- Key Lab of Plant Biotechnology in Universities of Shandong Province, College of Life Sciences, Qingdao Agricultural University, Qingdao, China
Heat shock transcription factors (Hsfs) are essential elements in plant signal transduction pathways that mediate gene expression in response to various abiotic stresses. Mungbean (Vigna radiata) is an important crop worldwide. The emergence of a genome database now allows for functional analysis of mungbean genes. In this study, we dissect the mungbean Hsfs using genome-wide identification and expression profiles. We characterized a total of 24 VrHsf genes and classified them into three groups (A, B, and C) based on their phylogeny and conserved domain structures. All VrHsf genes exhibit highly conserved exon-intron organization, with two exons and one intron. In addition, all VrHsf proteins contain 16 distinct motifs. Chromosome location analysis revealed that VrHsf genes are located on 8 of the 11 mungbean chromosomes, and that seven duplicated gene pairs had formed among them. Moreover, transcription patterns of VrHsf genes varied in different tissues, indicating their different roles in plant growth and development. We identified multiple stress related cis-elements in VrHsf promoter regions 2 kb upstream of the translation initiation codons, and the expression of most VrHsf genes was altered under different stress conditions, suggesting their potential functions in stress resistance pathways. These molecular characterization and expression profile analyses of VrHsf genes provide essential information for further function investigation.
Introduction
Plants often suffer from various abiotic stresses throughout their life cycles. Therefore, they have evolved complex defense mechanisms, such as morphological modulation and transcriptome adjustment, to protect themselves from adverse conditions (Blande et al., 2014; Zeng et al., 2016; Abdelrahman et al., 2018; Zandalinas et al., 2018). Likewise, we can alter gene expression to improve plant tolerance to different stress conditions (von Koskull Döring et al., 2007; Qu et al., 2013; Li S. et al., 2016; Ohama et al., 2017). Recently, numerous regulatory components in plants, such as transcription factors, have been identified to participate in multiple signal transduction pathways in response to various environmental stresses. Under unfavorable conditions, transcription factors are regulated to activate or suppress downstream target genes to sustain plant survival (Mittler et al., 2012; Xu et al., 2017; Zupin et al., 2017; Abdelrahman et al., 2018).
Heat shock transcription factors (Hsfs) are important regulatory elements in plants that play critical roles in signal transduction to mediate gene expression in response to multiple abiotic stresses, including cold, drought, salt, and heat stresses (Collins et al., 1995; Nover et al., 2001; Guo et al., 2016; Jacob et al., 2017). Hsf genes regulate the transcription of the molecular chaperones, heat shock proteins (Hsps), by recognizing heat shock elements (HSEs) within their promoter regions. Activation of Hsps protects cells from impairment under stress conditions (Akerfelt et al., 2010; Scharf et al., 2012; Guo et al., 2016). Hsf gene activity can be regulated by different conditions. Hsf proteins form cytoplasmic complexes with Hsp90/Hsp70 chaperones to maintain their inactive states under non-stress conditions, whereas under stress conditions, Hsf proteins are released and modified, allowing them to bind their target genes (Nover et al., 2001; Akerfelt et al., 2010; Scharf et al., 2012; Westerheide et al., 2012).
Recently, many Hsf gene families have been identified and analyzed in more than 20 plant species at a genome-wide scale (Nover et al., 2001; Fujimoto and Nakai, 2010; Lin et al., 2011; Giorno et al., 2012; Scharf et al., 2012; Guo et al., 2016; Wei et al., 2016). For example, there are 21 Hsf genes in Arabidopsis, 25 in rice, 24 in tomato, 52 in soybean, 40 in cotton, and 27 in potato (Li et al., 2014; Wang et al., 2014; Guo et al., 2016; Tang et al., 2016). Similar to other transcription factor families, plant Hsf proteins share a well conserved modular structure, such as a DNA binding domain (DBD) and hydrophobic heptad repeats (HR-A/B) (Baniwal et al., 2004). The Hsf N-terminal DBD is characterized by a conserved helix-turn-helix motif, containing one 3-helical bundle (α1, α2, and α3) and one 4-stranded β-sheet (β1, β2, β3, and β4). The DBD allows Hsf proteins to recognize HSEs in target promoter regions to regulate the downstream genes (Harrison et al., 1994; Scharf et al., 2012; Guo et al., 2016). The HR-A/B, which is also known as the oligomerization domain (OD), is connected to the DBD by a flexible peptide chain. This HR-A/B domain enables Hsf proteins to form homologous trimmers to efficiently bind Hsp promoters (Scharf et al., 2012; Guo et al., 2016). Moreover, some Hsf protein functional domains also consist of nuclear localization signal (NLS), nuclear export signal (NES), and transcriptional activation (AHA) motifs. The NLS motif, which is formed by a cluster of lysine and arginine residues, and the NES, which contains many leucine residues, are close to the C-terminus in some sub-classes of Hsf proteins. In the AHA motif, the “A,” “H,” and “A” represent different kinds of amino acids. Specifically, the first “A” indicates W, F, or Y, “H” indicates L, I, V, or M, and the last “A” represents D or E (Scharf et al., 2012; Liu et al., 2016; Guo et al., 2016; Jacob et al., 2017). Based on the characteristics of their flexible peptide chain and HR-A/B regions, Hsf proteins are generally classified into three groups, HsfA, HsfB, and HsfC (Scharf et al., 2012; Guo et al., 2016).
Mungbean is one of the most important crops and is commonly consumed by humans in many countries (Tang et al., 2014; Ganesan and Xu, 2017). Mungbean seeds and sprouts contain bioactive food compounds and abundant nutrients, and have potential health benefits for humans (Tang et al., 2014; Ganesan and Xu, 2017). However, sustainable mungbean production is challenged by various stresses, such as high temperature, cold, drought etc. (Blande et al., 2014; Fragkostefanakis et al., 2015). Therefore, it is very important to characterize mungbean stress resistant genes to allow for their modification for enhanced crop adaptability. In the current study, we report the molecular identification and characterization of VrHsf genes in mungbean, using a combination of approaches, including sequence alignment, evolutionary relationship investigation, gene duplication and motif analysis. Moreover, we investigate the expression patterns of VrHsf genes in various mungbean tissues, as well as their expression profiles under different stress treatments. Our findings provide a foundation for an improved understanding of the VrHsf gene family in mungbean, and will be useful for further characterization of VrHsf gene function.
Materials and Methods
Plant Materials and Growth Conditions
The sequenced mungbean genotype VC1973A (named Zhonglu in China) was used for all experiments (Kang et al., 2014). Mungbean seeds were germinated and grown in pots in a growth room, with 16 h 28°C light/8 h 22°C dark cycles. Humidity was maintained at approximately 30%.
Two weeks old mungbean seedlings were used for stress treatments. Cold stress treatment was performed by transferring plants to a 4°C chamber for 6 h. Heat stress was carried out by putting plants in a 40°C chamber for 6 h. For the drought stress treatment, plants were grown without watering for 6 days. For the salt stress treatment, plants were watered with 200 mmol salt solution, and tissues were collected 48 h after treatment. Shoots and roots were collected separately after stress treatments and then stored at -80°C before RNA extraction. Moreover, various tissues, such as roots, stems, leaves, flowers, pods and seeds, were also sampled for gene expression analysis. Roots, stems and leaves were collected from 3 weeks old seedlings, and flowers were sampled from 6 weeks old plants. The pods were sampled for analysis at the beginning of the pod stage. The seeds, at the full seed stage were used for gene expression analysis. Each sample was analyzed using three biological replicates.
Identification of VrHsf Members
The conserved amino acid sequence of DNA-binding domains (DBD, PF00447) was download from the protein family database Pfam1, and full-length amino acid sequences of Hsf proteins from Arabidopsis, soybean and potato were used as BLAST queries against the mungbean database2 and National Center for Biotechnology Information (NCBI). All output genes with default (Limit Expect Value 1e-5) were analyzed using the Pfam database (E = 1.0) and SMART3 to remove genes without conserved domain sequences. The classification of VrHsf proteins was performed using Heatster4. The theoretical iso-electric points, grand average of hydropathicity and protein molecular weight analyses were performed using the ProtParam tool5.
Phylogenetic Relationship Analysis
In total, 124 Hsf amino acid sequences from mungbean, soybean, Arabidopsis and potato were used for phylogenetic analysis (Nover et al., 2001; Chung et al., 2013; Tang et al., 2016). The N-proximal regions of Hsf proteins, from the start of the conserved DBD domain to the end of the HR-A/B region, were aligned using Clustal-X2. The alignment result was used to construct a phylogenetic tree using MEGA 6.0’s Neighbor-Joining method with pairwise deletion, 1000 bootstraps and a Poisson model (Tamura et al., 2013).
