- 1College of Pharmacy, Harbin Medical University, Harbin, China
- 2Pharmaceutical Experiment Teaching Center, College of Pharmacy, Harbin Medical University, Harbin, China
- 3Department of Colorectal Surgery, The Second Affiliated Hospital of Harbin Medical University, Harbin, China
- 4Department of Oral and Maxillofacial Surgery, The Second Affiliated Hospital of Harbin Medical University, Harbin, China
Aims: Chaperonin-containing TCP1 subunit (CCT) 6A is an oncogenic 6th subunit of the CCT family. Nevertheless, not much is documented regarding its function in colorectal cancer (COAD). This investigation seeks to explore the role of CCT6A in the prognosis of COAD.
Main Methods: Sequencing data from the Gene Expression Omnibus (GEO) and Cancer Genome Atlas database (TCGA) were employed to analyze the expression of CCT6A and its involvement in various regulatory networks behind COAD. Oncomine and Gene Expression Profiling Interactive Analysis (GEPIA) analyzed Levels of expression and survival rates, while GEPIA was used to uncover further the functional networks that involved CCT6A. Database for Annotation, Visualization, and Integrated Discovery (DAVID) tools were used to interpret Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathways. Evaluation of the expression levels of CCT6A in COAD samples was also verified via immunohistochemistry.
Key Findings: We found that the expression of CCT6A is up-regulated in COAD. CCT6A correlated with poor prognosis and decreased immune infiltrates such as CD4+ T cells, B cells, and dendritic cells. CCT6A is increased in COAD patients. CCT6A is associated with several gene networks related to the DDX family and mismatch repair pathways.
Significance: Our data showed that data mining was able to uncover data regarding levels of CCT6A and its involvement in genetic regulating pathways in COAD.
Introduction
COAD is the third most frequently encountered malignancies globally (Brody, 2015). Its incidence and mortality in China have been rising from 2000 to 2011 (Goldstein et al., 2016). There are no apparent symptoms in the early stage of colorectal cancer, with patients often presenting in advanced stages of the disease. Furthermore, symptoms of colorectal cancer manifest across a wide clinical spectrum depending on its primary origin. While advancements in treatment modalities have improved the survival rates of colorectal patients greatly, its prognosis is far from ideal, especially in those with advanced disease (Yu et al., 2018). However, the mechanisms underlying COAD have not been fully understood. Therefore, it is important to clarify COAD’s pathogenesis and uncover new biomarkers that may have therapeutic or prognostic value in patients with colorectal cancer.
Cancer immunotherapy (CI) has proved to be a useful treatment option in those with late-stage colorectal cancer (Zhang et al., 2019). Nevertheless, a small proportion of patients have failed to respond to this therapy, a phenomenon that may be attributed to interactions between the tumor and the immune system (Sarvaria et al., 2017). These immune markers may potentially be a source of prognostic predictors (Wagner and Roth, 2018; Pan et al., 2019). There is evidence that the immune microenvironment is a potential modulator of response to treatment and cancer progression (Dominguez et al., 2019; Leone et al., 2019; Wen et al., 2019). Tumor tissue infiltration by mononuclear immune cells has been reported to occur in several types of solid tumors, including COAD (Liu et al., 2017; Ren and Zhang, 2019). Yin et al. (2017) showed that the immune-microenvironment confers chemoresistance of COAD via IL6. Ye et al. (2019) reported tumor-infiltrating immune cells to play an essential role in COAD prognosis. These evidences highlight immune infiltration as a critical agent in COAD.
The CCT family is known to play oncogene roles in many cancers, especially CCT6A. Ma et al. (2018) showed that CCT6A was involved in the proteogenomic characterization of human colorectal cancer liver metastasis. Di Meo et al. (2019) reported that CCT6A was a potential prognostic biomarker in renal cell carcinoma. Klimczak et al. (2019) also suggested that CCT6A may be involved in imparting an unfavorable prognosis in breast cancer tumor progression. Despite this, the involvement of CCT6A in COAD has not been clarified.
The current investigation demonstrates the expression of CCT6A based on COAD patient data derived from the TCGA and other publicly accessible databases. Clinical data were correlated to the functional genetic networks related to CCT6A in COAD by using multi-dimensional analysis methods. A flowchart of our work is shown in Supplementary Figure 1. Our analysis may potentially uncover several novel therapeutic and prognostic treatment targets for COAD management.
Materials and Methods
Oncomine Analysis
The Oncomine 4.5 database was used to evaluate CCT6A mRNA expression in COAD. Oncomine1 is the most significant resource for oncogene chips and functions to host integrated data mining. It currently holds data from 86,733 cancer tissues and normal tissues with 715 gene expression data sets (Rhodes et al., 2007). Our analysis utilized data from a series of COAD studies (at Oct 2020), which included the Notterman colon (28 normal colon tissues vs. 50 COAD tissues), Kaiser colon (5 normal colon tissues vs. 10 COAD tissues), Skrzypczak colorectal (24 normal colon tissues vs. 36 COAD tissues), Ki colon (28 normal colon tissues vs. 50 COAD tissues), Hong colorectal (12 normal colon tissues vs. 70 COAD tissues), and Sabates-Bellver colon (32 normal colon tissues vs. 25 COAD tissues) studies. Detail information of these datasets were shown in Table 1. These datasets were collected and analyzed by Oncomine tool (at Oct 2020). COAD tissues were processed to quantify the CCT6A levels concerning its expression in normal tissue. Differences that had a p-value of less than 0.01 were considered significant.
UALCAN Analysis
UALCAN (v1.0)2 is an interactive internet-based platform for carrying out detailed analyses of TCGA genetic expression data and utilizes TCGA 3rd level of clinical and RNA-seq information derived from 31 types of cancer (Chandrashekar et al., 2017). We used UALCAN for quantification of a target gene(s)’ relative expression across different grades of tumors and stages of cancer, across healthy tissue and tumor tissue samples, and several other types of clinicopathological characteristics (at October 2020).
GEPIA Analysis
GEPIA (v1.0)3 is a comprehensive website that allows researchers to perform patient survival analysis, dimensionality reduction analysis, correlation analysis, profiling plotting, differential expression analysis, and detection of similar genes (Tang et al., 2019). The survival analyses of CCT6A and its associated COAD genes were performed using this tool (at October 2020).
TIMER Database Analysis
We utilized the TIMER (v1.0)4 database as a resource to analyze the various types of immune infiltrates in COAD (Li et al., 2016). The TIMER database is home to the immune profiles of 10,897 samples across 32 cancer types from TCGA. With this tool, we were able to determine the gene models of the molecular immune profile associated with CCT6A expression. Cells included in the analysis were dendritic, macrophage, neutrophil, B cells, CD4+ and CD8+ T cells (at October 2020).
Functional Enrichment Analysis
The functional enrichment analysis included the network analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment. The STRING online database (v2020)5 allowed us to build a protein-protein interaction network (PPI) network consisting of CCT6A related genes (Franceschini et al., 2013). DAVID (v6.8) bioinformatics6 was implemented in GO KEGG pathway analysis (at Oct 2020). The criteria were set as P < 0.05.
