- Laboratório de Biodiversidade Molecular e Conservação, Departamento de Genética e Evolução, Universidade Federal de São Carlos, São Paulo, Brazil
The megadiversity of the neotropical ichthyofauna has been associated to recent diversification processes, reflecting in subtle or lacking morphological differentiation between species, challenging the classical taxonomic identification. Leporinus friderici occurs in several river basins of South America, and its nominal taxonomic validity has been questioned. Its wide distribution within the Brazilian Shield suggests that this species could be genetically structured among the hydrographic basins, despite a sharp morphological similarity. In this study, we performed phylogenetic analyses, based on three nuclear (recombination activating gene 1, RAG1, recombination activating gene 2, RAG2, and myosin heavy chain 6 cardiac muscle alpha gene, Myh6) and two mitochondrial (COI and Cytochrome b, Cytb) markers, in specimens morphologically similar to L. friderici and related species from different hydrographic basins in South America. Our phylogenetic tree identified four well-supported clades, which point out to the existence of taxonomic inconsistencies within this fish group. A clade named L. cf. friderici sensu stricto included eight Molecular Operational Taxonomic Units recently diversified in the Brazilian Shield basins. These results were also confirmed by a single-gene species delimitation analysis. It is suggested that this clade includes a species complex, characterizing taxonomic uncertainties. Another clade recovered only L. friderici from the Suriname rivers, validating this nominal species in its type locality. A third no-named clade, characterized by deeper species divergence, recovered five different nominal species interleaved with other undescribed forms previously also recognized as L. cf. friderici, indicating taxonomic errors. The fourth clade only included L. taeniatus. Our results showed a complex scenario involving the morphotype L. cf. friderici and allowed us to address aspects related to evolutionary diversification of this fish group and historical processes involved with, highlighting the importance of revealing hidden biodiversity for the taxonomy and conservationist action plans of these fish.
Introduction
South America freshwater fish represent one-third of the world continental ichthyofauna (Reis et al., 2016). However, this huge biodiversity is relatively recent, mostly due to extensive speciation events during the last 10 Ma (Hubert et al., 2007; Albert and Reis, 2011). Several taxa diverged <1 Ma during the late Pleistocene because of the Quaternary activity that led to topographic changes such as recent drainage rearrangements (Ribeiro, 2006). These events probably reflected in subtle or lacking morphological differentiation between the emerged species, challenging the classical taxonomic identification.
Molecular analyses have been largely used to aid species identification and delimitation within neotropical fish (e.g., Carvalho et al., 2011; Pereira et al., 2011, 2013; Ramirez and Galetti, 2015; Machado et al., 2016), contributing in revealing hidden biodiversity (Pires et al., 2017; Ramirez et al., 2017a). Both DNA barcoding and phylogeny studies have provided important contributions for better understand the phylogenetic relationships within taxa, and historical and evolutionary processes involved in the species diversification (e.g., Hebert et al., 2003; Carvalho-Costa et al., 2011; Ramirez et al., 2017b). In this way, mitochondrial and nuclear DNA data can be used to define discrete genetic lineages, detecting reciprocal monophyly, and characterizing a Molecular Operational Taxonomic Unit (MOTU, cluster of orthologous sequences generated by an explicit algorithm), representing a monophyletic lineage that could or not correspond to a taxa (Blaxter et al., 2005; Jones et al., 2011).
Leporinus, from the Anostomidae family, is considered one of the richest genus within Characiformes, a predominant freshwater fish order in South America (Garavello and Britski, 2003). Recent morphological and molecular studies have shown Leporinus as a non-monophyletic genus (Sidlauskas and Vari, 2008; Ramirez et al., 2016), highlighting the need of a deep taxonomic revision on this group. Freshly, an integrated morphological, chromosomal, and molecular approach described the new genus Megaleporinus, gathering the largest body sized Leporinus species in a monophyletic clade (Ramirez et al., 2017b).
Among the remaining Leporinus species, Leporinus friderici Block (1794) has a wide geographic distribution occurring in most rivers of South America (Garavello et al., 1992). This large distribution has been investigated by several authors, who have found morphological (Géry et al., 1987; Renno et al., 1990, 1991; Garavello et al., 1992; Sidlauskas and Vari, 2012) and genetic (Renno et al., 1990, 1991) variations among populations, suggesting that this L. friderici morphotype may contain a species complex (Sidlauskas and Vari, 2012). While L. friderici from Suriname and French Guiana rivers has been recognized as the type species (Sidlauskas and Vari, 2012), the provisional nomenclature Leporinus cf. friderici has been used to refer to the remaining individuals of this morphotype. However, because of its wide distribution within the Brazilian Shield, one could expect some levels of genetic differentiation within the L. cf. friderici among hydrographic basins. Morphological variations in the eyes, body length, and color patterns have been already reported between L. cf. friderici from the Amazon and Paraná-Paraguay basins (Géry et al., 1987; Garavello et al., 1992).
In this study we tested the hypothesis that L. cf. friderici consists of a monophyletic group formed by different MOTUs currently separated in distinct basins within the Brazilian Shield. We performed a phylogenetic analysis using mitochondrial and nuclear markers to confirm L. cf. friderici as a monophyletic group, and a single-gene species delimitation analysis to characterize MOTUs. Finally, we infer on the evolutionary historical processes possibly responsible for the diversification within this group.
Materials and Methods
Ethics Statements
This research was conducted within the protocols approved by the Ethics Committee on Animal Experimentation (CEUA, Federal University of São Carlos, São Carlos, São Paulo, Brazil) and SISBIO-ICMBio (Authorization System and Biodiversity Information, Chico Mendes Institute for Biodiversity Conservation, Ministry of Environment, Brazil). The biological samples were obtained under CMBIO/MMA No. 32215 and CEUA No. 3893250615 permits following all legal requirements. Samples from Colombia and Peru were obtained by Jose Ariel Rodriguez and Hernan Ortega, respectively, who provided DNA aliquots for this study.
Biological Specimens and Data Sampling
Samples of fin, muscle, or liver tissues were collected of 53 L. cf. friderici specimens from eight hydrographic basins from the Brazilian Shield. Figure 1 illustrates the studied river basins. Samples of Leporinus agassizii Steindachner (1876), L. boehlkei Garavello (1988), and L. piau Fowler (1941), from Jaguaribe and São Francisco rivers and L. cf. parae Eigenmann and Ogle (1907), were also obtained. In addition, DNA sequences of Leporinus desmotes Fowler (1941), Leporinus fasciatus Block (1794), Leporinus lacustris Campos (1945), Leporinus octomaculatus Britski and Garavello (1993), Leporinus taeniatus Lütken (1875), Leporinus venerei Britski and Birindelli (2008), and Hypomasticus pachycheilus Britski (1976), were downloaded from GenBank (Ramirez et al., 2016). All these species were included in our study because they have been considered as closely related to L. friderici (Garavello, 1988; Britski and Birindelli, 2008; Ramirez et al., 2016). Sequences of L. friderici from Suriname, that is considered the type locality, were obtained from GenBank (Melo et al., 2016) as well. Information related to the specimens, vouchers, IDs, and site localities are recorded in the Supplementary Material (Supplementary Tables S1, S2).
