- 1Division of Anthropology, Institute of Organismic and Molecular Evolution, Faculty of Biology, Johannes Gutenberg University Mainz, Mainz, Germany
- 2Department of Virology, Biomedical Primate Research Centre, Rijswijk, Netherlands
Studies on the function of PRDM9 in model systems and its evolution during vertebrate divergence shed light on the basic molecular mechanisms of hybrid sterility and its evolutionary consequences. However, information regarding PRDM9-homolog, PRDM7, whose origin is placed in the primate evolutionary tree, as well as information about the fast-evolving DNA-binding zinc finger array of strepsirrhine PRDM9 are scarce. Thus, we aimed to narrow down the date of the duplication event leading to the emergence of PRDM7 during primate evolution by comparing the phylogenetic tree reconstructions of representative primate samples of PRDM orthologs and paralogs. To confirm our PRDM7 paralogization pattern, database-deposited sequences were used to test the presence/absence patterns expected from the paralogization timing. In addition, we extended the existing phylogenetic tree of haplorrhine PRDM9 zinc fingers with their strepsirrhine counterparts. The inclusion of strepsirrhine zinc fingers completes the PRDM9 primate phylogeny. Moreover, the updated phylogeny of PRDM9 zinc fingers showed distinct clusters of strepsirrhine, tarsier, and anthropoid degenerated zinc fingers. Here, we show that PRDM7 emerged on the branch leading to the most recent common ancestor of catarrhines; therefore, its origin is more recent than previously expected. A more detailed character evolutionary study suggests that PRDM7 may have evolved differently in Cercopithecoidea as compared to Hominoidea: it lacks the first four exons in Old World monkeys orthologs and exon 10 in Papionini orthologs. Dating the origin of PRDM7 is essential for further studies investigating why Hominoidea representatives need another putative histone methyltransferase in the testis.
Introduction
PRDM9 is a putative speciation gene consisting of the KRAB, SSXRD, and SET domains and a zinc finger array (Hayashi et al., 2005; Birtle and Ponting, 2006). Its expression is restricted to male and female germlines during meiosis I (Hayashi et al., 2005). PRDM9 has been studied in several haplorrhine primate species (Oliver et al., 2009; Auton et al., 2012; Groeneveld et al., 2012; Schwartz et al., 2014; Heerschop et al., 2016; Stevison et al., 2016). The zinc finger array of this gene has undergone rapid evolution with regard to the number of tandem repeats as well as its DNA-binding amino acids, and has been associated with positive selection (Oliver et al., 2009; Thomas et al., 2009; Myers et al., 2010). Phylogenies of PRDM9 show that the 5′-most zinc fingers of all studied primates are distinct from other canonical C2H2 zinc finger sequences (Schwartz et al., 2014; Heerschop et al., 2016).
During primate evolution, a duplication event led to the emergence of PRDM9′s homolog, PRDM7, presumably on the branch of Haplorrhini or Anthropoidea (Fumasoni et al., 2007; Vervoort et al., 2016). Due to the recency of this event, the two genes are highly similar in some aspects. While the PR/SET domains are having a 97% identical sequence, structural differences are present downstream of the PR domain (Blazer et al., 2016). PRDM7 shows an internal duplication of 89 bases in human exon 10 according to the transcript PRDM7-202 (Ensembl, transcript ID: ENST00000449207.7). This duplication is not based on repetitive elements, and leads to a frameshift and a new splicing site, which gives rise to some isoforms expressing four zinc fingers, while others do not express any zinc fingers (Fumasoni et al., 2007). While PRDM7 is mainly expressed in testis, expression patterns were observed in other tissues such as melanocytes (Hayashi et al., 2005; Fumasoni et al., 2007). Compared to PRDM9, PRDM7 exhibits a reduced catalytic function on H3K4me3 and has no catalytic function on H3K36me3. Three missense mutations in the PR domain of PRDM7, particularly the change from tyrosine to serine at position 357, might have led to these functional differences (Blazer et al., 2016). Fumasoni et al. (2007) suggested that PRDM9 is the ancestor gene of PRDM7 because its genomic surroundings and its gene and protein structures are more conserved in comparison to its paralog PRDM7.
The main goal of this paper is to narrow down the origin of PRDM7 with phylogenetic reconstructions and additional amplified sequences to support the findings. A second aim is to include strepsirrhine PRDM9 zinc fingers in an existing phylogeny to reveal if they form a cluster and, therefore, present a similar pattern of concerted evolution as haplorrhine zinc fingers.
Methods
Phylogeny of PRDM7 and PRDM9
Two strategies were applied to determine the origin of PRDM7 during primate divergence. The underlying sequences of the two phylogenetic reconstructions were obtained via BLASTN with normal search sensitivity, Ensembl version 102. The first one contains exon 7 to exon 9 including introns. Exon 7 and exon 9 of human PRDM9 were blasted within all primate species on Ensembl, and the sequences from the start of exon seven to the end of exon nine were downloaded. With BLASTN and “distant homologies” as search sensitivity, the homolog sequences of Bos taurus and Mus musculus were obtained as outgroups. The second phylogenetic reconstruction is based on the human PRDM9 3’ UTR which was also blasted within all primate species, extended to 1500 bases downstream and downloaded. In both cases, only BLAST results with 70% of the length and e-values below 0.0001 were accepted. The loci of the analyzed sequences and the genome assemblies can be found in Supplementary Data Sheets 1 and 2. If possible PRDM7 and PRDM9 were assigned with the help of syntenic regions on Ensembl. In cercopithecoid species, similar patterns in the BLAST results of PRDM7 exons (see section “Ensembl BLAST”) were used to distinguish the genes. The phylogenetic reconstruction was performed on ngphylogeny.fr (Dereeper et al., 2008; Lemoine et al., 2019) with the following workflow: MAFFT was used for the alignment (Katoh and Standley, 2013), Gblocks for the curation (Castresana, 2000; Talavera and Castresana, 2007), PhyML with 1000 bootstraps as support for the tree inference (Guindon et al., 2010; Lemoine et al., 2018) with default settings (substitution model: GTR + I + G) besides the bootstrap support and Newick for tree rendering (Junier and Zdobnov, 2010). Adjustments were made with the Interactive Tree of Life (iTOL) v4 (Letunic and Bork, 2019): nodes with bootstrap values below 500 were collapsed. B. taurus was set as outgroup in the phylogeny based on exons and introns.
