- 1Joint Laboratory for Translational Cancer Research of Chinese Medicine of the Ministry of Education of the People's Republic of China, International Institute for Translational Chinese Medicine, School of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, Guangzhou, China
- 2Key Laboratory of Bioactive Substances and Resources, Utilization of Chinese Herbal Medicine, Ministry of Education, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
- 3State Key Laboratory of Innovative Natural Medicine and TCM Injections, Jiangxi Qingfeng Pharmaceutical Co. Ltd., Ganzhou, China
- 4School of Chemistry, Chemical Engineering and Life Sciences, Wuhan University of Technology, Wuhan, China
Andrographolide, which is enriched in the leaves of Andrographis paniculata, has been known as “natural antibiotic” due to its pharmacological activities such as anti-inflammatory, antimicrobial and antioxidant effects. Several key enzymes in andrographolide biosynthetic pathway have been studied since the genome sequences were released, but its regulatory mechanism remains unknown. WRKY transcription factors proteins have been reported to regulate plant secondary metabolism, development as well as biotic and abiotic stresses. Here, WRKY transcription factors related to andrographolide biosynthesis were systematically identified, including sequences alignment, phylogenetic analysis, chromosomal distribution, gene structure, conserved motifs, synteny, alternative splicing event and Gene ontology (GO) annotation. A total of 58 WRKYs were identified in Chuanxinlian genome and phylogenetically classified into three groups. Moreover, nine WRKY genes underwent alternative splicing events. Furthermore, the combination of binding site prediction, gene-specific expression patterns, and phylogenetic analysis suggested that 7 WRKYs (ApWRKY01, ApWRKY08, ApWRKY12, ApWRKY14, ApWRKY19, ApWRKY20, and ApWRKY50) might regulate andrographolide biosynthesis. This study laid a foundation for understanding the regulatory mechanism of andrographolide biosynthesis and the improvement and breeding of Andrographis paniculata varieties.
Introduction
Andrographis paniculata (Burm.f.) Nees, a well-known traditional herb from the family Acanthaceae, has been used in eastern Asia for thousands of years (Lim et al., 2012). It has a broad spectrum of biological activities, such as anti-inflammatory, antimicrobial, antioxidant, hepatoprotective and hypoglycemic effects (Mishra et al., 2009; Yu et al., 2014). Previous studies have showed that the main active compounds of A. paniculata were ent-labdane-related diterpenes (ent-LRDs), including andrographolide (AD), neoandrographolide (NAD) and 14-deoxy-11,12-didehydroandrographolide (DDAD). Their contents varied significantly among different organs, with a marked increase in leaves and lowest in roots (Garg et al., 2015). Among the ent-LRDs, the most abundant constituent is the extremely bitter compound andrographolide, which has been used to treat upper respiratory tract infection, fever and diarrhea in clinical practice in many countries (Lim et al., 2012; Raina et al., 2013). Due to its marked anti-inflammatory and antimicrobial pharmacological activities, andrographolide has been recognized as a “natural antibiotic.” With the increasing clinical demand for andrographolide (Shao et al., 2015), how to efficiently obtain andrographolide by investigating the molecular mechanisms controlling its biosynthesis has received increasing attention. Recently, the genome of A. paniculata has been completely assembled, and some key genes for the biosynthesis of andrographolide have been discovered (Sun et al., 2019). However, there are no studies about the regulation of andrographolide content by transcription factors, which regulate secondary metabolism at the transcriptional level.
The WRKY TF family possesses ~60 amino acid binding domains and covers three classes (class I to class III) according to the WRKY domain number and the variety of zinc finger motifs (Hussain et al., 2019; Yan et al., 2019). It has been reported to have a significant role in plant secondary metabolism. Several WRKY TFs have been functionally identified as core regulators that regulate the biosynthesis of terpenoids. For example, AaWRKY1 positively regulates the biosynthesis of artemisinin by promoting the expression of the ADS, CYP71AV1 and DBR2 genes (Ma et al., 2009; Han et al., 2014; Jiang et al., 2016). Overexpression of a trichome-specific WRKY from Artemisia annua, named AaGSW1, significantly improves the contents of artemisinin and dihydroartemisinic acid by regulating CYP71AV1 and AaORA expression (Chen et al., 2017b). In addition, it has been reported that GaWRKY1 of Gossypium arboretum regulates sesquiterpene synthase in the biosynthesis pathway of sesquiterpene phytoalexins, including gossypol (Xu et al., 2004). Although some WRKY TFs have been identified in plants, there are no studies on WRKY TFs from A. paniculata. Andrographolide is a diterpenoid lactone derived from the terpene skeleton pathway. Therefore, we proposed that WRKY TFs may be involved in andrographolide biosynthesis.
The genome-wide identification of WRKY TFs has been documented in some plants, such as the model plant Arabidopsis thaliana (72); the important crops Oryza sativa (102), Zea mays (136), Glycine max (182), and Cucumis sativus (55); and the model medicinal plant Salvia miltiorrhiza (61) (Wu et al., 2005; Ling et al., 2011; Wei et al., 2012; Li et al., 2015). Although A. paniculata is a famous and widely used medicinal plant, its WRKY TFs has not been studied. This study aims to provide a more comprehensive analysis of the WRKY family in A. paniculata. In total, 58 WRKYs were identified in A. paniculata. Furthermore, analyses of chromosome locations, sequence alignment, the phylogenetic tree, gene structure, conserved motifs, GO annotation, synteny, and alternative splicing events were performed. To infer the function of WRKY genes regulating andrographolide biosynthesis, cis-element prediction, differentially expressed genes (DEGs) analysis, qRT-PCR detection, and phylogenetic analysis were applied in this study. Finally, we suggested seven WRKYs that may be involved in the biosynthesis of andrographolide, which laid the foundation for further regulatory research.
Materials and Methods
Plant Materials and Treatment
In this study, the plants of A. paniculata, which were pot cultivated in the laboratory at the School of Pharmaceutical Sciences of Guangzhou University of Chinese Medicine (China), were used. The healthy and full A. paniculata seeds were cultured in petri dishes for 10 days under 25°C until cotyledons grew. Then the seedlings were transplanted into flowerpots. For methyl jasmonate (MeJA) treatment, 20-day seedlings in flowerpots were used and each group (n = 3) was sprayed with 5 mM MeJA. The samples were collected at 0, 24, and 48 h after spaying from nine individual plants. For different organ samples, roots, stems, and leaves were collected from three plants grown for 45 days. All materials were immediately frozen in liquid nitrogen and stored at −80°C for analysis of the expression patterns of WRKY genes.
Database Search and Gene Identification
The WRKY sequences of A. thaliana were obtained from the Arabidopsis Information Resource (http://Arabidopsis.org/) and used as queries to search the A. paniculata genome database (Sun et al., 2019). Then, we corrected the sequences manually using the BLASTx algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi) by comparison with other plant WRKYs. All WRKY sequences were further confirmed with PROSITE (http://prosite.expasy.org/), and the conserved domains of each WRKY protein were obtained.
Phylogenetic, Gene Structure, Conserved Motif Analyses, and Chromosomal Locations
The WRKY domain amino acid sequences of A. paniculata and A. thaliana were aligned with ClustalX program by MEGA 7.0 software, and phylogenetic analysis was carried out using the neighbor-joining method with 1,000 bootstrap replicates. The molecular weights (MWs) and isoelectric points (pIs) of the putative WRKY proteins were calculated by the ExPASy proteomics server (https://web.expasy.org/protparam/). The gene exon-intron organization was constructed using Gene Structure Display Server (v2.0 http://gsds.cbi.pku.edu.cn/). Motifs in all ApWRKY protein sequences were identified by the MEME 5.0.1 online program (http://meme-suite.org/tools/meme) with the following parameters: number of repetitions, any; maximum number of motifs, 15; and the optimum width of each motif, between 20 and 50 residues. The figure was represented using TBtools software (Chen et al., 2020). The corresponding chromosomal locations of WRKY genes in the A. paniculata genome were downloaded from the A. paniculata genome database. The WRKY gene locations were then represented using Mapchart software.
Synteny Analysis, GO Annotation, Alternative Splicing Analysis, and Cis-element Prediction
The alignment was performed by LASTZ using CDS sequences of A. paniculata and four representative species (A. thaliana, Oryza sativa, Vitis vinifera, and Sesamum indicum). The syntenic block map was constructed by MCscan with cscore = 0.7. The gene ontology annotation of WRKY amino acid sequences was analyzed using Blast2GO (http://www.blast2go.com). The sequences were screened by BLASTp against the InterProScan database and NR database. After mapping to the GO term, the annotation was conducted with default parameters. Alternative splicing analysis was performed as in our previous study (Gao et al., 2019). The 2,000-bp promoter sequences of 56 genes encoding key enzymes in the andrographolide biosynthetic pathway were obtained from the A. paniculata genome database. Place (http://www.dna.affrc.go.jp/PLACE/signalscan.html) was used to identify the cis-elements by WRKY TFs.