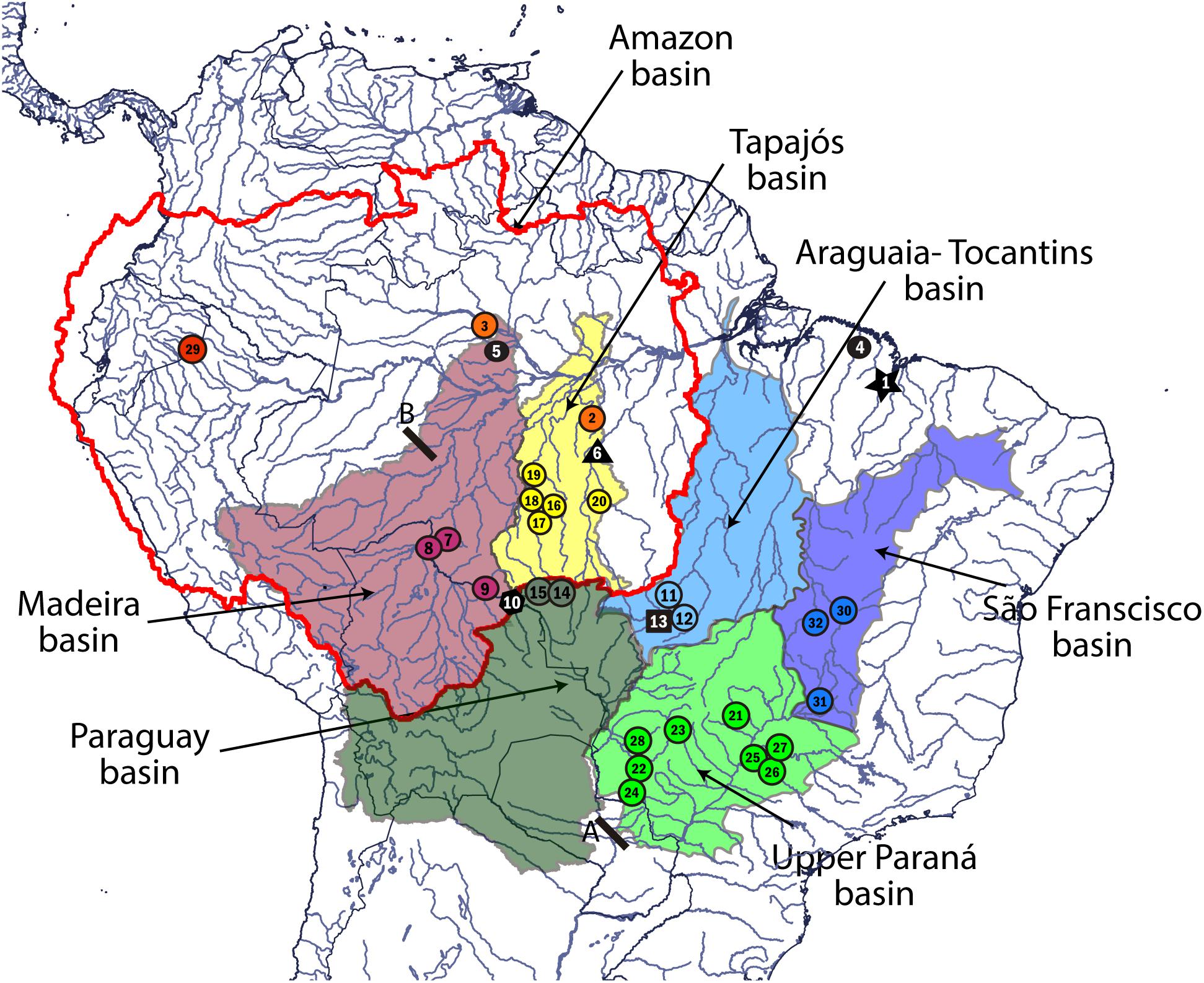
FIGURE 1. Collection sites and hydrographic basin of occurrence of Leporinus cf. friderici. Colored circles represent the MOTUs found in this study: L. agassizii (red), L. cf. friderici Amazon 1 (orange), L. cf. friderici Madeira 1 (purple), L. cf. friderici Paraná (light green), L. cf. friderici Paraguay (dark green), L. piau São Francisco (dark blue), L. cf. friderici Upper Tapajós (yellow), and L. cf. friderici Tocantins 1 (light blue). Black dot represents individuals collected as L. cf. friderici Amazon 2; black star represents L. cf. friderici Mearim; black square represents L. cf. friderici Tocantins 2; black polygon represents L. cf. friderici Madeira 2; black triangle represents L. cf. friderici Xingú. A, Prior Sete Quedas waterfalls region and Itaipú dam. B, Teotônio falls in the Madeira River. Map created by Q-GIS (http://www.qgis.org/). The collection site numbers are according to Supplementary Table S1.
DNA Extraction, Gene Amplification, and Sequencing
Total DNA was extracted using the conventional phenol–chloroform/proteinase K protocol (Sambrook et al., 1989). For the phylogenetic analysis at least one individual per each major sampled hydrographic basin was used. Cytochrome b (Cytb) and cytochrome oxidase subunit 1 (COI) mitochondrial regions were amplified according to Ramirez and Galetti (2015). The nuclear recombination activating gene 1 (RAG1), recombination activating gene 2 (RAG2), and myosin heavy chain 6 cardiac muscle alpha gene (Myh6) were amplified following Oliveira et al. (2011). PCR products obtained for both DNA strands were sequenced on an ABI 373xl sequencer (Applied Biosystems, Little Chalfont, United Kingdom).
Data Analysis
The obtained sequences were manually edited and aligned using Bioedit (Hall, 1999) and Clustal W (Thompson et al., 1994), respectively. All sequences were checked for indels and stop codons. The haplotypes of the nuclear genes were combined into a consensus sequence by coding polymorphic sites with the IUPAC ambiguity codes (IUPAC, 1974).
The phylogenetic analyses were conducted for concatenated sequences of all mitochondrial and nuclear genes using maximum parsimony (MP), implemented in PAUP∗4.0 (Swofford, 2003), with 1000 bootstrap replicates. We also performed maximum-likelihood (ML) analyses, using RAxML in XSEDE (Stamatakis, 2006; Stamatakis et al., 2008), through the web server CIPRES Science Gateway (Miller et al., 2010), and a partitioned model determined by PartitionFinder (Lanfear et al., 2012), under a GTR+G model, and 1000 bootstrap replicates.
A multilocus Bayesian species tree (BST) was estimated by ∗BEAST (Star-BEAST) (Heled and Drummond, 2010) using 150 million generations, sampled every 5000, and a burn-in of 300. A nucleotide substitution model selected was based on the Bayesian criterion, using JModeltest 2 (Darriba et al., 2012). The models chosen were HKY+I+G, HKY+G, K80+I, K80+I, and K80+I for COI, Cytb, Myh6, RAG1, and RAG2, respectively. The two mitochondrial gene tree topologies were linked and set to have an effective population size of one-quarter from that of nuclear genes. A lognormal relaxed clock was used for all partitions. Bayesian trees using all sequences were established for each gene, separately. These trees were estimated with 10 million of generations, sampling every 5000, and a burn-in of 10%. Yule speciation model was used and nucleotide substitution models followed the same criteria above cited (Supplementary Figures S1–S5). The convergence (sample size >200) and stationarity of the values were checked in TRACER v1.6 (Rambaut et al., 2014).
