- 1Microbial Sciences, Institute of Biology Leiden, Leiden University, Leiden, Netherlands
- 2Centre for Structural and Functional Genomics, Concordia University, Montreal, QC, Canada
- 3DSM Biosciences and Process Innovation, Center for Biotech Innovation, Delft, Netherlands
- 4DSM Food and Beverage, Center for Food Innovation, Delft, Netherlands
- 5Fungal Genetics and Technology Consultancy, Wageningen, AJ, Netherlands
The feruloyl esterase B gene (faeB) is specifically induced by hydroxycinnamic acids (e.g. ferulic acid, caffeic acid and coumaric acid) but the transcriptional regulation network involved in faeB induction and ferulic acid metabolism has only been partially addressed. To identify transcription factors involved in ferulic acid metabolism we constructed and screened a transcription factor knockout library of 239 Aspergillus niger strains for mutants unable to utilize ferulic acid as a carbon source. The ΔfarA transcription factor mutant, already known to be involved in fatty acid metabolism, could not utilize ferulic acid and other hydroxycinnamic acids. In addition to screening the transcription factor mutant collection, a forward genetic screen was performed to isolate mutants unable to express faeB. For this screen a PfaeB-amdS and PfaeB-lux613 dual reporter strain was engineered. The rationale of the screen is that in this reporter strain ferulic acid induces amdS (acetamidase) expression via the faeB promoter resulting in lethality on fluoro-acetamide. Conidia of this reporter strain were UV-mutagenized and plated on fluoro-acetamide medium in the presence of ferulic acid. Mutants unable to induce faeB are expected to be fluoro-acetamide resistant and can be positively selected for. Using this screen, six fluoro-acetamide resistant mutants were obtained and phenotypically characterized. Three mutants had a phenotype identical to the farA mutant and sequencing the farA gene in these mutants indeed showed mutations in FarA which resulted in inability to growth on ferulic acid as well as on short and long chain fatty acids. The growth phenotype of the other three mutants was similar to the farA mutants in terms of the inability to grow on ferulic acid, but these mutants grew normally on short and long chain fatty acids. The genomes of these three mutants were sequenced and allelic mutations in one particular gene (NRRL3_09145) were found. The protein encoded by NRRL3_09145 shows similarity to the FarA and FarB transcription factors. However, whereas FarA and FarB contain both the Zn(II)2Cys6 domain and a fungal-specific transcription factor domain, the protein encoded by NRRL3_09145 (FarD) lacks the canonical Zn(II)2Cys6 domain and possesses only the fungal specific transcription factor domain.
Introduction
Plant biomass represents an exploitable renewable carbon source from which to generate both fuels and chemicals. However, its robust nature generally prevents it from being utilized directly (Oliveira et al., 2015; Fatma et al., 2018). Extensive pretreatment of raw plant biomass is a general requirement for its exploitation, driving up the costs of the chemical exploitation process (Mosier et al., 2005; Carroll and Somerville, 2009; Sun et al., 2022). An alternative approach is to use enzymes to facilitate exploitation of raw biomass. Fungi are especially well equipped to deal with the robustness of plant biomass, being able to efficiently degrade plant cell walls through the secretion of cell wall degrading enzymes. Using complex regulatory responses, fungi are able to selectively secrete enzymes to utilize the nutrients available in their environment (Glass et al., 2013; Guerriero et al., 2015; Benocci et al., 2017; Niu et al., 2017). This property makes fungi ideal organisms for industrial exploitation of plant biomass. However, to optimize the exploitation process, the complex regulatory network regulating the expression of enzymes involved in the degradation process has to be understood thoroughly (see for reviews Huberman et al., 2016; Benocci et al., 2017; Alazi and Ram, 2018; Wu et al., 2020).
An important factor in acquiring robustness of plant biomass against fungal enzymatic degradation is the presence of lignin and other aromatic compounds including tannins and hydroxycinnamic acids such as ferulic acid. The presence of these aromatic compounds and the formation of covalent lignin-hydroxycinnamate-hemicellulose complexes together with the toxicity of these compounds prevent easy degradation by hemicellulolytic enzymes (Tabka et al., 2006; Selig et al., 2008; Wong et al., 2013). In this study, we have focused on the regulation of enzymes involved in the removal of ferulic acid from the plant cell wall and the subsequent metabolism of ferulic acid. Plants use ferulic acid to establish ester linkages between, for instance, arabinoxylans and pectins (Saulnier and Thibault, 1999; Mäkelä et al., 2015; Hatfield et al., 2017). Fungal degradation of ferulic acid from plant cell wall biomass is facilitated by ferulyol esterases. Several ferulyol esterases (FaeA-D) have been identified in A. niger, with expression of the genes being induced only under specific conditions. FaeA expression is induced on xylose, arabinose and several aromatics, whereas FaeB expression is only induced on aromatics (de Vries et al., 2002; Reijngoud et al., 2021). FaeC is generally lowly expressed (Dilokpimol et al., 2017; Oliveira et al., 2019; Lubbers et al., 2020) and no data are currently available for FaeD. The feruloyl esterases of A. niger display different substrate preferences and varying abilities to release ferulic acid or di-ferulic acids from plant biomass. FaeA is most active towards wheat arabinoxylan, whereas FaeB shows higher activity towards sugar beet pectin over arabinoxylan (de Vries et al., 2002). A more recent study indicated that FaeC shares the preference of FaeA to hydrolyse ferulic acid from xylan over pectin (Dilokpimol et al., 2017).
After the release of ferulic acid from the plant cell wall biomass, for further metabolism ferulic acid has to enter the cell via ferulic acid transporters. Moreover, it has recently been established that metabolism of ferulic acid involves genes from the peroxisomal CoA-dependent β-oxidative pathway (Lubbers et al., 2021a). Although in fungi the CoA-dependent β-oxidative pathway was mainly known for the degradation of fatty acids, plants have been shown previously to utilize the pathway for hydroxycinnamic acid degradation (Widhalm and Dudareva, 2015). Candidate genes involved in ferulic acid metabolism were identified in a comparative transcriptome analysis in combination with in silico functional predictions of strongly induced genes. These genes were subsequently deleted and the ability of the mutants to grow on different aromatics was analysed (Lubbers et al., 2021a). This led to the identification of HcsA (NRRL3_05989; putative hydroxycinnamate-CoA synthase), FoxA (NRRL3_00672; putative cinnamoyl-CoA hydratase/dehydrogenase) and KatA (NRRL3_05990; putative 3-ketoacyl CoA thiolase) that are required for growth on ferulic acid. The final step of the CoA-dependent oxidative pathway is proposed to be catalyzed by a thioesterase, with four thioesterases (TheA-D; NRRL3_00621, NRRL3_01539, NRRL3_02735 and NRRL3_06009) identified in the fungal genome. Individual disruption of these genes did not result in reduced growth on ferulic acid, suggesting that there is functional redundancy in this step of the pathway in A. niger (Lubbers et al., 2021a) (Figure 1). Subsequent genes involved in vanillic acid metabolism have been recently identified as well (Lubbers et al., 2021b).
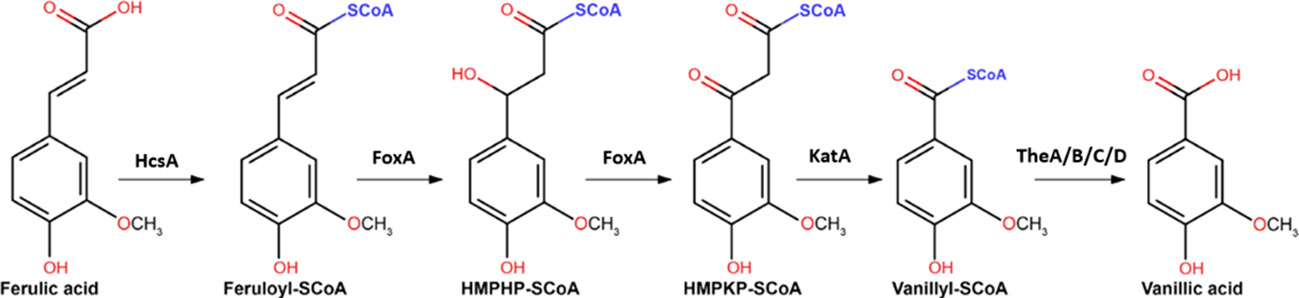
Figure 1 The proposed CoA-dependent β-oxidative pathway for the metabolism of ferulic acid to vanillic acid. Genes encoding the proposed enzymes include HcsA (NRRL3_05989; putative hydroxycinnamate-CoA synthase), FoxA (NRRL3_00672; putative cinnamoyl-CoA hydratase/dehydrogenase) and KatA (NRRL3_05990; putative 3-ketoacyl CoA thiolase), and TheA-TheD (NRRL3_00621, NRRL3_01539, NRRL3_02735 and NRRL3_06009) are indicated (Figure adapted from Lubbers et al., 2021a).
Both the genes encoding the enzymes involved in ferulic acid metabolism and the gene encoding feruloyl esterase B (FaeB) are induced in the presence of ferulic acid and other hydroxycinnamic acids (de Vries et al., 2002; Reijngoud et al., 2021; Lubbers et al., 2021a), suggesting that they might be controlled by the same transcription factor(s). We previously performed a screen to isolate A. niger mutants with constitutive expression of the faeB gene by using PfaeB-amdS and PfaeB-luciferase reporter strains (Reijngoud et al., 2021). More than 100 of these mutants were isolated and all were found to be mutated in the creA gene encoding the major player in carbon catabolite repression. The results from that study showed that loss of function of CreA resulted in constitutive expression of faeB. Unfortunately, this study did not yield other mutants in which a gain of function was introduced in the proposed transcriptional activator involved in the induction of faeB. Using the same screening principle but using other reporter strains, constitutive mutants in transcription factor involved in the induction of pectinases (GaaR), arabinases (AraR) and xylanases (XlnR) have been identified, showing the potential of this approach (Alazi et al., 2019; Reijngoud et al., 2019, Reijngoud and Ram, unpublished).
In this study, the search for transcription factors involved in faeB induction and ferulic acid metabolism was continued by using the same PfaeB-amdS and PfaeB-luciferase reporter strains (Reijngoud et al., 2021) but now using a new amdS counter selection strategy, and by constructing and screening a large collection of A. niger transcription factor deletion mutants. In the genome of A. niger approximately 660 predicted transcription factors, including approximately 440 transcription factors with a Zn(II)2Cys6 domain, are present. From the 660 genes encoding predicted transcription factors, 280 were selected for targeted deletion. These complementary approaches resulted in the identification of the FarA Zn(II)2Cys6 transcription factor as a transcription factor required not only for the metabolism of fatty acids, but also for the metabolism of hydroxycinnamic acids, including ferulic acid. The forward genetic screen identified, in addition to farA, a gene encoding a very peculiar protein (FarD) which is a new member of the Far-like transcription factor family. In contrast to FarA and FarB, FarD lacks the canonical Zn(II)2Cys6 domain.