Gene Expression Analysis and qRT-PCR
The RNA-seq reads from four organs (root, stem, leaf, flower, fruit) and MEJA-treated seedlings (0, 24, 48) were generated and the expression patterns of the WRKY genes were analyzed with TopHat and Cufflinks (Trapnell et al., 2012). According to the manufacturer's instructions, total RNA was extracted from each sample with an RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China). The quality of the RNA was detected by electrophoresis and a NanoDrop 2000C spectrophotometer. Total RNA was reverse transcribed into the first cDNA strand using a FastQuant RT Kit (Tiangen Biotech). The gene-specific primers were designed by Primer Premier 6.0, and the PCR product size was set between 130 and 200 bp (Supplementary Table 1). ApActin was used as the reference gene. qRT-PCR was performed using TaKaRa TB GreenTM Premix Ex TaqTM II (Dalian, China) on an Applied Biosystems 7500 Fast Real-Time PCR System. Three technical replicates were conducted for each sample. Statistical analysis was carried out to detect expression differences using IBM SPSS 20 software, and a P-value < 0.01 was considered extremely highly significant.
Results
Identification of the A. paniculata WRKY Family and Its Chromosomal Distribution
A total of 58 WRKY genes were identified from the genome database of A. paniculata, and the WRKY domain was further confirmed by PROSITE software. The gene chromosomal location analysis revealed that 57 WRKY genes were located on 21 chromosomes (Figure 1). The remaining 1 gene, WRKY55, was located at tig00000239. There was an average of 3 WRKY genes per chromosome, with the highest number of genes (five genes) located on chromosomes 1, 2, and 17. In addition, the lengths of the genomic DNA, cDNA, and the deduced protein; the domain sequence; the number of introns; and the physicochemical properties (molecular weight, isoelectric point, grand average of hydropathicity and aliphatic index) are summarized in Supplementary Table 2.
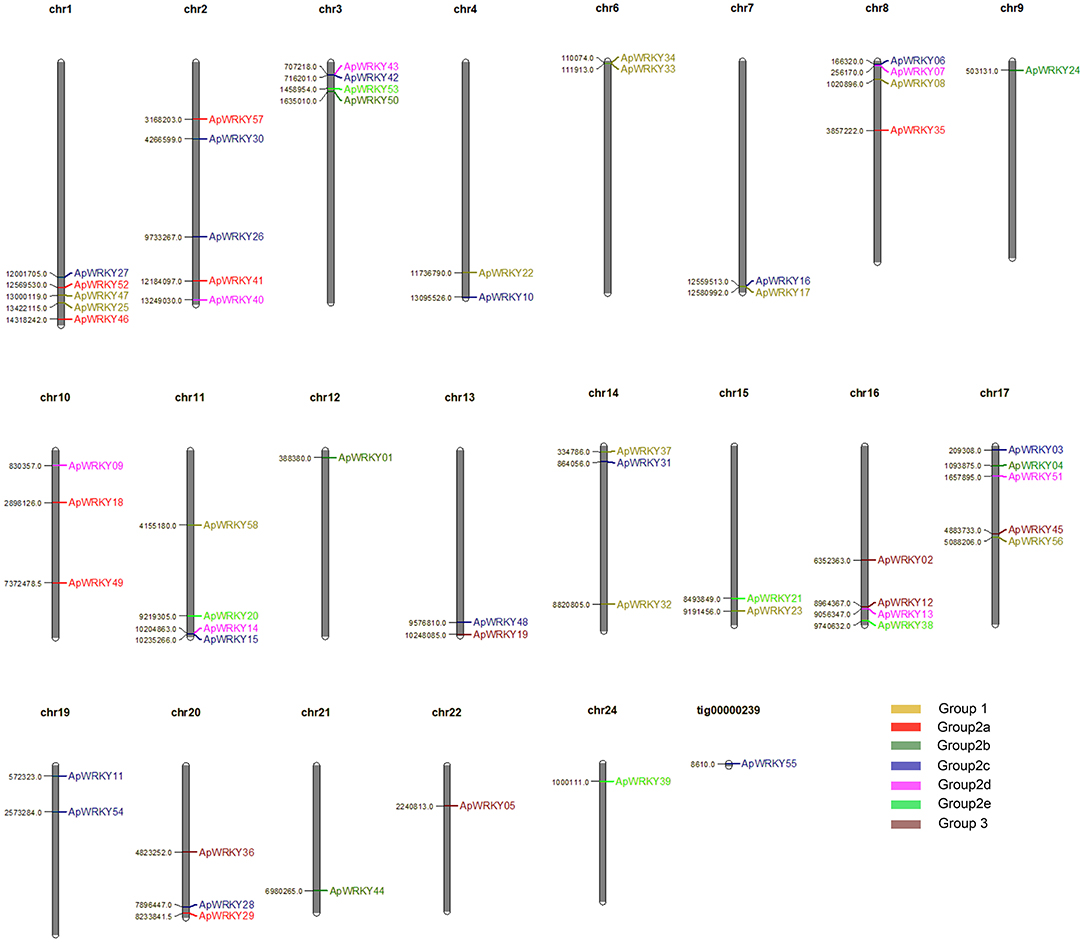
Figure 1. Distribution of 58 WRKY genes on the 21 Chuanxinlian chromosomes and one tig. Different group members are presented in various colors: yellow, Group 1; red, Group 2a; dark green, Group 2b; blue, Group 2c; pink, Group 2d; light green, Group 2e; brown, Group 3.
Classification of the WRKY Domains and Phylogenetic Analysis
The WRKY domain sequences of A. paniculata and A. thaliana were aligned with the default settings, and phylogenetic relationships were analyzed using the neighbor-joining algorithm by MEGA7.0 software with 1,000 bootstrap sampling steps (Figures 2, 3). The result of the WRKY domain sequence alignment indicated that each domain possessed ~60 residues, including the highly conserved motif WRKYGQK and a zinc finger motif. Among them, 57 domains contained the highly conserved sequence WRKYGQK, while only one had WRKYGKK (Figure 2). According to the classification of WRKYs from A. thaliana, the ApWRKYs were divided into three groups (Figure 3). Group I contained 12 members with two WRKY conserved domains, which were further designated the N-terminal WRKY domain (NTWD) and the C-terminal WRKY domain (CTWD). Forty members were clustered in Group II and possessed only one WRKY conserved domain. Based on the primary amino acid sequence, Group II proteins can be further divided into five subgroups (IIa-IIe). In addition, six WRKYs with one WRKY domain were identified in Group III. Group I and Group II harbored a C2H2 zinc finger motif, while Group III had the other zinc finger motif (C2HC).
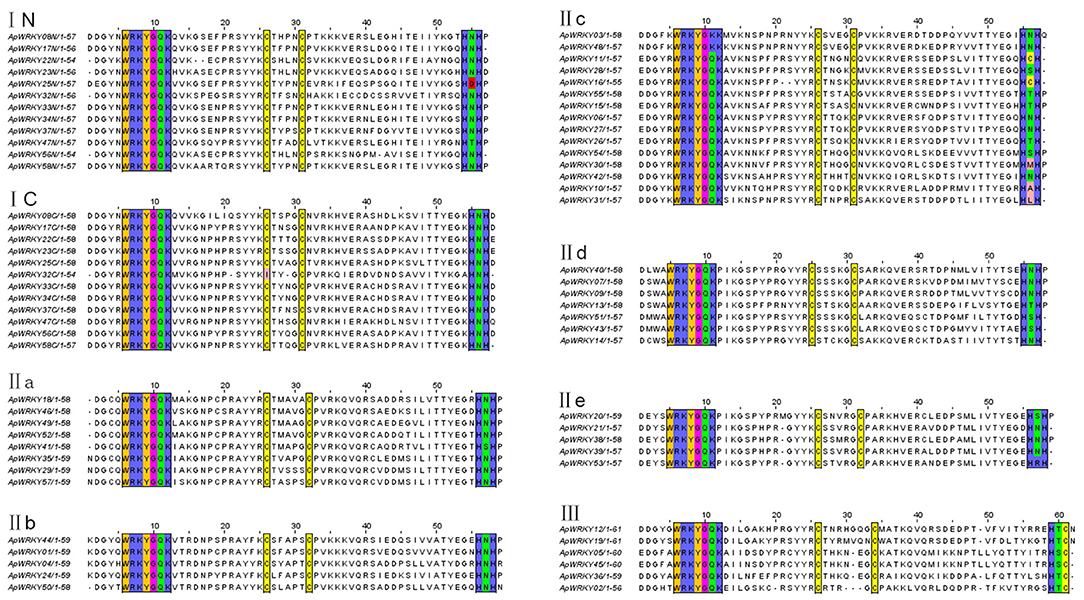
Figure 2. Comparison of deduced amino acid sequences of the WRKY domains from ApWRKYs. Black box indicates conserved WRKY amino acids and zinc-finger motif.
