- 1Department of Chemical and Biological Sciences, Japan Women’s University, Bunkyō, Japan
- 2National Institute for Basic Biology, Okazaki, Japan
- 3School of Life Sciences, The Graduate University for Advanced Studies (SOKENDAI), Okazaki, Japan
Common ancestors of vertebrates had four types of cone opsins: short-wavelength sensitive 1 (SWS1), SWS2, rhodopsin 2 (RH2), and long-wavelength sensitive (LWS) types. Whereas fish and birds retain all the types, mammals have lost two of them (SWS2 and RH2) possibly because of their nocturnal lifestyle during the Mesozoic Era. Considering that the loss of cone opsin types causes so-called color blindness in humans (e.g., protanopia), the ability to discriminate color by trichromatic humans could be lower than that in potentially tetrachromatic birds and fish. Behavioral studies using color-blind (cone opsin-knockout) animals would be helpful to address such questions, but it is only recently that the genome-editing technologies have opened up this pathway. Using medaka as a model, we introduced frameshift mutations in SWS2 (SWS2a and/or SWS2b) after detailed characterization of the loci in silico, which unveiled the existence of a GC–AG intron and non-optic expressed-sequence-tags (ESTs) that include SWS2a in part. Transcripts from the mutated SWS2 loci are commonly reduced, suggesting that the SWS2a/b-double mutants could produce, if any, severely truncated (likely dysfunctional) SWS2s in small amounts. The mutants exhibited weakened body color preferences during mate choice. However, the optomotor response (OMR) test under monochromatic light revealed that the mutants had no defect in spectral sensitivity, even at the absorbance maxima (λmax) of SWS2s. Evolutionary diversification of cone opsins has often been discussed in relation to adaptation to dominating light in habitats (i.e., changes in the repertoire or λmax are for increasing sensitivity to the dominating light). However, the present results seem to provide empirical evidence showing that acquiring or losing a type of cone opsin (or changes in λmax) need not substantially affect photopic or mesopic sensitivity. Other points of view, such as color discrimination of species-specific mates/preys/predators against habitat-specific backgrounds, may be necessary to understand why cone opsin repertories are so various among animals.
Introduction
Colors are virtual images evoked in the brain by light spectra received at the retina. Physically different spectra (e.g., yellow monochromatic light and red/green dichromatic light) make a person evoke an identical color, whereas an identical spectrum makes different people (e.g., monochromats, dichromats, and trichromats) evoke different colors. Animals possess various sets of photoreceptors in the retina. Thus, different animals may evoke different colors when looking at the same object, i.e., the world could differently be colored for different species or even individuals.
The mechanism for color perception in humans has been studied extensively (Solomon and Lennie, 2007; Conway, 2009; Neitz and Neitz, 2011). Under daylight, light is received by three types (or more precisely, two types with one subtype; see below) of visual pigments (cone opsins) and converted to an electronic signal of three channels (Young, 1802). This signal is then converted to a two-dimensional value defined by the red–green and blue–yellow axes (Hering, 1920) via complex neural networks of horizontal, bipolar, amacrine, and ganglion cells (Thoreson and Dacey, 2019), which is sent to the visual cortex of the brain where colors are evoked. However, this mechanism (still not fully understood, particularly the processing in the brain) will explain color perception only in a part of the Old World monkeys (Catarrhini). These animals, as do other mammalian species (except for monotremes), possess two types of cone opsins: short-wavelength sensitive 1 (SWS1) and long-wavelength sensitive (LWS) types (Bowmaker, 1998), which are often referred to as (ultra)violet and red opsins, respectively. Common ancestors of the catarrhine monkeys duplicated the LWS gene, accumulated missense substitutions, diversified absorbance maxima (λmax) of the proteins, and acquired so-called trichromacy. The cone cells in their retina are classified morphologically into two types (expressing SWS1 or either of the LWS subtypes), which are arranged largely at random (Viets et al., 2016).
Fish (and birds) possess two additional types of cone opsins, which mammals have lost during their nocturnal lifestyle in the Mesozoic Era: SWS2 and rhodopsin 2 (RH2), which are often referred to as blue and green opsins, respectively. These four types of cone opsins are expressed in four types of cone cells, which are regularly arranged in the retina (i.e., retinal mosaics; Tohya et al., 2003; Carleton et al., 2020). Although spatiotemporal patterns of cone opsin expression in fish are complex (Tsujimura et al., 2007; Dalton et al., 2014; Zimmermann et al., 2018), the retinal mosaic per se should suggest that the four types of cones could function coordinately for tetrachromacy, which has been suggested or actually demonstrated in some species (see Thoreson and Dacey, 2019 and references therein). Various attempts have been made to understand the vision of fish (e.g., spectral absorbance of opsin molecules, microspectrophotometry, electroretinography, theoretical modeling, and behavioral assays), but we still do not understand the mechanism or how differently their worlds would be colored in comparison with ours.
Recent genome-editing technologies have made it possible to establish color-blind animals by knocking out the cone opsin genes. Considering that color-blind patients have made great contributions to our understanding of the human trichromacy (Neitz et al., 1999; Neitz and Neitz, 2014), color-blind animals should provide promising opportunities for dissecting characteristics and mechanisms for color perception in animals. However, such resources are scarce at present. In mice, an SWS1-knockout strain is available (Greenwald et al., 2014), but dichromatic mice would not fit as a model for studying tetrachromacy. In zebrafish, there is a strain that specifically lacks the red cones, but the mutation is not in the cone opsin gene and the mutants are lethal at the larval stages (Taylor et al., 2005).
We recently established medaka strains that lack LWS and made some interesting findings by analyzing the mutants using a monochromatic light source, the Okazaki Large Spectrograph (OLS; Watanabe et al., 1982). For example, (i) whereas many previous studies at the molecular, cellular, physiological, behavioral, or theoretical levels ignored retinal inputs at λ > 700 nm, the wild-type medaka (as do some other fish species: M.M., Y.K., and S.F., in preparation) fully exhibit the optomotor response (OMR) at λ > 800 nm; (ii) the light-adapted, but not dark-adapted, lws mutants (LWSa/b-double knockouts) significantly reduced the OMR at λ ≥ 740 nm, demonstrating that the rod-dependent scotopic vision is dysfunctional in light-adapted medaka, and that not only LWSs (λmax at 561–562 nm) but also RH2s (λmax at 452–516 nm) can absorb and make medaka respond behaviorally to light at wavelengths > 200 nm longer than the λmax; (iii) an in-frame fusion of the tandemly located LWSa and LWSb loci, which are nearly identical in nucleotide sequence because of a recent gene conversion (i.e., a decreased copy number of the LWS genes), did not at all reduce red-light sensitivity, obscuring the necessity of their coexistence and coexpression; and (iv) a premating sexual isolation between body color variants, the color interfere (ci) mutant that lacks somatolactin alpha (SLα) and the actin beta (Actb)-SLα:green fluorescent protein (GFP) transgenic fish that overexpresses SLα, which can be observed convincingly under white, but not monochromatic lights (with half bandwidth of ± 5 nm), was significantly relaxed in the lws mutants under white light, suggesting the importance of LWSs in the color-dependent (not luminance-dependent) mate choice (Fukamachi et al., 2009a; Utagawa et al., 2016; Homma et al., 2017; Ikawa et al., 2017; Kamijo et al., 2018; Matsuo et al., 2018; Harada et al., 2019).
These kinds of reverse-genetic studies can provide direct empirical evidence for a causal relationship between cone opsin repertoire and animal behavior. Comparative studies using different species or heterospecific populations with different cone opsin repertoires could be another choice, but effects from other polymorphic genes (i.e., misidentification of a causal relationship) cannot be excluded. In this regard, the invention of genome-editing technologies provides a newly discovered and powerful strategy for assessing whether or not and to which degree each member of a cone opsin repertoire actually contributes to visual-dependent behaviors of animals.
In the present study, we focused on SWS2, whose ecological/evolutionary importance has been discussed in fish (Cortesi et al., 2015; Marques et al., 2017). Unlike LWSa and LWSb, the identity between SWS2a and SWS2b drops to 77.0% (271/352 amino acids) and their λmax is distinctly different (439 and 405 nm, respectively) (Matsumoto et al., 2006). Therefore, we established SWS2a-single, SWS2b-single, and SWS2a/b-double knockout medaka and assessed each phenotype using the methods we previously developed for assessing the lws mutants (Homma et al., 2017; Kamijo et al., 2018; Matsuo et al., 2018; Harada et al., 2019). Results of the mate-choice experiment and the OMR test will respectively tell us whether or not the sws2 mutants have defects (i.e., SWS2s play essential roles) in color discrimination and photosensitivity.
Results
The Medaka SWS2 Loci
Before the genome editing, we examined genomic sequences of the medaka SWS2a and SWS2b loci (Figure 1A) that are available at the GenBank (AB223056 and AB223057, respectively) and the UTGB (version 2.2.4)1 databases. The GenBank sequences (2,324 bp of SWS2a and 3,404 bp of SWS2b) were of the HNI strain, and the SWS2a sequence was identical to the whole-genome sequence of HNI in the UTGB, except that the fourth intron (249 bp) seemed to be of the Hd-rR strain for some unknown reason. We detected a total of six mismatches [five substitutions and one insertion/deletion (ins/del)] in SWS2b, one of which is located at the splice-donor site of the first intron (Figure 1A), i.e., GT in the GenBank, but GC in the UTGB.
