- 1Centre for Global Health, Zhejiang University School of Medicine, Hangzhou, China
- 2Department of Gastroenterology, The Third Xiangya Hospital, Central South University, Changsha, China
- 3Institute for Global Health, University College London, London, United Kingdom
Background: Whether a gluten-free diet (GFD) is a cause of irritable bowel syndrome (IBS) remains controversial. We aim at exploring the causal relationship between gluten intake and IBS within Mendelian randomization (MR) design.
Methods: We conducted a two-sample MR and selected single-nucleotide polymorphisms (SNPs) associated with GFD as instrumental variables (IVs). SNPs and genetic associations with GFD and IBS were obtained from the latest genome-wide association studies (GWAS) in Europeans (GFD: cases: 1,376; controls: 63,573; IBS: cases:1,121; controls: 360,073). We performed inverse variance weighting (IVW) as the primary method with several sensitivity analyses like MR-Egger and MR-PRESSO for quality control. The above analyses were re-run using another large dataset of IBS, as well as changing the p-value threshold when screening IVs, to verify the stability of the results.
Results: The final estimate indicated significant causal association [per one copy of effect allele predicted log odds ratio (OR) change in GFD intake: OR = 0.97, 95% confidence interval (CI) 0.96 to 0.99, p < 0.01] without heterogeneity statistically (Q = 2.48, p = 0.78) nor horizontal pleiotropy biasing the causality (p = 0.92). Consistent results were found in validation analyses. Results of MR Steiger directionality test indicated the accuracy of our estimate of the causal direction (Steiger p < 0.001).
Conclusion: GFD might be a protective factor of IBS. Therefore, we suggest taking a diet of lower gluten intake into account in IBS prevention and clinical practice.
Introduction
Irritable bowel syndrome (IBS) is a functional gastrointestinal disorder with recurrent abdominal pain associated with defecation and bowel habit change (Ford et al., 2020). Considered as the most common chronic functional gastrointestinal disease, it affects 5–10% of the world’s population, significantly deteriorating the quality of life (El-Salhy, 2015; Rej & Sanders, 2018; Flacco et al., 2019; Oka et al., 2020).
Since diet was assumed to be included in the pathophysiology of IBS and food ingestion triggered symptoms among up to 60% of IBS patients, controlling diets might be a primary prevention approach with low cost and high yield. Dietary modification like a gluten-free diet (GFD) was recommended to relieve their symptoms because of the overlap between IBS and gluten sensitivity (Lahner et al., 2013; De Giorgio et al., 2016); Verdu et al., 2009; Biesiekierski et al., 2011). However, when taking the confounding factors into account, the results of observational studies can be the opposite (Bowden et al., 2017). There is little high-quality evidence exploring the effect of specific diets on IBS (Ford et al., 2020), and whether a causal association between GFD and the development of IBS exists remains unanswered.
To investigate the causality of GFD and the lower risk of incident IBS, we conducted two-sample Mendelian randomization (MR). According to Mendel’s law of inheritance, genetic variants are randomly assigned prior to the onset of disease, and thus genetic variants are improbable to be downstream events of subsequently measured events. Thus, in the MR analysis, we selected appropriate genetic variants [single-nucleotide polymorphisms (SNPs)] as instrumental variables (IVs) for the exposure. In this way, it is possible to avoid reverse causality and confounding factors such as psychological and environmental factors (Saito et al., 2005; Bowden et al., 2017). Additionally, in the two-sample MR study, information on gene–outcome association and gene–exposure association were obtained from two separate samples of the same population. Based on existing data, it is more economical and efficient (Pierce & Burgess, 2013).
Materials and Methods
Study Design
In this MR analysis, we extracted genetic variants that are strongly associated with GFD from open genome-wide association studies (GWAS) as IVs and used IVs to replace GFD as new exposures to investigate the causal relationship between GFD and IBS. The extracted IVs must meet three core assumptions: 1) be strongly associated with the exposure; 2) be independent of confounders; and 3) influence the outcome exclusively via the exposure (details in Figure 1) (Bowden et al., 2015; Bowden & Holmes, 2019). The ratio of the gene–outcome association and gene–exposure association obtained from GWAS could assess the causal association between exposure and outcome (Lawlor et al., 2008). The total effect was calculated using the inverse variance weighting (IVW) method to summarize the estimate of every single IV. Sensitive analyses were conducted, including MR-Egger and MR-PRESSO to estimate horizontal pleiotropy, maximum likelihood (ML), weighted median (WME), robust adjusted profile score (RAPS), MRCIP, and MR-PRESSO for result validation. Furthermore, considering the small number of IBS cases, we used an outcome dataset with a larger number of cases (10,939) for validation. All the data we used were selected from publicly available databases with ethical approval in origin.

