- 1Laboratório de Genética e Evolução (LabGEv), Programa de Pós-Graduação em Biologia Evolutiva, Departamento de Biologia Estrutural, Molecular e Genética, Universidade Estadual de Ponta Grossa, Ponta Grossa, Brazil
- 2Postgraduate Program in Evolutionary Genetics and Molecular Biology, Federal University of São Carlos, São Paulo, Brazil
Cichlids constitute a diverse monophyletic group that have developed adaptive strategies to thrive in diverse environments. Andinoacara represents an example of diversification on the South American Andean uplift, providing a key model for understanding the evolution of biogeographic patterns. In this study, we analyzed the species Andinoacara latifrons using two mitochondrial markers (COI, cytb) and one nuclear marker (RAG1) in a populational level. Sequences were obtained through tissue collection and from the GenBank database. Populational analysis showed significant structuration among populations, also corroborated with population pairwise Fst results. Fu’s Fs and Tajima’s D results showed populations that seems to be under populational expansion. We identified 22 haplotypes using cytb. The population associations in the Cauca haplotype are related to the Momposina depression and the mixture of the Cauca-Magdalena river basins in the lower Cauca-Magdalena region. We constructed a new phylogenetic tree, which grouped mainly two A. latifrons lineages: (1) an upper Magdalena and Catatumbo clade and (2) an upper Cauca and upper Magdalena clade. Thus, A. latifrons represents a diverse entity that contributes to our understanding of the evolutionary history of northern South America. Our findings provide insight into devising public policies in determining refuges for the preservation of biodiversity in the lower Cauca and Magdalena regions in Colombia.
Introduction
The Neotropical region shows exceptional species richness and endemism due to diversifications associated with its unique geological history resulting in a complex topography and climatic history, but is still poorly understood (Bagley et al., 2016). The family Cichlidae, which occur in the region, are the most diverse group within the order Perciformes. The Neotropical region is estimated to have around 450 species and 50 genera of cichlids (Kullander, 2003). It represents a monophyletic clade with an incompletely understood phylogeny (López-Fernández et al., 2010) that includes species from Central America, the Antilles, and South America (Smith et al., 2008). Paleontological evidence suggests that this teleost group did not diversify into its recognizable modern families and genera until the Late Cretaceous or Paleogene (Reis et al., 2016). The north of South America shows one of the highest levels of species richness after Brazil, with the ichthyofauna of the Magdalena River located between the eastern and western cordilleras of the Colombian Andes long recognized as characteristic of this richness (Cala, 1987). Geologic processes involved in the formation of the trans-Andean region (it includes Panamá, Atrato-Pacific slope, Magdalena and Maracaibo basins) (Albert et al., 2006) contributed to the richness of this region, providing an important framework for addressing phylogeographic connections associated with the hydrographic history of the region (Rincon-Sandoval et al., 2019). In addition, the Andean uplift generated a biological dynamic that allows us to explain life histories that can be elucidated under evolutionary concepts of speciation, geographic isolation, vicariance, population dynamics, and others. Andinoacara represents a key example of these dynamics. The evolutionary history of cichlids could have originated from river-dwelling taxa, since they are the most diverse and geographically widespread and postulated to have maintained ancestral geographic patterns and distributions (Kullander, 1988).
In order to understand the Andinoacara species cluster, Musilová et al. (2008), confirmed that the polyphyletic group Aequidens pulcher rivulatus represents an unnamed genus with strong support through phylogenetic analysis. Subsequently, Musilová et al. (2009) designated this group as a new genus (Andinoacara) belonging to the tribe Cichlasomatini that includes eight valid species (Musilová et al., 2009). The processes of diversification within the Andinoacara group could have been governed by vicariance events, as is the case in many cichlids in the trans-Andean region. Thus, the distribution of Andinoacara ranges from the Magdalena Basin to the trans-Andean region of South America, including the northeast trans-Andean region (Maracaibo basin) and the south of Lima in central Perú (Musilová et al., 2015).
Andinoacara latifrons was originally described within Aequides, a genus consistently designated as a polyphyletic group (Musilová et al., 2008; Smith et al., 2008). Musilová et al. (2015) reconstructed the history of some trans-Andean cichlids using the mitochondrial gene cytochrome b (cytb), including several species in the genus Andinoacara. In this study, they found that A. latifrons form separate clades in Magdalena, Sinú, and Cauca, supporting allopatry by mtDNA. The synonyms of Andinoacara latifrons are Aequidens latifrons and Acara latifrons, and the type locality of the species is the Magdalena River, Colombia. The distribution of A. latifrons is centered in Colombia, in the Magdalena, Atrato, Sinú, Cauca, and San Juan basins (Kullander, 1998). The species reaches a maximum length of 17 cm. The conservation status of these species is least concern and there are no conservation measures in the habitat regions for this species. It does not occur in protected areas, but some areas of presence of this species are in risk of thread due to contamination of some industries like mining and petroleum. Mining projects have been developing in the region of the Upper Magdalena which in some years could generate an effect of loss of biodiversity, including the endemic fauna. This reason and the poor studies of the dynamic of this species, including reproduction, behavior, and the use of genetic and molecular markers for populational analysis allows designate to A. latifrons as a reference specie to preserve areas as the Depresión Momposina by anthropization damages.
To better understand the taxonomic and biogeographic evolutionary history of the species, it is important to understand and to analyze genetic patterns and the influence of the processes of diversification driven by geographic barriers. This is important insofar endemic species support the diversity of a region by their richness, also, population studies can help to recognize genetic patterns that show the level of mixture and structuration; it can be useful to track the states of endemic species like A. latifrons and avoid its extinction in some areas or habitat degradation.