Analyses of VrHsf Gene Structures, Protein Domains, Conserved Motifs, and cis-Elements
The CDS and genomic DNA sequences of mungbean Hsf genes were aligned with the Gene Structure Display Server program (GSDS)6 to illustrate exon/intron organization (Hu et al., 2015). VrHsf conserved domains were obtained using Heatster7, Pfam and SMART. The conserved motifs of VrHsf proteins were assessed via MEME tools8, and the parameters of the maximum number of motifs and the optimum motif widths were 16 and 6–50 amino acid residues, respectively. Promoter cis-elements were analyzed using the Plant cis-acting regulatory DNA elements (PLACE) database (Higo et al., 1999) and the distribution maps were constructed using iBS9.
Chromosomal Distribution and Duplication Analysis of VrHsf Genes
The MapInspect software10 was used for mapping VrHsf genes to mungbean chromosomes. Duplications of Hsf genes in mungbean were analyzed and marked as previously described (Zhang et al., 2015; Tang et al., 2016). The divergence time (T) of the duplicated genes was calculated as described (Lynch and Conery, 2000; Chen et al., 2014).
RNA Extraction and Quantitative Real-Time PCR Analysis
All mungbean RNA was extracted using Qiagen RNeasy mini kit following the instructions (Qiagen, United States)11. cDNA synthesis was conducted as previously described (Li et al., 2017). SuperScript II reverse transcriptase first-strand synthesis kit (Invitrogen) was used for the synthesis of the first strand cDNAs with 2 μg total RNA according to the instructions. The LightCycler 480 SYBR Green I Master Kit (Roche Diagnostics) was used for quantitative real-time PCR (qRT-PCR) using a LightCycler480 machine (Roche Diagnostics), according to the manufacturer’s instructions. The amplification program for qRT-PCR was performed as previously described (Li et al., 2017). For gene expression analysis, three biological replicates were used for each sample and gene expression was normalized to an Actin-expressing gene in mungbean (Vradi03g00210). All primers used for qRT-PCR analyses are listed in Supplementary Table 1.
Statistical Analysis
Statistical significance was analyzed by t-test using the SAS program (SAS Institute Inc.)12.
Results
Identification of VrHsf Genes in Mungbean
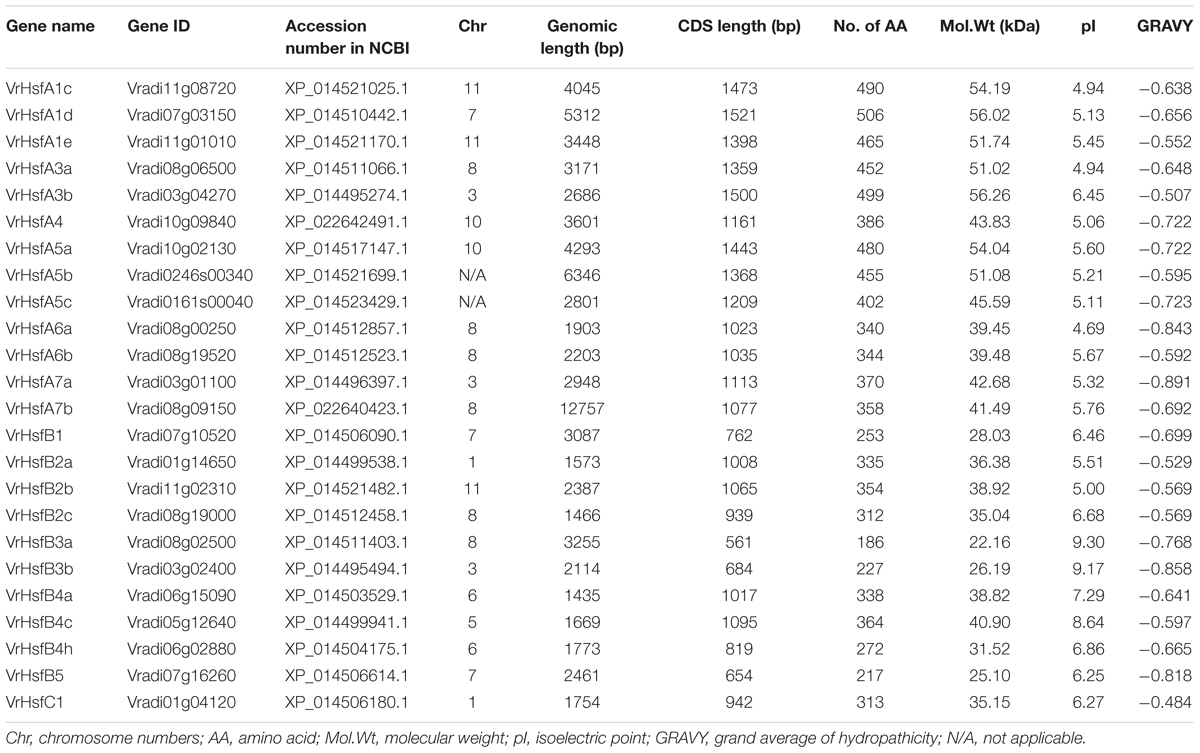
We used the conserved DBD amino acid sequence (PF00447) and the full-length amino acid sequences of Hsf proteins from Arabidopsis, soybean and potato as BLAST queries against the mungbean genome database and NCBI GenBank resources (Kang et al., 2014). We ultimately identified 24 VrHsf candidate genes in the mungbean genome with complete DBD and HR-A/B regions (Supplementary Figures 1, 2) and analyzed their genomic length, CDS length, number of amino acids, theoretical molecular weight, grand average of hydropathicity, and isoelectric point (Table 1). The genomic lengths of the VrHsf genes ranged from 1,435 bp (Vradi06g15090) to 12,757 bp (Vradi08g09150), the coding sequence sizes varied from 561 bp (Vradi08g02500) to 1,521 bp (Vradi07g03150), and the deduced number of amino acids ranged from 186 to 506. The grand averages of hydropathicity fell between -0.484 and -0.891, indicating that all the VrHsfs were predicted to be hydrophilic proteins. In addition, the predicted molecular weights ranged from 22.16 to 56.26 kDa, and the isoelectric points ranged from 4.69 to 9.30, indicating the structural diversity and functional variation of VrHsfs (Table 1).
Classification and Phylogenetic Analysis of VrHsf Genes
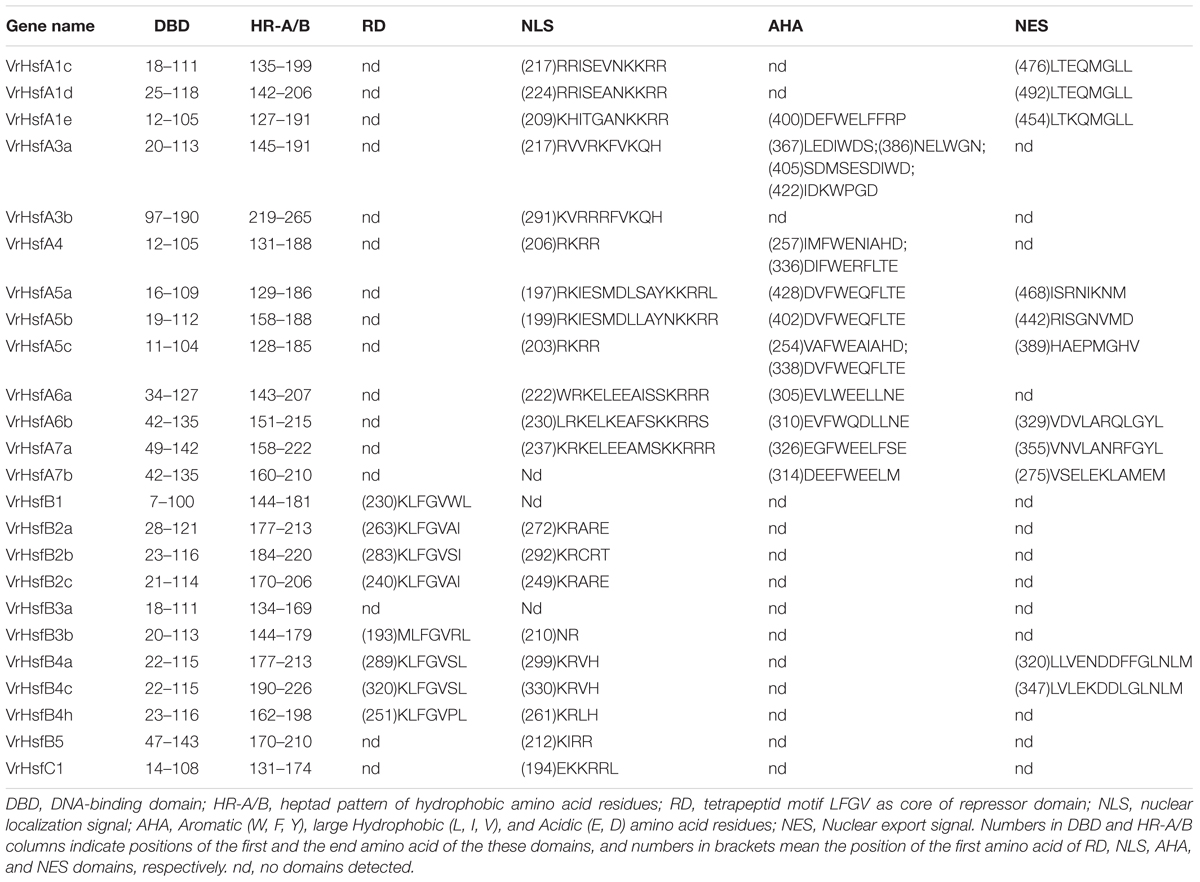
The VrHsf proteins are classified into three groups (A, B, and C) based on differences in the numbers of amino acids inserted between the HR-A and HR-B cores (Guo et al., 2016). Class A has a 21 amino acid insertion between the HR-A and HR-B regions, and Class C has a 7 amino acid insertion, whereas Class B Hsfs are more compact (Scharf et al., 2012). We aligned HR-A/B regions and used Heatster to classify VrHsfs into three groups. Among the 24 VrHsf members, 13 VrHsf proteins were grouped into class A, 10 VrHsf members belonged to class B and only 1 member was classified as class C (Table 1 and Supplementary Figure 2). We constructed a phylogenetic tree based on 124 Hsf amino acid sequences, including 24 mungbean Hsfs, 52 soybean Hsfs (Scharf et al., 2012), 21 Arabidopsis Hsfs (Scharf et al., 2012), and 27 potato Hsfs (Tang et al., 2016), using the conserved regions from the start of the conserved DBD domain to the end of the HR-A/B domain (Figure 1 and Supplementary Table 2) to investigate the evolutionary relationships among Hsf families and to gain insight into the potential function of VrHsfs from the well-studied Hsf families in other species. VrHsfs in class A were grouped into 6 distinct sub-classes (A1, A3–A7), and no VrHsf members were classified as A2, A8, or A9. VrHsfs in class B were sub-classified into five groups (B1-B5) (Table 1, Figure 1, and Supplementary Table 2). We also created a phylogenetic tree using only VrHsf N-proximal regions (Figure 2A). The class A members formed a single group in comparison with class B, which was divided into 3 sub-groups. VrHsfB1 sub-subclass had a distinct relationship with other VrHsfB sub-families, and VrHsfB5 had a much closer relationship with class A and class C families (Figure 2A).