PCRs of PRDM7 and PRDM9
For additional support, we used PCR to amplify the last exon of PRDM7 and PRDM9 (primer: 5′-ACATCTACCCTGACCA AAAAC-3′, 5′-CGGATTTGTTTAATCAGTTATTTC-3′) of Macaca mulatta (Tm: 52°C), Hylobates lar (Tm: 50°C), Pongo abelii (Tm: 52°C), Gorilla gorilla gorilla (Tm: 54°C), Pan troglodytes (Tm: 52°C), and Homo sapiens (Tm: 52°C). The differences in length were observed on an agarose gel with the Thermo Scientific GeneRuler 100 bp Plus DNA Ladder: PRDM7 gives a short amplicon and PRDM9 can give one or two longer amplicons due to length polymorphisms in the zinc finger array. We Sanger sequenced all three products of M. mulatta and used a BLAST routine to check whether the amplicons overlap with PRDM7 or PRDM9. Furthermore, we used PCR to amplify a sequence within exon 10 of PRDM7, containing a PRDM7-specific internal duplication for following species: Daubentonia madagascariensis, Cheirogaleus medius, Carlito syrichta, Callithrix jacchus, H. lar, P. abelii, G. g. gorilla, P. troglodytes, and H. sapiens (primer: 5′-AACTGTGCCCGGGATGAT-3′, 5′-ACTTGCTGCCCCACTTGAT-3′, Tm: 51°C). Samples from six non-related individuals per species (G. g. gorilla and P. troglodytes) were amplified. Human data on PRDM7’s internal duplication and PRDM9’s corresponding sequences were retrieved from the 1000 Genomes Project (Auton et al., 2015) by using an in-house shell-script. All exons were concatenated, and the unique sequences were selected with the help of in-house Python scripts (see Supplementary Data Sheet 1).
Ensembl BLAST
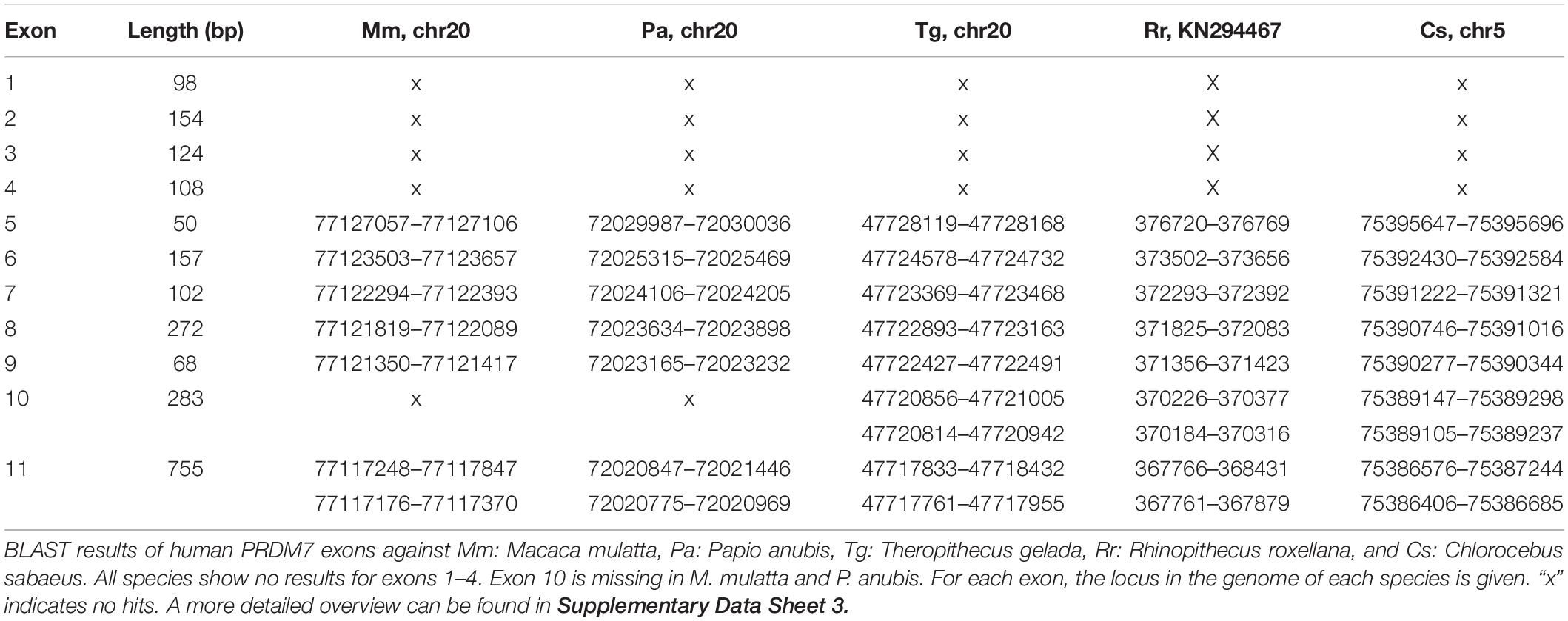
All human PRDM7_202 exons were blasted against the genomes of M. mulatta, Macaca fascicularis, Macaca nemestrina, Theropithecus gelada, Papio anubis, Cercocebus atys, Mandrillus leucophaeus, Chlorocebus sabaeus, Rhinopithecus roxellana, Rhinopithecus bieti, Piliocolobus tephrosceles, Nomascus leucogenys, P. abelii, G. g. gorilla, and P. troglodytes on Ensembl version 102, with default settings (Yates et al., 2020). For Cercopithecoidea, another BLAST with human PRDM9_201 exon 10 was performed. To evaluate the consequences of missing exons, human PRDM7 exons 5–11 were analyzed with ScanProsite (Sigrist et al., 2002; Castro et al., 2006) as well as human PRDM7 lacking exon 10 (based on the Papionini sequence).