Two different analyses, using a single-gene species delimitation approach based on COI sequences, were performed to determine the number of MOTUs within the clade widely distributed in the Brazilian Shield (L. cf. friderici sensu stricto, see below). First, the General Mixed Yule-Coalescent (GMYC) model (Pons et al., 2006) was used to determine the clustering of the COI haplotypes. This analysis was performed with a single threshold that was implemented in the SPLITS package using R 3.3.3 statistical software (R Core Team, 2017). For this analysis an ultrametric tree was generated using BEAST 2.3.2 (Bouckaert et al., 2014), with a lognormal relaxed clock, a birth and death model, and a HKY substitution model chosen by jModeltest 2 (Darriba et al., 2012). A total of 50 million MCMC generations and a burn-in of 10% were used. In second, a Bayesian analysis of genetic structure was implemented using BAPS software (Corander et al., 2008). The maximum number of genetically diverged groups (K) was firstly set up for 10 replicates, 10 times. The obtained groups containing samples from different basins were submitted to a second layer of analysis in BAPS using K = 1–3, replicated also 10 times. This hierarchical approach to DNA sequence clustering provides a useful way to increase statistical power and detect separated haplogroups that are assigned to conservative clusters (Cheng et al., 2013). The most likely K was chosen based on log (likelihood) and posterior probability values. Next, we consider the concordant groups between the two different analyses, and the groups presenting allopatry and reciprocal monophyly as the final number of MOTUs.
The genetic distances between MOTUS were calculated based on K2P model using MEGA 6.06 (Tamura et al., 2013). Finally, a haplotype network was generated using Median Joining (Bandelt et al., 1999) in the POPART software (Leigh and Bryant, 2015).
Results
We successful obtained a total of 127 sequences for the specimens studied herein [63 for the COI gene (557 bp) and 16 for each remaining amplified marker – Cytb (1005 bp), Myh6 (754 bp), RAG1 (1477 bp), and RAG2 (1023 bp)]. The GenBank accession numbers are shown in the Supplementary Material section (Supplementary Table S2).
The phylogenetic trees, generated by MP, ML, and BST analyses (Figure 2), strongly supported four clades with maximum support values. A clade recovered L. friderici from the Suriname basin alone. A monophyletic clade characterized by a recent diversification within the Brazilian Shield, named herein as L. cf. friderici sensu stricto, included specimens of L. cf. friderici from Amazonas (main channel), Madeira, Upper Tapajós, Tocantins, Paraguay, and Paraná basins, L. agassizii, and specimens of L. piau from São Francisco basin. A no-named clade, showing older diversification, joined specimens of L. cf. friderici, from Mearim, Tocantins, Turiaçu, Xingu, and Madeira basins, interleaved with L. boehlkei, L. lacustris, L. cf. parae, L. piau from Jaguaribe basin, and L. venerei. Lastly, a fourth clade recovered only L. taeniatus.
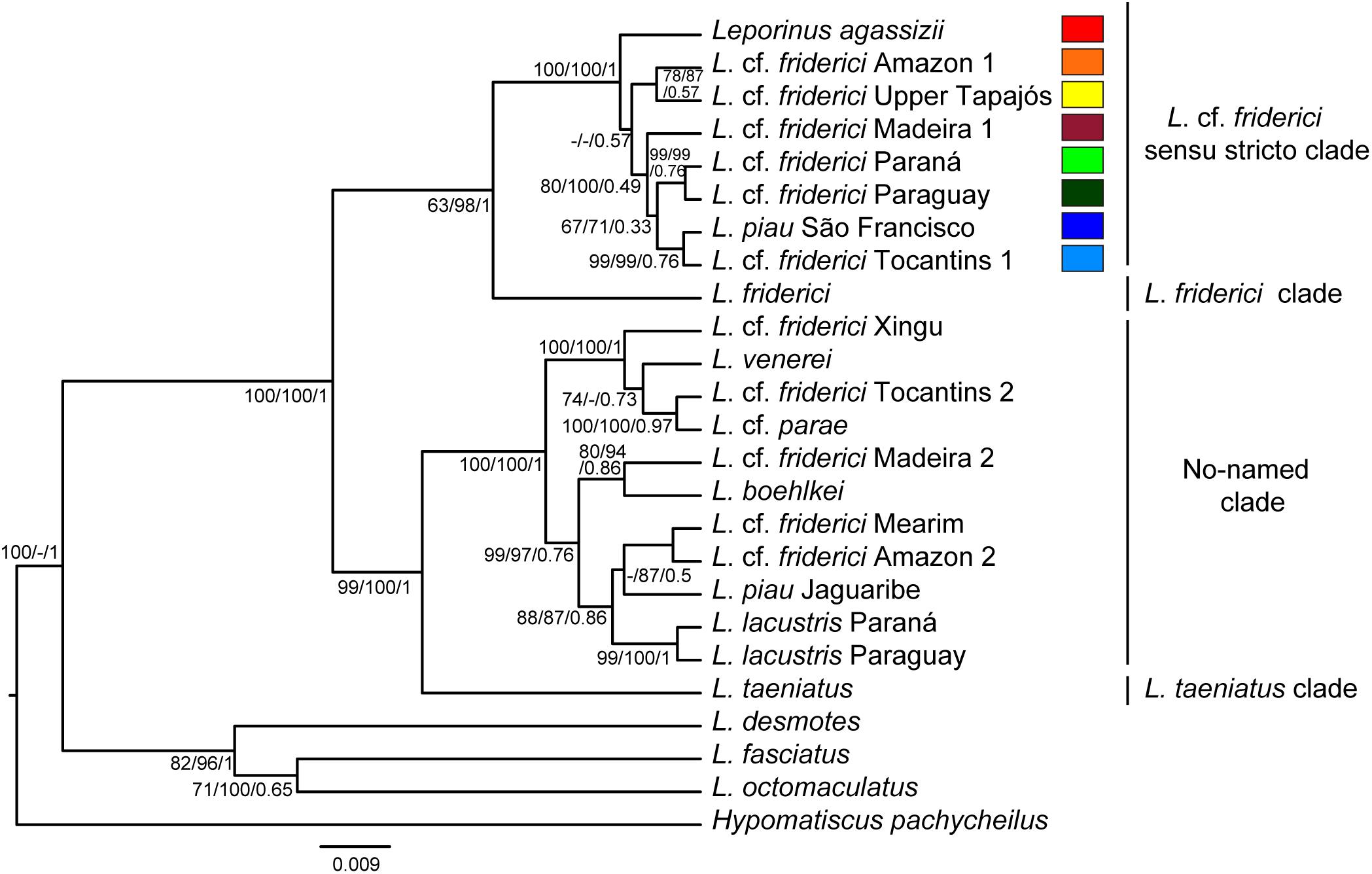
FIGURE 2. Species tree showing phylogenetic relationships of Leporinus cf. friderici. Trees were generated using five molecular markers (sequences with approximately 4900 bp). The topology corresponds to Bayesian tree. The numbers on the branches are bootstrap values for maximum parsimony and maximum likelihood, and posterior probability for Bayesian species tree. The scale bar indicates nucleotide substitutions per site.
The GMYC analysis, considering 23 parsimony-informative sites and no insertions or deletions within the COI sequences, identified seven MOTUs (CI: 6–7), with a significant likelihood ratio of 10.97 (P < 0.005) within L. cf. friderici sensu stricto. From these, six MOTUs corresponded to L. agassizii, L. cf. friderici Amazon 1, L. cf. friderici Madeira 1, L. cf. friderici Paraná, L. cf. friderici Paraguay, and L. cf. friderici Upper Tapajós. The seventh MOTU joined L. piau São Francisco and L. cf. friderici Tocantins 1 (Figure 3). The results of the first BAPS layer presented four as the most likely K with log (ml) = -554.0008 and 0.93 posterior probability values. In addition, the results of the second BAPS layer were similar to the GMYC MOTUs, but recovered L. piau São Francisco and L. cf. friderici Tocantins as two different MOTUs. The hierarchical analysis of BAPS could separate these populations, since information related to sample locations were given. Differently, GMYC method considers no prior information. Despite this little divergence in detecting MOTUs, both these lineages presenting recent divergence are reciprocally monophyletic and geographically isolated in nature.
