- 1Pediatric Intensive Care Unit, Beijing Children’s Hospital, Capital Medical University, National Center for Children’s Health, Beijing, China
- 2Research Unit of Critical Infection in Children, Chinese Academy of Medical Sciences (2019RU016), Beijing, China
- 3Beijing Key Laboratory for Genetics of Birth Defects, Beijing Pediatric Research Institute, MOE Key Laboratory of Major Diseases in Children, Beijing Children’s Hospital, Capital Medical University, National Center for Children’s Health, Beijing, China
Objectives: Whole exome sequencing (WES) has been widely used to detect genetic disorders in critically ill children. Relevant data are lacking in pediatric intensive care units (PICUs) of China. This study aimed to investigate the spectrum of monogenic disorders, the diagnostic yield and clinical utility of WES from a PICU in a large children’s hospital of China.
Methods: From July 2017 to February 2020, WES was performed in 169 critically ill children with suspected monogenic diseases in the PICU of Beijing Children’s Hospital. The clinical features, human phenotype ontology (HPO) terms, and assessment of clinical impact were analyzed.
Results: The media age of the enrolled children was 10.5 months (range, 1 month to 14.8 years). After WES, a total of 43 patients (25%) were diagnosed with monogenic disorders. The most common categories of diseases were metabolic disease (33%), neuromuscular disease (19%), and multiple deformities (14%). The diagnosis yield of children with “metabolism/homeostasis disorder” and “growth delay” or “ocular anomalies” was higher than that of children without these features. In addition, the diagnosis rate increased when more features were observed in children. The results of WES had an impact on the treatment for 30 cases (70%): (1) change of treatment (n = 11), (2) disease monitoring initiation (n = 18), (3) other systemic evaluation (n = 3), (4) family intervention (n = 2), and (5) rehabilitation and redirection of care toward palliative care (n = 12).
Conclusion: WES can be used as an effective diagnostic tool in the PICU of China and has an important impact on the treatment of patients with suspected monogenic conditions.
Introduction
Congenital genetic disease is frequent in pediatric intensive care units (PICUs) (French et al., 2019). At present, almost 7,000 monogenic diseases have been listed on the OMIM website1, and this number is growing at a rate of approximately 30–50 per month. Children with monogenic diseases who are admitted to PICU are often in critical condition and accompanied by multiple organ dysfunction (Sanford et al., 2019). Considering the genetic and clinical heterogeneity of monogenic disease, critical physicians cannot provide a rapid and clear diagnosis and make a targeted treatment plan in time. The condition of the patient may continue to progress, increasing the risk of death. Cunniff et al. (1995) reported that the mortality rate of children admitted to the hospital due to birth defects and genetic diseases was 4.5 times higher than that of children admitted for other reasons. Therefore, clinicians must diagnose critically ill children with suspected monogenic diseases in time to provide them with timely interventions, improve prognosis, reduce mortality, and minimize the burden on families and the national health system.
Next-generation sequencing provides a new strategy for diagnosing genetic disorders. In recent years, an increasing number of studies have shown that whole exome sequencing (WES) has a unique value in the diagnosis of monogenic diseases. WES could be used as a first-tier test for children with suspected monogenic disorders (Stark et al., 2016; Hu et al., 2018). More than 20% of patients could be diagnosed using WES (Yang et al., 2013; Meng et al., 2017; French et al., 2019). Farnaes et al. (2018) reported that WES could reduce mortality and hospitalization costs. This may be due to the fact that after obtaining a definite diagnosis, the best clinical care can be provided for the affected children, promoting the transition from empirical treatment to definite treatment of identified diseases where feasible.
Previous studies of WES mainly included infants in neonatal intensive care units (NICUs) (Meng et al., 2017; Farnaes et al., 2018; Petrikin et al., 2018; Swaggart et al., 2019; Gubbels et al., 2020). However, studies about PICUs either had a relatively small number of cases or were reported jointly with NICUs (Mestek-Boukhibar et al., 2018; French et al., 2019; Sanford et al., 2019; Australian Genomics Health Alliance Acute Care et al., 2020; Wang et al., 2020a). The present study focused on the clinical application of WES in critically ill patients in the PICU. The diagnostic yield, the monogenic disease spectrum of patients, and effect of WES on clinical management were systematically analyzed for the first time in a PICU of China.
Materials and Methods
Study Design and Clinical Samples
We retrospectively analyzed the demographics and clinical characteristics of critically ill patients in the PICU who received WES, the diagnostic yield of WES, and the clinical management of diagnosed patients.
From July 2017 to February 2020, 3,484 patients were admitted to the PICU of Beijing Children’s Hospital. A total of 169 patients who met the inclusion criteria were enrolled in this study (Figure 1).
