- 1Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, University of Oxford, Oxford, United Kingdom
- 2National Institute for Health Research Oxford Comprehensive Biomedical Research Centre, Botnar Research Centre, Nuffield Orthopaedic Centre, Oxford, United Kingdom
- 3Wellcome Centre for Human Genetics, University of Oxford, Oxford, United Kingdom
- 4Division of Rheumatology and Clinical Immunology, Humanitas Clinical and Research Center - IRCCS, Rozzano, Italy
Background: Ankylosing Spondylitis (AS) is a common form of inflammatory spinal arthritis with a complex aetiology and high heritability, involving more than 100 genetic associations. These include several AS-associated single nucleotide polymorphisms (SNPs) upstream of RUNX3, which encodes the multifunctional RUNT-related transcription factor (TF) 3. The lead associated SNP rs6600247 (p = 2.6 × 10−15) lies ∼13kb upstream of the RUNX3 promoter adjacent to a c-MYC TF binding-site. The effect of rs6600247 genotype on DNA binding and chromosome looping were investigated by electrophoretic mobility gel shift assays (EMSA), Western blotting-EMSA (WEMSA) and Chromosome Conformation Capture (3C).
Results: Interrogation of ENCODE published data showed open chromatin in the region overlapping rs6600247 in primary human CD14+ monocytes, in contrast to the Jurkat T cell line or primary human T-cells. The rs6600247 AS-risk allele is predicted to specifically disrupt a c-MYC binding-site. Using a 50bp DNA probe spanning rs6600247 we consistently observed reduced binding to the AS-risk “C” allele of both purified c-MYC protein and nuclear extracts (NE) from monocyte-like U937 cells. WEMSA on U937 NE and purified c-MYC protein confirmed these differences (n = 3; p < 0.05). 3C experiments demonstrated negligible interaction between the region encompassing rs6600247 and the RUNX3 promoter. A stronger interaction frequency was demonstrated between the RUNX3 promoter and the previously characterised AS-associated SNP rs4648889.
Conclusion: The lead SNP rs6600247, located in an enhancer-like region upstream of the RUNX3 promoter, modulates c-MYC binding. However, the region encompassing rs6600247 has rather limited physical interaction with the promoter of RUNX3. In contrast a clear chromatin looping event between the region encompassing rs4648889 and the RUNX3 promoter was observed. These data provide further evidence for complexity in the regulatory elements upstream of the RUNX3 promoter and the involvement of RUNX3 transcriptional regulation in AS.
Introduction
Background
Ankylosing Spondylitis (AS) is a form of inflammatory spondyloarthritis predominantly affecting the axial skeleton, which is characterised pathologically by enthesitis (Bridgewood et al., 2020). Extra-skeletal manifestations are also common in AS; these include inflammation of the gut (ranging from low-grade sub-clinical inflammation of the terminal ileum to overt inflammatory bowel disease - IBD), skin (psoriasis), and uveal tract (Stolwijk et al., 2015; Rizzo et al., 2017). AS was one of the first complex diseases in which a specific genetic effect was identified when its strong association with the major histocompatibility complex (MHC) immune response gene HLA-B27 was described nearly 50 years ago (Brewerton et al., 1973) (Schlosstein et al., 1973). However, it is clearly polygenic (Brown et al., 1997); even the MHC association is attributable to several alleles at more than one locus (Cortes et al., 2015) and more than 100 non-MHC genetic associations have now been suggested by genome-wide association studies (Burton et al., 2007; Reveille et al., 2010; Evans et al., 2011; Cortes et al., 2013). Shared genetic susceptibility factors undoubtedly contribute to the excess occurrence of psoriasis, IBD and uveitis not only in individuals with AS but also their relatives (Ellinghaus et al., 2016) (Robinson et al., 2016). One of the strongest non-HLA associations with AS is with the RUNX3 (Runt-related transcription factor (TF) 3) locus. RUNX3 is involved in T-cell function and plays a key role in the development of CD8+ T-cells (Egawa et al., 2007). It also influences many other cells, including helper T-cells, innate lymphoid, tissue resident, mucosa and gut cells (Ebihara et al., 2015; Behr et al., 2018). We have recently demonstrated that AS-associated non-coding single nucleotide polymorphisms (SNPs) in an enhancer-like region upstream of RUNX3 affect the binding of different factors: in particular the repressive nucleosome remodelling and deacetylase (NuRD) complex binds preferentially to the risk allele, while conversely interferon regulatory factor (IRF) five to the protective allele (Vecellio et al., 2021). However, the functional effects of these changes on gene transcription are still to be precisely determined. Our earlier observations were made in T-cells, but here we describe some of the functional effects of the lead AS-associated SNP in the vicinity of RUNX3 (rs6600247, p = 2.6 × 10−15 (Cortes et al., 2013) that are more obvious in CD14+ monocyte-like cells than CD8+ T-cells. First, we evaluate the chromatin landscape surrounding rs6600247 using the ENCODE database (https://genome.ucsc.edu/ENCODE/). Second, we demonstrate differential allelic binding of rs6600247 to the c-MYC TF. Finally, we investigate the chromosomal architecture and physical interactions between AS-associated sequences in the enhancer-like region upstream of RUNX3 and its promoter, showing a probable role of chromosome looping in the regulation of RUNX3.
