- 1Department of Thoracic Surgery, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, China
- 2Department of Thoracic Surgery, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China
- 3Department of Pathology, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China
Introduction: Lung adenocarcinoma with CTNNB1 mutation is relatively uncommon, and its clinicopathologic characteristics, disease course, and prognosis have not been well-studied.
Methods: A total of 564 lung adenocarcinoma patients were enrolled in this study. The relationship between CTTNB1 mutational status and clinicopathologic parameters, the rates of relapse-free survival (RFS) and overall survival (OS), and the mutational status of other genes commonly mutated in lung adenocarcinoma were analyzed.
Results: Of 564 lung adenocarcinoma patients, 30 (5.3%) harbored CTNNB1 mutations. Univariate analyses revealed that gender, smoking history, pleural invasion, and histological subtype were all significant predictors of RFS and OS. Pleural invasion and histological subtype remained significant predictors of RFS and OS in a multivariate analysis. There were no significant differences in RFS (p = 0.504) or OS (p = 0.054) between lung adenocarcinoma patients with CTNNB1 mutation and those without CTNNB1 mutation. However, patients with CTNNB1 mutation tended to have a worse OS.
Conclusions: Female patients and nonsmokers are likely to harbor CTNNB1 mutation and primary lung adenocarcinoma with mutated CTNNB1 has a poor prognosis.
Introduction
Lung cancer, which has the highest incidence of all cancers and the highest rate of disease-related fatalities, is the main cause of cancer-related death worldwide (Torre et al., 2015; Gu et al., 2017a; Gu et al., 2018). Lung adenocarcinoma is the most common pathological subtype, accounting for nearly 70% of all lung tumors (Sun et al., 2010). With the introduction of low-dose computed tomography, which enables earlier detection, the incidence of lung cancer, especially early-stage lung cancer, has risen sharply in recent years (Field et al., 2012).
With the advent of genomics, molecular or genetic variants affecting disease risk can be identified (Field, 2008). Mutations in the gene encoding β-catenin (CTNNB1) have been detected in numerous human malignancies, including lung cancer (Woenckhaus et al., 2008), malignant mesothelioma (Shigemitsu et al., 2001), desmoid tumors (Colombo et al., 2013), colon cancer (Akyol et al., 2019), and others. Woenckhaus et al. (2008) identified a number of differentially expressed genes in smoke-exposed bronchial epithelium and nonsmall cell lung cancers (NSCLCs), they found in adenocarcinomas, the cytoplasmic expression of beta-catenin was associated with shorter survival (p = 0.012). Shigemitsu et al. (2001) found CTNNB1 is infrequently mutated in lung cancer. Akyol et al. (2019) defined an immunohistochemical algorithm to dissect Wnt pathway alterations in formalin-fixed and paraffin-embedded neoplastic tissues and found all six colon adenomas of the 126 total adenomas studied for the altered/mutant β-catenin staining pattern had presumptively pathogenic point mutations or deletions in CTNNB1. The N-terminus of β-catenin, with contains conserved phosphorylated threonine/serine amino acid residues, is the most frequent location of cancer-related CTNNB1 mutations (Dar et al., 2017). The level of free β-catenin in the cytoplasmic pool is regulated by ubiquitination and proteasomal degradation (Akyol et al., 2019). β-catenin is a member of the Wnt signaling cascade and is associated with cadherin-mediated cell–cell adhesion systems (Woenckhaus et al., 2008). In lung tumors, the immunohistologic loss of β-catenin membrane staining along with a corresponding increase cytoplasmic or nuclear staining has been reported (Nozawa et al., 2006).
Although CTNNB1 mutation occurs in many tumors types, it has not been well-studied in the context of lung adenocarcinoma, and the clinicopathologic characteristics and prognosis of lung adenocarcinoma with mutated CTNNB1 has not been described. Therefore, we compared the clinicopathologic characteristics of 30 lung adenocarcinomas with CTNNB1 mutations with those of 534 lung adenocarcinomas with wild-type CTNNB1.
Materials and Methods
From July 2008 to April 2013, resected primary lung adenocarcinomas were collected at the Department of Thoracic Surgery of Shanghai Chest Hospital, Shanghai Jiaotong University. To confirm the diagnosis of primary lung cancer, all the patients received thorough preoperative testing at our hospital, including physical exams, serological tests, pulmonary function tests, chest/brain computed tomography (CT), technetium bone scanning, and abdominal ultrasound. Biopsies were done by bronchoscopy or endobronchial ultrasound-guided transbronchial needle aspiration, and in some cases, positron emission tomography CT was used to exclude mediastinal lymph node metastases (Gu et al., 2017b). The lung adenocarcinoma subtype was determined by light microscopy intraoperatively, using frozen sections, and confirmed postoperatively, using paraffin-embedded sections. All surgical samples had at least 5% tumor content. Each case was reviewed by at least two junior pathologists and a senior pathologist to confirm the histologic subtype of resected lung neoplasms. The combination of routine preoperative examination and intra-/postoperative pathological diagnosis is recommended to make an exact lung cancer diagnosis.