Patients
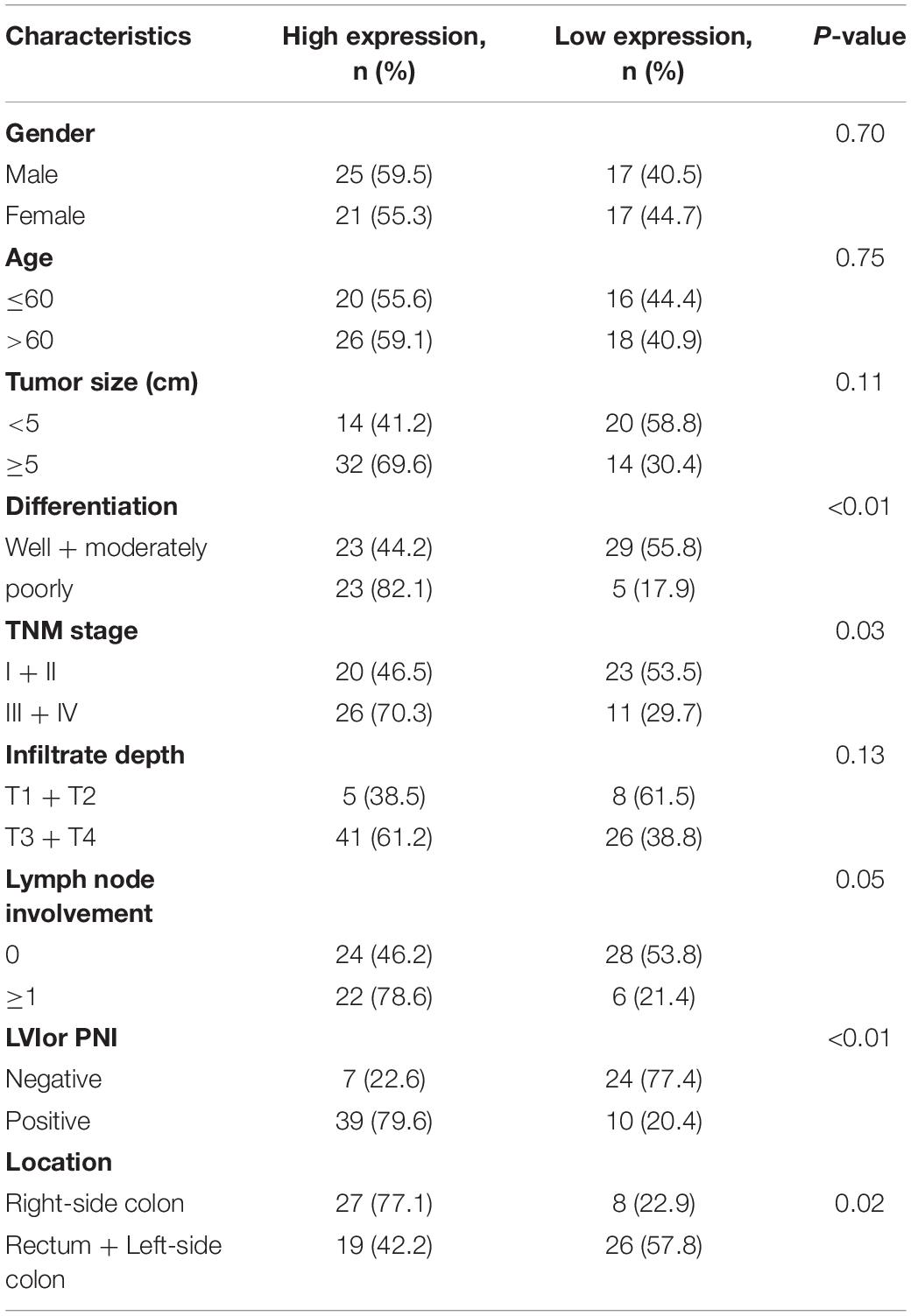
Eighty pairs of COAD tissue were harvested from patients who received surgery at the Department of Colorectal Cancer Surgery, Second Affiliated Hospital of Harbin Medical University (Harbin, China) between January 2015 and December 2019. The age of the patients was 49 years (range, 28–76 years). Additionally, normal COAD tissue samples were acquired by endoscopy from non-tumor areas from the patients with COAD enrolled in the study. All tissue specimens used in the current study were obtained after getting written informed consent from all participants. Study protocols were reviewed and passed by the Ethics Committee of Harbin Medical University. All the patients’ information was shown in Table 2.
Immunohistochemistry (IHC)
IHC was performed following standard procedures as described previously. Anti-CCT6A rabbit polyclonal antibodies (Cat. no. ab191951; 1:100 dilution; Abcam, Cambridge, United Kingdom) was added to the tissue sections before they were left to incubate overnight at 4°C. Following PBS rinse, samples were further incubated with biotinylated secondary antibody (cat. no. 111-035-003; 1:1,500 dilution; Jackson ImmunoResearch, United States) for 30 min at room temperature before being exposed to diaminobenzidine for 5 min at room temperature. The entire experiment was repeated with PBS replacing the primary antibody to produce a negatively staining control specimen.
Statistical Analysis
The data was a compilation from a minimum of 3 to 6 independent experiments as the mean ± SD (standard deviation). For the clinical tissue test, the data were appraised by a paired Student’s t-test, where P < 0.05 was indicative of statistical significance. Statistical analyses were performed by SPSS (version 19.0; United States) software and illustrated with GraphPad Prism (version 7.0; United States).
Results
CCT6A Expression in COAD
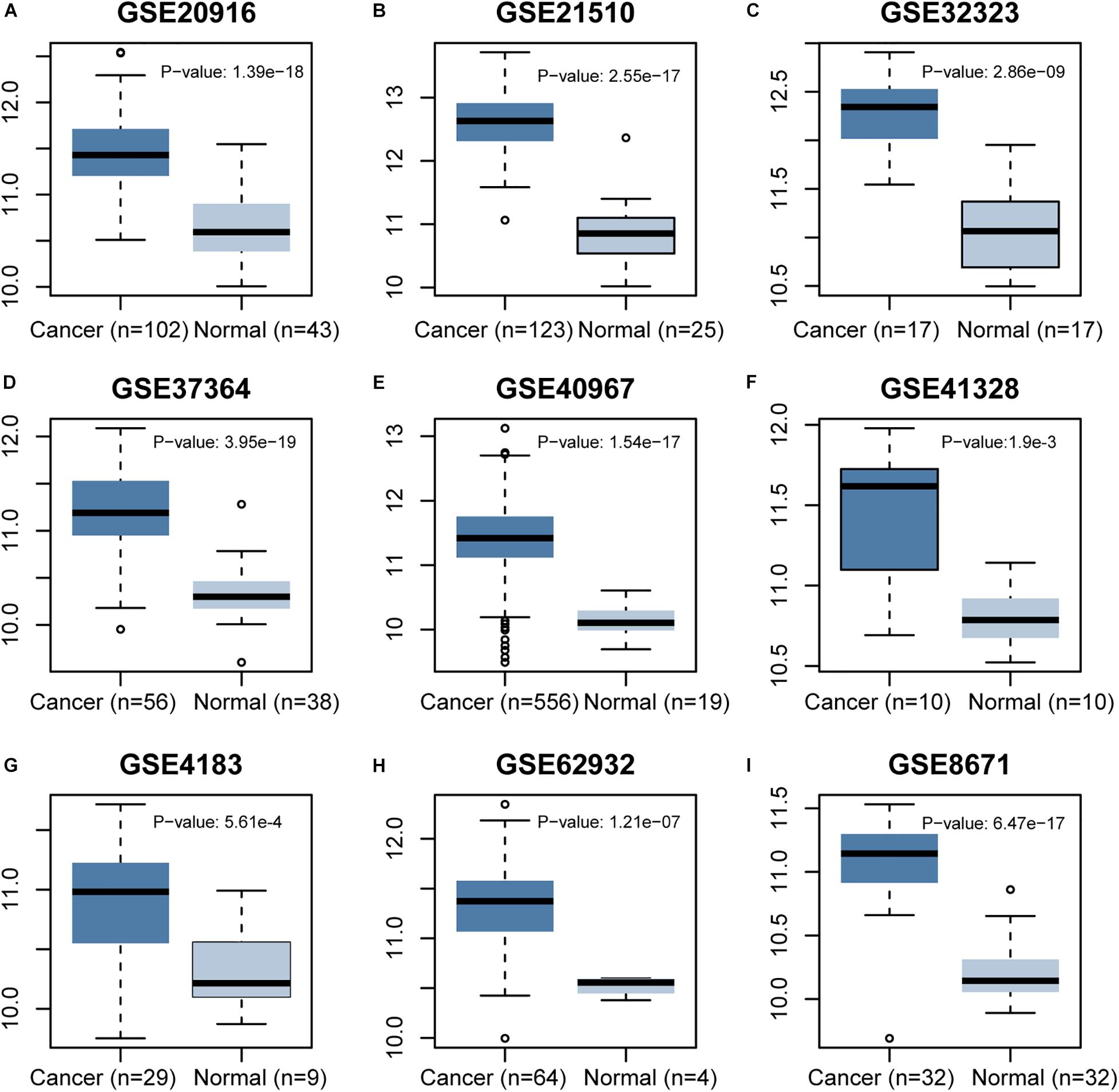
The CCT6A transcription levels across several studies on COAD studies extracted from the GEO database were evaluated. Based on the Oncomine 4.5 database, COAD tissues possessed a markedly raised CCT6A mRNA expression profile compared to healthy samples (p < 0.01) (Figure 1). The UALCAN database contained sub-group analyses data of COAD based on different clinicopathological shapes and also demonstrated that COAD tissues had higher CCT6A levels in contrast to normal samples (Figure 2A). CCT6A was differentially expressed in subgroup analyses according to tumor grade, disease stage, histological subtypes, and age, but not gender (Figures 2B–F). We collected more gene expression datasets of COAD from the GEO database, including GSE20916, GSE21510, GSE32323, GSE37364, GSE40967, GSE41328, GSE4183, GSE62932, and GSE8671. Based on these datasets, we have explored the expression variation status of the CCT6A gene between COAD and normal tissues. We found the expression levels of the CCT6A gene were significantly higher in COAD tissues than in normal tissues (Figures 3A–I). We have also performed differential expression analysis of the CCT6A gene in pan-cancers by using the GENET2 (v2.0) web tool7 (Park et al., 2019). We found that the CCT6A gene was significantly higher in some other cancer tissues than normal tissues (Figure 4). Based on this line of evidence, CCT6A may be a potential diagnostic tool in COAD.