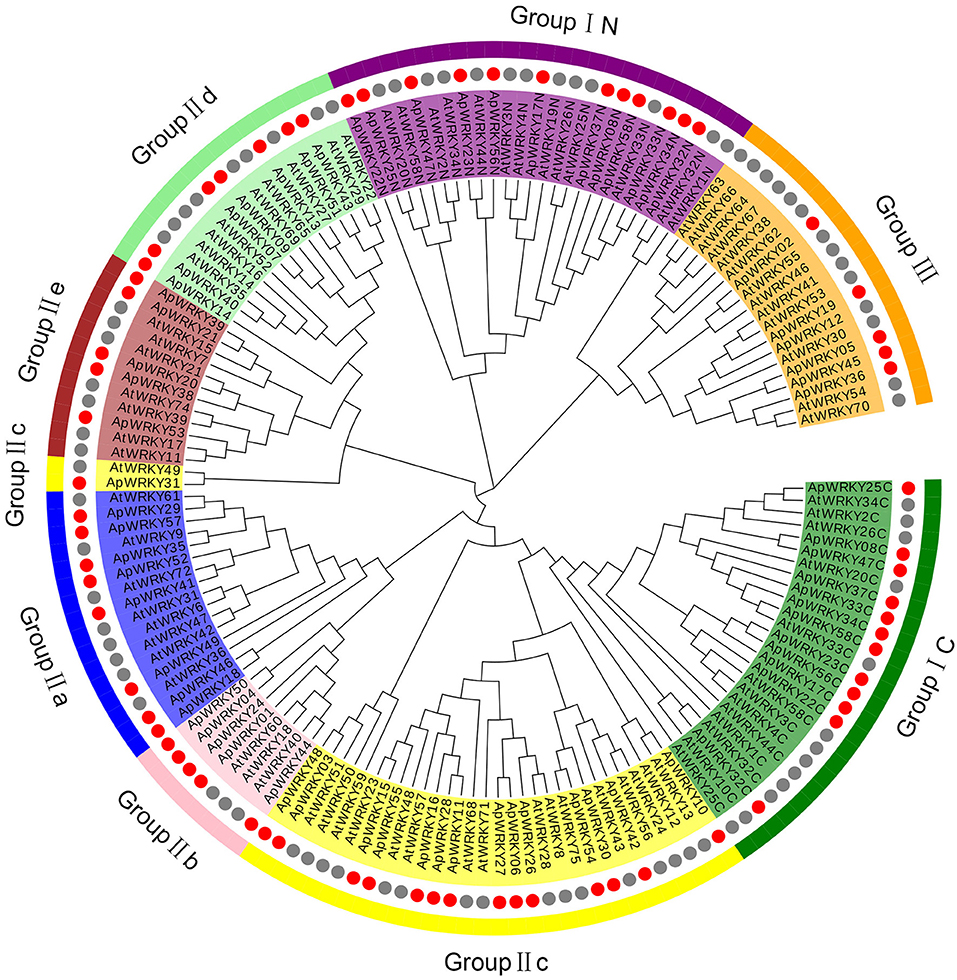
Figure 3. Phylogenetic relationships of WRKY proteins from A. paniculata and Arabidopsis using WRKY domain sequences. Groups are highlighted.
Gene Structure and Conserved Motifs of WRKYs
Conserved motifs are sometimes associated with specific functions such as transcription repression of EAR motif and R/KLFGV motif; transcriptional activation of EDLL motif and WRKY-protein interaction of VQ motif (Ohta et al., 2001; Ikeda and Ohme-Takagi, 2009; Tiwari et al., 2012; Chi et al., 2013). In this study, we used the MEME tool to predict the conserved motifs within the WRKY protein sequences, and a total of 15 motifs were obtained (Figure 4B and Supplementary Table 3). Some motifs were highly conserved and were distributed in all the proteins, while some specific motifs appeared only in certain groups (Figures 4A,B). For example, all WRKYs had motif-1, motif-2, and motif-3. However, motif-11, motif-14 and motif-15 existed only in Group I. Motif-7 and motif-9 were found only in Group III, and only Group IIa possessed motif-13. Moreover, motif-12 specifically belonged to Group IIe and Group IId. The information of these particular conserved motifs provided clues for further study on protein structure, function and evolution.
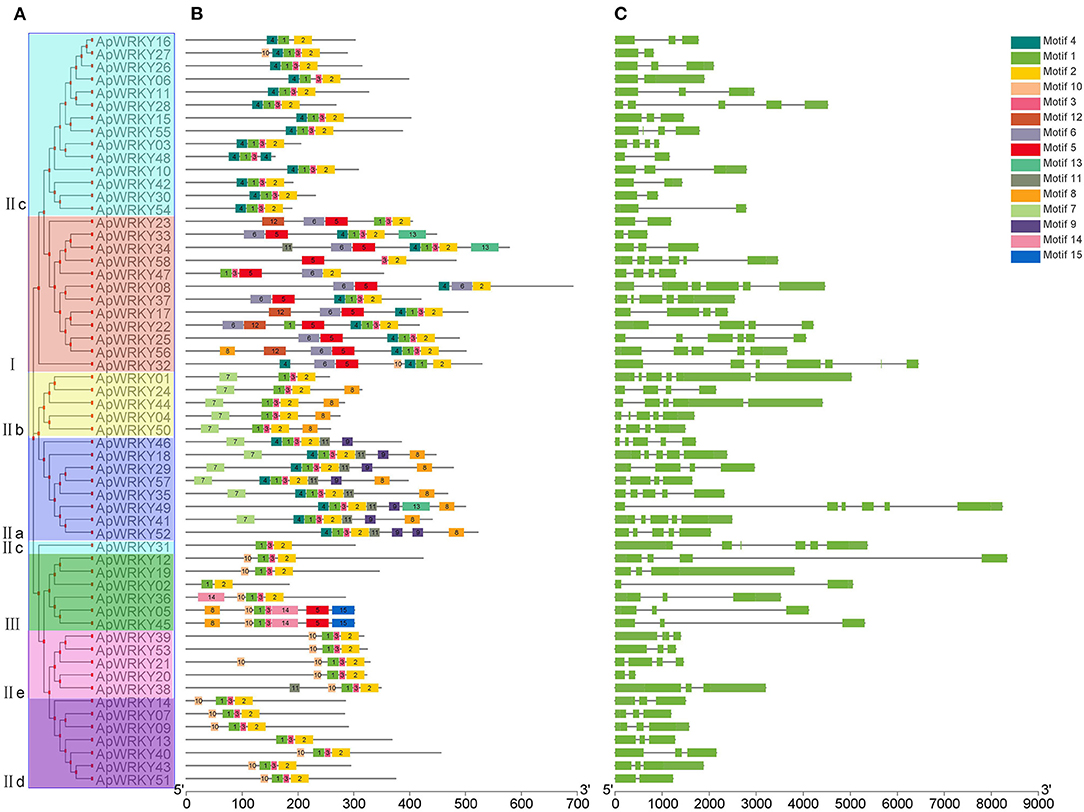
Figure 4. The gene structure and the distribution of conserved motifs within each ApWRKY in A. paniculata. (A) Phylogenetic tree of ApWRKY proteins. (B) The distribution of conserved motifs within each ApWRKY protein. (C) The gene structures of ApWRKYs. Exons were represented by green boxes and introns were represented by black lines.
The gene structure map showed that the number of introns in WRKY genes of A. paniculata varied from 1 to 5, and only two genes (ApWRKY06 and ApWRKY30) from Group IIc were intronless (Figure 4C). Members of Group I generally had more introns than others; for example, ApWRKY25, ApWRKY32, ApWRKY33, and ApWRKY58 contained up to five introns.
Go Ontology Annotation and Synteny Analysis of WRKY Genes
Blast2GO (http://www.blast2go.com) was used to annotate all the WRKYs. Gene Ontology consists of three categories: molecular function, biological process and cell component. The “binding” term, belonging to molecular function, appeared the most frequently, and 24 WRKYs were annotated with it (Supplementary Figure 1). In the biological process category, only three sequences were assigned to “response to stimulus.” Moreover, the remaining WRKYs were evenly distributed across the molecular function, biological process and cellular component categories with 12–13 sequences.