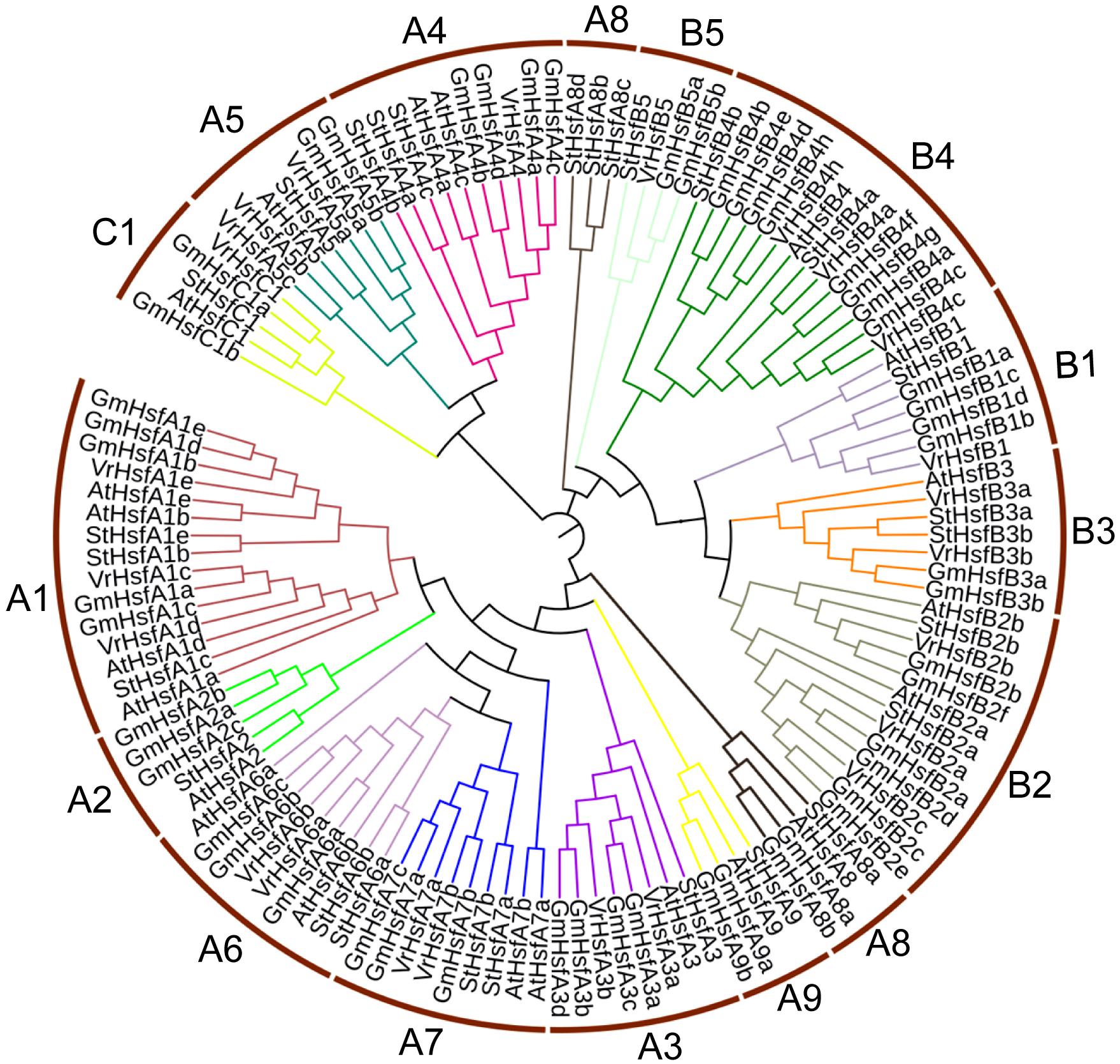
Figure 1. Evolutionary relationship analysis of VrHsf proteins. The N-proximal regions (from the start of the conserved DBD domain to the end of the HR-A/B region) of Hsf proteins from mungbean, soybean, Arabidopsis and potato were used to construct the phylogenetic tree using MEGA 6.0 with the Neighbor-Joining method.
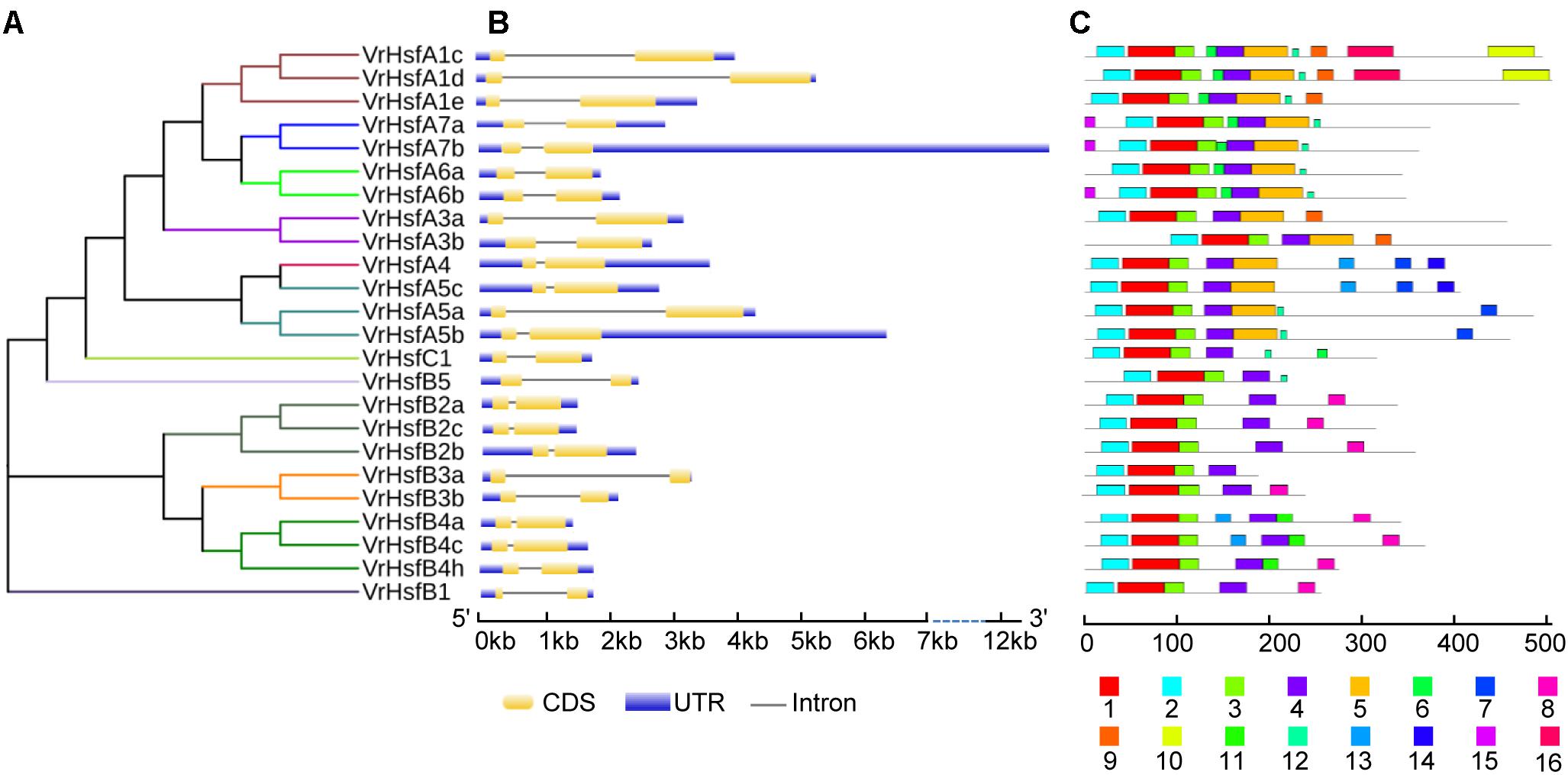
Figure 2. Phylogenetic and structure analysis of VrHsf genes. (A) The conserved domains of VrHsf proteins were used to construct the phylogenetic tree. (B) The exon/intron distribution of VrHsf genes were analyzed by comparing coding sequences with genomic sequences. The UTRs, exons and introns are represented by blue, yellow and black lines, respectively. (C) Motifs in VrHsf proteins identified by MEME. The motifs, numbered 1–16, are exhibited in different colored boxes.