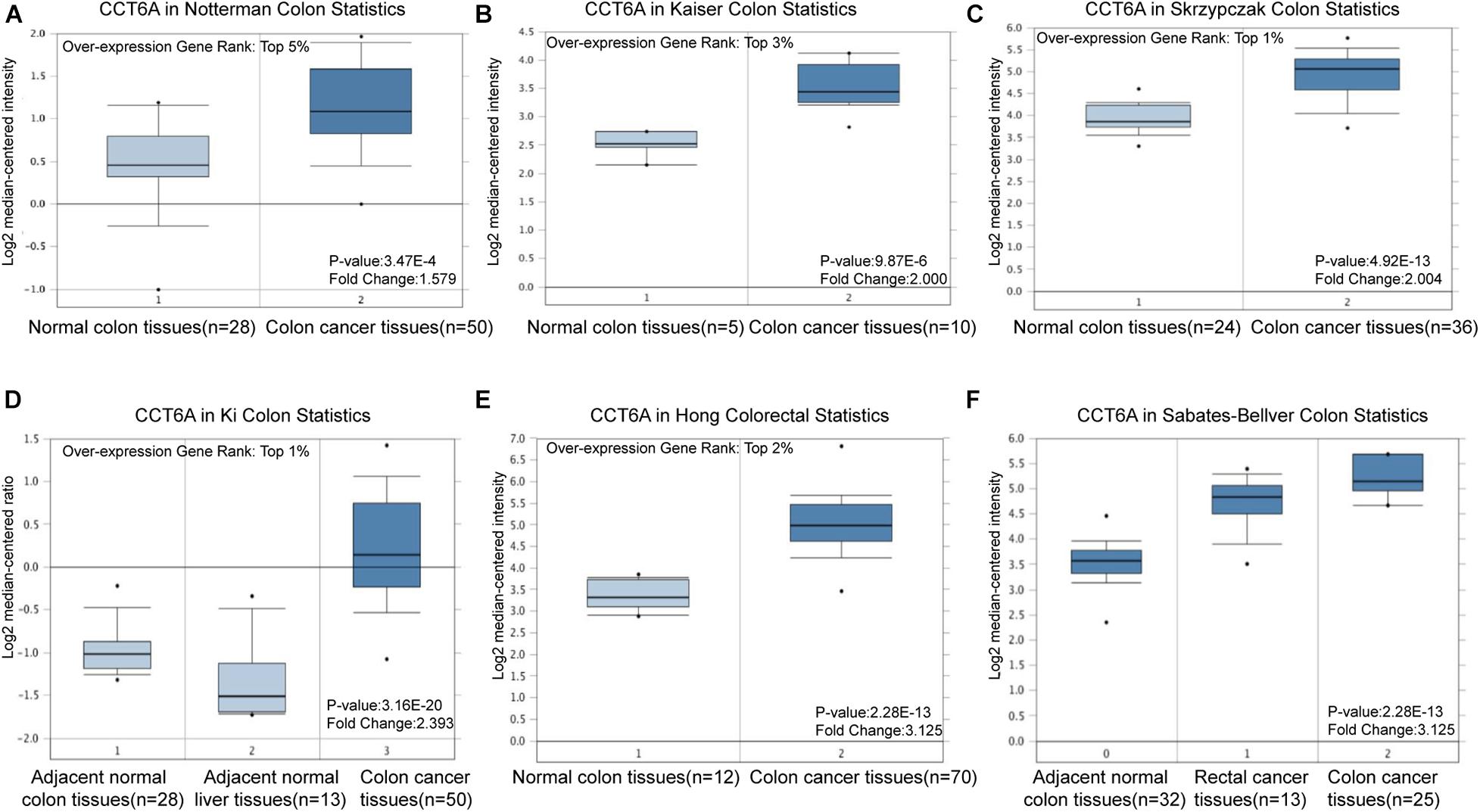
Figure 1. CCT6A transcription in COAD (Oncomine). Levels of CCT6A mRNA was significantly higher in COAD than that in normal tissue. The fold change, associated p-values, and overexpression gene rank, are shown based on Oncomine 4.5 analysis. (A–F) The box plot shows CCT6A mRNA levels in the Notterman Colon, Kaiser Colon, Skrzypczak Colon Ki Colon, Hong Colorectal and Sabates-Beliver Colon datasets.
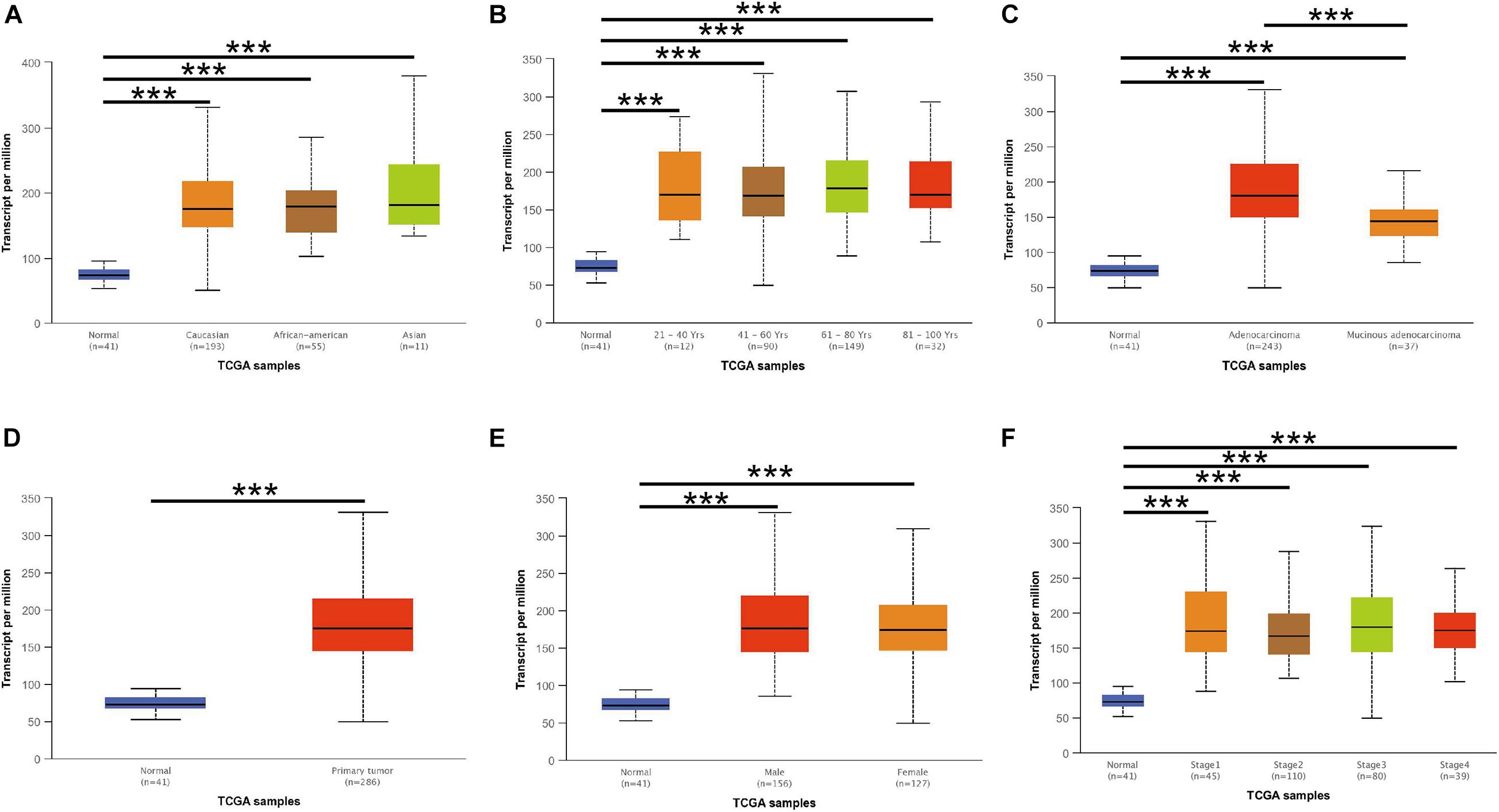
Figure 2. CCT6A transcription in subgroups of patients with COAD, stratified based on race, age, and other criteria (UALCAN). (A) Boxplot showing relative expression of CCT6A in normal and COAD samples. (B) Boxplot showing relative expression of CCT6A in normal individuals of either gender or male or female COAD patients. (C) Boxplot showing relative expression of CCT6A in normal individuals of any age or COAD patients aged 21–40, 41–60, 61–80, or 81–100 year. (D) Boxplot was showing relative expression of CCT6A in normal individuals of any ethnicity or COAD patients of Caucasian, African-American or Asian ethnicity. (E) Boxplot showing relative expression of CCT6A in normal individuals or COAD patients in stages 1, 2, 3, or 4. (F) Boxplot showing relative expression of CCT6A in normal individuals or COAD patients with grade 1, 2, 3, or 4 tumors. Data are mean ± SE. ***P < 0.001.