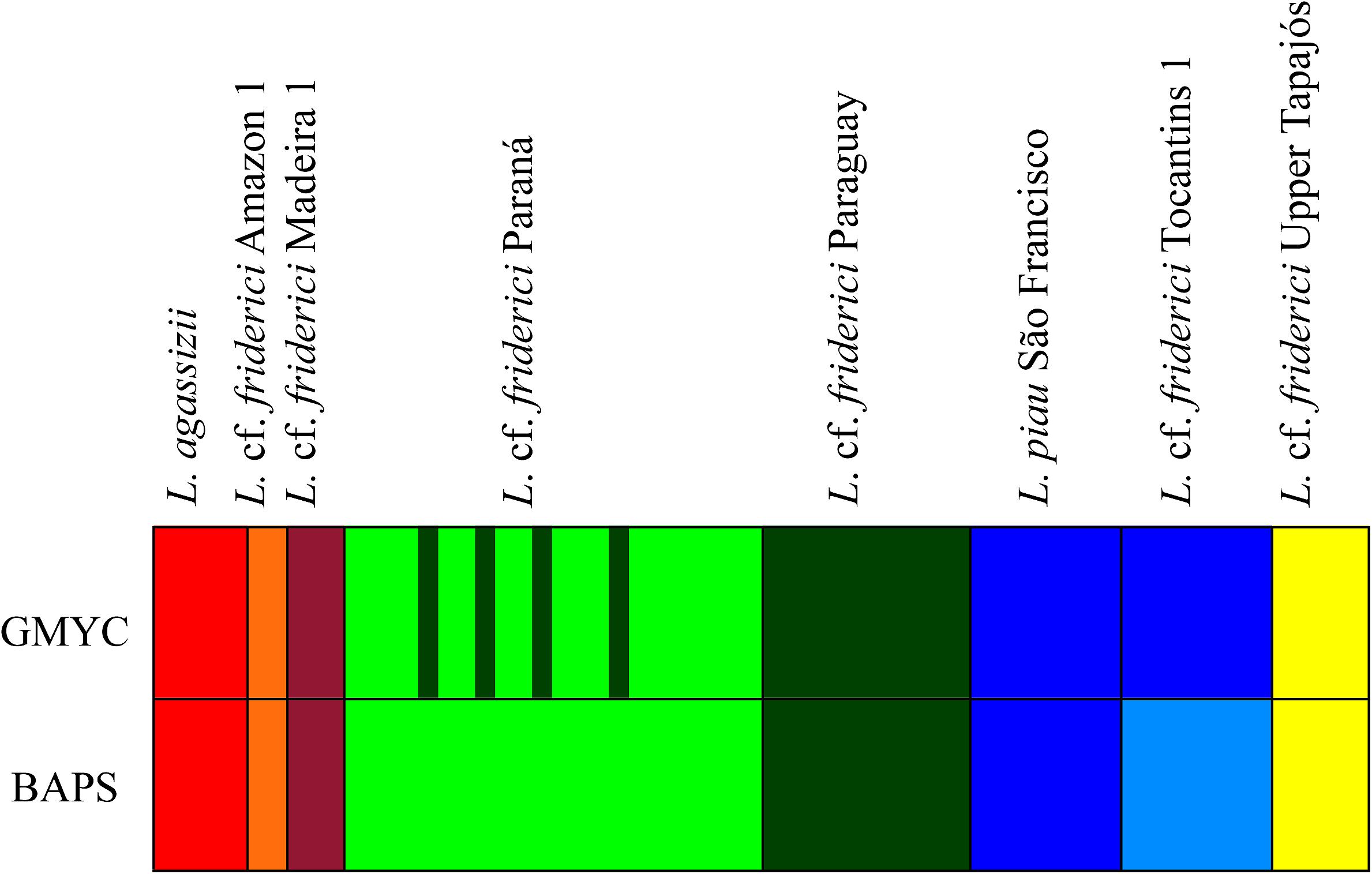
FIGURE 3. MOTUs identified based on the GMYC and BAPS analyses for Leporinus cf. friderici sensu stricto clade.
The mean genetic distance COI K2P values among the MOTUs ranged from 0.4 to 2.4%. The maximum intra-MOTU distance (0.5%) was observed in L. agassizii, while the minimum inter-MOTU distance (0.4%) was between L. cf. friderici Tocantins and L. piau São Francisco (Supplementary Table S3).
A total of 27 haplotypes was obtained within L. cf. friderici sensu stricto, in which each MOTU was represented by a haplogroup, except for the MOTUs from Paraná and Paraguay that shared one haplotype. L. piau São Francisco and L. cf. friderici Upper Tapajós were separated by only one mutational step, while the other haplogroups were connected by at least two mutational steps (Figure 4).
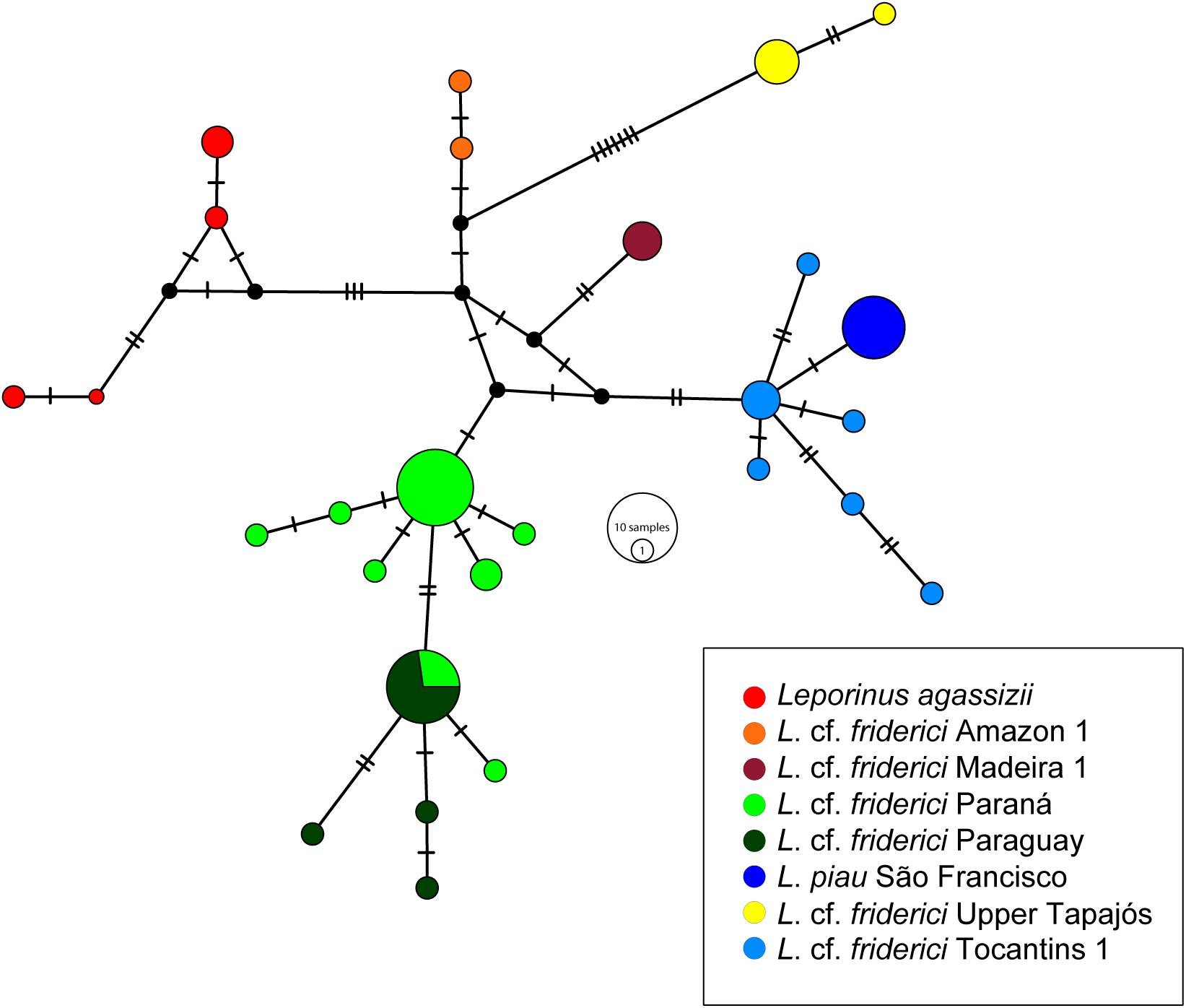
FIGURE 4. The Median Joining haplotype network of Leporinus cf. friderici sensu stricto clade using COI gene data. The small dark dots indicate the missing or not sampled haplotypes. The haplotypes are shown in different colors according to each MOTU in legend.
