- 1NHC Key Laboratory of Reproduction Regulation (Shanghai Institute of Planned Parenthood Research), Fudan University, Shanghai, China
- 2Reproductive Medical Center, Changhai Hospital of Shanghai, Shanghai, China
- 3Department of Obstetrics and Gynecology, International Peace Maternity and Child Health Hospital, Shanghai Jiao Tong University, Shanghai, China
- 4Huzhou Key Laboratory of Molecular Medicine, Huzhou Central Hospital, Zhejiang, China
- 5Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders, Ministry of Education, Shanghai Jiao Tong University, Shanghai, China
- 6Shanghai Institute of Planned Parenthood Research Hospital, Shanghai, China
In this study, we examined whether smoking and drinking affect sperm quality and the DNA methylation of the repetitive element LINE-1, MEST, P16, H19, and GNAS in sperm. Semen samples were obtained from 143 male residents in a minority-inhabited district of Guizhou province in southwest China. Quantitative DNA methylation analysis of the samples was performed using MassARRAY EpiTYPER assays. Sperm motility was significantly lower in both the nicotine-exposed (P = 0.0064) and the nicotine- and alcohol-exposed (P = 0.0008) groups. Follicle-stimulating hormone (FSH) levels were higher in the nicotine-exposed group (P = 0.0026). The repetitive element LINE-1 was hypermethylated in the three exposed groups, while P16 was hypomethylated in the alcohol and both the alcohol and nicotine exposure groups. Our results also show that alcohol and nicotine exposure altered sperm cell quality, which may be related to the methylation levels of MEST and GNAS. In addition, MEST, GNAS, and the repetitive element LINE1 methylation was significantly associated with the concentration of sperm as well as FSH and luteinizing hormone levels.
Introduction
Infertility is a major public health concern that affects 10%–20% of married couples attempting to conceive, and male infertility is the only or a common factor (Sharlip et al., 2002; Dada et al., 2008). Recent epidemiological studies have provided plenty of pieces of evidence that environmental exposure, lifestyle, and DNA methylation are closely related to male infertility (Kobayashi et al., 2017; Laqqan et al., 2017; Nasri et al., 2017; Santi et al., 2017; Donkin and Barres, 2018; Siddeek et al., 2018). Our studies before had shown associations of aberrant DNA methylation of several genes in spermatozoa with male infertility, but the study before chose only an asthenozoospermia patient, and other environmental elements such as smoking and drinking were not examined (Xu et al., 2016). It has been shown that long-term alcohol consumption and tobacco use have adverse effects on fertility (Besingi and Johansson, 2014; Hamad et al., 2018). Tobacco exposure is associated with impaired fecundability (Chavarro et al., 2009), slightly lower sperm viability, and reduced ejaculate volume, and spermatocyte apoptosis and disruption of the seminiferous tubules were observed (Kunzle et al., 2003; Gunes et al., 2018). Although cigarette smoking and alcohol consumption have been shown to affect DNA methylation patterns of human sperm, be related to semen quality, and have effects on endocrine control of reproductive and sexual functions, their effects on semen parameters is controversial (Gaur et al., 2010; Hansen et al., 2012; Povey et al., 2012; Chang et al., 2017; Laqqan et al., 2017; Alkhaled et al., 2018; Al Khaled et al., 2018).
Several studies have shown that smoking and drinking have similar effects on oxidative stress and DNA methylation in males of reproductive age and animal models, as has been observed for other chemicals, such as cadmium and bisphenol A (BPA) (Besingi and Johansson, 2014; Miao et al., 2014; Pierron et al., 2014; Jenkins et al., 2017; McCarthy et al., 2018). In imprinted genes, the methylation level present at pro-meiosis should be maintained throughout the male gamete development (Yaman and Grandjean, 2006; Marques et al., 2011). Nicotine exposure may alter the methylation of imprinted and non-imprinted genes in sperm that are associated with oligozoospermia and asthenozoospermia (Dai et al., 2017; Dong et al., 2017; Laqqan et al., 2017). Studies have also shown that chronic paternal alcohol exposure induced behavioral abnormalities in offspring due to alterations in the methylation of imprinted genes in sperm (Kim et al., 2014; Liang et al., 2014; Chastain and Sarkar, 2017).
Mesoderm-specific transcript (MEST) and GNAS are two maternally imprinted genes that are expressed from the paternal allele. The germ-line differentially methylated regions (DMRs) in MEST and GNAS exhibit differences in methylation levels between sperm and egg. During spermatogenesis, sperm genomic imprinting (especially in germ-line DMRs) is vulnerable to environmental factors (Marques et al., 2008). In addition, alcohol exposure could cause hypomethylation of H19 in the sperm of offspring and reduce the mean sperm concentration (Marques et al., 2011), and the methylation levels of H19 are related to sperm parameters, sperm chromatin, and DNA integrity (Montjean et al., 2015; Darbandi et al., 2018).
The promoter of long interspersed nucleotide element (LINE-1), which is used as a surrogate for global methylation, is enriched with methylated CpG dinucleotides and is usually silenced in normal tissues (Rangwala et al., 2009). However, the methylation levels of the LINE-1 DMRs were lower in BPA-exposed spermatozoa and asthenozoospermia (Miao et al., 2014; Xu et al., 2016). In addition, the P16 protein may inhibit mitosis in spermatogonia and is related to a loss of testicular function (Xin-Chang et al., 2002; Jeong et al., 2017), and we showed that increased methylation defects in the P16 DMR may be associated with low sperm motility (Xu et al., 2016). The imprint and non-imprint methylation marks at these DMRs are established during gametogenesis and affected by environmental exposures (Li et al., 2016). P16 methylation is strongly associated with smoking in different pathological conditions, including lung cancer and cervical cancer (Han et al., 2016; Han et al., 2017; Wang et al., 2017). However, the relationship between these genes, tobacco/alcohol exposure, and male infertility has not yet been elucidated.
To investigate the methylation modifications that occur under exposure to alcohol and nicotine, we performed a cohort study of the methylation at the repetitive element LINE-1 and four genes (MEST, P16, H19, and GNAS) in 143 subjects. The aim of this study was to assess whether the DNA methylation of these five genes is associated with the risk of male infertility under tobacco/alcohol exposure.
Methods
Subjects and Clinical Data
This study included 143 male residents from a minority-inhabited district in Sandu county of Guizhou province. All participants were interviewed by trained Chinese-speaking researchers and were asked about their demography, disease history, and lifestyle factors, including tobacco use and alcohol consumption. The standards for smokers and drinkers were as we described before (Liang et al., 2017; Yang et al., 2017). We defined smoking as consuming at least one cigarette per day for more than 6 months and drinking as consuming an alcohol beverage (beer, wine, and liquor) at least once a week for more than 6 months (Witkiewitz et al., 2017; Pang et al., 2018). As per the standards of the World Health Organization, semen samples were collected after 2 days of abstinence. Sperm counts and motility were assessed by a computer-aided sperm analysis system (Cyto-S; Alpha Innotech Corp., San Leandro, CA, USA) at 37°C. The remainder of the semen sample was stored at −80°C until further examination and DNA extraction. Our study was approved by the Ethics Committee of Shanghai Institute of Planned Parenthood Research, and the local approval of Guizhou province was not required. The individuals included in this study gave written informed consent before participating. All procedures were carried out in accordance with the approved guidelines and local regulations.
