco et al., 2022), and although it decreased in proportion in the silages, it still tended to occupy the second position of most abundant phyla in the silages. Shifts of Proteobacteria and Firmicutes from raw material to silages was the same as observed by Keshri et al. (2018) when studying maize silages and Franco et al. (2022) studying timothy grass silages. The silages treated with FPA showed a significantly lower presence of Firmicutes as compared to CONT (P=0.04; FDR=0.03) and other additive treatments. In addition, FPA silages had significantly higher abundances of Proteobacteria, as well as Actinobacteriota (P=0.04; FDR=0.03), even if in smaller amounts than the other phyla (Supplementary Table S1).
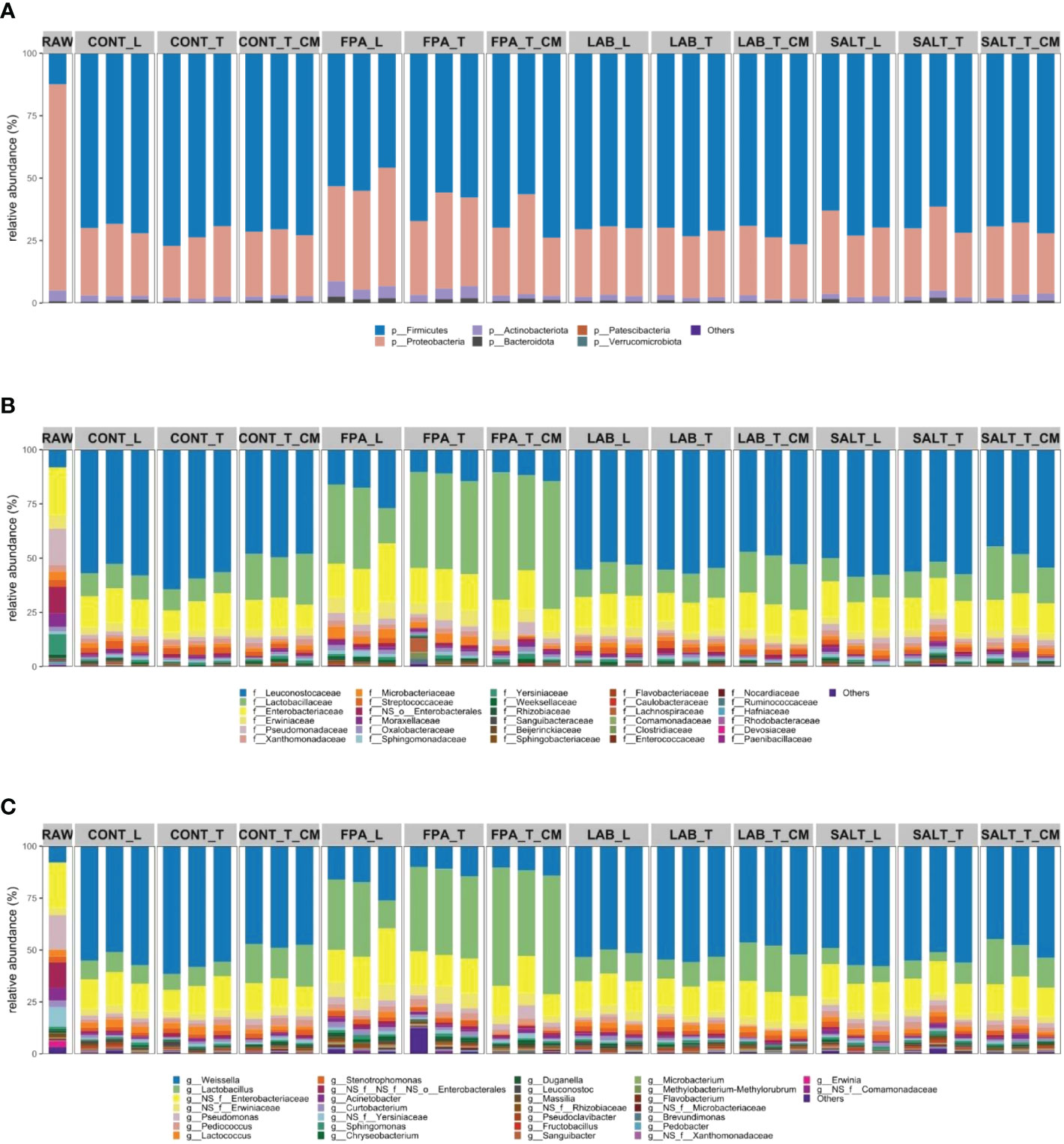
Figure 2 Relative abundance of bacterial communities at phylum (A), family (B) and genus (C) taxonomic levels of red clover dominated silages treated with additives under different compaction and soil contamination levels. RAW: raw material prior ensiling; CONT, Control; FPA, Formic and propionic acid-based additive; LAB, Lactic acid bacteria inoculant; SALT, Salt-based additive; L, loosely compacted silages; T, tightly compacted silages; CM, tightly compacted and contaminated silages.
