- 1Department of Biochemistry and Molecular Biology, University of Nevada, Reno, NV, United States
- 2Department of Agriculture, Veterinary, and Rangeland Sciences, University of Nevada, Reno, NV, United States
Insulin-like peptides (ILPs) have been identified in several invertebrates, particularly insects, and work on these ILPs has revealed many roles including regulation of energy homeostasis, growth, development, and lifespan to name a few. However, information on arthropod ILPs outside of insects is sparse. Studies of Ixodid tick ILPs are particularly scarce, despite their importance as vectors of infectious agents, most notably Lyme disease. The recent publication of the genome of the black-legged tick, Ixodes scapularis, has advanced opportunities to study this organism from a molecular standpoint, a resource sorely needed for an organism with challenging life history requirements for study in the laboratory, such as a long life cycle and obligate, prolonged, blood-feeding at each life stage. Through bioinformatics searches of the tick genome and other available I. scapularis databases, we identified four putative ILP sequences. Full-length sequences of these ILP transcripts were confirmed, and quantitative RT-PCR was used to examine expression levels of these ILPs in different life stages, feeding states, and adult tissues. This work serves as an initial characterization of ILP expression in ticks and provides the foundation for further exploration of the roles of ILPs in these important arthropod vectors.
Introduction
Insulin-like peptides (ILPs) are recognized to have several important functions in arthropods. This has been most extensively characterized in the dipterans Drosophila melanogaster and Aedes aegypti (1–4). Insects have multiple ILPs, ranging from as few as four in planthoppers (5), to eight in Ae. aegypti and D. melanogaster (2, 6, 7), to over 30 in Bombyx mori (8). The functions of these ILPs in most arthropods are largely unexplored, but, where investigated, ILPs have demonstrated pleiotropic roles, regulating metabolism, growth, reproduction, longevity, and larval molt, among other functions (9–11). Tissue expression of ILPs is also variable, with many produced in nervous tissue for neurohemal release. Others are produced in the midgut, fat body, or imaginal discs (7, 12, 13).
ILPs are variable in amino acid sequence, but are united in sharing conserved cysteine residues consistently spaced throughout the molecule. These cysteines form disulfide bridges, folding the peptide into a domain structure required for insulin receptor (IR) activation. The synthesis and post-translational processing of ILPs also is conserved. A prepropeptide is translated with B, C, and A chains (14). After disulfide bonds form, the C-chain is processed at dibasic cleavage sites, leaving behind the joined B and A chains. Some speculate that the C-chain is left intact in certain arthropod ILPs as is the case with human Insulin-like Growth Factor I (IGF-I) (2). However, to date, few native arthropod ILPs have been isolated and none of these examples retain the C-peptide (15).
Rapid genome and transcriptome sequencing, and associated bioinformatics processing of such data, have opened up previously unavailable resources for the study of neuropeptides and peptide hormones in arthropods. Our initial understanding of insect ILP structure, function, and signaling dynamics is the result of the isolation of bombyxin from B. mori (16) and Locusta Insulin-Related Peptide (LIRP) from Locusta migratoria (17). Characterization of individual ILPs through painstaking genetic methods such as PCR amplification with degenerate primers eventually gave way to available genomic and transcriptome databases. The availability of these sequences has permitted the use of genetic techniques such as RNAi and quantitative PCR to advance our knowledge of specific ILP functions. However, extraction, quantification, and synthesis of the ILP molecules themselves for study has remained a challenge for many arthropods.
The above mentioned research on ILPs is dominated by more easily studied model insects, not to mention the exhaustive vertebrate literature on insulin, IGFs, and relaxin, and little information on neuropeptide signaling in arachnids exists. In ticks, immunocytochemical evidence for the presence of ILPs has been reported in Ornithodoros parkerii (18) and Rhipicephalus appendiculatus (19) using an antibody to bombyxin. Immunostaining in Dermacentor variabilis using an anti-insulin antibody demonstrated a decrease in immunoreactivity in the synganglion after blood-feeding and mating (20). A partial fragment of an apparent A-chain of a D. variabilis ILP was detected by 454 pyrosequencing of the transcriptome (21), and a subsequent study showed a decrease in expression of this ILP in partially fed and replete ticks in comparison to unfed ticks (22). In Amblyomma americanum, upregulation of an ILP was detected in pathogen-challenged ticks (23), and knockdown of Insulin-like Growth Factor Binding Protein (IGFBP) in this tick prevented blood-feeding females from feeding to repletion (24).
In the United States, ticks are responsible for 95% of vector borne diseases (25). The publication of the genome sequence of Ixodes scapularis, the black-legged tick, has provided a wealth of information on this important vector (26). This was a much needed resource in our efforts to prevent the debilitating diseases transmitted by these ticks and others, including Lyme disease, babesisosis, anaplasmosis, ehrlichiosis, Rocky Mountain Spotted Fever, Southern Tick-Associated Rash Illness, Tick-Borne Relapsing Fever, tularemia, Colorado tick fever, Powassan encephalitis, and Q fever (25). In addition to its utility as a way to study vector-pathogen interactions, the I. scapularis genome and transcriptomes provide a unique opportunity to examine neuropeptides in a non-model arthropod. Therefore, in this study, we characterized the sequence and expression dynamics of ILPs in I. scapularis, to provide a basis for further explorations of these signaling molecules in ticks.
Methods
Bioinformatics Identification of Ixodes scapularis ILPs
Amino acid sequences from previously identified ILPs from Ae. aegypti, Anopheles stephensi, An. gambiae, D. melanogaster, and vertebrate ILPs [Mus musculus insulin and insulin-like growth factor I (IGF-I)] were used as queries in tBLASTn searches of the I. scapularis genome and expressed sequence tag (EST) databases, and I. ricinus transcriptome (TSA) databases through the National Center for Biotechnology Information (NCBI) website. To ensure detection of ILPs that might deviate from canonical insect ILP sequences, these queries were further searched against all available arthropods, from whole genome sequencing reads, transcriptomes, and EST databases. From these results, putative ILP amino acid sequences from representative arthropods outside of Hexapods were selected with an emphasis on arachnids (e.g., Aranae, Acari, Scorpiones, Opilionies, Amblypygi, Pseudoscorpiones, Solfugae), but also including the outgroup Merostomata (Atlantic Horseshoe Crab, Limulus polyphemus) (Supplementary Table 1). These were then leveraged as queries in searches against the I. scapularis genome and EST database to identify ILP signatures potentially unique to non-Hexapod arthropods.
Analyses of signal peptide sequences within putative prepropeptides were conducted using both the Neural networks and the Hidden Markov models contained in the SignalP program [http://www.cbs.dtu.dk/services/SignalP/ (27)].