Synteny is part of the important reference information for investigating the evolutionary history of gene families and exploring functional genes (Ksiazkiewicz et al., 2017; Raboanatahiry et al., 2017; Zhao et al., 2017; Zhang et al., 2018; Hardigan et al., 2020). In this study, we used three dicotyledonous plants comprising A. thaliana, Vitis vinifera and Sesamum indicum and one monocotyledonous O. sativa to construct a synteny relationship map with A. paniculata (Figure 5). The figure indicates that the collinear relationship between A. paniculata and S. indicum and V. vinifera was the most significant, while the gene pairs between A. paniculata and O. sativa were the least significant. These results provide clues for examining the relationship between functional genes.
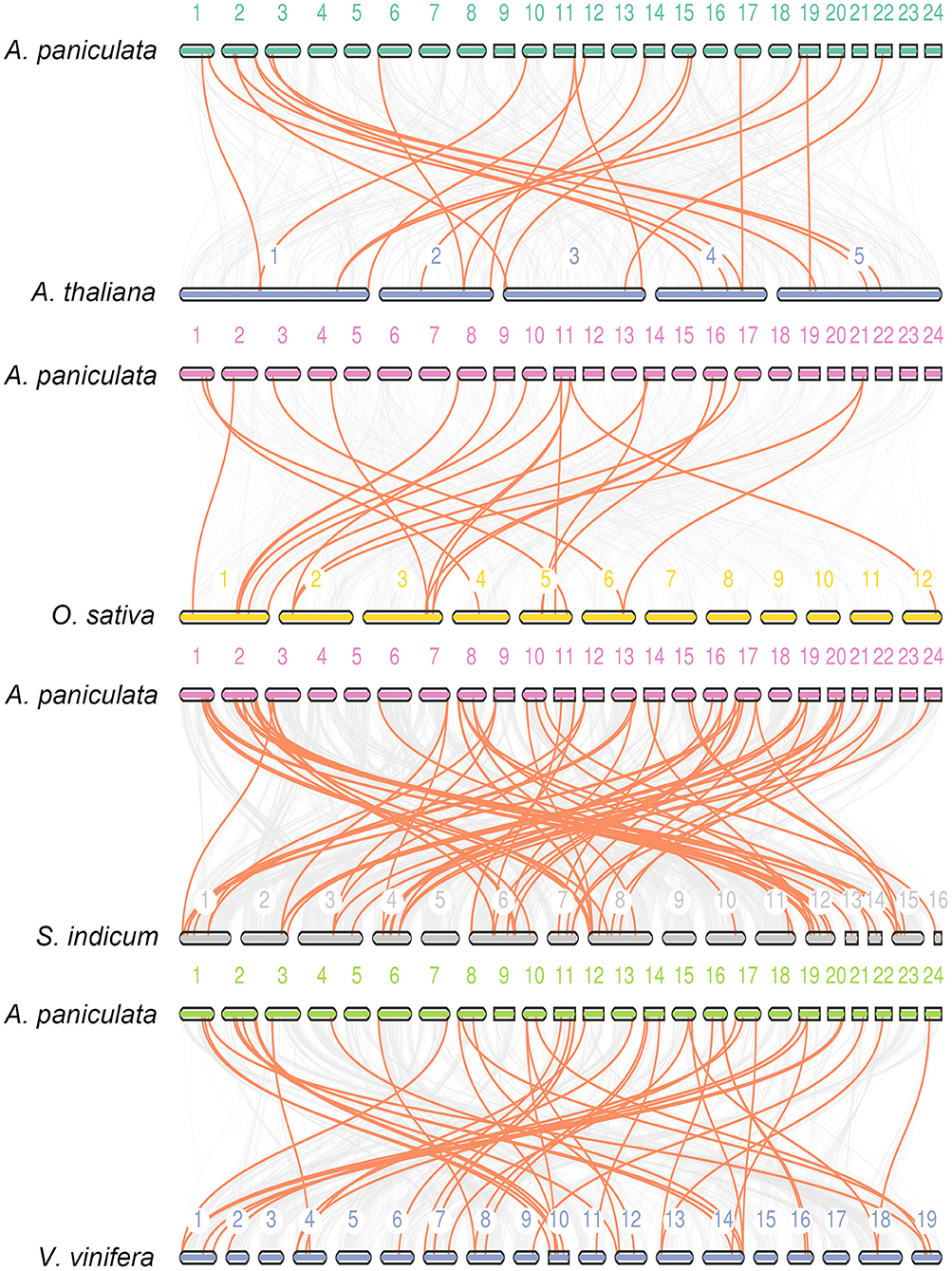
Figure 5. Synteny analysis of WRKY genes. Synteny between A. paniculata and Arabidopsis, O. sativa, S. indicum and V. vinifera is shown. Colored lines denote syntenic regions between Chuanxinlian chromosomes and others.
Analysis of Alternative Splicing Events
The main types of alternative splicing events include exon skipping, intron retention, mutually exclusive exons, A5SS (alternative 5' splice site) and A3SS (alternative 3' splice site) (Chaudhary et al., 2019). Based on the isoform detection and prediction (IDP) results of this study, 22 splicing events of 9 WRKY genes were identified (Figure 6). The splicing types included 12 IR isoforms, six non-IR isoforms, and nine ref isoforms. Five genes, including ApWRKY21, ApWRKY44, ApWRKY50, ApWRKY53, and ApWRKY58, had two splicing types. The remaining four genes presented three splicing types. After treatment with MeJA for 24 h, the expression levels of the isoforms from most genes were upregulated. For example, IR isoforms and ref isoforms produced by ApWRKY04, ApWRKY14, and ApWRKY50 were all induced. Moreover, after MeJA treatment for 48 h, the expression levels of various alternative splicing types varied. The expression of IR isoforms from ApWRKY21 and ApWRKY58 constantly increased within 48 h. However, the expression levels of the reference and IR isoforms of ApWRKY50 and ApWRKY04 were decreased at 48 h compared with 24 h. Alternative splicing is widespread in eukaryotic organisms, which leads to polymorphisms in the structure and function of proteins (Chaudhary et al., 2019). The complexity of these alternative splicing events may play an important regulatory role in the Chuanxinlian development-mediated MeJA pathway.
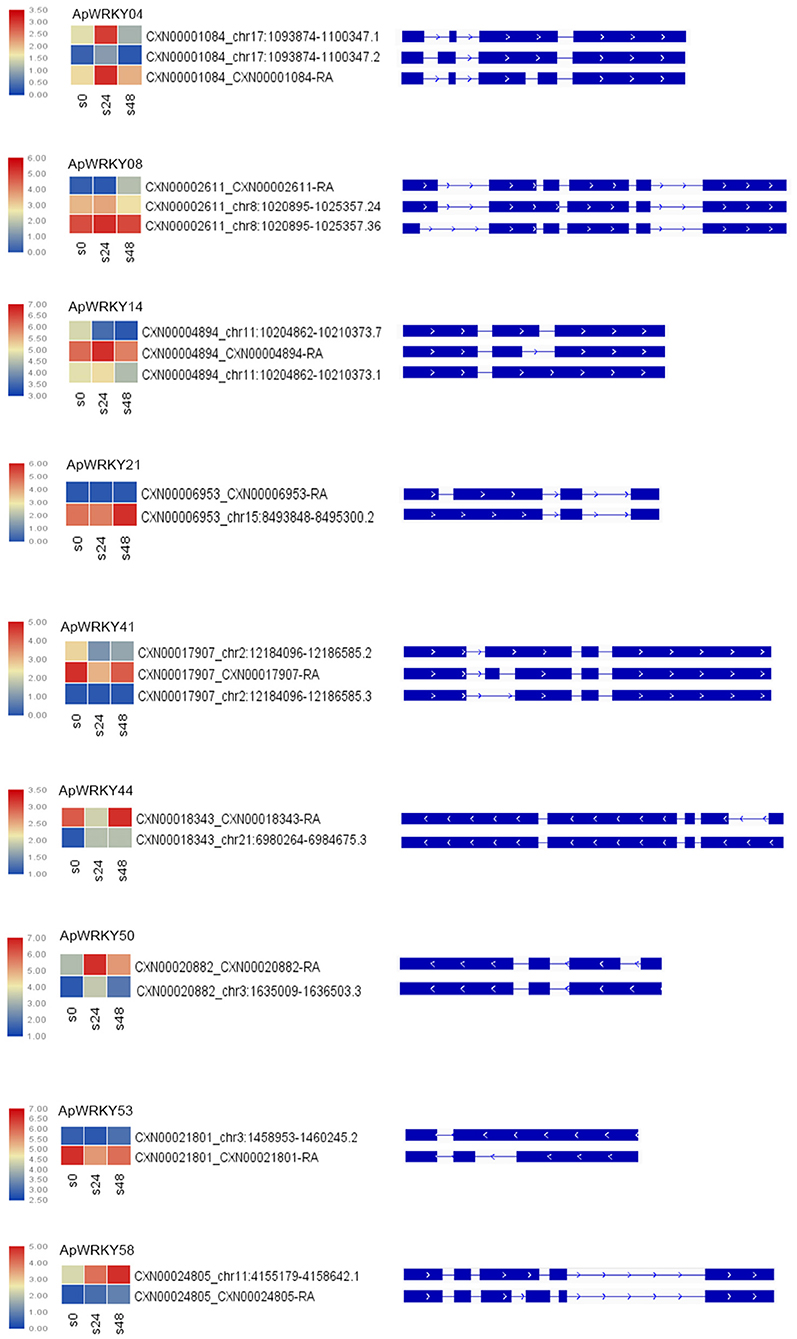
Figure 6. Alternative splicing isoforms of nine ApWRKY genes. Heatmap of the isoforms showed changes in the gene expression of various isoforms under MeJA stress.