Exon-Intron Organization and Conserved Motifs of VrHsf Genes
We constructed the exon-intron organization of the 24 VrHsf genes using the genomic and coding sequences. The gene structures showed that all the VrHsf genes exhibited a highly conserved exon-intron organization, with two exons and one intron (Figure 2B). The intron lengths varied, similar to Arabidopsis (Nover et al., 2001). We used MEME to predict putative motifs to further reveal the conservation and diversity of VrHsf genes (Figures 2C, 3 and Table 2), and we identified 16 distinct motifs among mungbean Hsf proteins (Figure 3). All the VrHsf members contain motifs 1, 2, 3, and 4. Motif 1 is closely connected with motifs 2 and 3. Combined, motifs 1, 2, and 3 represent the most conservative domain, the DBD domain. We considered motifs 4 and 5 the HR-A/B regions (Figure 2C). Some motifs were only found in specific VrHsf proteins, for example, motif 5 is specific to the class A family, and most class B members have motif 8, except for VrHsfB5 and VrHsfB3a (Figure 2C). Moreover, we identified NLS motifs in 21 VrHsf proteins and NES motifs in 9 class A and 2 class B VrHsfs. The class A Hsfs have short activator peptide motifs (AHA) and class B Hsfs contains the repressor domain tetrapeptide (Scharf et al., 2012). We detected AHA motifs, which may work as transcriptional activators, in 10 class A VrHsfs, while VrHsfA1c, VrHsfA1d, and VrHsfA3b have no AHA domains (Table 2). Eight of the 10 class B VrHsf genes contain the tetrapeptide LFGV, the RD domain (Table 2), and therefore may function as repressors in stress response pathways (Scharf et al., 2012). In addition, VrHsfC1 has no RD or AHA domains. In sum, these observations indicate the functional divergence among VrHsf genes.

Figure 3. Sequence logos of 16 motifs in VrHsf proteins. The “sites” indicate the number of VrHsf proteins containing each motif. The “width” indicates the amino acid number of each motif.
Chromosomal Location and Duplication Analysis of VrHsf Genes
The VrHsf gene family members have some common functions, and some members may have evolved new functions related to the original genes after gene duplication. The chromosomal locations of VrHsf genes can represent gene distributions after duplication. To map the locations of VrHsf genes on different chromosomes, we determined the distribution of VrHsf genes based on the mungbean genome database. Two candidate genes, VrHsfA5b and VrHsfA5c, were discarded due to the lack of chromosome information (Table 1). VrHsf genes were located on 8 of the 11 mungbean chromosomes, with no VrHsfs found on chromosomes 2, 4, or 9 (Figure 4). Chromosome 8 contained the most VrHsf genes, with four class A and two class B genes (VrHsfA3a, VrHsfA6a, VrHsfA6b, VrHsfA7b, VrHsfB2c, and VrHsfB3a), followed by chromosomes 3, 7, and 11, with three genes each (Figure 4). To investigate the duplication events, which may have occurred during mungbean genome evolution, we used the MpInspect software to analyze mungbean Hsf genes. The analysis identified six interchromosomal duplications and one intrachromosomal duplication (Figure 4). Class A proteins contained four duplication events, class B contained three duplication events and class C had no duplicated genes, and these gene duplications indicated similar functions for the duplicated gene pairs. In addition, chromosome 8 contained the most duplicated gene pairs, with five duplication events (VrHsfB2a/VrHsfB2c, VrHsfB3a/VrHsfB3b, VrHsfA3a/VrHsfA3b, VrHsfA6a/VrHsfA6b, and VrHsfA7a/VrHsfA7b) and all VrHsf members located on chromosome 8 had duplicated genes, indicating that chromosome 8 contained the original genes of many duplicated VrHsf genes. In contrast, genes on chromosome 10 had no duplication events (Figure 4).
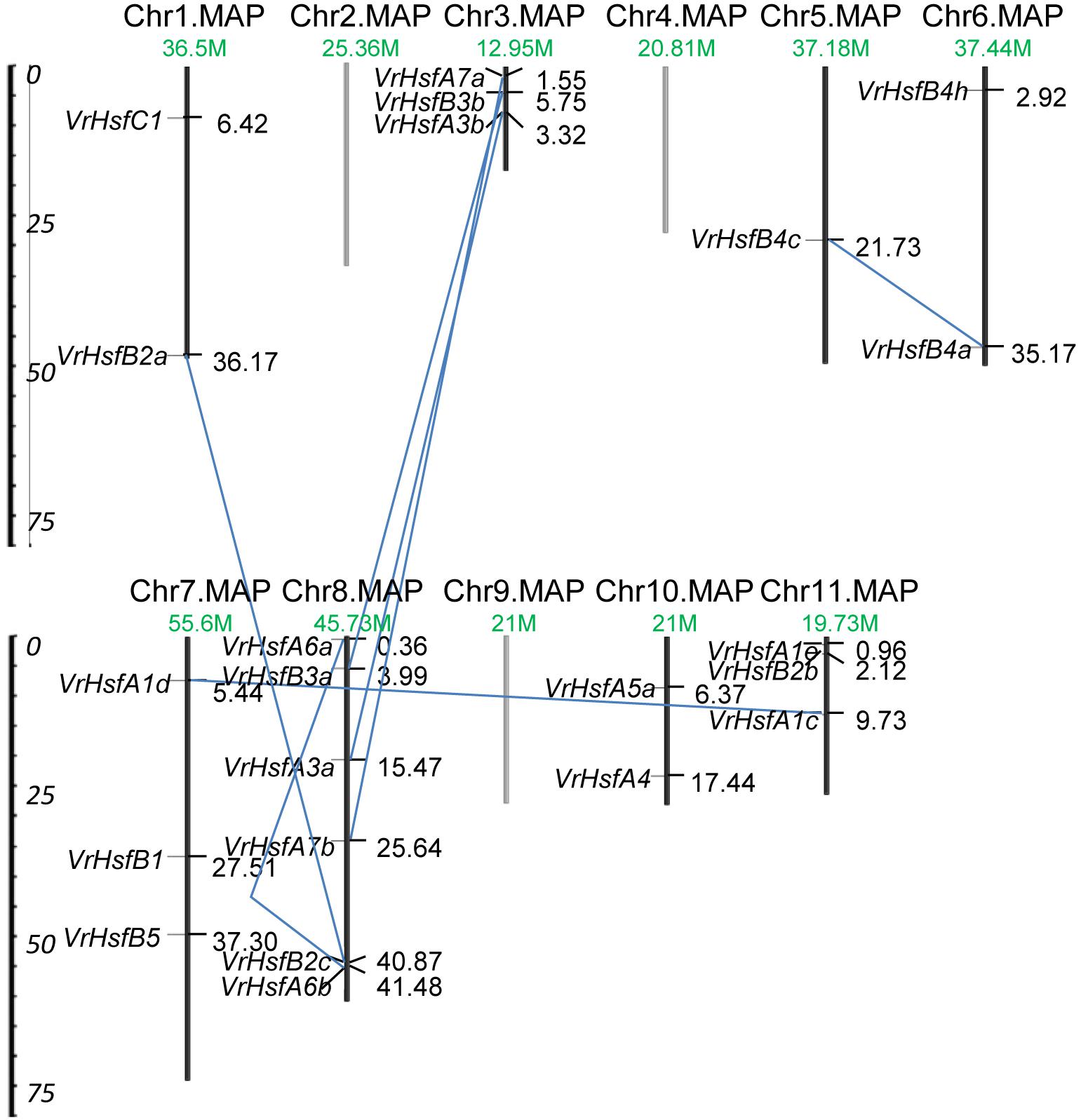
Figure 4. Chromosomal location and duplication analysis of VrHsf genes. Chromosome number and length are represented. The positions for each VrHsf gene are shown on the chromosome. The blue lines indicate segmental duplications.