I. scapularis Colony
A colony of I. scapularis was maintained at 20°C under 16:8 light:dark conditions at 95% humidity. Larvae and nymphs were fed on mice as described previously (28). Adults were fed on rabbit (29). All animals were treated humanely according to the guidelines of the Institutional Animal Care and Use Committee (IACUC protocol # 00682).
Sample Collection and RNA Extraction
RNA was extracted from three separate biological cohorts of I. scapularis eggs (~500), unfed larvae (10 individuals), nymphs (10 individuals), adult female (2 individuals), and adult male (5 individuals) ticks by placing them separately into 1.7 ml tubes, and extracting with TRIzol™ reagent (Invitrogen), following a slightly modified protocol from the manufacturer's instructions. The modified protocol is as follows: I. scapularis were collected into 1.7 ml tubes and were ground in liquid nitrogen with autoclaved plastic pestles. These samples were further homogenized in 100 μl of TRIzol™ with a tissumizer, then brought to 800 μl with additional TRIzol™. Solid tissue debris was pelleted by centrifugation at 12,000 x g at 4°C for 10 min, and the supernatant was transferred to a fresh tube. Thereafter, the manufacturer's protocol was followed for extraction of total RNA.
The resulting RNA pellet was resuspended in 20 μl of RNase/DNase free water incubated at 55–60°C for 10 min. RNA was treated with DNase I from the Direct-zolTM RNA MiniPrep Kit (Zymo Research) at RT for 30 min. Columns from this kit were pre-wet with RNA Wash Buffer, briefly centrifuged, then the DNase-treated RNA was added directly to the columns. Manufacturer protocols were followed for subsequent RNA recovery. Total RNA quantity was measured with Nanodrop spectrophotometer. Purity was determined by 260/280 and 260/230 ratios, with acceptable values in the ~1.8 and 2.0–2.2 range, respectively (all samples collected met purity standards). RNA samples were immediately stored at −80°C.
Sequencing Confirmation of I. scapularis ILPs
To confirm sequences, primers were designed to the identified I. scapularis ILP sequences using Primer3 (30, 31) and estimated optimal annealing temperatures were determined using Integrated DNA Technologies' online Oligoanalyzer tool (IDT) (Table 1). Synthesis of I. scapularis cDNA was accomplished using the SMARTer cDNA synthesis kit according to manufacturer's instructions (Takara Bio USA, Mountain View, CA) and 1 μg total RNA obtained from nymphs. These cDNA template and primers were used in polymerase chain reactions (PCR) to amplify sections of I. scapularis cDNA using Biotool Taq polymerase (Biotool, Ely, UK) with the following program: 40 cycles at 94°C, 30 s; 60°C, 30 s; 72°C, 1 min. This included an initial 5 min denaturing step at 94°C, and a final 10 min, 72°C extension step. These reactions were separated with gel electrophoresis and bands of the expected size were extracted from the gel and sent for Sanger sequencing (Genewiz, South Plainfield, NJ). Resulting sequences were aligned with the predicted sequences.

Table 1. Primer sequences used to amplify IsILP sequences for confirmation by Sanger sequencing or for relative quantification by qRT-PCR.
To confirm 5′ and 3′ ends of ILP transcripts not accessible to primer design, 5′ and 3′ rapid amplification of cDNA ends (RACE) was performed. For 3′ RACE, the anchor primer NotI d(T)2 (Table 1) was used to initiate cDNA synthesis using SMARTer cDNA synthesis kit and 1 μg total RNA from blood-fed nymphs. This template was used in nested PCR reactions as above, using gene-specific reverse primers and the NotI anchor primer. For 5′ RACE, specific ILP reverse primers were used with the SMARTer cDNA synthesis kit for generation of cDNA. These cDNA strands were purified and the 3′ end was polyadenylated (A) using Terminal Deoxynucleotidyl Transferase (TdT) and dATP (Thermo Fisher Scientific Inc.). Poly-A tailed cDNA was again isolated and used as template in nested PCR reactions employing NotI d(T)2, NotI and gene specific reverse primers. Resulting products were separated by gel electrophoresis and subcloned using the TOPO™ TA Cloning™ Kit for Subcloning (Invitrogen, Waltham, MA, USA). Plasmids were isolated from successful transformants and were Sanger sequenced as above.
Expression of I. scapularis ILPs
ILP expression was characterized using quantitative RT-PCR (qRT-PCR). The iScriptTM cDNA Synthesis Kit was used to convert 500 ng−1 μg I. scapularis total RNA samples from eggs, larvae, nymphs, and male and female adults (see above) into cDNA following manufacturer's protocols (BioRad, Hercules, CA, USA). Briefly, cDNA of ticks from different life stages was diluted 5x with deionized H2O before using it as a template in qRT-PCR experiments. One microliter cDNA was used in each 10 μl qRT-PCR reaction. Four different housekeeping genes were tested: β-tubulin, ubiquitin, ribosomal protein 4 (rps4), and I13A. β-tubulin was selected for final experiments because of its consistent expression in all life stages and tissues. Sequences of specific primers for IsILPs and the housekeeping gene, β-tubulin, are listed in Table 1. Each sample was run in triplicate wells of 96-well plate. qRT-PCR was performed on CFX touch Real-Time PCR Detection system using SYBR green master mix (BioRad, Hercules, CA, USA). All reactions were performed with initial 5 min at 95°C, followed by 40 cycles of 10 s at 95°C, 15 s at 64°C, and 15 s at 72°C, and a melt curve was analyzed at 70–95°C. Relative expression was calculated using 2−ΔΔCt method. Experiments were replicated two to four times with different tick cohorts.
Statistical Analysis
All data were analyzed using GraphPad Prism 7 software (La Jolla, CA, USA). Relative expression values of IsILP expression in different developmental stages and tissues were compared with one-way ANOVA, followed by Dunnett's multiple comparison test.
Results
I. scapularis ILP Identification
Four sequences with ILP characteristics were identified through bioinformatics searches of I. scapularis genome, transcriptome, and EST databases. The use of mosquito ILPs as queries in tBLASTn searches was sufficient to detect these sequences and no additional sequences were found with subsequent searches employing other arthropod ILPs. Sequences containing complete features of ILPs (start and stop codons, B and A chains, and signal peptide) were found for three I. scapularis ILPs. For IsILP4, only a partial sequence was obtained for I. scapularis which contained no stop codon. However, a highly similar transcriptome sequence from I. ricinus (accession: GANP01013821) was found that did contain a stop codon and untranslated regions further downstream. Reverse primers designed to these regions in the I. ricinus sequence were capable of amplifying PCR products in combination with forward primers from I. scapularis.
