- 1Centre for Microbial Diseases and Immunity Research, Department of Microbiology and Immunology, University of British Columbia, Vancouver, BC, Canada
- 2NanoMedicines Innovation Network (NMIN), Vancouver, BC, Canada
- 3ASEP Medical Holdings, Victoria, BC, Canada
Antibiotic failure can be defined as any clinical situation where treatment with antibiotics fails to cure the patient and remove the infection. Genetically-determined antibiotic resistance certainly contributes to antibiotic failure in the clinic, but this is not the only reason why antibiotics fail and it is likely not the most common cause of antibiotic failure. In this perspective article, we outline several widespread examples of situations where antibiotic treatment fails, even in the absence of formal resistance, including biofilm associated-infections (65% of all infections) as well as infections in sepsis (19.7% of all deaths) and immune compromised individuals. We then discuss various strategies that are being employed to address the issue of antibiotic failure and emphasize that antibiotic failure should be given increased awareness and resources to address this underappreciated but critical issue.
Introduction
Antibiotics are arguably the most important medical advancement of all time. Since their emergence in medical practice nearly a century ago, their astounding ability to treat infections has dramatically reduced the scourge of infectious diseases that plagued mankind. In the early part of the 20th century, infectious diseases like wound infections, cholera, diptheria, pneumonia, typhoid fever, plague, tuberculosis and syphilis were widespread and contributed to high morbidity and mortality (Shaw-Taylor, 2020). Beyond their ability to treat infections, antibiotics form the foundation upon which much of our current medical infrastructure is built. Antibiotics are essential for major and minor surgeries, organ transplants and early-term births. Antibiotics, together with improvements in public health, disease prevention, food safety, and sanitation, have helped to alleviate the burden on society of these debilitating diseases, so much so that the life expectancy of someone born today is nearly 30 years longer than someone born a century ago (Hutchings et al., 2019).
Unfortunately, antimicrobial resistance is (AMR) rapidly becoming a serious issue around the world. A recent article found that AMR contributes to nearly 5-million deaths worldwide and is the direct cause of >1-million deaths/year (Murray et al., 2022). Some have termed the AMR crisis “the slow-motion pandemic” (Sanderson and Thompson, 2021) with estimates of 10 million deaths/year by 2050 (O’Neill, 2016), a number likely exacerbated by the widespread use of antibiotics during the COVID-19 global pandemic (Subramanya et al., 2021). Of great concern is that our arsenal of effective antibiotics is dwindling at an alarming rate, as extensively discussed elsewhere (Howard et al., 2013; Årdal et al., 2020; Bhavnani et al., 2020). Intriguingly however, formal AMR is not the major contributor of infections to mortality as sepsis is directly associated with 11-million deaths annually (Rudd et al., 2020).
Here we wish to highlight common situations where antibiotics fail, have likely failed even during the gold age of antibiotic discovery, and will continue to fail even if new antibiotic compounds are discovered and introduced into the clinic. Specifically, we will discuss three widespread situations where antibiotics fail: biofilm-associated infections, infections in immunocompromised patients and the dysfunctional immune response to an infection known as sepsis. In all three cases, the primary course of treatment is to administer more antibiotics, but this strategy ignores the complexities of these serious medical conditions and does not address the true cause of antibiotic failure, often with poor outcomes for the afflicted patients.
Antibiotic Resistance vs. Antibiotic Failure
The term antibiotic resistance is commonplace in society due to the growing awareness of widespread AMR around the world. The mechanisms underlying AMR include acquisition of a (usually-mobile) genetic element (e.g., plasmids, transposons, etc.) encoding a resistance gene, or a chromosomal mutation developed under selective pressure (Munita and Arias, 2016). This can lead to production of an enzyme that chemically modifies/inactivates the antibiotic itself, alteration of the binding site on the antibiotic’s target or alteration in net uptake across of the bacterial envelope (Munita and Arias, 2016). The emergence and spread of AMR genes throughout populations reduces the effectiveness of antibiotics over time, resulting in increased or prolonged illness and greater morbidity and mortality.
Antibiotic failure can be defined as any situation where bacteria survive antibiotic treatment and the clinical symptoms of the infection persist. Certainly, AMR could be considered as one situation where antibiotics fail; however, there are many more situations where antibiotics frequently fail even in the absence of a genetically encoded resistance mechanism (Figure 1). Below, we highlight examples where antibiotic treatment often fails and emphasize that these situations are already widespread and physicians have limited treatment options to address them.
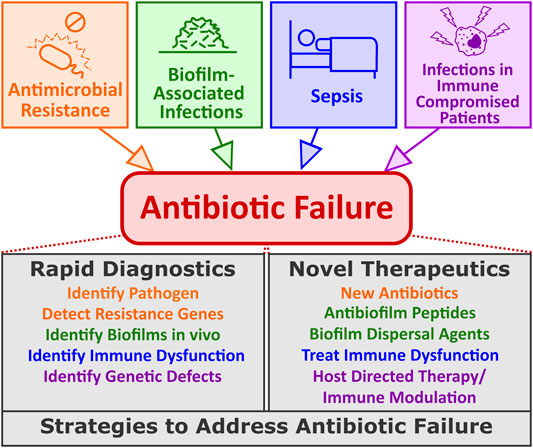
FIGURE 1. Common causes of antibiotic failure and potential strategies to address them. Antibiotic failure can be thought of as any clinical situation where administration of antibiotics fails to improve patient outcomes. Antibiotic failure may be due to AMR but is also common with biofilm-associated infections, infections in immunocompromised patients or in the serious condition known as sepsis. Several potential strategies have been proposed to deal with these different issues including advancements in new diagnostics and specialized therapeutics designed specifically for scenarios where antibiotics fail.
