- 1Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
- 2College of Forestry, Nanjing Forestry University, Nanjing, China
Hyphantria cunea (Drury) is a globally important forest pest. We found that the Serratia marcescens Bizio strain SM1 had insecticidal activity against H. cunea, but the transcriptomic response of H. cunea to SM1 were not clear. Therefore, we performed full-length sequencing of the transcriptomes of H. cunea larvae infected with SM1 and the control group. A total of 1,183 differentially expressed genes (DEGs) were identified by comparing the group infected with SM1 and the control group, including 554 downregulated genes and 629 upregulated genes. We found many downregulated genes in metabolic pathways. Furthermore, some of these downregulated genes were involved in cellular immunity, melanization, and detoxification enzymes, which showed that SM1 weakened H. cunea immunity. In addition, genes in the juvenile hormone synthesis pathway were upregulated, which was detrimental to the survival of H. cunea. This research analyzed the transcriptomic response of H. cunea to SM1 by high-throughput full-length transcriptome sequencing. The results provide useful information to explore the relationship between S. marcescens and H. cunea, and theoretical support for the application of S. marcescens and the control of H. cunea in the future.
1 Introduction
Hyphantria cunea (Drury) originated in North America, was introduced into east Asia and central Europe in the early 1940s, and then spread to Eurasia. At present, it is found in more than 30 countries and is an important forest pest worldwide (Ge et al., 2019). In 1979, it invaded the Dandong area of China’s Liaoning Province and gradually spread throughout China (Ji et al., 2003). Studies have shown that H. cunea has adapted to local climate conditions following its invasion and range expansion (Zhao et al., 2021). H. cunea has more than 600 host plants, including a variety of forestry and fruit trees, shrubs, herbs, and crops (Edosa et al., 2019). Due to its variety of host plants, large food intake, and strong reproductive and transmission ability, its invasion has seriously undermined the ecological environment, landscape, agriculture, forestry and many other aspects in China and caused great economic and environmental losses to China (Zhao, 2005). As an important pest control method, pathogenic microorganisms have been studied for their interactions with various pests (Xu et al., 2018; Bai et al., 2022). To control H. cunea, scientists have explored the use of various pathogenic microorganisms such as Bacillus thuringiensis, Beauveria bassiana, and H. cunea nucleopolyhedrovirus (HcNPV) (Zibaee et al., 2010; Aker & Tuncer, 2016; Sun et al., 2019).
Serratia marcescens Bizio (Enterobacterales: Yersiniaceae) can be used as a biocontrol bacterium to control some insect pests and plant pathogenic fungi (Nehme et al., 2007; Babashpour et al., 2012; Wang et al., 2013). Chen et al. (2001) isolated a strain of S. marcescens from dead Helicoverpa armigera (Hübner) and found that the strain had strong virulence on several insects, including Spodoptera exigua (Hübner), Pieris rapae (Linnaeus) and H. armigera. In addition, S. marcescens has also been found to be pathogenic to pests such as Diaphorina citri Kuwayama, Phthorimaea operculella (Zeller) and Spodoptera litura (Fabricius) (Aggarwal et al., 2017; Hu et al., 2018; Su et al., 2020). In our laboratory, we isolated a S. marcescens strain SM1 (hereafter refer to as SM1), and this strain had insecticidal activity against the larvae of H. cunea, indicating that SM1 has biocontrol potential against H. cunea (Feng et al., 2021).
We have previously showed that SM1 can induce strong immune responses of H. cunea by Solexa sequencing using an Illumina NovaSeq. Specifically, three signaling pathways, melanization and several cellular immune responses were activated, besides, several immune-related genes were also upregulated, including cytochrome P450s and uridine diphosphate-glycosyltransferases (Feng et al., 2021; Wang et al., 2021a). In addition to the induction of immune reactions, nonetheless, previous studies showed that many pathogenic bacteria can produce detrimental chemical toxins to affect a series of other genes expression. Thus, a comparative analysis of the transcriptomic response in H. cunea against SM1 using full-length single molecule real-time (SMRT) transcriptome sequencing may facilitate the understanding of the interaction between the pest and the bacterial pathogen. As a third-generation high-throughput sequencing technology, SMRT sequencing technology has been applied to transcriptome sequencing and analysis of many species, such as Agasicles hygrophila (Selman and Vogt), Bactrocera dorsalis (Hendel), Odontotermes formosanus (Shiraki), Rhopalosiphum padi (Linnaeus), Rhynchophorus ferrugineus (Olivier), and Sogatella furcifera (Horvath) (Jia et al., 2018; Chen et al., 2020; Feng et al., 2020; Yang et al., 2020; Ouyang et al., 2021; Wang et al., 2021b). Compared with the Sanger method and next-generation sequencing (NGS) technologies, SMRT sequencing technology has the advantages of being PCR-free, having a high speed, having long read lengths, and being capable of directly detecting epigenetic modifications, so it is very suitable for de novo genomic sequencing and high-quality assemblies of small genomes (Liu et al., 2015). In this study, the transcriptomic response on the metabolism, immunity and hormones of H. cunea to the infection of SM1 were analyzed using SMRT sequencing technology, which will provide the theoretical basis for the application of SM1 in the biocontrol of H. cunea.
2 Materials and methods
2.1 Insects and pathogenic bacteria
We collected H. cunea larvae in Huai’an, Jiangsu Province, China. In our laboratory, H. cunea larvae were fed with fresh poplar leaves in clear plastic boxes (20 cm×14 cm×10 cm) at an ambient temperature of 26 ± 1 °C with a light condition of 16 h light:8 h dark.
SM1 was isolated and stored in 25% glycerol at -80 °C.
2.2 Sample processing
SM1 was cultured on solid bacterial basal medium in the dark for 12 h at 30 °C to produce single colonies. A single colony of SM1 was proliferated with 50 mL seed culture medium at 200 r/min at 30 °C for 12 h. An appropriate amount of seed solution was added to 200 mL fermentation medium at 200 r/min at 30 °C for 36 h.
Healthy third instar H. cunea larvae were selected as experimental materials. Fresh poplar leaves of similar size were soaked in SM1 fermentation medium for 10-15 s, then two leaves with dry surface and 20 H. cunea larvae were placed into the triangular bottle (treatment group, SM_HC); leaves were soaked in sterile fermentation medium as the control group (CK). Live H. cunea larvae treated for 70 h were collected and stored at -80 °C for transcriptome sequencing. The test was repeated three times.
2.3 RNA sample preparation
We uesd the TRIzol method to extract total RNA from the samples. A Nanodrop 2000 spectrophotometer was used to determine the concentration and purity, and the integrity of RNA was detected by using agarose gel electrophoresis.
2.4 Library preparation and SMRT sequencing
The main process of library construction was as follows: The Clonetech SMARTerTM PCR cDNA Synthesis Kit was used to synthesize full-length cDNA of mRNA. Primer with Oligo dT was used for A-T base pairing with the polyA tail at the 3’ terminal of mRNA as primer for reverse synthesis of cDNA, and primer was added to the terminal of full-length cDNA synthesized in reverse. Full-length cDNA was obtained using PCR, PB magnetic beads were used to purify the amplified full-length cDNA, and small fragments of cDNA less than 1 kb were removed. The terminus of the full-length cDNA was repaired and connected to the SMRT dumbbell adapter. The fragments that were not connected to the adaptor were digested by exonuclease. PB magnetic beads were used to purify the fragments, and the sequencing library was obtained. After library construction, Qubit 3.0 was used for accurate quantification, and Agilent 2100 was used to detect the size of the library. After qualified detection, a PacBio sequencer was used to perform full-length transcriptome sequencing. The raw sequence generated by the PacBio sequencer totaled 86.4 Gbp and was deposited into the NCBI Sequence Read Archive (SRA) with accession number SRR22263802.
2.5 Sequel data output and quality control
SMRTlink software was used to preprocess the raw sequencing data based on the following main parameters: minimum predicted accuracy = 0.99, minimum number of passes = 3, maximum subread length = 15,000, minimum subread length = 50, and full-length transcript sequences were obtained by using the Iso-Seq analysis process. Subreads were obtained by splitting the single-molecule polymerase reads, and the subreads obtained from the same polymerase reads were self-corrected to form circular consensus sequences (CCS). CCSs were classified and full-length non-concatemer (FLNC) sequences were found by detecting chimeric sequences, primer sequences and 3′ poly-A sequences. The FLNC sequences were clustered, and redundancy was eliminated by the iterative clustering and error correction (ICE) tool of SMRTlink software and then further corrected by the arrow algorithm in SMRTlink, by which the obtained sequences were considered polished transcripts. Cd-hit software was used to cluster and remove redundancy (Li and Godzik, 2006), and the final full-length transcript sequence was used for subsequent analysis.
2.6 Functional annotation of transcripts
The basic functional annotations of full-length transcripts include NCBI nonredundant protein sequences (NR), Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Clusters of Orthologous Groups/Eukaryotic Orthologous Groups (COG/KOG) and Swiss-Prot Protein Sequence Database (Swiss-Prot) annotations. Isoform sequences were aligned to the NR, KEGG, COG/KOG and Swiss-Prot databases by diamond blastx, and protein IDs with high sequence similarity were obtained to gain the protein functional annotation information of the isoforms (Buchfink et al., 2015). According to the annotation results of five databases, the annotation status of all transcripts was statistically summarized, and the transcripts with ambiguous or contradictory annotations in the initial analysis were further verified by the BLAST function of the NCBI website (https://www.ncbi.nlm.nih.gov/).
2.7 Digital gene expression library preparation and analysis
The isoforms that had redundancy removed were used as the reference transcriptome sequences, and bowtie2 software was used to compare the clean reads of each sample with the references (Langmead, 2010). The comparison results of bowtie2 were counted by using RSEM, the number of reads from each sample to each transcript was obtained, and fragments per kilobase per million bases (FPKM) conversion was performed (Li and Dewey, 2011). The paired-end reads from the same fragment were counted as one fragment, and the expression levels of isoforms and transcripts were obtained. DESeq2 was used for differential expression analysis (Love et al., 2014). The threshold of the P-value in multiple tests was determined by using the false discovery rate (FDR) method. We set a threshold of FDR < 0.05 and an absolute value of log2fold change (FC) > 1 or log2FC < -1 to judge the significance of the difference in gene expression. Then, the genes expressed at different levels in the samples were further annotated by performing GO enrichment analysis and KEGG pathway enrichment analysis.
3 Results
3.1 Overview of the full-length transcriptome database
3.1.1 Sequencing data output and transcript clustering analysis
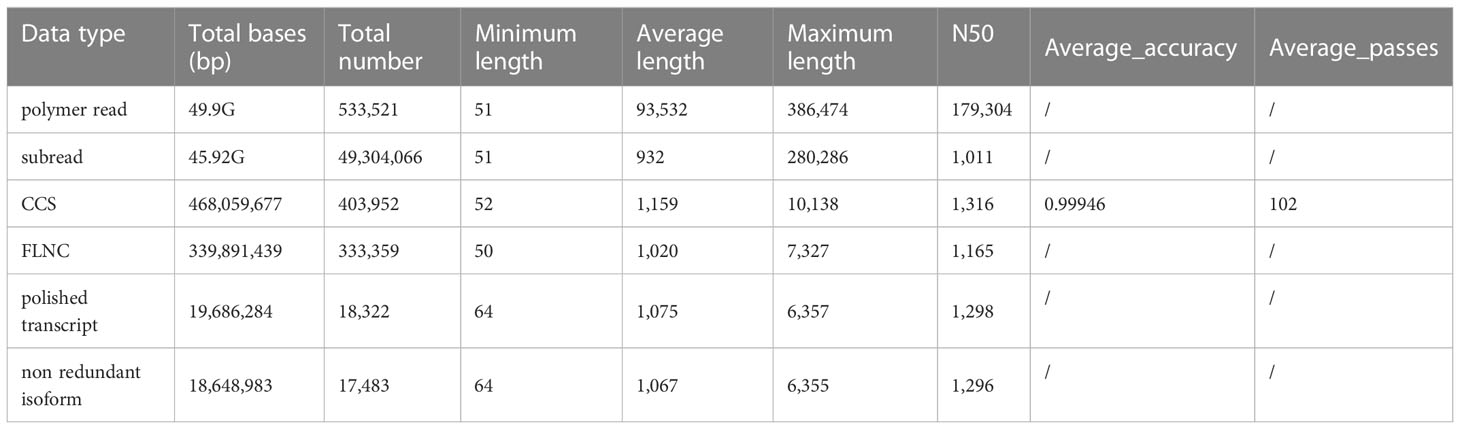
A total of 533,521 polymerase reads were generated by using PacBio SMRT sequencing technology. After removing the adapter sequences of the polymerase reads, the remaining sequence fragments are called subreads. 45.92 Gbp of subreads were obtained after preprocessing. CCSs are sequences with low error rates obtained by correcting sequencing errors among multiple sequencing results. In total, 403,952 CCSs were obtained. By classifying CCSs, a total of 333,359 FLNC sequences were identified. By clustering and correcting FLNC sequences using SMRTlink software, 18,322 polished transcripts were assembled. Using cd-hit software to cluster and eliminate redundancy, a total of 17,483 isoforms were assembled for further study. The above data details can be seen in Table 1.