Complete cDNA sequences of four ILPs were confirmed by Sanger sequencing of PCR products and 3′ and 5′ RACE products. These sequences contain features consistent with ILPs in other animals, including start and stop codons, signal peptides, conserved cysteine residues, and putative dibasic cleavage sites (Figures 1, 2). IsILP1 was named as such considering it was the first I. scapularis ILP noted in the literature (21). The remaining ILPs (3, 4, and 5) were named according to their amino acid similarity to An. gambiae ILPs, particularly ILP5 which contains additional amino acids between key Cys residues of the A-chain in comparison to other ILPs (Figure 2 and see section Discussion).
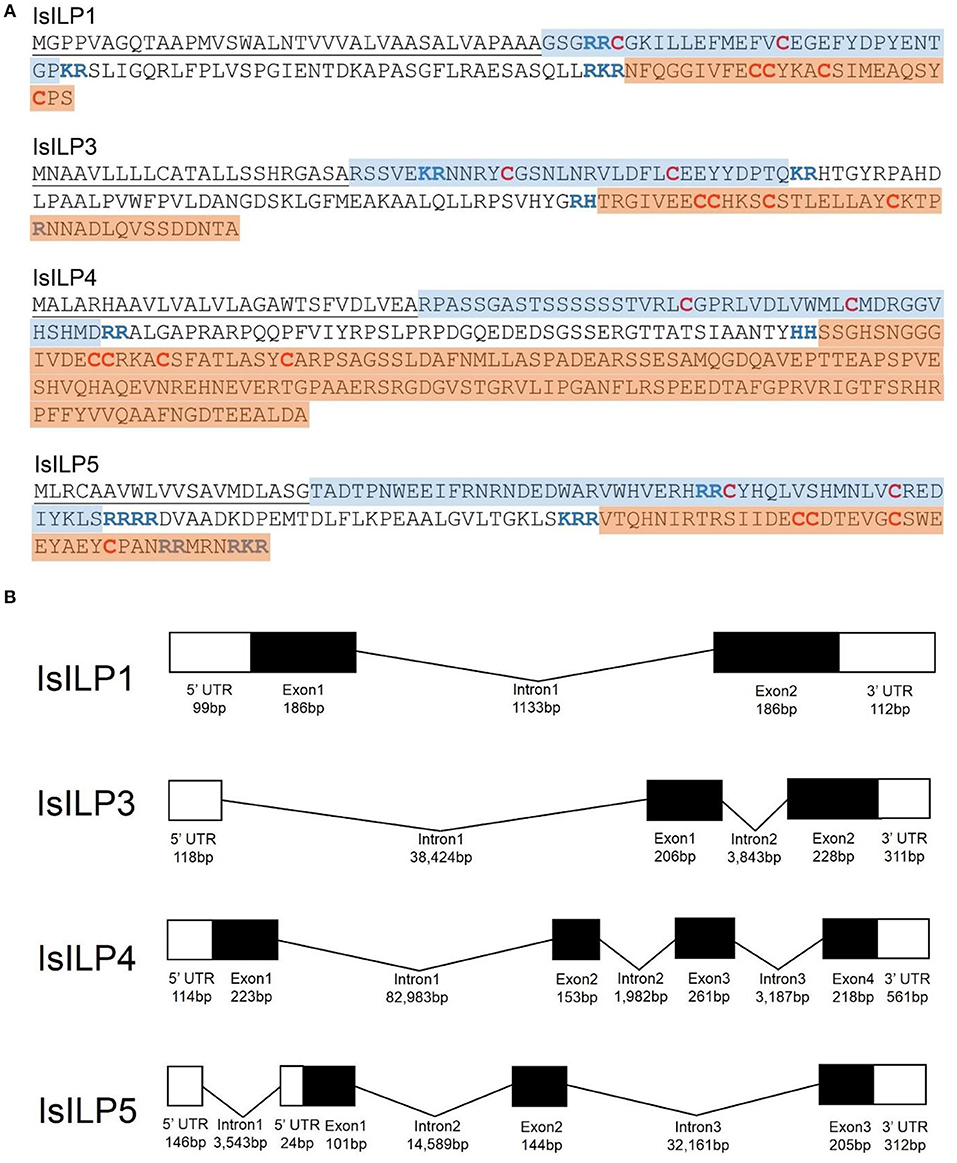
Figure 1. Structural features of I. scapularis ILPs. (A) Amino acid translations and structural predictions of I. scapularis ILP sequences confirmed by Sanger sequencing. Cysteine disulfide bridges in bold red text. Predicted di-basic cleavage sites in bold blue text; B-chain: Blue highlight; A-chain: Orange highlight; Signal peptide underlined. Genbank accession numbers: IsILP1: MK649814; IsILP3: MK649815; IsILP4: MK649816; IsILP5: MK649817; (B) Predicted gene models of IsILPs based on alignment of confirmed sequences to the I. scapularis genome (IsILP1, IsILP3: Contig: PKSA02008623, IsILP4: Contig: PKSA02013804, IsILP5: Contig: PKSA02002782). Intron and exon lengths in the diagram are not proportional.
