- 1Food and Feed Safety Research, Southern Regional Research Center, Agricultural Research Service, United States Department of Agriculture (USDA), New Orleans, LA, United States
- 2Food Processing and Sensory Quality Research, Southern Regional Research Center, Agricultural Research Service, United States Department of Agriculture (USDA), New Orleans, LA, United States
Aspergillus fungi produce mycotoxins that are detrimental to human and animal health. Two sections of aspergilli are of particular importance to cereal food crops such as corn and barley. Aspergillus section Flavi species like A. flavus and A. parasiticus produce aflatoxins, while section Circumdati species like A. ochraceus and A. sclerotiorum produce ochratoxin A. Mitigating these toxins in food and feed is a critical and ongoing worldwide effort. We have previously investigated biosynthetic gene clusters in Aspergillus flavus that are linked to fungal virulence in corn. We found that one such cluster, asa, is responsible for the production of aspergillic acid, an iron-binding, hydroxamic acid-containing pyrazinone metabolite. Furthermore, we found that the asa gene cluster is present in many other aflatoxin- and ochratoxin-producing aspergilli. The core gene in the asa cluster encodes the small nonribosomal peptide synthetase-like (NRPS-like) protein AsaC. We have swapped the asaC ortholog from A. sclerotiorum into A. flavus, replacing its native copy, and have also cloned both asaC orthologs into Saccharomyces cerevisiae. We show that AsaC orthologs in section Flavi and section Circumdati, while only containing adenylation-thiolation-reductase (ATR) domains, can selectively biosynthesize distinct pyrazinone natural products: deoxyaspergillic acid and flavacol, respectively. Because pyrazinone natural products and the gene clusters responsible for their production are implicated in a variety of important microbe-host interactions, uncovering the function and selectivity of the enzymes involved could lead to strategies that ultimately benefit human health.
Introduction
Aspergillus fungi infect crops and can contaminate food and feed with mycotoxins (Taniwaki et al., 2018). A. flavus and many other section Flavi species produce aflatoxins, potent carcinogens, that are regulated in the food supply worldwide (Frisvad et al., 2019). Aflatoxins are hepatotoxic and can cause acute illness as well as liver cancer with prolonged exposure (Wogan, 1966; Groopman et al., 1988). Aspergillus section Circumdati species, such as A. ochraceus, produce ochratoxin A, a nephrotoxin, mutagen, and potential carcinogen (Visagie et al., 2014). Ochratoxin A is found in many food and feed products, particularly cereals and grapes, and can even contaminate processed meat if the animal consumed infected grain. Aspergillus infection of crops, and subsequent mycotoxin contamination of the food supply, can occur preharvest or postharvest (Agriopoulou et al., 2020). Identifying key plant-pathogen interactions could lead to mitigating preharvest infection by toxin-producing fungi. In addition to known toxins, Aspergillus species also have the capacity to produce many other secondary metabolites, some of which may be involved in infection (Kjaerbolling et al., 2018; Romsdahl and Wang, 2019). For example, cyclopiazonic acid has been identified as an A. flavus pathogenicity factor during corn infection that results in ear rot and subsequent aflatoxin contamination (Chalivendra et al., 2017). The biosynthetic gene clusters responsible for secondary metabolite production are abundant in Aspergillus fungi, however a majority of these clusters are uncharacterized (Uka et al., 2020; Robey et al., 2021). Characterizing these clusters and determining the function of their metabolite products could lead to preharvest strategies that target fungal metabolites essential to the infection process (Bignell et al., 2016).
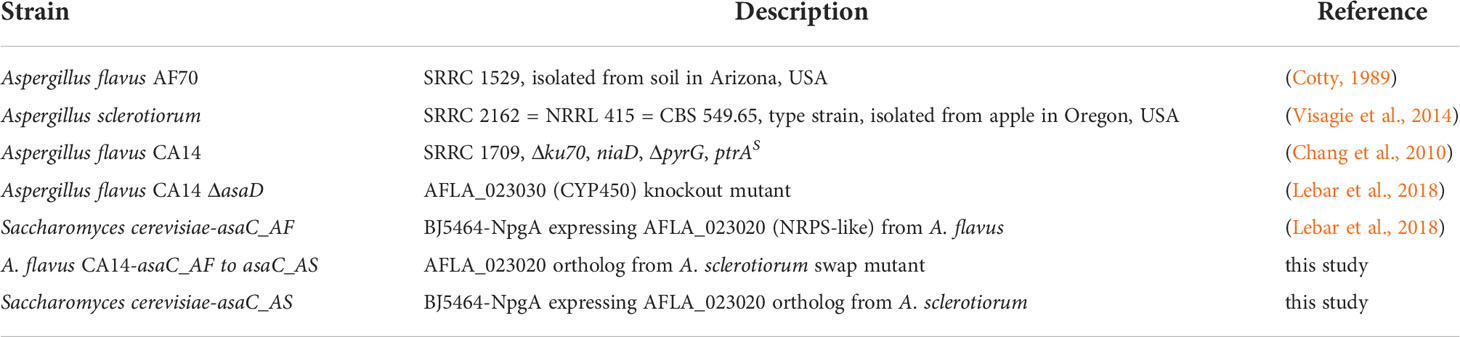
We have previously characterized the biosynthetic gene cluster asa in A. flavus and found it responsible for aspergillic acid production (Lebar et al., 2018). Aspergillic acid is a hydroxamic acid containing pyrazinone formed from leucine and isoleucine precursors that binds iron to form an Fe(III) trimer complex called ferriaspergillin (MacDonald, 1961; Pitt et al., 1983). The ability to produce aspergillic acid has been linked to fungal virulence in a corn kernel assay using asa knock out and genetically complimented strains (Lebar et al., 2018). The asa cluster in A. flavus encodes several biosynthetic proteins: a nonribosomal peptide synthetase-like (NRPS-like) enzyme (AsaC), a CYP450 oxidoreductase (AsaD), and a hydroxylase (AsaB), as well as an MFS transporter (AsaE), an Ankyrin domain protein (AsaA), and a zinc finger transcription factor (AsaR) (Figure 1A). We also previously reported that asa cluster orthologs were present in many aflatoxin-producing species (Aspergillus section Flavi) as well as ochratoxin-producing species (Aspergillus section Circumdati) (Lebar et al., 2019). The section Circumdati strains do not produce aspergillic acid but a similar analog named neoaspergillic acid, which is formed from two leucine residues (Micetich and MacDonald, 1965).
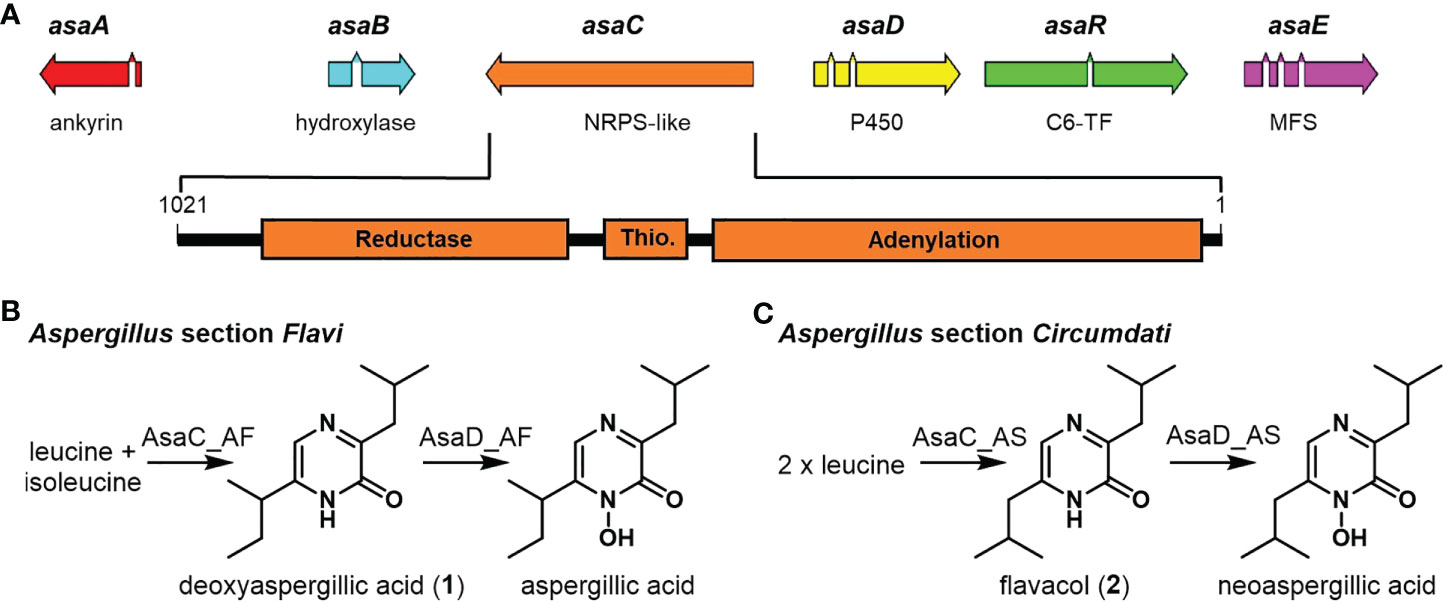
Figure 1 The pyrazinone precursors to aspergillic acids are formed by the NRPS-like enzyme AsaC in Aspergillus spp. (A) Schematic of the asa biosynthetic gene cluster responsible for aspergillic acid production contains the NRPS-like core gene asaC, which is comprised of Adenylation-Thiolation-Reductase (ATR) domains. Introns are shown as gaps in the genes. (B) Species in Aspergillus section Flavi such as A. flavus produce aspergillic acid from leucine and isoleucine through a deoxyaspergillic acid (1) intermediate. (C) Species in Aspergillus section Circumdati such as A. sclerotiorum produce neoaspergillic acid from two leucine residues from a flavacol (2) intermediate.