Material and methods
Strains, media and growth conditions
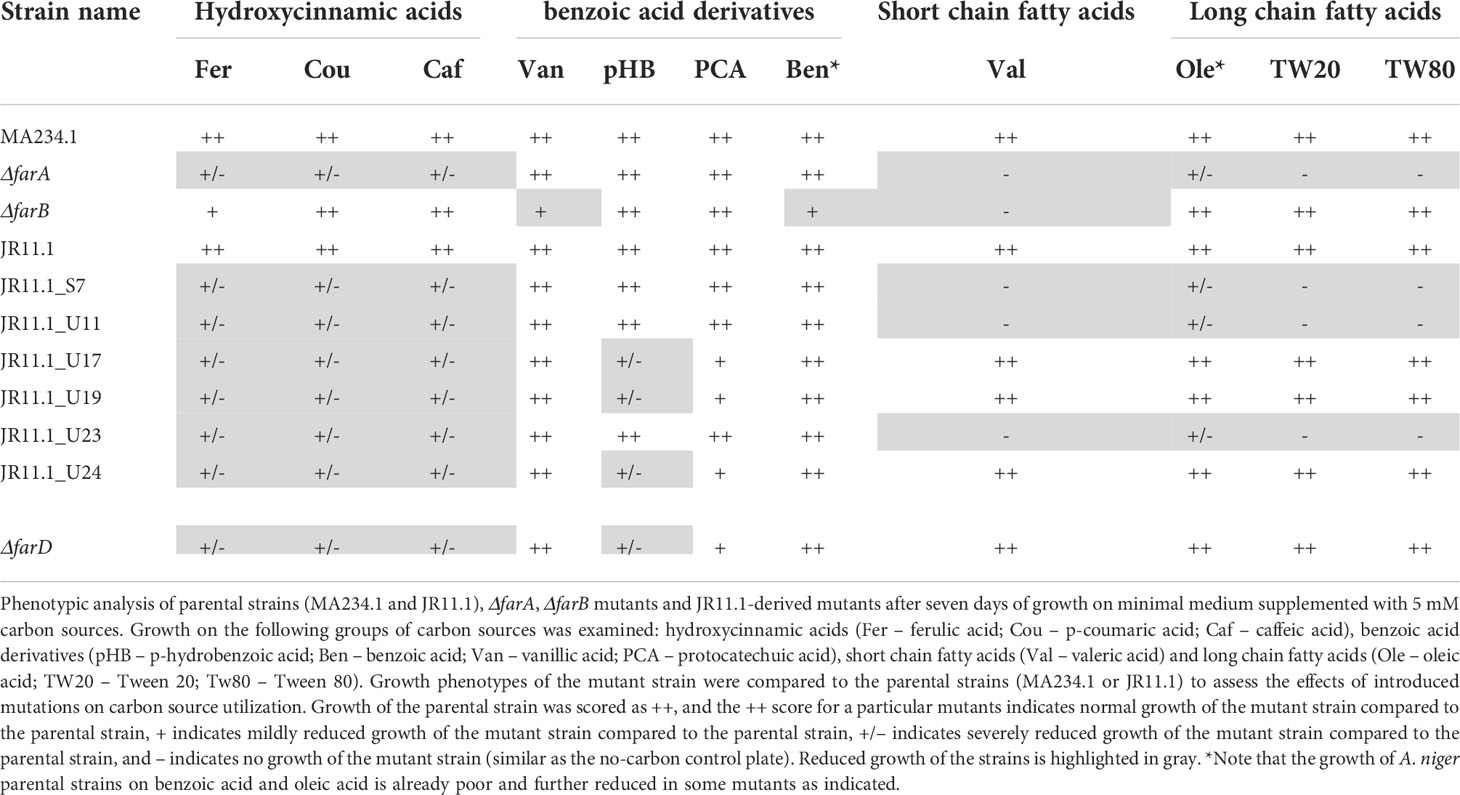
The A. niger strains used in this study are listed in Table 1. A. niger strain JR11.1 contains two reporter genes (PfaeB-amdS and PfaeB-lux613) to monitor the activity of the faeB promoter via the ability to grow on acetamide plates or via luciferase assay (Reijngoud et al., 2021). Strains were grown on liquid or solidified (containing 1.5% (w/v) Scharlau agar (Scharlau, Barcelona, Spain)) minimal medium (MM) or on complete medium (CM) MM contains 7 mM KCl, 8 mM KH2PO4, 70 mM NaNO3, and 2 mM MgSO4 (pH adjusted to 5.5) and spore elements as described (Arentshorst et al., 2012). MM was supplemented with 50 mM carbon source. Complete medium (CM) was also used and consists of MM supplemented with 0.1% casamino acids and 0.5% w/v yeast extract and 50 mM glucose. Transformants of A. niger were isolated and purified as described (Arentshorst et al., 2012) using a final concentration of 100 μg/mL hygromycin (In vivoGen, Toulouse France) or 100 μg/mL phleomycin (In vivoGen, Toulouse France). Mycelial growth assays were performed on MM plate, supplemented with 5 mM of the aromatic compounds or with 10 mM fatty acids. Aromatic compounds and fatty acids compounds were weighed, dissolved in sterile water, and mixed with an equal volume of 2 x concentrated MM-agar. Growth of strains was assayed by point inoculation of 5 μL filtered conidium suspension (1 x 106 conidia/mL) in the center of a MM medium containing plate and incubated for 5 to 10 days in the dark at 30°C. Growth characteristics of the strain was analyzed by comparison of the growth of the colony (radial extension, thickness of the mycelium, and the ability to form conidia) over time. Growth assays of different strains on the same substrate were always performed in the same growth experiments for proper comparisons. Growth experiments of the strains were repeated on the different substrate at least twice with consistent results. Growth phenotypes of the mutant strains were compared to the parental strains (MA234.1 or JR11.1) to assess the effects of introduced mutations on carbon source utilization. Growth of the parental strain was scored as ++, and the ++ score for a particular mutants indicates normal growth of the mutant strain compared to the parental strain, + indicates mildly reduced growth of the mutant strain compared to the parental strain, +/– indicates severely reduced growth of the mutant strain compared to the parental strain, and – indicates no growth of the mutant strain (similar as the no-carbon control plate). Escherichia coli DH5α was used for plasmid propagation and cultured at 37°C in Lysogeny broth (LB) medium, with ampicillin (100 μg/mL).
Molecular techniques
PCR amplifications were performed using either Phire (diagnostic PCR and bipartite flank amplification) or Phusion High-Fidelity DNA Polymerase (whole gene amplification) (Thermo Fisher Scientific, Carlsbad, CA, USA) according to the instructions provided. All primers are listed in Supplementary Table 1. DNA fragments were purified using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific, Carlsbad, CA, USA) and ligations were carried out using the CloneJET PCR Cloning Kit and the Rapid DNA Ligation Kit (Thermo Fisher Scientific, Carlsbad, CA, USA). DNA sequencing of specific genes was performed by Macrogen (The Netherlands).
Generation of transcription factor mutant library in A. niger
The general strategy for constructing and verifying the TF deletion strains is shown in Supplementary Figure 1. Primers were designed by Biomax, based on the A. niger CBS513.88 genome sequence and available upon request. Deletion constructs with the hygromycin selection marker were generated by PCR and assembled by either Gibson assembly (Gibson assembly cloning Kit, New England Biolabs) or fusion PCR. Gene deletion constructs were transformed into A. niger strain MA234.1 (Alazi et al., 2016). When available, two transformants were purified by streaking conidia on MM-hygromycin plates. Gene deletion mutants that gave viable colonies were purified once more on MM-hygromycin plates. For each strain, conidia were isolated from a twice purified clone and used to inoculated cultures to obtain mycelium for genomic DNA isolation and diagnostic PCR. Verification of correct integration was performed using primers as schematically depicted in Supplementary Figure 1. Correct integration of the bipartite fragments should lead to the amplification of a PCR fragment as the outside gene specific primer is not present on the bipartite fragment. Initially, 284 transcription factors were selected for gene disruption based on expression data and genomic clustering of transcription factors with potential target genes. In total 239 gene deletion mutants were identified in which at least one transformant was PCR positive for both flanks. Conidia of these transformants were counted and the concentration of conidia was adjusted to 1 x 106 conidia/ml and two glycerol stocks were made for each strain and stored at -80°C. Conidia from the glycerol stock were directly spotted on ferulic acid plates and poor growing strains were retested for growth on ferulic acid. The primers to delete the farA, farB and farD genes via the same method are listed in Supplementary Table 1.
UV mutagenesis
UV mutagenesis experiments were performed as described previously (Damveld et al., 2008) using JR11.1 (PfaeB-amdS, PfaeB-lux613) as the starting strain (Reijngoud et al., 2021). After determining the killing rate of the UV treatment, conidia with 60-70% survival were plated out on fluoro-acetamide plates with 0.05% ferulic acid. On each plate about 5*105 conidia (50 µL of 1*107 conidia/mL suspension) were plated. In total, around 30 plates for JR11.1 were prepared and incubated for 7 days at 30°C. Two plates were included on which non-mutagenized conidia were plated out. Mutants growing and sporulating on the fluoro-acetamide plates were purified twice. In total, six mutants resistant to fluoro-acetamide were isolated and analyzed in more detail for growth on different carbon sources (sugars, aromatics and fatty acids) and in luciferase assays.
Luciferase assays
Luciferase (lux) assays were performed as follows: 176 μL of MM (containing 1% (w/v) fructose and 0.003% (w/v) yeast extract), 4 μL 25 mM luciferin (Promega, E1605, Leiden, The Nethrlands) and 20 μL conidium suspension (1*106 conidia/mL) were pipetted together (in triplicate) into a well of a white, clear bottom, 96- well plate (Greiner Bio-One, ref. 655095, Alphen aan den Rijn, The Netherlands) and incubated for 24 h at 30°C in the Spark 10M Multimode Microplate reader (Tecan) while measuring luciferase activity and OD600 every 15 min. MM containing 0.1% (w/v) ferulic acid was prepared by dissolving 0.25 g ferulic acid in 250 mL MM, followed by filter sterilization. MM with lower concentrations of ferulic acid was prepared by diluting MM containing 0.1% ferulic acid with appropriate volumes of MM.
Sequencing of farA
To sequence the farA gene in JR11.1 derived mutants, the farA gene was amplified using Phire polymerase (high fidelity DNA polymerase, Thermo Fisher Scientific, Carlsbad, CA, USA) from genomic DNA of the respective mutant using primers FarAP1 and FarAP18 (Supplementary Table 1). The amplified 5.6- kb farA gene including ~1-kb flanking regions was sequenced using primers listed in Supplementary Table 1. Sequence reads were aligned to the farA gene of NRRL3 retrieved from gb.fungalgenomics.ca/portal/and analyzed in DNAman.
Genome sequencing of JR11.1 and JR11.1_U17, JR11.1_U19, and JR11.1_U24
Genomic DNA of A. niger strains was isolated as described (Arentshorst et al., 2012). The genomic DNA was further column-purified (NucleoSpin Plant II, Macherey-Nagel) for whole genome sequencing. The genomes of were sequenced at the McGill University Quebec Innovation Centre (Montreal, Canada) using the Illumina HiSeqX platform to about 125-fold coverage. The raw DNA reads, as received from the sequencing centre, were mapped to the Aspergillus niger NRRL3 v1 assembly with bowtie2 (Langmead and Salzberg, 2012). Variants were called with pilon (Walker et al., 2014) version 1.22 in –variants mode. The VCF files produced by pilon were filtered with vcffilter (Garrison et al., 2021) to keep only variants at locations covered by 6 or more reads, with no more than two genotypes present, and with more than 80% of the covering reads bearing the variant genotype. Variants at the same locations in the filtered files from all samples were tabulated with a custom Python script. Variants inside the coding sequence of a predicted gene and their effect on the predicted protein sequence were identified with another Python script. The Sequence Read Archive accession numbers for the JR11.1 parental strain and derived mutants JR11.1_U17, JR11.1_U19, and JR11.1_U24 are SRR18932555, SRR18932554, SRR18932553, and SRR18932552, respectively.
Complementation analysis of farA and farD mutants
Primers for PCR were designed to amplify complete expression cassettes for farA and farD open reading frames, including a ~1000 base pair promoter region and a ~500 base pair terminator region. PCR fragments were ligated into pJet1.2 (Thermo Fisher Scientific, Carlsbad, CA, USA) and sequenced. pJet1.2 plasmids containing the farA and farD genes were co-transformed to JR11.1_U11 (farA) and JR11.1_U17 (farD) with pAN8.1 (Punt and van den Hondel, 1992) which allows selection on phleomycin. Transformants were purified on solidified MM-phleomycin plates (100 μg/mL phleomycin) and single colonies were propagated on CM to harvest conidia. Conidia were spotted on MM-ferulic acid plates as described above. FarA and FarD primers (Supplementary Table 1) were used to amplify respective genes and PCR fragments were directly sequenced to confirm the presence of both the wild type and mutant allele in the respective complemented strains.