The Segmental Duplication Database (Bailey et al., 2001, 2002; She et al., 2004a,b) was searched to see if PRDM7 and PRDM9 are listed. To check if PRDM9 and PRDM7 differ in their predicted loss of function variants, both genes were scanned on Genome Aggregation Database (gnomAD), v2.1.1 (Karczewski et al., 2020).
Phylogeny of PRDM9 Zinc Fingers
The reconstruction of a PRDM9 zinc finger phylogeny is based on sequence data from Heerschop et al. (2016) and data from Schwartz et al. (2014). It encompasses zinc fingers of nine strepsirrhine species, including Microcebus murinus, C. medius, Eulemur coronatus, Prolemur simus, Varecia variegata, Propithecus coquereli, D. madagascariensis, Nycticebus coucang, and Otolemur garnettii and one degenerated zinc finger of the mouse and cow as outgroups. Sequences of C. medius, E. coronatus, V. variegata, D. madagascariensis, and N. coucang were obtained by sequencing PCR products amplified with the following primers: 5′-GCCCAGAACAGGCCAGA-3′ and 5′-TTCTGGTCTCTTTACACTCTTGG-3′ (Tm: 50°C, D. madagascariensis and E. coronatus); 5′-GGGATCAGAATCAGGAGCAG-3′ and 5′-CTCTAGTCAT GAAAGTAGAGGATTTG-3′ (Tm: 54°C, C. medius, V. variegata, and N. coucang). The shorter alleles of D. madagascariensis and E. coronatus were separated by gel electrophoresis and extracted using the QIAquick® Gel Extraction Kit of Qiagen. One allele of V. variegata was isolated by ligation into a pGEM T-vector and transfected into TOP10 electrocompetent cells. PRDM9 exon 11 zinc finger array sequences of C. medius, D. madagascariensis, E. coronatus, and V. variegata have been deposited in GenBank under the accession numbers MT862444–MT862447. The sequence of N. coucang can be found in Supplementary Data Sheet 1. Data on M. murinus (PRDM9, ENSMICG00000037613), P. simus (PRDM7, ENSPSMG00000016627), P. coquereli (PRDM7, ENSPCOG00000013284), and O. garnettii (GL873654:27557–28889) were extracted by looking for human PRDM9 exon 11 orthologs using BLAST on the Ensembl database, version 102. Although listed as PRDM7, we assumed that the sequences of P. coquereli and O. garnettii represent PRDM9. A zinc finger alignment was prepared manually, and DNA binding codons were removed. The phylogenetic reconstruction was performed on ngphylogeny.fr (Dereeper et al., 2008; Lemoine et al., 2019) with following workflow: MAFFT was used for the alignment (Katoh and Standley, 2013), BMGE for the curation (Criscuolo and Gribaldo, 2010), PhyML with SH-like support for the tree inference (Guindon et al., 2010; Lemoine et al., 2018) with default settings (substitution model: GTR + I + G) beside the support and Newick for tree rendering (Junier and Zdobnov, 2010). Adjustments were made with iTOL v4 (Letunic and Bork, 2019): nodes with SH-like values below 0.5 were collapsed. B. taurus was set as outgroup. A pruned phylogenetic tree of primate zinc fingers was constructed by merging the zinc fingers of Anthropoidea and Tarsiers, and editing the Newick file manually.
Results
The Origin of PRDM7
Phylogenetic reconstructions of PRDM7 and PRDM9 orthologs were performed to determine when PRDM7 arose. PCRs of the last exon should confirm the results. The basis of the presented phylogenetic reconstructions is represented by sequencing data obtained from the Ensembl genome browser, version 102. Both phylogenetic reconstructions yield similar topological results (Figure 1). PRDM7 of diverse catarrhine species forms a monophylum in the 3′ phylogeny. In the pylogeny including exons and introns, PRDM7 of Cercopithecoidea and Hominoidea cluster separately as sister taxa to PRDM9 of Catarrhini. PRDM7 branches off in the phylogenies following the Platyrrhini–Catarrhini split. The phylogenetic pattern representing PRDM9 in Hominoidea, Cercopithecoidea, Platyrrhini, and Strepsirrhini resembles the topology of the widely accepted species tree. The BLAST for exons 7 and 9 shows two results in Cebus capucinus imitator and they form sister taxa in the phylogeny. In the phylogeny based on the 3′ UTR, the second PRDM9 was not integrated because of unassembled regions.
