- 1Department of Plant Sciences, North Dakota State University, Fargo, ND, United States
- 2Edward T. Schafer Agricultural Research Center, Agricultural Research Service, United States Department of Agriculture, Fargo, ND, United States
Commercial hybrid seed production in sunflower currently relies on a single cytoplasmic male sterility (CMS) source, PET1 and the major fertility restoration gene, Rf1, leaving the crop highly vulnerable to issues with genetic bottlenecks. Therefore, having multiple CMS/Rf systems is important for sustainable sunflower production. Here, we report the identification of a new fertility restoration gene, Rf7, which is tightly linked to a new downy mildew (DM) resistance gene, Pl34, in the USDA sunflower inbred line, RHA 428. The Rf7 gene was genetically mapped to an interval of 0.6 cM on the lower end of linkage group (LG) 13, while Pl34 was mapped 2.1 cM proximal to the Rf7. Both the genes are located in a cluster of Rf and Pl genes. To gain further insights into the distribution of Rf genes in the sunflower breeding lines, we used a genome-wide association study (GWAS) approach to identify markers associated with the fertility restoration trait in a panel of 333 sunflower lines genotyped with 8,723 single nucleotide polymorphism (SNP) markers. Twenty-four SNP markers on the lower end of LG13 spanning a genomic region of 2.47 cM were significantly associated with the trait. The significant markers were surveyed in a world collection panel of 548 sunflower lines and validated to be associated with the Rf1 gene. The SNP haplotypes for the Rf1 gene are different from Rf5 and the Rf7gene located in the Rf gene cluster on LG13. The SNP and SSR markers tightly flanking the Rf7 gene and the Pl34 gene would benefit the sunflower breeders in facilitating marker assisted selection (MAS) of Rf and Pl genes.
Introduction
Cytoplasmic male sterility (CMS) is a maternally transmitted trait carried by the mitochondrial genome that prevents hermaphrodite plants from developing viable pollen, resulting in male-sterile plants (Chase, 2007). CMS provides a very useful mechanism to produce large numbers of female plants for commercial hybrid seed production (Harvey, 2004). CMS is a common phenomenon observed in over 150 diverse plant species (Laser and Lersten, 1972; Schnable and Wise, 1998; Fujii et al., 2011). In populations with CMS, one or more nuclear genes known as restorer-of-fertility (Rf) genes can suppress the expression of the aberrant mitochondrial CMS genes and restore viable pollen production (Chase, 2007). The CMS/Rf system has been an indispensable resource for commercial hybrid seed production in many crops including sunflower (Bohra et al., 2016).
The first CMS source of sunflower, PET1, was derived from an interspecific cross between Helianthus petiolaris subsp. petiolaris Nutt. and cultivated sunflower (Leclercq, 1969). Soon after, Kinman (1970) discovered a single dominant gene, Rf1, in the sunflower line T660006-2-1 that restores the fertility of CMS PET1 (Supplementary Figure S1). Since then, the Rf1 gene has been introduced from T660006-2-1 to many public and private sunflower inbred lines (Korell et al., 1992). A second dominant gene, Rf2, complementary to Rf1 was discovered from a restorer line MZ-1398 (Vranceanu and Stoenescu, 1971, 1978). However, Rf2 was described to be ubiquitous in nearly all sunflower inbred lines, including maintainer lines of PET1 (Horn et al., 2003; Serieys, 2005); therefore, Rf1 is the most important gene to track for fertility restoration of CMS PET1. Since the first discovery of CMS PET1, there have been over 70 CMS sources reported in sunflower (Serieys, 2005). Many of these CMS sources do not have a known complementary Rf gene or have an unstable cytoplasm (Seiler and Rieseberg, 1997).
To date, only seven Rf genes have been characterized and mapped in sunflower genome. The Rf1 gene from the sunflower lines RHA 266, RHA 271, RHA325, and RHA 439 was mapped to LG13 by different researchers (Gentzbittel et al., 1995; Berry et al., 1997; Horn et al., 2003; Kusterer et al., 2005; Yue et al., 2010). Gentzbittel et al. (1995, 1999) reported the presence of a distinct fertility restoration locus, Msc1, for the CMS PET1 cytoplasm and mapped it to LG12 on the restriction fragment length polymorphism (RFLP) map, which corresponds to LG7 of the public map. The Rf3 gene mapped on LG7 in the inbred lines RHA 280 and RHA 340 is equally capable of restoring fertility of PET1 CMS lines (Jan and Vick, 2007; Abratti et al., 2008; Liu et al., 2012). Feng and Jan (2008) identified a new dominant restorer gene, Rf4 originally from the diploid perennial wild species H. maximiliani, and mapped it to LG3 of the sunflower genome. The Rf4 gene restored the pollen fertility of the CMS GIG2 cytoplasm derived from wild H. giganteus. Schnabel et al. (2008) mapped the Rf-PEF1 gene for CMS PEF1 cytoplasm and demonstrated that Rf-PEF1 is not located on LG13 where the Rf1 gene resides. A restorer gene designated as Rf5 derived from wild H. annuus was mapped to the lower end of LG13 close to the Rf1gene (Qi et al., 2012). The most recent Rf gene described in sunflower, Rf6, was derived from H. angustifolius and is required to restore the male fertility of CMS 514A developed recently with H. tuberosus cytoplasm (Liu et al., 2013). Rf6 was mapped to LG3 of the public sunflower map.
While Rf genes have been mapped throughout the genome in different crop species, clustering of multiple Rf genes in the genome is also common across species (Melonek et al., 2016). For example, Rf1 and Rf5 genes were mapped in similar position on LG13 in sunflower (Yue et al., 2010; Qi et al., 2012), and in rice Rf1a (Rf5), Rf1b, Rf4, and Rf6(t) were mapped in proximity on chromosome 10 (Huang et al., 2000; Liu et al., 2004; Ahmadikhah and Karlov, 2006; Wang et al., 2006; Hu et al., 2012). In sunflower, the PET1 cytoplasm and its corresponding Rf1 gene has been solely used by the commercial seed industries around the globe for large-scale hybrid seed production (Dimitrijevic and Horn, 2018). Use of a single CMS source in sunflower hybrid production carries the risks associated with genetic bottlenecks in crops. The southern corn leaf blight epidemic of T-cytoplasmic maize revealed the dangers of hybrid-seed production using a single source of CMS (Levings, 1990; Harvey, 2004). Use of additional CMS/Rf sources would diversify the gene pool of the crop and reduce genetic vulnerability (Leclercq, 1969; Jan and Vick, 2007). The discovery of new CMS sources and corresponding Rf genes remains a goal of sunflower breeding.
Downy mildew (DM), caused by Plasmopara halstedii (Farlow) Berlese & de Toni, is a major disease affecting sunflower production globally (Gascuel et al., 2015). P. halstedii is a biotrophic oomycete pathogen commonly attacks sunflower during seedling stage (Meliala et al., 2000). Host resistance has been considered the best management approach to control the disease. Resistance against P. halstedii in sunflower has been described as the typical gene-for-gene interaction (Flor, 1971), where the host resistance R-gene(s) recognize and respond to the effector proteins produced by the compatible avirulence (Avr) genes of specific pathotypes (Chisholm et al., 2006; Viranyi and Spring, 2011). The DM resistance genes, designated as Pl, have long been deployed in elite sunflower lines (Vranceanu and Stoenescu, 1970; Zimmer and Kinman, 1972). However, due to dynamic changes in the pathogen population, new virulent races frequently evolve, which often overcome the effectiveness of the existing Pl genes. Fortunately, wild annual sunflower species have proven to be a reliable source of Pl genes for DM resistance (Seiler et al., 2017). To date, thirty-four Pl genes have been reported in sunflowers, namely Pl1–Pl33, and PlArg (for review see Ma et al., 2017; Dimitrijevic and Horn, 2018; Liu et al., 2018; Pecrix et al., 2018a,b). The ambiguity associated with persistent durability of R-gene mediated DM resistance compels the sunflower breeders to discover and deploy new Pl genes for sustained sunflower production.
Genetic linkage mapping based on biparental populations has been proven as a robust tool for detecting rarely occurring alleles that have a large effect on the phenotype (Nordborg and Weigel, 2008). Numerous dominant genes in many crop species have been successfully mapped using biparental linkage mapping. In contrast, genome wide association studies (GWAS) are a powerful approach to locate common alleles associated with phenotypes with much higher resolution than linkage mapping because they reflect historical recombination events in broad-based diversity panels (Nordborg and Weigel, 2008). In this study, we report the mapping of the two genes in a sunflower biparental mapping population, a fertility restoration gene, Rf7, and a DM resistance gene, Pl34. We also identified single nucleotide polymorphism (SNP) markers associated with the fertility restoration trait in sunflower lines using a GWAS approach. Finally, the detected significant SNP markers were surveyed in a larger population, which constitutes a global sunflower collection, to identify haplotypes associated with the Rf1 gene. The findings of this study could be a useful resource for identifying new Rf gene(s) and help alleviate the potential genetic vulnerability posed due to the exclusive use of a single CMS/Rf system in sunflower hybrid production.
Materials and Methods
Plant Materials
Bi-Parent Mapping Population
The F2 and F2-derived F3 populations were developed from two F1 plants derived from the cross RHA 428/HA 234 to map the Rf and Pl genes. HA 234 (PI 599778) is an oilseed sunflower maintainer line susceptible to DM, that was released by the USDA-ARS and Texas Agricultural Experiment Station in 1971. RHA 428 (PI 619206) is a male fertility restorer oilseed inbred line resistant to DM, which was selected from the cross RHA 801//RHA 365/PI 413157. PI 413157 is a wild H. annuus accession collected in New Mexico in 1974. RHA 428 was released by the USDA-ARS and the North Dakota Agricultural Experiment Station in 2000 as a DM resistant inbred line (Miller et al., 2002). DM tests indicated that RHA 428 is resistant to P. halstedii races 334, 700, 710, 714, 730, 734, 735, but susceptible to races 737 and 774 (Gulya, personal communication; Gilley et al., 2016). A total of 408 F2 plants were grown in the greenhouse in 2011. Because RHA 428 with CMS PET1 was used as a female parent in the initial cross, 126 F2 plants were male sterile, while 328 F2 plants were fertile and advanced to F3 generation for subsequent phenotypic evaluation for DM resistance and male fertility.