Promoter Structures of VrHsf Genes
We next investigated putative cis-elements in the promoter region 2 kb upstream of the translation initiation codons of VrHsf members. We obtained many cis-elements in the promoter regions including five known abiotic stress response elements (Figure 5). The stress related elements, Heat Stress Element (HSE), Low Temperature Responsive Element (LTRE), Dehydration-Responsive Element (DRE), C-Repeat Binding Factor (CBF), and ABA Responsive Element (ABRE) were characterized in VrHsf promoter regions (Figure 5). Distribution analysis of these cis-elements showed that all the VrHsf promoters contain HSE, CBF, ABRE, and DRE elements, and 17 of the 24 VrHsf promoter regions contain LTRE elements. All genes had multiple DREs and ABREs, pointing to their key roles in response to drought stress and ABA response pathways (Figure 5). These observations of the cis-elements in promoter regions imply that the VrHsf genes exhibit functional diversity and might be responsive to many different abiotic stresses.
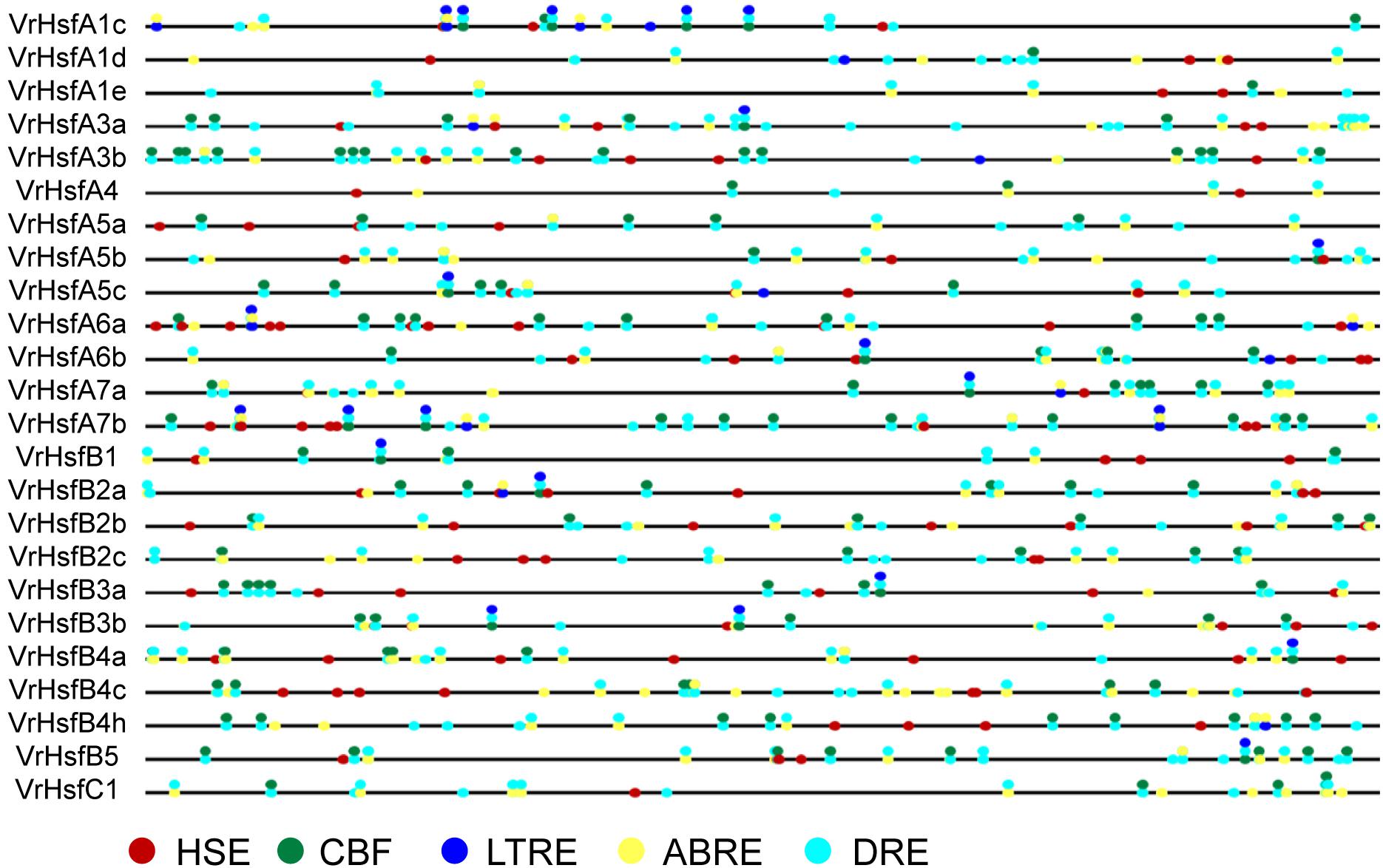
Figure 5. Cis-element analysis of VrHsf promoters 2 kb upstream of the translation initiation codons. Different colored circles indicate different response elements. HSE, Heat Stress Element; CBF, C-Repeat Binding Factor; ABRE, ABA Responsive Element; DRE, Dehydration-Responsive Element; LTRE, Low Temperature Responsive Element.
VrHsf Gene Expression Analysis in Multiple Tissues
To address the potential functions of VrHsf genes in different tissues, we used qRT-PCR to analyze their transcription patterns in various tissues, including the root, stem, leaf, flower, pod and seed (Figure 6). The expression patterns of each VrHsf gene varied in different tissues, indicating their potential functions in these tissues. Among the class A family, VrHsfA1c, VrHsfA1d, and VrHsfA5a were expressed at relatively high levels in all tissues, while VrHsfA4 was expressed at low levels (Figure 6). Moreover, VrHsfA1c, VrHsfA1d, and VrHsfA5a were more highly expressed in the root than in other tissues, indicating their critical roles in roots. Among the VrHsfB family, VrHsfB2a, VrHsfB2b, and VrHsfB4c were also expressed at high abundances in all tissues. In contrast, VrHsfB3a, VrHsfB3b, and VrHsfB5 were expressed at low levels, and were not expressed in the pod, seed, and leaf (Figure 6). In addition, the VrHsfC1 gene showed constitutively low expression in all tissues (Figure 6).
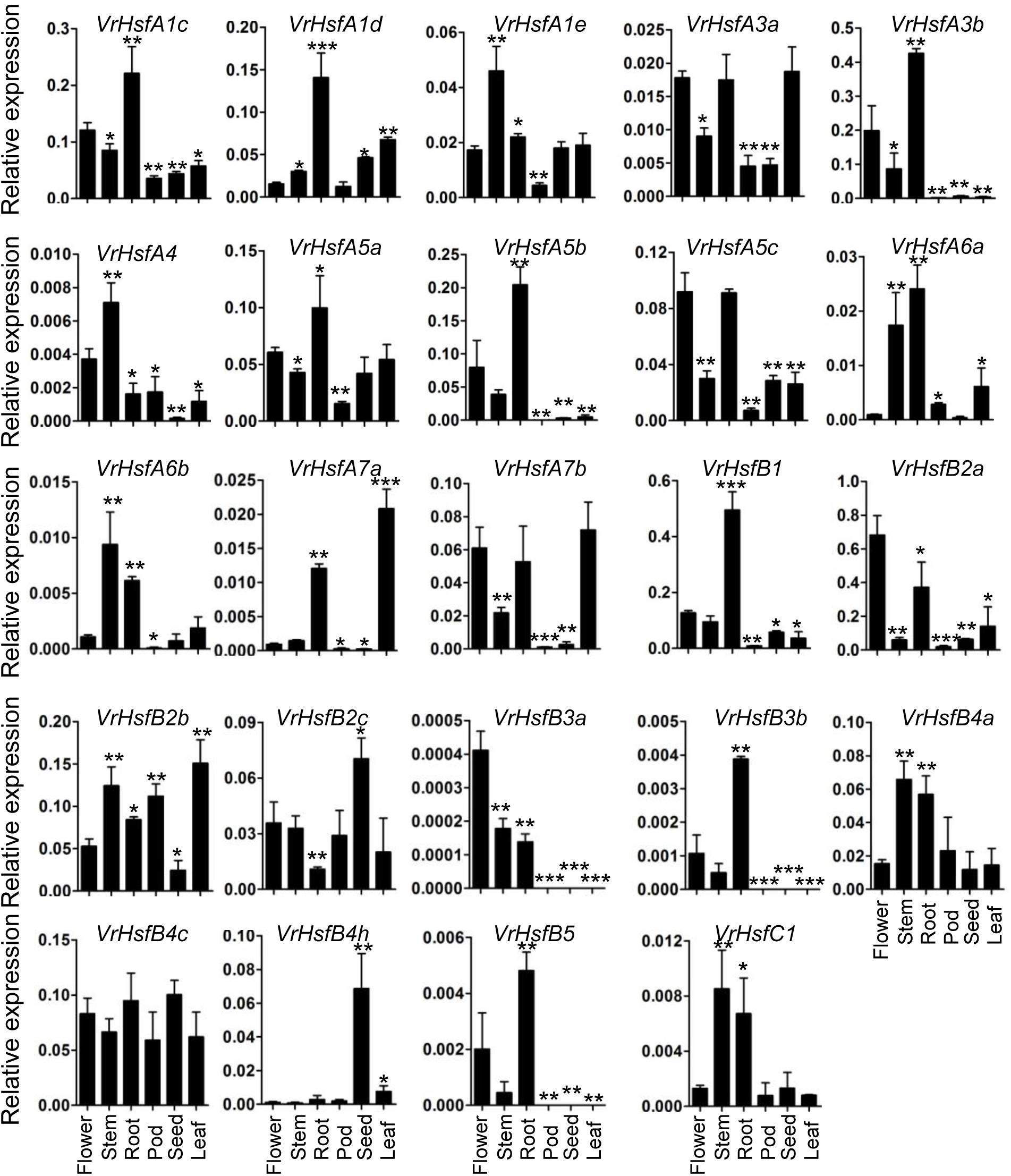
Figure 6. Expression analysis of VrHsf genes in different tissues. Tissues, such as flower, stem, root, leaf pod and seed were used for analysis. Each sample was analyzed using three biological replicates and normalized to an Actin-expressing gene in mungbean (Vradi03g00210). ∗∗∗, ∗∗, and ∗ are significantly different at P < 0.001, P < 0.01, and P < 0.05, respectively, compared with flower.