DNA Methylation Assay
DNA was extracted from the semen samples by using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) stored at −80°C. Bisulfite conversion of DNA was carried out using the Epitect Bisulfite Kit (Qiagen). Quantitative analysis of DNA methylation was performed using MassARRAY EpiTYPER assays (Sequenom, San Diego, CA, USA) according to a published protocol (He et al., 2013). Primers used in this study were designed using Methprimer (http://epidesigner.com; Table 1). CpG units that yielded data in >90% of the samples passed the initial quality control assessment. Epigenetic changes at DMRs are important in controlling the levels of gene expression. Thus methylation was measured at 76 CpG dinucleotides in the DMRs at the repetitive element LINE-1 and four genes (LINE-1: 20 CpG sites; MEST: 7q32, 130486175–130506297, 12 CpG sites; P16: 9p21, 21967752–21995043, 18 CpG sites; H19: 11p15.5, 142575532–142578146, 14 CpG sites; and GNAS: 20q13.3, 58839740–58911195, 12 CpG sites; Table 1).
Statistical Analysis
All data were analyzed with peak picking spectra interpretation tools to generate the ratios of methyl CpG/total CpG. EpiTyper software (Sequenom, San Diego, CA, USA) was used to quantify the methylated fraction of all CpG units. Statistics 18.0 software (SPSS, Inc., Somers, NY, USA) was used to perform all the statistical analyses in this study. Pearson’s correlation coefficient test and analysis of variance followed by Dunnett’s post hoct test were used to compare the categorical variables and the differences in the mean values of continuous variables between the two groups. All tests were two-tailed. Principal component analysis (PCA) was performed to identify underlying factors. Kaiser–Meyer–Olkin (KMO) value and Bartlett’s test of sphericity were checked to confirm that the data were suitable for factor analysis. The criterion of eigenvalue >1.0 was applied to determine the number of factors retained. Items were included in the factor on which they loaded highest (minimum accepted 0.4).
Results
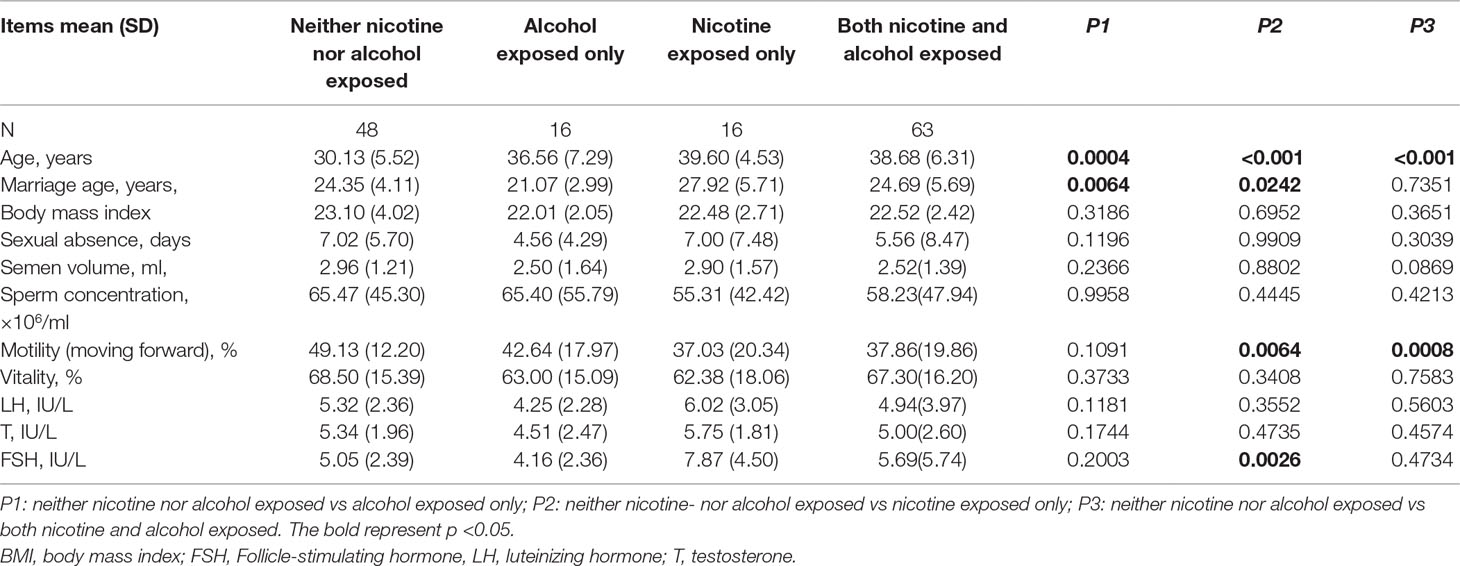
The results of the sperm motility assessment and the levels of follicle-stimulating hormone (FSH), luteinizing hormone (LH), and testosterone (T) are shown in Table 2. Sperm motility was significantly lower in the nicotine-exposed (P = 0.0064) and the nicotine- and alcohol-exposed (P = 0.0008) groups than in the control group. FSH levels were higher in the nicotine-exposed group (P = 0.0026). Methylation of each CpG site and adjusted linear regression of alcohol and nicotine exposure are shown in Suppl 1 and Suppl 2.
As shown in Figure 1, the average methylation levels of the repetitive element LINE-1 and four assessed genes in sperm from 143 minority male residents were compared. In general, the methylation levels in the repetitive element LINE-1 were higher in the three exposed groups (P < 0.001, P = 0.017, and P < 0.001, respectively) than in the control group, whereas methylation levels were lower in P16 in the nicotine-exposed group and in the nicotine- and alcohol-exposed group after correction of multiple testing (P < 0.001, Figure 1). Individual CpG sites within the same gene showed similar trends in methylation level. Compared to the controls, the methylation levels of nine CpG sites in the repetitive element LINE-1 (sites 2, 4.5.6, 7, 9, 14, 19, 20, 23, 25.26, and 27) were higher in the alcohol-exposed group, while the methylation levels of three CpG sites in the repetitive element LINE-1 (sites 7, 8, and 9) were higher in the nicotine-exposed group, and the levels of 14 CpG sites were significantly higher in the nicotine- and alcohol-exposed group (Figure 2).