The raw material showed a great variation in the abundance of families with no single-family dominating (Figure 2B). Some of the most abundant families in the raw material were Leuconostocaceae, Enterobacteriaceae, Erwiniaceae and Pseudomonadaceae. Although the Lactobacillaceae family was not identified in the raw material, it dominated fermentation in silages treated with FPA, regardless of compaction or contamination. Other families that were also present in FPA silages were Leuconostocaceae, Enterobacteriaceae and Erwiniaceae. The CONT, LAB and SALT silages were very similar to each other at the family level, with the most abundant communities being Leuconostocaceae, Lactobacillaceae, Enterobacteriaceae and Erwiniaceae.
The genus Weissella was identified in low abundance in the raw material (Figure 2C), but this genus dominated the fermentation of CONT, LAB and SALT silages. CONT, LAB and SALT silages were very similar in their bacterial composition at genus level, with only Lactobacillus being more abundant in LAB (P=0.004, FDR=0.4) as compared to CONT and Lactococcus being more abundant in CONT (P=0.003, FDR=0.2) as compared to SALT silage. Other genera that contributed to the fermentation in these silages in a similar way regardless of management conditions were Lactobacillus, Enterobacteriaceae sp. and Erwiniaceae sp. The silages treated with FPA were the most different from the remaining silages, regardless of management conditions. In comparison to CONT, the silages treated with FPA showed significantly greater relative abundance of Lactobacillus, Curtobacterium, Pseudomonas, and Erwiniaceae sp. (P=0.004, FDR=0.03). All the silages contained Lactobacillus, even though some of them in smaller amount, which is a community highly correlated with an excellent fermentation process. He et al. (2020) stated that high relative abundance of Lactobacillus has been linked to good fermentation quality of alfalfa silage. It is worth emphasizing that according to Wuyts et al. (2018), the identification of microbial composition through 16S rRNA sequencing may not differentiate between viable communities and dead bacterial cells. This deficiency may possibly distort the composition of active bacterial communities in the silages.
3.4. Correlations between relative abundance of bacterial communities and silage fermentation characteristics
To identify the relationships between the bacterial communities at genus level and parameters related to the silage fermentation characteristics, a Spearman’s correlation was performed (Figure 3). These results refer exclusively to non-contaminated loosely and tightly compacted silages. Although the Flavobacterium, Chryseobacterium, Methylobacterium-Methylorubrum, Acinetobacter, Rhizobiaceae sp., Yersiniaceae sp. and Enterobacterales spp. were present at abundance above filtering threshold, they did not present any correlation with the silage fermentation characteristics, and therefore were not shown in Figure 3. In a similar way, ratio between lactic acid and total fermentation acids, and ensiling losses were excluded because they were not correlated with the 28 bacterial taxonomic groups investigated here.