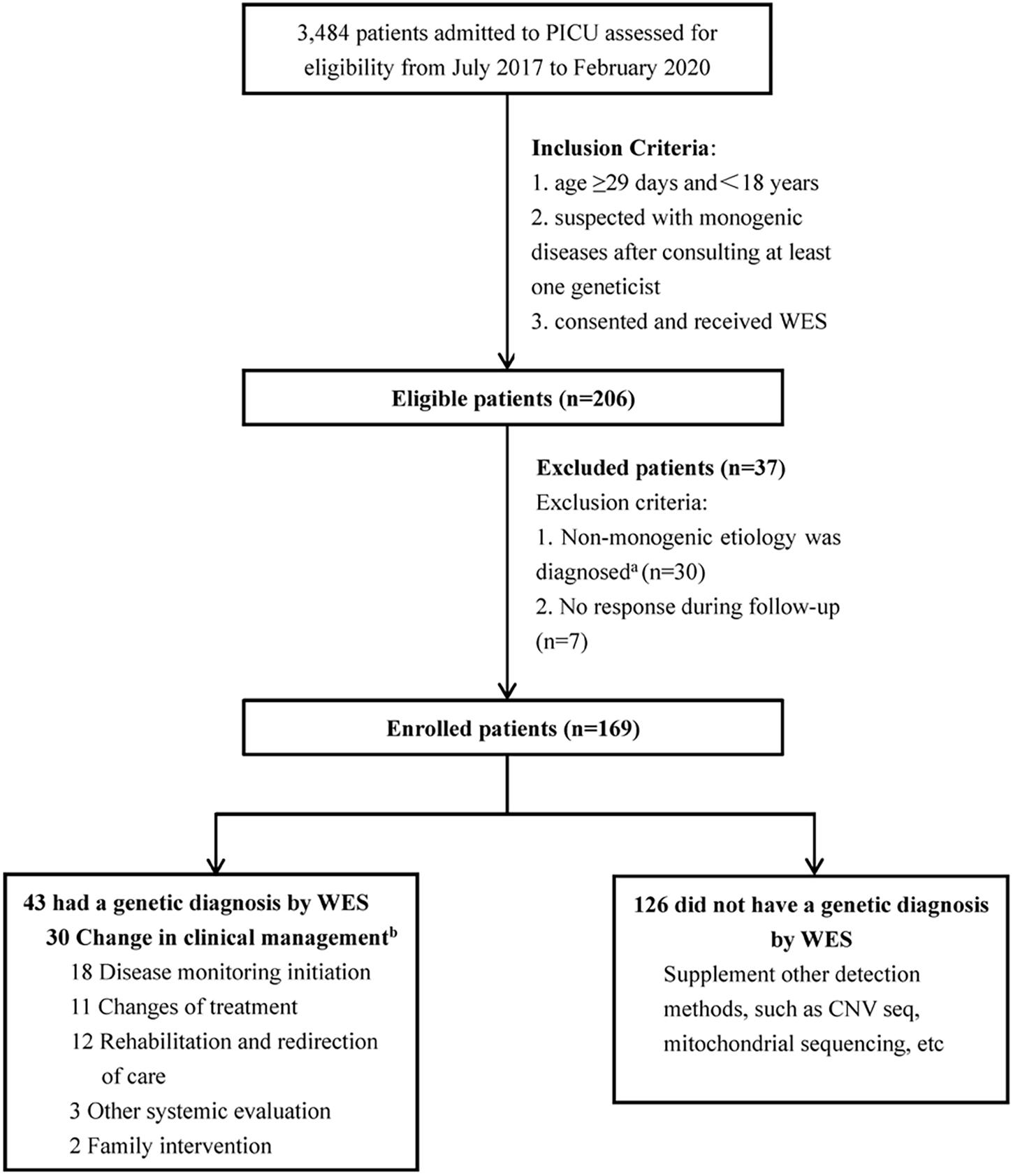
Figure 1. Flow diagram of the PICU patients who were enrolled, received genetic diagnosis by WES, and clinical management. aThe etiology of thirty patients was non-monogenic diseases, including: 19 acute necrotizing encephalopathy; 7 febrile infection-related epilepsy syndrome; 1 acute lymphoblastic leukemia; 1 poisoning; 1 heat stroke; 1 hemophagocytic syndrome. bSome patients had changed the clinical management more than 1 category.
The inclusion criteria were: (1) age ≥29 days and <18 years, (2) suspected with monogenic diseases after consulting at least one geneticist, and (3) consented and received WES (trio-WES or proband WES). The exclusion criteria were as follows: (1) diagnosed with non-monogenic etiology and (2) no response during follow-up. This study was approved by the institutional review board of Beijing Children’s Hospital (No. 2020-Z-098).
WES and Data Analysis
DNA was extracted from EDTA blood samples obtained from the probands and their parents (when available) using the Gentra Puregene Blood Kit (QIAGEN, Hilden, Germany). The SureSelect Human All Exon Kit V6 (Agilent Technologies, Santa Clara, United States) was used for whole exome capture. The target regions were sequenced on NovaSeq 6000 (Illumina, San Diego, United States). Raw reads were filtered using FastQC to remove low-quality reads. Clean reads were mapped to the reference genome GRCh37. Quality control information included the following: average read depth of >100×, accurate mapping rate of >98%, base capture rate of >55%, 20× mean depth coverage rate of >96%, and duplication rate of <25%. Single nucleotide variants were annotated and filtered by TGex2. Variants with a frequency over 1% in the gnomAD, ESP, or 1000G databases were excluded. Variants that lacked segregation in family members were also filtered. Variants were classified following the American College of Medical Genetics and Genomics and the Association for Molecular Pathology interpretation standards and guidelines (Richards et al., 2015). The genetic results were reviewed by a multidisciplinary team which included geneticists, pediatric intensive care physicians, neurologists as required, and compared with the phenotypes, related biochemical tests, or imaging examinations of the patients to make the final diagnosis (Supplementary Table 1).
Phenotype Analysis
The clinical features and the impact on patient prognosis were evaluated by reviewing electronic medical records. The clinical features were translated into human phenotype ontology (HPO) terms through the HPO website3. The top-level branching of HPO phenotypes was analyzed to ensure adequate counts of participants. The diagnostic rates of patients with and without HPO terms, the relationship between the phenotype complexity and expected diagnostic yield were analyzed.
Assessment of Clinical Impact
The changes in clinical management were determined from the medical records and follow-up by telephone until June 2020. These changes were grouped into five categories: (1) changes of treatment, (2) disease monitoring initiation, (3) other systemic evaluation, (4) family intervention, and (5) rehabilitation and redirection of care toward palliative care.
Statistical Analysis
All analyses were conducted in SPSS 22.0. Categorical variables were expressed as frequencies and percentages, continuous variables as median (interquartile range). Differences between groups were determined by Student t-test, Mann-Whitney U test, chi-square, if the expected counts were less than 5, Fisher’s exact test. p-values of less than 0.05 were considered to indicate statistical significance.