Many duplicated gene pairs displayed similar expression patterns in some tissues (Figure 6). For example, VrHsfA1c and VrHsfA1d showed similar expression levels in the root, leaf and seed. VrHsfA6a and VrHsfA6b expression patterns closely resembled each other in the flower and stem. VrHsfB4a and VrHsfB4c displayed similar expression patterns in the stem. Moreover, expression of VrHsfB3a conformed to VrHsfB3b in the pod, seed and leaf, indicating closely related functions in these tissues for these duplicated genes. In contrast, duplicated genes showed distinct expression levels in some tissues (Figure 6), indicating that they may have evolved new functions compared to the original genes.
Expression Analysis of VrHsf Genes in Stress Responses
To investigate the potential functions of VrHsf genes in response to different stresses, we analyzed VrHsf gene expression in mungbean shoots and roots under cold, drought, heat and salt conditions. The expression of most VrHsf genes changed in roots or shoots under stress treatments (Figure 7). The expression patterns of each VrHsf gene varied under different stress treatments. Expression of VrHsfA1d, VrHsfA7b, VrHsfB2b, and VrHsfB4a increased sharply under cold treatment in the shoot. Under drought stress, VrHsfA6a and VrHsfA6b exhibited a more than 10-fold increase in expression level in the root, compared with plants grown under normal conditions. The expression of many genes increased more than 10-fold under heat stress, including VrHsfA4, VrHsfA7a, VrHsfA7b, VrHsfB2c, VrHsfB4c, VrHsfB4h, VrHsfB5, and VrHsfC1 (Figure 7). Under salt treatment, VrHsfA6b showed the greatest change in expression with more than an 11-fold increase. The expression alterations of these VrHsf genes suggest their potential roles in response to the related stresses. In addition, the duplicated gene pairs displayed similar expression patterns under certain stresses (Figure 7), such as VrHsfA3a/VrHsfA3b, VrHsfA7a/VrHsfA7b, and VrHsfB2a/VrHsfB2c in both root and shoot under heat stress, VrHsfA6a/VrHsfA6b in root under drought stress, suggesting the functional redundancy of these duplicated genes.
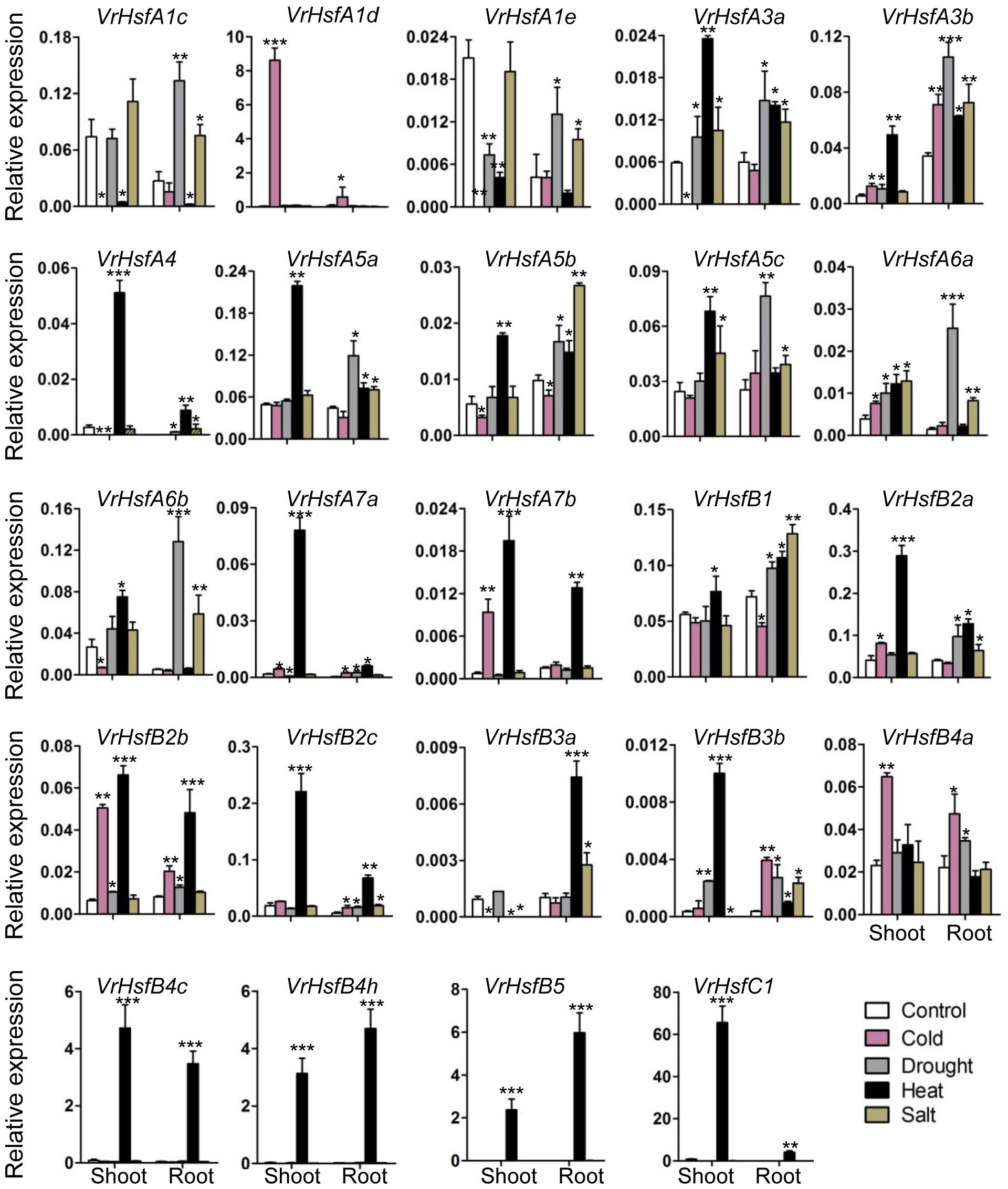
Figure 7. Expression analysis of VrHsf genes in response to different stresses. Two weeks old plants were used for various stress treatments. Each sample was analyzed using three biological replicates and normalized to an Actin-expressing gene in mungbean (Vradi03g00210). ∗∗∗, ∗∗, and ∗ are significantly different at P < 0.001, P < 0.01, and P < 0.05, respectively, compared with the relative control.
Discussion
During the past decades, the identification of Hsf genes in many species has greatly increased our knowledge of the molecular mechanisms of plant developmental and defense processes (Scharf et al., 2012; Guo et al., 2016; Wang et al., 2017). Mungbean is an important crop in the world and the emergence of its genome database allows functional analysis of mungbean genes (Kang et al., 2014). In this study, we identified 24 VrHsf genes and investigated their characteristics using the mungbean genome database.
