- NUS Environmental Research Institute, National University of Singapore, Singapore, Singapore
Rice has the natural morphological adaptation to germinate and elongate its coleoptile under submerged flooding conditions. The phenotypic deviation associated with the tolerance to submergence at the germination stage could be due to natural variation. However, the molecular basis of this variation is still largely unknown. A comprehensive understanding of gene regulation of different genotypes that have diverse rates of coleoptile elongation can provide significant insights into improved rice varieties. To do so, publicly available transcriptome data of five rice genotypes, which have different lengths of coleoptile elongation under submergence tolerance, were analyzed. The aim was to identify the correlation between promoter architecture, associated with transcriptional and hormonal regulation, in diverse genotype groups of rice that have different rates of coleoptile elongation. This was achieved by identifying the putative cis-elements present in the promoter sequences of genes upregulated in each group of genotypes (tolerant, highly tolerant, and extremely tolerant genotypes). Promoter analysis identified transcription factors (TFs) that are common and unique to each group of genotypes. The candidate TFs that are common in all genotypes are MYB, bZIP, AP2/ERF, ARF, WRKY, ZnF, MADS-box, NAC, AS2, DOF, E2F, ARR-B, and HSF. However, the highly tolerant genotypes interestingly possess binding sites associated with HY5 (bZIP), GBF3, GBF4 and GBF5 (bZIP), DPBF-3 (bZIP), ABF2, ABI5, bHLH, and BES/BZR, in addition to the common TFs. Besides, the extremely tolerant genotypes possess binding sites associated with bHLH TFs such as BEE2, BIM1, BIM3, BM8 and BAM8, and ABF1, in addition to the TFs identified in the tolerant and highly tolerant genotypes. The transcriptional regulation of these TFs could be linked to phenotypic variation in coleoptile elongation in response to submergence tolerance. Moreover, the results indicate a cross-talk between the key TFs and phytohormones such as gibberellic acid, abscisic acid, ethylene, auxin, jasmonic acid, and brassinosteroids, for an altered transcriptional regulation leading to differences in germination and coleoptile elongation under submergence. The information derived from the current in silico analysis can potentially assist in developing new rice breeding targets for direct seeding.
Introduction
Rice is one of the major cereal crops and staple food in Southeast Asian countries. In most of these countries, people sow seeds in the flooded rice fields through direct seeding that helps in reducing cost and manpower. During this process, rice seeds encounter flooding/submergence stress as they are exposed to hypoxia and even anoxia. Rice has the natural morphological adaptation to germinate and elongate its coleoptile under submerged flooding condition and also even in anoxic conditions. This coleoptile reaches the surface of water to get O2 for aerobic respiration to get sufficient energy for the development of roots and shoots (Narsai et al., 2015). However, there is phenotypic variation in coleoptile elongation and the tolerance to submergence at the germination stage among different genotypes and which could be due to natural variation. Tolerant genotypes germinate and elongate their coleoptile and hydrolyze starch to sugars for glycolysis and enhanced ethanolic fermentation to generate ATP (Lee et al., 2009). Besides, the plant hormone ethylene plays a major role during this adaptation (Ma et al., 2010; Steffens et al., 2012). Although a number of genetic studies provide information that the extent of tolerance to flooding in rice at the germination stage is due to natural variation (Septiningsih et al., 2013; Baltazar et al., 2014; Hsu and Tung, 2015), the molecular mechanism behind this phenotypic variation is largely unknown. A comprehensive understanding of the gene regulation of different genotypes tolerant to submergence/flooding can provide a significant insight into breeding of improved tolerant rice varieties for direct seeding cultivation.
A number of microarray and RNA-sequencing studies performed on rice germination under anoxia/hypoxia have shown the up-/downregulation of a number of stress-responsive genes (Lasanthi-Kudahettige et al., 2007; Narsai et al., 2009; Shingaki-Wells et al., 2011; Narsai et al., 2015; Hsu and Tung, 2017; Wu and Yang, 2020). During such stress conditions, transcription factors (TFs) play a key role in regulating the expression of genes that support them to survive through tolerance mechanism. The molecular mechanism of submergence has been studied on the rice genotype FR13A, which is able to survive 2 weeks as it has Submergence 1 (SUB1) locus that encodes genes such as SUB1A, SUB1B, and SUB1C that belong to the ethylene-response factor (ERF) subgroup VII (Xu et al., 2006). Among those three genes, SUB1A helps rice plants to tolerate submergence (Fukao and Bailey-Serres, 2008a, b). In addition, it has also been shown that SNORKELs genes such as SNORKEL 1 (SK1) and SNORKEL 2 (SK2) can also induce tolerance in rice plants (deep-water floating rice) by inducing GA for rapid internode elongation to rescue the plants from drowning (Hattori et al., 2009). These SNORKELs also belong to ERF subgroup VII. In rice coleoptiles, a number of ERFs such as ERF60, ERF67, and ERF68 genes have been shown to be upregulated under anoxic conditions and also belong to the ERF subgroup VII (Lin et al., 2019). These data favor the understanding that rice coleoptile elongation is promoted by ethylene during submerged conditions. Besides, it has been shown that auxin also plays a key role in coleoptile elongation during submergence (Ishizawa and Esashi, 1983; Wu and Yang, 2020). Auxin-dependent differential growth in rice coleoptile is shown to be due to the effect of the TF auxin response factor 1 (OsARF1) (Waller et al., 2002). Likewise, TF WRKY is also involved in the regulation of tolerance to rice under submergence (Viana et al., 2018). High accumulation of WRKY in both shoots and roots was observed in response to submergence. The expression of OsWRKY11 and OsWRKY56 was more than 100-fold in comparison to controls in the root tissues. These results support the importance of TFs in the regulation of tolerance of rice plants under submerged/flooding conditions. Recent studies of the roles of auxin in rice coleoptile elongation suggest that it plays a key role in the cell division and tropism of rice coleoptiles under submergence (Wu and Yang, 2020).
For such transcriptional regulation, the expression of a specific TF can regulate the expression of a number of specific sets of stress-responsive genes by binding to their cognate cis-acting elements in the promoters of specific genes (Mizoi et al., 2012). However, there are very limited studies performed on the cis-element enrichment analysis in the promoter of such stress-responsive genes. The studies so far revealed the identification of a number of putative cis-acting elements that can be potentially associated with TF families such as ARF, ERF, MYB, WRKY, bZIP, E2F, and ZnF (Mohanty et al., 2012; Lakshmanan et al., 2014; Sharma et al., 2018). This association of TFs also provides useful potential links with different hormonal signaling as it regulates the downstream genes through interactions with other regulatory molecules of signaling pathways (Song et al., 2005).
The genome-wide gene expression profile generated by microarray/transcriptome analyses on rice germination and coleoptile elongation under hypoxia/anoxia have revealed the involvement of a common molecular mechanism that is associated with carbohydrate metabolism, fermentation, hormone induction, cell division, and expansion (Lasanthi-Kudahettige et al., 2007; Shingaki-Wells et al., 2011; Narsai et al., 2015; Hsu and Tung, 2017). However, the level of tolerance to submergence/flooding during germination and coleoptile elongation could be due to the natural variation. The molecular basis of this variation due to transcriptional regulation is unknown. Hence, the aim was to analyze and identify the specific promoter architecture of gene expression data of different groups of diverse rice genotypes that display variation in the rate of coleoptile elongation under submerged conditions (Hsu and Tung, 2017). These analysis can provide a hypothesis on the key TFs that are associated with genetic variation of different genotypes of rice germination and coleoptile elongation. Further, it can be linked to common and/or unique transcriptional modules associated with different genotypes.
Materials and Methods
Extraction of Promoter Sequences of Common Genes Upregulated in Different Groups of Diverse Genotypes of Rice Tolerant to Submergence
Common upregulated genes in the coleoptiles of different genotype groups of rice under submerged conditions were identified from published RNA-sequencing data (Hsu and Tung, 2017). The five diverse rice genotypes-the japonica variety Nipponbare, two recombinant inbred lines F291 and F274-2a generated from a cross between Nipponbare and IR64, and two natural genotypes originated from Southeast Asia [8391 from Laos (IRGC 94599) and 8753 from Indonesia (IRGC 54313)]-were used for the promoter architecture analysis. To identify the promoter cis-element content of the common upregulated genes of different rice genotypes based on their extent of tolerance in terms of elongation of coleoptiles, common genes upregulated in different groups of genotypes (grouping done by Hsu and Tung, 2017) were extracted. The analysis was performed for all five tolerant genotypes (Nipponbare, F291, F274-2a, 8391, and 8753), which have 23 upregulated genes (Suppplementary Table 1); four highly tolerant genotypes (F291, F274-2a, 8391, and 8753), which have 16 upregulated genes (Suppplementary Table 2); and two extremely tolerant natural genotypes (8391 and 8753), which have 27 upregulated genes (Suppplementary Table 3). The promoter sequences [-1000, +200 nucleotide] relative to the experimentally verified Transcription Start Site (TSS) were extracted for these common upregulated genes from our in-house rice promoter sequence database (Mohanty et al., 2012).
To validate the identification of promoter architecture analysis of the three groups of genotypes that have varying degrees of tolerance to submergence, two additional analyses were performed. In one of the analysis that belongs to the intermediate tolerant group, two highly tolerant genotypes (F291 and F274-2a) that have 24 upregulated genes (Suppplementary Table 4) were analyzed to compare with the group that has four highly tolerant genotypes (F291, F274-2a, X8391, and X8753). In the other analysis, two genotypes (Nipponbare and F274-2a) that have 84 upregulated genes (Suppplementary Table 5) were analyzed for comparison with the group that has five tolerant genotypes (Nipponbare, F291, F274-2a, X8391, and X8753) from different backgrounds. The comparisons of the two groups of genotypes with respective similar groups of genotypes will support the analysis.
Identification of Putative cis-Elements and Associated TFs
Putative cis-elements were identified in the promoter regions of each set of common upregulated genes by the “The Dragon Motif Builder program” having EM2 option with a threshold value of 0.875 (Huang et al., 2005), which is similar to our earlier detection (Mohanty et al., 2012, 2016; Lakshmanan et al., 2014). The program identified a total of 30 motifs having a length of 8–10 nucleotides for each set of common genes. Motifs were selected having a threshold value of 10–3 and more than 50% occurrence. The biological significance of these motifs was verified by Transcription Factor Binding databases such as TRANSFAC (Matys et al., 20031), PLACE database (Higo et al., 19992), AGRIS (Davuluri et al., 2003; Yilmaz et al., 20113), and PlantPAN 3.0 (Chow et al., 20194). Putative cis-elements in the promoters of the upregulated genes were identified based on their sites for different plant TFs present in plant genes with a minimum sequence length of five nucleotides. The cutoffs for core and matrix similarities were more than 75%. TF genes, significantly upregulated, were identified from the RNA-sequencing data (Hsu and Tung, 2017) and annotated based on the RAP genome annotations (Itoh et al., 2007; Tanaka et al., 20085). The methods used to analyze the data have been summarized and presented in Figure 1.
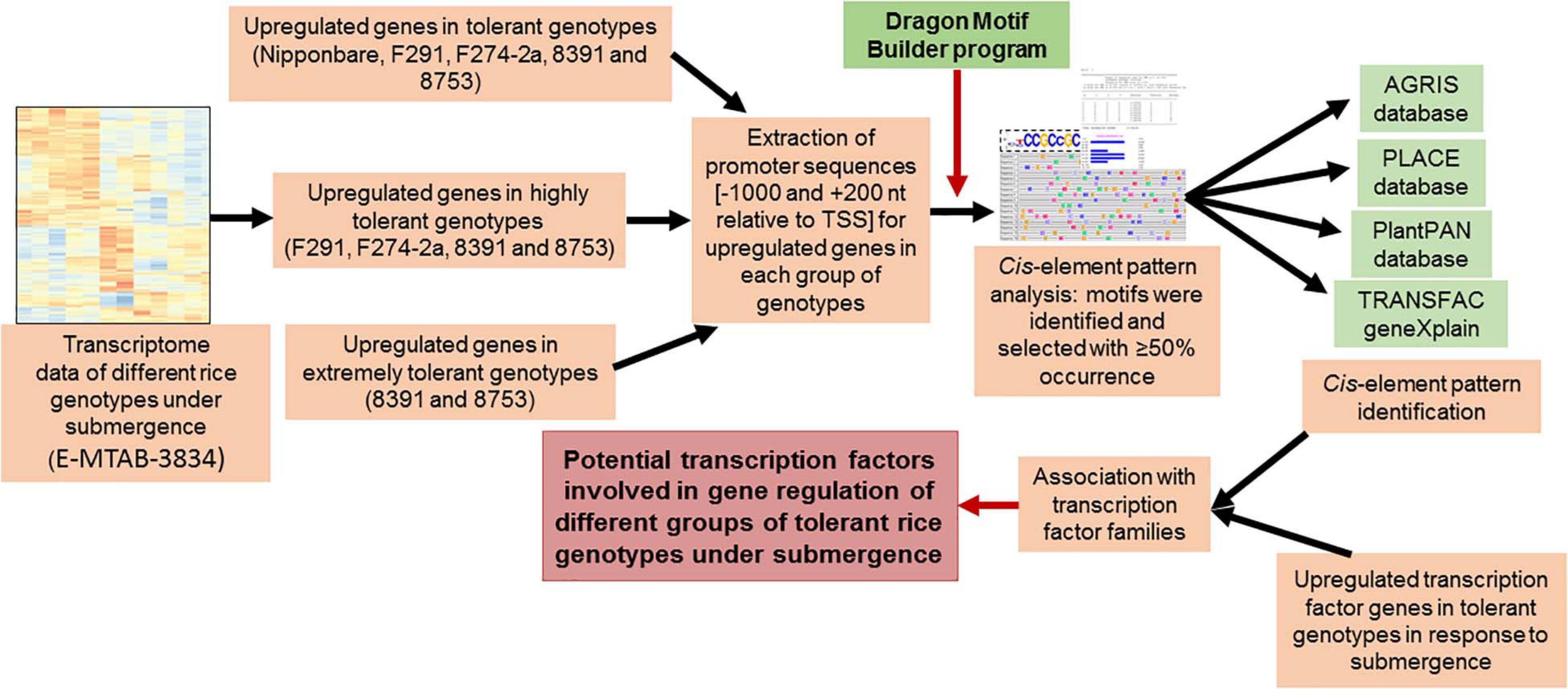
Figure 1. Summary of the methods used for the identification of the potential transcription factors in different groups of diverse rice genotypes tolerant to submergence. Promoter sequences of upregulated genes in different groups of genotypes that have different degrees of tolerance to submergence were extracted from our in-house rice promoter database, followed by identification and analysis of putative cis-elements by examining their presence in different TF binding databases. Potential TFs were identified by linking putative cis-elements with their associated transcription factors.
Results
Identification of Putative cis-Elements in the Upregulated Genes of Tolerant, Highly Tolerant, and Extremely Tolerant Diverse Rice Genotypes in Response to Submergence
Promoter regions of common genes upregulated in diverse genotypes of rice germination and coleoptile elongation in response to submergence tolerance contain cis-elements/TF binding sites that are responsible for the regulation of genes associated with various hormone signaling as well as metabolic pathways. Our analysis identified a number of common putative cis-elements in the promoter regions of the upregulated genes in tolerant (Nipponbare, F291, F274-2a, 8391, and 8753), highly tolerant (F291, F274-2a, 8391, and 8753), and extremely tolerant (8391 and 8753) genotypes. Putative cis-elements that are present in all genotypes are found to be associated with many known TFs such as MYB, bZIP, AP2/ERF, ARF, WRKY, ZnF, MADS-box, NAC, AS2, DOF, E2F, ARR-B, and HSF (Tables 1-3). Interestingly, both highly tolerant and extremely tolerant genotypes, in addition to the above binding sites for the associated TFs, also possess unique binding sites that are associated with TFs such as HY5 (bZIP), GBF3, 4, and 5 (bZIP), DPBF-3 (bZIP), ABF2, ABI5, bHLH (basic helix-loop-helix), and BES/BZR (BR-responsive TFs) (Table 2). Moreover, the extremely tolerant genotypes that have highly elongated coleoptiles comprise more specific binding sites that are associated with bHLH TFs such as BEE2, BIM1, BIM3, BM8, and BAM8 and bZIP TF ABF1 (Table 3). Hence, the following TF regulatory modules are identified as transcriptional activators/repressors that could be involved in controlling the regulation of germination and variation in coleoptile elongation of diverse rice genotypes in response to submergence tolerance.