Biofilm Associated Infections
Biofilms are communities of microorganisms that grow on surfaces. Clinically, biofilms are a major concern since biofilms are the cause of 65% of all infections (Sharma et al., 2019; Hancock et al., 2021). Common biofilm-associated infections in humans include skin and soft tissue infections, catheter-associated infections and dental plaque while more serious medical conditions include infections of implanted devices/prosthetics, endocarditis, and lung infections in cystic fibrosis patients (Wu et al., 2015; Vestby et al., 2020). Critically, biofilms are 10- to 1,000-fold more resistant to most antibiotics but this is usually adaptive multi-drug resistance due to their altered growth state, and reverts when the bacteria disperse from the biofilm. There are no approved biofilm treatments; instead, high doses or combinations of antibiotics are often used but treatments frequently fail (Ciofu et al., 2017). For example, high levels of antibiotics are used to treat biofilm-like abscess infections, cystic fibrosis lung infections, bronchiectasis, and chronic rhinosinusitis, but antibiotic effectiveness remains relatively poor and treatment still requires invasive procedures such as surgical debridement. Biofilms are an adaptive growth state of bacteria resulting in the differential expression of hundreds of genes, cf. planktonic bacteria (Dötsch et al., 2012). This undoubtedly contributes to multidrug and immune-response resistance together with other factors such as reduced metabolism and lowered bacterial growth rate (Vestby et al., 2020).
Sepsis
Sepsis is serious and life-threatening condition caused by immune dysfunction (Singer et al., 2016). Sepsis arises in response to an infection, which is usually bacterial but can include fungal or viral pathogens, and especially COVID-19 (Li et al., 2020); with antibiotics being the primary tool in sepsis management. However, antibiotics are frequently unsuccessful, as revealed by the death of 23% of severe sepsis patients in 2017 (49-million cases, 11-million deaths), representing 19.7% of all deaths globally (Rudd et al., 2020). Although this number eclipses the predicted 10-million deaths by 2050 due to antibiotic resistance (O’Neill, 2016), it has not been highlighted with the same level of alarm as the growing AMR crisis. There is great urgency to initiate treatment since for every hours delay in initiating antibiotic treatment results in a 7.6% increase in the chance of death (Kumar et al., 2006). Unfortunately, the early symptoms with which patients present are quite non-specific and identification of the offending pathogen takes time, so treatment is frequently delayed. Also, physicians are forced to treat patients empirically, often by prescribing broad spectrum antibiotics, in an attempt to control the source of infection as fast as possible. Therefore, sepsis could be considered a situation where antibiotic treatment often fail, and illustrates a situation where the underlying condition of a patient is likely as important as the pathogen itself.
Infections in Immunocompromised Patients
Immune compromised patients encompass a sizeable portion of the total population and include patients from different backgrounds and from a wide range of medical conditions. It is estimated that nearly 3% of all US adults (∼10 million individuals) have some type of immunosuppression (Harpaz et al., 2016). Many of these patients lead normal lives, but their impaired immune response makes them more susceptible to infections and if they do become ill, they have a higher chance of developing serious complications or dying. Furthermore, very young children, including premature babies, and the elderly tend to have weaker immune responses when compared to healthy adults (Dorshkind et al., 2009; PrabhuDas et al., 2011), representing sections of the population that are more vulnerable to infections.
Immunocompromised patients can be subdivided into two groups, those with primary immunodeficiencies in which a component of the host’s immune response are impaired due to a genetic mutation (McCusker et al., 2018) and those with secondary immunodeficiencies in which extrinsic factors impair the immune response (Chinen and Shearer, 2010). Nearly 270 different genetic disorders associated with primary immunodeficiencies have been identified impacting on various cellular processes related to cellular and humoral immunity, production of deficient antibodies, overall immune dysregulation or altered innate immunity, among others (Picard et al., 2015). Secondary immunodeficiencies are more common and may be due to a variety of causes including environmental stresses, metabolic disorders, malnutrition, the presence of particular chronic infections (e.g., HIV), or treatment of medical conditions with immune suppressive drugs (e.g., corticosteroids and treatments for cancer and autoimmune disorders, etc.) (Chinen and Shearer, 2010). All immunocompromised patients rely on a readily available arsenal of antibiotics should they get sick as their own immune response is hindered from dealing with the pathogen. Often these patients are treated aggressively to ensure that the infection is cleared as quickly as possible to prevent long term consequences associated with chronic infections but treatment is hampered by the lack of a supportive immune system (Ankomah and Levin, 2014).
Potential Solutions to Antibiotic Failure
Antibiotic failure is not due to a single cause and the medical consequences of such failure are diverse and potentially far-reaching. Therefore, it is likely that multi-pronged approaches will be required to address these issues in the coming years. In the following section, we focus on emerging strategies to address antibiotic failure, examining emerging technology from both a diagnostic perspective as well as a therapeutic standpoint (Figure 1) with a particular emphasis on the antibiotic failure scenarios described above.