Genome-Wide Association Study Panel
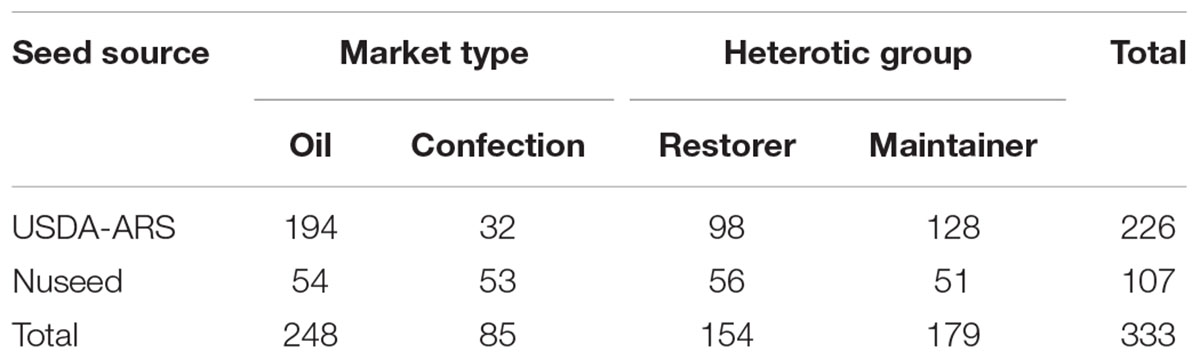
A GWAS population of 333 sunflower lines comprised of inbred and advanced breeding lines from the USDA-ARS breeding program and Seeds 2000, now Nuseed Americas Inc., was used to map male fertility restoration. This population includes lines from both oil and confection types and from the two major heterotic groups in cultivated sunflower (Table 1). Among the sunflower lines, 226 were USDA-ARS inbred lines released from 1970 to 2011 and the remaining 107 lines were from Nuseed.
Sunflower Evaluation Panel
A total of 548 sunflower genotypes were used as an evaluation panel to investigate the distribution of SNP markers associated with the male fertility restoration genes (Supplementary Table S1). These lines include 238 USDA-ARS released inbred lines (126 maintainer lines and 112 male fertility restoration lines), 63 germplasm lines and 247 plant introduction (PI) lines originally collected from 32 countries, which together capture a large portion of the global diversity present in cultivated sunflower. A total of 222 of the USDA-ARS released sunflower lines were common between the GWAS panel and the sunflower evaluation panel.
Phenotypic Characterization
Male Fertility Restoration
Male fertility evaluation was conducted for the bi-parental population. One hundred seventy-two fertile F2:3 families were grown in rows of 30 plants in the field at Fargo, ND, in June 2013. Evaluation of male fertility was conducted at the flowering stage. Plants that produced anthers and shed pollen were considered fertile, whereas those without anthers or pollen were considered sterile. Data on fertility restoration of the GWAS lines was obtained from breeder’s records.
Downy Mildew Resistance
Evaluation of DM resistance was conducted in the F2-derived fertile F3 families. A total of 172 F3 families along with the parental lines, RHA 428 and HA 234, were evaluated for resistance to DM in the greenhouse under controlled conditions in June 2013. Thirty seedlings of each F2:3 family were inoculated with the North American (NA) P. halstedii race 734, a virulent race identified in the United States in 2010 (Gulya et al., 2011) following the method described by Gulya et al. (1991). Sunflower seedlings infected by DM display white sporulation on cotyledons and true leaves in greenhouse tests. Resistant plants exhibit no sporulation.
The results of the F3 family tests were used to infer the genotypes of F2 plants at the DM resistance locus and male fertility restoration locus. A Chi-squared (χ2) analysis was performed to verify whether the observed ratios of segregation for the DM resistance and male fertility in the F3 population fit expected models.
DNA Extraction and Genotyping
Bi-Parental Mapping Population
Genomic DNA of the parents and 172 F2 progenies derived from the cross RHA 428/HA 234 was extracted from lyophilized tissues using the DNeasy 96 Plant Kit (Qiagen, Valencia, CA, United States). DNA concentrations were measured using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States), and were adjusted to 5 ng μl-1 for all samples for polymerase chain reaction (PCR) amplification.
A total of 860 simple sequence repeat (SSR) markers were selected to screen polymorphisms between the two parents (Tang et al., 2002; Yu et al., 2003). Bulked segregant analysis was conducted using polymorphic SSR markers (Michelmore et al., 1991). SSR markers associated with male fertility restoration and DM resistance were assessed in the 172 F2 individuals for linkage analysis and mapping. PCR of SSR markers was performed according to Qi et al. (2011), and PCR products were detected using an IR2 4300/4200 DNA Analyzer with denaturing polyacrylamide gel electrophoresis (LI-COR, Lincoln, NE, United States).
Additionally, a total of 58 SNP markers previously mapped to the lower end of LG13 were chosen for marker saturation in the region where male fertility restoration and DM resistance genes were mapped. Twenty-two of these SNPs were selected from Talukder’s map (hereafter referred to as NSA SNP) covering a region of 23.79 cM (Supplementary Table S2, Talukder et al., 2014), and 36 SNPs were selected from Bowers’s map (hereafter referred to as SFW SNP) covering a region of 26.62 cM (Supplementary Table S2, Bowers et al., 2012). Genotyping of most of the selected NSA SNP markers were conducted as described below, while genotyping of the SFW SNPs and a few NSA SNPs was performed using a strategy of converting SNPs into length polymorphism markers (Qi et al., 2015; Long et al., 2017). The primer sequences of the 15 polymorphic SFW SNPs and seven NSA SNPs are presented in Supplementary Table S3. The conditions of the SNP PCR reactions were described by Qi et al. (2015), and PCR products were detected using an IR2 4300/4200 DNA Analyzer with denaturing polyacrylamide gel electrophoresis (LI-COR, Lincoln, NE, United States).
GWAS and Sunflower Evaluation Panels
Total genomic DNA was extracted from 40 mg lyophilized young leaves of each sunflower line with the DNeasy 96 Plant Kit (Qiagen Inc., Valencia, CA, United States) using a modified protocol (Talukder et al., 2014). DNA was quantified using the PicoGreen kit (Molecular Probes) according to the kit instructions. Genotyping was carried out at BioDiagnostics, Inc., River Falls, WI, United States, with the custom-built Illumina Infinium chip (Illumina Inc., San Diego, CA, United States) containing 8,723 bi-allelic SNP markers developed by the National Sunflower Association (NSA) SNP consortium (Pegadaraju et al., 2013; Talukder et al., 2014). Automated SNP calling was performed using the cluster algorithm implemented in GenomeStudio v1.0 software (Illumina Inc., San Diego, CA, United States). All data was visually inspected and manually rescored if any errors were evident in the genotype calling.
Linkage Analysis and Mapping of Rf and Pl Genes
Construction of LG13 genetic map for RHA 428/HA 234 F2 population associated with the Rf and Pl genes were performed using JoinMap 4.1 software with a maximum likelihood mapping algorithm and Kosambi’s mapping function (Van Ooijen, 2006). A minimum likelihood of odds (LOD) ≥ 3.0 and a maximum distance of ≤ 50 centimorgans (cM) were used to test linkage among markers. The graphical representation of the linkage map was drawn using MapChart 2.2 (Voorrips, 2002).
Genome-Wide Association Study of Male Fertility Restoration
Population Structure and Kinship
The population structure of the GWAS panel was estimated using 681 SNP markers randomly selected from all 17 LGs of the sunflower genome that are spaced at least 1 cM apart. Subpopulation membership of each sunflower line was estimated using STRUCTURE v2.3.4 (Pritchard et al., 2000). An ancestry model that allows population admixture with no a priori information was used with a burn-in period of 100,000 iterations followed by 200,000 Markov Chain Monte Carlo (MCMC) iterations for subpopulation numbers (K) ranging from 1 to 10. Five runs for each K value were performed and the posterior probability [LnP(D)] was determined for each run. The optimum number of subpopulations was determined from ΔK, the rate of change in LnP(D) between successive K values, as proposed by Evanno et al. (2005). Kinship relationships among the lines of the GWAS population (K matrix) were derived using 4,630 SNP markers with minor allele frequency (MAF) of ≥ 0.05. The SPAGeDi software v1-5a (Hardy and Vekemans, 2002) was used to estimate a mean kinship coefficient (Loiselle et al., 1995) from SNP marker data, where negative kinship values between lines were set to 0 (Yu et al., 2006).
Association Mapping Analysis
Out of the 8,723 SNP markers, data from 5,019 markers were selected to run GWAS analyses because of their known map position in the sunflower genome (Talukder et al., 2014). Imputation of missing genotypes was performed using fastPHASE v1.3 software (Scheet and Stephens, 2006), assuming K = 38 clusters with the default settings of the EM algorithm. All marker-trait association tests were run using TASSEL v3.0 standalone (Bradbury et al., 2007). The SNP markers with MAF of ≤ 0.05 were removed from the analyses. Four different GWAS models were tested: first, we examined the association between the phenotype and SNP genotypes in a naïve analysis using the general linear model (GLM), y = Xα + e; second, a GLM analysis was performed that accounted for population structure as a cofactor (GLMQ), y = Xα + Qβ + e; third, a mixed linear model (MLM) analysis was performed that considered only kinship relatedness in the model, y = Xα + Kμ + e; and finally, an MLM analysis was performed that accounted for both population structure and kinship relatedness in the model (MLMQ), y = Xa + Qβ + Kμ + e (Yu et al., 2006). In the equations, y is the phenotype, X is the SNP genotype matrix, α is the vector of marker effects, Q is the population membership assignment matrices for subpopulations in the STRUCTURE analysis, β is the vector of subpopulation effects, K is the relative kinship matrix determined from the marker data, μ is the vector of kinship effects and, e is the vector of residual effects. Xα and Qβ represent fixed effects, and Kμ and e represent random effects. Quantile-quantile plots of estimated -log10(P) were produced for each model using the R statistical package (R Core Team, 2017) by plotting observed p-values of marker–trait associations against the expected p-values from the assumption that no association exists between markers and trait. The best fitting GWAS model was chosen by assessing the extent to which the analysis produced more significant results than expected by chance. Genome-wide marker–trait association p-values was corrected for multiple testing using 5% false discovery rate (FDR) (Benjamini and Hochberg, 1995). Given the distribution of empirical p-values of 4,630 markers, the FDR significance level cut-off corresponded to the p-value of 1.57-04, which was employed as the threshold for significant marker-trait associations in the GWAS analysis.
