- 1School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
- 2State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 3College of Plant Health and Medicine, Qingdao Agricultural University, Qingdao, China
- 4Agricultural Research Service, U.S. Department of Agriculture, Crop Genetics and Breeding Research Unit, University of Georgia – Tifton Campus, Tifton, GA, United States
- 5Department of Entomology and BIO5 Institute, University of Arizona, Tucson, AZ, United States
Helicoverpa armigera is a globally-important crop pest with a WZ (female)/ZZ (male) sex chromosome system. The absence of discernible sexual dimorphism in its egg and larval stages makes it impossible to address any sex-related theoretical and applied questions before pupation unless a W-specific sequence marker is available for sex diagnosis. To this end, we used one pair of morphologically pre-sexed pupae to PCR-screen 17 non-transposon transcripts selected from 4855 W-linked candidate reads identified by mapping a publicly available egg transcriptome of both sexes to the male genome of this species and detected the read SRR1015458.67499 only in the female pupa. Subsequent PCR screenings of this read and the previously reported female-specific RAPD (random amplified polymorphic DNA) marker AF18 with ten more pairs of pre-sexed pupae and different annealing positions and/or temperatures as well as its co-occurrence with the female-specific transcript splicing isoforms of doublesex gene of H. armigera (Hadsx) and amplification and sequencing of their 5′ unknown flanking sequences in three additional pairs of pre-sexed pupae verified that SRR1015458.67499 is a single copy protein-coding gene unique to W chromosome (named GUW1) while AF18 is a multicopy MITE transposon located on various chromosomes. Test application of GUW1 as a marker to sex 30 neonates of H. armigera yielded a female/male ratio of 1.14: 1.00. Both GUW1 and Hadsx splicing isoforms assays revealed that the H. armigera embryo cell line QB-Ha-E-1 is a male cell line. Taken together, GUW1 is not only a reliable DNA marker for sexing all stages of H. armigera and its cell lines, but also represents the first W-specific protein-coding gene in lepidopterans.
Introduction
Sex has profound effects on almost all aspects of animal life. This is manifested by the ubiquity of sexually-dimorphic morphological, physiological, behavioral, and life-history traits (Allen et al., 2011; Mowrey and Portman, 2012). Most notably, males and females differ in a broad variety of morphological traits that are involved in feeding, mate location, dispersal, escape from natural enemies, and/or oviposition. These include wing size and shape, size and color of pigment patches (female- or male-limited mimicry) and UV-reflective regions on the wings, size ratio of body parts (e.g., thorax to abdomen size ratio), color and density of body hairs, size and shape of sensory structures (e.g., antennae, eyes, auditory organs), size and shape of genitalia, and pheromone-releasing structures (Allen et al., 2011). For night-flying insects, female is usually the signaling sex which releases sex pheromone at night to attract and call the signal-receiving male which has more elaborate antennae. For day-flying species, male is the signaling sex which displays its vivid UV-reflective wings to attract the visual signal-receiving females for mating (Allen et al., 2011). In general, males are often smaller than females in body size but emerge earlier than the latter (male protandry) (Fischer and Fiedler, 2001; Esperk et al., 2007; Stillwell et al., 2010; Teder, 2014). The sexual size dimorphism is resulted from sex differnce in devlopemt time or even number of larval instar (larger sex has a longer development time or even more number of larval instar) (Esperk et al., 2007; Teder, 2014). In most insects, larger females also have a longer lifespan than smaller males (Fox et al., 2003). Less universal dimorphic traits include higher tolerance to insecticides (Li et al., 1992), heat stress (Gruntenko et al., 2016), and pathogen infection (Vincent and Sharp, 2014) as well as lower tendency to enter diapuse and shorter post-diapause period (Shimizu and Fujisaki, 2002) in females. Some of these secondary sexually-dimorphic traits, such as larger females (greater fecundity) and male protandry (better chance to mate with females), are largely associated with different reproductive roles, whereas others have a mix of sexual and non-sexual functions [e.g., arctiid moth auditory apparatus (Weller et al., 1999)] or function exclusively in non-reproductive contexts [e.g., female-limited mimetic color patterns (Kunte, 2009; Stillwell et al., 2010; Allen et al., 2011)].
Although sex of an individual insect is determined genetically upon fertilization and developed early in embryogenesis (Salz, 2011; Bopp et al., 2014; Biedler and Tu, 2016), most of the aforementioned sexual differences are uncovered in adult and/or pupal stages. This is because most insects do not exhibit conspicuous sexual dimorphism before pupation, and thus their eggs and larvae are often morphologically indistinguishable. Yet, characterization of the upstream diverged primary signal, master binary switch gene and autoregulatory gene of the sex determination pathway in various insects necessitates sex identification of eggs and young larvae/nymphs. Having a reliable method to sex eggs and larvae/nymphs of insects is also required for many other theoretical and applied research, such as molecular mechanisms of sexual trait development (Prakash and Monteiro, 2016) and genetic control of insect pests (San Andres et al., 2007). While flow cytometry measurement of DNA content (Nakamura et al., 1990; Aron et al., 2003), microscopic observation of sex chromatin (Traut and Marec, 1996; Fuková et al., 2005), and quantitative PCR (qPCR) measurement of the copy number (two copies in males vs. one copy in females) of the conserved Z chromosome gene in Lepidoptera (Belousova et al., 2019) can be used to determine sex of morphologically indistinguishable stages of some species, PCR gel analysis of sex-specific DNA markers revealed by RAPD (random amplified polymorphic DNA; Abe et al., 1998; Niu et al., 2007) assays or sex-specific chromosome (Y or W)-unique DNA sequences identified by comparison of male vs. female genome sequences (Krzywinski et al., 2004; Carvalho and Clark, 2013; Hall et al., 2013; Koerich et al., 2016) offers a more straightforward and reliable tool for sex identification. Such PCR-based sexing method is available for Ceratitis capitata (Douglas et al., 2004; San Andres et al., 2007; Gabrieli et al., 2010), Cydia pomonella (Fuková et al., 2009), Tribolium castaneum (Lagisz et al., 2010), and Gnatocerus cornutus (Gotoh et al., 2016), but has yet to be developed in many other crop pests.
Helicoverpa armigera is one of the most destructive polyphagous pests with hundreds of host plants belong to over 40 plant families (Zalucki et al., 1994; Zhao et al., 1998). Like Bombyx mori and most of lepidopterans (Fujii and Shimada, 2007; Traut et al., 2007), H. armigera has a WZ sex determination system, in which males are homogametic sex (ZZ) and females are heterogametic sex (WZ; Zhao et al., 2005; Niu et al., 2007). Male and female adults of H. armigera can be reliably sexed by their external genitalia and wing markings (Siverly, 1947; Zhao et al., 2005). At the pupal stage, the relative distance between the reproductive hole and the excretion hole can be used to sex male and female pupae (Zhao et al., 2005). For larvae older than the 3rd instar, one may recognize male and female larvae by the dorsal line, valve line, plane, and/or male testes (Jiang and Hu, 1995; Chen et al., 2004). However, sexing larvae older then the 3rd instar not only takes significant amount of time, practice and expertise, but also has a high error rate. Moreover, there are no morphological characters that can differentiate male from female eggs and larvae younger than the 3rd instar. Therefore, it is necessary to develop a PCR-based sexing method using W chromosome-unique DNA sequence for identification of male and female eggs, larvae and even pupae.
So far, no W-linked sequences for H. armigera are presently characterized, and only one female-specific RAPD marker (named AF18 hereafter) has been reported for this species (Niu et al., 2007). In this study, we first identified 4855 cDNA sequence reads putatively transcribed from the W chromosome of female eggs by mapping a publicly available egg transcriptome of both sexes (NCBI SRA dataset accession number SRP031603, Anantanawat, 2013) to the male genome of this species (Li et al., unpublished data). We then PCR-screened one pair of morphologically pre-sexed pupae for the presence of 17 non-transposon reads selected from the 4855 W-linked candidate sequences and detected the read SRR1015458.67499 in the female pupa, but not in the male pupa. We further PCR-screened ten more pairs of pre-sexed pupae with different annealing positions and/or temperatures (62 or 58°C) and found that varying the annealing position and/or temperature altered the female specificity of the previously reported RAPD marker AF18, but not the female specificity of SRR1015458.67499. We also used three additional pairs of pre-sexed pupae to check the co-occurrence of SRR1015458.67499 (one sequence read in SRP031603, Anantanawat, 2013) with the female-specific transcript splicing isoforms of H. armigera doublesex gene (Hadsx, Wang et al., 2014) and clone the 5′-flanking unknown sequences of SRR1015458.67499 and AF18. Finally, we tested the utility of SRR1015458.67499 as a marker to sex one embryo cell line and 30 neonate larvae of H. armigera. These experiments revealed that AF18 is a MITE (miniature inverted-repeat transposable elements) transposon located in multiple loci on both W and other chromosomes, whereas SRR1015458.67499 represents a single copy protein-coding gene unique to W chromosome (called GUW1) and thus can be used as a reliable DNA marker for sexing all growth stages of H. armigera.
Materials and Methods
H. armigera Strain and Cell Line
The laboratory colony of H. armigera used in this study was established with about 1200 larvae collected in tobacco fields in Xuchang (Henan, China) in June 2016 The colony had been maintained in a growth chamber at 26 ± 0.5°C with a photoperiod of 16 h light: 8 h dark and a relative humidity of 75 ± 5% (for adults) or 50 ± 5% (for larvae). Larvae of the colony had been reared on wheat germ-containing artificial diets (Waldbauer et al., 1984), whereas the adult moths had been fed with 10% honey water.
We pre-sexed 14 pairs of pupae based on the relative distance between the excretion and reproduction holes of each pupa according to Zhao et al. (2005). The pre-sexed pupae and a “tester set” of 30 unsexed neonate larvae were flash-frozen in liquid nitrogen and stored at −80°C for subsequent DNA and RNA extraction.
The embryo cell line QB-Ha-E-1 of H. armigera sexed in this study was established from another population (Zheng et al., 2010). QB-Ha-E-1 was seeded at 5 × 105 per mL in 15 mL flask and cultured at 28°C with Grace’s Insect Medium (Gibco/Life Technologies, New York, NY, United States) supplemented with 10% fetal bovine serum (Gibco/Life Technologies, New York, NY, United States), 50 U/mL penicillin and 50 μg/mL streptomycin (HyClone, Thermo Fisher Scientific, Logan, UT, United States). After three days, the cells were pelleted down by centrifuge at 300 × g, flash-frozen in liquid nitrogen and stored at −80°C for subsequent DNA and RNA extraction.
DNA/RNA Extraction and cDNA Synthesis
Genomic DNA (gDNA) was extracted from each of the 30 unsexed neonates as described (Li et al., 2002). Each of the 14 pairs of sexed pupae was individually grounded to powder in liquid nitrogen. Half of the powder was used to extract gDNA (Li et al., 2002). The other half was used to extract total RNA with the Trizol Regent (Life Technologies, Grand Island, NY, United States) according to the manufacturer’s manual. Likewise, half of the QB-Ha-E-1 cell pellet was used to extract gDNA and the other half for extraction of total RNA. The obtained DNA pellet from each neonate, pupa or the QB-Ha-E-1 cells was dissolved in the double distilled water (ddH2O), quantified using a NanoDrop 1000 instrument (NanoDrop, Wilmington, DE, United States), and then stored at −20°C.