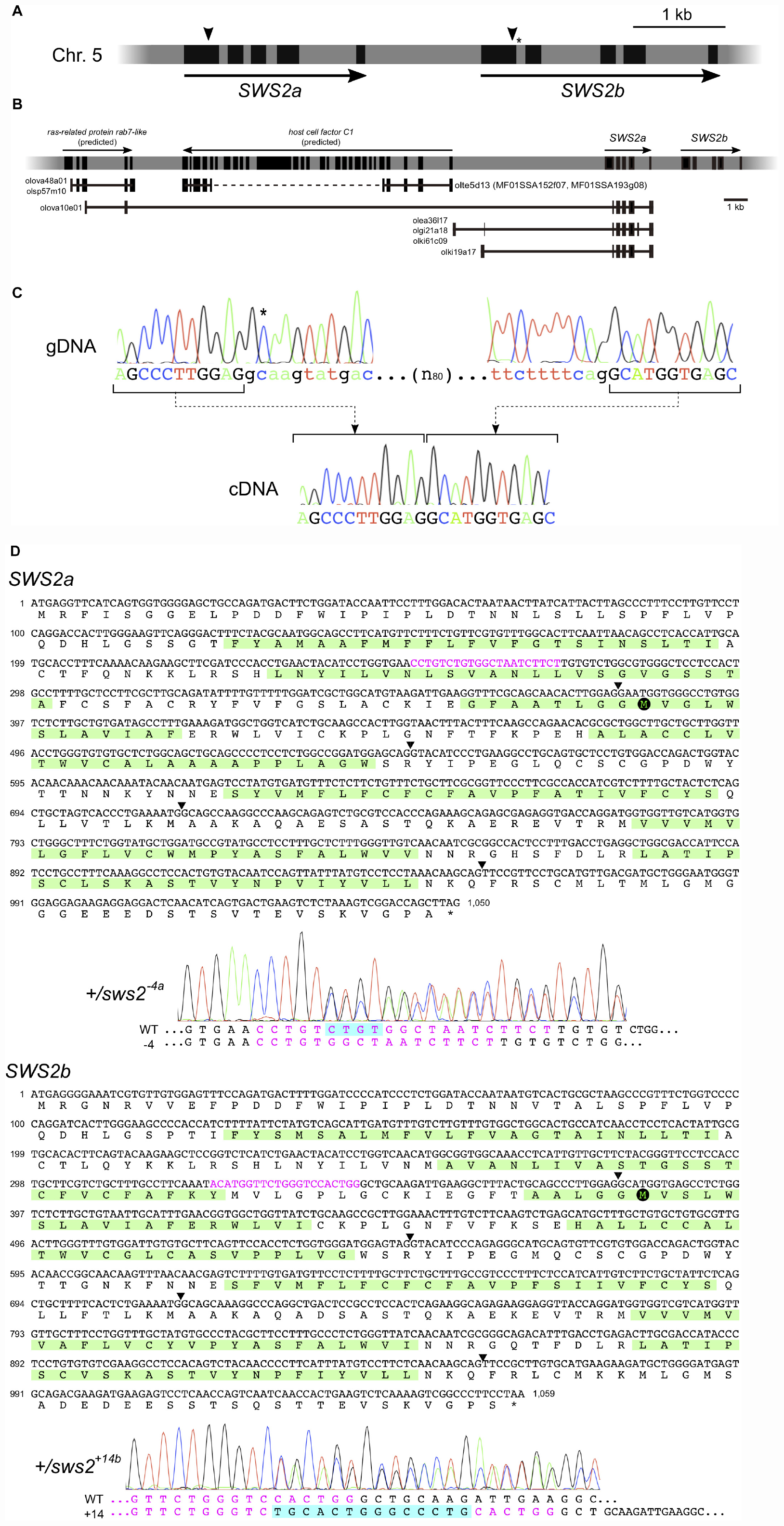
Figure 1. The medaka short-wavelength sensitive (SWS)2a and SWS2b loci. (A) A diagram of the SWS2 loci on the chromosome 5. The paralogs are concatenated tandemly in a head-to-tail manner. Black boxes represent coding regions (from the start codon to the stop codon). Arrowheads are approximate positions of target sequences for CRISPR/Cas9. An asterisk shows a position of the splice donor of the GC–AG intron (the first intron of SWS2b). (B) The expressed-sequence-tag (EST) clones containing a part of SWS2a. A horizontal gray bar with black boxes represents a part of chromosome 5. Black boxes indicate transcribed regions including untranslated regions (UTRs) (those of SWS2s do not include UTRs as A). Horizontal black lines with black boxes are ESTs found in the database at the National Bioresource Project (NBRP) medaka. ras-related protein rab7-like (XM_020703466 and XM_020703467) and host cell factor C1 (XM_011475416) have been registered as predicted genes in GenBank, and corresponding ESTs (olova48a01, olsp57m10, olte5d13, MF01SSA152f07, and MF01SSA193g08) are found presently existing in the database. A dotted line is the region not sequenced in the ESTs. A total of five EST clones (olova10e01, olea36l17, olgi21a18, olki61c09, and olki19a17) contained a part of SWS2a. Three of the five clones (olea36l17, olgi21a18, and olki61c09) additionally contain upstream intergenic regions and a part of the fourth intron. olki19a17 possesses a different intergenic region, and olova10e01 has a part of exons of the upstream ras-related protein rab7-like gene. (C) The GC–AG intron. Electropherograms of the genomic (top) and complementary (bottom) DNA sequences are shown. Uppercase indicates the first and second exons of SWS2b, and lowercase indicates the first intron. The C residue with an asterisk does not follow the GT–AG rule, but the intron is spliced out. (D) mRNA sequences of SWS2a and SWS2b and the sws2 mutations. Translated amino acid sequences are also shown, with the transmembrane domains (predicted by TMHMM; http://www.cbs.dtu.dk/services/TMHMM/) highlighted by green. Arrowheads are positions of introns. Target sequences are shown in magenta, and downstream methionines (potential initiation sites of translation) are highlighted by black circles. Electropherograms obtained for the sws2– 4a and sws2+14b heterozygotes are shown. Note that peaks are doubled from the position where the frameshift mutation is introduced. The sequences of mutated alleles (bottom) are determined by subtracting the known sequences of the wild type (top) from the doubled electropherogram.
This substitution seemed not to be negligible because it may cause mis-splicing. Hence, we screened the expressed-sequence-tag (EST) database provided by the National Bioresource Project (NBRP) Medaka2 to confirm the exon–intron boundaries. However, no EST (of 730,259 entries) was identical to SWS2b. This database search identified some interesting clones containing a part of the first and the entire second to fifth exons of SWS2a, which are connected to an upstream gene or intergenic regions (Figure 1B). Whether these strange ESTs were artifacts during library construction or indeed expressed and function in the organs from which they were isolated (i.e., the kidney, ovary, or gill) remains unknown.
Given that the examinations in silico, as described above, do not confirm the open-reading frames (ORFs) of SWS2a or SWS2b, we experimentally determined the ORFs using the ci strain, whose genome was going to be edited in this study. Direct sequencing of RT–PCR products revealed that the ORFs of SWS2a and SWS2b consist of 1,047 and 1,056 bp (excluding the stop codon), respectively, as reported for HNI in the GenBank database. These ORFs are split into five exons with conserved exon–intron boundaries, meaning that the first intron of SWS2b is indeed spliced out. Then, we examined the splice donor of the first intron by direct sequencing of genomic PCR products and found that GC is correct, as reported in the UTGB database (Figure 1C). Hence, although the first intron of SWS2b does not follow the GT–AG rule, it functions as an GC–AG intron, which has been reported from various species, including humans (Thanaraj, 2001).
Frameshift Mutations on the SWS2 Genes
The unusual ESTs in Figure 1B suggest that a frameshift mutation for knocking out SWS2a needs to be induced on the first exon (otherwise, these potentially functional ESTs could also be knocked out). We designed gRNAs that target either SWS2a or SWS2b [their ORFs were 75.1% (793/1,056) identical] for the CRISPR/Cas9 system and microinjected either or both of the gRNAs with the Cas9 mRNA into fertilized eggs of the ci and Actb-SLα:GFP strains.
Among a total of 29 G0 adults that successfully passed ins/del mutations to their F1s, we used eight G0s to obtain a total of 77 F1s, 15, 11, and 13 of which were heterozygous for the SWS2a-single, SWS2b-single, and SWS2a/b-double mutations, respectively (Tables 1–3). These ins/del mutations were classified into 19 haplotypes, four, four, and five of which were SWS2a-single, SWS2b-single, and SWS2a/b-double frameshift mutations, respectively. The F1 fish with the frameshift mutation/s were basically backcrossed with ci or Actb-SLα:GFP because F1s possessing an identical haplotype were scarce and insufficient for intercrossing (e.g., only one F1 for sws2+29a, three F1s for sws2+14b, but only male fish, etc.; see Tables 1–3). These backcrosses should basically reduce the risk of off targets.
Considering positions of the CRISPR/Cas9 target sequences and downstream codons for methionine (i.e., potential sites for translational initiation), SWS2 proteins translated from the frameshifted mRNA must severely be truncated losing multiple transmembrane domains, which should likely be dysfunctional as a G protein-coupled receptor (Figure 1D).
The sws2 Mutants
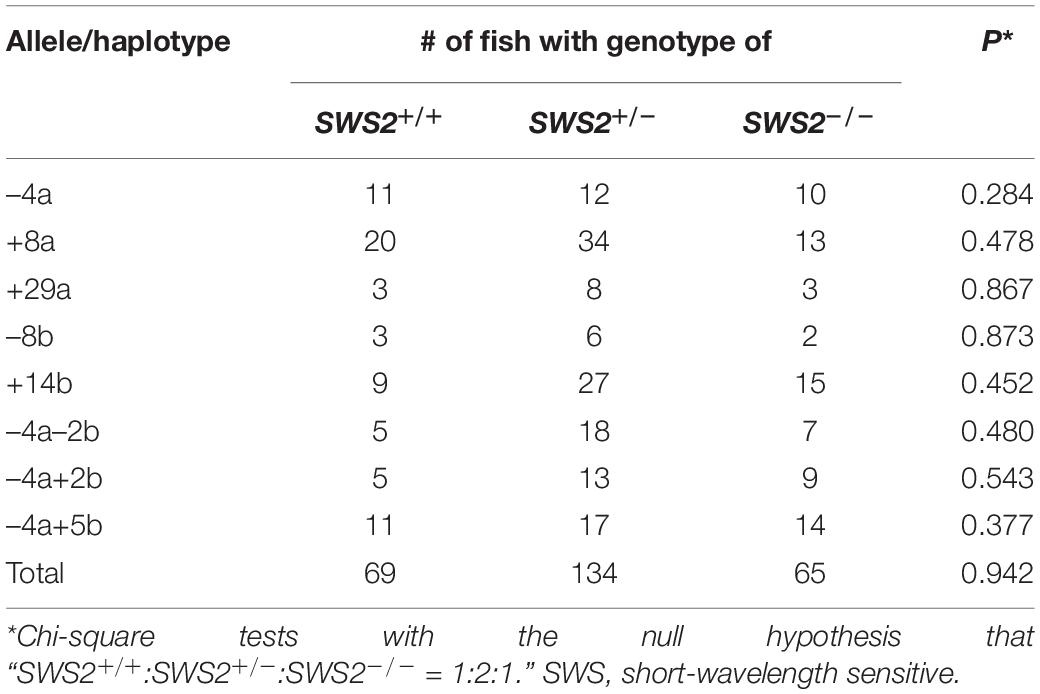
When the heterozygous F1s are intercrossed, an expected genotype ratio among their F2 siblings is SWS2+/+:SWS2+/–:SWS2–/– = 1:2:1, which was indeed observed for all the eight haplotypes (three SWS2a-single, two SWS2b-single, and three SWS2a/b-double frameshift mutations; Table 4). For example, we obtained a total of 33 adults by intercrossing the sws2–4a (a four-base deletion on SWS2a) heterozygotes and the genotype ratio was 11:12:10, which was not significantly different from the expected 8:17:8 (P = 0.284, chi-square test). Although the number of offspring per family (i.e., 11–67 F2s) was not always sufficiently large enough for statistical analysis, the overall ratio became 69:134:65, which is very close to the expected 67:134:67 (P = 0.942). Body colors of the sws2-mutant siblings are not distinguishable from those of the wild-type or heterozygous siblings on either ci or Actb-SLα:GFP background, at least for humans.