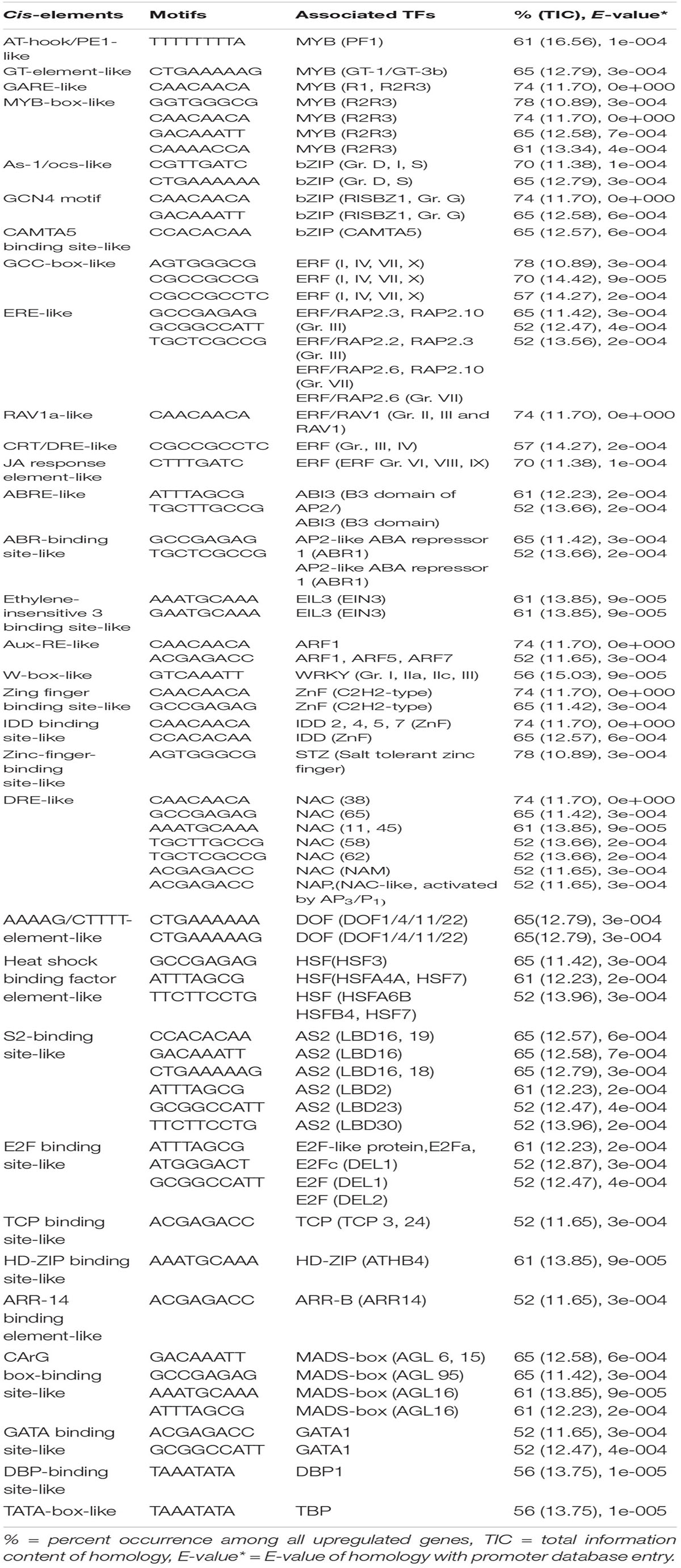
Table 1. Potential putative cis-elements identified in the promoters of upregulated genes in tolerant genotypes (Nipponbare, F291, F274-2a, 8391, and 8753).

Table 2. Potential putative cis-elements identified in the promoters of upregulated genes in highly tolerant genotypes (F291, F274-2a, 8391, and 8753).
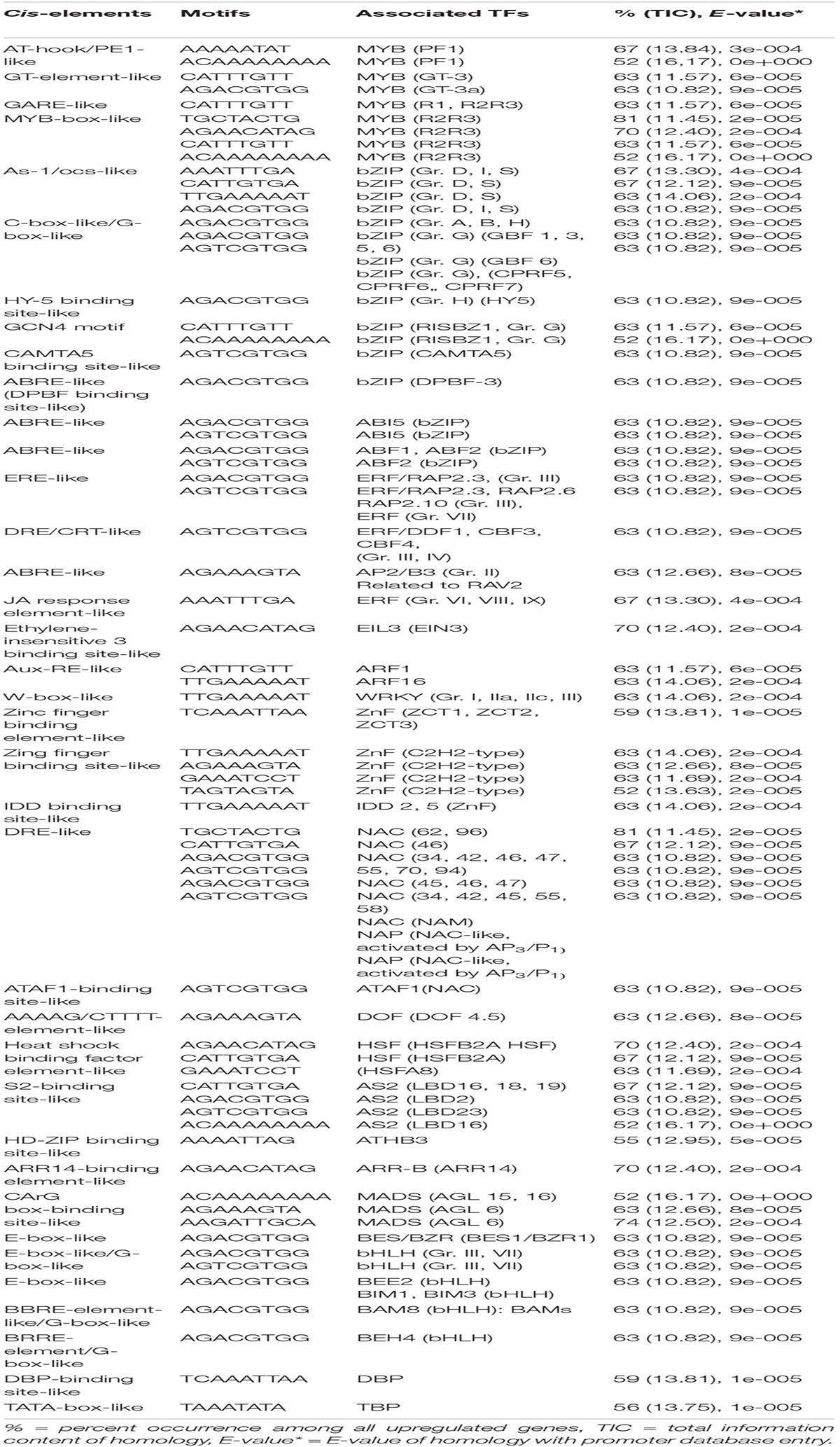
Table 3. Potential putative cis-elements identified in the promoters of upregulated genes in extremely tolerant genotypes (8391 and 8753).
MYB Regulatory Module
The promoter analysis of common upregulated genes in the tolerant, highly tolerant, and extremely tolerant genotypes identified high enrichment of a number of MYB-associated putative cis-elements such as AT-hook/PE1-like, GT-element-like, gibberellic acid response element (GARE)-like, and MYB-box-like. The MYB-box-like elements were highly enriched in the highly tolerant genotypes and, besides the promoters, also possess MYB-box related-like elements associated with MYB-like (CDC5) and MYB-like (CCA1, CDA-1) TFs (Tables 1-3). These elements are most likely associated with the upregulation of a number of MYB genes (Table 4) such as Os12g0125000 (MYB-like DNA-binding domain-containing protein), Os11g0128500 (MYB-like DNA-binding domain-containing protein), Os05g0553400 (Similar to MYB-related TF), Os01g0298400 (Putative typical P-type R2R3 MYB protein), Os12g0567300 (MYB TF domain-containing protein, R2R3 MYB), Os01g0874300 (Putative MYB-related protein, MYB2), and Os05g0140100 (R2R3 MYB TF) (Table 4).
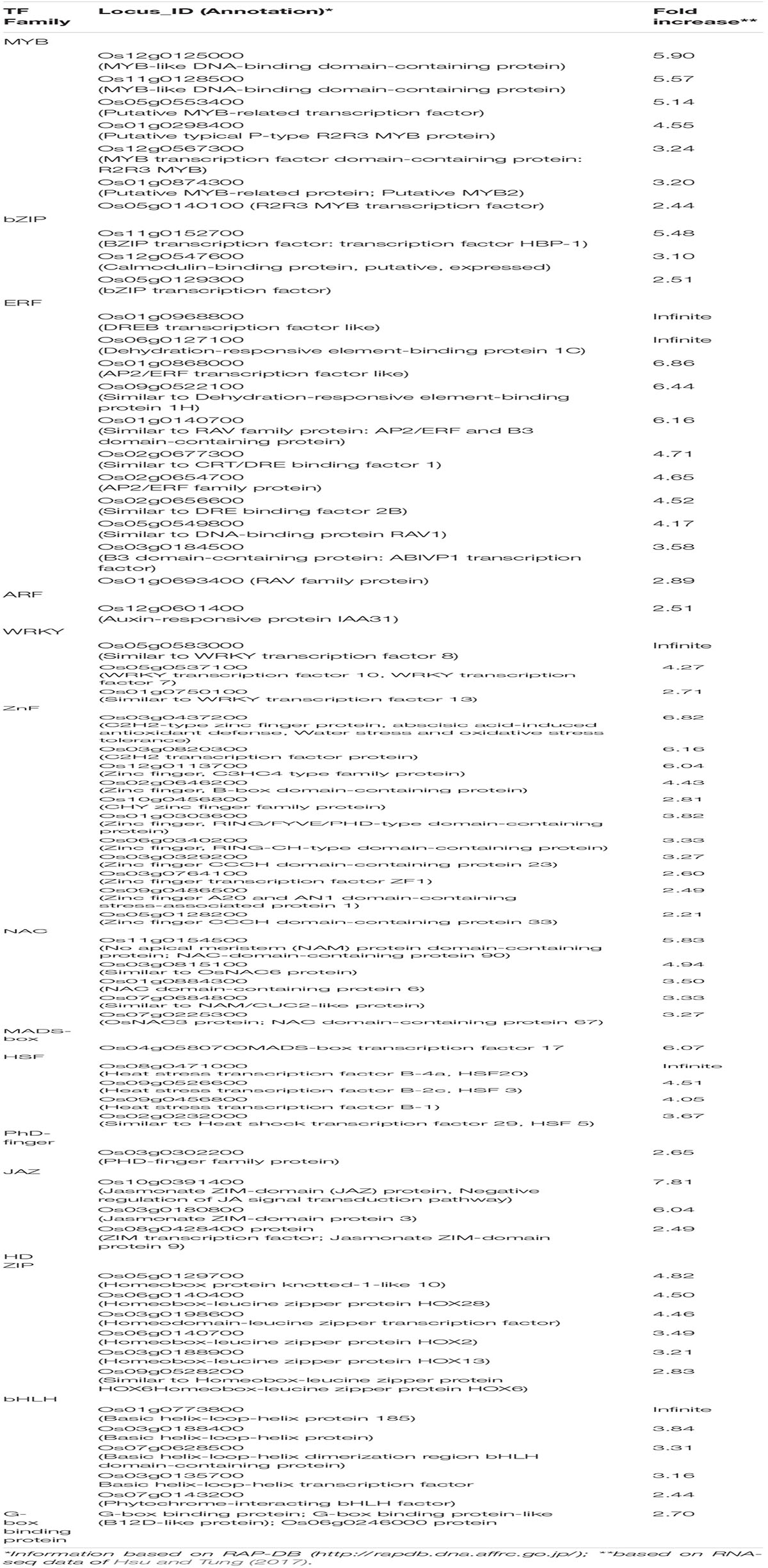
Table 4. List of upregulated transcription factors with potential significance to the identified putative cis-elements among upregulated genes in five genotypes from diverse background in response to submergence tolerance.
It has been highlighted that MYB TFs represent a major protein in rice and are involved in the transcriptional regulation of many developmental processes as well as abiotic and biotic stress conditions (Katiyar et al., 2012). Additionally, it has already been shown both experimentally and in silico analysis that MYB TFs that bind to MYB-box/GT-element-like elements play a key role in the transcriptional regulation of rice during germination under submergence and anoxia (Dolferus et al., 2003; Mohanty et al., 2012; Lakshmanan et al., 2014). In this analysis, MYB-box-like elements are more highly enriched compared to GARE-like elements. Under flooding/submergence/anoxic conditions, the plant hormone gibberellic acid (GA) plays a major role in rice and other plants such as barley (Gubler et al., 2002; Kaneko et al., 2002). It activates the endosperm reserve to aleurone layers for the induction of enzyme α-amylases for the hydrolysis of starch, protein, and cell wall reserve (Woodger et al., 2003; Lee et al., 2014). Under submerged/flooding conditions, α−amylases play a major role in providing sugar substrates by hydrolyzing starch for glycolysis and alcohol fermentation to generate ATP. Recently, Abdulmajid et al. (2020) have screened favorable rice genotypes for coleoptile elongation length sensitivity to exogenous gibberellin under submerged conditions. However, in this analysis, pyrimidine-box-like elements and GARE-like elements are less enriched compared to MYB-box-like elements, which are associated with MYB (R1, R2R3). A similar pattern was also identified in our previous analysis for the transcriptional regulation of coleoptile germination and elongation of the japonica rice under anoxia (Mohanty et al., 2012). It appears that submergence tolerance for rice germination may not be completely mediated by GA. Recently, it has been shown that two MYB TFs that bind to the same cis-element regulate the on/off switch of α-Amy expression (Chen et al., 2019). MYBS1 activates the expression of α-Amy during sugar starvation and promotes nuclear import of MYBS1, whereas MYBS2 behaves in the opposite manner during sugar provision. They have also shown that there is no enhancement of submergence tolerance in rice seedlings when the expression of MYBS2 was reduced. However, α-Amy is necessary for growth of rice seedlings under submergence as reduced seedling growth was observed in MYBS2-Ox lines under submergence. High enrichment of MYB-box-like element associated with R2R3 MYB and upregulation of R2R3 MYB genes in the tolerant genotypes suggest that abscisic acid (ABA) could also be playing a role together with other hormones in the germination and coleoptile elongation of rice under submergence.
bZIP Regulatory Module
As-1/ocs-like, GCN4 motif, and CAMTA5 binding site-like were highly enriched in the upregulated genes of tolerant, highly tolerant, and extremely tolerant genotypes (Tables 1-3). However, upregulated genes of highly tolerant and extremely tolerant genotypes possess specific ABRE-like elements associated with bZIP (Gr. A), bZIP (Gr. G) (GBF 3, 5, and 6), bZIP (DPBF-3), ABI5 (bZIP), and ABF2 (bZIP) and C-box-like/G-box-like and HY-5 binding site-like elements associated with GBF and HY5, respectively (Tables 2, 3). These binding sites could be associated with the upregulation of genes such as Os12g0547600 (Calmodulin-binding protein, putative, expressed), Os11g0152700 (bZIP transcription factor 79; transcription factor HBP-1), and Os05g0129300 (bZIP transcription factor) (Table 4). bZIP TFs play a key role in plant growth, development, and abiotic and biotic stress conditions in Arabidopsis and rice (Zhang et al., 2015; Yang et al., 2019). Although the role of bZIP in response to abiotic stresses has been well studied in Arabidopsis, a few cases have been characterized in rice (Tang et al., 2012, 2016; Zhang et al., 2017; Yang et al., 2019). Moreover, substantial enrichment of bZIP-associated elements suggest the significance of ABRE-like/CAMTA5/C-box/G-box-like elements in response to ABA signaling in response to submergence. The role of bZIP in response to submergence/flooding stress in rice is not well studied yet. In rice, OsbZIP45, an ortholog of maize GBF1, was shown to be induced by hypoxia (de Vetten and Ferl, 1995). Later, OsABF1, an ABA-responsive element (ABRE) binding bZIP TF, has been shown to be induced during different abiotic stresses, such as anoxia, drought, cold, salinity, and ABA in rice seedlings (Hossain et al., 2010). ABA plays a key role in plant physiology, development, seed maturation, dormancy, and responses to a number of abiotic stress conditions such as drought, cold, and salt (Dar et al., 2017). Promoter regions of many genes possess ABRE-responsive elements that are responsible for ABA-dependent regulation. These elements interact with various ABA-responsive TFs that regulate ABA response particularly (Kim et al., 2004). Recently, induction of bZIP in response to short- and long-term hypoxia in tomato root has been observed (Safavi-Rizi et al., 2020). In the previous analysis, we identified both high enrichment of As1/ocs-like element, ABRE-like, G-box-like, GCN4-like, and CAMTA5-like in the promoters of genes upregulated in anoxia (Mohanty et al., 2012), glycolysis and fermentation (Lakshmanan et al., 2014), and in the wild-type rice cultivar (Kinmaze) during germination and coleoptile elongation (Mohanty et al., 2016). In rice, all genotypes tolerant to submergence also showed upregulation of bZIP TF and a calmodulin-binding protein. Identification of CAMTA5 binding site-like elements associated with CAMTA5 TF proposes that it could be involved in regulating auxin transport and homeostasis (Galon et al., 2010). High enrichment of ABA-regulated bZIP TFs suggests a cross-talk between sugar and ABA-signaling during germination and coleoptile elongation under submergence. Besides, there could be some mutual enhancement between ABA and ethylene and ethylene could repress ABA activity depending on the requirement.
