- 1GenCology Research Centre, University of the Sunshine Coast, Maroochydore, QLD, Australia
- 2Department of Science, Technology and Environment, Ministry of Agriculture and Rural Development, Hanoi, Vietnam
- 3Research Institute for Aquaculture, Nha Trang, Vietnam
Selective breeding for marine finfish is challenging due to difficulties in reproduction, larval rearing, and on-growth in captive environments. The farming of Asian seabass (Lates calcarifer) has all these problems and our knowledge of the quantitative genetic information (heritability and correlations) of traits necessary for commercial exploitation is poor. The present study was conducted to address this knowledge gap and to provide information that can be applied to sea bass and other aquaculture species. We carried out a comprehensive genetic evaluation for three traits (body weight, total length, and survival) collected from a breeding population for Asian seabass over an eight-year period from 2010 to 2017. Statistical analysis was carried out on 4,567 adult fish at 105, 180, 270, 360, 450, and 570 days post-hatch (dph). The heritabilities (h2) estimated for body weight and length using linear mixed model were moderate to high (0.12 to 0.78 and 0.41 to 0.85, respectively) and they differed between the measurement periods. Survival during grow-out phase was analyzed using threshold logistic and probit models. The heritability estimates for survival rate on the underlying liability scale (hL2) varied from 0.05 to 0.21. When the observed heritability obtained from the linear mixed model was back-transformed to the liability scale, they were similar but not significant. In addition, we examined effects of genotype by environment (G × E) interaction on body traits. The genetic correlation for body weight between tank and sea cage cultures were high (0.91–0.94) in the first and second rearing periods (180 and 270 dph) but the correlation was decreased to 0.59 ± 0.33 at 360 dph. This suggests that the genotype by environment interaction is important for body traits in this population. Furthermore, the genetic correlations of body weights between different measurement periods were moderate but different from one. This suggests that body weights measured at different time points may be different traits and selection for improved early weight may not capture all genetic expressions in subsequent rearing periods in Asian seabass. Selection of the nucleus in sea cages may produce genotypes that do not perform equally well in tanks, although this deserves further studies to determine a suitable selection environment and optimize the breeding program. This paper discusses challenges encountered during implementation of the selection program for L. calcarifer.
Introduction
Aquaculture of Asian seabass or barramundi (Lates calcarifer, Bloch) has been growing in South East Asia and Australia. Regional production of this species in major producing countries increased 350% from ~20,000 tons in 1998 to 90,000 tons in 2017 (FAO Statistics). Asian seabass is highly fecund, euryhaline, and it has a rapid growth rate in both fresh and salt water. In addition, barramundi has good market acceptance and high economic values in many countries (Lawley, 2010; Robinson et al., 2010). Induced spawning is known to be difficult in this species and mass spawning (i.e., keeping broodfish in the same tank with a mating ratio of one female to one or two males) is normally practiced in commercial hatcheries (Evelyn et al., 2013). This practice does not allow the retention of progeny pedigrees unless DNA markers are used for parentage assignment and thus under commercial settings where genetic analysis is not practiced, the contribution made by individual broodstock to future generation is unknown. As a consequence, genetic variation in hatchery populations has declined and this has led to a severe loss in genetic diversity in many cultured stocks (Frost et al., 2006; Loughnan et al., 2015).
Selective breeding is a powerful tool to increase commercial production, and the success of selective breeding programs depends on a systematic approach involving several steps from the establishment of a base population to the development of the breeding objectives and selection strategies (Nguyen, 2016). In aquaculture, quantitative genetics, and selective breeding are in the early stages of development but substantial productivity improvement has already been achieved in fish and shellfish (Gjedrem and Rye, 2016). The principal breeding goals for aquaculture species are growth rate, disease resistance and product quality. The first selection experiments were conducted as long ago as 1919 (Embody and Hayford, 1925) to increase survival to furunculosis (due to Aeromonas salmonicida) in brook trout (Salvelinus fontinalis). Since then, several selection programs with the aim of improving growth rate and disease resistance have been reported for a range of fish (Knibb et al., 2016), crustacean (Hung et al., 2013), and mollusc (In et al., 2017). Improvement in growth rates of 5–15% (average 10%) per generation have been achieved—depending on species and testing environments (Gjedrem et al., 2012). It is estimated that in 2010 ~8.2% of global aquaculture production was based on genetically improved stocks. A typical example is Atlantic salmon (Salmo salar) where 97% production in Norway has been using improved stocks from artificial selection (Gjedrem, 2012). Hence, there is a strong need to develop an improved genetic line for Asian seabass in order to meet the growing demand for high quality seeds and breeding stock in aquaculture.
In any selective breeding programme, estimation of genetic parameters (heritability and correlations) is needed to understand the genetic basis of quantitative traits (Falconer and Mackay, 1996). They are also required for the evaluation of breeding candidates in order to estimate their genetic merit. These estimates are affected by several factors including genetic resource, sample size, culturing conditions, and the number of generations in experiments. The heritability has been reported for a range of traits in commercial aquaculture species, such as resistance to the bacterial disease columnaris in Rainbow trout (Evenhuis et al., 2015); growth related traits in tilapia (Charo-Karisa et al., 2006; Trọng et al., 2013), common carp (Vandeputte et al., 2004), and barramundi (Wang et al., 2008b; Domingos et al., 2013; Ye et al., 2017). To date, there has been limited genetic parameter estimates in Asian seabass and the published information that exists relates only to body traits recorded in the early stage of growth development, such as at 62 dph (Domingos et al., 2013), and 90 and 270 dph (Ye et al., 2017). Wang et al. (2008b) also analyzed the correlation of body weights between 90 and 270 dph while Ye et al. (2017) reported moderate heritability for growth traits at those measurement periods. Generally, the heritability and genetic correlations for body traits during growth trajectory from tagging to sexual maturity (about 3 kg) are rare in the literature.
The inheritance of survival characteristics during the grow-out phase has not been documented in Asian seabass, although it is one of the two traits (along with growth) that determines economic return for aquaculture enterprises. In contrast, numerous studies have showed existence of the additive genetic components for survival in tilapia (Luan et al., 2008; Thodesen et al., 2013; Ninh et al., 2014; Thoa et al., 2015), salmonids (Standal and Gjerde, 1987; Rye et al., 1990; Vehviläinen et al., 2010, 2012), shrimp (Vu et al., 2017), and molluscs (Dégremont et al., 2005; Liu et al., 2015). The results suggest that these species will respond positively to selection for survival. While systematic environmental effects and management options have been extensively investigated to improve survival rate in L. calcarifer, there are no reports of heritability for this trait especially survival rates during the grow-out phase in sea cages. Our study, for the first time, reports quantitative genetic basis of survival of Asian seabass (L. calcarifer).
Another important factor that merits a thorough evaluation in selective breeding programmes is the relationship between genotype and environmental interaction (G × E). The G × E effect is important when the selection and commercial production systems differ so as to develop a genetic line that can tolerate a range of environmental conditions. Understanding the G × E effect on traits of economic importance would aid in the design and optimization of a broadly applicable selective breeding programme for Asian seabass. It has been shown in red tilapia that selection response of genotypes to diverse environmental conditions can vary (Nguyen et al., 2017). Culture systems for Asian seabass are diverse and may involve different water sources (fresh or saline water) or different rearing systems (ponds, tanks, and floating sea cages). Studies in other species, such as Atlantic cod (Kolstad et al., 2006), rainbow trout (Kause et al., 2003), European seabass (Dupont-Nivet et al., 2008, 2010), common carp, and Pacific oysters (In et al., 2017) report low G × E effects. In Asian seabass, Domingos et al. (2013) did not find significant G × E interactions for early growth traits (62 dph) when comparing tank and pond or fresh and sea water environments. However, there is a lack of information about the G × E interactions for L. calcarifer grown in tanks and sea cages.