The core NRPS-like enzyme (AsaC), comprised of only Adenylation-Thiolation-Reductase (ATR) domains, is thought to be responsible for pyrazinone formation utilizing two amino acid residues. NRPS reductase domains liberate the NRP from the enzyme forming an aldehyde product (Forseth et al., 2013; Mullowney et al., 2018). AsaC is a noncanonical NRPS (“NRPS-like”) as it lacks a condensation domain (C), which is the domain that typically forms peptide bonds in NRPs. The pyrazinone precursors to aspergillic acid and neoaspergillic acid differ only by the location of one methyl group on one of the pendant aliphatic side chains (sec-butyl and isobutyl, respectively) (Figures 1B, C). Because the amino acid progenitors of these side chains are likely selected by the NRPS-like protein, we focused this study on determining the products of AsaC orthologs in both A. flavus and A. sclerotiorum. Investigating the selectivity of these small, very similar, noncanonical NRPSs would demonstrate their unique activity and could lead to bioengineering strategies to easily generate differently substituted pyrazinones and their iron-binding, hydroxamic acid analogs.
Materials and methods
Genomic analysis
The A. flavus 3357 asa biosynthetic gene cluster sequence (AFLA_023000 – AFLA_023050: asaA, asaB, asaC, asaD, asaR, and asaE, respectively) (Nierman et al., 2015; Lebar et al., 2018) was identified in other genomes using TBLASTN of the six proteins against all fungal genomes at JGI and all Pezizomycotina genomes from NCBI. The best hit for each protein was merged with the other best hits if they were within 2kb from each other, and then filtered for a total length of at least 10kb using a custom R script. The “annotate from” tool in Geneious Prime (version 2021.2) was used to annotate the six asa genes in each genome from their encoded proteins. The nonribosomal codes (A1-A10) of the AsaC orthologs were identified by aligning the GrsA-PheA to the Aspergillus spp. protein sequences using Muscle (Edgar, 2004). The phylogenetic tree of AsaC amino acid sequences was created with IQ-TREE (Nguyen et al., 2015) and the LG substitution matrix (Le and Gascuel, 2008) using the ultrafast bootstrapping approximation option. Geneious Prime was used to generate the identity matrix for the nucleotide and protein alignments.
Strains and culture conditions
All strains used in this study are described in Table 1. A. flavus AF70 (SRRC 1529) and A. sclerotiorum (SRRC 2162 = NRRL 415 = CBS 549.65) wild-type strains are housed in the Southern Regional Research Center (SRRC) permanent culture collection in New Orleans, USA. A. flavus CA14 Δku70, niaD, ΔpyrG, ptrAS (SRRC 1709, referred to herein as CA14) was used as host for transformation and is sensitive to pyrithiamine (ptrAS) (Chang et al., 2010). The control strain was CA14 transformed with vector pPTRI (Takara Bio Inc., Shiga, Japan) that contains the pyrithiamine (ptrA) resistance gene (Chang et al., 2010). The A. flavus CA14 asaD (AFLA_023030) gene knock out mutant was generated from CA14 using pyrithiamine selection as described previously (Lebar et al., 2018). Fungal spore stocks were generated from cultures point inoculated on double strength V8 agar (50 mL V8 juice, 40 g agar, pH 5.2 per liter of medium) supplemented with 3.0 g ammonium sulfate and 1 mg/ml uracil (2X V8 ASU) and incubated at 30°C in the light (Chang et al., 1993). Generation of the S. cerevisiae strain expressing the A. flavus asaC NRPS-like gene (AFLA_023020, herein called asaC_AF) was described previously (Lebar et al., 2018). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise noted.
Vector construction and fungal transformation of the CA14 asaC_AF to asaC_AS swap mutant
The A. flavus CA14 asaC_AF to asaC_AS gene swap mutant was generated from CA14 using pyrG selection. A. flavus CA14 (PTS) was transformed using an asaC_AS vector constructed by NEBuilder HiFi DNA Assembly method according to the manufacturer’s protocol (NEB E2621S, Ipswich, MA, USA). Briefly, the full length NRPS-like gene from A. sclerotiorum (asaC_AS) as well as the asaC_AF NRPS-like gene’s (AFLA_023020) native promotor and terminator were amplified by PCR using Q5 High-Fidelity DNA Polymerase (NEB M0492S). A. sclerotiorum gDNA was the template for amplification of the NRPS of asaC_AS using the primer pair Asc_NRPS_F and Asc_NRPS_R (Table 2). A. flavus 3357 gDNA served as the template for amplification of the native asaC_AF promotor and terminator regions using two primer pairs, AF_NRPS_prom_F/prom_R and AF_NRPS_Term_F/Term_R (Table 2). The three amplified PCR fragments were then assembled and cloned into a linearized pUC_pyrG plasmid (digest of NotI and BamHI) with the pyrG selection marker (Figure S1). PCR using the primer pair Asc_NRPS_RP nest_F/nest_R (Table 2) was used to screen and confirm the correct clone (Figure S2) as well as BamHI restriction enzyme digestion (Figure S3). Recombinant plasmids were linearized with NotI and used to transform protoplasts of A. flavus CA14 that were generated as described in Cary et al. (2006). Conidia were inoculated in Czapek-Dox (CZ; BD Difco, Franklin Lakes, NJ, USA) broth supplemented with 10 mM ammonium sulfate and 1 mg/ml uracil (CZ-ASU). Transformants were regenerated on CZ-AS without uracil agar plates. Correct transformants were confirmed by PCRs using the primer pair Asc_NRPS_F1 and Asc_NRPS_R1 (Table 2; Figure S4) and Asc_NRPS_F12 and pyrG-R (300) (Table 2; Figure S5).
Yeast expression system
Saccharomyces cerevisiae strain BJ5464-NpgA (MATα ura3-52 his3-Δ200 leu2-Δ1 trp1 pep4::HIS3 prb1Δ1.6R can1 GAL) was used as the yeast expression host (Mootz et al., 2002; Ma et al., 2019). The full length asaC NRPS-like gene (AFLA_023020) homolog from A. sclerotiorum (asaC_AS) was PCR amplified from A. sclerotiorum genomic DNA according to the manufacturer’s protocol using Q5 High-Fidelity DNA Polymerase (NEB M0492S) and the primer pair Asc_023020_F and Asc_023020_R (Table 2; Figure S6). The asaC_AS PCR product was subsequently cloned into the yeast 2 μm expression plasmid (a kind gift from Yi Tang, University of California, Los Angeles) XW55 (SpeI-SwaI digested) using the In-Fusion HD Cloning Kit (Takara Bio, Mountain View, CA, USA) to generate plasmid XW55-asaC_AS (Figure S7). The yeast expression plasmid was used to transform Stellar competent E. coli cells (Takara Bio) according to the manufacturer’s protocol. Cells were spread onto LB plates containing ampicillin (100μg/ml) and incubated at 37°C overnight. XW55-asaC_AS plasmid DNA was isolated from transformants using the Zyp-py™ Plasmid Miniprep Kit (ZymoResearch, Irvine, CA, USA). The yeast expression vectors were sequenced to confirm the correctness of the cloned genes. Two vacuolar proteases PEP4 and PRB1 were inactivated in the BJ5464-NpgA yeast host, which is critical to minimize proteolysis of the large recombinant proteins. The XW55-asaC_AS expression plasmid was transformed into S. cerevisiae BJ5464-NpgA by using S. c. EasyComp™ Transformation Kit (Thermo Fisher Scientific, Waltham, MA, USA). The presence of the XW55-asaC_AS plasmid DNA in transformed yeast was confirmed by KpnI and BamHI restriction enzyme digestion of isolated plasmid DNA (Figure S8) and by sequencing of the 3063 bp asaC_AS gene region amplified from isolated DNA using Q5 High-Fidelity DNA Polymerase. Yeast transformants harboring the XW55-asaC_AS vector were used for further biochemical analyses.
