- Department of Molecular Infection Biology, Helmholtz Centre for Infection Research, Braunschweig, Germany
The genus Yersinia includes three human pathogenic species, Yersinia pestis, the causative agent of the bubonic and pneumonic plague, and enteric pathogens Y. enterocolitica and Y. pseudotuberculosis that cause a number of gut-associated diseases. Over the past years a large repertoire of RNA-based regulatory systems has been discovered in these pathogens using different RNA-seq based approaches. Among them are several conserved or species-specific RNA-binding proteins, regulatory and sensory RNAs as well as various RNA-degrading enzymes. Many of them were shown to control the expression of important virulence-relevant factors and have a very strong impact on Yersinia virulence. The precise targets, the molecular mechanism and their role for Yersinia pathogenicity is only known for a small subset of identified genus- or species-specific RNA-based control elements. However, the ongoing development of new RNA-seq based methods and data analysis methods to investigate the synthesis, composition, translation, decay, and modification of RNAs in the bacterial cell will help us to generate a more comprehensive view of Yersinia RNA biology in the near future.
Introduction
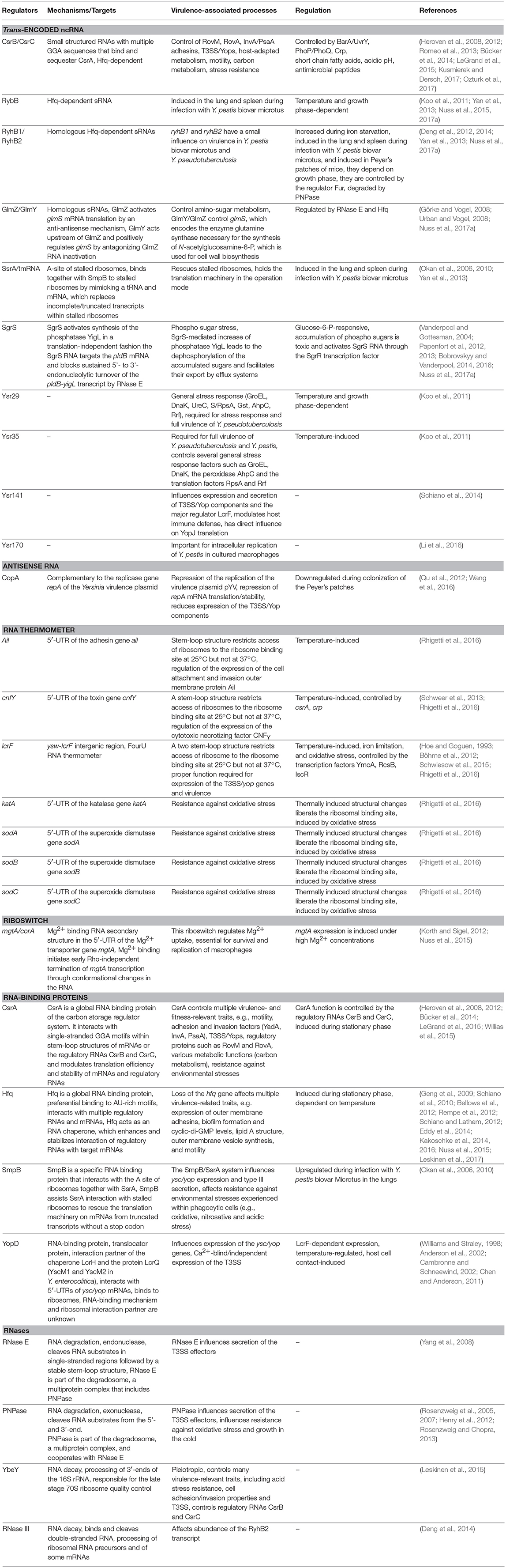
For a long time it was thought that the RNA make-up and biology of bacteria is rather simple and constitutes mainly of mRNAs, tRNAs, and rRNAs and a few specific RNAs such as the tmRNA/SsrA of the ribosome rescue system. However, over the past 10 years, research on bacterial RNA molecules has proven this to be incorrect, since bacteria possess a huge variety of RNAs. A large repertoire of cis- and trans-acting non-coding RNAs has been identified and a plethora of different RNA-based regulatory mechanisms and functions has been unveiled mainly in Enterobacteriaceae, but lately also in many other prokaryotes. This showed that RNA-based gene expression control is highly complex and global, and almost as diverse and multifarious as transcriptional control. Multiple structured RNA elements have been identified in the 5′-untranslated regions of mRNAs sensing temperatures (RNA thermometers) or metabolites (riboswitches) in order to modulate mRNA translation and/or stability (Kortmann and Narberhaus, 2012; Sherwood and Henkin, 2016; McCown et al., 2017). Moreover, a remarkably large number of small regulatory RNAs (sRNAs) exist in bacteria (i.e., 200–300 in Enterobacteriaceae, Barquist and Vogel, 2015; Nuss et al., 2015), which control mRNA expression and decay often in concert with RNA-binding proteins. Among the most prominent RNA-binding proteins are the RNA chaperone Hfq and the carbon storage regulator (Csr) protein CsrA, as well as RNases. Clever variations of different global RNA-sequencing (RNA-seq)-based techniques (Figure 1) used to define the overall RNA-protein interactome and the sRNA-target networks demonstrated that almost 50% of the bacterial mRNAs are subjected to sRNA-mediated regulation (Melamed et al., 2016; Hör and Vogel, 2017; Waters et al., 2017).
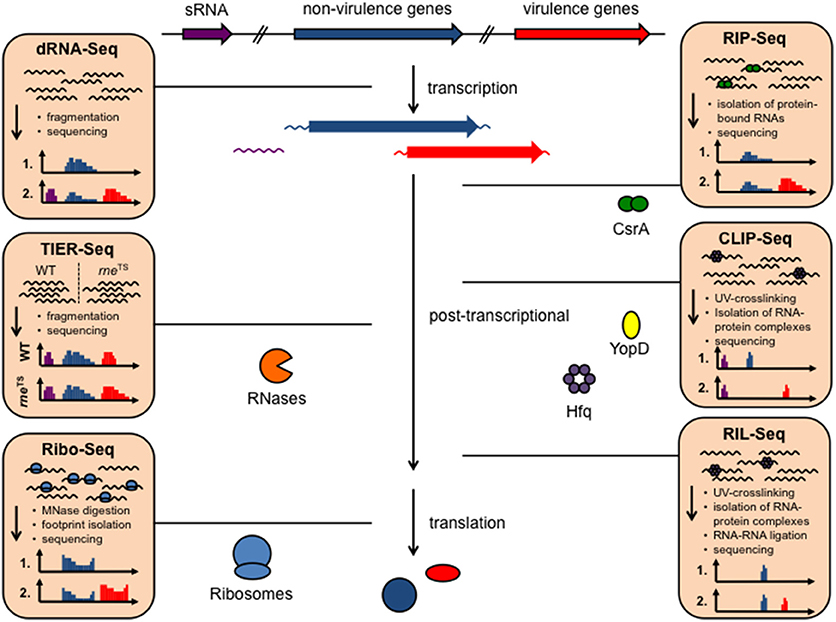
Figure 1. Available RNA-seq derived techniques, which can be used to study RNA-based control mechanisms in Yersinia. Different RNA-seq based techniques allow rapid and global investigation of transcript expression, transcription start sites, promoter structures, 5′- and 3′-UTR properties (dRNA-Seq), as well as binding of RNA-binding proteins to RNAs, such as CsrA, Hfq, and YopD (RIP-seq, CLIP-seq), RNA-RNA interactions facilitated by protein interaction (RIL-Seq), translation (Ribo-seq), and RNA processing and decay by heat-sensitive RNase E (rneTS) (TIER-seq).
The global analysis of RNA-based control strategies of virulence genes was started with a relatively small number of model bacteria. This included human pathogenic Yersinia species, such as Y. pestis, the causative agent of bubonic plague and the two enteric pathogens Y. pseudotuberculosis and Y. enterocolitica causing gut-associated diseases (Yersiniosis), i.e., ranging from enteritis, watery diarrhea, mesenteric lymphadenitis to post-infectious extraintestinal sequelae (Bottone, 1997; Koornhof et al., 1999; Smego et al., 1999; Valentin-Weigand et al., 2014). System level RNA-seq has the advantage that the entire transcriptome of an organism with all included RNA functions can be studied in a cellular and particular environmental (e.g., infection, virulence conditions) context. This allowed a single-nucleotide resolution and a dynamic view of the pathogen's gene expression, which are summarized and discussed in the present review. The establishment of comprehensive RNA maps of Y. pseudotuberculosis highlighted not only the breadth and large variety of RNA species, it also revealed new species-specific and conserved principles of RNA-based, post-transcriptional control strategies that contribute to the dynamic, spatial, and fine-tuned control of bacterial pathogenicity.
Global Transcriptional Analysis to Elucidate Fitness-Relevant Traits of Yersinia
In order to understand how the functional status of Yersinia is modulated in response to environmental conditions sensed outside or inside their hosts, detailed knowledge about the bacterial gene expression profile in different surroundings is required. RNA-seq is extremely well-suited for this task due to the high sensibility, high resolution and the possibility for high-throughput analysis of multiple in vitro and in vivo conditions (Kröger et al., 2013; Nuss et al., 2015, 2017a; Figures 1, 2). In particular transition from exponential to stationary phase and shifts from moderate temperatures to 37°C, mimicking host entry, were found to induce reprogramming of a large set of virulence genes. This includes colonization factors and immune defense mechanisms, encoded on the chromosome and the virulence plasmid, but also many catabolic/energy production genes. For instance, this uncovered the existence of a thermo-regulated “acetate switch,” which seems to prime the bacteria for growth in the digestive tract. This physiological switch occurs when bacteria shift from rapid growth in which they produce acetate from acetogenic carbon sources, e.g., glucose, to a metabolic program of slower growth, which is facilitated by the import and utilization of acetate (Nuss et al., 2015). This is accompanied by a thermo-induced up-regulation of multiple transport and catabolic genes for simple sugars. As the mammalian intestine is rich in short-chain fatty acids, in particular acetate, produced by the intestinal microbiota through consumption of available polysaccharides from diet and host-derived mucus (Cummings and Macfarlane, 1997), flipping the switch may facilitate ultilization of these microbiota-derived degradation products.