Discussion
Our phylogenetic analyses showed that specimens morphologically identified as L. friderici constitute a polyphyletic group, widely distributed along the South America (Figure 2). The individuals collected as L. friderici across the Brazilian Shield basins are not conspecific with L. friderici from the type locality, representing a different species. L. cf. friderici sensu stricto constitutes a monophyletic species complex distributed in Amazon, Madeira, Upper Tapajós, Tocantins, São Francisco, Paraná, and Paraguay river basins. This finding can represent a typical situation of recent diversification forming a strictly related group composed of potential cryptic species, revealing typical taxonomic uncertainties (Ramirez et al., 2017a). On the other hand, in the no-named clade the five nominal species L. boehlkei, L. cf. parae, L. lacustris, L. piau Jaguaribe, and L. venerei were interleaved with individuals that morphologically fit with the description of L. cf. friderici. In this clade, characterized by an older diversification, the use of the term L. cf. friderici hides undescribed cryptic species. All species from this clade share discriminative morphological general pattern with L. friderici (i.e., one–three spots on the body along the lateral-line, and dental formulae 4/4, except L. venerei that has 4/3, unique in Anostomidae), hindering the identification of these cryptic species. Moreover, three (L. lacustris, L. parae – L. cf. parae –, and L. venerei) of the five nominal species of this clade have been already considered as very similar in morphology due to their deep body, terminal mouth, anal fin long and dark, and three blotches on the lateral line (Britski and Birindelli, 2008).
Overall, the results obtained for L. cf. friderici sensu stricto confirm our hypothesis that there are different MOTUs within L. cf. friderici, currently separated in distinct river basins, but not all provisionally recognized as L. cf. friderici, that can be joined in a single monophyletic group.
Within the clade L. cf. friderici sensu stricto, L. agassizii is clearly recognized as a valid species different from L. friderici, mainly due to the presence of a longitudinal stripe, extending from dorsal fin to just before the caudal fin (Birindelli et al., 2013). L. agassizii was firstly described for the Iça river, Upper Amazon basin (Steindachner, 1876), and posteriorly it was also found in the Tefe lake, Nanay, Negro, and Branco rivers (Birindelli et al., 2013), from the same basin. This species has been described as restricted to the Upper Amazon basin. A parsimony analysis of endemism in the South America reported the Upper Amazon as a separate clade from the Amazon drainages (Hubert and Renno, 2006), suggesting that uplift of the paleoarches has promoted allopatric divergence in the ichthyofauna of this region, which was enhanced by marine incursions.
In our study, all individuals collected in rivers above Madeira river falls were joined in the single MOTU L. cf. friderici Madeira 1, while individuals downstream the Amazon basin were recovered in the MOTU L. cf. friderici Amazon 1 (Figure 3), with the exception of individuals from Upper Tapajós (see below), indicating these falls as possible barrier that limits the fish species distribution in the region. Previous studies had already reported evidences of structuring along the Amazon basin (Goulding, 1979; de Queiroz et al., 2013). This subdivision was attributed to geomorphological agents that allowed allopatric fragmentation and diversification events (Albert and Reis, 2011). In the Madeira river, the Teotônio fall seems to play a relevant role on the ichthyofauna diversification of the Amazon basin. This fall, besides other rapids, has been keeping apart the rivers from Upper and Lower Madeira, and has been considered a geographic barrier by limiting the fish species distribution in the region (Zanata and Toledo-Piza, 2004; Hubert et al., 2007; Torrente-Vilara et al., 2011).
Barrier effects can also explain the presence of L. cf. friderici Upper Tapajós joining individuals caught in the Juruena – Teles Pires sub-basin, in the Upper Tapajós (Supplementary Tables S1, S2). Waterfalls and rapids along the Tapajós river, and in its tributaries seem acting as barriers to fish dispersal (Britski and Garavello, 2005; Britski and Lima, 2008; Dagosta and de Pinna, 2017). The region above the Juruena – Teles Pires confluence river has been characterized by an endemic ichthyofauna different from other Amazon rivers (Carvalho and Bertaco, 2006; Britski and Lima, 2008), which could account to the separation of L. cf. friderici Upper Tapajós.
In turn, the Tocantins basin is considered an independent system from the Amazon basin, since its waters flow directly into the Atlantic Ocean (Albert and Reis, 2011). This fact was reflected in our analyses, in which the Tocantins individuals corresponded to a different genetic group (L. cf. friderici Tocantins 1). The final establishment of modern course of Tocantins (1.8 Ma) separated definitively this basin from Amazon (Rossetti and Valeriano, 2007), and the differentiation of the Tocantins ichthyofauna has been often associated to the rise of Gurupá arch, the Tucurui rapids, or the limited connectivity (Hubert et al., 2007; Hrbek et al., 2014).
The relationship between L. piau São Francisco and L. cf. friderici Tocantins 1 observed here can be accounted for a biogeographic history between the Tocantins and São Francisco basins, and the low genetic divergence (0.4%) between them likely represents a recent diversification. These two hydrographic basins share an extensive watershed, where the Sapão river (São Francisco basin) shares headwaters with the Galheiros river, Tocantins basin (Lima and Caires, 2011). The existence of these common headwaters can allow a fauna exchange between these basins. Geological evidence shows that the western border of Serra Geral from the Goiás plateau has been gradually eroded and could have potentially promoted headwater capture events between the São Francisco and Tocantins rivers (Lima and Caires, 2011). Geodispersal events (i.e., headwater capture) from Amazon river to eastern basins of the Brazilian Shield (as São Francisco river) have been already claimed in studies using molecular approaches (Hubert et al., 2007; Ramirez et al., 2017b).
While the L. piau specimens from São Francisco was linked to the L. cf. friderici sensu strito clade, L. piau from Jaguaribe was grouped in the no-named clade (Figure 2), revealing a clear taxonomic inconsistency. Fowler (1941) claimed the Salgado river (Jaguaribe basin) from Ceará state as type locality of L. piau, and included one paratype from Jatobá river (São Francisco basin). Consequently, specimens from São Francisco river have been usually cited as L. piau (Garavello and Britski, 2003; Carvalho et al., 2011). Our results pointed that specimens from São Francisco basin indeed constitute a different species from the L. piau from the Jaguaribe river, the type basin.