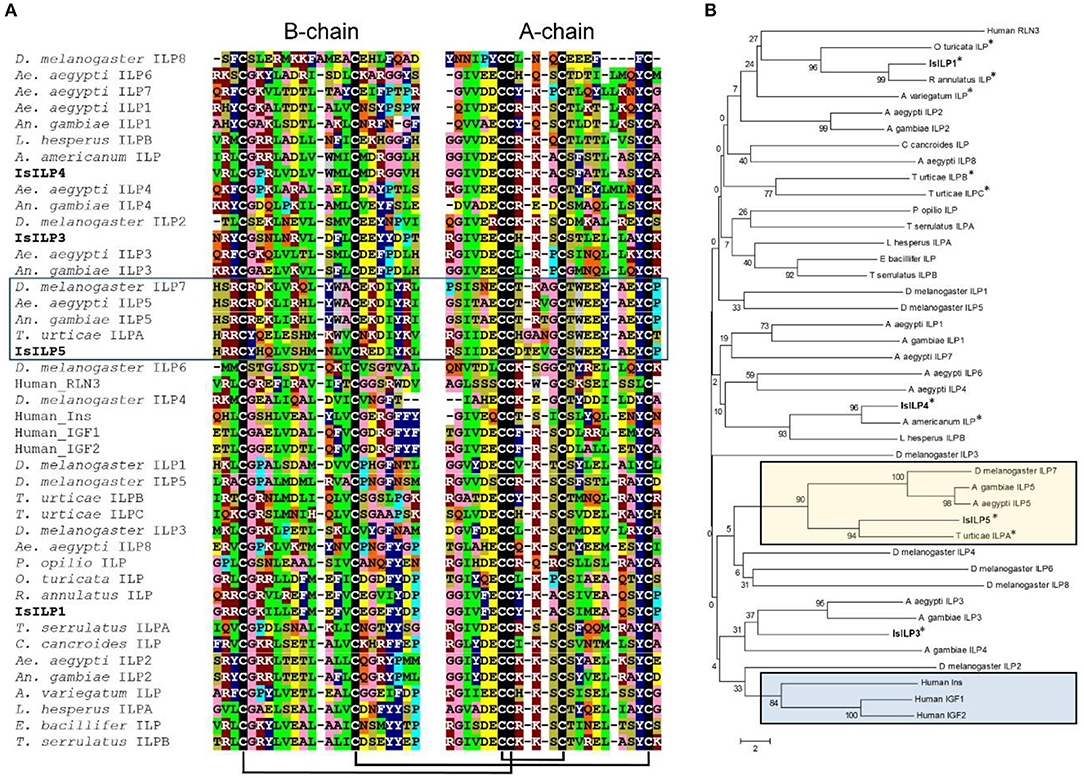
Figure 2. Comparison of IsILP amino acid sequences to human ILPs and selected arthropods. (A) Alignment of I. scapularis B- and A-chain ILP amino acid sequences (truncated around core Cys resides for the sake of space) with selected arachnid, insect, and human ILPs. Colors indicate amino acids with similar properties: green = hydrophobic; dark blue = aromatic; pink = small; dark red = basic; yellow = acidic, tan = hydroxyl, gray = tryptophan, light blue = proline, black = cysteine, orange = amide. Predicted disulfide bridges are connected by black bars. Box: ILP5 sub-group. Alignments performed in Clustal Omega (32) and colored manually. (B) Neighbor-joining tree of mosquito ILP A and B chain amino acid sequences comparing selected arachnid, insect, and human ILPs. Acari ILPs indicated by “*”. Tan box: ILP5-related sequences; Blue box: Human ILPs. Tree was constructed using MEGA version 6 (33). Accession numbers of sequences used for this figure can be found in Supplementary Table 2.
We encountered but were unable to amplify a putative additional I. scapularis ILP sequence that contained spacing of cysteines characteristic of an ILP A-chain: ILVFFSVKPDKKPAFSSKCCDGDCTKSVWKGSKISFHVFLPAPFVRVRE (accession: ABJB010579748). This may represent a pseudogene or an unrelated sequence that was picked up in our bioinformatics search by chance, or genome assembly in this region is of insufficient quality to design functional primers.
I. scapularis ILP Developmental Expression
ILP expression varied markedly between the developmental stages sampled. For fold difference calculations, eggs samples were used as a baseline (control). IsILP1 was significantly more expressed in the unfed larvae and adult females compared to eggs, nymphs, and adult males. In contrast, IsILP3 and IsILP5 expression was significantly higher in unfed larvae as compared to other life stages. IsILP4 was significantly more expressed in adult females, followed by larvae and nymphs having significantly more expression than adult males and eggs. For most IsILPs, there was little to no expression in adult males and eggs (Figures 3A–D).
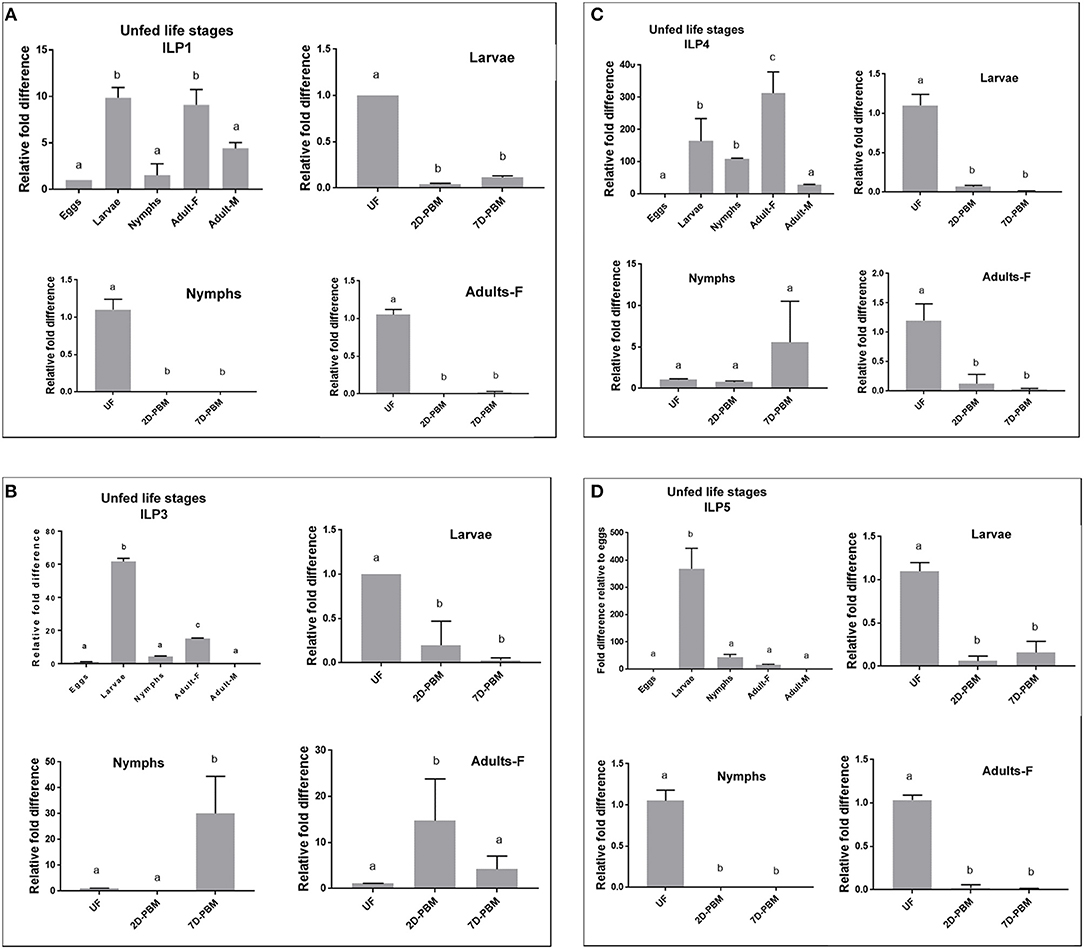
Figure 3. Expression of IsILP transcripts in I. scapularis life stages, as determined by quantitative RT-PCR. F, female; M, male; UF, unfed; 2D-PBM, 2 days post engorgement and host detachment; 7D-PBM, 7 days post engorgement and host detachment. Fold expression levels comparing life stages were scaled to expression in eggs. Feeding trials were examined within life stages by scaling expression to the unfed state. Treatments were examined by one-way ANOVA followed by Dunnett's multiple comparison test. (A) Expression of IsILP1 in unfed developmental stages and larvae, nymphs, and adult females post detachment. All unfed stages, F(4,5) = 29.27, P = 0.001; Larvae: F(2,3) = 4583, P < 0.0001; Nymphs: F(2,3) = 121, P = 0.001; Adults: F(2,3) = 410.2, P = 0.0002. (B) Expression of IsILP3 in unfed developmental stages and larvae, nymphs, and adult females post detachment. All unfed stages, F(4,5) = 1968, P < 0.0001; Larvae: F(2,3) = 21.46, P = 0.01; Nymphs: F(2,9) = 16.9, P = 0.0001; Adults: F(2,9) = 6.774, P = 0.01. (C) Expression of IsILP4 in unfed developmental stages and larvae, nymphs, and adult females post detachment. All unfed stages, F(4,5) = 16.5, P = 0.004; Larvae: F(2,3) = 111, P < 0.001; Nymphs: F(2,8) = 3.002, P = 0.10; Adults: F(2,3) = 24.25, P = 0.01. (D) Expression of IsILP5 in unfed developmental stages and larvae, nymphs, and adult females post detachment. All unfed stages, F(4,15) = 101.9, P < 0.0001; Larvae: F(2,6) = 99.08, P < 0.0001; Nymphs: F(2,9) = 264.6, P < 0.0001; Adults: F(2,6) = 661.4, P < 0.0001.