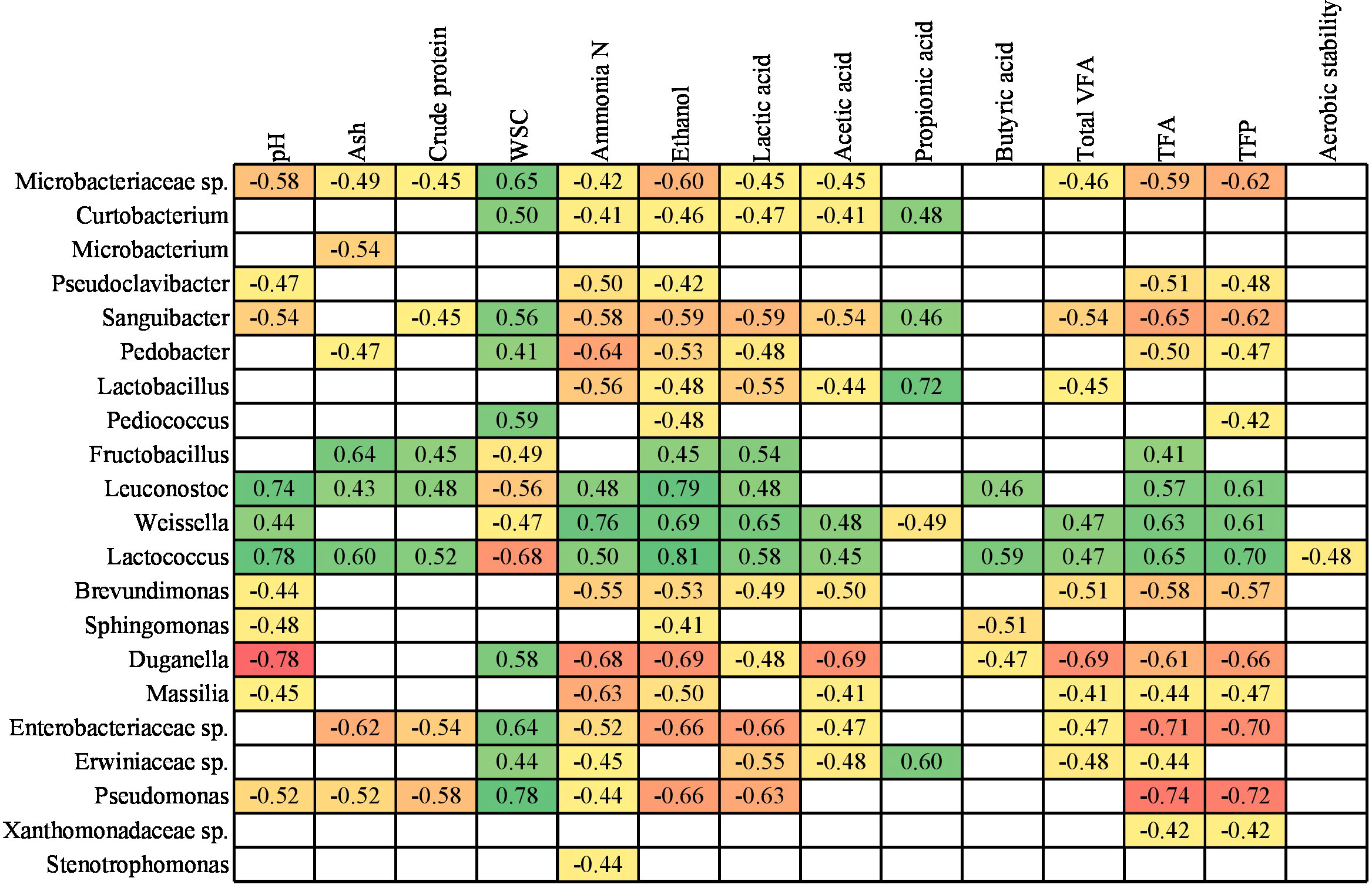
Figure 3 Spearman correlations between bacterial communities and silage parameters of red clover dominated silages. WSC, Water soluble carbohydrates; VFA, Volatile fatty acids; TFA, Total fermentation acids; TFP, Total fermentation products. From green (strong and positive correlation) to red colour (strong and negative correlation), while empty cells indicate non-significant correlation.