Phylogenetic analysis
Protein sequences from the A. niger NRRL3 strain encoding FarA (NRRL3_00665), FarB NRRL3_06630 and FarD (NRRL3_1945) were initially retrieved from https://gb.fungalgenomics.ca/portal/. The protein sequence from the A. flavus FarC protein (Aspfl2_3|2227896) was retrieved from https://mycocosm.jgi.doe.gov/mycocosm/home. Proteins sequences were used as queries for BLASTP searches at https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins. Hits were manually analyzed and ordered in their respective Far-family. For the 12 fungal species included in the phylogenetic tree, protein sequences were retrieved from https://mycocosm.jgi.doe.gov/mycocosm/home. Retrieved protein sequences were aligned in Clustal omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Upon the suspicion of misalignment due to structural annotation issues (false prediction of start/stop codon or intron/exon bounderies), genomic sequences were retrieved from the JGI website (https://mycocosm.jgi.doe.gov/mycocosm/home) and manually inspected and altered to improve the genemodel and thereby improving aligment of the protein sequences. The final phylogenic tree included in this study is based on those improved gene models.
Results
The FarA and FarB Zn(II)2Cys6transcription factors are involved in ferulic acid metabolism in A. niger
To identify transcription factors (TFs) required for growth on ferulic acid, a collection of 239 TF mutants was tested for growth on 0.2% (10.3 mM) ferulic acid. The genes encoding the TFs that were deleted are listed in Supplementary Table S2. The screen yielded two TF mutants with a reduced growth phenotype on ferulic acid, namely a deletion mutant in which the gene encoding the AcuK transcription factor was deleted (ΔacuK), and a deletion mutant in which the gene encoding the FarA transcription factor was deleted (ΔfarA). AcuK is required for the expression of genes related to gluconeogenesis (Suzuki et al., 2012). Because aromatics like ferulic acid are likely to be metabolized via TCA cycle intermediates, gluconeogenesis is key in sustaining growth using these compounds. The strongly reduced growth of the ΔacuK mutant on ferulic acid is therefore most likely not directly related to ferulic acid metabolism itself and the ΔacuK mutant was not studied further. The FarA transcription factor has an established function in fatty acid metabolism in A. nidulans (Hynes et al., 2006) and since ferulic acid metabolism was recently found to be dependent on the peroxisomal CoA-dependent β-oxidative genes (Lubbers et al., 2021a) the possible link of farA to ferulic acid metabolism was further investigated.
A second fatty acid regulator present in aspergilli is the FarB transcription factor. In A. nidulans, FarB has been shown to be required for the metabolism of short chain fatty acids (Hynes et al., 2006). Since a correct farB deletion mutant was not present in the original collection, a ΔfarB deletion mutant was made. The correct deletion of farA and farB in the respective mutants was confirmed by diagnostic PCRs (Supplementary Figures 2 and 3). The growth analysis on ferulic acid showed that deletion of farA strongly reduced growth on ferulic acid, while deletion of farB showed a partially reduced growth and reduced sporulation phenotype on ferulic acid (Figure 2). A more detailed growth analysis of the ΔfarA and ΔfarB mutants in addition to other mutants and other substrates will be presented below. A third Far-like transcription factor (FarC) has been identified in a number of Aspergilli, including Aspergillus flavus (Luo et al., 2016) but a FarC homolog is not present in the genome of A. niger (see below).
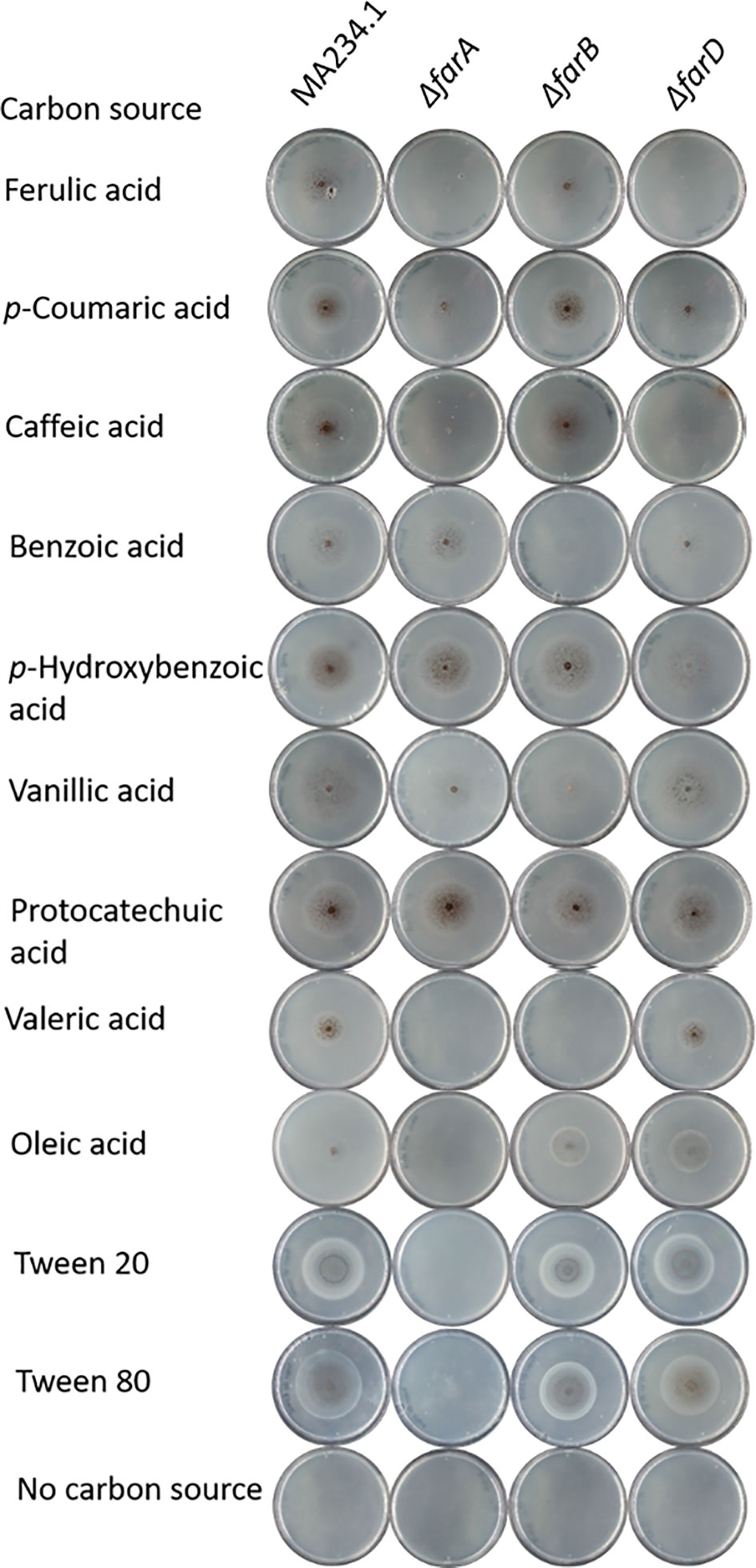
Figure 2 Phenotypic analysis of ΔfarA, ΔfarB and ΔfarD mutants. Mutant were grown for seven days on minimal medium with 5mM hydroxycinnamic acids, 5 mM benzoic acid derivatives, 1 mM valeric acid and 5 mM long chain fatty acids. Strain phenotypes were compared to the MA234.1 strains to assess the effects of introduced mutations on carbon source utilization.
Isolation of trans-acting mutants unable to induce faeB expression under inducing conditions
To identify additional regulatory mutants unable to induce the ferulic acid esterase B gene (faeB) in response to the presence of ferulic acid, a positive forward genetic screen was set up. The screen is based on the use of a PfaeB-amdS, PfaeB-lux613 dual reporter strain (Reijngoud et al., 2021) to isolate mutants unable to induce the faeB gene. The rationale of the screen is that ferulic acid will induce expression of the PfaeB-amdS reporter and that expression of the amdS gene will result in lethality on plates containing fluoro-acetamide. Mutants unable to induce the amdS gene from the PfaeB promoter will be resistant to fluoro-acetamide. The principle of the screen was assessed by analyzing the growth of PfaeB-amdS-containing reporter strains JR10.2 and JR11.1 (Reijngoud et al., 2021) on fluoro-acetamide plates in the absence and presence of ferulic acid. Induction of the PfaeB-amdS reporter in strain JR11.1 and to a lesser extent in JR10.2 by ferulic acid resulted in reduced growth on fluoro-acetamide plates (Figure 3). The growth reduction of reporter strain JR10.2 on fluoro-acetamide was less severe than of JR11.1 and is probably due to the presence of multiple copies of the pfaeB-amdS reporter in JR11.1. Because of the more reduced growth of JR11.1 on fluoro-acetamide plates containing ferulic acid, this strain was used for the isolation of mutants. To obtain mutants JR11.1 conidia were UV-mutagenized and plated out on MM medium plates containing fructose and nitrate as carbon and nitrogen sources respectively and 0.2% fluoro-acetamide and 0.05% ferulic acid. In addition, conidia that were not mutagenized were also plated out to isolate spontaneous mutants. From the initial screening plates 29 colonies were purified twice on MM plates containing fluoro-acetamide and ferulic acid, which resulted in the isolation of six mutants that were resistant to fluoro-acetamide in the presence of ferulic acid. One spontaneous mutant of JR11.1 was obtained (JR11.1_S7) and five mutants were obtained after UV mutagenesis (JR11.1_U11, JR11.1_U17, JR11.1_U19, JR11.1_U23 and JR11.1_U24. The ability to grow on fluoro-acetamide plates in the presence of 0.05% ferulic acid of these six JR11.1-derived UV mutants is shown in Figure 3.