IsILP1 and IsILP5 expression was significantly higher in unfed stages (larvae, nymphs, and adults) compared to the levels at 2 and 7 days after detachment from the host (Figures 3A,D). For IsILP3, expression was highest in unfed larvae compared to detachment from the host (Figure 3B); whereas in nymphs and adult females expression increased after detachment from the host and was higher at 2-day post detachment in adults and 7 day post detachment in the nymphs. For IsILP4, expression was significantly higher in unfed larvae and adults, whereas there was no difference in expression before and after a blood meal in the nymphal stage (Figure 3C).
I. scapularis ILP Adult Female Tissue Expression
Synganglion, salivary gland, midgut, and ovaries from unfed females were dissected to understand baseline tissue expression profiles of IsILPs. IsILP1 and IsILP5 had significantly higher expression in synganglion. IsILP5 expression was limited to synganglion. IsILP3 and IsILP4 expression was significantly higher in salivary glands. However, both expressed in all tissues examined.
IsILP1 expression was reduced after a blood meal in all tissues except for ovaries where no difference was noted in expression before or after feeding. IsILP3 expression was highest in unfed females in all tissues examined whereas IsILP4 expression was higher in midgut 7 days post detachment from the host. IsILP5 expression was highest in the synganglion of the unfed females. We noted some expression in ovaries; however, the Ct values were too high to accurately measure expression (36–38 out of 40 cycles) suggesting very little transcript was available. IsILP5 expression was higher in a few midgut and salivary gland samples at 7 day post detachment (Figure 4).
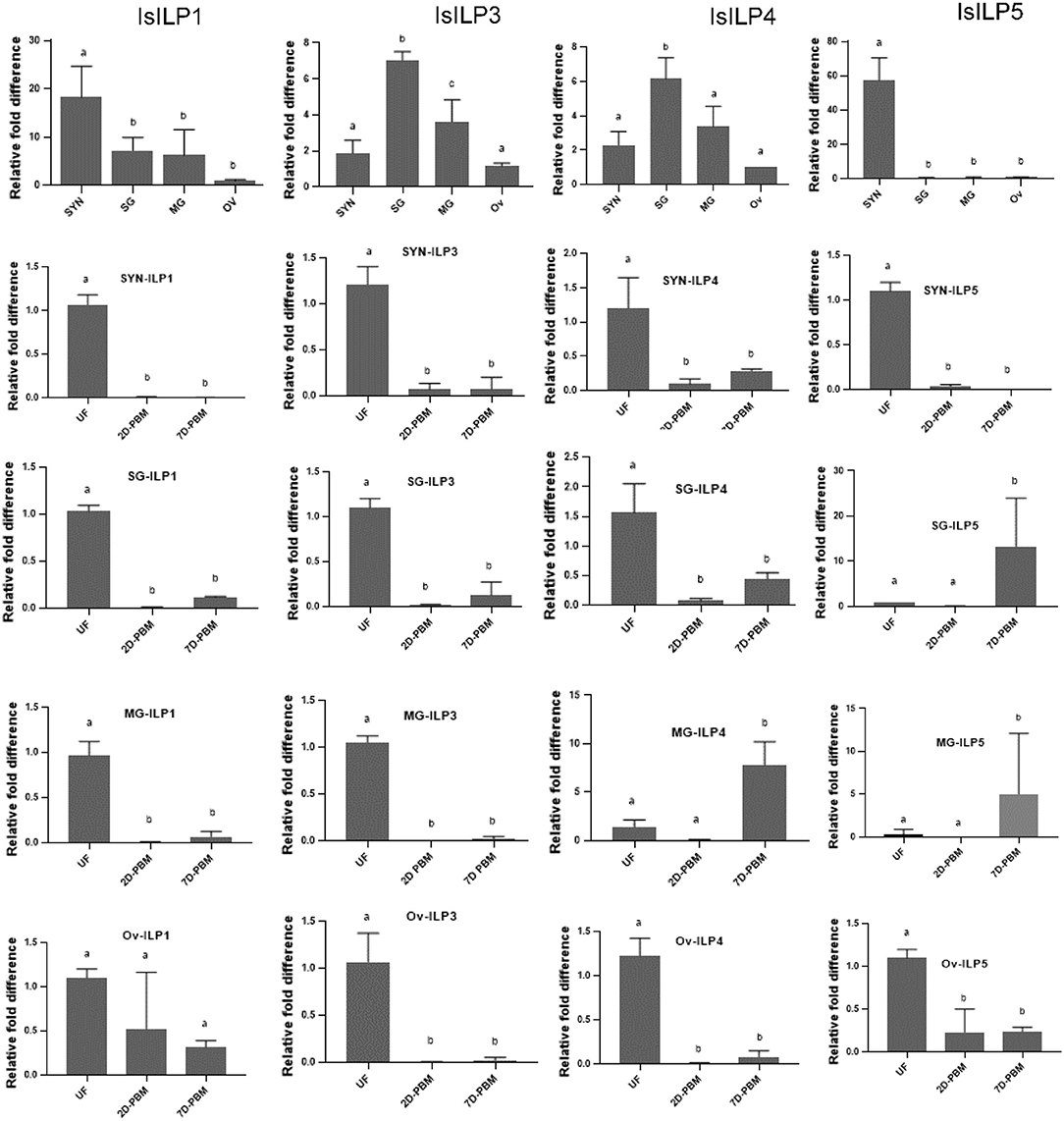
Figure 4. Expression of IsILP transcripts in adult female I. scapularis tissues, as determined by quantitative RT-PCR. SYN, synganglion; SG, salivary gland; MG, midgut; Ov, ovaries; UF, unfed; 2D-PBM, 2 days post engorgement and host detachment; 7D-PBM, 7 days post engorgement and host detachment. Top row: Expression of individual IsILPs in unfed adult females compared by body tissue. Fold expression levels were scaled to expression in ovaries and were examined by one-way ANOVA followed by Dunnett's multiple comparison test. IsILP1 unfed tissues, F(3,8) = 8.156, P = 0.008; IsILP1-Synganglion: F(2,6) = 252.1, P < 0.0001; IsILP1-salivary glands: F(2,6) = 836.8, P < 0.0001; IsILP1-midgut: F(2,6) = 97.47, P < 0.0001; IsILp1-Ovaries: F(2,6) = 3.491, P = 0.09. IsILP3 unfed tissues, F(3,7) = 25.42, P = 0.0004; IsILP3-Synganglion: F(2,6) = 68.23, P < 0.0001; IsILP3-salivary glands: F(2,6) = 98.0.8, P < 0.0001; IsILP3-midgut: F(2,6) = 374.6, P = 0.0003; IsILP3-ovaries: F(2,6) = 35.26, P = 0.0005. IsILP4 unfed tissues, F(3,7) = 14.26, P = 0.002; IsILP4-Synganglion: F(2,6) = 15.17, P = 0.004; IsILP4-salivary glands: F(2,6) = 21.64, P = 0.001, IsILP4-midgut: F(2,5) = 28.4, P = 0.001; IsILP4-ovaries: F(2,6) = 94.22, P < 0.0001. IsILP5 unfed tissues, F(3,8) = 57.74, P < 0.0001; IsILP5-Synganglion: F(2,6) = 340.3, P < 0.0001; IsILP5-salivary glands: F(2,7) = 5.176, P = 0.04, IsILP5-midgut: F(2,5) = 1.73, P = 0.2; IsILP5-ovaries: F(2,6) = 26.32, P < 0.001.
Discussion
To date, little examination has been made of ILPs in arachnids and our characterization of ILPs in I. scapularis provides a foundation for further exploration of the roles of these neuropeptides in this medically important arthropod. The discovery of four ILPs is consistent with the pattern seen in insects of multiple ILPs. Some of the I. scapularis ILP sequences we characterized have been previously noted as ILPs in the literature or as automated annotations in online databases. For instance, putative transcripts for IsILP1 were noted from I. scapularis (34) that were highly similar to an A-chain fragment from D. variabalis (21). However, this