Production, extraction, and analysis of hydroxamic acid trimer-iron(III) complexes
Ferriaspergillin and ferrineoaspergillin production by A. flavus (AF70, CA14 control, and the asaC_AF to asaC_AS swap mutant) and A. sclerotiorum strains was assessed by single point inoculation of spore suspensions on Aspergillus flavus and parasiticus agar (AFPA) medium [20 g yeast extract, 10 g bacto peptone, 0.5 g ferric ammonium citrate, 20 g agar, pH 5.0 per liter of medium, (Pitt et al., 1983)] incubated for 10 days at 30°C in the dark. The fungal colonies were excised from the medium. The fungus was lyophilized, pulverized, then extracted thrice (24 h, shaking at 200 RPM) with ethyl acetate containing 0.1% formic acid. The red-orange extract was filtered through filter paper (Whatman 1, GE Healthcare Life Sciences, USA) and concentrated in vacuo. The dried extracts were dissolved in methanol at 1 mg/ml and centrifuged to remove particulate before analysis on a Waters (Milford, MA, USA) ACQUITY UPLC system using PDA UV and QDa nominal mass detection as reported previously (Lebar et al., 2019). Separations were achieved with a BEH C18 1.7µm, 2.1 x 50 mm column and the following gradient solvent system: (0.5 ml/min, solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile); 5% B (0-1.25 min), gradient to 25% B (1.25-1.5 min), gradient to 100% B (1.5-5.0 min), 100% B (5.0-7.5 min), then column equilibration 5% B (7.6-10.1 min). UV absorbance was recorded at λ = 200-500 nm and measured at λ = 310 nm.
Production and extraction of substituted pyrazinones in Aspergilli and yeast
Conidia (106/ml) of A. flavus CA14 ΔasaD (AFLA_023030) and A. sclerotiorum were used to inoculate 100 ml YE-glycerol broth [2% w/v yeast extract (BD, Sparks, MD), 1% v/v glycerol, pH = 6.5) (Woodward, 1947)]. Static cultures were incubated at 30°C for 14 days. After removal of the mycelial mat, the spent broth was adjusted to pH = 3.0 with 6 N hydrochloric acid then extracted with chloroform (100 ml). The organic layer was removed and concentrated on a rotary evaporator. A portion of the extract was redissolved in methanol at 100 µg/ml then centrifuged (20,000 × g, 2 min) to remove particulates for HRMS analysis. Saccharomyces cerevisiae strains transformed with asaC_AF and asaC_AS genes were grown in yeast extract-peptone-dextrose broth [1% yeast extract (BD, Sparks, MD), 2% peptone, 2% dextrose; YPD] as described previously (Cacho and Tang, 2016). YPD (5 ml) was inoculated with a single colony of S. cerevisiae BJ5464-NpgA grown on selective media and incubated at 28°C for 3 days. The cultures were centrifuged (20,000 × g, 2 min). The supernatant was transferred to a clean vial and extracted with ethyl acetate containing 0.1% formic acid (5 ml), concentrated in vacuo, and then dissolved in methanol (200 µl) for HRMS analysis.
HRMS analysis of substituted pyrazinones
Analyses of deoxyaspergillic acid and flavacol in organic solvent extracts were conducted on a Waters ACQUITY UPLC system using photo diode array (PDA) UV and quadrupole time-of-flight (qTOF) mass detection with the following chromatographic conditions on a Waters BEH C18 1.7 μm, 2.1 mm × 50 mm column: 0.5 ml/min, solvent A (0.1% formic acid in water); solvent B (0.1% formic acid in acetonitrile); 25% B (0–1.5 min), gradient to 50% B (1.5–5.0 min), gradient to 100% B (5.0–5.1 min), 100% B (5.1–7.5 min), then column equilibration 5% B (7.6–10.0 min). High resolution mass (HRM) data were collected on a Waters Xevo G2-XS qTOF spectrometer equipped with a Z-spray ionization source running in ESI+ mode using Waters MassLynx 4.2 software. The qTOF conditions allowed for metabolite ionization (source temperature: 100°C; desolvation temperature: 250°C; desolvation gas flow: 600 L/h; cone gas flow: 50 L/h; capillary voltage: 3.0 kV; sampling cone voltage: 40 V). Analyses were performed in sensitivity and continuum mode, with a mass range of m/z 50–1200 and a scan time of 0.1 s. A data-independent acquisition method with elevated collision energy (MSE) was used with the following eV settings: 6 eV low energy and a high energy ramp from 10−45 eV. Mass calibration was performed with sodium formate. Lock mass data was acquired in MassLynx using leucine enkephalin as a reference at 20 sec. intervals with 3 scans to average. Data were analyzed on Waters UNIFI 1.9.4 software using the “accurate mass screening on MSE data” analysis method with lock mass corrected by UNIFI.
Protein modelling
The Aspergillus NRPS-like enzymes’ adenylation domains were modeled using the crystal structure of the Brevibacillus brevis gramicidin synthase-1 phenylalanine activation domain (PheA) complexed with cyclic-AMP and phenylalanine as a template (Conti et al., 1997). The PheA structure was obtained from the Protein Data Bank (pdb 1AMU) (Berman et al., 2000; Burley et al., 2017). The A. flavus and A. sclerotiorum adenylation domain models were generated using the homology modeler function within the Molecular Operating Environment (MOE 2020.0901, Chemical Computing Group, Montreal, QC, Canada) software. Structures were prepared using Protonate3D and QuickPrep within MOE to fill in missing atoms, build small loops, cap termini, form disulfide bonds, allow flipping of terminal amides, sulfonamide and imidazole groups, add hydrogens, and determine ionization state to optimize the hydrogen bond network. Substitution of isoleucine for leucine in the enzyme binding pocket was accomplished using the protein builder function in MOE and the orientation of the amino acid was optimized with QuickPrep in MOE as described above.
Results
The asa biosynthetic gene cluster is present in many Aspergillus section Flavi and section Circumdati species
TBLASTN queries of protein sequences in the asa cluster identified 19 Aspergillus section Flavi and 23 Aspergillus section Circumdati genomes that contained the entire cluster (Figure 2, Table S1). The identified clusters were quite similar. Although our query was designed to find the clustered genes in any order, all asa cluster orthologs that we found were ordered identically to asa in A. flavus 3357, the organism in which asa was first identified and characterized (Lebar et al., 2018). Aspergillus section Flavi asa cluster orthologs have mean pairwise nucleotide sequence identity of 79.0% to each other and 84.7% to A. flavus 3357 asa. Aspergillus section Circumdati asa cluster orthologs have a mean pairwise nucleotide sequence identity of 78.2% to each other and 50.7% to A. flavus 3357. The NRPS-like protein AsaC had similar homology between and within sections. Figure 2 shows a phylogenetic tree comparing all AsaC orthologs encoded in asa clusters. Aspergillus section Flavi AsaC orthologs have a mean pairwise amino acid sequence identity of 93.1% to each other and 95.3% to A. flavus 3357 AsaC, while section Circumdati AsaC orthologs have a mean pairwise amino sequence identity of 94.6% to each other and 71.6% to A. flavus 3357.
