- 1Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia
- 2A. N. Severtsov Institute of Ecology and Evolution, Russian Academy of Sciences, Moscow, Russia
- 3National Medical Research Radiological Center, Ministry of Health of the Russian Federation, Moscow, Russia
- 4National Medical Research Center for Obstetrics, Gynecology and Perinatology Named After Academician V.I. Kulakov, Ministry of Health of the Russian Federation, Moscow, Russia
The NETO2 gene (neuropilin and tolloid-like 2) encodes a protein that acts as an accessory subunit of kainate receptors and is predominantly expressed in the brain. Upregulation of NETO2 has been observed in several tumors; however, its role in tumorigenesis remains unclear. In this study, we investigated NETO2 expression in breast, prostate, and colorectal cancer using quantitative PCR (qPCR), as well as the effect of shRNA-mediated NETO2 silencing on transcriptome changes in colorectal cancer cells. In the investigated tumors, we observed both increased and decreased NETO2 mRNA levels, presenting no correlation with the main clinicopathological characteristics. In HCT116 cells, NETO2 knockdown resulted in the differential expression of 17 genes and 2 long non-coding RNAs (lncRNAs), associated with the upregulation of circadian rhythm and downregulation of several cancer-associated pathways, including Wnt, transforming growth factor (TGF)-β, Janus kinase (JAK)-signal transducer and activator of transcription (STAT), mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathways. Furthermore, we demonstrated the possibility to utilize a novel model organism, short-lived fish Nothobranchius furzeri, for evaluating NETO2 functions. The ortholog neto2b in N. furzeri demonstrated a high similarity in nucleotide and amino acid sequences with human NETO2, as well as was characterized by stable expression in various fish tissues. Collectively, our findings demonstrate the deregulation of NETO2 in the breast, prostate, and colorectal cancer and its participation in the tumor development primarily through cellular signaling.
Introduction
NETO2 (neuropilin and tolloid-like 2) was first described in 2001 as a gene demonstrating significant similarity to NETO1 (Stohr et al., 2002). Both genes encode putative transmembrane proteins containing two CUB domains, followed by a low-density lipoprotein class A (LDLa) module in the extracellular region and conserved FXNPXY-like motif in the cytoplasmic region. This structure of NETO proteins suggested their possible role in intracellular signaling pathways. Furthermore, a series of reports have shown the expression of NETO2 in the brain, as well as its involvement in the modulation of most kainate receptors (KARs) (Straub et al., 2011) and N-methyl-D-aspartate (NMDA) receptors (Wyeth et al., 2014). Additionally, the interaction of NETO2 with other neuronal proteins, including glutamate receptor-interacting protein (GRIP) (Tang et al., 2012) and K(+)-Cl(−) cotransporter (KKC2), has been reported (Ivakine et al., 2013). Currently, NETO2 and its paralog NETO1 are widely recognized as the main auxiliary subunits of KARs.
More recently, NETO2 was shown to be involved in carcinogenesis. Increased expression of the NETO2 gene has been observed in proliferating infantile hemangiomas (Calicchio et al., 2009), hepatocellular (Villa et al., 2016) and nasopharyngeal carcinomas (He et al., 2019), as well as in renal (Snezhkina et al., 2018), lung (Oparina et al., 2012), colorectal (Hu et al., 2015; Fedorova et al., 2017), gastric (Liu et al., 2019), and pancreatic (Li et al., 2019) cancers. Hu et al. (2015) have revealed that NETO2 overexpression significantly correlates with advanced tumor stage, invasion, and metastasis, as well as increases the risk of patient death in colorectal cancer. In patients with gastric and pancreatic cancers, a similar correlation was observed between NETO2 expression and tumor progression and worse overall survival (Li et al., 2019; Liu et al., 2019). Reportedly, NETO2 is related to five-gene transcriptomic signatures predicting rapidly growing tumors and survival in patients with hepatocellular carcinoma (Villa et al., 2016). Furthermore, NETO2 has been observed in a macrophage-related gene signature predicting resistance to targeted therapeutics, as well as survival, in glioma patients (Sun et al., 2019). Additionally, NETO2 expression is reportedly associated with the deletion of the PTEN (Phosphatase and tensin homolog) gene and high intratumor heterogeneity in prostate cancer (Yun et al., 2019).
In this study, we analyzed NETO2 expression in prostate, breast, and colorectal cancer samples obtained from Russian patients. In these sample sets, both increased and decreased NETO2 expression were observed, with no significant correlation between its mRNA levels and primary clinicopathological tumor characteristics. Additionally, we performed shRNA-mediated NETO2 knockdown in the colorectal cancer cell line, HCT116, resulting in the differential expression of 17 genes and 2 long non-coding RNAs (lncRNAs), as well as deregulation of several cellular pathways, including tumor-associated Wnt, transforming growth factor (TGF)-β, Janus kinase (JAK)-signal transducer and activator of transcription (STAT), mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathways. Moreover, we revealed a high similarity between human NETO2 and its ortholog in fish Nothobranchius furzeri, which demonstrate the shortest captive lifespan for a vertebrate (3 months), a novel model for investigating aging and aging-related pathologies, including cancer (Terzibasi et al., 2007). Additionally, we evaluated gene expression in different tissues of N. furzeri using quantitative PCR (qPCR). These results can help to generate unique fishes with gene overexpression/downregulation in the target tissues to establish its role in tumorigenesis [for example, using CRISPR activation (CRISPRa) or interference (CRISPRi) tools].
Materials and Methods
Patients and Tumor Samples
In total, 74 colorectal, 40 prostate and 32 breast tumors, with paired adjacent normal tissues, were collected at the National Medical Research Radiological Center, Ministry of Health of the Russian Federation. No patient received chemotherapy, radiation, targeted therapy, and/or immunotherapy before surgery. Postoperative tumor and normal tissues were immediately frozen in liquid nitrogen and store at −80°C. This study was approved by the Ethics Committee of P.A. Hertsen Moscow Cancer Research Institute, Ministry of Health of the Russian Federation. All experiments were performed in strict accordance with the principles outlined in the Declaration of Helsinki (1964). The patients provided written informed consent to participate in this study. The clinicopathological characteristics of the patients and tumors are presented in Supplementary Tables S1–S3. Additionally, colorectal cancer samples were genetically characterized by mutations in KRAS, NRAS, and BRAF, as well as the microsatellite instability (MSI) status. Analysis of mutations in the “hot spots” of the KRAS, NRAS, and BRAF genes was performed in Evrogen (Russia) using the qPCR with following validation of the results with Sanger sequencing. The MSI detection was carried out in the same company using PCR amplification of microsatellite markers (BAT25, BAT26, D2S123, D5S346, and D17S250).