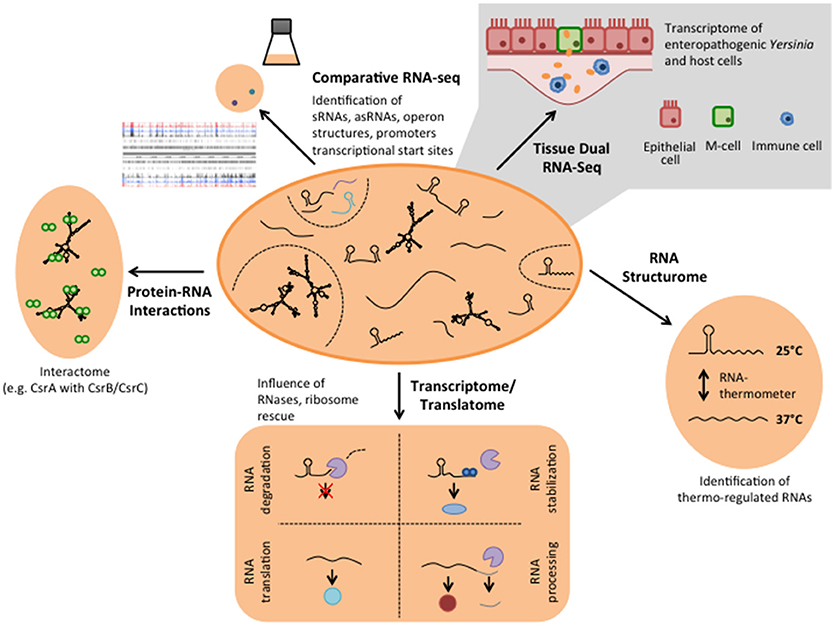
Figure 2. Different RNA-based regulatory mechanisms that control virulence in Yersinia. Global RNA biology approaches have been used to identify sRNAs, asRNAs, and sensory RNAs (RNA thermometer and riboswitches) as well as important RNA-binding proteins implicated in Yersinia virulence control.
The RNA-seq approach also allowed us to elucidate the regulatory architecture linking nutritional status to virulence. We identified a massive remodeling of the CRP-controlled network in response to temperature and discovered CRP as a transcriptional master regulator of numerous conserved and newly identified non-coding RNAs which participate in this process (Nuss et al., 2015). This finding highlighted a new level of complexity of the regulatory network in Yersinia. The concerted action of several transcriptional regulators and multiple non-coding RNAs controlled by CRP adjusts Yersinia fitness and virulence to the requirements of their environmental and virulent life-styles. This discovery showed that CRP is an integral component of a regulatory network that controls life-style switching and highlighted the power of comparative RNA-seq analysis.
Parallel analysis of the transcriptomes of Y. pseudotuberculosis during mouse infection (Tissue Dual RNA-seq, Figure 2) has further allowed us to study host-pathogen interaction. It enabled us to correlate gene expression changes of the pathogen with those occurring in the host and revealed genes and regulatory RNAs that are predominantly expressed or repressed during the infection (Avican et al., 2015; Nuss et al., 2015, 2017a). Numerous alterations of host transcripts associated with IL-6-triggered inflammatory and acute phase responses (Mmp8 induction), coagulative activities, metal ion sequestration, tissue repair and damage highlighted that the immune response in the gut-associated lymphoid follicles (Peyer's patches) upon a Y. pseudotuberculosis infection is dominated by infiltration of neutrophils and dendritic cells, and elicits a mixed TH1/TH17 response (Nuss et al., 2017a; Heine et al., 2018). In response, Yersinia increases the gene and expression dose of the virulence plasmid-encoded Ysc type III secretion system (T3SS) and the anti-phagocytic Yop effector proteins (Yops) to prevent the phagocytic attack. In addition, the bacteria induce high-affine ion (Fe2+ and Zn2+) sequestration systems, diverse stress response, and nutrient uptake systems to counteract ion deprivation, radical/oxidative stress, anti-microbial peptides, and nutrient restraints (Nuss et al., 2017a). Tissue Dual RNA-seq further showed that the outcome of the observed pathogen-host interactions could vary significantly when expression of certain pathogenicity factors is reduced or abrogated. The cytotoxic necrotizing factor Y (CNFY) expressed by some Y. pseudotuberculosis strains, e.g., YPIII and IP2666, is one of these factors. Suppression of CNFY triggers interferon-γ mediated responses fostering non-inflammatory bactericidal activities. Moreover, it promotes Ido1- and Arg1-mediated Tregs activation and proliferation and other immune suppressing and tolerogenic mechanisms that avoid systemic inflammation, prevent tissue destruction and drive this Y. pseudotuberculosis strain into persistency (Heine et al., 2018). This process is accompanied by a preterm reprogramming of the pathogen's transcriptome. Anaerobic and multiple acid and oxidative stress resistance genes (e.g., arcA, fnr, frdA, and wrbA) are already induced in the acute phase. It is assumed that this priming gives the bacteria a fitness edge against the host immune defense and facilitates establishment of a persistent microbiota-type life style (Heine et al., 2018). In parallel, the expression profile of many virulence genes changes toward a pattern seen at moderate temperature under in vitro growth conditions. Up-regulation of flagella and adhesion/invasion genes (e.g., invA) was observed during the establishment of persistence, whereas genes encoding important components of the T3SS and the Yops were down-regulated. This is accompanied by a change in the expression of the transcription factors of the Crp-CsrA-RovM-RovA regulatory cascade, indicating that these control factors determine the expression switch (Avican et al., 2015; Heine et al., 2018). Our findings suggest a life-style model for Y. pseudotuberculosis in which the bacteria change their gene expression profile from a virulent to an adapted phenotype, capable of persisting and spreading by fecal shedding. This further illustrates that the developed Tissue Dual RNA-seq approach is well suited to decipher the complexity of host-pathogen interactions during different stages of the infection. The comparative analysis between different strain isolates or mutants of potential virulence-relevant factors and regulators will help to decipher the complex interplay between of Y. pseudotuberculosis and the host response within distinct host niches.
Annotation and Characterization of Yersinia Transcripts
Since RNA-seq provided a single-nucleotide resolution transcriptional landscape of Y. pseudotuberculosis, it was used to map all genes of the chromosome and virulence plasmid (Nuss et al., 2015, 2017a; Figure 2). This led to the identification of new, so far not annotated and previously overlooked open reading frames (ORFs) and revealed a large set of antisense RNAs and small cis- and trans-encoded regulatory RNAs (Koo et al., 2011; Nuss et al., 2015, 2017a). Some of the identified antisense RNAs are long and cover whole genes or multiple genes of the virulence plasmid (Nuss et al., 2015). However, their individual impact on gene expression is still unclear. Moreover, certain variations of the RNA-seq protocols (e.g., treatment of the RNA with the terminator 5′-phosphate-dependent exonuclease short TEX), led to an enrichment of primary transcripts with 5′-triphosphate ends. Comparative analysis between the transcriptomes obtained with or without TEX treatment, called differential RNA-seq (dRNA-seq, Sharma and Vogel, 2014, Figures 1, 2), allowed global mapping of the transcription start sites of all transcripts. This led to the identification of promoter elements and the definition of operon structures, and improved the annotation of the Yersinia genome and virulence plasmid (Nuss et al., 2015).
The Yersinia RNA Structurome
In contrast to double-stranded DNA, RNA is a single-stranded molecule. However, it can undergo significant intramolecular base-pairing to take in an individual three-dimensional structure. It is known for quite some time that the intrinsic structure of an RNA impacts many cellular processes, most frequently by modulating its translation and/or decay. Protein synthesis is mainly affected by RNA structures, which influence the accessibility of the ribosome binding site (RBS). Based on the whole-genome transcriptomics study of Y. pseudotuberculosis several potential cis-regulatory RNA elements have been identified in long 5′-untranslated regions (5′-UTRs; >200 nt) of virulence, stress adaptation and metabolic genes in Y. pseudotuberculosis (Nuss et al., 2015). A bioinformatics approach identified 19 riboswitch-like elements (RLEs) among the long 5′-UTRs. Riboswitches are complex RNA structures that undergo structural changes in response to binding of small molecules, typically metabolites or ions, altering the expression of their downstream gene on the transcriptional or post-transcriptional level (Breaker, 2011; Serganov and Nudler, 2013). These features make them ideal to sense small molecule concentrations, e.g., nutrient and ion availability. Five of these RLEs showed features homologous to the threonine operon leader, the Moco (Molybdenum cofactor biosynthesis), FMN and the cobalamin riboswitch of E. coli and a cation-responsive riboswitch upstream of an Mg2+ transporter gene (corA) similar to mgtA of Salmonella (Nuss et al., 2015). However, the function of the majority of the riboswitch-like elements remains uncovered.