Results
General Information
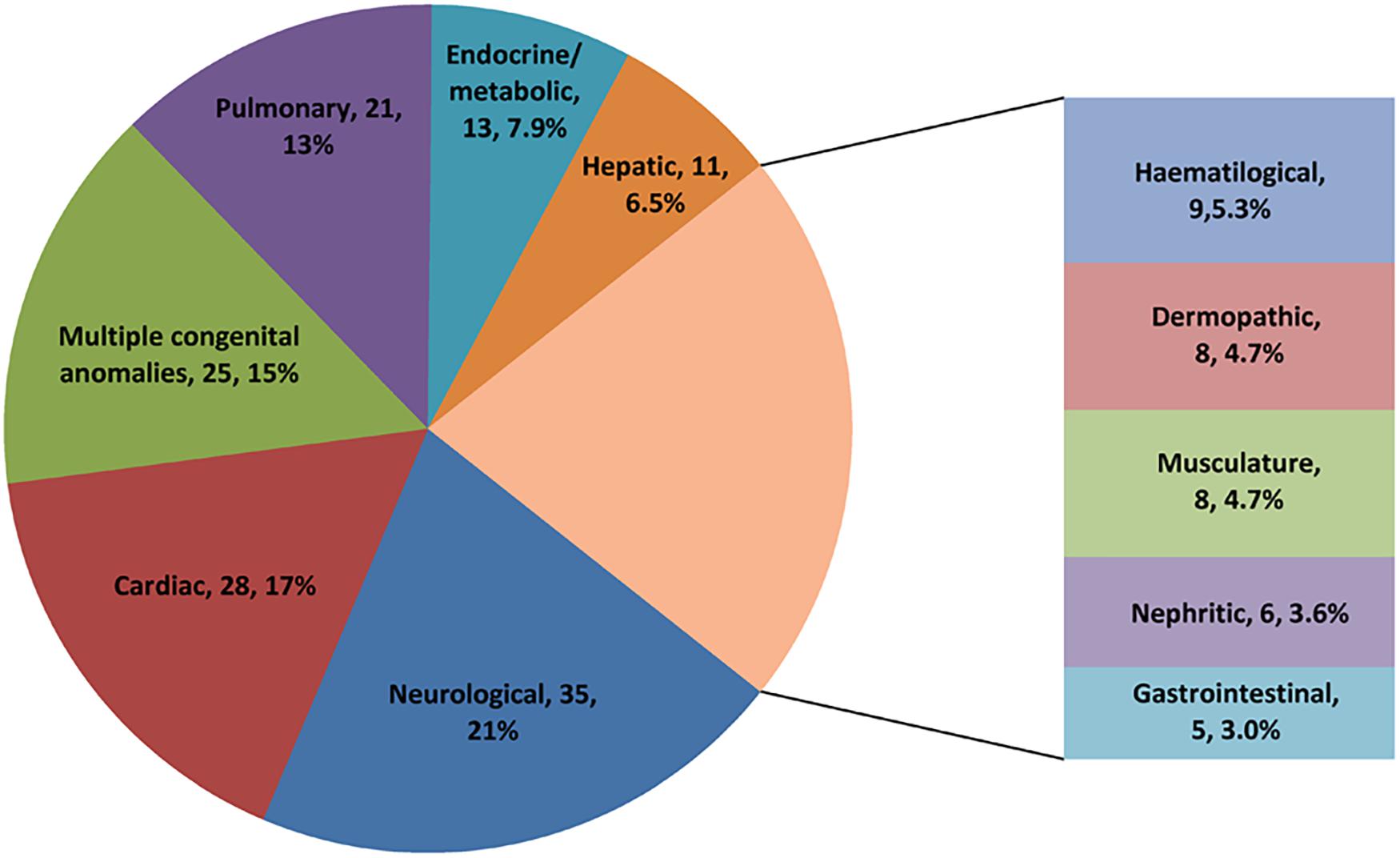
We enrolled for WES analysis 169 patients, 96 (57%) males, and 73 (43%) females. The ages of patients ranged from 1 month to 14.8 years, and the median age was 10.5 months (4 months, 47.5 months). The most common primary manifestation was neurological (21%), followed by cardiac (17%) and multiple congenital anomalies (15%) (Figure 2). The conditions of affected patients were severe with 82% of patients requiring mechanical ventilation and 62% of patients needing endotracheal intubation. In addition, more than 20% of patients received inotropic support or continuous renal replacement therapy. The mortality rate at discharge was 12%, and the mortality at 120 days after discharge was 32%.
Diagnostic Yield of WES in PICU
Among the 169 enrolled patients, trio WES was performed on 160 (95%) patients, while proband-only WES was performed on the remaining nine (5.3%) patients on the basis of the availability of parental samples. Forty-three cases had monogenic disorders, and the diagnosis rate was 25%.
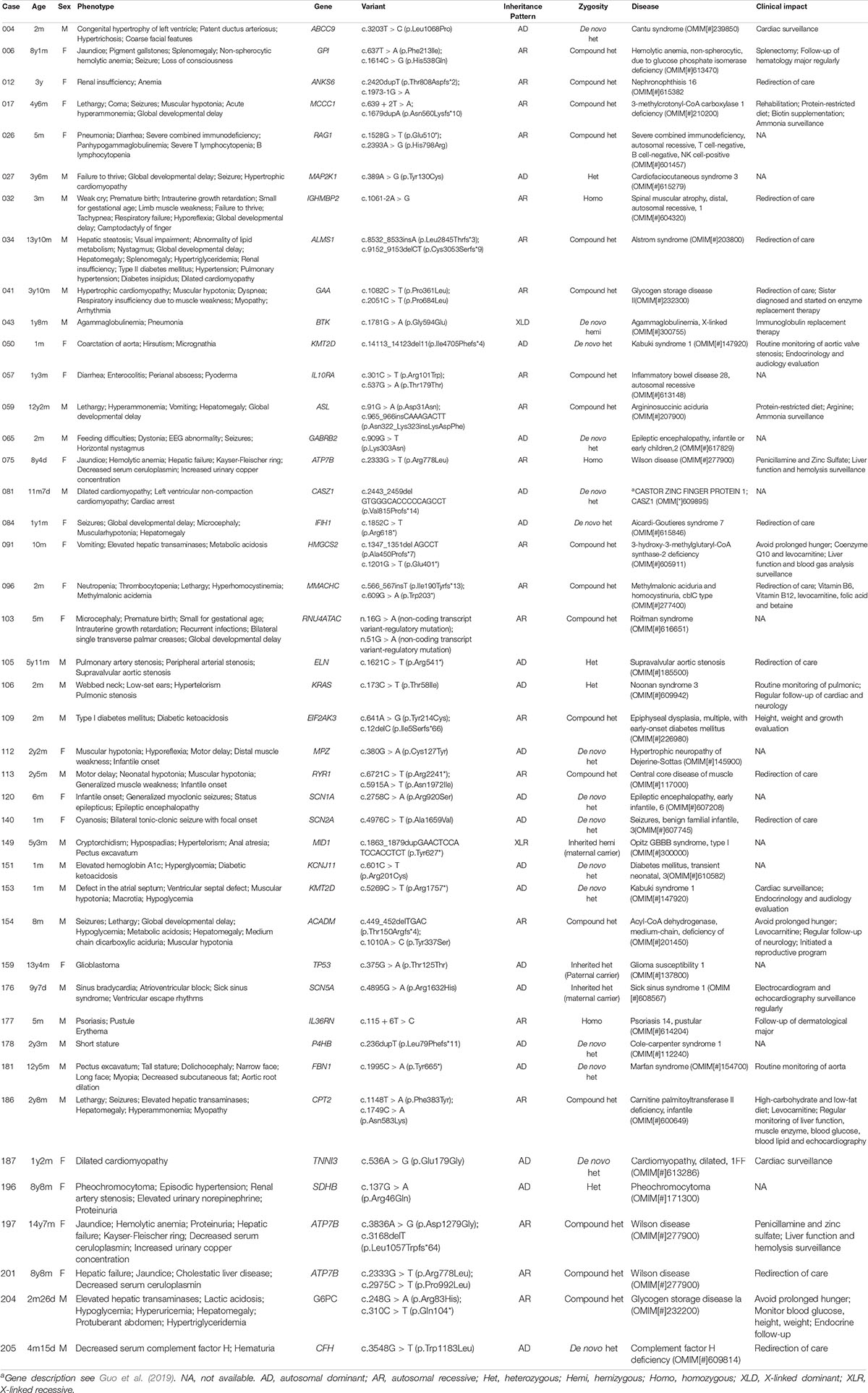
In the 43 diagnosed cases, 40 monogenic diseases were detected. The most common type was metabolic diseases (14/43, 33%), followed by neurological and muscular diseases (8/43, 19%), multiple malformations (6/43, 14%), cardiac diseases (4/43, 9.3%), immunodeficiency diseases (2/43, 4.7%), nephritic diseases (2/43, 4.7%), and other diseases (7/43, 16%) (Table 1 and Supplementary Table 1). ATP7B-related Wilson disease and KMT2D-related Kabuki syndrome were observed in 3 (7.0%) and 2 (4.7%) patients, respectively. The three