RNA Isolation and cDNA Synthesis
Tumor and normal tissues were homogenized in a lysis buffer using the MagNA Lyser Instrument (Roche, Switzerland). RNA was isolated from these tissues, as well as from cell cultures, using MagNA Pure Compact RNA Isolation Kit (Roche) on a MagNA Pure Compact System (Roche). The isolated RNA was quantified using a fluorometer, Qubit 2.0 (Thermo Fisher Scientific, United States). RNA quality was measured on an Agilent Bioanalyzer 2100 (Agilent Technologies, United States). Reverse transcription was performed from 1 mg of RNA using Mint Reagent Kit (Evrogen, Russia).
qPCR
Quantitative PCR was performed using TaqMan Gene Expression Assay (Thermo Fisher Scientific), primers and probes, for the NETO2 gene (Hs00983152_m1). GAPDH, GUSB, and B2M were used as reference genes for prostate, colorectal, and breast cancer samples, respectively. Primers and probes for reference genes are shown in Supplementary Table S4. qPCR was performed on an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific) according to the scheme described in Krasnov et al. (2015). Each reaction was performed in triplicate. Data obtained by qPCR were analyzed using the ddCt method and the original ATG software (Kudryavtseva et al., 2018). NETO2 expression was considered meaningful if at least a 2-fold expression was determined owing to potential variations in the reference gene expression.
Cell Culture
The human colorectal cancer HCT116 cell line was cultured in DMEM (Dulbecco’s Modified Eagle’s medium; PanEco, Russia) supplemented with 10% FBS (fetal bovine serum; Thermo Fisher Scientific) and 100 units of penicillin/streptomycin (Thermo Fisher Scientific) in a humidified incubator at 37°C and 5% CO2.
Cell Transfection
shRNA targeting the nucleotide residues 509–529 of the human NETO2 gene (NCBI Gene ID: 81831, location 16q12.1) protein-coding region was designed and synthesized as follows: Sense, 5′-GATCCGCGCCAAATTATCCTGACTCATCACGTGATGAG TCAGGATAATTTGGCGTTTTTG-3′, and antisense, 5′-AATTC AAAAACGCCAAATTATCCTGACTCATCACGTGATGAGTC AGGATAATTTGGCGCG-3′. This pair of oligonucleotides and non-specific control scrambled (SCR) shRNA were cloned into BamH and EcoR sites of pLSLP plasmid to establish the pLSLP-anti-NETO2 lentiviral vector. Lentiviral constructs were transfected into 293T cells using Lipofectamine LTX reagent (Thermo Fisher Scientific). Virus-containing supernatants were collected 24 h after the transfection and were used to infect target HCT116 cells in triple repeats with the addition of 5 μg/mL polybrene (Sigma, United States). Infected cells were selected using the regular growth medium containing 1 μg/mL puromycin (Sigma).
Western Blot Analysis
Western blot analysis was performed as previously described (Kudryavtseva et al., 2016). For NETO2 detection, the primary recombinant antibody EPR3497 (Abcam, United States) was used.
Transcriptome Sequencing and Analysis
For cDNA library preparation, we utilized RNA isolated from cell cultures with RIN (RNA integrity number) values ≥8. Libraries were prepared with TruSeq Stranded mRNA Library Prep Kit (Illumina, United States) according to the manufacturer’s recommendations. Transcriptome sequencing was performed on a NextSeq 500 System (Illumina) using 76 base pair single-end reads. At least 30M reads were obtained for each sample. The primary analysis of raw sequences was performed as previously described (Pudova et al., 2019). Then, data as transcripts per million (TPM) were imported in the R environment. Differential gene expression was analyzed using the DESeq2 package of Love et al. (2014). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used for pathway enrichment analysis.
Analysis of Orthologs
Nucleotide and amino acid sequences of NETO2 in human samples and fishes were aligned using Blastn and Blastp, respectively. Gene expression data were obtained from the Bgee database1.
Neto2b Expression Analysis in N. furzeri
The eggs of fish N. furzeri GRZ were obtained from a commercial supplier and were bred in the Aquatic Housing System (Aquaneering, United States) at the Center for Precision Genome Editing and Genetic Technologies for Biomedicine at the Engelhardt Institute of Molecular Biology. The study was approved by the Ethics Committee of the A.N. Severtsov Institute of Ecology and Evolution Russian Academy of Sciences (approval no. 27, 9.10.2019).
Eight tissue samples (brain, intestines, heart, head kidney, liver, stomach, muscles, and skin) from a female N. furzeri were subjected to RNA isolation using QIAzol lysis reagent (Qiagen, Germany), subsequently treated with DNase I (Thermo Fisher Scientific). Quantification of the isolated RNA was performed using the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Reverse transcription was carried out with 500 ng of RNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). A pair of primers (forward: CCACCCAACAAGGAGTGTGT and reverse: CCCGTGGAGGTAACAAGACC) was designed for neto2b gene detection by qPCR, performed on the Rotor-Gene Q 5 plex HRM (Qiagen) using the following scheme: 95°C for 10 min, 40 cycles of 95°C for 15 s, and 62°C for 60 s.
Statistical Analysis
The significance of differences observed between two groups (tumors/normal tissues and control cells/treated cells) was assessed using the non-parametric Mann-Whitney U test. Correlation analysis was performed using Spearman’s rank correlation coefficient (rs). For transcriptome analysis, the Benjamini-Hochberg method was used to calculate the adjusted p-values [the false discovery rate (FDR)]. Differences and correlations were considered significant at a p-value of < 0.05.