Still within L. cf. friderici sensu stricto, a well-supported differentiation between specimens from the Upper Paraná and Paraguay basins was also observed, although some individuals from the Upper Paraná showed haplotypes from the Paraguay basin (Figures 3, 4). It is possible that both L. cf. friderici Paraná and L. cf. friderici Paraguay reached their current distribution through ancient geodispersal events, as headwater captures between Amazon rivers and the Paraná and Paraguay basins. In the modern river basin landscape, the Paraguay basin has a watershed with the Guaporé, Tapajós, and Xingu rivers, while the Upper Paraná shared a watershed at the headwaters of the Tocantins basin (Albert and Reis, 2011). These hydrographic systems have experienced a long history of major capture events and formation of semipermeable barriers (Lundberg et al., 1998) that can support this hypothesis. The Upper Paraná ichthyofauna was separated from the Lower Paraná by the Sete Quedas Falls, a natural geographic barrier which no longer occurs. In the past this barrier isolated the Upper Paraná, where the ichthyofauna has been diverging, as already reported in Megaleporinus (Ramirez et al., 2017b) and Salminus (Machado et al., 2016). The shared haplotypes between L. cf. friderici Paraná and L. cf. friderici Paraguay are probably resulting of the removal of the natural barrier when the Itaipu hydroelectric was built. The resulted dam flooded an extensive area, including the no longer existent Sete Quedas Falls, allowing the connection between both ichthyofauna from Lower and Upper Paraná facilitating contact between mitochondrial lineages since the formation of Sete Quedas Falls (Júlio et al., 2009; Prioli et al., 2012).
Conclusion
Our study showed that L. cf. friderici as provisionally used hides at least two major situations. First, L. cf. friderici sensu stricto, a monophyletic clade joining eight MOTUs, potentially includes a true species complex, characterized by recent diversification across the Brazilian Shield basins. According to our initial expectations, L. cf. friderici sensu stricto is genetically structured along the Brazilian shield basins, and this structure appears to be related to geomorphological agents, determining the current hydrographic structure. Its taxonomic significance is an open question, requesting complementary studies for resolving this typical situation of taxonomic uncertainties. Second, a no-named clade, characterized by relatively older diversification, in our opinion, hides undescribed cryptic species under L. cf. friderici denomination, likely due to the morphology similarities that characterize the clades here studied (except L. taeniatus clade). However, this new MOTUs show deep phylogenetic divergence and they are interleaved with other nominal valid species (L. venerei, L. boehlkei, L. lacustris, L. piau, and L. cf. parae), supporting them as potential new species.
Overall, our results have important significance for the taxonomy and evolutionary knowledge of this fish group as well as for its conservation. Moreover, this scenario indicates that L. cf. friderici sensu stricto can constitute an excellent phylogeographic model in studying evolutionary and speciation processes acting in the South America basins. Despite its migratory behavior, L. cf. friderici cannot be considered as a single genetic stock even within the same basin (i.e., Amazon basin) and needs to be well known for having its whole diversity considered in any conservation effort. For a more complete understanding, the taxonomic status of each MOTU that was revealed herein needs to be evaluated using preferentially morphological and molecular data in an integrative approach.
Author Contributions
RS-S and JR collected the data, reviewed the literature, and achieved the bioinformatic analyses. All authors contributed to design the research, article writing and discussion, and approved the final version of the manuscript.
Funding
This study was supported by Conselho Nacional de Desenvolvimento Cientifíco e Tecnológico (CNPq, 304440/2009-4 and 473474/2011-5), SISBIOTA-Brazil Program (CNPq, 563299/2010-0; FAPESP, 10/52315-7), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, número to RS-S), and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2011/21836-4 to JR).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to C. Cramer, C. Doria, D. Carvalho, H. Ortega, J. A. Rodriguez, J. C. Riofrio, P. Venere, W. Troy, and U. Lopes for help to obtain part of the tissue or DNA samples and ICMBIO/MMA for sampling fish authorization (32215-1). We also thank H. Britski, P. Venere, J. Zuanon, N. Priorski, and H. Ortega for the specimen identification and the two reviewers for their comments and suggestions, which have improved the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00047/full#supplementary-material
FIGURE S1 | Bayesian tree for the cytochrome oxidase subunit 1 (COI) gene. Values on nodes represent the posterior probability.
FIGURE S2 | Bayesian tree for the cytochrome b (Cytb) gene. Values on nodes represent the posterior probability.
FIGURE S3 | Bayesian tree for the myosin heavy chain 6 cardiac muscle alpha (Myh6) gene. Values on nodes represent the posterior probability.
FiGURE S4 | Bayesian tree for the recombination activating gene 1 (RAG1) gene. Values on nodes represent the posterior probability.
FiGURE S5 | Bayesian tree for the recombination activating gene 2 (RAG2) gene. Values on nodes represent the posterior probability.
TABLE S1 | Vouchers and collection sites for all analyzed Leporinus cf. friderici.
TABLE S2 | GenBank accession numbers for all analyzed species.
TABLE S3 | Pairwise mean genetic distances values inter and intra-MOTU (in bold) Leporinus cf. friderici sensu stricto clade using K2p model. Values as percentage.
References
Albert, J. S., and Reis, R. E. (eds). (2011). “Major biogeographic and phylogenetic patterns,” in Historical Biogeography of Neotropical Freshwater Fishes, (Los Angeles, CA: University of California Press). doi: 10.1525/california/9780520268685.001.0001
Bandelt, H.-J. J., Forster, P., Röhl, A., and Rohl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Birindelli, J. L. O., Britski, H. A., and Lima, F. C. T. (2013). New species of Leporinus from the Rio Tapajós Basin, Brazil, and redescription of L. moralesi (Characiformes: Anostomidae). Copeia 2013, 238–247. doi: 10.1643/CI-12-081
Blaxter, M., Mann, J., Chapman, T., Thomas, F., Whitton, C., Floyd, R., et al. (2005). Defining operational taxonomic units using DNA barcode data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 1935–1943. doi: 10.1098/rstb.2005.1725
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C.-H., Xie, D., et al. (2014). BEAST 2: a software platform for bayesian evolutionary analysis. PLOS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Britski, H. A. (1976). Sobre uma nova espécie Leporinus da Amazônia. Acta Amaz. 6, 87–89. doi: 10.1590/1809-43921976064s087
Britski, H. A., and Birindelli, J. L. O. (2008). Description of a new species of the genus Leporinus Spix (Characiformes: Anostomidae) from the rio Araguaia, Brazil, with comments on the taxonomy and distribution of L. parae and L. lacustris. Neotrop. Ichthyol. 6, 45–51. doi: 10.1590/S1679-62252008000100005
Britski, H. A., and Garavello, J. C. (1993). Descrição de duas espécies novas de Leporinus da bacia do Tapajós (Pisces, Characiformes). Comun. Mus. Ciênc. Tecnol. PUCRS Sér. Zool. 6, 29-40.
Britski, H. A., and Garavello, J. C. (2005). Uma nova espécie de Leporinus Agassiz, 1829, da bacia Amazônica (Ostariophysi: Characiformes: Anostomidae). Comun. do Mus. Ciências e Tecnol. da PUCRS Série Zool. 18, 75–83.