children with Wilson disease had an initial symptom of acute liver failure, and then they were admitted to the PICU. Of all diagnosed patients, 20 (46%) were autosomal dominant (with 14 de novo, 2 inherited, and 4 inherited unknown), 21 (49%) were autosomal recessive (with 18 compound heterozygous and 3 homozygous), one was X-linked dominant (with de novo), and one was X-linked recessive (with maternal carrier).
In this study, the diagnostic rates were compared among patients with different phenotypes represented by HPO terms. We found that specific clinical features were associated with a molecular diagnosis. For example, patients with neurological and cardiovascular anomalies received diagnosis rates higher than 30%. Patients with HPO terms of “Abnormality of metabolism/homeostasis” (HP: 0001939), “Growth abnormality” (HP: 0001507), and “Abnormality of the eye” (HP: 0000478) had a higher diagnostic yield than those without the terms (Table 2) (P < 0.05). Moreover, we observed the relationship between the complexity of patient’s phenotype (reflected by the number of HPO terms) and the expected diagnostic rate (Figure 3A). The diagnostic yield of cases with only one HPO term was 17%. When more than three HPO terms were given, the diagnostic rate will increase and exceed 30%. For HPO terms greater than 6, the diagnosis rate will reach as high as 50%. However, no statistical difference was observed in the diagnosis yield between children with different HPO terms (P = 0.24).
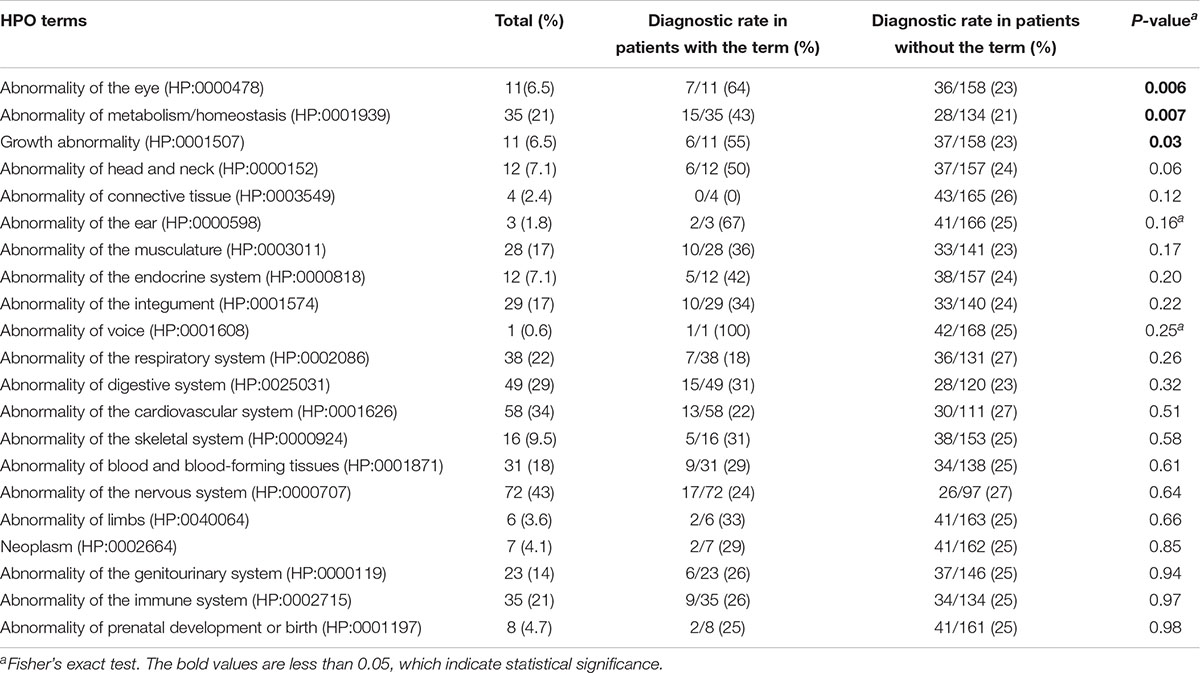
Table 2. Comparison of diagnostic rate by whole exome sequencing in groups with and without the phenotype.