Biofilm Specific Therapies
Current treatment options for biofilm associated infections include debridement of the infected site to try and remove the damaged tissue plus biofilm or infected implant followed by the prescription of a course of antibiotics to try and control the infection at the infected site as well as prevent dissemination to other parts of the body (Koo et al., 2017). Depending on the degree to which the biofilm infection has penetrated into the underlying tissue and the damage and inflammation present in the surrounding area, this type of biofilm treatment regimen can be quite invasive and still leaves the patient with a serious wound that may be susceptible to re-infection. In addition, due to the strong possibility that the biofilm persists, aggressive treatment with last resort antibiotics is often employed (Wu et al., 2015), further compounding the overuse of antibiotics. As mentioned above, no antibiofilm specific therapeutic has been approved for clinical use.
One of the simplest ways of addressing biofilms on implanted medical devices is to prevent them from developing in the first place. Numerous strategies have been employed to develop various bio-compatible coatings for these devices. Examples include the incorporation of metals such as silver (Wang et al., 2016; Ishihama et al., 2021) or copper (Gomes et al., 2020), incorporation of synthetic antimicrobial peptides (Riool et al., 2017; Yu et al., 2017) or encapsulating antimicrobial agents that are slowly released over time (Stærk et al., 2021) to prevent biofilm attachment and proliferation.
Targeting the extracellular matrix of biofilms may be possible through the use of exopolysaccharide degrading enzymes (Baker et al., 2016), proteases to target proteins within the extracellular biofilm matrix (Baidamshina et al., 2017) or DNases to break down extracellular DNA (Okshevsky et al., 2015). The idea is that dispersing the cells within a biofilm would return them to their susceptible planktonic state thereby making them susceptible to the effects of antibiotics. This is why dispersal agents would be best used in combination with some type of antibiotic. Chemical compounds may also be used to stimulate biofilm dispersal such as nitric oxide (Howlin et al., 2017) or quorum-sensing inhibitors (Brackman and Coenye, 2015).
Host defence peptides (HDPs) are natural antimicrobial compounds produced by virtually all complex life forms consisting of short cationic and amphipathic polypeptide chains that have both antimicrobial and immunomodulatory functions in vivo (Haney et al., 2019; Mookherjee et al., 2020). HDPs have recently garnered significant interest as a potential source of biofilm specific therapeutics stemming from the observation that a human HDP, LL-37, prevented biofilm formation in Pseudomonas aeruginosa at sub-inhibitory concentrations (Overhage et al., 2008). Since that observation, many natural and synthetic peptides have been identified that also exert antibiofilm specific activity that is mechanistically different from the effects of HDPs on planktonic bacteria (de la Fuente-Núñez et al., 2014; Haney et al., 2018). Importantly, many antibiofilm peptides have demonstrated efficacy in various animal models of biofilm-associated infections including infections of the skin (Pfalzgraff et al., 2018), sinuses (Alford et al., 2021) and abscesses (Mansour et al., 2016; Pletzer et al., 2018) as well as in human organoid models (Wu et al., 2021). The mechanism of action used by antibiofilm HDPs to kill biofilm cells is likely complex (Hancock et al., 2021) but has been shown to involve the targeting of guanosine tetraphosphate which is an important cell signalling molecule involved in the stringent stress response across many bacterial species (de la Fuente-Núñez et al., 2014). Overall, antibiofilm peptides represent a promising broad-spectrum therapeutic to treat biofilm-associated infections in the clinic.
Host Directed Therapies
Host directed therapies are treatments targeted to a host specific mechanism that can limit infection and may include excitement of a protective innate or adaptive immune response, inhibition of a host pathway that is required for infection, repairing or complementing a host defect (e.g., mutation) that leads to enhance susceptibility to infection, or restoring healthy microbial balance. As such, host directed therapies for infectious diseases are being increasingly explored as a way to mitigate antibiotic failure in immunocompromised patients and/or limit the general overuse of antibiotics. Certainly, among patients with primary immunodeficiencies, developing treatments aimed at mitigating or curing the causative genetic defect would have a tremendous impact on preventing infections in immunocompromised patients. Indeed, recent advances in gene therapy and the prospect of gene editing offered by CRISPR/Cas9 provides significant optimism for reversing many of these conditions in the near future (Booth et al., 2019). However, gene therapy may not be possible in all situations and in many cases the condition may not afflict a large enough population of patients, thereby making it difficult to attract the research and development efforts of pharmaceutical companies. Furthermore, patients with secondary immunodeficiencies likely have multiple immune pathways that are impacted by the presence of an infection or some immune suppressing drug regimen that genetic therapies alone are likely insufficient to completely address their underlying condition while adequately protecting against invading pathogens. Recently, we described a strategy whereby screening of a CRISPR/Cas9 mutant library in THP1 monocytes for deficiencies in uptake of Salmonella, led to the description of 183 potential targets for host directed therapy to mitigate such infections (Yeung et al., 2019).