Besides the eight haplotypes, we crossed female homozygotes for the sws2+1a+14b mutation (one-base and 14-base insertions on SWS2a and SWS2b, respectively) with heterozygous male fish. A genotype ratio of their siblings at adult stages was SWS2+/–:SWS2–/– = 7:6, which is not significantly different from the expected 6.5:6.5 (P = 0.782).
These results demonstrate that the lack of either or both SWS2s does not affect the viability of medaka, at least under laboratory conditions. All the nine lines are available as frozen sperm at the NBRP Medaka as the following names and IDs: sws2–4a (MT1158), sws2+8a (MT1159), sws2+29a (MT1160), sws2–8b (MT1161), sws2+14b (MT1162), sws2–4a–2b (MT1163), sws2+1a+14b (MT1178), sws2–4a+5b (MT1179), and sws2–4a+2b (MT1180).
Cone Opsin Expressions in the sws2 Mutants
Using two SWS2a-single (sws2+8a and sws2+29a), two SWS2b-single (sws2–8b and sws2+14b), and two SWS2a/b-double mutants (sws2–4a–2b and sws2–4a+2b) as representatives (n = 1 each), we compared the expression of full-length (more precisely, all-coding exon-containing) SWS2 mRNA with that of the wild-type (not SWS2-mutated) fish (n = 2) by stepwise RT–PCR. This experiment using eight adults was performed on two different genomic backgrounds, ci and Actb-SLα:GFP (i.e., n = 16 in total). No matter whether single or double, all the frameshift mutations commonly reduced the sws2 transcripts on both backgrounds (Figures 2A,B), likely reflecting the nonsense-mediated mRNA decay (NMD) (Lykke-Andersen and Jensen, 2015). Thus, the sws2 mutants should translate severely truncated SWS2 (Figure 1D) in small amounts (if any), which could further support the successful knockout of the SWS2.
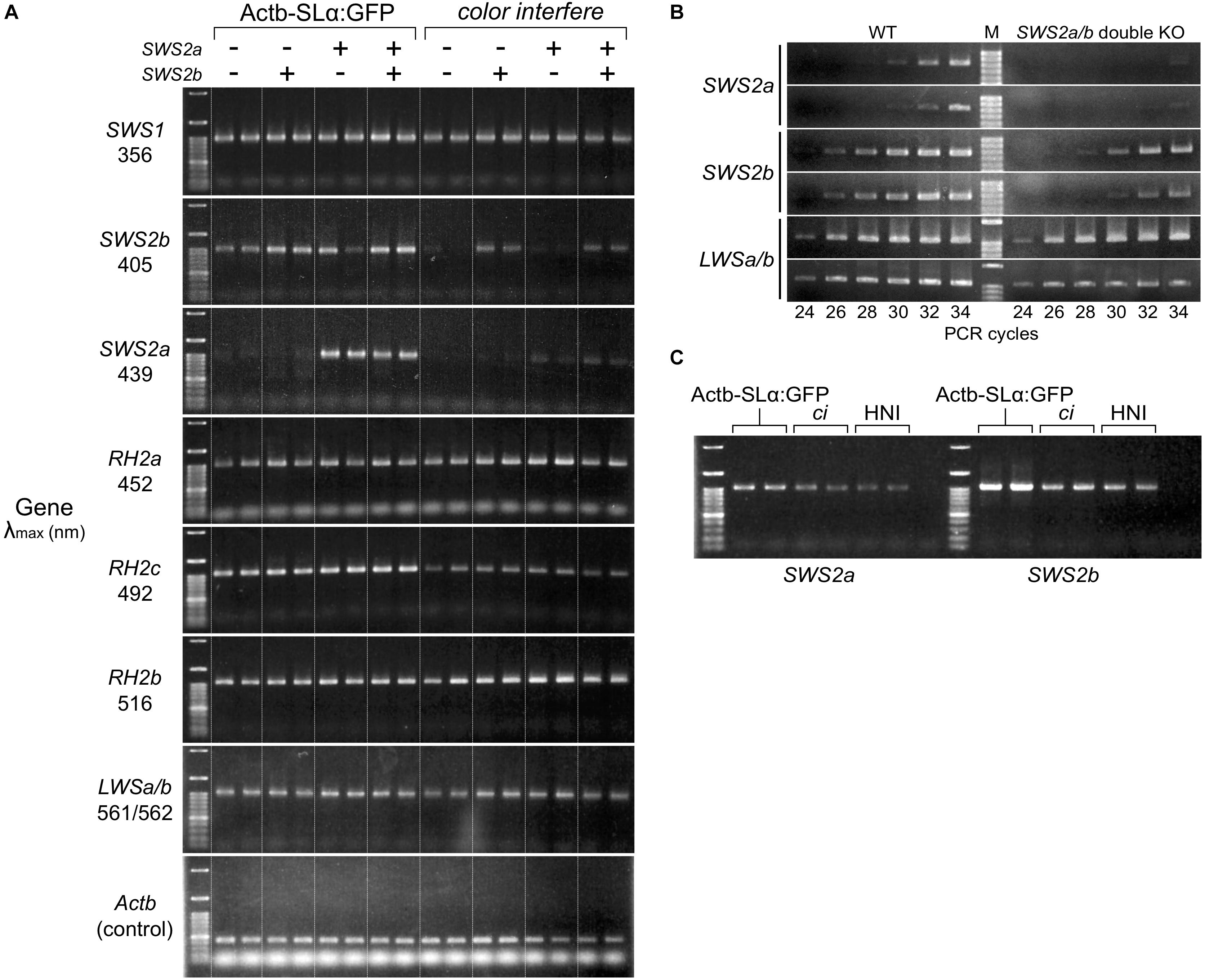
Figure 2. Expression of the cone opsin genes in the short-wavelength sensitive (sws)2 mutants. (A) RT-PCR of all the cone opsin genes in medaka. Gene names and λmax of the proteins are shown on the left. Long-wavelength sensitive (LWS)a and LWSb could not be analyzed separately because of their high sequence identity. Genomic backgrounds [actin beta (Actb)-somatolactin alpha (SLα):green fluorescent protein (GFP) or color interfere (ci)] and genotypes for the SWS2 loci [ + (homozygotes of the wild-type allele) or – (homozygotes of the mutated allele)] are shown on the top. From the left lane in each genomic background, results of the sws2– 4a+2b, sws2– 4a– 2b, sws2+29a, sws2+8a, sws2– 8b, sws2+14b, and two wild-type fish are shown. Note that expression is largely the same in the 16 individuals in terms of SWS1, rhodopsin (RH)2a, RH2b, and LWSa/b (that of RH2a may fluctuate slightly). Apparent differences are found in the expression of SWS2b, SWS2a, and RH2c. In RH2c, the expression is commonly higher in Actb-SLα:GFP than the expression in ci. In SWS2b and SWS2a, the expression is also higher in Actb-SLα:GFP than it is in ci (see results in the wild type). Additionally, the SWS2 expression differs depending on the genotype, i.e., when mutated, the expression is decreased on both genomic backgrounds. The number of PCR cycles was 30 for SWS2s, 26 for SWS1, RH2s, and LWSs, and 20 for Actb. (B) Examples of the stepwise RT-PCR for determining the appropriate number of PCR cycles (i.e., before plateau) for (A). Here, results at every two cycles between 24 and 34 cycles of two wild type and two SWS2a/b-double mutants (sws2– 4a+2b and sws2– 4a– 2b) with the ci background are shown for three genes (SWS2a, SWS2b, and LWSa/b). Note delayed amplifications in the sws2 mutants for SWS2a and SWS2b, but not LWSa/b. We used forward primers different from those listed in Table 6 (namely, f: 5′-AACAAGAAGCTTCGATCCCA for SWS2a and f: 5′-TTGTTGCTTCTACGGGTTCC for SWS2b), which is why products shorter than those in (A) were amplified. (C) RT-PCR of the SWS2 genes in three wild-type (i.e., with no mutations on any cone opsin genes) strains that express SLα differently. The Actb-SLα:GFP, ci, and HNI strains excessively, never, and ordinarily express SLα, respectively (Fukamachi et al., 2004, 2009b). The expression of SWS2a and SWS2b is stronger in Actb-SLα:GFP than in ci as in (A). Expression in HNI seems to be more similar to that in ci than to that in Actb-SLα:GFP, suggesting that the expression of SWS2 is enhanced in Actb-SLα:GFP (rather than suppressed in ci; see section “Discussion”).
Unexpectedly, SWS2a and SWS2b were expressed more strongly in Actb-SLα:GFP than they were in ci (Figure 2A). Until we noticed this, we repeated this assay without distinguishing the genomic backgrounds [as in our previous study for LWSs (Homma et al., 2017; Harada et al., 2019); note that LWS expressions in Figures 2A,B are similar between the backgrounds] being confused by apparently fluctuating or non-reproducible bands. Before we came to this conclusion, we had hypothesized the causes to include age (Valen et al., 2018), sex (Shao et al., 2014), and time of killing (Halstenberg et al., 2005). We might find a circadian fluctuation in SWS2 expression (data not shown) but did not further assess genetic or environmental factors that could affect the cone opsin expression.
The SWS2 expression in the standard wild-type strain that ordinarily expresses SLα (i.e., HNI) seemed to be more similar to that in ci than in Actb-SLα:GFP (Figure 2C). Thus, the SWS2 expression seemed to be enhanced in Actb-SLα:GFP rather than suppressed in ci (discussed further below).
Expressional assays for the other cone opsin genes (Figure 2A) revealed that: (1) as SWS2s, RH2c is expressed more strongly in Actb-SLα:GFP than in ci; (2) the expression of SWS1, RH2a, RH2b, and LWSa/b (LWSa and LWSb are too similar to be analyzed separately) between ci and Actb-SLα:GFP is similar; and (3) the loss of blue opsin seems to not induce upregulation of violet, green, or red opsins on either genomic background.
Mate Choice of the sws2 Mutants
ci and Actb-SLα:GFP are body color variants of medaka (pale gray and dark orange, respectively, Fukamachi et al., 2004, 2009b) that strongly prefer to mate within the strain under white light when they were separately reared from hatching (Fukamachi et al., 2009a; Ikawa et al., 2017). These sexual preferences cannot be observed under monochromatic light (Utagawa et al., 2016) and are significantly weakened when the LWS genes are knocked out (Kamijo et al., 2018).