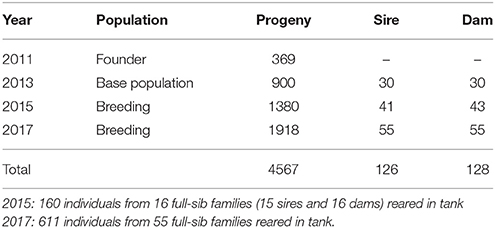
The current study, therefore, aimed to address two major issues: (i) the lack of genetic parameters for body traits and survival during growth trajectory of Asian seabass and (ii) the G × E interaction between tank and sea cage. The study used data collected from the breeding program for L. calcarifer at the Research Institute for Aquaculture No III (RIA 3), Vietnam between 2010 and 2017. The founder stocks used to form the base population for selection were collected in 2010 and 2011 from four wild and four hatcheries in the north, central and southern Vietnam. To obtain preliminary information about genetic diversity of these populations, seven microsatellite markers: Lca287, Lca371, Lca154, Lca178, LcaE22, Lca234, and Lca148 (Wang et al., 2008a) were used to analyse polymorphisms of 329 samples including eight wild and domesticated Asian seabass stocks (Phuong et al., 2013). Subsequent generations were produced in 2013, 2015, and 2017. In these years, siblings of each family were cultured in both/either sea cages and/or tanks over a period from 105 to 570 dph. A total of 4,567 fish from 128 full- and half-sib families were performance tested and morphometric measurements for body weight and total length were collected. A full pedigree traced back to the base population was used to estimate genetic parameters for growth traits in L. calcarifer. Our results showed that there is potential for genetic improvement to increase both growth and survival in the present Asian seabass population. However, there are challenges that need to be addressed in order to improve the effectiveness of future breeding program for this species.
Materials and Methods
Ethical Statement
All the methods and experimental protocols of this study were performed in accordance with guidelines and regulations approved by the animal ethics committee of the University of the Sunshine Coast, Australia (approval number ANE1613).
Experimental Location
The breeding program for Asian seabass, including breeding, rearing and grow-out experiments, was conducted at the National Mariculture Research and Development Center (MRDC) and Center for Environment and Disease Monitoring in Aquaculture (CEDMA), associated with the Research Institute for Aquaculture No.3 (RIA3) in Khanh Hoa, Central Vietnam (latitude 12.25, longitude 109.19 and it is situated at elevation 10 meters above sea level).
Origin of the Founder Stocks
The founder stock comprised four wild and four hatchery fish populations collected from different geographical regions in Vietnam: North (Hai Phong), Central (Khanh Hoa) and South (Vung Tau and Kien Giang) (Supplementary Table S1). They were collected in 2010 and transferred and reared under a common culture environment at the National Mariculture Research and Development Center in Nha Trang.
The fish were initially cultured in 20 m3 hatchery tanks and tagged before stocking in sea cages. Tagged fish were assigned randomly to 24 seacages (4 × 4 × 2.5 m) with equal representation of populations in each cage (20 fish per population per cage). After 17 months of culture in seacages, body measurements were made and the growth performance of the eight different populations were evaluated. Regardless of collection locations, wild populations had significant faster growth than those from hatcheries (Supplementary Table S2). The inferior performance of domestictaed stocks was likely resulted from poor management practices of hatcheries that may have caused inbreeding issues. The survival was high (85%) in all stock. There was a significant difference in polymorphism between the wild and domesticated populations. The allele numbers in each locus varied from 11 to 21 alleles/locus in the wild and from 3 to 8 alleles/locus in domesticated populations. The observed level of heterozygosity was also greater in the wild than hatchery sstocks (0.77–0.94 vs. 0.31–0.59). However, there was no clear distinction among populations, Fst value close to zero (Phuong et al., 2013). Based on these results (allele number >6 and observed heterozygosity > 0.6) together with the phenotypic performance, superior animals (400 fastest growth) from the four best stocks (Wild Vung Tau, Wild Khanh Hoa, hatchery fish and wild fish from Kien Giang) were used to form the base population.
Base Population
Parental fish were checked fornightly to confirm that they had reached sexual maturation. Mature broodstock were transferred from seacage to the breeding hatchery and were kept in 20 m3 tanks.
In 2011, a partial diallel cross involving fish from the four populations: Vung Tau and Khanh Hoa (both originating from the wild) and hatchery and wild fish from Kien Giang was carried out, following a single pair mating design. Thirty families (one male mated to one female) was produced after 15–20 days. Parental fish were injected using LHRHa and kept in tanks to allow natural spawning. Fish larvea were nursed until fingerling size (28.2 ± 1.2 g, L = 15.3 ± 0.2 cm). A random sample of 30 fish per family was tagged by using Passive Integrated Transponders (PIT). After tagging, the fingerlings were conditioned in tanks for another week before they were transferred into the seacage for grow-out over a period of 17 months. At harvest, body trait data were recorded and genetic evaluation was conducted to estimate breeding values (EBVs) for all individual fish in the pedigree. Based on EBV ranking and relationship of indviduals in the pedigree, the best performing (highest EBVs) individuals were selected to form the base population to produce subsequent generations for selection.
Family Production in Subsequent Generations in 2014/15 and 2017
In late 2014 and early 2015 the first generation (G1) were produced, involving 45 males and 45 female broodstocks. Mating was based not only on the EBVs but also the genetic relationships with other individuals in the pedigree. Basically, the selection and mate allocation involved three main steps: (1) We ordered families and individuals within each family on genetic merit, (2) Selected best male from best family and assigned to it the best female from best family, and (3) Checked for inbreeding (F) in potential progeny; if F = zero, proceeded to the mating, if not, assigned to best female from second best family; checked for F, and so on. Closely related matings, including full- and half-sibs, were not permitted.
The brood fish chosen for maturation assessment were healthy and normal in body shape. When assessing a prospective female, a catheter with diameter of 1.2 mm and 30 cm long was inserted into the genital pore about 5–10 cm and eggs were taken for microscopic examination. Females were considered ready to spawn when eggs showed uniform size (diameter 0.4–0.5 mm), were separated from each other, and were yellow in color. To assess maturity of males, gentle handling of the fish and slight pressure were applied on their abdomen near the genital pore. This resulted in the expulsion of sperm and, if it was white and viscous, the fish were considered to be ready for breeding. A total of 45 single pair matings were conducted in separate 7–10m3 hatching tanks.
Spawning usually occurred about 36 h after hormone admission. During this period, water temperature and salinity in the spawning tanks were maintained at 28.5–30.5°C and 32.0–33.0 ppt, respectively. Twelve hours after spawning, the fertilized (floating) eggs were collected and transferred to 500 L incubation tanks for hatching (each family in a different tank). The collection date of each egg was recorded.
Nursing, Rearing, and Grow-Out Testing
From day 2 until day 15, the larvae were fed rotifer (Brachionus plicatilis). Artemia nauplii were introduced at day 10 and fed until day 30. Subsequently a commerical pellet feed (composition 55% protein; 9% lipid, <9% fiber, <8% moisture) was used when larvae was about 23–25 days. During the larval rearing water temperature ranged from 25.2 to 29.4°C (yearly average water temperatures were 27.3°C in the morning and 28.2°C in the afternoon). Salinity was 33.4 g L-1 (range 31.4–34.5 g L−1), pH 8.2 (range 7.9–8.4), DO >5.1 mg/L (range 4.7–5.4 mg L−1), and Secchi clarity > 1.8 m. When the fish reached a weight of 28 ± 18 g (Length = 13 ± 2 cm), 60 indviduals per family were marked using Passive Intergrated Transponder (PIT) tags for indvidual identifcation. After tagging, fish of all the families were pooled and conditioned in large size tanks for 1 week. The survival rate from hatching to 30 days averaged 38.4%, but from day 30 to fingerling stage (about 60 days) it was high (average 91%). Communal grow-out of all families was conducted in 24 seacages (4 × 4 × 2.5 m) with similar number of fish per family in each cage (200 fish per cage). Once the fish were transferred to the sea cage system, they were fed a formulated diet for 2 weeks and then trash fish twice daily at a feeding rate of ~10% body weight in the first 2 months (~200 g per fish), 5% of body weight when fish weight ranged from 200 to 500 g and 3% of body weight when weight exceeded 500 g per fish. In addition to sea cages, representatives of each family were tested in 200 m3 tank (water depth of 1.5 m) at an initial stocking density of 800 fish per tank. Only commercial pellet feed containing 45% protein and 12% fat (Uni-President Ltd) was used in tanks. Water quality was monitored once a week.