Rapid Diagnostic Tests
When it comes to treating infections, particularly when sepsis is suspected, speed is of the essence. As mentioned above, there is a stiff penalty for every hour’s delay in initiation of appropriate therapy, e.g., potent antibiotics. Conversely, we do not want to use our most important and potent antibiotics in patients who are not at risk for sepsis, due to the potential for antibiotic resistance development. Conventional microbiological methods often take >24 h to pinpoint the causative bacteria species and identify an antibiotic that could be used for empirical treatment (Reller et al., 2009). An additional factor that is often underappreciated is that antibiotic susceptibility may be altered due to the use of in vitro growth conditions that are not reflective of in vivo conditions (Ersoy et al., 2017; Belanger et al., 2020), potentially leading to inappropriate antibiotic use with limited clinical benefit. For these reasons, significant efforts are underway to develop rapid diagnostics that can characterize the bacterial, fungal or viral source of an infections to enable application of appropriate therapeutics. Molecular diagnostic tests using PCR or mass spectrometry based technology are now available and can detect the presence of multiple pathogens in blood samples in a few hours, with some even detecting specific antimicrobial resistance genes (Afshari et al., 2012; Sullivan and Dien Bard, 2019).
Since sepsis is caused by a dysfunctional immune response, several groups have begun to examine whether the immune response itself could serve as a basis for a molecular diagnostic for sepsis. Early work from our research group identified a 31-gene expression signature, that was present in sepsis patients in the emergency room and reflected an immune dysfunction due to cellular reprogramming (Pena et al., 2014); it was recently reduced to 8 genes (Baghela et al., 2022). Likewise, other molecular signatures of sepsis in ICU patients have been reported in the literature (Scicluna et al., 2015; Sweeney et al., 2016) and may serve as a basis to develop novel sepsis diagnostics. Indeed, one test, SeptiCyte LAB, is already approved for use on sepsis patients in the ICU (Miller et al., 2018). More recent work has revealed the presence of endotypes of disease severity in individual sepsis patients, that can be assessed in the ER (Baghela et al., 2022). These endotypes reflect novel underlying mechanisms driving sepsis, differ in severity, and can be diagnosed using 2-gene expression signatures, which could be used as the basis to stratify sepsis patients into different (personalized) treatment groups.
Immune Modulators
An alternative host-directed strategy is to selectively modulate innate immunity to boost protective immunity, a tactic that has been used extensively in anti-viral therapy and to a lesser extent for bacterial infections (Hancock et al., 2012). Many strategies are being developed, including HDPs, that modulate the immune response due to an infection in a fairly unique way. Natural HDPs have the ability to influence a range of innate immune responses in vivo (Hancock et al., 2016). In general, HDPs promote immune functions which aid in the clearance of a pathogen such as upregulating the production of various chemokines that attract white blood cells to an infection site while at the same time, suppressing the potentially harmful effects of inflammation. These immunomodulatory effects can be specifically optimized in synthetic peptide sequences (Haney et al., 2015) and the immunomodulatory effects of synthetic HDPs can provide protection against a range of bacterial infections (Scott et al., 2007; Mansour et al., 2015). While not specifically explored in the context of immune compromised patients, it is conceivable that this patient population could potentially benefit from such a therapeutic approach, as would the general population.
Although discussed in the context of microbiota displacement therapy, probiotic organisms are also immune modulators (Cosseau et al., 2008; Cristofori et al., 2021). Indeed, combinations of respiratory microbes are a known anti-infective strategy used for lower respiratory infections in Latin America (Del-Rio-Navarro et al., 2012). Also, probiotics represent a strategy that is effective in decreasing the rate of sepsis in pre-term infants (Athalye-Jape and Patole, 2019).
Conclusion
AMR remains one of the most significant issues facing mankind and continued research into antibiotic discovery and clinical development of new antimicrobials should be prioritized and accelerated in the coming years. However, while providing new chemicals capable of killing resistant bacterial pathogens may alleviate the pressure arising from circulating AMR genes, such molecules do not address the root cause of why many antibiotics already fail, and antibiotic failure will continue to place a significant burden on global health.
There are substantial benefits to addressing antibiotic failure. Developing appropriate therapies to specifically target biofilm-associated infections would reduce the use of broad-spectrum antibiotics currently used to treat such infections, thereby reducing selective pressure for AMR development. Likewise, the development of rapid diagnostic tests capable of identifying specific bacterial or viral pathogens would provide clinicians with important information to initiate appropriate drug regimens that are tailored to each patient and reduce the use of antibiotics when unnecessary. Additionally, the costs associated with treating chronic (often biofilm) infections (Nussbaum et al., 2018) and severe sepsis (Paoli et al., 2018) are among the highest of all healthcare costs, so the financial incentives to solving antibiotic failure go well beyond the potential costs associated with developing new treatment strategies.
Indeed, antibiotic failure is not an issue that will go away simply by expanding our available chemical arsenal against pathogenic microbes. Addressing antibiotic failure will require an improved understanding of how microbes infect their hosts and understanding the complexities of the immune response when challenged with a pathogen. In addition, innovations in therapeutics and diagnostics that address fundamental processes related to antibiotic failure, beyond AMR, are required to reduce the incidence of antibiotic failure and provide clinicians with a full arsenal of tools to treat infectious diseases.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
RH gratefully acknowledges funding from the Canadian Institutes for Health Research (FDN-154287). This work was also supported by NMIN (the NanoMedicines Innovation Network), a member of the Networks of Centres of Excellence Canada program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Canadian Institutes for Health Research or NMIN. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Canadian Institutes for Health Research.