Here, we similarly examined sexual preferences of the SWS2-mutated ci and Actb-SLα:GFP under white light (Figure 3). In terms of the wild-type (not SWS2-mutated) ci and Actb-SLα:GFP (n = 10 and 8, respectively), only 23.9% ± 6.5% and 15.7% ± 2.8% (mean ± 95% confidence interval) of male courtships were directed to female fish of the other strain, respectively. These proportions were significantly increased in the sws2-mutated ci and Actb-SLα:GFP (six strains in total). Namely, the proportions were 40.4% ± 5.1%, 42.3% ± 12.6%, and 48.0% ± 10.7% in the sws2+1a+14b, sws2+29a, and sws2+14b with the ci background (n = 18, 4, and 4, respectively) (P < 0.05, one-way ANOVA followed by a Dunnett post hoc test using the wild-type ci as a control) and 42.3% ± 10.5%, 41.2% ± 13.1%, and 35.9% ± 13.4% in the sws2–4a–2b, sws2+29a, and sws2+14b with the Actb-SLα:GFP background (n = 6, 4, and 4, respectively) (P < 0.05, one-way ANOVA followed by a Dunnett post hoc test using the wild-type Actb-SLα:GFP as a control). We note that all the mutant strains showed a common characteristic, i.e., whereas the wild-type male medaka never preferred female fish of the other strain, the mutant male fish did occasionally prefer the other strain. Thus, although the sample sizes per strain per background (particularly, those for the single mutants) were not very big, not only the SWS2a/b-double mutants (n = 24 in total) but also the single mutants (n = 16 in total) have weakened the body color preferences.
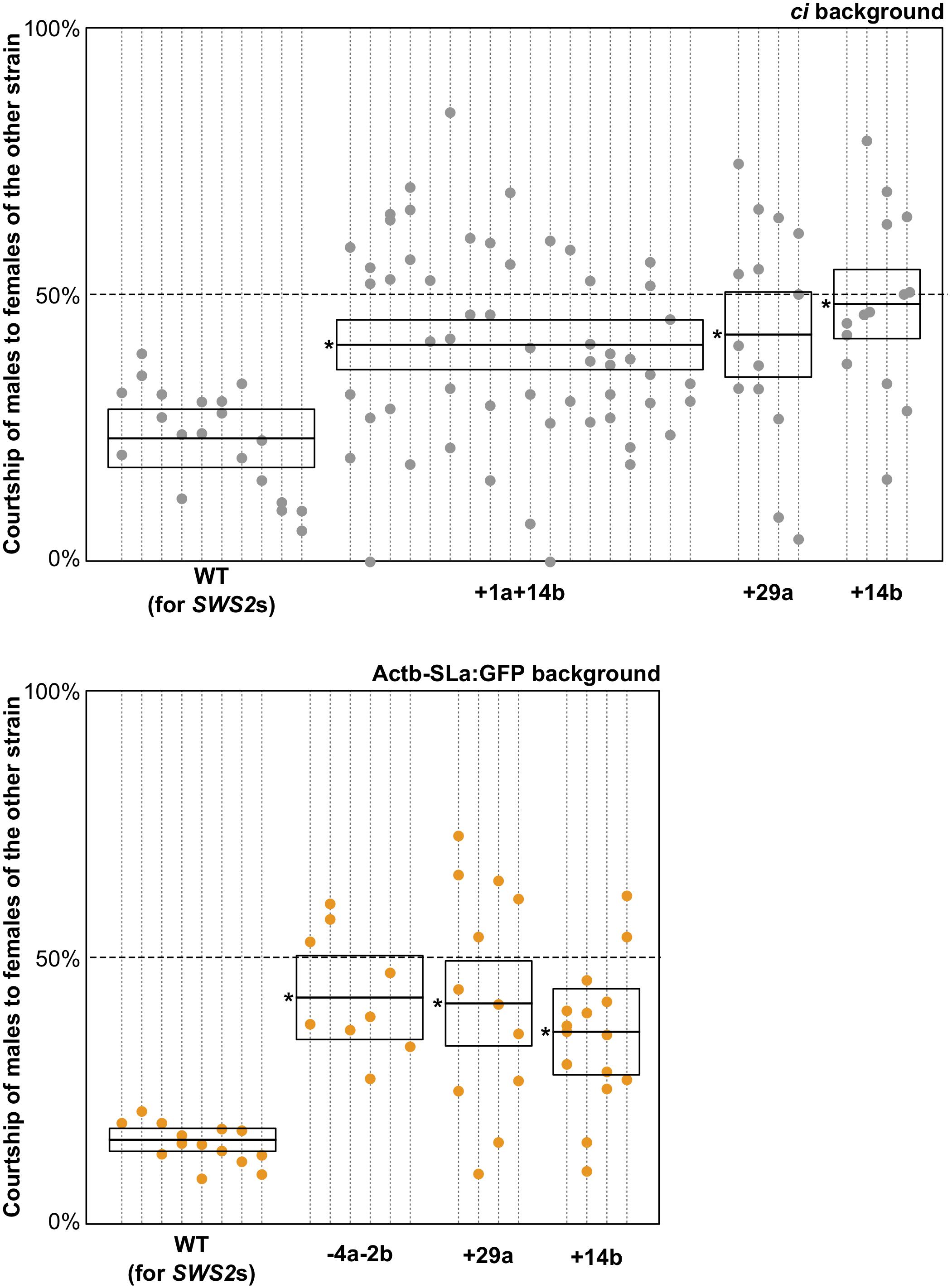
Figure 3. Mate choice of the short-wavelength sensitive (sws)2 mutants. Male fish of wild type, sws2+1a+14b, sws2– 4a– 2b, sws2+29a, and sws2+14b with the color interfere (ci) or actin beta (Actb)-somatolactin alpha (SLα):green fluorescent protein (GFP) background (shown by gray or orange, respectively) were given a choice between ci and Actb-SLα:GFP female fish. The cone opsin genotypes of the choice females were identical to those of the test males, except that we presented ci females with the sws2+1a+14b mutation and Actb-SLα:GFP females with the sws2+29a mutation to the wild-type males. Note that the wild-type males still exhibited strong preferences toward females of the same strain, indicating that body colors of the wild type and the cone opsin mutants are indistinguishable for not only humans but also medaka. Each dotted vertical line represents a male fish, and each circle on it is a result in a mate-choice trial (any trial with less than 10 courtships was ignored and is not shown). A box with a horizontal line shows the mean and 95% confidence interval of the ratio of courtship of male fish to female fish of the other strain (i.e., a relative sexual preference of the male fish). A one-way ANOVA followed by a Dunnett post hoc test revealed significant increases (shown by asterisks) in all the sws2 mutants by comparison with the wild type (P < 0.05) on both genomic backgrounds.
Behavioral Photosensitivity of the sws2 Mutants
As a lack of LWS clearly reduced sensitivity to red light (Homma et al., 2017; Matsuo et al., 2018), a lack of SWS2 may reduce sensitivity to blue light. Alternatively, the sensitivity may not be reduced because blue light could be absorbed by the neighboring violet and green opsins, whereas red (near infrared) light could not be absorbed by the neighboring green opsin. To address these hypotheses, we tested the blue light sensitivity of the sws2 mutants. The OMR of medaka is so conspicuous that we could manually distinguish the wild-type and lws-mutant fish based on their behavior under monochromatic light at λ = 760 nm, i.e., all the 48 fish (of 77 with unknown genotype) we judged to be OMR-positive were indeed either LWS+/+ or LWS+/– (Homma et al., 2017). That is, our manual assessments never included a false positive (but included false negatives because fish sometimes ignore/resist the rotating stripes).
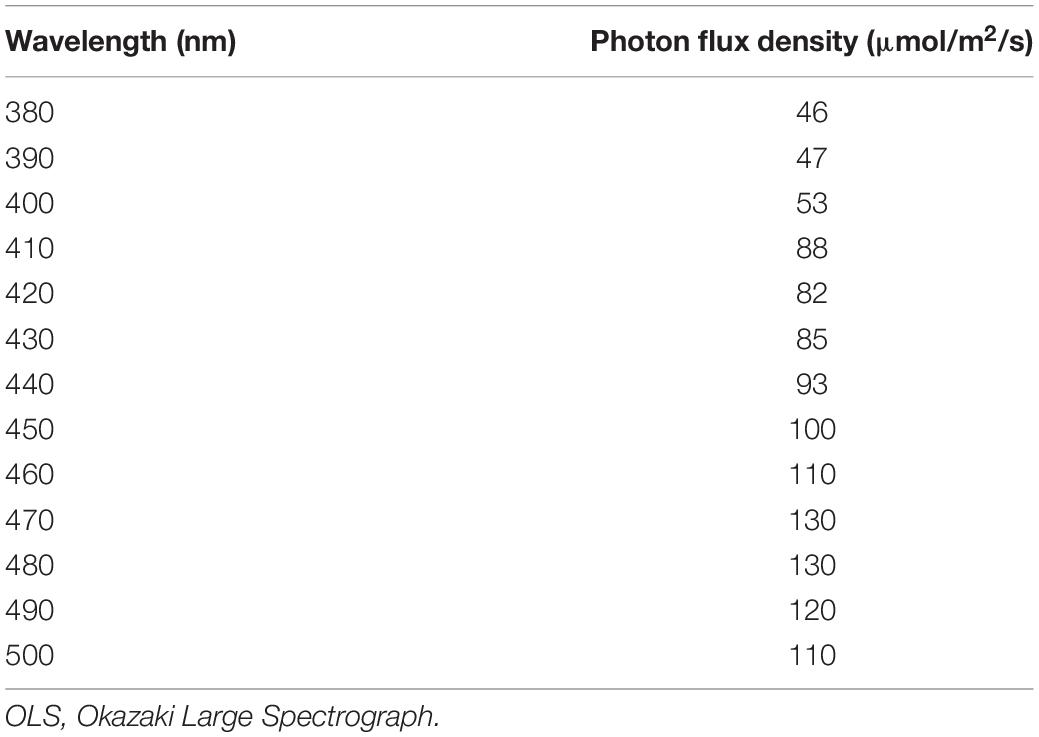
Using the OLS (Watanabe et al., 1982), we performed the OMR test at wavelengths of every 10 nm between 380 and 450 nm (see Table 5 for photon flux density) using the wild type (n = 5) and the sws2–4a, sws2+8a, sws2+29a, sws2–8b, sws2+14b, sws2–4a–2b, and sws2–4a+2b mutants (n = 5, 5, 5, 5, 5, 3, and 1, respectively). For the wild type and the double mutants (sws2–4a–2b and sws2–4a+2b), we additionally tested at every 10 nm between 460 and 500 nm. In all the strains at all the wavelengths, we manually assessed the fish to be OMR-positive.
The manual assessments might overlook quantitative reduction of the OMR in the sws2 mutants. Thus, we additionally performed quantitative OMR tests (Matsuo et al., 2018) focusing on λ = 440 nm for the SWS2a-single mutants (sws2–4a, sws2+8a, and sws2+29a; n = 5 each) and λ = 400 nm for the SWS2b-single mutants (sws2–8b and sws2+14b; n = 5 each) and the SWS2a/b-double mutants (sws2+1a+14b, sws2–4a–2b, and sws2–4a+2b; n = 5 each). We chose these wavelengths because λmax of SWS2a and SWS2b are 439 and 405 nm, respectively, and those of the neighboring SWS1 and RH2a are 356 and 452 nm, respectively (Matsumoto et al., 2006). Their genomic backgrounds were ci for sws2+29a, sws2+14b, sws2–4a–2b, and sws2–4a+2b and Actb-SLα:GFP for sws2–4a, sws2+8a, sws2–8b, sws2–4a+5b, and sws2+1a+14b. We used ci as the wild-type control (n = 5). As shown in Figures 4A–C, all the sws2 mutants responded to the rotating stripes as quickly (Figure 4A) and for as long (Figure 4B) as the wild type, which resulted in the similar overall swimming distances per OMR test (Figure 4C; P > 0.05, one-way ANOVA followed by a Dunnett post hoc test).