Methods
Genotyping
DNA was extracted using the Qiagen AllPrep DNA/RNA Mini Kit (Qiagen Ltd., Manchester, United Kingdom) and genotyped for rs6600247 using TaqMan SNP assay (custom order by Life Technologies, Paisley, United Kingdom), for the cells (obtained by the buffy coat) used in the functional studies.
In Silico Investigation
We used the UCSC genome browser build hg19 and the Roadmap database [https://genome.ucsc.edu/ENCODE/] to investigate the epigenetic landscape of rs6600247 upstream of the RUNX3 promoter, which is strongly associated with AS (p = 4.2 × 10–15) (Cortes et al., 2013). Histone modifications and GeneHancer (a database of human regulatory elements and their inferred target genes) tracks were selected to evaluate regulatory elements and chromosome looping between promoters and enhancer regions (Fishilevich et al., 2017).
Cell Lines, Culture and Primary Human Cell Isolation
Blood samples were obtained from AS patients with ethical approval (COREC 06/Q1606/139) and informed patient consent. CD8+ T-cells and CD14+ monocytes were isolated from AS patients’ peripheral blood mononuclear cells (PBMCs) using a CD8+ T-cell or a CD14+ monocyte isolation kit (Miltenyi, Bisley, Surrey, United Kingdom), respectively. Jurkat, U937, CD8+ and CD14+ cells were resuspended at 1×106/ml in pre-warmed Roswell Park Memorial Institute medium supplemented with 10% fetal bovine serum, penicillin/streptomycin and l-glutamine, and rested overnight. Cells were then harvested for experiments.
Electrophoretic Mobility Gel Shift Assay
The impact of rs6600247, which lies in a c-MYC binding-site (Figure 2A), was assessed by EMSA. We designed DNA probes including either the protective T or the AS-risk variant C to evaluate the disruption of a c-MYC consensus motif. The DNA probes used in EMSAs (50-bp single-stranded biotinylated DNA probe incorporating rs6600247) were mixed and annealed at room temperature for 1 h. Probes were then incubated for 20 min with nuclear extracts (NE) obtained either from primary CD8+ T-cells or a monocyte cell line from histiocytic lymphoma (U937) stimulated with phorbol-12-myristate-13-acetate (PMA). The sequences of the synthetic single-stranded oligonucleotides are listed below:
C* s (sense): 5′-CTCCATGACGCAATTTGGGCTCCGTTATGAGTCAGCTCAAGTAA-3′; T* s: 5′-CTCCATGACGCAATTTGGGCTCTGTTATGAGTCAGCTCAAGTAA-3′; C* as (antisense): 5′-TTACTTGAGCTGACTCATAACGGAGCCCAAATTGCGTCATGGAG-3′; T* as: 5′-TTACTTGAGCTGACTCATAACAGAGCCCAAATTGCGTCATGGAG-3′.
(Underlined base highlights the position of rs6600247).