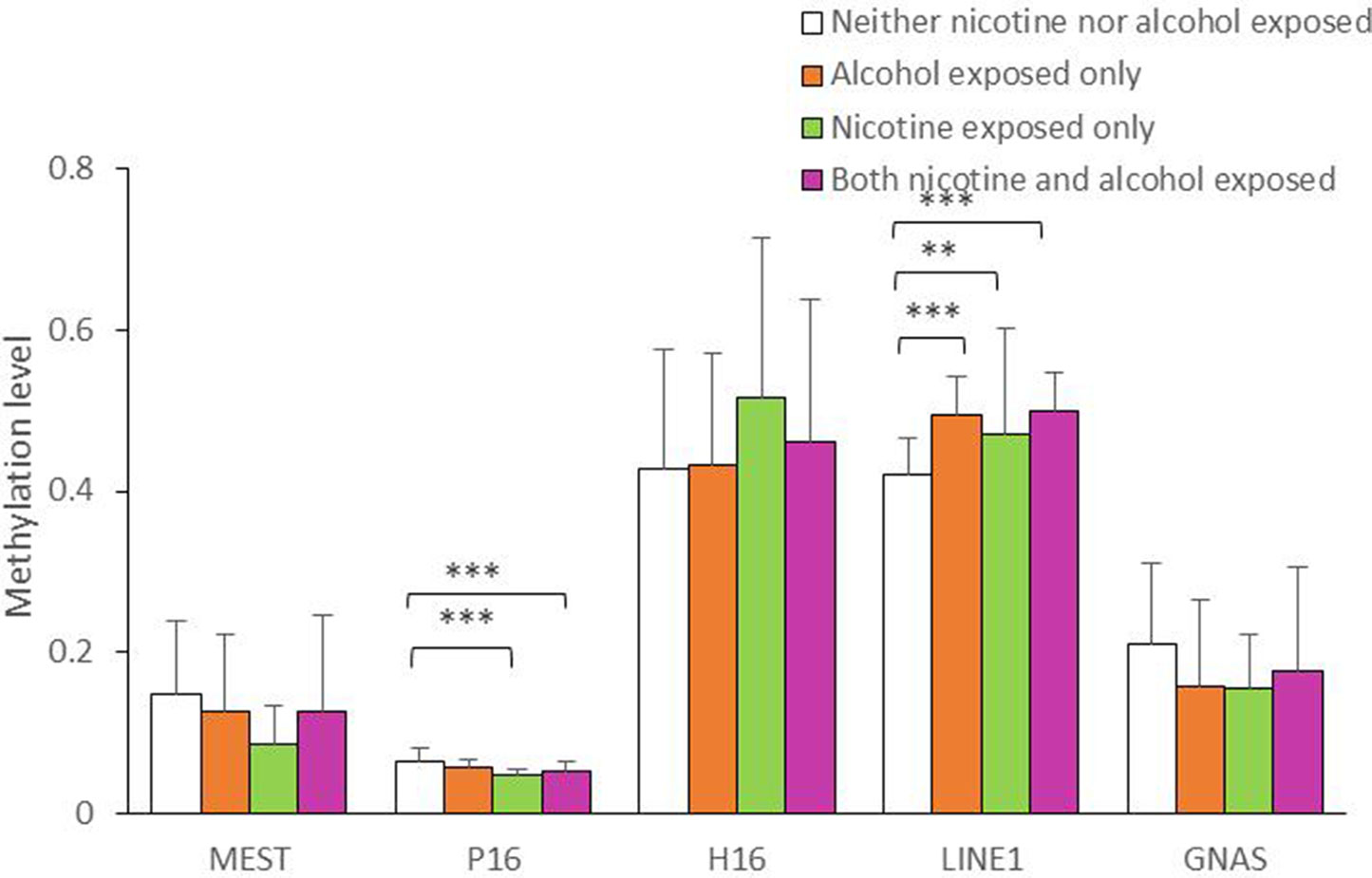
Figure 1 Average methylation level of the five genes in all the subjects. The comparison of average methylation levels at MEST, P16, H19, LINE1, and GNAS in human sperms of 143 minority male residents, respectively. Data are means ± SD. Statistically significant differences are represented with asterisks: **P < 0.01, ***P < 0.001. Not significant, P > 0.05.
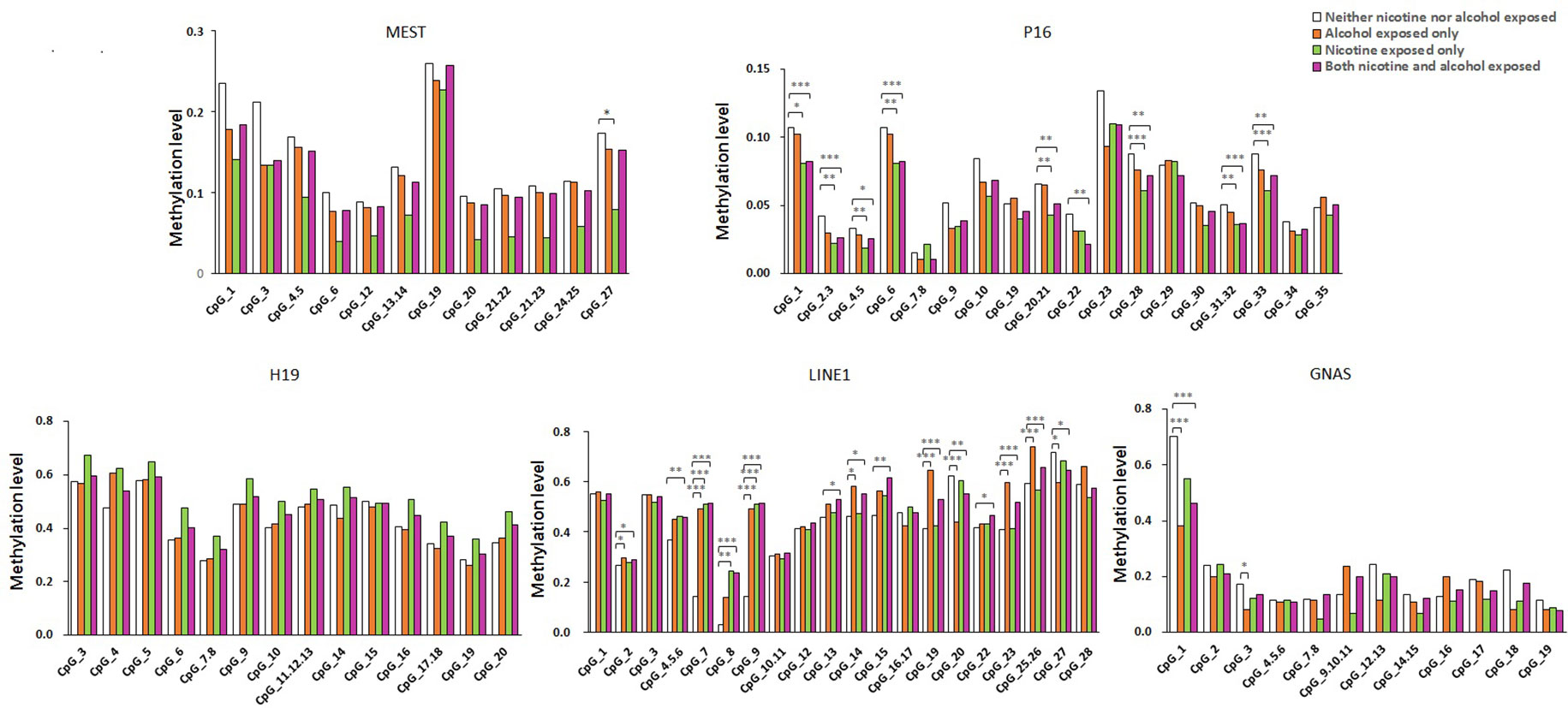
Figure 2 Methylation level of the five genes in all the subjects. The comparison of site-specific methylation levels at MEST, P16, H19, LINE1, and GNAS DMR of the neither nicotine- nor alcohol-exposed controls (N = 48) with alcohol-exposed only cases (N = 16), nicotine-exposed only cases (N = 16), both nicotine- and alcohol-exposed cases (N = 63), respectively. The data are the geometric mean ratios of methylation levels at each CpG site. *P < 0.05, **P < 0.01, ***P < 0.001. Not significant, P > 0.05
Among the imprinted genes, we found that the methylation levels of two CpG sites in the GNAS DMR (sites 1 and 3) were significantly lower in the alcohol-exposed and the nicotine- and alcohol-exposed groups, and only one site (site 11) in the MEST was lower in the nicotine-exposed group. However, the methylation levels of most CpGs in H19 did not show obvious differences among the three groups (Figure 2).
Our results showed that the methylation levels of eight CpGs in P16 in the nicotine-exposed and nicotine- and alcohol-exposed groups were lower than in the controls. In contrast, there were only one CpG sites with lower methylation levels in the alcohol-exposed group when compared to the control group (Figure 2).