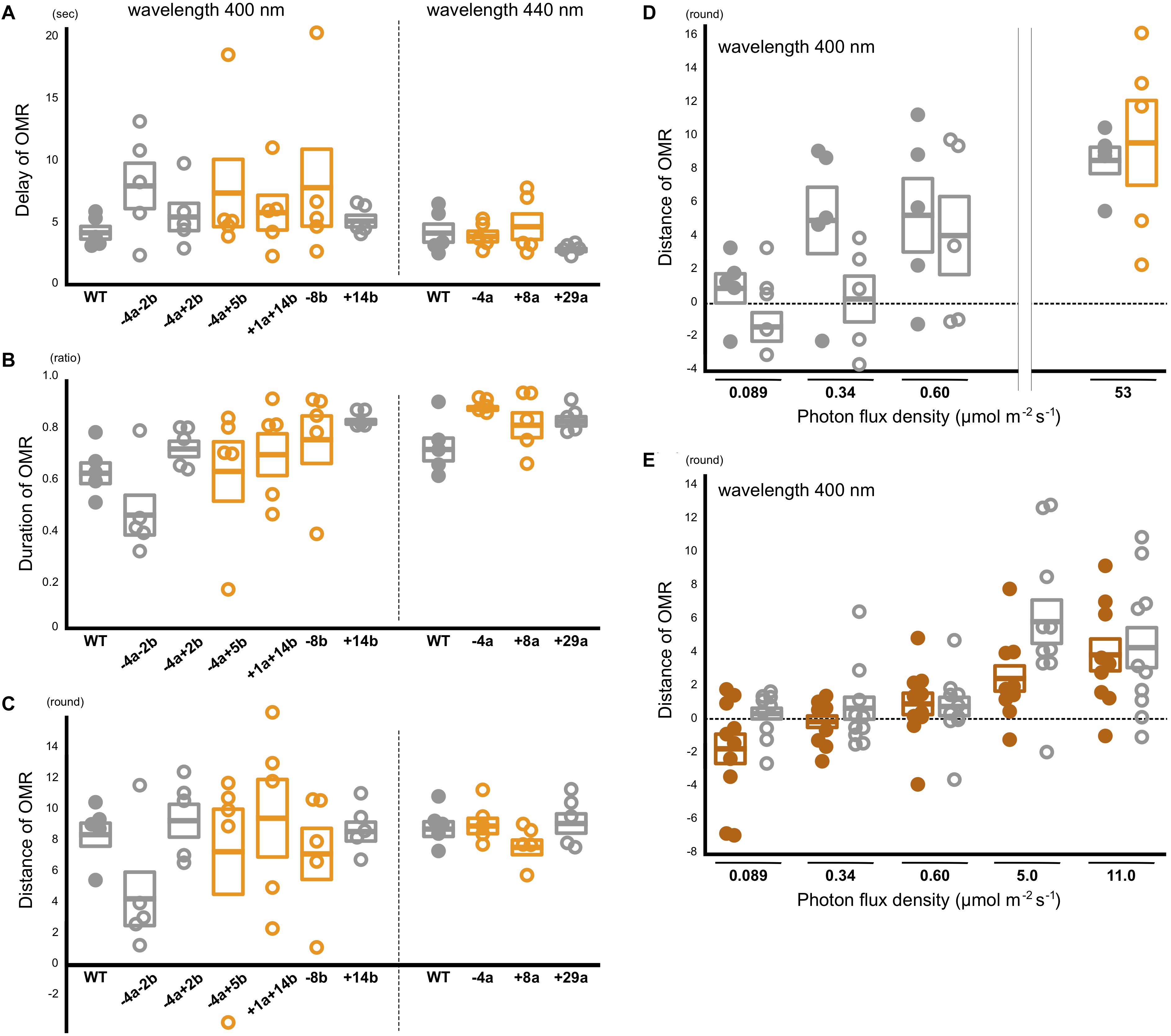
Figure 4. Behavioral photosensitivity of the short-wavelength sensitive (sws)2 mutants. The optomotor response (OMR) of adult fish was tested individually under monochromatic light at λ = 400 nm for the wild type, SWS2a/b-double, and SWS2b-single mutants (left) and λ = 440 nm for the wild type and SWS2a-single mutants (right). Each circle represents the result for one fish (n = 5 for each strain). Closed circle, wild type; open circle, sws2 mutants. Colors represent genomic backgrounds of the strains [gray, color interfere (ci); orange, actin beta (Actb)-somatolactin alpha (SLα):green fluorescent protein (GFP)]. The OMR was quantified by three parameters: (A) delay: the time elapsed until the fish started the OMR after switching the direction of stripe rotation, (B) duration: the proportion of time the fish was exhibiting the OMR, and (C) distance: the distance the fish swam in the direction of stripe rotation during the test (Matsuo et al., 2018). Mean and standard error of the mean is shown as a box with a horizontal line. No significant difference was detected between the wild type and the sws2 mutants in any of the six comparisons (i.e., the delay, duration, or distance at λ = 400 or 440 nm) (P > 0.05; one-way ANOVA followed by a Dunnett post hoc test using the wild type as a control). (D) The OMR tests under mesopic conditions. The results at 53 μmol/m2/s (photopic condition) are those in (C). Closed circle, wild type; open circle, sws2+1a+14b (n = 5 each). See panel (C) for other symbols. The results at 0.089 μmol/m2/s (i.e., approximately –4 to + 4 rounds per individual and –2 to + 2 rounds in average) seem to indicate OMR-negative. From this standpoint, the OMR at 0.34 μmol/m2/s might be negative in the mutants, but positive in the wild type, although the difference is not significant (P > 0.05, Student’s t-test). (E) Further examination of the OMR under mesopic conditions. As the wild type (closed brown circles), we used the HNI strain (n = 9–10; one fish died during the tests). As the sws2 mutants (open gray circles), we used the sws2+1a+14b with the ci background (n = 10). The differences are not significant at any photon flux density (P > 0.05, Student t test). The results in panels D,E may be somewhat inconsistent (e.g., the response at 0.60 μmol/m2/s seems to be higher in D than it is in E), which would reflect, for example, experiments on different days using different fish, experiments at different times of the day (i.e., diurnal fluctuation of cone opsin expression), potential differences in light angles or rotation speeds, or deterioration of the xenon arc lamp [we adjusted the photon flux densities using neutral density filters and the slits of the Okazaki Large Spectrograph (OLS)].
We further hypothesized that a difference in the OMR (i.e., blue light sensitivity) could not be detected in the experiments described above because the monochromatic light from the OLS was too strong (i.e., bright enough even for the sws2 mutants to fully recognize the rotating stripes). Thus, we performed the quantitative OMR tests at decreased photon flux densities (i.e., at mesopic conditions) using the wild type and the sws2+1a+14b mutants with the ci background (n = 5 each). Whereas both the strains swam for about nine rounds per OMR test at 53 μmol/m2/s (Figures 4C,D), the distance dropped to four to five rounds at 0.60 μmol/m2/s in both strains. At 0.089 μmol/m2/s, the OMR seemed to be substantially negative in both strains (i.e., 1.1 ± 0.9 and –1.2 ± 0.9 rounds, respectively; it should be noted that these are distances for which fish followed the rotating stripes in 2 min in a tank of 19 cm diameter; see section “Materials and Methods”). At 0.34 μmol/m2/s, the wild type might better respond than the sws2+1a+14b mutant, although the difference was not significant (P = 0.095, Student’s t-test).
To assess further the potential difference in behavioral photosensitivity under the mesopic condition, we repeated the OMR tests using different individuals. We used the standard HNI strain (n = 10) and the sws2+1a+14b mutant with the ci background (n = 10; Figure 4E). The OMR seems to be positive at 5.0 and 11.0 μmol/m2/s and negative at 0.089 μmol/m2/s in both strains. At neither 0.34 nor 0.60 μmol/m2/s an apparent or statistically significant difference in the OMR could be detected (P > 0.05, Student’s t-test). A difference might be detected at different photon flux densities or wavelengths or by methods other than the OMR test. However, effects of the SWS2 loss on behavioral blue light sensitivity would in any case be subtle, or might even be absent, while the defects in color-dependent mate choice were relatively apparent (Figure 3).
Discussion
Apparent Effects of the sws2 Mutations on Mate Choice but Not the Optomotor Response
Although we did not confirm it at the protein level (antibodies specific for the medaka SWS2a or SWS2b are not available), the frameshift mutations (Tables 1–3 and Figure 1D) and the suppressed mRNA expressions (Figures 2A,B) strongly suggest that the SWS2a/b-double mutants lack functional blue opsins. On losing one of the four types of cone opsins in the retina, the mutants had been expected to have a defect in color vision, and reduced body color preferences were actually detected (Figure 3). The similar reduction in body color preferences had also been observed in the LWSa/b-double mutants (Kamijo et al., 2018). Therefore, we suspect that these cone opsin knockouts could not distinguish the body colors of ci and Actb-SLα:GFP as clearly as the wild type, and this visual limitation caused the weakly biased color preferences, as reported in other fish using turbid water (Engström-Öst and Candolin, 2007; Sundin et al., 2010; Ehlman et al., 2018). To know to what extent the abilities for color discrimination are actually reduced in the sws2 and lws mutants, further experiments using different colors, possibly based on learning (e.g., Siebeck et al., 2014; Escobar-Camacho et al., 2017), are necessary.
Four types of cone cells are regularly arranged in the fish retina forming the retinal mosaic (Nishiwaki et al., 1997; Allison et al., 2010), but expressions of cone opsins are rather complex; for example, different types of opsins could be coexpressed in a single cone and the expression can change relying on growth stages, ambient light, or visual angles (Dalton et al., 2014; Sakai et al., 2018; Zimmermann et al., 2018). In our studies, proximate consequences of the SWS2 or LWS loss (e.g., effects on the retinal mosaic or electrophysiological responses in the retina or downstream cascades) remain unknown, which are inevitable (and complex) subjects for unveiling the mechanism for color perception (Thoreson and Dacey, 2019). However, one solid conclusion at present is that our reverse-genetic studies demonstrated causal relationships (not simply associations) between a cone opsin repertoire and a color-dependent social behavior (Figure 3; Kamijo et al., 2018).