The total RNA sample obtained from each pupa or the QB-Ha-E-1 cells was treated with DNase I (Promega, Madison, WI, United States) and RNase inhibitor (Thermo Fisher Scientific, Wilmington, DE, United States) for 40 min to remove potential gDNA contamination, cleaned with phenol/chloroform extraction, and dissolved in diethyl pyrocarbonate (DEPC) treated-H2O. The concentration of each cleaned RNA sample was determined using a NanoDrop 1000 instrument. We transcribed 1 μg of each RNA sample into cDNA using a mixture of random hexamer primers and oligo(dT)20 and Quant Reverse transcriptase (Tiangen Biotech, Beijing, China).
Genomic PCR and RT-PCR Gel Analyses
We designed a pair of primers for each of the 17 selected putative W-linked transcript 454 reads (see the corresponding primers in Supplementary Table S1) and the reference gene elongation factor 1 alpha (EF-1α; present in both males and females; Supplementary Table S2) with Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, United States) for the initial genomic PCR (gPCR) verification of their W chromosome uniqueness. For SRR1015458.67499 (i.e., GUW1), the only verified W-specific transcript in the initial PCR screen, we designed another pair of primers (GUW1-118-F and GUW1-118-R in Supplementary Table S2) for further verification of its W chromosome uniqueness and utility as a female specific molecular marker, and one gene-specific reverse primer GUW1-118-R (Supplementary Table S2 and Figure 5A) for amplification of its 5′-flanking sequences by genome walking. For the previously-reported female-specific RAPD marker AF18 (Niu et al., 2007), we used the same AF18-F and AF18-R primers (Supplementary Table S2) reported in Niu et al. (2007) for its initial verification. We also designed another pair of primers AF18-436-F3 and AF18-436-R3 for its further verification and another gene-specific reverse primer AF-18-GSP (see these primers in Supplementary Table S2) for amplification of its 5′-flanking sequences by genome walking. All the primers that we designed have a Tm value of 60°C and thus the related gPCR reactions for the 17 putative W-linked transcript 454 reads and EF-1α used the same PCR set up and cycling conditions (see below) except for the genome working PCRs and the PCR reactions of AF18. For RT-PCR detection of sex-specific transcript splicing isoforms of the doublesex gene, we used the primer pair Hadsx-F and Hadsx-R (Supplementary Table S2) and cycling conditions as reported in Wang et al. (2014).
The gPCR reactions for all of the tested putative W-linked transcripts, AF18, and EF-1α as well as RT-PCR reactions for the doublesex gene were initiated in a 25 μL reaction containing 1 μL template, 0.25 μmol/L of both the forward and reverse primers for each gene, 2.5 μL 2.5 mM dNTP, 5 μL PCR buffer with Mg2+ (Takara, Dalian) and 5 U ExTaq DNA polymerase. The PCR or RT-PCR reactions were completed in an Eppendorf Mastercycler® nexus (Eppendorf, Borsdorf, Leipzig, Germany) with a cycling program of denaturation at 95°C for 10 min, followed by 35 cycles of 95°C for 30 s, 60°C (62°C and 58°C for amplification of AF18 with AF-18F and AF-18R) for 30 s and 72°C for 1 min, and a final extension of 72°C for 5 min. The resultant gPCR or RT-PCR products were separated at 200 V for about 15 min on 2% agarose gels in the 1 × TAE buffer, visualized by ethidium bromide staining, and documented using a ChampGel (Sagecreation, Beijing, China).
Genome Walking
We used six gDNA samples extracted from three pairs of male and female pupae of H. armigera to clone the 5′-flanking sequences of GUW1 and AF18 by genome walking. As diagramed in Figure 5A, we digested 2.5 μg of each of the six gDNA samples with 2 μL of 5 U/μL MspI (New England Biolab, Boston, MA, United States) in a 100 μL reaction in a 37°C water bath for 12 h. The digested gDNA samples were purified by TIANquick Midi Purification Kit (Tiangen Biotech). We then ligated 200 ng of each MspI-digested DNA sample to the MspI adapter at 16°C for 16 h in a 20 μL reaction containing 1 × T4 ligation buffer and 2 U T4 ligases (New England Biolab, Boston, MA, United States). The MspI adapter was formed by annealing 1:1 ratio of the oligos MspI-a and MspI-b (Supplementary Table S2) by a 5-step descending heating program of 90, 70, 50, 30, and 16°C for 30 min, respectively.
We used the ligated products of the six gDNA samples as the templates to PCR-amplify the 5′-flanking sequences of AF18 and GUW1 with the general forward primer MspI-a and the gene-specific reverse primers AF18-GSP and GUW1-118-R (Supplementary Table S2 and Figure 5A), respectively. The genome walking PCR reactions were carried out in a 25 μL reaction mixture containing 2.5 μL of PCR buffer, 0.5 μL of dNTP (10 mM), 0.25 μL of primestar GXL polymerase (5 U/mL), 0.5 μL of the specific primer pairs (20 mM), and 1 μL of the template DNA. The PCR cycling conditions consisted of denaturation at 95°C for 5 min, followed by five cycles of 95°C denaturation for 1min, annealing for 30 s from 65 to 60°C at a declining rate of 1°C/cycle, extension at 72°C for 1 min, 25 cycles of 95°C denaturation for 1 min, annealing at 60°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 5 min. The PCR products were electrophoresed on a 2% agarose gel in the 1 × TAE buffer and visualized by ethidium bromide staining. The obtained bands of both AF18 and GUW1 were cloned into the pGEM®-T Easy Vector (Promega, Madison, WI, United States) and sequenced by Beijing Genomic Institute (BGI; Beijing, China).
Data Analyses
The egg 454 sequence data were downloaded from NCBI SRA datasets (the accession number was SRP031603 and the run number was SRR1013685, SRR1015459, and SRR1015457) (Anantanawat, 2013). The downloaded SRA data were process into fastq format by the fastq-dump software, which is a part of SRA Toolkit. Then we mapped the egg transcriptome data to our unpublished H. armigera male pupa genome (including mitochondrion genome) using the NCBI local BLAST 2.2.26. The unmapped sequences were putatively derived from the W chromosome and 17 of them were selected for further verification.
AF18 and GUW1 as well as the 5′ flanking sequences obtained by genome walking were further analyzed by Blast to NCBI databases and H. armigera male genome to annotate them and verify their W chromosome uniqueness. Chi-square (χ2) test was used to determine if the sex ratio of H. armigera neonates determined by the GUW1 marker is significantly different from the expected 1:1 sex ratio.
Results
Identification of W-Linked Candidate Transcripts
A total of 394,630 raw 454 sequence reads of H. armigera eggs (≤24 h post oviposition) of both sexes (Anantanawat, 2013) were downloaded from the NCBI website and then mapped to an unpublished male nuclear and mitochondrial genome of this species (Li et al., unpublished data). This transcriptome to genome blast revealed that 98.77% of the raw 454 sequence reads mapped at least one time to the male genomes. The remaining 4855 (1.23%) sequence reads failed to hit any scaffolds of the male genome and thus were putatively transcribed from the W chromosome of female eggs.
Characterization of a Gene Unique to W Chromosome
In order to characterize one or more transcripts unique to W chromosome for development of PCR-based DNA markers for sexing male vs. female eggs, larvae, pupae, or even cells, we selected 24 non-transposon transcripts of >200 base pairs (bps) from the 4855 unmapped candidate sequences for designating a pair of PCR primers with Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA, United States). The software Primer Premier 5.0 found suitable primers (Supplementary Table S1) for 17 out of the 24 transcripts, but no suitable primers were found for the other 7 transcripts.
We prepared two gDNA samples from a pair of pupae sexed by the relative distance between their reproduction and excretion holes, and used the two gDNA samples as the templates to PCR-amplify the above 17 transcripts and EF-1α, a reference gene known to exist in both male and female individuals. As expected, EF-1α was detected in both the male and female pupae (Figure 1). No expected bands were detected for 15 out of the 17 transcripts in both the male and female pupae (Figure 1). One of the two remaining transcripts, SRR1017685.5288, like the reference gene EF-1α, was present in both the male and female pupae (Figure 1), and therefore is not a W-specific gene. By contrast, another remaining transcript, SRR1015458.67499, was detected only in the female pupa (Figure 1), and thus is probably a gene unique to W chromosome [here after named gene unique to W 1 (GUW1)] and can be used to differentiate female vs. male cells and individuals at all developmental stages. Furthermore, Blast search of GUW1 against the published male genome of the Australian population of H. armigera (Pearce et al., 2017) also failed to hit any scaffolds.
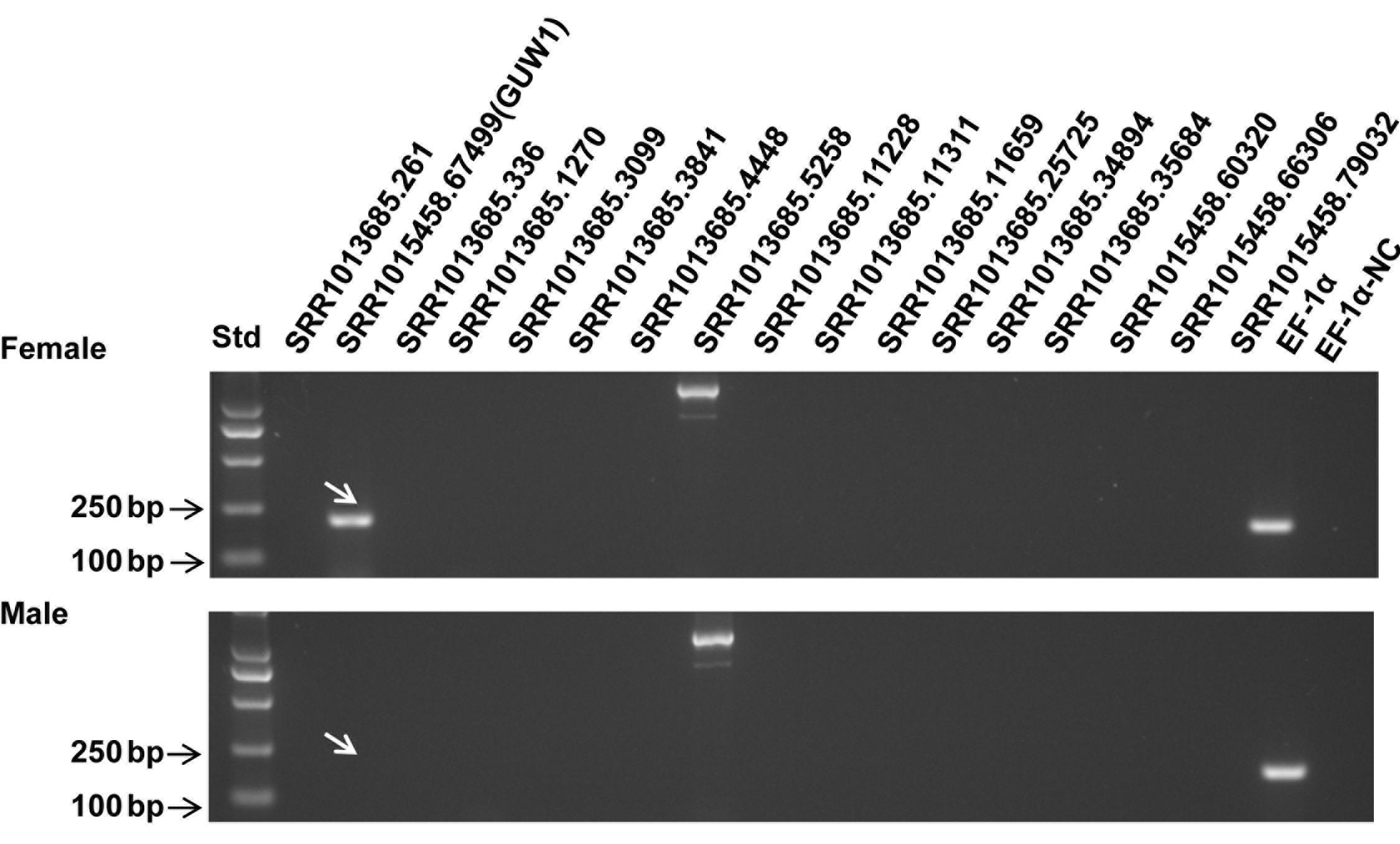
Figure 1. PCR detection of 17 putative W-linked sequences in a pair of H. armigera pupae. Genomic DNA extracted from one female (top panel) and one male (bottom panel) pupae of H. armigera were used as the templates to PCR-amplify EF-1α (as a positive control for autosome genes) and 17 selected putative W-linked sequences using the primers (Supplementary Table S1) designed based on their transcripts. The lane names starting with “SRR101” represent the PCR products of the 17 putative W-linked sequences. Std, molecular weight marker. EF-1α-NC, EF-1α negative control (no template control).