Interestingly, binding sites associated with bZIPs such as HY5, DPBF-3, and GBF 3, 5, and 6 were identified in both highly tolerant and extremely tolerant rice genotypes. HY5 is a key integrating factor for light and ABA pathways, and it stimulates ABA signaling pathway by binding to the promoter of ABI5 (Chen et al., 2008). It also regulates cell elongation and proliferation besides its other roles in plant growth and development (Jing et al., 2013). The presence of high enrichment of ABRE-like elements associated with different bZIP TFs especially in both highly tolerant and extremely tolerant genotypes suggests a possible temporal role of ABA signaling in response to longer coleoptile elongation.
ERF Regulatory Module
A number of putative cis-elements associated with different groups of ERF TFs were identified in the promoter regions of the upregulated genes in tolerant, highly tolerant, and extremely tolerant genotypes in response to submergence (Tables 1-3). Genes in tolerant genotypes were highly enriched with GCC-box−like elements associated with Groups VI, VIII, and IX ERF TFs; ERE-like elements associated with Groups I, IV, and VII TFs; jasmonic acid response element (JARE)-like elements associated with Gr. VI, VIII, and IX ERF TFs; and RAV-1-like element associated with ERF/RAV1 (Gr. II) TFs (Table 1). There is also enrichment of ABRE-like elements associated with Group II ABI3 AP2/B3 related to RAV TF, CRT/DRE-like elements associated with Group IV ERF TF and ABRE-binding site-like associated with ABR1 TF (Table 1). The enrichment of cis-elements related to the ERF group is less compared to their presence in tolerant genotypes (Tables 2, 3). GCC-box-like elements are also absent in these groups (Tables 2, 3). However, these genes possess binding sites associated with ERF (Gr. VI, VIII, and IX), ABI3 AP2/B3 related to RAV and ERF/RAP 2.4, and ERF/RAP 2.3, 2.6, and 2.10 (Tables 2, 3). The enrichment pattern of these elements could be correlated with the upregulation of a number of ERF genes such as Os01g0968800 (DREB transcription factor-like), Os06g0127100 (Dehydration-responsive element-binding protein 1C), Os01g0868000 (AP2/ERF transcription factor-like), Os09g0522100 (Dehydration-responsive element-binding protein 1H), Os01g0140700 (Similar to RAV2: AP2/ERF and B3 domain-containing protein), Os02g0677300 (Similar to CRT/DRE binding factor 1), Os02g0654700 (AP2/ERF family protein, abiotic stress response), Os02g0656600 (DRE binding factor 2B), Os03g0184500 (B3 domain-containing protein, ABIVP1 transcription factor), and Os01g0693400 (RAV family protein) (Table 4).
In rice, ERF VII plays a key role in flooding, hypoxia, and submergence conditions (Bailey-Serres et al., 2012; Bui et al., 2015; Gibbs et al., 2015). SUBMERGENCE 1A (Sub1A), an ERF-type TF, regulates submergence/flooding tolerance in rice. Although the japonica cultivar “Nipponbare” lacks this gene (Fukao et al., 2006; Xu et al., 2006), it germinates and elongates its coleoptile under anoxia and submergence conditions. Hence, this shows that there could be some other mechanism that is independent of SUB1A in these genotypes to tolerate anoxia and submergence (Lee et al., 2009). An earlier evidence by Ishizawa and Esashi (1988) showed that ethylene is important for the transport of sucrose from the scutellum to the coleoptile during germination and coleoptile elongation of rice seeds. Recently, it has been reported that ethylene plays an important role in the signal transduction pathway dependent on ethylene and oxygen under hypoxia/submergence conditions. In contrast, two ERFs such as SK1 and SK2 are involved in submergence adaptation in deep-water rice by rapidly elongating the internodes through the action of GA response (Hattori et al., 2009).
In Arabidopsis, members of ERF-VIIs such as ERF71/HRE2, ERF72/RAP2.3, ERF73/HRE1, ERF74/RAP2.12, and ERF75/RAP2.2 are induced under limited oxygen conditions to regulate many hypoxia-induced genes involved in fermentation, sugar metabolism, and ethylene biosynthesis. Besides, it has been shown that RAP2.12 is present in the plasma membrane in an inactive state in the presence of oxygen and then moves to the nucleus under hypoxia/submergence conditions (Kosmacz et al., 2015). This mechanism shows the fast response of the TF to oxygen shortage in order to protect plant cells. In Arabidopsis, a higher survival rate under low oxygen condition was observed by overexpressing RAPs (RAP2.2, RAP2.3, and RAP2.12) (Papdi et al., 2015; Yao et al., 2017). In this analysis, GCC-box-like elements associated with ERF (I, IV, VII, and X) and ERE-like elements associated with ERF (Gr. III) such as RAP2.2, RAP2.3, RAP2.6, and RAP2.10 were identified. However, identification of moderate enrichment of RAV1a-like elements can be associated with the expression of Os05g0549800 (AP2/ERF and B3 domain-containing protein: similar to DNA-binding protein RAV1). In Arabidopsis, during seed germination and early seedling development, RAV1 plays a key role in ABA signaling by repressing the expression of ABI3, ABI4, and ABI5 by binding to the 5′−CAACA−3′ motif within the promoter of ABI3, ABI4, and ABI5. These results suggest that RAV1 could also be acting as a repressor of ABA signaling. In addition to ABA inhibition, ethylene also inhibits the biosynthesis of jasmonate (JA) during coleoptile elongation of etiolated rice seedlings (Xiong et al., 2017). Although the role of ERF-VIIs has been well studied in response to submergence, it is still a complex process and the regulatory mechanism is still not clearly understood yet.
The upregulated genes in the tolerant genotypes are highly enriched with ABR1 (AP2-like ABA repressor 1) binding-site-like motifs associated with ABR1 TF belonging to AP2-domain-containing protein group X in rice. TF ABR1 functions as a negative regulator of ABA response during seed germination. The expression of this TF is induced during various abiotic stresses, such as drought, cold, and salt in Arabidopsis (Pandey et al., 2005). In rice, the group X OsERFs are also closely related to the ortholog of ABR1 in Arabidopsis (Mishra et al., 2013) and show significant expression during different developmental stages and response to various abiotic stresses. Interestingly, the expression of AtERF#111/ABR1 in Arabidopsis was shoot specific and induced under submergence and hypoxia (van Veen et al., 2016; Yeung et al., 2018). Later, the regulation of AtERF#111 expression was suggested to be related to mechanical stress during submergence as it was regulated by WRKY18, 33, and 40 (Birkenbihl et al., 2017; Bäumler et al., 2019). Significant induction of TFs WRKY18, WRKY33, and WRKY40 is reported to be induced under both submergence and anoxia as well as wounding stress (Hsu et al., 2013; Tsai et al., 2014; Wang et al., 2015). This shows that ABR1 related to wounding/pathogen response was also induced during submergence to stimulate the immune response against the threat of wounding or pathogen infection after flooding (Bäumler et al., 2019). This TF could be involved in the immune response mechanism during rice germination and coleoptile elongation in tolerant genotypes in response to submergence.
EIN3/EIL1 Regulatory Module
The promoter analysis of the upregulated genes in all tolerant, highly tolerant, and extremely tolerant genotypes identified the presence of putative EIN3 binding-like elements in 50–70% of the genes (Tables 1-3). This binding site can be associated with EIN3/EIL1 TF. However, there was no expression of this TF in tolerant genotypes. This TF is the master transcriptional regulator of ethylene signaling as it is required for the activation of the ethylene pathway. It regulates the transcription of ethylene-responsive genes under different environmental and spatiotemporal conditions in Arabidopsis (An et al., 2010; Dolgikh et al., 2019). Besides, EIN3/EIL1 controls multiple transcriptional cascades. It also targets genes such as ERF1, PIF3, and CBF1/2/3, which are important regulators during different abiotic stress conditions and developmental processes (Zhang et al., 2011; Shi et al., 2012; Zhong et al., 2012). It has been reported that EIN3 is involved in the regulation of a secondary transcriptional ethylene response that includes TFs such as AP2/ERFs: ERF1, ERF5, WRKY14/47, PIF3, NAC6, and RAP2.2 (Chang et al., 2013). EIN3 binding also modulates feedback regulation of the ethylene signaling pathway and integrates between different hormone-mediated pathways. These results suggest that it could be playing a key role in rice germination and coleoptile elongation mainly to activate ethylene signaling and ERF TFs and could also act as a mediator in regulating other hormone signaling and TFs.
ARF Regulatory Module
Aux-RE-like elements associated with ARF TFs are identified in all tolerant, highly tolerant, and extremely tolerant genotypes, and this could be associated with the expression of ARF gene such as Os12g0601400 (Auxin-responsive protein IAA31) (Tables 1-4). The phytohormone auxin plays a key role in plant growth and development. It has been suggested that the elongation of rice coleoptile under submerged conditions could be cooperatively regulated by the endogenous concentration of ethylene and auxin (Ishizawa and Esashi, 1983). Moreover, it was shown that external IAA addition had a positive effect on the initial elongation of coleoptile (Breviario et al., 1992), whereas ethylene enhanced the later stage of elongation (Hoson et al., 1992). Auxin mainly promotes cell division. It has been shown that coleoptile in rice elongates rapidly in response to auxin treatment (Perrot-Rechenmann, 2010). OsARF, a rice homolog of the ARF, was found to be positively correlated with auxin-dependent differential growth in rice coleoptiles (Waller et al., 2002). Recently, Wu and Yang (2020) have revealed the involvement of auxin signaling in regulating rice coleoptile elongation as well as regulation of carbohydrate metabolism and secondary metabolism under submergence. Besides, auxin plays a key role in cell division during coleoptile elongation in the japonica rice under submergence (Nghi et al., 2020). Differences in auxin transport determine the length of coleoptile while availability of higher auxin level determines the final length of coleoptile under submerged conditions. They have also claimed that the long coleoptile in rice under submergence is due to an increase in auxin transport by the influx carrier AUX1. Besides, it also regulates the expression of other TFs. The presence of Aux-RE-like elements in the promoters of upregulated genes present in all groups and significant expression of ARF gene suggest a role of ARF and auxin signaling in coleoptile elongation under submergence.
WRKY Regulatory Module
Promoter analysis of the upregulated genes identified enrichment of W-box-like elements associated with WRKY TF in all tolerant, highly tolerant, and extremely tolerant genotypes, and those could be associated with the upregulation of WRKY genes such as Os05g0583000 (Similar to WRKY transcription factor 8), Os05g0537100 (WRKY transcription factor 10, WRKY transcription factor 7), and Os01g0750100 (WRKY transcription factor 13) (Tables 1-4). Identification of W-box-like elements also coincides with the previous identification in the genes upregulated during germination and coleoptile elongation in rice (Mohanty et al., 2012; Lakshmanan et al., 2014). WRKY plays a key role in plant growth and development, secondary metabolite biosynthesis, and response to a number of abiotic and biotic stresses (Phukan et al., 2016). Increase in the expression of WRKY has also been shown by microarray data in Arabidopsis (Loreti et al., 2005) and rice (Lasanthi-Kudahettige et al., 2007) in response to oxygen deficiency. The role of WRKY in the regulation of submergence tolerance in rice has also been reported by Viana et al. (2018). Also, in transgenic Arabidopsis expressing a sunflower WRKY, HaWRKY76 was found to be tolerant to flooding (complete submergence) by preserving carbohydrates, mainly sucrose and starch, through the repression of fermentation pathways (Raineri et al., 2015). Upregulation of WRKYs at different time points of submergence stress was also observed in maize (Campbell et al., 2016). Similar observations were also identified in alcohol dehydrogenase 1 (ADH1)-deficient mutant of rice where WRKY could be playing a key role in cell survival rather than elongation (Mohanty et al., 2016). Interestingly, W-box elements have been identified in the promoter of upregulated WRKY genes such as OsWRKY11, OsWRKY56, and OsWRKY62 during submergence (Viana et al., 2018). This shows the self-regulation of WRKY in response to submergence stress. Overall, it shows that WRKY could be playing a role by self-regulating through an on–off switch depending on the condition in response to submergence.
A number of WRKY TFs are known to be ABA responsive and are involved in ABA signaling pathways. However, in Arabidopsis, three WRKY genes such as AtWRKY18, AtWRKY40, and AtWRKY60 negatively regulate ABA signaling. Among them, AtWRKY 40 binds to the promoter of ABI4 and ABI5 and represses the expression of ABA-responsive genes (Shang et al., 2010). ABI4 itself and WRKY 9 positively regulate ABI4 expression in Arabidopsis (Chen et al., 2013), whereas ABI3/VP1 (Feng et al., 2014) and Basic Pentacysteine (BPC) negatively regulate ABI4 (Mu et al., 2017). However, the TF WRKY 6 induces the expression of RAV1 by binding to the W-box motif present in its promoter and this represses the expression of ABI3, ABI4, and ABI5 (Huang et al., 2016).
In addition to the high enrichment of putative cis-elements associated with WRKY TF, high enrichment of putative cis-elements associated with AB3/VP1 and ABI5 was also identified. Recently, the role of WRKYs in hypoxia tolerance has been demonstrated in Arabidopsis (Tang et al., 2021). Through genetic and molecular experiments, it has been shown that WRKYs synergistically (WRKY33 interacts with WRKY12) increase the activation of TF RAP2.2 to increase the hypoxia tolerance in Arabidopsis and support the role of WRKY in hypoxia tolerance. The analysis of promoter architecture suggests its self-regulation depending on its requirement to activate other TFs to enhance submergence tolerance.
ZnF Regulatory Module
The promoter analysis results identified a high enrichment of zinc-finger binding element-like associated with ZnF TF in all tolerant, highly tolerant, and extremely tolerant genotypes (Tables 1-3). These elements are probably associated with the upregulation of different ZnF genes such as Os03g0437200 (C2H2-type zinc finger protein, ABA-induced antioxidant defense, water stress, and oxidative stress tolerance), Os03g0820300 (C2H2 transcription factor), Os12g0113700 (Zinc finger, C3HC4-type family protein), Os02g0646200 (Zinc finger, B-box domain-containing protein), Os10g0456800 (CHY zinc finger family protein), Os01g0303600 (Zinc finger, RING/FYVE/PHD-type domain-containing protein), Os06g0340200 (Zinc finger, RING-CH-type domain-containing protein), Os03g0329200 (Zinc finger CCCH domain-containing protein 23), Os03g0764100 (Zinc finger transcription factor ZF1), Os09g0486500 (Zinc finger A20 and AN1 domain-containing stress-associated protein 1), and Os05g0128200 (Zinc finger CCCH domain-containing protein 33) (Table 4). The enrichment of zinc finger binding-like elements are higher in extremely tolerant genotypes compared to tolerant genotypes. This suggests a key role of ZnF in submergence tolerance although the molecular mechanism is not known yet. ZnF family TFs are known to be involved in important transcriptional regulation of plant responses to different abiotic stresses, such as drought, temperature, light, and salt (Wang et al., 2018). In rice, ZFP and C2H2 ZnF proteins are induced by a number of abiotic stresses including cold, drought, and salt (Jin et al., 2018). In our previous analysis, we identified high enrichment of zinc finger binding element-like elements associated with ZnF TFs as well as upregulation of a number of C2H2 and other ZnF proteins in the upregulated genes related to anaerobic metabolism in rice under anoxia (Lakshmanan et al., 2014). We also found upregulation of the expression of ZnF TF in wild-type rice under complete submergence (Mohanty et al., 2016). Besides, it has been reported that ZnF TF was upregulated in both rice and Arabidopsis in response to hypoxia/anoxia (Loreti et al., 2005; Lasanthi-Kudahettige et al., 2007; Pandey and Kim, 2012). In rice, a CCCH-type zinc finger protein was significantly induced by hypoxia/submergence stress indicating a key role during hypoxia/submergence stress in rice (Pandey and Kim, 2012). A B-box type zinc finger protein displayed significantly higher expression in soybean tolerant genotypes in response to flooding treatment compared to sensitive genotypes (Yu et al., 2019).
NAC Regulatory Module
High enrichment of DRE-like elements associated with NAC TFs is identified in the promoters of upregulated genes in all tolerant, highly tolerant, and extremely tolerant genotypes (Tables 1-3). This high enrichment can be associated with the activities of several NAC genes such as Os11g0154500 [No apical meristem (NAM) protein domain-containing protein; NAC-domain-containing protein 90], Os03g0815100 (Similar to OsNAC6 protein), Os01g0884300 (NAC domain-containing protein 6), Os07g0684800 (Similar to NAM/CUC2-like protein), and Os07g0225300 (OsNAC3 protein) (Table 4). Members of NAC family TFs play a major role in regulating abiotic and biotic stresses in Arabidopsis and many other crops (Nakashima et al., 2012; Shao et al., 2015; Yuan et al., 2019a, b). In many cases, NAC TF is regulated through the ABA-dependent signal transduction pathway (Chen et al., 2014). In addition, there is evidence regarding the interaction of NAC and JA signaling pathway. Regulation of NAC through both ABA and JA to abiotic stress tolerance has been elucidated in Arabidopsis and other plants (Bu et al., 2008; Yoshii et al., 2010). It also maintains membrane integrity during abiotic stresses in Arabidopsis (Yong et al., 2019). It could be regulating germination and coleoptile elongation to maintain membrane integrity in cross-talk with other hormones such as ABA and JA. Moreover, reactive oxygen and nitrogen species (ROS and RNS) that accumulate during submergence/hypoxia activate a number of TFs including Heat Shock Factor (HSFs) and NAC families to control the homeostasis of the harmful molecules (Gonzali et al., 2015).