3.1.2 Functional annotation of transcripts
In total, 11,912 transcripts were annotated in the NR database; 3,581 transcripts were annotated in the GO database; 6,580 transcripts were annotated in the KEGG database; 7,855 transcripts were annotated in the KOG database; and 8,761 transcripts were annotated in the SwissProt database. Additionally, 2,467 transcripts were annotated in all of these databases, and 5,564 transcripts were not annotated in any of these databases.
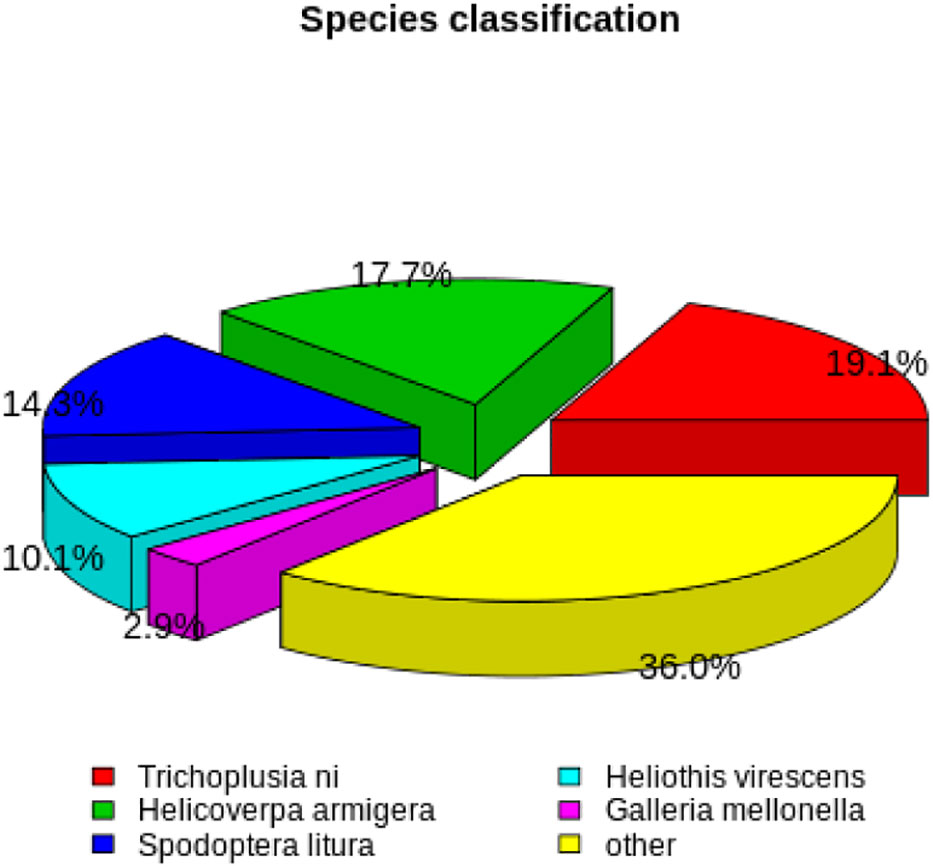
Based on NR annotations, 19.1% of the H. cunea sequences were aligned to Trichoplusia ni (Hübner), followed by H. armigera (17.7%), S. litura (14.3%), Heliothis virescens (Fabricius) (10.1%), Galleria mellonella (Linnaeus) (2.9%), and others (36.0%) (Figure 1).
Based on GO annotations, the transcriptome of H. cunea was distributed to 46 GO terms of the three main functional processes. In the biological process (BP) category, cellular process (1411) and metabolic process (1268) had the most transcripts. With respect to the cellular component (CC) category, the most abundant terms were cell (1506) and cell part (1469), followed by organelle (1222). Regarding the molecular function (MF) category, binding (1662) and catalytic activity (1609) were the subcategories with the most transcripts (Figure 2).
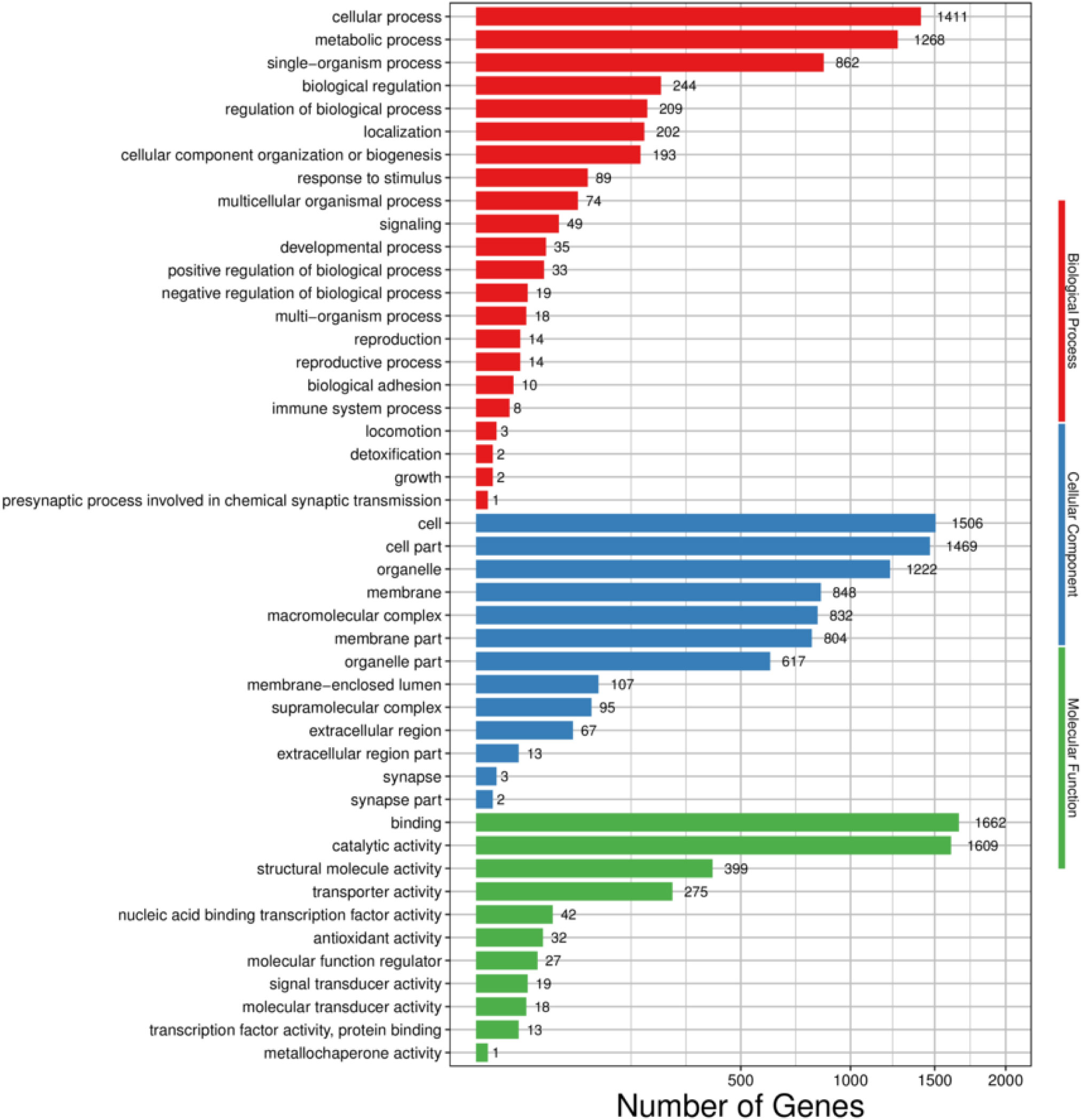
Figure 2 GO functional classifcations of H. cunea transcripts. Red, blue and green represent biological process, cellular component and molecular function, respectively. The x-axis represents the number of transcripts; the y-axis represents GO categories.
A total of 6,580 transcripts were annotated into 275 KEGG pathways. These KEGG pathways can be divided into five large branches, including cellular processes, environmental information processing, genetic information processing, metabolism and organismal systems, and further divided into 34 smaller branches. For cellular processes, transport and catabolism (868) had the largest numbers of transcripts. For environmental information processing, signal transduction (875) was the main subcategory. For genetic information processing, translation (783) was the most abundant term. For metabolism, energy metabolism (634) was the term with the largest numbers of transcripts. For organismal systems, the largest number of transcripts was assigned to the endocrine system (655) (Figure 3).
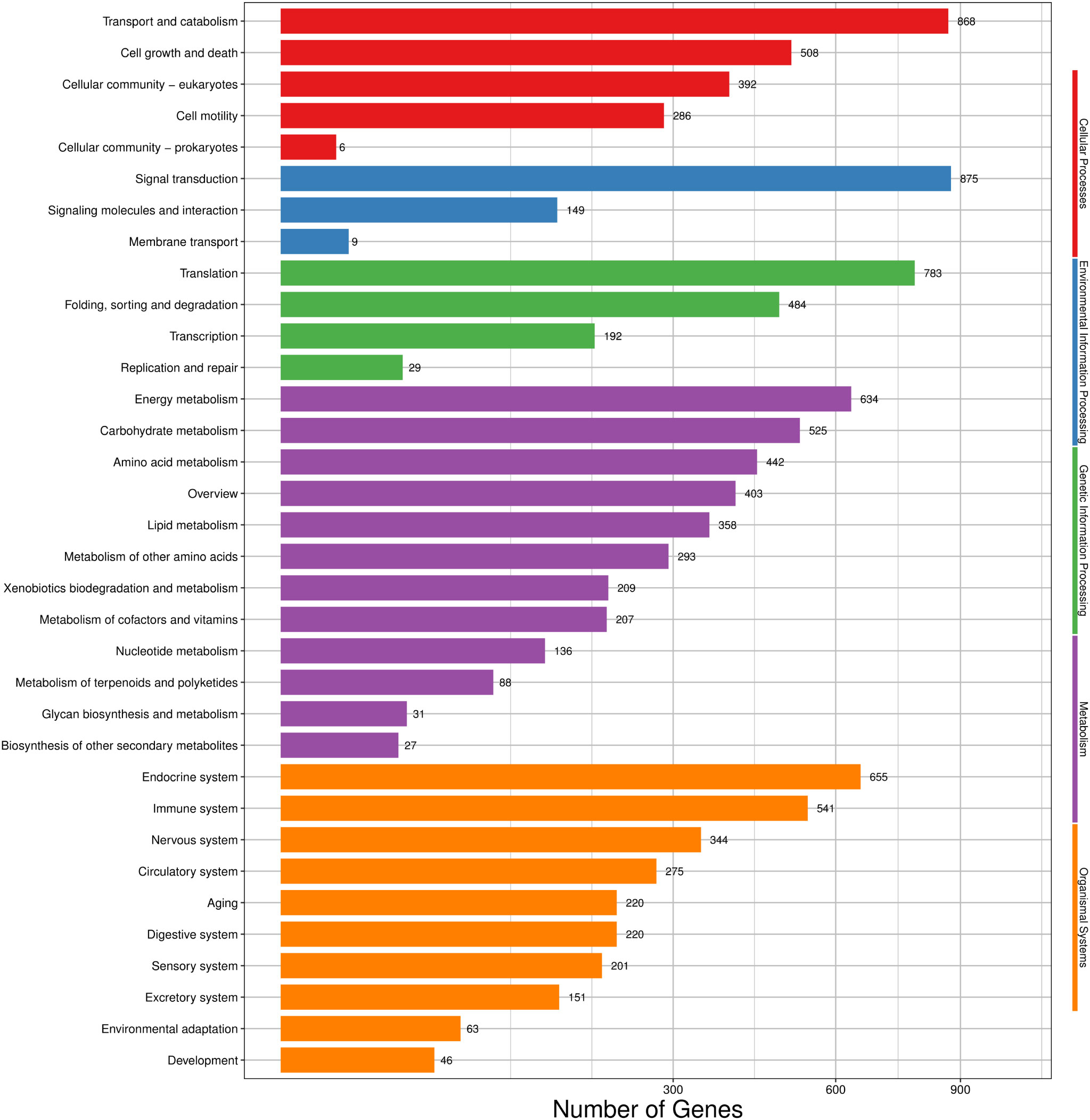
Figure 3 KEGG classification of H. cunea transcripts. Red, blue, green, purple and orange represent cellular processes, environmental information processing, genetic information processing, metabolism and organismal systems, respectively. The x-axis represents the number of transcripts; the y-axis represents KEGG pathway categories.