In total, 601 patients with primary lung adenocarcinoma were identified. Of these, 17 and 20 patients were excluded because they received neoadjuvant chemotherapy or were lost to follow-up, respectively. The remaining 564 patients were enrolled in this study.
Informed consent was given by all patients or their legal representatives. The study was initiated after obtaining Institutional Review Board approval. The medical records for all patients were reviewed to collect corresponding clinicopathologic data, including sex, age, smoking status, pathologic tumor, node, and metastasis (TNM) stage [according to the staging system of the 7th edition of the American Joint Committee on Cancer (Edge et al., 2010)], thyroid transcription factor-1 status, and treatment information. Data on disease recurrence and survival were obtained from follow-up clinic visits or by telephone.
Bioinformatics Analysis
Data of The Cancer Genome Atlas (TCGA) were analyzed by Gene Expression Profiling interactive Analysis (http://gepia.cancer-pku.cn/) and Kaplan–Meier Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=lung). Gene CTNNB1 were further analyzed by Gene Expression Profiling interactive Analysis and the survival curves were draw and compared by Kaplan–Meier Plotter.
Mutational Analysis
The mutational status of EGFR, KRAS, and CTNNB1 was determined by targeted sequencing and verified by DNA sequencing analysis. Relevant primers were designed to amplify all known ALK fusion variants by quantitative real-time reverse transcriptase PCR of cDNA. ALK fluorescent in situ hybridization was used to confirm the presence of ALK gene fusions (Wang et al., 2012).
Statistical Analysis
Clinicopathologic data was analyzed using the SPSS 22.0 software package (SPSS Inc, Chicago, IL). Relapse-free survival (RFS) and overall survival (OS) were estimated by the Kaplan–Meier method, and differences were compared by log-rank testing using Prism 6 (GraphPad Software, La Jolla, CA). A p value of <0.05 was considered statistically significant.
Results
Mutational Status of Lung Adenocarcinomas
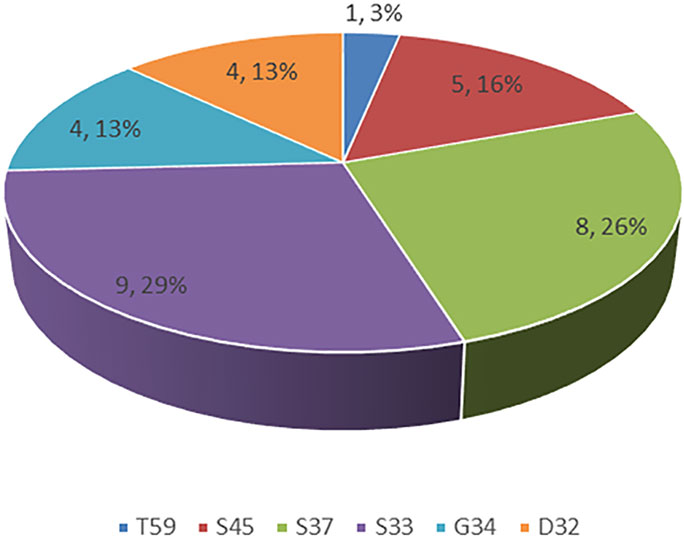
Of the 564 lung adenocarcinoma patients examined, 30 (5.3%) harbored CTNNB1 mutations (Table 1). The distributions of specific mutation types are shown in Figure 1.
Relationship Between Clinicopathologic Factors and CNTTB1 Mutational Status
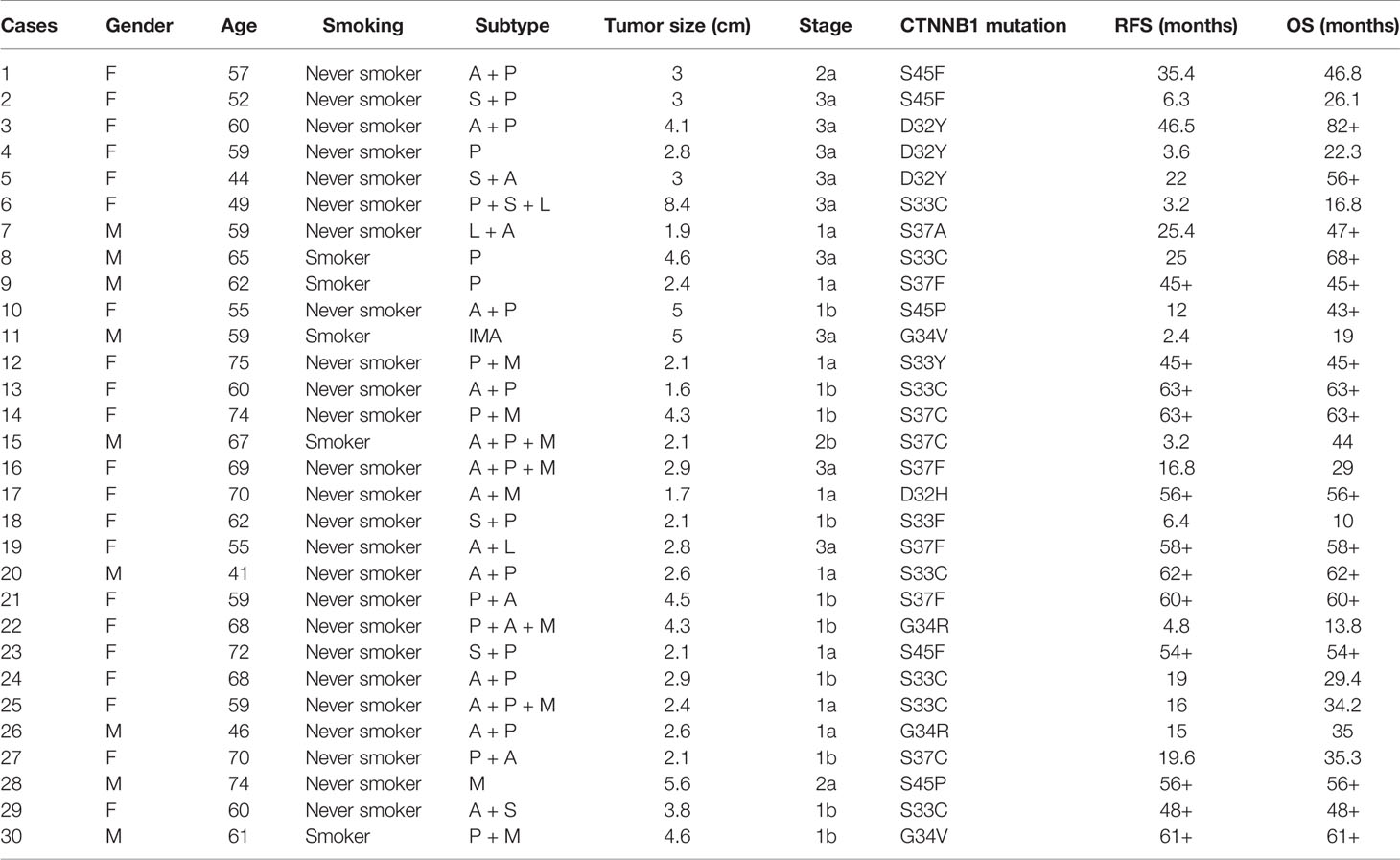
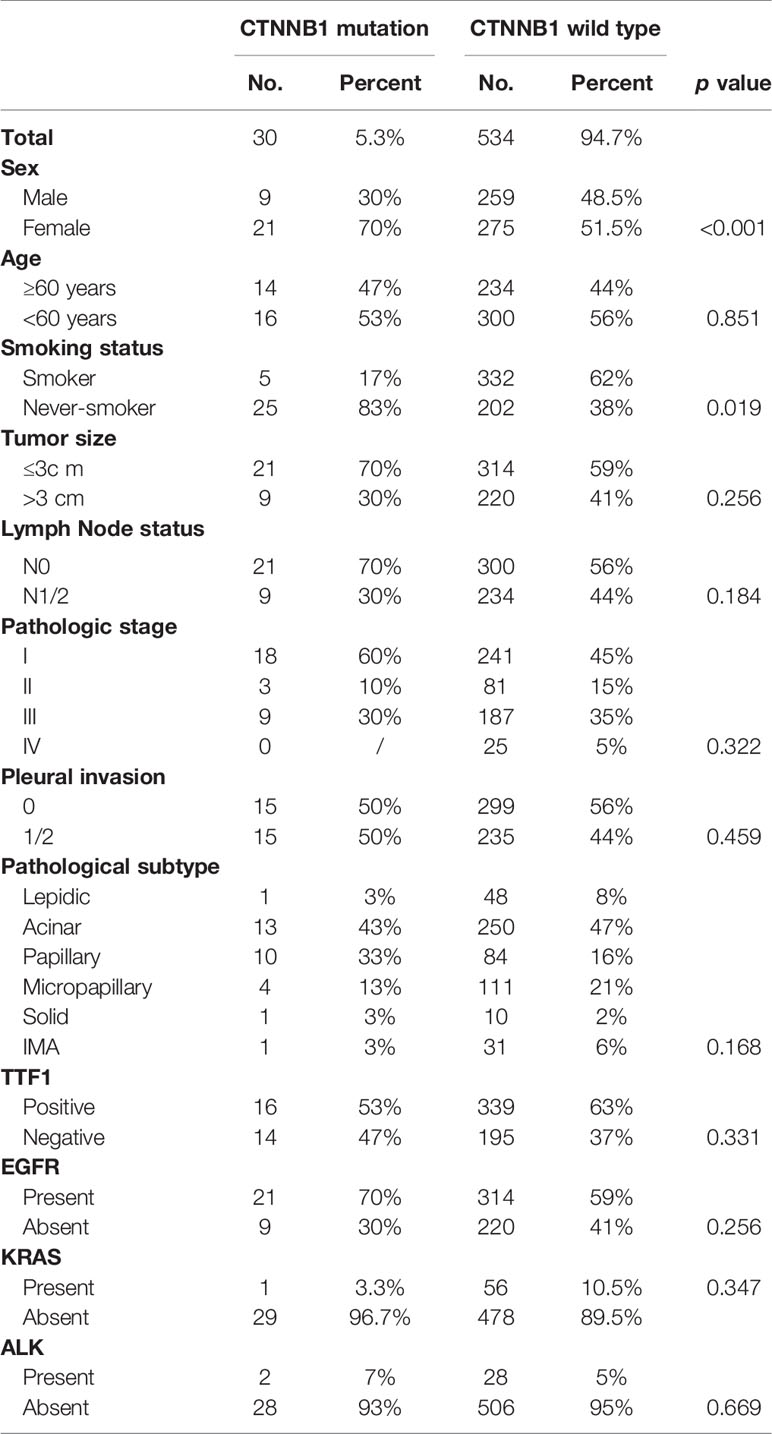