Results
NETO2 Expression in Breast, Prostate, and Colorectal Cancer
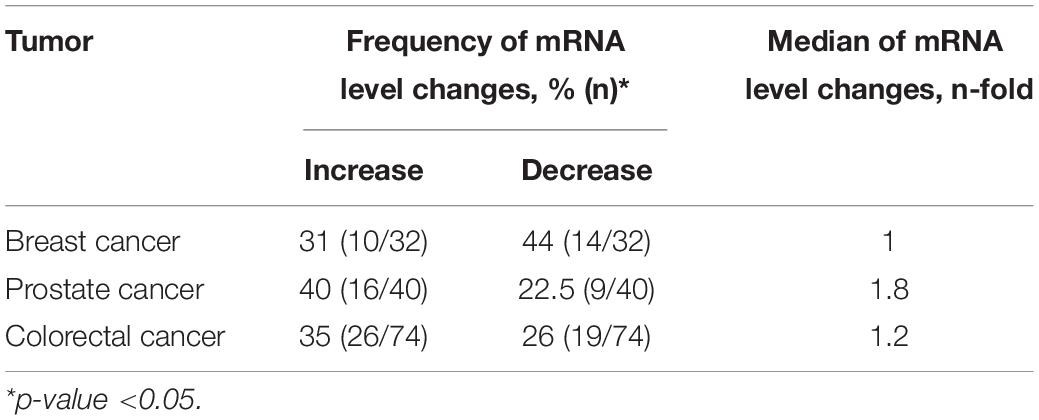
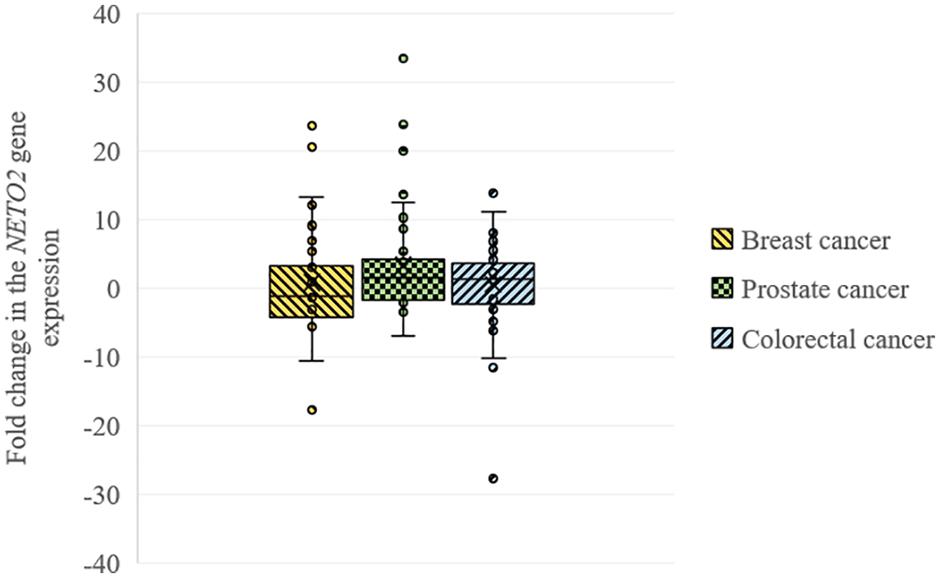
Using qPCR, we analyzed the relative NETO2 mRNA level in sets of breast (n = 32), prostate (n = 40), and colorectal (n = 74) cancer samples. In breast cancer, NETO2 gene expression was increased in 31% of cases (2–24-fold), with decreased mRNA levels detected in 44% of samples (2–18-fold) (Table 1). NETO2 expression was characterized by a 2–33-fold average increase in 40% of prostate cancer samples and a 2–7-fold average decrease in 22.5% of cases. In colorectal cancer, NETO2 mRNA levels were increased in 35% (2–14-fold) of investigated samples and decreased in 26% of cases (2–28-fold). The highest median value of altered gene expression was determined in prostate cancer (Table 1). In general, a similar trend of NETO2 expression was observed in investigated cancer types (Figure 1).
Furthermore, we performed a correlation analysis between several clinicopathological characteristics of tumors (stage for all cancers, differentiation for prostate and colorectal cancer, mutation, and MSI status for colorectal cancer) and NETO2 expression. The highest correlation coefficients were observed between NETO2 expression and pathological stage for breast cancer (rs = 0.15) and prostate cancer (rs = 0.25), tumor differentiation for prostate cancer (rs = −0.16), and BRAF mutation status (rs = 0.18) for colorectal cancer. Correlation coefficients of less than 0.1 were observed between the NETO2 mRNA level and other investigated clinicopathological characteristics.
Stable Knockdown of NETO2 in HCT116
The pLSLP lentiviral vector expressing shRNA targeting NETO2 (pLSLP-anti-NETO2) and the vector expressing non-targeting SCR shRNA were transfected into HCT116 in triplicate. Protein and mRNA levels of the NETO2 gene were measured in experimental and control HCT116 cells using western blotting and qPCR (Figure 2). The results obtained confirmed the NETO2 knockdown.