Conflict of Interest
EH and RH have invented and filed for patent protection on related antibiofilm peptide sequences. This patent has been assigned to their employer, the University of British Columbia, and has been licensed to ABT Innovations Inc., in which both have an ownership position. RH has filed patents related to sepsis gene expression signatures and licenced this to Sepset Biosciences Inc., a Vancouver company in which he has a significant ownership position. ABT Innovations Inc. and Sepset Biosciences Inc. are subsidiaries of ASEP Medical Holdings. EH is employed by the ASEP and receives salary while RH holds an executive position and is on the Board of ASEP.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Afshari, A., Schrenzel, J., Ieven, M., and Harbarth, S. (2012). Bench-to-Bedside Review: Rapid Molecular Diagnostics for Bloodstream Infection - A New Frontier? Crit. Care 16, 222. doi:10.1186/cc11202
Alford, M. A., Choi, K.-Y. G., Trimble, M. J., Masoudi, H., Kalsi, P., Pletzer, D., et al. (2021). Murine Model of Sinusitis Infection for Screening Antimicrobial and Immunomodulatory Therapies. Front. Cel. Infect. Microbiol. 11, 621081. doi:10.3389/fcimb.2021.621081
Ankomah, P., and Levin, B. R. (2014). Exploring the Collaboration between Antibiotics and the Immune Response in the Treatment of Acute, Self-Limiting Infections. Proc. Natl. Acad. Sci. U.S.A. 111, 8331–8338. doi:10.1073/pnas.1400352111
Årdal, C., Balasegaram, M., Laxminarayan, R., McAdams, D., Outterson, K., Rex, J. H., et al. (2020). Antibiotic Development - Economic, Regulatory and Societal Challenges. Nat. Rev. Microbiol. 18, 267–274. doi:10.1038/s41579-019-0293-3
Athalye-Jape, G., and Patole, S. (2019). Probiotics for Preterm Infants - Time to End All Controversies. Microb. Biotechnol. 12, 249–253. doi:10.1111/1751-7915.13357
Baghela, A., Pena, O. M., Lee, A. H., Baquir, B., Falsafi, R., An, A., et al. (2022). Predicting Sepsis Severity at First Clinical Presentation: The Role of Endotypes and Mechanistic Signatures. EBioMedicine 75, 103776. doi:10.1016/j.ebiom.2021.103776
Belanger, C. R., Lee, A. H.-Y., Pletzer, D., Dhillon, B. K., Falsafi, R., and Hancock, R. E. W. (2020). Identification of Novel Targets of Azithromycin Activity Against Pseudomonas aeruginosa Grown in Physiologically Relevant Media. Proc. Natl. Acad. Sci. 117, 33519–33529. doi:10.1073/pnas.2007626117
Baidamshina, D. R., Trizna, E. Y., Holyavka, M. G., Bogachev, M. I., Artyukhov, V. G., Akhatova, F. S., et al. (2017). Targeting Microbial Biofilms Using Ficin, a Nonspecific Plant Protease. Sci. Rep. 7, 46068. doi:10.1038/srep46068
Baker, P., Hill, P. J., Snarr, B. D., Alnabelseya, N., Pestrak, M. J., Lee, M. J., et al. (2016). Exopolysaccharide Biosynthetic Glycoside Hydrolases Can Be Utilized to Disrupt and Prevent Pseudomonas A Biofilms. Sci. Adv. 2, e1501632. doi:10.1126/sciadv.1501632
Bhavnani, S. M., Krause, K. M., and Ambrose, P. G. (2020). A Broken Antibiotic Market: Review of Strategies to Incentivize Drug Development. Open Forum Infect. Dis. 7, ofaa083. doi:10.1093/ofid/ofaa083
Booth, C., Romano, R., Roncarolo, M. G., and Thrasher, A. J. (2019). Gene Therapy for Primary Immunodeficiency. Hum. Mol. Genet. 28, R15–R23. doi:10.1093/hmg/ddz170
Brackman, G., and Coenye, T. (2015). Quorum Sensing Inhibitors as Anti-Biofilm Agents. Curr. Pharm. Des. 21, 5–11. doi:10.2174/1381612820666140905114627
Chinen, J., and Shearer, W. T. (2010). Secondary Immunodeficiencies, Including HIV Infection. J. Allergy Clin. Immunol. 125, S195–S203. doi:10.1016/j.jaci.2009.08.040
Ciofu, O., Rojo-Molinero, E., Macià, M. D., and Oliver, A. (2017). Antibiotic Treatment of Biofilm Infections. APMIS 125, 304–319. doi:10.1111/apm.12673
Cosseau, C., Devine, D. A., Dullaghan, E., Gardy, J. L., Chikatamarla, A., Gellatly, S., et al. (2008). The Commensal Streptococcus salivarius K12 Downregulates the Innate Immune Responses of Human Epithelial Cells and Promotes Host-Microbe Homeostasis. Infect. Immun. 76, 4163–4175. doi:10.1128/IAI.00188-08
Cristofori, F., Dargenio, V. N., Dargenio, C., Miniello, V. L., Barone, M., and Francavilla, R. (2021). Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation: A Door to the Body. Front. Immunol. 12, 578386. doi:10.3389/fimmu.2021.578386