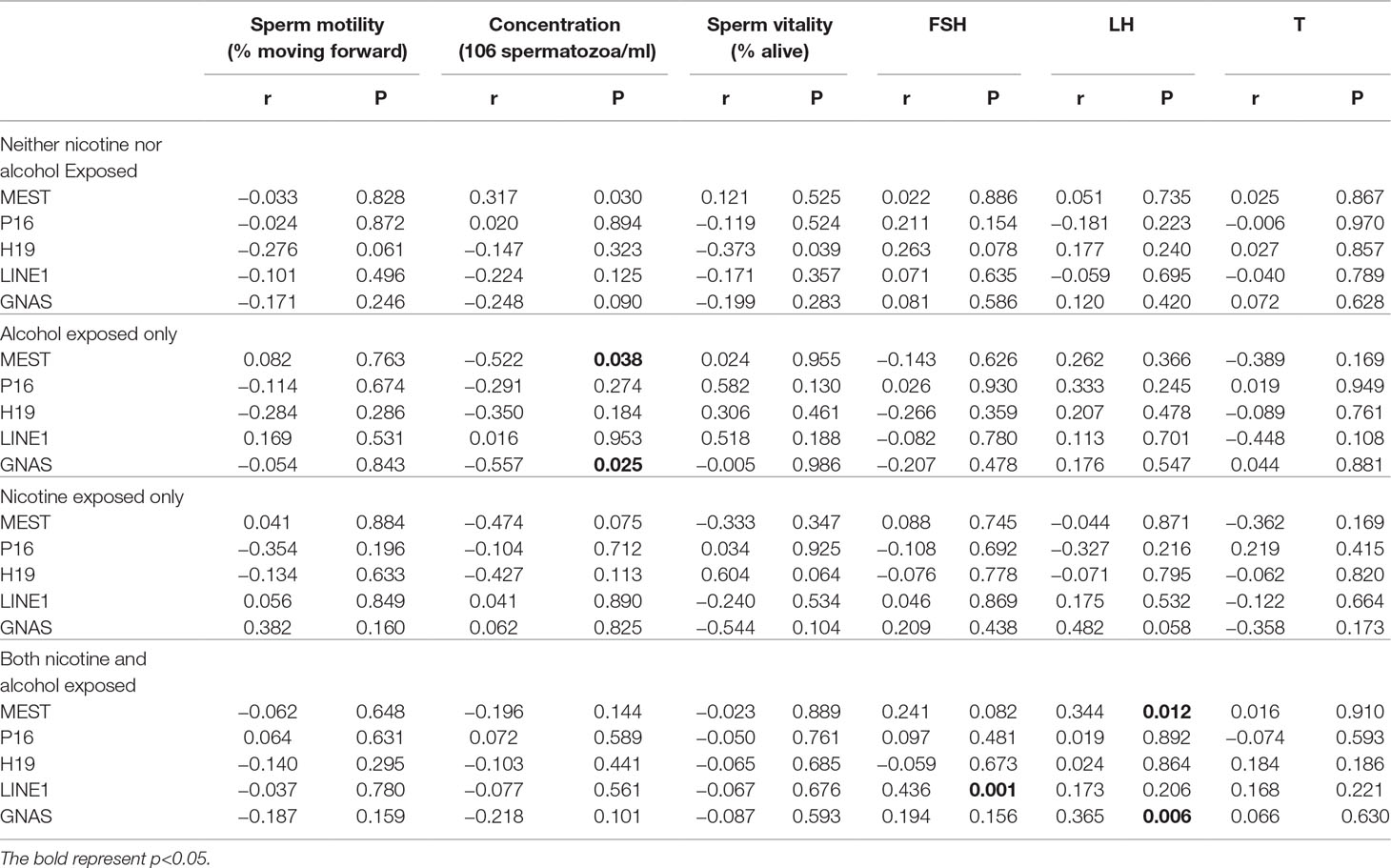
The data were checked for normal distributions using the Shapiro–Wilk test. The correlations in methylation between loci were analyzed using the Pearson’s test. The methylation between MEST and P16, and MEST and GNAS were positively correlated, respectively (Suppl 3). The correlations between the average methylation levels and seminological parameters or hormones were analyzed using the Pearson’s (normal distributions) or Spearman’s correlation (abnormal distributions), respectively. Correlation tests for gene modulation levels and phenotypic indices showed that the average methylation levels of MEST and GNAS were inversely correlated with sperm concentration [r = −0.522 (P = 0.038) and r = −0.557 (P = 0.025), respectively; Figure 3A] in the alcohol-exposed group. The average methylation levels of MEST and GNAS were positively correlated with LH levels [r = 0.344 (P = 0.012) and r = 0.365 (P = 0.006), respectively], and the methylation of the repetitive element LINE1 was positively correlated with FSH level (r = 0.436, P = 0.001; Figure 3B) in the nicotine- and alcohol-exposed group. However, no association between gene methylation and the phenotypic indices was observed in the nicotine-exposed group (P > 0.05, Table 3).The multivariate correlation pattern between the variables was investigated using PCA. The KMO measure was 0.669, indicating that sufficient correlation existed between these variables to proceed with factor analysis. Five components were extracted by factor analysis using PCA (Suppl 4). Variables located near each other such as MEST, GNAS, FSH, and LH were strongly correlated (Figure 4).
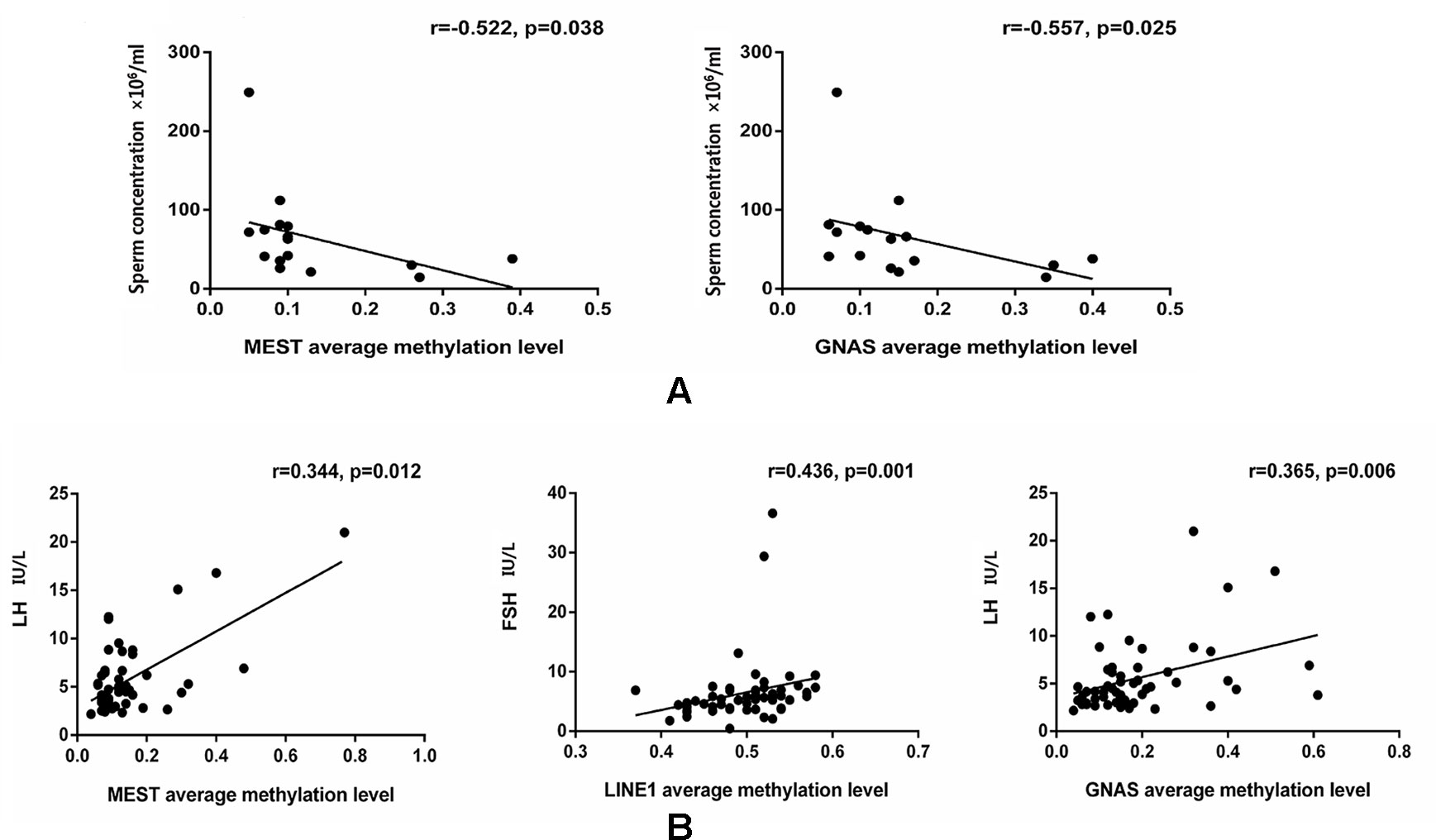
Figure 3 Comparison of the gene methylation levels and semen quality/hormone level. (A) The significant correlations between the average methylation levels, seminological parameters and hormones in the alcohol exposed only group. (B) The significant correlations between the average methylation levels, seminological parameters and hormones in the both nicotine and alcohol exposed group. The dots are the intersection points between the average methylation levels, seminological parameters and hormones.