Structural alteration of a sensory RNA segment can also be triggered by temperature shifts. For instance, a thermo-controlled melting process of a double-stranded region, like a molecular zipper, is used to modulate gene expression of several bacterial heat shock and virulence-relevant genes (Kortmann and Narberhaus, 2012). One of these thermo-sensitive RNA elements (RNA thermometer) was identified upstream of lcrF (Table 1), the virulence plasmid-encoding gene for the master regulator of the T3SS/Yop machinery. The lcrF RNA thermometer is highly conserved between the human pathogenic Yersinia species and consists of a two stem-loop structure of which the second hairpin sequesters and hides the ribosome binding site (RBS) within its stem region at moderate temperatures (20–30°C). Elevated temperatures (37°C) lead to partial unfolding of the stem-loop, which renders the RBS accessible for the small subunit of the ribosome, leading to translation initiation (Hoe and Goguen, 1993; Böhme et al., 2012).
To investigate whether additional RNA thermometers or thermo-sensitive RNA elements are encoded on the Y. pseudotuberculosis genome, a structure RNA profile was recently generated of an entire transcriptome (Figure 2). For this purpose total RNA of the bacteria were isolated, incubated at 25, 37, and 42°C, and subsequently treated with single-strand and double-strand specific RNases. RNA-seq analysis of the resulting RNA fragments (PARS: parallel analysis of RNA structures) from 1,750 transcripts identified numerous temperature-sensitive RNAs (Rhigetti et al., 2016). This revealed that the RNA structurome of this pathogen is highly dynamic and encodes many more RNA thermometers. In total, 16 new RNA thermometers were discovered in 5′-UTRs of metabolic, heat stress, and virulence-relevant genes. Among them are many oxidative stress genes (sodA, sodB, sodC, and katA), and two very important virulence genes, encoding the cell adhesion and invasion factor Ail, contributing to host cell colonization and host resistance, and the CNFY toxin, triggering host tissue inflammation and damage (Rhigetti et al., 2016; Table 1). Besides the putative RNA thermometer, several small regulatory RNAs and tRNAs were also shown to be thermo-sensitive in this study. It will be interesting to investigate whether they promote and/or participate in the thermo-regulation of their target mRNA(s), and whether they are exploited by the pathogen to modify bacterial metabolism, physiology or pathogenicity upon entering a warm-blooded host, similar to the RNA thermometers.
Control by Regulatory RNAs
Small regulatory RNAs (sRNAs) are central regulators that play an essential role in various physiological and virulence associated processes in bacteria (Papenfort and Vogel, 2010, 2014; Heroven et al., 2017; Quereda and Cossart, 2017; Westermann, 2018). sRNAs are involved in the fine-tuning of these processes, allowing a rapid adaptation of pathogens to new environments. They have unique regulatory characteristics and control target gene expression post-transcriptionally through base-pairing with their transcripts. Such interaction can alter translation initiation or mRNA stability (Fröhlich and Vogel, 2009; Storz et al., 2011; Gorski et al., 2017). Functional analysis of sRNAs in different microorganisms revealed that often several sRNAs are part of the same regulatory cascade and sRNA-target network. This network typically includes multiple feedback loops and mixed regulatory circuits, illustrating the true complexity of RNA-based control system (Nitzan et al., 2017; Hör et al., 2018).
A myriad of sRNAs have been identified in the human pathogenic Yersinia species in independent high-throughput RNA-seq studies and are mainly referred to as Ysr (Yersinia small RNAs) (Koo et al., 2011; Koo and Lathem, 2012; Qu et al., 2012; Beauregard et al., 2013; Yan et al., 2013; Nuss et al., 2015, 2017a). The Ysrs are presumed Yersinia-specific sRNAs that were identified in an sRNA search in Y. pseudotuberculosis (Koo et al., 2011). A detailed list and the characteristics of these sRNAs are summarized in two recent reviews (Martínez-Chavarría and Vadyvaloo, 2015; Nuss et al., 2017b) (Figure 2, Table 1). Two aspects that are particularly striking are that (i) many sRNAs are directly regulated by the cAMP receptor protein (Crp) in response to growth phase and temperature, and that (ii) the majority of sRNAs are specific for Yersinia. This indicates that they are important players for the biological fitness and adaptation in their individual environmental and host niches, and reservoirs. A drawback of the latter feature is that most bioinformatic algorithms to identify potential targets of sRNAs rely on comparative genomics, i.e., functional and network analysis depending on sequence conservation between species (Wright et al., 2013), and are thus not suitable for the analysis of most Ysr sRNAs. Consequently, very little information is available about their molecular function and role for the pathogen. Nonetheless, the RNA chaperone Hfq, which mediates annealing of many sRNAs to their target mRNAs and protects them from ribonuclease cleavage or facilitates sRNA turnover or activity, was found to be required for virulence (Table 1). An hfq mutant of Y. pestis is impaired in its ability to resist phagocytosis and survive within macrophages at the initial stage of infection, and is highly attenuated in mice after subcutaneous or intravenous injection (Geng et al., 2009). Moreover, Hfq was found to regulate biofilm gut blockage that facilitates flea-borne transmission of Y. pestis (Rempe et al., 2012). Hfq of Y. pseudotuberculosis and Y. enterocolitica modulates the expression of multiple virulence-relevant cell surface structures such as adhesins/invasins (e.g., Ail, OmpX, MyfA pili, YadA, InvA) and expression of the T3SS/yop genes (Schiano et al., 2010, 2014; Schiano and Lathem, 2012; Kakoschke et al., 2014, 2016). Moreover, several Hfq-dependent alterations in the lipid A structure, motility, and biofilm formation defects, and changes in outer membrane vesicle synthesis were identified, which also impact virulence (Eddy et al., 2014; Leskinen et al., 2017). This strongly indicates that Hfq acts by controlling the expression of virulence-associated genes, probably in conjunction with small non-coding RNAs. However, up to now only very little is known about virulence-associated sRNAs and whether they are Hfq-dependent.
An initial analysis with a few Ysr sRNAs showed that Ysr29, Ysr35, and Ysr141 seem to play a role in pathogenesis (Koo et al., 2011; Schiano et al., 2014; Table 1). The specific target of Ysr35 remains unknown, but expression of multiple proteins was found to be affected in an ysr35 mutant. Among them are several general stress response factors such as the chaperones GroEL and DnaK, the peroxidase AhpC, the translational factors S1 (RpsA) and the ribosome recycling factor Rrf (Koo et al., 2011). The virulence plasmid-encoded sRNA Ysr141 was found to enhance the synthesis of LcrF and many effector proteins (YopJ, YopE, YopK, YpkA), whereby only a direct influence of Ysr141 on YopJ translation was documented (Schiano et al., 2014). Recent work has also identified multiple sRNAs of Y. pestis, which are upregulated within macrophages. Among them is Ysr170, which was shown to be important for intracellular replication (Li et al., 2016). Much more is known about conserved sRNAs and their impact on pathogenicity of Yersinia. The most prominent influence on pathogenesis has been shown for the global carbon storage regulator system (Csr) (Heroven et al., 2012; Romeo et al., 2013; Vakulskas et al., 2015; Kusmierek and Dersch, 2017; Nuss et al., 2017b). The Csr system of Yersinia consists of the small RNA-binding protein CsrA, which is highly conserved among many bacterial species, and the two non-coding sRNAs CsrB and CsrC. The CsrA protein typically affects the translation and/or stability of target mRNAs by binding to GGA motifs within the mRNA. Activity of CsrA is controlled and antagonized by the sRNAs CsrB and CsrC as these are able to sequester multiple CsrA molecules, which prevents binding of target transcripts (Romeo et al., 1993, 2013; Liu and Romeo, 1997; Liu et al., 1997; Romeo, 1998; Babitzke and Romeo, 2007; Romeo and Babitzke, 2018). In Yersinia, similar to other Enterobacteriaceae, the Csr system constitutes an important link between metabolic, stress, and virulence gene regulation by sRNAs, and plays a crucial role in virulence (Heroven et al., 2012; Kusmierek and Dersch, 2017; Nuss et al., 2017b). It has been found to control expression of the flagella master regulator FlhDC, the colonization factors InvA and PsaA, and multiple fitness and virulence-relevant cellular processes. These are e.g., motility/chemotaxis, biofilm formation, resistance against various stresses, including antibiotics and oxygen radicals, and adaptation of the central carbon metabolism by regulating the pyruvate-acetyl-CoA-tricarboxylate acid cycle flux (Heroven et al., 2008; Bücker et al., 2014; LeGrand et al., 2015; Willias et al., 2015; Kusmierek and Dersch, 2017). A very recent study of our group further demonstrated that the abundance of both CsrB and CsrC sRNAs in Y. pseudotuberculosis is reduced during the colonization of Peyer's patches compared to growth under different in vitro conditions (Nuss et al., 2017a). This indicates that more “free” CsrA is required during the infection process. In fact, loss of CsrA was shown to reduce the synthesis and completely abolish the secretion of the Yop effector proteins in Y. pseudotuberculosis (Nuss et al., 2017a). Most interestingly, CsrA also influences Yop secretion in Y. enterocolitica (Ozturk et al., 2017), although the overall outcome is quite different. This might be explained by the different growth conditions used for the Yop secretion assay or differences between the species.
Furthermore, RNA-seq approaches used to profile gene expression of Y. pseudotuberculosis and Y. pestis during host colonization revealed several other conserved sRNAs that are induced during infection. Among these sRNAs are RyhB1 and RyhB2—two sRNAs that are induced during iron starvation experienced in host tissue, SgrS—an sRNA upregulated under glucose-phosphate stress, and GlmY involved in cell wall synthesis (Table 1) (Massé and Gottesman, 2002; Vanderpool and Gottesman, 2004; Reichenbach et al., 2008; Urban and Vogel, 2008; Wadler and Vanderpool, 2009; Deng et al., 2012; Papenfort et al., 2012, 2013; Yan et al., 2013; Bobrovskyy and Vanderpool, 2014, 2016; Nuss et al., 2017a). However, lack of these sRNAs had only a mild effect on host colonization by the pathogens, indicating that they are implicated in the fine-tuning of virulence-relevant fitness processes.