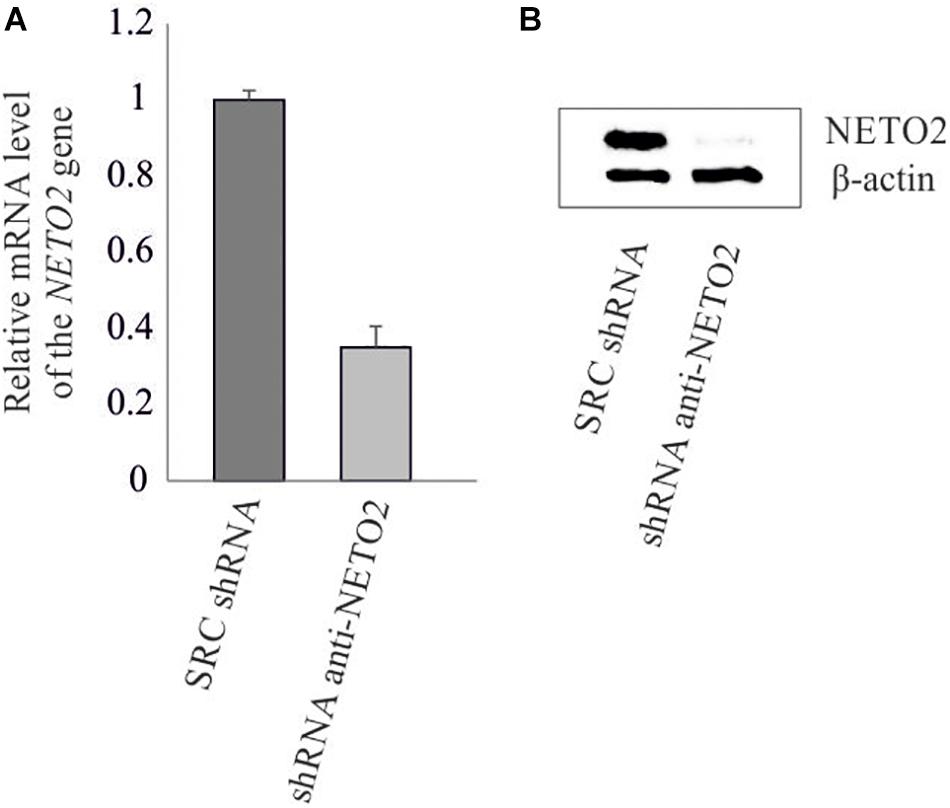
Figure 2. shRNA-mediated knockdown of NETO2 in the HCT116 cell line. Relative NETO2 expression in HCT116 cells transfected with targeting shRNA and non-specific SCR shRNA was measured using qPCR at mRNA level (A) and western blotting (B) at the protein level.
Effects of NETO2 Knockdown on Gene Expression
Seventeen protein-coding differently expressed genes (DEGs) and two lncRNAs (DE lncRNAs) were revealed through the RNA-Seq analysis (Log2FC ≥ 1, Log2FC ≤ −1, CPM ≥ 1, FDR < 0.05) in treated HCT116 cells when compared with the control group. We detected the upregulation of eight genes (CYP2U1, NR1D1, ZNF804A, SEMA3C, DBP, NT5E, NAV3, and KDM5D) and one lncRNA (LINC02043), as well as the downregulation of nine genes (MUC16, KRT6A, ACVRL1, PTP4A1, HYAL1, CLIC3, TEX19, DES, and KCNK3) and one lncRNAs (AL355075.4) (Figure 3A).
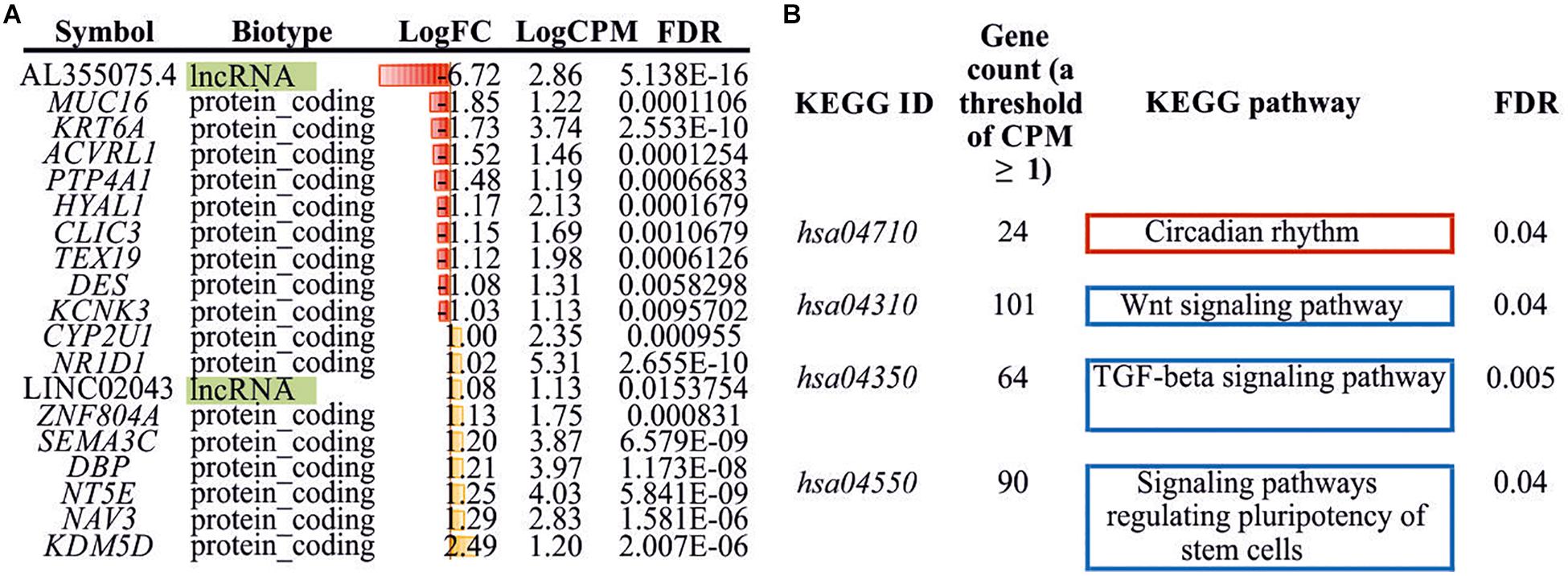
Figure 3. Differently expressed genes/lncRNAs and enriched KEGG pathways. (A) DEGs/DE lncRNAs (Log2FC ≥ 1, Log2FC ≤ −1, CPM ≥ 1, FDR < 0.05) identified between experimental and control HCT116 cells. (B) Enriched KEGG pathways (FDR < 0.05) with DEGs (CPM ≥ 1, FDR < 0.05). KEGG, Kyoto Encyclopedia of Genes and Genomes; DEG, differently expressed genes.
Additionally, we performed KEGG pathway enrichment analysis using all DEGs with FDR < 0.05. Three significantly enriched pathways for downregulated genes and one pathway for upregulated genes were identified (Figure 3B). These included “Wnt signaling pathway,” “TGF-beta signaling pathway,” “signaling pathways regulating pluripotency of stem cells,” and “circadian rhythm.”