During the grow-out period of 17–18 months, fish were measured six times (one measurement every 3–4 months). One to 2 days prior to the measurements, the fish were conditioned in cages without food. Three main traits were recorded: growth related characteristics (body weight, total length), survival, and sexual maturity. A detailed description of the measurement methods is given in sections Family Production in Subsequent Generations in 2014/15 and 2017 and Other Measurements. The individual tag number, deformity and health conditions were also recorded. Digital scale and ruler were used for measuring weight and length. Survival was recoded as a binary trait and coded as 1 if the fish were still alive as 0 if the fish were absent at the final measurement and this trait was also expressed as percent difference in the number of fish at stocking and final harvest (570 dph).
Other Measurements
Maturity
Maturity of females was determined by biopsy as described above (section Family Production in Subsequent Generations in 2014/15 and 2017). The maturity of males was determined by stripping method. They were recorded in the form of presence and absence (coded as 1 and 0, respectively).
Deformity
Morphological deformity included a range of measures, namely lower jaw, nasal erosion, abnormal skeleton and deformed operculum (Nguyen et al., 2016, 2018a).
Food Conversion Ratio (FCR)
Total amount of feed provided in each net cage consumed by the fish was recorded to calculate FCR. This trait was not included in analysis because it was not possible to collect the amount of uneaten feed in sea cages.
Traits Used for Genetic Analysis
Due to the limited data records and details for sexual maturity, deformity and health condition, they were not included in our genetic analysis. This study focussed on three main traits: body weight, total length and survival. Body weight and length were measured at six different time points: 105, 180, 270, 360, 450, and 570 dph. These body traits showed continuous variation and were analyzed using linear mixed model. However, survival data was treated as a binary trait and analyzed using generalized mixed model as below (section Statistical Analysis).
Statistical Analysis
Linear Mixed Model
Heritability (the observed/measurable variations that are due to genetic inheritance) for traits showing continuous expressions (body weight and length) was estimated using Restricted Maximum Likelihood Method (REML) in a uni- or multi-variate mixed model (Henderson, 1975). Numerator relationship matrix was calculated from the pedigree that included both full- and half-sib families. The number of half-sib families in generations 2015 and 2017 was 12 and 22, respectively. In mathematical notation, the mixed model is written as:
where yijklm is the vector of observations for traits studied, Yi is the systematic fixed effect of spawning year (2011, 2013, 2015, and 2017), and Ej is the fixed effect of testing environments. The non-significant effect of sex (P > 005) was omitted from the final model. The random factors in the model were sire (sk) and dam (dl). The dam component () is most likely a combination of maternal and common environmental effects (thus, , referred to as ) caused by the separate rearing of full-sib families until individuals reached a suitable size for physical tagging. The log likelihood ratio test (LTR) showed that the common full-sib effect was significant (P < 0.05). The term eijklm signifies residual errors.
Heritabilities for body traits were estimated from a univariate model (Equation 1). The genetic variance () was calculated as 4 where is sire variance (Falconer and Mackay, 1996). The dam variance component (), in this case, was a combination of the maternal, dominant and common environmental effects, also named as common full-sib effects (). The “and(dam)” option used in ASReml assumed equal sire and dam variances ()(Ninh et al., 2014). The phenotypic variance () was calculated as the sum of the additive genetic sire variance
(), the dam variance (), the common full-sib () and the residual variance (), as or . The heritability was calculated as and the common environmental effect was calculated as . Genetic and phenotypic correlations were estimated from a two-trait sire and dam model with the same fixed effects as shown in Equation (1). The correlations were calculated as the covariance divided by the product of the standard deviations of traits: where was the estimated additive genetic or phenotypic covariance between the two traits, and and are the additive genetic or phenotypic variances of traits 1 and 2, respectively.
To examine the interaction between genotype and environment (G × E), the expressions in tank and cage were treated as different traits. A multi-trait model was applied to estimate the between-environment genetic correlations through a numerator relationship matrix obtained from the pedigree. In this analysis, Model 1 was used and all the effects were the same as shown in Equation (1), except that the effect of environment was excluded. Due to differences in standard deviations between the two culture systems, body weight, and length were square root transformed to estimate the between-environment genetic correlations. As the phenotypes were measured on different animals in tank and pond, there is no phenotypic correlation between trait expressions between the two environments.
Generalised Threshold Model
In this study, survival was treated as a binary character (0 = dead/or missing and 1 = alive). In addition to the linear mixed model (1), this trait was also analyzed using a threshold sire and dam model. The threshold models assume that the data follow a binominal distribution and logit (model 2) and probit (model 3) functions were used.
With the threshold logistic sire (sm) and dam model (2), heritability for survival was calculated using the variance of the logit link function, which implies a correction of the residual variance by factor π2/3.
where is sire variance, is dam variance, and
Under model 3 (probit threshold model), the heritability for survival was calculated as:
where is sire variance, is dam variance, and
For binomial observations, estimates of h2 on the observed scale (0/1) were transformed to the liability scales (logit and probit) using the formula of Robertson and Lerner (1949):
where is the heritability on the observed (0/1) scale, is the estimated heritability on the liability (logit or probit) scale, p is a proportion of a given survival rate in the data, and z is the height of the ordinate of normal distribution corresponding to a truncation point applied to p proportion of survival.
The REML and mixed model approach have been implemented in ASReml version 4.0 (Gilmour et al., 2009). ASReml provides flexibility to specify different co-variance structures or different fixed and random effects to avoid any possible bias associated with the genetic parameter estimates.
Results
Basic Statistics and Characteristics of the Data
The body weight and length measured six times during the grow-out phase from 105 to 570 days post-hatch (dph) are given in Table 1. The weight and length of the fish increased steadily until 270 dph after which there was a rapid increase in growth rate until the final harvest. At 570 days, the mean body weight was 2.3 kg and mean length was 53.9 cm. The coefficients of variation for body traits were greater in the earlier (105, 180, and 270 dph) than later phase of growth development (360, 450, and 570 dph). The growth curve of the experimental fish at six different grow-out periods is presented in Supplementary Figure 1. Pedigree structure of the data is shown in Table 2.
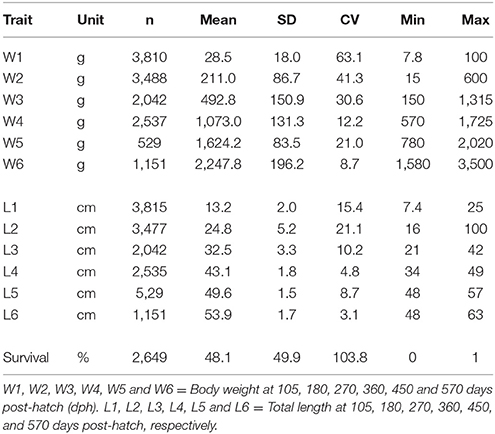
Table 1. Number of observations (n), mean, standard deviation (SD), coefficient of variation (CV, %), minimum, and maximum values for body weight and length during the growth trajectory.