Verification and Annotation of GUW1 as a W-Specific Gene and of AF18 as a Multicopy Transposon
Three additional experiments were conducted to verify if GUW1 and the previously identified female-specific DNA marker AF18 (Niu et al., 2007) are W-specific sequences. First, we individually extracted gDNAs from 10 more pairs of morphologically pre-sexed pupae and tried to PCR-amplify both GUW1 and AF18 from the 10 pairs of pupae. Gel analysis of the PCR products showed the presence of GUW1 (Figure 2A) and AF18 (Figure 2B) in all 10 female pupae but not in all 10 male pupae. When the annealing temperature for PCR amplification of AF18 decreased from 62°C (Figure 2B) to 58°C (Figure 2C), the expected AF18 band occurred not only in the 10 female pupae but also in 4 (No. 2, 3, 5, and 9) out of the 10 male pupae (Figure 2C). Besides, 9 out of the 10 male pupae also had 1 or 2 obvious bands larger than the expected AF18 band. When the annealing position of the reverse primer was moved only 13 bp downstream (compare the primers AF18-436-R3 and AF18-R in Supplementary Table S2), the expected AF18 band appeared not only in the 10 female pupae but also in the 10 male pupae (Figure 3A). By contrast, shifting the forward primer 7 bp downstream (GUW1-118-F in Supplementary Table S2) and the reverse primer 81 bp upstream (GUW1-118-R in Supplementary Table S2) did not change the sex specificity of GUW1 band in any of the 10 pairs of pupae (Figure 3B).
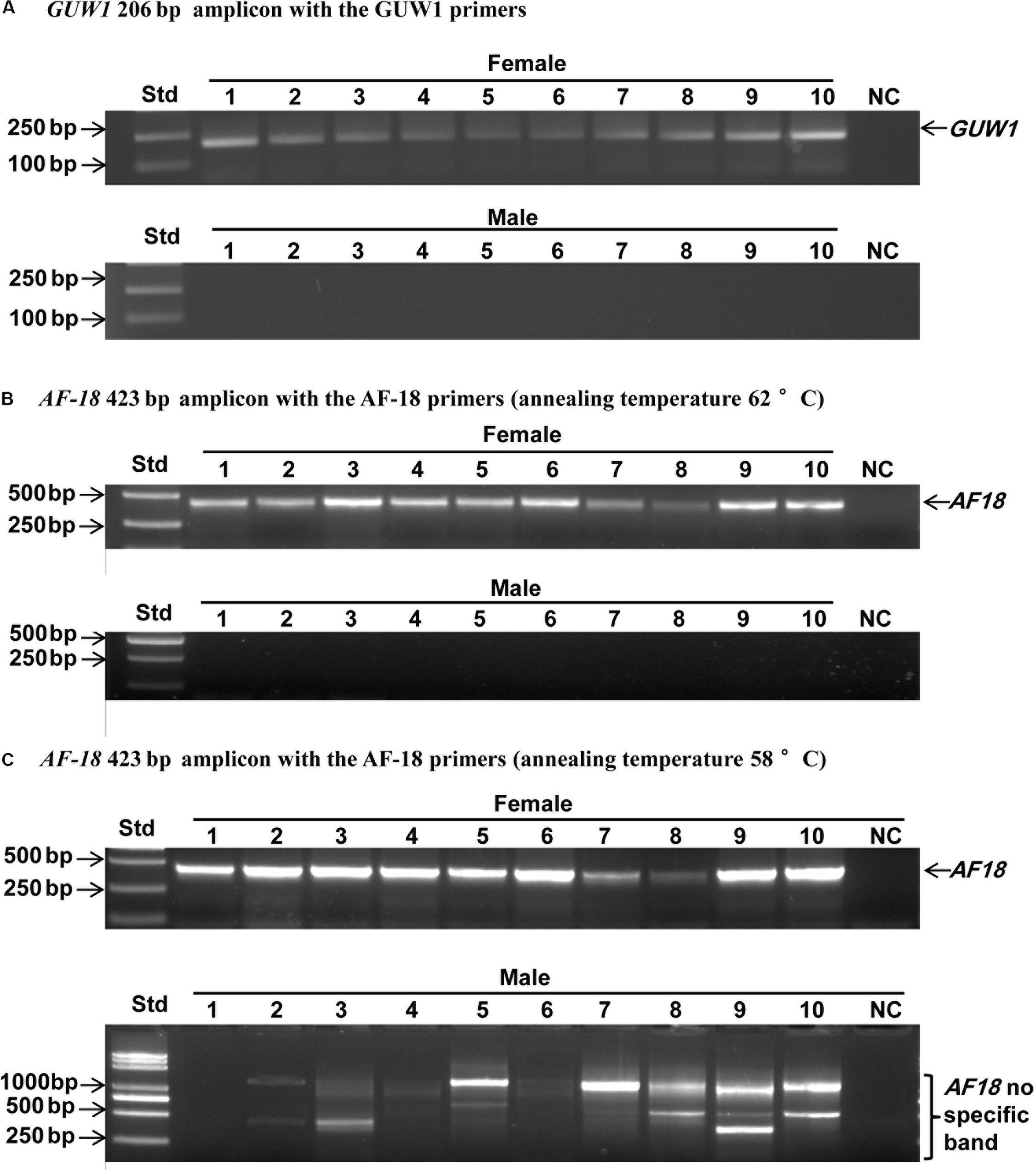
Figure 2. Sex identification of ten pairs of morphologically pre-sexed H. armigera pupae with the 206 bp GUW1 amplicon (A) and the 423 bp AF18 amplicon obtained at the annealing temperature of 62°C (B) and 58°C (C), respectively. Genomic DNA samples were individually extracted from ten pairs of morphologically pre-sexed pupae and used as the templates to PCR-amplify GUW1 with the primer pair GUW1-F and GUW1-R (Supplementary Table S2) and AF18 with the primer pair AF18-F and AF18-R (Supplementary Table S2). Lanes numbered with 1–10 indicate the 10 female (top panel) or male (bottom panel) pupae in each figure. Std, molecular weight marker. NC, negative control (no template control) for GUW1 (A) or AF18 (B–C).
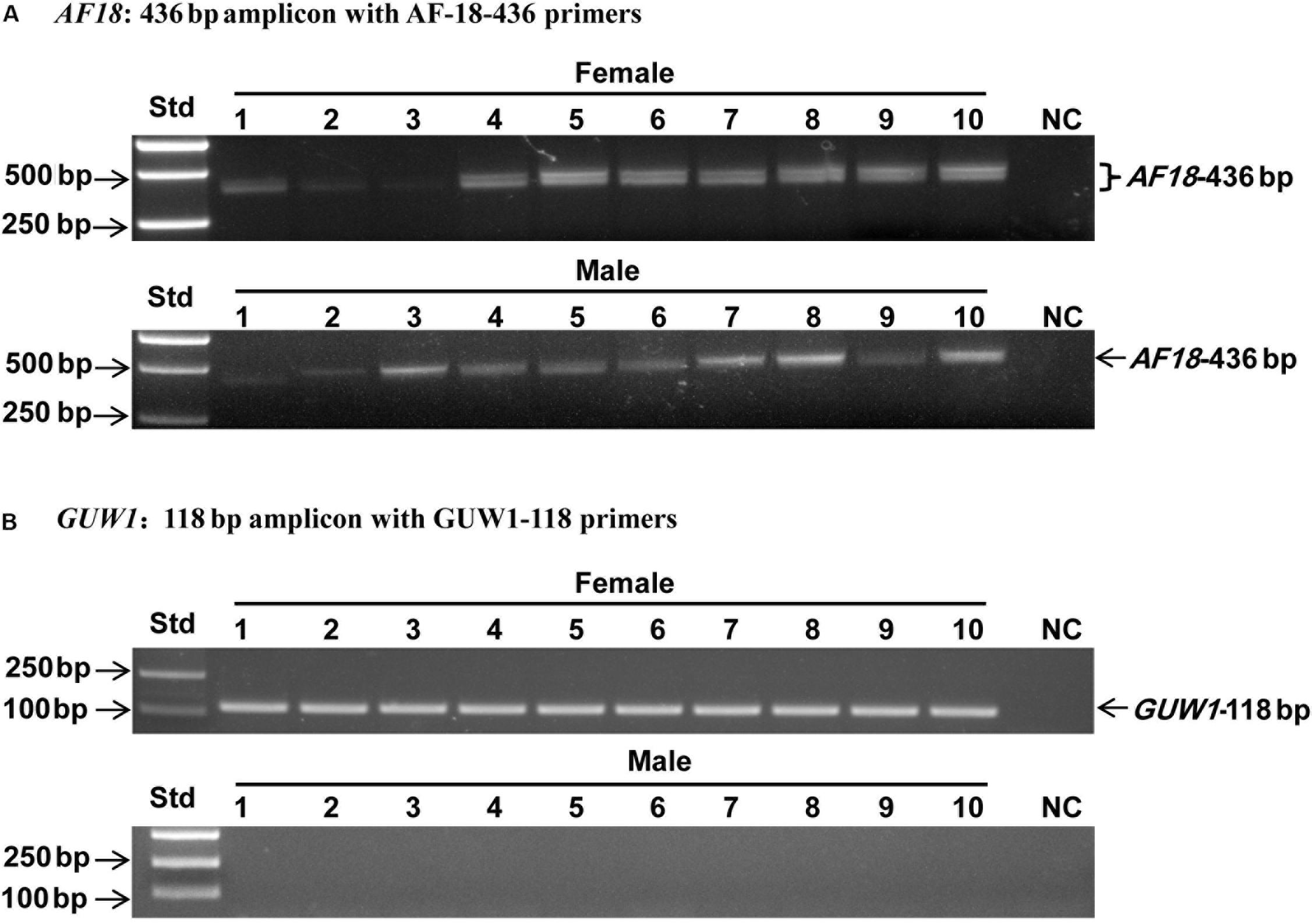
Figure 3. Sex identification of ten pairs of morphologically pre-sexed H. armigera pupae with the 436 bp AF18 amplicon (A) and the 118 bp GUW1 amplicon (B). Genomic DNA samples from the same ten pairs of pupae as in Figure 2 were used as the templates to PCR-amplify GUW1 with the primer pair GUW1-118-F and GUW1-118-R (Supplementary Table S2) and AF18 with AF18-436-F3 and AF18-436-R3 (Supplementary Table S2). Lanes numbered with 1–10 indicate the 10 female (top panel) or male (bottom panel) pupae in each figure. Std, molecular weight marker. NC, negative control (no template control) for AF18 (A) and GUW1 (B).