NETO2 Gene Ortholog in Fish N. furzeri
We analyzed the NETO2 ortholog in the short-lived fish N. furzeri, termed neto2b (NCBI Gene ID: 107381994, location sgr07). Alignment of cDNA sequences demonstrated 69% of identity, whereas amino acid sequence alignment revealed higher similarity with 85% of conservative substitutions (positives) and only 2% of gaps (Supplementary Figure S1). In comparison, 71% of nucleotide sequences (cDNA) and 74% of amino acid sequence (6% of gaps) similarities were observed in Danio rerio (Supplementary Figure S2). Using qPCR, we measured neto2b expression in different tissues of N. furzeri (female) to estimate its potential use as a fish model presenting gene tissue-specific overexpression/downregulation. As observed in humans, neto2b demonstrated the highest mRNA levels in the fish brain. The expression of neto2b in other investigated tissues is presented in Figure 4.
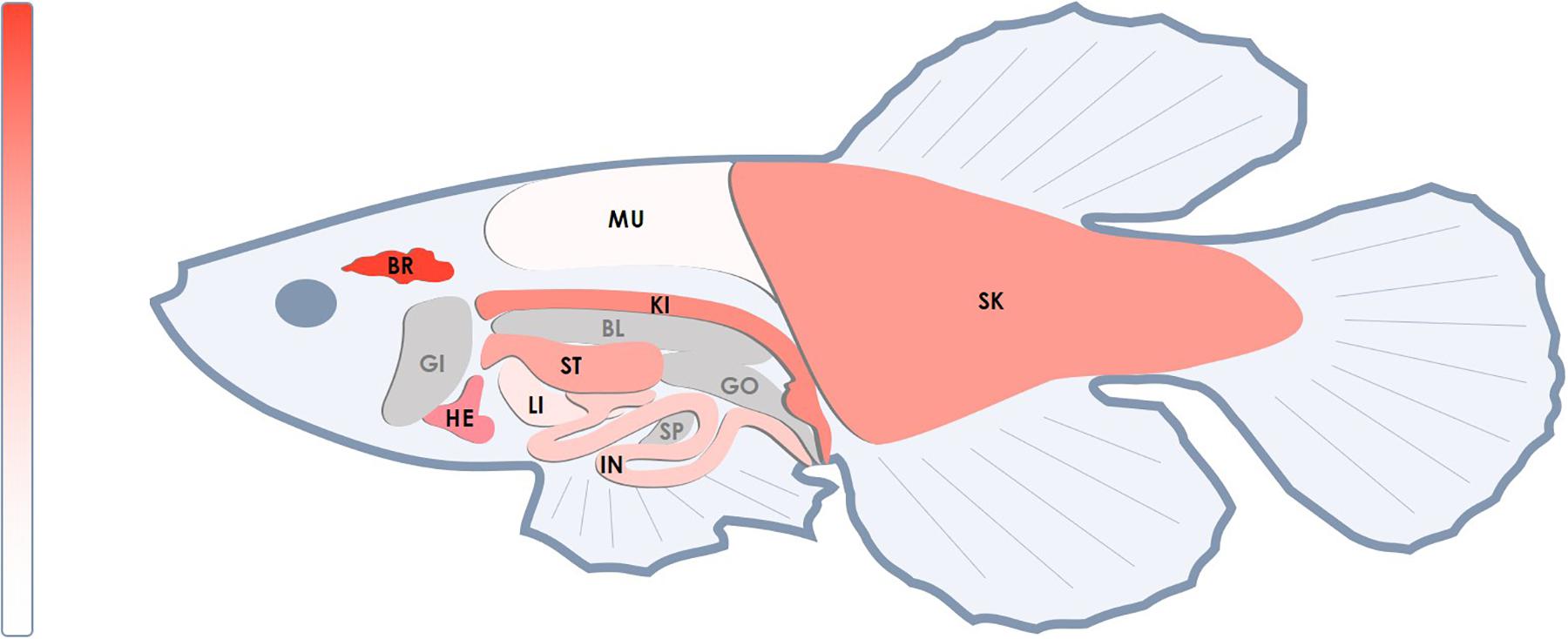
Figure 4. Relative mRNA levels of the neto2b gene in different tissues of N. furzeri. The highest mRNA level of neto2b was detected in the brain tissue and was taken as 1. The expression of neto2b in other studied tissues was measured relatively this value. Darker red indicates higher expression. Gray color indicates tissues where the gene expression has not been measured. BR, brain; HE, heart; ST, stomach; SK, skin; IN, intestines; LI, liver; KI, head kidney; MU, muscles; SP, spleen; BL, bladder; GI, gills; GO, gonads.
Discussion
A series of studies aimed at investigating the biological functions of NETO2 has reported its crucial role in neural glutamate signaling. However, NETO2 was also found to be upregulated in several tumors. Our research group was one of the first to reveal NETO2 overexpression in solid tumors (renal and lung cancer) (Oparina et al., 2012). Recently, it has been reported that elevated NETO2 expression was associated with tumor progression, poor prognosis, and reduced survival in cancer patients (Hu et al., 2015; He et al., 2019). In this study, we first demonstrated the deregulation of NETO2 expression in breast and prostate cancer. The NETO2 mRNA level was predominantly downregulated in breast cancer (44%), with elevated gene expression observed in 31% of samples. In prostate cancer, NETO2 mRNA levels were upregulated in 40% of cases, with a 1.8 median change. Negligible correlations were observed between the NETO2 expression and stage of breast and prostate cancer, as well as differentiation of prostate tumors. For colorectal cancer, we expanded a set of samples from a previous study (Fedorova et al., 2017) and confirmed that NETO2 expression was increased in about one-third of these tumors (41 and 35% in previous and current studies, respectively). Nevertheless, a study evaluating Chinese patients with colorectal cancer has revealed the upregulation of NETO2 in 52.6% of cases, reporting an association between expression and advanced tumor stage and invasion, poor differentiation, lymph node metastasis, and unfavorable prognosis in patients (Hu et al., 2015). In contrast, we failed to observe any significant correlations between the NETO2 mRNA level and tumor stage, as well as differentiation that can be explained by the difference of the studied cohorts. Also, no association of NETO2 expression with KRAS, NRAS, and BRAF mutations or MSI status were found.