In this study, we analyzed the genetic and haplotype diversity of A. latifrons at the population level using mitochondrial [cytochrome oxidase 1 (COI) and cytb] and nuclear [recombination activating gene (RAG1)] markers. As A. latifrons is an important model species, evolutionarily significant units (ESUs) (Ryder, 1986) could be established for conservation purposes, as well as to elucidate evolutionary processes that reflect the history of drainage basin formation.
Materials and Methods
Sample Collection
The material used and amplified for posterior analysis was acquired from the Ichthyological section of the Zoological Collections of the University of Tolima [Colección Zoológica Universidad del Tolima, Ictiología, Ibagué, Colombia (CZUT-IC)]. We also used cytb sequences from the GenBank database (Clark et al., 2016). Detailed information of specimens (including region, authors, accession number, and gene) are summarized in Supplementary Material 1.
DNA Extraction and Amplification
DNA was extracted from muscle tissue preserved in 96% ethanol through the phenol-chloroform method (Sambrook and Russell, 2006). To perform molecular systematics analysis, the mitochondrial genes COI and cytb, and the nuclear gene RAG1 were used. To amplify ca. 610 pb COI we used the primers FishF1 5′-TCAACCAACCACAAAGACATTGGCAC-3′ and Fish R1 5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′ (Ward et al., 2005). The PCR conditions in 25 μL were as follows: 2.5 μL of Buffer (10X), 1.25 μL of MgCl2 (50 mM), 0.5 μL of mix dNTPs (10 mM), 0.5 μL of each primer (10 mM), 0.2 μL of Polymerase Taq (1U), 17.3 μL of ultrapure water and 2.5 of DNA template (50 ng/μL). The thermal profile consisted of an initial denaturation of 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 45 s at 52°C and 1 min at 72°C, and a final extension of 10 min at 72°C. The fragment of 940 pb of the gene cytb was amplified with primers Fishcytb-F 5′-ACCACCGTTGTTATTCAACTACAAGAAC-3′ and Truccytb-R 5′-CCGACTTCCGGATTACAAGACCG-3′ (Sevilla et al., 2007). The PCR conditions in 25 μL were: 2.5 μL of Buffer (10X), 0.8 μL of MgCl2 (50 mM), 0.5 μL of mix dNTPs (10 mM), 0.5 μL of each primer (10 mM), 0.2 μL of Polymerase Taq (1U), 18.1 μL of ultrapure water and 2.5 of DNA template (50 ng/μL). The thermal profile consisted of an initial denaturation of 2 min at 94°C, followed by 36 cycles of 1 min of denaturation at 94°C, 1 min at 55°C, 1 min at 72°C, and a final extension of 8 min at 72°C. The nuclear gene RAG1 was amplified with the primers forward 5′-CTGAGCTGCAGTCAGTACCATAAGATGT-3′ and reverse 5′-CTGAGTCCTTGTGAGCTTCCATRAAYTT-3′ (Grande et al., 2004). The conditions and the thermal profile to amplify the 1481 pb of PCR product were the same as those used for the cytb gene. The PCR products were purified with the Kit Illumina GFXTM PCR DNA and Gel Band Purification (GE HealthCare), following the manufacturer’s instructions. PCR products were sequenced by Macrogen Inc. (Korea) and ACTGene Análises Moleculares (Rio Grande do Sul, Brazil). After sequencing, each electropherogram was verified manually.
Populational Analysis
We selected the cytb marker to perform haplotype and genetic analysis due to its large number of sequences, we use 55 sequences for analysis (see Supplementary Material 1). We performed an analysis of molecular variance (AMOVA) considering seven groups according to proximity in basins [Group 1 (loMg, Nechi, Jorge, loCau); Group 2 (Sinu); Group 3 (Cesar, SNSM); Group 4 (UpCau); Group 5 (UpMg); Group 6 (Atrato); Group 7 (Catatumbo)], population pairwise Fst, and Fu’s Fs and Tajima’s D statistics using the Arlequin software (Excoffier et al., 2007). A haplotype network was constructed using the TCS method (Clement et al., 2000) implemented in the software PopArt v.1.7 (Leigh and Bryant, 2015). Populations were labeled as: loMg, lower Magdalena; Nech, Nechí; Sinu, Sinu; Ces, Cesar; UpCau, Upper Cauca River; Jorge, San Jorge River; UpMg, Upper Magdalena; Atr, Atrato; Cat, Catatumbo River; SNSM, coastal rivers of Sierra Nevada de Santa Marta; loCau, lower Cauca.
Phylogenetic Analysis
A Bayesian tree was constructed using a concatenated matrix from the genes COI, cytb, and RAG1 with a sample of 15 sequences and a total of 2,778 pair bases (see Supplementary Material 1). We ran 500,000 MCMC simulations, discarding a 20% burn-in. The Akaike Information Criterion, corrected (AICc) was used to select the model of molecular evolution (COI = HKY and cytb = TN93 + 5 gamma categories; RAG1 = K2 + 5 gamma categories) using the jModelTest software (Posada, 2008). Mr. Bayes v.3.2.6 (Huelsenbeck and Ronquist, 2001) was used to construct the tree. Phylogenetic trees from cytb, COI, and RAG1 were calculated and also documented in Supplementary Material 2. Clade visualization and editing was performed in Fig Tree v1.4.2 (Rambaut, 2012). Adobe Photoshop CS6 (v 13.0) was used to improve the trees output from the analyses for use in subsequent figures.