Western Blotting - Electrophoretic Mobility Gel Shift Assay
DNA probes as for EMSA (above) were incubated with nuclear extract obtained from U937, CD8+ T-cells or purified c-MYC human recombinant protein (Abcam, ab169901 Cambridge, United Kingdom) as previously described (Allen et al., 2017) and separated on DNA retardation gels at 100 V on ice. The samples were transferred on to nitrocellulose membranes for Western blotting (WB), then blocked with 5% milk in Tris Buffer Saline +0.1% Tween (TBST) for 1 h at room temperature (RT) before incubating overnight at 4°C with the primary antibody for c-MYC (Santa Cruz Biotechnology sc-40, Dallas, Texas United States). Secondary goat anti-rabbit antibody (1:10,000 dilution) was added (1 h RT) and the membranes washed before Horse Radish peroxidase substrate (Thermo Fisher Scientific, Waltham, Massachusetts, United States) added for imaging. ImageJ (NIH) was used for quantifying WEMSA bands (Schneider et al., 2012).
Chromosome Conformation Capture
Chromosome conformation capture (3C) was performed as previously described (Miele et al., 2006). Briefly, libraries were prepared as follows: 1.5 × 107 of U937 or Jurkat cells were cross-linked with formaldehyde at 1% of the final volume. Glycine [0.125M] was used to quench cross-linking and cells were lysed in cold lysis buffer on ice using a Dounce homogenizer (Sigma Aldrich, Gillingham, United Kingdom). Cells were resuspended in specific restriction enzyme buffer (10 μL were kept as undigested control). The remaining samples were digested overnight at 37°C with 500 units of Sac1 (New England Biolabs, Hitchin, United Kingdom). Digestion was stopped by the addition of 10% sodium dodecyl sulfate incubated at 65°C for 30 min. T4 ligase (Ambion, Thermo Fisher Scientific, Waltham, Massachusetts, United States) was used to perform ligation for 4 h at 16°C. Proteinase K was added prior to reversal of cross-linking at 65°C overnight. Proteinase K was added to the undigested and digested controls saved earlier. DNA was purified using phenol-chloroform extraction, followed by ethanol precipitation. 3C template was resuspended in 500 ul H2O, while undigested and digested controls in 50 ul. The quality of the chromatin samples was assessed on agarose gels. Bacterial Artificial Chromosome (BAC) preparations were performed similarly as genomic controls. 3C PCR primers were designed along the same strand and same orientation to accomplish specific amplification across 3C ligation junctions. Full list of primers is available in Supplementary Table S1 and their genomic position relative to RUNX3 is shown in Figure 3
We interrogated a genomic region upstream the RUNX3 distal promoter, including few AS-associated SNPs in U937 (monocyte-like) and Jurkat (T-lymphocyte-like) cell lines. The bait was placed at the distal promoter (P2) with amplification primers at the AS-associated SNPs rs6600247 and rs4648889 along with three intergenic regions.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA from CD8+ and CD14+ cells was isolated with TRIzol (Invitrogen, Paisley, United Kingdom) and reverse transcribed with Superscript III (Invitrogen, Thermo Fisher Scientific, 168 Third Avenue, Waltham, Massachusetts, United States) to synthesise cDNA as previously described (Vecellio et al., 2018). The specific primers were: RUNX3 sense (s): 5′-ACTCAG CAC CAC AAG CCA CT-3′; RUNX3 antisense (as): 5′-GTC GGA GAA TGG GTT CAG TT-3′. Quantitative PCR was performed in triplicate and the 2−ΔCt method was used to calculate the expression of RUNX3 relative to β-actin (ID Assay qHsaCED0036269, Bio-Rad Laboratories, Kidlington, United Kingdom).
Historical Controls and RUNX3 Expression
RUNX3 transcription in AS cases and controls was evaluated from previously published data derived from RNA-seq in PBMCs from 72 AS cases and 62 healthy controls and stratified for rs6600247 (Li et al., 2017).
Results
Genomic Landscape Interrogation Suggests a Regulatory Role for rs6600247
Figure 1A shows the genomic landscape at the RUNX3 locus, with the lead AS-associated SNP rs6600247 lying ∼13kb upstream of the distal promoter while the regulatory SNP rs4648889 is physically closer to the promoter. SNP rs6600247 is situated within a region of open chromatin, defined by a peak for dnase I hypersensitvity (DHS - indicative of regions of open chromatin) (Figure 1B), and a peak of H3K4Me1 histone modification. This sequence also binds the transcription factor c-MYC (ENCODE Factorbook (http://www.factorbook.org/human/chipseq/tf/) (Figure 1B). Taken together, these data suggest an enhancer-type element surrounding rs6600247, so we sought to determine a regulatory role of this SNP. The DHS peak overlapping rs6600247 is seen specifically in CD14+ monocytes (Figure 1B). For this reason, we conducted our functional experiments in U937 cells, a pro-monocytic, human myeloid leukaemia cell line, exhibiting monocyte-like features.