FIGURE 1. Diagram of the Mendelian randomization assumptions supporting a two-sample Mendelian randomization analysis of the causal effect of gluten-free diet on irritable bowel syndrome (the dashed lines indicate possible causal effects between variables which may be against the Mendelian randomization assumptions).
Data Source
GWAS provided large-sized, repeatedly checked samples, which made our studies available and reliable. We obtained genetic predictors and genetic associations with GFD from the latest and biggest GWAS of GFD (Elsworth et al., 2020). The study ascertained GFD standards with the Oxford WebQ (an online questionnaire reporting the condition of food intaking), and participants who follow a GFD or wheat-free diet routinely were considered as GFD cases. The summary data covered the range of 64,949 European individuals (cases: 1,376, controls: 63,573) (Hemani et al., 2018; Liu et al., 2011).
Genetic associations with IBS were acquired from the latest United Kingdom Biobank GWAS study. United Kingdom Biobank collects the deep genetic and traits data of 488,377 individuals, including 361,194 individuals with IBS genetic information (cases: 1,121, controls: 360,073) (Bycroft et al., 2018). IBS cases refer to participants who have recurrent abnormal pain or discomfort for at least 3 days per month during the last 3 months accompanied by two or more of the following conditions: 1) symptom ease after defecation, 2) changes in defecation frequency during seizures, and 3) seizure with stool changes (Spiller, 2003; Enck et al., 2016). The GWAS used BOLT-LMM106 to adjust for sex and age, and the first 20 ancestor principal components (PCs) as covariates (Wu et al., 2021).
The dataset for validation was a large GWAS based on the UK biobank, 2021 (cases: 10,939, controls: 451,994) (Elsworth et al., 2020). In this study, the outcome was self-reported IBS, obtained from verbal interviews by trained nurses.
Selection of Instrumental Variables
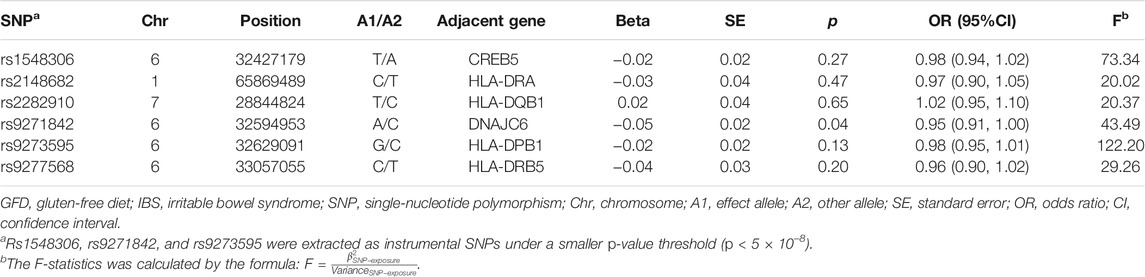
Appropriate SNPs must be linked to GFD strongly (p < 5 × 10–8) with low linkage disequilibrium (LD) (r2 < 0.001) and high minor allele frequency (MAF
To reduce the interference from confounders, each instrumental SNP was estimated for the possible connections with confounders including age when full-time education was completed (226,899 individuals), the year full-time education ended (112,569 individuals), circulating leptin levels adjusted for body mass index (BMI) (33,987 individuals), average total household income before tax (397,751 individuals), age of smoking initiation (341,427 individuals), and alcoholic drinks per week (335,394 individuals) (details in Supplementary Figure S1 and Table S2) (Kilpeläinen et al., 2016; Bowden et al., 2017; Liu et al., 2019).
Statistical Analysis
All statistical analyses were performed in R 4.0.3 with R package “TwoSampleMR” and “MRPRESSO.”
Primary Statistical Analysis
IVW was chosen as the primary analysis, as it is the most efficient method to estimate the causal effect when the included IVs are all valid (satisfied the InSIDE assumption) (Bowden et al., 2015) and the intercept of the IVW regression equation is assumed to be 0 (Bowden et al., 2017).
The causal association between exposure and outcome
When genetic variants were not related, we estimated an overall effect combining the estimated ratio of each variant, called IVW estimator. IVW estimator refers to a consistent estimate of the causal association if IVs meet all IVs assumptions [
Sensitive Analyses
The MR analysis was also conducted in different methods to check the robustness of the results. Other analysis methods that were exerted included MR Egger, ML, WME, RAPS, MRCIP, and MR-PRESSO. Each method has its own characteristics. MR Egger contains the square of SE as its weight, and the intercept is taken into consideration (Bowden et al., 2017). The probability of each nucleotide substitution is considered in ML aiming to find a phylogenetic tree that can engender observation data in a higher possibility (Luque-Fernandez et al., 2018). WME is robust to outliners focusing on the median of the distribution function estimated by IVW to analyze (Bowden et al., 2016a). RAPS can downweight outliers, while MRCIP is a new robust MR method that can handle correlated and idiosyncratic pleiotropy at the same time through the PRW-EM algorithm combined with expectation–maximization (EM) algorithm (Zhao et al., 2020; Xu et al., 2021). MR-PRESSO was conducted to estimate the horizontal pleiotropy and the total effect after removing outliers (Verbanck et al., 2018).
