- Faculty of Biology, Technion—Israel Institute of Technology, Haifa, Israel
Gonadotropin-releasing hormone (GnRH) stimulates the expression of multiple genes in the pituitary gonadotropes, most notably to induce synthesis of the gonadotropins, luteinizing hormone (LH), and follicle-stimulating hormone (FSH), but also to ensure the appropriate functioning of these cells at the center of the mammalian reproductive endocrine axis. Aside from the activation of gene-specific transcription factors, GnRH stimulates through its membrane-bound receptor, alterations in the chromatin that facilitate transcription of its target genes. These include changes in the histone and DNA modifications, nucleosome positioning, and chromatin packaging at the regulatory regions of each gene. The requirements for each of these events vary according to the DNA sequence which determines the basal chromatin packaging at the regulatory regions. Despite considerable progress in this field in recent years, we are only beginning to understand some of the complexities involved in the role and regulation of this chromatin structure, including new modifications, extensive cross talk, histone variants, and the actions of distal enhancers and non-coding RNAs. This short review aims to integrate the latest findings on GnRH-induced alterations in the chromatin of its target genes, which indicate multiple and diverse actions. Understanding these processes is illuminating not only in the context of the activation of these hormones during the reproductive life span but may also reveal how aberrant epigenetic regulation of these genes leads to sub-fertility.
Introduction
Gonadotropin-releasing hormone (GnRH) regulates the expression of multiple gonadotropic genes [e.g., Ref. (1–4)], to control population size (5–7), differentiation (8), morphology, and migration (9–11) as well as response to other regulatory hormones [e.g., Ref. (12–15)]. The GnRH receptor (GnRHR)-induced activation of MAP- and other kinase pathways (16–18), culminates in expression and/or activation of gene-specific transcription factors [e.g., Ref. (19–25)], allowing them to bind the DNA and stimulate transcription, often via recruitment of coactivators which catalyze chromatin modifications [e.g., (19, 26, 27)]. However, some MAPKs are associated with the chromatin, where they phosphorylate histones (28, 29), and GnRH also targets several chromatin and DNA-modifying genes directly [Ref. (8, 19, 30, 31); Figure 1], indicating much broader mechanisms for moderating chromatin organization.
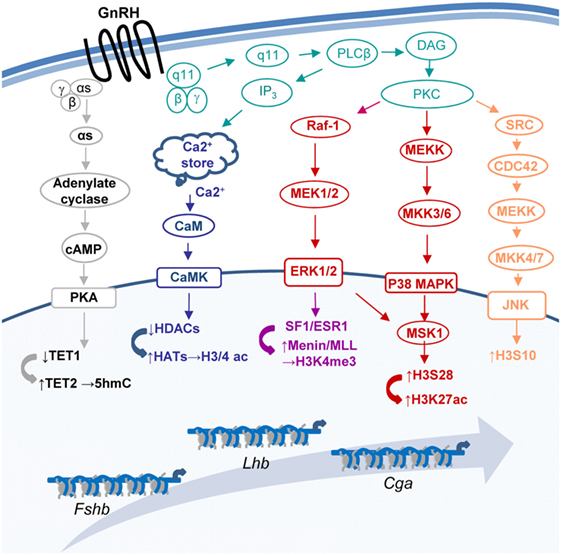
Figure 1. Some of the pathways through which gonadotropin-releasing hormone (GnRH) modifies the chromatin at the three gonadotropin subunit genes. GnRH binds its receptor (GnRHR) to activate a number of pathways that modify the chromatin and lead to changes in expression of the genes encoding the common gonadotropin α-subunit (Cga) and the hormone specific β-subunits of luteinizing hormone (Lhb) and follicle-stimulating hormone (Fshb).
Although the nucleosome is usually highly stable, chromatin structure is dynamic, and this plays a role in determining the accessibility of regulatory DNA, via chromatin modifications which alter nucleosome behavior (32). Many histone modifications occur on the N-terminal tails; some affect contact with DNA through altering histone charge, while others “write” a signal which is recognized by protein effectors [“readers” (33, 34)]. However, chromatin-modifying complexes often comprise multiple components with activities to both “read” and “write” various modifications including those on DNA, as well as ATP-dependent remodeling enzymes that reposition or reorganize the nucleosomes to facilitate transcription initiation and also transition of RNAPII through nucleosomes. Such a diversity of distinct enzymes in a single complex allows for sophisticated dialog and cross talk (33, 35, 36).
This short review will highlight the multiple ways through which GnRH targets the chromatin at the gonadotropin genes which, in addition to clarifying the regulation of these genes during development, should lead to greater understanding of how aberrant epigenetic regulation of these genes might underlie fertility problems.
Histone Acetylation and Deacetylation
By neutralizing the positive charge of lysines on histone N-terminal tails, acetylation at this residue disrupts histone–DNA interactions to make chromatin more accessible, and is thus commonly found at active regions of the genome. Accordingly, basal expression levels of the three gonadotropin genes in partially differentiated gonadotrope-precursor αT3-1 cells closely correlate with levels of H3 acetylation and inversely with H3 occupancy (31). Although H3 and H4 undergo acetylation at various lysines, their differential acetylation at the N-terminus may have a redundant role in transcription such that, in certain contexts, the cumulative charge neutralization influences the transcriptional outcome of a gene more than the acetylation of any specific lysine (37, 38). However, acetylated lysines can be recognized by bromodomain proteins, including multiple chromatin-modifying and remodeling enzymes, such that this modification may well function as a specific recognition site for additional transcriptional activators (39). GnRH increases gonadotrope H3 acetylation, seen both globally and at the 5′ end of the Cga gene which encodes the gonadotropin common α subunit, indicating that this comprises part of the regulatory mechanism of GnRH-induced upregulation of gene expression (31). Histone acetylation is catalyzed by the histone acetyl transferase (HAT) activity of several common transcriptional coactivators, some of which have been shown to mediate hormonally-induced expression of the gonadotropin genes (19, 26, 40).