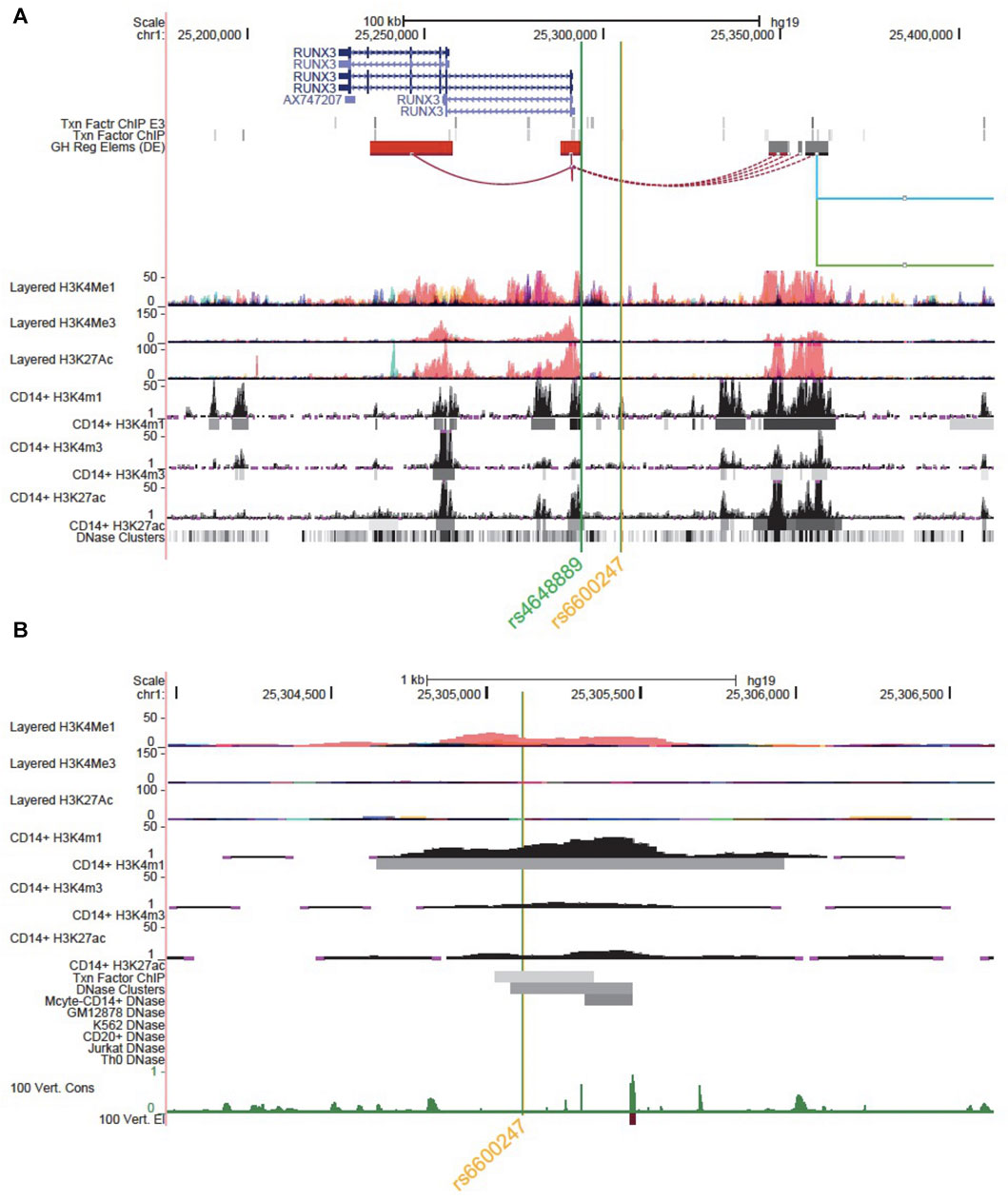
FIGURE 1. Genomic landscape interrogation suggests a regulatory role for rs6600247. UCSC Genome Browser analysis of the RUNX3 locus at chr1:25,177,797–25,409,807. The green and orange vertical lines show the location of SNP rs4648889 and rs6600247. RUNX3 gene location (blue) is from the UCSC genes database. Two tracks show transcription factor binding by ChIP (Txn Facto ChIP E3 and Txn Factor ChIP). GeneHancer track shows human regulatory elements and their inferred target genes. ENCODE layered H3K27ac, H3K4Me1 and H3K4me3 tracks (from 7 cell lines) show enrichment of these marks, indicating promoter and enhancer regions. CD14+ H3K4me1, CD14+ H3K4me3 and CD14+ H3K27ac show enrichment and peaks called specifically in CD14+ monocytes (ENCODE). Dnase clusters show DNase-I hypersensitivity clusters (ENCODE); (B) Zoomed Genome Browser view (chr1:25,303,971–25,306,641) upstream of the promoter of RUNX3 showing a peak for H3K4Me1 enrichment overlapping rs6600247 (vertical line; Layered tracks and ENCODE histone tracks as in (A). Additional datasets displayed are ENCODE transcription factor Chip-seq peaks and DNaseI HS peaks that directly overlap rs6600247, ENCODE dnase I HS for CD14+ monocytes and vertebrate conservation.
rs6600247 AS-Risk C Allele Alters c-MYC Binding to Deoxyribonucleic Acid