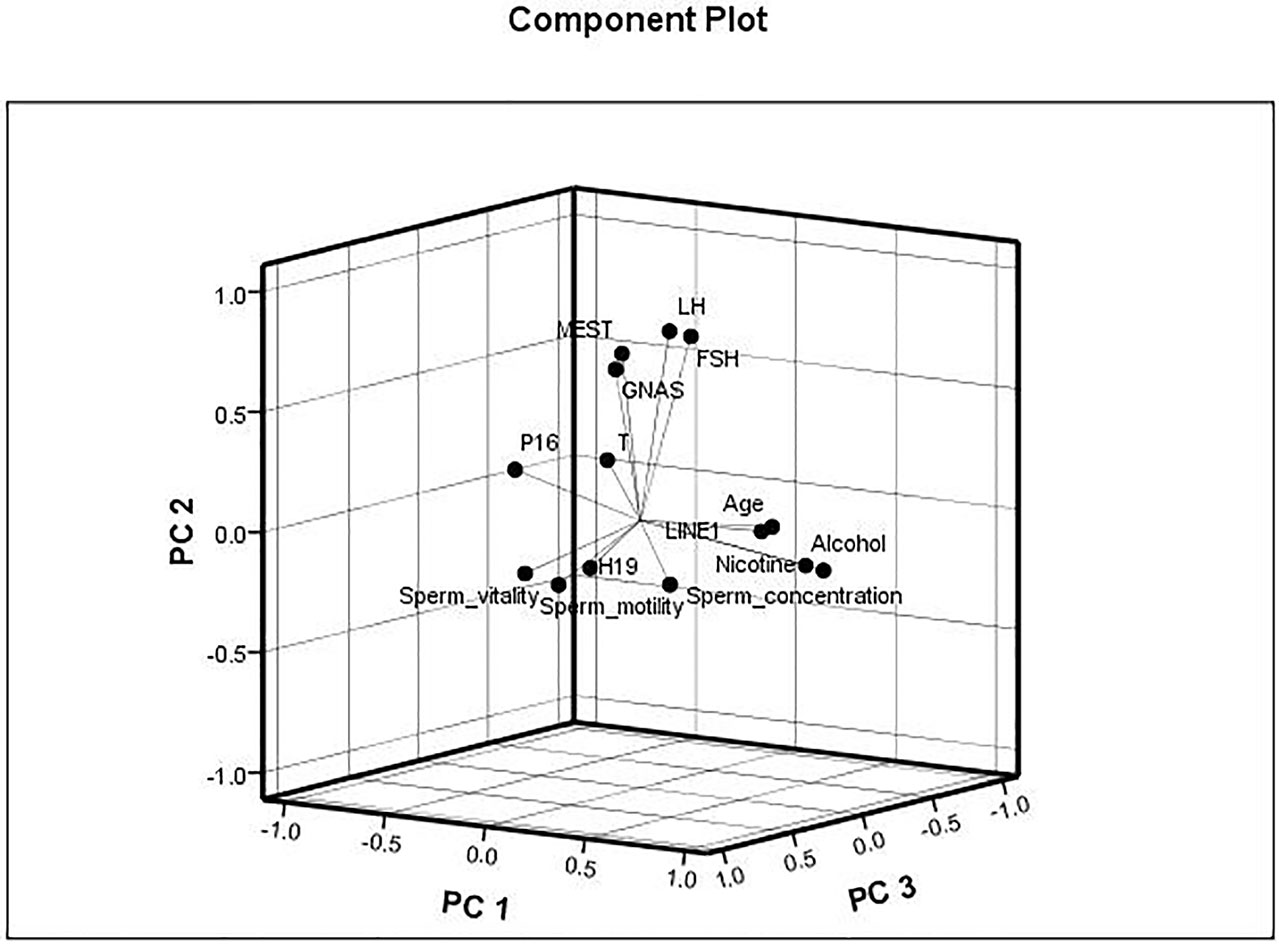
Figure 4 Multivariate correlation patterns. The Kaiser–Meyer–Olkin measure was 0.669, indicating that sufficient correlation existed between these variables to proceed with factor analysis. Three components were extracted by factor analysis using principal component analysis. Factor 1 labeled as “PC1” contains nicotine exposed, alcohol exposed, age, LINE1, P16, and sperm motility and had loadings of 0.852, 0.829, 0.678, 0.578, −0.516, and −0.403, respectively. This factor explained 20.0% of the total variance. Factor 2 labeled as “PC2” contains MEST, GNAS, follicle-stimulating hormone (FSH), and luteinizing hormone (LH) and had loadings of 0.672, 0.629, 0.775, and 0.789, respectively, explaining 17.5% of the total variance. Factor 3 labeled as “PC3” contains sperm vitality, H19, and sperm concentration and had loadings of 0.746, and 0.632, and −0.516, respectively, explaining 9.3% of the total variance.
Discussion
In this study, we observed that alcohol and nicotine exposure altered sperm cell quality, which may be related to the methylation levels of MEST and GNAS. The methylation levels of MEST, GNAS, and the repetitive element LINE1 were significantly associated with sperm concentration and FSH and LH levels.
Recent studies have shown that tobacco use and alcohol consumption may increase the risk of global aberrant DNA methylation (Hamid et al., 2009; Semmler et al., 2015). Aberrant methylation of LINE-1 has been reported in many recent studies on aging (Cho et al., 2015) and cancer (Pattamadilok et al., 2008; Suzuki et al., 2013; Suter et al., 2004), and LINE-1 methylation status could reflect the influence of environmental conditions or lifestyle habits on the genome (Schernhammer et al., 2010). High tobacco use might lead to a high risk of LINE-1 hypermethylation-related cancers in men (Andreotti et al., 2014; Karami et al., 2015). To the best of our knowledge, our study showed, for the first time, that hypermethylation of the LINE-1 gene in sperm was associated with alcohol and nicotine exposure. However, the association between LINE-1 methylation level and alcohol and tobacco exposure in sperm needs to be further explored in future studies.