Based on the KOG database, 7,855 transcripts were annotated and grouped into 26 KOG groups. General function prediction only (1080) was the largest group in the 26 KOG groups, followed by Cytoskeleton (934) and Posttranslational modification, protein turnover, chaperones (837) (Figure 4).
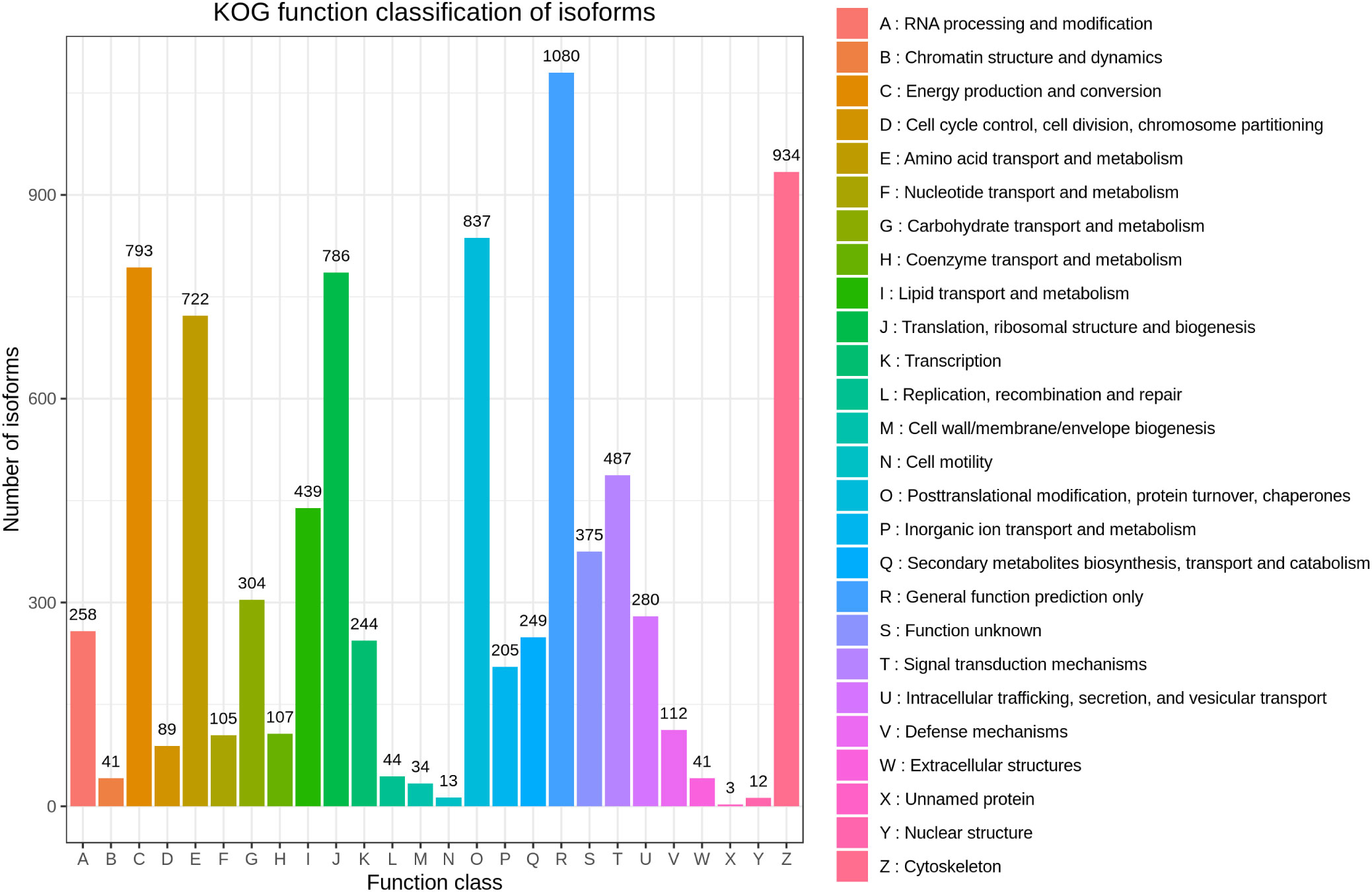
Figure 4 KOG annotation of H. cunea transcripts. The x-axis represents KOG categories; the y-axis represents the number of transcripts.
3.1.3 DEGs in H. cunea in response to SM1 infection
We attempted to identify the upregulated and downregulated DEGs in H. cunea infected with SM1 by using PacBio Sequel sequencing to investigate the transcriptomic response of H. cunea. To improve the accuracy of expression measurements, we combined the data from three biological replicates. Using the combined data, FPKM values were computed, and the results were compared between the replicate SM_HC and CK groups. When FDR < 0.05 and log2FC > 1 or log2FC < -1, DEGs were considered to be significantly different between the SM_HC and CK groups (Figure 5A). In total, 1,183 DEGs were identified, including 629 upregulated DEGs and 554 downregulated DEGs (Figure 5B).
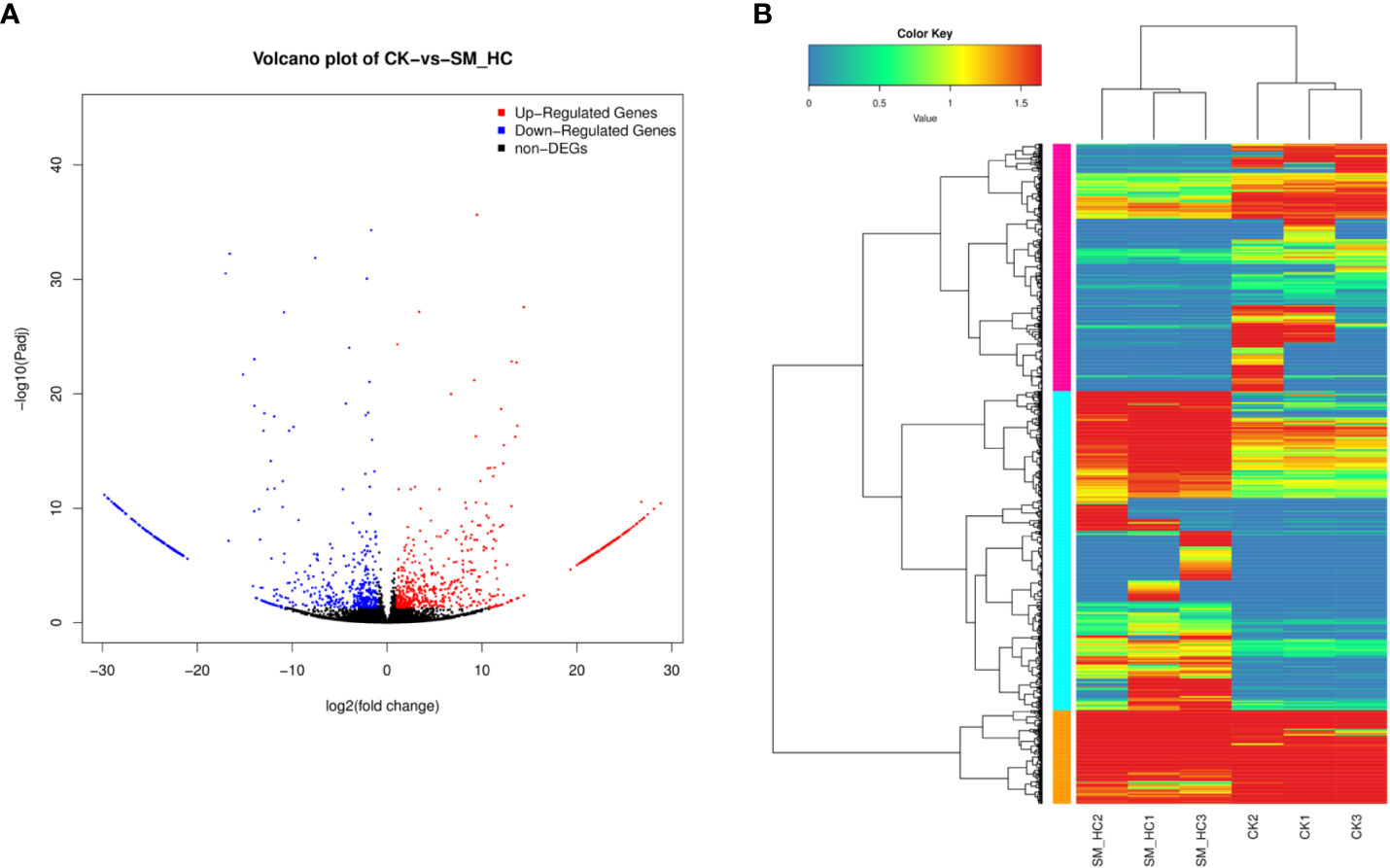
Figure 5 Overview of DEGs. (A) Comparison of DEGs between the CK library and SM_HC library. Red spots represent signifcantly upregulated genes and blue spots represent significantly downregulated genes. Black spots indicate no significant diferences in gene expression. (B) Heatmaps illustrating diferences in normalized log signal intensity for the identified H cunea genes. S. marcescens treatment groups were labeled as SM_HC1, SM_HC2 and SM_HC3, and control groups were labeled as CK1, CK2 and CK3. Red indicates genes expressed at high levels and blue indicates genes expressed at low levels. The colors from blue to red indicate gradually increasing expression.
Based on GO enrichment analysis, the functions of the 244 DEGs in H. cunea were classified into 34 groups of three main categories. In the BP category, cellular process (77) and metabolic process (76) were the most abundant terms, followed by single-organism process (61). With regard to the CC category, cell (88), cell part (87) and organelle (70) were highly enriched. In terms of the MF category, the main subcategories were catalytic activity (127) and binding (116) (Figure 6).
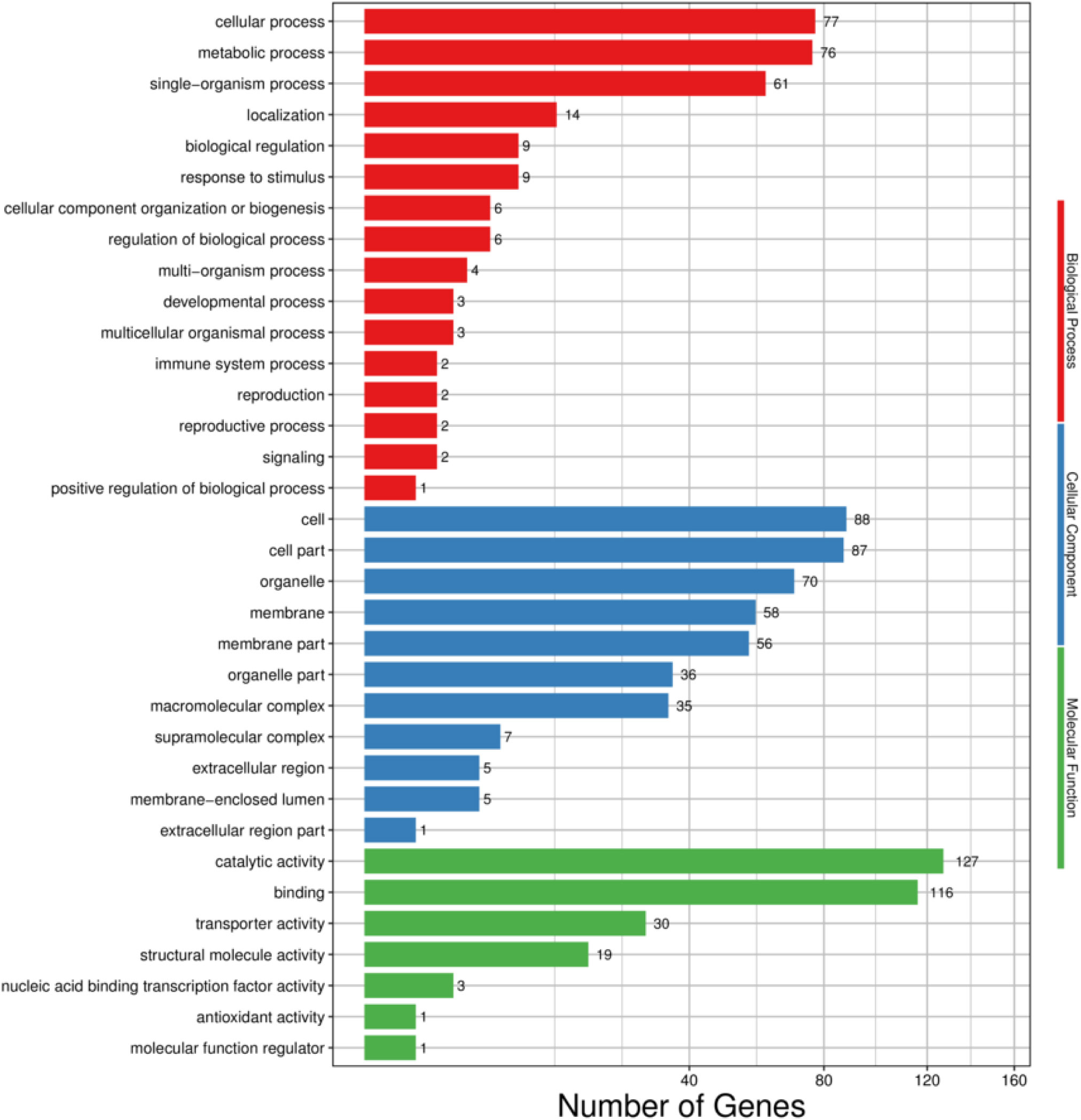
Figure 6 GO functional classifcation of DEGs in H. cunea. Red, blue and green represent biological process, cellular component and molecular function. The x-axis represents the number of transcripts; the y-axis represents GO categories.