is the first study to specifically focus on IsILPs, and IsILPs 3 and 4 are identified for the first time in our study. It is of interest that only IsILP5 (ISCW002549, partial fragment annotated as ILP4), and not IsILP1, was initially annotated in the I. scapularis genome (26). Also, an ILP sequence was recently recognized from A. americanum that is highly similar to IsILP4, yet this did not map to the I. scapularis genome (23). This highlights the large and scattered nature of the current I. scapularis genome and the challenge of assembling and annotating a genome with extensive repetitive sequences and expansive intron regions.
The IsILPs we identified fit the characteristic hallmarks of ILPs in other organisms, containing signal peptides, B and A chains, and dibasic cleavage sites for excision of the C-peptide. Conserved cysteine residues are predicted to form disulfide bridges between B and A chains or internally in the A chain (Figures 1A, 2A). Insulin-like growth factors (IGFs) have a similar structure, yet retain a short C-peptide that is not cleaved (11). None of the IsILPs in this study appear to have a short C-peptide indicative of an IGF-like structure. Some other commonalities for ILPs include basic (Arg, Lys, and His) and amino acids with aromatic rings (Tyr and His) upstream of the first Cys of the B-chain, and consistently spaced hydrophobic (particularly Leu) or charged residues (frequently Glu, but also Asp). IsILP5, AgILP5, AaILP5, and DmILP7 are unusual in that they have additional amino acids between Cys 2 and 3 of the A-chain compared to other ILPs, where there are consistently three amino acids (Figure 2A). These particular ILPs share considerable amino acid sequence conservation in comparison to other ILPs, particularly in the A-chain. Outside of this particular sub-group, IsILPs vary greatly in their similarity to each other and to those of other arthropods (Figure 2). As has been the case with other invertebrate ILPs, amino acid sequence similarity alone provides only limited information as to their potential functions when comparing between species. It is likely that the secondary structure imposed by disulfide bonds and as yet unverified key amino acid residues may preserve the functionality of these molecules in binding to their cognate IRs. Unfortunately, the chemical synthesis of invertebrate ILPs has been a challenge and has so far limited the study of structure-activity relationships with techniques such as alanine screens (3, 15).
The IsILPs we identified in this study had from two to four exons (Figure 1B), comparable to other arthropod examples (A. gambiae: 1–3 exons; Ae. aegypti: 2–3 exons; D. melanogaster: 1–3 exons) (35, 36). Curiously, the ILP with the most conserved amino acid structure among the arthropods we examined, ILP5 (DmILP7), exhibited high variability in intron number between organisms (AgILP5: 0 introns; AaILP5: 1 intron; DmILP7: 2 introns; IsILP5: 3 introns). The gene structure of IsILP1 was consistent with previous predictions in I. scapularis of a single intron (21). IsILPs 3, 4, and 5 are predicted to have massive introns, ranging from 32,161 to 82,983 bp. However, this is not inconsistent with observed intron lengths in other organisms such as IGF1 in humans, which has a 55,952 bp intron (35). Both IsILP1 and IsILP3 were found on the same contig (PKSA02008623), but curiously the entire IsILP1 gene sequence was found within intron 1 of the IsILP3 sequence. We encountered another contig containing both IsILP1 and IsILP3 (PKSA02013879) with a similar gene structure, but with different intron lengths for IsILP3 (Intron 1: 35,843 bp; Intron 2: 3,773 bp). It is unclear whether the current genome assembly accurately reflects the gene structure of these IsILPs. This confusion may be clarified with newer sequencing technologies permitting longer reads allowing more robust genome assemblies.