In our study, compared to the control group, hypomethylation of P16 was observed in the tobacco-exposed group. In addition, we found that GNAS methylation was decreased in the alcohol-exposed group, and P16 methylation was decreased in the nicotine- and alcohol-exposed group. Previous studies have suggested that chronic paternal alcohol exposure might contribute to mental deficits in offspring via abnormal methylation of imprinted genes (such as H19 and Peg3) in sperm (Liang et al., 2014) and that methylation levels could be easily modified by air pollution, heavy metals, and other environmental factors, both in vivo and in vitro (Baccarelli and Bollati, 2009; Haggarty, 2013). An association between nicotine/alcohol exposure and methylation has been demonstrated in pregnant women (Barua and Junaid, 2015; Lee et al., 2015). In this study, we studied the influence of nicotine/alcohol exposure in male residents from Guizhou province, a population with a low fertility rate. The relationship between nicotine/alcohol exposure and the methylation of these five genes in male residents of Guizhou has not been previously reported. Abnormal DNA methylation in spermatozoa seems to be involved in environmental factor-induced transgenerational disruptive spermatogenesis (Anway et al., 2005; Anway et al., 2006). The impact of environmental factors on the epigenetic phenotype might affect offspring through abnormal spermatozoa methylation (Anway and Skinner, 2006). Some evidence has suggested that abnormal DNA methylation of imprinted genes may be associated with spermatogenesis failure (Boissonnas et al., 2010), and an observable decrease in the concentration of sperm was reported in patients with H19 hypomethylation (Stouder et al., 2011). In addition, aberrant MEST DNA methylation has been shown to be significantly associated with increased FSH levels (Klaver et al., 2013). However, we found that alcohol exposure altered sperm cell quality and was related to the hypomethylation of MEST and GNAS. MEST and GNAS methylation levels were significantly associated with increased LH levels, and LINE1 methylation was significantly associated with increased FSH levels. However, further studies are needed to explore the mechanisms underlying the association between chronic nicotine and alcohol exposure and aberration methylation of MEST, GNAS, and LINE1 with sperm quality and abnormal FSH and LH levels.
Our study has several limitations. First, our study had a small sample size, which restricts the generalizability of our results. Therefore, our results must be verified in larger cohorts using different techniques. In addition, further studies are needed to explore how changes in methylation due to smoking and drinking affect fertility.
In conclusion, our results show that the different methylation levels of four genes, MEST, P16, LINE-1, and GNAS, alter the sperm cells of patients who consume alcohol and use nicotine. Both smoking and drinking impair sperm/semen quality and hormone levels. Thus, methylation of MEST, GNAS, and LINE1 may be associated with sperm concentration and FSH and LH levels.
Ethics Statement
Our study was approved by the Ethics Committee of Shanghai Institute of Planned Parenthood Research (SIPPR) and the local approval of Guizhou province was not required.
Author Contributions
Designed and coordinated the study: WZ, AZ, HS, JD. Performed the experiments: FS, ZZ, YS, JX, ML. Analyzed the data: XX, JL, XS, MM, JD. Contributed reagents/materials/analysis tools: YY, FS, BW. Wrote the paper: WZ, FS, JD. Helped in critical revision of the manuscript for important intellectual content: DJ, XH, ML.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Chunli Zhong, Yuan Yang, Kanglian Chen, and Jiang Zhu for help in collecting samples from the Population and Family Planning Institute of Guizhou Province. We would like to acknowledge research support from the National Nature Science Foundation of China (Nos. 81571503, 81270744, and 81771655) and the Major State Basic Research Development Program of China (973 Program, 2014CB943104).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01001/full#supplementary-material
References
Al Khaled, Y., Tierling, S., Laqqan, M., Lo Porto, C., Hammadeh, M. E. (2018). Cigarette smoking induces only marginal changes in sperm DNA methylation levels of patients undergoing intracytoplasmic sperm injection treatment. Andrologia 50. doi: 10.1111/and.12818
Alkhaled, Y., Laqqan, M., Tierling, S., Lo Porto, C., Amor, H., Hammadeh, M. E. (2018). Impact of cigarette-smoking on sperm DNA methylation and its effect on sperm parameters. Andrologia. doi: 10.1111/and.12950
Andreotti, G., Karami, S., Pfeiffer, R. M., Hurwitz, L., Liao, L. M., Weinstein, S. J., et al. (2014). LINE1 methylation levels associated with increased bladder cancer risk in pre-diagnostic blood DNA among US (PLCO) and European (ATBC) cohort study participants. Epigenetics 9, 404–415. doi: 10.4161/epi.27386
Anway, M. D., Skinner, M. K. (2006). Epigenetic transgenerational actions of endocrine disruptors. Endocrinology 147, S43–S49. doi: 10.1210/en.2006-0640
Anway, M. D., Cupp, A. S., Uzumcu, M., Skinner, M. K. (2005). Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308, 1466–1469. doi: 10.1126/science.1108190
Anway, M. D., Memon, M. A., Uzumcu, M., Skinner, M. K. (2006). Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J. Androl. 27, 868–879. doi: 10.2164/jandrol.106.000349
Baccarelli, A., Bollati, V. (2009). Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 21, 243–251. doi: 10.1097/MOP.0b013e32832925cc
Barua, S., Junaid, M. A. (2015). Lifestyle, pregnancy and epigenetic effects. Epigenomics 7, 85–102. doi: 10.2217/epi.14.71
Besingi, W., Johansson, A. (2014). Smoke-related DNA methylation changes in the etiology of human disease. Hum Mol. Genet. 23, 2290–2297. doi: 10.1093/hmg/ddt621
Boissonnas, C. C., Abdalaoui, H. E., Haelewyn, V., Fauque, P., Dupont, J. M., Gut, I., et al. (2010). Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur. J. Hum. Genet. 18, 73–80. doi: 10.1038/ejhg.2009.117
Chang, R. C., Skiles, W. M., Chronister, S. S., Wang, H., Sutton, G. I., Bedi, Y. S., et al. (2017). DNA methylation-independent growth restriction and altered developmental programming in a mouse model of preconception male alcohol exposure. Epigenetics 12, 841–853. doi: 10.1080/15592294.2017.1363952
Chastain, L. G., Sarkar, D. K. (2017). Alcohol effects on the epigenome in the germline: role in the inheritance of alcohol-related pathology. Alcohol 60, 53–66. doi: 10.1016/j.alcohol.2016.12.007
Chavarro, J. E., Rich-Edwards, J. W., Rosner, B. A., Willett, W. C. (2009). Caffeinated and alcoholic beverage intake in relation to ovulatory disorder infertility. Epidemiology 20, 374–381. doi: 10.1097/EDE.0b013e31819d68cc
Cho, Y. H., Woo, H. D., Jang, Y., Porter, V., Christensen, S., Hamilton, R. F., Jr., et al. (2015). The Association of LINE-1 Hypomethylation with Age and Centromere Positive Micronuclei in Human Lymphocytes. PLoS One 10, e0133909. doi: 10.1371/journal.pone.0133909
Dada, R., Kumar, R., Shamsi, M. B., Tanwar, M., Pathak, D., Venkatesh, S., et al. (2008). Genetic screening in couples experiencing recurrent assisted procreation failure. Indian J. Biochem. Biophys. 45, 116–120.