Results
DNA sequences of 907 pb of the mitochondrial cytb gene were used from a total of 55 sequences (16 sequences from this study and 35 sequences from Musilová et al., 2009), a fragment of 436 pb of the mitochondrial gene COI (16 sequences), and 1436 pb from a nuclear gene RAG1. The nucleotide diversity of the entire dataset of each gene were cytb = 0.021, COI = 0.20, and RAG1 = 0.0024.
Populational Analysis
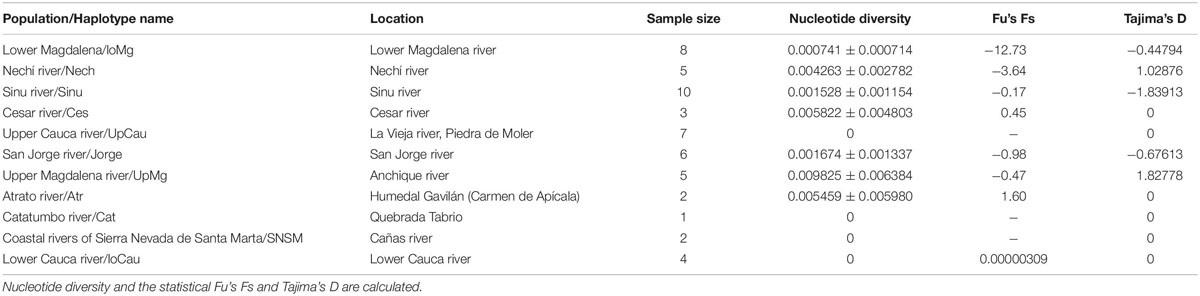
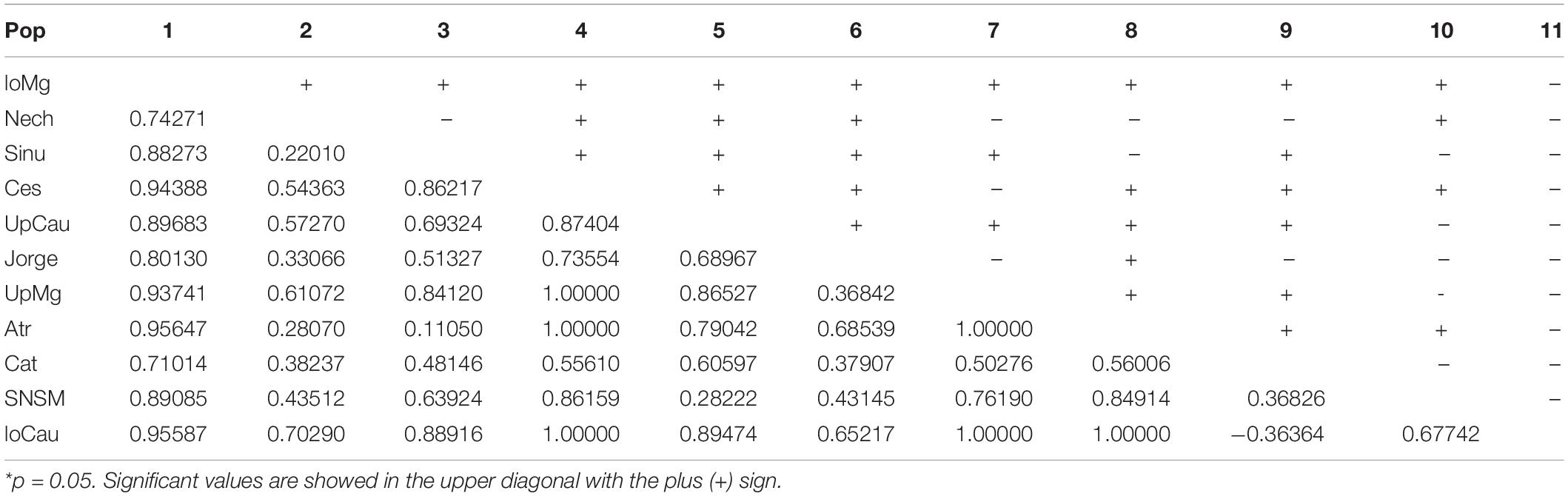
The statistical results of nucleotide diversity, Fu’s Fs and Tajima’s D indexes for the cytb gene are summarized in Table 1. Negative values for the Fu’s Fs index show purifying selection or populational expasion (lower Magdalena, Nechí, Sinu, San Jorge, Upper Magdalena river), while positive values (Ces and Atr) represent deficiency of alleles; non-calculated values in some populations can be related with presence of unique sequences. The Tajima’ D also reinforce the populational expansion in the lower Magdalena, Sinú, and San Jorge populations. The AMOVA showed the highest variation among population and groups (64.49%) (Table 2); also, fixation indexes were more significant among and within populations. Population pairwise Fst results are showed in the Table 3, the highest values between populations are related with the higher distances among rivers in the different basins.
For the mitochondrial gene cytb, we found 84 segregating sites, and 66 parsimony-informative sites; the Tajima’s D of the entire dataset showed a result of D = -0.393, indicating an excess of low frequency of polymorphic sequences, it could signify possible expansion or purifying events, as showed specifically in Table 1 for each population. Likewise, we found 22 different cytb haplotypes. Haplotypes results indicate little unique share haplotypes between populations (Figure 1), and slight differences with the UpCauca + San Jorge + Nechí + Cesar + Sinu + UpMg haplotypes constitute an important structure belonging to the Cauca River basin, and could be considered the main ancestor haplotype (the dotted haplotype; Figure 1). It means that the region of the Upper Cauca located on the Western Cordillera and the Upper Magdalena located on the Eastern Cordillera represent an ancient haplogroup with colonizations to the downstream in the past. In addition, direct associations of the populations UpMag and Catatumbo are related to the history of the two basins, due to the uplifting of the central and eastern Cordilleras. The Catatumbo basin produced a more isolated group, induced by a higher rate of mutational steps, mainly explained by distance isolation.