Second, we used the gDNA and RNA samples from 3 pairs of pupae as the templates to amplify GUW1 and the sex-unique transcript isoforms of Hadsx gene, respectively. According to Wang et al. (2014), the primer pairs we used for Hadsx (Hadsx-F and Hadsx-R in Supplementary Table S2) should yield four bands of 668/683 bp (not separable on gels) and 797/812 bp (not separable on gels) in females, but one band of 419 bp in males. Gel analysis of the GUW1 and Hadsx PCR or RT-PCR products showed that all the 3 pupae with the female-unique transcript isoforms of Hadsx had the expected GUW1 band, whereas those with the male-unique transcript isoform of Hadsx lacked the GUW1 band (Figure 4).
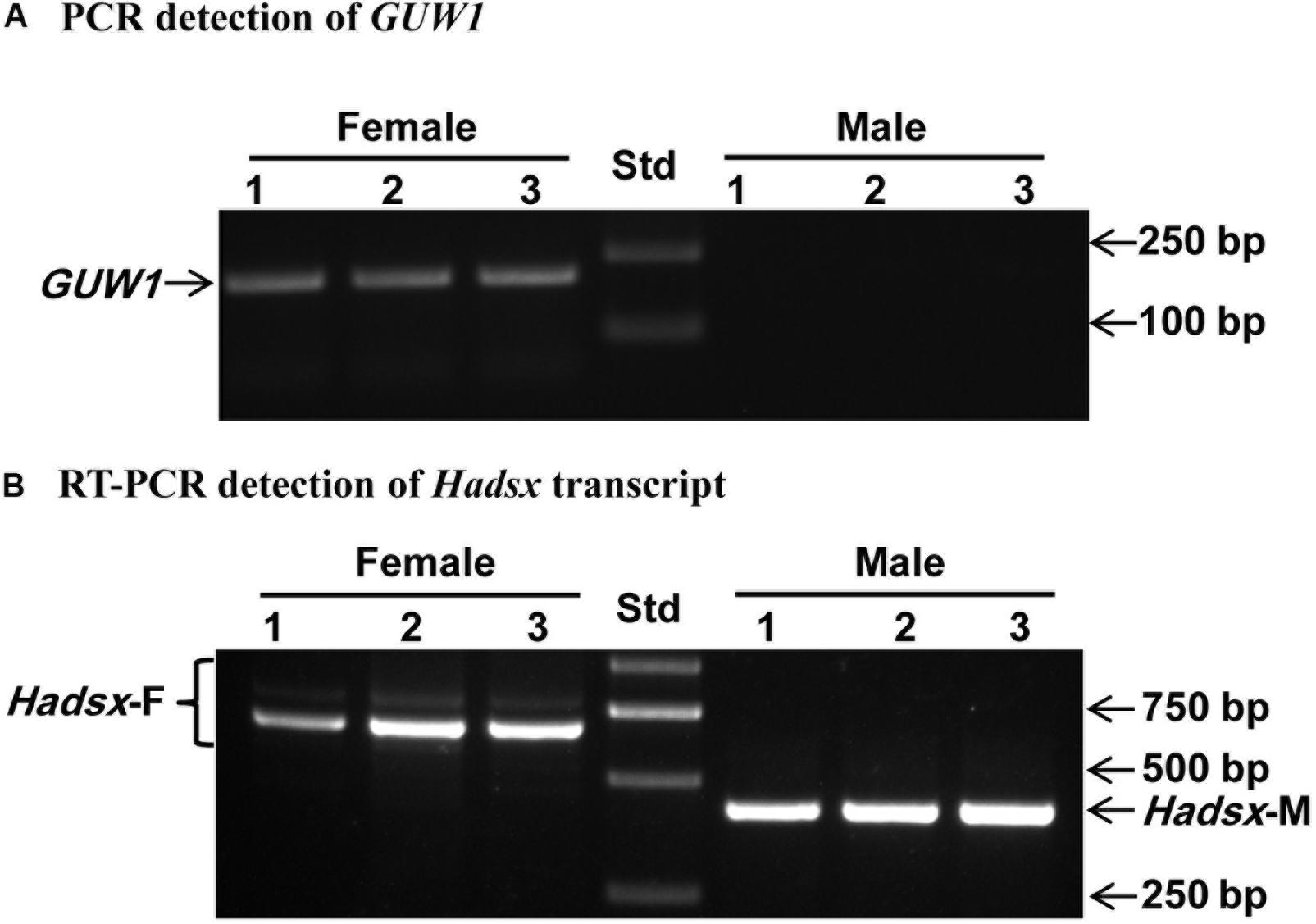
Figure 4. Sex identification of three pairs of morphologically pre-sexed H. armigera pupae with the 206 bp GUW1 amplicon (A) and Hadsx transcript (B). Genomic DNA and total RNA samples individually extracted from three pairs of male and female pupae were used as the templates to PCR-amplify GUW1 with the primers GUW1-118-F and GUW1-118-R (Supplementary Table S2) and RT-PCR-amplify Hadsx transcripts with the primers Hadsx-F and Hadsx-R (Supplementary Table S2), respectively. Lanes numbered with 1–3 indicate the three female (top panel) or male (bottom panel) pupae in each figure. Std, molecular weight marker.
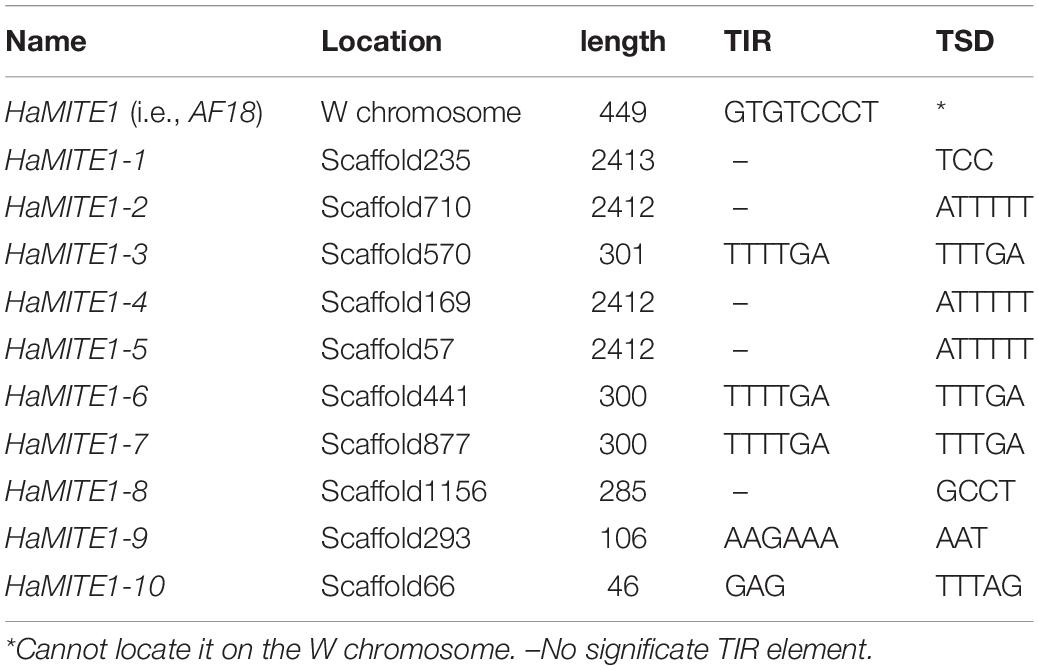
We also conducted genome walking to amplify the 5′ flanking sequences of AF-18 and GUW1 from the above three pairs of pupae (Figure 5A). Gel analysis of the genome walking products revealed one band of 635 bp for GUW1 only in the three female pupae, but two bands of 158 and 290 bp for AF18 in both the three female and three male pupae (Figure 5B). Sequencing of the two AF18 bands reveals that they not only miss the first 11 (band 1) and 45 bp (band 2) of the reported AF18 sequence respectively, but also share no significant similarity in their 5′ flanking sequences (Figure 5C and Supplementary Figure S1). These results indicate that the two bands represent two different copies of AF18 located on Z chromosome or autosome, whereas the previously reported female-specific copy (Niu et al., 2007) locates on W chromosome. Careful examination of the reported 449 bp AF18 sequence (Niu et al., 2007; Figure 5C) confirms that it is a MITE (Miniature inverted repeat transposable element; named MITE1_Har) because it has an AT-rich internal sequence (65% = 35%A + 30%T), a pair of 8-bp terminal inverted repeats (TIRs, left TIR = GTGTCCCT), and lacks coding potential. Blast search with MITE1-Har as the query yielded 137 significant hits (e-value ≤ 1.0e-10) to the H. armigera male genome scaffolds (Supplementary Table S3), indicating this transposon has multiple copies present on both Z chromosome and autosomes. We listed 10 representative copies of MITE1_Har in Table 1.
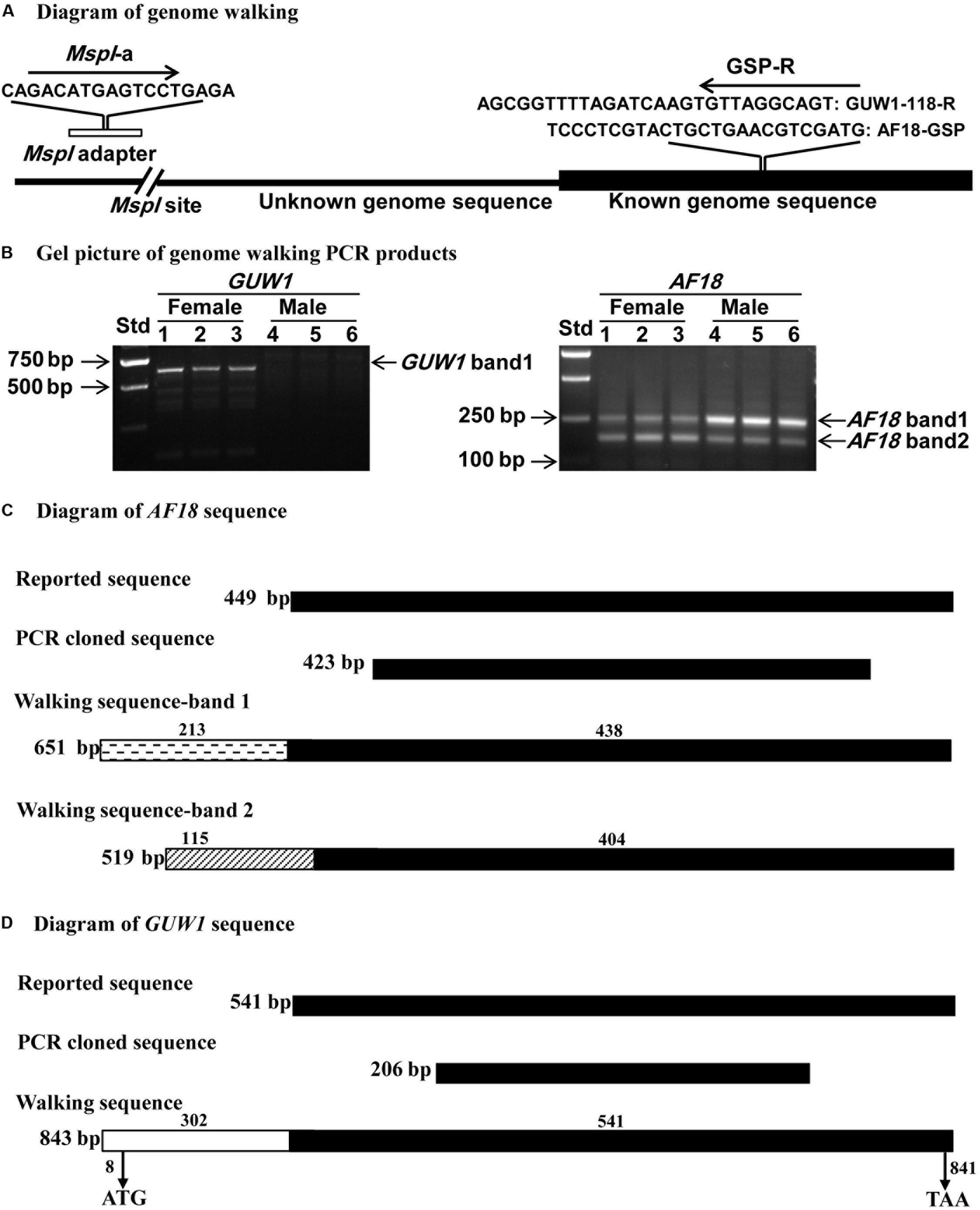
Figure 5. Schematic alignments of GUW1 and AF18 with their 5′ flanking sequence obtained by genome walking. Genome DNA samples from the same three pairs of male and female pupae as in Figure 4A were completely digested by MspI and ligated to MspI adaptor. The ligation mixtures were used as the templates to PCR-amplify the 5′ unknown flanking sequences of GUW1 and AF18 using the general primer MspI-a and the gene-specific primers GUW1-118-R and AF18-GSP (Supplementary Table S2), respectively (A). The obtained genome walking PCR products of GUW1 (left panel in B) and AF18 (right panel in B) from the three pairs of pupae were separated on 2% agarose gels (B). The two genome walking bands of AF18 from each of the six pupae and the single walking band of GUW1 from the three female pupae were sequenced and schematically aligned with the reported and cloned AF18 (C) and GUW1 sequences (D), respectively. Filled black boxes represent the known (reported) sequences of AF18 or GUW1, whereas clear (GUW1), cross-striped (AF18 band 1) or dashed (AF18 band 2) boxes represent the 5′ flanking sequences of the two markers identified by our genome walking experiment. The lengths of the 5′ flanking and reported (known) parts of the three walking sequence are placed above each box. The total length of each sequence is shown on the right side of each box. The positions of start and stop codons of the GUW1 open reading frame are shown below the walking sequence box. Std, molecular weight marker.