Results
Molecular Mapping of Rf and DM Resistance Genes in RHA 428
Phenotypic Assessments
A total of 408 F2 plants from the cross RHA 428/HA 234 were grown in the greenhouse in 2011. The F2 population segregated in 328 male fertile: 126 male sterile, fitting a single gene model of 3 male fertile: 1 homozygous male sterile (χ2 = 1.8355, df = 1, P = 0.1755). No seed was obtained from the 126 male sterile F2 plants. The F3 family tests were performed to identify the F2 plants homozygous for DM resistance and male fertility. A total of 172 F3 families with good seed set were selected and evaluated in the field for male fertility in the summer of 2013. Fifty-six families were all fertile with no segregation, while 116 families showed segregation for fertility restoration. The segregation ratio fit an expected ratio of 1 homozygous male fertile: 2 heterozygous male fertile (χ2 = 0.046512, df = 1, P = 0.8292). These data indicated that the male fertility restoration in RHA 428 is controlled by a single dominant gene, designated as Rf7.
DM tests of 172 F3 families with P. halstedii race 734 indicated co-segregation with male fertility restoration. Out of the 56 F3 families homozygous for male fertility restoration, 52 were homozygous for DM resistance and four F3 families were homozygous for susceptibility. Among the 116 F3 families segregating for male fertility, 114 were segregating for DM resistance, while only two were homozygous for DM resistance. The results suggested that the DM resistant gene is linked to the male fertility restoration gene. Since RHA 428 shows a differential specificity against P. helianthi races other than the known DM R genes (Gilley et al., 2016), this gene in RHA 428 was named as Pl34.
Linkage Map Construction of the Rf and Pl Genes
Simple sequence repeat markers were used for initial linkage map construction and to study marker-trait association. Out of 860 SSR markers screened for polymorphism between the parents RHA 428 and HA 234, a total of 293 SSR markers (34%) showed polymorphism between two parents. Bulked segregant analysis with polymorphic SSR markers revealed that the male fertility restoration trait was associated with markers on LG13. Seven LG13 specific polymorphic SSR markers were used to screen the F2 population, and linkage analysis mapped the Rf7 gene on LG13 (Figure 1C). The SSR marker, ORS511 mapped at 0.9 cM distal and two co-segregating SSR markers, ORS191 and ORS316 mapped at 1.2 cM proximal to the Rf7 gene on LG13. The Pl34 DM resistance gene was mapped only 2.1 cM downstream of the Rf gene. Two co-segregating SSR markers, ORS191 and ORS316 were 0.9 cM distal to Pl34, while HT382 was 3.4 cM proximal to Pl34 (Figure 1C).
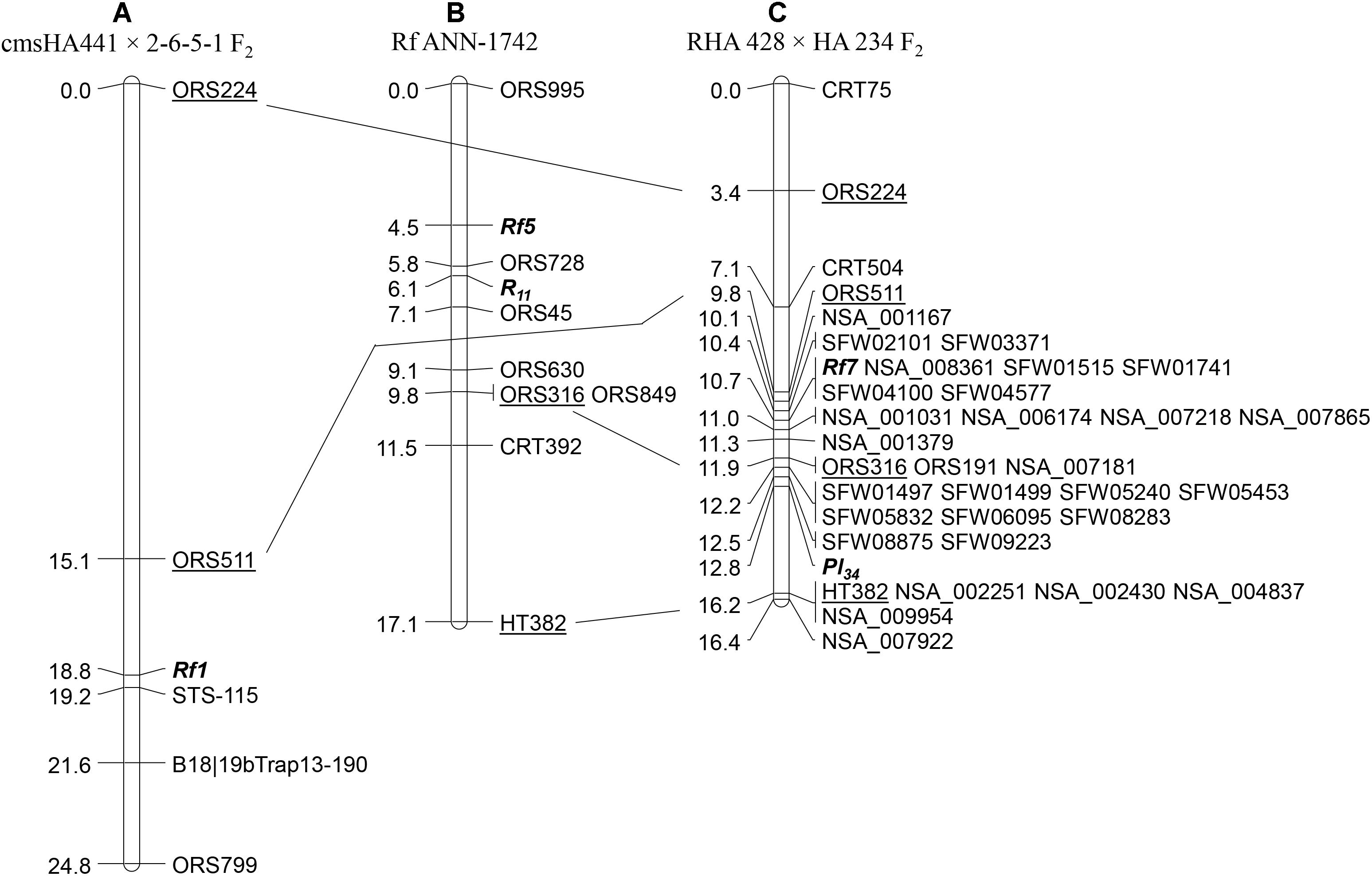
Figure 1. Comparison of the male fertility restoration genes mapped on linkage group 13 in different studies: (A) maps showing the location of Rf1 gene (Yue et al., 2010); (B) the location of Rf5 gene (Qi et al., 2012); and (C) the position of Rf7 gene mapped in the current study. The common markers across different maps are underlined.
SNP Marker Saturation of the Gene Region
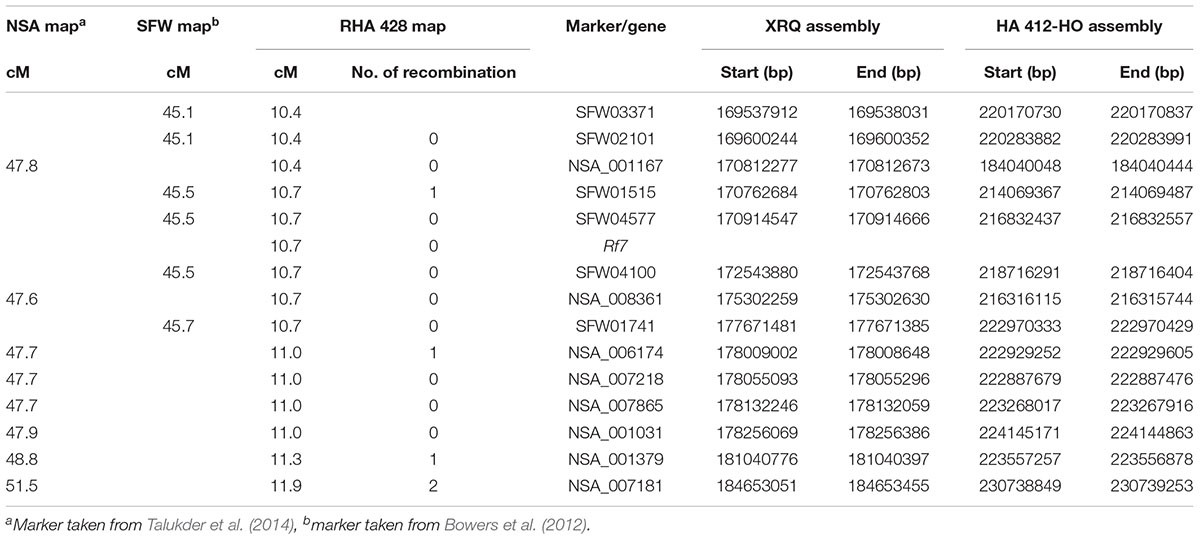
Of the 58 SNP markers tested, 28 showed polymorphism between the two parents, and were assigned to the LG13 map (Figure 1C). Thirteen SNPs were positioned to the Rf7 gene interval between ORS511 and ORS316, while 14 SNPs were mapped to the Pl34 gene interval between ORS316 and HT382. One was mapped at the end of LG13 (Figure 1C). Out of 13 SNP markers mapped in the Rf7 gene interval, five of them were co-segregating with the gene. These five markers spanned 6.9 and 8.9 Mb on chromosome 13 of the XRQ and HA412-HO sunflower genome assemblies, respectively (Table 2).
Comparison of Other Rf Genes Mapped on LG13
Two other male fertility restoration genes, Rf1 and Rf5, have been previously mapped at the lower end of LG13 near the genomic region of the Rf7gene mapped in this study (Yue et al., 2010; Qi et al., 2012). The common SSR markers, ORS511 and ORS316 in the Rf7map, were separately mapped to the Rf1 and Rf5 genetic maps. ORS511 is distal to Rf1 and Rf7 with 3.7 and 0.9 cM in their maps, while ORS316 is proximal to Rf5 and Rf7 with 5.3 and 1.2 cM in their maps, indicating Rf7 resides in a Rf gene cluster region at the lower end of LG13 (Figure 1).