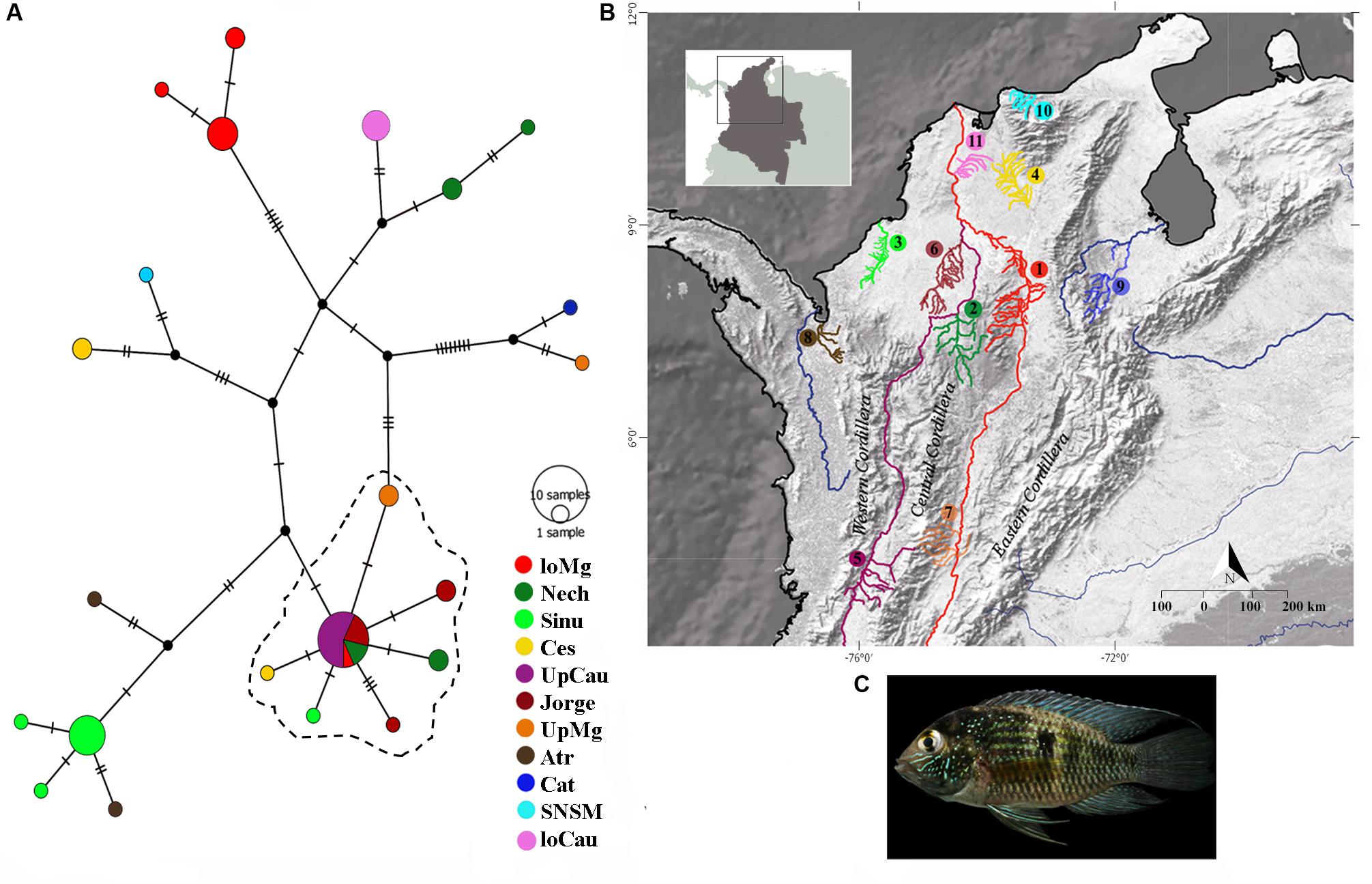
Figure 1. Haplotype distributions of Andinoacara latifrons constructed using the mitochondrial gene cytochrome b (cytb). (A) Haplotype network with main localities; dotted lines indicate most similar haplotypes. (B) Geographic distribution of samples. loMag, lower Magdalena; Nech, Nechí; Sinu, Sinu; Ces, Cesar; UpCau, Upper Cauca river; Jorge, San Jorge river; UpMg, Upper Magdalena; Atr, Atrato; Cat, Catatumbo river; SNSM, coastal rivers of Sierra Nevada de Santa Marta; loCau, lower Cauca. (C) Photo: Andinoacara latifrons.
Phylogenetic Analysis
The concatenated tree grouped two main clades with the highest supporting values (Figure 2): (1) a UpMg + Cat clade and (2) a clade from UpCau + UpMg, representing mostly the Magdalena and Cauca river basin, respectively. Separate trees from cytb, COI, and RAG1 are in Supplementary Material 2, showing similar topologies among them. Even without a large number of sequences, the combination of mitochondrial (cytb + COI) and nuclear (RAG1) genes highlighted these main clades that correspond to the geological and biogeographic history of the region and shows that the evolution of A. latifrons was driven by the geological history of the drainage basin, corroborating the proposal of Musilová et al. (2008). The two main clades described can be lineages seen as evolutionary units which represent important river basins, principally the Magdalena-Cauca river basin that is constantly under contamination and pollution caused by human actions.