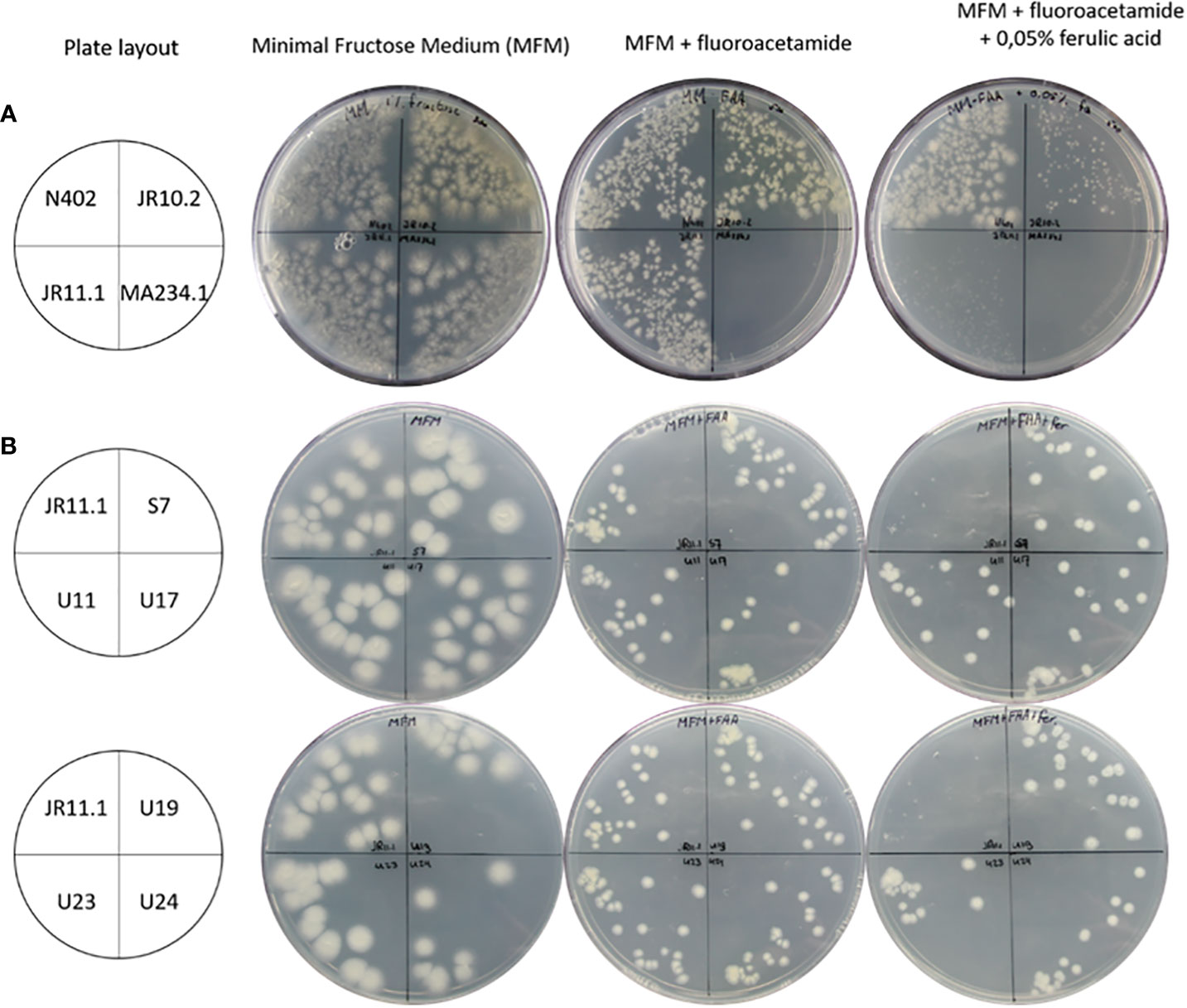
Figure 3 Growth analysis of reporter strains and reporter strain derived mutants. (A) Analysis of strains JR10.2 and JR11.1 for susceptibility to 5-fluoro-acetamide in the presence of the ferulic acid inducer. Growth of the reporter strains was compared to N402 and MA234.1 (amdS gene expressed from the constitutive gpdA promoter) on minimal fructose medium (MFM), MFM supplemented with 0.2% 5-fluoro-acetamide and MFM supplemented with 0.2% 5-fluoro-acetamide and 0.05% (2.6 mM) ferulic acid. Roughly 500 conidia of each strain were spread on the plates in the indicated quadrants. (B) Screen for UV mutants in the JR11.1 background that are resistant to fluoro-acetamide in the presence of the ferulic acid inducer. Growth of six UV mutants was compared to the JR11.1 parental strain on minimal fructose medium (MFM), MFM supplemented with 0.2% fluoro-acetamide and MFM supplemented with 0.2% fluoro-acetamide and 0.05% (2.6 mM) ferulic acid. Roughly 20 conidia of each strain were spread on the plates in the indicated quadrants.
The JR11.1 strain also carries the PfaeB-luciferase (PfaeB-lux613) reporter construct. To determine if the mutations that allow these six mutants in the JR11.1 background to grow on fluoro-acetamide were trans-acting, luciferase assays under non-inducing and inducing conditions were performed. Expression of the pfaeB-lux613 reporter is low under non-inducing conditions and was not induced by 5 mM ferulic acid in the six mutants while ferulic acid strongly induces luciferase activity in the JR11.1 reporter strain (Figure 4). This indicates that the mutations introduced in the fluoro-acetamide resistant mutants work in trans; the mutations affect both the faeB promoter upstream of the amdS gene and the faeB promoter upstream of the lux613 gene.
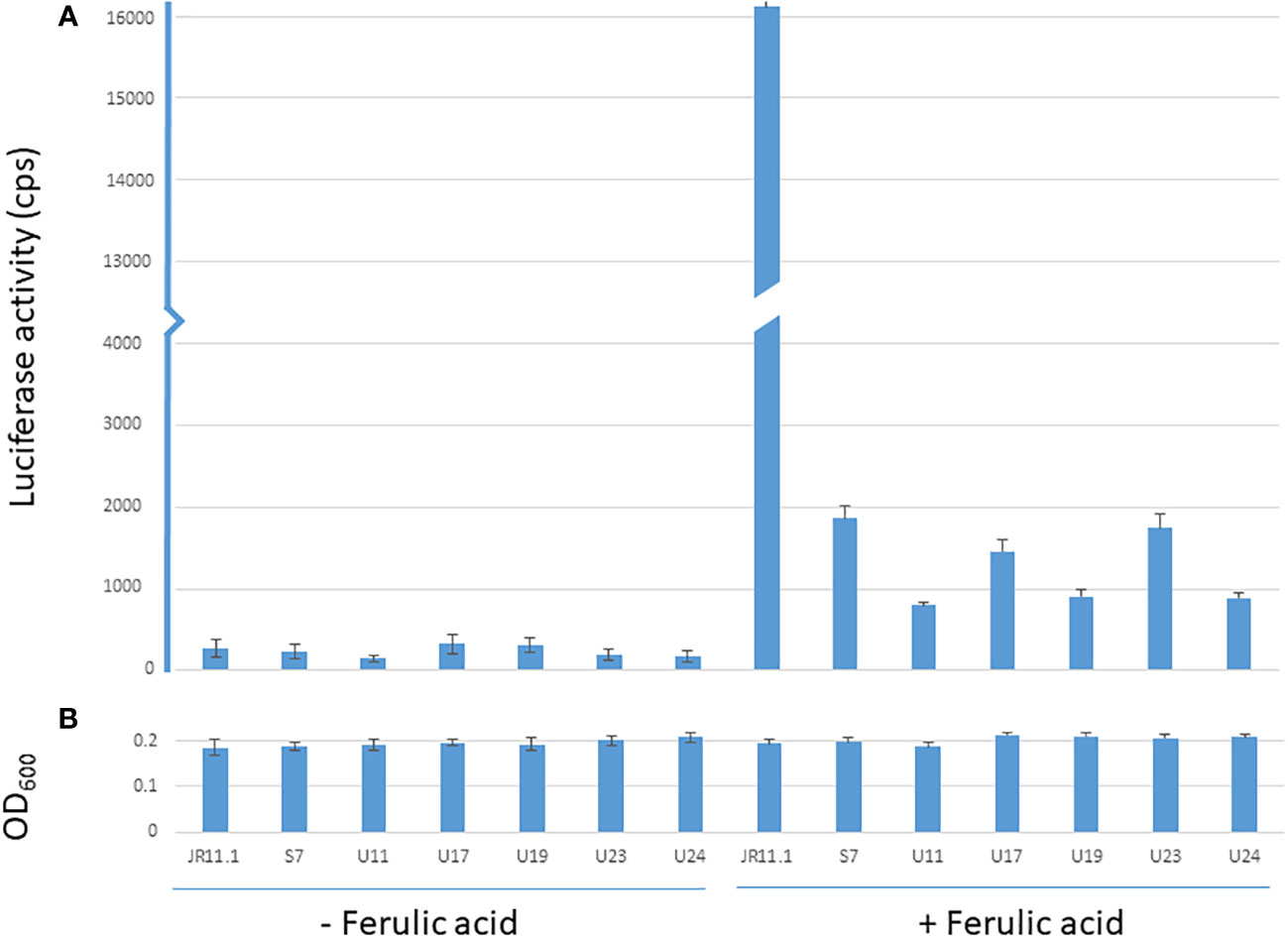
Figure 4 Luciferase and OD600 values of the reporter strain and mutants under non-inducing and inducing conditions. Luminescence values (cps) of reporter strain JR11.1 and mutants derived from JR11.1 (S7, U11, U17, U19, U23 and U24) were compared to parental strain JR11.1. Strains were grown in MM with D-fructose as carbon source without or with 5 mM ferulic acid as inducer. Luciferase activity (A) and OD600 values (B) after 20 hours of growth at 30 °C are shown.
Growth analysis of ΔfarA, ΔfarB and the six trans-acting fluoro-acetamide resistant mutants for growth on aromatics and fatty acid carbon sources
With the confirmation that the six fluoro-acetamide resistant mutants contained mutations functioning in trans, a detailed growth analysis on a variety of carbon sources including hydroxycinnamic acids, benzoic acid-derivatives, and long- and short fatty acids was performed with the ΔfarA and ΔfarB mutants (Figure 1 and Table 2) and the six mutants (Table 2). All six mutants displayed a reduced growth phenotype on ferulic acid, caffeic acid and coumaric acid, similar to the ΔfarA strain. The ΔfarB strain showed a reduced growth phenotype on ferulic acid compared to the parental strain, but the growth defect was less severe than that found for the ΔfarA strain.
The ΔfarA and ΔfarB strains and the six fluoro-acetate resistant mutants were tested for growth on the benzoic acid relatives of hydroxycinnamic acids. The benzoic acid derivatives have the same ring structure as hydroxycinnamic acids, but lack their C3-moiety (prop-2-enoic acid (-C=C-COOH)). Growth on these carbon sources revealed two groups within the six mutants. The first group consists of the ΔfarA mutant and mutants JR11.1_S7, JR11.1_U11 and JR11.1_U23, which were able to grow on all tested benzoic acid derivatives (p-hydrobenzoic acid and benzoic acid, vanillic acid and protocatechuic acid) whereas the second group, consisting of JR11.1_U17, JR11.1_U19 and JR11.1_U24, showed a reduced growth phenotype on p-hydroxybenzoic acid. The ΔfarB strain showed a growth reduction on benzoic acid (Figure 1 and Table 2).
The growth analysis of the different mutants on short- and long chain fatty acids revealed that mutants JR11.1_S7, JR11.1_U11 and JR11.1_U23 showed a reduced growth phenotype on short fatty acids (valeric acid) and long chain fatty acids (oleic acid, Tween 20 and Tween 80) similar to the ΔfarA mutant. Mutants JR11.1_U17, JR11.1_U19 and JR11.1_U24 were able to grow on short- and long chain fatty acids, indicating that these mutants phenotypically differ from ΔfarA mutants. The ΔfarB mutant did not grow on the short fatty acid tested, but grew well on long fatty acids (Figure 1 and Table 2), indicating that mutants JR11.1_U17, JR11.1_U19 and JR11.1_U24 were also different from farB mutants.
Genetic characterization of ferulic acid non-utilizing mutants
Since the growth phenotypes of three of the six fluoro-acetamide mutants was identical to the growth phenotype of the ΔfarA mutant, the farA gene was amplified and sequenced for these three mutants (JR11.1_S7, JR11.1_U11 and JR11.1_U23) as well as for the remaining three mutants (JR11.1_U17, JR11.1_U19 and JR11.1_U24). Sequencing analysis revealed that the three farA-like mutants (JR11.1_S7, JR11.1_U11 and JR11.1_U23) contain mutations in the farA gene, while no mutations were found in the farA gene of the three other mutants (JR11.1_U17, JR11.1_U19 and JR11.1_U24). Mutant JR11.1_S7 has a T to G mutation, which leads to nonsense mutation TAT (Tyrosine) to TAG (stop codon) at nucleotide position 2063 in the farA ORF. The resulting truncated FarA protein has only 443 amino acids of the 912 amino acids remaining. JR11.1_U11 has a T to A mutation leading to a nonsense mutation TTA (Leucine) to TAA (stop codon) at nucleotide position 3102 in the farA ORF, resulting in a truncated protein of 578 amino acids. In mutant JR11.1_U23 a T has been inserted at nucleotide position 1092, resulting in a frameshift and an immediate nonsense mutation CCT AAG (Proline and Lysine) to CCT TAA (Proline and Stop codon) at amino acid position 163. The presence of a nonsense mutation in the farA gene of all three of the mutants and the fact that all three mutants have the same phenotype suggests a loss-of-function mutation of farA, which corresponds with the observed ΔfarA-like phenotype. FarA contains two PFAM domains including a Zn(II)2Cys6 binuclear cluster domain (PF00172.18) comprising amino acids 49-88 and a fungal specific transcription factor domain (PF04082.18) comprising amino acids 242-484. The position of these domains as well as the positions of nonsense mutations in the mutants is shown in Figure 5. The loss-of-function mutation in JR11.1-U11, leading to a C-terminally truncated version of FarA, indicates that the Zn(II)2Cys6 binuclear cluster domain and the fungal specific transcription factor are not sufficient for a functional FarA protein.