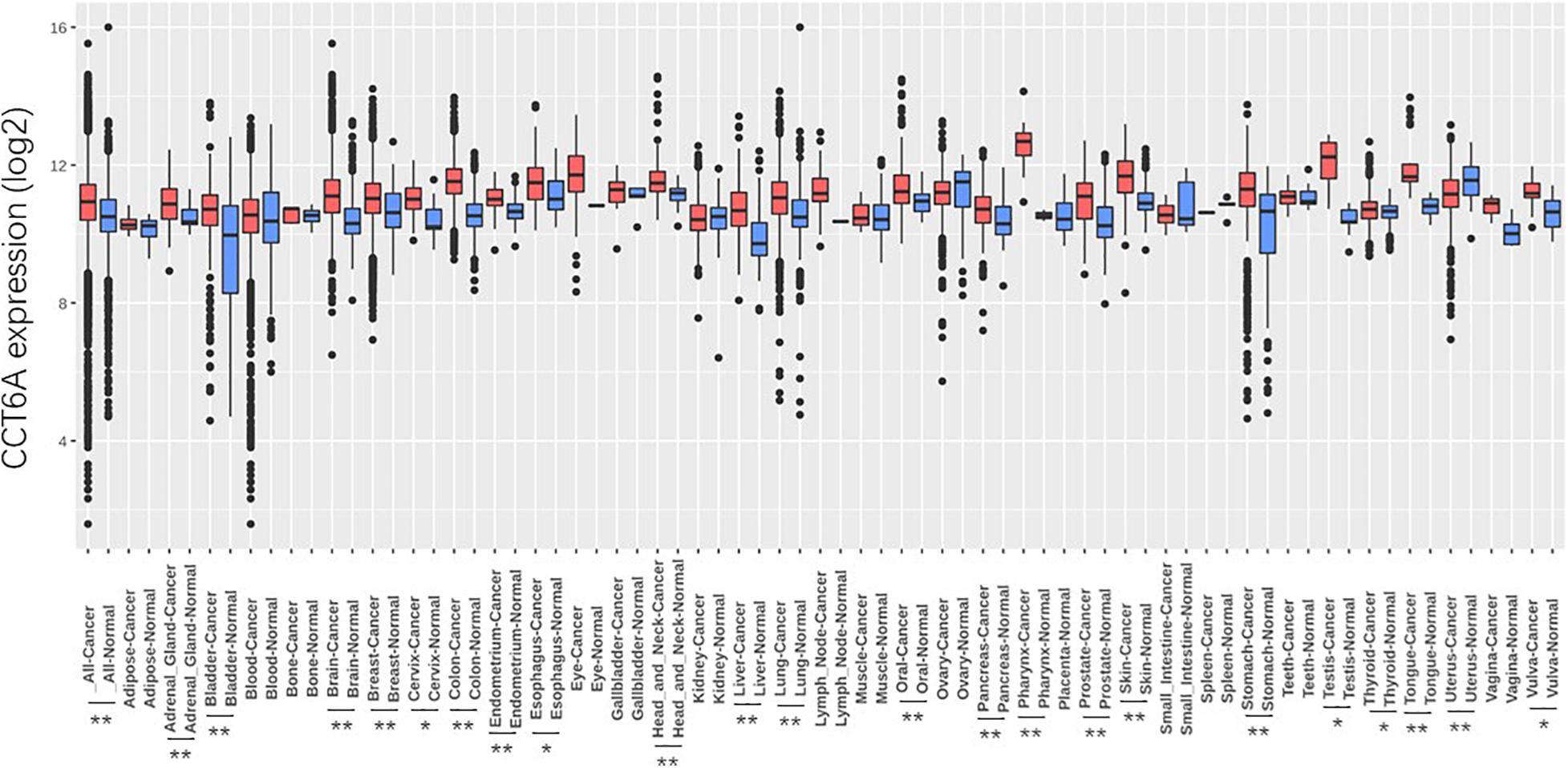
Figure 4. Differential expression analysis of CCT6A gene in pan-cancers. Data are mean SE. *P < 0.05 and **P < 0.01.
CCT6A Is Up-Regulated in COAD Patients
To determine whether CCT6A expression may correlate with COAD, immunohistochemistry was performed. CCT6A expression was detected primarily in the cytoplasm in the perinuclear region (Figure 5). According to our data, we found that the protein level of CCT6A in COAD is up-regulated in comparison to normal intestinal tissue. Our data suggest that CCT6A expression is increased in COAD patients and detected in the cytomembrane.
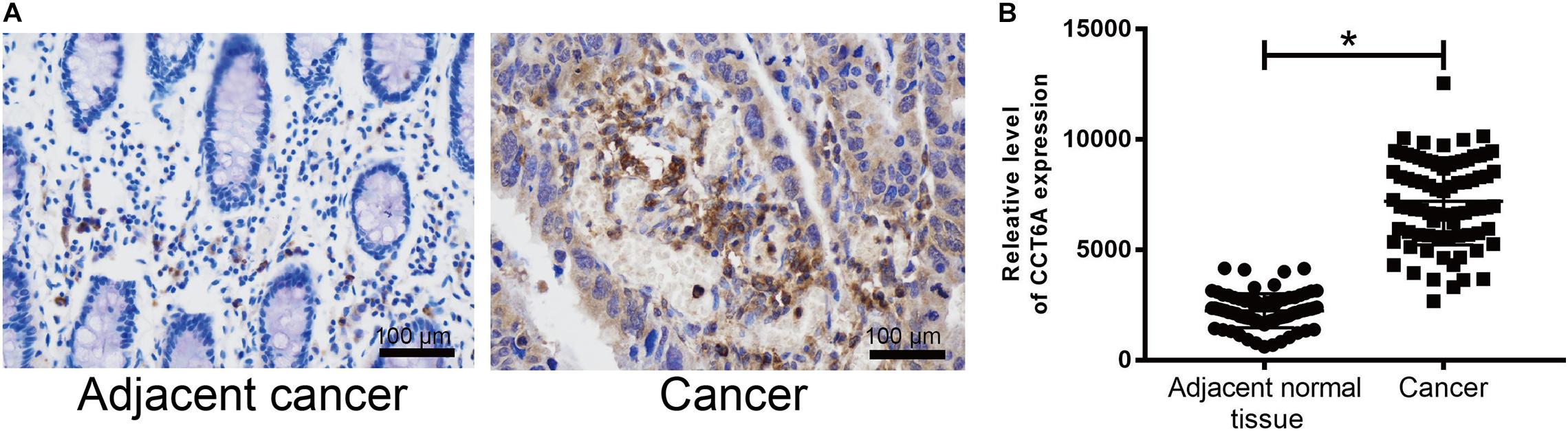
Figure 5. CCT6A expression is up-regulated in COAD patients. (A,B) Immunohistochemistry was employed to test the protein level of CCT6A in COAD patients. Data are mean ± SE. *P < 0.05 vs. normal group.