Genome-Wide Association Study of the Rf Gene
Association Analysis
Genome-wide association study analysis was performed using four different models. The best fitting model for this GWAS panel was identified using quantile-quantile plots constructed from the observed vs expected -log10(p) values of each models (Supplementary Figure S2). As expected, the highest number of significant markers (FDR < 0.05) was observed for the GLM model, with many suspected as false positives. The number of significant markers were dramatically reduced with the inclusion of structure (with K = 3 subpopulations; Figures 2, 3) and/or kinship covariates in the model. The deviation of observed p-values from the expected p-values was minimal for the MLMQ model, which accounted both a structure variable and kinship as a random factor. We concluded that this was the best fitting model for our GWAS panel. A total of 24 significant SNP markers were associated with the fertility restoration trait with p < 1.57-04 (Figure 4). All 24 markers were located from 46.40 to 48.87 cM on LG13 of the NSA sunflower map (Table 3). We performed a blastn search using sequences of the 24 significant SNP markers on both XRQ and HA412-HO sunflower genome assemblies to locate the physical position of these markers1. Out of 24 significant SNP markers, no sequence homology was found for NSA_000112 and NSA_007865 on LG13 in either of the sunflower genome assemblies. The remaining 22 significant SNP markers were found between 169361232 and 181040776 bp on chromosome 13 of the XRQ genome (Table 3). This corresponds with 215330128 to 224803649 bp on chromosome 13 of the HA412-HO assembly except for NSA_001167, which was found at the 184040048 bp position on chromosome 13 about 31.29 Mb from the rest of the SNP markers (Table 3).
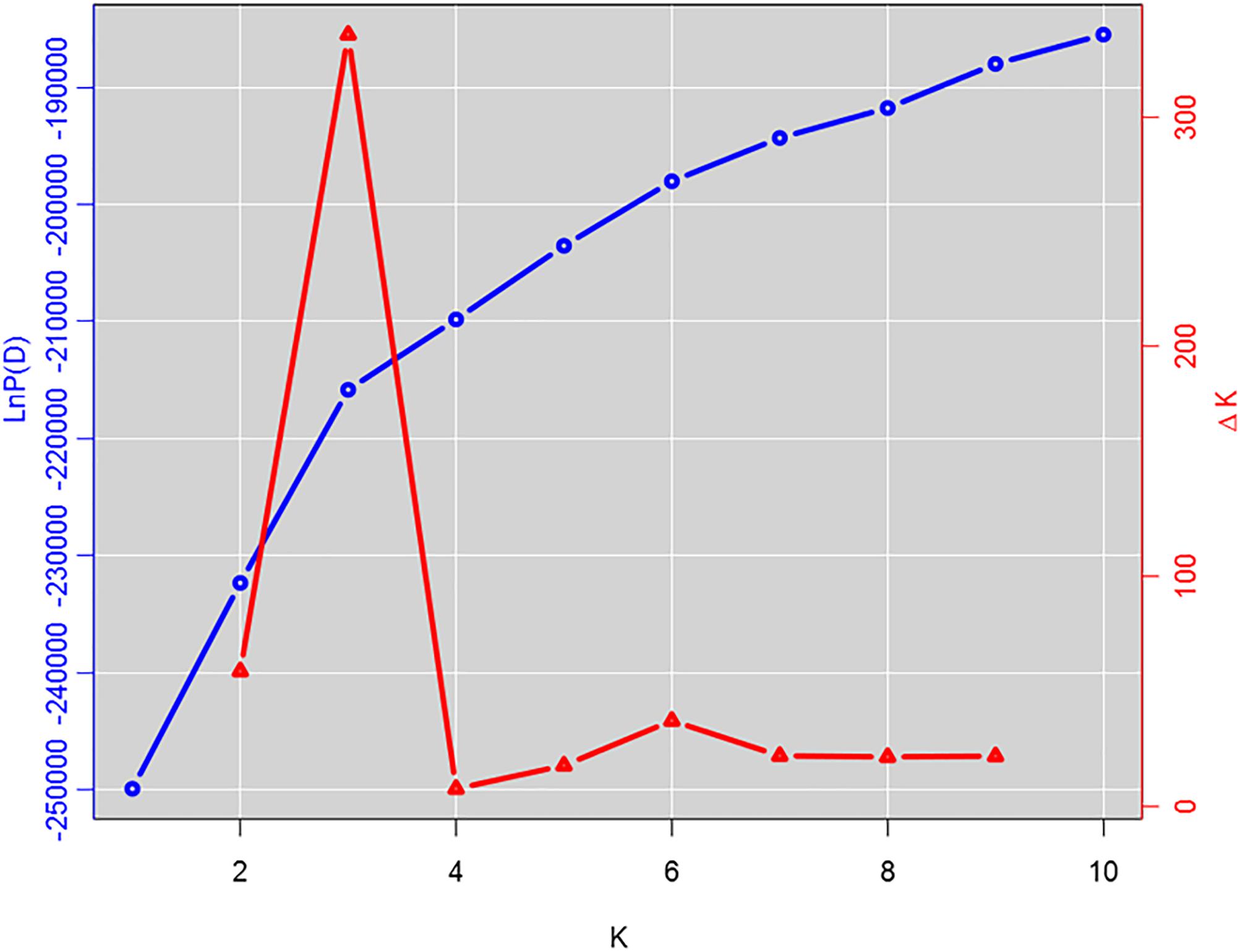
Figure 2. Estimation of number of subpopulations in the genome-wide association study (GWAS) panel. The log probability, LnP(D) for each value of K (K = 1–10) averaged over 5 runs of STRUCTURE analysis with 100,000 burn-in steps and 200,000 simulation steps is plotted with the ΔK values for each of the successive K runs.
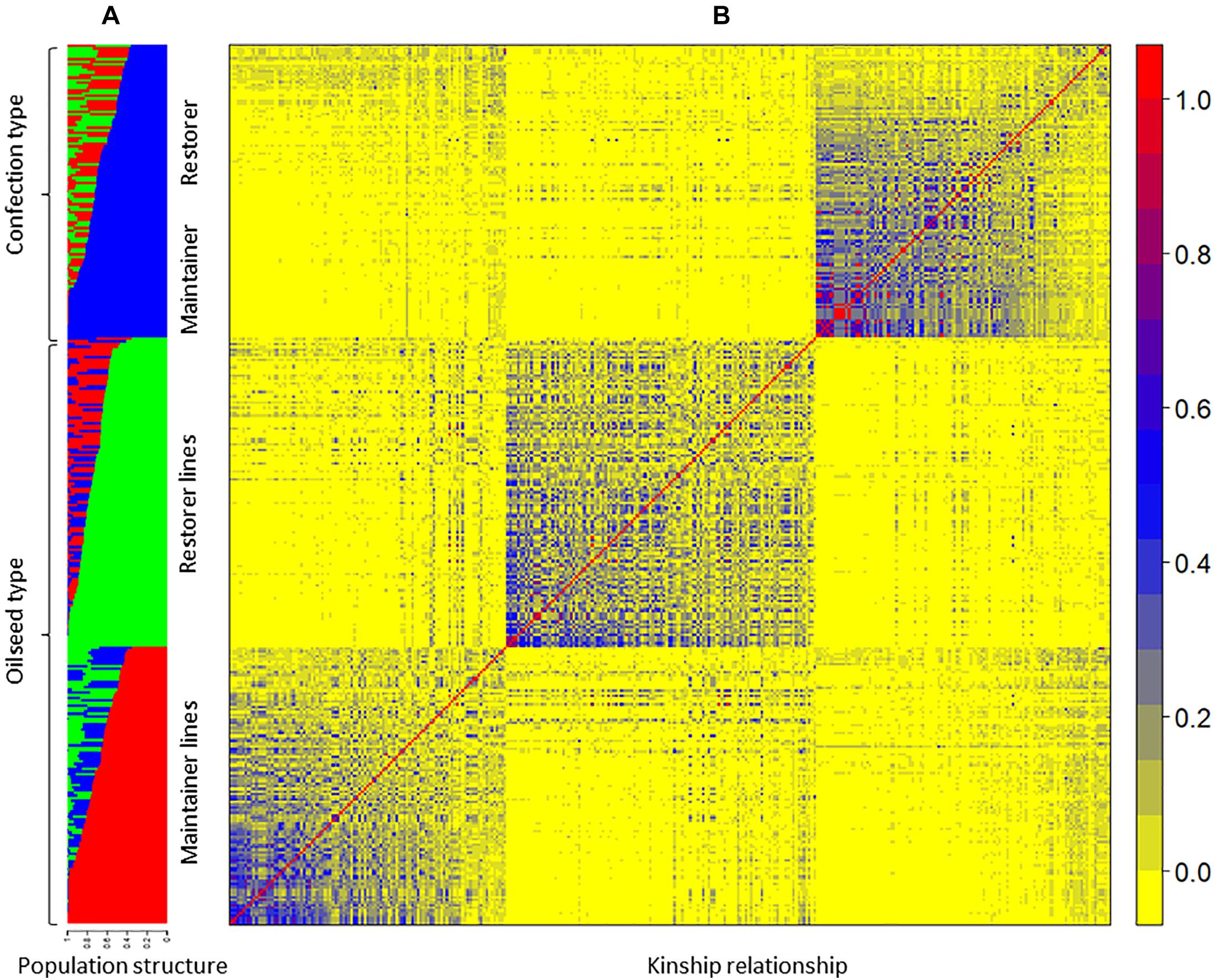
Figure 3. (A) Bar plot of the STRUCTURE analysis. Each of the 333 genotypes is represented by a vertical bar, which is partitioned into K colored segments that represent the individual’s estimated membership to the K clusters, (B) heat map of relative kinship matrix among the sunflower lines used in the genome-wide association study panel.