Of the 30 patients with CTNNB1 mutations, there were 21 (70%) female patients and 9 (30%) male patients, ranging in age from 22 to 81 years (median, 59 years). Histologically, 263 of the tumors were acinar-predominant (47%), 115 were micropapillary-predominant (20%), 94 were papillary-predominant (17%), 49 were lepidic-predominant (9%), 32 were mucinous adenocarcinoma-predominant (5%), and 11 were solid-predominant (2%). Most of the patients had early-stage lung cancer in both the CNTTB1 mutation group (stage I: 18/30, 60%) and the CNTTB1 wild-type group (stage I: 241/534, 45%). Age (p = 0.851), tumor size (p = 0.256), lymph node status (p = 0.184), pathologic stage (p = 0.322), and the presence of pleural invasion (p = 0.459) were similar between lung adenocarcinomas with CTNNB1 mutation and lung adenocarcinomas without CTNNB1 mutation, but the former group tended to have more female patients (p < 0.001) and more smokers (p = 0.019) (Table 2).
Relationship Between CNTTB1 Mutational Status and Survival
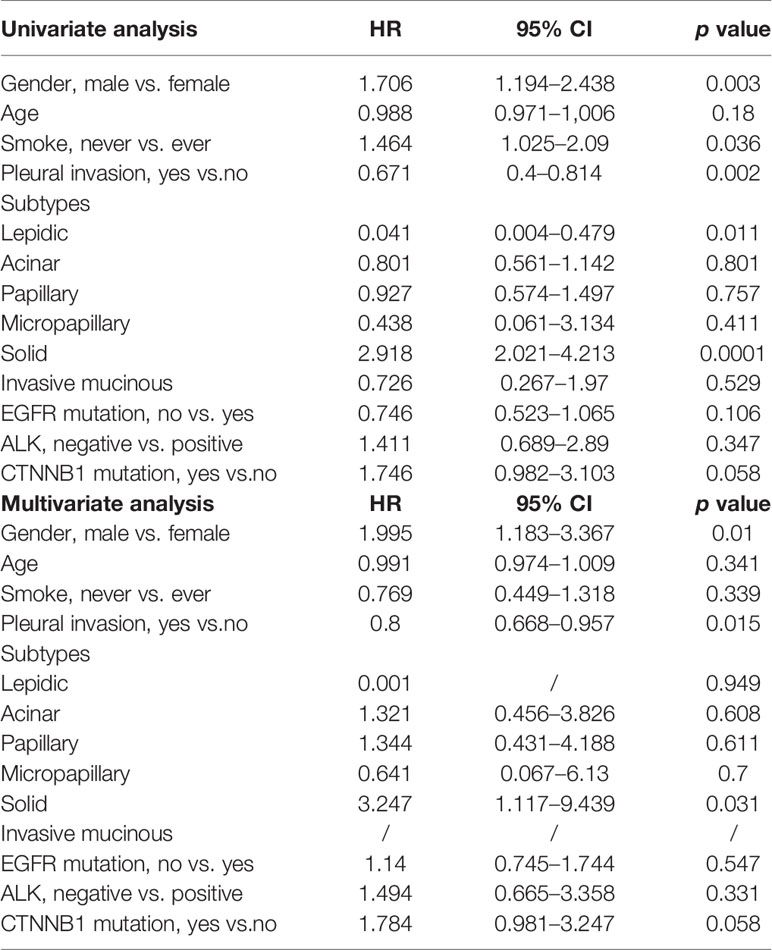
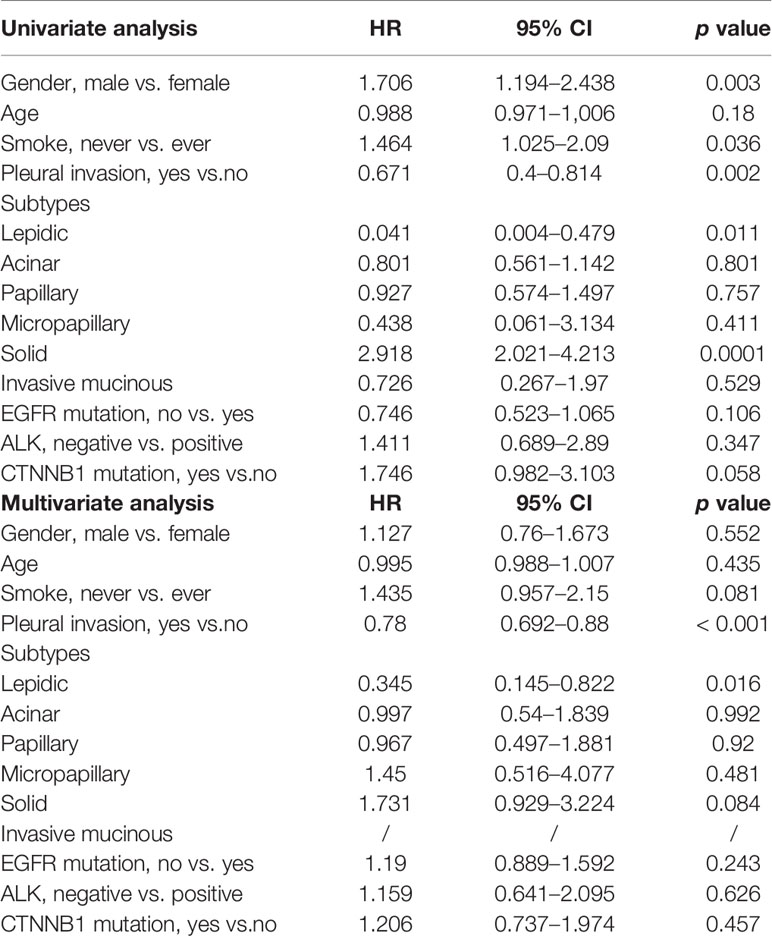
Univariate analysis revealed that gender, smoking history, pleural invasion, and histological subtype were all significant predictors of RFS and OS (Table 3). Pleural invasion and histological subtype were still significant predictors of RFS and OS in a multivariate analysis (Table 4).