High CCT6A Expression Impacts the Prognosis of COAD Patients
The association and impact of CCT6A in COAD were further explored by investigating its effect on patient survival. This was performed using the GEPIA databases. A higher level of CCT6A correlated with a shorter overall survival time in COAD (P = 0.0071), indicating that raised CCT6A expressions were a risk factor for poor survival in COAD patients (Figure 6A). Further, we evaluated the prognostic efficiency of CCT6A in an independent dataset of 62 COAD cohorts (GSE12945 from the GEO database). Based on this dataset, Kaplan-Meier survival analysis was performed using an online tool, ProgScan, which employs a minimum P-value approach to the optimal cut-off point in continuous gene expression (Mizuno et al., 2009). Using this method, we divided the 62 patients into different groups according to the minimum P-value and found that CCT6A successfully divided patients into different risk groups (Figures 6B,C).
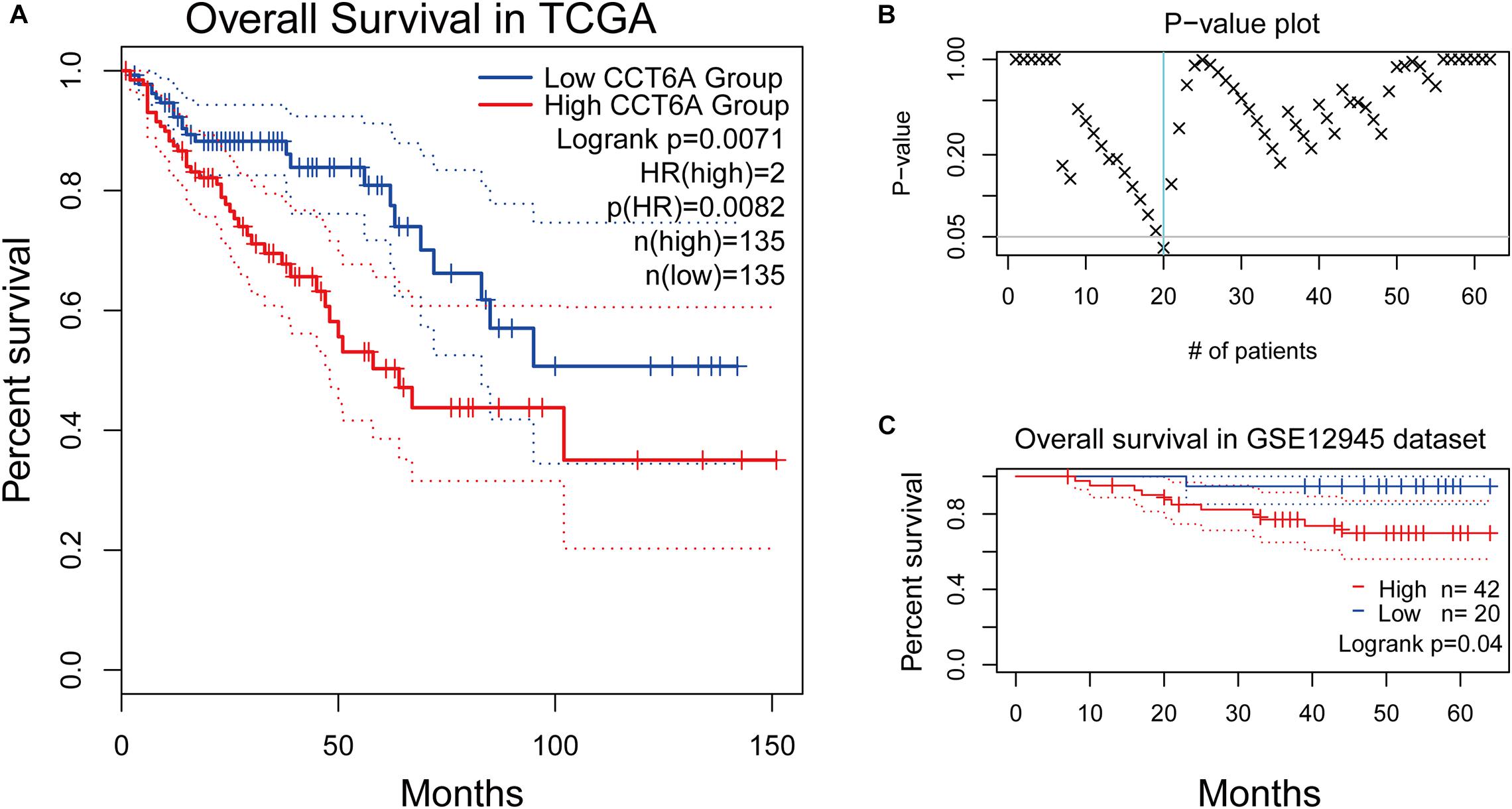
Figure 6. Survival analysis of CCT6A in different datasets. (A) Kaplan-Meier survival curves comparing the high and low expression of CCT6A in different types of cancer using the GEPIA. (B) The p-value plot of survival analysis result by ProgScan. (C) Kaplan-Meier survival curves comparing the high and low expression of CCT6A in the GSE12945 dataset.
CCT6A Expression Correlated With Immune-Cell Infiltration in COAD
Tumor-infiltrating lymphocytes have been found to have prognostic value in various tumors (Fu et al., 2019). Therefore, we explored if the expression of CCT6A was linked to levels of immune infiltrates in COAD using the TIMER resource. We found that CCT6A levels correlated negatively with levels of infiltrating B cells, CD4+T cells, neutrophils, and dendritic cells but positively associated with tumor purity (Figure 7), suggesting a potentially immune-suppressed role of CCT6A.