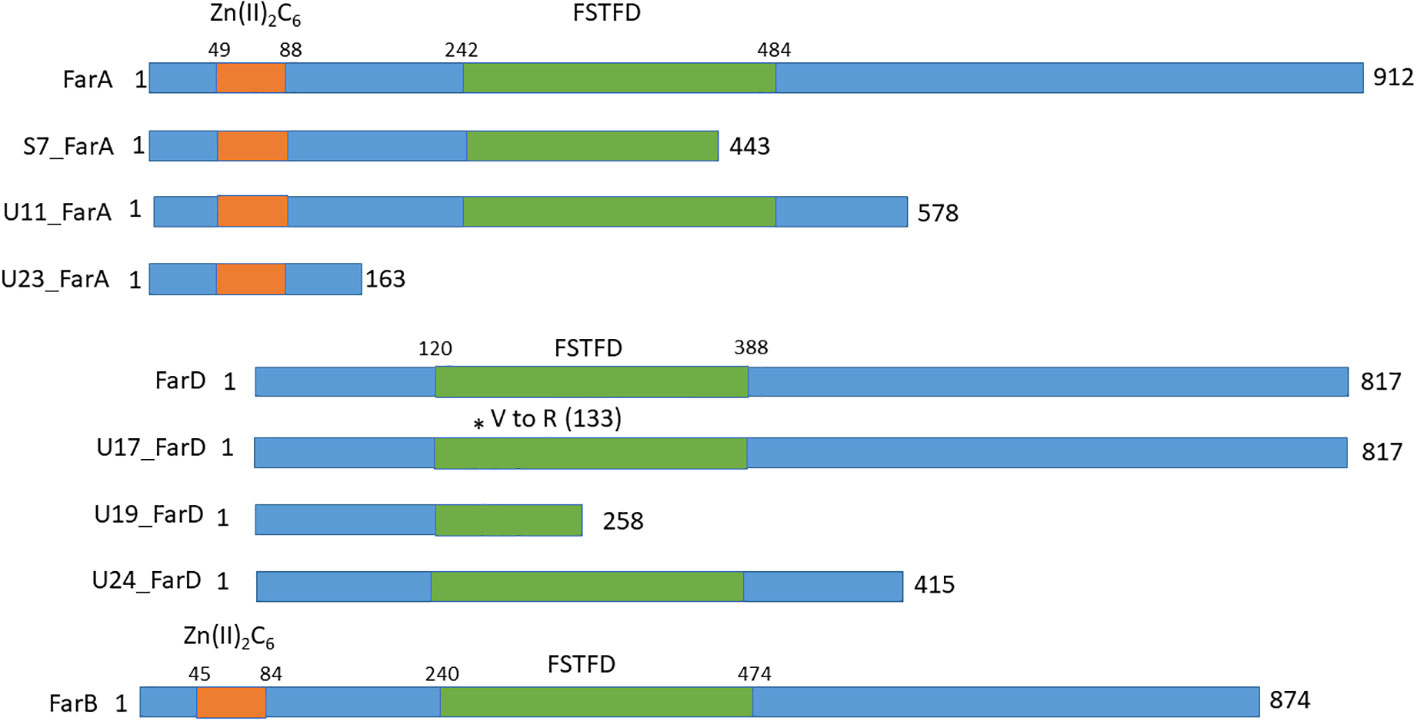
Figure 5 Schematic overview of the domains and the positions of nonsense mutations in FarA, FarD and FarB identified in the fluoro-acetamide resistant mutants. The Zn(II)2Cys6 binding domain in FarA and FarB are indicated by the orange box and the Fungal Specific Transcription Factor Domain (FSTFD) is indicated in green. * indicates the position of the mutation.
To identify the gene(s)s responsible for the reduction of growth on ferulic acid shown for the three remaining mutants (JR11.1_U17, JR11.1_U19 and JR11.1_U24), whole genome sequencing was performed. All three mutants showed only a limited number of SNPs in open reading frames, with JR11.1_U17, JR11.1_U19 and JR11.1_U24 having 6, 9, and 6 SNPs respectively in protein-encoding regions (Supplementary Table 3). All three mutants contain a mutation in the gene NRRL3_09145, which is predicted to encode a protein of 817 amino acids. A PFAM analysis showed that the protein encoded by NRRL3_09145 contains a fungal specific transcription factor domain (PF04082.18) comprising amino acids 120-388. Usually this domain is accompanied by a Zn(II)2Cys6 binuclear cluster domain. However, in the case of NRRL3_09145, this domain was absent. Incorrect annotation of the predicted region was carefully checked by computationally and manually analyzing the DNA sequence and mRNA transcripts of NRRL3_09145. The mRNA reads corresponding to the NRRL3_09145 from several growth conditions showed no evidence of alternative splicing; e.g. an unusually long intron, that could explain the absence of a Zn(II)2Cys6 binuclear cluster domain in the so far annotated gene model (data not shown). The Zn(II)2Cys6 binuclear cluster domain was absent in A. niger and other filamentous fungi that possess a NRRL3_09145 orthologue (see below). Using NRRL3_09145 as a query for a BLASTP search to look for homologous proteins in A. niger revealed that NRRL3_09145 is most similar to the FarB transcription factor and more distantly related to FarA. The FarB protein shows 24% identity over the entire protein sequence to FarD, while FarA and FarD show 16% identity towards each other. The identity between FarB and FarD is mostly found in the fungal specific transcription factor domain (29%) and FarA and FarD show 17% identity in the fungal specific transcription factor domain. Far A and FarB show 16 and 19% sequence identity between the entire protein sequence and the fungal specific transcription factor domain, respectively. Since the name FarC was already given to a FarA/B like protein found in A. flavus and some other Aspergilli, and because NRRL3_09145 is clearly different from the FarC proteins, the protein encoded by the NRRL3_09145 gene is called FarD.
At least two of the mutations found in farD suggest that the observed phenotype of the mutants is due to a loss of function of farD (Figure 5). JR11.1_U17 has a C to T mutation at position 1402 in the farD ORF, leading to a nonsense mutation CAA (Q) to TAG (Stop codon) and resulting in a truncated protein of 451 amino acids. JR11.1_U19 has GT to AG mutations leading to a missense mutation GTG (Valine) to AGG (Arginine) at nucleotide position 396/397 in the farD ORF, resulting in an amino acid change at position 133. JR11.1_U24 has an A to T mutation at position 826 in the farD ORF, leading to a nonsense mutation AAG (Leucine) to TAA (Stop codon) resulting in a truncated protein of 258 amino acids.
To confirm that the observed phenotypes of the two identified groups of UV mutants were indeed caused by loss-of-function mutations in farA or farD, mutants JR11.1_U11 (farA) and JR11.1_U17 (farD) were complemented by transformation with a full length version of farA or farD gene expressed from their endogenous promoter. Complemented strains were able to grow on ferulic acid indicating that a loss-of-function of farA or farD is responsible for the ferulic acid related phenotypes observed in JR11.1_U11 and JR11.1_U17, respectively.
Targeted deletion of farD confirms its role in ferulic acid metabolism
To further confirm the role of farD for growth on ferulic acid as well as on other substrates, the farD gene was deleted in MA234.1 using the hygromycin split marker. Proper deletion of the farD gene was confirmed by diagnostic PCR (Supplementary Figure 4). The phenotypic growth defect of the ΔfarD mutant was compared to the growth phenotype of the ΔfarA and ΔfarB mutants as well as the farD mutants resulting from the UV mutagenesis. As expected, deletion of farD led to the growth phenotype first observed for the farD UV mutants (Figure 1 and Table 2).
Growth phenotypes of the ΔfarA, ΔfarB and ΔfarD strains on various substrates are shown in Figure 1 and summarized in Table 2. From these results it is clear that both FarA and FarD are important for growth on ferulic acid, coumaric acid and caffeic acid. Importantly, while the function of FarA and FarD is required for growth on these hydroxycinnamic acids, FarA and FarD seem to have distinct functions in the metabolism of short and long chain fatty acids. As shown, FarA is required for fatty acid metabolism, while FarD is fully dispensable for growth on valeric acid, oleic acid, Tween20 and Tween80 (Figure 2). FarB is specifically required for growth on the short chain fatty acid valeric acid, but not for growth on long chain fatty acids (Figure 2).
Phylogenetic analysis of the family of Far transcription factors
Several Far transcription factors have been previously described and studied. FarA and FarB are relatively well studied in a few species (mainly A. nidulans, A. oryzae, A. flavus and Fusarium oxysporum) (Hynes et al., 2006, Rocha et al., 2008; Garrido et al., 2012; Luo et al., 2016; Bermúdez-García et al., 2019) In A. flavus, another homolog of FarA and FarB was identified and named FarC (Luo et al., 2016). The gene model of A. flavus FarC was updated compared to the initial publication and predicted to consist of 527 amino acids (Supplemental Data 1). In this study, an additional member of the Far transcription factor family was identified and named FarD.
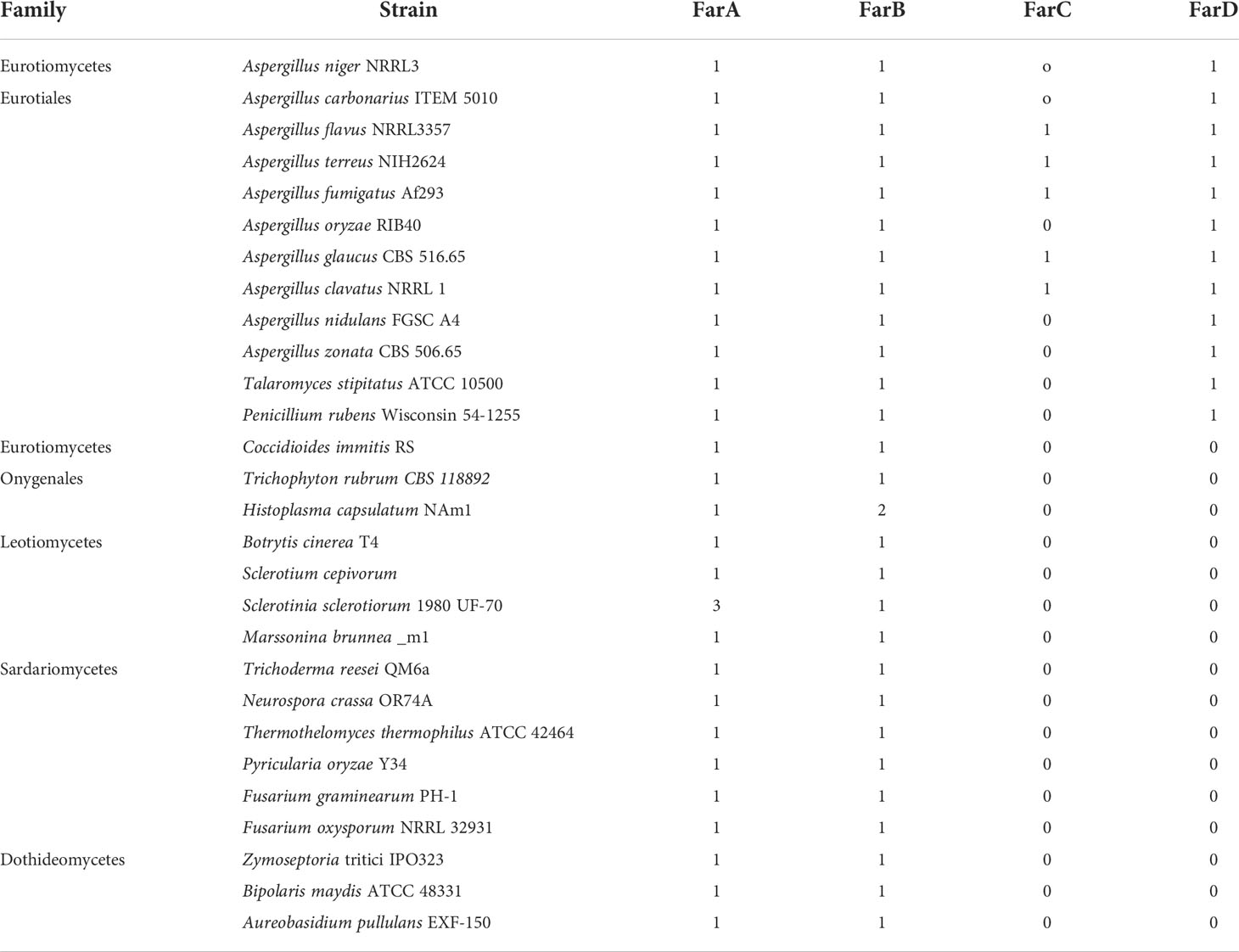
The farD gene of A. niger is predicted to encode a 817 amino acid long protein. Protein BLAST searches using the A. niger FarD protein (NRRL3_09145) as a query revealed that the protein is conserved among fungal species within the order Eurotiales. Fungi belonging to the same class of Eurotiomycetes, but a different order (Onygenales) which include Coccidioides immitis, Trichophyton rubrum, and Histoplasma capsulatum, do not contain a FarD ortholog. More distantly related Pezizomycetes fungi belonging to the classes of Leotiomycetes, Sordariomycetes, and Dothideomycetes also do not have a FarD homolog. In Table 3, we have summarized the presence or absence of FarD homologs in a selection of 28 fungal species representing the Pezizomycotina group. None of the FarD orthologs identified contained a Zn(II)2Cys6 DNA binding domain, but all contained the fungal specific transcription factor domain (PFAM PF04082). The gene model of A. oryzae, the only aberrant model, is predicted to be 163 amino acids longer at the N-terminus of the protein. However, this predicted protein sequence also does not contain a Zn(II)2Cys6 DNA binding domain and is likely a result of a mispredicted gene model.