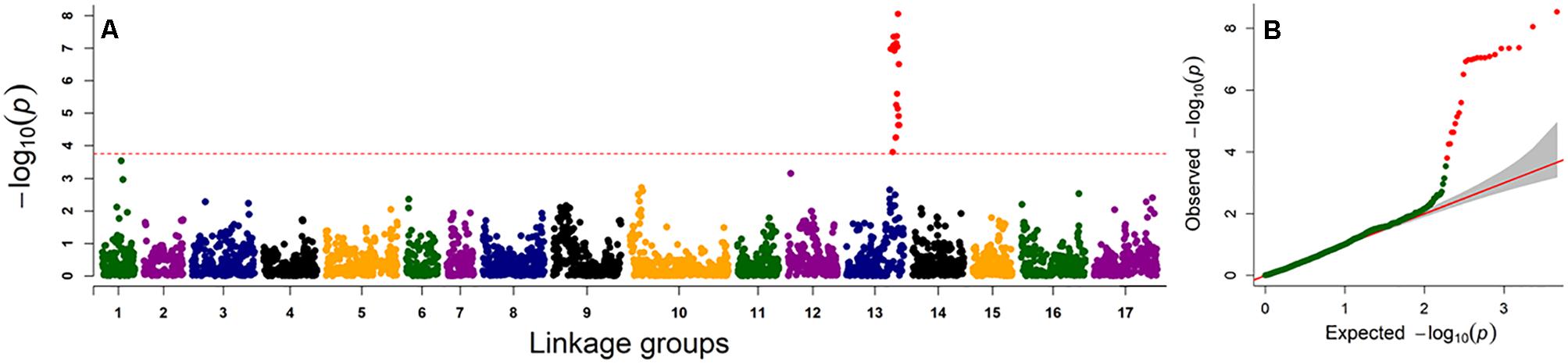
Figure 4. Genome-wide association scan for male fertility restoration trait in sunflower. (A) Manhattan plot for male fertility restoration using mixed linear model (MLMQ). The dashed horizontal line represents the FDR0.05-adjusted significance threshold (p < 1.57-04), (B) Quantile–quantile plots of male fertility restoration trait for MLMQ model.
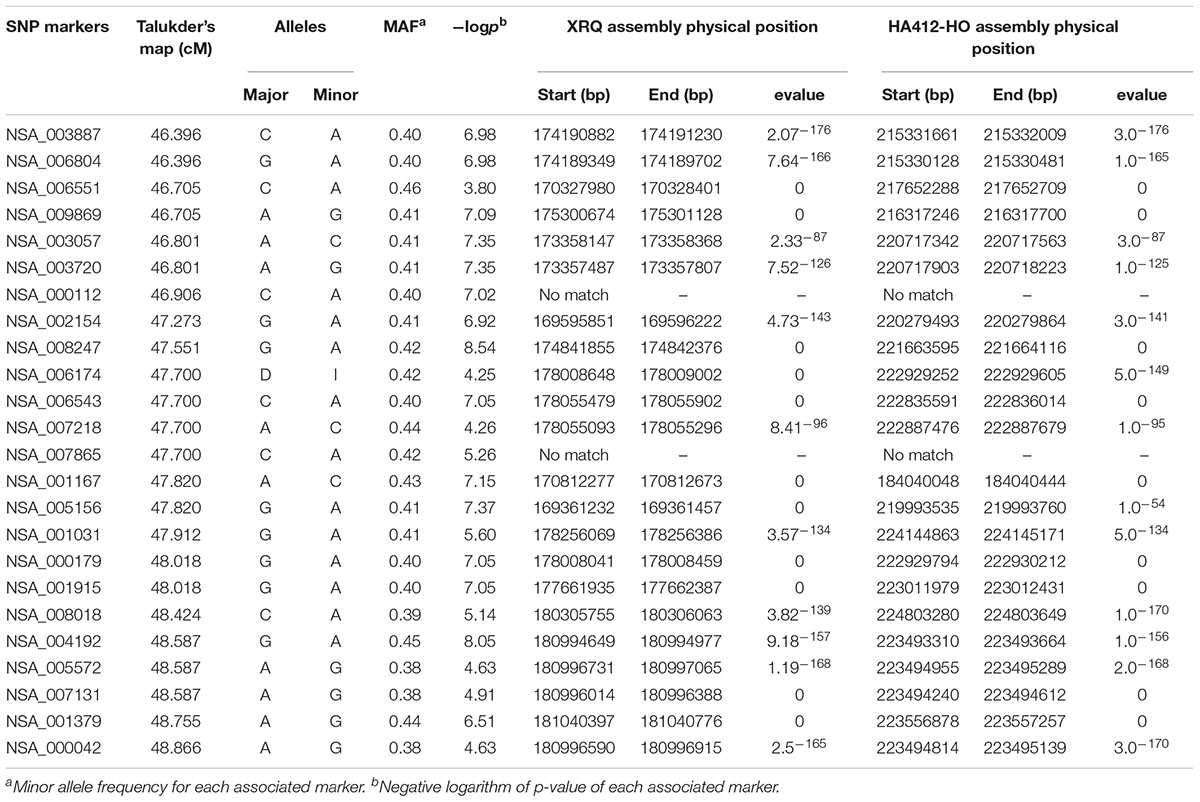
Table 3. Linkage and physical positions of the 24 significant SNP markers associated with the Rf1 gene identified using genome-wide association mixed model analysis in a panel of 333 sunflower lines.
Survey of Significant SNP Marker Alleles in the Sunflower Evaluation Panel
Twenty-four significant SNP markers identified in the GWAS analysis were surveyed in the evaluation panel comprised of 548 sunflower lines from the fertility restorer and maintainer heterotic groups, and unassigned PIs. A total of 133 sunflower lines shared all 24 significant SNP marker alleles, of which 92 were USDA released restorer inbred lines, 19 were USDA germplasm lines and another 19 were PI accessions (Table 4 and Supplementary Table S4). Surprisingly, three inbred maintainer lines, HA 452, HA 821 (LP-1), and HA 821 (LS-1), which do not restore fertility in sunflower, also shared all 24 significant SNP marker alleles. Seven restorer lines, RHA 266, RHA 271, RHA 273, RHA 274, RHA 296, RHA 325, and RHA 439, known to possess the Rf1 gene on LG13, belong to this group of 133 sunflower lines, suggesting that these SNPs are associated with the Rf1gene (Supplementary Table S4). In the present study, we mapped the Rf7 gene from the restorer line RHA 428 on LG13, which shared only seven significant SNP marker alleles detected in the GWAS analysis (Supplementary Table S4). The two inbreed lines, RHA 801 and RHA 365, used as parents in the RHA 428 pedigree shared 24 and 17 significant SNP marker alleles, respectively, different from those of RHA 428 (Supplementary Table S4). Also, the restorer line HA-R9 possessing Rf5 on LG13, shared only five alleles out of 24 significant SNP marker alleles (Supplementary Table S4). Two restorer inbred lines, RHA 280 and RHA 340, known to possess the Rf3 gene on LG7, shared only 3–4 significant SNP marker alleles (Supplementary Table S4). Eighteen USDA released restorer inbred lines and three germplasm lines known to restore fertility in sunflower with unknown Rf genes shared 2–18 of the significant SNP markers (Table 4 and Supplementary Table S4).
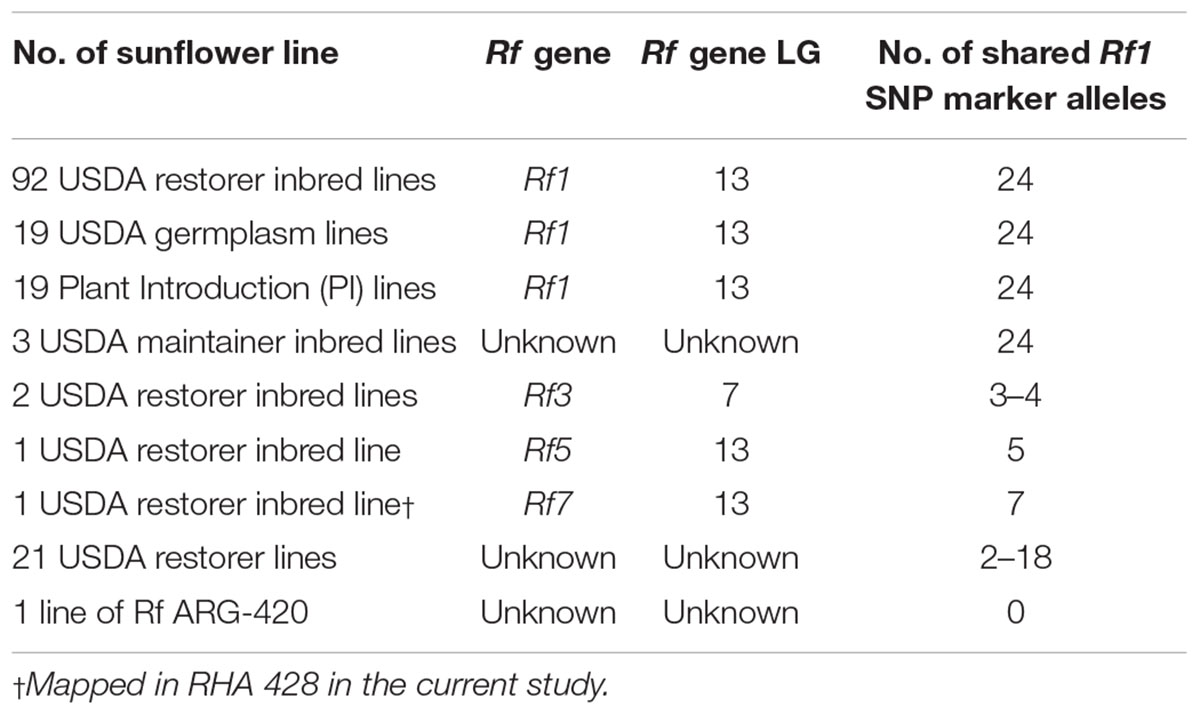
Table 4. Distribution of the fertility restoration (Rf) gene in the world sunflower collection panel of 548 lines.
Discussion
Crop wild relatives (CWR) of sunflower have revolutionized sunflower by providing many genes of utmost economic value, for example, CMS/Rf genes for commercial hybrid production, disease resistance genes for rust, DM, Sclerotinia wilt and rot, Phomopsis stem canker, Verticillium wilt, Alternaria leaf spot, and herbicide resistance genes (for review see Seiler et al., 2017). In the present study, we utilized a biparental linkage mapping approach to map the Rf7 gene in RHA 428, and an association mapping approach to discover the Rf1 haplotype and compare it to this and other Rf loci. RHA 428 is a progeny derived from the cross involving a wild H. annuus accession (PI 413157). Both the male fertility restoration and the DM resistance genes in RHA 428 are derived from PI 413157. Linkage analysis using SSR markers mapped the Rf7 gene to the lower end of LG13, only 2.1 cM from the Pl34 DM resistance gene (Figure 1C). A pair of co-segregating SSR markers, ORS316 and ORS191, were mapped in between the two genes at 1.2 cM proximal to Rf7 and 0.9 cM distal to Pl34 (Rf7/ORS316 and ORS191/Pl34). Additional SNP markers selected from two high density SNP maps (Bowers et al., 2012; Talukder et al., 2014) saturated the genomic region around both genes and delimit the genes within even narrower intervals. Five of these SNP markers were co-segregating with the new Rf7 gene, a useful resource for MAS breeding of fertility restoration (Figure 1C).