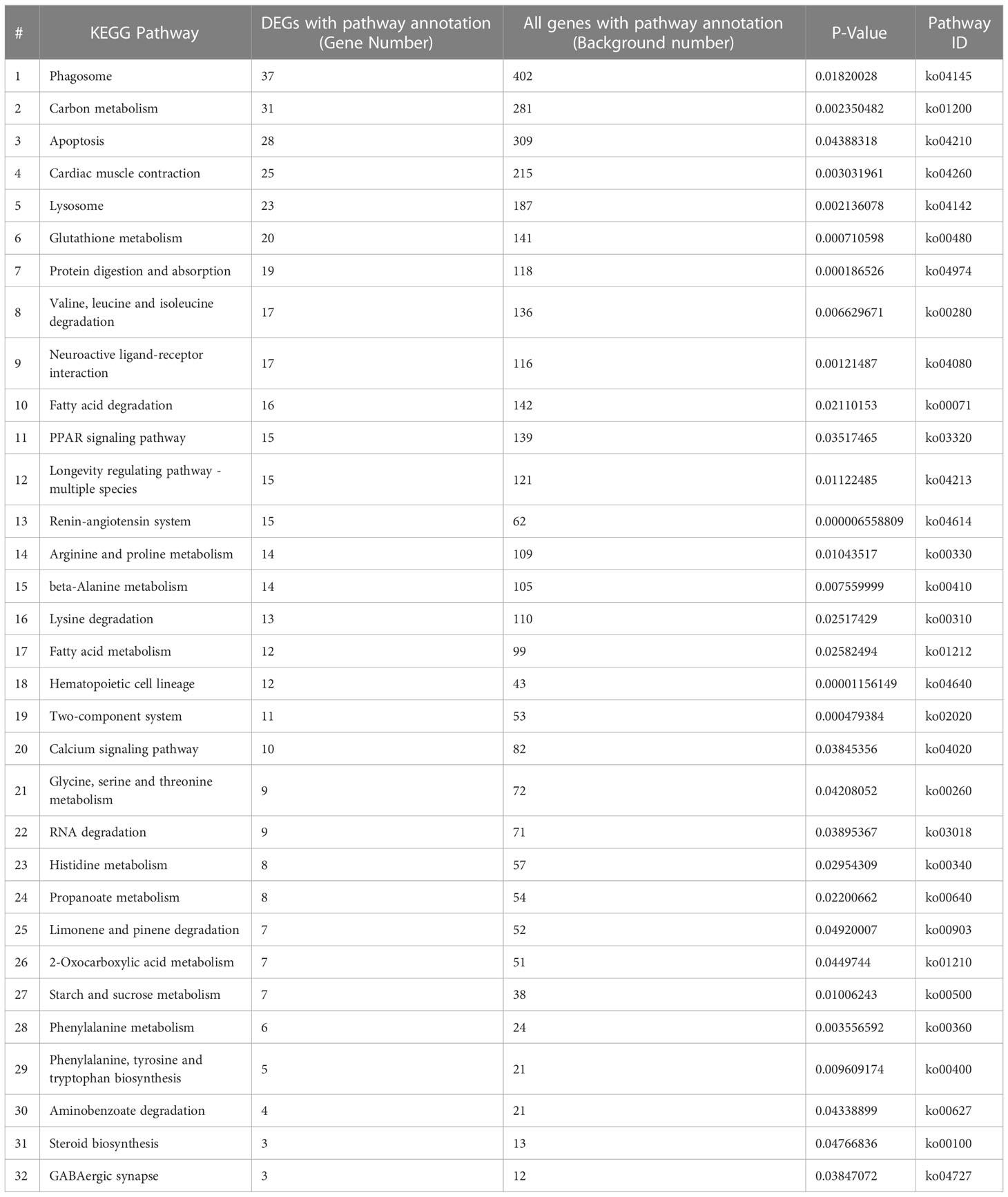
DEGs were mapped to typical KEGG pathways, identifying biological pathways that respond to SM1 treatment, and a total of 319 DEGs were distributed into 170 KEGG pathways (Supplementary Table S1). The pathways were considered highly enriched when the P-values were <0.05 (Table 2). Based on DEG enrichment analysis, the highly enriched pathway with the most unigenes was phagosome (37), followed by carbon metabolism (31), apoptosis (28) and cardiac muscle contraction (25).
3.2 Transcriptomic response of H. cunea to SM1 infection
3.2.1 SM1 suppressed the expression of metabolism-related genes of H. cunea
Metabolism of organisms is an important link for maintaining normal physiological activities. We searched the KEGG database and filtered out genes related to metabolic pathways of H. cunea, which could be divided into energy metabolism, metabolism of cofactors and vitamins, amino acid metabolism, metabolism of terpenoids and polyketides, nucleotide metabolism, xenobiotic biodegradation and metabolism, carbohydrate metabolism, and lipid metabolism. The results indicated that SM1 infection regulated a large number of genes in H. cunea, and many of them were downregulated. For example, a total of 20 genes were downregulated in amino acid metabolism (Figure 7A) and 13 genes were downregulated in the oxidative phosphorylation pathway (Figure 7B) (Supplementary Table S2). In this study, downregulated genes in amino acid metabolism were distributed in histidine-, glycine-, serine-, threonine-, alanine-, aspartate-, glutamate-, tryptophan-, tyrosine-, cysteine-, methionine-, valine-, leucine-, isoleucine-, lysine-, phenylalanine-, arginine- and proline-related metabolic pathways. Oxidative phosphorylation is an important part of biological energy metabolism. In our results, the downregulated genes in the oxidative phosphorylation pathway accounted for most of the total downregulated genes related to energy metabolism. A total of 16 genes were downregulated in energy metabolism, of which 13 genes were located in the oxidative phosphorylation pathway. The 13 genes included NADH dehydrogenase (NDUFAB1, NDUFB5, ND1 and ND5), cytochrome b (CYTB), cytochrome c oxidase (COX1), succinate dehydrogenase cytochrome b560 subunit (SDHC), Rieske iron-sulfur protein (RISP) and ATPase (ATPeVS1, ATPeV1E, ATPeF1A, ATPeF0B and ATPeF1B). In addition, V-type proton ATPase (ATPeVS1 and ATPeV1E) genes appeared not only in the oxidative phosphorylation pathway but also in cellular immune-related pathways. In summary, SM1 infection led to downregulation of some metabolism-related genes in H. cunea.
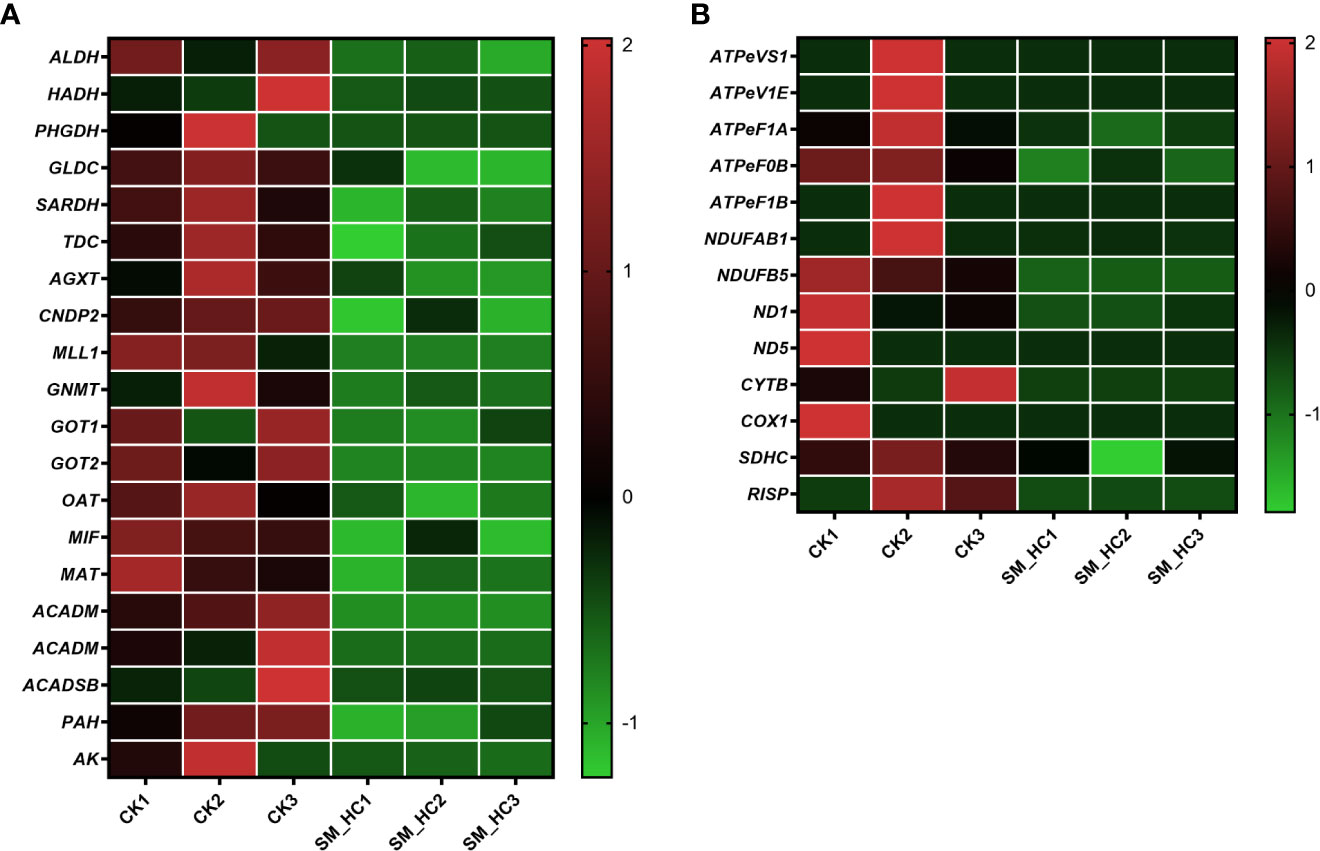
Figure 7 Heatmap analysis of downregulation genes in the metabolic pathways in H cunea infected by SM1. The control groups were labeled as CK1, CK2 and CK3. H cunea groups infected with SM1 were labeled as SM_HC1, SM_HC2 and SM_HC3. (A) Amino acid metabolism. (B) Oxidative phosphorylation.
3.2.2 SM1 suppressed the immunity of H. cunea
3.2.2.1 SM1 inhibited the expression of cellular immune-related genes in H. cunea
We detected many cellular immune-related genes, including genes in the apoptosis, lysosome, autophagy, endocytosis and phagosome pathways. Some of these genes were downregulated in H. cunea infected with SM1 compared with uninfected H. cunea (Figure 8A and Supplementary Table S2). Actin and tubulin are two kinds of proteins associated with the cytoskeleton. In H. cunea larvae infected with SM1, 9 actin genes, 2 α-tubulin genes and 3 β-tubulin genes were downregulated. Actin and α-tubulin genes appear in the apoptosis pathway and phagosome pathway, and β-tubulin is involved in the phagosome pathway. V-type proton ATPase subunit E (ATPeV1E) and V-type proton ATPase subunit S1 (ATPeVS1) each had one downregulated gene. These 2 genes are present in the phagosome pathway, while ATPeVS1 gene is also present in the lysosome pathway. Both cathepsin L (CTSL) and cathepsin D (CTSD) are involved in apoptosis, lysosome and autophagy pathways, and cathepsin L is also involved in phagocytosis. Each of the 2 cathepsins had one downregulated gene. In addition, 1 sphingomyelin phosphodiesterase (SMPD) gene, 2 CD53 antigen (CD53-1 and CD53-2) genes, 1 CD63 antigen (CD63) gene and 1 Niemann-Pick C2 protein (NPC2) gene were downregulated in the lysosome pathway, and 1 gene of stromal membrane-associated protein (SMAP) was downregulated in the endocytosis pathway. In short, SM1 infection downregulated many genes related to cellular immunity in H. cunea, which destroyed the cellular immune response.