When conducting MR analysis, the three core assumptions above may be violated due to the horizontal pleiotropy, leading to inaccurate results. Pleiotropy includes vertical and horizontal pleiotropy. Since eliminating the horizontal pleiotropy would make the result more reliable while removing the vertical one might distort the causal estimation, we only test horizontal pleiotropy (Bowden et al., 2017). The test methods were comparing the intercept of the MR Egger function with 0 and applying MR-PRESSO. We also search the GWAS Catalog to avoid more specific pleiotropic paths (Buniello et al., 2019). Besides, the heterogeneity was quantified by the p-value derived from Cochran’s Q test and checked deeper by the leave-one-out sensitivity test.
In our design, we assumed that IVs subsequently affect outcomes by influencing exposure. Therefore, we used the MR Steiger directionality test to examine whether the results we found followed the direction in our hypothesis (Hemani et al., 2017). The Steiger p-value of less than 0.05 was considered to show the correct direction.
Result
According to the criteria mentioned above, we extracted three SNPs that met our inclusion criteria (rs1548306, rs9271842, and rs9273595) and three more SNPs under the lower p-value threshold (p < 1 × 10–5) (rs2148682, rs2282910, and rs9277568) as IVs to conduct MR. As rs9271842 was not included in our outcome database; rs9271847 was used as a proxy to gain the gene–outcome association. F-statistics ranging from 20.02 to 122.20 indicated less likely bias caused by weak IVs (details in Table 1; Supplementary Table S1).
The results of IVW with three IVs illustrated a suggestive effect of GFD on the risk of IBS [per one copy of effect allele predicted log odds ratio (OR) change in GFD intake: OR = 0.97, 95% confidence interval (CI) 0.95 to 0.99, p < 0.05]. A consistent result was found when six IVs were included into the analysis (IVW, OR = 0.97, 95%CI 0.96 to 0.99, p < 0.01).
Given the insufficient number of three IVs to complete only partial sensitivity analyses, we completed sensitivity analyses primarily when six IVs were available. In sensitive analyses, results in the other six methods (MR Egger, ML, WME, RAPS, MRCIP, and MR-PRESSO) all went along well with what we supposed despite some small deviations. The scatter plot also showed that the curves fitted by the various methods were generally consistent across parameters (details in Figure 2; Supplementary Figure S2). We found no statistically significant horizontal pleiotropy [MR Egger (intercept): p = 0.51 (3 IVs), p = 0.92 (6 IVs); MR-PRESSO: p = 0.85 (6 IVs, three IVs not enough for this method)] or heterogeneity in the IVW method [Q = 0.95, p = 0.63 (3 IVs); Q = 2.48, p = 0.78 (6 IVs)] (additional funnel plot in Supplementary Figure S3), and no obvious factors were found when searching the GWAS Catalog that could cause possible pleiotropy. Furthermore, we conducted leave-one-out sensitivity estimations by extracting one of the six SNPs out each time. The result of leave-one-out sensitivity estimations illustrated that every SNP of the six hardly biases the outcome (details in Supplementary Figure S4). Results of the MR Steiger directionality test indicated the accuracy of our estimate of the causal direction (Steiger p < 0.001).
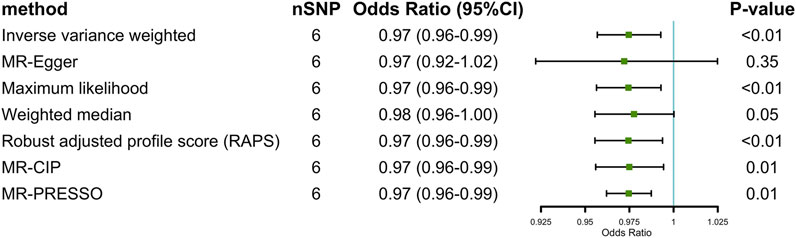
FIGURE 2. Two-sample Mendelian randomization estimations of the causal effect of gluten-free diet on irritable bowel syndrome with IVW, MR Egger, ML, WME, RAPS, MRCIP, and MR PRESSO (at p < 1 × 10–5).