Different numbers of Hsf genes have been found in different species. Mungbean contains 24 VrHsfs that are similar to Arabidopsis, tomato, rice and potato, but different from the legume plants peanut and soybean (Scharf et al., 2012; Tang et al., 2016; Wang et al., 2017). This difference is most likely due to the fact that soybean and peanut had two duplications during evolution, while only one duplication occurred in mungbean (Chung et al., 2013; Kang et al., 2014; Tang et al., 2016). Although gene numbers increased due to double duplications in a variety of species, some duplicated genes lost functions in the evolution process (Wang et al., 2015). Therefore, there may be many more non-functional Hsf genes in soybean and peanut than in mungbean. The sub-classes A2, A8, and A9, which have been identified in many species (Scharf et al., 2012; Wang et al., 2017), were not found in mungbean (Figure 1 and Table 1). Most of the sub-classes are shared among many species, but some clusters were lost during evolution in some plants. For example, peanut does not contain the A3, A6a, A7, B3, and B4 sub-classes (Wang et al., 2017). These observations imply the functional conservation and divergence of Hsf genes among different plants. Moreover, sub-classes of mungbean A family Hsf genes were closely clustered, compared with B family Hsf genes, which include three clades. And VrHsfB5 displayed a closer relationship with A family genes (Figure 2A), indicating the functional diversity of mungbean B family Hsf genes. In conclusion, since many plant species grow in different conditions, the evolved diversity of Hsf genes may contribute significantly to the plants’ survival and adaption to the environment.
Although most of the Hsf genes play critical roles in response to abiotic stresses, Hsf genes show remarkable functional diversification in Arabidopsis, for example, AtHsfA3 works as part of drought stress signaling and AtHsfA9 controls Hsp expression during seed development (Scharf et al., 2012). VrHsf genes were classified into three classes based on the gene structure diversity (Figure 1), indicating their function diversity for different classes of VrHsfs genes. In addition, some VrHsf genes classified into the same sub-classes contained different conserved domains, such as VrHsfA1c and VrHsfA1e (Figure 3 and Table 2), which suggested that they might have different functions and cannot replace each other in the stress response pathway.
The involvement of Hsf genes in plant growth and development has been revealed in past decades, and homologous genes exhibit either functional conservation or divergence in different species (Scharf et al., 2012; Guo et al., 2016). Expression profiles of VrHsf genes in different tissues may be closely related to their functions in organ development. For example, the expression of VrHsfB4h is higher in seeds than any other tested tissues (Figure 6), indicating its potential function in seed development. However, the expression pattern of AtHsfB4, the homologous gene in Arabidopsis, is distinct from VrHsfB4h (Figure 1). Overexpression of AtHsfB4 in Arabidopsis induces specific effects on root development (Begum et al., 2013). Although homologous genes often have similar functions in different species, the evolution of plants enables gene function diversification. AtHsfB4 has three homologous genes in mungbean, VrHsfB4a, VrHsfB4c, and VrHsfB4h (Figure 1). In addition to the low expression of VrHsfB4h in root, VrHsfB4a and VrHsfB4c might have some functions in common with AtHsfB4 in root development.
The Hsf genes play critical roles in protecting plants from stresses (Scharf et al., 2012; Guo et al., 2016). Most VrHsf genes contain multiple HSE, CBF, ABRE, DRE, and LTRE cis-elements (Figure 5), which suggest that VrHsf genes could be involved in various stress responses, such as heat and drought. Accordingly, most of the VrHsf gene expression patterns change under stress conditions in roots or shoots (Figure 7). The numbers and types of cis-elements are varied among the VrHsf promoters, resulting in different expression profiles of VrHsf genes in the tested tissues and under different stresses (Figures 6, 7), exemplifying the functional diversity of these VrHsf genes. However, for some VrHsf genes, although they contained these cis-elements in the promoter regions, their expression levels did not change under certain stresses. For example, VrHsfA1c had all five cis-elements in its promoter region, but did not show changes in expression in shoots under drought conditions. Moreover, the expression level of VrHsfA3a in roots under cold stresses is similar to that in normal conditions (Figures 5, 7). One possibility for this might be that the treatment time was not sufficient to activate VrHsfA3a gene expression. In addition, epigenetic and somatic genome variations also play important roles in stress responses (Li X. et al., 2016), although these VrHsf genes did not show expression alterations, they may have some epigenetic and somatic genome variations, which require further investigation.
Gene duplication is particularly prevalent in plants and is an essential source for evolution. The duplicated genes can be lost, pseudogenized or become novel genes (Kondrashov et al., 2002; Baskaran et al., 2017). We identified seven duplicated gene pairs in the mungbean genome (Figure 4), all of which were generated between 73.88 and 91.01 MYA (Lynch and Conery, 2000; Chen et al., 2014). However, mungbean experienced one ancient whole-genome duplication (WGD) 58 MYA (Kang et al., 2014). Thus, the duplication of VrHsfs occurred early before the WGD. The seven duplicated gene pairs showed either similar or divergent expression levels (Figures 6, 7). The Hsf duplicated genes also displayed functional divergence in plants (Zhang et al., 2016). In mungbean, some duplicated gene pairs also showed functional divergence during evolution (Figures 6, 7). For example, VrHsfB2a exhibited higher expression levels than its duplicated gene VrHsfB2c in flower and root (Figure 6). Moreover, VrHsfB3b showed increased expression under heat stress in shoot, while its duplicated gene VrHsfB3a displayed decreased transcription level under heat stress in shoot (Figure 7). And also VrHsfB3a and VrHsfB3b showed different cis-element array in the promoter region (Figure 5), which may be responsible for their expression alterations under stresses conditions. Transposable elements (TEs) can move genes or gene fragments to new chromosomal positions, and create new duplications (Jiang et al., 2004; Lai et al., 2005). Among the duplicated genes, VrHsfA7b contained an HSF-type DNA-binding domain and a plant transposase domain, belonging to the Ptta/En/Spm gene family (Supplementary Figure 3). Thus, VrHsf7b may not only have Hsf protein functions, but may also work as a transposase that may have produced its duplicated gene VrHsf7a (Figure 4). The transposition of the En/Spm family is regulated through interacting autoregulatory or epigenetic mechanisms (Fedoroff, 1999; Staginnus et al., 2001), which might explain why VrHsf7a and VrHsf7b are located on different chromosomes (Table 1).
In summary, we have characterized Hsf genes in mungbean, and shown that their expression profile changes in response to stresses. VrHsf gene modification might improve abiotic stress tolerance of mungbean plants. However, much work remains to fully understand the mechanisms of VrHsf gene functions in stress responses.
Author Contributions
SL and CC conceived and designed the research. SL, RW, YD, and HJ conducted the experiments and analyzed the data. SL wrote the manuscript with the input of RW. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31601372), the Qingdao Applied Basic Research Program (Grant No. 15-9-1-45-jch), and the Qingdao Agricultural University Scientific Research Foundation (Grant Nos. 6631115039 and 6631113304).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00736/full#supplementary-material
Footnotes
- ^http://pfam.xfam.org/search
- ^http://plantgenomics.snu.ac.kr/mediawiki-1.21.3/index.php/Main_Page
- ^http://smart.embl-heidelberg.de/
- ^http://www.cibiv.at/services/hsf/
- ^https://web.expasy.org/protparam/
- ^http://gsds.cbi.pku.edu.cn/index.php
- ^http://www.cibiv.at/services/hsf/
- ^http://meme.sdsc.edu/meme/intro.html
- ^http://ibs.biocuckoo.org/
- ^http://www.mybiosoftware.com/mapinspect-compare-display-linkage-maps.html
- ^http://www.qiagen.com
- ^http://www.sas.com
References
Abdelrahman, M., Jogaiah, S., Burritt, D. J., and Tran, L. S. P. (2018). Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant Cell Environ. 41, 1972–1983. doi: 10.1111/pce.13123
Akerfelt, M., Morimoto, R. I., and Sistonen, L. (2010). Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555. doi: 10.1038/nrm2938
Baniwal, S. K., Bharti, K., Chan, K. Y., Fauth, M., Ganguli, A., Kotak, S., et al. (2004). Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29, 471–487. doi: 10.1007/BF02712120
Baskaran, P., Jaleta, T. G., Streit, A., and Rodelsperger, C. (2017). Duplications and positive selection drive the evolution of parasitism-associated gene families in the nematode strongyloides papillosus. Genome Biol. Evol. 9, 790–801. doi: 10.1093/gbe/evx040