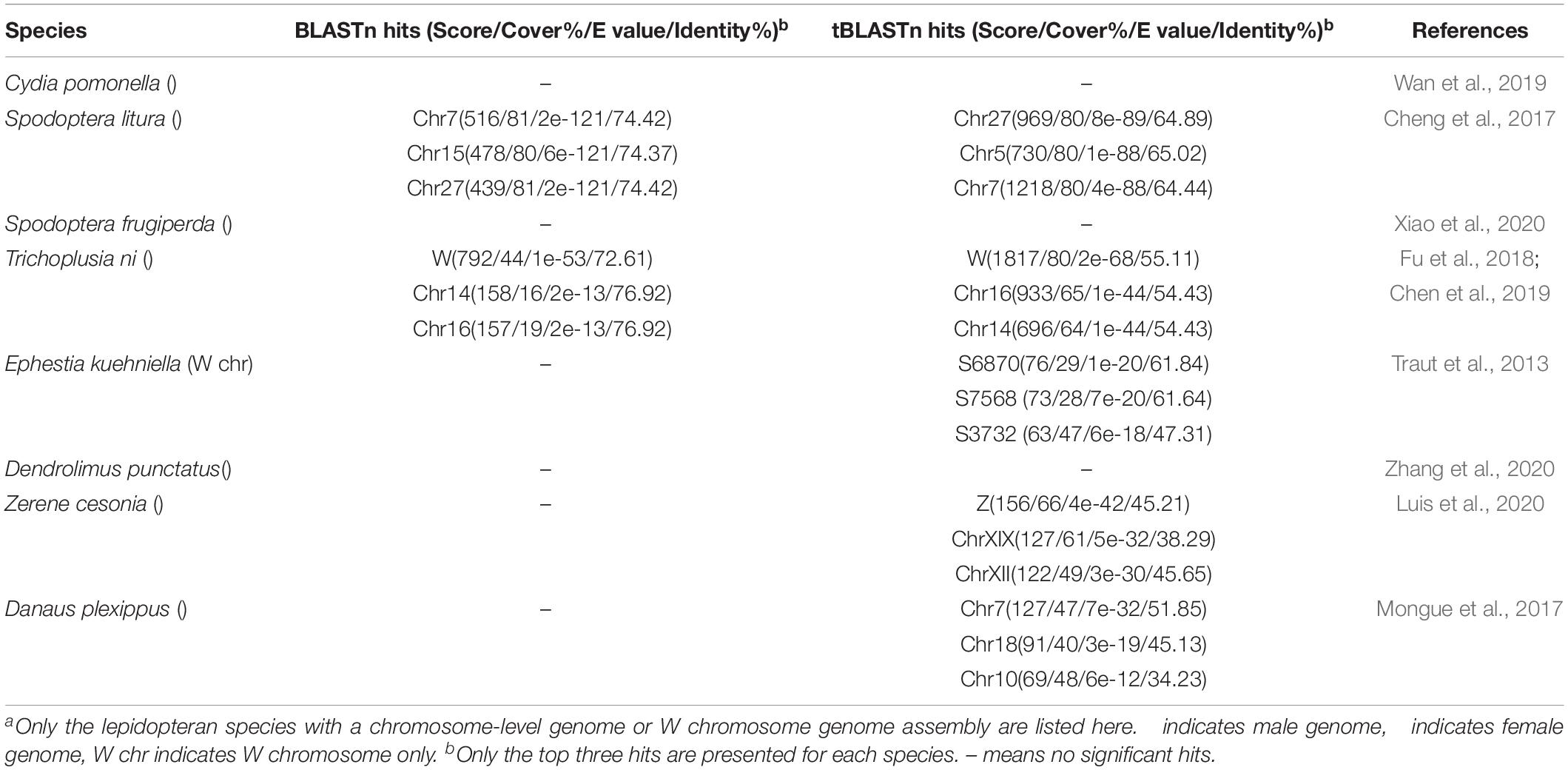
In contrast, sequencing of the only genome walking band for GUW1 from female pupae shows no 5′ end missing of the reported transcript (SRR1015458.67499) sequence (Figures 5D, 6), suggesting that the band is originated from the same locus with SRR1015458.67499 on W chromosome. Consistent with the absence of a genome walking band for GUW1 in the male pupae, BLASTn search with the assembled/edited sequence of the reported transcript plus the genome walking band (Figure 6) as the query produced no significant hits to the male H. armigera genome scaffolds. The assembled/edited GUW1 sequence has an open reading frame encoding a protein of 277 amino acids (aa) (Figure 6). BLASTn search with GUW1 as the query against the NCBI nucleotide collection found no similar sequences. BLASTp search against the NCBI protein database showed that the predicted GUW1 protein shares the highest amino acid sequence identity (69%) with the hypothetical protein RR48_05862 (GenBank accession# KPJ14768, 446 aa) predicted from Papilio machaon male adult genome (Li et al., 2015) in their first N-terminal 234 amino acids (query coverage 84%, e-value = 3e-108). BLASTn search against the publicly available chromosome-level genomes of eight lepidopteran species detected five partial alignments (in the 5′ 683 bp) with an identity of =0.74% and a coverage of =0.81% (2 on autosome 7, 2 on autosome 15, and 1 on autosome 27) in the genome of Spodoptera litura, 10 short partial alignments (in the 5′ 376 bp) with an identity of =0.73% and a coverage of =0.44% (4 on W chromosome, 1 on autosome 14, 1 on autosome 16, 1 on autosome 5, 1 on autosome 7, 1 on autosome 18 and 1 on autosome 19) in the genome of Trichoplusia ni, and no significant hits in the genomes of C. pomonella (female), Spodoptera frugiperda (female), Ephestia kuehniella (W chromosome only), Dendrolimus punctatus (female), Zerene cesonia (male), and Danaus plexippus (female) (Table 2). tBLASTn search against the genomes of the 8 lepidopteran species revealed multiple 5′ partial alignments (in the N-terminal 225 aa) with a coverage of =0.80% and an identity of =65%. in the genomes of S. litura, T. ni, E. kuehniella, Z. cesonia, and D. plexipus but no hits in the other three species (Table 2).

Figure 6. Nucleotide and deduced amino acid sequence of GUW1. The merged sequence of the reported GUW1 transcript (upright) and its 5′ flanking sequence (italicized) has an open reading frame (ORF) encoding a protein of 277 amino acids. The start and stop codons of the ORF are underlined. The annealing direction and position of the gene-specific primer GUW1-118R (Supplementary Table S2) used for genome walking of GUW1 are depicted with an arrowed line.
Application of GUW1-Based PCR Gel Analysis to Sex Young H. armigera Larvae and Cell Line
To test the utility of the W-specific GUW1 gene for sexing young larvae, we randomly selected 30 neonates of H. armigera and extracted gDNA from each of the 30 larvae. PCR gel analysis with the gene-specific primers for the W-specific marker gene GUW1 (GUW1-118-F and GUW1-118-R in Supplementary Table S2) and the autosomal gene EF-1α (EF1α-F and EF1α-R in Supplementary Table S2) showed the presence of the EF-1α band in all the 30 first-instar larvae (Figure 7). By contrast, the GUW1 band was detected only in 16 out of the 30 neonates. Chi-square (χ2) test confirmed that the sex ratio (16 female: 14 male = 1.14:1.0) obtained with the GUW1 marker was not significantly different from the expected 1:1 sex ratio (χ2 value = 0.067, P = 0.80).
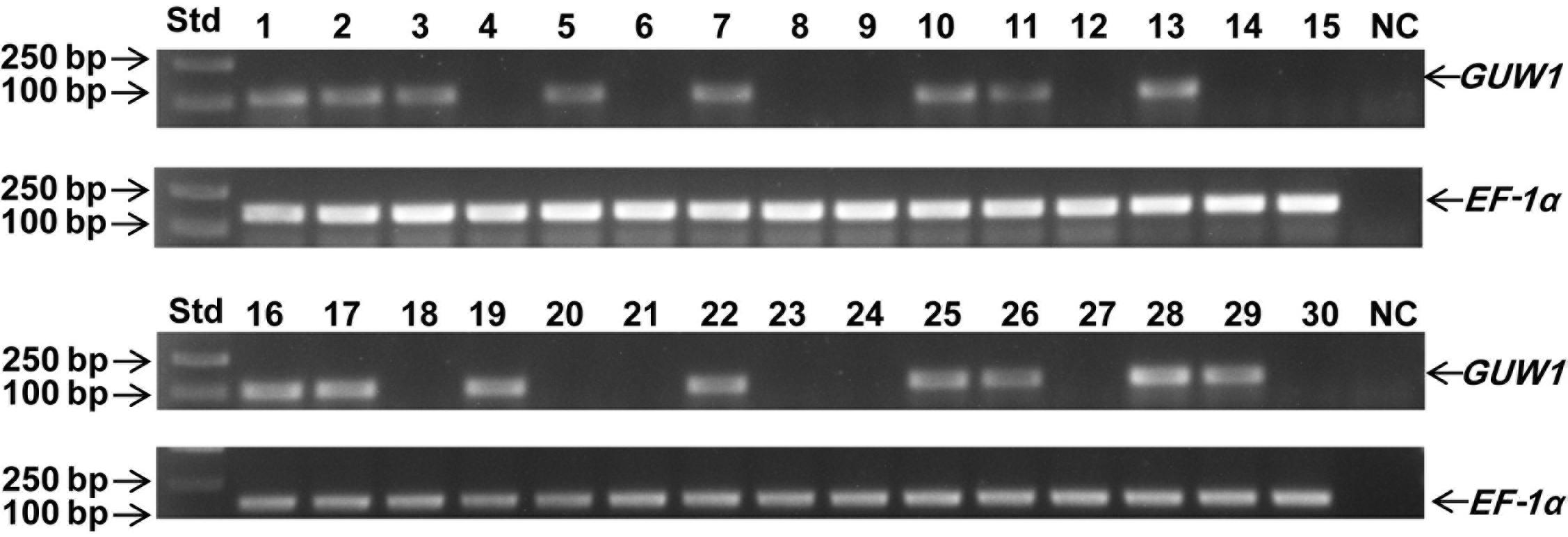
Figure 7. Test application of the 118 bp GUW1 amplicon for sex identification of 30 neonates of H. armigera. Genomic DNA samples individually extracted from 30 neonates of <5 h were used as the templates to PCR-amplify the W-linked GUW1 with the primer pair GUW1-118-F and GUW1-118-R (Supplementary Table S2) and the autosomal gene EF-1α with the primer pair EF1α-F and EF1α-R (Supplementary Table S2). Lanes numbered with 1–30 represent the 30 neonates. Std, molecular weight marker. NC, negative control (no template control) for GUW1 and EF-1α.