Expression Patterns of WRKYs in A. paniculata
Different expression profiles of genes in various organs and stresses can supply essential information for screening for functional genes. The expression levels of WRKYs in A. paniculata were calculated using RNA-seq data (Figure 7 and Supplementary Table 4). ApWRKY02 and ApWRKY13 were not expressed in all organs (FPKM value >1), and other genes were expressed at different levels among the organs of flowers, fruits, roots, stems and leaves. A total of 16 genes were most highly expressed in leaves (FPKM value >10); and 5, 12, and 5 genes in fruits, roots, and stems were most highly expressed (FPKM value >10), respectively. These genes may participate in organ development in A. paniculata. The expression profiles of 58 WRKYs in MeJA-treated seedlings were also observed. After MeJA treatment for 24 h, the expression of 12 WRKYs was upregulated compared with the expression at 0 h. After 48 h of MeJA treatment, 4 WRKYs were induced consistently within 24 h. For example, the expression profile of ApWRKY58 increased from 7.12 to 41.57. These results provide transcriptome reference information for us to reasonably speculate on genes that may be responsible for the development and tolerance regulation of A. paniculata.
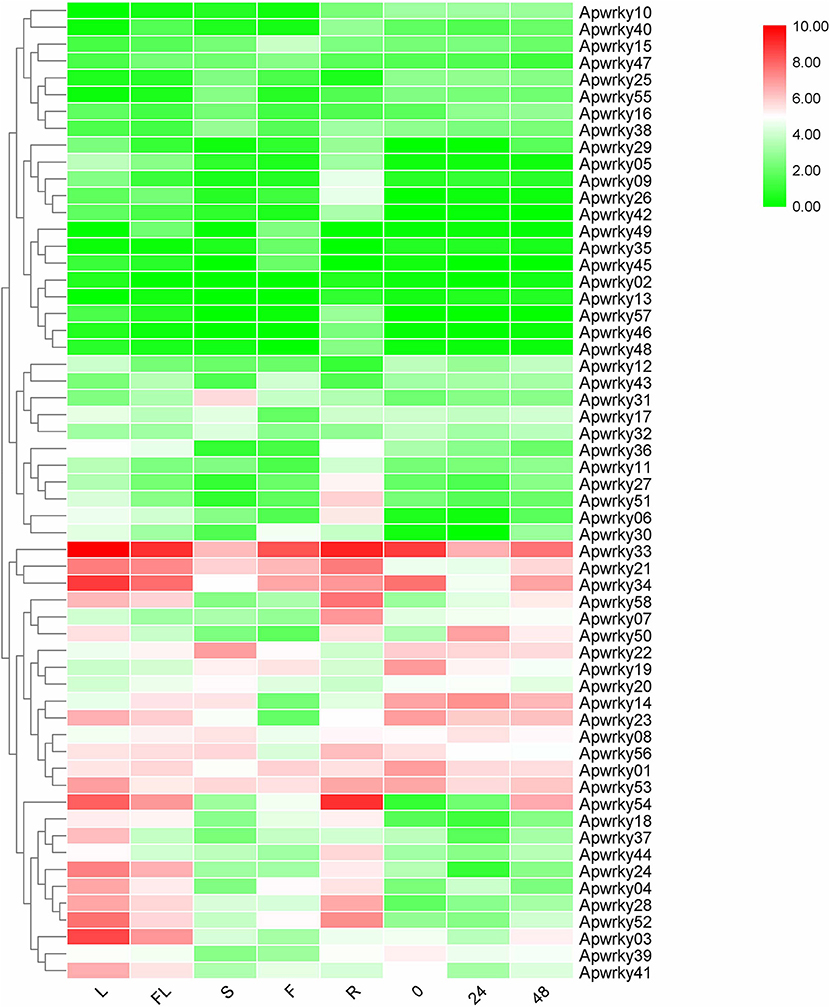
Figure 7. Heatmaps representing the expression profiles of Chuanxinlian ApWRKY genes in leaf, root, stem, flower, fruit and MeJA-treated seedings.
WRKY Transcription Factors Associated With Andrographolide Biosynthesis
Four approaches, including WRKY binding site prediction, differential gene expression analysis, coexpression analysis, and phylogenetic analysis, were performed to predict genes involved in andrographolide biosynthesis. WRKY transcription factors can specifically bind to the W-box element [(T) TGAC (C)] located in the target gene promoter (Cheng et al., 2019; Liu et al., 2019; Zhu et al., 2019; Lv et al., 2020). In our study, the 2.0 kb promoter sequences of 56 key enzyme genes in the andrographolide biosynthetic pathway were analyzed, and a total of six binding sites were identified, of which WRKY71OS was possessed by all key enzyme gene promoters (Supplementary Table 5). The promoters of various genes possessed different binding sites. HMGR1 possessed six types of binding sites, and HMGS1 possessed three types of binding sites. These results imply that WRKYs may bind directly to W-box elements to regulate the expression of key enzyme genes and thereby affect andrographolide biosynthesis in A. paniculata.
Based on the expression level of WRKYs derived from the RNA-seq data, we preliminarily selected 24 WRKY genes, and their expression was further confirmed using qRT-PCR (Figures 8, 9). The qRT-PCR results showed that six genes (ApWRKY01, ApWRKY08, ApWRKY12, ApWRKY14, ApWRKY19, and ApWRKY20) were both most highly expressed in leaves and MeJA-treatment seedlings. Notably, another gene ApWRKY50 was most highly expressed in leaves, followed by stems, and lowest in roots. The expression level of ApWRKY50 was nearly 40 times higher than in roots, similar to that of the key enzyme gene CPS2. Though it was downregulated by MeJA, it seemed that ApWRKY50 maybe participate in the biosynthesis of andrographolide in leaves. A phylogenetic tree was further constructed using protein sequences of the candidates and the functional WRKYs known to regulate secondary metabolism. The seven WRKYs were all clustered with the functional WRKYs that regulated terpene biosynthesis. For instance, ApWRKY12 and ApWRKY19 were clustered with AaWRKY1, which regulate artemisinin from A. annua (Ma et al., 2009; Han et al., 2014). ApWRKY01 and ApWRKY50 belonged to Clade 2 with GaWRKY01 from Gossypium arboreum, AtWRKY40 and AtWRKY18 from A. thaliana (Xu et al., 2004; Alfieri et al., 2018). Therefore, based on the sensitive and tissue specific gene expression manner and evolutionary relationship of WRKYs, we suggested that the seven candidates are likely to participate in the biosynthesis of andrographolide.