After changing the outcome dataset for validation, we still observed that GFD was associated with a lower risk of IBS (IVW, OR = 0.93, 95%CI 0.89 to 0.97, p < 0.01) without statistically significant horizontal pleiotropy (p = 0.51) or heterogeneity (Q = 5.43, p = 0.37).
Discussion
In this study, we found that GFD was associated with a lower risk of IBS, and this causal effect provided evidence for IBS prevention and was valuable for future investigations and clinical practice.
Accumulated previous studies proved that specific food, especially gluten, might play a key role in inducing and worsening some symptoms of IBS (Zar et al., 2001; Atkinson et al., 2004; Spencer et al., 2014). With regard to the putative mechanism, some studies have pointed out that gluten might change the bowel barrier and increase the permeability of the small bowel, evoking inflammation and higher sensitivity (Vazquez-Roque et al., 2013; Shahbazkhani et al., 2015). As the initial stimulator of hypersensitivity, bowel inflammation could cause lasting mild inflammation and bowel irritability, which in turn generate or exacerbate IBS (Neal et al., 1997; Spiller et al., 2000; Gwee et al., 2003). Additionally, epithelial T lymphocytes and mast cells proliferated slightly under the colonic biopsies further verified the role of inflammation in the onset of IBS (Barbara et al., 2004; Ohman & Simrén, 2010). Besides, it was interesting to note that except for bowel reactions, gluten also showed systemic effects including the sense of fatigue (Biesiekierski et al., 2013; Shahbazkhani et al., 2015). Since psychological factors are crucial to the pathogenesis of IBS, the protective effect of GFD on IBS may also be explained in this way (Drossman, 1999).
To our knowledge, this was the first MR study providing insights on the preventive effect of GFD, while the long-term adherence to GFD was inversely associated with IBS development. It is widely acknowledged that strict diet control was hardly a long-term intervention, which makes related randomized control trials (RCT) evidence insufficient. Meanwhile, our MR study could overcome the susceptible bias of confounders and reverse causality shared by traditional observational studies.
Although the current MR study was performed rigorously and had plenty of strengths, some limitations exist. Firstly, a spurious causal relationship may be caused by the possible difference of frequency in people with diverse genetic backgrounds. To minimize this bias, our IVs were all selected from genetic variants from sources of European ancestry and genomic control research. Nevertheless, our results in the European group might not generalize to other races (Garcia-Mazcorro et al., 2018). Secondly, in MR, it was a challenge to eliminate the interferences of potential confounders and horizontal pleiotropy. Therefore, we applied six other methods besides IVW and found consistent results. Besides, MR Egger and MR-PRESSO were employed to assess horizontal pleiotropy and found unlikely to exhibit such bias. Furthermore, before conducting MR analysis, we had estimated associations between some confounders and SNPs and found that the SNPs all met IV assumption two which ensured higher accuracy and reliability (details in Supplementary Table S2). Thirdly, as the self-administered questionnaire of GFD from United Kingdom Biobank only presents binary choices, Yes/No, the effect of GFD on preventing IBS could be only described qualitatively rather than quantitatively. Detailed investigation on gluten intake might show more information, but we were not aware of any available large GWAS related at present. Fourthly, the limited number of cases and coefficient of determination make the statistical power relatively insufficient. To minimize this effect, we verified our hypothesis in another larger dataset. Considering the limited statistical power, larger and higher-quality GWAS studies or well-designed trials should be conducted to validate this conclusion. Last but not least, the health benefit of following a GFD among the population without gastrointestinal diseases like IBS remains a core controversial issue for public health. Further studies are necessary because of the current lack of experimental evidence (Staudacher and Gibson, 2015; Dionne et al., 2018; Melini & Melini, 2019). Thus, we still need to be vigilant about potential dietary change risks and inducing nutrition deficiency while commencing and adhering to a GFD.
Furthermore, our results demonstrated the protective effect of GFD, but GFD is not exactly the same as just reducing gluten intake, especially when there were some other components in the gluten-containing food. Fructans, another key ingredient found in gluten-containing foods demonstrated adverse effects not only on the bowel but also on mental problems. An RCT directly clarified that fructan rather than gluten was regarded as a culprit (Skodje et al., 2018). Several RCTs also compared FODMAPs (low fermentable, oligosaccharides, disaccharides, monosaccharides, and polyols) with GFD and found positive results, since fructan was an oligosaccharide in FODMAPs (Halmos et al., 2014; Dionne et al., 2018; Krieger-Grübel et al., 2020). Therefore, whether gluten was a crucial step in GFD to develop IBS or merely an inducer still needs further investigation on other ingredients in gluten-containing foods.