de la Fuente-Núñez, C., Reffuveille, F., Haney, E. F., Straus, S. K., and Hancock, R. E. W. (2014). Broad-Spectrum Anti-Biofilm Peptide that Targets a Cellular Stress Response. PLoS Pathog. 10, e1004152. doi:10.1371/journal.ppat.1004152
Del-Rio-Navarro, B. E., Espinosa-Rosales, F. J., Flenady, V., and Sienra-Monge, J. J. (2012). Cochrane Review: Immunostimulants for Preventing Respiratory Tract Infection in Children. Evid.-Based Child. Health 7, 629–717. doi:10.1002/ebch.1833
Dorshkind, K., Montecino-Rodriguez, E., and Signer, R. A. J. (2009). The Ageing Immune System: Is it Ever Too Old to Become Young Again? Nat. Rev. Immunol. 9, 57–62. doi:10.1038/nri2471
Dötsch, A., Eckweiler, D., Schniederjans, M., Zimmermann, A., Jensen, V., Scharfe, M., et al. (2012). The Pseudomonas Aeruginosa Transcriptome in Planktonic Cultures and Static Biofilms Using RNA Sequencing. PLoS One 7, e31092. doi:10.1371/journal.pone.0031092
Ersoy, S. C., Heithoff, D. M., Barnes, L., Tripp, G. K., House, J. K., Marth, J. D., et al. (2017). Correcting a Fundamental Flaw in the Paradigm for Antimicrobial Susceptibility Testing. EBioMedicine 20, 173181. doi:10.1016/j.ebiom.2017.05.026
Gomes, I. B., Simões, M., and Simões, L. C. (2020). Copper Surfaces in Biofilm Control. Nanomaterials 10, 2491. doi:10.3390/nano10122491
Hancock, R. E. W., Alford, M. A., and Haney, E. F. (2021). Antibiofilm Activity of Host Defence Peptides: Complexity Provides Opportunities. Nat. Rev. Microbiol. 19, 786–797. doi:10.1038/s41579-021-00585-w
Hancock, R. E. W., Haney, E. F., and Gill, E. E. (2016). The Immunology of Host Defence Peptides: Beyond Antimicrobial Activity. Nat. Rev. Immunol. 16, 321–334. doi:10.1038/nri.2016.29
Hancock, R. E. W., Nijnik, A., and Philpott, D. J. (2012). Modulating Immunity as a Therapy for Bacterial Infections. Nat. Rev. Microbiol. 10, 243–254. doi:10.1038/nrmicro2745
Haney, E. F., Brito-Sánchez, Y., Trimble, M. J., Mansour, S. C., Cherkasov, A., and Hancock, R. E. W. (2018). Computer-Aided Discovery of Peptides that Specifically Attack Bacterial Biofilms. Sci. Rep. 8, 1871. doi:10.1038/s41598-018-19669-4
Haney, E. F., Mansour, S. C., Hilchie, A. L., de la Fuente-Núñez, C., and Hancock, R. E. W. (2015). High Throughput Screening Methods for Assessing Antibiofilm and Immunomodulatory Activities of Synthetic Peptides. Peptides 71, 276–285. doi:10.1016/j.peptides.2015.03.015
Haney, E. F., Straus, S. K., and Hancock, R. E. W. (2019). Reassessing the Host Defense Peptide Landscape. Front. Chem. 7, 43. doi:10.3389/fchem.2019.00043
Harpaz, R., Dahl, R. M., and Dooling, K. L. (2016). Prevalence of Immunosuppression Among US Adults, 2013. JAMA 316, 2547–2548. doi:10.1001/jama.2016.16477
Howard, S. J., Catchpole, M., Watson, J., and Davies, S. C. (2013). Antibiotic Resistance: Global Response Needed. Lancet Infect. Dis. 13, 1001–1003. doi:10.1016/S1473-3099(13)70195-6
Howlin, R. P., Cathie, K., Hall-Stoodley, L., Cornelius, V., Duignan, C., Allan, R. N., et al. (2017). Low-Dose Nitric Oxide as Targeted Anti-biofilm Adjunctive Therapy to Treat Chronic Pseudomonas Aeruginosa Infection in Cystic Fibrosis. Mol. Ther. 25, 2104–2116. doi:10.1016/j.ymthe.2017.06.021
Hutchings, M. I., Truman, A. W., and Wilkinson, B. (2019). Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 51, 72–80. doi:10.1016/j.mib.2019.10.008
Ishihama, H., Ishii, K., Nagai, S., Kakinuma, H., Sasaki, A., Yoshioka, K., et al. (2021). An Antibacterial Coated Polymer Prevents Biofilm Formation and Implant-Associated Infection. Sci. Rep. 11, 3602. doi:10.1038/s41598-021-82992-w
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P., and Hall-Stoodley, L. (2017). Targeting Microbial Biofilms: Current and Prospective Therapeutic Strategies. Nat. Rev. Microbiol. 15, 740–755. doi:10.1038/nrmicro.2017.99
Kumar, A., Roberts, D., Wood, K. E., Light, B., Parrillo, J. E., Sharma, S., et al. (2006). Duration of Hypotension before Initiation of Effective Antimicrobial Therapy Is the Critical Determinant of Survival in Human Septic Shock. Crit. Care Med. 34, 1589–1596. doi:10.1097/01.CCM.0000217961.75225.E9
Li, H., Liu, L., Zhang, D., Xu, J., Dai, H., Tang, N., et al. (2020). SARS-CoV-2 and Viral Sepsis: Observations and Hypotheses. The Lancet 395, 1517–1520. doi:10.1016/S0140-6736(20)30920-X