Despite several studies indicating the involvement of NETO2 in tumorigenesis, the underlying mechanism remains unclear. Li et al. (2019) have investigated NETO2 functions in pancreatic cancer, and demonstrated that NETO2 knockdown reduced the proliferative capacity of pancreatic cancer cells and suppressed tumor growth in vivo; NETO2 overexpression conversely stimulated cell proliferation, invasion, and migration via the activation of the STAT3 pathway. Similar observations were reportedly documented following NETO2 knockdown in nasopharyngeal carcinoma cells (He et al., 2019). The depletion of NETO2 expression results in decreased proliferation, invasion, and migration of tumor cells and induced apoptosis via activating Caspase-3 signaling. Additionally, NETO2 overexpression promotes the invasion and metastasis of gastric cancer cells by inducing epithelial-mesenchymal transition (EMT) by upregulating TNFRSF12A, which mediates the activation of the PI3K/AKT/NF-κB/Snail axis (Liu et al., 2019). In the present work, we demonstrated that stable NETO2 knockdown in HCT116 cells resulted in the downregulation of Wnt and TGF-β signaling pathways, as well as signaling pathways regulating the pluripotency of stem cells. In the KEGG database, the “signaling pathways regulating pluripotency of stem cells” group comprises the Jak-STAT, MAPK, PI3K-Akt, Wnt, and TGF-β signaling pathways. All these stem cell-related pathways are extensively implicated in tumorigenesis (Dreesen and Brivanlou, 2007). Thus, our results demonstrated the participation of NETO2 in the deregulation of cell signaling in tumors and confirmed its relation to STAT and PI3K-Akt signaling as previously reported. Conversely, NETO2 reduction leads to an upregulation of the circadian rhythm pathway in colorectal cancer cells. Circadian rhythm is closely associated with the cell cycle and has been implicated in DNA-damage response (Hunt and Sassone-Corsi, 2007), with the disruption of this pathway considered a risk factor for carcinogenesis (Straif et al., 2007). According to literature, NETO2 is related to cell proliferation and apoptosis in tumor cells (He et al., 2019; Li et al., 2019). These processes could be connection points between the functions of NETO2 and the circadian rhythm pathway.
Furthermore, we determined 17 genes presenting more than a 2-fold change in expression following NETO2 knockdown. Increased expression was observed in genes involved in cell metabolism [CYP2U1 (arachidonic acid metabolism) and NT5E (metabolism of nucleotides)], circadian rhythm (NR1D1 and DBP), regulation of developmental processes (SEMA3C and NAV3), histone demethylation (KDM5D), and transcriptional regulation (ZNF804A). Decreased mRNA levels were noted for genes related to cell adhesion (MUC16), differentiation (TEX19), proliferation, and migration (ACVRL1, PTP4A1, and HYAL1), as well as cytoskeleton organization (KRT6A and DES) and ion transmembrane transport (CLIC3 and KCNK3). Most genes (CYP2U1, SEMA3C, NT5E, KRT6A, NAV3, ACVRL1, KCNK3, MUC16, PTP4A1, and HYAL1) were previously found to be implicated in the development of colorectal cancer. Interestingly, NETO2 is reportedly associated with the neuron navigator 3 (NAV3) gene, which is also predominantly expressed in the nervous tissue. Copy number changes in the NAV3 gene have been observed in colorectal cancer (Carlsson et al., 2012). Moreover, NAV3 is reportedly involved in the p73-mediated tumor suppression, as well as in Jak-STAT and GnRH signaling in colorectal cancer cells (Carlsson et al., 2012; Uboveja et al., 2020). Furthermore, the participation of NETO2 has been revealed in the Jak-STAT signaling pathway during cancer. Additionally, NETO2 silencing resulted in the altered expression of two lncRNAs, LINC02043, and AL355075.4, for which the functions and related cellular pathways have not been established. However, lncRNA LINC02043 was previously reported as a prognostic marker of recurrence-free survival in hepatocellular carcinoma (Luo et al., 2020). Notably, regulation of NETO2 expression with microRNAs and microRNA-lncRNA interaction has been recently shown in esophageal cancer. NETO2 was determined as a target of miR-206 and miR-143-5p, and participation of NETO2 tumor progression and angiogenesis through overexpression of lncRNA FAM225A absorbed miR-206 was revealed (Wada et al., 2020; Zhang et al., 2020). LncRNAs found in our study can also be potential regulators of NETO2 expression in colorectal cancer.
Although a wide range of studies has proposed increased NETO2 expression in several tumors, we revealed its deregulation in breast, prostate, and colorectal cancers (both decreased and increased mRNA levels). The function of NETO2 in tumorigenesis remains unclear; however, our findings and those obtained in previous studies indicate its participation in cancer-related signaling pathways. Moreover, all identified cancer-related signaling pathways (Wnt, TGF-β, STAT, MAPK, and PI3K-Akt) have previously been shown to be associated with breast, prostate, and colorectal cancer (Dhillon et al., 2007; Massagué, 2008; Pencik et al., 2016; Zhan et al., 2016; Jiang et al., 2020).
Finally, we analyzed the ortholog of the human NETO2 gene in the unique animal model, the short-lived fish N. furzeri. This fish is characterized by an extremely short captive lifespan of 3 months. The use of N. furzeri for genetic studies allows for the rapid generation of transgenic lines and short-term experiments. We showed that the ortholog neto2b had the high similarity with human NETO2 in nucleotide (69%) and amino acid (85%) sequences (even more than those for D. rerio), as well as stable expression in the majority of the fish tissues. These results confirm that NETO2 is a very conservative gene among vertebrates and indicate the possibility to use N. furzeri for the study of the gene function and its role in tumorigenesis at the organism level.
Data Availability Statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/PRJNA655834.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of P.A. Hertsen Moscow Cancer Research Institute, Ministry of Health of the Russian Federation. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Bioethics Commission from the A. N. Severtsov Institute of Ecology and Evolution, Russian Academy of Sciences.