Britski, H. A., and Lima, F. C. T. (2008). A new species of Hemigrammus from the upper Rio Tapajós Basin in Brazil (Teleostei: Characiformes: Characidae). Copeia 2008, 565–569. doi: 10.1643/CI-07-134
Campos, A. A. (1945). Contribuição ao estudo das espécies brasileiras do gênero Leporinus. Pap. Avulsos Dept. Zool. 5, 141–158.
Carvalho, D. C., Oliveira, D. A. A., Pompeu, P. S., Leal, C. G., Oliveira, C., and Hanner, R. (2011). Deep barcode divergence in Brazilian freshwater fishes: the case of the São Francisco River basin. Mitochondrial DNA 22, 80–86. doi: 10.3109/19401736.2011.588214
Carvalho, T. P., and Bertaco, V. A. (2006). Two new species of Hyphessobrycon (Teleostei: Characidae) from upper rio Tapajós basin on Chapada dos Parecis, central Brazil. Neotrop. Ichthyol. 4, 301–308. doi: 10.1590/S1679-62252006000300001
Carvalho-Costa, L. F., Piorski, N. M., Willis, S. C., Galetti, P. M. Jr., and Ortí, G. (2011). Molecular systematics of the neotropical shovelnose catfish genus Pseudoplatystoma Bleeker 1862 based on nuclear and mtDNA markers. Mol. Phylogenet. Evol. 59, 177–194. doi: 10.1016/j.ympev.2011.02.005
Cheng, L., Connor, T. R., Siren, J., Aanensen, D. M., and Corander, J. (2013). Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 30, 1224–1228. doi: 10.1093/molbev/mst028
Corander, J., Marttinen, P., Sirén, J., and Tang, J. (2008). Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9:539. doi: 10.1186/1471-2105-9-539
Dagosta, F. C. P., and de Pinna, M. C. C. (2017). Biogeography of Amazonian fishes: deconstructing river basins as biogeographical units. Neotrop. Ichthyol. 15, 1–24. doi: 10.1590/1982-0224-20170034
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
de Queiroz, L. J., Torrente-Vilara, G., Ohara, W. M., Pires, T., Zuanon, J. A., and Doria, C. R. (eds). (2013). Peixes do Rio Madeira. São Paulo: Dialeto Latin American Documentary.
Eigenmann, C. H., and Ogle, F. (1907). An Annotated list of characin fishes in the United States National Museum and the Museum of Indiana University, with descriptions of new species. Proc. U.S. Natl. Mus. 33, 1–36.
Fowler, H. W. (1941). Academy of natural sciences A collection of fresh-water fishes obtained in Eastern Brazil by Dr. Rodolpho Von Ihering. Proc. Acad. Nat. Sci. Phila. 93, 123–199.
Garavello, J. C. (1988). A new species of the genus Leporinus Spix from the Rio Meta, Colombia, South America (Pisces, Ostariophysi, Anostomidae). Proc. Acad. Nat. Sci. Phila. 140, 143–149.
Garavello, J. C., and Britski, H. A. (2003). “Family anostomidae,” in Check List of the Freshwater Fishes of South and Central America, eds R. E. Reis, S. O. Kullander, and C. J. Ferraris Jr. (Porto Alegre: EDIPUCRS), 71–84.
Garavello, J. C., Dos Reis, S. F., and Strauss, R. E. (1992). Geographic variation in Leporinus friderici (Bloch) (Pisces: Ostariophysi: Anostomidae) from the Paraná-Paraguay and Amazon River basins. Zool. Scr. 21, 197–200. doi: 10.1111/j.1463-6409.1992.tb00320.x
Géry, J., Mahnert, V., and Dlouhy, C. (1987). Poisson characoïdes non characidae du paraguay (Pisces, Ostariophysi). Rev. Suisse Zool. 94, 357–464. doi: 10.5281/zenodo.46287
Goulding, M. (1979). Ecologia da Pesca do rio Madeira. Manaus: Instituto Nacional de Pesquisas da Amazônia.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hebert, P. D. N., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Heled, J., and Drummond, A. J. (2010). Bayesian inference of species trees from multilocus data. Mol. Biol. Evol. 27, 570–580. doi: 10.1093/molbev/msp274
Hrbek, T., da Silva, V. M. F., Dutra, N., Gravena, W., Martin, A. R., and Farias, I. P. (2014). A new species of river dolphin from Brazil or: how little do we know our biodiversity. PLOS ONE 9:e83623. doi: 10.1371/journal.pone.0083623
Hubert, N., Duponchelle, F., Nuñez, J., Garcia-Davila, C., Paugy, D., and Renno, J.-F. (2007). Phylogeography of the piranha genera Serrasalmus and Pygocentrus: implications for the diversification of the Neotropical ichthyofauna. Mol. Ecol. 16, 2115–2136. doi: 10.1111/j.1365-294X.2007.03267.x
Hubert, N., and Renno, J.-F. (2006). Historical biogeography of South American freshwater fishes. J. Biogeogr. 33, 1414–1436. doi: 10.1111/j.1365-2699.2006.01518.x
IUPAC (1974). Abbreviations and symbols for Nucleic Acids, Polynucleotides and their constituents. Int. Union Pure Appl. Chem. Int. Union Biochem. 40, 277–331.
Jones, M., Ghoorah, A., and Blaxter, M. (2011). JMOTU and taxonerator: turning DNA barcode sequences into annotated operational taxonomic units. PLOS ONE 6:e19259. doi: 10.1371/journal.pone.0019259
Júlio, H. F. Jr., Tós, C. D., Agostinho,Ǎ. A., and Pavanelli, C. S. (2009). A massive invasion of fish species after eliminating a natural barrier in the upper rio Paraná basin. Neotrop. Ichthyol. 7, 709–718. doi: 10.1590/S1679-62252009000400021
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Leigh, J. W., and Bryant, D. (2015). POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Lima, F. C. T., and Caires, R. A. (2011). Peixes da Estação Ecológica Serra Geral do Tocantins, bacias dos Rios Tocantins e São Francisco, com observações sobre as implicações biogeográficas das “águas emendadas” dos Rios Sapão e Galheiros. Biota Neotrop. 11, 231–250. doi: 10.1590/S1676-06032011000100024
Lundberg, J. G., Marshall, L. G., Guerrero, J., Horton, B., Malabarba, M. C. S. L., and Wesselingh, F. P. (1998). “The stage for Neotropical fish diversification: a history of tropical South American rivers,” in Phylogeny and Classification of Neotropical Fishes, eds L. R. Malabarba, R. E. Reis, R. P. Vari, Z. M. S. Lucena, and C. A. S. Lucena (Porto Alegre: EDIPUCRS), 13–48.
Lütken, C. F. (1875). Characinae novae Brasiliae centralis a clarissimo J. Reinhardt in provincia Minas-Geraes circa oppidulum Lagoa Santa in lacu ejusdem nominis, flumine Rio das Velhas et rivulis affluentibus collectae, secundum characteres essentiales breviter descriptae. Oversigt over det Kongelige Danske Videnskabernes Selskabs Forhandlinger og dets Medlemmers Arbeider (Kjøbenhavn) 1874, 127–143.