Of the 80 antisense RNAs (asRNAs) that have been discovered to be part of the Y. pseudotuberculosis transcriptome, multiple are encoded on the virulence plasmid. Several were complementary to transcripts of the T3SS/Yop genes, although their influence on the expression of these virulence genes, and the influence on Yersinia virulence has not yet been investigated. However, one asRNA, CopA/incRNA, has been shown to be extremely important for the virulence process. CopA is implicated in the RNA-based control of the copy number of the virulence plasmid. The copy number of the Yersinia virulence plasmid, an IncFII plasmid, is controlled by the replicase RepA, which is tightly controlled and negatively regulated at the transcriptional and the post-transcriptional level. Under non-secretion conditions (e.g., in the absence of host cells), synthesis of RepA is repressed by (i) CopB, a transcriptional repressor that interacts with the repA promoter, and (ii) the asRNA CopA, that binds to the 5′ end of the longer of two repA transcripts preventing translation of a short leader peptide whose translation is coupled with and required for repA translation (Blomberg et al., 1990, 1992; Nordström, 2006; Wang et al., 2016; Pilla and Tang, 2018). Despite other IncFII plasmids, transcription of copA and copB of the Yersinia virulence plasmid are not constitutive. They are temperature-regulated and repressed during infection of the Peyer's patches, leading to a reduction of the ratio of the CopA asRNA and the repA transcript level, which results in an increase from 1–4 to up to 4–12 plasmid copies per bacterial cell during host tissue colonization (Wang et al., 2016). It could be demonstrated that up-regulation of the virulence plasmid copy number during the infection is crucial for virulence and that one of the secreted T3SS substrates (YopD) is involved. However, the molecular mechanism coupling copy number control to host cell binding still needs to be elucidated.
In summary, the identification of regulatory sRNAs and asRNAs using RNA-seq in the context of an infection process highlights that this approach is a powerful tool for the discovery of new infection-relevant regulatory processes. Application of newly developed RNA-seq technologies such as pulse-chase-expression global target searches (Massé et al., 2005; Papenfort et al., 2006; Westermann et al., 2016), coupling of RNA-seq with ribosome profiling (Guo et al., 2014; Wang et al., 2015), M2-tagged sRNA affinity purification coupled with RNA-seq (Lalaouna et al., 2015; Tomasini et al., 2017), RNA interaction by ligation and sequencing—short RIL-Seq (Melamed et al., 2016), or global small non-coding RNA target identification by ligation and sequencing—short GRIL-seq (Han et al., 2016) could help to identify direct targets of the regulatory RNAs and their role in Yersinia biology.
Translational Control
During translation mRNA is decoded for protein synthesis in a multi-step process consisting of initiation, elongation, termination, and recycling (Melnikov et al., 2012). This process is performed by the ribosome and several other translation associated components like tRNAs, initiation factors (IF), elongation factors (EF), and recycling factors (RF and RRF) (Melnikov et al., 2012). In addition to the complex transcriptional control strategies, gene expression of Yersinia is extensively regulated on the level of translation. Regulation of translation initiation occurs by changing the accessibility of the RBS for the small ribosomal subunit (30S, small subunit). As previously described, this can be achieved by masking the RBS with an RNA binding protein such as CsrA (Heroven et al., 2008, 2012) or an RNA thermo-loop (Baba et al., 1991; Böhme et al., 2012).
Other important riboregulators can modulate sRNA-mRNA interaction and influence the function of the translation machinery. For instance, the trans-translation control system or ribosome rescue system is composed of a small RNA-binding protein SsrB which transfers the polypeptide chain of ribosomes that are stalled on damaged or incomplete transcripts to the small, stable sRNA SsrA/tmRNA (Okan et al., 2006). The fact that SsrA/tmRNA is strongly upregulated in Y. pestis during the colonization of the lung and spleen and an ssrA mutant strain is strongly attenuated for virulence in a mouse infection model, strongly indicated that ribosome rescue is crucial for virulence (Okan et al., 2010; Yan et al., 2013). Reduced stress resistance and influence of the expression of the major virulence regulator LcrF in an ssrA/ssrB mutant strain could be the reason for this drastic phenotype (Okan et al., 2006, 2010).
One additional and very special translational control pathway of Yersinia virulence is mediated by the T3SS protein YopD, which, together with YopB, builds up the translocon pore in the eukaryotic membrane. Under non-secretion conditions, YopD is present in the bacterial cell and interacts with its chaperone LcrH, which prevents its degradation (Francis and Wolf-Watz, 1998; Edgren et al., 2012; Dewoody et al., 2013). Alone or bound to its chaperone LcrH, YopD seems to use sequences within the AU-rich regions in the proximity of the RBS of multiple yop-encoding mRNAs. This hinders translation initiation and reduces Yop protein expression (Williams and Straley, 1998; Anderson et al., 2002; Chen and Anderson, 2011). However, other control factors seem to contribute to YopD-mediated translational repression, as the identified 5′-UTR regions are important, but do not seem to be sufficient to promote this process. In fact, the secreted factor LcrQ (LcrM1 and LcrM2 in Y. enterocolitca) was found to contribute to post-transcriptional repression by YopD (Cambronne and Schneewind, 2002). Moreover, YopD was found to interact with the small 30S subunits of the ribosome. This indicates the presence of a different population of ribosomes (ribosome heterogeneity, Byrgazov et al., 2013), which negatively influences translation initiation complex formation (Kopaskie et al., 2013). How and to what extent these interactions affect translation of virulence-encoded genes is still unknown. YopD-binding could block the anti-RBS in the 16S rRNA from binding with the RBS. However, this alone would not explain why in particular translation of yop mRNAs is abrogated.
RNA Processing and Decay
Fast adaptation to changing surroundings is crucial for bacterial pathogens, such as Yersinia cycling between various environmental reservoirs, mammals, and humans in order to efficiently colonize their respective host and thrive in an occupied niche. Essential for this process is a controlled degradation of transcripts, which is mediated by a complex set of ribonucleases (RNases). The amount and type of RNases, their essentiality and collaboration in RNA metabolism is strongly depending on the investigated pathogen (e.g., Yersinia pseudotuberculosis > 15 RNases) (Arraiano et al., 2010; Lawal et al., 2011; Hui et al., 2014). The different RNases vary to a great extend in their mode of action, specificity for certain RNA features, and the nature of their targets in general (Arraiano et al., 2010; Lawal et al., 2011; Hui et al., 2014). Most bacteria, including all human pathogenic Yersinia species, contain low-specificity single-stranded endonucleases (e.g., RNase E), double-stranded endonucleases (e.g., RNAse III) and 3′-exonucleases (e.g., PNPase) (Mohanty and Kushner, 2016). RNA degradation occurs typically by consecutive endonucleolytic cleavage followed by exonucleolytic degradation from the 3′-end. The 5′-end is generally more stabilized due to the triphosphate end, but also the 3′-end can be rendered more stable toward RNases, e.g., by the presence of hairpin structures, RNA modification/polyadenylation or coverage by translating ribosomes. In addition to maturation and activation of certain transcripts (e.g., rRNA, tRNA), several conserved RNases were found to play a crucial role in global and/or targeted mRNA and sRNA turnover and also virulence in many pathogens (Clements et al., 2002; Ygberg et al., 2006; Lawal et al., 2011; Rosenzweig and Chopra, 2013).
In order to determine global mRNA decay rates, the bacterial culture is generally treated with the RNA polymerase inhibitor rifampicin prior to isolation of total RNA. To identify RNase targets and decay pathways, RNA-seq approaches with a transiently inactivated endoribonuclease followed by RNA-seq—short TIER-seq (Chao et al., 2017, Figure 1) or with RNase-deficient mutants were recently used to quantify transcript steady-state levels in Leskinen et al. (2015) and Chen et al. (2016).
In Yersinia, especially the RNases PNPase and RNase E were found to occupy well-established roles in the regulation of virulence factors and growth under several infection-relevant conditions (oxidative and cold stress) (Rosenzweig et al., 2007; Yang et al., 2008; Rosenzweig and Chopra, 2013). PNPase forms together with the RNase E, the helicase RhlB and the glycolytic enzyme enolase a multi protein complex, called the degradosome (Rice and Vanderpool, 2011). RNase E represents the scaffolding enzyme for this complex hyperstructure and while some of the affected virulence-relevant properties were associated with an intact buildup of the degradosome, others only require RNase E or PNPase activity and RNA binding ability (Henry et al., 2012; Rosenzweig and Chopra, 2013). A Yersinia pnp mutant is significantly less virulent in the mouse compared to the isogenic wild-type strain. It was shown that PNPase is required for optimal functioning of the T3SS. Yet, this did not seem to depend on its ribonuclease activity, but requires its S1 domain. As the pnp mutant strain also expressed similar or even enhanced levels of T3SS-encoded transcripts, it is most likely that its virulence attenuation is due to a limiting T3SS activity (Rosenzweig et al., 2005, 2007). Besides the RNases of the degradosome, there is an increasing amount of information that additional RNases contribute to virulence. For instance, the single-strand specific RNase YbeY, which is important for the processing of the 3′-end of the 16S rRNAs, was recently shown to influence the expression of many virulence genes (colonization factors, T3SS, and the amount of both Csr regulatory RNAs) (Leskinen et al., 2015). How precisely YbeY influences all these processes is still unclear, but the large number of differentially regulated fitness and virulence-relevant genes in the YbeY-deficient mutant correlates with the severe phenotype and illustrates a crucial role in cell homeostasis and virulence. Besides these RNA processing and decay pathways, other RNA degrading enzymes were found to be differentially regulated in response to various virulence-related growth conditions in vitro and during infection in vivo, indicating that more RNases may play a role in mRNA stability regulation. The exploitation of advanced RNA-seq combined with new target search approaches by coupling a UV crosslinking reaction to covalently link the RNase of interest to its target RNA will elucidate the role of RNases in various cellular processes (Waters et al., 2017). Determination of the cleavage sites and profiling of cleavage products of RNases, as established for other pathogens (Linder et al., 2014; Pobre and Arraiano, 2015; Redko et al., 2016; Chao et al., 2017; Le Rhun et al., 2017), will enable us to obtain genome-wide cleavage maps of different RNases of yersiniae under virulence-relevant conditions and will facilitate unraveling their complex RNA degradation network.