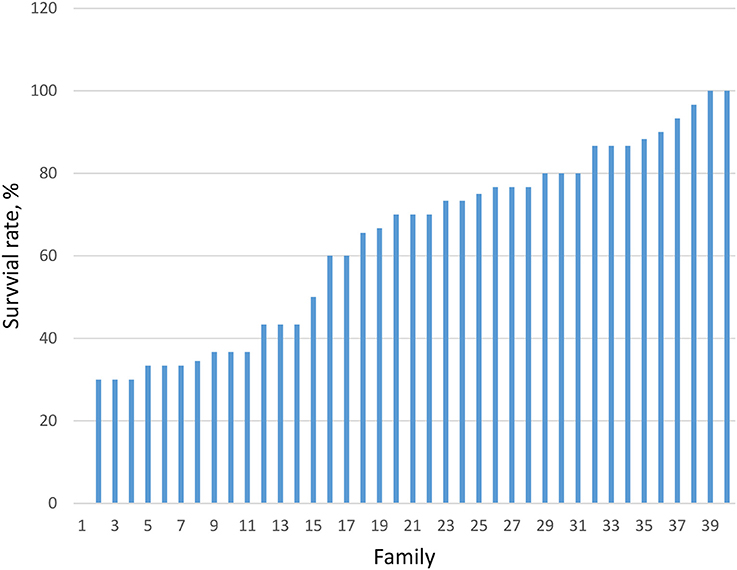
Survival trait during grow-out phase (105 to 360 days) ranged from 30 to 100% among 30 families produced in 2013 and 40 families in 2015 (averaging 48.1%). Significant (P < 0.05) differences in the survival rates among the families in 2015 is presented in Figure 1.
Effects of Culture Environments
A prevailing commercial production system for Asian seabass is sea cage, but tank culture is also increasingly practiced. In this study, growth traits were investigated in two different culture systems: tanks and sea cages. The fish grew significantly faster (P < 0.001) in sea cages than in tanks (Figure 2). After 270 days, average weight of fish stocked in sea cages was 698.5 g (373 mm length) and in tanks 231.6 g (291 mm length). The differences in weight and length between the two environments varied during the growing period and were significantly (p < 0.001) greater after 270 dph than during the early phase of growth (66% for W3 vs. 21% for W1).
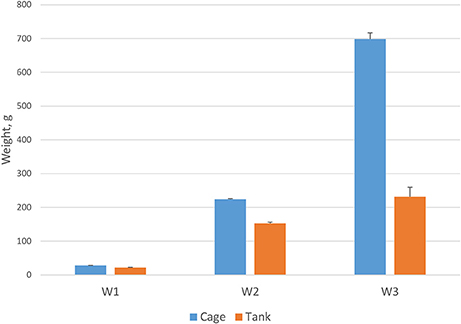
Figure 2. Fish body weight in cage (n = 2058) and tank (n = 576) over different measurement periods at 105, 220, and 270 days post-hatch (P < 0.001).
Sex Differences in Growth Traits
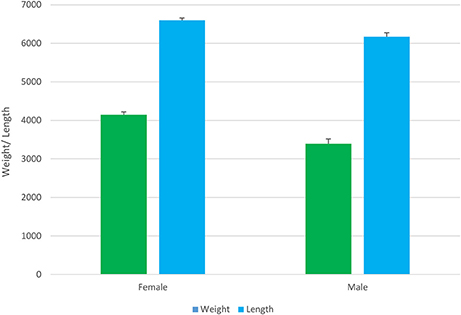
Between-sex differences in growth was examined in parental candidates prior to breeding. In this population of Asian seabass, the females had significantly (P < 0.001) greater body weight than the males (4.2 vs. 3.4 kg; Figure 3). The sexual differences in body weight were not significant during the grow-out phase either in tanks or cages. However, the between-sex differences in growth-related traits are widely observed in aquaculture, such as tilapia and prawn (Hung et al., 2014).
Heritability and Common Full-Sib Effects for Growth Traits
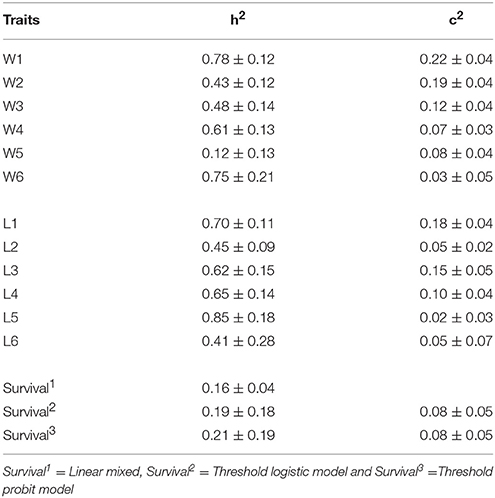
The estimated heritabilities (h2) for body weight and length at different times of measurement were moderate to high (Table 3). The heritability estimates ranged from 0.12 to 0.78 for body weight and 0.41–0.85 for total length. Except for W5 (h2 = 0.12) that was likely due to measurement errors, the heritability for both traits was higher at 105 to 450 dph of on-grown than at other measurement periods. Across all measurements, the heritability estimates for weight and length were significant.
The common full-sib (c2) effects accounted from 2 to 22% of the total variation for weight and length. The c2 effects were larger for W1-W3 than those obtained for W4-W6 (16–22% vs. 2–10% for body weight).
Heritability Estimates for Survival Using Different Statistical Models
Three models were used to estimate heritability for survival. The heritability estimate from linear mixed model (model 1) was = 0.05. Threshold probit model (model 3) gave the highest value at = 0.21, whereas the estimate obtained from threshold logistic model (model 2) was intermediate ( 0.19). When the observed heritability obtained from the linear mixed model (1) was back-transformed to the underlying liability scale, the estimates were generally similar between the linear and threshold models (2 and 3). However, across the three statistical models used, the heritabilities for survival on both observed scale (model 1) and underlying liability (models 2 and 3) were not different from zero due to their high standard errors (Table 3). The common full-sib effect on survival was low (8%).
Phenotypic and Genetic Correlations
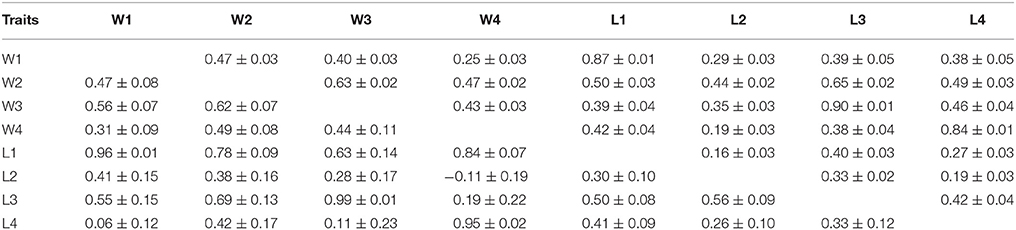
Phenotypic and genetic correlation of body weight between different times points are presented in Table 4. In general, genetic correlation of weight traits estimated for four measurements were all positive (0.31 to 0.62) and significantly differed from one. The genetic correlation (rg) between weight and length at the same age was high (0.95 to 0.99), except for the moderate estimate between W2 and L2 (rg = 0.38). However, the rg values between weight and length at different ages were low or moderate, of which two estimates between W1 and L4 or between W4 and L2 were not different from zero, due to their high standard errors. Overall, the genetic correlations of traits between successive rearing periods were stronger than those that are further apart. The phenotypic correlations were all positive and significant.
Genotype by Environment (G×E) Interaction
Table 5 shows the between-environment genetic correlations (rg) for weight traits. Due to the limited data records at 450 and 570 dph in both cages and tanks, the between-environment genetic correlations were estimated for body weights at three ages: 180 (W2), 270 (W3), and 360 dph (W4). The genetic correlation between tank and cage culture was high (0.91–0.94) for W2 and W3. The genetic correlation estimate (0.84 ± 0.12) was similar for total length. However, the between-environment genetic correlation decreased as growth progressed (rg = 0.59 ± 0.33 for W4, 360 dph). Our results suggest that the effect of G×E interaction is potentially important for growth traits measured in later stages of development in Asian seabass. This is particularly significant when the commercial production system (e.g., tank) is incompatible with a controlled maintenance of broodstock in the selection nucleus (e.g., cage).