Author Contributions
AS and AVK conceived and designed the work. NV clinically characterized tumor tissue samples. AL designed and maintained knockout cell line. MF, VP, and AAK performed the qPCR experiments. ZG, EP, MF, II, and MS performed the RNA isolation, library preparation, and western blot analysis. GK, DT, and VP carried out bioinformatics analysis. TD performed experimental procedures with the fish. AS, GS, and AVK wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Grant 075-15-2019-1660 from the Ministry of Science and Higher Education of the Russian Federation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the National Medical Research Radiological Center, Ministry of Health of the Russian Federation for tissue samples, National Medical Research Center for Obstetrics, Gynecology and Perinatology named after Academician V.I. Kulakov, Ministry of Health of the Russian Federation, and A. N. Severtsov Institute of Ecology and Evolution for assistance in the data analysis. This work was performed using the equipment(s) at EIMB RAS “Genome” center (http://www.eimb.ru/rus/ckp/ccu_genome_c.php).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.594933/full#supplementary-material
Footnotes
References
Calicchio, M. L., Collins, T., and Kozakewich, H. P. (2009). Identification of signaling systems in proliferating and involuting phase infantile hemangiomas by genome-wide transcriptional profiling. Am. J. Pathol. 174, 1638–1649. doi: 10.2353/ajpath.2009.080517
Carlsson, E., Ranki, A., Sipila, L., Karenko, L., Abdel-Rahman, W. M., Ovaska, K., et al. (2012). Potential role of a navigator gene NAV3 in colorectal cancer. Br. J. Cancer 106, 517–524. doi: 10.1038/bjc.2011.553
Dhillon, A. S., Hagan, S., Rath, O., and Kolch, W. (2007). MAP kinase signalling pathways in cancer. Oncogene 26, 3279–3290. doi: 10.1038/sj.onc.1210421
Dreesen, O., and Brivanlou, A. H. (2007). Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev. 3, 7–17.
Fedorova, M. S., Snezhkina, A. V., Pudova, E. A., Abramov, I. S., Lipatova, A. V., Kharitonov, S. L., et al. (2017). Upregulation of NETO2 gene in colorectal cancer. BMC Genet. 18(Suppl. 1):117. doi: 10.1186/s12863-017-0581-8
He, A. R., Zhu, Q., and Gao, S. (2019). Reducing NETO2 expression prevents human nasopharyngeal carcinoma (NPC) progression by suppressing metastasis and inducing apoptosis. Biochem. Biophys. Res. Commun. 513, 494–501. doi: 10.1016/j.bbrc.2019.03.061
Hu, L., Chen, H. Y., Cai, J., Yang, G. Z., Feng, D., Zhai, Y. X., et al. (2015). Upregulation of NETO2 expression correlates with tumor progression and poor prognosis in colorectal carcinoma. BMC Cancer 15:1006. doi: 10.1186/s12885-015-2018-y
Hunt, T., and Sassone-Corsi, P. (2007). Riding tandem: circadian clocks and the cell cycle. Cell 129, 461–464. doi: 10.1016/j.cell.2007.04.015
Ivakine, E. A., Acton, B. A., Mahadevan, V., Ormond, J., Tang, M., Pressey, J. C., et al. (2013). Neto2 is a KCC2 interacting protein required for neuronal Cl- regulation in hippocampal neurons. Proc. Natl. Acad. Sci. U.S.A. 110, 3561–3566. doi: 10.1073/pnas.1212907110
Jiang, N., Dai, Q., Su, X., Fu, J., Feng, X., and Peng, J. (2020). Role of PI3K/AKT pathway in cancer: the framework of malignant behavior. Mol. Biol. Rep. 47, 4587–4629. doi: 10.1007/s11033-020-05435-1
Krasnov, G. S., Dmitriev, A. A., Sadtritdinova, A. F., Fedorova, M. S., Snezhkina, A. V., Melnikova, N. V., et al. (2015). [Evaluation of gene expression of Hexokinases in colorectal cancer with the use of bioinformatics methods]. Biofizika 60, 1050–1056.
Kudryavtseva, A. V., Fedorova, M. S., Zhavoronkov, A., Moskalev, A. A., Zasedatelev, A. S., Dmitriev, A. A., et al. (2016). Effect of lentivirus-mediated shRNA inactivation of HK1, HK2, and HK3 genes in colorectal cancer and melanoma cells. BMC Genet. 17(Suppl. 3):156. doi: 10.1186/s12863-016-0459-1
Kudryavtseva, A. V., Nyushko, K. M., Zaretsky, A. R., Shagin, D. A., Sadritdinova, A. F., Fedorova, M. S., et al. (2018). [Suppression of NR0B2 gene in clear cell renal cell carcinoma is associated with Hypermethylation of Its promoter]. Mol. Biol. 52, 482–488. doi: 10.7868/S0026898418030114
Li, Y., Zhang, Y., and Liu, J. (2019). NETO2 promotes pancreatic cancer cell proliferation, invasion and migration via activation of the STAT3 signaling pathway. Cancer Manag. Res. 11, 5147–5156. doi: 10.2147/CMAR.S204260
Liu, J. Y., Jiang, L., He, T., Liu, J. J., Fan, J. Y., Xu, X. H., et al. (2019). NETO2 promotes invasion and metastasis of gastric cancer cells via activation of PI3K/Akt/NF-kappaB/Snail axis and predicts outcome of the patients. Cell Death Dis. 10:162. doi: 10.1038/s41419-019-1388-5
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Luo, Y., Ye, J., Wei, J., Zhang, J., and Li, Y. (2020). Long noncoding RNAbased risk scoring system predicts prognosis of alcoholrelated hepatocellular carcinoma. Mol. Med. Rep. 22, 997–1007. doi: 10.3892/mmr.2020.11179
Oparina, N. Y., Sadritdinova, A. F., Snezhkina, A. V., Dmitriev, A. A., Krasnov, G. S., Senchenko, V. N., et al. (2012). Increase in NETO2 gene expression is a potential molecular genetic marker in renal and lung cancers. Russ. J. Genet. 48, 506–512. doi: 10.1134/S1022795412050171