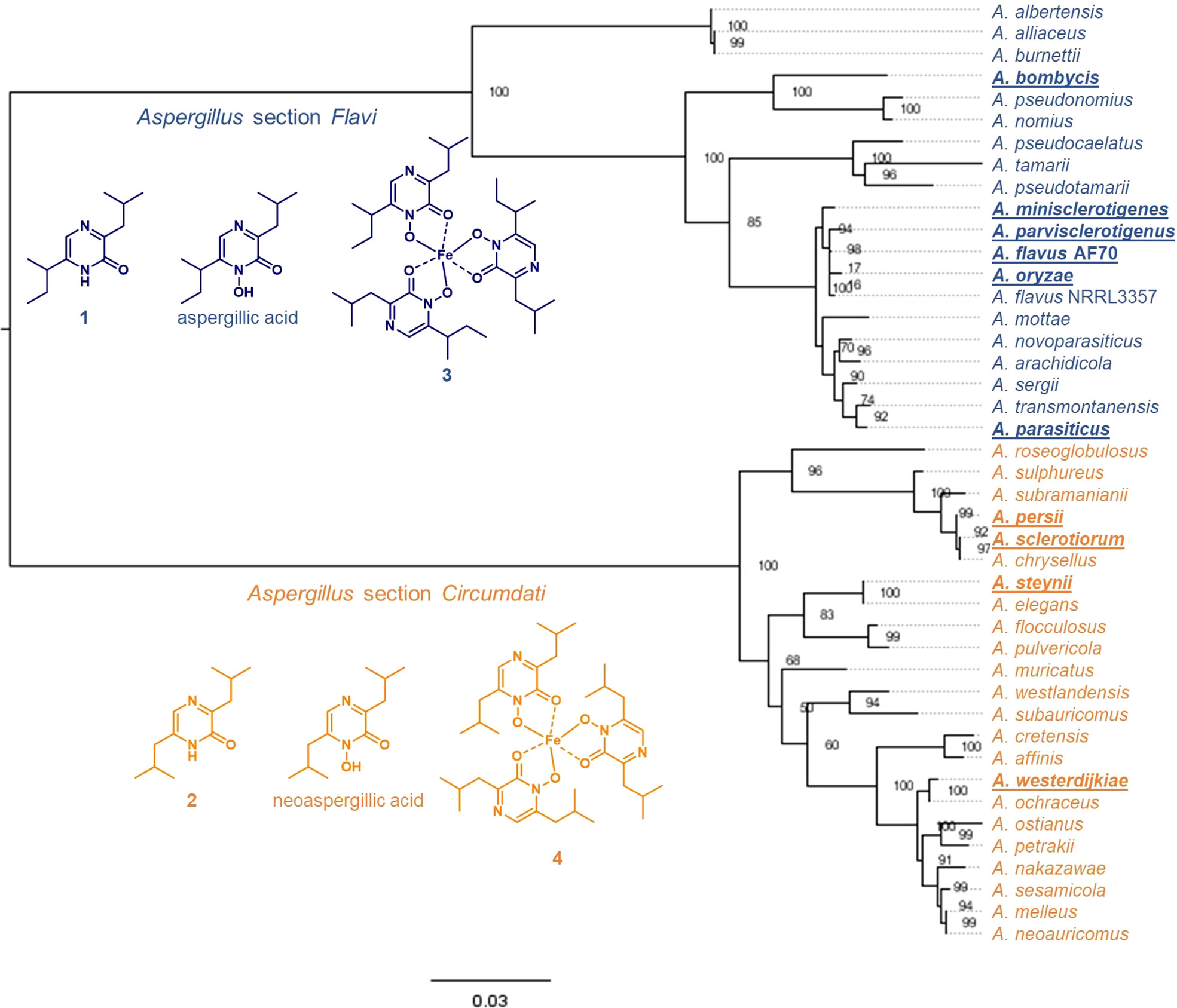
Figure 2 Sequence homology of AsaC in Aspergillus section Flavi is distinct from section Circumdati species. Phylogenetic tree of AsaC NRPS-like orthologs in Aspergillus section Flavi (blue) and Circumdati (orange) species. Chemical confirmation of the substituted pyrazinones and their derivatives produced by these species are in underlined bold font (Lebar et al., 2019). Blue: predominantly deoxyaspergillic acid-derived (1), including aspergillic acid and ferriaspergillin (3); orange: exclusively flavacol-derived (2), including neoaspergillic acid and ferrineoaspergillin (4). The node values represent the branch bootstrap values using the ultrafast bootstrap approximation.
A. flavus asaC_AS swap mutant produces ferrineoaspergillin
We have previously shown that many species from Aspergillus section Flavi, including A. flavus produce mainly ferriaspergillin (from aspergillic acid, See Figure 2, blue bolded font) and species from Aspergillus section Circumdati, including A. sclerotiorum, produce ferrineoaspergillin (Figure 2, orange bolded font) (Lebar et al., 2019). To further investigate the role of asaC in pyrazinone biosynthesis, the asaC_AF gene naturally occurring in A. flavus was swapped with the asaC ortholog from A. sclerotiorum (asaC_AS). Wild-type A. flavus and the A. flavus CA14 control produce the same mix of ferriaspergillin (3) and its analogs (Figures 3A, B) on AFPA medium (see also Figure S9, S10). A. sclerotiorum produces ferrineoaspergillin (4, Figure 3C and Figure S11). The A. flavus strain swapped with the asaC_AS ortholog produces ferrineoaspergillin (Figure 3D and Figure S12) and a small shoulder peak with similar retention time to 3. However, the identity of this shoulder peak could not be verified. These findings suggest that the two amino acid precursor residues forming the pyrazinone intermediate – and, subsequently, (neo)aspergillic acid and ferri(neo)aspergillin – are dependent only on AsaC.
Figure 3 A. flavus containing asaC from A. sclerotiorum produces an iron(III)-bound trimer found naturally only in Aspergillus section Circumdati species. (A–D) UV chromatograms of Aspergillus extracts grown on AFPA measured in relative absorbance units (AU) at λ = 310 nm.
AsaC is solely responsible for the biosynthesis of aspergillic acid precursor pyrazinones
To further validate our observation that only the NRPS-like enzyme AsaC is necessary to selectively biosynthesize pyrazinones, we cloned asaC_AF and asaC_AS into yeast host strains and compared the yeast extracts to extracts of A. flavus ΔasaD and A. sclerotiorum. We previously reported that A. flavus ΔasaD, a knockout mutant of the P450 oxidase responsible for the conversion of deoxyaspergillic acid to aspergillic acid, produces an abundance of deoxyaspergillic acid (Lebar et al., 2018). Both deoxyaspergillic acid and flavacol share the molecular formula C12H20N2O and their [M+H]+ ion is 209.16484 m/z. The extracted ion chromatogram (EIC, m/z = 209.165) of A. flavus ΔasaD grown in yeast extract (YE) glycerol broth confirms that deoxyaspergillic acid (1) is present (Figure 4A, left and Figure S13), as is a small amount of flavacol (2, Figure S14). High energy MS data shows a distinct fragment peak at 137 m/z for deoxyaspergillic acid (Figure 4A, right). The extracted ion chromatogram (EIC, m/z = 209.165) of A. sclerotiorum grown in YE glycerol shows only one peak, flavacol (2), which has a distinct fragment at 123 m/z in its high energy MS spectrum (Figure 4B and Figure S15). The extract prepared from S. cerevisiae expressing asaC from A. flavus (asaC_AF) shows deoxyaspergillic acid (1) as the main product (Figure 4C and Figure S16), as well as a small amount of flavacol (2, Figure S17), while S. cerevisiae expressing asaC from A. sclerotiorum (asaC_AS) contains only flavacol (2, Figure 4D and Figure S18), mirroring their cognate fungal extracts. It has been previously shown that deoxyaspergillic acid is formed from one leucine and one isoleucine residue (MacDonald, 1961) and flavacol is formed from two leucine residues (Micetich and MacDonald, 1965). We herein show that AsaC selectively dictates which amino acids the pyrazinone products are derived from. AsaC_AF predominately selects isoleucine and leucine, while AsaC_AS selects two leucine residues. Previously reported NRPS-like enzymes that are comprised of one ATR module produce products comprised of two of the same amino acid (Forseth et al., 2013; Morgan et al., 2021), much like AsaC_AS. AsaC_AF is unique in that it makes a product with two distinct amino acids from one ATR module. It is also interesting to note that the pattern of relative amounts of deoxyaspergillic acid, flavacol, and a minor flavacol analog (RT = 3.32 min) in the A. flavus ΔasaD strain and the yeast expressing asaC_AF are nearly identical (Figure 4A, C, see also Figures S19, S20). We can infer that this ratio is entirely controlled by the AsaC enzyme and is not dependent on any other genes in the asa cluster.