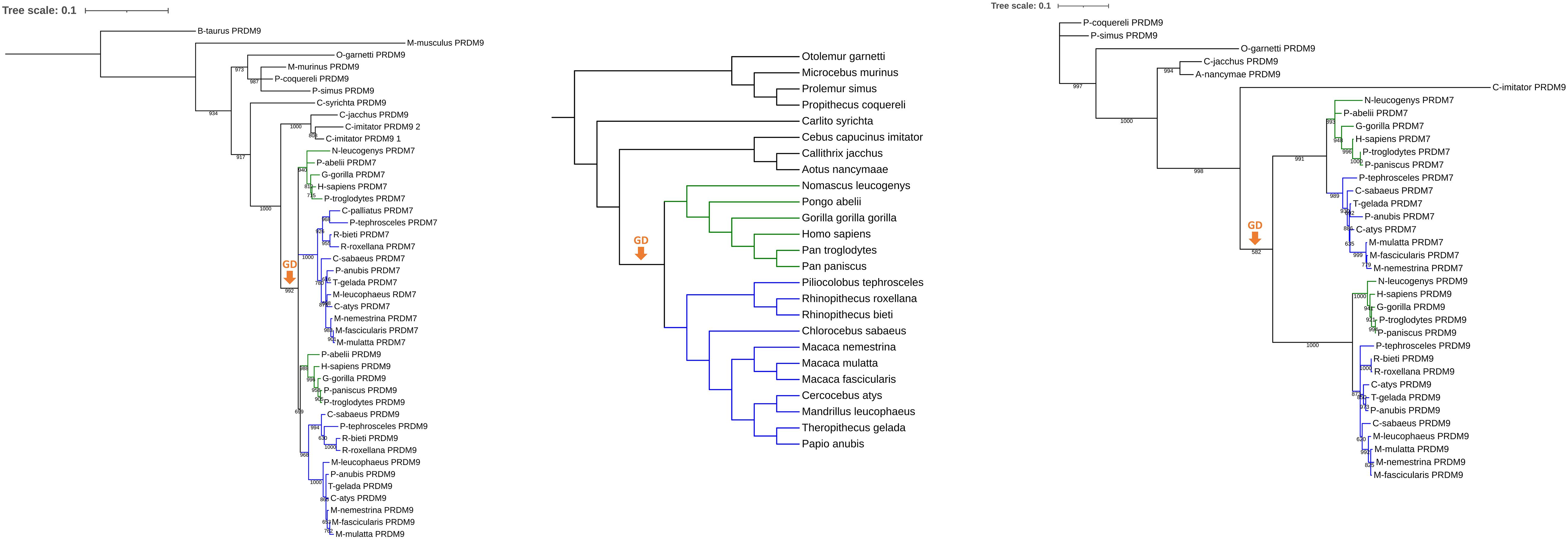
Figure 1. The origin of PRDM7. The phylogenetic reconstructions on primate PRDM7 and PRDM9 sequences based on exon seven to nine (left) and the 1500 bp downstream of the stop codon (right) show the gene duplication event, marked with an orange arrow. In both cases, PRDM9 sequences of Catarrhini form a cluster and PRDM7 sequences of Cercopithecoidea (blue) and Hominoidea (green) are the sister taxa (left) or taxon (right). That suggests that PRDM7 originated on the branch of catarrhine before the split of Cercopithecoidea and Hominoidea. PRDM9 of Bos taurus was chosen as the outgroup in the phylogeny based on exons 7–9. The bootstrap values of nodes are displayed. The scale bar indicates the mean number of nucleotide substitutions per site. The phylogeny in the middle displays a species tree that was created based on Geissmann (2003) and Perelman et al. (2011).
The PCR amplification of exon 11 of M. mulatta, H. lar, P. abelii, G. g. gorilla, P. troglodytes, and H. sapiens exhibited two or three products representing PRDM7 and PRDM9 (Figures 2C,D). The presence of two PRDM9 alleles with length polymorphisms explains the amplification of a third product. Sequencing of these M. mulatta products and subsequent BLASTs indicate that the shortest sequence represents the PRDM7 and the two longer sequences represent the PRDM9 alleles.
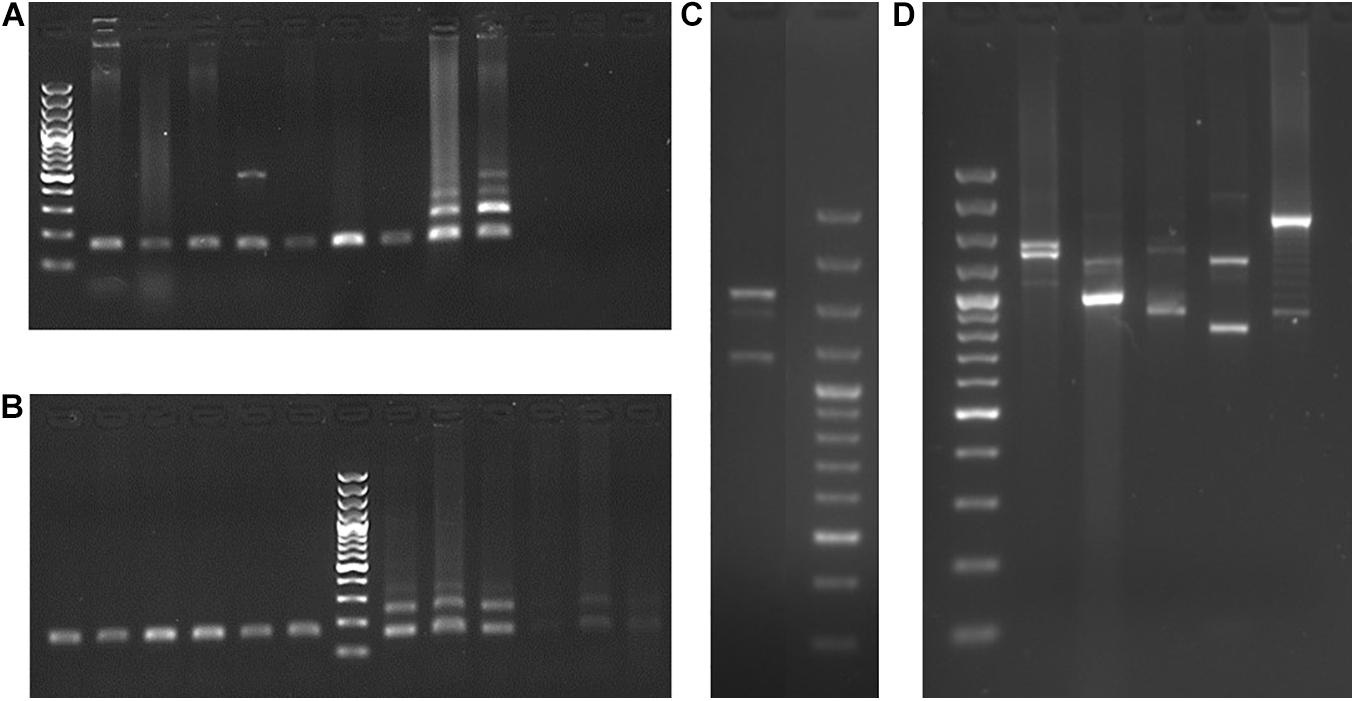
Figure 2. Characterization of PRDM7 and PRDM9 in primates. PCR products of PRDM7 and PRDM9 for (A) exon 10 of D. madagascariensis, C. medius, C. syrichta, C. jacchus, H. lar, P. abelii, G. g. gorilla, P. troglodytes, and H. sapiens show that the internal duplication in PRDM7 is exclusive to chimp and human. (B) Exon 10 of several, mostly unrelated gorillas (left side of the ladder) and chimpanzees (right side of the ladder) reveals that there is no incomplete lineage sorting regarding the internal duplication in PRDM7. (C) Exon 11 of M. mulatta. (D) Exon 11 of H. lar, P. abelii, G. g. gorilla, P. troglodytes, and H. sapiens. The last two gels show both, PRDM7 and PRDM9 amplicons in these anthropoid species. The Thermo Scientific GeneRuler 100 bp Plus DNA Ladder was used.