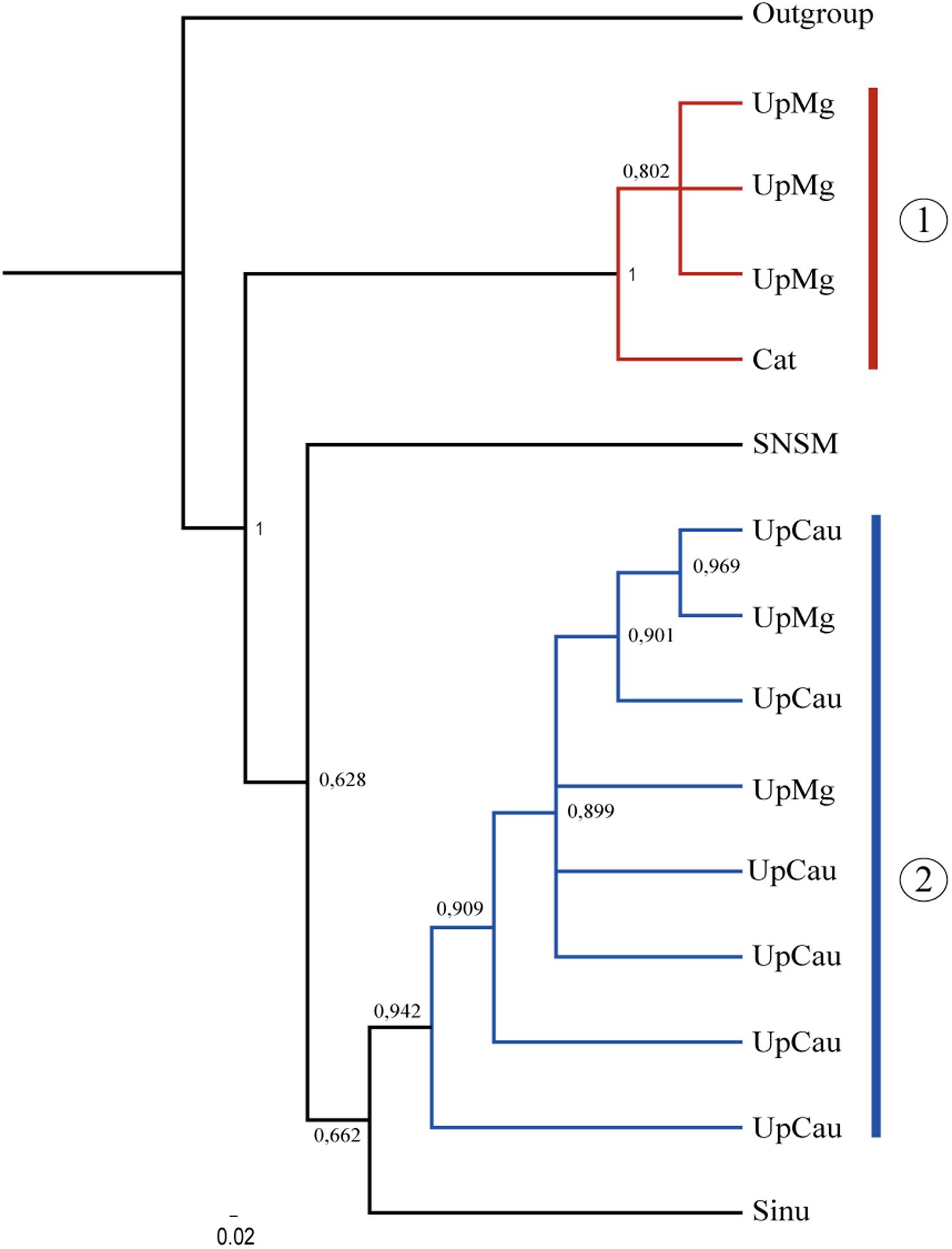
Figure 2. Phylogenetic tree of A. latifrons constructed with the mitochondrial genes: cytb + COI and the nuclear rag1. Branch support values are represented in Arabic numbers. Sinu, sinu; UpMg, Upper Magdalena; Cat, Catatumbo; SNSM, Coastal rivers of Sierra Nevada de Santa Marta; loCau, lower Cauca; UpCau, Upper Cauca; Outgroup, Geophagus steindachneri. The two main clades are shown.
Discussion
The trans-Andean species A. latifrons, as type species of the genus Andinoacara (Musilová et al., 2009), represents an endemic entity with distribution in the Magdalena, Cauca, and Caribbean basins, as well as the Catatumbo River. The haplotype diversity of the cytb gene from the Colombian populations have proven to be congruent with the geological history of this region. Thus, different haplotypes allow us locate each population in a determined region where the higher structuration appears in the farthest places which the genetic flux is lower (e.g., haplotype of the SNSM and Cat) of the ancestral haplotypes of UpMg and UpCau as corroborated with Fst values (Table 3). This framework can help to inform public policies for the conservation of Colombian biomes through evidences of some species like A. latifrons, where the most isolated haplotypes constitute important diversity units of this species.
In this study, the populations of the upper Magdalena-Cauca had an association with the Caribbean basins, but not too strong which is explained by the configuration and presence of Serranias, Sierras, and mountains, isolated from the Colombian Andes. This association allow us to draw explanations on aspects of the biological evolution of these populations (Castaño-Uribe, 1999). The well supported UpMg + Cat and UpCau + UpMg clades (Figure 2) in the Bayesian tree may explain precisely this configuration around the natural history of the Andes and the dynamics of freshwater fauna highlighting also, an ancient lineage that converge with the original description of A. latifrons from Magdalena river basin.