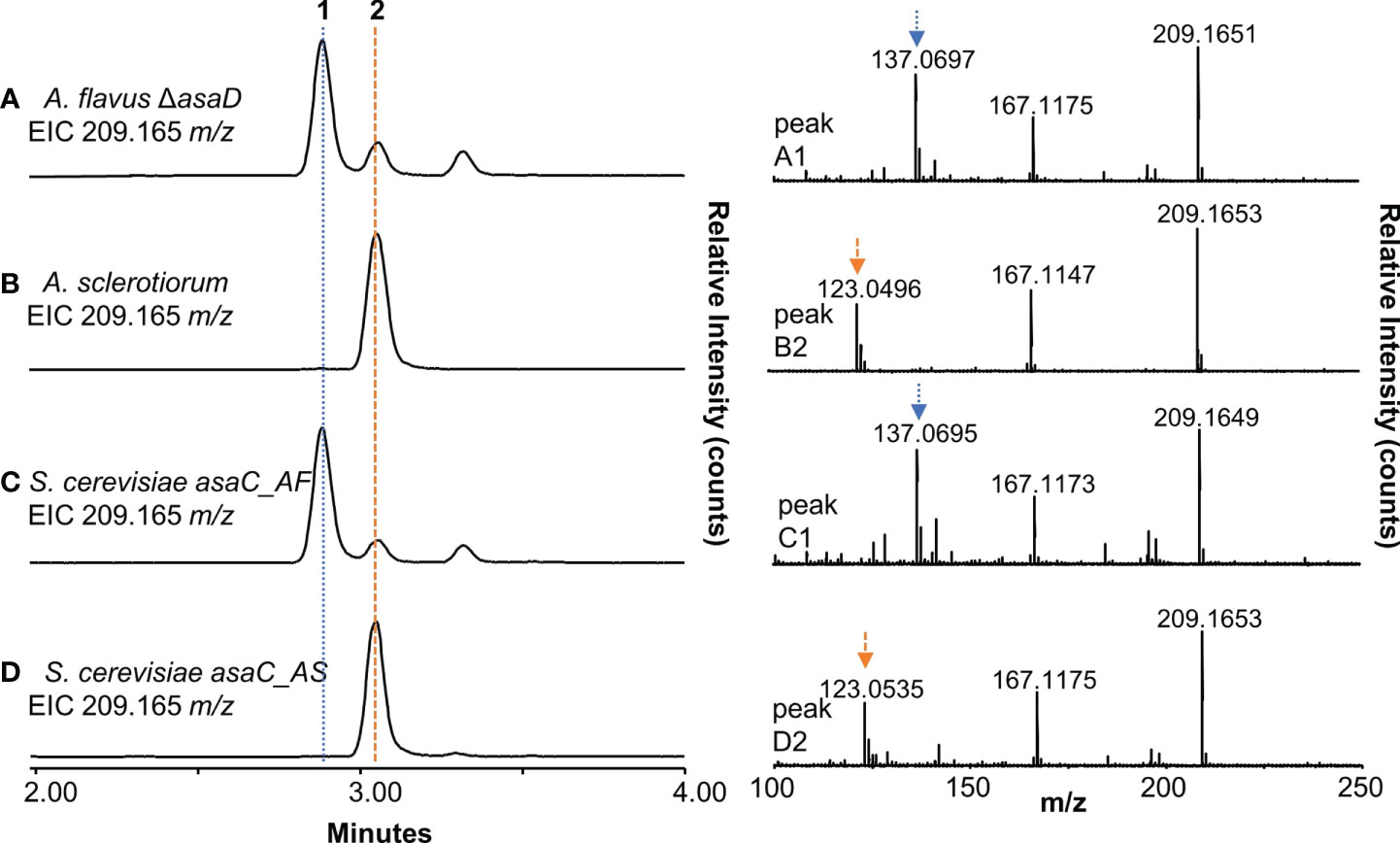
Figure 4 AsaC forms aspergillic acid precursor pyrazinones. (A), left) Extracted ion chromatogram (EIC) of A. flavus ΔasaD. (A), right) High energy mass spectra of peak 1 from A. flavus ΔasaD extract. (B), left) EIC of A. sclerotiorum extract. (B), right) High energy mass spectra of peak 2 from A. sclerotiorum extract. (C), left) EIC of S. cerevisiae expressing asaC from A. flavus (asaC_AF) extract. (C), right) High energy mass spectra of peak 1 from S. cerevisiae asaC_AF extract. (D), left) EIC of S. cerevisiae expressing asaC from A. sclerotiorum (asaC_AS) extract. (D), right) High energy mass spectra of peak 2 from S. cerevisiae asaC_AS extract. 1: deoxyaspergillic acid; 2: flavacol.
The nonribosomal codes of AsaC in Aspergillus section Flavi and section Circumdati differ by only one residue
The adenylation (A) domains of NRPS enzymes selectively activate amino acids, which are then tethered to the thiolation (T) domain. The selectivity of the adenylation domain has been investigated extensively in bacteria and to a lesser extent, in fungi (Kudo et al., 2019). A seminal study reporting the crystal structure of the adenylation domain of the NRPS responsible for phenylalanine activation in gramicidin S biosynthesis (GrsA-PheA) identified ten key residues that are responsible for the selectivity (Conti et al., 1997). These ten residues became known as the “nonribosomal code” and are designated A1-A10. Alignment of the GrsA-PheA to the Aspergillus spp. AsaC protein sequences revealed that Aspergillus section Flavi and section Circumdati species share 9 of the 10 nonribosomal code residues (Figure 5A, Figures S21, S22). All section Flavi species contain isoleucine at A8, while all section Circumdati species contain serine at this position. To highlight the ten key amino acids in the A domains of AsaC_AF and AsaC_AS, model structures were prepared using the PheA (1AMU pdb) crystal as a template (Conti et al., 1997). The AsaC_AF and AsaC_AS models complexed with leucine or isoleucine were prepared by exchanging the substrate phenylalanine in the PheA crystal and preserving the orientation of the respective substrate amino acids. The models predict that both the highly conserved A1 aspartic acid and the A10 lysine residues lie proximal to the substrate Leu or Ile residues in AsaC_AF and AsaC_AS (Figures 5B–D). In each model, ionic interactions are predicted to form between the substrate leucine or isoleucine amino acid and the A1 aspartic acid consistent with interactions observed in the PheA (1AMU pdb) template. The highly conserved A10 lysine, along with glycine 275 (A6) and isoleucine 307 (A8) in AsaC_AF and glycine 274 (A6) and serine 306 (A8) in AsaC_AS are also predicted to interact with the substrate amino acids, but these interactions are predicted to be relatively much weaker in this set of models (Figures 5B–D).
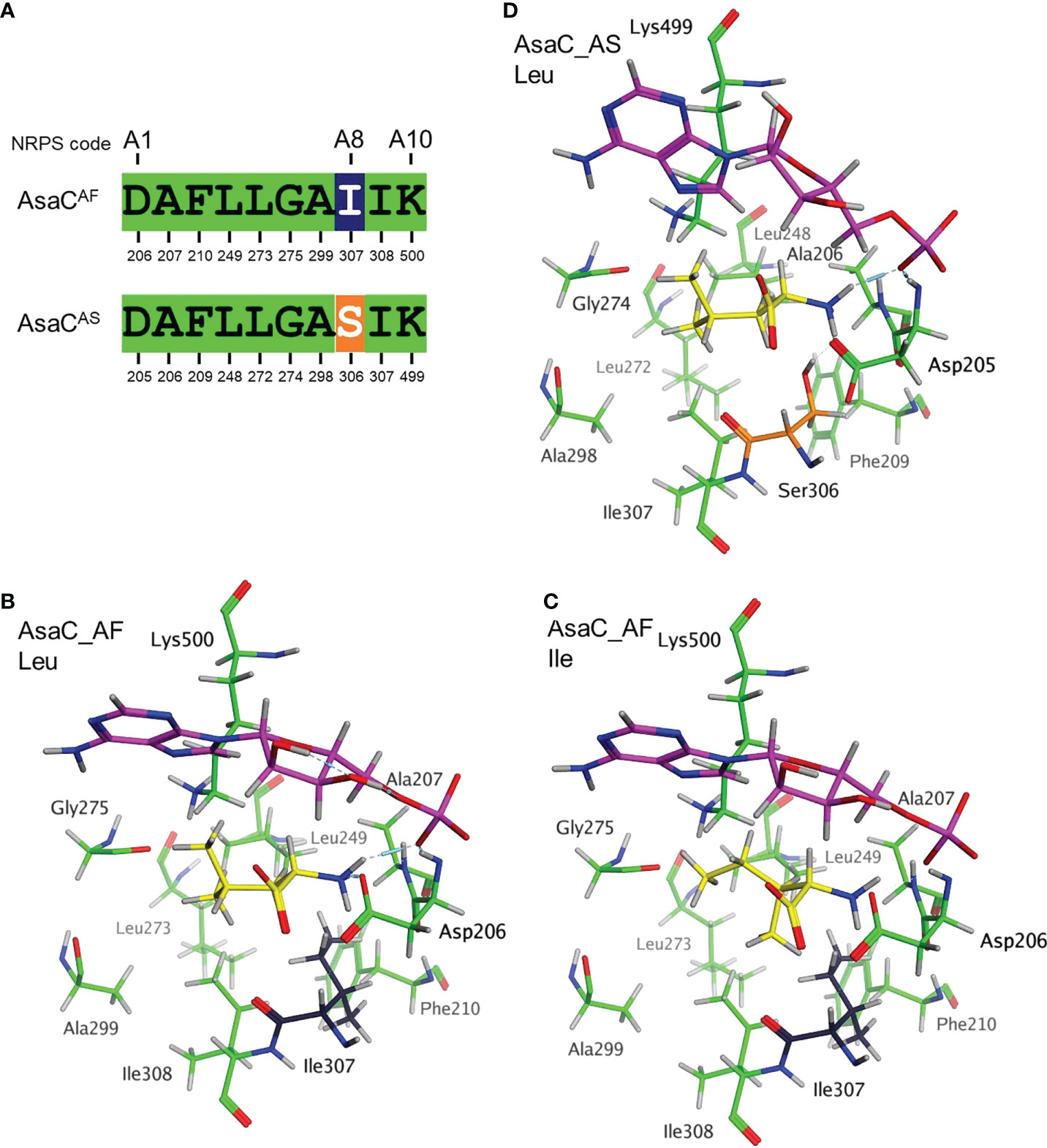
Figure 5 The nonribosomal codes of AsaC in Aspergillus section Flavi and section Circumdati differ by only one residue. (A) Alignment of the 10 key amino acid residues in the nonribosomal code using GrsA-PheA as a reference. AsaC_AF represents the code from A. flavus as well as AsaC from all Aspergillus section Flavi species sequenced to date (see Figure 1). AsaC_AS represents the code from A. sclerotiorum and AsaC from all Aspergillus section Circumdati species sequenced to date (see Figure 1). Stick rendering model of the 10 key amino acids in AsaC_AF (B, C) and AsaC_AS (D) enzymes using the PheA crystal structure (pdb 1AMU) as a template. Key amino acid colors are consistent with the alignment in panel (A). Complexed amino acids leucine (B, D) or isoleucine (C) are colored yellow, and cAMP (in magenta) is orientated similarly near the top of each panel.