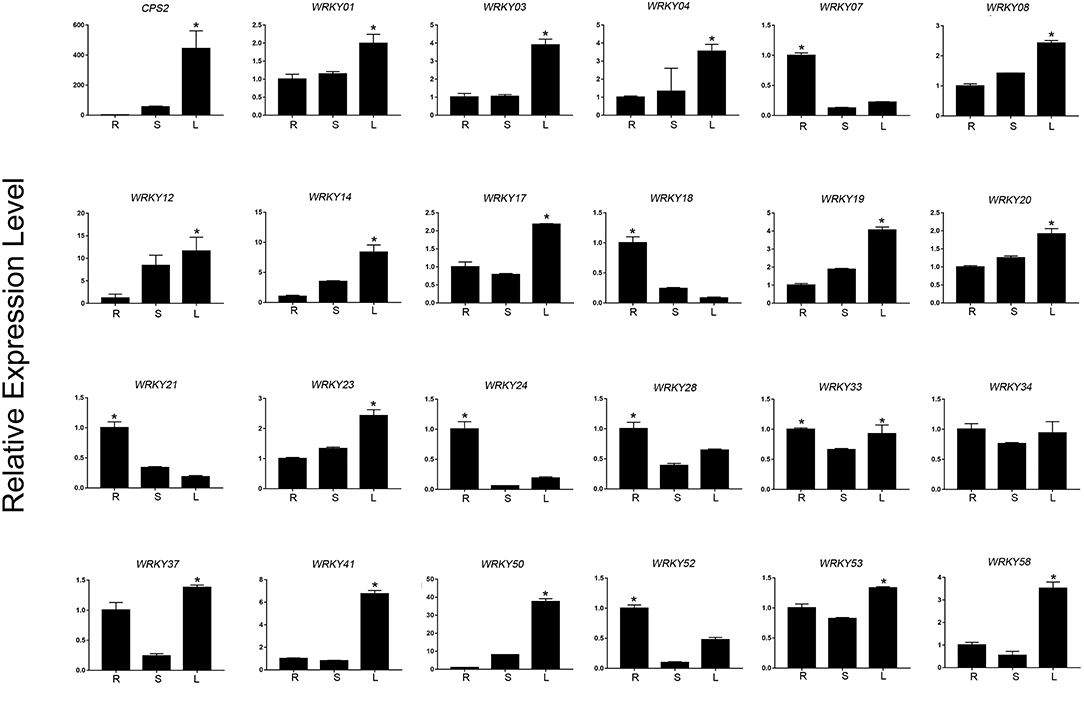
Figure 8. qRT-PCR analysis of ApWRKYs with putative roles in andrographolide biosynthesis in different organs (root, stem, leaf) in A. paniculata. The actin gene was tested as an internal reference. CPS2 was used as the positive control. The asterisks (*) represent significant differences (P < 0.05).
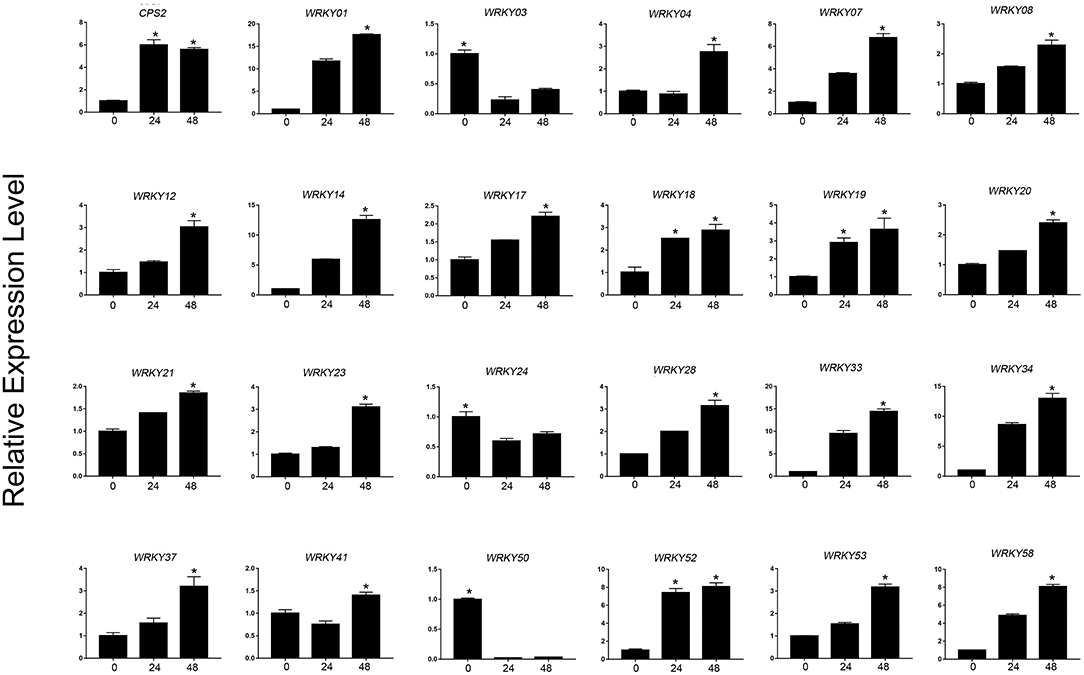
Figure 9. qRT-PCR analysis of ApWRKYs with putative roles in andrographolide biosynthesis in MeJA-treated seedlings (MeJA-24 and MeJA-48, respectively) in A. paniculata. The actin gene was tested as an internal reference. CPS2 was used as the positive control. The asterisks (*) represent significant differences (P < 0.05).
Discussion
Medicinal herbs have been widely used clinically due to their natural origins, rich variety, and low toxicity. A. paniculata is a well-known Chinese medicine in China and South Asian countries, and andrographolide, as the main bioactive component of A. paniculata, has many pharmacological activities. As the demand for andrographolide has grown over the past years, obtaining higher contents of andrographolide by controlling the biosynthesis pathway of andrographolide has attracted researchers' attention. Transcription factors are important regulators that can activate biosynthetic enzyme genes at the transcription level to promote active compound production (Bai et al., 2018; Cao et al., 2018; Pan et al., 2019). In A. paniculata, transcription factors have rarely been studied. Therefore, we report work relevant to the WRKY TFs in the regulation of andrographolide biosynthesis. This represents a first major step for the further identification of WRKYs involved in andrographolide biosynthesis.
Some special motifs may provide certain reference information for investigating genes function. The conserved motif distribution in all 58 WRKYs was obtained using MEME online software. Motif 1, motif 2 and motif 3, motif 5, motif 6 and motif 14 were all within the WRKY domain region, among which motif 1 was the core WRKY motif (WRKYGQK). Motif 2, motif 3 and motif 4 were assigned to a complete C2H2-type zinc finger structure. In the 58 ApWRKYs, 16 proteins have the EAR (ERF-associated amphiphilic repression) motif, which appears in the form of LxLxL or DLNxxP, was the first active repression motif reported in plants with diverse biological functions (Kagale and Rozwadowski, 2011; Shyu et al., 2012; Rakesh et al., 2014). Among the 16 ApWRKY proteins, only 3 ApWRKYs have DLNxxP-type EAR motifs. The remaining 13 proteins all possessed the LXLXL-type EAR motif. Moreover, 8 ApWRKY proteins (ApWRKY5, ApWRKY6, ApWRKY24, ApWRKY39, ApWRKY42, ApWRKY45, ApWRKY50, and ApWRKY57) contain LxxLL motifs, which may participate in protein-protein interactions that can activate or repress transcription. In Arabidopsis, AtERF11 with an EAR motif plays a key role in affecting ethylene biosynthesis by negatively regulating the expression of AtACS2/5 (Li et al., 2011). In Salvia miltiorrhiza, SmJAZ8 has an LXLXL-type EAR motif at the N terminus and negatively regulates the biosynthesis of salvianolic acids and tanshinones by reducing the expression of the enzyme genes of both biosynthetic pathways (Pei et al., 2018). The ApWRKYs possessed these specific motifs may have similar functions in development, stress response as well as secondary metabolism.
Considerable studies demonstrate that “multi-omics”-based method was an efficient tool to discovery candidate genes involving plant development, stress-response, and specialized metabolism. For example, metabolome and transcriptome data of 20 tomato tissues and stages were integrated; and a new transcription factor SlERF.G3-like was co-expressed with flavonoid compounds and biosynthetic genes. Further investigation revealed SlERF.G3-like positive regulated flavonoid biosynthesis in tomato (Li et al., 2020). In our previous study, the AP2/ERF transcription factor family of Salvia miltiorrhiza was analyzed via a genome-wide survey (Ji et al., 2016). A candidate gene SmERF128 was obtained and identified as a positive regulator promoting the production of tanshinone, which was the famous active compound treating cardiovascular diseases (Zhang et al., 2019). Andrographolide, the main medicinal component of A. paniculata, exhibits site specificity, which is reflected in its accumulation being most abundant in leaves and least abundant in roots (Garg et al., 2015). In addition, the andrographolide content in seedlings was significantly increased after MeJA treatment (Sharma et al., 2015; Sun et al., 2019). Previous studies have shown that many enzyme genes and transcription factors are involved in the formation of multiple secondary metabolites, and their expression patterns are significantly correlated with the distribution of active components (Lu et al., 2013; Ji et al., 2016, 2017; Huang et al., 2019; Li et al., 2019; Zhang et al., 2019). In this study, 7 WRKYs were proposed as candidates in andrographolide biosynthesis. The expression of these genes was all most highly in leaves, followed by stems and lowest in roots; and six of them were induced by MeJA. Though the expression of ApWRKY50 did not increase after MeJA treatment, its expression in leaves was much higher than in roots. Therefore, they were likely to be involved in andrographolide biosynthesis. In addition, the phylogenetic analysis indicated that the 7 WRKYs were all grouped with the functionally characterized WRKYs, which were involved in terpenoid biosynthesis. ApWRKY19 and ApWRKY12 were in Clade 1 with AaWRKY1 from A. annua and AtWRKY46 from A. thaliana. In A. annua, AaWRKY1 transcription factor binds W-box of ADS promoter to promote the production of artemisinin; and in A. thaliana, AtWRKY46 is the positive regulator in the BR (brassinosteroids) pathway (Ma et al., 2009; Han et al., 2014; Jiang et al., 2016; Chen et al., 2017a). In addition, ApWRKY1 was closely related to GaWRKY1 from cotton, which participates in regulation of sesquiterpene biosynthesis (Xu et al., 2004). Moreover, ApWRKY14 and ApWRKY20 were clustered with PqWRKY1 from Panax quinquefolius and OsWRKY13 from rice. PqWRKY1 is a positive regulator related to triterpene ginsenoside biosynthesis, while overexpression of OsWRKY13 downregulate Momilactone A biosynthesis (Qiu et al., 2008; Sun et al., 2013). These results indicated our candidates may be involved in the biosynthesis of andrographolide in A. paniculata. Further functional verification experiments based on these genes are yet to be completed.