Despite the apparent effects on mate choice, blue light sensitivity of the sws2 mutants was equivalent to that of the wild type (Figure 4). By contrast, in our previous study, an apparent reduction in photopic red light sensitivity could be detected in the lws mutants (Homma et al., 2017; Matsuo et al., 2018). These potentially controversial results are not at all surprising because whereas red light (λ > 740 nm) could only be absorbed by LWSs (λmax = 561–562 nm), blue light tested in this study (λ = 380–500 nm) could likely be absorbed by not only SWS2s (λmax = 405–439 nm) but also the neighboring SWS1 (λmax = 356 nm) and RH2s (λmax = 452–516 nm) (Matsumoto et al., 2006). Indeed, we previously found that the wild-type and LWSa/b-knockout medaka could exhibit the OMR at λ ≤ 830 and ≤ 740 nm, respectively (Homma et al., 2017; Matsuo et al., 2018). That is, LWSs and RH2s can absorb and make medaka respond behaviorally to light at wavelengths > 200 nm longer than the λmax. The absorption of a wide range of light might be the same for SWS2s and SWS1, which enabled compensation of the SWS2 loss by SWS1 (and maybe also RH2 and LWS, although we still do not know how widely the cone opsins cover wavelengths shorter than the λmax). The present results also indicate that the compensation does not require upregulation of SWS1, RH2, or LWS (Figure 2A). Compensation by RH1 is unlikely because the rod vision has been shown to be dysfunctional in light-adapted medaka, i.e., only the dark-adapted, but not light-adapted, lws mutants could show the OMR at λ ≥ 750 nm (Homma et al., 2017).
We note that SWS2 also plays only a dispensable role for blue light sensitivity in zebrafish larvae, i.e., when the SWS2-expressing cones are chemically ablated, the ability to detect contrasts between dark red and blue on an RGB monitor is temporarily reduced, but quickly recovered within 24 h without regenerating the SWS2 cones (Hagerman et al., 2016). Thus, the absence of SWS2 seems to have little effect on photopic blue light sensitivity in both medaka adults and zebrafish larvae.
Adaptive Evolution of Cone Opsin Repertoire
Divergent repertoires and λmax of cone opsins in fish have often been discussed in relation to adaptation to various underwater light conditions and body colors of mating partners; for example, in blue light-dominating clear water, blue light sensitivity is increased by increased SWS2 expression or blue-shift of λmax, which promotes sexual selection of a blue nuptial coloration. Although this evolutionary scenario (i.e., sensory drive) sounds attractive and many researchers are reporting results that support it (see Cummings and Endler, 2018 and references therein), the reverse-genetic evidence in medaka and zebrafish demonstrating little difference in behavioral blue light sensitivity regardless of the presence or absence of SWS2 even at a wavelength close to its λmax (Figure 4; Hagerman et al., 2016) seems to suggest that the “dominant-light” hypothesis needs to be reconsidered carefully.
Increased photosensitivity to a dominating wavelength should increase not only a signal from an object of interest but also noise from the background. A study reporting a depth-dependent shift in λmax of SWS2, but away from a dominating wavelength in cottoid fishes (whereas RH1 for scotopic vision did shift toward the dominating wavelength) (Cowing et al., 2002), is implicative in that the authors suggested that the shift is to increase the signal-to-noise ratio (the “noise reduction” hypothesis). In Catarrhini, the functional importance of the duplicated and diversified λmax in LWSs is theoretically (Lewis and Li, 2006) and ecologically (Melin et al., 2017) interpreted for discrimination of red objects (e.g., fruits) against green background (e.g., leaves). Animals other than Catarrhini should also have a particular color (or luminance) that needs to be discriminated against background for efficient survival and reproduction in nature, which could drive the diversifications of repertoire, λmax, or expression of cone opsins (the “color discrimination” hypothesis).
Plastic Expression of the Cone Opsin Genes
Another interesting finding in this study is the apparent increase in the expression of SWS2a, SWS2b, and RH2c in Actb-SLα:GFP in comparison with that in ci (Figure 2). The expression of cone opsins is plastic depending on, for example, light conditions (Fuller et al., 2010; Hofmann et al., 2010; Sakai et al., 2016) and circadian rhythms (Halstenberg et al., 2005; Li, 2005; Johnson et al., 2013), but all the fish used in Figure 2 were reared under identical light conditions and killed for RNA extraction at the same time. The Actb-SLα:GFP strain is established by introducing a transgene that expresses SLα and Renilla GFP (hrGFP II; Agilent Technologies) into ci (Fukamachi et al., 2009b). Therefore, nothing else but SLα and/or Renilla GFP must be the cause for the differences in expression of SWS2a, SWS2b, and RH2c between ci and Actb-SLα:GFP.
The Renilla GFP in Actb-SLα:GFP is not visible under white light. However, it does exist in all cells that express the gene for Actb, including in the lenses of the eyes (Fukamachi et al., 2009b). According to the datasheet provided by the manufacturer, absorption/emission spectra of Renilla GFP are nearly symmetrical with absorption/emission peaks at 500/506 nm, respectively (Figure 5A). This indicates that, using 503 nm as a border, a part of light at shorter wavelengths (blue light) is absorbed and converted to light at longer wavelengths (green light) before reaching the retina of Actb-SLα:GFP (Figure 5B). Some of the green light would directly go out of the eyes because the direction of emission by Renilla GFP molecules must be random. Thus, the retina of Actb-SLα:GFP would mostly receive less blue light (and more green light) in comparison with that of ci, i.e., ci and Actb-SLα:GFP should effectively live in different light conditions.
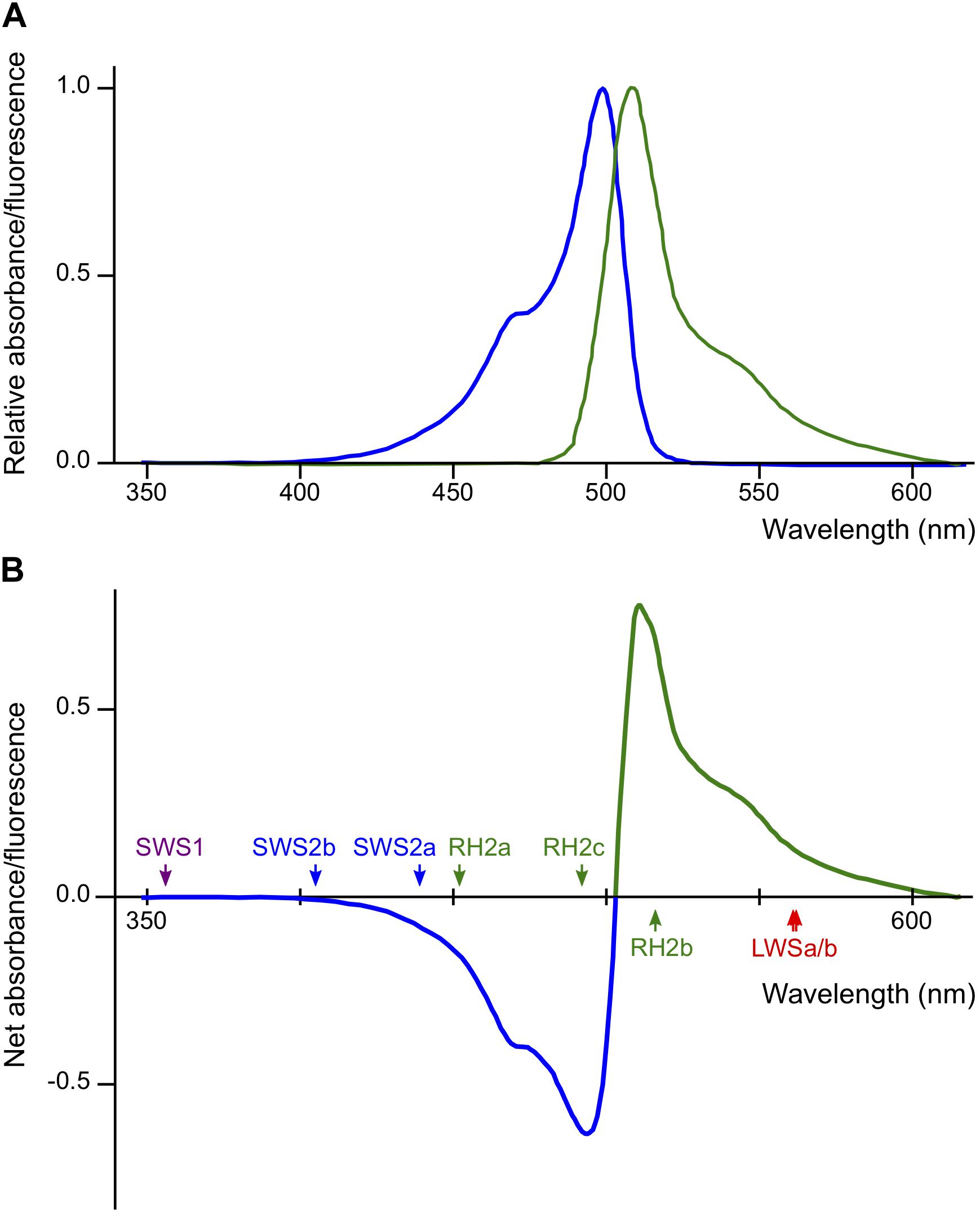
Figure 5. Potential effects of Renilla green fluorescent protein (GFP) on the vision of actin beta (Actb)-somatolactin alpha (SLα):GFP. (A) Excitation (blue) and emission (green) spectra of Renilla GFP (hrGFPII; Agilent Technologies) (Ward and Cormier, 1979). Excitation and emission peaks are 500 and 506 nm, respectively. The spectra are largely symmetrical using 503 nm as a border. (B) Net spectral absorbance (minus values) and fluorescence (plus values) by Renilla GFP based on the data in (A). This graph indicates that a part of the blue light (λ < 503 nm) impinging on the eyes of Actb-SLα:GFP (through the cornea and lens that weakly express Renilla GFP; Fukamachi et al., 2009b) would be absorbed, converted to green light (λ > 503 nm), and scattered into all directions before reaching the retina. The λmax of the medaka cone opsins are shown by colored arrows.
However, again, this interpretation becomes inconsistent with the “dominant-light” hypothesis because λmax of the increased SWS2a, SWS2b, and RH2c (439, 405, and 492 nm, respectively) are within the range of decreased ambient light (λ = 400–500 nm). We interpreted this result that the decreased blue light impinging on the retina (decreased signals from blue light-absorbing cone cells) was compensated for by the increased expression of SWS2a, SWS2b, and RH2c, i.e., the biased light spectrum was corrected by counter-biased expression of the cone opsins (the “white-balance” hypothesis). However, this interpretation does not explain the lack of increase in RH2a with λmax at 452 nm in Actb-SLα:GFP (Figure 2A).
An alternative interpretation is that SLα directly (or indirectly via body color recognition; Ikawa et al., 2017) enhanced the SWS2a/SWS2b/RH2c expression. Considering that the expression of SWS2s between SLα-expressing HNI and SLα-deficient ci (Figure 2C) is similar, Renilla GFP would more likely be the cause. Reverse genetics to remove the Renilla GFP from Actb-SLα:GFP or rearing ci in Renilla GFP solution, for example, may address these possibilities.