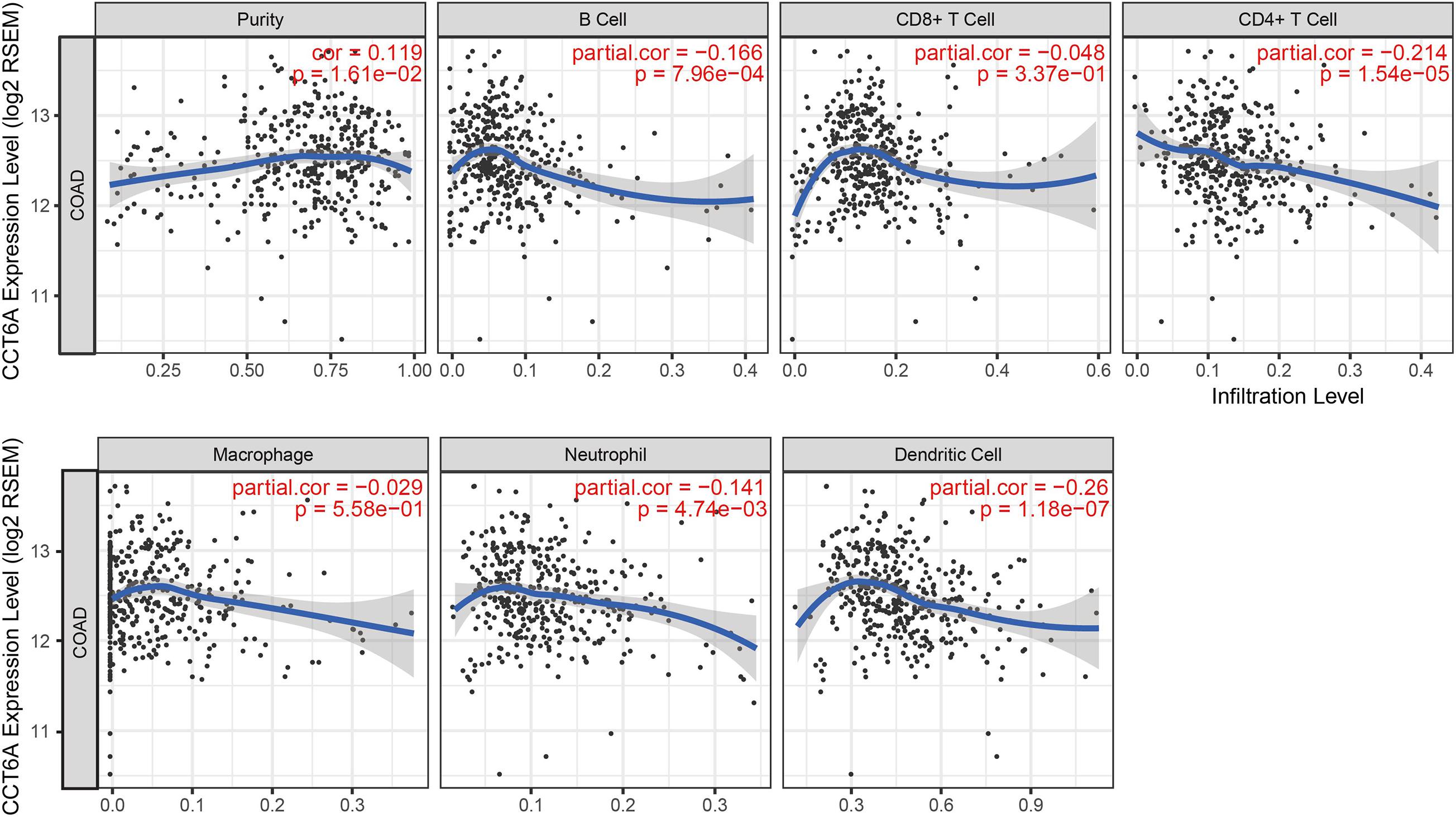
Figure 7. Correlation of CCT6A expression with immune infiltration level in COAD (colon adenocarcinoma).
Enrichment Analysis of CCT6A Functional Networks in COAD
To further investigate the potential target genes of CCT6A in COAD, we identified the top 100 associated genes of CCT6A in COAD using the GEPIA database. We uploaded all the 100 genes to DAVID to determine overrepresented KEGG pathways and GO categories. Data analysis with GO demonstrated that the following biological processes (BP) had enriched genes: ribosome biogenesis and cellular macromolecule metabolic process (Figure 8A and Table 3). For cell component (CC), the up-regulated DEGs were increased in intracellular organelle lumen and intracellular part (Figure 8B). Furthermore, molecular function (MF) analysis also indicated enrichment in RNA binding (Figure 8C). Figure 8D contains the most significantly enriched KEGG pathways of these genes, including Mismatch repair and Homologous recombination.
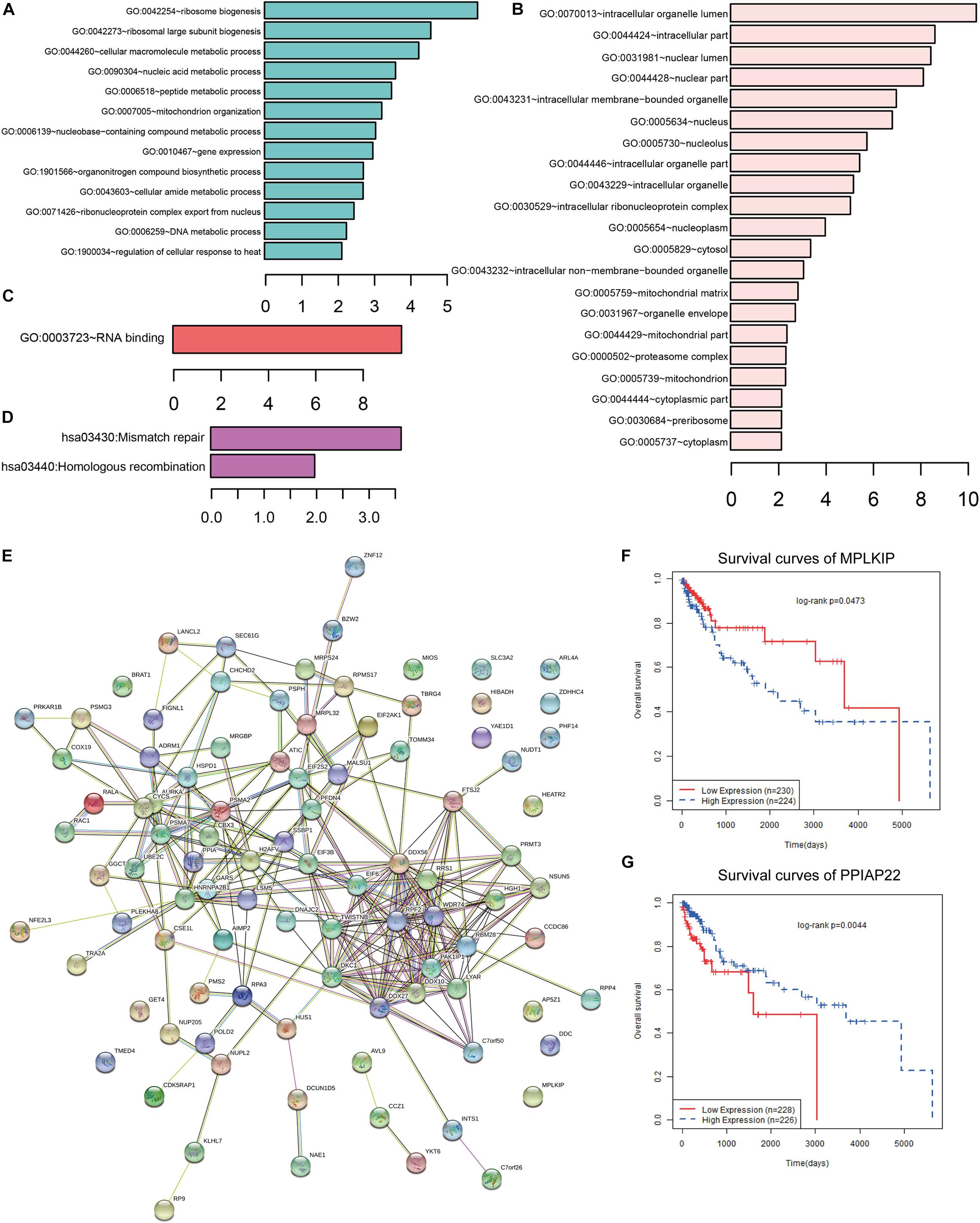
Figure 8. Functional analysis of CCT6A in COAD. (A) GO BP, (B) GO CC, (C) GO MF and (D) KEGG pathway enrichment analysis of CCT6A associated genes. (E) The PPI network of CCT6A related genes using the STRING online database. (F,G) Kaplan-Meier survival curves of MPLKIP and PPIAP22 in COAD patients.
Using the STRING online database (see text footnote 5), 95 out of the 100 CCT6A associated genes were screened into the PPI network complex (Figure 8E). Some network nodes were of a higher degree and displayed hubs features (For example, DDX27, DDX56, DDX10, and RPF2). We also found that several genes such as MPLKIP and PPIAP22 in this network were significantly associating with COAD patients’ survival (Figures 8F,G).