FarA and FarB orthologs are found in all 28 species analyzed (Table 3). Using the newly predicted gene model of A. flavus FarC as a query, only a few species, all belonging to the order Eurotiales, contained a ortholog of FarC (Table 3). Aspergillus flavus FarC as well as any of the other FarC homologs do not contain a Zn(II)2Cys6 domain but all contain the fungal specific transcription factor domain (PFAM PF04082).
For the 12 species belonging to the Eurotiales that are mentioned in Table 3, a phylogenetic tree was constructed using Clustal Omega for the FarA, FarB, FarC and FarD proteins (when present). To optimize protein alignments, gene models were optimized to include e.g. a Zn(II)2Cys6 domain or to adjust intron/exon bouderies if appropriate. The protein sequences used to construct the phylogenetic tree are listed in Supplementary Data 1. The phylogenetic analysis shows that the Far protein form distinct phylogenetic (Figure 6).
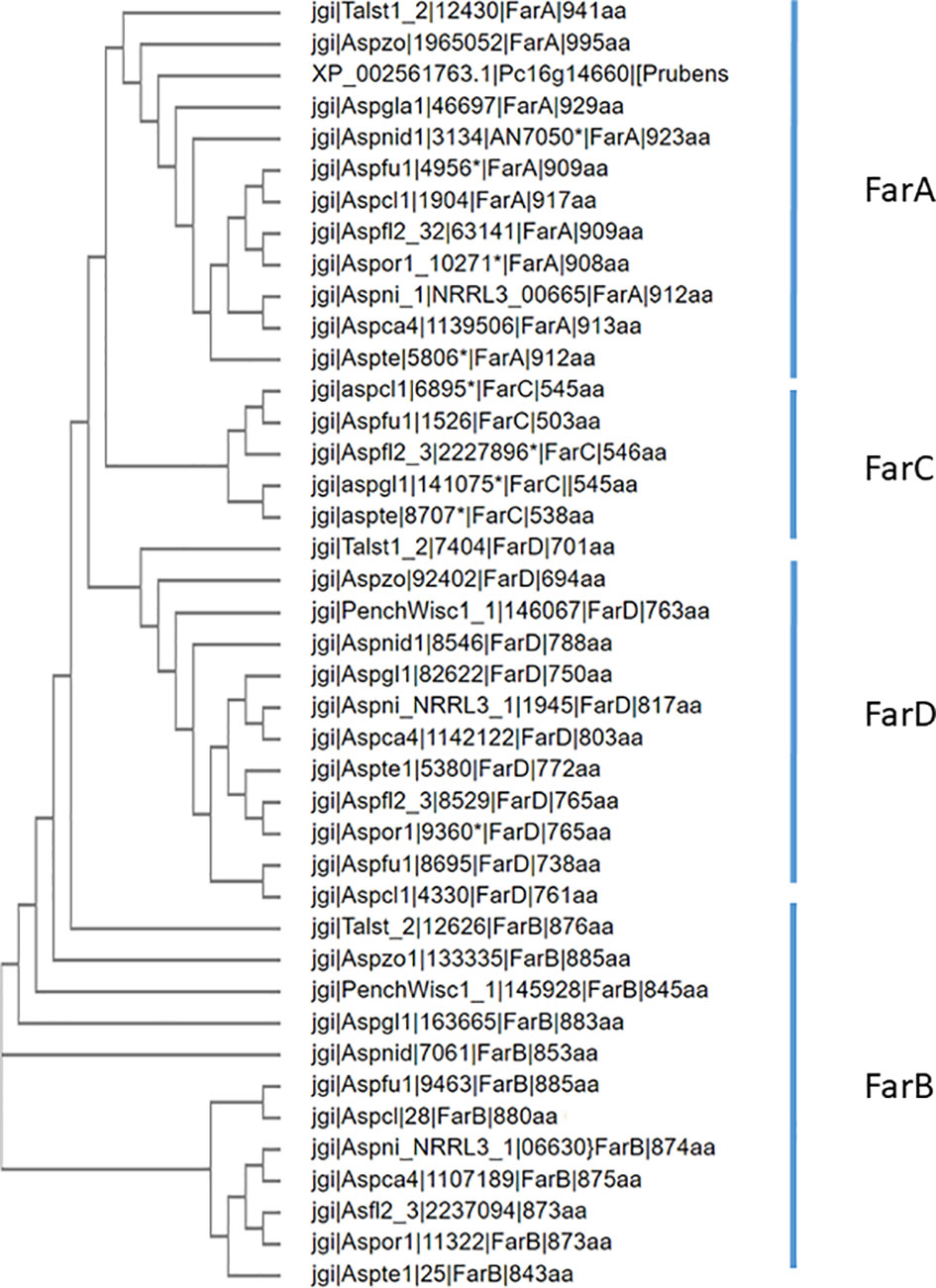
Figure 6 Phylogenetic analysis of Far transcription factors and Far-like transcription factor in the genomes of 12 representative Eurotiales fungal genomes. Using Far proteins from A. niger (FarA, FarB, FarD, NRRL3_11483 and NRRL3_07314 or A. flavus (FarC) as queries, orthologous Far-like transcription factors were identified. Clustal omega was used to construct the phylogenetic tree.
Discussion
The interest in fungal metabolic pathways required for the degradation of plant derived aromatic compounds has increased a lot in the last decade, but detailed knowledge is still limited (see for recent reviews Mäkelä et al., 2015; Lubbers et al., 2019a). One of the reasons for the interest is related to the transition from a fossil-based economy to a bio-based economy using plant cell wall carbohydrates for the production of biofuels and chemicals. Plant cell walls are mostly composed of polysaccharides (cellulose, xylan, arabinanan and pectins) but also contain significant amounts of lignin (Liu et al., 2018). The amount of lignin in the plant cell wall varies among species and the plant tissue and varies between 1 and 24% of lignin of total cell wall residues (Neutelings, 2011) Lignin is generally a polymer composed of three main types of monolignol building blocks (sinapyl alcohol, coniferyl alcohol, and p-coumaryl alcohol) present in plant secondary cell walls (see for review Liu et al., 2018). Although A. niger does not possess the enzyme spectrum to fully degrade lignin, it can grow on and metabolize the aromatics of which lignin consists. Apart from lignin, plant cell walls contain other aromatic compounds such as tannins and hydroxycinnamic acids like ferulic acid which A. niger can metabolize (van Diepeningen et al., 2004; Lubbers et al., 2019b; Lubbers et al., 2021a; Arentshorst et al., 2021). Another reason for the interest in improving knowledge about enzymes involved in the metabolism of aromatic compounds is to make valuable compounds from the left-over lignins or other plant cell associated aromatics (Kamimura et al., 2019; Lubbers et al., 2021b) obtained from biorefinery processes. In the last few years several metabolic pathways for aromatics have been (partially) elucidated in Aspergilli, including salicylic acid (Martins et al., 2015; Lubbers et al., 2021c), protocatechuic acid (Martins et al., 2015; Lubbers and de Vries, 2021), ferulic acid (Lubbers et al., 2021a) and gallic acid (Arentshorst et al., 2021). Ferulic acid represents an important aromatic compound in relation to efficient conversion of plant cell wall biomass into fermentable sugars. The pathway of ferulate metabolism has been recently studied and several genes involved in the peroxisomal CoA-dependent β-oxidative pathway were found to be required for the degradation of hydoxycinnamic acids, including ferulic acid, p-coumaric acid and caffeic acid (Lubbers et al., 2021a). The peroxisomal CoA-dependent β-oxidative pathway is wellknown to be required for the metabolism of fatty acids in fungi (Hiltunen et al., 2003; Hynes et al., 2008) and is also essential for hydroxycinnamic acid catabolism.
The transcriptional control of genes involved in metabolic degradation of aromatics is largely unknown. Only a few transcription factors that are involved in regulation of genes encoding the enzymes involved the degradation of aromatics have been identified. These include the Zn(II)2Cys6 transcription factor SdrA which is required for the metabolism of cinnamic acid and sorbic acid (Plumridge et al., 2010; Lubbers et al., 2019b, Seekles et al., 2022) and the TanR Zn(II)2Cys6 transcription factor required for the degradation of tannin into gallic acid and subsequent metabolism of gallic acid (Arentshorst et al., 2021). Here we show that deletion of FarA, a well described transcription factor required for the induction of genes involved in peroxisomal degradation of fatty acids (Hynes et al., 2006; Hynes et al., 2008), is also required for the metabolism of hydroxycinnamic acids, including ferulic acid, p-coumaric acid and caffeic acid. The degradation of cinnamic acid was less affected in our mutants (results not shown), because its metabolism uses a different pathway controlled by the SdrA transcription factor (Lubbers et al., 2019b). The FarA target genes required for the utilization of ferulic acid, p-coumaric acid and caffeic acid as well as fatty acids are foxA and katA. Based on homology with the FoxA protein of A. nidulans (Maggio-Hall and Keller, 2004), the A. niger FoxA (NRRL3_00672) protein was identified and considered to act as a candidate β-oxidation hydratase/dehydrogenase, and KatA (NRRL3_5990) is predicted to be a 3-keto-acyl CoA thiolase. Deletion of foxA or katA in A. niger results in reduced growth on hydroxycinnamic acids (ferulic acid, p-coumaric acid and caffeic acid) as well as on fatty acids (Lubbers et al., 2021a). In this study, we show that deletion of the FarA transcription factor leads to similar phenotypes indicating that the expression of foxA and katA s is probably controlled by FarA. The presence of putative FarA transcription factor binding sites (5’-CCTCGG) (van den Berg et al., 2010) in the promoters of A. niger foxA (at nucleotide position –225) and katA (at nucleotide position –400) is further in support that FarA is directly controlling their expression.
The first step in the oxidative peroxisomal degradation pathway of ferulic acid is catalyzed by hydroxycinnamate-CoA synthase encoded by hcsA (NRRL3_05989). Deletion of hcsA results in strongly reduced growth on several hydroxycinnamic acids but not on cinnamic acid (see above). However, growth on fatty acids was not affected (Lubbers et al., 2021a). Interestingly, also in the hcsA promoter a putative FarA binding site is present (at nucleotide position -871 before the start codon). Whether FarA is indeed directly binding to the promoter of these genes and required for the induced expression of ferulic acid requires further studies.