To test the utility of the W-specific GUW1 gene for sexing H. armigera cell lines, we extracted gDNA and total RNA from the embryo cell line QB-Ha-E-1 of H. armigera (Zheng et al., 2010) and gDNA from a pair of female and male pupae. We then used the obtained gDNA and RNA samples as the templates to PCR-amplify GUW1 and RT-PCR-amplify Hadsx transcripts, respectively. Gel analysis of the gDNA PCR products detected the GUW1 band in the female pupa, but not in the embryo cell line QB-Ha-E-1 and male pupa (see the left panel in Figure 8). Consistent with the absence of GUW1 gene in QB-Ha-E-1, gel analysis of the RT-PCR products showed that QB-Ha-E-1 had the male-unique transcript isoform of Hadsx (see the right panel in Figure 8).
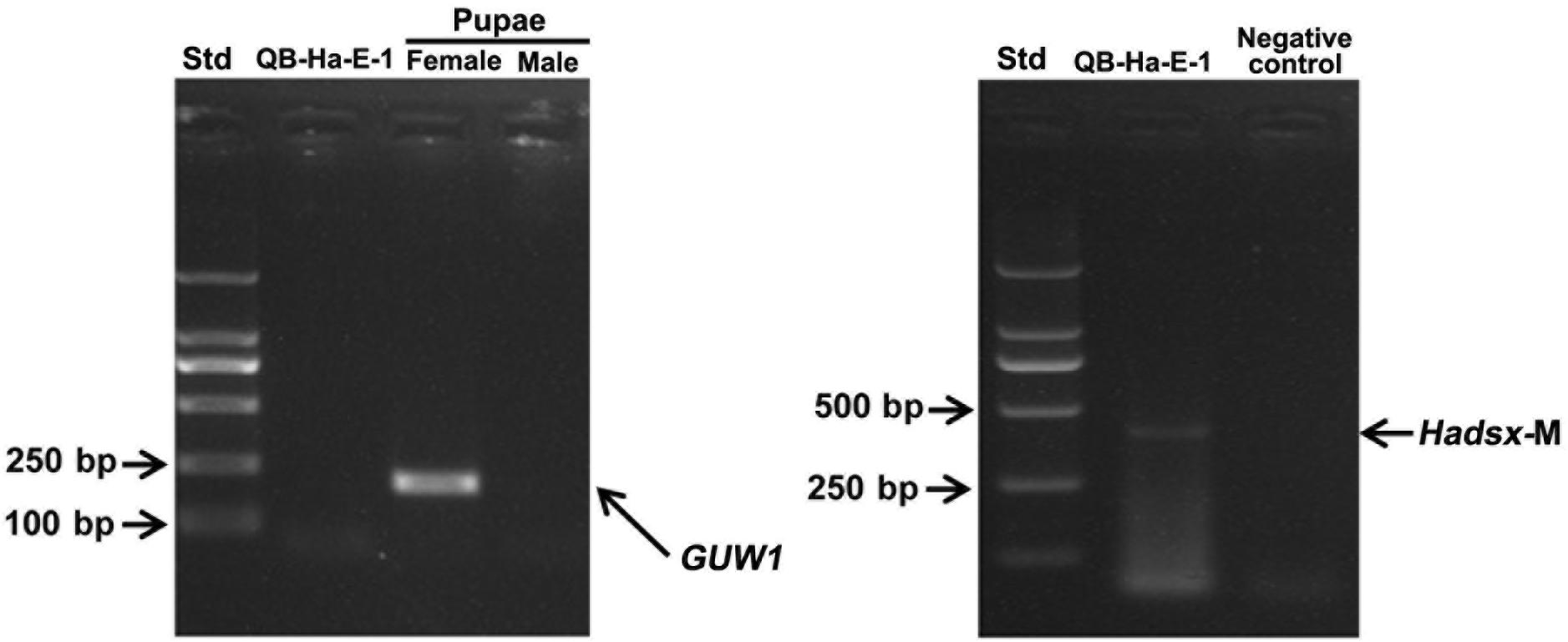
Figure 8. Sex identification of H. armigera cell line QB-Ha-E-1 with the 206 bp GUW1 amplicon (left panel) and Hadsx transcript (right panel). The gDNA samples extracted from a pair of pre-sexed pupae as well as the gDNA and total RNA samples from the H. armigera cell line QB-Ha-E-1 were used as the templates to PCR-amplify GUW1 (left panel) with the primer pair GUW1-F and GUW1-R (Supplementary Table S2) and RT-PCR-amplify Hadsx transcripts (right panel) with the primers Hadsx-F and Hadsx-R (Supplementary Table S2), respectively. Std, molecular weight marker.
Discussion
The absence of conspicuous sexual dimorphism prior to pupation makes it necessary to identify W chromosome-linked genes or sequences for accurate and reliable determination of the gender of individual eggs and larvae of H. armigera. Recent studies have demonstrated that characterization of Y or W chromosome-linked sequences or genes can be achieved through segregation and linkage analysis of transcriptomic alleles from parents to their F1 male and female progenies (Bergero and Charlesworth, 2011; Chibalina and Filatov, 2011), comparison of male vs. female transcriptomes of an inbred line (Muyle et al., 2012), and contrast of male vs. female genomes of a given species (Portela et al., 2010; Chen et al., 2012; Koerich et al., 2016; Fraïsse et al., 2017). Unlike the above three omics approaches that rely on either transcriptome-transcriptome contrast (Bergero and Charlesworth, 2011; Chibalina and Filatov, 2011; Muyle et al., 2012) or genome-genome subtraction (Portela et al., 2010; Chen et al., 2012; Koerich et al., 2016; Fraïsse et al., 2017), here we identified 4855 putative W-linked transcript sequences of H. armigera by mapping an egg transcriptome of both sexes (Anantanawat, 2013) to a male genome of this species (Li et al., unpublished data). This approach, like the other three omics approaches, is also based on the fact that sequences unique to Y or W chromosome are not present in the genome of a female or male.
Genomic PCR analysis of a small set of 17 non-transposon transcripts selected from the 4855 putative W-linked transcript sequences found only 1 W-linked sequence (i.e., the true positive GUW1; detected only in the female pupa) but 15 negatives (not detected in pupae of both sexes) and 1 false positive (detected in pupae of both sexes) (Figure 1). Such a high ratio (15/17) of negatives is probably caused largely by the repeat-rich introns commonly present in the sex-specific chromosome-linked genes (Gatti and Pimpinelli, 1983; Carvalho et al., 2000; Reugels et al., 2000). These giant introns may locate within one primer or between the primer pairs of the 15 negatives, leading to the failure of the corresponding PCR reactions. Alternatively, some of the 15 negative sequences could be derived from the microorganisms (symbionts or parasites) that lived only in the eggs used for generation of the egg transcriptome (Anantanawat, 2013) and thus were not detected in both male and female pupae. Lastly, inter-population differences of H. armigera could be the third possible reason, whereby some of the 15 negative sequences present in the Bt resistant Australian population employed for production of the egg transcriptome (Anantanawat, 2013) might be deleted in the Chinese population used in this study.
Several additional experiments and analyses were conducted to verify the W chromosome uniqueness and identities of the only positive GUW1 identified here and the previously-reported female-specific RAPD marker AF18 (Niu et al., 2007). Detection of the AF18 band in both male and female pupae at a lower annealing temperature (from 62 to 58°C; Figure 2C) or at a different annealing position for the reverse primer (13 bp downstream; Figure 3A), observation of two genome walking bands in both male and female pupae (Figure 5B), and lack of sequence similarity between the 5′ flanking sequences of the two genome walking bands and the 5′-end of the reported AF18 sequence (Niu et al., 2007; Figure 5C) strongly suggest that AF18 has multiple copies in the genome and some of its copies also sit on Z chromosome and/or autosomes. Consistent with this inference, our sequence analyses confirm that AF18 sequence is a MITE (named MITE1_Har) with 137 significant hits (e-value ≤ 1.0e-10) to the H. armigera male genome scaffolds (Table 1 and Supplementary Table S3). This is not unexpected as the lepidopteran W chromosomes are rich in transposons (Abe et al., 2005; Fuková et al., 2007; Traut et al., 2013).
In contrast, multiple evidence including detection of GUW1 only in the female pupae regardless of the annealing positions of the two primers (Figures 2A, 3B), coupling of GUW1 with female-unique splicing of Hadsx transcript (Figure 4), appearance of a single genome walking band only in the female pupae (Figures 5B,D), reliable sexing of a “tester set” of 30 neonates with GUW1 (Figure 7) and one H. armigera cell line (Figure 8), and its lack of no significant hits to the male genomes of both the Chinese (Li et al., unpublished data) and Australian (Pearce et al., 2017) populations confirm that GUW1 is indeed a single copy sequence unique to W chromosome. Translation analysis reveals that GUW1 encodes a protein of 277 amino acids (Figure 6), whose first N-terminal 234 amino acids has multiple tBLASTn hits of 34-65% similarity on several autosomes of S. litura, D. plexippus, T. ni, and Z. cesonia, Z chromosome of Z. cesonia, and W chromosome of T. ni and E. kuehniella (Table 2), and is 69% identical to the N-terminal half (446 amino acids in total) of the hypothetical protein RR48_05862 of P. machaon. However, none of the tBLASTn hits on the W chromosome of T. ni and E. kuehniella represents an intact protein-coding gene (Traut et al., 2013; Fu et al., 2018; Chen et al., 2019). And RR48_05862 is not a W chromosome-unique protein-coding gene since it was annotated from the male adult genome of P. machaon (Li et al., 2015). Further experiments are required to decipher the functions and evolutionary relationship between GUW1 and RR48_05862.