Discussion
Heritability for Survival
The heritability estimate for survival during the grow out phase from 105 days post-hatch (dph) to 570 dph was low (<12%) in this study. This is as expected for binary characters that often require a larger sample size to obtain reliable heritability estimates (Davies et al., 2015; Nguyen et al., 2018b). Further, survival as well as fitness-related traits are known to be influenced by many environmental factors, such as nutrition, feeding regimes, stocking density, or social competition (Thoa et al., 2016). This is also because fitness-related traits exhibit small additive genetic components and thus, heritabilities are low for these characters. Despite the low and non-significance of the heritability estimates, our study showed that there are heritable genetic components determining survival during grow-out. This is supported by the highly significant differences in survival rate among families studied (Figure 1). Our results are consistent with observations in other aquaculture species which show that the heritability for survival traits during grow-out phase was low (0.05–0.17), such as tilapia (Hamzah et al., 2017a), common carp (Dong et al., 2015), rainbow trout (Vehviläinen et al., 2012) and giant freshwater prawn (Vu et al., 2017). Some other studies, however, reported moderate to high heritability (h2 = 0.53) for the survival trait (Ninh et al., 2014). A meta-analysis of 31 studies across aquaculture species (Nguyen, submitted) showed that the weighted mean heritability for survival rate was 0.14 ± 0.03, suggesting that improving this character by selection, although possible, may be slow due to environmental factors.
Heritability for Growth Traits
There is limited information regarding the additive genetic variability for growth trajectory in Asian seabass. In this study, we report, for the first time, the heritability for growth traits at six different time points (105–570 dph) in a selected population of Asian seabass. The moderate to high heritabilities obtained for body weight and length across six growth stages indicate that there is substantial genetic variation in growth related traits (weight and length). This suggests that selection for growth should be effective in this population. By contrast, previous research reported heritability for juvenile fish 90 dph (≈18 g or ≈10 cm in length). The published h2 estimates ranged from 0.12 to 0.24 for weight, length and condition factors (Wang et al., 2008b) or 0.15 to 0.22 in 62 dph fish (Domingos et al., 2013). These h2 estimates generally had high standard errors (0.09 to 0.22) and may have been overestimated because the common full-sib effects were not accounted for in statistical models. The significant heritability values obtained from our study indicate that selection to improve body traits should be effective in this population. The heritability reported for growth traits were moderate to high in other species, such as kingfish (Premachandra et al., 2017), salmon or giant freshwater prawn (Hung and Nguyen, 2014).
Common Full-Sib Effects
The common full-sib effects (c2) observed in the present study was due to separate rearing of each family in different tanks before tagging. The c2 effect accounted for between 3 and 22% of the total variation for weight and length. Our c2 estimate is in good agreement with those reported in other marine (Falica et al., 2017) and freshwater (Hamzah et al., 2017b) fish species. The magnitude of the c2 effect generally diminished with the growth time (Table 3) and the c2 estimate was significantly larger for growth traits during the early phase of growth development than those at harvest (Oliveira et al., 2016). Strategies to reduce the c2 effect in selective breeding programs can include: (i) shortening the spawning and nursing time for full- and half-sib families; (ii) physical tagging of the fingerlings at an early age as possible; or (iii) early communal rearing after birth using molecular techniques for parentage assignment. In breeding programs where these factors can be well managed, the c2 effects were small and there may be little improvement by including them in the evaluation of breeders (Winkelman and Peterson, 1994; Martinez et al., 1999; Pante et al., 2002; Gjerde et al., 2004; Kjøglum et al., 2005). For this barramundi population, inclusion of the c2 effect in statistical models is deemed essential in order to minimize possible biases in genetic parameter estimates and predicted breeding values that may lead to reductions in selection accuracy and hence, a reduction in genetic gain.
Correlations at Different Measurements
Genetic correlations of body weight between different time points are all positive. However, the estimates differed significantly from one, suggesting that body traits at different growth phases in this population are genetically different. Hence, selection in the early stage of growth development may not capture all genetic expression at later stages of growth or at the time of commercial harvest. This contrasts with results reported in other aquaculture species. For instance, Ninh et al. (2013) showed high genetic correlations (0.85–0.99) for body weight at 3, 7, and 12 months of age in common carp. A similar correlation was also found in giant freshwater prawn (Hung et al., 2014).
Consistent with the results reported for other species, the genetic correlations between weight and length measured at the same time/age were high (0.88–0.99) in this population of barramundi. The genetic correlation between weight and length in our study were close to one as were those reported in Atlantic salmon and rainbow trout (Gjerde and Gjedrem, 1984), tilapia (Nguyen et al., 2010), carp (Ninh et al., 2011), or shrimp (Nguyen et al., 2014). This suggests that these growth traits are under control by a similar or same set of genes. Our results indicate that either body weight or length can be used as selection criterion in aquaculture species. However, measurement of length at an early age may not be a good predictor of body weight at other growth periods because in our research the genetic correlations between these traits differed significantly from one.
Genotype by Environment (G×E) Interaction
Multi-variate assessment of genotype by environment (G×E) interaction suggests that the G×E effect was not important in the first year of on-growth; however, it had significant impact on body traits in adult fish. The longer the fish were grown, the stronger the G×E effects had on body traits. Thus, selection of breeding candidates in sea cages may not be effective when the fish are cultured in tanks. However, in a study with barramundi, Domingos et al. (2013) reported close to one genetic correlations (0.87–0.99) for homologous body traits between fresh and sea water (62 dph) and between tank (343 dph) and pond (469 dph) environments. G×E interactions between tank and cage have not been reported for traits of economic importance in adult barramundi and no comparison is possible with the results obtained from the present study. During sea cage culture, a range of environmental factors may be influential including: ambient temperature, water parameters, feeds and feeding, culture system, and management and husbandry practices. For example, Asian seabass brooders raised in a sea cage net had better growth performance than those raised in tanks (Wang et al., 2008b) and those reared in intensive tanks outperformed their family counterparts in a semi-intensive pond (Domingos et al., 2013). Our results also showed that the fish grew faster in sea cages than in tanks.
The G×E interaction effects have been examined in many aquaculture species (Nguyen, 2016; Sae-Lim et al., 2016) and have shown that, when the environments are “similar” (e.g., freshwater pond vs. freshwater cage), the G×E effects may not be important for growth traits. However, when the selection and production environments differed greatly (e.g., sea cages vs. freshwater tanks) the G×E is of biological significance. G×E effects resulted in a reduced genetic gain and lowered genetic parameter estimates in red tilapia (Nguyen et al., 2017). In these cases, the G×E interaction effects must be accounted for in genetic improvement programs or, when the economic losses are greater than the cost of running a new breeding program, separate genetic lines should be developed for each culture environment. Continuing accumulation of the growth data in future generations would enable the better assessment of the G×E effects in this population of barramundi.