Figure 8 Heatmap analysis of downregulation immune-related genes in H cunea infected by SM1. The control groups were labeled as CK1, CK2 and CK3. H cunea groups infected with SM1 were labeled as SM_HC1, SM_HC2 and SM_HC3. (A) Cellular immune. (B) Melanization. (C) Detoxification enzyme.
3.2.2.2 SM1 inhibited the melanization in H. cunea
Melanization is an important defense against pathogens in many arthropods. We searched the transcriptome data for genes associated with melanization. The results showed that SM1 infection weakened melanization of H. cunea larvae by regulating some genes (Figure 8B and Supplementary Table S2). As part of the protease cascade of melanization, serine proteases (SPs) promote the melanization process. In our study, two SPs (SPs1 and SPs2) genes were downregulated in H. cunea larvae infected with SM1. Trypsin, a class of serine proteases, also participates in and promotes melanization. In our data, 8 trypsin-like protein (TLP) genes and 1 trypsin-like serine protease (Tryp)-related gene were downregulated by SM1. As a negative regulator of the serine protease cascade pathway, two serine protease inhibitor (SPn1 and SPn2) genes were upregulated. Cuticular protein is a component of the melanic color pattern, and 2 genes related to cuticular protein (CPR and LCP) were downregulated in H. cunea larvae infected with SM1. In addition, as negative regulator of melanization, angiotensin converting enzyme (ACE) was upregulated after SM1 infection. In conclusion, SM1 infection inhibited the melanization in H. cunea.
3.2.2.3 SM1 caused the downregulation of detoxification enzyme genes in H. cunea
In insect detoxification, P450s (CYPs) and glutathione S-transferases (GSTs) both play important roles. Our results showed that the expression of 3 CYPs and 3 GSTs were downregulated in H. cunea infected with SM1 (Figure 8C and Supplementary Table S2). After sequence alignment and naming by the cytochrome P450 nomenclature committee, these three CYPs were determined to be CYP4M86, CYP341B54 and CYP6AE183, respectively. The three downregulated GSTs all belonged to the sigma class of cytosolic GSTs (GSTs1, GSTs2 and GSTs3). In this study, SM1 infection caused the downregulation of some detoxification enzyme genes in H. cunea.
3.2.3 SM1 induced juvenile hormone synthesis-related genes in H. cunea
Juvenile hormone (JH) is an important hormone regulating insect growth, development and reproduction. In our study, after SM1 infection, there were 5 DEGs in the JH synthetic pathway of H. cunea, and 4 genes were upregulated (Figure 9 and Supplementary Table S2). The expression of 1 gene of farnesyl diphosphate synthase (FDPS) that catalyzes isopentenyl diphosphate to form farnesyl diphosphate was upregulated. Two genes for farnesal dehydrogenase (FALDH1 and FALDH2), an enzyme that converts farnesal to farnesoic acid, were upregulated. One gene for juvenile hormone acid methyltransferase (JHAMT), which converts juvenile hormone acid to JH, was upregulated. In this study, most DEGs in the JH synthesis pathway were upregulated, indicating that JH synthesis in H. cunea larvae was induced to increase after SM1 infection.
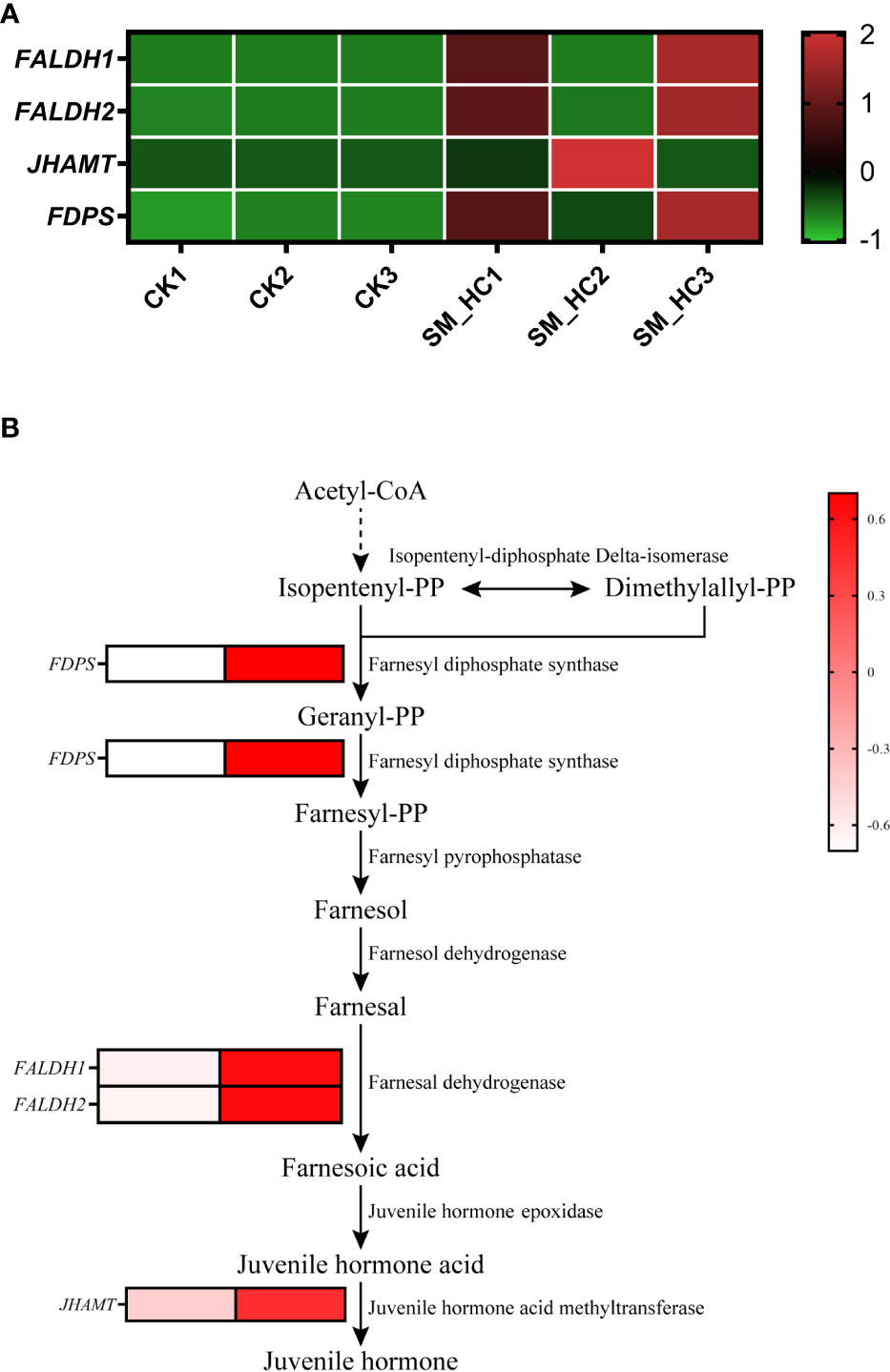
Figure 9 Changes in JH synthesis in H cunea infected by SM1. (A) Heatmap analysis of upregulation genes in JH synthesis using transcriptome data. The control groups were labeled as CK1, CK2 and CK3. H cunea groups infected with SM1 were labeled as SM_HC1, SM_HC2 and SM_HC3. (B) Diagram of genes related to JH synthesis in H cunea infected by SM1.
4 Discussion
In our study, the metabolic pathways affected by SM1 infection can be divided into 8 categories: energy metabolism, metabolism of cofactors and vitamins, amino acid metabolism, xenobiotic biodegradation and metabolism, nucleotide metabolism, carbohydrate metabolism, lipid metabolism, and metabolism of terpenoids and polyketides. Oxidative phosphorylation is an important pathway in energy metabolism and the main source of energy in organisms: it can produce ATP and provide energy for various life activities of organisms (Ruiz-Pesini et al., 2004; D’Elia et al., 2006). In this study, 13 genes in the oxidative phosphorylation pathway of H. cunea were downregulated after SM1 infection. In Epiphyas postvittana (Walker), some genes in the oxidative phosphorylation pathway were downregulated after infection with NPV (Gatehouse et al., 2007). A similar situation was observed in a Musca domestica Linnaeus study, after 48 h of bacterial infection, the genes related to oxidative phosphorylation were downregulated, indicating that energy homeostasis and mitochondrial function were disrupted in the late stage of infection (Tang et al., 2014). In Ceracris kiangsu Tsai, when oxidative phosphorylation was destroyed, the production of ATP was blocked, resulting in an imbalance between the supply and demand of energy metabolism and the failure to maintain the normal life activities of cells, ultimately leading to death (Zhao et al., 2004). Amino acids are important molecules in every organism, and various amino acids play different functions in the organism. Huang et al. (2009) found that after Bombyx mori (Linnaeus) was infected with Bacillus bombyseptieus Hartman, most genes related to metabolic pathways, including amino acid metabolism, were upregulated. This was because B. mori needed to meet the basic material and energy requirements for the growth and reproduction of B. bombyseptieus during infection. On the one hand, amino acids may be directly used by pathogens; on the other hand, when the body needs a large amount of energy, amino acids can be used as materials for energy supply, such as proline (Auerswald et al., 1998; Scaraffia and Wells, 2003). In addition, it may be related to the involvement of amino acids in the production of immune-related substances or cells (Xu et al., 2015). However, in our results, approximately half of the DEGs related to amino acid metabolism pathways were downregulated. We speculate that this situation may be related to the infection time and proliferation of the pathogen. With the aggravation of pathogen infection, the host is more severely damaged, and the proportion of upregulation and downregulation in amino acid metabolism pathway genes may gradually change. This needs to be verified in our future studies. According to our current results, SM1 infection caused metabolic dysfunction in larvae of H. cunea.
Cellular immunity is a defense response mediated by blood cells (hemocytes), including phagocytosis, encapsulation, nodulation, etc (Ali Mohammadie Kojour et al., 2020). The cellular uptake of particulates (>0.5 μ) within a plasma-membrane envelope is called phagocytosis, which is closely related to and partly overlaps the endocytosis of soluble ligands by fluid-phase macropinocytic and receptor pathways (Gordon, 2016). Actin can interact with many proteins and participate in many important biological processes, including immunity (Pollard and Cooper, 2009; Dominguez & Holmes, 2011). Phagocytosis is associated with the reorganization of the actin cytoskeleton, and actin can mediate phagocytosis and direct killing of bacteria (May and Machesky, 2001; Sandiford et al., 2015). When Scylla paramamosain Estampador lacked actin, they had higher morbidity and mortality after infection with Vibrio alginolyticus (Miyamoto et al.) Sakazaki or white spot syndrome virus (WSSV) (Sun et al., 2017b). V-ATPase is a class of important transporters that play an important role in plasma membrane proton transport in various cell types and are important for cellular processes (Wagner et al., 2004; Beyenbach and Wieczorek, 2006; Forgac, 2007; Hinton et al., 2009). Studies on many insects, including D. citri, Sphenophorus levis Vaurie, and Locusta migratoria (Linnaeus), have found that the interference of V-ATPase gene will affect the growth and survival of insects (Mohan et al., 2021; Guo et al., 2022; Liu et al., 2022). Studies have shown that RNAi knockdown of V-ATPase in Monochamus alternatus Hope inhibited the expression of immunity-related genes, including lysozyme, TAK1, and pelle (Li et al., 2022). In our results, there were downregulated genes related to the phagocytosis pathway in both actin and V-ATPase. This suggested that the relevant immune response was affected by SM1. Cathepsin is associated with various physiological processes such as immunity, aging, development and tissue remodeling (Conus, 2010; Stoka et al., 2016; Vidak et al., 2019), and its inadequate processing or activation can lead to cell death (Chwieralski et al., 2006). In studies of B. mori and Procambarus clarkii (Girard), CTSL was an important protease involved in the innate immune response (Dai et al., 2017; Sun et al., 2021). CTSL in the hepatopancreas of Litopenaeus vannamei (Boone) was downregulated after infection with WSSV (Zhai et al., 2021). CTSD has also been found to be associated with immune responses in invertebrates (Li et al., 2010; Menike et al., 2013; Yu et al., 2020). After knockout of CTSD from Eriocheir sinensis (H. Milne Edwards) with RNAi, the expression levels of many immune-related genes were decreased, and the mortality was increased after infection with Spiroplasma eriocheiris Wang (Ning et al., 2018). In this study, both CTSL and CTSD genes were downregulated after SM1 infection. The CTSL gene was not only located in the apoptosis, autophagy, and lysosome pathways similar to CTSD but also involved in the phagosome pathway. This suggested that these pathways were affected to some extent by SM1 infection. Lysosomes are the main degradation regions in cells. Lysosomes obtain substrates through phagocytosis, endocytosis, and autophagy and degrade them (Sagné et al., 2001; Saftig and Klumperman, 2009; Rong et al., 2011; Jézégou et al., 2012; Liu et al., 2012). In this study, in addition to CTSL and CTSD, downregulated genes in the lysosome pathway also included CD53, CD63, NPC2 and SMPD. In studies of other arthropods, these genes have indeed been classified as immune-related genes (Wang et al., 2019; Lawrie et al., 2020; Liao et al., 2020). Studies have reported that stromal membrane-associated protein is involved in the regulation of endocytosis (Tanabe et al., 2006), and its gene was downregulated in our study. In conclusion, the cellular immune response of H. cunea larvae was suppressed or destroyed by SM1 infection.