Conclusion
Our result provided suggestive evidence of a protective causal effect of GFD on the IBS incident. Therefore, we suggested taking a diet of reducing gluten intake into account in terms of IBS prevention in clinical practice.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
Conceptualization: JC and XW; methodology: YS, JC, and XC; writing—original draft preparation: YS, SW, XC, MD, and YX; writing—review and editing: XW, JC, and TH; visualization: YS and XC; supervision: XW and JC; funding acquisition: XW and MD. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (81970494) and Key Research and Development Program of Hunan Province (2019SK 2041).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank MRC Integrative Epidemiology Unit and Neale Lab for providing publicly available summary data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.684535/full#supplementary-material
References
Atkinson, W., Sheldon, T. A., Shaath, N., and Whorwell, P. J. (2004). Food Elimination Based on IgG Antibodies in Irritable Bowel Syndrome: a Randomised Controlled Trial. Gut 53 (10), 1459–1464. doi:10.1136/gut.2003.037697
Barbara, G., Stanghellini, V., De Giorgio, R., Cremon, C., Cottrell, G. S., Santini, D., et al. (2004). Activated Mast Cells in Proximity to Colonic Nerves Correlate with Abdominal Pain in Irritable Bowel Syndrome. Gastroenterology 126 (3), 693–702. doi:10.1053/j.gastro.2003.11.055
Biesiekierski, J. R., Newnham, E. D., Irving, P. M., Barrett, J. S., Haines, M., Doecke, J. D., et al. (2011). Gluten Causes Gastrointestinal Symptoms in Subjects without Celiac Disease: a Double-Blind Randomized Placebo-Controlled Trial. Am. J. Gastroenterol. 106 (3), 508–514. doi:10.1038/ajg.2010.487
Biesiekierski, J. R., Peters, S. L., Newnham, E. D., Rosella, O., Muir, J. G., and Gibson, P. R. (2013). No Effects of Gluten in Patients with Self-Reported Non-celiac Gluten Sensitivity after Dietary Reduction of Fermentable, Poorly Absorbed, Short-Chain Carbohydrates. Gastroenterology 145 (2), 320–328. e83. doi:10.1053/j.gastro.2013.04.051
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian Randomization with Invalid Instruments: Effect Estimation and Bias Detection through Egger Regression. Int. J. Epidemiol. 44 (2), 512–525. doi:10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016b). Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 40 (4), 304–314. doi:10.1002/gepi.21965
Bowden, J., Del Greco M., F., Minelli, C., Davey Smith, G., Sheehan, N. A., and Thompson, J. R. (2016a). Assessing the Suitability of Summary Data for Two-Sample Mendelian Randomization Analyses Using MR-Egger Regression: the Role of the I2 Statistic. Int. J. Epidemiol. 45 (6), dyw220–1974. doi:10.1093/ije/dyw220
Bowden, J., Del Greco M, F., Minelli, C., Davey Smith, G., Sheehan, N., and Thompson, J. (2017). A Framework for the Investigation of Pleiotropy in Two-Sample Summary Data Mendelian Randomization. Statist. Med. 36 (11), 1783–1802. doi:10.1002/sim.7221
Bowden, J., and Holmes, M. V. (2019). Meta‐analysis and Mendelian Randomization: A Review. Res. Syn Meth 10 (4), 486–496. doi:10.1002/jrsm.1346
Buniello, A., MacArthur, J. A. L., Cerezo, M., Harris, L. W., Hayhurst, J., Malangone, C., et al. (2019). The NHGRI-EBI GWAS Catalog of Published Genome-wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res. 47 (D1), D1005–D1012. doi:10.1093/nar/gky1120
Burgess, S., Butterworth, A., and Thompson, S. G. (2013). Mendelian Randomization Analysis with Multiple Genetic Variants Using Summarized Data. Genet. Epidemiol. 37 (7), 658–665. doi:10.1002/gepi.21758
Burgess, S., Dudbridge, F., and Thompson, S. G. (2016). Combining Information on Multiple Instrumental Variables in Mendelian Randomization: Comparison of Allele Score and Summarized Data Methods. Statist. Med. 35 (11), 1880–1906. doi:10.1002/sim.6835