Experience and Challenges
Despite some initial achievements in this study (including a new set of genetic parameters and identifying the G×E interaction effects for growth traits), practical implementation of the breeding program for Asian seabass is faced with several challenges. The biggest problem is achieving synchronized spawning of mating pairs in order to produce many families within a reasonable time interval (2–3 weeks). In the population we studied, the fish generally reached maturity after two years of age although at this stage the sex ratio was uneven with a much larger proportion of males (43.1%) than females (8.2%). The proportion of fish whose sex was not identified was 48.7%. Furthermore, females after harvest (average 2.3 kg) were not ready to spawn and the breeding failure rate was high. In the base population (G0), parental fish were injected using LHRHa and kept in tanks to allow natural spawning. With this method, the spawning rate was only 66%. Since G1, artificial insemination (i.e., stripping of eggs and collection of sperm after HCG admission) increased spawning success to 78% although fertilization and hatching rates were low (53%) compared with natural spawning in tanks (66%). Due to these problems, selection had to be made by using lower EBV candidates and thus, affecting genetic parameters and genetic progress achieved for growth traits in this population. In addition, larval and post-larval rearing showed unwanted cannibalism. To overcome this problem, “shooters”/or cannibals (unusual large size fish) were removed from rearing tanks twice a week. Grading was not used to minimize possible bias in genetic parameter estimates. As a result, fingerling weight/size varied greatly among families studied and affected growth performance in both tanks and sea cages. The common full-sib effects were also important for body traits (2–22% of total variance) in this population. Changing environments in some years had noticeable negative impacts on the growth and health of the breeding population as a result of parasitic and bacterial diseases. Main diseases included the protozoal disease Trichodiniasis, fish louse due to a parasitic crustacean, and a bacterial disease Vibrosis and generally caused reductions in growth and maturation rates in on-grow fish and breeding candidates. In 2013, significant loss (70% of parental candidates) occurred when the water temperature dropped below 15°C during the season before winter. Further, operculum deformity (opaque eyes) due to parasites or environmental factors occurred in 60% of the on-grow fish even though a freshwater bath using KMnO4 was applied once a week. Obviously, morphological deformity and parasite diseases (e.g., skin fluke) are crucial to Asian seabass and should be recorded and included in genetic analyses. A fourth problem was that in the first 2 years of the breeding program there were not enough spawning/rearing tanks to accommodate all the families produced for subsequent performance testing in sea cages and tanks. Small size of sea cages also affected growth rate of the experimental fish. Finally, live foods, such as scad or other trash fish, were not always available and this affected the spawning rate and reproductive performance of brood fish. Alternative diets are sought to overcome these problems. As a consequence, the number of families expected to produce in each generation (at least 60) was not always achieved. Mass spawning together with parentage assignment techniques using DNA markers to enable early communal rearing of all families soon after birth may be an option to be considered in the future breeding program for Asian seabass.
Conclusions
There are heritable genetic variations for growth traits that can be exploited by genetic selection in selective breeding programs for barramundi. However, selection for growth in the early stages (e.g., 105, 180, or 270 dph) may not capture all the genetic variation necessary in later rearing periods. This is because genetic correlations of body weight at different ages, although positive and moderate, differed significantly from one. The genetic correlation for body weight expressions between tank and sea cage culture decreased as growth progressed especially after 360 dph, suggesting that the relationship between genotype and environment is important for body traits including weight and length in this population of Asian seabass. In conclusion, selection for growth traits should be made at (or close to) harvest to maximize commercial production. Continuing collection of growth data in different environments is needed to ascertain the G×E interaction effect and if it is significant there may be a call to have separate breeding programs for tank and cage environments. Expansion of the breeding objectives by including new traits, namely improved disease resistance and reduced cannibalism would maximize productivity and revenue for Asian seabass aquaculture.
Author Contributions
PK, TP, ND, WK, and NN conceived and designed the experiments, analyzed the data, and prepared and approved the manuscripts.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was funded by Ministry of Agriculture and Rural Development, Vietnam (project number: KHCN2013). We extend our sincere thanks to Dr. Nguyen Huu Ninh, Director of RIA3 for his support and to staff of RIA3 for their kindly assistance in the management of the animals. Prof. Richard Burns at University of the Sunshine Coast provided valuable comments to improve the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00191/full#supplementary-material
References
Charo-Karisa, H., Komen, H., Rezk, M. A., Ponzoni, R. W., van Arendonk, J. A., and Bovenhuis, H. (2006). Heritability estimates and response to selection for growth of Nile tilapia (Oreochromis niloticus) in low-input earthen ponds. Aquaculture 261, 479–486. doi: 10.1016/j.aquaculture.2006.07.007
Davies, S. W., Scarpino, S. V., Pongwarin, T., Scott, J., and Matz, M. V. (2015). Estimating trait heritability in highly fecund species. G3 Genes Genomes Genet. 5, 2639–2645. doi: 10.1534/g3.115.020701
Dégremont, L., Bédier, E., Soletchnik, P., Ropert, M., Huvet, A., Moal, J., et al. (2005). Relative importance of family, site, and field placement timing on survival, growth, and yield of hatchery-produced Pacific oyster spat (Crassostrea gigas). Aquaculture 249, 213–229. doi: 10.1016/j.aquaculture.2005.03.046
Domingos, J. A., Smith-Keune, C., Robinson, N., Loughnan, S., Harrison, P., and Jerry, D. R. (2013). Heritability of harvest growth traits and genotype–environment interactions in barramundi, Lates calcarifer (Bloch). Aquaculture 402, 66–75. doi: 10.1016/j.aquaculture.2013.03.029
Dong, Z., Nguyen, N. H., and Zhu, W. (2015). Genetic evaluation of a selective breeding program for common carp Cyprinus carpio conducted from 2004 to 2014. BMC Genet. 16:94. doi: 10.1186/s12863-015-0256-2
Dupont-Nivet, M., Karahan-Nomm, B., Vergnet, A., Merdy, O., Haffray, P., Chavanne, H., et al. (2010). Genotype by environment interactions for growth in European seabass (Dicentrarchus labrax) are large when growth rate rather than weight is considered. Aquaculture 306, 365–368. doi: 10.1016/j.aquaculture.2010.05.025
Dupont-Nivet, M., Vandeputte, M., Vergnet, A., Merdy, O., Haffray, P., Chavanne, H., et al. (2008). Heritabilities and GxE interactions for growth in the European sea bass (Dicentrarchus labrax L.) using a marker-based pedigree. Aquaculture 275, 81–87. doi: 10.1016/j.aquaculture.2007.12.032
Embody, G. C., and Hayford, C. O. (1925). The advantage of rearing brook trout fingerlings from selected breeders. Trans. Am. Fish. Soc. 55, 135–148. doi: 10.1577/1548-8659(1925)55[135:TAORBT]2.0.CO;2
Evelyn, G. D. J.-A., Felix, G. A., and Valentin, T. (2013). “Early development and seed production of Asian seabass, Lates calcarifer,” in Biology and Culture of Asian Seabass Lates Calcarifer, ed D. R. Jerry (Boca Raton, FL: CRC Press), 17–30.
Evenhuis, J., Leeds, T., Marancik, D., LaPatra, S., and Wiens, G. (2015). Rainbow trout () resistance to columnaris disease is heritable and favorably correlated with bacterial cold water disease resistance. J. Anim. Sci. 93, 1546–1554. doi: 10.2527/jas.2014-8566
Falica, B. K., Lehnert, S. J., Pitcher, T. E., Heath, D. D., and Higgs, D. M. (2017). Ontogentic shifts in genetic and maternal effects on length and survival in Chinook salmon (Oncorhynchus tshawytscha). Aquaculture 468, 218–225. doi: 10.1016/j.aquaculture.2016.10.003
Frost, L. A., Evans, B. S., and Jerry, D. R. (2006). Loss of genetic diversity due to hatchery culture practices in barramundi (Lates calcarifer). Aquaculture 261, 1056–1064. doi: 10.1016/j.aquaculture.2006.09.004
Gilmour, A. R., Gogel, B., Cullis, B., Thompson, R., and Butler, D. (2009). ASReml User Guide Release 3.0. Hemel Hempstead, UK: VSN International Ltd.