The opposing activity is executed by histone deacetylases (HDACs) which repress expression of the gonadotropin β-subunit genes in gonadotrope precursor cells (22, 41). Exposure of these cells to GnRH allows de-repression of the Lhb and Fshb genes as a result of activation of calmodulin-dependent kinases, which phosphorylate class II HDACs associated with the gene promoters, leading to their nuclear export (17, 22, 41). A similar mechanism may be responsible for the repression of Fshb in the more fully differentiated LβT2 cell line, as GnRH or an HDAC inhibitor facilitated its expression quite specifically, indicating repression by HDACs, which is overcome by GnRH (42, 43).
Both HAT and HDAC enzymes are characteristically found in large multiprotein complexes whose recruitment may follow other chromatin modifications, while they often also recruit additional modifying enzymes to these loci to provide elaborate cross talk (44). In partially differentiated gonadotropes, SIN3A and SMRT corepressors were found at the Fshb gene promoter, together with class I and class II HDACs. GnRH treatment caused loss of SIN3A and HDAC association, and various components were displaced following knockdown of HDAC4 or either of the co-repressors, suggesting their central roles in this complex (22). The nature of the repressive HDAC complex at the Lhb gene is less clear, but as this gene is regulated by DNA methylation and by TET1 (8), additional modifying enzymes are clearly involved: HDAC-containing complexes are often recruited by methylated DNA-binding proteins (MBPs) or DNA methyl transferases (DNMTs), and the HDAC-containing PRC2 complex contains also EZH2 which represses transcription by catalyzing H3K27me3 (35, 45).
On the other hand, HATs are often associated with other chromatin-activating enzymes, including additional HATs and chromatin-remodeling enzymes, as well as chromodomain proteins that bind H3K4me3. The chromodomain-helicase–DNA-binding domain protein 1 (CHD1) in yeast SAGA/SLIK HAT complexes is recruited to gene promoters in this way, thereby facilitating HAT activity (46) while also altering nucleosomal stability or turnover. Notably, CHD1 is found at the active Cga promoter, but its levels were significantly reduced after disruption of enhancer function, and it was not found at the Lhb gene promoter (47, 48). This drop in CHD1 association with the Cga gene was accompanied by a drop in histone acetylation and H3K4me3 levels which suggests a common pathway/complex of recruiting these enzymes, although the exact mechanism has still to be elucidated. We did not measure histone phosphorylation in this context, but histone acetylation has also been linked to phosphorylation, since discovery that the HAT GCN5 binds preferentially phosphorylated H3S10 which couples these modifications in EGF-induced transcription (49–51).
Histone Phosphorylation
Given that GnRHR signaling involves activation of several MAPKs, it is not surprising that GnRH induces histone phosphorylation, seen globally and at the gonadotropin promoters (31). Some of the MAPKs activated by GnRH, including JNK, can target histone phosphorylation directly (28, 29) and this kinase is responsible for H3S10p at the Cga promoter. The phosphorylation of H3S28 at the Cga 5′end is via GnRH-activation of MSK1, which is a downstream target of ERK and p38 MAPK (31).
By introducing a positive charge, this phosphorylation would be expected to destabilize the DNA–histone interactions and thus aide in passage through the nucleosome (32). However, the role of H3 phosphorylation in transcriptional regulation is still not fully understood, as it is seen at both repressed and active genes (52, 53). Notably, phosphorylation of H3S10 on the Cga gene 5′ end/promoter, even though it is increased in response to GnRH, appears to have little function on on-going and/or hormonally upregulated Cga transcription, as no effect was noted when its basal levels were reduced by over 90%. However, H3S28p at the first nucleosome in the transcribed region appears to play a role in elongation, presumably facilitating RNAPII transition through this nucleosome, which is likely aided by GnRH-induced H3K27ac (31).
H3S10 or S28 phosphorylation may also alter the affinity of additional chromatin binding proteins to their targets, as reported above for GCN5 (49, 54). At the Cga gene, H3K9ac appeared independent of H3S10p, while inhibition of H3S28p was accompanied by a drop in global levels of H3K27ac, although H3S18ac was unaffected (31). In other contexts, H3S10p plays a more crucial role in the activation of repressed genes, as it can trigger the displacement of the HP1γ repressor, and facilitate recruitment of the SWI/SNF chromatin-remodeling enzyme Brg1 and RNAPII (55, 56). The interpretation of histone phosphorylation thus appears to be highly context specific, involving cross talk with neighboring histone residues to determine the precise outcome.