Pencik, J., Pham, H. T. T., Schmoellerl, J., Javaheri, T., Schlederer, M., Culig, Z., et al. (2016). JAK-STAT signaling in cancer: from cytokines to non-coding genome. Cytokine 87, 26–36. doi: 10.1016/j.cyto.2016.06.017
Pudova, E. A., Lukyanova, E. N., Nyushko, K. M., Mikhaylenko, D. S., Zaretsky, A. R., Snezhkina, A. V., et al. (2019). Differentially expressed genes associated with prognosis in locally advanced lymph node-negative prostate cancer. Front. Genet. 10:730. doi: 10.3389/fgene.2019.00730
Snezhkina, A. V., Nyushko, K. M., Zaretsky, A. R., Shagin, D. A., Sadritdinova, A. F., Fedorova, M. S., et al. (2018). Transcription factor SAP30 is involved in the activation of NETO2 gene expression in clear cell renal cell carcinoma. Mol. Biol. 52, 385–392. doi: 10.7868/S0026898418030072
Stohr, H., Berger, C., Frohlich, S., and Weber, B. H. (2002). A novel gene encoding a putative transmembrane protein with two extracellular CUB domains and a low-density lipoprotein class A module: isolation of alternatively spliced isoforms in retina and brain. Gene 286, 223–231. doi: 10.1016/s0378-1119(02)00438-9
Straif, K., Baan, R., Grosse, Y., Secretan, B., El Ghissassi, F., Bouvard, V., et al. (2007). Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. 8, 1065–1066. doi: 10.1016/S1470-2045(07)70373-X
Straub, C., Zhang, W., and Howe, J. R. (2011). Neto2 modulation of kainate receptors with different subunit compositions. J. Neurosci. 31, 8078–8082. doi: 10.1523/JNEUROSCI.0024-11.2011
Sun, X., Liu, X., Xia, M., Shao, Y., and Zhang, X. D. (2019). Multicellular gene network analysis identifies a macrophage-related gene signature predictive of therapeutic response and prognosis of gliomas. J. Transl. Med. 17:159. doi: 10.1186/s12967-019-1908-1
Tang, M., Ivakine, E., Mahadevan, V., Salter, M. W., and McInnes, R. R. (2012). Neto2 interacts with the scaffolding protein GRIP and regulates synaptic abundance of kainate receptors. PLoS One 7:e51433. doi: 10.1371/journal.pone.0051433
Terzibasi, E., Valenzano, D. R., and Cellerino, A. (2007). The short-lived fish Nothobranchius furzeri as a new model system for aging studies. Exp. Gerontol. 42, 81–89. doi: 10.1016/j.exger.2006.06.039
Uboveja, A., Satija, Y. K., Siraj, F., Sharma, I., and Saluja, D. (2020). p73 - NAV3 axis plays a critical role in suppression of colon cancer metastasis. Oncogenesis 9:12. doi: 10.1038/s41389-020-0193-4
Villa, E., Critelli, R., Lei, B., Marzocchi, G., Camma, C., Giannelli, G., et al. (2016). Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular carcinomas and worst survival. Results from a prospective study. Gut 65, 861–869. doi: 10.1136/gutjnl-2014-308483
Wada, M., Goto, Y., Tanaka, T., Okada, R., Moriya, S., Idichi, T., et al. (2020). RNA sequencing-based microRNA expression signature in esophageal squamous cell carcinoma: oncogenic targets by antitumor miR-143-5p and miR-143-3p regulation. J. Hum. Genet. 65, 1019–1034. doi: 10.1038/s10038-020-0795-x
Wyeth, M. S., Pelkey, K. A., Petralia, R. S., Salter, M. W., McInnes, R. R., and McBain, C. J. (2014). Neto auxiliary protein interactions regulate kainate and NMDA receptor subunit localization at mossy fiber-CA3 pyramidal cell synapses. J. Neurosci. 34, 622–628. doi: 10.1523/JNEUROSCI.3098-13.2014
Yun, J. W., Lee, S., Ryu, D., Park, S., Park, W. Y., Joung, J. G., et al. (2019). Biomarkers associated with tumor heterogeneity in prostate cancer. Transl. Oncol. 12, 43–48. doi: 10.1016/j.tranon.2018.09.003
Zhan, T., Rindtorff, N., and Boutros, M. (2016). Wnt signaling in cancer. Oncogene 36, 1461–1473. doi: 10.1038/onc.2016.304
Keywords: NETO2, breast cancer, prostate cancer, colorectal cancer, fish model
Citation: Fedorova MS, Snezhkina AV, Lipatova AV, Pavlov VS, Kobelyatskaya AA, Guvatova ZG, Pudova EA, Savvateeva MV, Ishina IA, Demidova TB, Volchenko NN, Trofimov DY, Sukhikh GT, Krasnov GS and Kudryavtseva AV (2020) NETO2 Is Deregulated in Breast, Prostate, and Colorectal Cancer and Participates in Cellular Signaling. Front. Genet. 11:594933. doi: 10.3389/fgene.2020.594933
Received: 14 August 2020; Accepted: 19 November 2020;
Published: 10 December 2020.
Edited by:
Yuriy L. Orlov, I.M. Sechenov First Moscow State Medical University, RussiaReviewed by:
Stanislav Rybtsov, University of Edinburgh, United KingdomElvira Galieva, Novosibirsk State University, Russia
Copyright © 2020 Fedorova, Snezhkina, Lipatova, Pavlov, Kobelyatskaya, Guvatova, Pudova, Savvateeva, Ishina, Demidova, Volchenko, Trofimov, Sukhikh, Krasnov and Kudryavtseva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anastasiya V. Snezhkina, bGVmdGdlckByYW1ibGVyLnJ1; Anna V. Kudryavtseva, cmhpemFtb2ViYUBtYWlsLnJ1
†These authors have contributed equally to this work
 Maria S. Fedorova
Maria S. Fedorova Anastasiya V. Snezhkina
Anastasiya V. Snezhkina Anastasiya V. Lipatova
Anastasiya V. Lipatova Vladislav S. Pavlov1
Vladislav S. Pavlov1 Zulfiya G. Guvatova
Zulfiya G. Guvatova Elena A. Pudova
Elena A. Pudova George S. Krasnov
George S. Krasnov Anna V. Kudryavtseva
Anna V. Kudryavtseva