The regions chr5: 23504220–23528686 and ch16: 90123418–90147718 are listed in the Segmental Duplication Database and match with the loci of PRDM9 (chr5: 23507724–23528706) and PRDM7 (chr16: 90122974–90142338) of the human genome assembly GRCh37 on which the Segmental Duplication Database is based.
According to gnomAD, the expected and observed predicted losses of function variants are similar for both genes, with values of 1.01 for PRDM9 and 1.02 for PRDM7.
Internal Duplication of PRDM7 Is Exclusively Found in Human and Chimp
PRDM7 displays an internal duplication in human. Our aim is to check whether this is a distinct marker to distinguish PRDM7 from PRDM9 in primates. The PCR amplification of the region encompassing PRDM7’s internal duplication showed that only the P. troglodytes and H. sapiens samples displayed products of two distinct sizes, thus differing from a variety of strepsirrhine and haplorrhine species of primates (Figure 2A). To further support this observation, we amplified this region in five non-related gorillas and four non-related chimpanzees to exclude the possibility of incomplete lineage sorting of ancestral polymorphisms. The gorilla samples showed one product, while for the chimpanzee samples, we observed at least two products (Figure 2B). Human data obtained from the 1000 Genomes Project confirmed our PCR results; while 246 non-identical PRDM7 sequences showed internal duplication in exon 10, the 74 non-identical PRDM9 sequences showed the shorter version of exon 10.
Absent PRDM7 Exons in Catharrhini
A BLAST search of human PRDM7 (PRDM7_202) exons, examining the genome assemblies of 11 Cercopithecoidea and four Hominoidea representatives from the Ensembl database, was performed to screen for differences in these taxa. It generated two hits with the expected length, indicating the high sequence similarity between PRDM7 and PRDM9. No hit was obtained for PRDM7 exons 1–4 in the 11 Cercopithecoidea and Gibbon genomes. Furthermore, exons 1–6 of chimpanzee PRDM7 were not listed in the BLAST results. The sequences upstream of exons 5 and 7 contain unassembled sites in the cases of C. sabaeus, R. roxellana, T. gelada, N. leucogenys, and P. troglodytes. Nevertheless, the BLAST analysis of P. abelii and G. g. gorilla yielded all exons. In addition, macaques and P. anubis lack exon 10, while the sequence between exons 9 and 11 of both species revealed no unassembled sections. The lengths of these sequences were 1718 bp in P. anubis, 3502 bp in M. mulatta, and 2917 bp in H. sapiens. The BLAST results for Cercopithecoidea showed two PRDM7 exon 10 hits (Table 1), which are due to the insert of 89 bp in exon 10 (found in chimpanzees and humans). N. leucogenys lacks exon 7 and 10 in its PRDM9 locus. The BLAST query sequence with human PRDM9 exon 10 resulted in a better fit with one hit. Furthermore, two hits for exon 11 were obtained for Cercopithecoidea. This might indicate their origin from rudimentary zinc finger repeats.
A ScanProsite analysis of manually truncated human PRDM7 amino acid sequences revealed that exons 2–4 encode the KRAB domain and exon 10 partly encodes the SET domain.
Missing Cluster of Strepsirrhine Zinc Fingers
We integrated strepsirrhine PRDM9 zinc finger sequences into a previously published data set to see if they form a distinct cluster like Tarsiers and Anthropoids do. The phylogeny of PRDM9 zinc fingers (Figure 3) showed three clusters of 5′ end degenerated zinc fingers belonging to the strepsirrhine, tarsier, and anthropoid taxa with a C2H2 zinc finger of N. coucang as sister taxa of O. garnettii degenerated zinc finger. In contrast, the classical zinc fingers of Strepsirrhini do not form a cluster. C2H2 zinc fingers of O. garnettii and N. coucang form outgroups to all other C2H2 zinc fingers. With the exceptions of one D. madagascariensis, one E. coronatus, and two of P. simus orthologs, Lemuriformes zinc fingers branch off from the lineage leading to haplorrhine C2H2 zinc fingers. Tarsier classical zinc fingers form a monophylum in the haplorrhine cluster.