During follow-up, 19 (63.3%) patients with lung adenocarcinomas with mutated CTNNB1 and 259 (48.5%) patients with lung adenocarcinomas with wild-type CTNNB1 experienced a relapse, and 10 (33.3%) and 111 (20.8%) patients died, respectively. There were no statistically significant differences in RFS (p = 0.504) or OS (p = 0.054) between patients with CTNNB1 mutation and patients without CTNNB1 mutation (Figure 2). However, patients with CTNNB1 mutation tended to have a worse OS.
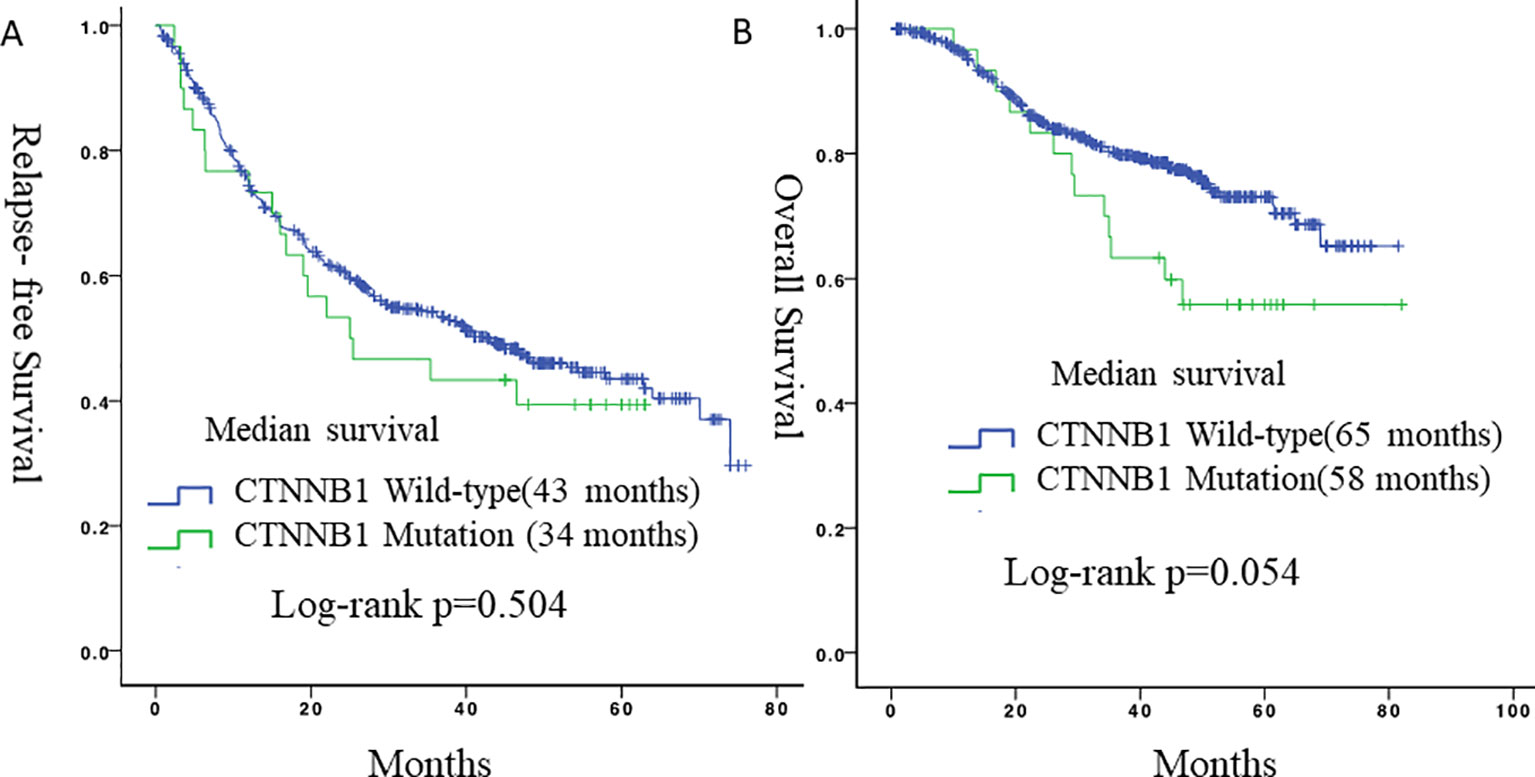
Figure 2 Survival curves for relapse-free survival and overall survival according to CTNNB1 status. (A) Relapse-free survival between the two groups. (B) Overall survival between the two groups.