Histone Methylation
The mono-, di- or trimethylation of specific lysines by histone methyltransferases and demethylases distinguishes transcriptionally active from inactive chromatin domains [reviewed by Ref. (57)]. At the gonadotropin genes, levels of H3K4 trimethylation (H3K4me3) which marks the 5′ ends and/or promoters of all actively transcribed genes and is essential for transcription initiation (58), correlate well with basal expression levels and increase following GnRH exposure (27). Mammals have six distinct complexes capable of catalyzing this modification, with multiple subunits affording different mechanisms of recruitment and regulation (59, 60). The mixed-lineage leukemia (MLL)-COMPASS-like complex 1/2 is recruited to the gonadotropin genes during their upregulation by GnRH and is responsible for the GnRH-induced increase in H3K4me3 at all three gonadotropin promoters (27). Unique among the Set1/COMPASS-like complexes, the MLL1/2 complex contains menin (60), which interacts with various gene-specific transcription factors including ERα (61, 62) and Sf-1 (27). The GnRH-induced association of menin with the β-subunit genes, is dependent on Sf-1. Sf-1 recruits ERα to the Lhb promoter, after GnRH-induced modification of both factors. Both of these factors appear to play roles in the recruitment of the MLL1/2 complex and thus also H3K4me3 at these genes (12, 21, 27).
The GnRH-induction of H3K4me3 at the gonadotropin genes alters nucleosomal occupancy, and there was increased association of H3 at the promoters following inhibition of menin which was not overcome by exposure to GnRH (27). H3K4me3 was previously reported to play a role in maintaining low nucleosomal occupancy, likely due to its ability to bind chromatin-remodeling enzymes such as CHD1 and ISWI (46, 63, 64). As described above, it also helps recruit HAT complexes (65, 66) and TFIID (67). However, whether the menin-dependent loss of H3 at the gonadotropin gene promoters following GnRH exposure involves such a mechanism is not yet clear.
Trimethylation of H3K36 (H3K36me3) in the coding regions of all three gonadotropin genes also increases following exposure to GnRH (27). This is likely a direct consequence of increased transcription rates as it is catalyzed by Set2 which is recruited by the elongating S2p form of RNAPII [reviewed by Ref. (68)], and the increased level of this modification at the gonadotropin genes correlates with elevated association of RNAPII (27). H3K36me3 is reported to have a number of roles including regulating histone exchange (69), suppression of initiation through recruitment of DNMT3b and intragenic DNA methylation (70), RNA processing (71), chromatin organization (72), and others, which have yet to be explored in this context.
H2B Ubiquitination
Monoubiquitinated H2B at lysine 120 (H2BK120ub) is generally associated with actively transcribed genes, as first reported in yeast, although its precise function in mammals has been controversial. It is reportedly required for recruitment of the Set1/COMPASS complex to gene promoters due to its recognition by one of the complex subunits. This ubiquitin is later removed by a component of the SAGA HAT complex to allow recruitment of the kinase that phosphorylates RNAPII at S2, thus signaling promoter escape and elongation (68, 73). However, it appears that for many mammalian genes H2BK120ub is found primarily in the transcribed region where it plays a role in the reassembly of nucleosomes in the wake of elongating RNAPII (74–76). The presence and requirement of H2BK120ub at mammalian gene promoters has been disputed, perhaps in part due to the different chromatin organization in the various genes studied and the associated diverse transcription dynamics (see below), and possibly also to earlier experimental protocols which mapped its exact genomic location with poorer resolution.
The apparent lack of requirement for H2BK120ub at some mammalian promoters is likely due to the more numerous complexes that can catalyze H3K4me3 than found in yeast. As described above, these complexes contain distinct subunits which allow recruitment of the lysine methyl transferase (KMT) complex through diverse proteins including transcription factors. Accordingly, there is no apparent correlation between levels of promoter H2BK120ub and expression levels of the three gonadotropin genes, nor with levels of H3K4me3 at their promoters. However, GnRH induces a major increase in H2BK120ub levels globally, and specifically in the coding regions of the gonadotropin genes, in keeping with the changes in coding region H3K36me3 described above (27). The fact that this elevation in H2BK120ub was also noted globally suggests that it is a common event at the multiple genes upregulated by GnRH.
Histone Citrullination
Histone citrullination, in which histone tail arginine residues are converted by peptidylarginine deiminase (PAD) enzymes to citrulline has been observed but is still poorly understood. The citrullination is thought to induce chromatin decondensation, and was recently shown to be particularly crucial for transcriptional activation during early embryonic development (77, 78). Notably, PAD family members are highly expressed in female reproductive tissues, have been indicated to play a critical role in female reproduction, and a correlation was seen particularly between PAD2 expression levels, and stages of the estrous cycle (79). Moreover, GnRH was reported to induce PAD2 nuclear localization in gonadotropes, where it stimulates citrullination of H3 at R2, R8, and R17. The inhibition of this activity was seen to blunt the GnRH stimulatory effect on the gonadotropin β-subunit genes, suggesting that histone citrullination mediates part of the GnRH effect, although it has yet to be shown whether it targets the gonadotropin genes directly and the exact mechanism involved (30).
The DNA-Modifying TET Enzymes
DNA methylation is generally considered a stable mechanism to repress gene expression, often in concert with repressive histone modifications. However, since the discovery of the TET family of enzymes (80, 81), it has become clear that methylated cytosines (5mC) can be hydroxylated to 5hmC, often imparting a very different function as the 5hmC modified-DNA is not recognized similarly by some of the 5mC-binding proteins (82) which can thus lead to de-repression through “functional demethylation.” The maintenance DNMT, DNMT1 also binds 5hmC DNA with much lower affinity than to 5mC DNA, leading to a passive demethylation in replicating cells (83, 84). More recently, the TET enzymes were noted to catalyze additional modification of 5hmC, to bases that are quickly removed by the base-excision repair mechanism, in a form of active demethylation (85, 86). Despite this potential of the TET enzymes to relieve inhibition of gene expression, TET1 is also sometimes found in repressor complexes in association with other inhibitory chromatin-modifying enzymes (87–89).