DOF Regulatory Module
Sequence motif “AAAAG/CTTTT”-element-like was identified in the promoters of all tolerant, highly tolerant, and extremely tolerant upregulated genes, which can be associated with DOF (DNA binding with one finger) TFs. These TFs are mainly plant-specific (Noguero et al., 2013) and are involved in the regulation of various processes in plant metabolism, seed germination, phytochrome response, and various developmental processes (Noguero et al., 2013; Wu et al., 2015). However, the biological function of this TF is not well studied in rice yet. Experiments on OsDof3 suggested that it interacts with GAMYB to induce the expression of RAmy1A to facilitate GA signaling during the germination of rice seeds (Washio, 2003). In this analysis, DOF could be interacting with GA and other plant hormones at the initial stage of germination to activate the expression of stress-responsive genes to tolerate submergence during rice germination and coleoptile elongation.
HSF Regulatory Module
Enrichment of heat shock binding factor element-like in the promoter regions of upregulated genes of all tolerant, highly tolerant, and extremely tolerant genotypes could potentially be correlated with the upregulation of HSF genes such as Os08g0471000 (Heat stress transcription factor B-4a, HSF20), Os09g0526600 (Heat stress transcription factor B-2c, HSF 3), Os09g0456800 (Heat stress transcription factor B-1), and Os02g0232000 (Similar to Heat shock transcription factor 29, HSF 5) (Tables 1-4). HSFs are shown to be involved in heat stress and also other abiotic stresses (Nishizawa et al., 2008). Induction of a number of HSPs and HSTs in response to anoxia/hypoxia has been reported in Arabidopsis and rice (Loreti et al., 2005; Lasanthi-Kudahettige et al., 2007). HSF proteins play a major role in protecting cellular responses under stress conditions by preventing the misfolding and denaturation of proteins (Liberek et al., 2008) and also in the activation of HSP pathway during either anoxia or heat stress to cope with the production of ROS.
AS2/LBD Regulatory Module
Identification of a high enrichment of S2-binding-like elements in all tolerant, highly tolerant, and extremely tolerant genotypes proposes their possible association with Lateral organ boundaries domain (LBD) family TF AS2 (Tables 1-4). However, we did not see any expression of this gene in any of the tolerant genotypes. This TF plays a key role in different abiotic stress conditions. It is one of the hypoxia-induced TFs in Arabidopsis during flooding stress (Licausi et al., 2010). ARF7 and ARF19 TFs are known to regulate the expression of LBD16 and LBD29 for lateral organ development in Arabidopsis (Okushima et al., 2007). Recently, it has been shown that these TF genes could be regulated by auxin signaling under submergence (Wu and Yang, 2020), although the function is not known yet. This element could be present in the upregulated genes of all groups for lateral organ development later once the coleoptile reaches the surface of water for O2 availability.
E2F Regulatory Module
A moderate enrichment of E2F-binding site-like elements associated with E2F TFs was identified in the upregulated genes of the tolerant genotypes (Table 1). E2F TF plays a role in the regulation of cell division in rice coleoptile elongation under anoxia (Kosugi and Ohashi, 2002). The presence of E2F-binding site-like elements associated with E2F TFs was also detected in the promoters of upregulated genes under anoxia in rice coleoptile elongation and in wild-type rice demonstrating coleoptile elongation under anoxia (Mohanty et al., 2012, 2016).
TCP Regulatory Module
TCP binding site-like elements associated with TCP TF are enriched in the promoters of upregulated genes in tolerant and highly tolerant genotypes (Tables 1, 2). This TF plays a key role in cell growth, different hormone response pathways, and abiotic stress responses (Li et al., 2005; Danisman, 2016). The role of TCP in submergence tolerance is not known yet. However, in Arabidopsis, the activity of AtTCP14 is necessary for seed germination and there is a functional relationship between this TF and GA (Tatematsu et al., 2008). Subsequently, both TCP14 and TCP15 were reported to regulate cell proliferation (Kieffer et al., 2011) and required for GA-dependent regulation of seed germination in Arabidopsis (Resentini et al., 2015). Identification of TCP-binding sites in tolerant and highly tolerant genotypes suggests their roles in initial cell proliferation of coleoptile during germination under submergence in rice.
HD-ZIP (ATHB4) Regulatory Module
Promoter analysis identified the presence of HD-ZIP binding site-like elements associated with HD-ZIP TFs in the promoters of all upregulated genes of all tolerant, highly tolerant, and extremely tolerant genotypes (Tables 1-3). The enrichment of this element is most probably associated with the activities of a number of HD-ZIP genes such as Os06g014040 (Homeobox protein knotted-1-like 10), Os05g0129700 (Homeobox-leucine zipper protein HOX28), Os03g0198600 (Homeodomain-leucine zipper transcription factor), Os06g0140700 (Homeobox-leucine zipper protein HOX2), Os03g0188900 (Homeobox-leucine zipper protein HOX13), and Os09g0528200 (Similar to Homeobox-leucine zipper protein HOX6) (Table 4). HD-ZIP is involved in the regulation of many developmental processes and response to different abiotic stresses in many plants (Annapurna et al., 2016; Tang et al., 2019; Zhao et al., 2019). However, the role of HD-ZIP in response to submergence stress is not known yet. In Arabidopsis seedlings, AtHB4 gene was shown to regulate both shade avoidance and hormone-mediated development, particularly BR (Sorin et al., 2009). AtHB4 acts downstream of P1F1 and was involved in the activation of hypocotyl cell wall composition and elongation in response to short day plants (Capella et al., 2015). The presence of this binding site in highly tolerant and extremely tolerant genotypes could be playing a role in coleoptile elongation by mediating the BR signaling pathway.
ARR-B Regulatory Module
ARR-B binding element-like associated with type-B ARRs was identified among upregulated genes in all tolerant, highly tolerant, and extremely tolerant genotypes (Tables 1-3). Type-B ARRs such as ARR1-ARR2, ARR10-ARR14, and ARR18-ARR21 play a key role as positive regulators of cytokinin signaling (Argyros et al., 2008). Besides, they also play an important role in the downregulation of ABA activity in response to cytokinin. Moreover, the binding sites of ARR-B TFs are known to be associated with metabolism of BR, BR signaling, and transcriptional regulation by TFs such as BZR2; BEH1, 2, 3, and 4; BIM1; and MYB30 (Zubo and Schaller, 2020). Genes that belong to ARR-B are also involved in the induction of GA biosynthesis and the reduction of GA perception (Marín-de la Rosa et al., 2015). These binding sites were identified in the genes associated with ethylene biosynthesis and signal transduction. Since genes in all genotypes possess these binding sites, it suggests that they could be playing a key role together with other hormones to regulate the elongation and germination of coleoptile in response to submergence tolerance.
MADS-box (AGL) Regulatory Module
High percentage of CArG box-binding site-like elements corresponding to MADS box (AGL) TF were identified in the upregulated genes of all tolerant, highly tolerant, and extremely tolerant genotypes (Tables 1-3), and this could be due to the induction of Os04g0580700 (MADS-box transcription factor 17) (Table 4). MADS box (AGL) TFs are key regulators of many developmental processes in plants (Smaczniak et al., 2012). However, their role in seed germination is unknown except for a few MADS-box genes such as AGL25, AGL67, and AGL 21 (Chiang et al., 2009; Bassel et al., 2011; Yu et al., 2017). AGL25 is reported to be involved in temperature-dependent seed germination by inducing the GA biosynthetic pathway and the ABA catabolic pathway, whereas AGL67 and AGL21 act as negative regulators of seed germination. AGL21 incorporates both hormone signal and environmental signal to ABA signaling by balancing ABI5 to prevent germination under adverse conditions (Yu et al., 2017).
BES/BZR Regulatory Module
Interestingly, E-box-like elements associated with BES/BZR TFs involved in Brassinosteroid (BR) signaling pathway were identified only in the upregulated genes of highly tolerant and extremely tolerant genotypes that had longer coleoptile elongation in response to submergence (Tables 2, 3). BR is one of the most important plant steroidal hormones that regulate a wide range of plant growth, development including cell elongation and seed germination, and responses to biotic and abiotic stresses (Guo et al., 2013; Tong et al., 2014; Zhang et al., 2014; Li et al., 2016; Ahammed et al., 2020). It has also been shown earlier that BRs can increase coleoptile length in rice (Yamamuro et al., 2000). However, the molecular mechanism of BR that controls the germination and coleoptile elongation of rice seeds is not well known yet. High level of BR inactivates BIN2 and activates dephosphorylation of TFs BZR1 and BZR2/BES1 by protein phosphatase for their accumulation in the nucleus to increase their DNA-binding activity for the expression of BR-responsive genes (Wang et al., 2012). BZR1 and BZR2/BES1 regulate a large number of structural and metabolic genes as well as cell wall biogenesis and regulatory genes (Yu et al., 2011). Gene expression profiles have also revealed the induction of the expression of many cell wall extension and loosening enzymes and expansins (Guo et al., 2009). Several studies have reported that rice has a well-preserved BR signaling pathway like Arabidopsis (Li et al., 2009; Tong and Chu, 2012; Tong et al., 2014). It also regulates developmental processes through a cross-talk interaction with other signaling pathways. In lowland rice plants, both BRs and GA act antagonistically in response to submergence tolerance (Schmitz et al., 2013). They have indicated that BR induces GA catabolic genes and also DELLA proteins to limit GA level during submergence, and the cross-talk between BR and GA depends on tissues and hormone levels in rice (Tong et al., 2014). Although ABA and BR signaling are known to be antagonist of each other, recently it has been reported that both BR and ABA co-regulate to enhance seed germination in Arabidopsis. BES1, an important TF of the BR signaling pathway, assists in seed germination by weakening ABA signaling pathway induction by interfering the transcriptional activity of ABI5 (Zhao et al., 2019).
BR also activates different stress adaptive signaling pathways by directly or indirectly regulating different stress-responsive TFs such as bZIP, MYB, WRKY, NAC, DREB, etc. through BIN2 and key BZR1/BES1 TFs (Sharma et al., 2017). Genetic analysis shows that BZR1 and PIF, a bHLH TF, directly interact and promote cell elongation and etiolation (Oh et al., 2012). Moreover, BZR1 and PIF4 along with ARF6 promote genes that are involved in cell expansion (Oh et al., 2014). Both E-box motifs and G-box (CACGTG) motifs are associated with TF BZR1/2 and PIFs, and these are highly enriched in ARF6 binding regions. BEE2 is a bHLH TF, which is shown to be involved in the regulation of cell elongation by BR (Friedrichsen et al., 2002). BEE2 also interacts with ARF6 like PIF6 to promote cell elongation (Oh et al., 2014). These findings suggest a key role of BR-responsive TFs in cell elongation.
bHLH Regulatory Module
E-box-like/G-box-like elements associated with bHLH (Gr. III and VII) including PIFs were identified in both highly tolerant and extremely tolerant genotypes (Tables 2, 3). Interestingly, a set of E-box-like and BBRE-element-like/G-box-like elements associated with TFs such as BEE2, BIM1, BIM3, BAM8, and BEH4 that belong to bHLH were identified only in the upregulated genes of the two extremely tolerant genotypes. These elements could be associated with the induction of different bHLH TF genes such as Os01g0773800 (bHLH protein 185), Os03g0188400 (bHLH protein), Os07g0628500 (bHLH dimerization region bHLH domain-containing protein), Os03g0135700 (bHLH transcription factor), and Os07g0143200 (Phytochrome-interacting bHLH factor) (Table 4). bHLH TFs are involved in the regulation of many cellular processes (Zhao et al., 2020) such as seed germination (Penfield et al., 2005), light signaling (Leivar et al., 2008), hormone signaling (Fernández-Calvo et al., 2011), responses to wounding, drought, salt, and low temperature (Sun et al., 2018). They also play a key role in BR-responsive gene expression to support coleoptile elongation, which has already been discussed earlier.
Phytochrome-interacting factors (PIFs) are bHLH TFs. Besides the bHLH domain, they have active phytochrome A/B binding domains. They are involved in various physiological processes such as seed germination, photomorphogenesis, shade responses, flowering time, and leaf senescence (Leivar and Quail, 2011; Casal, 2013; Sakuraba et al., 2014). Besides, PIFs promote cell elongation under low Red:far Red light conditions by inducing the transcription of genes related to growth (Paik et al., 2017). Involvement of PIFs in signaling responses (GH3, IAA, and ARF), cell wall modification, and elongation has been studied (Zhang et al., 2013; Pfeiffer et al., 2014). In addition, they are involved in a variety of hormone-response pathways such as GA, BR, JA, ethylene, and nitric oxide (Mazzella et al., 2014; Paik et al., 2017). However, the study of phototropism of rice coleoptile under submergence is not known yet. Interestingly, it was suggested that submerged coleoptiles exhibited only a slight R-induced growth inhibition (Pjon and Furuya, 1974). Studies show that they are less phototropic compared to other gramineae coleoptiles such as maize and oats (Neumann and Iino, 1997). Experiments on the coleoptile photomorphogenesis 1 (cpm1) mutant suggest that this gene has a role in phytochrome-mediated inhibition of coleoptile growth. Later studies on phototropism rice coleoptile demonstrates that auxin is involved in this process (Haga et al., 2005).
Identification of Putative cis-Elements in the Upregulated Genes of Intermediately Tolerant Diverse Rice Genotypes in Response to Submergence Tolerance
To compare and validate the identification of cis-elements in tolerant, highly tolerant, and extremely tolerant genotype groups, upregulated genes in the intermediate group, which has two highly tolerant genotypes, F291 and F274-2a, were also analyzed (Table 5). The analysis results identified highly enriched significant cis-elements that are associated with MYB, bZIP, ABI5, different groups of AP2/ERF, AP2/B3, EIL, ARF, WRKY, ZnF, NAC, DOF, HSF, AS2, TCP, ARR-B, MADS box, GATA, BES/BZR, bHLH, and BPC2 (Table 5). Interestingly, the analysis results for the intermediate group are exactly similar to the identification of cis-elements and their associated TFs in highly tolerant genotypes (Table 2). In addition, it is slightly different in terms of promoter architecture content from the analysis that was performed for extremely tolerant genotypes (Table 3). Hence, the analysis results for both intermediately tolerant genotypes and highly tolerant genotypes suggest that they have a common gene regulatory mechanism that allows longer coleoptile elongation of rice seeds during germination, in response to submergence tolerance.

Table 5. Potential putative cis-elements identified in the promoters of upregulated genes in two intermediately tolerant genotypes (F291 and F274-2a).
Similarly, to validate the identification results, analysis was performed for a set of upregulated genes that have the moderate genotype Nipponbare and highly tolerant genotype F274-2a. The analysis results identified cis-elements that are associated with TFs such as MYB, bZIP, ERF, ABI3, AP2/B3, ARF, ZnF, NAC, HSF, AS2, TCP, ARR-B, MADS box, GATA, DBP1, and TBP (Table 6). Remarkably, the potential TFs that were identified also show similar promoter architecture with results obtained for tolerant genotypes (Table 1). The analysis results also did not identify specific cis-elements that are associated with BES/BZR and bHLH TFs present in highly tolerant and extremely tolerant genotypes (Tables 2, 3).