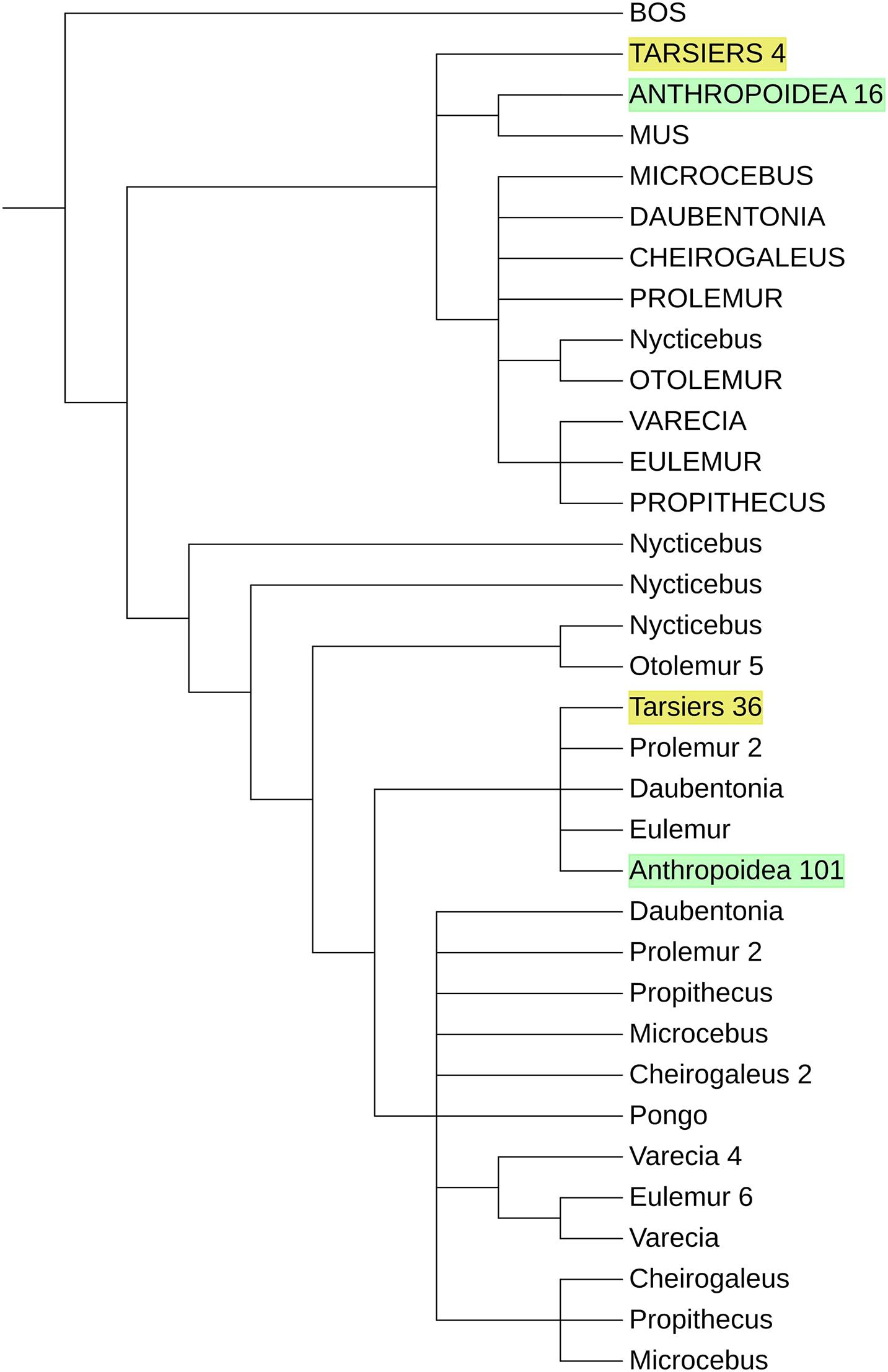
Figure 3. Schematic phylogeny of primate PRDM9 zinc fingers. The degenerated zing fingers of Strepsirrhini, Tarsier, and Anthropoids form clusters. The C2H2 zinc fingers of the strepsirrhine primates are the sister taxa of the other primates, but they do not show a distinct clustering. Sequences of Anthropoidea and Tarsiers are merged with the number of zinc fingers indicated at the end of the identifier. Degenerated zinc fingers are displayed in capital letters, and the zinc fingers of Anthropoidea and Tarsiers are marked in green and yellow, respectively. See Supplementary Data Sheet 1 for a more detailed version.
Discussion
By including strepsirrhine data to the phylogeny of PRDM9 zinc fingers previously published by Heerschop et al. (2016), we herein show that strepsirrhines contrast the haplorrhine primates by not forming a cluster as Tarsiers or Anthropoids do. Therefore, the realization of a concerted evolution of the zinc fingers as proposed by Schwartz et al. (2014) for the anthropoid primates does not apply for the PRDM9 zinc fingers of Strepsirrhini, especially regarding C2H2 zinc fingers of the two Lorisiformes representatives, O. garnettii and N. coucang. They branch off earlier in the phylogeny than those of the other strepsirrhine primates. This may indicate different patterns and regimes governing PRDM9 zinc finger evolution in the two branches (Lemurs and Loris) representing the deepest split of Strepsirrhini.
The date of PRDM7’s origin is narrowed down to the lineage leading to extant Catarrhini, after the platyrrhines split off. The well-supported topology obtained from the phylogenetic reconstructions is corroborated by presence/absence analyses. Accordingly, no signature of a paralogization event leading to PRDM7 was observed in platyrrhine species following a BLAST query of the genomes of several representatives of these species. The corresponding period dates back to an estimated 32–43 million years ago, and since then several segmental duplications arose in catarrhine primates as indicated by the analysis of the human genome (Lander et al., 2001; Samonte and Eichler, 2002; Perelman et al., 2011). These segmental duplications are not evenly distributed throughout chromosomes, thus being predominantly located in the pericentromeric and subtelomeric regions (Bailey et al., 2001; Lander et al., 2001). Due to PRDM7’s position at the end of chromosome 16q and its putative emergence during a period of segmental duplication, it is reasonable to speculate that this gene probably originated from the aforementioned rearrangements. These assumptions support the previously established hypothesis that portrays PRDM9 as its ancestral gene. Fumasoni et al. (2007) concluded that PRDM9 is the predecessor of PRDM7 based on the conserved genomic flanks and the high similarity of rodent co-orthologs.
The internal duplication of PRDM7 is an exclusive characteristic of chimpanzees and humans and is therefore not suitable for distinguishing PRDM7 from PRDM9 in other species. Nevertheless, the effects of this frameshifting rearrangement on the function of PRDM7 with and without the duplication remain to be determined. The BLAST searches of human PRDM7 exons querying catarrhine genomes revealed that cercopithecine primates and N. leucogenys might lack the first four exons, while P. troglodytes might lack six exons. It remains unclear whether these exons are missing or the inability to detect them is due to poorly assembled segments. Furthermore, PRDM7 exon 10 is missing in macaques and P. anubis. T. gelada, M. leucophaeus, and C. atys are more closely related to P. anubis than they all are to macaques. However, the BLAST results of exon 10 show a match in the sister taxa of P. anubis. Here is yet to be determined if information is lacking or if there were two separate events leading to the loss of exon 10 in macaques and P. anubis. Exons 2–4 are coding for the KRAB domain, while exon 10 codes partially for the SET domain. Therefore, PRDM7 is putatively experiencing a loss of functional constraints on the cercopithecine branch.