Outlook: RNA Interactome and Regulatory Networks in Yersinia
The discovery of novel RNA-based regulatory strategies anticipates many exciting new insights into the complex post-transcriptional control network of virulence gene expression. Yet, determining the individual targets and interaction partners (protein and RNA) of the multitude of mRNAs, asRNAs, and sRNAs identified in Y. pseudotuberculosis during recent RNA-seq approaches is a time-consuming and very labor-intensive task. However, new approaches have been successfully established to generate a snapshot of the interactome of RNAs that bind to a particular RNA-binding protein, e.g., CsrA, Hfq, or YopD. Such RNA interactomes can be obtained by RNA immunoprecipitation followed by RNA-seq (RIP-seq) (Figure 2), by covalently linked protein-RNA interactions followed by RNA-seq (CLIP-seq) (Figure 2), RNA interaction by ligation and sequencing (RIL-seq), or cross-linking, ligation and sequencing of hybrids (CLASH) (Bilusic et al., 2014; Holmqvist et al., 2016; Melamed et al., 2016; Waters et al., 2017). These techniques cannot only be used to profile the individual targets and entire binding sites (positions and sequences) of an RNA-binding protein, they will also allow us to improve sRNA target prediction, and facilitate the discovery of new sRNA control circuits and regulatory pathways (Dugar et al., 2016; Heidrich et al., 2017; Michaux et al., 2017). Similarly, RNA-RNA hybrids can be enriched or cross-linked and then sequenced to obtain a genome-wide Yersinia RNA-RNA network as done for E. coli (Liu et al., 2017).
Other open questions concerning the RNA-based control network of Yersinia virulence factors are: How are they conserved or remodeled during evolution between different strains and species, and how do acquired variations change virulence gene expression and pathogenicity? Numerous species- and strain-specific sensory and regulatory RNAs and many intra-species variations of certain sRNAs have been discovered in human pathogenic Yersiniae over the past years. For instance the PhoP regulator, controlling expression of the CsrC sRNA is not expressed in Y. pseudotuberculosis strain YPIII, but in IP32953 (Pisano et al., 2014). Moreover, the stability of the CsrC RNAs of strain YPIII and IP32953 differs significantly due to a 20 nt insertion in CsrC of IP32953, which renders the transcript more susceptible to degradation (Nuss et al., 2014). Based on the fact that the Csr system is crucial for T3SS/Yop expression and virulence (Bücker et al., 2014; Nuss et al., 2017a), it seems likely that even small variations (patho-specific alterations) between closely related strains could have a significant influence on the pathogen's potential to readjust and adapt to different hosts, host niches and reservoirs.
Another future task will be to elucidate the precise molecular mechanism how the RNA-binding translocator protein YopD controls translation of yop mRNAs. One technique to investigate ribosome-associated regulation, is ribosome profiling (Ribo-seq, Figure 1). With this method, all in vivo positions of extracted ribosomes on mRNAs (polysomes) can be identified enabling a dynamic view of ribosome movement along all mRNAs of a bacterial cell at a certain growth condition (Li et al., 2012; Ingolia, 2014). The function of ribosome subpopulations, such as those bound by YopD could be investigated by crosslinking of the ribosomes to the mRNA followed by affinity purification of the resulting YopD-ribosomal complex.
Ongoing characterization of the identified RNA-based control mechanisms of Yersinia will also have to address how the different riboregulators and RNAs contribute to the colonization of different host niches and whether and how they are implicated in the reprogramming of the pathogen, e.g., Y. pseudotuberculosis during the switch from the acute to the persistent infection mode. As regulatory RNAs, such as CsrB and CsrC, are implicated in the control of the regulator RovA, which forms a bistable switch, leading to high- and low-invasive subpopulations (Nuss et al., 2016), it is important to investigate the impact of riboregulators and non-coding RNAs on the formation of heterogeneous subpopulation with different virulence-relevant features. A major advance for this endeavor would be the development of single-cell RNA-seq approaches, in which the transcriptome of multiple individual bacterial cells could be profiled in different host niches and hosts during the entire course of an infection. So far single-cell RNA-seq has mainly been used for eukaryotic cells, however when applied to single bacteria this approach still suffers from technical limitations, which will have to be overcome in the future. This includes the establishment of more robust analyses and a more effective single-cell lysis and cDNA synthesis (Zhang et al., 2018). The next challenge will then be to analyze and compare the huge amount of generated data in an optimized and standardized integrative manner. This demands the establishment of (i) high quality, and free accessible standardized data-bases and (ii) the development of innovative bioinformatics protocols for data analysis and interpretation that allow integration of different technical and experimental approaches (e.g., different RNA-seq techniques, different OMICs, strains, tissues, and time-points of infection). Once achieved, this will give a comprehensive view on the complex RNA biology of Yersinia and will open up a new level of our understanding of virulence control of bacterial pathogens.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
Work on this topic was supported by Deutsche Forschungsgemeinschaft (DE616/6, DE616/7), and the Helmholtz Society. PD is member and supported by the Germany Center for Infection Research (DZIF) under grant number DZIF-TTU 06.801.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Anderson, D. M., Ramamurthi, K. S., Tam, C., and Schneewind, O. (2002). YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J. Bacteriol. 184, 1287–1295. doi: 10.1128/JB.184.5.1287-1295.2002
Arraiano, C. M., Andrade, J. M., Domingues, S., Guinote, I. B., Malecki, M., Matos, R. G., et al. (2010). The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34, 883–923. doi: 10.1111/j.1574-6976.2010.00242.x
Avican, K., Fahlgren, A., Huss, M., Heroven, A. K., Beckstette, M., Dersch, P., et al. (2015). Reprogramming of Yersinia from virulent to persistent mode revealed by complex in vivo RNA-seq analysis. PLoS Pathog. 11:e1004600. doi: 10.1371/journal.ppat.1004600
Baba, K., Takeda, N., and Tanaka, M. (1991). Cases of Yersinia pseudotuberculosis infection having diagnostic criteria of Kawasaki disease. Contrib. Microbiol. Immunol. 12, 292–296.
Babitzke, P., and Romeo, T. (2007). CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 10, 156–163. doi: 10.1016/j.mib.2007.03.007
Barquist, L., and Vogel, J. (2015). Accelerating discovery and functional analysis of small RNAs with new technologies. Annu. Rev. Genet. 49, 367–394. doi: 10.1146/annurev-genet-112414-054804
Beauregard, A., Smith, E. A., Petrone, B. L., Singh, N., Karch, C., Mcdonough, K. A., et al. (2013). Identification and characterization of small RNAs in Yersinia pestis. RNA Biol. 10, 397–405. doi: 10.4161/rna.23590
Bellows, L. E., Koestler, B. J., Karaba, S. M., Waters, C. M., and Lathem, W. W. (2012). Hfq-dependent, co-ordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol. Microbiol. 86, 661–674. doi: 10.1111/mmi.12011
Bilusic, I., Popitsch, N., Rescheneder, P., Schroeder, R., and Lybecker, M. (2014). Revisiting the coding potential of the E. coli genome through Hfq co-immunoprecipitation. RNA Biol. 11, 641–654. doi: 10.4161/rna.29299
Blomberg, P., Nordström, K., and Wagner, E. G. (1992). Replication control of plasmid R1: RepA synthesis is regulated by CopA RNA through inhibition of leader peptide translation. EMBO J. 11, 2675–2683. doi: 10.1002/j.1460-2075.1992.tb05333.x
Blomberg, P., Wagner, E. G., and Nordström, K. (1990). Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 9, 2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x
Bobrovskyy, M., and Vanderpool, C. K. (2014). The small RNA SgrS: roles in metabolism and pathogenesis of enteric bacteria. Front. Cell. Infect. Microbiol. 4:61. doi: 10.3389/fcimb.2014.00061
Bobrovskyy, M., and Vanderpool, C. K. (2016). Diverse mechanisms of post-transcriptional repression by the small RNA regulator of glucose-phosphate stress. Mol. Microbiol. 99, 254–273. doi: 10.1111/mmi.13230
Böhme, K., Steinmann, R., Kortmann, J., Seekircher, S., Heroven, A. K., Berger, E., et al. (2012). Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518. doi: 10.1371/journal.ppat.1002518
Bottone, E. J. (1997). Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10, 257–276.