As for lung adenocarcinomas from TCGA, there was no significant differences in the distributions of CTNNB1 mRNA expression among different lung adenocarcinoma stages (Supplementary Figure 1). Besides, between lung adenocarcinoma patients with and without CTNNB1 mutation, there was no significant differences in RFS (p = 0.49), while significant differences were found in OS (p = 8.9e−05). (Supplementary Figures 2 and 3).
Discussion
Lung cancer remains the leading cause of cancer-related death worldwide (Gu et al., 2017). Low-dose computed tomography screening reduces the mortality of lung cancer by as much as 20% in high-risk patients (National Lung Screening Trial Research Team, 2011). Early detection and diagnosis increases the number of patients who are newly diagnosed with lung cancer while it is still early-stage, improving the prognosis of lung cancer patients as a whole. For individuals at a high risk of developing lung cancer, periodic screening could have a survival benefit. Recently, some lung cancer risk prediction models have been constructed to make lung cancer screening more efficient (Spitz et al., 2007; Raji et al., 2012). With the development of gene mutation testing, targeted therapy has changed the treatment strategy for lung cancer. In this study, we describe the clinicopathological characteristics of lung adenocarcinoma with CTNNB1 mutation.
β-catenin is important for the establishment and maintenance of the epithelial layer and is a key downstream component of the canonical Wnt signaling pathway. The WNT/β-catenin pathway is involved in cancer and pluripotent stem cell signaling, which may suggest the mechanism underlying cancer stem cells. In this study, out of 564 patients, 30 (5.3%) patients with CTNNB1 mutations were identified. Kase et al. (2000) conducted an immunohistochemical analysis of 331 lung cancer specimens and reported that β-catenin expression was reduced in 122 (37%) of the samples, which was associated with significantly worse patient survival. Similarly, Woenckhaus et al. (2008) reported that reduced membrane staining of β-catenin and its abnormal accumulation in the cytoplasm and/or nuclei of lung adenocarcinoma cells was associated with shorter survival (p = 0.012). Another study also suggested that reduced β-catenin expression in surgically resected non-small cell lung cancer specimens was associated with lymph node metastasis and a poor prognosis (Retera et al., 1998). These studies suggest that decreased expression of β-catenin is associated with an unfavorable prognosis in lung cancer.
In our study, during follow-up, 19 patients (63.3%) with lung adenocarcinomas with CTNNB1 mutations and 259 patients (48.5%) with lung adenocarcinomas with wild-type CTNNB1 relapsed, and 10 (33.3%) and 111 (20.8%) patients died, respectively. Patients with CTNNB1 mutations therefore tended to have a worse prognosis, although this did not reach statistical significance. When compare with data from TCGA, patients with CTNNB1 mutation in TCGA also had worse OS. Our findings therefore correspond well to the results of previous studies and common directory (Sunaga et al., 2001).
In Cox proportional hazards models, univariate analyses revealed that gender, smoking history, the presence of pleural invasion, and histological subtype were all significant predictors of RFS and OS. Pleural invasion and histological subtype remained significant predictors of RFS and OS in a multivariate analysis. With respect to histological subtype, adenocarcinoma patients with micropapillary or solid subtypes, which are defined as high-risk subtypes in the 2011 classification proposed by the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society (Travis et al., 2011), had significantly worse prognosis. As for pleural invasion, pleural invasion, as well as visceral invasion, is considered an aggressive and invasive factor in NSCLC and has been included in the TNM staging system as a factor that should upstage the T factor (Rami-Porta et al., 2007; Travis et al., 2008; Butnor and Travis, 2012). Shimizu et al. (2005) demonstrated that velopharyngeal insufficiency (VPI) is a significant and independent predictor of a poor prognosis regardless of tumor size or N status, and as a result, VPI is a good indicator of the degree of invasion and aggressiveness of NSCLC. As more early-staged lung neoplasms are detected, whether VPI has impact on survival of patients with early-staged lung cancer is unknown. Therefore, Jiang et al. (2015) published a meta-analysis and found VPI together with tumor size has a synergistic effect on survival in patients with N0 disease. Patients with stage IB NSCLC and larger tumor size with VPI might be considered for adjuvant chemotherapy after surgical resection and need careful preoperative evaluation and postoperative follow-up.