We analysed the DNA sequence at rs6600247 and found that the SNP lies within a c-MYC consensus binding motif (Figure 2A). We hypothesised that binding of c-MYC protein to a DNA sequence containing the risk allele C would be reduced. The results of EMSA assessing the relative c-MYC protein binding to the C or T alleles are shown in Figure 2. We first incubated probes with recombinant c-MYC purified protein, and observed a specific DNA/protein complex with both alleles but markedly less to the AS-risk allele C than the protective T allele. (Figure 2B; lane 3-4, n = 3). We then incubated the same probes with NE from U937 (monocyte-like cells) and observed a major protein/DNA complex binding to the protective T allele, but none with the C allele (Figure 2C, lanes 3-4, n = 3). In both cases, successful competition with a 100-fold excess of unlabelled probe confirmed the specificity of the complex (Figure 2B, lane 5-6 and Figure 2C lane 5–6). We next used WEMSA to quantitate the relative binding of c-MYC to each allele of rs6600247. Markedly less c-MYC enrichment was seen with the C risk vs T allele using either c-MYC purified protein or U937 NE (Figures 2D,E, relative band intensities p = 0.01 and p = 0.05, respectively, two-sample t test). We also repeated these experiments using CD8+ T-cells and Jurkat NE, showing no differential binding between the two alleles (Supplementary Figure S1).
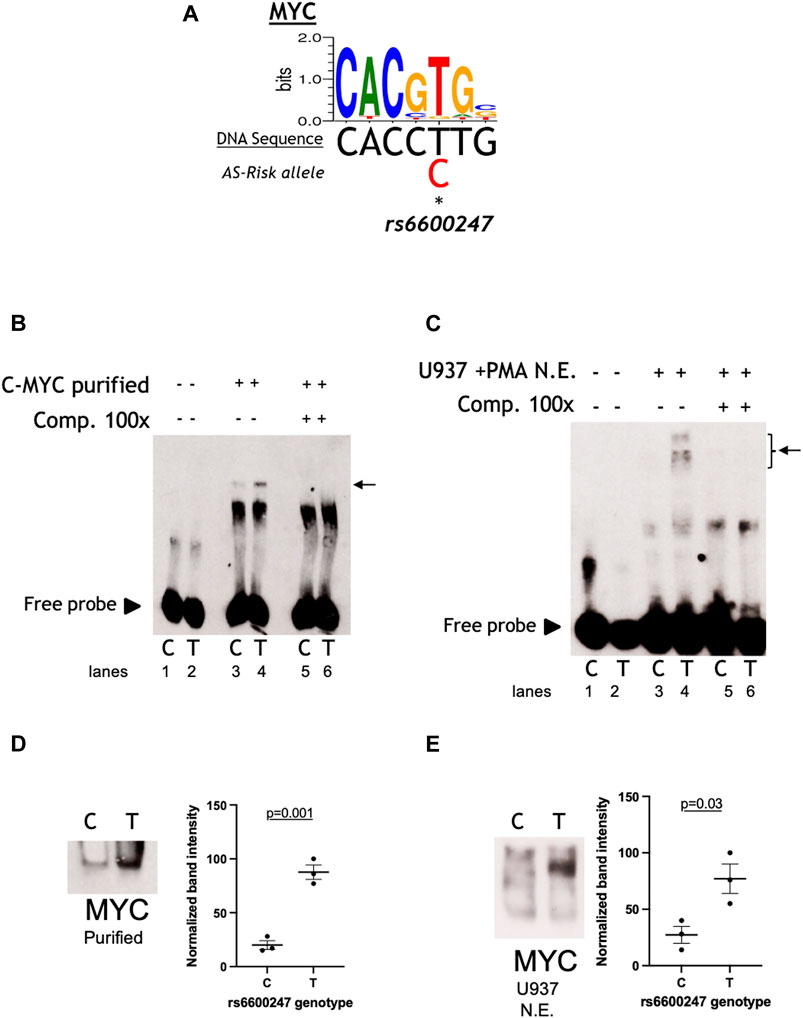
FIGURE 2. rs6600247 risk allele affects C-Myc binding. (A) C-Myc binding motif analyzed using the MEME program and the location of rs6600247 risk allele; (B) EMSA using c-MYC purified protein with or without specific competitor (Comp 100x), n = 4, allele (C or T) of rs6600247 included in the 50bp biotinylated