Rf Gene Cluster in Sunflower and Other Crops
The lower end of LG13 is very significant for sunflower breeders as many genes of economic importance have been reported to cluster at this genomic region. Qi et al. (2012) mapped a fertility restoration gene, Rf5 in the wild H. annuus-derived Rf ANN-1742 sunflower line, which is tightly linked to the sunflower rust resistance gene R11 (Figure 1B). In the current study, the SSR marker ORS316 mapped at 1.2 cM proximal to Rf7 also mapped at 5.3 cM proximal to the Rf5 gene in the Qi et al. (2012) map. The most used male fertility restorer gene, Rf1, was also mapped to the lower end of LG13 (Gentzbittel et al., 1995, 1999; Berry et al., 1997; Horn et al., 2003; Yu et al., 2003; Kusterer et al., 2005; Yue et al., 2010). Earlier authors used other DNA markers, including RFLP, amplified fragment length polymorphism (AFLP), random-amplified polymorphism DNA (RAPD) or target region amplification polymorphism (TRAP) markers, to map the Rf1 gene. Because these markers are not routinely used and can have unclear results, they could not be used for comparative mapping. However, the latest effort of Rf1 gene mapping (Yue et al., 2010) revealed that an SSR marker, ORS511, mapped at 3.7 cM distal to Rf1 gene (Figure 1A). The same marker in the current study maps 0.9 cM distal to Rf7 gene (Figure 1C). Coincident genomic locations of Rf1, Rf5, and Rf7 are a strong indication that these genes are clustered on the lower end of LG13. Clustering of Rf genes has been observed in other species, for example, four fertility restorers, Fr, Fr2, FrPI207228, and FrXR235 were mapped to the same linkage group in common bean (Jia et al., 1997), in rice Rf1a, Rf1b, Rf4 and Rf5, which encode a PPR protein, are clustered on chromosome 10 near the Rf1 locus (Zhang et al., 2002; Akagi et al., 2004; Komori et al., 2004; Wang et al., 2006; Fujii et al., 2008; Hu et al., 2012; Kazama and Toriyama, 2014; Melonek et al., 2016). PPR-gene clusters have also been reported at petunia Rf (Bentolila et al., 2002), radish Rfo/Rfk1 (Brown et al., 2003; Koizuka et al., 2003) and, Rf1 and Rf2 of monkeyflower (Mimulus guttatus) on LG7 (Barr and Fishman, 2010).
Relationship of the Rf Genes in the LG13 Cluster
The male fertility restorer gene Rf1 was discovered in the sunflower line T66006-2-1, derived from a cross involving a wild H. annuus accession from Texas (Supplementary Figure S1). The other two Rf genes, Rf5 and Rf7, mapped in the same LG13 were derived from sunflower lines developed using two independent wild H. annuus accessions. The H. annuus accession of the Rf5 gene (PI 613748) was collected from Oklahoma, while the wild accession of the Rf7 gene (PI 413157) in our study was a collection from New Mexico, United States. In the present study, 24 significant SNP markers were identified to be associated with the Rf1 gene. Of the 159 restorer lines in an evaluation panel of 548 sunflower lines, 130 lines retained all 24 SNP alleles. This finding is consistent with previous reports of widespread introduction of the dominant nuclear restorer Rf1 gene in the sunflower breeding materials (Korell et al., 1992; Serieys, 2005; Jan and Vick, 2007). It seems that 24 significant SNP alleles associated with Rf1 were transmitted as a haplotype for over five decades (1970–2011) of breeding and the development of 92 restorer lines (Table 4 and Supplementary Table S4). In addition, 19 restorer germplasm lines with restoration from different wild species and 19 PI lines collected from 10 different countries also retained 24 significant SNP alleles. However, in the comparison of 24 SNP marker alleles associated with the Rf1 gene to HA-R9 (Rf5) and RHA 428 (Rf7), only five and seven SNPs retained the Rf1 alleles in HA-R9 and RHA 428, respectively (Supplementary Table S4). In addition, one NSA SNP marker, NSA_008361 co-segregating with Rf7 identified in the present study was not found to be associated with Rf1 in GWAS analysis (Table 3). Rf5 is linked to a rust R gene R11, while Rf7 is linked to a DM R gene Pl34 (Figures 1B,C). Taken together, it indicated that the Rf5 and Rf7 genes are different in genomic composition than the Rf1 gene. However, we cannot rule out the possibility that Rf7 could potentially be an alternate Rf1 source to that of the original wild H annuus in Texas. Further characterization of these closely linked genes would elucidate the evolutionary relationships among Rf1, Rf5, and Rf7 by additional fine mapping combined with a whole genome resequencing approach.
Due to the size and the complexity of the sunflower genome, cloning of sunflower Rf genes has not been successful yet. Owens et al. (2018) recently reported a candidate gene, HanXRQChr13g0419821 which encodes an aldehyde dehydrogenase gene for Rf1. HanXRQChr13g0419821 is located at the 174,082,899 bp position within the interval of 24 significant SNPs associated with Rf1 (Table 3). Meanwhile, map and sequence-based analysis of the Rf5 gene region on HA412-HO and XRQ genome assemblies identified two candidate genes for Rf5, which encodes PPR proteins (Qi et al., 2018).
Surprisingly, three USDA-ARS released inbred lines, HA 452, HA 821 (LP-1) and HA 821 (LS-1) also shared all 24 SNP marker alleles associated with Rf1. These inbred lines are maintainer lines with no Rf allele. HA 452 is an F6 derived F7 line selected from the cross of two maintainer lines, HA 335/HA 412 (Miller et al., 2006). HA 821 (LP-1) and HA 821 (LS-1) were derived from mutagenesis of a maintainer line, HA 821 (Miller and Vick, 1999). While the pedigree of these sunflower lines clearly suggests that these are maintainer lines, the exact reason for the presence of positive SNP alleles associated with Rf1 gene is not known.
A Strategy for Using Pl34 With Rf7 in Sunflower Breeding
In this study, we also mapped a DM resistance gene, Pl34, at the lower end of LG13 that is tightly linked to the Rf7 fertility gene, at a genetic distance of 2.1 cM. Three additional DM resistance genes, Pl5, Pl8 and Pl21, have been previously reported to map in this genomic region (Bert et al., 2001; Radwan et al., 2003; Bachlava et al., 2011; Vincourt et al., 2012; Qi et al., 2017). Comparative analysis of map location implies that Pl34, Pl8, Pl5, and Pl21 are positioned at 0.9, 1.2, 5.1, and 13.2 cM proximal to the common SSR marker, ORS316 (Vincourt et al., 2012; Qi et al., 2017), suggesting that Pl21 is little farther away from the remaining three-gene in the cluster. The Pl5 and Pl8 genes originated from wild H. tuberosus and H. argophyllus, respectively, while the Pl34 was derived from wild H. annuus. In addition to their diverse origin, Pl5, Pl8, and Pl34 also showed differential response against a recent collection of 185 P. halstedii isolates in the United States (Gilley et al., 2016). Two diagnostic SNP markers, NSA_000423 and NSA_002220, for Pl8 were not mapped in the Pl34 map (Qi et al., 2017). Taken together, this indicates that Pl34 is a different gene from Pl5 and Pl8. It appears that Pl34 might not be a good choice as a sole defense against the damage caused by the DM but could be a good candidate for pyramiding Pl resistance in sunflower lines that could preferentially be transferred along with the tightly linked Rf7 fertility restoration gene identified in this study. The Rf1 gene has been extensively used in sunflower breeding over four decades for hybrid seed production. The use of the new Rf7 gene would potentially diversify the genetic makeup of the hybrids. The high-throughput amenable SNP markers co-segregating with the Rf7 gene will expedite MAS and transfer of the Rf allele into elite breeding lines through conventional breeding.
Data Availability
The datasets generated for this study can be found in the publicly accessible repository: figshare (https://doi.org/10.6084/m9.figshare.7754378.v1 and https://doi.org/10.6084/m9.figshare.7754420.v1).
Ethics Statement
The experiments were performed in compliance with current laws of the United States.
Author Contributions
LQ, BH, and C-CJ conceived and designed the experiments. All authors performed the experiments. ZT and LQ analyzed the data and wrote the paper. BH, GM, and C-CJ commented on the manuscript before submission.
Funding
This research was supported by the USDA-ARS CRIS Project No. 3060-21000-043-00D. Mention of trade names or commercial products in this report is solely for the purpose of providing specific information and does not imply recommendations or endorsement by the US Department of Agriculture. The USDA is an equal opportunity provider and employer.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge Nuseed Americas for providing phenotypic and genotypic data for our analyses. The authors would like to thank Angelia Hogness for her assistance in the laboratory, greenhouse, and field.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00216/full#supplementary-material
FIGURE S1 | Pedigree of the Rf1 gene origin (taken from Korell et al., 1992).
FIGURE S2 | Quantile–quantile plots of observed vs. expected –log10 (p) values from four different genome-wide association models: (i) general linear model (GLM), (ii) general linear model with population structure used as a cofactor (GLMQ), (iii) mixed linear model that accounted only kinship relatedness (MLM), and (iv) a mixed linear model that accounted for both population structure and kinship relatedness in the analysis (MLMQ). The red lines represent the expected values under the null distribution.
TABLE S1 | List of sunflower inbred/germplasm lines used for single nucleotide polymorphism marker validation associated with male fertility restoration Rf1 gene. The detailed information of each line can be found in the USDA National Plant Germplasm Systerm: https://www.ars-grin.gov/npgs/.
TABLE S2 | Map position and sequences of SNP markers on LG13 selected from published sunflower linkage maps used for map saturation in the present study.
TABLE S3 | Allele-specific polymerase chain reaction (PCR) primer sequences of polymorphic SNP markers mapped in this study.
TABLE S4 | Distribution of 24 significant SNP markers in the sunflower evaluation panel identified using GWAS analysis of fertility restoration trait. The black font color indicates SNP allele associated with fertility restoration, while the red font color indicates the alternate SNP allele.