Gjedrem, T. (2012). Genetic improvement for the development of efficient global aquaculture: a personal opinion review. Aquaculture 344, 12–22. doi: 10.1016/j.aquaculture.2012.03.003
Gjedrem, T., Robinson, N., and Rye, M. (2012). The importance of selective breeding in aquaculture to meet future demands for animal protein: a review. Aquaculture 350, 117–129. doi: 10.1016/j.aquaculture.2012.04.008
Gjedrem, T., and Rye, M. (2016). Selection response in fish and shellfish: a review. Rev. Aquaculture 10, 168–179. doi: 10.1111/raq.12154
Gjerde, B., and Gjedrem, T. (1984). Estimates of phenotypic and genetic parameters for carcass traits in Atlantic salmon and rainbow trout. Aquaculture 36, 97–110. doi: 10.1016/0044-8486(84)90057-7
Gjerde, B., Terjesen, B. F., Barr, Y., Lein, I., and Thorland, I. (2004). Genetic variation for juvenile growth and survival in Atlantic cod (Gadus morhua). Aquaculture 236, 167–177. doi: 10.1016/j.aquaculture.2004.03.004
Hamzah, A., Mekkawy, W., Khaw, H. L., Nguyen, N. H., Yee, H. Y., Abu Bakar, K. R., et al. (2017a). Genetic parameters for survival during the grow-out period in the GIFT strain of Nile tilapia (Oreochromis niloticus) and correlated response to selection for harvest weight. Aquaculture Res. 48, 47–55. doi: 10.1111/are.12859
Hamzah, A., Thoa, N. P., and Nguyen, N. H. (2017b). Genetic analysis of a red tilapia (Oreochromis spp.) population undergoing three generations of selection for increased body weight at harvest. J. Appl. Genet. 58, 509–519. doi: 10.1007/s13353-017-0411-8
Henderson, C. R. (1975). Best linear unbiased estimation and prediction under a selection model. Biometrics 31, 423–447. doi: 10.2307/2529430
Hung, D., and Nguyen, H. N. (2014). Genetic inheritance of female and male morphotypes in giant freshwater prawn Macrobrachium rosenbergii. PLoS ONE 9:e90142. doi: 10.1371/journal.pone.0090142
Hung, D., Nguyen, N. H., Hurwood, D. A., and Mather, P. B. (2014). Quantitative genetic parameters for body traits at different ages in a cultured stock of giant freshwater prawn (Macrobrachium rosenbergii) selected for fast growth. Mar. Freshw. Res. 65, 198–205. doi: 10.1071/MF13111
Hung, D., Vu, N. T., Nguyen, N. H., Ponzoni, R. W., Hurwood, D. A., and Mather, P. B. (2013). Genetic response to combined family selection for improved mean harvest weight in giant freshwater prawn (Macrobrachium rosenbergii) in Vietnam. Aquaculture 412, 70–73. doi: 10.1016/j.aquaculture.2013.07.015
In, V. V., Sang, V. V., O'Connor, W., Van, P. T., Dove, M., Knibb, W., et al. (2017). Are strain genetic effect and heterosis expression altered with culture system and rearing environment in the Portuguese oyster (Crassostrea angulata)? Aquac. Res. 48, 4058–4069. doi: 10.1111/are.13227
Kause, A., Ritola, O., Paananen, T., Mäntysaari, E., and Eskelinen, U. (2003). Selection against early maturity in large rainbow trout Oncorhynchus mykiss: the quantitative genetics of sexual dimorphism and genotype-by-environment interactions. Aquaculture 228, 53–68. doi: 10.1016/S0044-8486(03)00244-8
Kjøglum, S., Grimholt, U., and Larsen, S. (2005). Non-MHC genetic and tank effects influence disease challenge tests in Atlantic salmon (Salmo salar). Aquaculture 250, 102–109. doi: 10.1016/j.aquaculture.2005.03.021
Knibb, W., Miller, A., Quinn, J., D'Antignana, T., and Nguyen, N. H. (2016). Comparison of lines shows selection response in kingfish (Seriola lalandi). Aquaculture 452, 318–325. doi: 10.1016/j.aquaculture.2015.11.015
Kolstad, K., Thorland, I., Refstie, T., and Gjerde, B. (2006). Genetic variation and genotype by location interaction in body weight, spinal deformity and sexual maturity in Atlantic cod (Gadus morhua) reared at different locations off Norway. Aquaculture 259, 66–73. doi: 10.1016/j.aquaculture.2005.12.022
Luan, T. D., Olesen, I., Ødegård, J., Kolstad, K., and Dan, N. C. (2008). “Genotype by environment interaction for harvest body weight and survival of Nile tilapia (Oreochromis niloticus) in brackish and fresh water ponds”, in Proceedings from the Eighth International Symposium on Tilapia Aquaculture (Cairo) 231–240.
Lawley, D. (2010). Repositioning Australian Farmed Barramundi: Online Consumer Survey Findings. University of Sunshine Coast, Sunshine Coast, QLd.
Liu, J., Lai, Z., Fu, X., Wu, Y., Bao, X., Hu, Z., et al. (2015). Genetic parameters and selection responses for growth and survival of the small abalone Haliotis diversicolor after four generations of successive selection. Aquaculture 436, 58–64. doi: 10.1016/j.aquaculture.2014.10.046
Loughnan, S. R., Smith-Keune, C., Jerry, D. R., Beheregaray, L. B., and Robinson, N. A. (2015). Genetic diversity and relatedness estimates for captive barramundi (Lates calcarifer, Bloch) broodstock informs efforts to form a base population for selective breeding. Aquaculture Res. 47, 3570–3584. doi: 10.1111/are.12807
Martinez, V. c., Neira, R., and Gall, G. A. E. (1999). Estimation of genetic parameters from pedigreed populations: lessons from analysis of alevin weight in Coho salmon (Oncorhynchus kisutch). Aquaculture 180, 223–236. doi: 10.1016/S0044-8486(99)00203-3
Nguyen, H. N. (2016). Genetic improvement for important farmed aquaculture species with a reference to carp, tilapia and prawns in Asia: achievements, lessons and challenges. Fish Fish. 17, 483–506. doi: 10.1111/faf.12122
Nguyen, N. H., Hamzah, A., and Ngo, T. P. (2017). Effects of genotype by environment interaction on genetic gain and population genetic parameters in Red tilapia (Oreochromis spp). Front. Genet. 8:82. doi: 10.3389/fgene.2017.00082
Nguyen, N. H., Ponzoni, R. W., Abu-Bakar, K. R., Hamzah, A., Khaw, H. L., and Yee, H. Y. (2010). Correlated response in fillet weight and yield to selection for increased harvest weight in genetically improved farmed tilapia (GIFT strain), Oreochromis niloticus. Aquaculture 305, 1–5. doi: 10.1016/j.aquaculture.2010.04.007
Nguyen, N. H., Premachandra, H., Kilian, A., and Knibb, W. (2018a). Genomic prediction using DArT-Seq technology for yellowtail kingfish Seriola lalandi. BMC Genomics 19:107. doi: 10.1186/s12864-018-4493-4.