Breaker, R. R. (2011). Prospects for riboswitch discovery and analysis. Mol. Cell 43, 867–879. doi: 10.1016/j.molcel.2011.08.024
Bücker, R., Heroven, A. K., Becker, J., Dersch, P., and Wittmann, C. (2014). The pyruvate-tricarboxylic acid cycle node: a focal point of virulence control in the enteric pathogen Yersinia pseudotuberculosis. J. Biol. Chem. 289, 30114–30132. doi: 10.1074/jbc.M114.581348
Byrgazov, K., Vesper, O., and Moll, I. (2013). Ribosome heterogeneity: another level of complexity in bacterial translation regulation. Curr. Opin. Microbiol. 16, 133–139. doi: 10.1016/j.mib.2013.01.009
Cambronne, E. D., and Schneewind, O. (2002). Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5' untranslated region of yop mRNA. J. Bacteriol. 184, 5880–5893. doi: 10.1128/JB.184.21.5880-5893.2002
Chao, Y., Li, L., Girodat, D., Förstner, K. U., Said, N., Corcoran, C., et al. (2017). In vivo cleavage map illluminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell 65, 39–51. doi: 10.1016/j.molcel.2016.11.002
Chen, R., Weng, Y., Zhu, F., Jin, Y., Liu, C., Pan, X., et al. (2016). Polynucleotide phosphorylase regulates multiple virulence factors and the stabilities of small RNAs RsmY/Z in Pseudomonas aeruginosa. Front. Microbiol. 7:247. doi: 10.3389/fmicb.2016.00247
Chen, Y., and Anderson, D. M. (2011). Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol. Microbiol. 80, 966–980. doi: 10.1111/j.1365-2958.2011.07623.x
Clements, M. O., Eriksson, S., Thompson, A., Lucchini, S., Hinton, J. C., Normark, S., et al. (2002). Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. U.S.A. 99, 8784–8789. doi: 10.1073/pnas.132047099
Cummings, J. H., and Macfarlane, G. T. (1997). Role of intestinal bacteria in nutrient metabolism. J. Parenter. Enteral Nutr. 21, 357–365. doi: 10.1177/0148607197021006357
Deng, Z., Liu, Z., Bi, Y., Wang, X., Zhou, D., Yang, R., et al. (2014). Rapid degradation of Hfq-free RyhB in Yersinia pestis by PNPase independent of putative ribonucleolytic complexes. Biomed Res. Int. 2014:798918. doi: 10.1155/2014/798918
Deng, Z., Meng, X., Su, S., Liu, Z., Ji, X., Zhang, Y., et al. (2012). Two sRNA RyhB homologs from Yersinia pestis biovar microtus expressed in vivo have differential Hfq-dependent stability. Res. Microbiol. 163, 413–418. doi: 10.1016/j.resmic.2012.05.006
Dewoody, R. S., Merritt, P. M., and Marketon, M. M. (2013). Regulation of the Yersinia type III secretion system: traffic control. Front. Cell. Infect. Microbiol. 3:4. doi: 10.3389/fcimb.2013.00004
Dugar, G., Svensson, S. L., Bischler, T., Wäldchen, S., Reinhardt, R., Sauer, M., et al. (2016). The CsrA-FliW network controls polar localization of the dual-function flagellin mRNA in Campylobacter jejuni. Nat. Commun. 7:11667. doi: 10.1038/ncomms11667
Eddy, J. L., Gielda, L. M., Caulfield, A. J., Rangel, S. M., and Lathem, W. W. (2014). Production of outer membrane vesicles by the plague pathogen Yersinia pestis. PLoS ONE 9:e107002. doi: 10.1371/journal.pone.0107002
Edgren, T., Forsberg, A., Rosqvist, R., and Wolf-Watz, H. (2012). Type III secretion in Yersinia: injectisome or not? PLoS Pathog. 8:e1002669. doi: 10.1371/journal.ppat.1002669
Francis, M. S., and Wolf-Watz, H. (1998). YopD of Yersinia pseudotuberculosis is translocated into the cytosol of HeLa epithelial cells: evidence of a structural domain necessary for translocation. Mol. Microbiol. 29, 799–813. doi: 10.1046/j.1365-2958.1998.00973.x
Fröhlich, K. S., and Vogel, J. (2009). Activation of gene expression by small RNA. Curr. Opin. Microbiol. 12, 674–682. doi: 10.1016/j.mib.2009.09.009
Geng, J., Song, Y., Yang, L., Feng, Y., Qiu, Y., Li, G., et al. (2009). Involvement of the post-transcriptional regulator of Hfq in Yersinia pestis virulence. PLoS ONE 4:e6213. doi: 10.1371/journal.pone.0006213
Görke, B., and Vogel, J. (2008). Noncoding RNA control of the making and breaking of sugars. Genes Dev. 22, 2914–2925. doi: 10.1101/gad.1717808
Gorski, S. A., Vogel, J., and Doudna, J. A. (2017). RNA-based recognition and targeting: sowing the seeds of specificity. Nat. Rev. Mol. Cell Biol. 18, 215–228. doi: 10.1038/nrm.2016.174
Guo, M. S., Updegrove, T. B., Gogol, E. B., Shabalina, S. A., Gross, C. A., and Storz, G. (2014). MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 28, 1620–1634. doi: 10.1101/gad.243485.114
Han, K., Tjaden, B., and Lory, S. (2016). GRIL-seq provides a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation. Nat. Microbiol. 2:16239. doi: 10.1038/nmicrobiol.2016.239
Heidrich, N., Bauriedl, S., Barquist, L., Li, L., Schoen, C., and Vogel, J. (2017). The primary transcriptome of Neisseria meningitidis and its interaction with the RNA chaperone Hfq. Nucleic Acids Res. 45, 6147–6167. doi: 10.1093/nar/gkx168
Heine, W., Beckstette, M., Heroven, A. K., Thiemann, S., Heise, U., Nuss, A. M., et al. (2018). Loss of CNFY toxin-induced inflammation drives Yersinia pseudotuberculosis into persistency. PLoS Pathog. 14:e1006858. doi: 10.1371/journal.ppat.1006858
Henry, A., Shanks, J., Van Hoof, A., and Rosenzweig, J. A. (2012). The Yersinia pseudotuberculosis degradosome is required for oxidative stress, while its PNPase subunit plays a degradosome-independent role in cold growth. FEMS Microbiol. Lett. 336, 139–147. doi: 10.1111/j.1574-6968.12000.x
Heroven, A., Böhme, K., Rohde, M., and Dersch, P. (2008). A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol. 68, 1179–1195. doi: 10.1111/j.1365-2958.2008.06218.x
Heroven, A. K., Böhme, K., and Dersch, P. (2012). The Csr/Rsm system of Yersinia and related pathogens: a post-transcriptional strategy for managing virulence. RNA Biol. 9, 379–391. doi: 10.4161/rna.19333
Heroven, A. K., Nuss, A. M., and Dersch, P. (2017). RNA-based mechanisms of virulence control in Enterobacteriaceae. RNA Biol. 14, 471–487. doi: 10.1080/15476286.2016.1201617
Hoe, N. P., and Goguen, J. D. (1993). Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175, 7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993
Holmqvist, E., Wright, P. R., Li, L., Bischler, T., Barquist, L., Reinhardt, R., et al. (2016). Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35, 991–1011. doi: 10.15252/embj.201593360
Hör, J., Gorski, S. A., and Vogel, J. (2018). Bacterial RNA biology on a genome scale. Mol. Cell 70, 785–799. doi: 10.1016/j.molcel.2017.12.023
Hör, J., and Vogel, J. (2017). Global snapshots of bacterial RNA networks. EMBO J. 36, 245–247. doi: 10.15252/embj.201696072
Hui, M. P., Foley, P. L., and Belasco, J. G. (2014). Messenger RNA degradation in bacterial cells. Annu. Rev. Genet. 48, 537–559. doi: 10.1146/annurev-genet-120213-092340
Ingolia, N. T. (2014). Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 15, 205–213. doi: 10.1038/nrg3645
Kakoschke, T., Kakoschke, S., Magistro, G., Schubert, S., Borath, M., Heesemann, J., et al. (2014). The RNA chaperone Hfq impacts growth, metabolism and production of virulence factors in Yersinia enterocolitica. PLoS ONE 9:e86113. doi: 10.1371/journal.pone.0086113
Kakoschke, T. K., Kakoschke, S. C., Zeuzem, C., Bouabe, H., Adler, K., Heesemann, J., et al. (2016). The RNA chaperone Hfq is essential for virulence and modulates the expression of four adhesins in Yersinia enterocolitica. Sci. Rep. 6:29275. doi: 10.1038/srep29275
Koo, J. T., Alleyne, T. M., Schiano, C. A., Jafari, N., and Lathem, W. W. (2011). Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia-specific small, noncoding RNAs required for virulence. Proc. Natl. Acad. Sci. U.S.A. 108, E709–E717. doi: 10.1073/pnas.1101655108
Koo, J. T., and Lathem, W. W. (2012). Global discovery of small noncoding RNAs in pathogenic Yersinia species. Adv. Exp. Med. Biol. 954, 305–314. doi: 10.1007/978-1-4614-3561-7_38
Koornhof, H. J., Smego, R. A. Jr., and Nicol, M. (1999). Yersiniosis. II: The pathogenesis of Yersinia infections. Eur. J. Clin. Microbiol. Infect. Dis. 18, 87–112. doi: 10.1007/s100960050237
Kopaskie, K. S., Ligtenberg, K. G., and Schneewind, O. (2013). Translational regulation of Yersinia enterocolitica mRNA encoding a type III secretion substrate. J. Biol. Chem. 288, 35478–35488. doi: 10.1074/jbc.M113.504811
Korth, M. M., and Sigel, R. K. (2012). Unusually high-affinity Mg(2+) binding at the AU-rich sequence within the antiterminator hairpin of a Mg(2+) riboswitch. Chem. Biodivers. 9, 2035–2049. doi: 10.1002/cbdv.201200031
Kortmann, J., and Narberhaus, F. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265. doi: 10.1038/nrmicro2730