The W chromosome in Lepidoptera is replete with transposons but poor in protein-coding genes (Sahara et al., 2003, 2012; Abe et al., 2005; Fuková et al., 2007; Traut et al., 2013). Comprehensive gene-based surveys have not found a single protein-coding gene on the W chromosome in Heliconius melpomene (Pringle et al., 2007), Bicyclus anynana (Beldade et al., 2009), Plutella xylostella (Baxter et al., 2011), C. pomonella (Wan et al., 2019), S. litura (Cheng et al., 2017), S. frugiperda (Xiao et al., 2020), and T. ni (Fu et al., 2018; Chen et al., 2019). High-throughput sequencing of the W chromosome of the flour moth (E. kuehniella) has not detected a single protein-coding gene either (Traut et al., 2013). Even in the most extensively-studied lepidopteran B. mori, no protein-coding gene but a piRNA gene (Fem) has been identified recently on the W chromosome (Kiuchi et al., 2014; Fujii et al., 2015; Zhang et al., 2018). The prediction of 14 protein-coding loci on the putative W-linked scaffolds of D. plexippus (Mongue et al., 2017) represents one exception, but the W chromosome of this species is probably a neo-W chromosome—fusion of ancient W with an autosome, rather than a typical fully-degenerated W chromosome (Mongue et al., 2017). The other two exceptions found so far are a truncated W homolog of the Z-linked period gene in Antheraea pernyi (Gotter et al., 1999) and a W homolog of the Z-linked laminin A gene in the peppered moth (Biston betularia; Van’t Hof et al., 2013). These two W homologs might have resulted from the Z-linked period or laminin A gene by transposon-mediated ectopic recombination between the Z and W chromosomes (Van’t Hof et al., 2013). By contrast, GUW1 characterized and verified at the transcription level in this study is a W-specific protein coding gene and is not present on the Z or autosome in H. armigera. Thus, GUW1 is the first W-specific protein coding gene found in Lepidoptera and represents a breakthrough that provides new insights into the evolution of the W chromosome in butterflies and moths.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
XL and ZD conceived and designed the experiments, and analyzed the data. ZD and YZ performed the experiments. XL, ZD, MZ, YZ, XN, CL, and JH wrote the manuscript. All authors have read and approved the manuscript for publication.
Funding
This work was supported by USDA National Institute of Food and Agriculture (hatch grant ARZT-1360890-H31-164 and multi state grant ARZT-1370400-R31-168) and Zhengzhou University Talent Discipline Project (32310206).
Disclaimer
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00649/full#supplementary-material
References
Abe, H., Kanehara, M., Terada, T., Ohbayashi, F., Shimada, T., Kawai, S., et al. (1998). Identification of novel random amplified polymorphic DNAs (RAPDs) on the W chromosome of the domesticated silkworm, Bombyx mori, and the wild silkworm, B. mandarina, and their retrotransposable element-related nucleotide sequences. Genes Genet. Syst. 73, 243–254. doi: 10.1266/ggs.73.243
Abe, H., Mita, K., Yasukochi, Y., Oshiki, T., and Shimada, T. (2005). Retrotransposable elements on the W chromosome of the silkworm, Bombyx mori. Cytogenet. Genome Res. 110, 144–151. doi: 10.1159/000084946
Allen, C. E., Zwaan, B. J., and Brakefield, P. M. (2011). Evolution of sexual dimorphism in the lepidoptera. Annu. Rev. Entomol. 56, 445–464.
Anantanawat, J. K. (2013). Effects of Inducible Tolerance to Bacillus Thuringiensis on the Egg Transciptomes and Egg Parasitism in Helicoverpa Armigera. Ph.D. dissertation, University of Adelaide, Adelaide.
Aron, S., de Menten, L., and Van Bockstaele, D. R. (2003). Brood sex ratio determination by flow cytometry in ants. Mol. Ecol. Notes 3, 471–475. doi: 10.1046/j.1471-8286.2003.00488.x
Baxter, S. W., Davey, J. W., Johnston, J. S., Shelton, A. M., Heckel, D. G., Jiggins, C. D., et al. (2011). Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS One 6:e19315. doi: 10.1371/journal.pone.0019315
Beldade, P., Saenko, S. V., Pul, N., and Long, A. D. (2009). A gene-based linkage map for Bicyclus anynana butterflies allows for a comprehensive analysis of synteny with the lepidopteran reference genome. PLoS Genet. 5:e1000366. doi: 10.1371/journal.pgen.1000366
Belousova, I., Ershov, N., Pavlushin, S., Ilinsky, Y., and Martemyanov, V. (2019). Molecular sexing of Lepidoptera. J. Insect Physiol. 114, 53–56. doi: 10.1016/j.jinsphys.2019.02.005
Bergero, R., and Charlesworth, D. (2011). Preservation of the Y transcriptome in a 10-million-year-old plant sex chromosome system. Curr. Biol. 21, 1470–1474. doi: 10.1016/j.cub.2011.07.032
Bopp, D., Saccone, G., and Beye, M. (2014). Sex determination in insects: variations on a common theme. Sex. Dev. 8, 20–28. doi: 10.1159/000356458
Carvalho, A. B., and Clark, A. G. (2013). Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 23, 1894–1907. doi: 10.1101/gr.156034.113
Carvalho, A. B., Lazzaro, B. P., and Clark, A. G. (2000). Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc. Natl. Acad. Sci. U.S.A. 97, 13239–13244. doi: 10.1073/pnas.230438397
Chen, D., Niu, B., and Weng, H. (2004). Study on larval sexual character and spermatogenesis of cotton bollworm (Helicoverpa armigera). Acta Agric. Zhejiangensis 6, 354–357.
Chen, N., Bellott, D. W., Page, D. C., and Clark, A. G. (2012). Identification of avian W-linked contigs by short-read sequencing. BMC Genomics 13:183. doi: 10.1186/1471-2164-13-183
Chen, W. B., Yang, X. W., Tetreau, G., Song, X. Z., Coutu, C., Hegedus, D., et al. (2019). A high-quality chromosome-level genome assembly of a generalist herbivore, Trichoplusia ni. Mol. Ecol. Resour. 19, 485–496. doi: 10.1111/1755-0998.12966
Cheng, T., Wu, J., Wu, Y., Chilukuri, R. V., Huang, L., Yamamoto, K., et al. (2017). Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat. Ecol. Evol. 1, 1747–1756. doi: 10.1038/s41559-017-0314-4
Chibalina, M. V., and Filatov, D. A. (2011). Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr. Biol. 21, 1475–1479. doi: 10.1016/j.cub.2011.07.045
Douglas, L. J., Untalan, P. M., and Haymer, D. S. (2004). Molecular sexing in the Mediterranean fruit fly, Ceratitis capitata. Insect Biochem. Mol. Biol. 34, 159–165. doi: 10.1016/j.ibmb.2003.08.006
Esperk, T., Tammaru, T., Nylin, S., and Teder, T. (2007). Achieving high sexual size dimorphism in insects: females add instars. Ecol. Entomol. 32, 243–256. doi: 10.1111/j.1365-2311.2007.00872.x
Fischer, K., and Fiedler, K. (2001). Sexual differences in life-history traits in the butterfly Lycaena tityrus: a comparison between direct and diapause development. Entomol. Exp. Appl. 100, 325–330. doi: 10.1046/j.1570-7458.2001.00879.x
Fox, C. W., Dublin, L., and Pollitt, S. J. (2003). Gender differences in lifespan and mortality rates in two seed beetle species. Funct. Ecol. 17, 619–626. doi: 10.1046/j.1365-2435.2003.00781.x
Fraïsse, C., Picard, M. A. L., and Vicoso, B. (2017). The deep conservation of the Lepidoptera Z chromosome suggests a non-canonical origin of the W. Nat. Commun. 8:1486. doi: 10.1038/s41467-017-01663-5
Fu, Y., Yang, Y. J., Zhang, H., Farley, G., Wang, J. L., Quarles, K. A., et al. (2018). The genome of the Hi5 germ cell line from Trichoplusia ni, an agricultural pest and novel model for small RNA biology. eLife 7:e31628.
Fujii, T., Abe, H., Kawamoto, M., Banno, Y., and Shimada, T. (2015). Positional cloning of the sex-linked giant egg (Ge) locus in the silkworm, Bombyx mori. Insect. Mol. Biol. 24, 213–221. doi: 10.1111/imb.12150
Fujii, T., and Shimada, T. (2007). Sex determination in the silkworm, Bombyx mori: a female determinant on the W chromosome and the sex-determining gene cascade. Semin. Cell Dev. Biol. 18, 379–388. doi: 10.1016/j.semcdb.2007.02.008
Fuková, I., Neven, L. G., Bárcenas, N. M., Gund, N. A., Dalíková, M., Marec, F., et al. (2009). Rapid assessment of the sex of codling moth Cydia pomonella (Linnaeus) (Lepidoptera: Tortricidae) eggs and larvae. J. Appl. Entomol. 133, 249–261.
Fuková, I., Nguyen, P., and Marec, F. (2005). Coding moth cytogenetics: karyotype, chromosomal location of rDNA, and molecular differentiation of sex chromosomes. Genome 48, 1083–1092. doi: 10.1139/g05-063
Fuková, I., Traut, W., Vítková, M., Nguyen, P., Kubčèková, S., Marec, F., et al. (2007). Probing the W chromosome of the codling moth, Cydia pomonella, with sequences from microdissected sex chromatin. Chromosoma 116, 135–145. doi: 10.1007/s00412-006-0086-0
Gabrieli, P., Falaguerra, A., Siciliano, P., Gomulski, L. M., Scolari, F., Zacharopoulou, A., et al. (2010). Sex and the single embryo: early deveiopment in the Mediterranean fruit fly, Ceratitis capitata. BMC Dev. Biol. 10:12. doi: 10.1186/1471-213X-10-12
Gatti, M., and Pimpinelli, S. (1983). Cytological and genetic analysis of the Y chromosome of Drosophila melanogaster. Chromosoma 88, 349–373. doi: 10.1007/bf00285858
Gotoh, H., Ishiguro, M., Nishikawa, H., Morita, S., Okada, K., Miyatake, T., et al. (2016). Molecular cloning and functional characterization of the sex-determination gene doublesex in the sexually dimorphic broad-horned beetle Gnatocerus cornutus (Coleoptera, Tenebrionidae). Sci. Rep. 6:29337. doi: 10.1038/srep29337
Gotter, A. L., Levine, J. D., and Reppert, S. M. (1999). Sex-linked period genes in the silkmoth, Antheraea pernyi: implications for circadian clock regulation and the evolution of sex chromosomes. Neuron 24, 953–965. doi: 10.1016/s0896-6273(00)81042-9
Gruntenko, N. E., Karpova, E. K., Burdina, E. V., Adonyeva, N. V., Andreenkova, O. V., Alekseev, A. A., et al. (2016). Probable mechanism of sexual dimorphism in insulin control of Drosophila heat stress resistance. Physiol. Entomol. 41, 59–66. doi: 10.1111/phen.12125
Hall, A. B., Qi, Y. M., Timoshevskiy, V., Sharakhova, M. V., Sharakhov, I. V., Tu, Z., et al. (2013). Six novel Y chromosome genes in Anopheles mosquitoes discovered by independently sequencing males and females. BMC Genomics 14:273. doi: 10.1186/1471-2164-14-273
Jiang, Y., and Hu, M. (1995). Study on male and female differentiation of Helicoverpa armigera larvae. J. Appl. Insects 4, 230–239.