Perhaps of greater value toward understanding IsILP functions are the expression patterns of ILPs. In specific body tissues, IsILP5 was almost exclusively expressed in the synganglion suggesting a neurotransmitter or neurohemal role. IsILP1 was also expressed at significantly higher levels in the synganglion compared to other tissues. In contrast, IsILPs 3 and 4 were expressed in all tissues examined with highest expression in the salivary glands. Tick salivary glands undergo a growth phase during feeding in order to increase saliva production and then greatly reduce in size after host detachment. As some ILPs act as growth factors, one might speculate higher expression in these tissues may indicate synthesis and storage of these ILPs in preparation for initiation of a feeding event, where their release subsequently stimulates salivary gland growth.
Expression of most ILPs decreased after a blood meal which is similar to a previous study where expression of an ILP (the homolog of IsILP1) dropped during and after a blood meal in D. variabilis females (22). In our study, this occurred in most tissues and in most life stages. The importance of a reduction in IsILP transcripts in response to feeding is, as yet, unclear, but may be tied to regulation of energy storage as is the case for ILPs in other organisms. Also of interest are exceptions to this trend, such as the increase in expression of IsILP4 and IsILP5 in the midgut of adult females 7 days post blood meal. The increase in expression in this tissue after a blood meal may suggest a role of IsILPs 4 and 5 in blood digestion, a situation similar to that seen in Ae. aegypti where AaILP3 secreted from the brain stimulates the midgut to produce the digestive enzyme trypsin following a blood meal (4). In addition, nymphs increased expression of IsILPs 3 and 4 after detaching from hosts, possibly suggesting a developmental role unique to this life stage. A greater number of timepoints over a longer period may further elucidate the role of IsILPs before, during, and after blood feeding, and during development.
In adult I. scapularis males, IsILP expression was low for all ILPs except IsILP1. The primary activity for adult males of this species is to survive long enough to find and mate with a female. While this activity is most successful if the males cling to and search a vertebrate host where a prospective female is likely to be attached for feeding, the males themselves do not feed. Considering the major functions of ILPs identified in other animals include storage of nutrients, endocrine cascades resulting in yolk protein synthesis, and growth, the lack of IsILP expression in males may make sense in that they do not engage in these processes in the adult stage.
In addition to ILP expression levels in ticks, binding proteins, which were not investigated in this study, may also play a role in insulin signaling dynamics. For instance, knockdown of IGFBP in A. americanum prevented blood-feeding females from feeding to repletion (24). Although they are not ILPs, IGFBPs do interact with ILPs and can regulate binding dynamics with the insulin receptor by altering degradation rates and binding kinetics. Exploration of the expression dynamics of IGFBPs and their impact on IsILP stability in I. scapularis will provide further information on the roles of these signaling molecules in tick physiology. ILPs also have been implicated in immune responses, and expression of an ILP (homolog of IsILP1) was upregulated in A. americanum exposed to Ehrlichia chaffeensis, the causative agent of human monocytic ehrlichiosis (23). Whether similar expression patterns occur in I. scapularis in response to pathogens remains to be explored, but it is a subject of particular interest given the role of insulin signaling in immunity in other blood-feeding arthropods (37), and the importance of I. scapularis as a vector of disease.
Conclusions
Ticks represent an extreme lifestyle compared to many animals. They feed exclusively on vertebrate blood, and Ixodid ticks feed only three times in their entire life cycle. From these scant meals, they can endure many months between feedings, yet still manage to deal with the harsh realities of the environment as other arthropods do, but without the luxury of regular food intake. No doubt absorption and conservation of energy is critical to their survival and ILPs are likely to play a key role in this process. We are eager to uncover the role of these signaling molecules with regard to the extreme physiology of these arachnids, and this study provides an important first step in the characterization and role of these neuropeptides.
Author Contributions
AN and MG-N wrote the draft manuscript, conceived the experiments, and wrote the final manuscript. RP and PV amplified and sequenced the ILP transcripts. AS and BG carried out the insulin transcript expression study. MG-N performed the statistical analysis.
Funding
This project was funded in part through an NIH-NIAID R21AI128393 to MG-N. The UNR Cellular and Molecular Imaging Core Facility, funded through NIH NIGMS P20 GM103650, was used, in part, for data collection.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2019.00292/full#supplementary-material
References
1. Géminard C, Arquier N, Layalle S, Bourouis M, Slaidina M, Delanoue R, et al. Control of metabolism and growth through insulin-like peptides in Drosophila. Diabetes. (2006) 55(Suppl. 2): S5–8. doi: 10.2337/db06-S001
2. Riehle MA, Fan Y, Cao C, Brown MR. Molecular characterization of insulin-like peptides in the yellow fever mosquito, Aedes aegypti: expression, cellular localization, and phylogeny. Peptides. (2006) 27:2547–60. doi: 10.1016/j.peptides.2006.07.016
3. Brown MR, Clark KD, Gulia M, Zhao Z, Garczynski SF, Crim JW, et al. An insulin-like peptide regulates egg maturation and metabolism in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. (2008) 105:5716–21. doi: 10.1073/pnas.0800478105
4. Gulia-Nuss M, Robertson AE, Brown MR, Strand MR. Insulin-like peptides and the target of rapamycin pathway coordinately regulate blood digestion and egg maturation in the mosquito Aedes aegypti. PLoS ONE. (2011) 6:e20401. doi: 10.1371/journal.pone.0020401
5. Xu H-J, Xue J, Lu B, Zhang XC, Zhuo JC, He SF, et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature. (2015) 519:464–7. doi: 10.1038/nature14286
6. Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. (2001) 20:213–21. doi: 10.1016/S0960-9822(01)00068-9
7. Colombani J, Andersen DS, Léopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. (2012) 336: 582–5. doi: 10.1126/science.1216689
8. Aslam AFM, Kiya T, Mita K, Iwami M. Identification of novel bombyxin genes from the genome of the silkmoth Bombyx mori and analysis of their expression. Zool Sci. (2011) 28:609–16. doi: 10.2108/zsj.28.609
9. Antonova Y, Arik AJ, Moore W, Riehle MA, Brown MR. Insulin-like peptides: structure, signaling, and function. Insect Endocrinol. (2012) 2:63–92. doi: 10.1016/B978-0-12-384749-2.10002-0
10. Strand MR, Brown MR, Vogel KJ. Mosquito peptide hormones: diversity, production, and function. Adv Insect Physiol. (2016) 51:145–88. doi: 10.1016/bs.aiip.2016.05.003
11. Nässel DR, Broeck JV. Insulin/IGF signaling in Drosophila and other insects: factors that regulate production, release and post-release action of the insulin-like peptides. Cell Mol Life Sci. (2016) 73:271–90. doi: 10.1007/s00018-015-2063-3
12. Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. (2012) 336:579–82. doi: 10.1126/science.1216735
13. Nässel DR. Insulin-producing cells and their regulation in physiology and behavior of Drosophila. Can J Zool. (2012) 90:476–88.