Conclusion
In this study, a total of 58 WRKY genes were identified in A. paniculata genome. ApWRKYs can be divided into group I, group II (subgroup a-e) and group III supported by phylogeny, additional protein motifs, and intron/exon structures. Based on the gene expression pattern, qRT-PCR detection, and phylogenetic analyses, we obtained seven candidate genes that may participate in andrographolide biosynthesis. Therefore, based on the mentioned results, seven candidate WRKYs were inferred to be genes regulating andrographolide biosynthesis. The function of these genes is worth further exploration and may increase the production of andrographolide in metabolic engineering.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
AJ and LD conceived the research. RZ, ZC, LZ, and WY performed the experiment, data analysis, and wrote the paper. ZX, BL, YM, and HG contributed to transcriptome data analysis and discussion. AJ and CJ revised the paper. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (Grant no. 82003895); the Science and Technology Project of Guangdong Province (Grant no. 2019A1515110594); and the National Science and Technology Major Project for Significant New Drugs Development (2019ZX09201005-006-003).
Conflict of Interest
YM and CJ were employed by the company Jiangxi Qingfeng Pharmaceutical Co. Ltd., Ganzhou, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.601689/full#supplementary-material
Supplementary Figure 1. GO classification of the WRKYs in A. paniculata.
Supplementary Figure 2. Phylogenetic analysis of candidate WRKYs and functional WRKYs from other species.
Supplementary Table 1. Primer sequences of the candidate WRKYs used for qRT-PCR analyses.
Supplementary Table 2. Complete list of WRKY transcription factors identified in the A. paniculata genome.
Supplementary Table 3. Conserved motifs identified in WRKY transcription factors from A. paniculata by MEME analysis.
Supplementary Table 4. The average FPKM values of 58 WRKY genes in different organs and MeJA-treated seedlings.
Supplementary Table 5. Regulatory elements targeted by WRKY TFs in the promoters of the enzyme-coding genes of the andrographolide biosynthesis pathway.
References
Alfieri, M., Vaccaro, M., Cappetta, E., Ambrosone, A., De, T., and Leone, A. (2018). Coactivation of MEP-biosynthetic genes and accumulation of abietane diterpenes in Salvia sclarea by heterologous expression of WRKY and MYC2 transcription factors. Sci. Rep. 8:11009. doi: 10.1038/s41598-018-29389-4
Bai, Z., Li, W., Jia, Y., Yue, Z., Jiao, J., Huang, W., et al. (2018). The ethylene response factor SmERF6 co-regulates the transcription of SmCPS1 and SmKSL1 and is involved in tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. Planta 248, 243–255. doi: 10.1007/s00425-018-2884-z
Cao, W., Wang, Y., Shi, M., Hao, X., Zhao, W., Wang, Y., et al. (2018). Transcription factor SmWRKY1 positively promotes the biosynthesis of tanshinones in Salvia miltiorrhiza. Front. Plant Sci. 9:544. doi: 10.3389/fpls.2018.00554
Chaudhary, S., Jabre, I., Reddy, A., Staiger, D., and Syed, N. (2019). Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci. 24, 496–506. doi: 10.1016/j.tplants.2019.02.006
Chen, C., Chen, H., Zhang, Y., Thomas, H., Frank, M., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J., Nolan, T., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017a). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Chen, M., Yan, T., Shen, Q., Lu, X., Pan, Q., Huang, Y., et al. (2017b). GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 214, 304–316. doi: 10.1111/nph.14373
Cheng, X., Zhao, Y., Jiang, Q., Yang, J., Zhao, W., Taylor, I., et al. (2019). Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain. Nucleic Acids Res. 47, 4308–4318. doi: 10.1093/nar/gkz113
Chi, Y., Yang, Y., Zhou, Y., Zhou, J., Fan, B., Yu, J., et al. (2013). Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6, 287–300. doi: 10.1093/mp/sst026
Gao, H., Li, F., Xu, Z., Huang, C., Xiong, C., Jiang, C., et al. (2019). Genome-wide analysis of methyl jasmonate-regulated isoform expression in the medicinal plant Andrographis paniculata. Ind. Crops Prod. 135, 39–48. doi: 10.1016/j.indcrop.2019.04.023
Garg, A., Agrawal, L., Misra, R., Sharma, S., and Ghosh, S. (2015). Andrographis paniculata transcriptome provides molecular insights into tissue-specific accumulation of medicinal diterpenes. BMC Genomics 16:659. doi: 10.1186/s12864-015-1864-y
Han, J., Wang, H., Lundgren, A., and Brodelius, P. (2014). Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry 102, 89–96. doi: 10.1016/j.phytochem.2014.02.011
Hardigan, M., Feldmann, M., Lorant, A., Bird, K., Famula, R., Acharya, C., et al. (2020). Genome synteny has been conserved among the octoploid progenitors of cultivated strawberry over millions of years of evolution. Front. Plant Sci. 10:1789. doi: 10.3389/fpls.2019.01789
Huang, Q., Sun, M., Yuan, T., Wang, Y., Shi, M., Lu, S., et al. (2019). The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem. 274, 368–375. doi: 10.1016/j.foodchem.2018.08.119
Hussain, A., Noman, A., Khan, M., Zaynab, M., Aqeel, M., Anwar, M., et al. (2019). Molecular regulation of pepper innate immunity and stress tolerance: an overview of WRKY TFs. Microb. Pathog. 135:103610. doi: 10.1016/j.micpath.2019.103610
Ikeda, M., and Ohme-Takagi, M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50, 970–975. doi: 10.1093/pcp/pcp048
Ji, A., Jia, J., Xu, Z., Li, Y., Bi, W., Ren, F., et al. (2017). Transcriptome-guided mining of genes involved in Crocin Biosynthesis. Front. Plant Sci. 8:518. doi: 10.3389/fpls.2017.00518
Ji, A., Luo, H., Xu, Z., Zhang, X., Zhu, Y., Liao, B., et al. (2016). Genome-wide identification of the AP2/ERF gene family involved in active constituent biosynthesis in Salvia miltiorrhiza. Plant Genome 9, 1–11. doi: 10.3835/plantgenome2015.08.0077
Jiang, W., Fu, X., Pan, Q., Tang, Y., Shen, Q., Lv, Z., et al. (2016). Overexpression of AaWRKY1 leads to an enhanced content of artemisinin in Artemisia annua. Biomed. Res. Int. 2016:7314971. doi: 10.1155/2016/7314971
Kagale, S., and Rozwadowski, K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146. doi: 10.4161/epi.6.2.13627
Ksiazkiewicz, M., Nazzicari, N., Yang, H., Nelson, M., Renshaw, D., Rychel, S., et al. (2017). A high-density consensus linkage map of white lupin highlights synteny with narrow-leafed lupin and provides markers tagging key agronomic traits. Sci. Rep. 7:15335. doi: 10.1038/s41598-017-15625-w
Li, C., Li, D., Shao, F., and Lu, S. (2015). Molecular cloning and expression analysis of WRKY transcription factor genes in Salvia miltiorrhiza. BMC Genomics 16:200. doi: 10.1186/s12864-015-1411-x
Li, Y., Chen, Y., Zhou, L., You, S., Deng, H., Chen, Y., et al. (2020). MicroTom Metabolic Network: rewiring tomato metabolic regulatory network throughout the growth cycle. Mol Plant 13, 1203–1218. doi: 10.1016/j.molp.2020.06.005
Li, Z., Liu, C., Zhang, Y., Wang, B., Ran, Q., and Zhang, J. (2019). The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J. Exp. Bot. 70, 5471–5486. doi: 10.1093/jxb/erz307
Li, Z., Zhang, L., Yu, Y., Quan, R., Zhang, Z., Zhang, H., et al. (2011). The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J. 68, 88–99. doi: 10.1111/j.1365-313X.2011.04670.x