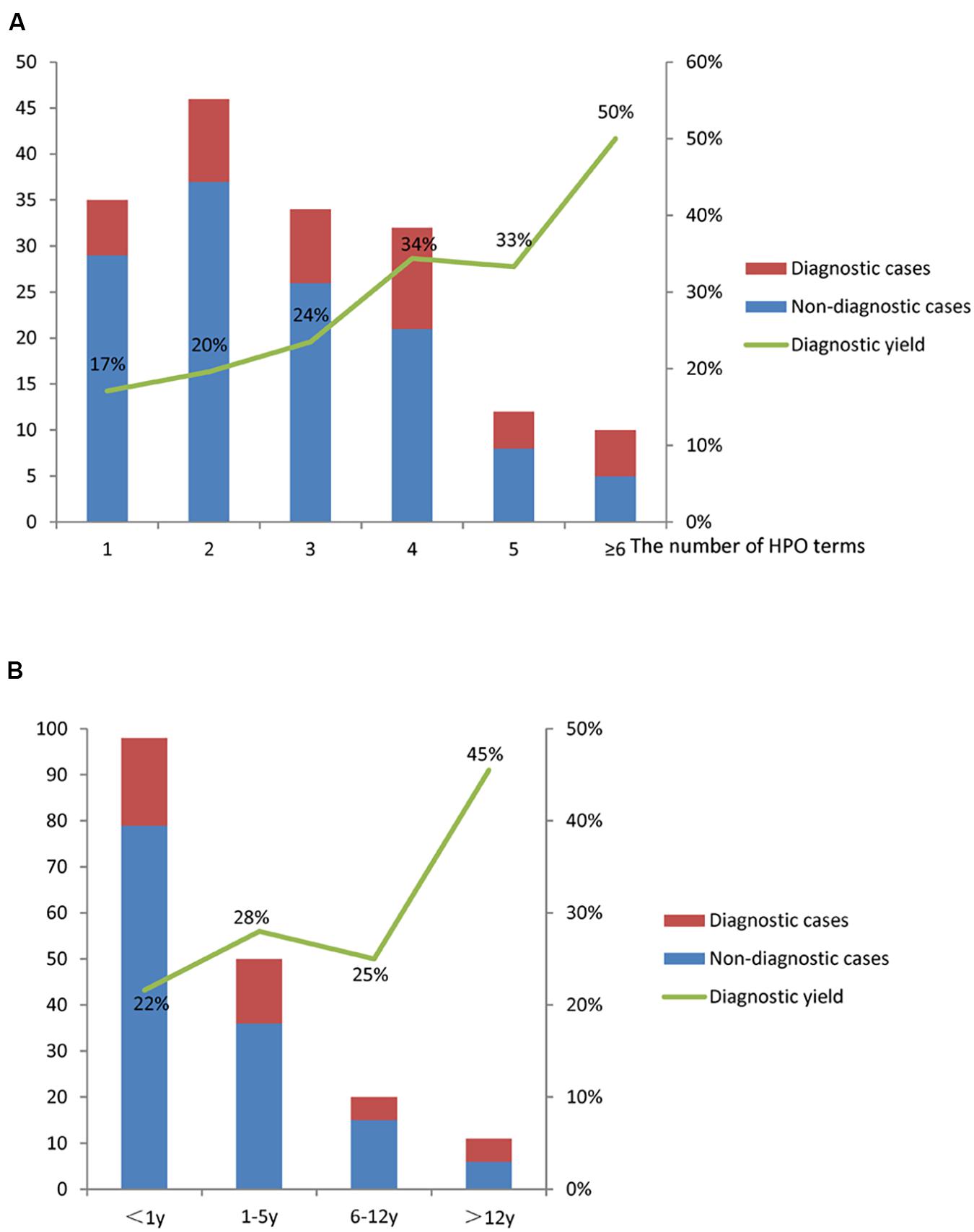
Figure 3. The diagnostic yield of WES in the PICU. (A) The diagnostic yield increased with the number of HPO terms; (B) the diagnostic yields for different age groups were different.
The diagnostic yield of patients aged more than 12 years was the highest (45%), and that of patients aged less than 1 year was relatively low (22%) (Figure 3B). No statistical difference was observed in the diagnosis rate between children of different age groups (P = 0.36).
The media turnaround time of WES in diagnosed patients was 11 days, of which 44% of patients received definite diagnosis before discharge.
Clinical Impact of WES Diagnosis
In this study, 30 patients (70%) were affected by the clinical management. We evaluated the impact of WES on clinical treatment through the following aspects (Table 1 and Supplementary Table 1).
Changes of Treatment
Treatment changes affected 11 patients. Due to infection-induced “hemolytic crisis” with seizure and impairment of consciousness, case 006 was admitted to the PICU. In addition, the child was hospitalized many times because of repeated jaundice. This patient was diagnosed as GPI-related non-spherocytic hemolytic anemia (OMIM: 613470) through WES and discharged from the hospital after laparoscopic cholecystectomy and splenectomy. Her condition was normal, and her jaundice was significantly alleviated.
Moreover, some metabolic diseases required diet therapy in addition to drug treatment. For example, a patient (case 059) with argininosuccinic aciduria (OMIM: 207900) received a protein-restricted diet, another child (case 186) with carnitine palmitoyltransferase II deficiency (OMIM: 600649) needed a high-carbohydrate and low-fat diet, while other children should avoid prolonged hunger.
Disease Monitoring Initiation
Eighteen patients underwent further disease monitoring after diagnosis. For example, regular electrocardiogram monitoring and echocardiography surveillance were required for case 176 with sick sinus syndrome to monitor for arrhythmias. Liver function and blood gas monitoring was critical for case 91 with 3-hydroxy-3-methylglutaryl-CoA synthase-2 deficiency to understand the metabolic state of the body and provide timely intervention. Routine neurological examinations were required for patients with brain damage.
Other Systemic Evaluation
After the diagnosis of WES, three patients were subjected to other systemic evaluation: cases 050 and 153 had KMT2D-related Kabuki syndrome 1 (OMIM: 147920) and underwent endocrinology and audiology evaluations. Case 109 had EIF2AK3-related multiple epiphyseal dysplasia with early-onset diabetes mellitus (OMIM: 226980). In addition to ongoing diabetes treatment, height, weight, growth, and development were evaluated and monitored.
Family Intervention
In addition to the effect on medical services, WES also had a potential influence on family members. Case 041 was diagnosed with glycogen storage disease II (OMIM: 232300). The child’s 9-year-old sister had normal activity endurance but could not tolerate activities such as dancing and running. She underwent relevant genetic testing and was subsequently diagnosed. Fortunately, she was started on enzyme replacement therapy, and lives a normal life at present. Another family initiated a reproductive program through genetic counseling.