Conclusion
The present study established SWS2-knockout medaka and demonstrated a defect in color-dependent mating behaviors (Figure 3). However, its defect in behavioral blue light sensitivity is subtle, if not absent (Figure 4). These results could possibly be interpreted that, whereas a signal from the SWS2-expressing cones (together with those from the SWS1/RH2/LWS-expressing cones, i.e., a signal of four channels) is essential for the normal (tetrachromatic) color vision, the blue channel is dispensable and the remaining violet/green/red channels are sufficient for detecting blue light (although the three-channel signal may not be sensed as blue). This kind of a clear-cut evidence demonstrating a causal relationship (not an association) between genotype and phenotype can only be obtained by reverse (or forward) genetics. The recent genome-editing technologies have brought animal-vision studies to a new era. Although the diversification in cone opsins has often exclusively been discussed in relation to the sensory-drive hypothesis, the present results strongly suggest that increased transcription or even acquiring cone opsin types/subtypes does not necessarily increase (i.e., adapt) behavioral photosensitivity to a certain wavelength in photopic or mesopic conditions. Other viewpoints are necessary to understand the evolution of color vision in animals.
Materials and Methods
We essentially adopted the same procedures which were used to establish and characterize the lws mutants (Homma et al., 2017; Kamijo et al., 2018; Matsuo et al., 2018; Harada et al., 2019). Here, we outline the strategies in brief.
Targeted Mutagenesis of the SWS2 Genes by the CRISPR/Cas9 System
The ci and Actb-SLα:GFP strains were used as hosts for genome editing. ci has a mutation on the gene for SLα (Fukamachi et al., 2004), and Actb-SLα:GFP is a transgenic ci strain that ectopically overexpresses SLα driven by the promotor for actin beta (Actb) (Fukamachi et al., 2009b). The transgene also expresses Renilla GFP under the control of the internal ribosome entry site (IRES). We have been using these strains to establish color-blind medaka because of their unique color-dependent mate-choice behaviors (Fukamachi et al., 2009a; Utagawa et al., 2016; Ikawa et al., 2017).
Double-strand oligonucleotides complementary to target sequences (5′-CCTGTCTGTGGCTAATCTTCT-3′ for SWS2a and 5′-ACATGGTTCTGGGTCCACTGG-3′ for SWS2b) were inserted into a DR274 vector (Addgene) from which guide RNA (gRNA) were transcribed using an AmpliScribe T7-flash Transcription Kit (Epicentre). The Cas9 mRNA was synthesized from an hCas9 vector (Addgene) using an mMessage mMachine SP6 kit (Life Technology). The RNAs were purified using an RNeasy mini Kit (Qiagen) and microinjected into one-cell-stage embryos using GD-1 glass capillaries (Narishige) and an IM-9b microinjector (Narishige) under an SZX16 stereomicroscope (Olympus).
Ins/del mutations induced on the target sequences were detected using a heteroduplex mobility assay (HMA) on 12% polyacrylamide gels using the primers shown in Table 6. The microinjected adults that possess an ins/del mutation (G0) were crossed with the wild type to obtain heterozygotes of the ins/del mutation (F1). Ins/del mutations causing a frameshift were screened by genomic PCR and direct sequencing using the primers shown in Table 6. Heterozygous fish with an identical frameshift mutation on SWS2a and/or SWS2b were intercrossed to obtain homozygotes for the mutation/s.
Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted from the eyes of fully matured adults using Isogen II reagent (Nippon Gene), incubated with deoxyribonuclease (RT Grade) for Heat Stop (Nippon Gene), and used as templates for reverse transcription by ReverTra Ace (Toyobo) and polyT primers. PCR primers for each cone opsin gene are listed in Table 6. Temperature conditions were: 96°C for 1 min; 20–30 cycles (we performed stepwise PCR for each gene and stopped the reaction before the amplification plateaued) of 98°C for 20 s, 60°C for 1 min, 72°C for 1 min; and 72°C for 10 min. The products were electrophoresed on a 1% agarose gel and detected by ethidium bromide staining and UV transillumination (BioDoc-It Imaging System, UVP).
Mate-Choice Experiments
The medaka spawn every morning. We put one male and two female fish in a tank (20 cm × 13 cm with a water level of about 5 cm), let them mate freely for 30 min, and manually counted courtships (approaching behaviors) to each female fish. The body length was strictly equalized between choice females; i.e., the difference was less than 1 mm. A sexual preference of the male fish in the trial was calculated as a ratio of the courtships. If a male fish courted less than 10 times in a trial, we abolished the datum. This trial was repeated for two or four times in consecutive 2 or 4 days presenting different female individuals, and an overall preference of the male was calculated as an average in the trials. A preference of a strain was calculated as an average of all the male fish in the strain, and the values were compared between strains by a one-way analysis of variance (ANOVA) followed by a Dunnett post hoc test using IBM SPSS Statistics (ver. 25) for Mac (IBM Corp.).
The Optomotor Response Test
Bright-adapted adult medaka were put into a cylindrical glass tank (19 cm in diameter). The tank was placed in a drum (24 cm in diameter), inside of which vertical stripes were made using Indian ink-painted plastic paper and aluminum foil (because ordinary white paper and black ink fluoresce under blue light). The entire drum was irradiated from the top using monochromatic light from an OLS (Watanabe et al., 1982). In the experiments in Figures 4D,E, we simultaneously irradiated infrared light (λ = 940 nm) using Hololight (PiPhotonics) because the blue light was not sufficiently strong enough for video recording. This infrared light should not affect the OMR (i.e., should be invisible for medaka) because the OMR was negative at λ > 840 nm in our previous experiments (Homma et al., 2017; Matsuo et al., 2018).
After 30 s of acclimation, the stripes were rotated in the clockwise, counterclockwise, clockwise, and counterclockwise directions in 10 rpm for 30 s each. Behaviors were video-recorded using an A10FHDIR (Kenko) or an ORCA-R2 digital CCD camera (Hamamatsu Photonics). Because the ORCA-R2 camera was not very sensitive to light at λ < 400 nm, some image processing was needed to analyze the movies recorded at λ = 400 nm, which are summarized in Supplementary Movie S1.
The position (x–y coordinates) of fish in each video frame was extracted using UMATracker software (Yamanaka and Takeuchi, 2018), from which we calculated three parameters to quantify the OMR: delay, duration, and distance (Matsuo et al., 2018). The delay and duration were calculated as averages, whereas the distance was a sum, in four rotations. The values in each parameter were compared at each wavelength using a one-way ANOVA followed by a Dunnett post hoc test using the wild type as a control (Figures 4A–C). For Figures 4D,E, we applied a Student’s t-test at each photon flux density.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Ethics Statement
The animal study was reviewed and approved by the Animal Experiment Committee of Japan Women’s University.
Author Contributions
YH introduced the sws2 mutations. NK and MG established and characterized the sws2 mutants (the OMR and RT-PCR). CT and YS performed the mate choice. MM quantitatively analyzed the OMR. TU and YK provided technical advice on the OLS. SF examined the SWS2 loci in silico, designed and supervised the wet experiments, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by a Grant-in-Aid for Scientific Research (C) (#17K07506) from the Japan Society for the Promotion of Science (JSPS), a grant for Joint Research (#01111904) by the National Institutes of Natural Sciences (NINS), and research funds from JWU to SF. This study was also supported as the Priority Collaborative Research Project (#16-101) and Collaborative Experiments using the Large Spectrograph (#15-602, #16-601, and #17-703) in NIBB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mone Nagayama and Hinako Sato of JWU for their contributions to the mutant establishment and the mate-choice experiments, respectively. We also thank Haruna Ono and Mana Sakakibara of JWU for their contributions to the OMR tests under mesopic conditions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00801/full#supplementary-material
MOVIE S1 | Summary of retouch processes for tracking movies recorded at λ = 400 nm.
Footnotes
References
Allison, T. W., Barthel, L. K., Skebo, K. M., Takechi, M., Kawamura, S., and Raymond, P. A. (2010). Ontogeny of cone photoreceptor mosaics in zebrafish. J. Comp. Neurol. 518, 4182–4195. doi: 10.1002/cne.22447
Bowmaker, J. K. (1998). Evolution of colour vision in vertebrates. Eye 12, 541–547. doi: 10.1038/eye.1998.143
Carleton, K. L., Escobar-Camacho, D., Stieb, S. M., Cortesi, F., and Marshall, N. J. (2020). Seeing the rainbow: mechanisms underlying spectral sensitivity in teleost fishes. J. Exp. Biol. 223:jeb193334. doi: 10.1242/jeb.193334
Conway, B. R. (2009). Color vision, cones, and color-coding in the cortex. Neuroscientist 15, 274–290. doi: 10.1177/1073858408331369
Cortesi, F., Musilová, Z., Stieb, S. M., Hart, N. S., Siebeck, U. E., Malmstrøm, M., et al. (2015). Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proc. Natl. Acad. Sci. U.S.A. 112, 1493–1498. doi: 10.1073/pnas.1417803112
Cowing, J. A., Poopalasundaram, S., Wilkie, S. E., Bowmaker, J. K., and Hunt, D. M. (2002). Spectral tuning and evolution of short wave-sensitive cone pigments in cottoid fish from Lake Baikal. Biochemistry 41, 6019–6025. doi: 10.1021/bi025656e
Cummings, M. E., and Endler, J. A. (2018). 25 Years of sensory drive: the evidence and its watery bias. Curr. Zool. 64, 471–484. doi: 10.1093/cz/zoy043
Dalton, B. E., Loew, E. R., Cronin, T. W., and Carleton, K. L. (2014). Spectral tuning by opsin coexpression in retinal regions that view different parts of the visual field. Proc. R. Soc. B Biol. Sci. 281, 1–9. doi: 10.1098/rspb.2014.1980
Ehlman, S. M., Martinez, D., and Sih, A. (2018). Male guppies compensate for lost time when mating in turbid water. Behav. Ecol. Sociobiol. 72:46. doi: 10.1007/s00265-018-2468-8