14. De Meyts P, Gauguin L, Svendsen AM, Sarhan M, Knudsen L, Nøhr J. Structural basis of allosteric ligand–receptor interactions in the insulin/relaxin peptide family. Ann NY Acad Sci. (2009) 1160:45–53. doi: 10.1111/j.1749-6632.2009.03837.x
15. Nuss AB, Brown MR. Isolation of an insulin-like peptide from the Asian malaria mosquito, Anopheles stephensi, that acts as a steroidogenic gonadotropin across diverse mosquito taxa. Gen Comp Endocrinol. (2018) 258:140–8. doi: 10.1016/j.ygcen.2017.05.007
16. Ishizaki H, Suzuki A. The brain secretory peptides that control moulting and metamorphosis of the silkmoth, Bombyx mori. Int J Dev Biol. (1994) 38:301–10.
17. Hetru Ch, Li KW, Bulet P, Lagueux M, Hoffmann JA. Isolation and structural characterization of an insulin-related molecule, a predominant neuropeptide from Locusta migratoria. Eur J Biochem. (1991) 201:495–9. doi: 10.1111/j.1432-1033.1991.tb16308.x
18. Zhu XX, Oliver JH. Immunocytochemical localization of an insulin-like substance in the synganglion of the tick Ornithodoros parkeri (Acari: Argasidae). Exp Appl Acarol. (1991) 13:153–9.
19. Šimo L, Slovák M, Park Y, Žitňan D. Identification of a complex peptidergic neuroendocrine network in the hard tick, Rhipicephalus appendiculatus. Cell Tissue Res. (2009) 335:639–55. doi: 10.1007/s00441-008-0731-4
20. Davis HH, Dotson EM, Oliver JH. Localization of insulin-like immunoreactivity in the synganglion of nymphal and adult Dermacentor variabilis (Acari: Ixodidae). Exp Appl Acarol. (1994) 18:111–22.
21. Donohue KV, Khalil SMS, Grozinger CM, Sonenshine DE, Roe RM. Neuropeptide signaling sequences identified by pyrosequencing of the American dog tick synganglion transcriptome during blood feeding and reproduction. Insect Biochem Mol Biol. (2010) 40:79–90. doi: 10.1016/j.ibmb.2009.12.014
22. Bissinger BW, Donohue KV, Khalil SM, Grozinger CM, Sonenshine DE, Zhu J, et al. Synganglion transcriptome and developmental global gene expression in adult females of the American dog tick, Dermacentor variabilis (Acari: Ixodidae). Insect Mol Biol. (2011) 4:465–91. doi: 10.1111/j.1365-2583.2011.01086.x
23. Van Horn TR, Adalsteinsson SA, Westby KM, Biro E, Myers JA, et al. Landscape physiognomy influences abundance of the lone star tick, Amblyomma americanum (Ixodida: Ixodidae), in Ozark forests. J Med Entomol. (2018) 55:982–8. doi: 10.1093/jme/tjy038
24. Mulenga A, Khumthong R. Silencing of three Amblyomma americanum (L.) insulin-like growth factor binding protein-related proteins prevents ticks from feeding to repletion. J Exp Biol. (2010) 231:1153–61. doi: 10.1242/jeb.035204
25. Adams DA, Thomas KR, Jajosky R, Foster L, Sharp P, Onweh DH, et al. Summary of notifiable infectious diseases and conditions - United States 2014. MMWR Morb Mortal Wkly Rep. (2016) 63:1–152. doi: 10.15585/mmwr.mm6354a1
26. Gulia-Nuss M, Nuss AB, Meyer J, Sonenshine DE, Roe RM, Waterhouse RM, et al. (2016). Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 7:10507. doi: 10.1038/ncomms10507
27. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. (2011) 8:785–6. doi: 10.1038/nmeth.1701
28. Nuss AB, Mathew MG, Gulia-Nuss M. Rearing Ixodes scapularis, the black-legged tick: feeding immature stages on mice. J Vis Exp. (2017). e55286. doi: 10.3791/55286
29. Ibelli AMG, Kim TK, Hill CC, Lewis LA, Bakshi M, Miller S, et al. A blood meal-induced Ixodes scapularis tick saliva serpin inhibits trypsin and thrombin, and interferes with platelet aggregation and blood clotting. Int J Parasitol. (2014) 44:369–79. doi: 10.1016/j.ijpara.2014.01.010
30. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3 - new capabilities and interfaces. Nucleic Acids Res. (2012) 40:e115. doi: 10.1093/nar/gks596
31. Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. (2007) 23:1289–91. doi: 10.1093/bioinformatics/btm091
32. Chojnacki S, Cowley A, Lee J, Foix A, Lopez R. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. (2017) 45:W550–553. doi: 10.1093/nar/gkx273
33. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. (2013) 30:2725–9. doi: 10.1093/molbev/mst197
34. Egekwu N, Sonenshine DE, Bissinger BW, Roe RM. Transcriptome of the female synganglion of the black-legged tick Ixodes scapularis (Acari: Ixodidae) with comparison between illumina and 454 systems. PLoS ONE. (2014) 9:e102667. doi: 10.1371/journal.pone.0102667
35. Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. Ensemble 2018. Nucleic Acids Res. (2018) 46:D754–61. doi: 10.1093/nar/gkx1098
36. Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. (2015) 43:D707–13. doi: 10.1093/nar/gku1117
Keywords: insulin, tick, ILPs, peptide, blood-feeding
Citation: Sharma A, Pooraiiouby R, Guzman B, Vu P, Gulia-Nuss M and Nuss AB (2019) Dynamics of Insulin Signaling in the Black-Legged Tick, Ixodes scapularis. Front. Endocrinol. 10:292. doi: 10.3389/fendo.2019.00292
Received: 30 November 2018; Accepted: 23 April 2019;
Published: 21 May 2019.
Edited by:
María Jesús Delgado, Complutense University of Madrid, SpainReviewed by:
Ian Orchard, University of Toronto Mississauga, CanadaAlejandro Cabezas-Cruz, Institut National de la Recherche Agronomique (INRA), France
Petr Kopacek, Institute of Parasitology (ASCR), Czechia
Copyright © 2019 Sharma, Pooraiiouby, Guzman, Vu, Gulia-Nuss and Nuss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrew B. Nuss, nuss@cabnr.unr.edu
Monika Gulia-Nuss, mgulianuss@unr.edu
†These authors have contributed equally to this work
 Arvind Sharma
Arvind Sharma Rana Pooraiiouby
Rana Pooraiiouby Blanca Guzman1†
Blanca Guzman1† Monika Gulia-Nuss
Monika Gulia-Nuss Andrew B. Nuss
Andrew B. Nuss