In this context, TET1, which is highly expressed in gonadotrope-precursor cells, was found to repress the Lhb gene in association with promoter H3K27 methylation, possibly playing a role in recruitment of the KMT enzyme to this locus. Tet1 expression is downregulated to allow the precursor gonadotropes to complete their differentiation, following exposure to GnRH and also in response to estrogens or androgens via steroid receptors that bind the Tet1 promoter (8). With developmental or experimentally-induced downregulation of TET1, it is replaced at the Lhb gene promoter by TET2 which hydroxymethylates the methylated CpGs on this gene promoter so, in concert with the GnRH-activated transcription factors, facilitating Lhb expression (8). In this way, the exposure of the partially differentiated gonadotrope precursors to GnRH promotes their final differentiation via downregulation of Tet1 and the ensuing elevation in gonadal steroids provides the feedback to keep Tet1 repressed.
Nucleosomal Organization and Remodeling
For all three gonadotropin genes, nucleosome levels drop following GnRH treatment, reflecting histone displacement (27, 31). However, the organization of the chromatin at the Cga and Lhb gene promoters in functional gonadotropes differs markedly, in accordance with their distinct expression levels and means of regulation. In the gonadotropes, the Cga proximal promoter, similar to that of many other highly expressed genes but unlike its state in non-gonadotropes, is exposed and accessible to transcription factors, such that transcription may be initiated quite easily (48). Upregulation of Cga expression by GnRH is, therefore, likely directed primarily at the level of RNAPII promoter escape and elongation such that after exposure to GnRH it is quickly released from the promoter (27, 48). This is reflected in much higher levels of RNAPII at the Cga than Lhb promoter in unstimulated mature gonadotropes, and the fact that the first nucleosome in the coding region, characteristic of genes with high RNAPII occupancy (90), is positioned 10 bp further downstream than at the Lhb gene. RNAPII transition through this nucleosome is clearly facilitated by the incorporation of a histone H2A variant which allows greater nucleosomal mobility (48), and likely further enhanced by GnRH-induced modifications targeting the histones in this nucleosome (Figure 2).
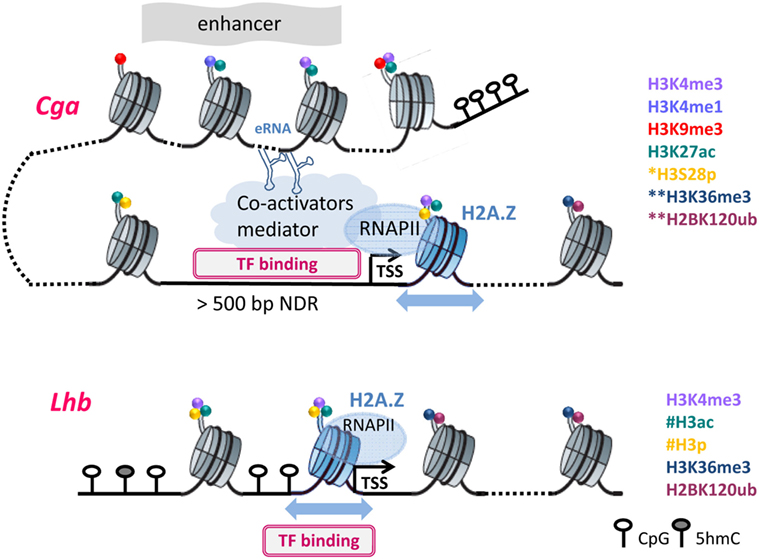
Figure 2. The different chromatin organization necessitates distinct modifications to facilitate transcription: Chromatin organization at Cga and Lhb regulatory regions are shown in their active states, with DNA and histone modifications as noted. *S28p was examined only at the proximal region of Cga; **at the downstream nucleosomes, only H3K36me3 and H2K120Bub were assessed; #histone acetylation and phosphorylation are inferred but were not measured directly on the Lhb promoter. Furthermore, the nucleosomal density on the Lhb proximal promoter precluded precise mapping of H3K4me3. Dotted lines represent DNA packaged into additional nucleosomes that are not shown, and the blue arrows represent nucleosome mobility due to H2A.Z incorporation. Further details can be found in the text.
This organization of the Cga gene differs fundamentally from that of Lhb whose proximal promoter is packaged into a nucleosome (Figure 2) that encompasses binding sites of Sf-1, Pitx-1, and Egr-1, which activate this gene (91, 92). The initiation of transcription must thus require reorganization to allow access of these factors to the DNA, likely facilitated by the incorporation of H2A.Z at the Lhb promoter (48, 93). The binding of TFs to sites that are buried inside the nucleosome is thought to be modulated by the thermally driven spontaneous “breathing” (94). This involves partial wrapping and unwrapping of the DNA at the entry and exit to nucleosome which can expose the binding site, such that its ability to bind is also a function of the distance of the binding site from the nucleosome dyad (95, 96). These fluctuations, which are typically much faster than rates of nucleosome repositioning, likely work together with increased nucleosome mobility to facilitate the initial access of Sf-1 and/or Pitx-1 to their binding sites (93). Binding of either of these “pioneer” factors would destabilize the nucleosome both through the binding itself and the recruitment of the histone-modifying enzymes described above, as well as quite possibly ATP-dependent chromatin-remodeling enzymes. Clearly there is much more work to be done in order to understand the various components and their intricate roles in the activation of this gene, and also whether the nucleosomes at the Fshb gene promoter are similarly organized and modified following GnRH exposure.