The convergence zone in the Depresión Momposina (Herrera et al., 2001) allows some admixture of populations from the lower Cauca-Magdalena basin with populations around this depression (San Jorge, Nechí and lower Magdalena, Figure 1) with populations located more upstream. It constitutes a hybridization area that can validate some of the Andean orogeny and the previous age of populations of A. latifrons. However, population structuring is seen in many populations when compared to each other (Table 3). This conclusion was drawn from the comparison of the ages of the last orogenic processes in the Andean Cordillera (12–10 Ma) and the vicariance of A. latifrons and A. pulcher (∼3.3 Ma).
The Sierra Nevada de Santa Marta (SNSM) is surrounded by the Caribbean Sea to the north, the Cesar River Valley to the southeast, and the Magdalena River to the southwest (Macdonald and Hurley, 1969). This geographic setting helps explain the shared histories of the populations of Ces and SNSM (Figure 1) and also, the structuration of the SNSM and UpMg populations. The continental margin of the Caribbean has been exposed to a wide variety of orogenic processes (Reyes et al., 2004) that have invoked changes in water bodies, consequently influencing the associated fauna. Based on a range of information on organism dynamics, the use of whole-genome sequence (WGS) data offers a high molecular resolution but is costly (Cushman et al., 2018). In order to generate an approximation of evolutionary histories and genetic inferences, we selected to use mitochondrial and nuclear genetic markers that offered a base line and good resolution of haplotype histories in A. latifrons. This is useful not only in ecological terms but also to subsequently understand behavioral, reproductive, morphological and phylogenetic traits of different populations. Thus, as a first approach to understand the complexity of this species and its behavior as an endemic species, it was quite efficient to know at least a little, from the genetic and phylogenetic point of view, the distribution and possible expansion events of A. latifrons. In addition, it reinforces the idea of the richness in the Colombian basins and how they support the existing biodiversity.
Implications for Conservation
Developing conservation strategies to establish priority units has historically been difficult. Genetic studies are constantly changing what we know about evolutionary processes. For this reason, the International Union for Conservation of Nature (IUCN, 2020) considers genetic variability an important factor in planning conservation programs (McNeely et al., 1991). It is estimated that the world has more genetic variability than we currently know of, and therefore taxonomic units have to be conserved considering these aspects.
Frankham (2010) highlights three contribution levels made by genetics and molecular biology to conservation genetics. The first level is the conservation objective. Secondly, genetic factors contribute to the risk of extinction through inbreeding depression, loss of genetic diversity, and loss of evolutionary potential. Thirdly, genetic diversity impacts the survival of ecosystems, their function, and their diversity. When we think about these considerations, we need to discuss the units to be conserved, so that ESUs are considered as stocks that deserve separate management and have a high conservation priority. To preserve evolutionary processes, management must preserve natural networks of genetic connections between populations, rather than just as isolated populations within that network (as we point in the Colombian Depresión Momposina). Genetic techniques are also essential, because they provide estimates of genetic flow between populations and this could guide efforts to maintain historical levels of genetic exchange between populations (Crandall et al., 2000).
Genomic data confer better results than microsatellite data to delineate Conservation Units (CUs) since genomic data can quantify adaptive variation (Funk et al., 2012). However, more work is needed in the development of new analyses to delineate CUs and to test adaptive differentiation. For example, an ideal analysis could delineate the ESUs and Management Units (MUs), and quantify the adaptive differentiation between these different hierarchical CUs in a single analytical framework, analogous to the current Bayesian MCMC approaches developed to simultaneously infer phylogenies, time of divergence and historical demographic parameters of the given sequence (Drummond et al., 2012). This focus could be important in delineating conservation strategies in the Magdalena-Cauca and Caribbean basins of Colombia mainly by the increasing of mining.
Fish living in high energy and highly oxygenated mountain rivers, such as Astroblepus (Astroblepididae), Chaetostoma (Loricariidae), and Trichomycterus (Trichomycteridae) (Reis et al., 2016), are ecologically important in the upper part of the Magdalena basin. Andinoacara latifrons is a species targeted by artisanal fishermen, although the IUCN does not consider it as at risk. However, the IUCN states that the species could be affected by sedimentation and pollution related to deforestation (logging, land use by agriculture, and livestock) and to a lower degree, mining (through associated pollution and sedimentation) that could also affect the drainage of the Magdalena at medium and low altitudes. Just below the Magdalena, in the Momposina basin, ∼80% of the Ciénagas are concentrated in depressions in the rocky base of the Late Cretaceous deposits, with stagnant or river-dependent bodies of water accumulating sediments. The muddy plumes of the Magdalena River and the Canal del Dique greatly affect the quality of the water, and the combination of mud and reduced salinity may have caused serious damage to coral reefs near where the river flows into the ocean. Data indicate that the Magdalena River contributes ∼10% of the total sediment load discharged from the east coast of South America (Restrepo et al., 2006).
Morphological adaptations of Andean fishes have helped them to colonize and survive at different altitudes (Anderson and Maldonado-Ocampo, 2011). This contributes to the higher richness of Andean fishes found in Colombia. Although cichlids are not the largest teleost group in the region, they are of importance to conservation; in addition, their evolutionary history in South America is correlated to the orogenic history of Andes, this they are considered the ideal model animals for evolutionary study. The high diversity of cichlids and recent environmental degradation in this region suggest that separate conservation strategies are needed for different basins (Figure 1). Andinoacara latifrons is found on flood plains and in water that is directly influenced by sedimentation and oxygenation processes. These factors have the greatest impact in the low Magdalena-Cauca region with important repercussions occurring in the Caribbean, including corals reef systems. Andinoacara latifrons is an abundant species that highlights the evolutionary history of the Colombian Andes, and that is being affected by environmental contamination. The niche preference of this non-migratory species makes its conservation important and suggests the need for implementation of public policies that promote good practice in mineral extraction.