Based on our results, the FarB transcription factor in A. niger plays also a role in relation to degradation of aromatics and fatty acids. Its role in ferulic acid degradation seems less important than that of FarA, because growth of the ΔfarB strain is less affected than of the ΔfarA strain. An unexpected finding was the reduced growth of ΔfarB on benzoic acid and vanillic acid. The growth of the ΔfarB strain was as expected reduced on the short chain fatty acid valeric acid but not on long chain fatty acids. As in A. flavus, the FarB transcription factor in A. niger seems to be mostly involved in the regulation of enzymes involved in metabolism of short chain fatty acids (Luo et al., 2016).
In this study, we identified FarD as a new regulatory factor involved in the metabolism of hydroxycinnamic acids. Our results show that the absence of FarD reduces ability to grow on ferulic acid and other hydroxycinnamic acids. The farD gene was identified in a screen for mutants that were no longer able to induce the faeB gene, indicating that FarD, directly or indirectly, controls the induction of faeB in response to hydroxycinnamic acids. The mutant screen also yielded FarA loss-of-function mutants. The surprising finding is that the FarD protein lacks any known DNA binding motif, but contains the fungal specific transcription factor domain (PFAM04082) that is normally found in Zn(II)2Cys6 transcription factors. The fungal specific transcription factor domain is around 240 amino acids long and overlaps with the CDD (conserved domain database) cd12148 known as the fungal transcription factor homology domain (Fungal_TF_MHR). The cd12148 domain is generally around 400 amino acids in length. This domain is thought to play a role in regulating the transcriptional activity of these factors (Schjerling and Holmberg, 1996; MacPherson et al., 2006). To the best of our knowledge this is the first report that describes functional analysis of a gene encoding a protein with a transcription factor homology domain, but lacking the canonical Zn(II)2Cys6 function. The mechanism by which FarD exerts its function is unknown.
Besides a role in the induction of genes required for peroxisomal beta-oxidation, FarA has also been implicated in the induction of expression of genes encoding cutinases in Fusarium solani, F. oxysporum, A. oryzae and A. nidulans (Kämper et al., 1994; Li and Kolattukudy, 1997, Rocha et al., 2008, Garrido et al., 2012; Bermúdez-García et al., 2019). For F. solani it was shown that Ctf1 (FarA homolog) binds directly to the cut1 promoter (Li and Kolattukudy, 1997). So far, a role for FarA (or any other Far protein) in directly inducing FaeB (or other feruroyl esterases) had not been described. The observation that in farA and farD mutants the PfaeB-amdS and the PfaeB-lux reporter gene are not induced in response to ferulic acid suggest that FarA (and/or FarD) are directly involved in the induction. A putative FarA binding site at position -474 upstream of the translational start site supports the possibility that FarA binds directly to its target effector genes.
The amdS selection method has been successfully used in several studies to identify regulatory mutants. Such a screen requires a gene which is specifically induced under a certain growth condition. The promoter of that gene is cloned in front of the amdS reporter gene which allows the selection of mutants that express the amdS gene under non-inducing conditions. Screens have been performed using genes specifically induced by galacturonic acid (exo-polygalacturonase X (pgaX)), arabinose (arabinofuranosidase A (abfA)) or ferulic acid (feruloyl esterase B (faeB)) respectively (Niu et al., 2017; Reijngoud et al., 2019; Reijngoud et al., 2021). In all these studies, the aim was to identify mutants resulting in constitutive expression of the target gene. In this study by using a counter selection strategy of amdS, we aimed to isolate mutants unable to induce faeB expression in order to find regulatory mutants. To screen for those mutants, we used the possibility to counterselect for the expression of amdS using fluoro-acetamide. In an initial experiment, it was confirmed that the induction of the amdS gene via the faeB promoter with ferulic acid resulted in lethality on fluoro-acetamide (Figure 3) which allowed the selection of mutants. To the best of our knowledge it is the first time that the a promoter-amdS reporter is used in a counter selection strategy to identify mutants defective in induction. Our results show that it is a powerful method to obtain and identify both gain- and loss of function mutations in trans-acting factors.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR18932555 https://www.ncbi.nlm.nih.gov/, SRR18932554 https://www.ncbi.nlm.nih.gov/, SRR18932553 https://www.ncbi.nlm.nih.gov/, SRR18932552.
Author contributions
MA: Investigation, conceptualization, validation, data analysis, methodology, visualization. JR: Investigation, conceptualization, data analysis, methodology, data analysis. DT: Investigation, conceptualization, validation, data analysis, methodology, visualization, first draft manuscript writing. IR: DNA sequencing, data analysis, manuscript writing. YA: investigation, data analysis. HP: Conceptualization, data analysis, funding. NP: Conceptualization, data analysis, funding. JV: Conceptualization, data analysis, funding manuscript writing. PP: Conceptualization, data analysis, manuscript writing. AT: Conceptualization, data analysis, manuscript writing, funding. AR: Conceptualization, data analysis, original draft writing, funding acquisition, overall supervision. All authors agree to be accountable for the content of the work.
Funding
This project was partially funded by the Genome Canada and Génome Québec and partially funded the Kluyver Centre for Genomics of Industrial Fermentation.
Conflict of interest
Authors YA, HP, NP were employed by the company DSM Netherlands.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2022.978845/full#supplementary-material
References
Alazi E., Niu J., Kowalczyk J. E., Peng M., Aguilar Pontes M. V., Kan J. A. L., et al. (2016). The transcriptional activator GaaR of Aspergillus niger is required for release and utilization of D-galacturonic acid from pectin. FEBS Lett. 590 (12), 1804–1815. doi: 10.1002/1873-3468.12211
Alazi E., Ram A. F. J. (2018). Modulating transcriptional regulation of plant biomass degrading enzyme networks for rational design of industrial fungal strains. Front. Bioeng. Biotech. 6. doi: 10.3389/fbioe.2018.00133
Alazi E., Niu J., Otto S. B., Arentshorst M., Pham T. T. M., Tsang A., et al. (2019). W361R mutation in GaaR, the regulator of D-galacturonic acid-responsive genes, leads to constitutive production of pectinases in Aspergillus niger. Microbiol. Open 8 (5), e00732. doi: 10.1002/mbo3.732
Arentshorst M., Di Falco M., Moisan M. C., Reid I. D., Spaapen T. O. M., van Dam J., et al. (2021). Identification of a conserved transcriptional activator-repressor module controlling the expression of genes involved in tannic acid degradation and gallic acid utilization in Aspergillus niger. Front. Fungal Biol. 2, 681631. doi: 10.3389/ffunb.2021.681631
Arentshorst M., Ram A. F., Meyer V. (2012). Using non-homologous end-joining-deficient strains for functional gene analyses in filamentous fungi. Methods Mol. Biol. 835, 133–150. doi: 10.1007/978-1-61779-501-5_9
Benocci T., Aguilar-Pontes M. V., Zhou M., Seiboth B., de Vries R. P. (2017). Regulators of plant biomass degradation in ascomycetous fungi. Biotechnol. Biofuels 10, 1–25. doi: 10.1186/s13068-017-0841-x
Bermúdez-García E., Peña-Montes C., Martins I., Pais J., Pereira C. S., Sánchez S., et al. (2019). Regulation of the cutinases expressed by Aspergillus nidulans and evaluation of their role in cutin degradation. Appl. Microbiol. Biotechnol. 103 (9), 3863–3874. doi: 10.1007/s00253-019-09712-3
Bos C. J., Debets A. J., Swart K., Huybers A., Kobus G., Slakhorst S. M. (1988). Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14, 437–443. doi: 10.1007/BF00521266
Carroll A., Somerville C. (2009). Cellulosic biofuels. Annu. Rev. Plant Biol. 60, 165–182. doi: 10.1146/annurev.arplant.043008.092125
Carvalho N. D., Arentshorst M., Jin Kwon M., Meyer V., Ram A. F. (2010). Expanding the ku70 toolbox for filamentous fungi: Establishment of complementation vectors and recipient strains for advanced gene analyses. Appl. Microbiol. Biotechnol. 87 (4), 1463–1473. doi: 10.1007/s00253-010-2588-1
Damveld R. A., Franken A., Arentshorst M., Punt P. J., Klis F. M., van den Hondel C. A., et al. (2008). A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 178 (2), 873–881. doi: 10.1534/genetics.107.073148
de Vries R. P., vanKuyk P. A., Kester H. C., Visser J. (2002). The Aspergillus niger faeB gene encodes a second feruloyl esterase involved in pectin and xylan degradation and is specifically induced in the presence of aromatic compounds. Biochem. J. 363 (Pt 2), 377–386. doi: 10.1042/bj3630377
Dilokpimol A., Mäkelä M. R., Mansouri S., Belova O., Waterstraat M., Bunzel M., et al. (2017). Expanding the feruloyl esterase gene family of Aspergillus niger by characterization of a feruloyl esterase FaeC. Nat. Biotechnol. 37 (Pt B), 200–209. doi: 10.1016/j.nbt.2017.02.007
Fatma S., Hameed A., Noman M., Ahmed T., Shahid M., Tariq M., et al. (2018). Lignocellulosic biomass: A sustainable bioenergy source for the future. Protein Pept. Lett. 25 (2), 148–163. doi: 10.2174/0929866525666180122144504
Garrido S. M., Kitamoto N., Watanabe A., Shintani T., Gomi K. (2012). Functional analysis of FarA transcription factor in the regulation of the genes encoding lipolytic enzymes and hydrophobic surface binding protein for the degradation of biodegradable plastics in Aspergillus oryzae. Biosc. Bioeng. 113 (5), 549–555. doi: 10.1016/j.jbiosc.2011.12.014
Garrison E., Kronenberg Z. N., Dawson E. T., Pedersen B. S., Prins P. (2021). Vcflib and tools for processing the VCF variant call format. BioRxiv. doi: 10.1101/2021.05.21.445151
Glass N. L., Schmoll M., Cate J. H., Coradetti S. (2013). Plant cell wall deconstruction by ascomycete fungi. Annu. Rev. Microbiol. 67, 477–498. doi: 10.1146/annurev-micro-092611-150044
Guerriero G., Hausman J. F., Strauss J., Ertan H., Siddiqui K. S. (2015). Destructuring plant biomass: Focus on fungal and extremophilic cell wall hydrolases. Plant Sci. 234, 180–193. doi: 10.1016/j.plantsci.2015.02.010
Hatfield R. D., Rancour D. M., Marita J. M. (2017). Grass cell walls: A story of cross-linking. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02056
Hiltunen J. K., Mursula A. M., Rottensteiner H., Wierenga R. K., Kastaniotis A. J., Gurvitz A. (2003). The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 27 (1), 35–64. doi: 10.1016/S0168-6445(03)00017-2
Huberman L. B., Liu J., Qin L., Glass N. L. (2016). Regulation of the lignocellulolytic response in filamentous fungi. Fun. Biol. Rev. 30, 101–111. doi: 10.1016/j.fbr.2016.06.001
Hynes M. J., Murray S. L., Duncan A., Khew G. S., Davis M. A. (2006). Regulatory genes controlling fatty acid catabolism and peroxisomal functions in the filamentous fungus Aspergillus nidulans. Eukaryotic Cell 5 (5), 794–805. doi: 10.1128/EC.5.5.794-805.2006
Hynes M. J., Murray S. L., Khew G. S., Davis M. A. (2008). Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics 178 (3), 1355–1369. doi: 10.1534/genetics.107.085795
Kamimura N., Sakamoto S., Mitsuda N., Masai E., Kajita S. (2019). Advances in microbial lignin degradation and its applications. Curr. Opin. Biotechnol. 56, 179–186. doi: 10.1016/j.copbio.2018.11.011
Kämper J. T., Kämper U., Rogers L. M., Kolattukudy P. E. (1994). Identification of regulatory elements in the cutinase promoter from Fusarium solani f. sp. pisi (Nectria haematococca). J. Biol. Chem. 269 (12), 9195–9204.