Positive and strong correlations were found between Leuconostoc and Lactococcus with ensiling parameters, such as pH, ammonia N, ethanol, butyric acid, total fermentation acids and total fermentation products (Figure 3). Weissella, which was one of the most abundant genera throughout the silages, was positively correlated with pH, ammonia N, ethanol, acetic acid, total VFA, total fermentation acids and total fermentation products, but it was negatively correlated with propionic acid. Some of these results agree with Zheng et al. (2022), as these authors also found a positive correlation between Weissella and pH and acetic acid, and at the same time a negative correlation between this genus and WSC and propionic acid. Franco et al. (2022) also identified positive correlation of Weissella with pH, however contradictory to the current study, no correlation was found with acetic acid and WSC, while a positive correlation with propionic acid.
Interestingly, the Fructobacillus, Leuconostoc, Weissella and Lactococcus genera were positively correlated with lactic acid production (Figure 3), and consequently showed a negative correlation with WSC, since they are used as the main substrate in fermentation that produces organic acids. Aerobic stability had the lowest correlation with bacterial communities, since only Lactococcus was negatively correlated to it.
Ideally, for optimal silage fermentation, the pH is expected to be low concomitantly with a low presence of ammonia N, ethanol and VFAs, which are indicative of quality and the lower the better. Thus, the bacterial taxonomic groups negatively correlated with these parameters, implies that the more of these bacteria, the lower the contents for these parameters. Some of the genera negatively correlated with these parameters that can potentially be linked to good silage fermentation characteristics were Pseudoclavibacter, Sanguibacter, Brevundimonas, Duganella, Massilia and Pseudomonas. However, contradictorily, some of these genera (e.g., Sanguibacter, Brevundimonas, Duganella and Pseudomonas) were also negatively correlated with the lactic acid production, although it would be preferable in terms of silage preservation.
Li et al. (2022) found that Pediococcus were negatively correlated with pH and concomitantly positively correlated with the lactic acid concentration in lucerne silages. However, these findings were not consistent with our study, as this genus did not show a significant correlation with pH and lactic acid. Stenotrophomonas, among the many possible correlations with several silage fermentation characteristics, showed a negative correlation solely with ammonia production (Figure 3). This result corroborates with Zeng et al. (2020), who studying mixed maize and soybean silages also found ammonia N as the only significant negative correlation with Stenotrophomonas. This fact indicates that these bacteria might block the production of ammonia N or vice versa, in case it is inhibited by ammonia N.
Lactobacillus, which was one of the most abundant genus especially in FPA treated silages (Figure 2C), was negatively correlated to ammonia N, ethanol and acetic acid (Figure 3). However, this genus was also negatively correlated to lactic acid, and positively correlated to propionic acid, which might be unexpected, as this can be related to poor fermentation quality. The Enterobacteriaceae and Erwiniaceae families, also among the most abundant in the silage fermentation process (Figures 2B, C), showed a negative correlation with the production of ammonia N, lactic acid and acetic acid, and as a possible consequence there was a positive correlation with WSC (Figure 3).
High concentrations of ammonia N in silage indicate protein breakdown generally as result of clostridial fermentation or extensive plant protease activity. Pseudomonas was among the five most commonly found genera in these silages and showed negative correlation with most silage fermentation characteristics (Figure 3), including pH, ammonia N and ethanol. Thus, the presence of Pseudomonas in silages can be a positive factor implying that the pH of the silage will tend to be low, concomitantly with low ammonia N and ethanol concentrations as also found by Ogunade et al. (2018). Considering our results, some other bacterial genera may also be involved in decreasing protein breakdown because the relative abundances of Curtobacterium, Pedobacter and Microbacteriaceae sp. were negatively correlated with ammonia N.