Discussion
COAD is the most common intestinal tumor worldwide, especially in women (Hassanlou et al., 2019), and has been reported to be associated with many oncogenes (Yin et al., 2017). Existing literature reports CCT6A to possess oncogenic features in a various human cancer (Di Meo et al., 2019; Huang et al., 2019). This study of bioinformatics analysis of publically available genetic sequencing data was performed to gain further insight into the role of CCT6A in the genetic regulation of COAD. We used a panel of bioinformatics tools to perform systematic analysis of CCT6A across different COAD datasets. Oncomine is the biggest resource and tool for oncogene chips data collection and integrated data mining. It currently holds data from 86,733 cancer tissues and normal tissues with a total of 715 gene expression data sets. UALCAN is an online platform to perform detailed analyses of gene expression data on the level of clinical and RNA-seq information of TCGA datasets. TIMER web server is a comprehensive resource for systematical analysis of immune infiltrates across diverse cancer types. TIMER web server allows users to input function-specific parameters, with resulting figures dynamically displayed to conveniently access the tumor immunological, clinical, and genomic features. David provides a comprehensive set of functional annotation tools for investigators to understand biological meaning behind large list of genes. Transcriptional sequencing data derived from hundreds of clinical samples from TCGA and GEO revealed that COAD tissues possessed higher CCT6A mRNA levels in contrast to healthy colon tissue (Figures 1–4).
To ensure if the level of CCT6A correlates from the onset of COAD. Immunohistochemistry analyses were employed to test the expression of CCT6A in COAD patients. Our data indicate that CCT6A was up-regulated in COAD patients and detected in the cytomembrane (Figure 5). These results suggested that aberrantly increased CCT6A levels are a feature of COAD, and may benefit from clinical validation studies to determine its value as a potential prognostic biomarker. There was a consistent correlation between CCT6A expression and patient prognosis, indicating that higher levels of this gene were indicative of poorer prognosis (Figure 6A). Based on an independent COAD dataset, CCT6A could also successfully divided patients into different risk groups (Figures 6B,C).
This study also sought to clarify the association between tumor immune cell infiltrates and CCT6A expression in COAD. Lower CCT6A expression was associated with higher infiltrating B cells, CD4+ T cells, neutrophils, and dendritic cells, but positively correlated with tumor purity (Figure 7). A significant proportion of immune cell infiltrated in COAD constituted of B cells, specifically terminally differentiated memory B cells or plasma cells, and highlights the presence of a specific immune response in this condition (Shimabukuro-Vornhagen et al., 2014). In the early stages of Crohn’s-like lymphoid reaction development, there is clustering of mature antigen presenting dendritic cells and CD4+ T-cells. As Crohn’s-like lymphoid reaction matures, lymphoid follicles are created as a result of increased recruitment of B-cells, as well as follicular dendritic cells (Maoz et al., 2019). Increased macrophage infiltration in colorectal cancer tissues was significantly associated with increased chemoresistance and poor prognosis (Yin et al., 2017). Patients who underwent resection of colorectal cancer have been reported to possess a novel neutrophil phenotype comprising increased phagocytosis, less apoptosis, and lowered NET formation (Richardson et al., 2017)Dendritic cells’ presence dictates a tumor’s response to immunotherapy, effector T cell trafficking to the tumor site, and T cell anti-tumor immunity (Hope et al., 2017). When interpreted as a whole, CCT6A may possess a role in polarization of tumor-associated macrophages (TAM).
To further gain insight into CCT6A regulated network, we identified the top 100 related genes of CCT6A in COAD using the GEPIA database. We constructed a PPI network of these related genes using the STRING database. The results showed that the DDX27, DDX10, and DDX56 were hub genes of the CCT6A regulated network. DDX (DEAD-box helicase) is the biggest RNA helicase family responsible for short RNA duplex unwinding, a key regulator of RNA biogenesis. Human DDX3X and its yeast ortholog Ded1p are a DDX related closely to the human DDX4 and fly Vasa subfamily. As with all other DDXs, Like all DDXs, DDX3X comprises two RecA-like domains (D1D2) which makes up its helicase core. It contains 12 highly conserved sequence motifs (Figure 1A). The D1D2 core of DDX3X is flanked by largely unstructured N- or C-terminal tails, but have motifs responsible for the subfamily’s unique functions. Yang C et al. showed that DDX27 possessed an oncogenic role in COAD by regulating the stem cell-like activity of COAD cells (Liu et al., 2019). Shi et al. (2019) suggested that higher DDX10 expression was deletion inhibiting key cellular activities by MAPK signaling pathway (Shi and Hao, 2019). Kouyama et al. (2019) reported DDX56 regulated COAD cell proliferation and cell cycle both in vitro and in vivo by promoting WEE1. Pathway and GO enrichment analysis revealed that the top 100 related genes of CCT6A were mainly related to ribosome biogenesis and mismatch repair (Figures 8A–D). We also found that several genes such as MPLKIP and PPIAP22 in this network were significantly associating with COAD patients’ survival (Figures 8F,G).
In summary, CCT6A mRNA levels were markedly raised in COAD in contrast to normal colon tissue and correlates with poor prognosis. We also found that expression level of CCT6A is related to decreased immune infiltrates of B, CD4+ T and dendritic cells. Finally, the correlated network and functional annotation analysis revealed that CCT6A was related to the DDX family. This study provides several supporting lines of evidence that highlight the critical role of CCT6A in COAD and its potential as a prognostic marker.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
Study protocols were reviewed and passed by the Ethics Committee of Harbin Medical University (KY-2016-036). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
HS and YW: conception and design. YW, HS, and H-YJ: administrative support. X-YY, X-XS, J-HZ, Y-XY, and LY: provision of study materials. YW, HS, LG, and LY: collection and assembly of data. H-YJ, X-YY, X-XS, J-HZ, Y-XY, LG, and LY: data analysis and interpretation. All authors: manuscript writing and final approval of manuscript.
Funding
This research was supported by the Special Funds for the Construction of High-level Universities and Advantageous Disciplines, Basic Scientific Research in Colleges and Universities, Post-doctor of Heilongjiang Province (LBH-Z16164), Undergraduate Student Innovation Project (201910226350), and Medical Wisdom Research Fund by the Heilongjiang Sunshine Health Foundation (H21L0802).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.629856/full#supplementary-material
Supplementary Figure 1 | The flow chart of our analysis steps.