Kröger, C., Colgan, A., Srikumar, S., Händler, K., Sivasankaran, S. K., Hammarlof, D. L., et al. (2013). An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 14, 683–695. doi: 10.1016/j.chom.2013.11.010
Kusmierek, M., and Dersch, P. (2017). Regulation of host-pathogen interactions via the post-transcriptional Csr/Rsm system. Curr. Opin. Microbiol. 41, 58–67. doi: 10.1016/j.mib.2017.11.022
Lalaouna, D., Carrier, M. C., Semsey, S., Brouard, J. S., Wang, J., Wade, J. T., et al. (2015). A 3' external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol. Cell 58, 393–405. doi: 10.1016/j.molcel.2015.03.013
Lawal, A., Jejelowo, O., Chopra, A. K., and Rosenzweig, J. A. (2011). Ribonucleases and bacterial virulence. Microb. Biotechnol. 4, 558–571. doi: 10.1111/j.1751-7915.2010.00212.x
Le Rhun, A., Lécrivain, A. L., Reimegard, J., Proux-Wera, E., Broglia, L., Della Beffa, C., et al. (2017). Identification of endoribonuclease specific cleavage positions reveals novel targets of RNase III in Streptococcus pyogenes. Nucleic Acids Res. 45, 2329–2340. doi: 10.1093/nar/gkw1316
LeGrand, K., Petersen, S., Zheng, Y., Liu, K. K., Ozturk, G., Chen, J. Y., et al. (2015). CsrA impacts survival of Yersinia enterocolitica by affecting a myriad of physiological activities. BMC Microbiol. 15:31. doi: 10.1186/s12866-015-0343-6
Leskinen, K., Pajunen, M. I., Varjosalo, M., Fernández-Carrasco, H., Bengoechea, J. A., and Skurnik, M. (2017). Several Hfq-dependent alterations in physiology of Yersinia enterocolitica O:3 are mediated by derepression of the transcriptional regulator RovM. Mol. Microbiol. 103, 1065–1091. doi: 10.1111/mmi.13610
Leskinen, K., Varjosalo, M., and Skurnik, M. (2015). Absence of YbeY RNase compromises the growth and enhances the virulence plasmid gene expression of Yersinia enterocolitica O:3. Microbiology 161, 285–299. doi: 10.1099/mic.0.083097-0
Li, G. W., Oh, E., and Weissman, J. S. (2012). The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541. doi: 10.1038/nature10965
Li, N., Hennelly, S. P., Stubben, C. J., Micheva-Viteva, S., Hu, B., Shou, Y., et al. (2016). Functional and structural analysis of a highly-expressed Yersinia pestis small RNA following infection of cultured macrophages. PLoS ONE 11:e0168915. doi: 10.1371/journal.pone.0168915
Linder, P., Lemeille, S., and Redder, P. (2014). Transcriptome-wide analyses of 5'-ends in RNase J mutants of a gram-positive pathogen reveal a role in RNA maturation, regulation and degradation. PLoS Genet. 10:e1004207. doi: 10.1371/journal.pgen.1004207
Liu, M. Y., Gui, G., Wei, B., Preston, J. F. III., Oakford, L., Yüksel, U., et al. (1997). The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272, 17502–17510. doi: 10.1074/jbc.272.28.17502
Liu, M. Y., and Romeo, T. (1997). The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 179, 4639–4642. doi: 10.1128/jb.179.14.4639-4642.1997
Liu, T., Zhang, K., Xu, S., Wang, Z., Fu, H., Tian, B., et al. (2017). Detecting RNA-RNA interactions in E. coli using a modified CLASH method. BMC Genomics 18:343. doi: 10.1186/s12864-017-3725-3
Martínez-Chavarría, L. C., and Vadyvaloo, V. (2015). Yersinia pestis and Yersinia pseudotuberculosis infection: a regulatory RNA perspective. Front. Microbiol. 6:956. doi: 10.3389/fmicb.2015.00956
Massé, E., and Gottesman, S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. doi: 10.1073/pnas.032066599
Massé, E., Vanderpool, C. K., and Gottesman, S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005
McCown, P. J., Corbino, K. A., Stav, S., Sherlock, M. E., and Breaker, R. R. (2017). Riboswitch diversity and distribution. RNA 23, 995–1011. doi: 10.1261/rna.061234.117
Melamed, S., Peer, A., Faigenbaum-Romm, R., Gatt, Y. E., Reiss, N., Bar, A., et al. (2016). Global mapping of small RNA-target interactions in bacteria. Mol. Cell 63, 884–897. doi: 10.1016/j.molcel.2016.07.026
Melnikov, S., Ben-Shem, A., Garreau De Loubresse, N., Jenner, L., Yusupova, G., and Yusupov, M. (2012). One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567. doi: 10.1038/nsmb.2313
Michaux, C., Holmqvist, E., Vasicek, E., Sharan, M., Barquist, L., Westermann, A. J., et al. (2017). RNA target profiles direct the discovery of virulence functions for the cold-shock proteins CspC and CspE. Proc. Natl. Acad. Sci. U.S.A. 114, 6824–6829. doi: 10.1073/pnas.1620772114
Mohanty, B. K., and Kushner, S. R. (2016). Regulation of mRNA decay in bacteria. Annu. Rev. Microbiol. 70, 25–44. doi: 10.1146/annurev-micro-091014-104515
Nitzan, M., Rehani, R., and Margalit, H. (2017). Integration of bacterial small RNAs in regulatory networks. Annu. Rev. Biophys. 46, 131–148. doi: 10.1146/annurev-biophys-070816-034058
Nordström, K. (2006). Plasmid R1–replication and its control. Plasmid 55, 1–26. doi: 10.1016/j.plasmid.2005.07.002
Nuss, A. M., Beckstette, M., Pimenova, M., Schmühl, C., Opitz, W., Pisano, F., et al. (2017a). Tissue dual RNA-seq: a fast discovery path for infection-specific functions and riboregulators shaping host-pathogen transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 114 E791–E800. doi: 10.1073/pnas.1613405114
Nuss, A. M., Heroven, A. K., and Dersch, P. (2017b). RNA regulators: formidable modulators of yersinia virulence. Trends Microbiol. 25, 19–34. doi: 10.1016/j.tim.2016.08.006
Nuss, A. M., Heroven, A. K., Waldmann, B., Reinkensmeier, J., Jarek, M., Beckstette, M., et al. (2015). Transcriptomic profiling of Yersinia pseudotuberculosis reveals reprogramming of the Crp regulon by temperature and uncovers Crp as a master regulator of small RNAs. PLoS Genet. 11:e1005087. doi: 10.1371/journal.pgen.1005087
Nuss, A. M., Schuster, F., Kathrin Heroven, A., Heine, W., Pisano, F., and Dersch, P. (2014). A direct link between the global regulator PhoP and the Csr regulon in Y. pseudotuberculosis through the small regulatory RNA CsrC. RNA Biol. 11, 580–593. doi: 10.4161/rna.28676
Nuss, A. M., Schuster, F., Roselius, L., Klein, J., Bücker, R., Herbst, K., et al. (2016). A precise temperature-responsive bistable switch controlling Yersinia virulence. PLoS Pathog. 12:e1006091. doi: 10.1371/journal.ppat.1006091
Okan, N. A., Bliska, J. B., and Karzai, A. W. (2006). A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2:e6. doi: 10.1371/journal.ppat.0020006
Okan, N. A., Mena, P., Benach, J. L., Bliska, J. B., and Karzai, A. W. (2010). The smpB-ssrA mutant of Yersinia pestis functions as a live attenuated vaccine to protect mice against pulmonary plague infection. Infect. Immun. 78, 1284–1293. doi: 10.1128/IAI.00976-09
Ozturk, G., Legrand, K., Zheng, Y., and Young, G. M. (2017). Yersinia enterocolitica CsrA regulates expression of the Ysa and Ysc type 3 secretion system in unique ways. FEMS Microbiol. Lett. 364:fnx204. doi: 10.1093/femsle/fnx204
Papenfort, K., Pfeiffer, V., Mika, F., Lucchini, S., Hinton, J. C., and Vogel, J. (2006). SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62, 1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x
Papenfort, K., Podkaminski, D., Hinton, J. C., and Vogel, J. (2012). The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc. Natl. Acad. Sci. U.S.A. 109, E757–E764. doi: 10.1073/pnas.1119414109
Papenfort, K., Sun, Y., Miyakoshi, M., Vanderpool, C. K., and Vogel, J. (2013). Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153, 426–437. doi: 10.1016/j.cell.2013.03.003
Papenfort, K., and Vogel, J. (2010). Regulatory RNA in bacterial pathogens. Cell Host Microbe 8, 116–127. doi: 10.1016/j.chom.2010.06.008
Papenfort, K., and Vogel, J. (2014). Small RNA functions in carbon metabolism and virulence of enteric pathogens. Front. Cell. Infect. Microbiol. 4:91. doi: 10.3389/fcimb.2014.00091
Pilla, G., and Tang, C. M. (2018). Going around in circles: virulence plasmids in enteric pathogens. Nat. Rev. Microbiol. 16, 484–495. doi: 10.1038/s41579-018-0031-2
Pisano, F., Heine, W., Rosenheinrich, M., Schweer, J., Nuss, A. M., and Dersch, P. (2014). Influence of PhoP and intra-species variations on virulence of Yersinia pseudotuberculosis during the natural oral infection route. PLoS ONE 9:e103541. doi: 10.1371/journal.pone.0103541
Pobre, V., and Arraiano, C. M. (2015). Next generation sequencing analysis reveals that the ribonucleases RNase II, RNase R and PNPase affect bacterial motility and biofilm formation in E. coli. BMC Genomics 16:72. doi: 10.1186/s12864-015-1237-6
Qu, Y., Bi, L., Ji, X., Deng, Z., Zhang, H., Yan, Y., et al. (2012). Identification by cDNA cloning of abundant sRNAs in a human-avirulent Yersinia pestis strain grown under five different growth conditions. Future Microbiol. 7, 535–547. doi: 10.2217/fmb.12.13