Rehabilitation and Redirection of Care Toward Palliative Care
Considering the severity and poor prognosis of the diseases, parents may make different decisions after the diagnosis of WES. Case 017 had 3-methylcrotonyl-CoA carboxylase 1 deficiency (OMIM: 210200). After treatment in the PICU, the child’s vital signs gradually stabilized, but consciousness did not return. The parents chose to leave the PICU for rehabilitation training and accompany the child after diagnosis. The child is now conscious and able to walk. However, 11 patients’ family members chose to stop invasive surgery or life support equipment to alleviate the pain of their children. Most of the children suffered from severe disorders or life-limiting conditions.
Discussion
WES has been increasingly used to diagnose genetic diseases. Previous studies on WES mainly focused on NICUs (Smith et al., 2015; Willig et al., 2015; Wang et al., 2020b). In the present study, we first reported the use of WES in a relatively large number of critically ill children in a PICU from China. The age of patients in this study ranged from 1 month to 14.8 years, with an average age of 2.8 years, which was older than those in previous similar studies (Mestek-Boukhibar et al., 2018; French et al., 2019; Australian Genomics Health Alliance Acute Care et al., 2020; Wang et al., 2020a). The most common presentation was neurological (21%), cardiac (17%), and multiple congenital anomalies (15%), which were similar to those in previous studies (Farnaes et al., 2018; Wang et al., 2020a).
The diagnosis yield varied with different phenotypes, and increased with the phenotypic complexity. Stark et al. (2016) reported that infants with characteristics of strongly suggestive monogenic diseases, such as neurometabolic status and skeletal dysplasia, received WES, and the diagnosis yield was 57%. In the present study, the indication for clinical WES that was assessed by HPO phenotype analysis as having high diagnosis value included (1) “abnormality of metabolism/homeostasis,” such as metabolic acidosis; (2) “growth abnormality,” such as failure to thrive; and (3) “abnormality of the eye,” such as hypertelorism. Therefore, the strict screening of phenotypes can be used to improve the diagnosis yield. However, a previous study had shown that restricting the recruitment to these specific phenotypes would miss many important diagnoses (French et al., 2019). In the future work, more stringent screening should be carried out on the conditions for joining the group to improve the diagnosis rate. Interestingly, Trujillano et al. (2017) reported that complex phenotypes including several symptoms tend to have a higher diagnostic yield, which is consistent with our findings. This might suggest that patients’ phenotypes with several specific symptoms have a higher probability to be explained by monogenic reasons. Therefore, we recommend physicians who request WES to include as many symptoms as possible in their clinical description, but to mark those symptoms that seem to be specific to a particular patient. WES can find the complex manifestations of monogenic diseases. With its wide application in clinical practice, it can be used to easily diagnose patients with overlapped symptoms and mixed phenotypes.
Combined with other detection methods or information supplementation, the diagnosis yield could be improved. Hu et al. (2020) reported that through parallel tests of WES and copy number variant sequencing, the diagnosis yield could be increased from 37 to 53%. In the present study, for patients who were strongly suspected of monogenic diseases but could not be diagnosed after WES, pediatric intensive care physicians and geneticists supplemented the patient’s phenotype or made suggestions to add other detection methods (e.g., copy number variant sequencing, mitochondrial sequencing) through clinical/laboratory rounds and online communication. In this way, two patients were diagnosed. Case 148 with muscular hypotonia and progressive dyspnea received a diagnosis by mitochondrial genome sequencing. He was diagnosed with Leigh syndrome (OMIM: 256000). Case 206 with development delay was diagnosed through copy number variant sequencing (8q24.3 dup 3.6 Mb; 18q22.2-q23 del 9.9 Mb). In the future work, we will continue to strengthen clinical–laboratory interaction so as to improve the efficiency of diagnosis.
For the uncertain significance variants in this study, the following methods usually were taken to help improve the judgment result: (1) Supplement related tests, such as case 006, the final diagnosis was confirmed through supplementation of glucose phosphate isomerase activity test; (2) Test relatives of the proband with the same symptoms, such as case 041, the WES test was performed on the sister of the patient with the same symptoms, and there was a same variant, then the final diagnosis was given; (3) Perform functional verification (not covered in this article).
In addition, the diagnosis yield was also affected by socioeconomic and age factors. In this study, the diagnosis yield of children aged >12 years was the highest (45%), whereas that of children under 1 year was 22%, which was lower than that previously reported (Meng et al., 2017; Wang et al., 2020a). This may be due to the fact that more than half of the enrolled patients in this study were less than 1 year old and were treated in the PICU at the first onset, and their phenotypes were often atypical, mostly congenital heart disease and convulsions. Their parents were willing to find the etiology of the disease through relevant tests. Therefore, in this age group, the percentage of patients who received WES was high, but the diagnosis yield was relatively low. On the other hand, children who were >12 years old often had obvious phenotypes when they were hospitalized and experienced the diagnostic odyssey outside. Thus, they had a higher chance to be diagnosed by WES.
Among the critically ill children, 48–81% of the diagnosed children had changes in clinical management (Willig et al., 2015; Meng et al., 2017; Wu et al., 2019; Wang et al., 2020a). In this study, the diagnosis of WES affected 30 cases (70%) in various aspects. First, WES had an impact on medical treatment: (1) Implement any method, including drugs, surgery, and diet, while avoiding other ineffective or potentially harmful treatments. For example, in case 006 who was diagnosed with GPI-related non-spherocytic hemolytic anemia, she was hospitalized many times in the past because of repeated jaundice. Splenectomy was performed after definite diagnosis. The available data showed that splenectomy was a common and effective treatment (Beutler et al., 1974; Fermo et al., 2019). Thereafter, the need for blood transfusions could be significantly reduced, and the quality of life of patients could be greatly improved. Although the prognosis is serious for some metabolic diseases, earlier diagnosis could lead to better results through diet control (Erez et al., 2011). (2) Disease surveillance and evaluation of the system involved. Single gene diseases often cause multi-system damage. After a clear diagnosis, through targeted disease surveillance, the affected organs could be identified. Once problems occur, interventions should be made in advance to achieve the purpose of long-term survival. For example, case 176 was diagnosed with sick sinus syndrome 1, and electrocardiogram and echocardiography should be monitored regularly. The genetic diagnosis also has impacts on the family: (1) Change of attitude to treatment. For patients with definite diagnosis, if the disease could be treated (including surgery, medicine, diet), parents are more confident to cooperate with doctors for treatment and regular re-examination. Even for patients with unclear diagnosis, some genetic diseases may be excluded. Thus, parents are urged to actively seek further diagnosis and treatment. In our study, roughly 20% of parents chose positive treatment. (2) Withdrawing treatment. For serious diseases that cannot be cured, parents have also found the causes of their children’s illness. Some parents could consider earlier overall hospice care to alleviate their children’s pain in the final stages of life and accompany them through the last days of their lives (Oberender and Tibballs, 2011). (3) Familial genetic counseling. Understanding the molecular cause of the patients greatly facilitated further genetic counseling, including follow-up reproductive planning and screening of siblings. Case 041 was diagnosed with glycogen storage disease II. His sister was later diagnosed, started on enzyme replacement therapy, and lives a normal life at present.