Mansour, S. C., de la Fuente-Núñez, C., and Hancock, R. E. W. (2015). Peptide IDR-1018: Modulating the Immune System and Targeting Bacterial Biofilms to Treat Antibiotic-Resistant Bacterial Infections. J. Pept. Sci. 21, 323–329. doi:10.1002/psc.2708
Mansour, S. C., Pletzer, D., de la Fuente-Núñez, C., Kim, P., Cheung, G. Y. C., Joo, H.-S., et al. (2016). Bacterial Abscess Formation Is Controlled by the Stringent Stress Response and Can Be Targeted Therapeutically. EBioMedicine 12, 219–226. doi:10.1016/j.ebiom.2016.09.015
McCusker, C., Upton, J., and Warrington, R. (2018). Primary Immunodeficiency. Allergy Asthma Clin. Immunol. 14, 61. doi:10.1186/s13223-018-0290-5
Miller, R. R., Lopansri, B. K., Burke, J. P., Levy, M., Opal, S., Rothman, R. E., et al. (2018). Validation of a Host Response Assay, SeptiCyte LAB, for Discriminating Sepsis from Systemic Inflammatory Response Syndrome in the ICU. Am. J. Respir. Crit. Care Med. 198, 903–913. doi:10.1164/rccm.201712-2472OC
Mookherjee, N., Anderson, M. A., Haagsman, H. P., and Davidson, D. J. (2020). Antimicrobial Host Defence Peptides: Functions and Clinical Potential. Nat. Rev. Drug Discov. 19, 311–332. doi:10.1038/s41573-019-0058-8
Munita, J. M., and Arias, C. A. (2016). Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 4 (2). doi:10.1128/microbiolspec.VMBF-0016-2015
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Aguilar, G. R., Gray, A., et al. (2022). Global burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. The Lancet 399, 629–655. doi:10.1016/S0140-6736(21)02724-0
Nussbaum, S. R., Carter, M. J., Fife, C. E., DaVanzo, J., Haught, R., Nusgart, M., et al. (2018). An Economic Evaluation of the Impact, Cost, and Medicare Policy Implications of Chronic Nonhealing Wounds. Value in Health 21, 27–32. doi:10.1016/j.jval.2017.07.007
Okshevsky, M., Regina, V. R., and Meyer, R. L. (2015). Extracellular DNA as a Target for Biofilm Control. Curr. Opin. Biotechnol. 33, 73–80. doi:10.1016/j.copbio.2014.12.002
O’Neill, J. (2016). Home | AMR Review. Review on Antimicrobial Resistance. Available at: http://amr-review.org/ (Accessed September 6, 2016).
Overhage, J., Campisano, A., Bains, M., Torfs, E. C. W., Rehm, B. H. A., and Hancock, R. E. W. (2008). Human Host Defense Peptide LL-37 Prevents Bacterial Biofilm Formation. Infect. Immun. 76, 4176–4182. doi:10.1128/iai.00318-08
Paoli, C. J., Reynolds, M. A., Sinha, M., Gitlin, M., and Crouser, E. (2018). Epidemiology and Costs of Sepsis in the United States-An Analysis Based on Timing of Diagnosis and Severity Level*. Crit. Care Med. 46, 1889–1897. doi:10.1097/CCM.0000000000003342
Pena, O. M., Hancock, D. G., Lyle, N. H., Linder, A., Russell, J. A., Xia, J., et al. (2014). An Endotoxin Tolerance Signature Predicts Sepsis and Organ Dysfunction at Initial Clinical Presentation. EBioMedicine 1, 64–71. doi:10.1016/j.ebiom.2014.10.003
Pfalzgraff, A., Brandenburg, K., and Weindl, G. (2018). Antimicrobial Peptides and Their Therapeutic Potential for Bacterial Skin Infections and Wounds. Front. Pharmacol. 9, 281. doi:10.3389/fphar.2018.00281
Picard, C., Al-Herz, W., Bousfiha, A., Casanova, J.-L., Chatila, T., Conley, M. E., et al. (2015). Primary Immunodeficiency Diseases: an Update on the Classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J. Clin. Immunol. 35, 696–726. doi:10.1007/s10875-015-0201-1
Pletzer, D., Mansour, S. C., and Hancock, R. E. W. (2018). Synergy between Conventional Antibiotics and Anti-biofilm Peptides in a Murine, Sub-Cutaneous Abscess Model Caused by Recalcitrant ESKAPE Pathogens. Plos Pathog. 14, e1007084. doi:10.1371/journal.ppat.1007084
PrabhuDas, M., Adkins, B., Gans, H., King, C., Levy, O., Ramilo, O., et al. (2011). Challenges in Infant Immunity: Implications for Responses to Infection and Vaccines. Nat. Immunol. 12, 189–194. doi:10.1038/ni0311-189
Reller, L. B., Weinstein, M., Jorgensen, J. H., and Ferraro, M. J. (2009). Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices. Clin. Infect. Dis. 49, 1749–1755. doi:10.1086/647952
Riool, M., de Breij, A., Drijfhout, J. W., Nibbering, P. H., and Zaat, S. A. J. (2017). Antimicrobial Peptides in Biomedical Device Manufacturing. Front. Chem. 5, 63. doi:10.3389/fchem.2017.00063