Machado, C. D. B., Ishizuka, T. K., Freitas, P. D. D., Valiati, V. H., and Galetti, P. M. (2016). DNA barcoding reveals taxonomic uncertainty in Salminus (Characiformes). Syst. Biodivers 15, 372–382. doi: 10.1080/14772000.2016.1254390
Melo, B. F., Sidlauskas, B. L., Hoekzema, K., Frable, B. W., Vari, R. P., and Oliveira, C. (2016). Molecular phylogenetics of the Neotropical fish family Prochilodontidae (Teleostei: Characiformes). Mol. Phylogenet. Evol. 102, 189–201. doi: 10.1016/j.ympev.2016.05.037
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), (New Orleans, LA: IEEE), 1–8. doi: 10.1109/GCE.2010.5676129
Oliveira, C., Avelino, G. S., Abe, K. T., Mariguela, T. C., Benine, R. C., Ortí, G., et al. (2011). Phylogenetic relationships within the speciose family characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol. Biol. 11:275. doi: 10.1186/1471-2148-11-275
Pereira, L. H. G., Hanner, R., Foresti, F., and Oliveira, C. (2013). Can DNA barcoding accurately discriminate megadiverse Neotropical freshwater fish fauna? BMC Genet. 14:20. doi: 10.1186/1471-2156-14-20
Pereira, L. H. G., Pazian, M. F., Hanner, R., Foresti, F., and Oliveira, C. (2011). DNA barcoding reveals hidden diversity in the Neotropical freshwater fish Piabina argentea (Characiformes: Characidae) from the Upper Paraná Basin of Brazil. Mitochondrial DNA 22(Suppl. 1), 87–96. doi: 10.3109/19401736.2011.588213
Pires, A. A., Ramirez, J. L., Galetti, P. M., Troy, W. P., and Freitas, P. D. (2017). Molecular analysis reveals hidden diversity in Zungaro (Siluriformes: Pimelodidade): a genus of giant South American catfish. Genetica 145, 335–340. doi: 10.1007/s10709-017-9968-8
Pons, J., Barraclough, T., Gomez-Zurita, J., Cardoso, A., Duran, D., Hazell, S., et al. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609. doi: 10.1080/10635150600852011
Prioli, A. J., Carlo, V. A., Soria, T. V., Prioli, S. M., Pavanelli, C. S., Prioli, R. A., et al. (2012). Mitochondrial D-loop nucleotide diversity in Astyanax (Osteichthyes, Characidae) from the upper Paraná and upper Paraguay River basins. Genet. Mol. Res 11, 1064–1074. doi: 10.4238/2012.April.27.5
R Core Team (2017). R: A Language and Environment for Statistical Computing. Available at: https://www.r-project.org/
Rambaut, A., Suchard, M. A., Xie, D., and Drummond, A. J. (2014). Tracer v1.6. Available at: http://tree.bio.ed.ac.uk/software/tracer/
Ramirez, J. L., Birindelli, J. L., Carvalho, D. C., Affonso, P. R. A. M., Venere, P. C., Ortega, H., et al. (2017a). Revealing hidden diversity of the underestimated neotropical ichthyofauna: DNA barcoding in the recently described genus Megaleporinus (Characiformes: Anostomidae). Front. Genet. 8:149. doi: 10.3389/fgene.2017.00149
Ramirez, J. L., Birindelli, J. L. O., and Galetti, P. M. (2017b). A new genus of anostomidae (Ostariophysi: Characiformes): diversity, phylogeny and biogeography based on cytogenetic, molecular and morphological data. Mol. Phylogenet. Evol. 107, 308–323. doi: 10.1016/j.ympev.2016.11.012
Ramirez, J. L., Carvalho-Costa, L. F., Venere, P. C., Carvalho, D. C., Troy, W. P., and Galetti, P. M. (2016). Testing monophyly of the freshwater fish Leporinus (Characiformes. Anostomidae) through molecular analysis. J. Fish Biol. 88, 1204–1214. doi: 10.1111/jfb.12906
Ramirez, J. L., and Galetti, P. M. Jr. (2015). DNA barcode and evolutionary relationship within Laemolyta Cope 1872 (Characiformes: Anostomidae) through molecular analyses. Mol. Phylogenet. Evol 93, 77–82. doi: 10.1016/j.ympev.2015.07.021
Reis, R. E., Albert, J. S., Di Dario, F., Mincarone, M. M. M., Petry, P. L., and Rocha, L. R. (2016). Fish biodiversity and conservation in South America. J. Fish Biol. 89, 12–47. doi: 10.1111/jfb.13016
Renno, J.-F., Berrebi, P., Boujard, T., and Guyomard, R. (1990). Intraspecific genetica differentiation of Leporinus friderici (Anostomidae, Pisces) in French Guiana and Brazil: a genetic approach to the refuge theory. J. Fish Biol. 36, 85–95. doi: 10.1111/j.1095-8649.1990.tb03522.x
Renno, J.-F., Machordom, A., Blanquer, A., and Boursot, P. (1991). Polymorphism of mitochondrial genes in populations of Leporinus friderici (Block, 1794): intraspecific structure and zoogeography of the Neotropical fish. Genetica 84, 137–142. doi: 10.1007/BF00116554
Ribeiro, A. C. (2006). Tectonic history and the biogeography of the freshwater fishes from the coastal drainages of eastern Brazil: an example of faunal evolution associated with a divergent continental margin. Neotrop. Ichthyol. 4, 225–246. doi: 10.1590/S1679-62252006000200009
Rossetti, D. F., and Valeriano, M. M. (2007). Evolution of the lowest amazon basin modeled from the integration of geological and SRTM topographic data. Catena 70, 253–265. doi: 10.1016/j.catena.2006.08.009
Sambrook, J., Fritish, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. NewYork, NY: Cold Spring Harbor Laboratory Press.
Sidlauskas, B. L., and Vari, R. P. (2008). Phylogenetic relationships within the South American fish family Anostomidae (Teleostei, Ostariophysi,Characiformes). Zool. J. Linn. Soc. 154, 70–210. doi: 10.1111/j.1096-3642.2008.00407.x
Sidlauskas, B. L., and Vari, R. P. (2012). Diversity and distribution of anostomoid fishes (Teleostei: Characiformes) throughout the Guianas. Cybium 36, 71–103.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Swofford, D. L. (2003). PAUP∗. Phylogenetic Analysis Using Parsimony (∗ and Other Methods). Version 4. Sunderland, MA: Sinauer Associates.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Torrente-Vilara, G., Zuanon, J., Leprieur, F., Oberdorff, T., and Tedesco, P. A. (2011). Effects of natural rapids and waterfalls on fish assemblage structure in the Madeira River (Amazon Basin). Ecol. Freshw. Fish 20, 588–597. doi: 10.1111/j.1600-0633.2011.00508.x
Keywords: neotropical fish, MOTUs, fish phylogeny, taxonomic uncertainties, cryptic species
Citation: Silva-Santos R, Ramirez JL, Galetti PM Jr and Freitas PD (2018) Molecular Evidences of a Hidden Complex Scenario in Leporinus cf. friderici. Front. Genet. 9:47. doi: 10.3389/fgene.2018.00047
Received: 30 October 2017; Accepted: 31 January 2018;
Published: 15 February 2018.
Edited by:
Rodrigo A. Torres, Universidade Federal de Pernambuco, BrazilReviewed by:
Izeni Pires Farias, Federal University of Amazonas, BrazilMarcelo Ricardo Vicari, Ponta Grossa State University, Brazil
Copyright © 2018 Silva-Santos, Ramirez, Galetti and Freitas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rosane Silva-Santos, cm9zYW5lc2FudG9zLmdlbkBnbWFpbC5jb20=
 Rosane Silva-Santos
Rosane Silva-Santos Jorge L. Ramirez
Jorge L. Ramirez Pedro M. Galetti Jr.
Pedro M. Galetti Jr. Patrícia D. Freitas
Patrícia D. Freitas