The most common result of a gene duplication is the pseudogenization of one copy (Ohno, 1972). While PRDM7 might be a pseudogene in Old World monkeys, its putative H3K4 methyltransferase activity, shown in a catalytic assay and in cells with human PRDM7 cDNA (Blazer et al., 2016), and the homologous structure in gorilla and orangutan could indicate that there is a sub- or neofunctionalization of this young PRDM member in Hominoidea family. However, due to similar branch lengths during both PRDM7- and PRDM9-divergence, both positive selection and loss of functional constraints apparently did not play a major role after paralogization and early evolution of the duplicate. Similarly, character evolution as revealed by the relative proportion of functional substitutions obtained by querying the gnomAD database for both human PRDM7 and PRDM9 does not point toward signatures of selection on the population level, too. PRDM9 has an essential function in meiosis by determining hotspot loci and is a putative speciation gene (Mihola et al., 2009; Baudat et al., 2010; Myers et al., 2010). However, some mammals do not have functional PRDM9 copies, like dogs (Oliver et al., 2009; Muñoz-Fuentes et al., 2011). In human, loss of function did not result in sterility as shown in a case of a mother (Narasimhan et al., 2016).
The available data on primate genomes are limited. Most primate genome assemblies are on a level of scaffolds, for example, the C. imitator assembly. There is a possibility that with new and improved data and coverage of primate diversity, the outcome may differ from ours. However, the results shown in this article are strong evidence that PRDM7 originated on the branch of Catarrhini. Therefore, genes annotated as PRDM7 in non-catarrhine species are probably PRDM9, exemplified in P. coquereli on Ensembl. We also suggest that PRDM7 in ruminants (Ahlawat et al., 2016) is a different PRDM member and possibly resulted from another duplication event.
Based on the data and interpretations herein, species with a putative functional PRDM7 can be reasonably well defined and further studies can focus to elucidate the function of this recent PRDM member.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SH performed the experiments and the analysis and wrote the manuscript. SH and HZ planned the study and discussed the results. ZF extracted the DNA of the non-related chimpanzees and gorillas. ZF and EV provided samples. EV and HZ revised the manuscript. All authors read and approved the final manuscript.
Funding
SH was funded with a doctoral scholarship by the German Academic Scholarship Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jessica Ries for preliminary work on strepsirrhine sequences, Christine Driller and Juergen Brosius for Tarsier samples, and the Department of Anthropology, University of Zurich, for Daubentonia samples. We would like to thank Editage (www.editage.com) for English language editing. This publication uses data collected within the framework of the Ph.D. thesis “Genomweite Charakterisierung der putativen Rekombinationshotspots zweier Makakenarten—PRDM9 in der Primatenevolution” of SH published in 2020 at the Faculty of Biology, Johannes Gutenberg University Mainz.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.593725/full#supplementary-material
Supplementary Data Sheet 1 | Partial sequences of PRDM9 and PRDM7 exon 11 of Macaca mulatta as well as the zinc finger array of PRDM9 of Nycicebus coucang. A detailed phylogeny on the PRDM9 zinc fingers. A table on primate species, the corresponding genome assemblies and loci of PRDM7 and PRDM9. In-house scripts to work with data of the 1000 genomes project.
Supplementary Data Sheet 2 | Blasted Loci for the phylogenies on PRDM7 and PRDM9.
Supplementary Data Sheet 3 | Ensembl BLAST results of human PRDM7 exons.
References
Ahlawat, S., Sharma, P., Sharma, R., Arora, R., and De, S. (2016). Zinc finger domain of the PRDM9 gene on chromosome 1 exhibits high diversity in ruminants but its paralog PRDM7 contains multiple disruptive mutations. PLoS One 11:e0156159. doi: 10.1371/journal.pone.0156159
Auton, A., Abecasis, G., Altshuler, D., Kang, H. M., Jan, O. K., Shane, M., et al. (2015). A global reference for human genetic variation. Nature 526, 68–74. doi: 10.1038/nature15393
Auton, A., Fledel-Alon, A., Pfeifer, S., Venn, O., Ségurel, L., Street, T., et al. (2012). A fine-scale chimpanzee genetic map from population sequencing. Science 336, 193–198. doi: 10.1126/science.1216872
Bailey, J. A., Gu, Z., Clark, R. A., Knut, R., Rhea, V. S., Stuart, S., et al. (2002). Recent segmental duplications in the human genome. Science 297, 1003–1007. doi: 10.1126/science.1072047
Bailey, J. A., Yavor, A. M., Massa, H. F., Trask, B. J., and Eichler, E. E. (2001). Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 11, 1005–1017. doi: 10.1101/gr.gr1871r
Baudat, F., Buard, J., Grey, C., Fledel-Alon, A., Ober, C., Przeworski, M., et al. (2010). PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840. doi: 10.1126/science.1183439
Birtle, Z., and Ponting, C. P. (2006). Meisetz and the birth of the KRAB motif. Bioinformatics 22:2845. doi: 10.1093/bioinformatics/btl498
Blazer, L. L., Lima-Fernandes, E., Gibson, E., Eram, M. S., Loppnau, P., Arrowsmith, C. H., et al. (2016). PR domain containing protein 7 (PRDM7) is a histone 3 lysine 4 trimethyltransferase. J. Biol. Chem. 291, 13509–13519. doi: 10.1074/jbc.M116.721472
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Castro, E. D, Sigrist, C. J. A., Gattiker, A., Bulliard, V., Langendijk-Genevaux, P. S., Gasteiger, E., et al. (2006). ScanProsite: detection of prosite signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365. doi: 10.1093/nar/gkl124
Criscuolo, A., and Gribaldo, S. (2010). BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10:210. doi: 10.1186/1471-2148-10-210
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Fumasoni, I., Meani, N., Rambaldi, D., Scafetta, G., Alcalay, M., and Ciccarelli, F. D. (2007). Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol. Biol. 7:187. doi: 10.1186/1471-2148-7-187
Geissmann, T. (2003). Vergleichende Primatologie [Comparative Primatology]. New York, NY: Springer-Verlag.