Discussion
Typical NRPS enzymes are modular; each module (usually ATC) adds one distinct amino acid to the peptide chain (Schwarzer et al., 2003; Evans, 2016). Because pyrazinone metabolites arise from two amino acids, a dimodular NRPS would be predicted to form these products. Pyrazinone metabolites have been reported from bacteria, such as the aureusimines (=phevalin, tyrvalin) produced by Staphylococcus aureus (Wyatt et al., 2010; Zimmermann and Fischbach, 2010). AusA (=PzaA), a dimodular NRPS responsible for the biosynthesis of aureusimines, is comprised of the domains ATCATR. Each module (ATC and ATR) is responsible for activating a distinct amino acid, while the C forms a peptide bond between the two activated amino acids. The AusA reductase domain (R) liberates the dipeptide as an aldehyde and, after spontaneous cyclization and oxidation, the substituted pyrazinones are formed (Figure 6A) (Wilson et al., 2013).
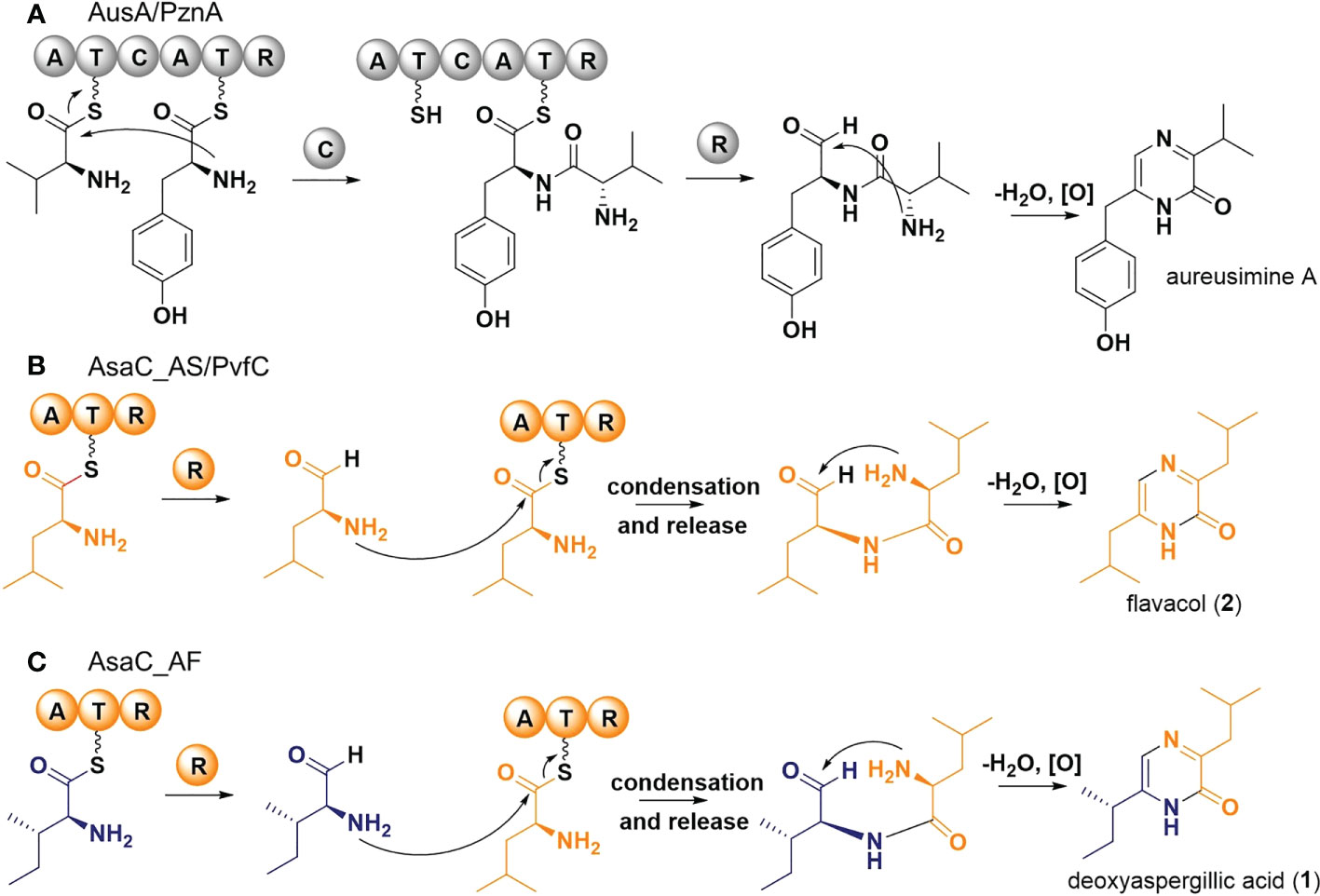
Figure 6 Proposed biosynthetic pathways of pyrazinone metabolites. (A) Schematic of the biosynthesis of aureusimine from valine and tyrosine by Staphylococcus aureus AusA. Adapted from (Wyatt et al., 2010; Zimmermann and Fischbach, 2010). (B) Schematic of the biosynthesis of flavacol (2) from two leucines by PvfC in Pseudomonas fluorescens and AsaC_AS in Aspergillus sclerotiorum. Adapted from (Morgan et al., 2021). (C) Schematic of the biosynthesis of deoxyaspergillic acid (1) from isoleucine and leucine by AsaC_AF in A flavus.
Unexpectedly, and in contrast to the dimodular NRPS AusA (ATCATR) from Staphylococcus, the unimodular NRPS-like enzyme AsaC (ATR) forms pyrazinones in Aspergillus species. A unimodular ATR enzyme with similar activity from Pseudomonas fluorescens has been recently characterized. A detailed mechanistic investigation was conducted on PvfC by Li and coworkers (Morgan et al., 2021). PvfC is the core biosynthesis gene in the Pseudomonas virulence factor (pvf) biosynthetic gene cluster. PvfC was shown to make flavacol and a related substituted imidazole from two leucine residues. AsaC_AS likely functions similarly to the proposed mechanism for PvfC (Figure 6B): 1. A leucine residue is activated and tethered to the thiolation domain (T); 2. The reductase domain (R) liberates the residue as an aldehyde (NH2-Leu-CHO); 3. Another leucine residue is activated and tethered to T; 4. The amine from NH2-Leu-CHO formed in step 2 is condensed with the tethered leucine, liberating a dipeptide aldehyde (NH2-Leu-Leu-CHO); 5. Spontaneous cyclisation and oxidation then form flavacol (2). How the condensation in step 4 is accomplished without a canonical condensation domain requires further investigation.