Bycroft, C., Freeman, C., Petkova, D., Band, G., Elliott, L. T., Sharp, K., et al. (2018). The UK Biobank Resource with Deep Phenotyping and Genomic Data. Nature 562 (7726), 203–209. doi:10.1038/s41586-018-0579-z
De Giorgio, R., Volta, U., and Gibson, P. R. (2016). Sensitivity to Wheat, Gluten and FODMAPs in IBS: Facts or Fiction. Gut 65 (1), 169–178. doi:10.1136/gutjnl-2015-309757
Dionne, J., Ford, A. C., Yuan, Y., Chey, W. D., Lacy, B. E., Saito, Y. A., et al. (2018). A Systematic Review and Meta-Analysis Evaluating the Efficacy of a Gluten-free Diet and a Low FODMAPs Diet in Treating Symptoms of Irritable Bowel Syndrome. Am. J. Gastroenterol. 113 (9), 1290–1300. doi:10.1038/s41395-018-0195-4
Drossman, D. A. (1999). Review Article: an Integrated Approach to the Irritable Bowel Syndrome. Aliment. Pharmacol. Ther. 13 (Suppl. 2), 3–14. doi:10.1046/j.1365-2036.1999.0130s2003.x
El-Salhy, M. (2015). Recent Developments in the Pathophysiology of Irritable Bowel Syndrome. Wjg 21 (25), 7621–7636. doi:10.3748/wjg.v21.i25.7621
Elsworth, B. L., Lyon, M. S., Alexander, T., Liu, Y., Matthews, P., Hallett, J., et al. (2020). The MRC IEU Open GWAS Data Infrastructure. bioRxiv. 244293v1. doi:10.1101/2020.08.10.244293
Enck, P., Aziz, Q., Barbara, G., Farmer, A. D., Fukudo, S., Mayer, E. A., et al. (2016). Irritable Bowel Syndrome. Nat. Rev. Dis. Primers. 2, 16014. doi:10.1038/nrdp.2016.14
Flacco, M. E., Manzoli, L., De Giorgio, R., Gasbarrini, A., Cicchetti, A., Bravi, F., et al. (2019). Costs of Irritable Bowel Syndrome in European Countries with Universal Healthcare Coverage: a Meta-Analysis. Eur. Rev. Med. Pharmacol. Sci. 23 (7), 2986–3000. doi:10.26355/eurrev_201904_17580
Ford, A. C., Sperber, A. D., Corsetti, M., and Camilleri, M. (2020). Irritable Bowel Syndrome. The Lancet 396 (10263), 1675–1688. doi:10.1016/S0140-6736(20)31548-8
Garcia-Mazcorro, J., Noratto, G., and Remes-Troche, J. (2018). The Effect of Gluten-free Diet on Health and the Gut Microbiota Cannot Be Extrapolated from One Population to Others. Nutrients 10 (10), 1421. doi:10.3390/nu10101421
Gwee, K.-A., Collins, S. M., Read, N. W., Rajnakova, A., Deng, Y., Graham, J. C., et al. (2003). Increased Rectal Mucosal Expression of Interleukin 1beta in Recently Acquired post-infectious Irritable Bowel Syndrome. Gut 52 (4), 523–526. doi:10.1136/gut.52.4.523
Halmos, E. P., Power, V. A., Shepherd, S. J., Gibson, P. R., and Muir, J. G. (2014). A Diet Low in FODMAPs Reduces Symptoms of Irritable Bowel Syndrome. Gastroenterology 146 (1), 67–75. e5. doi:10.1053/j.gastro.2013.09.046
Hemani, G., Tilling, K., and Davey Smith, G. (2017). Orienting the Causal Relationship between Imprecisely Measured Traits Using GWAS Summary Data. Plos Genet. 13 (11), e1007081. doi:10.1371/journal.pgen.1007081
Hemani, G., Zheng, J., Elsworth, B., Wade, K. H., Haberland, V., Baird, D., et al. (2018). The MR-Base Platform Supports Systematic Causal Inference across the Human Phenome. eLife 7, e34408. doi:10.7554/eLife.34408
Johnson, T. (2014) Efficient Calculation for multi‐SNP Genetic Risk Scores. Technical Report the Comprehensive R Archive Network. online, Available at: http://cran.r-project.org/web/packages/gtx/vignettes/ashg2012.pdf [accessed 2014 11 19]
Kilpeläinen, T. O., Carli, J. F. M., Skowronski, A. A., Sun, Q., Kriebel, J., Feitosa, M. F., et al. (2016). Genome-wide Meta-Analysis Uncovers Novel Loci Influencing Circulating Leptin Levels. Nat. Commun. 7, 10494. doi:10.1038/ncomms10494
Krieger-Grübel, C., Hutter, S., Hiestand, M., Brenner, I., Güsewell, S., and Borovicka, J. (2020). Treatment Efficacy of a Low FODMAP Diet Compared to a Low Lactose Diet in IBS Patients: A Randomized, Cross-Over Designed Study. Clin. Nutr. ESPEN 40, 83–89. doi:10.1016/j.clnesp.2020.09.020