Concluding Comments
The organization of the chromatin at the gonadotropin genes and its GnRH-induced modifications that facilitate transcription is crucial in understanding how these genes are activated during the reproductive lifespan, but may also have implications in non-pituitary GnRHR-expressing cancer cells. There is increasing indication, however, that much larger distal genomic regions function to determine expression of specific genes, as shown for the eRNA that regulates Cga chromatin (47, 97). The likelihood that genes are regulated by a variety of distal enhancers in various scenarios (97), points to highly complex gene-regulation in distinct developmental, hormonally activated and pathological contexts, while emphasizing the importance of the chromatin architecture in extensive genomic regions. As the epigenome is susceptible to external perturbations, elucidation of the full complement of elements that regulate gonadotropin gene expression and their chromatin organization will further our understanding of abnormal gonadotropin levels and the ensuing pituitary-origin reproductive disorders, while opening the way for epigenetic targeting as a basis for fertility drug development and treatment.
Author Contributions
All authors contributed to this review, and all have read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past coauthorship with one of the authors PM.
Funding
This work was supported by funding from The Israel Science Foundation (grant 840/12 and 1850/17 to PM).
References
1. Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. Gonadotropin-releasing hormone receptor-coupled gene network organization. J Biol Chem (2001) 276:47195–201. doi:10.1074/jbc.M108716200
2. Kakar SS, Winters SJ, Zacharias W, Miller DM, Flynn S. Identification of distinct gene expression profiles associated with treatment of LβT2 cells with gonadotropin-releasing hormone agonist using microarray analysis. Gene (2003) 308:67–77. doi:10.1016/S0378-1119(03)00446-3
3. Ruf F, Park MJ, Hayot F, Lin G, Roysam B, Ge YC, et al. Mixed analog/digital gonadotrope biosynthetic response to gonadotropin-releasing hormone. J Biol Chem (2006) 281:30967–78. doi:10.1074/jbc.M606486200
4. Zhang H, Bailey JS, Coss D, Lin B, Tsutsumi R, Lawson MA, et al. Activin modulates the transcriptional response of LβT2 cells to gonadotropin-releasing hormone and alters cellular proliferation. Mol Endocrinol (2006) 20:2909–30. doi:10.1210/me.2006-0109
5. Savulescu D, Feng J, Ping YS, Mai O, Boehm U, He B, et al. Gonadotropin-releasing hormone-regulated prohibitin mediates apoptosis of the gonadotrope cells. Mol Endocrinol (2013) 27:1856–70. doi:10.1210/me.2013-1210
6. Feng J, Lawson MA, Melamed P. A proteomic comparison of immature and mature mouse gonadotrophs reveals novel differentially expressed nuclear proteins that regulate gonadotropin gene transcription and RNA splicing. Biol Reprod (2008) 79:546–61. doi:10.1095/biolreprod.108.068106
7. Miles LE, Hanyaloglu AC, Dromey JR, Pfleger KD, Eidne KA. Gonadotropin-releasing hormone receptor-mediated growth suppression of immortalized LbetaT2 gonadotrope and stable HEK293 cell lines. Endocrinology (2004) 145:194–204. doi:10.1210/en.2003-0551
8. Yosefzon Y, David C, Tsukerman A, Pnueli L, Qiao S, Boehm U, et al. An epigenetic switch repressing Tet1 in gonadotropes activates the reproductive axis. Proc Natl Acad Sci U S A (2017) 114:10131–6. doi:10.1073/pnas.1704393114
9. Rahamim-Ben Navi L, Tsukerman A, Feldman A, Melamed P, Tomić M, Stojilkovic SS, et al. GnRH induces ERK-dependent bleb formation in gonadotrope cells, involving recruitment of members of a GnRH receptor-associated signalosome to the blebs. Front Endocrinol (2017) 8:113. doi:10.3389/fendo.2017.00113
10. Navratil AM, Knoll JG, Whitesell JD, Tobet SA, Clay CM. Neuroendocrine plasticity in the anterior pituitary: gonadotropin-releasing hormone-mediated movement in vitro and in vivo. Endocrinology (2007) 148:1736–44. doi:10.1210/en.2006-1153
11. Godoy J, Nishimura M, Webster NJ. Gonadotropin-releasing hormone induces miR-132 and miR-212 to regulate cellular morphology and migration in immortalized LbetaT2 pituitary gonadotrope cells. Mol Endocrinol (2011) 25:810–20. doi:10.1210/me.2010-0352
12. Luo M, Koh M, Feng J, Wu Q, Melamed P. Cross talk in hormonally regulated gene transcription through induction of estrogen receptor ubiquitylation. Mol Cell Biol (2005) 25:7386–98. doi:10.1128/MCB.25.16.7386-7398.2005
13. Kotitschke A, Sadie-Van Gijsen H, Avenant C, Fernandes S, Hapgood JP. Genomic and nongenomic cross talk between the gonadotropin-releasing hormone receptor and glucocorticoid receptor signaling pathways. Mol Endocrinol (2009) 23:1726–45. doi:10.1210/me.2008-0462