double-stranded DNA probe is given below the image; horizontal arrow indicates specific protein-DNA complex formation; (C) EMSA using nuclear extract (N.E.) from U937 cells stimulated with phorbol 12-myristate 13-acetate (PMA) with or without specific competitor (Comp 100x), n = 4; rs6600247 allele and complex formation indicated as in (B,D) WEMSA using C-Myc purified protein and blotted with an antibody against C-Myc; (E) WEMSA using U937 nuclear extract and blotted with an antibody against C-Myc. The blot is representative of n = 3 experiments. Binding in (D,E) was quantified using ImageJ software and is representative of three different experiments, demonstrating that the risk allele for rs6600247 shows fewer binding properties for C-Myc.
The RUNX3 Promoter Interacts With the rs4648889 Region Rather Than rs6600247
We used 3C to test plausible chromosome looping interactions between the AS-associated SNP rs6600247 and the RUNX3 distal promoter. Figure 3A shows the RUNX3 genomic region interrogated, the location of the primers and Sac1 restriction sites. Baits were designed to capture Sac1 fragments containing rs6600247, three intergenic fragments with H3K4me1 enrichment, and additionally with a previously-studied AS-associated SNP rs4648889. There was very low interaction frequency between rs6600247 and the distal RUNX3 promoter, either in U937 or Jurkat cells. A stronger interaction frequency was observed between the distal promoter and the region encompassing the AS-associated SNP rs4648889 (Figure 3B) confirming its functional role.