Lahner, E., Bellentani, S., Bastiani, R. D., Tosetti, C., Cicala, M., Esposito, G., et al. (2013). A Survey of Pharmacological and Nonpharmacological Treatment of Functional Gastrointestinal Disorders. United Eur. Gastroenterol. j. 1 (5), 385–393. doi:10.1177/2050640613499567
Lawlor, D. A., Harbord, R. M., Sterne, J. A. C., Timpson, N., and Davey Smith, G. (2008). Mendelian Randomization: Using Genes as Instruments for Making Causal Inferences in Epidemiology. Statist. Med. 27 (8), 1133–1163. doi:10.1002/sim.3034
Liu, B., Young, H., Crowe, F. L., Benson, V. S., Spencer, E. A., Key, T. J., et al. (2011). Development and Evaluation of the Oxford WebQ, a Low-Cost, Web-Based Method for Assessment of Previous 24 H Dietary Intakes in Large-Scale Prospective Studies. Public Health Nutr. 14 (11), 1998–2005. doi:10.1017/S1368980011000942
Liu, M., Jiang, Y., Jiang, Y., Wedow, R., Li, Y., Brazel, D. M., et al. (2019). Association Studies of up to 1.2 Million Individuals Yield New Insights into the Genetic Etiology of Tobacco and Alcohol Use. Nat. Genet. 51 (2), 237–244. 23 and Me Research Team, HUNT All-In Psychiatry. doi:10.1038/s41588-018-0307-5
Luque-Fernandez, M. A., Schomaker, M., Rachet, B., and Schnitzer, M. E. (2018). Targeted Maximum Likelihood Estimation for a Binary Treatment: A Tutorial. Stat. Med. 37 (16), 2530–2546. doi:10.1002/sim.7628
Machiela, M. J., and Chanock, S. J. (2015). LDlink: a Web-Based Application for Exploring Population-specific Haplotype Structure and Linking Correlated Alleles of Possible Functional Variants: Fig. 1. Bioinformatics 31 (21), 3555–3557. doi:10.1093/bioinformatics/btv402
Melini, V., and Melini, F. (2019). Gluten-Free Diet: Gaps and Needs for a Healthier Diet. Nutrients 11 (1), 170. doi:10.3390/nu11010170
Neal, K. R., Hebden, J., and Spiller, R. (1997). Prevalence of Gastrointestinal Symptoms Six Months after Bacterial Gastroenteritis and Risk Factors for Development of the Irritable Bowel Syndrome: Postal Survey of Patients. BMJ 314 (7083), 779. doi:10.1136/bmj.314.7083.779
Öhman, L., and Simrén, M. (2010). Pathogenesis of IBS: Role of Inflammation, Immunity and Neuroimmune Interactions. Nat. Rev. Gastroenterol. Hepatol. 7 (3), 163–173. doi:10.1038/nrgastro.2010.4
Oka, P., Parr, H., Barberio, B., Black, C. J., Savarino, E. V., and Ford, A. C. (2020). Global Prevalence of Irritable Bowel Syndrome According to Rome III or IV Criteria: a Systematic Review and Meta-Analysis. Lancet Gastroenterol. Hepatol. 5 (10), 908–917. doi:10.1016/S2468-1253(20)30217-X
Pierce, B. L., and Burgess, S. (2013). Efficient Design for Mendelian Randomization Studies: Subsample and 2-sample Instrumental Variable Estimators. Am. J. Epidemiol. 178 (7), 1177–1184. doi:10.1093/aje/kwt084
Rej, A., and Sanders, D. (2018). Gluten-Free Diet and its ‘Cousins' in Irritable Bowel Syndrome. Nutrients 10 (11), 1727. doi:10.3390/nu10111727
Saito, Y. A., Petersen, G. M., Locke, G. R., and Talley, N. J. (2005). The Genetics of Irritable Bowel Syndrome. Clin. Gastroenterol. Hepatol. 3 (11), 1057–1065. doi:10.1016/s1542-3565(05)00184-9
Shahbazkhani, B., Sadeghi, A., Malekzadeh, R., Khatavi, F., Etemadi, M., Kalantri, E., et al. (2015). Non-Celiac Gluten Sensitivity Has Narrowed the Spectrum of Irritable Bowel Syndrome: A Double-Blind Randomized Placebo-Controlled Trial. Nutrients 7 (6), 4542–4554. doi:10.3390/nu7064542
Skodje, G. I., Sarna, V. K., Minelle, I. H., Rolfsen, K. L., Muir, J. G., Gibson, P. R., et al. (2018). Fructan, rather Than Gluten, Induces Symptoms in Patients with Self-Reported Non-celiac Gluten Sensitivity. Gastroenterology 154 (3), 529–539. e2. doi:10.1053/j.gastro.2017.10.0410.1053/j.gastro.2017.10.040