14. Chen J, An BS, So WK, Cheng L, Hammond GL, Leung PC. Gonadotropin-releasing hormone-I-mediated activation of progesterone receptor contributes to gonadotropin alpha-subunit expression in mouse gonadotrophs. Endocrinology (2010) 151:1204–11. doi:10.1210/en.2009-0816
15. Thackray VG, Mellon PL, Coss D. Hormones in synergy: regulation of the pituitary gonadotropin genes. Mol Cell Endocrinol (2010) 314:192–203. doi:10.1016/j.mce.2009.09.003
16. Naor Z. Signaling by G-protein-coupled receptor (GPCR): studies on the GnRH receptor. Front Neuroendocrinol (2009) 30:10–29. doi:10.1016/j.yfrne.2008.07.001
17. Melamed P, Savulescu D, Lim S, Wijeweera A, Luo Z, Luo M, et al. Gonadotrophin-releasing hormone signalling downstream of calmodulin. J Neuroendocrinol (2012) 24:1463–75. doi:10.1111/j.1365-2826.2012.02359.x
18. Mugami S, Dobkin-Bekman M, Rahamim-Ben Navi L, Naor Z. Differential roles of PKC isoforms (PKCs) in GnRH stimulation of MAPK phosphorylation in gonadotrope derived cells. Mol Cell Endocrinol (2018) 463:97–105. doi:10.1016/j.mce.2017.04.004
19. Melamed P, Kadir MN, Wijeweera A, Seah S. Transcription of gonadotropin beta subunit genes involves cross-talk between the transcription factors and co-regulators that mediate actions of the regulatory hormones. Mol Cell Endocrinol (2006) 252:167–83. doi:10.1016/j.mce.2006.03.024
20. Lim S, Pnueli L, Tan JH, Naor Z, Rajagopal G, Melamed P. Negative feedback governs gonadotrope frequency-decoding of gonadotropin releasing hormone pulse-frequency. PLoS One (2009) 4:e7244. doi:10.1371/journal.pone.0007244
21. Luo Z, Wijeweera A, Oh Y, Liou YC, Melamed P. Pin1 facilitates the phosphorylation-dependent ubiquitination of SF-1 to regulate gonadotropin beta-subunit gene transcription. Mol Cell Biol (2010) 30:745–63. doi:10.1128/MCB.00807-09
22. Lim S, Luo M, Koh M, Yang M, Bin Abdul Kadir MN, Tan JH, et al. Distinct mechanisms involving diverse histone deacetylases repress expression of the two gonadotropin beta-subunit genes in immature gonadotropes, and their actions are overcome by gonadotropin-releasing hormone. Mol Cell Biol (2007) 27:4105–20. doi:10.1128/MCB.00248-07
23. Pnueli L, Luo M, Wang S, Naor Z, Melamed P. Calcineurin mediates the gonadotropin-releasing hormone effect on expression of both subunits of the follicle-stimulating hormone through distinct mechanisms. Mol Cell Biol (2011) 31:5023–36. doi:10.1128/MCB.06083-11
24. Ely HA, Mellon PL, Coss D. GnRH induces the c-Fos gene via phosphorylation of SRF by the calcium/calmodulin kinase II pathway. Mol Endocrinol (2011) 25:669–80. doi:10.1210/me.2010-0437
25. Thompson IR, Ciccone NA, Xu S, Zaytseva S, Carroll RS, Kaiser UB. GnRH pulse frequency-dependent stimulation of FSHβ transcription is mediated via activation of PKA and CREB. Mol Endocrinol (2013) 27:606–18. doi:10.1210/me.2012-1281
26. Mouillet JF, Sonnenberg-Hirche C, Yan X, Sadovsky Y. p300 regulates the synergy of steroidogenic factor-1 and early growth response-1 in activating luteinizing hormone-beta subunit gene. J Biol Chem (2004) 279:7832–9. doi:10.1074/jbc.M312574200
27. Wijeweera A, Haj M, Feldman A, Pnueli L, Luo Z, Melamed P. Gonadotropin gene transcription is activated by menin-mediated effects on the chromatin. Biochim Biophys Acta (2015) 1849:328–41. doi:10.1016/j.bbagrm.2015.01.001
28. Tiwari VK, Stadler MB, Wirbelauer C, Paro R, Schübeler D, Beisel C. A chromatin-modifying function of JNK during stem cell differentiation. Nat Genet (2011) 44:94–100. doi:10.1038/ng.1036
29. Klein AM, Zaganjor E, Cobb MH. Chromatin-tethered MAPKs. Curr Opin Cell Biol (2013) 25:272–7. doi:10.1016/j.ceb.2013.01.002
30. Khan SA, Edwards BS, Muth A, Thompson PR, Cherrington BD, Navratil AM. GnRH stimulates peptidylarginine deiminase catalyzed histone citrullination in gonadotrope cells. Mol Endocrinol (2016) 30:1081–91. doi:10.1210/me.2016-1085
31. Haj M, Wijeweera A, Rudnizky S, Taunton J, Pnueli L, Melamed P. Mitogen and stress-activated protein kinase 1 is required for gonadotropin-releasing hormone-mediated activation of gonadotropin α subunit expression. J Biol Chem (2017) 292:20720–31. doi:10.1074/jbc.M117.797845
32. Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol (2013) 20:259–66. doi:10.1038/nsmb.2470
33. Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem (2011) 80:473–99. doi:10.1146/annurev-biochem-061809-175347
34. Patel DJ, Wang Z. Readout of epigenetic modifications. Annu Rev Biochem (2013) 82:81–118. doi:10.1146/annurev-biochem-072711-165700
35. Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications – writers that read. EMBO Rep (2015) 16:1467–81. doi:10.15252/embr.201540945
36. Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet (2016) 32:42–56. doi:10.1016/j.tig.2015.10.007
37. Martin AM, Pouchnik DJ, Walker JL, Wyrick JJ. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics (2004) 167:1123–32. doi:10.1534/genetics.104.026674