Footnotes
- ^ www.oncomine.org
- ^ http://ualcan.path.uab.edu
- ^ http://gepia.cancer-pku.cn/
- ^ https://cistrome.shinyapps.io/timer/
- ^ http://string-db.org
- ^ https://david.ncifcrf.gov/
- ^ http://gent2.appex.kr/gent2/
References
Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B., et al. (2017). UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19, 649–658. doi: 10.1016/j.neo.2017.05.002
Di Meo, A., Batruch, I., Brown, M. D., Yang, C., Finelli, A., Jewett, M. A., et al. (2019). Searching for prognostic biomarkers for small renal masses in the urinary proteome. Int. J. Cancer 146, 2315–2325. doi: 10.1002/ijc.32650
Dominguez, C. X., Muller, S., Keerthivasan, S., Koeppen, H., Hung, J., Gierke, S., et al. (2019). Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer. Discov. 10, 232–253. doi: 10.1158/2159-8290.CD-19-0644
Franceschini, A., Szklarczyk, D., Frankild, S., Kuhn, M., Simonovic, M., Roth, A., et al. (2013). STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815. doi: 10.1093/nar/gks1094
Fu, Q., Chen, N., Ge, C., Li, R., Li, Z., Zeng, B., et al. (2019). Prognostic value of tumor-infiltrating lymphocytes in melanoma: a systematic review and meta-analysis. Oncoimmunology 8:1593806. doi: 10.1080/2162402X.2019.1593806
Goldstein, D. A., Zeichner, S. B., Bartnik, C. M., Neustadter, E., and Flowers, C. R. (2016). Metastatic Colorectal Cancer: A Systematic Review of the Value of Current Therapies. Clin. Colorectal Cancer 15, 1–6. doi: 10.1016/j.clcc.2015.10.002
Hassanlou, M., Soltani, B. M., Medlej, A., Kay, M., and Mowla, S. J. (2019). Hsa-miR-6165 downregulates insulin-like growth factor-1 receptor (IGF-1R) expression and enhances apoptosis in SW480 cells. Biol. Chem. 401, 477–485. doi: 10.1515/hsz-2018-0421
Hope, C., Emmerich, P. B., Papadas, A., Pagenkopf, A., Matkowskyj, K. A., Van De Hey, D. R., et al. (2017). Versican-Derived Matrikines Regulate Batf3-Dendritic Cell Differentiation and Promote T Cell Infiltration in Colorectal Cancer. J. Immunol. 199, 1933–1941. doi: 10.4049/jimmunol.1700529
Huang, K., Zeng, Y., Xie, Y., Huang, L., and Wu, Y. (2019). Bioinformatics analysis of the prognostic value of CCT6A and associated signalling pathways in breast cancer. Mol. Med. Rep. 19, 4344–4352. doi: 10.3892/mmr.2019.10100
Klimczak, M., Biecek, P., Zylicz, A., and Zylicz, M. (2019). Heat shock proteins create a signature to predict the clinical outcome in breast cancer. Sci. Rep. 9:7507. doi: 10.1038/s41598-019-43556-1
Kouyama, Y., Masuda, T., Fujii, A., Ogawa, Y., Sato, K., Tobo, T., et al. (2019). Oncogenic splicing abnormalities induced by DEAD-Box Helicase 56 amplification in colorectal cancer. Cancer Sci. 110, 3132–3144. doi: 10.1111/cas.14163
Leone, R. D., Zhao, L., Englert, J. M., Sun, I. M., Oh, M. H., Sun, I. H., et al. (2019). Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 366, 1013–1021. doi: 10.1126/science.aav2588
Li, B., Severson, E., Pignon, J. C., Zhao, H., Li, T., Novak, J., et al. (2016). Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 17:174. doi: 10.1186/s13059-016-1028-7
Liu, H., Ren, G., Chen, H., Liu, Q., Yang, Y., and Zhao, Q. (2019). Predicting lncRNA-miRNA interactions based on logistic matrix factorization with neighborhood regularized. Knowledge Based Syst. 191:105261. doi: 10.1016/j.knosys.2019.105261
Liu, X., Wu, S., Yang, Y., Zhao, M., Zhu, G., and Hou, Z. (2017). The prognostic landscape of tumor-infiltrating immune cell and immunomodulators in lung cancer. Biomed. Pharmacother. 95, 55–61. doi: 10.1016/j.biopha.2017.08.003
Ma, Y. S., Huang, T., Zhong, X. M., Zhang, H. W., Cong, X. L., Xu, H., et al. (2018). Proteogenomic characterization and comprehensive integrative genomic analysis of human colorectal cancer liver metastasis. Mol. Cancer 17:139. doi: 10.1186/s12943-018-0890-1
Maoz, A., Dennis, M., and Greenson, J. K. (2019). The Crohn’s-Like Lymphoid Reaction to Colorectal Cancer-Tertiary Lymphoid Structures With Immunologic and Potentially Therapeutic Relevance in Colorectal Cancer. Front. Immunol. 10:1884. doi: 10.3389/fimmu.2019.01884
Mizuno, H., Kitada, K., Nakai, K., and Sarai, A. (2009). PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics 2:18. doi: 10.1186/1755-8794-2-18
Pan, J. H., Zhou, H., Cooper, L., Huang, J. L., Zhu, S. B., Zhao, X. X., et al. (2019). LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front. Immunol. 10:6. doi: 10.3389/fimmu.2019.00006
Park, S. J., Yoon, B. H., Kim, S. K., and Kim, S. Y. (2019). GENT2: an updated gene expression database for normal and tumor tissues. BMC Med. Genomics 12:101. doi: 10.1186/s12920-019-0514-7
Ren, X., and Zhang, Z. (2019). Understanding tumor-infiltrating lymphocytes by single cell RNA sequencing. Adv. Immunol. 144, 217–245. doi: 10.1016/bs.ai.2019.08.004
Rhodes, D. R., Kalyana-Sundaram, S., Mahavisno, V., Varambally, R., Yu, J., Briggs, B. B., et al. (2007). Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166–180. doi: 10.1593/neo.07112
Richardson, J. J. R., Hendrickse, C., Gao-Smith, F., and Thickett, D. R. (2017). Characterization of systemic neutrophil function in patients undergoing colorectal cancer resection. J. Surg. Res. 220, 410–418e411. doi: 10.1016/j.jss.2017.07.036
Sarvaria, A., Madrigal, J. A., and Saudemont, A. (2017). B cell regulation in cancer and anti-tumor immunity. Cell Mol. Immunol. 14, 662–674. doi: 10.1038/cmi.2017.35
Shi, J. H., and Hao, Y. J. (2019). DDX10 overexpression predicts worse prognosis in osteosarcoma and its deletion prohibits cell activities modulated by MAPK pathway. Biochem. Biophys. Res. Commun. 510, 525–529. doi: 10.1016/j.bbrc.2019.01.114
Shimabukuro-Vornhagen, A., Schlosser, H. A., Gryschok, L., Malcher, J., Wennhold, K., Garcia-Marquez, M., et al. (2014). Characterization of tumor-associated B-cell subsets in patients with colorectal cancer. Oncotarget 5, 4651–4664. doi: 10.18632/oncotarget.1701
Tang, Z., Kang, B., Li, C., Chen, T., and Zhang, Z. (2019). GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560. doi: 10.1093/nar/gkz430
Wagner, D. C., and Roth, W. (2018). [Prognostic significance of immune cell infiltrates in tumor pathology]. Pathologe 39, 532–538. doi: 10.1007/s00292-018-0541-8
Wen, Y., Chen, X., Zhu, X., Gong, Y., Yuan, G., Qin, X., et al. (2019). Photothermal-Chemotherapy Integrated Nanoparticles with Tumor Microenvironment Response Enhanced the Induction of Immunogenic Cell Death for Colorectal Cancer Efficient Treatment. ACS Appl. Mater Interf. 11, 43393–43408. doi: 10.1021/acsami.9b17137
Ye, L., Zhang, T., Kang, Z., Guo, G., Sun, Y., Lin, K., et al. (2019). Tumor-Infiltrating Immune Cells Act as a Marker for Prognosis in Colorectal Cancer. Front. Immunol. 10:2368. doi: 10.3389/fimmu.2019.02368
Yin, Y., Yao, S., Hu, Y., Feng, Y., Li, M., Bian, Z., et al. (2017). The Immune-microenvironment Confers Chemoresistance of Colorectal Cancer through Macrophage-Derived IL6. Clin. Cancer Res. 23, 7375–7387. doi: 10.1158/1078-0432.CCR-17-1283
Yu, W., Jiang, J., Xie, L., Li, B., Luo, H., Fu, Y., et al. (2018). Mortality Trends in Colorectal Cancer in China During 2000-2015: A Joinpoint Regression and Age-Period-Cohort Analysis. Prev. Chronic Dis. 15:E156. doi: 10.5888/pcd15.180329
Zhang, R., Zhu, Z., Lv, H., Li, F., Sun, S., Li, J., et al. (2019). Immune Checkpoint Blockade Mediated by a Small-Molecule Nanoinhibitor Targeting the PD-1/PD-L1 Pathway Synergizes with Photodynamic Therapy to Elicit Antitumor Immunity and Antimetastatic Effects on Breast Cancer. Small 15:e1903881. doi: 10.1002/smll.201903881
Keywords: chaperonin-containing TCP1 subunit 6A, colorectal cancer, immune infiltrates, prognosis, functional network analysis
Citation: Sun H, Wang Y, Jing H-Y, Yang X-Y, Shi X-X, Zhang J-H, Yu Y-X, Gao L, Wang X-Y, Li W-H and Yu L (2021) Chaperonin-Containing TCP1 Subunit 6A Is a Prognostic Potential Biomarker That Correlates With the Presence of Immune Infiltrates in Colorectal Cancer. Front. Genet. 12:629856. doi: 10.3389/fgene.2021.629856
Received: 16 November 2020; Accepted: 23 March 2021;
Published: 04 May 2021.
Edited by:
Shibiao Wan, St. Jude Children’s Research Hospital, United StatesReviewed by:
Qiqi Xie, Lanzhou University, ChinaSherry Bhalla, Mount Sinai Hospital, United States
Bo Feng, Shanghai Jiao Tong University, China
Copyright © 2021 Sun, Wang, Jing, Yang, Shi, Zhang, Yu, Gao, Wang, Li and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Yu, YWxsZW5nbDA2MDFAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Hui Sun1,2†
Hui Sun1,2† Hao-Yu Jing
Hao-Yu Jing Wan-Hong Li
Wan-Hong Li Lei Yu
Lei Yu