Footnotes
References
Abratti, G., Bazzalo, M. E., and León, A. (2008). “Mapping a novel fertility restoration gene in sunflower,” in Proceedings of the 17th International Sunflower Conference, ed. L. Velasco (Paris: International Sunflower Association),617–621.
Ahmadikhah, A., and Karlov, G. (2006). Molecular mapping of the fertility-restoration gene Rf4 for WA-cytoplasmic male sterility in rice. Plant Breed. 25, 363–367. doi: 10.1111/j.1439-0523.2006.01246.x
Akagi, H., Nakamura, A., Yokozeki-Misono, Y., Inagaki, A., Takahashi, H., Mori, K., et al. (2004). Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 108, 1449–1457. doi: 10.1007/s00122-004-1591-2
Bachlava, E., Radwan, O. E., Abratti, G., Tang, S., Gao, W., Heesacker, A. F., et al. (2011). Downy mildew (Pl8 and Pl14) and rust (RAdv) resistance genes reside in close proximity to tandemly duplicated clusters of non-TIR-like NBS-LRR encoding genes on sunflower chromosomes 1 and 13. Theor. Appl. Genet. 122, 1211–1221. doi: 10.1007/s00122-010-1525-0
Barr, C. M., and Fishman, L. (2010). The nuclear component of a cytonuclear hybrid incompatibility in Mimulus maps to a cluster of pentatricopeptide repeat genes. Genetics 184, 455–465. doi: 10.1534/genetics.109.108175
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 289–300.
Bentolila, S., Alfonso, A., and Hanson, M. (2002). A pentatricopeptide repeat containing gene restores male sterility to male-sterile plants. Proc. Natl. Acad. Sci. U.S.A. 99, 10887–10892. doi: 10.1073/pnas.102301599
Berry, S. T., Leon, A. J., Peerbolte, R., Challis, C., Livini, C., Jones, R., et al. (1997). “Presentation of the Advanta sunflower RFLP linkage map for public research,” in Proceedings of the 19th Sunflower Research Forum (Mandan, ND: National Sunflower Association), 113–118.
Bert, P. F., Tourvieille de Labrouhe, D., Philippon, J., Mouzeyar, S., Jouan, I., Nicolas, P., et al. (2001). Identification of a second linkage group carrying genes controlling resistance to downy mildew (Plasmopara halstedii) in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 103, 992–997. doi: 10.1007/s001220100660
Bohra, A., Jha, U. C., Adhimoolam, P., Bisht, D., and Singh, N. P. (2016). Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep. 35, 967–993. doi: 10.1007/s00299-016-1949-3
Bowers, J. E., Bachlava, E., Brunick, R. L., Knapp, S. J., and Burke, J. M. (2012). Development of a 10,000 locus genetic map of the sunflower genome based on multiple crosses. G3 2, 721–729. doi: 10.1534/g3.112.002659
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Brown, G. G., Formanova, N., Jin, H., Wargachuk, R., Dendy, C., Patil, P., et al. (2003). The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35, 262–272. doi: 10.1046/j.1365-313X.2003.01799.x
Chase, C. D. (2007). Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23, 81–90. doi: 10.1016/j.tig.2006.12.004
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Dimitrijevic, A., and Horn, R. (2018). Sunflower hybrid breeding: from markers to genomic selection. Front. Plant Sci. 8:2238. doi: 10.3389/fpls.2017.02238
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Feng, J., and Jan, C. C. (2008). Introgression and molecular tagging of Rf4, a new male fertility restoration gene from wild sunflower Helianthus maximiliani L. Theor. Appl. Genet. 117, 241–249. doi: 10.1007/s00122-008-0769-4
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Fujii, S., Bond, C. S., and Small, I. D. (2011). Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. U.S.A. 108, 1723–1728. doi: 10.1073/pnas.1007667108
Fujii, S., Kazama, T., and Toriyama, K. (2008). “Molecular studies on cytoplasmic male sterility-associated genes and restorer genes in rice,” in Rice Biology in the Genomics Era, eds H. Y. Hirano, A. Hirai, Y. Sano, and T. Sasaki (Berlin, Germany: Springer Publisher), 205–216.
Gascuel, Q., Martinez, Y., Boniface, M.-C., Vear, F., Pichon, M., and Godiard, L. (2015). The sunflower downy mildew pathogen Plasmopara halstedii. Mol. Plant Path. 16, 109–122. doi: 10.1111/mpp.12164
Gentzbittel, L., Mestries, E., Mouzeyar, S., Mazeyrat, F., Badaoui, S., Vear, F., et al. (1999). A composite map of expressed sequences and phenotypic traits of the sunflower (Helianthus annuus L.) genome. Theor. Appl. Genet. 99, 218–234. doi: 10.1007/s001220051228
Gentzbittel, L., Vear, F., Zhang, Y. X., and Berville, A. (1995). Development of a consensus linkage RFLP map of cultivated sunflower (Helianthus annuus L.). Theor. Appl. Genet. 90, 1079–1086. doi: 10.1007/BF00222925
Gilley, M. A., Misar, C. G., Gulya, T. J., and Markell, S. G. (2016). “Prevalence and virulence of Plasmopara halstedii (downy mildew) in sunflowers,” in Proceeding of the 38th Sunflower Research Forum (Mandan, ND: National Sunflower Association).
Gulya, T. J., Markell, S., McMullen, M., Harveson, B., and Osborne, L. (2011). “New virulent races of downy mildew: distribution, status of DM resistant hybrids, and USDA sources of resistance,” in Proceedings of the 33th Sunflower Research Forum (Mandan, ND: National Sunflower Association).
Gulya, T. J., Miller, J. F., Viranyi, F., and Sackston, W. E. (1991). Proposed internationally standardized methods for race identification of Plasmopara halstedii. Helia 14, 11–20.
Hardy, O. J., and Vekemans, X. (2002). SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620. doi: 10.1046/j.1471-8286.2002.00305.x
Harvey, M. (2004). “The use of cytoplasmic male sterility for hybrid seed production,” in Molecular Biology and Biotechnology of Plant Organelles, eds H. Daniell and C. Chase (Dordrecht: Springer Publishers), 617–628.
Horn, R., Kusterer, B., Lazarescu, E., Prufe, M., and Friedt, W. (2003). Molecular mapping of the Rf1 gene restoring pollen fertility in PET1-based F1 hybrids in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 106, 599–606. doi: 10.1007/s00122-002-1078-y
Hu, J., Wang, K., Huang, W., Liu, G., Gao, Y., Wang, J., et al. (2012). The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine rich protein GRP162. Plant Cell 24, 109–122. doi: 10.1105/tpc.111.093211
Huang, Q., He, Y., Jing, R., Zhu, R., and Zhu, Y. (2000). Mapping of the nuclear fertility restorer gene for HL cytoplasmic male sterility in rice using microsatellite markers. Chin. Sci. Bull. 45, 430–432. doi: 10.1007/BF02884944
Jan, C. C., and Vick, B. A. (2007). Inheritance and allelic relationships of fertility restoration genes for seven new sources of male-sterile cytoplasm in sunflower. Plant Breed. 126, 213–217. doi: 10.1111/j.1439-0523.2007.01350.x
Jia, M. H., He, S., Vanhouten, W., and Mackenzie, S. (1997). Nuclear fertility restorer genes map to the same linkage group in cytoplasmic male-sterile bean. Theor. Appl. Genet. 95, 205–210. doi: 10.1007/s001220050549
Kazama, T., and Toriyama, K. (2014). A fertility restorer gene, Rf4, widely used for hybrid rice breeding encodes a pentatricopeptide repeat protein. Rice 7:28. doi: 10.1186/s12284-014-0028-z
Kinman, M. L. (1970). “New development in the USDA and state experiment station sunflower breeding programs,” in Proceedings of the 4th International Sunflower Conference (Paris: International Sunflower Association), 181–183.
Koizuka, N., Imai, R., Fujimoto, H., Hayakawa, T., Kimura, Y., Kohno-Murase, J., et al. (2003). Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 34, 407–415. doi: 10.1046/j.1365-313X.2003.01735.x
Komori, T., Ohta, S., Murai, N., Takakura, Y., Kuraya, Y., Suzuki, S., et al. (2004). Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). Plant J. 37, 315–325. doi: 10.1046/j.1365-313X.2003.01961.x
Korell, M., Mösges, G., and Friedt, W. (1992). Construction of a sunflower pedigree map. Helia 15, 7–16.
Kusterer, B., Horn, R., and Friedt, W. (2005). Molecular mapping of the fertility restoration locus Rf1 in sunflower and development of diagnostic markers for the restorer gene. Euphytica 143, 35–43. doi: 10.1007/s10681-005-1795-9
Laser, K. D., and Lersten, N. R. (1972). Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot. Rev. 38, 425–454. doi: 10.1007/BF02860010
Leclercq, P. (1969). Une stérilité mâle cytoplasmique chez le tournesol. Ann. Amélior. Plantes 19, 99–106.
Levings, C. S. (1990). The Texas cytoplasm of maize: cytoplasmic male sterility and disease susceptibility. Science 250, 942–947. doi: 10.1126/science.250.4983.942
Liu, X. Q., Xu, X., Tan, Y. P., Li, S. Q., Hu, J., Huang, J. Y., et al. (2004). Inheritance and molecular mapping of two fertility-restoring loci for Honglian gametophytic cytoplasmic male sterility in rice (Oryza sativa L.). Mol. Genet. Genomics 276, 586–594. doi: 10.1007/s00438-004-1005-9
Liu, Z., Mulpuri, S., Feng, J., Vick, B. A., and Jan, C. C. (2012). Molecular mapping of the Rf3 fertility restoration gene to facilitate its utilization in breeding confection sunflower. Mol. Breed. 29, 275–284. doi: 10.1007/s11032-011-9563-0
Liu, Z., Wang, D. M., Feng, J., Seiler, G. J., Cai, X., and Jan, C. C. (2013). Diversifying sunflower germplasm by integration and mapping of a novel male fertility restoration gene. Genetics 193, 727–737. doi: 10.1534/genetics.112.146092/-/DC1
Liu, Z., Zhang, L., Ma, G. J., Seiler, G. J., Jan, C. C., and Qi, L. L. (2018). Molecular mapping of the downy mildew and rust resistance genes in a sunflower germplasm line TX16R. Mol. Breed. 39:19. doi: 10.1007/s11032-018-0921-z
Loiselle, B. A., Sork, V. L., Nason, J., and Graham, C. (1995). Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 82, 1420–1425. doi: 10.2307/2445869
Long, Y. M., Chao, W. S., Ma, G. J., Xu, S. S., and Qi, L. L. (2017). An innovative SNP genotyping method adapting multiple platforms and throughputs. Theor. Appl. Genet. 130, 597–607. doi: 10.1007/s00122-016-2838-4
Ma, G. J., Markell, S. G., Song, Q. J., and Qi, L. L. (2017). Genotyping-by-sequencing targeting of a novel downy mildew resistance gene Pl20 from wild Helianthus argophyllus for sunflower (Helianthus annuus L.). Theor. Appl. Genet. 130, 1519–1529. doi: 10.1007/s00122-017-2906-4
Meliala, C., Vear, F., and Tourvieille de Labrouhe, D. (2000). Relation between date of infection of sunflower downy mildew (Plasmopara halstedii) and symptoms development. Helia 23, 35–44.