The mechanism of AsaC_AF from A. flavus is more interesting since the pyrazinone product is formed from two different amino acids. We propose AsaC_AF first activates an isoleucine residue. After reductive release forming NH2-Ile-CHO, a leucine is then activated and tethered to T. In a similar mechanism to AsaC_AS, AsaC_AF likely facilitates condensation and release forming NH2-Leu-Ile-CHO, which affords deoxyaspergillic acid (1) after cyclisation and oxidation (Figure 6C). The initial activation step of AsaC_AF appears to be more promiscuous than the activation of leucine by AsaC_AS because we observed both deoxyaspergillic acid and a small amount of flavacol (from NH2-Leu-Ile-CHO and NH2-Leu-Leu-CHO, respectively) in AsaC_AF experiments, but only flavacol in the AsaC_AS experiments. Deoxyaspergillic acid is the predominant AsaC_AF product, however, indicating the enzyme is surprisingly selective as each of the two distinct amino acids must be selected in the appropriate order to afford the mixed disubstituted product. This pyrazinone product contains an alpha-isobutyl group and a sec-butyl adjacent to the amide NH. If the proposed mechanism is correct and the order of activations were switched, the positions of the butyl groups would also be switched. Further investigation is required to determine the mechanism by which AsaC_AF incorporates different amino acids at different steps of deoxyaspergillic acid biosynthesis with only one ATR module.
The modeling presented here provides a coarse prediction of the arrangement of the crucial ten amino acids in the Aspergillus AsaC_AF and AsaC_AS enzymes thought to be involved in substrate recognition based upon the adenylation domain in the PheA-GrsA template structure. In the PheA structure, the highly conserved A1 (Asp235) and A10 (Lys517) residues interact directly with the substrate phenylalanine (Conti et al., 1997). For example, the A1 (Asp235) residue forms electrostatic interactions with the amino group of the substrate phenylalanine while the A10 (Lys517) residue interacts with substrate phenylalanine carboxylic acid. This is not necessarily the case in the Aspergillus AsaC models. In both Aspergillus AsaC enzymes the A1 (Asp205 or Asp206) residue is oriented so that it would be predicted to interact favorably with the amino group of the substrate amino acid. In contrast, the A10 (Lys499 or Lys500) is positioned in manner so that it generates electrostatic interactions with Gly274, but the carboxylic acid of the substrate amino acid is rotated away from the A10 lysine (Figure 5). As discussed above, the AsaC_AF and AsaC_AS enzymes have different substrate amino acid preferences. The A8 residues, Ile307 and Ser306, that differ between the two Aspergillus enzymes may play a role in substrate recognition (leucine or isoleucine) and therefore which pyrazinone products are produced. Ile307 from AsaC_AF and the Ser306 from AsaC_AS are positioned just below the respective A1 residues (Asp205 or Asp206) in the substrate pocket. These A8 residues are predicted to interact with the A1 residue(s) and this may allow them some influence on substrate specificity. Site-directed mutagenesis combined with further modeling of the Aspergillus enzymes will help to elucidate the structure-function relationship of specific amino acids for these ATR enzymes.
While the exact function of the pyrazinone natural products produced by Aspergillus species with respect to virulence is unknown, Dolezal et al. (2013) reported that asaC was upregulated during A. flavus infection of corn. We were later able to show that an A. flavus mutant in which asaC was knocked out was less virulent in a corn kernel infection model when compared to the control strain that harbored an intact asaC gene. Little is currently known about the function of these metabolites in A. sclerotiorum and other ochratoxin A-producing aspergilli. Because the downstream metabolites in the asa biosynthesis pathways are hydroxamic acid-containing pyrazinones that bind iron (e.g. aspergillic acid and neoaspergillic acid), it is quite possible that these molecules are acting as siderophores, thus contributing to Aspergillus virulence (Haas, 2014). Flavacol can be isolated from both A. flavus and A. sclerotiorum and could be acting as a signaling molecule for virulence, which has been reported in some bacteria. The Pseudomonas virulence factor (pvf) gene cluster, which includes the flavacol-producing ATR enzyme PvfC, is responsible for the production of unidentified autoinducers with many functions (Kretsch et al., 2021). PVF autoinducers were found to regulate a variety of secreted molecules, including toxins and proteins, that play a key role in bacterial virulence. Many pyrazinone-producing NRPSs have also been found in gut bacteria and appear to be involved in host colonization. NRPS-like enzymes have been engineered to make complex and diverse small molecules that resemble natural products (van Dijk et al., 2016; van Dijk and Wang, 2018).
Conclusion
We have shown that AsaC NRPS-like enzymes with only an ATR module selectively produce disubstituted pyrazinones. Both AsaC homologs from A. flavus and A. sclerotiorum lack a C domain but are able to condense two amino acids to produce substituted pyrazinones. The selectivity of AsaC_AF from A. flavus is especially interesting in that it selectively forms the dipeptide product from two different amino acids while harboring only one A domain. Because these small genes produce functional enzymes when cloned into yeast, their selectivity can be further probed and, ideally, modified to produce a wide variety of substituted pyrazinone products. Further harnessing the biosynthetic machinery that produces these simple yet abundant and important pyrazinone natural products could lead to treatment strategies that have the potential to encourage beneficial microbes or inhibit pathogenic microbes in a variety of hosts.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ML and JC designed the research. ML, JC, BM, CC-W, QW, and CM performed experiments and analysed data. All authors wrote the manuscript and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2022.1029195/full#supplementary-material
References
Agriopoulou S., Stamatelopoulou E., Varzakas T. (2020). Advances in occurrence, importance, and mycotoxin control strategies: Prevention and detoxification in foods. Foods 9 (2), 137. doi: 10.3390/foods9020137
Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., et al. (2000). The protein data bank. Nucleic Acids Res. 28 (1), 235–242. doi: 10.1093/nar/28.1.235
Bignell E., Cairns T. C., Throckmorton K., Nierman W. C., Keller N. P. (2016). Secondary metabolite arsenal of an opportunistic pathogenic fungus. Philos. Trans. R Soc. Lond B Biol. Sci. 371 (1709), 20160023. doi: 10.1098/rstb.2016.0023
Burley S. K., Berman H. M., Kleywegt G. J., Markley J. L., Nakamura H., Velankar S. (2017). Protein data bank (PDB): The single global macromolecular structure archive. Methods Mol. Biol. 1607, 627–641. doi: 10.1007/978-1-4939-7000-1_26
Cacho R. A., Tang Y. (2016). Reconstitution of fungal nonribosomal peptide synthetases in yeast and In vitro. Methods Mol. Biol. 1401, 103–119. doi: 10.1007/978-1-4939-3375-4_7
Cary J. W., Ehrlich K. C., Bland J. M., Montalbano B. G. (2006). The aflatoxin biosynthesis cluster gene, aflX, encodes an oxidoreductase involved in conversion of versicolorin a to demethylsterigmatocystin. Appl. Environ. Microbiol. 72 (2), 1096–1101. doi: 10.1128/AEM.72.2.1096-1101.2006
Chalivendra S. C., DeRobertis C., Chang P. K., Damann K. E. (2017). Cyclopiazonic acid is a pathogenicity factor for aspergillus flavus and a promising target for screening germplasm for ear rot resistance. Mol. Plant Microbe Interact. 30 (5), 361–373. doi: 10.1094/MPMI-02-17-0026-R
Chang P.-K., Cary J. W., Bhatnagar D., Cleveland T. E., Bennett J. W., Linz J. E., et al. (1993). Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl. Environ. Microbiol. 59 (10), 3273–3279. doi: 10.1128/aem.59.10.3273-3279.1993
Chang P.-K., Scharfenstein L. L., Wei Q., Bhatnagar D. (2010). Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 81 (3), 240–246. doi: 10.1016/j.mimet.2010.03.010