4. Conclusions
This study indicated that practical guidelines of good silage production management, such as the use of additives, thorough compaction and avoiding soil contamination, are of fundamental importance. Use of a formic and propionic acid-based additive improved fermentative quality of red clover dominated grass that was ensiled under different management conditions, but effects of a salt-based additive and particularly a lactic acid bacteria inoculant were minor under the conditions of the current experiment, although the salt-based additive was effective in improving the aerobic stability of the silages. The limited responses to compaction may be related to the use of airtight laboratory silos. Contamination of the silage with soil stimulated an undesired type of fermentation with higher amounts of ethanol, acetic and propionic acids and also greater pH.
The formic and propionic acid treated silages presented a greater diversity of bacterial communities than silages treated with other additives. Proteobacteria were the most abundant in raw material, followed by Firmicutes, and major shifts happened in these communities after the silage fermentation process. Weissella was found in small amounts in the raw material prior to ensiling but was the genus that mostly dominated the fermentation in the silages. The most abundant communities in the silage, such as Weissella, Lactobacillus and Pseudomonas, were correlated with several parameters of silage fermentation quality, in a mixed way between both ideal and poor fermentation quality directions. This requires further investigations in order to understand the cause-and-effect relationships between these factors. Thus, studies like this that combine parameters related to silage quality and bacterial communities provide a deeper comprehension of silage fermentation processes. This approach offers new knowledge that allows us to manipulate the silage fermentation mechanisms, which consequently contribute to the improvement of the quality and use of the silages.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found at: NCBI; PRJNA886493.
Author contributions
MF contributed to conceptualization, methodology, formal analysis, visualization, data curation and writing original draft, review and editing. IT contributed to methodology, formal analysis, visualization, data curation and review and editing manuscript. AH contributed to conceptualization, methodology, review and editing, and funding acquisition. MR contributed to conceptualization, methodology, review and editing, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by MiMi project funded by the Academy of Finland (grant number 322827) and NurmiNauta project funded by the Centre for Economic Development, Transport and the Environment for South Ostrobothnia, Finland (grant number 67424/2018).
Acknowledgments
We thank the Finnish Functional Genomics Centre supported by University of Turku, Åbo Akademi University and Biocenter Finland for sequencing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2022.1080535/full#supplementary-material
References
Bai J., Xu D., Xie D., Wang M., Li Z., Guo X. (2020). Effects of antibacterial peptide-producing Bacillus subtilis and Lactobacillus buchneri on fermentation, aerobic stability, and microbial community of alfalfa silage. Bioresour Technol. 315, 123881. doi: 10.1016/j.biortech.2020.123881
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Franco M., Jalava T., Stefanski T., Kuoppala K., Timonen P., Winquist E., et al. (2018). “Grass silage for biorefinery - effect of additives on silage quality and liquid solid separation of timothy and red clover silages,” in Proc. 9th Nordic Feed Science Conference, 49–54 (Uppsala, Sweden: Swedish University of Agricultural Sciences (SLU).
Franco M., Tapio I., Pirttiniemi J., Stefański T., Jalava T., Huuskonen A., et al. (2022). Fermentation quality and bacterial ecology of grass silage modulated by additive treatments, extent of compaction and soil contamination. Fermentation 8, 156. doi: 10.3390/fermentation8040156
Fu Z., Sun L., Hou M., Hao J., Lu Q., Liu T., et al. (2022). Effects of different harvest frequencies on microbial community and metabolomic properties of annual ryegrass silage. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.971449
Guo X. S., Ke W. C., Ding W. R., Ding L. M., Xu D. M., Wang W. W., et al. (2018). Profiling of metabolome and bacterial community dynamics in ensiled Medicago sativa inoculated without or with Lactobacillus plantarum or Lactobacillus buchneri. Sci. Rep. 8, 357. doi: 10.1038/s41598-017-18348-0
He L., Lv H., Xing Y., Wang C., You X., Chen X., et al. (2020). The nutrients in Moringa oleifera leaf contribute to the improvement of stylo and alfalfa silage: Fermentation, nutrition and bacterial community. Bioresour Technol. 301, 122733. doi: 10.1016/j.biortech.2020.122733
Holmes B., Bolsen K. (2009). “What’s new in silage management?,” in XV International Silage Conference. 61–76 (Madison: University of Wisconsin).