Melonek, J., Stone, J. D., and Small, I. (2016). Evolutionary plasticity of restorer of-fertility-like proteins in rice. Sci. Rep. 6:35152. doi: 10.1038/srep35152
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Miller, J. F., Gulya, T. J., and Seiler, G. J. (2002). Registration of five fertility restorer sunflower germplasms. Crop Sci. 42, 989–991. doi: 10.2135/cropsci2002.9890
Miller, J. F., Gulya, T. J., and Vick, B. A. (2006). Two maintainer and three restorer Sclerotinia-tolerant oilseed sunflowers. Crop Sci. 46, 2727–2728. doi: 10.2135/cropsci2006.06.0436
Miller, J. F., and Vick, B. A. (1999). Registration of three low palmitic acid and five low stearic acid sunflower genetic stocks. Crop Sci. 39, 305–306. doi: 10.2135/cropsci1999.0011183X003900010080x
Nordborg, M., and Weigel, D. (2008). Next-generation genetics in plants. Nature 456, 720–723. doi: 10.1038/nature07629
Owens, G. L., Baute, G. J., Hubner, A., and Rieseberg, L. H. (2018). Genomic sequences and copy number evolution during hybrid crop development in sunflowers. Evol. Appl. 11, 1–12. doi: 10.111/eva.12603
Pecrix, Y., Buendia, L., Penouilh-Suzette, C., Maréchaux, M., Legrand, L., Bouchez, Q., et al. (2018a). Sunflower resistance to multiple downy mildew pathotypes revealed by recognition of conserved effectors of the oomycete Plasmopara halstedii. Plant J. 97, 730–748. doi: 10.1111/tpj.14157
Pecrix, Y., Penouilh-Suzette, C., Stéphane Muños, S., Felicity Vear, F., and Godiard, L. (2018b). Ten broad spectrum resistances to downy mildew physically mapped on the sunflower genome. Front. Plant Sci. 9:1780. doi: 10.3389/fpls.2018.01780
Pegadaraju, V., Nipper, R., Hulke, B. S., Qi, L. L., and Schultz, Q. (2013). De novo sequencing of the sunflower genome for SNP discovery using the RAD (Restriction site Associated DNA) approach. BMC Genomics 14:556. doi: 10.1186/1471-2164-14-556
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155,945–959.
Qi, L. L., Gulya, T., Seiler, G. J., Hulke, B. S., and Vick, B. A. (2011). Identification of resistance to new virulent races of rust in sunflowers and validation of DNA markers in the gene pool. Phytopathology 101, 241–249. doi: 10.1094/PHYTO-06-10-0162
Qi, L. L., Long, Y. M., Jan, C. C., Ma, G. J., and Gulya, T. J. (2015). Pl17 is a novel gene independent of known downy mildew resistance genes in the cultivated sunflower (Helianthus annuus L.). Theor. Appl. Genet. 128, 757–767. doi: 10.1007/s00122-015-2470-8
Qi, L. L., Long, Y. M., Talukder, Z. I., Ma, G. J., Song, Q. J., and Seiler, G. J. (2018). “Map and sequence-based candidate gene analysis of the male fertility restoration gene Rf5 in sunflower,” in Proceedings of the International Symposium “Sunflower and Climate Change” (Paris: International Sunflower Association), 32.
Qi, L. L., Seiler, G. J., Vick, B. A., and Gulya, T. J. (2012). Genetics and mapping of the R11 gene conferring resistance to recently emerged rust races, tightly linked to male fertility restoration, in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 125, 921–932. doi: 10.1007/s00122-012-1883-x
Qi, L. L., Talukder, Z. I., Hulke, B. S., and Foley, M. E. (2017). Development and dissection of diagnostic SNP markers for the downy mildew resistance genes PlArg and Pl8 and maker assisted gene pyramiding in sunflower (Helianthus annuus L.). Mol. Genet. Genomics 292, 551–563. doi: 10.1007/s00438-017-1290-8
R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Radwan, O., Bouzidi, M. F., Vear, F., Philippon, J., Tourvieille de Labrouhe, D., Nicolas, P., et al. (2003). Identification of non-TIRNBS-LRR markers linked to the Pl5/Pl8 locus for resistance to downy mildew in sunflower. Theor. Appl. Genet. 106, 1438–1446. doi: 10.1007/s00122-003-1196-1
Scheet, P., and Stephens, M. (2006). A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78, 629–644. doi: 10.1086/502802
Schnabel, U., Engelmann, U., and Horn, R. (2008). Development of markers for the use of the PEF1-cytoplasm in sunflower hybrid breeding. Plant Breed. 127, 587–591.
Schnable, P. S., and Wise, R. P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180. doi: 10.1016/S1360-1385(98)01235-7
Seiler, G. J., Qi, L. L., and Marek, L. F. (2017). Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 57, 1–19. doi: 10.2135/cropsci2016.10.0856
Seiler, G. J., and Rieseberg, L. H. (1997). “Systematic, origin, and germplasm resources of the wild and domesticated sunflower,” in Sunflower Technology and Production, ed. A. A. Schneiter (Madison,WI: ASA), 21–66.
Serieys, H. (2005). “Identification, study and utilization in breeding programs of new CMS sources in the FAO subnetwork,” in Proceedings of the Sunflower Subnetwork Progress Report (Rome: FAO), 47–53.
Talukder, Z. I., Gong, L., Hulke, B. S., Pegadaraju, V., Song, Q., Schultz, Q., et al. (2014). A high-density SNP map of sunflower derived from RAD-sequencing facilitating fine-mapping of the rust resistance gene R12. PLoS One 9:e98628. doi: 10.1371/journal.pone.0098628
Tang, S., Yu, J. K., Slabaugh, M. B., Shintani, D. K., and Knapp, S. J. (2002). Simple sequence repeat map of the sunflower genome. Theor. Appl. Genet. 105, 1124–1136. doi: 10.1007/s00122-002-0989-y
Van Ooijen, J. W. (2006). JoinMap® 4.0: Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen: Kyazma BV.
Vincourt, P., As-sadi, F., Bordat, A., Langlade, N. B., Gouzy, J., Pouilly, N., et al. (2012). Consensus mapping of major resistance genes and independent QTL for quantitative resistance to sunflower downy mildew. Theor. Appl. Genet. 125, 909–920. doi: 10.1007/s00122-012-1882-y
Viranyi, F., and Spring, O. (2011). Advances in sunflower downy mildew research. Eur. J. Plant Pathol. 129, 207–220. doi: 10.1007/s10658-010-9683-0
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Vranceanu, A. V., and Stoenescu, F. M. (1970). Immunity to sunflower downy mildew due to a single dominant gene. Probl. Agric. 22, 34–40.
Vranceanu, A. V., and Stoenescu, F. M. (1971). Pollen fertility restorer gene from cultivated sunflower (Helianthus annuus L.). Euphytica 20, 536–541.
Vranceanu, A. V., and Stoenescu, F. M. (1978). Gene for pollen fertility restoration in sunflowers. Euphytica 27, 617–627. doi: 10.1007/BF00043193
Wang, Z., Zou, Y., Zhang, Q., Li, X., Chen, L., Wu, H., et al. (2006). Cytoplasmic male sterility of rice with Boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18, 676–687. doi: 10.1105/tpc.105.038240
Yu, J., Pressoir, G., Briggs, W. H., Vroh, B. I., Yamasaki, M., Doebley, J. F., et al. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. doi: 10.1038/ng1702
Yu, J. K., Tang, S., Slabaugh, M. B., Heesacker, A., Cole, G., Herring, M., et al. (2003). Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci. 43, 367–387. doi: 10.2135/cropsci2003.3670
Yue, B., Vick, B. A., Cai, X., and Hu, J. (2010). Genetic mapping for the Rf1 (fertility restoration) gene in sunflower (Helianthus annuus L.) by SSR and TRAP markers. Plant Breed. 129, 24–28. doi: 10.1111/j.1439-0523.2009.01661.x
Zhang, Q. Y., Liu, Y. G., Zhang, G. Q., and Mei, M. T. (2002). Molecular mapping of the fertility restorer gene Rf4 for WA cytoplasmic male sterility in rice. Acta Genet. Sin. 29, 1001–1004.
Keywords: sunflower, restorer-of-fertility, downy mildew, linkage mapping, genome-wide association study, gene cluster
Citation: Talukder ZI, Ma G, Hulke BS, Jan C-C and Qi L (2019) Linkage Mapping and Genome-Wide Association Studies of the Rf Gene Cluster in Sunflower (Helianthus annuus L.) and Their Distribution in World Sunflower Collections. Front. Genet. 10:216. doi: 10.3389/fgene.2019.00216
Received: 14 November 2018; Accepted: 27 February 2019;
Published: 14 March 2019.
Edited by:
Badri Padhukasahasram, Illumina, United StatesReviewed by:
Felicity Vear, INRA Centre Auvergne-Rhône-Alpes, FranceReza Darvishzadeh, Urmia University, Iran
Shuancang Yu, Beijing Vegetable Research Center, China
Copyright © 2019 Talukder, Ma, Hulke, Jan and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lili Qi, bGlsaS5xaUBhcnMudXNkYS5nb3Y=
 Zahirul I. Talukder
Zahirul I. Talukder Guojia Ma
Guojia Ma Brent S. Hulke
Brent S. Hulke Chao-Chien Jan2
Chao-Chien Jan2 Lili Qi
Lili Qi