Dai, J., Wang, Z., Xu, W., Zhang, M., Zhu, Z., Zhao, X., et al. (2017). Paternal nicotine exposure defines different behavior in subsequent generation via hyper-methylation of mmu-miR-15b. Sci. Rep. 7, 7286. doi: 10.1038/s41598-017-07920-3
Darbandi, M., Darbandi, S., Agarwal, A., Baskaran, S., Dutta, S., Sengupta, P., et al. (2018). Reactive oxygen species-induced alterations in H19-Igf2 methylation patterns, seminal plasma metabolites, and semen quality. J. Assist. Reprod. Genet. doi: 10.1007/s10815-018-1350-y
Dong, H., Wang, Y., Zou, Z., Chen, L., Shen, C., Xu, S., et al. (2017). Abnormal methylation of imprinted genes and cigarette smoking: assessment of Their association with the risk of male infertility. Reprod. Sci. 24, 114–123. doi: 10.1177/1933719116650755
Donkin, I., Barres, R. (2018). Sperm epigenetics and influence of environmental factors. Mol. Metabol. 14, 1–11. doi: 10.1016/j.molmet.2018.02.006
Gaur, D. S., Talekar, M. S., Pathak, V. P. (2010). Alcohol intake and cigarette smoking: impact of two major lifestyle factors on male fertility. Indian J Pathol Microbiol 53, 35–40. doi: 10.4103/0377-4929.59180
Gunes, S., Metin Mahmutoglu, A., Arslan, M. A., Henkel, R. (2018). Smoking-induced genetic and epigenetic alterations in infertile men. Andrologia 50, e13124. doi: 10.1111/and.13124
Haggarty, P. (2013). Epigenetic consequences of a changing human diet. Proc. Nutr. Soc. 72, 363–371. doi: 10.1017/S0029665113003376
Hamad, M. F., Dayyih, W. A. A., Laqqan, M., AlKhaled, Y., Montenarh, M., Hammadeh, M. E. (2018). The status of global DNA methylation in the spermatozoa of smokers and non-smokers. Reprod. Biomed. Online 37, 581–589. doi: 10.1016/j.rbmo.2018.08.016
Hamid, A., Wani, N. A., Kaur, J. (2009). New perspectives on folate transport in relation to alcoholism-induced folate malabsorption–association with epigenome stability and cancer development. FEBS J. 276, 2175–2191. doi: 10.1111/j.1742-4658.2009.06959.x
Han, J. C., Xu, F., Chen, N., Qi, G. B., Wei, Y. J., Li, H. B., et al. (2016). Promoter methylations of RASSF1A and p16 is associated with clinicopathological features in lung cancers. J. Cancer Res. Ther. 12, 340–349. doi: 10.4103/0973-1482.154926
Han, Y. D., Wang, X. B., Cui, N. H., Zhang, S., Wang, C., Zheng, F. (2017). Associations of P16INK4a promoter hypermethylation with squamous intra-epithelial lesion, cervical cancer and their clinicopathological features: a meta-analysis. Oncotarget 8, 1871–1883. doi: 10.18632/oncotarget.12202
Hansen, M. L., Thulstrup, A. M., Bonde, J. P., Olsen, J., Hakonsen, L. B., Ramlau-Hansen, C. H. (2012). Does last week’s alcohol intake affect semen quality or reproductive hormones? A cross-sectional study among healthy young Danish men. Reprod. Toxicol. 34, 457–462. doi: 10.1016/j.reprotox.2012.06.004
He, J., Zhang, A., Fang, M., Fang, R., Ge, J., Jiang, Y., et al. (2013). Methylation levels at IGF2 and GNAS DMRs in infants born to preeclamptic pregnancies. BMC Genomics 14, 472. doi: 10.1186/1471-2164-14-472
Jenkins, T. G., James, E. R., Alonso, D. F., Hoidal, J. R., Murphy, P. J., Hotaling, J. M., et al. (2017). Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology 5, 1089–1099. doi: 10.1111/andr.12416
Jeong, M. G., Song, H., Shin, J. H., Jeong, H., Kim, H. K., Hwang, E. S. (2017). Transcriptional coactivator with PDZ-binding motif is required to sustain testicular function on aging. Aging Cell 16, 1035–1042. doi: 10.1111/acel.12631
Karami, S., Andreotti, G., Liao, L. M., Pfeiffer, R. M., Weinstein, S. J., Purdue, M. P., et al. (2015). LINE1 methylation levels in pre-diagnostic leukocyte DNA and future renal cell carcinoma risk. Epigenetics 10, 282–292. doi: 10.1080/15592294.2015.1006505
Kim, P., Choi, C. S., Park, J. H., Joo, S. H., Kim, S. Y., Ko, H. M., et al. (2014). Chronic exposure to ethanol of male mice before mating produces attention deficit hyperactivity disorder-like phenotype along with epigenetic dysregulation of dopamine transporter expression in mouse offspring. J. Neurosci. Res. 92, 658–670. doi: 10.1002/jnr.23275
Klaver, R., Tuttelmann, F., Bleiziffer, A., Haaf, T., Kliesch, S., Gromoll, J. (2013). DNA methylation in spermatozoa as a prospective marker in andrology. Andrology 1, 731–740. doi: 10.1111/j.2047-2927.2013.00118.x
Kobayashi, N., Miyauchi, N., Tatsuta, N., Kitamura, A., Okae, H., Hiura, H., et al. (2017). Factors associated with aberrant imprint methylation and oligozoospermia. Sci. Rep. 7, 42336. doi: 10.1038/srep42336
Kunzle, R., Mueller, M. D., Hanggi, W., Birkhauser, M. H., Drescher, H., Bersinger, N. A. (2003). Semen quality of male smokers and nonsmokers in infertile couples. Fertil. Steril. 79, 287–291. doi: 10.1016/S0015-0282(02)04664-2
Laqqan, M., Tierling, S., Alkhaled, Y., LoPorto, C., Hammadeh, M. E. (2017). Alterations in sperm DNA methylation patterns of oligospermic males. Reprod. Biol. 17, 396–400. doi: 10.1016/j.repbio.2017.10.007
Laqqan, M., Tierling, S., Alkhaled, Y., Porto, C. L., Solomayer, E. F., Hammadeh, M. E. (2017). Aberrant DNA methylation patterns of human spermatozoa in current smoker males. Reprod. Toxicol. 71, 126–133. doi: 10.1016/j.reprotox.2017.05.010
Lee, B. Y., Park, S. Y., Ryu, H. M., Shin, C. Y., Ko, K. N., Han, J. Y., et al. (2015). Changes in the methylation status of DAT, SERT, and MeCP2 gene promoters in the blood cell in families exposed to alcohol during the periconceptional period. Alcohol. Clin. Exp. Res. 39, 239–250. doi: 10.1111/acer.12635
Li, Y., Xie, C., Murphy, S. K., Skaar, D., Nye, M., Vidal, A. C., et al. (2016). Lead exposure during early human development and DNA methylation of imprinted gene regulatory elements in adulthood. Environ. Health Perspect. 124, 666–673. doi: 10.1289/ehp.1408577
Liang, F., Diao, L., Liu, J., Jiang, N., Zhang, J., Wang, H., et al. (2014). Paternal ethanol exposure and behavioral abnormities in offspring: associated alterations in imprinted gene methylation. Neuropharmacology 81, 126–133. doi: 10.1016/j.neuropharm.2014.01.025
Liang, H., Xu, W., Chen, J., Shi, H., Zhu, J., Liu, X., et al. (2017). The Association between exposure to environmental bisphenol A and gonadotropic hormone levels among men. PLoS One 12, e0169217. doi: 10.1371/journal.pone.0169217
Marques, C. J., Costa, P., Vaz, B., Carvalho, F., Fernandes, S., Barros, A., et al. (2008). Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol. Hum. Reprod. 14, 67–74. doi: 10.1093/molehr/gam093