Lim, J., Chan, T., Ng, D., Sagineedu, S., Stanslas, J., and Wong, W. (2012). Andrographolide and its analogues: versatile bioactive molecules for combating inflammation and cancer. Clin. Exp. Pharmacol. Physiol. 39, 300–310. doi: 10.1111/j.1440-1681.2011.05633.x
Ling, J., Jiang, W., Zhang, Y., Yu, H., Mao, Z., Gu, X., et al. (2011). Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 12.:471. doi: 10.1186/1471-2164-12-471
Liu, Y., Yang, T., Lin, Z., Gu, B., Xing, C., Zhao, L., et al. (2019). A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 17, 1770–1787. doi: 10.1111/pbi.13099
Lu, X., Zhang, L., Zhang, F., Jiang, W., Shen, Q., Zhang, L., et al. (2013). AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 198, 1191–1202. doi: 10.1111/nph.12207
Lv, B., Wu, Q., Wang, A., Li, Q., Dong, Q., Yang, J., et al. (2020). A WRKY transcription factor, FtWRKY46, from Tartary buckwheat improves salt tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 147, 43–53. doi: 10.1016/j.plaphy.2019.12.004
Ma, D., Pu, G., Lei, C., Ma, L., Wang, H., Guo, Y., et al. (2009). Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50, 2146–2161. doi: 10.1093/pcp/pcp149
Mishra, K., Dash, A., Swain, B., and Dey, N. (2009). Anti-malarial activities of Andrographis paniculata and Hedyotis corymbosa extracts and their combination with curcumin. Malaria J. 8:26. doi: 10.1186/1475-2875-8-26
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/TPC.010127
Pan, Q., Wang, C., Xiong, Z., Wang, H., Fu, X., Shen, Q., et al. (2019). CrERF5, an AP2/ERF transcription factor, positively regulates the biosynthesis of bisindole alkaloids and their precursors in Catharanthus roseus. Front. Plant Sci. 10:931. doi: 10.3389/fpls.2019.00931
Pei, T., Ma, P., Ding, K., Liu, S., Jia, Y., Ru, M., et al. (2018). SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acids and tanshinones in Salvia miltiorrhiza hairy roots. J. Exp. Bot. 69, 1663–1678. doi: 10.1093/jxb/erx484
Qiu, D., Xiao, J., Xie, W., Liu, H., Li, X., Xiong, L., et al. (2008). Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol. Plant 1, 538–551. doi: 10.1093/mplant/ssn012
Raboanatahiry, N., Chao, H., Guo, L., Gan, J., Xiang, J., Yan, M., et al. (2017). Synteny analysis of genes and distribution of loci controlling oil content and fatty acid profile based on QTL alignment map in Brassica napus. BMC Genomics 18:776. doi: 10.1186/s12864-017-4176-6
Raina, A., Gupta, V., Sivaraj, N., and Dutta, M. (2013). Andrographis paniculata (Burm. f.) Wall. ex Nees (kalmegh), a traditional hepatoprotective drug from India. Genet. Resour. Crop Evol. 60, 1181–1189. doi: 10.1007/s10722-012-9953-0
Rakesh, K., Asmita, G., Sanjay, R., Ruchi, S., Uday, V., Pravendra, N., et al. (2014). The EAR Motif controls the early flowering and senescence phenotype mediated by over-Expression of SlERF36 and is partly responsible for changes in stomatal density and photosynthesis. PLoS ONE 9:e101995. doi: 10.1371/journal.pone.0101995
Shao, Y., Gao, J., Wu, X., Li, Q., Wang, J., Ding, P., et al. (2015). Effect of salt treatment on growth, isoenzymes and metabolites of Andrographis paniculata (Burm. f.) Nees. Acta Physiol. Plant 37, 1–12. doi: 10.1007/s11738-015-1787-x
Sharma, S., Jha, Z., Sinha, R., and Geda, A. (2015). Jasmonate-induced biosynthesis of andrographolide in Andrographis paniculata. Physiol. Plant 153, 221–229. doi: 10.1111/ppl.12252
Shyu, C., Figueroa, P., DePew, C., Cooke, T., Sheard, L., Moreno, J., et al. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24, 536–550. doi: 10.1105/tpc.111.093005
Sun, W., Leng, L., and Yin, Q. (2019). The genome of the medicinal plant Andrographis paniculata provides insight into the biosynthesis of the bioactive diterpenoid neoandrographolide (vol 97, pg 841, 2018). Plant J. 97, 996–996. doi: 10.1111/tpj.14162
Sun, Y., Niu, Y., Xu, J., Li, Y., Luo, H., Zhu, Y., et al. (2013). Discovery of WRKY transcription factors through transcriptome analysis and characterization of a novel methyl jasmonate-inducible PqWRKY1 gene from Panax quinquefolius. Plant Cell Tiss. Org. 114, 269–277. doi: 10.1007/s11240-013-0323-1
Tiwari, S., Belachew, A., Ma, S., Young, M., Ade, J., Shen, Y., et al. (2012). The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70, 855–865. doi: 10.1111/j.1365-313X.2012.04935.x
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Wei, K., Chen, J., Chen, Y., Wu, L., and Xie, D. (2012). Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 19, 153–164. doi: 10.1093/dnares/dsr048
Wu, K., Guo, Z., Wang, H., and Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Xu, Y., Wang, J., Wang, S., Wang, J., and Chen, X. (2004). Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol. 135, 507–515. doi: 10.1104/pp.104.038612
Yan, H., Li, M., Xiong, Y., Wu, J., da Silva, J., and Ma, G. (2019). Genome-wide characterization, expression profile analysis of WRKY family genes in Santalum album and functional identification of their role in abiotic stress. Int. J. Mol. Sci. 20:5676. doi: 10.3390/ijms20225676
Yu, B., Dai, C., Jiang, Z., Li, E., Chen, C., Wu, X., et al. (2014). Andrographolide as an anti-H1N1 drug and the mechanism related to retinoic acid-inducible gene-I-like receptors signaling pathway. Chin. J. Integr. Med. 20, 540–545. doi: 10.1007/s11655-014-1860-0
Zhang, J., Zhao, P., Zhao, J., and Chen, G. (2018). Synteny-based mapping of causal point mutations relevant to sand rice (Agriophyllum squarrosum) trichomeless1 mutant by RNA-sequencing. J. Plant Physiol. 231, 86–95. doi: 10.1016/j.jplph.2018.09.003
Zhang, Y., Ji, A., Xu, Z., Luo, H., and Song, J. (2019). The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza. Plant Mol. Biol. 100, 83–93. doi: 10.1007/s11103-019-00845-7
Zhao, T., Holmer, R., de Bruijn, S., Angenent, G., van den Burg, H., and Schranz, M. (2017). Phylogenomic synteny network analysis of mADS-Box transcription factor genes reveals lineage-specific transpositions, ancient tandem duplications, and deep positional conservation. Plant Cell 29, 1278–1292. doi: 10.1105/tpc.17.00312
Keywords: WRKY, Andrographis paniculata, andrographolide biosynthesis, genome-wide, expression patterns
Citation: Zhang R, Chen Z, Zhang L, Yao W, Xu Z, Liao B, Mi Y, Gao H, Jiang C, Duan L and Ji A (2021) Genomic Characterization of WRKY Transcription Factors Related to Andrographolide Biosynthesis in Andrographis paniculata. Front. Genet. 11:601689. doi: 10.3389/fgene.2020.601689
Received: 03 September 2020; Accepted: 22 December 2020;
Published: 18 January 2021.
Edited by:
Madhav P. Nepal, South Dakota State University, United StatesReviewed by:
Ethan Andersen, Francis Marion University, United StatesBharathi Raja Ramadoss, University of Saskatchewan, Canada
Copyright © 2021 Zhang, Chen, Zhang, Yao, Xu, Liao, Mi, Gao, Jiang, Duan and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lixin Duan, bmxpem5AZ3p1Y20uZWR1LmNu; Aijia Ji, YWpqaUBnenVjbS5lZHUuY24=
†These authors have contributed equally to this work
 Rongrong Zhang
Rongrong Zhang Zhenzhen Chen1†
Zhenzhen Chen1† Baosheng Liao
Baosheng Liao Lixin Duan
Lixin Duan Aijia Ji
Aijia Ji