Quereda, J. J., and Cossart, P. (2017). Regulating Bacterial Virulence with RNA. Annu. Rev. Microbiol. 71, 263–280. doi: 10.1146/annurev-micro-030117-020335
Redko, Y., Galtier, E., Arnion, H., Darfeuille, F., Sismeiro, O., Coppée, J. Y., et al. (2016). RNase J depletion leads to massive changes in mRNA abundance in Helicobacter pylori. RNA Biol. 13, 243–253. doi: 10.1080/15476286.2015.1132141
Reichenbach, B., Maes, A., Kalamorz, F., Hajnsdorf, E., and Gorke, B. (2008). The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 36, 2570–2580. doi: 10.1093/nar/gkn091
Rempe, K. A., Hinz, A. K., and Vadyvaloo, V. (2012). Hfq regulates biofilm gut blockage that facilitates flea-borne transmission of Yersinia pestis. J. Bacteriol. 194, 2036–2040. doi: 10.1128/JB.06568-11
Rhigetti, F., Nuss, A. M., Twittenhoff, C., Beele, S., Urban, K., Will, S., et al. (2016). Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U.S.A. 113, 7237–7242. doi: 10.1073/pnas.1523004113
Rice, J. B., and Vanderpool, C. K. (2011). The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 39, 3806–3819. doi: 10.1093/nar/gkq1219
Romeo, T. (1998). Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29, 1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x
Romeo, T., and Babitzke, P. (2018). Global regulation by CsrA and its RNA antagonists. Microbiol. Spectr. 6:RWR-0009-2017. doi: 10.1128/microbiolspec.RWR-0009-2017
Romeo, T., Gong, M., Liu, M. Y., and Brun-Zinkernagel, A. M. (1993). Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J. Bacteriol. 175, 4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993
Romeo, T., Vakulskas, C. A., and Babitzke, P. (2013). Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15, 313–324. doi: 10.1111/j.1462-2920.2012.02794.x
Rosenzweig, J. A., and Chopra, A. K. (2013). The exoribonuclease polynucleotide phosphorylase influences the virulence and stress responses of yersiniae and many other pathogens. Front. Cell. Infect. Microbiol. 3:81. doi: 10.3389/fcimb.2013.00081
Rosenzweig, J. A., Chromy, B., Echeverry, A., Yang, J., Adkins, B., Plano, G. V., et al. (2007). Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol. Lett. 270, 255–264. doi: 10.1111/j.1574-6968.2007.00689.x
Rosenzweig, J. A., Weltman, G., Plano, G. V., and Schesser, K. (2005). Modulation of Yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 280, 156–163. doi: 10.1074/jbc.M405662200
Schiano, C. A., Bellows, L. E., and Lathem, W. W. (2010). The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect. Immun. 78, 2034–2044. doi: 10.1128/IAI.01046-09
Schiano, C. A., Koo, J. T., Schipma, M. J., Caulfield, A. J., Jafari, N., and Lathem, W. W. (2014). Genome-wide analysis of small RNAs expressed by Yersinia pestis identifies a regulator of the Yop-Ysc type III secretion system. J. Bacteriol. 196, 1659–1670. doi: 10.1128/JB.01456-13
Schiano, C. A., and Lathem, W. W. (2012). Post-transcriptional regulation of gene expression in Yersinia species. Front. Cell. Infect. Microbiol. 2:129. doi: 10.3389/fcimb.2012.00129
Schweer, J., Kulkarni, D., Kochut, A., Pezoldt, J., Pisano, F., Pils, M. C., et al. (2013). The cytotoxic necrotizing factor of Yersinia pseudotuberculosis (CNFY) enhances inflammation and Yop delivery during infection by activation of Rho GTPases. PLoS Pathog. 9:e1003746. doi: 10.1371/journal.ppat.1003746
Schwiesow, L., Lam, H., Dersch, P., and Auerbuch, V. (2015). Yersinia type III secretion system master regulator LcrF. J. Bacteriol. 198, 604–614. doi: 10.1128/JB.00686-15
Serganov, A., and Nudler, E. (2013). A decade of riboswitches. Cell 152, 17–24. doi: 10.1016/j.cell.2012.12.024
Sharma, C. M., and Vogel, J. (2014). Differential RNA-seq: the approach behind and the biological insight gained. Curr. Opin. Microbiol. 19, 97–105. doi: 10.1016/j.mib.2014.06.010
Sherwood, A. V., and Henkin, T. M. (2016). Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374. doi: 10.1146/annurev-micro-091014-104306
Smego, R. A., Frean, J., and Koornhof, and, H. J. (1999). Yersiniosis I: Microbiological and clinicoepidemiological aspects of plague and non-plague Yersinia infections. Eur. J. Clin. Microbiol. Infect. Dis. 18, 1–15. doi: 10.1007/s100960050219
Storz, G., Vogel, J., and Wassarman, K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891. doi: 10.1016/j.molcel.2011.08.022
Tomasini, A., Moreau, K., Chicher, J., Geissmann, T., Vandenesch, F., Romby, P., et al. (2017). The RNA targetome of Staphylococcus aureus non-coding RNA RsaA: impact on cell surface properties and defense mechanisms. Nucleic Acids Res. 45, 6746–6760. doi: 10.1093/nar/gkx219
Urban, J. H., and Vogel, J. (2008). Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 6:e64. doi: 10.1371/journal.pbio.0060064
Vakulskas, C. A., Potts, A. H., Babitzke, P., Ahmer, B. M., and Romeo, T. (2015). Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol. Mol. Biol. Rev. 79, 193–224. doi: 10.1128/MMBR.00052-14
Valentin-Weigand, P., Heesemann, J., and Dersch, P. (2014). Unique virulence properties of Yersinia enterocolitica O:3 - an emerging zoonotic pathogen using pigs as preferred reservoir host. Int. J. Med. Microbiol. 304, 824–834. doi: 10.1016/j.ijmm.2014.07.008
Vanderpool, C. K., and Gottesman, S. (2004). Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54, 1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x
Wadler, C. S., and Vanderpool, C. K. (2009). Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res. 37, 5477–5485. doi: 10.1093/nar/gkp591
Wang, H., Avican, K., Fahlgren, A., Erttmann, S. F., Nuss, A. M., Dersch, P., et al. (2016). Increased plasmid copy number is essential for Yersinia T3SS function and virulence. Science 353, 492–495. doi: 10.1126/science.aaf7501
Wang, J., Rennie, W., Liu, C., Carmack, C. S., Prévost, K., Caron, M. P., et al. (2015). Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Res. 43, 10308–10320. doi: 10.1093/nar/gkv1158
Waters, S. A., McAteer, S. P., Kudla, G., Pang, I., Deshpande, N. P., Amos, T. G., et al. (2017). Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E. EMBO J. 36, 374–387. doi: 10.15252/embj.201694639
Westermann, A. J. (2018). Regulatory RNAs in virulence and host-microbe interactions. Microbiol. Spectr. 6:RWR-0002-2017. doi: 10.1128/microbiolspec.RWR-0002-2017
Westermann, A. J., Förstner, K. U., Amman, F., Barquist, L., Chao, Y., Schulte, L. N., et al. (2016). Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529, 496–501. doi: 10.1038/nature16547
Williams, A. W., and Straley, S. C. (1998). YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J. Bacteriol. 180, 350–358.
Willias, S. P., Chauhan, S., Lo, C. C., Chain, P. S., and Motin, V. L. (2015). CRP-mediated carbon catabolite regulation of Yersinia pestis biofilm formation Is enhanced by the carbon storage regulator protein, CsrA. PLoS ONE 10:e0135481. doi: 10.1371/journal.pone.0135481
Wright, P. R., Richter, A. S., Papenfort, K., Mann, M., Vogel, J., Hess, W. R., et al. (2013). Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. U.S.A. 110, E3487–E3496. doi: 10.1073/pnas.1303248110
Yan, Y., Su, S., Meng, X., Ji, X., Qu, Y., Liu, Z., et al. (2013). Determination of sRNA expressions by RNA-seq in Yersinia pestis grown in vitro and during infection. PLoS ONE 8:e74495. doi: 10.1371/journal.pone.0074495
Yang, J., Jain, C., and Schesser, K. (2008). RNase E regulates the Yersinia type 3 secretion system. J. Bacteriol. 190, 3774–3778. doi: 10.1128/JB.00147-08
Ygberg, S. E., Clements, M. O., Rytkönen, A., Thompson, A., Holden, D. W., Hinton, J. C., et al. (2006). Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 74, 1243–1254. doi: 10.1128/IAI.74.2.1243-1254.2006
Keywords: RNA thermometer, RNA stability, RNA processing, Csr/Rsm system, virulence, gene regulation, small regulatory RNAs
Citation: Knittel V, Vollmer I, Volk M and Dersch P (2018) Discovering RNA-Based Regulatory Systems for Yersinia Virulence. Front. Cell. Infect. Microbiol. 8:378. doi: 10.3389/fcimb.2018.00378
Received: 31 July 2018; Accepted: 05 October 2018;
Published: 25 October 2018.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Kai Papenfort, Ludwig-Maximilians-Universität München, GermanySatish Raina, Gdansk University of Technology, Poland
Erik Holmqvist, Uppsala University, Sweden
Copyright © 2018 Knittel, Vollmer, Volk and Dersch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Dersch, petra.dersch@helmholtz-hzi.de
† These authors have contributed equally to this work
 Vanessa Knittel
Vanessa Knittel Ines Vollmer†
Ines Vollmer† Petra Dersch
Petra Dersch