Marques, C. J., Joao Pinho, M., Carvalho, F., Bieche, I., Barros, A., Sousa, M. (2011). DNA methylation imprinting marks and DNA methyltransferase expression in human spermatogenic cell stages. Epigenetics 6, 1354–1361. doi: 10.4161/epi.6.11.17993
McCarthy, D. M., Morgan, T. J., Jr., Lowe, S. E., Williamson, M. J., Spencer, T. J., Biederman, J., et al. (2018). Nicotine exposure of male mice produces behavioral impairment in multiple generations of descendants. PLoS Biol. 16, e2006497. doi: 10.1371/journal.pbio.2006497
Miao, M., Zhou, X., Li, Y., Zhang, O., Zhou, Z., Li, T., et al. (2014). LINE-1 hypomethylation in spermatozoa is associated with Bisphenol A exposure. Andrology 2, 138–144. doi: 10.1111/j.2047-2927.2013.00166.x
Montjean, D., Zini, A., Ravel, C., Belloc, S., Dalleac, A., Copin, H., et al. (2015). Sperm global DNA methylation level: association with semen parameters and genome integrity. Andrology 3, 235–40. doi: 10.1111/andr.12001
Nasri, F., Gharesi-Fard, B., Namavar Jahromi, B., Farazi-Fard, M. A., Banaei, M., Davari, M., et al. (2017). Sperm DNA methylation of H19 imprinted gene and male infertility. Andrologia 49. doi: 10.1111/and.12766
Pang, Y., Holmes, M. V., Guo, Y., Yang, L., Bian, Z., Chen, Y., et al. (2018). Smoking, alcohol, and diet in relation to risk of pancreatic cancer in China: a prospective study of 0.5 million people. Cancer Med. 7, 229–239. doi: 10.1002/cam4.1261
Pattamadilok, J., Huapai, N., Rattanatanyong, P., Vasurattana, A., Triratanachat, S., Tresukosol, D., et al. (2008). LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int. J. Gynecol. Cancer 18, 711–717. doi: 10.1111/j.1525-1438.2007.01117.x
Pierron, F., Baillon, L., Sow, M., Gotreau, S., Gonzalez, P. (2014). Effect of low-dose cadmium exposure on DNA methylation in the endangered European eel. Environ. Sci. Technol. Lett. 48, 797–803. doi: 10.1021/es4048347
Povey, A. C., Clyma, J. A., McNamee, R., Moore, H. D., Baillie, H., Pacey, A. A., et al. (2012). Modifiable and non-modifiable risk factors for poor semen quality: a case-referent study. Hum. Reprod. 27, 2799–2806. doi: 10.1093/humrep/des183
Rangwala, S. H., Zhang, L., Kazazian, H. H., Jr (2009). Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 10, R100. doi: 10.1186/gb-2009-10-9-r100
Santi, D., De Vincentis, S., Magnani, E., Spaggiari, G. (2017). Impairment of sperm DNA methylation in male infertility: a meta-analytic study. Andrology 5, 695–703. doi: 10.1111/andr.12379
Schernhammer, E. S., Giovannucci, E., Kawasaki, T., Rosner, B., Fuchs, C. S., Ogino, S. (2010). Dietary folate, alcohol and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut 59, 794–799. doi: 10.1136/gut.2009.183707
Semmler, A., Heese, P., Stoffel-Wagner, B., Muschler, M., Heberlein, A., Bigler, L., et al. (2015). Alcohol abuse and cigarette smoking are associated with global DNA hypermethylation: results from the German Investigation on Neurobiology in Alcoholism (GINA). Alcohol 49, 97–101. doi: 10.1016/j.alcohol.2015.01.004
Sharlip, I. D., Jarow, J. P., Belker, A. M., Lipshultz, L. I., Sigman, M., Thomas, A. J., et al. (2002). Best practice policies for male infertility. Fertil Steril 77, 873–882. doi: 10.1016/S0015-0282(02)03105-9
Siddeek, B., Mauduit, C., Simeoni, U., Benahmed, M. (2018). Sperm epigenome as a marker of environmental exposure and lifestyle, at the origin of diseases inheritance. Mutat Res 778, 38–44. doi: 10.1016/j.mrrev.2018.09.001
Stouder, C., Somm, E., Paoloni-Giacobino, A. (2011). Prenatal exposure to ethanol: a specific effect on the H19 gene in sperm. Reprod. Toxicol. 31, 507–512. doi: 10.1016/j.reprotox.2011.02.009
Suter, C. M., Martin, D. I., Ward, R. L. (2004). Hypomethylation of L1 retrotransposons in colorectal cancer and adjacent normal tissue. Int J Colorectal Dis 19, 95–101. doi: 10.1007/s00384-003-0539-3
Suzuki, M., Shiraishi, K., Eguchi, A., Ikeda, K., Mori, T., Yoshimoto, K., et al. (2013). Aberrant methylation of LINE-1, SLIT2, MAL and IGFBP7 in non-small cell lung cancer. Oncol. Rep. 29, 1308–1314. doi: 10.3892/or.2013.2266
Wang, B. H., Li, Y. Y., Han, J. Z., Zhou, L. Y., Lv, Y. Q., Zhang, H. L., et al. (2017). Gene methylation as a powerful biomarker for detection and screening of non-small cell lung cancer in blood. Oncotarget 8, 31692–31704. doi: 10.18632/oncotarget.15919
Witkiewitz, K., Hallgren, K. A., Kranzler, H. R., Mann, K. F., Hasin, D. S., Falk, D. E., et al. (2017). Clinical validation of reduced alcohol consumption after treatment for alcohol dependence using the World Health Organization risk drinking levels. Alcohol. Clin. Exp. Res. 41, 179–186. doi: 10.1111/acer.13272
Xin-Chang, Z., Peng, W., Zhao-Yuan, H., Xiao-Bin, H., Ru-Jin, Z., Yi-Xun, L. (2002). Expression of P16(INK4a) in testis of rhesus monkey during heat stress and testosterone undecanoate induced azoospermia or oligozoospermia. Contraception 65, 251–255. doi: 10.1016/S0010-7824(01)00305-5
Xu, J., Zhang, A., Zhang, Z., Wang, P., Qian, Y., He, L., et al. (2016). DNA methylation levels of imprinted and nonimprinted genes DMRs associated with defective human spermatozoa. Andrologia 48, 939–947. doi: 10.1111/and.12535
Yaman, R., Grandjean, V. (2006). Timing of entry of meiosis depends on a mark generated by DNA methyltransferase 3a in testis. Mol. Reprod. Dev. 73, 390–397. doi: 10.1002/mrd.20430
Keywords: DNA methylation, nicotine/alcohol exposure, male infertility, imprint gene, sperm
Citation: Zhang W, Li M, Sun F, Xu X, Zhang Z, Liu J, Sun X, Zhang A, Shen Y, Xu J, Miao M, Wu B, Yuan Y, Huang X, Shi H and Du J (2019) Association of Sperm Methylation at LINE-1, Four Candidate Genes, and Nicotine/Alcohol Exposure With the Risk of Infertility. Front. Genet. 10:1001. doi: 10.3389/fgene.2019.01001
Received: 21 January 2019; Accepted: 20 September 2019;
Published: 18 October 2019.
Edited by:
Yun Liu, Fudan University, ChinaReviewed by:
Osman A. El-Maarri, University of Bonn, GermanyNa Zhu, Columbia University, United States
Peng Chen, Jilin University, China
Copyright © 2019 Zhang, Li, Sun, Xu, Zhang, Liu, Sun, Zhang, Shen, Xu, Miao, Wu, Yuan, Huang, Shi and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianliang Huang, aHhsNzQwMDY0MUAxNjMuY29t; Huijuan Shi, c2hpaHVpanVhbjIwMTFAMTYzLmNvbQ==; Jing Du, ZHVqaW5nNDJAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Wenjing Zhang1,2†
Wenjing Zhang1,2† Junwei Liu
Junwei Liu Aiping Zhang
Aiping Zhang Jianhua Xu
Jianhua Xu Jing Du
Jing Du