After receiving a definite diagnosis, some patients controlled their symptoms through drugs or diet and avoided ineffective treatment; through systemic assessment and regular monitoring of the disease state, some patients could be intervened in time to avoid life-threatening diseases, such as metabolic crisis; while some parents decided to withdraw the life support equipment. All of the above scenarios reduced the medical care costs from different aspects.
The limitation of this study is that the turnaround time of WES is relatively long. At present, some studies have reported the application of rapid WES in critically ill children (Carey et al., 2020; Freed et al., 2020; Wang et al., 2020b). In future works, we will continue to improve testing methods, optimize testing procedures, and reduce testing costs.
Conclusion
A retrospective analysis was performed on the clinical application of WES in a single PICU of China in the past 3 years. Approximately a quarter of critically ill children with suspected monogenic diseases were diagnosed by WES. In addition, 70% of the patients had a positive impact on medical treatment and families. WES played a key role in family decision-making and clinical treatment, and may improve the prognosis of some children and reduce the economic burden on families and the society. Therefore, we recommend that critically ill children with suspected monogenic diseases in PICUs be tested for WES. With the optimization of the testing process and the reduction of the cost, an increasing number of patients in PICUs are expected to benefit from WES.
Data Availability Statement
According to national legislation/guidelines, specifically the Administrative Regulations of the People’s Republic of China on Human Genetic Resources (http://www.gov.cn/zhengce/content/2019-06/10/content_5398829.htm, http://english.www.gov.cn/policies/latest_releases/2019/06/10/content_281476708945462.htm), no additional raw data is available at this time. Data of this project can be accessed after an approval application to the China National Genebank (CNGB, https://db.cngb.org/cnsa/). Please refer to https://db.cngb.org/, or email: Q05HQmRiQGNuZ2Iub3Jn for detailed application guidance. The accession code HRA001063 should be included in the application.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of the Beijing Children’s Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
YL designed the analysis plan, collected and analyzed the data, drafted the manuscript, and critically reviewed and revised the manuscript for important intellectual content. CH made substantial contributions to the design of the study, coordinated and supervised the data collection, and critically reviewed and revised the manuscript for important intellectual content. XH, RG, JG, and ZQ led the analysis of whole exome sequencing, interpreted results, and critically reviewed and revised the manuscript for important intellectual content. KL, HG, JZ, JL, ZL, and XJ led the clinical treatment of the patients, interpreted the results, and critically reviewed and revised the manuscript for important intellectual content. WL and SQ conceptualized and designed the study, coordinated and supervised the data collection, and critically reviewed the manuscript for important intellectual content. All authors approved the final manuscript as submitted and agreed to be accountable for all aspects of the work.
Funding
This work was funded by the Ministry of Science and Technology of China (2016YFC1000306), the CAMS Innovation Fund for Medical Sciences (2019-I2M-5-026), the Beijing Municipal Commission of Health and Family Planning Foundation (2018-2-1141 and 2020-4-1144), the Special Fund of the Pediatric Medical Coordinated Development Center of Beijing Hospitals Authority (XTCX201807), and the Beihang University and Capital Medical University Advanced Innovation Center for Big Data-Based Precision Medicine Plan (BHME-201905).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all the participating families in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.677699/full#supplementary-material
Footnotes
References
Australian Genomics Health Alliance Acute Care Flagship, Lunke, S., Eggers, S., Wilson, M., Patel, C., Barnett, C. P., et al. (2020). Feasibility of ultra-rapid exome sequencing in critically Ill infants and children with suspected monogenic conditions in the australian public health care system. JAMA 323, 2503–2511. doi: 10.1001/jama.2020.7671
Beutler, E., Sigalove, W. H., Muir, W. A., Matsumoto, F., and West, C. (1974). Glucosephosphate-isomerase (GPI) deficiency: GPI elyria. Ann. Intern. Med. 80, 730–732. doi: 10.7326/0003-4819-80-6-730
Carey, A. S., Schacht, J. P., Umandap, C., Fasel, D., Weng, C., Cappell, J., et al. (2020). Rapid exome sequencing in PICU patients with new-onset metabolic or neurological disorders. Pediatr. Res. 88, 761–768. doi: 10.1038/s41390-020-0858-x
Cunniff, C., Carmack, J. L., Kirby, R. S., and Fiser, D. H. (1995). Contribution of heritable disorders to mortality in the pediatric intensive care unit. Pediatrics 95, 678–681.