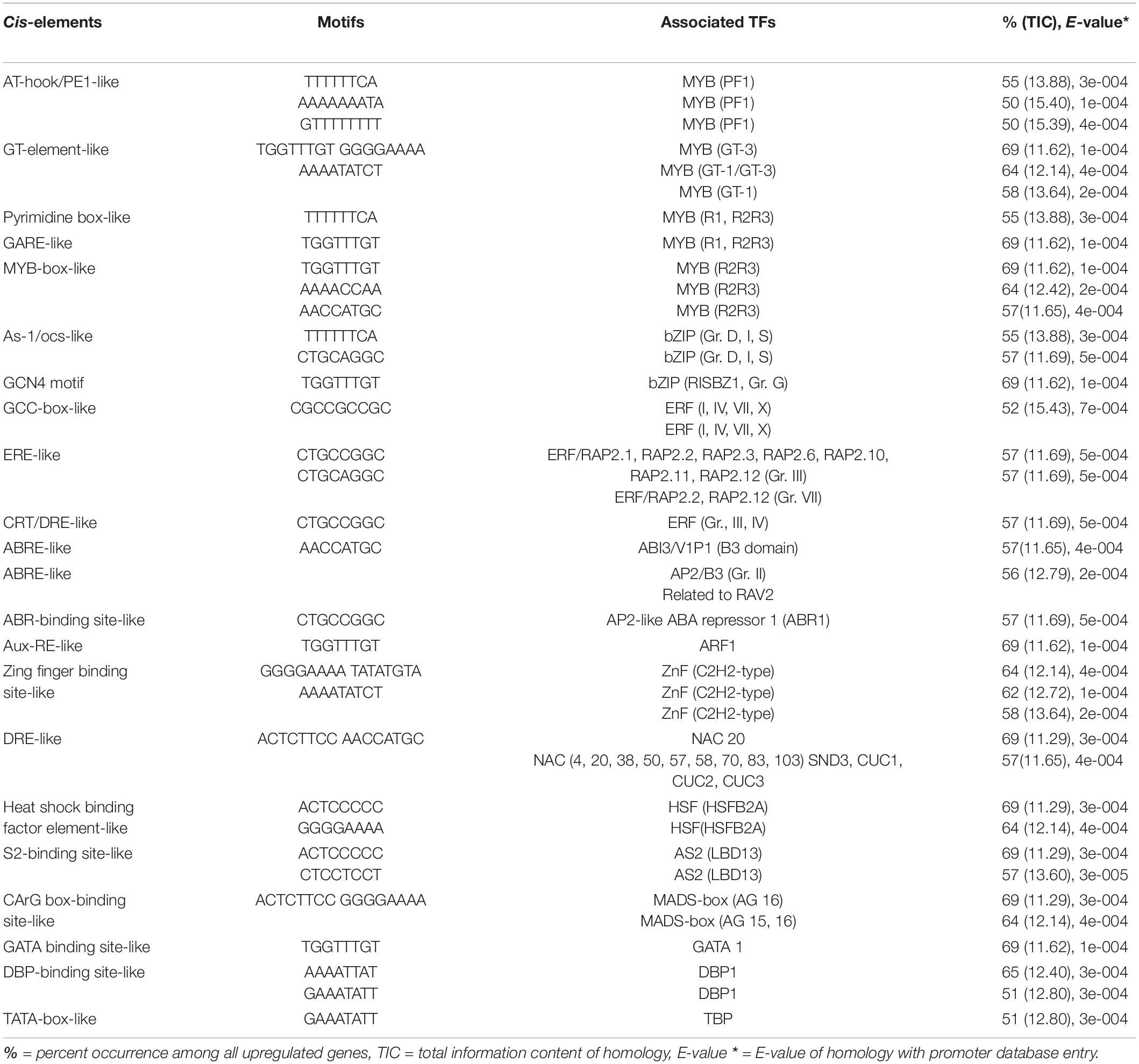
Table 6. Potential putative cis-elements identified in the promoters of upregulated genes in a group that has one tolerant genotype and one highly tolerant genotype (Nipponbare and F274-2a).
Discussion
The global climate variation causes extreme weather changes and often causes severe rain, which is harmful for rice seed germination and seedling growth. Although rice has the unique ability to tolerate such conditions, different genotypes of rice show different degrees of tolerance to submergence by elongating their coleoptiles. Hence, it is essential to understand the regulatory mechanism that helps in tolerating such adverse conditions. It is a complex process and involves interactive mechanisms of both metabolic and transcriptional regulation and hormonal signaling. A number of studies show that different types of hormones mainly activate transcriptional regulation that enables different plant metabolic processes to tolerate any adverse conditions due to climate change (Nemhauser et al., 2006). During rice germination and coleoptile elongation under submergence, although ethylene and ERF factors are known to be involved, the role of other TFs and hormonal signaling is not well understood yet. Therefore, the promoter architecture of upregulated genes of tolerant genotypes with different rates of coleoptile elongation could give some indication regarding the cis-elements content and their association with specific TFs. This information would support in providing a regulatory role of potential TFs and different hormonal signaling pathways involved in the transcriptional regulation of submergence tolerance mechanism. In this study, promoter architecture was analyzed for different sets of common genes associated with different degrees of submergence tolerance such as tolerant genotypes (Nipponbare, two recombinant inbred lines F291 and F274-2a, and two natural genotypes 8391 and 8753), highly tolerant genotypes with longer coleoptile elongation (F291, F274-2a, 8391, and 8753), and extremely tolerant natural genotypes with the longest coleoptile elongation (8391 and 8753). The promoter architecture analysis of upregulated genes for tolerant genotypes including O. sativa japonica identified the presence of putative cis-elements that are associated with TFs such as MYB, bZIP, AP2/ERF, ARF, EIN3, ABI3, ABR1,WRKY, ZnF, MADS-box, NAC, AS2, DOF, HD-ZIP, E2F, ARR-B, and HSF (Table 1). For the verification of these findings, promoters of the upregulated genes of a group that has tolerant genotypes, Nipponbare and F274-2a, were analyzed. The analysis identified most of the cis-elements that are associated with TFs identified in tolerant genotypes (Table 6). Interestingly, in addition to these TFs, there was identification of more specific binding sites associated with different specific TFs such as ABRE-binding site associated with bZIP, ABI5, ABF2, and bZIP (DPBF3); HY-5 binding site-like associated with HY5; G-box-like associated with GBF3, 5, and 6; E-box-like associated with BES/BZR TF; and E-box/G-box-like elements associated with bHLH (PIF7) in highly tolerant genotypes (Table 2). Moreover, the promoter architecture of the extremely tolerant genotypes (longest coleoptile elongation) is quite fascinating. They contain binding sites that are present in both tolerant and highly tolerant genotypes as well as higher enrichment of more binding sites such as ABRE-like associated with both ABF1 and ABF2 (bZIP), E-box-like/G-box-like and BBRE-element-like/G-box-like associated with bHLH, BEE2(bHLH), and BAM8 (bHLH) involved in BR-mediated signaling (Table 3). These binding sites are completely absent in the tolerant genotypes and less enriched or absent in highly tolerant genotypes. To support these findings, an additional promoter architecture analysis was performed by taking an intermediate genotype group that has two highly tolerant genotypes, F291 and F274-2a. The putative cis-element analysis of this group also identified less enrichment or complete absence of those specific binding sites that are present in the extremely tolerant genotypes (Table 5). These findings clearly show that the promoter architecture varies from genotype to genotype and the difference in tolerance mechanism and coleoptile elongation could be dependent on the presence of specific binding elements that are associated with specific transcriptional regulation by TFs in the promoter of genes upregulated during submergence.
Based on the promoter architecture of genes upregulated in all the three groups of different genotypes, the results suggest that there is involvement of both MYBs such as MYB (R1, R2R3) and MYB (R2R3) during the initial stage, i.e., imbibition and germination where both hormones ABA and GA play a key role. Upon imbibition of rice seed, the GA content gradually increases and breaks dormancy by inducing the secretion of hydrolytic enzymes and the endogenous ABA level decreases rapidly due to the induction of ABA catabolic genes (Figure 2). GAMYB is a GA-responsive R1, R2R3 TF that induces the expression of α-amylase gene (RAmy3D) in the aleurone layer for starch degradation to provide substrates for the germination and coleoptile elongation of rice seeds (Perata et al., 1997) under submergence. During this process, DOF TF interacts with GAMYB for induction of the expression of α-amylase to enable GA signaling (Zou et al., 2008; Figure 2). The high enrichment of MYB-box-like elements associated with MYB R2R3 TF and ABA signaling suggest that these TFs that are involved in seed maturation might be stored in rice seeds to initiate initial germination process during the early stage of germination (Sreenivasulu et al., 2008). High enrichment of ERF-VII TFs including RAP2.2, RAP2.3, RAP2.6, RAP2.10, and RAV1 shows their involvement and the importance of the hormone ethylene in germination and coleoptile elongation of rice seeds in response to submergence. The gaseous hormone ethylene is required for the transport of sucrose to the coleoptile (Ishizawa and Esashi, 1988). This is again supported by the higher expression of two ERF genes such as LOC_Os01g21120 in all genotypes [higher expression in tolerant genotypes compared to sensitive genotype (IR64) and higher expression of LOC_Os07g47790 in the tolerant genotypes] (Hsu and Tung, 2017). The role of ethylene in coleoptile elongation is also supported by the identification of EIN3-binding site elements in all tolerant, highly tolerant, and extremely tolerant genotypes (Tables 1 – 3). EIN3 becomes active in the dark and regulates the ethylene signaling pathway to transcribe ethylene-responsive genes. Since this TF has a unique role in regulating multiple transcriptional regulation, it could also be involved in feedback regulation by ethylene and regulation of other hormones and TFs (Dolgikh et al., 2019; Figure 2). Besides, high enrichment of ERE-like elements associated with ERF TF (Gr. III) such as RAP2.2, RAP2.3, RAP2.6, and RAP2.10 suggests their roles in fermentation and sugar metabolism in response to submergence (Tables 1-3). Identification of RAV-like elements associated with RAV1 implies their role in repressing the ABA signaling pathway by inhibiting the expression of ABI3, ABI4, and ABI5 by binding to the 5′−CAACA−3′ cis-element in their promoters (Feng et al., 2014; Figure 2). The enrichment of binding sites associated with ERF TFs including GCC-box-like elements associated with ERF (I, IV, VII, and X) was highest in tolerant genotypes and less enriched in the extremely tolerant genotypes. High enrichment of RAP2.3, RAP2.6, RAP2.10, and ERF (Gr. VII) could be associated with a common role of ethylene in all genotypes. The enrichment of ARF binding sites associated with TF ARF and the expression of ARF gene suggest the role of auxin in an auxin-dependent elongation of rice coleoptile through the interaction with ethylene (Ishizawa and Esashi, 1984; Breviario et al., 1992). Interaction of both hormones also inhibited root elongation in rice seedlings (Qin et al., 2017) and plays an important role in cell division and regulation of carbohydrate metabolism (Wu and Yang, 2020). Auxin upregulates GA biosynthesis genes and regulates the expression of GA metabolism genes through the action of Aux/IAA and ARF (Frigerio et al., 2006). Moreover, the co-regulation of both ethylene and auxin happens at the level of transcription as well as transport response. Besides AuxRE-like elements, there was identification of CAMTA5 binding site-like elements associated with bZIP (CAMTA5) TF. This TF could be involved in regulating auxin transport and homeostasis (Galon et al., 2010).
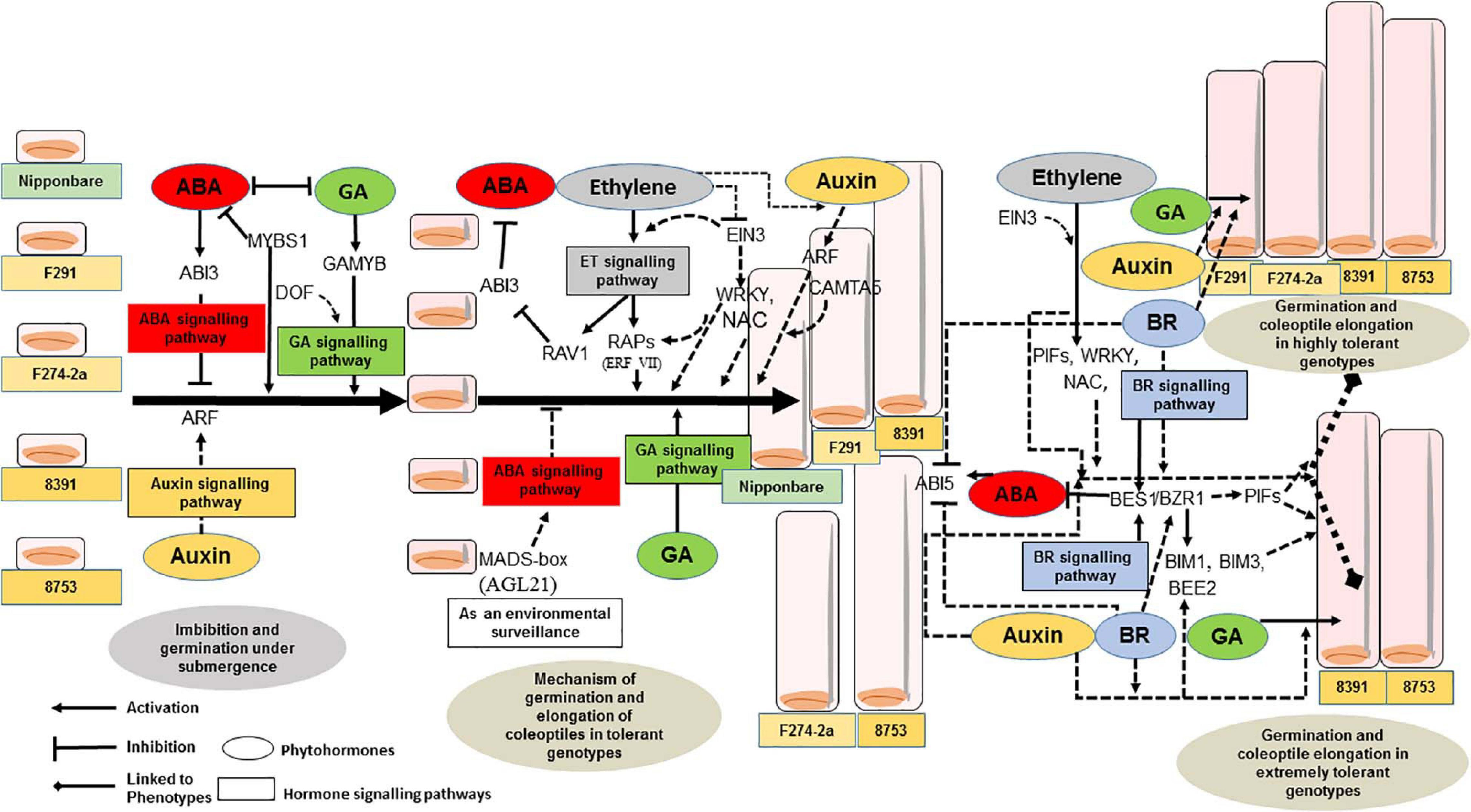
Figure 2. Hypothetical model representing transcriptional regulatory mechanisms leading to differences in germination and coleoptile elongation of diverse rice genotypes in response to submergence tolerance. TFs acting as activators/repressors along with hormonal signaling induce the synthesis of cell building blocks for elongation of coleoptiles in response to submergence in rice genotypes from diverse backgrounds. The differences in the length of coleoptiles could be due to altered transcriptional regulatory mechanisms involving bHLH and BES/BZR TFs together with other TFs and a cross-talk between BR and other signaling pathways. The dashed lines represent the hypothesis predicted from the gene expression data of tolerant, highly tolerant, and extremely tolerant genotypes that exhibit differences in coleoptile elongation in response to submergence tolerance.
It is known that bZIP 11 (Gr. S1) is induced by sucrose and can regulate sugar metabolism. It is most probably involved in adaptations to carbon starvation in Arabidopsis (Ma et al., 2011). bZIP also activates genes involved in trehalose metabolism and amino acid metabolism in response to stress (Ma et al., 2011; Weiste and Dröge-Laser, 2014). In rice, OsTPP7 encodes trehalose-6-phosphate phosphatase and is involved in the induction of starch mobilization during germination and coleoptile elongation (Kretzschmar et al., 2015). However, this gene was found to be upregulated in Nipponbare and the two RIL genotypes, F291 and F274-2a, but downregulated in two natural genotypes, and that could be due to structural variation (Hsu and Tung, 2017). Identification of as-1/ocs-like elements associated with bZIP (Gr. D, I, S) in tolerant, highly tolerant, and extremely tolerant genotypes propose their possible role in carbon and amino acid metabolism during coleoptile elongation in response to submergence. The majority of the bZIP TFs play a key role in ABA signaling pathways. G-box-related cis-elements associated with bZIPs are found in auxin-induced promoters and function as modulators of auxin-induced transcription in Arabidopsis (Weiste and Dröge-Laser, 2014). High enrichment of ABRE-like and G-box-like elements associated with bZIP TF could probably be involved in auxin-induced transcription for the longer elongation of coleoptiles in response to submergence in highly tolerant and extremely tolerant genotypes (Figure 2).
High enrichment of MYB-box-like elements associated with MYB R2R3 TF in the promoter regions of upregulated genes in all groups of genotypes indicates the involvement of pre-existing ABA not only at the beginning of germination but also at a later stage of germination through activation–inhibition machinery (Tables 1-3). It has been reported that ethylene–ABA interaction inhibits root growth in rice seedlings (Ma et al., 2014). In the dark, ethylene plays a double role in rice: stimulates coleoptile elongation and inhibits root growth (Ma et al., 2010, 2013; Yang et al., 2015), which is different from the response of ethylene in Arabidopsis (Bleecker and Kende, 2000). Moreover, it is motivating to find the presence of E-box-like/G-box-like elements associated with bHLH TFs such as PIF3 and PIF7 and BES/BZR TF in the promoters of upregulated genes in the highly tolerant and extremely tolerant genotypes (Tables 2, 3). Among them, the enrichment of the binding sites are much higher in the extremely tolerant natural genotypes. bHLH TFs are major players in phytochrome signal transduction and expression of BR-responsive genes (Duek and Fankhauser, 2005; Serna, 2007). PIFs and the hormone BR could be contributing in the regulation of longer cell elongation of rice coleoptiles under submergence (Paik et al., 2017) compared to moderate elongation in the O. sativa japonica genotype (Figure 2). Among the highly tolerant and extremely tolerant genotypes, E-box-like/G-box-like elements and BBRE-element-like/G-box-like element are more enriched in the extremely tolerant natural genotypes compared to the two recombinant inbred lines (RILs; F291 and F274-2a) derived from a cross between Nipponbare and IR64 (Tables 2, 3). BZR1 and PIF4 together with ARF4 induce genes that are involved in cell expansion (Oh et al., 2014). Moreover, BEE2 also regulates cell elongation by BR and interacts with ARF6 (Oh et al., 2014). These data indicate a potential involvement of BEE2, PIFs (bHLH), and BR-mediated signaling pathways contributing to the phenotypes of the two natural genotypes in regulating the highly elongated coleoptile growth compared to the RILs (Figure 2). Additionally, BR is reported to stimulate seed germination and inactivate the negative effect of ABA on seed germination by a negative feedback mechanism that modulates ABA signaling (Steber and McCourt, 2001; Xi et al., 2010).