Huhtanen P., Nousiainen J., Rinne M. (2006). Recent developments in forage evaluation with special reference to practical applications. Agric. Food Sci. 15, 293–323. doi: 10.2137/145960606779216317
Huhtanen P., Rinne M., Nousiainen J. (2007). Evaluation of the factors affecting silage intake of dairy cows: a revision of the relative silage dry-matter intake index. Animal 1, 758–770. doi: 10.1017/S175173110773673X
Jaakkola S., Rinne M., Heikkilä T., Toivonen V., Huhtanen P. (2006). Effects of restriction of silage fermentation with formic acid on milk production. Agric. Food Sci. 15, 200–218. doi: 10.2137/145960606779216290
Järvenranta K., Kuoppala K., Rinne M., Virkajärvi P. (2016). Legumes in ruminant production systems in European cold climates - limitations and opportunities. Legume Perspect. 12, 34–35.
Kaldmäe H., Kärt O., Olt A., Selge A., Keres I. (2009). Inoculant effects on red clover silage: fermentation products and nutritive value. Agron. Res. 7, 793–800.
Keshri J., Chen Y., Pinto R., Kroupitski Y., Weinberg Z. G., Sela S. (2018). Microbiome dynamics during ensiling of corn with and without Lactobacillus plantarum inoculant. Appl. Microbiol. Biotechnol. 102, 4025–4037. doi: 10.1007/s00253-018-8903-y
King C., McEniry J., O’Kiely P. (2012). A note on the fermentation characteristics of red clover silage in response to advancing stage of maturity in the primary growth. Irish J. Agr. Food Res. 51, 79–84. doi: 10.2307/41756847
Knicky M., Spörndly R. (2015). Short communication: Use of a mixture of sodium nitrite, sodium benzoate, and potassium sorbate in aerobically challenged silages. J. Dairy Sci. 98, 5729–5734. doi: 10.3168/jds.2015-9332
König W., König E., Elo K., Vanhatalo A., Jaakkola S. (2019). Effects of sodium nitrite treatment on the fermentation quality of red clover-grass silage harvested at two dry matter concentrations and inoculated with clostridia. Agric. Food Sci. 28, 155–164. doi: 10.23986/afsci.85114
Kung L. Jr. (2010). Aerobic stability of silage (Visalia, CA, USA) (Accessed Proceedings of the California Alfalfa and Forage Symposium and Crop/cereal Conference).
Kung L., Shaver R. D., Grant R. J., Schmidt R. J. (2018). Silage review: Interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 101, 4020–4033. doi: 10.3168/jds.2017-13909
Kuoppala K., Ahvenjärvi S., Rinne M., Vanhatalo A. (2009). Effects of feeding grass or red clover silage cut at two maturity stages in dairy cows. 2. dry matter intake and cell wall digestion kinetics. J. Dairy Sci. 92, 5634–5644. doi: 10.3168/jds.2009-2250
Li P., Lu Y., Zhao M., Chen L., Zhang C., Cheng Q., et al. (2021). Effects of phenyllactic acid, lactic acid bacteria, and their mixture on fermentation characteristics and microbial community composition of timothy silage. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.743433
Li P., Zhang Y., Gou W., Cheng Q., Bai S., Cai Y. (2019). Silage fermentation and bacterial community of bur clover, annual ryegrass and their mixtures prepared with microbial inoculant and chemical additive. Anim. Feed Sci. Technol. 247, 285–293. doi: 10.1016/j.anifeedsci.2018.11.009
Li P., Zhao W., Yan L., Chen L., Chen Y., Gou W., et al. (2022). Inclusion of abandoned rhubarb stalk enhanced anaerobic fermentation of alfalfa on the Qinghai Tibetan Plateau. Bioresour Technol. 347, 126347. doi: 10.1016/j.biortech.2021.126347
McAllister T. A., Dunière L., Drouin P., Xu S., Wang Y., Munns K., et al. (2018). Silage review: Using molecular approaches to define the microbial ecology of silage. J. Dairy Sci. 101, 4060–4074. doi: 10.3168/jds.2017-13704
McDonald P., Henderson A. R., Heron S. J. E. (1991). The biochemistry of silage. 2nd ed (Marlow, UK: Chalcombe Publications).