Groeneveld, L. F., Atencia, R., Garriga, R. M., and Vigilant, L. (2012). High diversity at PRDM9 in chimpanzees and bonobos. PLoS One 7:e39064. doi: 10.1371/journal.pone.0039064
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hayashi, K., Yoshida, K., and Matsui, Y. (2005). A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438, 374–378. doi: 10.1038/nature04112
Heerschop, S., Zischler, H., Merker, S., Perwitasari-Farajallah, D., and Driller, C. (2016). The pioneering role of PRDM9 indel mutations in tarsier evolution. Sci. Rep. 6:34618. doi: 10.1038/srep34618
Junier, T., and Zdobnov, E. M. (2010). The newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26, 1669–1670. doi: 10.1093/bioinformatics/btq243
Karczewski, K. J., Francioli, L. C., Tiao, G., Beryl, B. C., Laura, D. G., Daniel, P. B., et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443. doi: 10.1038/s41586-020-2308-7
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi: 10.1038/35057062
Lemoine, F., Correia, D., Lefort, V., Doppelt-Azeroual, O., Mareuil, F., Cohen-Boulakia, S., et al. (2019). NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 47, W260–W265. doi: 10.1093/nar/gkz303
Lemoine, F., Domelevo Entfellner, J. B., Wilkinson, E., Correia, D., Dávila Felipe, M., De Oliveira, T., et al. (2018). Renewing felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556, 452–456. doi: 10.1038/s41586-018-0043-0
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Mihola, O., Trachtulec, Z., Vlcek, C., Schimenti, J. C., and Forejt, J. (2009). A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323, 373–375. doi: 10.1126/science.1163601
Muñoz-Fuentes, V., Di Rienzo, A., and Vilà, C. (2011). Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One 6:e25498. doi: 10.1371/journal.pone.0025498
Myers, S., Bowden, R., Tumian, A., Bontrop, R. E., Freeman, C., MacFie, T. S., et al. (2010). Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327, 876–879. doi: 10.1126/science.1182363
Narasimhan, V. M., Hunt, K. A., Mason, D., Laura, S., Tee, L., Mark, T., et al. (2016). Health and population effects of rare gene knockouts in adult humans with related parents. Science 352, 474–477. doi: 10.1126/science
Oliver, P. L., Goodstadt, L., Bayes, J. J., Birtle, Z., Roach, K. C., Phadnis, N., et al. (2009). Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5:e1000753. doi: 10.1371/journal.pgen.1000753
Perelman, P., Johnson, W. E., Roos, C., Seuánez, H. N., Horvath, J. E., Moreira, M. A. M., et al. (2011). A molecular phylogeny of living primates. PLoS Genet. 7:e1001342. doi: 10.1371/journal.pgen.1001342
Samonte, R. V., and Eichler, E. E. (2002). Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3, 65–72. doi: 10.1038/nrg705
Schwartz, J. J., Roach, D. J., Thomas, J. H., and Shendure, J. (2014). Primate evolution of the recombination regulator PRDM9. Nat. Commun. 5:4370. doi: 10.1038/ncomms5370
She, X., Horvath, J. E., Jiang, Z., Wilson, R. K., Webb, M., Eva, E. E., et al. (2004a). The structure and evolution of centromeric transition regions within the human genome. Nature 430, 857–864. doi: 10.1038/nature02806
She, X., Jiang, Z., Clark, R. A., Cheng, Z., Church, D. M., Aaron, L. H., et al. (2004b). Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431, 927–930. doi: 10.1038/nature03062
Sigrist, C. J. A., Cerutti, L., Hulo, N., Gattiker, A., Falquet, L., Pagni, M., et al. (2002). PROSITE: A documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 3, 265–274. doi: 10.1093/bib/3.3.265
Stevison, L. S., Woerner, A. E., Kidd, J. M., Kelley, J. L., Veeramah, K. R., McManus, K. F., et al. (2016). The time scale of recombination rate evolution in great apes. Mol. Biol. Evol. 33, 928–945. doi: 10.1093/molbev/msv331
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Thomas, J. H., Emerson, R. O., and Shendure, J. (2009). Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One 4:e8505. doi: 10.1371/journal.pone.0008505
Vervoort, M., Meulemeester, D., Béhague, J., and Kerner, P. (2016). Evolution of Prdm genes in animals: insights from comparative genomics. Mol. Biol. Evol. 33, 679–696. doi: 10.1093/molbev/msv260
Keywords: PRDM7, PRDM9, gene duplication, paralogization, primate evolution
Citation: Heerschop S, Fagrouch Z, Verschoor EJ and Zischler H (2021) Pinpointing the PRDM9-PRDM7 Gene Duplication Event During Primate Divergence. Front. Genet. 12:593725. doi: 10.3389/fgene.2021.593725
Received: 11 August 2020; Accepted: 01 February 2021;
Published: 24 February 2021.
Edited by:
Badri Padhukasahasram, Illumina, United StatesReviewed by:
Federico Guillermo Hoffmann, Mississippi State University, United StatesJosé Arroyo, Santa Fe Institute, United States
Zdenek Trachtulec, Institute of Molecular Genetics (ASCR), Czechia
Copyright © 2021 Heerschop, Fagrouch, Verschoor and Zischler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sacha Heerschop, aGVlcnNjaG9wQHVuaS1tYWluei5kZQ==; Hans Zischler, emlzY2hsZXJAdW5pLW1haW56LmRl
 Sacha Heerschop
Sacha Heerschop Zahra Fagrouch
Zahra Fagrouch Ernst J. Verschoor
Ernst J. Verschoor Hans Zischler
Hans Zischler