During submergence/flooding stress, rice coleoptiles also suffer from low osmotic stress in addition to hypoxia/anoxia stress. A number of TFs such as AREB/ABFs are known to regulate ABA signaling during such stress conditions (Yoshida et al., 2010). In this promoter architecture analysis, identification of ABRE-like elements associated with ABF1 and ABF2 TFs could be involved in osmotic stress tolerance in the highly tolerant and extremely tolerant genotypes where the coleoptile elongation is higher compared to the moderate genotype O. sativa japonica. Among these two groups, there is high enrichment of ABF1 and ABF2 associated motifs in the extremely tolerant genotypes compared to highly tolerant genotypes (Tables 2, 3). As these TFs are associated with ABA signaling, the endogenous ABA level could be acting as an on–off switch having a spatiotemporal regulation to activate/repress other TFs and hormonal signaling pathways during the germination and coleoptile elongation under submergence (Figure 2). The enrichment of ABRE-like elements associated with both ABI3 and ABI5 TFs in all five tolerant genotypes is complicated as both are positive regulators of ABA signaling and involved in the arrest of seed germination (Lopez-Molina et al., 2002). They have shown that ABI5 is a bZIP TF, and it acts downstream of ABI3 to inhibit germination. A MADS-box TF AGL21 positively regulates the expression of ABI5 and responds to a number of environmental stresses and plant hormones during seed germination (Yu et al., 2017). It is reported that it could be acting as a surveillance integrator to incorporate external environmental signals and endogenous hormonal signals to ABA signaling for the regulation of seed germination and early post-germination growth. In this analysis, identification of putative cis-elements associated with ABI3 and ABI5 and high enrichment of MADS-box (AGL) TFs indicate a probable role of AGL as an environmental surveillance integrator to safeguard seed germination process by inducing ABA signaling through ABI5 TF (Figure 2). Although the putative cis-element associated with ABI3 is present in all groups, the elements associated with ABI5 are more enriched in highly tolerant and extremely tolerant genotypes. Also, during seed germination in Arabidopsis, JA enhances ABA activation to inhibit seed germination through JAZ repressors of the JA signaling pathway by regulating ABI3 and ABI5 TFs (Pan et al., 2020). Upregulation of the expression of JA ZIM-domain protein indicates their possible role in the activation of ABI3 and ABI5 for the spatiotemporal safeguard seed germination process.
The role of WRKY TFs in response to coleoptile elongation in rice under submergence is not well studied yet. However, studies on WRKY TFs reported that it plays a role in hypoxic stress response in persimmon (Zhu et al., 2019), rice (Shiono et al., 2014; Mohanty et al., 2016), and sunflower (Raineri et al., 2015), and submergence tolerance in rice (Viana et al., 2018). Recent studies on WRKY stated that a regulatory module composed of WRKY33 and WRKY12 together with RAP2.2 plays a key role in hypoxia tolerance in Arabidopsis (Tang et al., 2021). During submergence stress, interaction of WRKY 33 with WRKY12 upregulates the expression of RAP2.2, which acts downstream of both WRKY33 and WRKY12. RAP2.2 is an ethylene response TF that normally regulates genes associated with ethylene production, metabolism, and induction of genes encoding sugar metabolism and fermentation pathway enzymes (Hinz et al., 2010).
The identification of high enrichment of DRE-like elements associated with NAC TF in all groups of genotypes and expression of NAC genes suggest their role in regulating germination and coleoptile elongation together with other hormones to maintain membrane integrity. Similarly, the HD-ZIP (AtHB) gene acts downstream of P1F1 and regulates activation of hypocotyl cell wall composition and elongation through hormone-mediated development, particularly BR (Sorin et al., 2009; Capella et al., 2015). However, tolerant genotypes lack binding sites for BR-regulated TFs such as BES/BZR as well as bHLH (PIFs), whereas these are highly enriched particularly in highly tolerant and extremely tolerant genotypes (Tables 2, 3). In darkness, PIFs are normally active and involved in the regulation of gene expression to stimulate the skotomorphogenic response (Leivar et al., 2008). BZR1 and PIF4 interact with each other and regulate BR-induced gene expression (Martínez et al., 2018). There is also identification of ARR-B type binding site-like associated with ARR-B TFs. Type-B ARR binding sites are linked to the transcriptional regulation by TFs such as BZR2; BEH1, 2, 3, 4; BIM1; and MYB30 (Zubo and Schaller, 2020). MYB30 cooperates with BES1 and regulates the expression of BR-induced genes (Lee et al., 2009). Moreover, BR and GA are known to regulate cell elongation in rice, and BR regulates cell elongation by controlling GA metabolism (Tong et al., 2014). It seems that there is a cross-talk between BR, GA, and other hormones for the contribution of coleoptile elongation (Figure 2).
Interestingly, BBRE-element-like/G-box-like/E-box-like elements associated with TF BIM1, BIM3, and BEH1 are highly enriched in the extremely tolerant natural genotypes that have maximum coleoptile elongation (Figure 2 and Table 3). These genes may play a role in longer elongation of their coleoptiles compared to the highly tolerant genotypes. Besides the contribution of the BES1/BZR1 and PIF4 interaction for the longer elongation of coleoptiles in the extremely tolerant genotypes, BEE2 could also be involved in the BR signaling pathway for the maximum elongation of coleoptile in those two genotypes. Additionally, BAM8 (β-amylase–like proteins), a bHLH TF, could be acting as a metabolic sensor by interacting with the BR signaling pathway (Soyk et al., 2014).
Besides these binding sites, there were identifications of binding sites for TFs such as ABR1, HD-ZIP (ATHB4), TCP, ZNF, E2F, AS2 (LBD), NAC, and HSF (Tables 1-3, 5, 6). The roles of these TFs have not been studied in response to submergence stress. However, they could be involved in different protective roles during germination and coleoptile elongation in response to submergence such as HSF in protecting and preventing cellular responses (Liberek et al., 2008), ABR1TF in relation to mechanical stress, and NAC TF in controlling homeostasis of the cells. C2H2 zinc finger proteins could be involved in targeting the antioxidant genes associated with ROS scavenging (Han et al., 2020). TFs such as E2F and TCP could be involved in cell elongation and proliferation in a GA dependent pathway (Kieffer et al., 2011).
Conclusion
Germination and coleoptile elongation of diverse rice genotypes vary in response to submergence. Promoter architecture of upregulated genes associated with different tolerant genotypes suggests a fine-tuning at the transcriptional level that affects the phenotype of different genotypes. The germination and elongation of rice coleoptiles in response to submergence tolerance in all three genotype groups could be due to a combination of mechanisms involving different TFs such as MYB, bZIP, AP2/ERF, ARF, WRKY, ZnF, MADS-box, NAC, AS2, DOF, E2F, ARR-B, and HSF, and hormonal regulation by GA, ABA, ethylene, JA, and auxin. There could be re-balance between GA and ABA to activate other TFs and stress-responsive genes for the elongation of coleoptile to escape the submergence stress. Interestingly, the longer coleoptile elongation in the highly tolerant and extremely tolerant genotypes could be due to the involvement of additional TFs such as bHLH and BES/BZR along with BR signaling and a cross-talk with other signaling pathways. Moreover, the maximum elongation in the two extremely tolerant natural genotypes might be due to the additional transcriptional regulatory mechanism governed by TFs such as BEE2, BIM1, BIM3, and BAM8, which are not present in the other two genotype groups. The variation in coleoptile elongation in different groups of genotypes is certainly due to the difference in transcriptional regulatory mechanism controlled by specific TFs along with a synergistic cross-talk interaction between different hormones, which needs further experimental validation. This analysis provides a potential mechanism of transcriptional regulation across rice genotypes from diverse backgrounds, which may be helpful for rice breeding targets for direct seeding to improve rice production.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
BM designed and analyzed the data, and wrote the manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
BM would like to thank NUS Environmental Research Institute, National University of Singapore, Singapore, for providing the facility to do the work and Dr. Pallavi Panda and Miss Puravi Panda for proofreading the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.639654/full#supplementary-material
Abbreviations
ABA, Abscisic Acid; ABF1/2, ABA-responsive element binding factor 1/2; ABI3/4/5, Abscisic Acid-Insensitive 3/4/5; ABR1, AP2-like ABA repressor 1; ABRE, Abscisic Acid Response Element; AGL, Agamous-like; AP2/B3, APETALA2/B3; AP2/ERF, APETALA2/Ethylene-Responsive Element; ARR-B-Type-B Arabidopsis Response Regulator, ARF, Auxin Response Factor; ATHB4 Arabidopsis thaliana Homeobox 2; AUX1, Auxin influx carrier Auxin Resistant 1; BAM 8, BZR1-BAM 8; BAM1, β-amylase (BAM)–like 2; BEE, BR Enhanced Expression; BEH4, BES1/BZR1 Homolog 4; BES1, BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR 1; bHLH, basic Helix-Loop-Helix; BIM, BES1-Interacting MYC-Like; BIN2, Brassinosteroid Insensitive 2; BPC, Basic Penta Cysteine; BR, Brassinosteroid; bZIP, Basic Leucine Zipper; BZR1/2, Brassinazole-Resistant 1/2; CArG-box, ‘C-A rich-G-box; CAMTA, Calmodulin binding transcription factor; CBF, C-repeat Binding Factor; CCA1, circadian clock associated 1; C2H2: CCHH Zinc Finger; C3H, CCCH Zinc Finger; CCCH CPRF 5/6/7, Common Plant Regulatory Factor 5/6/7; cpm1, coleoptile photomorphogenesis 1; DOF, DNA-Binding with One Finger; DPBF-1/2, Dc3 Promoter Binding Factor 1/2; DREB, Dehydration-Responsive Element-Binding; E2F, E2 factor; EIL, Ethylene-Insensitive 3 (EIN3)-like protein3 (EIL3); EIN, Ethylene-Insensitive 3; ERF, Ethylene Response Factor; GA, Gibberellic Acid; GARE, Gibberellic Acid Response Element; GBF, G-Box Binding Factor; HD, ZIP-Homeodomain Leucine Zipper; HSF, Heat Shock Factor; HY5, Elongated Hypocotyl 5; IAA, Indole − 3 − Acetic Acid; JA, Jasmonate; JA, Jasmonic Acid; JARE, Jasmonic acid Response Element; JAZ, Jasmonate ZIM domain; LBD, Asymmetric Leaves 2 (ASL2)/Lateral organ boundaries domain; MADS-box, Minichromosome Maintenance Factor 1 (MCM1 from Saccharomyces cerevisiae), Agamous (AG; from Arabidopsis thaliana), Deficiens (DEF: from Antirrhinum majus), and Serum Response Factor (SRF; from Homo sapiens); MYB, Myeloblastosis; NAC, NAM, ATAF/, 2, and CUC2; NAM, No apical meristem; PIF1/3/4/7, Phytochrome Interacting Factor 1/3/4/7; RAP, Related to AP2; RAV, Related to ABI3/VP1; RNA, seq-RNA sequencing; ROS, Reactive oxygen species; SUB1, Submergence 1; SK1, SNORKEL1; SK2, SNORKEL2; TCP, TEOSINTE BRANCHED 1; TFs, Transcription Factors; TSS, Transcription Start Site; ZCT 1, 2, 3, Zinc Finger Catharanthus Transcription Factor 1, 2, and 3; ZnF, Zinc Finger.
Footnotes
- ^ www.gene-regulation.com
- ^ http://www.dna.affrc.go.jp/htdocs/PLACE/
- ^ http://arabidopsis.med.ohio-state.edu/
- ^ http://PlantPAN.itps.ncku.edu.tw
- ^ http://rapdb.dna.affrc.go.jp/
References
Abdulmajid, D., Ali, N., Eltahawy, M. S., Liu, E., Dang, X., and Mining, D. H. (2020). Mining favorable alleles for rice coleoptile elongation length sensitivity to exogenous gibberellin under submergence condition. J. Plant Growth Regul. doi: 10.1007/s00344-020-10196-z
Ahammed, G. J., Li, X., Liu, A., and Chen, S. (2020). Brassinosteroids in plant tolerance to abiotic stress. J. Plant Growth Regul. 39, 1451–1464. doi: 10.1007/s00344-020-10098-0
An, F., Zhao, Q., Ji, Y., Li, W., Jiang, Z., Yu, X., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22, 2384–2401. doi: 10.1105/tpc.110.076588
Annapurna, B., Khurana, J. P., and Mukesh, J. (2016). Characterization of rice homeobox genes, OsHOX22 and OsHOX24, and over-expression of OsHOX24 in transgenic Arabidopsis suggest their role in abiotic stress response. Front. Plant Sci. 7:627. doi: 10.3389/fpls.2016.00627
Argyros, R. D., Mathews, D. E., Chiang, Y. H., Palmer, C. M., Thibault, D. M., Etheridge, N., et al. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20, 2102–2116. doi: 10.1105/tpc.108.059584
Bailey-Serres, J., Fukao, T., Gibbs, D. J., Holdsworth, M. J., Lee, S. C., Licausi, F. et al. (2012). Making sense of low oxygen sensing. Trends Plant Sci. 17, 129–138. doi: 10.1016/j.tplants.2011.12.004
Baltazar, M. D., Ignacio, J. C. I., Thomson, M. J., Ismail, A. M., Mendioro, M. S., and Septiningsih, E. M. (2014). QTL mapping for tolerance of anaerobic germination from IR64 and the aus landrace Nanhi using SNP genotyping. Euphytica 197, 251–260. doi: 10.1007/s10681-014-1064-x
Bassel, G. W., Lan, H., Glaab, E., Gibbs, D. J., Gerjets, T., Krasnogor, N., et al. (2011). Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc. Natl. Acad. Sci. U.S.A. 108, 9709–9714. doi: 10.1073/pnas.1100958108
Bäumler, J., Riber, W., Klecker, M., Müller, L., Dissmeyer, N., Weig, A. R., et al. (2019). AtERF#111/ABR1 is a transcriptional activator involved in the wounding response. Plant J. 100, 969–990.
Birkenbihl, R. P., Kracher, B., and Somssich, I. E. (2017). Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 29, 20–38. doi: 10.1105/tpc.16.00681
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Breviario, D., Giani, S., Di Vietri, P., and Coraggio, I. (1992). Auxin and growth regulation of rice coleoptile segments: molecular analysis. Plant Physiol. 98, 488–495. doi: 10.1104/pp.98.2.488
Bu, Q., Jiang, H., Li, C.-B., Zhai, Q., Zhang, J., Wu, X., et al. (2008). Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 18, 756–767. doi: 10.1038/cr.2008.53
Bui, L. T., Giuntoli, B., Kosmacz, M., Parlanti, S., and Licausi, F. (2015). Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 236, 37–43. doi: 10.1016/j.plantsci.2015.03.008
Campbell, M. T., Proctor, C. A., Dou, Y., Schmitz, A. J., Phansak, P., Kruger, G. R., et al. (2016). Genetic and molecular characterization of submergence response identifies Subtol6 as a major submergence tolerance locus in maize. PLoS One 10:e0120385. doi: 10.1371/journal.pone.0120385
Capella, M., Ribone, P. A., Arce, A. L., and Chan, R. L. (2015). Arabidopsis thaliana HomeoBox 1 (AtHB1), a homedomain-leucine zipper I (HD-Zip I) transcription factor, is regulated by phytochrome-interacting factor 1 to promote hypocotyl elongation. New Phytol. 207, 669–682. doi: 10.1111/nph.13401
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. doi: 10.1146/annurev-arplant-050312-120221
Chang, K. N., Zhong, S., Weirauch, M. T., Hon, G., Pelizzola, M., Li, H., et al. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2:e00675.
Chen, H., Zhang, J., Neff, M. M., Hong, S. W., Zhang, H., et al. (2008). Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. U.S.A. 105, 4495–4500. doi: 10.1073/pnas.0710778105
Chen, L., Zhang, L., Li, D., Wang, F., and Yu, D. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 110, E1963–E1971.
Chen, X., Wang, Y., Lv, B., Li, J., Luo, L., Lu, S., et al. (2014). The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 55, 604–619. doi: 10.1093/pcp/pct204
Chen, Y. S., Ho, T. H. D., Liu, L., Lee, D. H., Lee, C. H., Chen, Y. R., et al. (2019). Sugar starvation-regulated MYBS2 and 14-3-3 protein interactions enhance plant growth, stress tolerance, and grain weight in rice. Proc. Natl. Acad. Sci. U.S.A. 116, 21925–21935. doi: 10.1073/pnas.1904818116
Chiang, G. C., Barua, D., Kramer, E. M., Amasino, R. M., and Donohue, K. (2009). Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 106, 11661–11666. doi: 10.1073/pnas.0901367106
Chow, C. N., Lee, T. Y., Hung, Y. C., Li, G. Z., Tseng, K. C., Liu, Y. H., et al. (2019). PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 8, D1155–D1163.