Conti E., Stachelhaus T., Marahiel M. A., Brick P. (1997). Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin s. EMBO J. 16 (14), 4174–4183. doi: 10.1093/emboj/16.14.4174
Cotty P. J. (1989). Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79 (7), 808. doi: 10.1094/Phyto-79-808
Dolezal A. L., Obrian G. R., Nielsen D. M., Woloshuk C. P., Boston R. S., Payne G. A. (2013). Localization, morphology and transcriptional profile of Aspergillus flavus during seed colonization. Mol. Plant Pathol. 14 (9), 898–909. doi: 10.1111/mpp.12056
Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi: 10.1093/nar/gkh340
Evans B. S. (2016). Nonribosomal peptide and polyketide biosynthesis. (New York, NY: Humana Press) 1401. doi: 10.1007/978-1-4939-3375-4
Forseth R. R., Amaike S., Schwenk D., Affeldt K. J., Hoffmeister D., Schroeder F. C., et al. (2013). Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew Chem. Int. Ed. Engl. 52 (5), 1590–1594. doi: 10.1002/anie.201207456
Frisvad J. C., Hubka V., Ezekiel C. N., Hong S. B., Novakova A., Chen A. J., et al. (2019). Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 93, 1–63. doi: 10.1016/j.simyco.2018.06.001
Groopman J. D., Cain L. G., Kensler T. W., Harris C. C. (1988). Aflatoxin exposure in human populations: Measurements and relationship to cancer. CRC Crit. Rev. Toxicol. 19 (2), 113–145. doi: 10.3109/10408448809014902
Haas H. (2014). Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Natural Product Rep. 31 (10), 1266–1276. doi: 10.1039/c4np00071d
Kjaerbolling I., Vesth T. C., Frisvad J. C., Nybo J. L., Theobald S., Kuo A., et al. (2018). Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc. Natl. Acad. Sci. United States America 115 (4), E753–E761. doi: 10.1073/pnas.1715954115
Kretsch A. M., Morgan G. L., Acken K. A., Barr S. A., Li B. (2021). Pseudomonas virulence factor pathway synthesizes autoinducers that regulate the secretome of a pathogen. ACS Chem. Biol. 16 (3), 501–509. doi: 10.1021/acschembio.0c00901
Kudo F., Miyanaga A., Eguchi T. (2019). Structural basis of the nonribosomal codes for nonproteinogenic amino acid selective adenylation enzymes in the biosynthesis of natural products. J. Ind. Microbiol. Biotechnol. 46 (3-4), 515–536. doi: 10.1007/s10295-018-2084-7
Lebar M. D., Cary J. W., Majumdar R., Carter-Wientjes C. H., Mack B. M., Wei Q., et al. (2018). Identification and functional analysis of the aspergillic acid gene cluster in Aspergillus flavus. Fungal Genet. Biol. 116, 14–23. doi: 10.1016/j.fgb.2018.04.009
Lebar M. D., Mack B. M., Carter-Wientjes C. H., Gilbert M. K. (2019). The aspergillic acid biosynthetic gene cluster predicts neoaspergillic acid production in aspergillus section circumdati. World Mycotoxin J. 12 (3), 213–222. doi: 10.3920/wmj2018.2397
Le S. Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25 (7), 1307–1320. doi: 10.1093/molbev/msn067
MacDonald J. C. (1961). Biosynthesis of aspergillic acid. J. Biol. Chem. 236, 512–514. doi: 10.1016/S0021-9258(18)64394-7
Ma W., Zhao L., Zhao W., Xie Y. (2019). (E)-2-Hexenal, as a potential natural antifungal compound, inhibits aspergillus flavus spore germination by disrupting mitochondrial energy metabolism. J. Agric. Food Chem. 67 (4), 1138–1145. doi: 10.1021/acs.jafc.8b06367
Micetich R. G., MacDonald J. C. (1965). Biosynthesis of neoaspergillic and neohydroxyaspergillic acids. J. Biol. Chem. 240, 1962–1695. doi: 10.1016/S0021-9258(18)97490-9
Mootz H. D., Schorgendorfer K., Marahiel M. A. (2002). Functional characterization of 4'-phosphopantetheinyl transferase genes of bacterial and fungal origin by complementation of Saccharomyces cerevisiae lys5. FEMS Microbiol. Lett. 213 (1), 51–57. doi: 10.1111/j.1574-6968.2002.tb11285.x
Morgan G. L., Li K., Crawford D. M., Aube J., Li B. (2021). Enzymatic synthesis of diverse heterocycles by a noncanonical nonribosomal peptide synthetase. ACS Chem. Biol. 16 (12), 2776–2786. doi: 10.1021/acschembio.1c00623
Mullowney M. W., McClure R. A., Robey M. T., Kelleher N. L., Thomson R. J. (2018). Natural products from thioester reductase containing biosynthetic pathways. Nat. Prod. Rep. 35 (9), 847–878. doi: 10.1039/c8np00013a
Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Nierman W. C., Yu J., Fedorova-Abrams N. D., Losada L., Cleveland T. E., Bhatnagar D., et al. (2015). Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome Announcements 3 (2), e00168–e00115. doi: 10.1128/genomeA.00168-15
Pitt J. I., Hocking A. D., Glenn D. R. (1983). An improved medium for the detection of Aspergillus flavus and A. parasiticus. J. Appl. Bacteriol. 54, 109–114. doi: 10.1111/j.1365-2672.1983.tb01307.x
Robey M. T., Caesar L. K., Drott M. T., Keller N. P., Kelleher N. L. (2021). An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Proc. Natl. Acad. Sci. U.S.A. 118 (19), e2020230118. doi: 10.1073/pnas.2020230118
Romsdahl J., Wang C. C. C. (2019). Recent advances in the genome mining of aspergillus secondary metabolites (covering 2012-2018). Medchemcomm 10 (6), 840–866. doi: 10.1039/c9md00054b
Schwarzer D., Finking R., Marahiel M. A. (2003). Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20 (3), 275. doi: 10.1039/b111145k
Taniwaki M. H., Pitt J. I., Magan N. (2018). Aspergillus species and mycotoxins: occurrence and importance in major food commodities. Curr. Opin. Food Sci. 23, 38–43. doi: 10.1016/j.cofs.2018.05.008
Uka V., Cary J. W., Lebar M. D., Puel O., De Saeger S., Diana Di Mavungu J. (2020). Chemical repertoire and biosynthetic machinery of the aspergillus flavus secondary metabolome: A review. Compr. Rev. Food Sci. Food Saf. 19 (6), 2797–2842. doi: 10.1111/1541-4337.12638
van Dijk J. W., Guo C. J., Wang C. C. (2016). Engineering fungal nonribosomal peptide synthetase-like enzymes by heterologous expression and domain swapping. Org. Lett. 18 (24), 6236–6239. doi: 10.1021/acs.orglett.6b02821
van Dijk J. W. A., Wang C. C. C. (2018). Expanding the chemical space of nonribosomal peptide synthetase-like enzymes by domain and tailoring enzyme recombination. Org. Lett. 20 (17), 5082–5085. doi: 10.1021/acs.orglett.8b01581
Visagie C. M., Varga J., Houbraken J., Meijer M., Kocsube S., Yilmaz N., et al. (2014). Ochratoxin production and taxonomy of the yellow aspergilli (Aspergillus section Circumdati). Stud. Mycol. 78, 1–61. doi: 10.1016/j.simyco.2014.07.001
Wilson D. J., Shi C., Teitelbaum A. M., Gulick A. M., Aldrich C. C. (2013). Characterization of AusA: A dimodular nonribosomal peptide synthetase responsible for the production of aureusimine pyrazinones. Biochemistry 52 (5), 926–937. doi: 10.1021/bi301330q
Wogan G. N. (1966). Chemical nature and biological effects of the aflatoxins. Bacteriol. Rev. 30 (2), 460–470. doi: 10.1128/br.30.2.460-470.1966
Woodward C. R. (1947). Production of aspergillic acid by surface cultures of Aspergillus flavus. J. Bacteriol. 54 (3), 375–379. doi: 10.1128/jb.54.3.375-379.1947
Wyatt M. A., Wang W., Roux C. M., Beasley F. C., Heinrichs D. E., Dunman P. M., et al. (2010). Staphylococcus aureus nonribosomal peptide secondary metabolites regulate virulence. Science 329 (5989), 294–296. doi: 10.1126/science.1188888
Keywords: mycotoxin, aspergillic acid, ATR, biosynthesis, aflatoxin, ochratoxin, flavacol
Citation: Lebar MD, Mack BM, Carter-Wientjes CH, Wei Q, Mattison CP and Cary JW (2022) Small NRPS-like enzymes in Aspergillus sections Flavi and Circumdati selectively form substituted pyrazinone metabolites. Front. Fungal Biol. 3:1029195. doi: 10.3389/ffunb.2022.1029195
Received: 26 August 2022; Accepted: 14 October 2022;
Published: 26 October 2022.
Edited by:
Selma Pascale Snini, UMR5503 Laboratoire de Génie Chimique (LGC), FranceReviewed by:
Wenbing Yin, Institute of Microbiology (CAS), ChinaMehdi Razzaghi-Abyaneh, Pasteur Institute of Iran (PII), Iran
Copyright © 2022 Lebar, Mack, Carter-Wientjes, Wei, Mattison and Cary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew D. Lebar, bWF0dGhldy5sZWJhckB1c2RhLmdvdg==
 Matthew D. Lebar
Matthew D. Lebar Brian M. Mack
Brian M. Mack Carol H. Carter-Wientjes
Carol H. Carter-Wientjes Qijian Wei1
Qijian Wei1 Christopher P. Mattison
Christopher P. Mattison Jeffrey W. Cary
Jeffrey W. Cary