Conclusion
In conclusion, our study highlights the importance of recognizing genetic diversity in endemic species, such as A. latifrons, not only for the conservation of these species, but also to better understand the taxonomy and historical pattern of geographical colonization. Our findings provide insight into devising public policies in determining refuges for the preservation of biodiversity, including regions of the Lower Cauca and Magdalena river basins in Colombia.
Data Availability Statement
The datasets generated for this study can be found in the GenBank MN514625, MN514615, MN514616, MN514613, MN514614, MN514612, MN514611, MN514623, MN514619, MN514621, MN514624, MN514609, MN514622, MN514620, MN514618, MN514610, MN514617, MN563598, MN563588, MN563590, MN563585, MN563586, MN563584, MN563583, MN563596, MN563592, MN563594, MN563597, MN563579, MN563595, MN563593, MN563591, MN563582, MN563599, MN514595, MN514592, MN514590, MN514597, MN514594, MN514600, MN514596, MN514599, MN514598, MN514602, MN514606, MN514591, MN514608, MN514601, MN514593, MN514607, and MN514589.
Ethics Statement
The animal study was reviewed and approved by the COEP (Comissão de Ética em Pesquisa, Sub-Comissão de Ética em Animais) Universidade Estadual de Ponta Grossa.
Author Contributions
LD performed the analysis under supervision of RA and MS. All authors helped in the writing and analysis of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledged to Colección Zoológica, Sección de Ictiología de la Universidad del Tolima, and his Curator Francisco Villa for donating tissues. Thanks to Christian Conde for identifying some material and Edison Duarte for collaboration in the field. Thanks to Jorge E. García-Melo (www.jorgegarciabiophoto.com) by providing a photo of A. latifrons. Thanks to CAPES and OEA for supporting the grant for LD. Thanks to CNPq for the financial support and special thanks to the team of the Laboratory of Evolutionary Biology (LabGEv).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00815/full#supplementary-material
References
Albert, J. S., Lovejoy, N. R., and Crampton, W. G. R. (2006). Miocene tectonism and the separation of cis- and trans-Andean river basins: evidence from Neotropical fishes. J. South Am. Earth Sci. 21, 14–27. doi: 10.1016/j.jsames.2005.07.010
Anderson, E. P., and Maldonado-Ocampo, J. A. (2011). A regional perspective on the diversity and conservation of tropical Andean fishes. Conserv. Biol. 25, 30–39. doi: 10.1111/j.1523-1739.2010.01568.x
Bagley, J. C., Matamoros, W. A., Mcmahan, C. D., Tobler, M., Chakrabarty, P., and Johnson, J. B. (2016). Phylogeography and species delimitation in convict cichlids (Cichlidae: Amatitlania): implications for taxonomy and Plio-Pleistocene evolutionary history in Central America. Biol. J. Linn. Soc. 120, 155–170. doi: 10.1111/bij.12845
Cala, P. (1987). Ictiofauna Dulceacuícolede Colombia. 84. Available online at: http://www.accefyn.org.co/revista/Volumen_16/62/69-84.pdf (accessed June 15, 2019).
Castaño-Uribe, C. (1999). Sierras y Serranías de Colombia. Cali: Colección Ecológica Banco de Occidente, 205.
Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Sayers, E. W. (2016). Genbank. Nucleic Acids Res. 44, D67–D72. doi: 10.1093/nar/gkv1276
Clement, M., Posada, D., and Crandall, K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Crandall, K. A., Bininda-emonds, O. R. P., Mace, G. M., and Wayne, R. K. (2000). Considering evolutionary processes in conservation biology. TREE 15, 290–295. doi: 10.1016/s0169-5347(00)01876-0
Cushman, S. A., Shirk, A. J., Howe, G. T., Murphy, M. A., Dyer, R. J., and Joost, S. (2018). Editorial: the least cost path from landscape genetics to landscape genomics: challenges and opportunities to explore NGS data in a spatially explicit context. Front. Genet. 9:215. doi: 10.3389/fgene.2018.00215
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Excoffier, L., Laval, G., and Schneider, S. (2007). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50.
Frankham, R. (2010). Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 143, 1919–1927. doi: 10.1016/j.biocon.2010.05.011
Funk, W. C., McKay, J. K., Hohenlohe, P. A., and Allendorf, F. W. (2012). Harnessing genomics for delineating conservation units. Trends Ecol. Evol. 27, 489–496. doi: 10.1016/j.tree.2012.05.012
Grande, T., Laten, H., and López, J. A. (2004). Phylogenetic relationships of extant Esocid species (Teleostei: Salmoniformes) based on mophological and molecular characters. Copeia 4, 743–757. doi: 10.1643/CG-04-007R1
Herrera, L. F., Sarmiento, G., Romero, F., Botero, P. J., and Berrio, J. C. (2001). Evolución ambiental de la depresión momposina (Colombia) desde el pleistoceno tardio a los paisajes actuales. Geol. Colomb. 26, 95–121.
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
IUCN (2020). The IUCN Red List of Threatened Species. Version 2020-1. Available online at: https://www.iucnredlist.org (accessed March 19, 2020).
Kullander, S. O. (1988). Teleocichla, a new genus of south american rheophilic Cichlid fishes with six new species (Teleostei: Cichlidae). Copeia 1988, 196–230. doi: 10.2307/1445938
Kullander, S. O. (1998). “A phylogeny and classification of the South American Cichlidae (Teleostei: Perciformes),” Phylogeny and Classification of Neotropical Fishes, eds Malabarba, L. R., Reis, R. E., Vari, R. P., Lucena, Z. M. S., and Lucena, C. A. S. (Porto Alegre: Edipucrs), 461–498.