Erez, A., Nagamani, S. C., and Lee, B. (2011). Argininosuccinate lyase deficiency-argininosuccinic aciduria and beyond. Am. J. Med. Genet. C Semin. Med. Genet. 157C, 45–53. doi: 10.1002/ajmg.c.30289
Farnaes, L., Hildreth, A., Sweeney, N. M., Clark, M. M., Chowdhury, S., Nahas, S., et al. (2018). Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom. Med. 3:10. doi: 10.1038/s41525-018-0049-4
Fermo, E., Vercellati, C., Marcello, A. P., Zaninoni, A., Aytac, S., Cetin, M., et al. (2019). Clinical and molecular spectrum of glucose-6-phosphate isomerase deficiency. Report of 12 new cases. Front. Physiol. 10:467. doi: 10.3389/fphys.2019.00467
Freed, A. S., Clowes Candadai, S. V., Sikes, M. C., Thies, J., Byers, H. M., Dines, J. N., et al. (2020). The impact of rapid exome sequencing on medical management of critically Ill children. J. Pediatr. 226, 202–212. doi: 10.1016/j.jpeds.2020.06.020
French, C. E., Delon, I., Dolling, H., Sanchis-Juan, A., Shamardina, O., Megy, K., et al. (2019). Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 45, 627–636. doi: 10.1007/s00134-019-05552-x
Gubbels, C. S., Vannoy, G. E., Madden, J. A., Copenheaver, D., Yang, S., Wojcik, M. H., et al. (2020). Prospective, phenotype-driven selection of critically ill neonates for rapid exome sequencing is associated with high diagnostic yield. Genet. Med. 22, 736–744. doi: 10.1038/s41436-019-0708-6
Guo, J., Li, Z., Hao, C., Guo, R., Hu, X., Qian, S., et al. (2019). A novel de novo CASZ1 heterozygous frameshift variant causes dilated cardiomyopathy and left ventricular noncompaction cardiomyopathy. Mol. Genet Genomic Med. 7:e828. doi: 10.1002/mgg3.828
Hu, X., Guo, R., Guo, J., Qi, Z., Li, W., and Hao, C. (2020). Parallel tests of whole exome sequencing and copy number variant sequencing increase the diagnosis yields of rare pediatric disorders. Front. Genet. 11:473. doi: 10.3389/fgene.2020.00473
Hu, X., Li, N., Xu, Y., Li, G., Yu, T., Yao, R. E., et al. (2018). Proband-only medical exome sequencing as a cost-effective first-tier genetic diagnostic test for patients without prior molecular tests and clinical diagnosis in a developing country: the China experience. Genet. Med. 20, 1045–1053. doi: 10.1038/gim.2017.195
Meng, L., Pammi, M., Saronwala, A., Magoulas, P., Ghazi, A. R., Vetrini, F., et al. (2017). Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 171:e173438. doi: 10.1001/jamapediatrics.2017.3438
Mestek-Boukhibar, L., Clement, E., Jones, W. D., Drury, S., Ocaka, L., Gagunashvili, A., et al. (2018). Rapid paediatric sequencing (RaPS): comprehensive real-life workflow for rapid diagnosis of critically ill children. J. Med. Genet. 55, 721–728. doi: 10.1136/jmedgenet-2018-105396
Oberender, F., and Tibballs, J. (2011). Withdrawal of life-support in paediatric intensive care–a study of time intervals between discussion, decision and death. BMC Pediatr. 11:39. doi: 10.1186/1471-2431-11-39
Petrikin, J. E., Cakici, J. A., Clark, M. M., Willig, L. K., Sweeney, N. M., Farrow, E. G., et al. (2018). The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genom. Med. 3:6. doi: 10.1038/s41525-018-0045-8
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17, 405–424. doi: 10.1038/gim.2015.30
Sanford, E. F., Clark, M. M., Farnaes, L., Williams, M. R., Perry, J. C., Ingulli, E. G., et al. (2019). Rapid whole genome sequencing has clinical utility in children in the PICU. Pediatr. Crit. Care Med. 20, 1007–1020. doi: 10.1097/PCC.0000000000002056
Smith, L. D., Willig, L. K., and Kingsmore, S. F. (2015). Whole-Exome sequencing and whole-genome sequencing in critically Ill neonates suspected to have single-gene disorders. Cold Spring Harb. Perspect. Med. 6:a023168. doi: 10.1101/cshperspect.a023168
Stark, Z., Tan, T. Y., Chong, B., Brett, G. R., Yap, P., Walsh, M., et al. (2016). A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet. Med. 18, 1090–1096. doi: 10.1038/gim.2016
Swaggart, K. A., Swarr, D. T., Tolusso, L. K., He, H., Dawson, D. B., and Suhrie, K. R. (2019). Making a genetic diagnosis in a level IV Neonatal intensive care unit population: who, when, how, and at what cost? J. Pediatr. 213:e214. doi: 10.1016/j.jpeds.2019.05.054
Trujillano, D., Bertoli-Avella, A. M., Kumar Kandaswamy, K., Weiss, M. E., Koster, J., Marais, A., et al. (2017). Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur. J. Hum. Genet. 25, 176–182. doi: 10.1038/ejhg.2016.146
Wang, H., Lu, Y., Dong, X., Lu, G., Cheng, G., and Qian, Y. (2020a). Optimized trio genome sequencing (OTGS) as a first-tier genetic test in critically ill infants: practice in China. Hum. Genet. 139, 473–482. doi: 10.1007/s00439-019-02103-8
Wang, H., Qian, Y., Lu, Y., Qin, Q., Lu, G., Cheng, G., et al. (2020b). Clinical utility of 24-h rapid trio-exome sequencing for critically ill infants. NPJ Genom. Med. 5:20. doi: 10.1038/s41525-020-0129-0
Willig, L. K., Petrikin, J. E., Smith, L. D., Saunders, C. J., Thiffault, I., Miller, N. A., et al. (2015). Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir. Med. 3, 377–387. doi: 10.1016/S2213-2600(15)00139-3
Wu, E. T., Hwu, W. L., Chien, Y. H., Hsu, C., Chen, T. F., Chen, N. Q., et al. (2019). Critical trio exome benefits in-time decision-making for pediatric patients with severe illnesses. Pediatr. Crit. Care Med. 20, 1021–1026. doi: 10.1097/PCC.0000000000002068
Keywords: whole exome sequence, pediatric intensive care unit, monogenic disorders, clinical application, effective
Citation: Liu Y, Hao C, Li K, Hu X, Gao H, Zeng J, Guo R, Liu J, Guo J, Li Z, Qi Z, Jia X, Li W and Qian S (2021) Clinical Application of Whole Exome Sequencing for Monogenic Disorders in PICU of China. Front. Genet. 12:677699. doi: 10.3389/fgene.2021.677699
Received: 08 March 2021; Accepted: 03 August 2021;
Published: 01 September 2021.
Edited by:
Natália Duarte Linhares, Genuity Science, IrelandReviewed by:
Ni-Chung Lee, National Taiwan University, TaiwanBaiba Lace, Biomedical Research and Study Centre, Latvia
Copyright © 2021 Liu, Hao, Li, Hu, Gao, Zeng, Guo, Liu, Guo, Li, Qi, Jia, Li and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Li, bGl3ZWlAYmNoLmNvbS5jbg==; Suyun Qian, c3lxaWFuMjAyMEAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share senior authorship
 Yingchao Liu
Yingchao Liu Chanjuan Hao
Chanjuan Hao Kechun Li
Kechun Li Xuyun Hu
Xuyun Hu Hengmiao Gao1,2
Hengmiao Gao1,2 Jiansheng Zeng
Jiansheng Zeng Ruolan Guo
Ruolan Guo Jun Guo
Jun Guo Xinlei Jia
Xinlei Jia Wei Li
Wei Li Suyun Qian
Suyun Qian