Rudd, K. E., Johnson, S. C., Agesa, K. M., Shackelford, K. A., Tsoi, D., Kievlan, D. R., et al. (2020). Global, Regional, and National Sepsis Incidence and Mortality, 1990-2017: Analysis for the Global Burden of Disease Study. The Lancet 395, 200–211. doi:10.1016/S0140-6736(19)32989-7
Sanderson, S., and Thompson, W. (2021). Can Antibiotic Prescribing Recover its Downward Trajectory during 2021? BDJ Pract. 34, 12–14. doi:10.1038/s41404-021-0710-y
Scicluna, B. P., Klein Klouwenberg, P. M. C., van Vught, L. A., Wiewel, M. A., Ong, D. S. Y., Zwinderman, A. H., et al. (2015). A Molecular Biomarker to Diagnose Community-Acquired Pneumonia on Intensive Care Unit Admission. Am. J. Respir. Crit. Care Med. 192, 826–835. doi:10.1164/rccm.201502-0355OC
Scott, M. G., Dullaghan, E., Mookherjee, N., Glavas, N., Waldbrook, M., Thompson, A., et al. (2007). An Anti-Infective Peptide that Selectively Modulates the Innate Immune Response. Nat. Biotechnol. 25, 465–472. doi:10.1038/nbt1288
Sharma, D., Misba, L., and Khan, A. U. (2019). Antibiotics versus Biofilm: an Emerging Battleground in Microbial Communities. Antimicrob. Resist. Infect. Control. 8, 76. doi:10.1186/s13756-019-0533-3
Shaw‐Taylor, L. (2020). An Introduction to the History of Infectious Diseases, Epidemics and the Early Phases of the Long‐Run Decline in Mortality†. Econ. Hist. Rev. 73, E1–E19. doi:10.1111/ehr.13019
Singer, M., Deutschman, C. S., Seymour, C. W., Shankar-Hari, M., Annane, D., Bauer, M., et al. (2016). The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 315, 801–810. doi:10.1001/jama.2016.0287
Stærk, K., Grønnemose, R. B., Palarasah, Y., Kolmos, H. J., Lund, L., Alm, M., et al. (2021). A Novel Device-Integrated Drug Delivery System for Local Inhibition of Urinary Tract Infection. Front. Microbiol. 12, 685698. doi:10.3389/fmicb.2021.685698
Subramanya, S. H., Czyż, D. M., Acharya, K. P., and Humphreys, H. (2021). The Potential Impact of the COVID-19 Pandemic on Antimicrobial Resistance and Antibiotic Stewardship. VirusDis. 32, 330–337. doi:10.1007/s13337-021-00695-2
Sullivan, K. V., and Dien Bard, J. (2019). New and Novel Rapid Diagnostics that Are Impacting Infection Prevention and Antimicrobial Stewardship. Curr. Opin. Infect. Dis. 32, 356–364. doi:10.1097/QCO.0000000000000565
Sweeney, T. E., Wong, H. R., and Khatri, P. (2016). Robust Classification of Bacterial and Viral Infections via Integrated Host Gene Expression Diagnostics. Sci. Transl. Med. 8, 346ra91. doi:10.1126/scitranslmed.aaf7165
Vestby, L. K., Grønseth, T., Simm, R., and Nesse, L. L. (2020). Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 9, 59. doi:10.3390/antibiotics9020059
Wang, J., Li, J., Guo, G., Wang, Q., Tang, J., Zhao, Y., et al. (2016). Silver-nanoparticles-modified Biomaterial Surface Resistant to staphylococcus: New Insight into the Antimicrobial Action of Silver. Sci. Rep. 6, 32699. doi:10.1038/srep32699
Wu, B., Catherine), , Haney, E. F., Akhoundsadegh, N., Pletzer, D., Trimble, M. J., et al. (2021). Human Organoid Biofilm Model for Assessing Antibiofilm Activity of Novel Agents. NPJ Biofilms Microbiomes 7, 1–13. doi:10.1038/s41522-020-00182-4
Wu, H., Moser, C., Wang, H.-Z., Høiby, N., and Song, Z.-J. (2015). Strategies for Combating Bacterial Biofilm Infections. Int. J. Oral Sci. 7, 1–7. doi:10.1038/ijos.2014.65
Yeung, A. T. Y., Choi, Y. H., Lee, A. H. Y., Hale, C., Ponstingl, H., Pickard, D., et al. (2019). A Genome-Wide Knockout Screen in Human Macrophages Identified Host Factors Modulating Salmonella Infection. mBio 10, e02169–19. doi:10.1128/mBio.02169-19
Keywords: antibiotics, antibiotic failure, antimicrobial resistance, biofilm, host defence peptides, sepsis
Citation: Haney EF and Hancock REW (2022) Addressing Antibiotic Failure—Beyond Genetically Encoded Antimicrobial Resistance. Front. Drug. Discov. 2:892975. doi: 10.3389/fddsv.2022.892975
Received: 09 March 2022; Accepted: 14 March 2022;
Published: 30 May 2022.
Edited by:
Stefania Galdiero, University of Naples Federico II, ItalyReviewed by:
William William Shafer, Emory University, United StatesCopyright © 2022 Haney and Hancock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert E. W. Hancock, Ym9iQGhhbmNvY2tsYWIuY29t
†These authors have contributed equally to this work
 Evan F. Haney
Evan F. Haney Robert E. W. Hancock
Robert E. W. Hancock