There are several limitations to this study. First, the sample size was relatively small. Contributing to the small sample size, there were several patients with CTNNB1 gene mutations who could not be included in the data analysis because of incomplete clinicopathological records. Finally, the patients’ outcomes could have been influenced by the use of different treatment strategies, which may confound the survival analysis.
In summary, our results suggest that female patients and nonsmokers are likely to harbor CTNNB1 mutation and primary lung adenocarcinoma with mutated CTNNB1 has a poor prognosis. Further research is needed to verify our results. However, these data suggest that β-catenin could be a potential therapeutic target for advanced-stage lung cancer.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics committee of Shanghai chest hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conception and design: CZ and CC. Development of methodology: CZ, WL, and CC. Acquisition of data (provided surgical samples, gene detection, pathological diagnosis, acquired and managed patients, provided facilities, etc.): CZ, WL, JS, JZ, and CC. Analysis and interpretation of data: CZ. Writing, review, and/or revision of the manuscript: CZ and CC.
Funding
This work was supported by Shanghai Pulmonary Hospital Innovation Team Program (fkcx1906).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01367/full#supplementary-material
References
Akyol, A., Güner, G., Özşeker, H. S., Işık, A., Atcı, O., Fearon, E. R., et al. (2019). An immunohistochemical approach to detect oncogenic CTNNB1 mutations in primary neoplastic tissues. Lab. Invest. 99 (1), 128. doi: 10.1038/s41374-018-0121-9
Butnor, K. J., Travis, W. D. (2012). Editorial comment: recent advances in our understanding of lung cancer visceral pleural invasion and other forms of minimal invasion: implications for the next TNM classification.
Colombo, C., Miceli, R., Lazar, A. J., Perrone, F., Pollock, R. E., Gronchi, A., et al. (2013). CTNNB1 45F mutation is a molecular prognosticator of increased postoperative primary desmoid tumor recurrence: an independent, multicenter validation study. Cancer 119 (20), 3696–3702. doi: 10.1002/cncr.28271
Dar, M. S., Singh, P., Mir, R. A., et al. (2017). Beta-catenin N-terminal domain: an enigmatic region prone to cancer causing mutations. Mutat. Res. Rev. Mutat. Res. 773, 122–133. doi: 10.1016/j.mrrev.2017.06.001
Edge, S. B., Byrd, D. R., Compton, C. C. (2010). Cancer staging manual. American Joint Committee on Cancer (AJCC). 7th ed. (New York: Springer).
Field, J. K., Smith, R. A., Aberle, D. R., Oudkerk, M., Baldwin, D. R., Yankelevitz, D., et al. (2012). International association for the study of lung cancer computed tomography screening workshop 2011 report. J. Thoracic Oncol. 7 (1), 10–19. doi: 10.1097/JTO.0b013e31823c58ab
Field, J. K. (2008). Lung cancer risk models come of age. Cancer Prev. Res. 1 (4), 226–228. doi: 10.1158/1940-6207.CAPR-08-0144
Gu, C., Wang, R., Pan, X., Huang, Q., Zhang, Y., Yang, J., et al. (2017a). Sublobar resection versus lobectomy in patients aged ≤ 35 years with stage IA non-small cell lung cancer: a SEER database analysis. J. Cancer Res. Clin. Oncol. 143 (11), 2375–2382. doi: 10.1007/s00432-017-2499-y
Gu, C., Pan, X., Chen, Y., Yang, J., Zhao, H., Shi, J., et al. (2017b). Short-term and mid-term survival in bronchial sleeve resection by robotic system versus thoracotomy for centrally located lung cancer. Eur. J. Cardio-Thoracic Surg. 53 (3), 648–655. doi: 10.1093/ejcts/ezx355
Gu, C., Wang, R., Pan, X., Huang, Q., Luo, J., Zheng, J., et al. (2017c). Comprehensive study of prognostic risk factors of patients underwent pneumonectomy. J. Cancer 8 (11), 2097. doi: 10.7150/jca.19454
Gu, C., Huang, Z., Dai, C., Wang, Y., Ren, Y., Chen, C., et al. (2018). Prognostic analysis of limited resection versus lobectomy in stage IA small cell lung cancer patients based on the surveillance, epidemiology, and end results registry database. Front. Genet. 9, 1–6. doi: 10.3389/fgene.2018.00568
Jiang, L., Liang, W., Shen, J., Chen, X., Shi, X., He, J., et al. (2015). The impact of visceral pleural invasion in node-negative non-small cell lung cancer: a systematic review and meta-analysis. Chest 148 (4), 903–911. doi: 10.1378/chest.14-2765
Kase, S., Sugio, K., Yamazaki, K., Okamoto, T., Yano, T., Sugimachi, K., et al. (2000). Expression of E-cadherin and β-catenin in human non-small cell lung cancer and the clinical significance. Clin. Cancer Res. 6 (12), 4789–4796.