Melanization is an important physiological process in insects that plays an important role in wound healing, cuticle tanning, immunity and so on (Nakhleh et al., 2017). Tryp and TLP are important enzymes in the process of melanization, and they have been reported to participate in immune defense reactions in a variety of animals (Zhou et al., 2012; Sun et al., 2017a; Yang et al., 2021). In E. sinensis, when Tryp was silenced, the expression of prophenoloxidase was downregulated, and the mortality rate of E. sinensis infected by S. eriocheiris increased (Gao et al., 2019). In this study, we found that 1 Tryp gene and 8 TLP genes were downregulated. In addition, 2 SPs genes with the Tryp domain were also downregulated. Obviously, SM1 inhibited the melanization of H. cunea. Cuticle proteins are important components of insect cuticles (Suderman et al., 2006; Arakane et al., 2012; Noh et al., 2016). Loss of cuticle protein genes blocks the deposition of melanin (Xiong et al., 2017). It has been found that the degree of melanism in silkworm larvae is positively correlated with the expression of cuticle protein genes (Qiao et al., 2020). In our results, two cuticle protein genes were downregulated, which obviously has a negative effect on melanization. Some studies have found that ACE and SPn are both negative regulators of melanization in different insects (Tong et al., 2005; Chu et al., 2015; Huybrechts and Coltura, 2018; Kausar et al., 2018). In this study, some genes in these negative regulators were upregulated. This represents an inhibition of melanism. In conclusion, our results indicated that SM1 infection inhibited the melanization in H. cunea larvae.
GSTs are a class of enzymes widely present in aerobic organisms. GSTs play an important role in the detoxification of exogenous and endogenous substances and are also involved in a variety of life functions (Enayati et al., 2005; Li et al., 2007). Insect CYPs function in several aspects, including feeding, growth, development, and protection against xenobiotics. Protection against xenobiotics includes resistance to pesticides and tolerance to plant toxins (Scott and Wen, 2001). When insects are infected with pathogens, detoxification enzymes might be inhibited. For example, studies have found that CYP activity in Dysdercus koenigii (Fabricius) was decreased after infection with Aspergillus niger van Tieghem, and a large number of GSTs in Drosophila melanogaster Meigen were downregulated in the overall response to fungi (Trienens et al., 2017; Kumar et al., 2019). Sun et al. (2019) found that different amounts of GST genes were inhibited in H. cunea larvae by the stress of different concentrations of HcNPV. Similar results were also found in our study, in which 3 CYP genes and 3 GST genes in H. cunea were downregulated by SM1 infection. This indicated that SM1 infection caused the inhibition of CYPs and GSTs, which would reduce the ability of H. cunea to metabolize various harmful substances including the metabolites of SM1.
When pathogens infect insects, immune-related genes are induced to improve the immune ability of insects to fight the pathogens. SM1 is a gram-negative bacteria. Lipopolysaccharide (LPS), a component of the cell wall of SM1, was a possible proinflammatory cytokine stimulant that could greatly induce the immune-related gene expression in some insects (Chappell et al., 2000). Therefore, we have reported that SM1 can induce the immunity of H. cunea, which upregulates many immune genes in H. cunea (Feng et al., 2021; Wang et al., 2021a). On the other hand, studies have shown that LPS was an important virulence factor of many pathogenic bacteria (Whitfield and Trent, 2014), which could help bacteria resist the active component of the host innate immune response and promote the survival of bacteria in the body; at the same time, it had the characteristics of endotoxin, causing cytokine storm and disease, and even leading to the death of the host (Anwar and Choi, 2014; Feng et al., 2023). However, there is no report at home and abroad about how SM1 destroys H. cunea. Therefore, we found that the immunity of H. cunea was inhibited after SM1 infection in this study. This study is of great significance to further study the interaction between SM1 and H. cunea.
JH is associated with the regulation of insect molting and metamorphosis and is an important hormone related to insect growth and development. In addition, JH also influences ovarian development, caste differentiation, orchestrating phase polyphenism, reproductive behavior and many other aspects (Hartfelder, 2000). In our study, most of the genes related to the JH synthesis pathway were upregulated after the H. cunea larvae were infected with SM1. This has also been found in studies involving B. mori. A study found that the expression of many JH-related genes was upregulated after B. mori infection with NPV and B. bombyseptieus, and it was speculated that the pathogens could prolong the larval period of hosts by activating JH-related genes to facilitate the pathogens to obtain more nutrients for reproduction (Huang et al., 2009). In the case of S. exigua and S. litura, studies have also found that JH analogue (JHA) treatment of host larva would extend the larval period, improve the food conversion rate and intake, and ultimately improve the pathogen proliferation efficiency and yield by improving the nutritional level of host larva (Liu et al., 2005; Lasa et al., 2007; Li et al., 2009). On the other hand, studies have shown that JH is also involved in insect immunity. Flatt et al. (2008) found that JH was an immunosuppressant that reduces the expression of antimicrobial peptide (AMP) genes in D. melanogaster. A similar situation was also found in Aedes aegypti (Linnaeus) (Chang et al., 2021). Studies have found that treatment with JH or JHA for Eurygaster integriceps Puton, Neobellieria bullata (Parker) and G. mellonella could reduce the number of hemocytes and inhibit the formation of nodules (Franssens et al., 2006; Zibaee et al., 2012; Sezer and Ozalp, 2015). In addition, JH was found to inhibit spreeding of plasmatocytes in S. exigua (Kim et al., 2008). In conclusion, the increase in JH not only benefits the proliferation of pathogens in terms of nutrition but also inhibits the immune function of insects. Our results showed that SM1 infection induced the upregulation of genes related to JH synthesis in H. cunea, which was detrimental to H. cunea.
5 Conclusions
In this study, we analyzed the metabolic, immune and hormonal damage caused by SM1 in H. cunea larvae using full-length SMRT transcriptome sequencing. In metabolic pathways, represented by amino acid metabolism and oxidative phosphorylation, many genes in the pathways were downregulated. In immunity, cellular immunity, melanism and detoxification enzymes were negatively affected to some extent. Among hormones, genes related to the juvenile hormone synthesis pathway were upregulate, which was unfavorable for H. cunea larvae. This study revealed the transcriptomic response of H. cunea to the infection of SM1 and provided theoretical support and a basis for the improvement of the control effect of SM1 and the biological control of H. cunea.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI as SRR22263802.
Author contributions
Conceptualization, FT. Methodology, LZ, FT and ZW. Software, LZ. Validation, LZ and ZW. Formal analysis, LZ and XT. Investigation, LZ and XT. Resources, FT. Data curation, LZ, FT, XT and ZW, Writing-original draft, LZ. Writing-review and editing, FT. Visualization, LZ. Supervision, FT. Project administration, FT. Funding acquisition, FT. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Priority Academic Program Development Fund of Jiangsu Higher Education Institutions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1093432/full#supplementary-material
References
Aggarwal, C., Paul, S., Tripathi, V., Paul, B., Khan, M. A. (2017). Characterization of putative virulence factors of Serratia marcescens strain SEN for pathogenesis in Spodoptera litura. J. Invertebr. Pathol. 143, 115–123. doi: 10.1016/j.jip.2016.12.004
Aker, O., Tuncer, C. (2016). Pathogenicity of Beauveria bassiana on larvae of fall webworm, Hyphantria cunea (Drury)(Lepidoptera: Arctiidae) at different temperatures. Int. J. Entomol. Res. 1 (6), 16–20.
Ali Mohammadie Kojour, M., Han, Y. S., Jo, Y. H. (2020). An overview of insect innate immunity. Entomol. Res. 50 (6), 282–291. doi: 10.1111/1748-5967.12437
Anwar, M., Choi, S. (2014). Gram-negative marine bacteria: Structural features of lipopolysaccharides and their relevance for economically important diseases. Mar. Drugs 12 (5), 2485–2514. doi: 10.3390/md12052485
Arakane, Y., Lomakin, J., Gehrke, S. H., Hiromasa, Y., Tomich, J. M., Muthukrishnan, S., et al. (2012). Formation of rigid, non-flight forewings (elytra) of a beetle requires two major cuticular proteins. PloS Genet. 8 (4), e1002682. doi: 10.1371/journal.pgen.1002682
Auerswald, L., Schneider, P., Gäde, G. (1998). Utilisation of substrates during tethered flight with and without lift generation in the African fruit beetle Pachnoda sinuata (Cetoniinae). J. Exp. Biol. 201 (15), 2333–2342. doi: 10.1242/jeb.201.15.2333
Babashpour, S., Aminzadeh, S., Farrokhi, N., Karkhane, A., Haghbeen, D. K. (2012). Characterization of a chitinase (Chit62) from Serratia marcescens B4A and its efficacy as a bioshield against plant fungal pathogens. Biochem. Genet. 50 (9), 722–735. doi: 10.1007/s10528-012-9515-3
Bai, J. Y., Cao, J. Y., Zhang, Y., Xu, Z., Li, L., Liang, L. W., et al. (2022). Comparative analysis of the immune system and expression profiling of Lymantria dispar infected by Beauveria bassiana. Pestic. Biochem. Phys. 187, 105212. doi: 10.1016/j.pestbp.2022.105212
Beyenbach, K. W., Wieczorek, H. (2006). The V-type h+ ATPase: Molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209 (4), 577–589. doi: 10.1242/jeb.02014
Buchfink, B., Xie, C., Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12 (1), 59–60. doi: 10.1038/nmeth.3176
Chang, M. M., Wang, Y. H., Yang, Q. T., Wang, X. L., Wang, M., Raikhel, A. S., et al. (2021). Regulation of antimicrobial peptides by juvenile hormone and its receptor, methoprene-tolerant, in the mosquito Aedes aegypti. Insect Biochem. Molec. 128, 103509. doi: 10.1016/j.ibmb.2020.103509
Chappell, V. L., Le, L. X., LaGrone, L., Mileski, W. J. (2000). Stat proteins play a role in tumor necrosis factor alpha gene expression. Shock 14 (3), 400–403. doi: 10.1097/00024382-200014030-00027
Chen, X. W., Fan, H., Chen, R., Yu, X. J., Lan, H. X., Sun, B. Z. (2001). Study of the pathogenicity on pests and the utility in fields of Serratia marcescens. J. Tianjin Agr. Coll. 8, 28–30.