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9 (4), 357–359. doi: 10.1038/nmeth.1923
Li D., Kolattukudy P. E. (1997). Cloning of cutinase transcription factor 1, a transactivating protein containing Zn2Cys6 binuclear cluster DNA-binding motif. J. Biol. Chem. 272 (19), 12462–12467. doi: 10.1074/jbc.272.19.12462
Liu Q., Luo L., Zheng L. (2018). Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 19 (2), 335. doi: 10.3390/ijms19020335
Lubbers R. J. M., de Vries R. P. (2021). Production of protocatechuic pcid from p-hydroxyphenyl (H) units and related aromatic compounds using an Aspergillus niger cell factory. mBio 29;12 (3), e0039121. doi: 10.1128/mBio.00391-21
Lubbers R. J. M., Dilokpimol A., Navarro J., Peng M., Wang M., Lipzen A., et al. (2019b). Cinnamic acid and sorbic acid conversion are mediated by the same transcriptional regulator in Aspergillus niger. Front. Bioeng. Biotechnol. 7. doi: 10.3389/fbioe.2019.00249
Lubbers R. J. M., Dilokpimol A., Nousiainen P. A., Cioc R. C., Visser J., Bruijnincx P. C. A., et al. (2021b). Vanillic acid and methoxyhydroquinone production from guaiacyl units and related aromatic compounds using Aspergillus niger cell factories. Microb. Cell Fact. 20 (1), 151. doi: 10.1186/s12934-021-01643-x
Lubbers R. J. M., Dilokpimol A., Visser J., de Vries R. P. (2021a). Aspergillus niger uses the peroxisomal CoA-dependent β-oxidative genes to degrade the hydroxycinnamic acids caffeic acid, ferulic acid, and p-coumaric acid. Appl. Microbiol. Biotechn. 105 (10), 4199–4211. doi: 10.1007/s00253-021-11311-0
Lubbers R. J. M., Dilokpimol A., Visser J., Hildén K. S., Mäkelä M. R., de Vries R. P. (2021c). Discovery and functional analysis of a salicylic acid hydroxylase from Aspergillus niger. Appl. Environ. Microbiol. 87 (6), e02701–e02720. doi: 10.1128/AEM.02701-20
Lubbers R. J. M., Dilokpimol A., Visser J., Mäkelä M. R., Hildén K. S., de Vries R. P. (2019a). A comparison between the homocyclic aromatic metabolic pathways from plant-derived compounds by bacteria and fungi. Biotechnol. Adv. 37, 107396. doi: 10.1016/j.biotechadv.2019.05.002
Lubbers R. J. M., Liwanag A. J., Peng M., Dilokpimol A., Benoit-Gelber I., de Vries R. P. (2020). Evolutionary adaptation of Aspergillus niger for increased ferulic acid tolerance. J. Appl. Microbiol. 128, 735–746. doi: 10.1111/jam.14505
Luo X., Affeldt K. J., Keller N. P. (2016). Characterization of the far transcription factor family in Aspergillus flavus. G3 (Bethesda Md.) 6 (10), 3269–3281. doi: 10.1534/g3.116.032466
MacPherson S., Larochelle M., Turcotte B. (2006). A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70 (3), 583–604. doi: 10.1128/MMBR.00015-06
Maggio-Hall L. A., Keller N. P. (2004). Mitochondrial beta-oxidation in Aspergillus nidulans. Mol. Microbiol. 54 (5), 1173–1185. doi: 10.1111/j.1365-2958.2004.04340.x.PID:15554960
Mäkelä M. R., Marinović M., Nousiainen P., Liwanag A. J., Benoit I., Sipilä J., et al. (2015). Aromatic metabolism of filamentous fungi in relation to the presence of aromatic compounds in plant biomass. Adv. Appl. Microbiol. 91, 63–137. doi: 10.1016/bs.aambs.2014.12.001
Martins T. M., Hartmann D. O., Planchon S., Martins I., Renaut J., Silva Pereira C. (2015). The old 3-oxoadipate pathway revisited: New insights in the catabolism of aromatics in the saprophytic fungus Aspergillus nidulans. Fung. Gen. Biol. 74, 32–44. doi: 10.1016/j.fgb.2014.11.002
Mosier N., Wyman C., Dale B., Elander R., Lee Y. Y., Holtzapple M., et al. (2005). Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresource Techn. 96 (6), 673–686. doi: 10.1016/j.biortech.2004.06.025
Neutelings G. (2011). Lignin variability in plant cell walls: Contribution of new models. Plant Sci. 181 (4), 379–386. doi: 10.1016/j.plantsci.2011.06.012
Niu J., Alazi E., Reid I. D., Arentshorst M., Punt P. J., Visser J., et al. (2017). An evolutionarily conserved transcriptional activator-repressor module controls expression of genes for d-galacturonic acid utilization in Aspergillus niger. Genetics 205, 169–183. doi: 10.1534/genetics.116.194050
Oliveira D. M., Finger-Teixeira A., Mota T. R., Salvador V. H., Moreira-Vilar F. C., Molinari H. B., et al. (2015). Ferulic acid: A key component in grass lignocellulose recalcitrance to hydrolysis. Plant Biotechnol. J. 13 (9), 1224–1232. doi: 10.1016/j.ophtha.2013.03.046
Oliveira D. M., Mota T. R., Oliva B., Segato F., Marchiosi R., Ferrarese-Filho O., et al. (2019). Feruloyl esterases: biocatalysts to overcome biomass recalcitrance and for the production of bioactive compounds. Bioresource Technol. 278, 408–423. doi: 10.1016/j.ophtha.2013.03.046
Plumridge A., Melin P., Stratford M., Novodvorska M., Shunburne L., Dyer P. S., et al. (2010). The decarboxylation of the weak-acid preservative, sorbic acid, is encoded by linked genes in aspergillus spp. Fungal Genet. Biol. 47 (8), 683–692. doi: 10.1016/j.fgb.2010.04.011
Punt P. J., van den Hondel C. A. (1992). Transformation of filamentous fungi based on hygromycin b and phleomycin resistance markers. Methods Enzymol. 216, 447–457. doi: 10.1016/0076-6879(92)16041-H
Reijngoud J., Arentshorst M., Ruijmbeek C., Reid I., Alazi E. D., Punt P. J., et al. (2021). Loss of function of the carbon catabolite repressor CreA leads to low but inducer-independent expression from the feruloyl esterase b promoter in Aspergillus niger. Biotechnol. Lett. 43, 1323–1336. doi: 10.1007/s10529-021-03104-2
Reijngoud J., Deseke M., Halbesma E. T. M., Alazi E., Arentshorst M., Punt P. J., et al. (2019). Mutations in AraR leading to constitutive expression of arabinolytic genes in Aspergillus niger under derepressing conditions. Appl. Microbiol. Biotechnol. 103 (10), 4125–4136. doi: 10.1007/s00253-019-09777-0
Rocha A.L., Di Pietro A., Ruiz-Roldán C., Roncero M.I. (2008). Ctf1, a transcriptional activator of cutinase and lipase genes in Fusarium oxysporum is dispensable for virulence. Mol. Plant Pathol. 9 (3), 293–304. doi: 10.1111/j.1364-3703.2007.00463.x
Saulnier L., Thibault J. F. (1999). Ferulic acid and diferulic acids as components of sugar-beet pectins and maize bran heteroxylans. J. Sci. Food Agric. 79, 396–402. doi: 10.1002/(SICI)1097-0010(19990301)79:3<396::AID-JSFA262>3.0.CO;2-B
Schjerling P., Holmberg S. (1996). Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24 (23), 4599–4607. doi: 10.1093/nar/24.23.4599
Seekles S.J., van Dam J., Arentshorst M., Ram A.F.J. (2022). Natural variation and the role of Zn2Cys6 Transcription Factors SdrA, WarA and WarB in sorbic acid resistance of Aspergillus niger. Microorganisms 10 (2), 221. doi: 10.3390/microorganisms10020221
Selig M. J., Knoshaug E. P., Adney W. S., Himmel M. E., Decker S. R. (2008). Synergistic enhancement of cellobiohydrolase performance on pretreated corn stover by addition of xylanase and esterase activities. Bioresource Technol. 99 (11), 4997–5005. doi: 10.1016/j.biortech.2007.09.064
Sun D., Lv Z. W., Rao J., Tian R., Sun S. N., Peng F. (2022). Effects of hydrothermal pretreatment on the dissolution and structural evolution of hemicelluloses and lignin: A review. Carbohydr. Polym. 281, 119050. doi: 10.1016/j.carbpol.2021.119050
Suzuki Y., Murray S. L., Wong K. H., Davis M. A., Hynes M. J. (2012). Reprogramming of carbon metabolism by the transcriptional activators AcuK and AcuM in Aspergillus nidulans. Mol. Microbiol. 84 (5), 942–964. doi: 10.1111/j.1365-2958.2012.08067.x
Tabka M. G., Herpoël-Gimbert I., Monod F., Asther M., Sigoillot J. C. (2006). Enzymatic saccharification of wheat straw for bioethanol production by a combined cellulase xylanase and feruloyl esterase treatment. Enz. Microb. Techn. 39 (4), 897–902. doi: 10.1016/j.enzmictec.2006.01.021
van den Berg R. A., Braaksma M., van der Veen D., van der Werf M. J., Punt P. J., van der Oost J., et al. (2010). Identification of modules in Aspergillus niger by gene co-expression network analysis. Fungal Genet. Biol. 47 (6), 539–550. doi: 10.1016/j.fgb.2010.03.005
van Diepeningen A. D., Debets A. J., Varga J., van der Gaag M., Swart K., Hoekstra R. F. (2004). Efficient degradation of tannic acid by black aspergillus species. Mycol. Res. 108 (Pt 8), 919–925. doi: 10.1017/s0953756204000747
Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. (2014). Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9 (11), e112963. doi: 10.1371/journal.pone.0112963
Widhalm J. R., Dudareva N. (2015). A familiar ring to it: Biosynthesis of plant benzoic acids. Mol. Plant 8 (1), 83–97. doi: 10.1016/j.molp.2014.12.001
Wong D. W., Chan V. J., Liao H., Zidwick M. J. (2013). Cloning of a novel feruloyl esterase gene from rumen microbial metagenome and enzyme characterization in synergism with endoxylanases. J. Ind. Microbiol. Biotechnol. 40 (3-4), 287–295. doi: 10.1007/s10295-013-1234-1
Keywords: transcriptional regulation, peroxisome, hydroxycinnamic acids, feruloyl esterase, fatty acids, CoA-dependent β-oxidative pathway
Citation: Arentshorst M, Reijngoud J, van Tol DJC, Reid ID, Arendsen Y, Pel HJ, van Peij NNME, Visser J, Punt PJ, Tsang A and Ram AFJ (2022) Utilization of ferulic acid in Aspergillus niger requires the transcription factor FarA and a newly identified Far-like protein (FarD) that lacks the canonical Zn(II)2Cys6 domain. Front. Fungal Biol. 3:978845. doi: 10.3389/ffunb.2022.978845
Received: 26 June 2022; Accepted: 17 October 2022;
Published: 08 November 2022.
Edited by:
Marcos Sergio Toledo, Federal University of São Paulo, BrazilReviewed by:
Taina Kristina Lundell, University of Helsinki, FinlandMao Peng, Westerdijk Fungal Biodiversity Institute, Netherlands
Copyright © 2022 Arentshorst, Reijngoud, van Tol, Reid, Arendsen, Pel, van Peij, Visser, Punt, Tsang and Ram. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arthur F. J. Ram, YS5mLmoucmFtQGJpb2xvZ3kubGVpZGVudW5pdi5ubA==
†Present address: Peter J. Punt, Gingko Bioworks NL, Netherlands
 Mark Arentshorst
Mark Arentshorst Jos Reijngoud1
Jos Reijngoud1 Ian D. Reid
Ian D. Reid Herman J. Pel
Herman J. Pel Jaap Visser
Jaap Visser Adrian Tsang
Adrian Tsang Arthur F. J. Ram
Arthur F. J. Ram