Danisman, S. (2016). TCP transcription factors at the interface between environmental challenges and the plant’s growth responses. Front. Plant Sci. 7:1930. doi: 10.3389/fpls.2016.01930
Dar, N. A., Amin, I., Wani, W., Wani, S. A., Shikari, A. B., Wani, S. H., et al. (2017). Abscisic acid: a key regulator of abiotic stress tolerance in plants. Plant Gene 11, 106–111. doi: 10.1016/j.plgene.2017.07.003
Davuluri, R. V., Sun, H., Palaniswamy, S. K., Matthews, N., Molina, C., Kurtz, M., et al. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinform. 4:25. doi: 10.1186/1471-2105-4-25
de Vetten, N. C., and Ferl, R. J. (1995). Characterization of a maize G-box binding factor that is induced by hypoxia. Plant J. 7, 589–601. doi: 10.1046/j.1365-313x.1995.7040589.x
Dolferus, R., Klok, E. J., Delessert, C., Wilson, S., Ismond, K. P., Good, A. G., et al. (2003). Enhancing the anaerobic response. Ann. Bot. 91, 111–117. doi: 10.1093/aob/mcf048
Dolgikh, V. A., Pukhovaya, E. M., and Zemlyanskaya, E. V. (2019). Shaping ethylene response: the role of EIN3/EIL1 transcription factors. Front. Plant Sci. 10:1030. doi: 10.3389/fpls.2019.01030
Duek, P. D., and Fankhauser, C. (2005). bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 10, 51–54. doi: 10.1016/j.tplants.2004.12.005
Feng, C. Z., Chen, Y., Wang, C., Kong, Y. H., Wu, W. H., and Chen, Y. F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80, 654–668.
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Friedrichsen, D. M., Nemhauser, J., Muramitsu, T., Maloof, J. N., Alonso, J., Ecker, J. R., et al. (2002). Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162, 1445–1456.
Frigerio, M., Alabadı, D., Perez-Gomez, J., Garcıa-Carcel, L., Phillips, A. L., Hedden, P., et al. (2006). Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142, 553–563. doi: 10.1104/pp.106.084871
Fukao, T., and Bailey-Serres, J. (2008a). Ethylene—a key regulator of submergence responses in rice. Plant Sci. 175, 43–51. doi: 10.1016/j.plantsci.2007.12.002
Fukao, T., and Bailey-Serres, J. (2008b). Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. U.S.A. 105, 16814–16819. doi: 10.1073/pnas.0807821105
Fukao, T., Xu, K., Ronald, P. C., and Bailey-Serres, J. (2006). A variable cluster of ethylene response factor–like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell Online 18, 2021–2034. doi: 10.1105/tpc.106.043000
Galon, Y., Aloni, R., Nachmias, D., Snir, O., Feldmesser, E., Scrase-Field, S., et al. (2010). Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 232, 165–178. doi: 10.1007/s00425-010-1153-6
Gibbs, D. J., Conde, J. V., Berckhan, S., Prasad, G., Mendiondo, G. M., and Holdsworth, M. J. (2015). Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 169, 23–31. doi: 10.1104/pp.15.00338
Gonzali, S., Loreti, E., Cardarelli, F., Novi, G., Parlanti, S., and Pucciariello, C. (2015). Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nat. Plants 1:15151.
Gubler, F., Chandler, P. M., White, R. G., Llewellyn, D. J., and Jacobsen, J. V. (2002). Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 129, 191–200. doi: 10.1104/pp.010918
Guo, H., Li, L., Aluru, M., Aluru, S., and Yin, Y. (2013). Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 16, 545–553. doi: 10.1016/j.pbi.2013.08.002
Guo, H. Q., Li, L., Ye, H. X., Yu, X. F., Algreen, A., and Yin, Y. H. (2009). Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl Acad. Sci. U.S.A. 106, 7648–7653. doi: 10.1073/pnas.0812346106
Haga, K., Takano, M., Neumann, R., and Iino, M. (2005). The rice COLEOPTILE PHOTOTROPISM 1 gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin. Plant Cell 17, 103–115. doi: 10.1105/tpc.104.028357
Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N., Qiu, N., et al. (2020). C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 11:115. doi: 10.3389/fpls.2020.00115
Hattori, Y., Nagai, K., Furukawa, S., Song, X.-J., Kawano, R., Sakakibara, H., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030. doi: 10.1038/nature08258
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hinz, M., Wilson, I. W., Yang, J., Buerstenbinder, K., Llewellyn, D., Dennis, E. S., et al. (2010). Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153, 757–772. doi: 10.1104/pp.110.155077
Hoson, T., Masuda, Y., and Pilet, P. E. (1992). Auxin content in air and water grown rice coleoptiles. J. Plant Physiol. 139, 685–689. doi: 10.1016/s0176-1617(11)81711-6
Hossain, M. A., Cho, J.-I. I., Han, M., Ahn, C. H., Jeon, J. S., An, G., et al. (2010). The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 167, 1512–1520. doi: 10.1016/j.jplph.2010.05.008
Hsu, F. C., Chou, M. Y., Chou, S. J., Li, Y. R., Peng, H. P., and Shih, M. C. (2013). Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell 25, 2699–2713. doi: 10.1105/tpc.113.114447
Hsu, S.-K., and Tung, C.-W. (2015). Genetic mapping of anaerobic germination associated QTLs controlling coleoptile elongation in rice. Rice 8:38.
Hsu, S.-K., and Tung, C.-W. (2017). RNA-Seq analysis of diverse rice genotypes to identify the genes controlling coleoptile growth during submerged germination. Front. Plant Sci. 8:762. doi: 10.3389/fpls.2017.00762
Huang, E., Yang, L., Chowdhary, R., Kassim, A., and Bajic, V. B. (2005). “An algorithm for ab-initio DNA motif detection,” in Information Processing and Living System. World Scientific, eds V. B. Bajic and T. W. Tan, (London: Imperial College Press), 611–614. doi: 10.1142/9781860946882_0004
Huang, Y., Feng, C. Z., Ye, Q., Wu, W. H., and Chen, Y. F. (2016). Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 12:e1005833. doi: 10.1371/journal.pgen.1005833
Ishizawa, K., and Esashi, Y. (1983). Cooperation of ethylene and auxin in the growth-regulation of rice coleoptile segments. J. Exp. Bot. 34, 74–82. doi: 10.1093/jxb/34.1.74
Ishizawa, K., and Esashi, Y. (1984). Osmoregulation in rice coleoptile elongation as promoted by cooperation between IAA and ethylene. Plant Cell Physiol. 25, 495–504.
Ishizawa, K., and Esashi, Y. (1988). Action mechanism of ethylene in the control of sugar translocation in relation to rice coleoptile growth. I. Sucrose metabolism. Plant Cell Physiol. 29, 1311–1341.
Itoh, T., Tanaka, T., Barrero, R. A., Yamasaki, C., Fujii, Y., Hilton, P. B., et al. (2007). Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 17, 175–183.
Jin, Y. M., Piao, R., Yan, Y. F., Chen, M., Wang, L., He, H., et al. (2018). Overexpression of a new zinc finger protein transcription factors OsCTZFP8 improves cold tolerance in rice. Int. J. Genomics. 2018, 1–13. doi: 10.1155/2018/5480617
Jing, Y., Zhang, D., Wang, X., Tang, W., Wang, W., Huai, J., et al. (2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25, 242–256. doi: 10.1105/tpc.112.105742
Kaneko, M., Itoh, H., Ueguchi, M., Ashikari, M., and Matsuoka, M. (2002). The α-Amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 128, 1264–1270. doi: 10.1104/pp.010785
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansa, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Kieffer, M., Master, V., Waites, R., and Davies, B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158. doi: 10.1111/j.1365-313x.2011.04674.x
Kim, S., Kang, J. Y., Cho, D. I., Park, J. H., and Kim, S. Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40, 75–87. doi: 10.1111/j.1365-313x.2004.02192.x
Kosmacz, M., Parlanti, S., Schwarzländer, M., Kragler, F., Licausi, F., and Van Dongen, J. T. (2015). The stability and nuclear localization of the transcription factor RAP2.12 are dynamically regulated by oxygen concentration. Plant Cell Environ. 38, 1094–1103. doi: 10.1111/pce.12493
Kosugi, S., and Ohashi, Y. (2002). E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 29, 45–59. doi: 10.1046/j.1365-313x.2002.01196.x
Kretzschmar, T., Pelayo, M. A., Trijatmiko, K. R., Gabunada, L. F., Alam, R., Jimenez, R., et al. (2015). A trehalose-6-phosphate phosphatase enhances anaerobic germination tolerance in rice. Nat. Plants 24:15124.
Lakshmanan, M., Mohanty, B., Lim, S.-H., Ha, S.-H., and Lee, D.-Y. (2014). Metabolic and transcriptional regulatory mechanisms underlying the anoxic adaptation of rice coleoptile. AoB PLANTS 6:Plu026.
Lasanthi-Kudahettige, R., Magneschi, L., Loreti, E., Gonzali, S., Licausi, F., Novi, G., et al. (2007). Transcript profiling of the anoxic rice coleoptile. Plant Physiol. 144, 218–231. doi: 10.1104/pp.106.093997
Lee, K. W., Chen, P. W., Lu, C. A., Chen, S., Ho, T. H., and Yu, S. M. (2009). Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2:ra61. doi: 10.1126/scisignal.2000333
Lee, K. W., Chen, P. W., and Yu, S. M. (2014). Metabolic adaptation to sugar/O2 deficiency for anaerobic germination and seedling growth in rice. Plant Cell Environ. 37, 2234–2244.
Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., et al. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823. doi: 10.1016/j.cub.2008.10.058
Leivar, P., and Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Li, C., Potuschak, T., Colón-Carmona, A., Gutiérrez, R. A., and Doerner, P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl Acad. Sci. U.S.A. 102, 12978–12983. doi: 10.1073/pnas.0504039102
Li, L., Yu, X., Thompson, A., Guo, M., Yoshida, S., Asami, T., et al. (2009). Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58, 275–286. doi: 10.1111/j.1365-313x.2008.03778.x
Li, Q. F., Xiong, M., Xu, P., Huang, L. C., Zhang, C. Q., and Liu, Q. Q. (2016). Dissection of brassinosteroid-regulated proteins in rice embryos during germination by quantitative proteomics. Sci. Rep. 6:34583. doi: 10.1038/srep34583
Liberek, K., Lewandowska, A., and Zietkiewicz, S. (2008). Chaperones in control of protein disaggregation. Embo J. 27, 328–335. doi: 10.1038/sj.emboj.7601970
Licausi, F., vanDongen, J. T., Giuntoli, B., Novi, G., Santaniello, A., Geigenberger, P., et al. (2010). HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 62, 302–315. doi: 10.1111/j.1365-313x.2010.04149.x
Lin, C. C., Chao, Y. T., Chen, W. C., Ho, H. Y., Chou, M. Y., Li, Y. R., et al. (2019). Regulatory cascade involving transcriptional and N-end rule pathways in rice under submergence. Proc. Natl Acad. Sci. U.S.A. 116, 3300–3309. doi: 10.1073/pnas.1818507116
Lopez-Molina, L., Mongrand, S., Mclachlin, D. T., Chait, B. T., and Chua, N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. doi: 10.1046/j.1365-313x.2002.01430.x
Loreti, E., Poggi, A., Novi, G., Alpi, A., and Perata, P. (2005). A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137, 1130–1138. doi: 10.1104/pp.104.057299
Ma, B., Chen, S. Y., and Zhang, J. S. (2010). Ethylene signaling in rice. Chin. Sci. Bull. 55, 2204–2210. doi: 10.1007/s11434-010-3192-2
Ma, B., He, S. J., Duan, K. X., Yin, C. C., Chen, H., Yang, C., et al. (2013). Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant 6, 1830–1848. doi: 10.1093/mp/sst087
Ma, B., Yin, C. C., He, S. J., Lu, X., Zhang, W. K., Lu, T. G., et al. (2014). Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings. PLoS Genet. 10:e1004701. doi: 10.1371/journal.pgen.1004701
Ma, J., Hanssen, M., Lundgren, K., Hernández, L., Delatte, T., Ehlert, A., et al. (2011). The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol. 191, 733–745. doi: 10.1111/j.1469-8137.2011.03735.x
Marín-de la Rosa, N., Pfeiffer, A., Hill, K., Locascio, A., Bhalerao, R. P., Miskolczi, P., et al. (2015). Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. PLoS Genet. 11:e1005337. doi: 10.1371/journal.pgen.1005337
Martínez, C., Espinosa-Ruíz, A., de Lucas, M., Bernardo-García, S., Franco-Zorrilla, J. M., and Prat, S. (2018). PIF4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. Embo J. 37:e99552.
Matys, V., Fricke, E., Geffers, R., Gossling, E., Haubrock, M., Hehl, R., et al. (2003). TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31, 74–378.
Mazzella, M. A., Casal, J. J., Muschietti, J. P., and Fox, A. R. (2014). Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 5:52. doi: 10.3389/fpls.2014.00052
Mishra, M., Kanwar, P., Singh, A., Pandey, A., Kapoor, S., and Pandey, G. K. (2013). Plant omics: genome-wide analysis of ABA Repressor1 (ABR1) related genes in rice during abiotic stress and development. Omics J. Int. Biol. 17, 439–450. doi: 10.1089/omi.2012.0074
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Mohanty, B., Hearth, V., Wijaya, E., Reyes, B. D., and Lee, D. Y. (2012). Patterns of cis-element enrichment reveal potential regulatory modules in the transcriptional regulation of anoxia response of japonica rice. Gene 511, 235–242. doi: 10.1016/j.gene.2012.09.048
Mohanty, B., Takahashi, H., de los Reyes, B. G., Wijaya, E., Nakazono, E., and Lee, D.-Y. (2016). Transcriptional regulatory mechanism of alcohol dehydrogenase 1-deficient mutant of rice for cell survival under complete submergence. Rice 9:51.
Mu, Y., Zou, M., Sun, X., He, B., Xu, X., Liu, Y., et al. (2017). BASIC PENTACYSTEINE proteins repress ABSCISIC ACID INSENSITIVE4 expression via direct recruitment of the Polycomb-Repressive Complex 2 in Arabidopsis root development. Plant Cell Physiol. 58, 607–621.
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103.
Narsai, R., Edwards, J. M., Roberts, T. H., Whelan, J., Joss, G. H., and Atwell, B. J. (2015). Mechanisms of growth and patterns of gene expression in oxygen deprived rice coleoptiles. Plant J. 82, 25–40. doi: 10.1111/tpj.12786
Narsai, R., Howell, K. A., Carroll, A., Ivanova, A., Millar, A. H., and Whelan, J. (2009). Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol. 151, 306–322. doi: 10.1104/pp.109.142026
Nemhauser, J. L., Hong, F., and Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475. doi: 10.1016/j.cell.2006.05.050
Neumann, R., and Iino, M. (1997). Phototropism of rice (Oryza sativa L.) coleoptiles: fluence-response relationships, kinetics and photogravitropic equilibrium. Planta 201, 288–292. doi: 10.1007/s004250050068
Nghi, K. N., Tagliani, A., Mariotti, L., Weits, D. A., Perata, P., and Pucciariello, C. (2020). Auxin is required for the long coleoptile trait in japonica rice under submergence. New Phytol. 229, 85–93. doi: 10.1111/nph.16781
Nishizawa, A., Yabuta, Y., and Shigeoka, S. (2008). Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 147, 1251–1263. doi: 10.1104/pp.108.122465
Noguero, M., Atif, R. M., Ochatt, S., and Thompson, R. D. (2013). The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
Oh, E., Zhu, J. Y., Bai, M. Y., Arenhart, R. A., Sun, Y., and Wang, Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3:e03031.