Chen, J., Yu, Y. Y., Kang, K., Zhang, D. W. (2020). SMRT sequencing of the full-length transcriptome of the white-backed planthopper Sogatella furcifera. PeerJ 8, e9320. doi: 10.7717/peerj.9320
Chu, Y., Zhou, F., Liu, Y., Hong, F., Wang, G. R., An, C. J. (2015). Ostrinia furnacalis serpin-3 regulates melanization cascade by inhibiting a prophenoloxidase-activating protease. Insect Biochem. Molec. 61, 53–61. doi: 10.1016/j.ibmb.2015.03.007
Chwieralski, C. E., Welte, T., Bühling, F. (2006). Cathepsin-regulated apoptosis. Apoptosis 11 (2), 143–149. doi: 10.1007/s10495-006-3486-y
Conus, S. (2010). Cathepsins and their involvement in immune responses. Swiss Med. Wkly. 140, 1–8. doi: 10.4414/smw.2010.13042
Dai, L. S., Chu, S. H., Yu, X. M., Li, Y. Y. (2017). A role of cathepsin l gene in innate immune response of crayfish (Procambarus clarkii). Fish Shellfish Immun. 71, 246–254. doi: 10.1016/j.fsi.2017.10.021
D’Elia, D., Catalano, D., Licciulli, F., Turi, A., Tripoli, G., Porcelli, D., et al. (2006). The MitoDrome database annotates and compares the OXPHOS nuclear genes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae. Mitochondrion 6 (5), 252–257. doi: 10.1016/j.mito.2006.07.001
Dominguez, R., Holmes, K. C. (2011). Actin structure and function. Annu. Rev. Biophys. 40, 169–186. doi: 10.1146/annurev-biophys-042910-155359
Edosa, T. T., Jo, Y. H., Keshavarz, M., Anh, Y. S., Noh, M. Y., Han, Y. S. (2019). Current status of the management of fall webworm, Hyphantria cunea: Towards the integrated pest management development. J. Appl. Entomol. 143 (1-2), 1–10. doi: 10.1111/jen.12562
Enayati, A. A., Ranson, H., Hemingway, J. (2005). Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 14 (1), 3–8. doi: 10.1111/j.1365-2583.2004.00529.x
Feng, K., Jiang, D. B., Luo, J., Tang, F. (2023). OfGNBP silencing enhances the toxicity of Serratia marcescens bizio (SM1) to Odontotermes formosanus (Shiraki). Pestic. Biochem. Phys. 189, 105306. doi: 10.1016/j.pestbp.2022.105306
Feng, K., Lu, X. Y., Luo, J., Tang, F. (2020). SMRT sequencing of the full-length transcriptome of Odontotermes formosanus (Shiraki) under Serratia marcescens treatment. Sci. Rep. 10 (1), 1–13. doi: 10.1038/s41598-020-73075-3
Feng, K., Luo, J., Ding, X., Tang, F. (2021). Transcriptome analysis and response of three important detoxifying enzymes to Serratia marcescens bizio (SM1) in Hyphantria cunea (Drury)(Lepidoptera: Noctuidae). Pestic. Biochem. Phys. 178, 104922. doi: 10.1016/j.pestbp.2021.104922
Flatt, T., Heyland, A., Rus, F., Porpiglia, E., Sherlock, C., Yamamoto, R., et al. (2008). Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J. Exp. Biol. 211 (16), 2712–2724. doi: 10.1242/jeb.014878
Forgac, M. (2007). Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Bio. 8 (11), 917–929. doi: 10.1038/nrm2272
Franssens, V., Smagghe, G., Simonet, G., Claeys, I., Breugelmans, B., De Loof, A., et al. (2006). 20-hydroxyecdysone and juvenile hormone regulate the laminarin-induced nodulation reaction in larvae of the flesh fly, Neobellieria bullata. Dev. Comp. Immunol. 30 (9), 735–740. doi: 10.1016/j.dci.2005.10.010
Gao, Q., Wei, P. P., Zhou, H. F., Hao, W. J., Hou, L. B., Ning, M. X., et al. (2019). The function of two trypsin-like serine proteases from Eriocheir sinensis involved in Spiroplasma eriocheiris infection. Aquaculture 501, 519–526. doi: 10.1016/j.aquaculture.2018.12.014
Gatehouse, H. S., Markwick, N. P., Poulton, J., Ward, V. K., Young, V., Wilson, S., et al. (2007). Effects of EppoNPV infection on gene expression in Epiphyas postvittana larvae. N. Z. Plant Prot. 60, 33–41. doi: 10.30843/nzpp.2007.60.4630
Ge, X. Z., He, S. Y., Zhu, C.Y., Wang, T., Xu, Z. C., Zong, S. X. (2019). Projecting the current and future potential global distribution of Hyphantria cunea (Lepidoptera: Arctiidae) using CLIMEX. Pest Manage. Sci. 75 (1), 160–169. doi: 10.1002/ps.5083
Gordon, S. (2016). Phagocytosis: an immunobiologic process. Immunity 44 (3), 463–475. doi: 10.1016/j.immuni.2016.02.026
Guo, C. F., Qiu, J. H., Hu, Y. W., Xu, P. P., Deng, Y. Q., Tian, L., et al. (2022). Silencing of V-ATPase-E gene causes midgut apoptosis of Diaphorina citri and affects its acquisition of huanglongbing pathogen. Insect Sci. 0, 1–13. doi: 10.1111/1744-7917.13146
Hartfelder, K. (2000). Insect juvenile hormone: From” status quo” to high society. Braz. J. Med. Biol. Res. 33, 157–177. doi: 10.1590/S0100-879X2000000200003
Hinton, A., Bond, S., Forgac, M. (2009). V-ATPase functions in normal and disease processes. Pflug. Arch. Eur. J. Phy. 457 (3), 589–598. doi: 10.1007/s00424-007-0382-4
Huang, L. L., Cheng, T. C., Xu, P. Z., Cheng, D. J., Fang, T., Xia, Q. Y. (2009). A genome-wide survey for host response of silkworm, Bombyx mori during pathogen Bacillus bombyseptieus infection. PloS One 4 (12), e8098. doi: 10.1371/journal.pone.0008098
Hu, W., Kuang, F., Lu, Z. J., Zhang, N., Chen, T. T. (2018). Killing effects of an isolated Serratia marcescens KH-001 on Diaphorina citri via lowering the endosymbiont numbers. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00860
Huybrechts, R., Coltura, L. (2018). Immune-induced angiotensin-converting enzyme assures the appearance of complementary peptides in Locusta migratoria for fine-tuning the innate immune response by inhibiting immune-activated phenoloxidase. Trends Entomol. 14, 11–16. doi: 10.31300/TENT.14.2018.11-16
Jézégou, A., Llinares, E., Anne, C., Kieffer-Jaquinod, S., O’Regan, S., Aupetit, J., et al. (2012). Heptahelical protein PQLC2 is a lysosomal cationic amino acid exporter underlying the action of cysteamine in cystinosis therapy. P. Natl. Acad. Sci. 109 (50), E3434–E3443. doi: 10.1073/pnas.1211198109
Jia, D., Wang, Y. X., Liu, Y. H., Hu, J., Guo, Y. Q., Gao, L. L., et al. (2018). SMRT sequencing of full-length transcriptome of flea beetle Agasicles hygrophila (Selman and vogt). Sci. Rep. 8 (1), 1–8. doi: 10.1038/s41598-018-20181-y
Ji, R., Xie, B. Y., Li, X. H., Gao, Z. X., Li, D. M. (2003). Research progress on the invasive species, Hyphantria cunea. Entomol. Knowl. 40 (1), 13–18.
Kausar, S., Abbas, M. N., Qian, C., Zhu, B., Gao, J., Sun, Y., et al. (2018). Role of Antheraea pernyi serpin 12 in prophenoloxidase activation and immune responses. Arch. Insect Biochem. 97 (2), e21435. doi: 10.1002/arch.21435
Kim, Y., Jung, S., Madanagopal, N. (2008). Antagonistic effect of juvenile hormone on hemocyte-spreading behavior of Spodoptera exigua in response to an insect cytokine and its putative membrane action. J. Insect Physiol. 54 (6), 909–915. doi: 10.1016/j.jinsphys.2008.03.012
Kumar, D., Kumari, S., Verma, D. (2019). Evaluation of Aspergillus niger as a biocontrol agent in the insect pest management of red cotton bug, Dysdercus koenigii (Heteroptera: Pyrrhocoridae). J. Sci. Res. 11 (2), 235–247. doi: 10.3329/jsr.v11i2.39286
Langmead, B. (2010). Aligning short sequencing reads with bowtie. Curr. Protoc. Bioinf. 32 (1), 11–17. doi: 10.1002/0471250953.bi1107s32
Lasa, R., Caballero, P., Williams, T. (2007). Juvenile hormone analogs greatly increase the production of a nucleopolyhedrovirus. Biol. Control 41 (3), 389–396. doi: 10.1016/j.biocontrol.2007.02.012
Lawrie, R. D., Mitchell, R. D., III, Deguenon, J. M., Ponnusamy, L., Reisig, D., Pozo-Valdivia, A. D., et al. (2020). Multiple known mechanisms and a possible role of an enhanced immune system in Bt-resistance in a field population of the bollworm, Helicoverpa zea: Differences in gene expression with RNAseq. Int. J. Mol. Sci. 21 (18)6528, 1–24. doi: 10.3390/ijms21186528
Liao, X. Z., Wang, C. G., Wang, B., Qin, H. P., Hu, S. K., Zhao, J. C., et al. (2020). Research into the hemocyte immune response of Fenneropenaeus merguiensis under decapod iridescent virus 1 (DIV1) challenge using transcriptome analysis. Fish Shellfish Immun. 104, 8–17. doi: 10.1016/j.fsi.2020.05.053
Li, C. B., Cai, Y. X., Ding, C. X., Wang, F. H., Li, G. H., Pang, Y. (2009). Effect of juvenile hormone analogues on multiplication of recombinant Spodoptera exigua nucleopolyhedrovirus (Sexd1). Chin. J. Biol. Control. 25 (3), 209–214. doi: 10.16409/j.cnki.2095-039x.2009.03.006
Li, B., Dewey, C. N. (2011). RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12 (1), 1–16. doi: 10.1186/1471-2105-12-323
Li, W. Z., Godzik, A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22 (13), 1658–1659. doi: 10.1093/bioinformatics/btl158
Li, X. C., Schuler, M. A., Berenbaum, M. R. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52, 231–253. doi: 10.1146/annurev.ento.51.110104.151104
Liu, B., Du, H. W., Rutkowski, R., Gartner, A., Wang, X. C. (2012). LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science 337 (6092), 351–354. doi: 10.1126/science.1220281
Liu, X. J., Liang, X. Y., Guo, J., Shi, X. K., Merzendorfer, H., Zhu, K. Y., et al. (2022). V-ATPase subunit a is required for survival and midgut development of Locusta migratoria. Insect Mol. Biol. 31 (1), 60–72. doi: 10.1111/imb.12738
Liu, Y. P., Wang, F. H., Su, Z. J., Li, G. H., Pang, Y. (2005). Effect of juvenile hormone analogues on multiplication of Spodoptera litura nucleopolyhedrovirus. Acta Entomol. Sin. 48 (6), 866. doi: 10.16380/j.kcxb.2005.06.008
Liu, Y. H., Wang, L., Yu, L. (2015). The principle and application of the single-molecule real-time sequencing technology. Hereditas 37 (3), 259–268. doi: 10.16288/j.yczz.14-323
Li, X. J., Yin, H. Y., Guo, W. L., Niu, X. X., Dong, G. P., Fang, J. M., et al. (2022). RNAi suppression of vacuolar ATPase subunit h inhibits immunity-related gene expression in pine sawyer beetle (Coleoptera: Cerambycidae). J. Entomol. Sci. 57 (2), 204–212. doi: 10.18474/JES21-33
Li, C. H., Zhang, H., Li, L., Song, L. S. (2010). Identification of a cathepsin d potentially involved in H2A cleavage from scallop Chlamys farreri. Mol. Biol. Rep. 37 (3), 1451–1460. doi: 10.1007/s11033-009-9534-2
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 1–21. doi: 10.1186/s13059-014-0550-8
May, R. C., Machesky, L. M. (2001). Phagocytosis and the actin cytoskeleton. J. Cell Sci. 114 (6), 1061–1077. doi: 10.1242/jcs.114.6.1061
Menike, U., Ariyasiri, K., Choi, J. Y., Lee, Y., Wickramaarachchi, W. D. N., Premachandra, H. K., et al. (2013). Manila Clam, Ruditapes philippinarum cathepsin d: Molecular analysis and immune response against brown ring disease causing Vibrio tapetis challenge. Korean J. Malacol. 29 (2), 155–161. doi: 10.9710/kjm.2013.29.2.155
Mohan, C., Shibao, P. Y. T., de Paula, F. F. P., Toyama, D., Vieria, M. A. S., Figuera, A., et al. (2021). hRNAi-mediated knock-down of Sphenophorus levis V-ATPase e in transgenic sugarcane (Saccharum spp interspecific hybrid) affects the insect growth and survival. Plant Cell Rep. 40 (3), 507–516. doi: 10.1007/s00299-020-02646-5
Nakhleh, J., El Moussawi, L., Osta, M. A. (2017). The melanization response in insect immunity. Adv. Insect Physiol. 52, 83–109. doi: 10.1016/bs.aiip.2016.11.002
Nehme, N. T., Liégeois, S., Kele, B., Giammarinaro, P., Pradel, E., Hoffmann, J. A., et al. (2007). A model of bacterial intestinal infections in Drosophila melanogaster. PloS Pathog. 3 (11), e173. doi: 10.1371/journal.ppat.0030173
Ning, M., Yuan, M., Liu, M., Gao, Q., Wei, P., Gu, W., et al. (2018). Characterization of cathepsin d from Eriocheir sinensis involved in Spiroplasma eriocheiris infection. Dev. Comp. Immunol. 86, 1–8. doi: 10.1016/j.dci.2018.04.018
Noh, M. Y., Muthukrishnan, S., Kramer, K. J., Arakane, Y. (2016). Cuticle formation and pigmentation in beetles. Curr. Opin. Insect. Sci. 17, 1–9. doi: 10.1016/j.cois.2016.05.004
Ouyang, H. L., Wang, X. Y., Zheng, X. L., Lu, W., Qin, F. P., Chen, C. (2021). Full-length SMRT transcriptome sequencing and SSR analysis of Bactrocera dorsalis (Hendel). Insects 12 (10), 938. doi: 10.3390/insects12100938
Pollard, T. D., Cooper, J. A. (2009). Actin, a central player in cell shape and movement. Science 326 (5957), 1208–1212. doi: 10.1126/science.1175862
Qiao, L., Yan, Z. W., Xiong, G., Hao, Y. J., Wang, R. X., Hu, H., et al. (2020) Excess melanin precursors rescue defective cuticular traits in stony mutant silkworms probably by upregulating four genes encoding RR1-type larval cuticular proteins. Insect Biochem. Mol. Biol. 119, 103315. doi: 10.1016/j.ibmb.2020.103315
Rong, Y. G., McPhee, C. K., Deng, S. S., Huang, L., Chen, L. L., Liu, M., et al. (2011). Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc. Natl. Acad. Sci. 108 (19), 7826–7831. doi: 10.1073/pnas.1013800108
Ruiz-Pesini, E., Mishmar, D., Brandon, M., Procaccio, V., Wallace, D. C. (2004). Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303 (5655), 223–226. doi: 10.1126/science.1088434
Saftig, P., Klumperman, J. (2009). Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10 (9), 623–635. doi: 10.1038/nrm2745
Sagné, C., Agulhon, C., Ravassard, P., Darmon, M., Hamon, M., El Mestikawy, S., et al. (2001). Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc. Natl. Acad. Sci. 98 (13), 7206–7211. doi: 10.1073/pnas.12118349
Sandiford, S. L., Dong, Y., Pike, A., Blumberg, B. J., Bahia, A. C., Dimopoulos, G. (2015). Cytoplasmic actin is an extracellular insect immune factor which is secreted upon immune challenge and mediates phagocytosis and direct killing of bacteria, and is a plasmodium antagonist. PloS Pathog. 11 (2), e1004631. doi: 10.1371/journal.ppat.1004631
Scaraffia, P. Y., Wells, M. A. (2003). Proline can be utilized as an energy substrate during flight of Aedes aegypti females. J. Insect Physiol. 49 (6), 591–601. doi: 10.1016/S0022-1910(03)00031-3
Scott, J. G., Wen, Z. (2001). Cytochromes P450 of insects: The tip of the iceberg. Pest Manage. Sci. 57 (10), 958–967. doi: 10.1002/ps.354
Sezer, B., Ozalp, P. (2015). Effects of pyriproxyfen on hemocyte count and morphology of Galleria mellonella. Fresenius Environ. Bull. 24 (2a), 621–625.