Spencer, M., Chey, W. D., and Eswaran, S. (2014). Dietary Renaissance in IBS: Has Food Replaced Medications as a Primary Treatment Strategy. Curr. Treat. Options. Gastro 12 (4), 424–440. doi:10.1007/s11938-014-0031-x
Spiller, R. C., Jenkins, D., Thornley, J. P., Hebden, J. M., Wright, T., Skinner, M., et al. (2000). Increased Rectal Mucosal Enteroendocrine Cells, T Lymphocytes, and Increased Gut Permeability Following Acute Campylobacter Enteritis and in post-dysenteric Irritable Bowel Syndrome. Gut 47 (6), 804–811. doi:10.1136/gut.47.6.804
Spiller, R. C. (2003). Postinfectious Irritable Bowel Syndrome1 1Abbreviations Used in This Paper: EC,_; IBS, Irritable Bowel Syndrome; PI, Postinfective. Gastroenterology 124 (6), 1662–1671. doi:10.1016/s0016-5085(03)00324-x
Staudacher, H. M., and Gibson, P. R. (2015). How Healthy Is a Gluten-free Diet. Br. J. Nutr. 114 (10), 1539–1541. doi:10.1017/S000711451500330X
UK biobank (2021) Comprehensive Genomic Analysis of Dietary Habits in UK Biobank Identifies Hundreds of Genetic Loci and Establishes Causal Relationships between Educational Attainment and Healthy Eating. online, Available at: https://www.biorxiv.org/content/10.1101/662239v1.full (accessed 6 Feb 2021)
Vazquez–Roque, M. I., Camilleri, M., Smyrk, T., Murray, J. A., Marietta, E., O'Neill, J., et al. (2013). A Controlled Trial of Gluten-free Diet in Patients with Irritable Bowel Syndrome-Diarrhea: Effects on Bowel Frequency and Intestinal Function. Gastroenterology 144 (5), 903–911. e3. doi:10.1053/j.gastro.2013.01.049
Verbanck, M., Chen, C.-Y., Neale, B., and Do, R. (2018). Detection of Widespread Horizontal Pleiotropy in Causal Relationships Inferred from Mendelian Randomization between Complex Traits and Diseases. Nat. Genet. 50 (5), 693–698. doi:10.1038/s41588-018-0099-7
Verdu, E. F., Armstrong, D., and Murray, J. A. (2009). Between Celiac Disease and Irritable Bowel Syndrome: The "No Man's Land" of Gluten Sensitivity. Am. J. Gastroenterol. 104 (6), 1587–1594. doi:10.1038/ajg.2009.188
Wu, Y., Murray, G. K., Byrne, E. M., Sidorenko, J., Visscher, P. M., and Wray, N. R. (2021). GWAS of Peptic Ulcer Disease Implicates Helicobacter pylori Infection, Other Gastrointestinal Disorders and Depression. Nat. Commun. 12 (1), 1146. doi:10.1038/s41467-021-21280-7
Xu, S., Fung, W. K., and Liu, Z. (2021). MRCIP: a Robust Mendelian Randomization Method Accounting for Correlated and Idiosyncratic Pleiotropy. Brief. Bioinformatics 22, bbab019. Advance online publication. doi:10.1093/bib/bbab019
Zar, S., Kumar, D., and Benson, M. J. (2001). Food Hypersensitivity and Irritable Bowel Syndrome. Aliment. Pharmacol. Ther. 15 (4), 439–449. doi:10.1046/j.1365-2036.2001.00951.x
Keywords: irritable bowel syndrome, gluten-free diet, mendelian randomization, causal association, genome-wide association studies
Citation: Sun Y, Chen X, Wang S, Deng M, Xie Y, Wang X, Chen J and Hesketh T (2021) Gluten-free Diet Reduces the Risk of Irritable Bowel Syndrome: A Mendelian Randomization Analysis. Front. Genet. 12:684535. doi: 10.3389/fgene.2021.684535
Received: 15 April 2021; Accepted: 18 October 2021;
Published: 09 November 2021.
Edited by:
Safarina G. Malik, Eijkman Institute for Molecular Biology, IndonesiaReviewed by:
Sharon Ross, National Cancer Institute (NCI), United StatesDing Ye, Zhejiang Chinese Medical University, China
Copyright © 2021 Sun, Chen, Wang, Deng, Xie, Wang, Chen and Hesketh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyan Wang, d2FuZ3hpYW95YW5AY3N1LmVkdS5jbg==; Jie Chen, bWVkX2NoZW5qaWVAemp1LmVkdS5jbg==
†These authors have contributed equally to this work and share the first authorship
 Yuhao Sun
Yuhao Sun Xuejie Chen2†
Xuejie Chen2† Shuyang Wang
Shuyang Wang Ying Xie
Ying Xie Xiaoyan Wang
Xiaoyan Wang Jie Chen
Jie Chen