Engström-Öst, J., and Candolin, U. (2007). Human-induced water turbidity alters selection on sexual displays in sticklebacks. Behav. Ecol. 18, 393–398. doi: 10.1093/beheco/arl097
Escobar-Camacho, D., Marshall, J., and Carleton, K. L. (2017). Behavioral color vision in a cichlid fish: metriaclima benetos. J. Exp. Biol. 220, 2887–2899. doi: 10.1242/jeb.160473
Fukamachi, S., Kinoshita, M., Aizawa, K., Oda, S., Meyer, A., and Mitani, H. (2009a). Dual control by a single gene of secondary sexual characters and mating preferences in medaka. BMC Biol. 7:64. doi: 10.1186/1741-7007-7-64
Fukamachi, S., Yada, T., Meyer, A., and Kinoshita, M. (2009b). Effects of constitutive expression of somatolactin alpha on skin pigmentation in medaka. Gene 442, 81–87. doi: 10.1016/j.gene.2009.04.010
Fukamachi, S., Sugimoto, M., Mitani, H., and Shima, A. (2004). Somatolactin selectively regulates proliferation and morphogenesis of neural-crest derived pigment cells in medaka. Proc. Natl. Acad. Sci. U.S.A. 101, 10661–10666. doi: 10.1073/pnas.0401278101
Fuller, R. C., Noa, L. A., and Strellner, R. S. (2010). Teasing apart the many effects of lighting environment on opsin expression and foraging preference in bluefin killifish. Am. Nat. 176, 1–13. doi: 10.1086/652994
Greenwald, S. H., Kuchenbecker, J. A., Roberson, D. K., Neitz, M., and Neitz, J. (2014). S-opsin knockout mice with the endogenous M-opsin gene replaced by an L-opsin variant. Vis. Neurosci. 31, 25–37. doi: 10.1017/S0952523813000515
Hagerman, G. F., Noel, N. C. L., Cao, S. Y., DuVal, M. G., Oel, A. P., and Allison, W. T. (2016). Rapid recovery of visual function associated with blue cone ablation in Zebrafish. PLoS One 11:e0166932. doi: 10.1371/journal.pone.0166932
Halstenberg, S., Lindgren, K. M., Samagh, S. P. S., Nadal-Vicens, M., Balt, S., and Fernald, R. D. (2005). Diurnal rhythm of cone opsin expression in the teleost fish Haplochromis burtoni. Vis. Neurosci. 22, 135–141. doi: 10.1017/S0952523805222022
Harada, Y., Matsuo, M., Kamei, Y., Goto, M., and Fukamachi, S. (2019). Evolutionary history of the medaka long-wavelength sensitive genes and effects of artificial regression by gene loss on behavioural photosensitivity. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-39978-6
Hofmann, C. M., O’Quin, K. E., Smith, A. R., and Carleton, K. L. (2010). Plasticity of opsin gene expression in cichlids from Lake Malawi. Mol. Ecol. 19, 2064–2074. doi: 10.1111/j.1365-294X.2010.04621.x
Homma, N., Harada, Y., Uchikawa, T., Kamei, Y., and Fukamachi, S. (2017). Protanopia (red color-blindness) in medaka: a simple system for producing color-blind fish and testing their spectral sensitivity. BMC Genet. 18:10. doi: 10.1186/s12863-017-0477-7
Ikawa, M., Ohya, E., Shimada, H., Kamijo, M., and Fukamachi, S. (2017). Establishment and maintenance of sexual preferences that cause a reproductive isolation between medaka strains in close association. Biol. Open 6, 244–251. doi: 10.1242/bio.022285
Johnson, A. M., Stanis, S., and Fuller, R. C. (2013). Diurnal lighting patterns and habitat alter opsin expression and colour preferences in a killifish. Proc. R. Soc. B Biol. Sci. 280:20130796. doi: 10.1098/rspb.2013.0796
Kamijo, M., Kawamura, M., and Fukamachi, S. (2018). Loss of red opsin genes relaxes sexual isolation between skin-colour variants of medaka. Behav. Processes 150, 25–28. doi: 10.1016/j.beproc.2018.02.006
Lewis, A., and Li, Z. (2006). Are cone sensitivities determined by natural color statistics? J. Vis. 6, 285–302. doi: 10.1167/6.3.8
Li, P. (2005). Circadian rhythms of behavioral cone sensitivity and long wavelength opsin mRNA expression: a correlation study in zebrafish. J. Exp. Biol. 208, 497–504. doi: 10.1242/jeb.01424
Lykke-Andersen, S., and Jensen, T. H. (2015). Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16, 665–677. doi: 10.1038/nrm4063
Marques, D. A., Taylor, J. S., Jones, F. C., Di Palma, F., Kingsley, D. M., and Reimchen, T. E. (2017). Convergent evolution of SWS2 opsin facilitates adaptive radiation of threespine stickleback into different light environments. PLoS Biol. 15:e2001627. doi: 10.1371/journal.pbio.2001627
Matsumoto, Y., Fukamachi, S., Mitani, H., and Kawamura, S. (2006). Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes). Gene 371, 268–278. doi: 10.1016/j.gene.2005.12.005
Matsuo, M., Ando, Y., Kamei, Y., and Fukamachi, S. (2018). A semi-automatic and quantitative method to evaluate behavioral photosensitivity in animals based on the optomotor response (OMR). Biol. Open 7:bio033175. doi: 10.1242/bio.033175
Melin, A. D., Chiou, K. L., Walco, E. R., Bergstrom, M. L., Kawamura, S., and Fedigan, L. M. (2017). Trichromacy increases fruit intake rates of wild capuchins (Cebus capucinus imitator). Proc. Natl. Acad. Sci. U.S.A. 114, 10402–10407. doi: 10.1073/pnas.1705957114
Neitz, J., and Neitz, M. (2011). The genetics of normal and defective color vision. Vision Res. 51, 633–651. doi: 10.1016/j.visres.2010.12.002
Neitz, J., Neitz, M., He, J. C., and Shevell, S. K. (1999). Trichromatic color vision with only two spectrally distinct photopigments. Nat. Neurosci. 2, 884–888. doi: 10.1038/13185
Neitz, M., and Neitz, J. (2014). Curing color blindness—mice and nonhuman primates. Cold Spring Harb. Perspect. Med. 4:a017418. doi: 10.1101/cshperspect.a017418
Nishiwaki, Y., Oishi, T., Tokunaga, F., and Morita, T. (1997). Three-dimensional reconstitution of cone arrangement on the spherical surface of the retina in the medaka eyes. Zoolog. Sci. 14, 795–801. doi: 10.2108/zsj.14.79
Sakai, Y., Kawamura, S., and Kawata, M. (2018). Genetic and plastic variation in opsin gene expression, light sensitivity, and female response to visual signals in the guppy. Proc. Natl. Acad. Sci. U.S.A. 115, 12247–12252. doi: 10.1073/pnas.1706730115
Sakai, Y., Ohtsuki, H., Kasagi, S., Kawamura, S., and Kawata, M. (2016). Effects of light environment during growth on the expression of cone opsin genes and behavioral spectral sensitivities in guppies (Poecilia reticulata). BMC Evol. Biol. 16:106. doi: 10.1186/s12862-016-0679-z
Shao, Y. T., Wang, F. Y., Fu, W. C., Yan, H. Y., Anraku, K., Chen, I. S., et al. (2014). Androgens increase lws opsin expression and red sensitivity in male three-spined sticklebacks. PLoS One 9:e100330. doi: 10.1371/journal.pone.0100330
Siebeck, U. E., Wallis, G. M., Litherland, L., Ganeshina, O., and Vorobyev, M. (2014). Spectral and spatial selectivity of luminance vision in reef fish. Front. Neural Circuits 8:118. doi: 10.3389/fncir.2014.00118
Solomon, S. G., and Lennie, P. (2007). The machinery of colour vision. Nat. Rev. Neurosci. 8, 276–286. doi: 10.1038/nrn2094
Sundin, J., Berglund, A., and Rosenqvist, G. (2010). Turbidity hampers mate choice in a pipefish. Ethology 116, 713–721. doi: 10.1111/j.1439-0310.2010.01787.x
Taylor, M. R., Kikkawa, S., Diez-Juan, A., Ramamurthy, V., Kawakami, K., Carmeliet, P., et al. (2005). The zebrafish pob gene encodes a novel protein required for survival of red cone photoreceptor cells. Genetics 170, 263–273. doi: 10.1534/genetics.104.036434
Thanaraj, T. A. (2001). Human GC-AG alternative intron isoforms with weak donor sites show enhanced consensus at acceptor exon positions. Nucleic Acids Res. 29, 2581–2593. doi: 10.1093/nar/29.12.2581
Thoreson, W. B., and Dacey, D. M. (2019). Diverse cell types, circuits, and mechanisms for color vision in the vertebrate retina. Physiol. Rev. 99, 1527–1573. doi: 10.1152/physrev.00027.2018
Tohya, S., Mochizuki, A., and Iwasa, Y. (2003). Difference in the retinal cone mosaic pattern between zebrafish and medaka: cell-rearrangement model. J. Theor. Biol. 221, 289–300. doi: 10.1006/jtbi.2003.3192
Tsujimura, T., Chinen, A., and Kawamura, S. (2007). Identification of a locus control region for quadruplicated green-sensitive opsin genes in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 104, 12813–12818. doi: 10.1073/pnas.0704061104
Utagawa, U., Higashi, S., Kamei, Y., and Fukamachi, S. (2016). Characterization of assortative mating in medaka: mate discrimination cues and factors that bias sexual preference. Horm. Behav. 84, 9–17. doi: 10.1016/j.yhbeh.2016.05.022
Valen, R., Karlsen, R., and Helvik, J. V. (2018). Environmental, population and life-stage plasticity in the visual system of Atlantic cod. J. Exp. Biol. 221:jeb165191. doi: 10.1242/jeb.165191
Viets, K., Eldred, K. C., and Johnston, R. J. (2016). Mechanisms of photoreceptor patterning in vertebrates and invertebrates. Trends Genet. 32, 638–659. doi: 10.1016/j.tig.2016.07.004
Ward, W. W., and Cormier, M. J. (1979). Energy transfer protein in coelenterate bioluminescence. J. Biol. Chem. 254, 781–788.
Watanabe, M., Furuya, M., Miyoshi, Y., Inoue, Y., Iwahashi, I., and Matsumoto, K. (1982). Design and performance of the okazaki large spectrograph for photobiological research. Photochem. Photobiol. 36, 491–498. doi: 10.1111/j.1751-1097.1982.tb04407.x
Yamanaka, O., and Takeuchi, R. (2018). UMATracker: an intuitive image-based tracking platform. J. Exp. Biol. 221:jeb182469. doi: 10.1242/jeb.182469
Young, T. (1802). The bakerian lecture: on the theory of light and colours. Philos. Trans. R. Soc. London 92, 12–48. doi: 10.1098/rstl.1802.0004
Keywords: medaka (Oryzias lapites), reverse genetics, color discrimination, spectral sensitivity, sensory drive, short wavelength sensitive gene
Citation: Kanazawa N, Goto M, Harada Y, Takimoto C, Sasaki Y, Uchikawa T, Kamei Y, Matsuo M and Fukamachi S (2020) Changes in a Cone Opsin Repertoire Affect Color-Dependent Social Behavior in Medaka but Not Behavioral Photosensitivity. Front. Genet. 11:801. doi: 10.3389/fgene.2020.00801
Received: 11 January 2020; Accepted: 06 July 2020;
Published: 12 August 2020.
Edited by:
Naoki Osada, Hokkaido University, JapanReviewed by:
Daisuke Kojima, The University of Tokyo, JapanIshwar Parhar, Monash University Malaysia, Malaysia
Copyright © 2020 Kanazawa, Goto, Harada, Takimoto, Sasaki, Uchikawa, Kamei, Matsuo and Fukamachi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shoji Fukamachi, ZnVrYW1hY2hpQGZjLmp3dS5hYy5qcA==
†These authors have contributed equally to this work
 Nodoka Kanazawa1†
Nodoka Kanazawa1† Tamaki Uchikawa
Tamaki Uchikawa Yasuhiro Kamei
Yasuhiro Kamei Shoji Fukamachi
Shoji Fukamachi