McEniry J., O’Kiely P., Clipson N. J. W., Forristal P. D., Doyle E. M. (2007). The relative impacts of wilting, chopping, compaction and air infiltration on the conservation characteristics of ensiled grass. Grass Forage Sci. 62, 470–484. doi: 10.1111/j.1365-2494.2007.00602.x
Muck R. E., Nadeau E. M. G., McAllister T. A., Contreras-Govea F. E., Santos M. C., Kung L. (2018). Silage review: Recent Adv. Future uses silage additives. J. Dairy Sci. 101, 3980–4000. doi: 10.3168/jds.2017-13839
Ni K., Wang F., Zhu B., Yang J., Zhou G., Pan Y., et al. (2017). Effects of lactic acid bacteria and molasses additives on the microbial community and fermentation quality of soybean silage. Bioresour Technol. 238, 706–715. doi: 10.1016/j.biortech.2017.04.055
Ogunade I. M., Jiang Y., Pech Cervantes A. A., Kim D. H., Oliveira A. S., Vyas D., et al. (2018). Bacterial diversity and composition of alfalfa silage as analyzed by Illumina MiSeq sequencing: Effects of Escherichia coli O157:H7 and silage additives. J. Dairy Sci. 101, 2048–2059. doi: 10.3168/jds.2017-12876
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2019). Vegan: Community ecology package. R Package Version 2.5-6, 2019.
Pahlow G., Muck R., Driehuis F., Oude Elferink S., Spoelstra S. F. (2003). “Microbiology of ensiling,” in Silage Science and Technology, 31–93. doi: 10.2134/agronmonogr42.c2
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ren H., Feng Y., Pei J., Li J., Wang Z., Fu S., et al. (2020). Effects of Lactobacillus plantarum additive and temperature on the ensiling quality and microbial community dynamics of cauliflower leaf silages. Bioresour Technol. 307, 123238. doi: 10.1016/j.biortech.2020.123238
Rinne M., Jalava T., Stefanski T., Kuoppala K., Timonen P., Winquist E., et al. (2018). “Optimizing grass silage quality for green biorefineries,” in Proc. of the 27th General Meeting of the European Grassland Federation, 820–822 (Cork, Ireland: Grassland Science in Europe).
Rinne M., Nykänen A. (2000). Timing of primary growth harvest affects the yield and nutritive value of timothy-red clover mixtures. Agric. Food Sci. 9, 121–134. doi: 10.23986/afsci.5654
Salo T., Eurola M., Rinne M., Seppälä A., Kaseva J., Kousa T. (2014). “The effect of nitrogen and phosphorus concentrations on nutrient balances of cereals and grass silage,” in MTT report vol. 147. Available at: http://urn.fi/URN:ISBN:978-952-487-541-7.
Seppälä A., Heikkilä T., Mäki M., Miettinen H., Rinne M. (2013). Controlling aerobic stability of grass silage-based total mixed rations. Anim. Feed Sci. Technol. 179, 54–60. doi: 10.1016/j.anifeedsci.2012.11.011
Seppälä A., Heikkilä T., Mäki M., Rinne M. (2016). Effects of additives on the fermentation and aerobic stability of grass silages and total mixed rations. Grass Forage Sci. 71, 458–471. doi: 10.1111/gfs.12221
Wang M., Gao R., Franco M., Hannaway D. B., Ke W., Ding Z., et al. (2021). Effect of mixing alfalfa with whole-plant corn in different proportions on fermentation characteristics and bacterial community of silage. Agriculture 11, 174. doi: 10.3390/agriculture11020174
Wang S., Li J., Zhao J., Dong Z., Dong D., Shao T. (2022). Dynamics of the bacterial communities and predicted functional profiles in wilted alfalfa silage. J. Appl. Microbiol. 132, 2613–2624. doi: 10.1111/jam.15417
Weissbach F. (1996). “New developments in crop conservation,” in Proceedings of the 11th Int. Silage Conference, 11–25 (Aberystwyth, UK: IGER Publ. Section).