Kiuchi, T., Koga, H., Kawamoto, M., Shoji, K., Sakai, H., Arai, Y., et al. (2014). A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509, 633–636. doi: 10.1038/nature13315
Koerich, L. B., Dupim, E. G., Faria, L. L., Dias, F. A., Dias, A. F., Trindade, G. S., et al. (2016). First report of Y-linked genes in the kissing bug Rhodnius prolixus. BMC Genomics 17:100. doi: 10.1186/s12864-016-2425-8
Krzywinski, J., Nusskern, D. R., Kern, M. K., and Besansky, N. J. (2004). Isolation and characterization of Y chromosome sequences from the African malaria mosquito Anopheles gambiae. Genetics 166, 1291–1302. doi: 10.1534/genetics.166.3.1291
Kunte, K. (2009). The diversity and evolution of batesian mimicry in Papilio Swallowtail butterflies. Evolution 63, 2707–2716. doi: 10.1111/j.1558-5646.2009.00752.x
Lagisz, M., Wilde, K. E., and Wolff, K. (2010). The development of PCR-based markers for molecular sex identification in the model insect species Tribolium castaneum. Entomol. Exp. Appl. 134, 50–59. doi: 10.1111/j.1570-7458.2009.00935.x
Li, X., Tan, F., and You, Z. (1992). Sexual difference of Pectinophora gossypiella larvae in tolerance to insecticides and its metabolic mechanism. J. Southwest Agric. Univ. 14, 303–306.
Li, X. C., Berenbaum, M. R., and Schuler, M. A. (2002). Cytochrome P450 and actin genes expressed in Helicoverpa zea and Helicoverpa armigera: paralogy/orthology identification, gene conversion and evolution. Insect Biochem. Mol. Biol. 32, 311–320. doi: 10.1016/s0965-1748(01)00092-3
Li, X. Y., Fan, D. D., Zhang, W., Liu, G. C., Zhang, L., Zhao, L., et al. (2015). Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat. Commun. 6:8212. doi: 10.1038/ncomms9212
Luis, R. C., Jennifer, F., Caleb, B., Steven, M. V., and Brian, A. C. (2020). Genome assembly of the dogface butterfly zerene. Genome Biol. Evol. 12, 3580–3585. doi: 10.1093/gbe/evz254
Mongue, A. J., Nguyen, P., Volenikova, A., and Walters, J. R. (2017). Neo-sex chromosomes in the monarch butterfly, Danaus plexippus. G3 7, 3281–3294. doi: 10.1534/g3.117.300187
Mowrey, W. R., and Portman, D. S. (2012). Sex differences in behavioral decision-making and the modulation of shared neural circuits. Biol. Sex Diff. 3:8. doi: 10.1186/2042-6410-3-8
Muyle, A., Zemp, N., Deschamps, C., Mousset, S., Widmer, A., Marais, G. A. B., et al. (2012). Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biol. 10:e1001308. doi: 10.1371/journal.pbio.1001308
Nakamura, D., Tiersch, T. R., Douglass, M., and Chandler, R. W. (1990). Rapid identification of sex in birds by flow cytometry. Cytogenet. Cell Genet. 53, 201–205. doi: 10.1159/000132930
Niu, B. L., Weng, H. B., Li-Hua, H. E., Shen, W. F., and Meng, Z. Q. (2007). Cloning and sequencing of two molecular markers located on the sex chromosomes of the cotton bollworm, Helicoverpa armigera. Acta Entomol. Sin. 50, 649–654.
Pearce, S. L., Clarke, D. F., East, P. D., Elfekih, S., Gordon, K. H. J., Jermiin, L. S., et al. (2017). Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 15:63. doi: 10.1186/s12915-017-0402-6
Portela, J., Grunau, C., Cosseau, C., Beltran, S., Dantec, C., Parrinello, H., et al. (2010). Whole-genome in-silico subtractive hybridization (WISH)–using massive sequencing for the identification of unique and repetitive sex-specific sequences: the example of Schistosoma mansoni. BMC Genomics 11:387. doi: 10.1186/1471-2164-11-387
Prakash, A., and Monteiro, A. (2016). Molecular mechanisms of secondary sexual trait development in insects. Curr. Opin. Insect Sci. 17, 40–48. doi: 10.1016/j.cois.2016.06.003
Pringle, E. G., Baxter, S. W., Webster, C. L., Papanicolaou, A., Lee, S. F., Jiggins, C. D., et al. (2007). Synteny and chromosome evolution in the lepidoptera: evidence from mapping in Heliconius melpomene. Genetics 177, 417–426. doi: 10.1534/genetics.107.073122
Reugels, A. M., Kurek, R., Lammermann, U., and Bunemann, H. (2000). Mega-introns in the dynein gene DhDhc7(Y) on the heterochromatic Y chromosome give rise to the giant threads loops in primary spermatocytes of Drosophila hydei. Genetics 154, 759–769.
Sahara, K., Yoshido, A., Kawamura, N., Ohnuma, A., Abe, H., Mita, K., et al. (2003). W-derived BAC probes as a new tool for identification of the W chromosome and its aberrations in Bombyx mori. Chromosoma 112, 48–55. doi: 10.1007/s00412-003-0245-5
Sahara, K., Yoshido, A., and Traut, W. (2012). Sex chromosome evolution in moths and butterflies. Chromosome Res. 20, 83–94. doi: 10.1007/s10577-011-9262-z
Salz, H. K. (2011). Sex determination in insects: a binary decision based on alternative splicing. Curr. Opin. Genet. Dev. 21, 395–400. doi: 10.1016/j.gde.2011.03.001
San Andres, V., Urbaneja, A., Sabater-Munoz, B., and Castanera, P. (2007). A novel molecular approach to assess mating success of sterile Ceratitis capitata (Diptera: Tephritidae) males in sterile insect technique programs. J. Econ. Entomol. 100, 1444–1449. doi: 10.1093/jee/100.4.1444
Shimizu, K., and Fujisaki, K. (2002). Sexual differences in diapause induction of the cotton bollworm, Helicoverpa armigera (Hb.) (Lepidoptera : Noctuidae). Appl. Entomol. Zool. 37, 527–533. doi: 10.1303/aez.2002.527
Siverly, R. E. (1947). A morphological study of the male and female genitalia of Heliothis armigera (Corn Earworm Moth). Am. Midland Nat. 38, 712–724.
Stillwell, R. C., Blanckenhorn, W. U., Teder, T., Davidowitz, G., and Fox, C. W. (2010). Sex differences in phenotypic plasticity affect variation in sexual size dimorphism in insects: from physiology to evolution. Annu. Rev. Entomol. 55, 227–245. doi: 10.1146/annurev-ento-112408-085500
Teder, T. (2014). Sexual size dimorphism requires a corresponding sex difference in development time: a meta-analysis in insects. Funct. Ecol. 28, 479–486. doi: 10.1111/1365-2435.12172
Traut, W., and Marec, F. (1996). Sex chromatin in Lepidoptera. Q. Rev. Biol. 71, 239–256. doi: 10.1086/419371
Traut, W., Sahara, K., and Marec, F. (2007). Sex chromosomes and sex determination in Lepidoptera. Sex. Dev. 1, 332–346. doi: 10.1159/000111765
Traut, W., Vogel, H., Glöckner, G., Hartmann, E., and Heckel, D. G. (2013). High-throughput sequencing of a single chromosome: a moth W chromosome. Chromosome Res. 21, 491–505. doi: 10.1007/s10577-013-9376-6
Van’t Hof, A. E., Nguyen, P., Dalíková, M., Edmonds, N., Marec, F., Saccheri, I. J., et al. (2013). Linkage map of the peppered moth, Biston betularia (Lepidoptera, Geometridae): a model of industrial melanism. Heredity 110, 283–295. doi: 10.1038/hdy.2012.84
Vincent, C. M., and Sharp, N. P. (2014). Sexual antagonism for resistance and tolerance to infection in Drosophila melanogaster. Proc. R. Soc. B Biol. Sci. 281:20140987. doi: 10.1098/rspb.2014.0987
Waldbauer, G. P., Cohen, R. W., and Friedman, S. (1984). An improved procedure for laboratory rearing of the corn earworm, Heliothis zea (Lepidoptera: Noctuidae). Great Lakes Entomol. 17, 113–118.
Wan, F., Yin, C., Tang, R., Chen, M., Wu, Q., Huang, C., et al. (2019). A chromosome-level genome assembly of Cydia pomonella provides insights into chemical ecology and insecticide resistance. Nat. Commun. 10:4237. doi: 10.1038/s41467-019-12175-9
Wang, X. Y., Zheng, Z. Z., Song, H. S., and Xu, Y. Z. (2014). Conserved RNA cis-elements regulate alternative splicing of Lepidopteran doublesex. Insect Biochem. Mol. Biol. 44, 1–11. doi: 10.1016/j.ibmb.2013.10.009
Weller, S. J., Jacobson, N. L., and Conner, W. E. (1999). The evolution of chemical defences and mating systems in tiger moths (Lepidoptera: Arctiidae). Biol. J. Linn. Soc. 68, 557–578. doi: 10.1111/j.1095-8312.1999.tb01188.x
Xiao, H., Ye, X., Xu, H., Mei, Y., Yang, Y., Chen, X., et al. (2020). The genetic adaptations of fall armyworm Spodoptera frugiperda facilitated its rapid global dispersal and invasion. Mol. Ecol. Resour. doi: 10.1111/1755-0998.13182
Zalucki, M. P., Murray, D. A. H., and Gregg, P. C. (1994). Ecology of Helicoverpa armigera (Hübner) and H. punctigera (Wallengren) in the inland of Australia: larval sampling and host plant relationships during winter and spring. Austral. J. Zool. 42, 329–346.
Zhang, S., Shen, S., Peng, J., Zhou, X., Kong, X., Ren, P., et al. (2020). Chromosome-level genome assembly of an important pine defoliator, Dendrolimus punctatus (Lepidoptera; Lasiocampidae). Mol. Ecol. Resour. doi: 10.1111/1755-0998.13169
Zhang, Z. J., Niu, B. L., Ji, D. F., Li, M. W., Li, K., James, A. A., et al. (2018). Silkworm genetic sexing through W chromosome-linked, targeted gene integration. Proc. Natl. Acad. Sci. U.S.A. 115, 8752–8756. doi: 10.1073/pnas.1810945115
Zhao, J. Z., Zhao, K. J., Lu, M. G., Fan, X. L., and Guo, S. D. (1998). Interactions between Helicoverpa armigera and transgenic Bt cotton in North China. Sci. Agric. Sin. 31, 1–6.
Zhao, X. C., Dong, J. F., Tang, Q. B., Yan, Y. H., Gelbič, I., Van Loon, J. J., et al. (2005). Hybridization between Helicoverpa armigera and Helicoverpa assulta (Lepidoptera: Noctuidae): development and morphological characterization of F1 hybrids. Bull. Entomol. Res. 95, 409–416. doi: 10.1079/ber2005372
Keywords: doublesex, egg transcriptome, sex identification, male genome, sexual dimorphism, transposon
Citation: Deng Z, Zhang Y, Zhang M, Huang J, Li C, Ni X and Li X (2020) Characterization of the First W-Specific Protein-Coding Gene for Sex Identification in Helicoverpa armigera. Front. Genet. 11:649. doi: 10.3389/fgene.2020.00649
Received: 14 June 2019; Accepted: 28 May 2020;
Published: 19 June 2020.
Edited by:
Jiannis Ragoussis, McGill University, CanadaCopyright © 2020 Deng, Zhang, Zhang, Huang, Li, Ni and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianchun Li, bHhjQGVtYWlsLmFyaXpvbmEuZWR1; bHhjQGFyaXpvbmEuZWR1
 Zhongyuan Deng
Zhongyuan Deng Yakun Zhang
Yakun Zhang Min Zhang
Min Zhang Jinyong Huang
Jinyong Huang Changyou Li
Changyou Li Xinzhi Ni
Xinzhi Ni Xianchun Li
Xianchun Li