Kullander, S. O. (2003). “Family Cichlidae (Cichlids),” in Check List of the Freshwater Fishes of South and Central America, eds R. E. Reis, S. O. Kullander, and C. J. Ferraris Jr. (Porto Alegre: EDIPUCRS), 605–654.
Leigh, J. W., and Bryant, D. (2015). POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
López-Fernández, H., Winemiller, K. O., and Honeycutt, R. L. (2010). Multilocus phylogeny and rapid radiations in Neotropical cichlid fishes (Perciformes: Cichlidae: Cichlinae). Mol. Phylogenet. Evol. 55, 1070–1086. doi: 10.1016/j.ympev.2010.02.020
Macdonald, W. D., and Hurley, P. M. (1969). Precambrian gneisses from northern Colombia, South America. Bull. Geol. Soc. Am. 80, 1867–1872.
McNeely, J. A., Miller, K. R., Reid, W. V., Mittermeier, R. A., and Werner, T. B. (1991). Conserving the World’s Biological Diversity. Gland: IUCN.
Musilová, Z., Říčan, O., Janko, K., and Novák, J. (2008). Molecular phylogeny and biogeography of the Neotropical cichlid fish tribe Cichlasomatini (Teleostei: Cichlidae: Cichlasomatinae). Mol. Phylogenet. Evol. 46, 659–672. doi: 10.1016/j.ympev.2007.10.011
Musilová, Z., Říčan, O., and Novák, J. (2009). Phylogeny of the neotropical cichlid fish tribe cichlasomatini (Teleostei: Cichlidae) based on morphological and molecular data, with the description of a new genus. J. Zool. Syst. Evol. Res. 47, 234–247. doi: 10.1111/j.1439-0469.2009.00528.x
Musilová, Z., Říčan, O., Říčanová, Š, Janšta, P., Gahura, O., and Novák, J. (2015). Phylogeny and historical biogeography of trans-Andean cichlid fishes (Teleostei: Cichlidae). Vertebr. Zool. 65, 333–350.
Posada, D. (2008). jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. doi: 10.1093/molbev/msn083
Rambaut, A. (2012). FigTree v. 1.4.0. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed September 20, 2018).
Reis, R. E., Albert, J. S., Di Dario, F., Mincarone, M. M. M., Petry, P. L., and Rocha, L. R. (2016). Fish biodiversity and conservation in South America. J. Fish Biol. 89, 12–47. doi: 10.1111/jfb.13016
Restrepo, J. D., Zapata, P., Diaz, J. M., Garzo, J., and García, C. B. (2006). Fluvial fluxes into the Caribbean Sea and their impact on coastal ecosystems: the Magdalena River. Colombia 50, 33–49. doi: 10.1016/j.gloplacha.2005.09.002
Reyes, H. A., Montenegro, B. M., and Gómez, P. D. (2004). Tecnoestatigrafía y evolución geológica. Boletín Geol. 26, 19–38.
Rincon-Sandoval, M., Betancur, R. R., and Maldonado-Ocampo, J. A. (2019). Comparative phylogeography of trans-Andean freshwater fishes based on genome-wide nuclear and mitochondrial markers. Mol. Ecol. 28, 1096–1115. doi: 10.1111/mec.15036
Ryder, O. A. (1986). Species conservation and systematics: the dilemma of the subspecies. Trends Ecol. Evol. 1, 9–10. doi: 10.1016/0169-5347(86)90059-5
Sambrook, J., and Russell, D. W. (2006). Purification of nucleic acids by extraction with Phenol:Chloroform. Cold Spring Harb. Protoc 2006:db.rot4455. doi: 10.1101/pdb.prot4455
Sevilla, R. G., Diez, A., Norén, M., Mouchel, O., Jérôme, M., Verrez-Bagnis, V., et al. (2007). Primers and polymerase chain reaction conditions for DNA barcoding teleost fish based on the mitochondrial cytochrome b and nuclear rhodopsin genes. Mol. Ecol. Notes 7, 730–734. doi: 10.1111/j.1471-8286.2007.01863.x
Smith, W. L., Chakrabarty, P., and Sparks, J. S. (2008). Phylogeny, taxonomy, and evolution of Neotropical cichlids (Teleostei: Cichlidae: Cichlinae). Cladistics 24, 625–641. doi: 10.1111/j.1096-0031.2008.00210.x
Keywords: cichlids, isolation, Cauca-Magdalena basin, distribution, conservation
Citation: De la Ossa-Guerra LE, Santos MH and Artoni RF (2020) Genetic Diversity of the Cichlid Andinoacara latifrons (Steindachner, 1878) as a Conservation Strategy in Different Colombian Basins. Front. Genet. 11:815. doi: 10.3389/fgene.2020.00815
Received: 28 November 2019; Accepted: 07 July 2020;
Published: 24 July 2020.
Edited by:
Badri Padhukasahasram, Illumina, United StatesReviewed by:
Silvia Perea, National Museum of Natural Sciences (MNCN), SpainVera Nisaka Solferini, State University of Campinas, Brazil
Copyright © 2020 De la Ossa-Guerra, Santos and Artoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Ferreira Artoni, cmZhcnRvbmlAZ21haWwuY29t
 Luz Elena De la Ossa-Guerra
Luz Elena De la Ossa-Guerra Mateus Henrique Santos
Mateus Henrique Santos Roberto Ferreira Artoni
Roberto Ferreira Artoni