Stoka, V., Turk, V., Turk, B. (2016). Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res. Rev. 32, 22–37. doi: 10.1016/j.arr.2016.04.010
Suderman, R. J., Dittmer, N. T., Kanost, M. R., Kramer, K. J. (2006). Model reactions for insect cuticle sclerotization: Cross-linking of recombinant cuticular proteins upon their laccase-catalyzed oxidative conjugation with catechols. Insect Biochem. Mol. Biol. 36 (4), 353–365. doi: 10.1016/j.ibmb.2006.01.012
Sun, Y. X., Chen, C., Xu, W. J., Abbas, M. N., Mu, F. F., Ding, W. J., et al. (2021). Functions of Bombyx mori cathepsin l-like in innate immune response and anti-microbial autophagy. Dev. Comp. Immunol. 116, 103927. doi: 10.1016/j.dci.2020.103927
Sun, Y., Wang, Y., Liu, W., Zhou, J. L., Zeng, J., Wang, X. H., et al. (2017a). Upregulation of a trypsin-like serine protease gene in Antheraea pernyi (Lepidoptera: Saturniidae) strains exposed to different pathogens. J. Econ. Entomol. 110 (3), 941–948. doi: 10.1093/jee/tox096
Sun, B. Z., Wang, Z., Wang, Z. Y., Ma, X. C., Zhu, F. (2017b). A proteomic study of hemocyte proteins from mud crab (Scylla paramamosain) infected with white spot syndrome virus or Vibrio alginolyticus. Front. Immunol. 8. doi: 10.3389/fimmu.2017.00468
Sun, L. L., Yin, J. J., Du, H., Liu, P., Cao, C. W. (2019). Characterisation of GST genes from the Hyphantria cunea and their response to the oxidative stress caused by the infection of Hyphantria cunea nucleopolyhedrovirus (HcNPV). Pestic. Biochem. Physiol. 163, 254–262. doi: 10.1016/j.pestbp.2019.11.019
Su, Z. T., Zhang, L. Y., Xu, T. M., Du, G. Z., Chen, B., Xiao, G. L. (2020). Isolation, identification and insecticidal activity of pathogenic Serratia marcescens from potato tuber moth larvae. Chin. J. Biol. Control. 36 (3), 361–370. doi: 10.16409/j.cnki.2095-039x.2020.03.003
Tanabe, K., Kon, S., Natsume, W., Torii, T., Watanabe, T., Satake, M. (2006). Involvement of a novel ADP-ribosylation factor GTPase-activating protein, SMAP, in membrane trafficking: Implications in cancer cell biology. Cancer Sci. 97 (9), 801–806. doi: 10.1091/mbc.E05-10-0909
Tang, T., Li, X., Yang, X., Yu, X., Wang, J. H., Liu, F. S., et al. (2014). Transcriptional response of Musca domestica larvae to bacterial infection. PloS One 9 (8), e104867. doi: 10.1371/journal.pone.0104867
Tong, Y. R., Jiang, H. B., Kanost, M. R. (2005). Identification of plasma proteases inhibited by Manduca sexta serpin-4 and serpin-5 and their association with components of the prophenol oxidase activation pathway. J. Biol. Chem. 280 (15), 14932–14942. doi: 10.1074/jbc.M500532200
Trienens, M., Kraaijeveld, K., Wertheim, B. (2017). Defensive repertoire of Drosophila larvae in response to toxic fungi. Mol. Ecol. 26 (19), 5043–5057. doi: 10.1111/mec.14254
Vidak, E., Javoršek, U., Vizovišek, M., Turk, B. (2019). Cysteine cathepsins and their extracellular roles: Shaping the microenvironment. Cells 8 (3), 264. doi: 10.3390/cells8030264
Wagner, C. A., Finberg, K. E., Breton, S., Marshansky, V., Brown, D., Geibel, J. P. (2004). Renal vacuolar h+-ATPase. Physiol. Rev. 84 (4), 1263–1314. doi: 10.1152/physrev.00045.2003
Wang, G. B., Na, S., Qin, L. (2019). Uncovering the cellular and humoral immune responses of Antheraea pernyi hemolymph to Antheraea pernyi nucleopolyhedrovirus infection by transcriptome analysis. J. Invertebr. Pathol. 166, 107205. doi: 10.1016/j.jip.2019.107205
Wang, Z. Q., Feng, K., Tang, F., Xu, M. (2021a). Activation of the host immune response in Hyphantria cunea (Drury)(Lepidoptera: Noctuidae) induced by Serratia marcescens bizio. Insects 12 (11), 1–17. doi: 10.3390/insects12110983
Wang, X., Xu, X., Ullah, F., Ding, Q., Gao, X. W., Desneux, N., et al. (2021b). Comparison of full-length transcriptomes of different imidacloprid-resistant strains of Rhopalosiphum padi (L.). Entomol. Gen. 41 (3), 289–304. doi: 10.1127/entomologia/2021/0972
Wang, K., Yan, P. S., Cao, L. X., Ding, Q. L., Shao, C., Zhao, T. F. (2013). Potential of chitinolytic Serratia marcescens strain JPP1 for biological control of Aspergillus parasiticus and aflatoxin. BioMed. Res. Int. 2013, 1–7. doi: 10.1155/2013/397142
Whitfield, C., Trent, M. S. (2014). Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83 (1), 99–128. doi: 10.1146/annurev-biochem-060713-035600
Xiong, G., Tong, X. L., Gai, T. T., Li, C. L., Qiao, L., Monteiro, A., et al. (2017). Body shape and coloration of silkworm larvae are influenced by a novel cuticular protein. Genetics 207 (3), 1053–1066. doi: 10.1534/genetics.117.300300
Xu, Y. J., Luo, F., Gao, Q., Shang, Y., Wang, C. (2015). Metabolomics reveals insect metabolic responses associated with fungal infection. Anal. Bioanal. Chem. 407 (16), 4815–4821. doi: 10.1007/s00216-015-8648-8
Xu, L. T., Zhang, Y. Q., Zhang, S. H., Deng, J. D., Lu, M., Zhang, L. W., et al. (2018). Comparative analysis of the immune system of an invasive bark beetle, Dendroctonus valens, infected by an entomopathogenic fungus. Dev. Comp. Immunol. 88, 65–69. doi: 10.1016/j.dci.2018.07.002
Yang, H., Ji, T. W., Xiong, H. R., Zhang, Y. Y., Wei, W. Z. (2021). A trypsin-like serine protease domain of masquerade gene in crayfish Procambarus clarkii could activate prophenoloxidase and inhibit bacterial growth. Dev. Comp. Immunol. 117, 103980. doi: 10.1016/j.dci.2020.103980
Yang, H. J., Xu, D. P., Zhuo, Z. H., Hu, J. M., Lu, B. Q. (2020). SMRT sequencing of the full-length transcriptome of the Rhynchophorus ferrugineus (Coleoptera: Curculionidae). PeerJ 8, e9133. doi: 10.7717/peerj.9133
Yu, X. M., Chen, J. L., Abbas, M. N., Gul, I., Kausar, S., Dai, L. S. (2020). Characterization of the cathepsin d in Procambarus clarkii and its biological role in innate immune responses. Dev. Comp. Immunol. 111, 103766. doi: 10.1016/j.dci.2020.103766
Zhai, Y. F., He, P. M., Jia, R. (2021). iTRAQ-based quantitative proteomic analysis of differentially expressed proteins in the hepatopancreas of Litopenaeus vannamei after WSSV infection. Dis. Aquat. Organ. 145, 51–61. doi: 10.3354/dao03594
Zhao, T. Z. (2005). Damage analysis and loss evaluation after hyphantria cunea (Drury)’s invading China (Beijing: Beijing Forestry University). [PhD thesis].
Zhao, J., Luo, X., Chen, D. H., Wang, J. D., Yang, Z. R. (2004). Study on the locusts energy metabolizability inhibited by the insecticidal protein purified from Pseudomonas pseudoalcaligenes. Acta Microbiol. Sin. 44 (3), 365–368. doi: 10.13343/j.cnki.wsxb.2004.03.021
Zhao, L. Q., Wang, W., Qiu, Y., Torson, A. S. (2021). Plasticity of nutrient accumulation patterns in diapausing fall webworm pupa. B. Entomol. Res. 116 (6), 637–644. doi: 10.1017/S0007485321000201
Zhou, L. M., Wu, S. G., Liu, D. C., Xu, B., Zhang, X. F., Zhao, B. S. (2012). Characterization and expression analysis of a trypsin-like serine protease from planarian Dugesia japonica. Mol. Biol. Rep. 39 (6), 7041–7047. doi: 10.1007/s11033-012-1535-x
Zibaee, A., Bandani, A. R., Malagoli, D. (2012). Methoxyfenozide and pyriproxifen alter the cellular immune reactions of Eurygaster integriceps puton (Hemiptera: Scutelleridae) against Beauveria bassiana. Pestic. Biochem. Physiol. 102 (1), 30–37. doi: 10.1016/j.pestbp.2011.10.006
Keywords: Hyphantria cunea (Drury), full-length transcriptome, SMRT sequencing, Serratia marcescens Bizio, transcriptomic response
Citation: Zhang L, Tang X, Wang Z and Tang F (2023) The transcriptomic response of Hyphantria cunea (Drury) to the infection of Serratia marcescens Bizio based on full-length SMRT transcriptome sequencing. Front. Cell. Infect. Microbiol. 13:1093432. doi: 10.3389/fcimb.2023.1093432
Received: 09 November 2022; Accepted: 24 January 2023;
Published: 16 February 2023.
Edited by:
Letian Xu, Hubei University, ChinaCopyright © 2023 Zhang, Tang, Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Tang, tangfang76@foxmail.com
 Ling Zhang
Ling Zhang Xinyi Tang1,2
Xinyi Tang1,2 Fang Tang
Fang Tang