Nguyen, N. H., Quinn, J., Powell, D., Elizur, A., Thoa, N. P., Nocillado, J., et al. (2014). Heritability for body colour and its genetic association with morphometric traits in Banana shrimp (Fenneropenaeus merguiensis). BMC Genet. 15:132. doi: 10.1186/s12863-014-0132-5
Nguyen, N. H., Fitzgibbon, Q. P., Quinn, J., Smith, G., Battaglene, S., and Knibb, W. (2018b). Can metamorphosis survival during larval development in spiny lobster Sagmariasus verreauxi be improved through quantitative genetic inheritance? BMC Genet. 19:27. doi: 10.1186/s12863-018-0621-z
Nguyen, N., Whatmore, P., Miller, A., and Knibb, W. (2016). Quantitative genetic properties of four measures of deformity in yellowtail kingfish Seriola lalandi Valenciennes, 1833. J. Fish Dis. 39, 217–228. doi: 10.1111/jfd.12348
Ninh, N. H., Ponzoni, R. W., Nguyen, N. H., Woolliams, J. A., Taggart, J. B., McAndrew, B. J., et al. (2011). A comparison of communal and separate rearing of families in selective breeding of common carp (Cyprinus carpio): estimation of genetic parameters. Aquaculture 322, 39–46. doi: 10.1016/j.aquaculture.2011.09.031
Ninh, N. H., Ponzoni, R. W., Nguyen, N. H., Woolliams, J. A., Taggart, J. B., McAndrew, B. J., et al. (2013). A comparison of communal and separate rearing of families in selective breeding of common carp (Cyprinus carpio): responses to selection. Aquaculture 408–409, 152–159. doi: 10.1016/j.aquaculture.2013.06.005
Ninh, N. H., Thoa, N. P., Knibb, W., and Nguyen, N. H. (2014). Selection for enhanced growth performance of Nile tilapia (Oreochromis niloticus) in brackish water (15–20ppt) in Vietnam. Aquaculture 428, 1–6. doi: 10.1016/j.aquaculture.2014.02.024
Oliveira, C. A. L., Ribeiro, R. P., Yoshida, G. M., Kunita, N. M., Rizzato, G. S., Oliveira, S. N., et al. (2016). Correlated changes in body shape after five generations of selection to improve growth rate in a breeding program for Nile tilapia Oreochromis niloticus in Brazil. J. Appl. Genet. 57, 487–493. doi: 10.1007/s13353-016-0338-5
Pante, M. J. R., Gjerde, B., McMillan, I., and Misztal, I. (2002). Estimation of additive and dominance genetic variances for body weight at harvest in rainbow trout, Oncorhynchus mykiss. Aquaculture 204, 383–392. doi: 10.1016/S0044-8486(01)00825-0
Phuong, T. H., Chi, L. V., and Nguyen, V. Q. (2013). Evaluating genetic variations of Asian seabass (Lates calcarifer) populations by using microsatellites. Sci. Technol. J. Agric. Rural Dev. (In Vietnamese) 3&4, 170–175.
Premachandra, H. K. A., Nguyen, N. H., Miller, A., D'Antignana, T., and Knibb, W. (2017). Genetic parameter estimates for growth and non-growth traits and comparison of growth performance in sea cages vs land tanks for yellowtail kingfish Seriola lalandi. Aquaculture 479, 169–175. doi: 10.1016/j.aquaculture.2017.05.043
Robertson, A., and Lerner, I. M. (1949). The heritability of all-or-none traits; viability of poultry. Genetics 34, 395–411.
Robinson, N. A., Schipp, G., Bosmans, J., and Jerry, D. R. (2010). Modelling selective breeding in protandrous, batch-reared Asian sea bass (Lates calcarifer, Bloch) using walkback selection. Aquac. Res. 41, e643–e655. doi: 10.1111/j.1365-2109.2010.02584.x
Rye, M., Lillevik, K. M., and Gjerde, B. (1990). Survival in early life of Atlantic salmon and rainbow trout: estimates of heritabilities and genetic correlations. Aquaculture 89, 209–216. doi: 10.1016/0044-8486(90)90126-8
Sae-Lim, P., Gjerde, B., Nielsen, H. M., Mulder, H., and Kause, A. (2016). A review of genotype-by-environment interaction and micro-environmental sensitivity in aquaculture species. Rev. Aquaculture 8, 369–393. doi: 10.1111/raq.12098
Standal, M., and Gjerde, B. (1987). Genetic variation in survival of Atlantic salmon during the sea-rearing period. Aquaculture 66, 197–207. doi: 10.1016/0044-8486(87)90106-2
Thoa, N. P., Knibb, W., Ninh, N. H., Van Dai, N., Nhat, P. H., and Nguyen, N. H. (2015). Genetic variation in survival of tilapia ( Oreochromis niloticus, Linnaeus, 1758) fry during the early phase of rearing in brackish water environment (5–10ppt). Aquaculture 442, 112–118. doi: 10.1016/j.aquaculture.2015.02.040
Thoa, N. P., Ninh, N. H., Knibb, W., and Nguyen, N. H. (2016). Does selection in a challenging environment produce Nile tilapia genotypes that can thrive in a range of production systems? Sci. Rep. 6:21486. doi: 10.1038/srep21486
Thodesen, J., Rye, M., Wang, Y.-X., Li, S.-J., Bentsen, H. B., and Gjedrem, T. (2013). Genetic improvement of tilapias in China: genetic parameters and selection responses in growth, pond survival and cold-water tolerance of blue tilapia (Oreochromis aureus) after four generations of multi-trait selection. Aquaculture 396, 32–42. doi: 10.1016/j.aquaculture.2013.02.010
Trọng, T. Q., Mulder, H. A., van Arendonk, J. A., and Komen, H. (2013). Heritability and genotype by environment interaction estimates for harvest weight, growth rate, and shape of Nile tilapia (Oreochromis niloticus) grown in river cage and VAC in Vietnam. Aquaculture 384, 119–127. doi: 10.1016/j.aquaculture.2012.12.022
Vandeputte, M., Kocour, M., Mauger, S., Dupont-Nivet, M., De Guerry, D., Rodina, M., et al. (2004). Heritability estimates for growth-related traits using microsatellite parentage assignment in juvenile common carp (Cyprinus carpio L.). Aquaculture 235, 223–236. doi: 10.1016/j.aquaculture.2003.12.019
Vehviläinen, H., Kause, A., Koskinen, H., and Paananen, T. (2010). Genetic architecture of rainbow trout survival from egg to adult. Genet. Res. 92, 1–11. doi: 10.1017/S0016672310000017
Vehviläinen, H., Kause, A., Kuukka-Anttila, H., Koskinen, H., and Paananen, T. (2012). Untangling the positive genetic correlation between rainbow trout growth and survival. Evol. Appl. 5, 732–745. doi: 10.1111/j.1752-4571.2012.00251.x
Vu, N. T., Trong, T. Q., and Nguyen, N. H. (2017). Effects of selection for fast growth on survival rate during grow-out phase in giant freshwater prawn (Macrobrachium rosenbergii). BMC Genet. 18:56. doi: 10.1186/s12863-017-0521-7
Wang, C., Lo, L., Feng, F., Zhu, Z., and Yue, G. (2008a). Identification and verification of QTL associated with growth traits in two genetic backgrounds of Barramundi (Lates calcarifer). Anim. Genet. 39, 34–39. doi: 10.1111/j.1365-2052.2007.01672.x
Wang, C. M., Lo, L. C., Zhu, Z. Y., Lin, G., Feng, F., Li, J., et al. (2008b). Estimating reproductive success of brooders and heritability of growth traits in Asian sea bass (Lates calcarifer) using microsatellites. Aquaculture Res. 39, 1612–1619. doi: 10.1111/j.1365-2109.2008.02034.x
Winkelman, A., and Peterson, R. (1994). Heritabilities, dominance variation, common environmental effects and genotype by environment interactions for weight and length in chinook salmon. Aquaculture 125, 17–30. doi: 10.1016/0044-8486(94)90278-X
Keywords: genetic improvement, selection, Barramundi, heritability, correlation and genotype by environment interaction
Citation: Khang PV, Phuong TH, Dat NK, Knibb W and Nguyen NH (2018) An 8-Year Breeding Program for Asian Seabass Lates calcarifer: Genetic Evaluation, Experiences, and Challenges. Front. Genet. 9:191. doi: 10.3389/fgene.2018.00191
Received: 19 March 2018; Accepted: 11 May 2018;
Published: 29 May 2018.
Edited by:
Peng Xu, Xiamen University, ChinaReviewed by:
Jesús Fernández, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), SpainJunhong Xia, Sun Yat-sen University, China
Copyright © 2018 Khang, Phuong, Dat, Knibb and Nguyen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pham Van Khang, S2hhbmcuUGhhbUByZXNlYXJjaC51c2MuZWR1LmF1
Nguyen Hong Nguyen, bm5ndXllbkB1c2MuZWR1LmF1
 Pham Van Khang
Pham Van Khang Truong Ha Phuong3
Truong Ha Phuong3 Nguyen Hong Nguyen
Nguyen Hong Nguyen