Begum, T., Reuter, R., and Schoffl, F. (2013). Overexpression of AtHsfB4 induces specific effects on root development of Arabidopsis. Mech. Dev. 130, 54–60. doi: 10.1016/j.mod.2012.05.008
Blande, J. D., Holopainen, J. K., and Ninemets, Ü (2014). Plant volatiles in polluted atmospheres: stress responses and signal degradation. Plant Cell Environ. 37, 1892–1904. doi: 10.1111/pce.12352
Chen, X., Chen, Z., Zhao, H., Zhao, Y., Cheng, B., and Xiang, Y. (2014). Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS One 9:e87156. doi: 10.1371/journal.pone.0087156
Chung, E., Kim, K. M., and Lee, J. H. (2013). Genome-wide analysis and molecular characterization of heat shock transcription factor family in Glycine max. J. Genet. Genom. 40, 127–135. doi: 10.1016/j.jgg.2012.12.002
Collins, G. G., Nie, X., and Saltveit, M. E. (1995). Heat shock proteins and chilling sensitivity of mung bean hypocotyls. J. Exp. Bot. 46, 795–802. doi: 10.1093/jxb/46.7.795
Fedoroff, N. V. (1999). The suppressor-mutator element and the evolutionary riddle of transposons. Genes Cells 4, 11–19. doi: 10.1046/j.1365-2443.1999.00233.x
Fragkostefanakis, S., Roth, S., Schleiff, E., and Scharf, K. D. (2015). Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 38, 1881–1895. doi: 10.1111/pce.12396
Fujimoto, M., and Nakai, A. (2010). The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 277, 4112–4125. doi: 10.1111/j.1742-4658.2010.07827.x
Ganesan, K., and Xu, B. (2017). A critical review on phytochemical profile and health promoting effects of mung bean (Vigna radiata). Food Sci. Hum. Wellness 7, 11–33. doi: 10.1016/j.fshw.2017.11.002
Giorno, F., Guerriero, G., Baric, S., and Mariani, C. (2012). Heat shock transcriptional factors in malus domestica: identification, classification and expression analysis. BMC Genomics 13:639. doi: 10.1186/1471-2164-13-639
Guo, M., Liu, J. H., Ma, X., Luo, D. X., Gong, Z. H., and Lu, M. H. (2016). The plant heat stress transcription sactors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7:114. doi: 10.3389/fpls.2016.00114
Harrison, C. J., Bohm, A. A., and Nelson, H. C. (1994). Crystal structure of the DNA binding domain of the heat shock transcription factor. Science 263, 224–227. doi: 10.1126/science.8284672
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jacob, P., Hirt, H., and Bendahmane, A. (2017). The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 15, 405–414. doi: 10.1111/pbi.12659
Jiang, N., Bao, Z., Zhang, X., Eddy, S. R., and Wessler, S. R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431, 569–573. doi: 10.1038/nature02953
Kang, Y. J., Kim, S. K., Kim, M. Y., Lestari, P., Kim, K. H., Ha, B. K., et al. (2014). Genome sequence of mungbean and insights into evolution within vigna species. Nat. Commun. 5:5443. doi: 10.1038/ncomms6443
Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3:RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008
Lai, J., Li, Y., Messing, J., and Dooner, H. K. (2005). Gene movement by helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. U.S.A. 102, 9068–9073. doi: 10.1073/pnas.0502923102
Li, P. S., Yu, T. F., He, G. H., Chen, M., Zhou, Y. B., Chai, S. C., et al. (2014). Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genomics 15:1009. doi: 10.1186/1471-2164-15-1009
Li, S., Ying, Y., Secco, D., Wang, C., Narsai, R., Whelan, J., et al. (2017). Molecular interaction between PHO2 and GIGANTEA reveals a new crosstalk between flowering time and phosphate homeostasis in Oryza sativa. Plant Cell Environ. 40, 1487–1499. doi: 10.1111/pce.12945
Li, S., Yue, W., Wang, M., Qiu, W., Zhou, L., and Shou, H. (2016). Mutation of osgigantea leads to enhanced tolerance to polyethylene glycol-generated osmotic stress in rice. Front. Plant Sci. 7:465. doi: 10.3389/fpls.2016.00465
Li, X., Xing, T., and Du, D. (2016). Identification of top-ranked proteins within a directional protein interaction network using the pagerank algorithm: applications in humans and plants. Curr. Issues Mol. Biol. 20, 29–46.
Lin, Y. X., Jiang, H. Y., Chu, Z. X., Tang, X. L., Zhu, S. W., and Cheng, B. J. (2011). Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12:76. doi: 10.1186/1471-2164-12-76
Liu, Z. W., Wu, Z. J., Li, X. H., Huang, Y., Li, H., Wang, Y. X., et al. (2016). Identification, classification, and expression profiles of heat shock transcription factors in tea plant Camellia sinensis) under temperature stress. Gene 576, 52–59. doi: 10.1016/j.gene.2015.09.076
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Mittler, R., Finka, A., and Goloubinoff, P. (2012). How do plants feel the heat? Trends Biochem. Sci. 37, 118–125. doi: 10.1016/j.tibs.2011.11.007
Nover, L., Bharti, K., Döring, P., Mishra, S. K., Ganguli, A., and Scharf, K. D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperons 6, 177–189.
Ohama, N., Sato, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. doi: 10.1016/j.tplants.2016.08.015
Qu, A. L., Ding, Y. F., Jiang, Q., and Zhu, C. (2013). Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 432, 203–207. doi: 10.1016/j.bbrc.2013.01.104
Scharf, K. D., Berberich, T., Ebersberger, I., and Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 1819, 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Staginnus, C., Huettel, B., Desel, C., Schmidt, T., and Kahl, G. (2001). A PCR-based assay to detect En/Spm-like transposon sequences in plants. Chromosom. Res. 9, 591–605. doi: 10.1023/A:1012455520353
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, D., Dong, Y., Ren, H., Li, L., and He, C. (2014). A review of phytochemistry, metabolite changes, and medicinal uses of the common food mung bean and its sprouts (Vigna radiata). Chem. Cent. J. 8:4. doi: 10.1186/1752-153X-8-4
Tang, R., Zhu, W., Song, X., Lin, X., Cai, J., Wang, M., et al. (2016). Genome-wide identification and function analyses of heat shock transcription factors in potato. Front. Plant Sci. 7:490. doi: 10.3389/fpls.2016.00490
von Koskull Döring, P., Scharf, K. D., and Nover, L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. doi: 10.1016/j.tplants.2007.08.014
Wang, J., Sun, N., Deng, T., Zhang, L., and Zuo, K. (2014). Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genomics 15:961. doi: 10.1186/1471-2164-15-961
Wang, P., Song, H., Li, C., Li, P., Li, A., Guan, H., et al. (2017). Genome-wide dissection of the heat shock transcription factor family genes in Arachis. Front. Plant Sci. 8:106. doi: 10.3389/fpls.2017.00106
Wang, Z., Zhou, Z., Liu, Y., Liu, T., Li, Q., Ji, Y., et al. (2015). Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 27, 323–336. doi: 10.1105/tpc.114.135103
Wei, Y., Hu, W., Xia, F., Zeng, H., Li, X., Yan, Y., et al. (2016). Heat shock transcription factors in banana: genome-wide characterization and expression profile analysis during development and stress response. Sci. Rep. 6:36864. doi: 10.1038/srep36864
Westerheide, S., Raynes, R., Powell, C., Xue, B., and Uversky, V. (2012). HSF transcription factor family, heat shock response, and protein intrinsic disorder. Curr. Protein Pept. Sci. 13, 86–103. doi: 10.2174/138920312799277956
Xu, H. X., Li, X. Y., and Chen, J. W. (2017). Comparative transcriptome profiling of freezing stress responses in loquat (Eriobotrya japonica) fruitlets. J. Plant Res. 130, 893–907. doi: 10.1007/s10265-017-0942-4
Zandalinas, S. I., Mittler, R., Balfagón, D., Arbona, V., and Gómez-Cadenas, A. (2018). Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 162, 2–12. doi: 10.1111/ppl.12540
Zeng, J. K., Li, X., Zhang, J., Ge, H., Yin, X. R., and Chen, K. S. (2016). Regulation of loquat fruit low temperature response and lignification involves interaction of heat shock factors and genes associated with lignin biosynthesis. Plant Cell Environ. 39, 1780–1789. doi: 10.1111/pce.12741
Zhang, J., Jia, H., Li, J., Li, Y., Lu, M., and Hu, J. (2016). Molecular evolution and expression divergence of the Populus euphratica Hsf genes provide insight into the stress acclimation of desert poplar. Sci. Rep. 6:30050. doi: 10.1038/srep30050
Zhang, J., Liu, B., Li, J., Zhang, L., Wang, Y., Zheng, H., et al. (2015). Hsf and Hsp gene families in populus: genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genomics 16:181. doi: 10.1186/s12864-015-1398-3
Keywords: mungbean, heat shock transcription factor, gene family, abiotic stress, gene expression
Citation: Li S, Wang R, Jin H, Ding Y and Cai C (2019) Molecular Characterization and Expression Profile Analysis of Heat Shock Transcription Factors in Mungbean. Front. Genet. 9:736. doi: 10.3389/fgene.2018.00736
Received: 16 July 2018; Accepted: 22 December 2018;
Published: 11 January 2019.
Edited by:
Ping Ma, University of Georgia, United StatesReviewed by:
Heng Zhang, Shanghai Institutes for Biological Sciences (CAS), ChinaYiwen Liu, University of Arizona, United States
Copyright © 2019 Li, Wang, Jin, Ding and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai Li, bGkyMDE0c2h1YWlAMTYzLmNvbQ== Chunmei Cai, Y2FpY2h1bm1laTA5MDJAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Shuai Li
Shuai Li Runhao Wang
Runhao Wang Hanqi Jin†
Hanqi Jin†