Weissbach F., Schmidt L., Hein E. (1974). “Method of anticipation of the run of fermentation in silage making based on the chemical composition of green fodder,” in Proceedings of the 12th International Grassland Congress. 663–673(Moscow, Russia).
Wilkinson J. M., Rinne M. (2018). Highlights of progress in silage conservation and future perspectives. Grass Forage Sci. 73, 40–52. doi: 10.1111/gfs.12327
Woolford M. K. (1975). Microbiological screening of the straight chain fatty acids (C1-C12) as potential silage additives. J. Sci. Food Agric. 26, 219–228. doi: 10.1002/jsfa.2740260213
Wright D. A., Gordon F. J., Steen R. W. J., Patterson D. C. (2000). Factors influencing the response in intake of silage and animal performance after wilting of grass before ensiling: a review. Grass Forage Sci. 55, 1–13. doi: 10.1046/j.1365-2494.2000.00198.x
Wuyts S., van Beeck W., Oerlemans E. F. M., Wittouck S., Claes I. J. J., de Boeck I., et al. (2018). Carrot juice fermentations as man-made microbial ecosystems dominated by lactic acid bacteria. Appl. Environ. Microbiol. 84, e00134–e00118. doi: 10.1128/AEM.00134-18
Wyngaarden S., Gaudin A., Deen W., Martin R. (2015). Expanding red clover (Trifolium pratense) usage in the corn–soy–wheat rotation. Sustainability 7, 15487–15509. doi: 10.3390/su71115487
Xu D., Ding W., Ke W., Li F., Zhang P., Guo X. (2019). Modulation of metabolome and bacterial community in whole crop corn silage by inoculating homofermentative Lactobacillus plantarum and heterofermentative Lactobacillus buchneri. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03299
Xu S., Yu G. (2021). MicrobiotaProcess: An R package for analysis, visualization and biomarker discovery of microbiome. R Package Version 1.6.1, 2021.
Yu Z., Morrison M. (2004). Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36, 808–812. doi: 10.2144/04365ST04
Zeng T., Li X., Guan H., Yang W., Liu W., Liu J., et al. (2020). Dynamic microbial diversity and fermentation quality of the mixed silage of corn and soybean grown in strip intercropping system. Bioresour Technol. 313, 123655. doi: 10.1016/j.biortech.2020.123655
Keywords: aerobic stability, animal feedstock, compaction, contamination, density, microbiome, silage additive, Trifolium pratense
Citation: Franco M, Tapio I, Huuskonen A and Rinne M (2022) Fermentation quality and bacterial ecology of red clover dominated silage modulated by different management factors. Front. Anim. Sci. 3:1080535. doi: 10.3389/fanim.2022.1080535
Received: 26 October 2022; Accepted: 29 November 2022;
Published: 15 December 2022.
Edited by:
Bianca Castiglioni, National Research Council (CNR), ItalyReviewed by:
Xianjun Yuan, Nanjing Agricultural University, ChinaMichael D. Flythe, Agricultural Research Service (USDA), United States
Anusorn Cherdthong, Khon Kaen University, Thailand
Copyright © 2022 Franco, Tapio, Huuskonen and Rinne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcia Franco, bWFyY2lhLmZyYW5jb0BsdWtlLmZp
 Marcia Franco
Marcia Franco Ilma Tapio
Ilma Tapio Arto Huuskonen2
Arto Huuskonen2