National Lung Screening Trial Research Team (2011). The national lung screening trial: overview and study design. Radiology 258 (1), 243–253. doi: 10.1148/radiol.10091808
Nozawa, N., Hashimoto, S., Nakashima, Y., Matsuo, Y., Koga, T., Sugio, K., et al. (2006). Immunohistochemical α-and β-catenin and E-cadherin expression and their clinicopathological significance in human lung adenocarcinoma. Pathol. Res. Pract. 202 (9), 639–650. doi: 10.1016/j.prp.2006.03.007
Raji, O. Y., Duffy, S. W., Agbaje, O. F., Baker, S. G., Christiani, D. C., Field, J. K., et al. (2012). Predictive accuracy of the Liverpool Lung Project risk model for stratifying patients for computed tomography screening for lung cancer: a case–control and cohort validation study. Ann. Internal Med. 157 (4), 242–250. doi: 10.7326/0003-4819-157-4-201208210-00004
Rami-Porta, R., Ball, D., Crowley, J., Giroux, D. J., Jett, J., Travis, W. D., et al. (2007). The IASLC Lung Cancer Staging Project: proposals for the revision of the T descriptors in the forthcoming (seventh) edition of the TNM classification for lung cancer. J. Thoracic Oncol. 2 (7), 593–602. doi: 10.1097/JTO.0b013e31807a2f81
Retera, J. M., Leers, M. P., Sulzer, M. A., Dar, M. J. (1998). The expression of beta-catenin in non-small-cell lung cancer: a clinicopathological study. J. Clin. Pathol. 51 (12), 891–894. doi: 10.1136/jcp.51.12.891
Shigemitsu, K., Sekido, Y., Usami, N., Mori, S., Sato, M., Horio, Y., et al. (2001). Genetic alteration of the β-catenin gene (CTNNB1) in human lung cancer and malignant mesothelioma and identification of a new 3p21. 3 homozygous deletion. Oncogene 20 (31), 4249. doi: 10.1038/sj.onc.1204557
Shimizu, K., Yoshida, J., Nagai, K., Nishimura, M., Ishii, G., Morishita, Y., et al. (2005). Visceral pleural invasion is an invasive and aggressive indicator of non-small cell lung cancer. J. Thoracic Cardiovasc. Surg. 130 (1), 160–165. doi: 10.1016/j.jtcvs.2004.11.021
Spitz, M. R., Hong, W. K., Amos, C. I., Wu, X., Schabath, M. B., Dong, Q., et al. (2007). A risk model for prediction of lung cancer. J. Natl. Cancer Institute 99 (9), 715–726. doi: 10.1093/jnci/djk153
Sun, Y., Ren, Y., Fang, Z., Li, C., Fang, R., Gao, B., et al. (2010). Lung adenocarcinoma from East Asian never-smokers is a disease largely defined by targetable oncogenic mutant kinases. J. Clin. Oncol. 28 (30), 4616. doi: 10.1200/JCO.2010.29.6038
Sunaga, N., Kohno, T., Kolligs, F. T., Fearon, E. R., Saito, R., Yokota, J. (2001). Constitutive activation of the Wnt signaling pathway by CTNNB1 (β-catenin) mutations in a subset of human lung adenocarcinoma. Genes Chromosomes Cancer 30 (3), 316–321. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1097>3.0.co;2-9
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., Jemal, A. (2015). Global cancer statistics, 2012. CA: Cancer J. Clinicians 65 (2), 87–108. doi: 10.3322/caac.21262
Travis, W. D., Brambilla, E., Rami-Porta, R., Vallières, E., Tsuboi, M., Rusch, V., et al. (2008). Visceral pleural invasion: pathologic criteria and use of elastic stains: proposal for the 7th edition of the TNM classification for lung cancer. J. Thoracic Oncol. 3 (12), 1384–1390. doi: 10.1097/JTO.0b013e31818e0d9f
Travis, W. D., Brambilla, E., Noguchi, M., Nicholson, A. G., Geisinger, K. R., Yatabe, Y., et al. (2011). IASLC/ATS/ERS international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 6 (2), 244–285. doi: 10.1097/JTO.0b013e318206a221
Wang, R., Pan, Y., Li, C., Hu, H., Zhang, Y., Li, H., et al. (2012). The use of quantitative real-time reverse transcriptase PCR for 5′ and 3′ portions of ALK transcripts to detect ALK rearrangements in lung cancers. Clin. Cancer Res. 18 (17), 4725–4732. doi: 10.1158/1078-0432.CCR-12-0677
Keywords: adenocarcinoma, lung cancer, CTNNB1, mutation, prognosis
Citation: Zhou C, Li W, Shao J, Zhao J and Chen C (2020) Analysis of the Clinicopathologic Characteristics of Lung Adenocarcinoma With CTNNB1 Mutation. Front. Genet. 10:1367. doi: 10.3389/fgene.2019.01367
Received: 17 October 2019; Accepted: 12 December 2019;
Published: 07 February 2020.
Edited by:
Tao Huang, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Liang Liu, Fudan University Shanghai Cancer Center, ChinaDeli Liu, Cornell University, United States
Copyright © 2020 Zhou, Li, Shao, Zhao and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Chen, Y2hlbnRob3JhY2ljQDE2My5jb20=
 Chao Zhou
Chao Zhou Wentao Li2
Wentao Li2 Jikai Zhao
Jikai Zhao Chang Chen
Chang Chen