Oh, E., Zhu, J. Y., and Wang, Z. Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14, 802–809. doi: 10.1038/ncb2545
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., and Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130. doi: 10.1105/tpc.106.047761
Paik, I., Kathare, P. K., Kim, J. I., and Huq, E. (2017). Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10, 1035–1046. doi: 10.1016/j.molp.2017.07.002
Pan, J., Hu, Y., Wang, H., Guo, Q., Chen, Y., Howe, G. A., et al. (2020). Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 32, 3846–3865. doi: 10.1105/tpc.19.00838
Pandey, D. M., and Kim, S. R. (2012). Identification and expression analysis of hypoxia stress inducible CCCH-type zinc finger protein genes in rice. J. Plant Biol. 55, 489–497. doi: 10.1007/s12374-012-0384-4
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li, L., and Luan, S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139, 1185–1193. doi: 10.1104/pp.105.066324
Papdi, C., Perez-Salamo, I., Joseph, M. P., Giuntoli, B., Bogre, L., Koncz, C., et al. (2015). The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12. RAP2.2 and RAP2.3. Plant J. 82, 772–784. doi: 10.1111/tpj.12848
Penfield, S., Josse, E. M., Kannangara, R., Gilday, A. D., Halliday, K. J., and Graham, I. A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15, 1998–2006. doi: 10.1016/j.cub.2005.11.010
Perata, P., Guglielminetti, L., and Alpi, A. (1997). Mobilization of endosperm reserves in cereal seeds under anoxia. Ann. Bot. 79, 49–56. doi: 10.1093/oxfordjournals.aob.a010306
Perrot-Rechenmann, C. (2010). Cellular responses to auxin: division versus expansion. Cold Spring Harb. Perspect. Biol. 2:a001446. doi: 10.1101/cshperspect.a001446
Pfeiffer, A., Shi, H., Tepperman, J. M., Zhang, Y., and Quail, P. H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7, 1598–1618. doi: 10.1093/mp/ssu087
Phukan, U. J., Jeena, G. S., and Shukla, R. K. W. R. K. Y. (2016). Transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Pjon, C. J., and Furuya, M. (1974). Phytochrome action in Oryza sativa L. VII. Effects of light and aeration on coleoptile growth under submerged conditions. Plant Cell Physiol. 15, 663–668. doi: 10.1093/oxfordjournals.pcp.a075051
Qin, H., Zhang, Z., Wang, J., Chen, X., Wei, P., and Huang, R. (2017). The activation of OsEIL1 on YUC8 transcription and auxin biosynthesis is required for ethylene-inhibited root elongation in rice early seedling development. PLoS Genet. 13:e1006955. doi: 10.1371/journal.pgen.1006955
Raineri, J., Ribichich, K. F., and Chan, R. L. (2015). The sunflower transcription factor HaWRKY76 confers drought and flood tolerance to Arabidopsis thaliana plants without yield penalty. Plant Cell Rep. 34, 2065–2080. doi: 10.1007/s00299-015-1852-3
Resentini, F., Felipo-Benavent, A., Colombo, L., Blázquez, M. A., Alabadí, D., and Masiero, S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8, 482–485. doi: 10.1016/j.molp.2014.11.018
Safavi-Rizi, V., Herde, M., and Stöhr, C. (2020). RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root. Sci. Rep. 10:1692.
Sakuraba, Y., Jeong, J., Kang, M. Y., Kim, J., Paek, N. C., and Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636.
Schmitz, A. J., Folsom, J. J., Jikamaru, Y., Ronald, P., and Walia, H. (2013). SUB1A-mediated submergence tolerance response in rice involves differential regulation of the brassinosteroid pathway. New Phytol. 198, 1060–1070.
Septiningsih, E. M., Ignacio, J. C., Sendon, P. M., Sanchez, D. L., Ismail, A. M., and Mackill, D. J. (2013). QTL mapping and confirmation for tolerance of anaerobic conditions during germination derived from the rice landrace Ma-Zhan Red. Theor. Appl. Genet. 126, 1357–1366.
Serna, L. (2007). bHLH proteins know when to make a stoma. Trends Plant Sci. 12, 483–485. doi: 10.1016/j.tplants.2007.08.016
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell, 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shao, H. B., Wang, H. Y., and Tang, X. L. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6:902. doi: 10.3389/fpls.2015.00902
Sharma, I., Kaur, N., and Pati, P. K. (2017). Brassinosteroids: a promising option in deciphering remedial strategies for abiotic stress tolerance in rice. Front. Plant Sci. 8:2151. doi: 10.3389/fpls.2017.02151
Sharma, N., Dang, T. M., Singh, N., Ruzicic, S., Mueller-Roeber, B., Baumann, U., et al. (2018). Allelic variants of OsSUB1A cause differential expression of transcription factor genes in response to submergence in rice. Rice 11:2.
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578–2595. doi: 10.1105/tpc.112.098640
Shingaki-Wells, R. N., Huang, S., Taylor, N. L., Carroll, A. J., Zhou, W., and Millar, A. H. (2011). Differential molecular responses of rice and wheat coleoptiles to anoxia reveal novel metabolic adaptations in amino acid metabolism for tissue tolerance. Plant Physiol. 156, 1706–1724. doi: 10.1104/pp.111.175570
Shiono, K., Yamauchi, T., Yamazaki, S., Mohanty, B., Malik, A. I., Nagamura, Y., et al. (2014). Microarray analysis of laser-microdissected tissues indicates the biosynthesis of suberin in the outer part of roots during formation of a barrier to radial oxygen loss in rice (Oryza sativa). J. Exp. Bot. 65, 4795–4806. doi: 10.1093/jxb/eru235
Smaczniak, C., Immink, R. G. H., Angenent, G. C., and Kaufmann, K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098. doi: 10.1242/dev.074674
Song, C. P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Sorin, C., Salla-Martret, M., Bou-Torrent, J., Roig-Villanova, I., and Martínez-García, J. F. (2009). ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 59, 266–277. doi: 10.1111/j.1365-313X.2009.03866.x
Soyk, S., Simková, K., Zürcher, E., Luginbühl, L., Brand, L. H., Vaughan, K. C., et al. (2014). The enzyme-like domain of arabidopsis nuclear b-amylases is critical for DNA sequence recognition and transcriptional activation. Plant Cell 26, 1746–1763. doi: 10.1105/tpc.114.123703
Sreenivasulu, N., Usadel, B., Winter, A., Radchuk, V., Scholz, U., Stein, N., et al. (2008). Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol. 146, 1738–1758. doi: 10.1104/pp.107.111781
Steber, C. M., and McCourt, P. (2001). A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 125, 763–769. doi: 10.1104/pp.125.2.763
Steffens, B., Kovalev, A., Gorb, S. N., and Sauter, M. (2012). Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. Plant Cell 24, 3296–3306. doi: 10.1105/tpc.112.101790
Sun, X., Wang, Y., and Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Bioph. Res. Co. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123
Tanaka, T., Antonio, B. A., Kikuchi, S., Matsumoto, T., Nagamura, Y., Numa, H., et al. (2008). The rice annotation project database (RAP-DB). Nucleic Acids Res. 36, D1028–D1033. doi: 10.1093/nar/gkm978
Tang, H., Bi, H., Liu, B., Lou, S., Song, Y., Tong, S., et al. (2021). WRKY33 interacts with WRKY12 protein to up-regulate RAP2.2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol. 229, 106–125. doi: 10.1111/nph.17020
Tang, N., Ma, S., Zong, W., Yang, N., Lv, Y., Yan, C., et al. (2016). MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28, 2161–2177. doi: 10.1105/tpc.16.00171
Tang, N., Zhang, H., Li, X., Xiao, J., and Xiong, L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. doi: 10.1104/pp.111.190389
Tang, Y., Wang, J., Bao, X., Liang, M., Lou, H., Zhao, J., et al. (2019). Genome-wide identification and expression profile of HD-ZIP genes in physic nut and functional analysis of the JcHDZ16 gene in transgenic rice. BMC Plant Biol. 19:298. doi: 10.1186/s12870-019-1920-x
Tatematsu, K., Nakabayashi, K., Kamiya, Y., and Nambara, E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52. doi: 10.1111/j.1365-313X.2007.03308.x
Tong, H., Xiao, Y., Liu, D., Gao, S., Liu, L., Yin, Y., et al. (2014). Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant cell 26, 4376–4393. doi: 10.1105/tpc.114.132092
Tong, H. N., and Chu, C. C. (2012). Brassinosteroid signaling and application in rice. J. Genet. Genomics 39, 3–9. doi: 10.1016/j.jgg.2011.12.001
Tsai, K. J., Lin, C. Y., Ting, C. Y., and Shih, M. C. (2014). Ethylene plays an essential role in the recovery of Arabidopsis during post-anaerobiosis reoxygenation. Plant Cell Environ. 37, 2391–2405. doi: 10.1111/pce.12292
van Veen, H., Vashisht, D., Akman, M., Girke, T., Mustroph, A., Reinen, E., et al. (2016). Transcriptomes of eight Arabidopsis thaliana accessions reveal core conserved, genotype- and organ-specific responses to flooding stress. Plant Physiol. 172, 668–689. doi: 10.1104/pp.16.00472
Viana, V. E., Marini, N., Busanello, C., Pegoraro, C., Fernando, J. A., Da Maia, L. C., et al. (2018). Regulation of rice responses to submergence by WRKY transcription factors. Biol. Plant 62, 551–560. doi: 10.1007/s10535-018-0806-3
Waller, F., Furuya, M., and Nick, P. (2002). OsARF1, an auxin response factor from rice, is auxin-regulated and classifies as a primary auxin responsive gene. Plant Mol. Biol. 50, 415–425. doi: 10.1023/A:1019818110761
Wang, C., Ding, Y., Yao, J., Zhang, Y., Sun, Y., Colee, J., et al. (2015). Arabidopsis Elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 83, 1019–1033. doi: 10.1111/tpj.12946
Wang, K., Ding, Y., Cai, C., Chen, Z., and Zhu, C. (2018). The role of C2H2 zinc finger proteins in plant responses to abiotic stresses. Physiol. Plan. 165, 690–700. doi: 10.1111/ppl.12728
Wang, Z. Y., Bai, M. Y., Oh, E., and Zhu, J. Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724. doi: 10.1146/annurev-genet-102209-163450
Washio, K. (2003). Functional dissections between GAMYB and DOF transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 133, 850–863. doi: 10.1104/pp.103.027334
Weiste, C., and Dröge-Laser, W. (2014). The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 5:3883. doi: 10.1038/ncomms4883
Woodger, F. J., Gubler, F., Pogson, B., and Jacobsen, J. V. (2003). The role of GAMYB transcription factors in GA-regulated gene expression. J. Plant Growth Regul. 22, 176–184. doi: 10.1007/s00344-003-0025-8
Wu, Q., Li, D., Li, D., Liu, X., Zhao, X., Li, X., et al. (2015). Overexpression of OsDof12 affects plant architecture in rice (Oryza sativa L.). Front. Plant Sci. 6:833. doi: 10.3389/fpls.2015.00833
Wu, Y. S., and Yang, C. Y. (2020). Comprehensive transcriptomic analysis of auxin responses in submerged rice coleoptile growth. Int. J. Mol. Sci. 21:1292. doi: 10.3390/ijms21041292
Xi, W., Liu, C., Hou, X., and Yu, H. (2010). MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22, 1733–1748. doi: 10.1105/tpc.109.073072
Xi, W., and Yu, H. (2010). MOTHEROFFTANDTFL1 regulates seed germination and fertility relevant to the brassinosteroid signaling pathway. Plant Signal. Behav. 5, 1315–1317. doi: 10.4161/psb.5.10.13161
Xiong, Q., Ma, B., Lu, X., Huang, Y.-H., He, S.-J., Yang, C., et al. (2017). Ethylene-inhibited jasmonic acid biosynthesis promotes mesocotyl/coleoptile elongation of etiolated rice seedlings. Plant Cell 29, 1053–1072. doi: 10.1105/tpc.16.00981
Xu, K., Xu, X., Fukao, T., Canlas, P., Maghirang-Rodriguez, R., Heuer, S., et al. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708. doi: 10.1038/nature04920
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1606. doi: 10.1105/tpc.12.9.1591
Yang, C., Lu, X., Ma, B., Chen, S. Y., and Zhang, J. S. (2015). Ethylene signaling in rice and Arabidopsis: conserved and diverged aspects. Mol. Plant 4, 495–505. doi: 10.1016/j.molp.2015.01.003
Yang, Y., Li, J., Li, H., Yang, Y., Guang, Y., and Zhou, Y. (2019). The bZIP gene family in watermelon: Genome-wide identification and expression analysis under cold stress and root-knot nematode infection. Peer J. 7:e7878. doi: 10.7717/peerj.7878
Yao, Y., He, R. J., Xie, Q. L., Zhao, X. H., Deng, X. M., He, J. B., et al. (2017). ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 213, 1667–1681. doi: 10.1111/nph.14278
Yeung, E., van Veen, H., Vashisht, D., Paiva, A. L. S., Hummel, M., Rankenberg, T., et al. (2018). A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc. Natl Acad. Sci. U.S.A. 115, E6085–E6094. doi: 10.1101/276519
Yilmaz, A., Mejia-Guerra, M. K., Kurz, K., Liang, X., Welch, L., and Grotewold, E. (2011). AGRIS: the Arabidopsis gene regulatory information server, an update. Nucleic Acids Res. 39, D1118–D1122. doi: 10.1093/nar/gkq1120
Yong, Y., Zhang, Y., and Lyu, Y. (2019). A stress-responsive NAC transcription factor from tiger lily (LlNAC2) interacts with LlDREB1 and LlZHFD4 and enhances various abiotic stress tolerance in Arabidopsis. Int. J. Mol. Sci. 20:3225. doi: 10.3390/ijms20133225
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3are master transcription factors that cooperatively regulate ABRE dependent ABA signalling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Yoshii, M., Yamazaki, M., Rakwal, R., Kishi-Kaboshi, M., Miyao, A., and Hirochika, H. (2010). The NAC transcription factor RIM1 of rice is a new regulator of jasmonate signaling. Plant J. 61, 804–815. doi: 10.1111/j.1365-313X.2009.04107.x
Yu, L. H., Wu, J., Zhang, Z. S., Miao, Z. Q., Zhao, P. X., Wang, Z., et al. (2017). Arabidopsis MADS-Box transcription factor AGL21 acts as environmental surveillance of seed germination by regulating ABI5 expression. Mol. Plant. 10, 834–845. doi: 10.1016/j.molp.2017.04.004
Yu, X., Li, L., Zola, J., Aluru, M., Ye, H., Foudree, A., et al. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646. doi: 10.1111/j.1365-313X.2010.04449.x
Yu, Z., Chang, F., Lv, W., Sharmin, R. A., Wang, Z., Kong, J., et al. (2019). Identification of QTN and candidate gene for seed-flooding tolerance in soybean [Glycine max (L.) Merr.] using Genome-Wide Association Study (GWAS). Genes 10:957. doi: 10.3390/genes10120957
Yuan, X., Wang, H., Cai, J., Bi, Y., Li, D., and Song, F. (2019a). Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 19:278. doi: 10.1186/s12870-019-1883-y
Yuan, X., Wang, H., Cai, J., Li, D., and Song, F. (2019b). NAC transcription factors in plant immunity. Phytopathol. Res. 1:3. doi: 10.1186/s42483-018-0008-0
Zhang, C., Bai, M. Y., and Chang, K. (2014). Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 33, 683–696. doi: 10.1007/s00299-014-1578-7
Zhang, C., Li, C., Liu, J., Lv, Y., Yu, C., Li, H., et al. (2017). The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J Exp. Bot. 68, 4695–4707. doi: 10.1093/jxb/erx260
Zhang, L., Li, Z., Quan, R., Li, G., Wang, R., and Huang, R. (2011). An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 157, 854–865. doi: 10.1104/pp.111.179028
Zhang, L., Xia, C., Zhao, G., Liu, J., Jia, J., and Kong, X. (2015). A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol. Plant 153, 538–554. doi: 10.1111/ppl.12261
Zhang, Y., Mayba, O., Pfeiffer, A., Shi, H., Tepperman, J. M., Speed, T. P., et al. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9:e1003244. doi: 10.1371/journal.pgen.1003244
Zhao, Q., Fan, Z., Qiu, L., Che, Q., Wang, T., Li, Y., et al. (2020). MdbHLH130, an Apple bHLH transcription factor, confers water stress resistance by regulating stomatal closure and ROS homeostasis in transgenic tobacco. Front. Plant Sci. 11:543696. doi: 10.3389/fpls.2020.543696
Zhao, X., Dou, L., Gong, Z., Wang, X., and Mao, T. (2019). BES1 hinders ABSCISIC ACID INSENSITIVE5 and promotes seed germination in Arabidopsis. New Phytol. 221, 908–918. doi: 10.1111/nph.15437
Zhong, S., Shi, H., Xue, C., Wang, L., Xi, Y., Li, J., et al. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22, 1530–1535. doi: 10.1016/j.cub.2012.06.039
Zhu, Q. G., Gong, Z. Y., Huang, J., Grierson, D., Chen, K. S., and Yin, X. R. (2019). High-CO2/hypoxia-responsive transcription factors DkERF24 and DkWRKY1 interact and activate DkPDC2 promoter. Plant Physiol. 180, 621–633. doi: 10.1104/pp.18.01552
Zou, X., Neuman, D., and Shen, Q. J. (2008). Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiol. 148, 176–186. doi: 10.1104/pp.108.123653
Keywords: rice, germination and coleoptile elongation, submergence tolerance, promoter cis-element, transcription factor, phytohormones, plant breeding
Citation: Mohanty B (2021) Promoter Architecture and Transcriptional Regulation of Genes Upregulated in Germination and Coleoptile Elongation of Diverse Rice Genotypes Tolerant to Submergence. Front. Genet. 12:639654. doi: 10.3389/fgene.2021.639654
Received: 09 December 2020; Accepted: 08 February 2021;
Published: 16 March 2021.
Edited by:
Deborah A. Triant, University of Virginia, United StatesReviewed by:
Nisha Singh, Cornell University, United StatesIshaan Gupta, Indian Institute of Science Education and Research, India
Copyright © 2021 Mohanty. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bijayalaxmi Mohanty, ZXJpdjk3QG51cy5lZHUuc2c=; YmlqYXlhbGF4bWkubW9oYW50eUBnbWFpbC5jb20=
 Bijayalaxmi Mohanty
Bijayalaxmi Mohanty