38. Anamika K, Krebs AR, Thompson J, Poch O, Devys D, Tora L. Lessons from genome-wide studies: an integrated definition of the coactivator function of histone acetyl transferases. Epigenetics Chromatin (2010) 3:18. doi:10.1186/1756-8935-3-18
39. Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett (2002) 513:124–8. doi:10.1016/S0014-5793(01)03309-9
40. Miller RS, Wolfe A, He L, Radovick S, Wondisford FE. CREB binding protein (CBP) activation is required for luteinizing hormone beta expression and normal fertility in mice. Mol Cell Biol (2012) 32:2349–58. doi:10.1128/MCB.00394-12
41. Melamed P. Histone deacetylases and repression of the gonadotropin genes. Trends Endocrinol Metab (2008) 19:25–31. doi:10.1016/j.tem.2007.10.008
42. Oride A, Kanasaki H, Mijiddorj T, Sukhbaatar U, Miyazaki K. Trichostatin A specifically stimulates gonadotropin FSHβ gene expression in gonadotroph LβT2 cells. Endocr J (2014) 61:335–42. doi:10.1507/endocrj.EJ13-0411
43. Mijiddorj T, Kanasaki H, Sukhbaatar U, Oride A, Ishihara T, Kyo S. Retinoic acid and retinaldehyde dehydrogenase are not involved in the specific induction of the follicle-stimulating hormone β subunit by trichostatin A, a selective inhibitor of histone deacetylase. Gen Comp Endocrinol (2017) 242:59–65. doi:10.1016/j.ygcen.2015.11.020
44. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res (2011) 21:381–95. doi:10.1038/cr.2011.22
45. Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet (2009) 10:295–304. doi:10.1038/nrg2540
46. Pray-Grant MG, Daniel JA, Schieltz D, Yates JR III, Grant PA. CHD1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature (2005) 433:434–8. doi:10.1038/nature03242
47. Pnueli L, Rudnizky S, Yosefzon Y, Melamed P. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin α-subunit gene. Proc Natl Acad Sci U S A (2015) 112:4369–74. doi:10.1073/pnas.1414841112
48. Rudnizky S, Bavly A, Malik O, Pnueli L, Melamed P, Kaplan A. H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat Commun (2016) 7:12958. doi:10.1038/ncomms12958
49. Sawicka A, Seiser C. Histone H3 phosphorylation – a versatile chromatin modification for different occasions. Biochimie (2012) 94:2193–201. doi:10.1016/j.biochi.2012.04.018
50. Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell (2000) 5:917–26. doi:10.1016/S1097-2765(00)80257-9
51. Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell (2000) 5:905–15. doi:10.1016/S1097-2765(00)80256-7
52. Baek SH. When signaling kinases meet histones and histone modifiers in the nucleus. Mol Cell (2011) 42:274–84. doi:10.1016/j.molcel.2011.03.022
53. Rossetto D, Avvakumov N, Côté J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics (2012) 7:1098–108. doi:10.4161/epi.21975
54. Sawicka A, Seiser C. Sensing core histone phosphorylation – a matter of perfect timing. Biochim Biophys Acta (2014) 1839:711–8. doi:10.1016/j.bbagrm.2014.04.013
55. Vicent GP, Ballaré C, Zaurin R, Saragüeta P, Beato M. Chromatin remodeling and control of cell proliferation by progestins via cross talk of progesterone receptor with the estrogen receptors and kinase signaling pathways. Ann N Y Acad Sci (2006) 1089:59–72. doi:10.1196/annals.1386.025
56. Winter S, Simboeck E, Fischle W, Zupkovitz G, Dohnal I, Mechtler K, et al. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J (2008) 27:88–99. doi:10.1038/sj.emboj.7601954
57. Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell (2012) 48:491–507. doi:10.1016/j.molcel.2012.11.006
58. Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol (2009) 29:6074–85. doi:10.1128/MCB.00924-09
59. Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol (2008) 20:341–8. doi:10.1016/j.ceb.2008.03.019
60. Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem (2012) 81:65–95. doi:10.1146/annurev-biochem-051710-134100
61. Dreijerink KM, Mulder KW, Winkler GS, Höppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res (2006) 66:4929–35. doi:10.1158/0008-5472.CAN-05-4461
62. Dreijerink KM, Lips CJ, Timmers HT. Multiple endocrine neoplasia type 1: a chromatin writer’s block. J Intern Med (2009) 266:53–9. doi:10.1111/j.1365-2796.2009.02115.x
63. Sims RJ III, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem (2005) 280:41789–92. doi:10.1074/jbc.C500395200
64. Nikolov M, Stützer A, Mosch K, Krasauskas A, Soeroes S, Stark H, et al. Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Mol Cell Proteomics (2011) 10:M110.005371. doi:10.1074/mcp.M110.005371
65. Petty E, Pillus L. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet (2013) 29:621–9. doi:10.1016/j.tig.2013.06.006
66. Wozniak GG, Strahl BD. Hitting the ‘mark’: interpreting lysine methylation in the context of active transcription. Biochim Biophys Acta (2014) 1839:1353–61. doi:10.1016/j.bbagrm.2014.03.002
67. Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell (2007) 13:58–69. doi:10.1016/j.cell.2007.08.016
68. Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem (2006) 75:243–69. doi:10.1146/annurev.biochem.75.103004.142422
69. Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature (2012) 489:452–5. doi:10.1038/nature11326
70. Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature (2017) 543:72–7. doi:10.1038/nature21373
71. Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science (2010) 327:996–1000. doi:10.1126/science.1184208
72. Ulianov SV, Khrameeva EE, Gavrilov AA, Flyamer IM, Kos P, Mikhaleva EA, et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res (2016) 26:70–84. doi:10.1101/gr.196006.115
73. Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell (2008) 29:653–63. doi:10.1016/j.molcel.2008.02.014
74. Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell (2007) 131:1084–9. doi:10.1016/j.cell.2007.09.046
75. Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell (2008) 31:57–66. doi:10.1016/j.molcel.2008.04.025
76. Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol (2008) 10:483–8. doi:10.1038/ncb1712
77. Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol (2009) 184:205–13. doi:10.1083/jcb.200806072
78. Zhang X, Liu X, Zhang M, Li T, Muth A, Thompson PR, et al. Peptidylarginine deiminase 1-catalyzed histone citrullination is essential for early embryo development. Sci Rep (2016) 6:38727. doi:10.1038/srep38727
79. Horibata S, Coonrod SA, Cherrington BD. Role for peptidylarginine deiminase enzymes in disease and female reproduction. J Reprod Dev (2012) 58:274–82. doi:10.1262/jrd.2011-040
80. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (2009) 324:930–5. doi:10.1126/science.1170116
81. Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature (2010) 466:1129–33. doi:10.1038/nature09303
82. Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol (2010) 11:607–20. doi:10.1038/nrm2950
83. Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res (2012) 40:4841–9. doi:10.1093/nar/gks155
84. Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell (2013) 152:1146–59. doi:10.1016/j.cell.2013.02.004
85. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5carboxylcytosine. Science (2011) 333:1300–3. doi:10.1126/science.1210597
86. He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science (2011) 333:1303–7. doi:10.1126/science.1210944
87. Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature (2011) 473:343–8. doi:10.1038/nature10066
88. Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature (2011) 473:389–93. doi:10.1038/nature09934
89. Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, et al. Mbd3/NURD complex regulates expression of 5hydroxymethylcytosine marked genes in embryonic stem cells. Cell (2011) 147:1498–510. doi:10.1016/j.cell.2011.11.054
90. Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell (2008) 132:887–98. doi:10.1016/j.cell.2008.02.022
91. Tremblay JJ, Drouin J. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone beta gene transcription. Mol Cell Biol (1999) 19:2567–76. doi:10.1128/MCB.19.4.2567
92. Dorn C, Ou Q, Svaren J, Crawford PA, Sadovsky Y. Activation of luteinizing hormone beta gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J Biol Chem (1999) 274:13870–6. doi:10.1074/jbc.274.20.13870
93. Rudnizky S, Malik O, Bavly A, Pnueli L, Melamed P, Kaplan A. Nucleosome mobility and the regulation of gene expression: insights from single-molecule studies. Protein Sci (2017) 26:1266–77. doi:10.1002/pro.3159
94. Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol (1995) 254:130–49. doi:10.1006/jmbi.1995.0606
95. Tims HS, Gurunathan K, Levitus M, Widom J. Dynamics of nucleosome invasion by DNA binding proteins. J Mol Biol (2011) 411:430–48. doi:10.1016/j.jmb.2011.05.044
96. Moyle-Heyrman G, Tims HS, Widom J. Structural constraints in collaborative competition of transcription factors against the nucleosome. J Mol Biol (2011) 412:634–46. doi:10.1016/j.jmb.2011.07.032
Keywords: gonadotropin-releasing hormone, gonadotrope, luteinizing hormone, follicle-stimulating hormone, chromatin, histone, transcription, gene
Citation: Melamed P, Haj M, Yosefzon Y, Rudnizky S, Wijeweera A, Pnueli L and Kaplan A (2018) Multifaceted Targeting of the Chromatin Mediates Gonadotropin-Releasing Hormone Effects on Gene Expression in the Gonadotrope. Front. Endocrinol. 9:58. doi: 10.3389/fendo.2018.00058
Received: 01 November 2017; Accepted: 09 February 2018;
Published: 27 February 2018
Edited by:
Zvi Naor, Tel Aviv University, IsraelReviewed by:
Gregoy Y. Bedecarrats, University of Guelph, CanadaSimon J. Rhodes, Indiana University—Purdue University Indianapolis, United States
Copyright: © 2018 Melamed, Haj, Yosefzon, Rudnizky, Wijeweera, Pnueli and Kaplan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philippa Melamed, philippa@tx.technion.ac.il
 Philippa Melamed
Philippa Melamed Majd Haj
Majd Haj Sergei Rudnizky
Sergei Rudnizky Ariel Kaplan
Ariel Kaplan