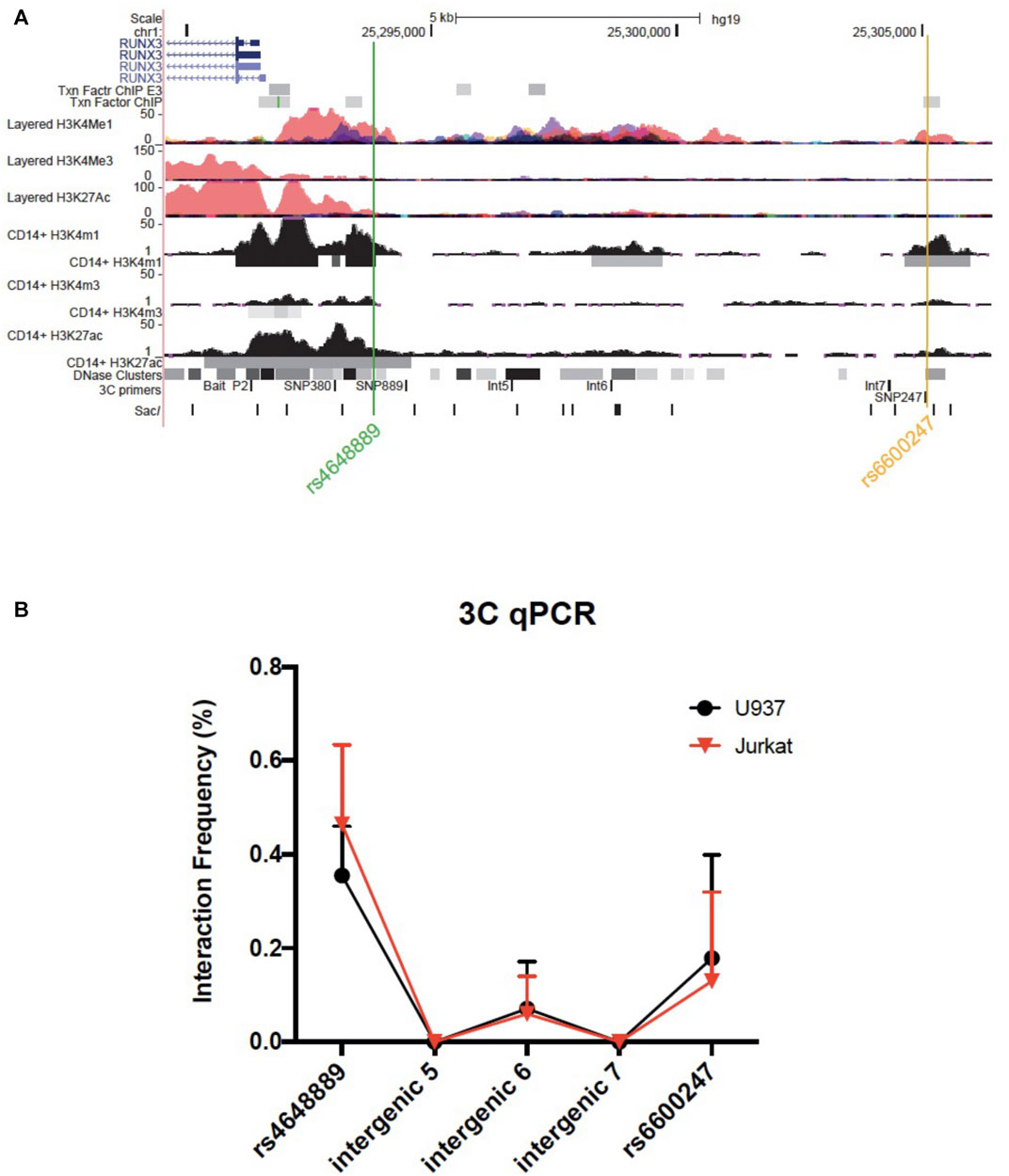
FIGURE 3. Chromosome looping investigation demonstrates interaction between the RUNX3 promoter and region encompassing rs4648889 rather than rs6600247. (A) Location of the RUNX3 genomic region chr1:25,289,567–25,306,400. Tracks shown as in Figure 1, with the addition of 3C-qPCR primers and SacI enzyme cutting sites. Bait fragment is located at RUNX3 distal promoter (P2); AS-associated SNPs primers used in these experiments are named as follows: SNP889 (for rs4648889), Int5, Int6, Int7 (for intergenic regions 5, 6 and 7) and SNP247 (for rs6600247); (B) Results of the 3C-qPCR analysis showing Increased relative interaction frequency between RUNX3 P2 and the region encompassing rs4648889, with a modest interaction with the rs6600247 region. Theses interactions were seen in both U937 (red) and Jurkat (black) cell lines.
rs6600247 Genotype has No Effect on RUNX3 Expression
Primary CD14+ monocytes and CD8+ T-cells from AS patients were used to evaluate RUNX3 mRNA expression stratified on rs6600247 genotype (n = 5 each genotype) (Figures 4A,B). There was a non-significant trend for lower expression in CD14+ monocytes with the AS-risk CC genotype compared to protective TT and heterozygous TC genotypes (TT vs CC: 4.6 ± 1.8 vs 2.0 ± 0.4; TT vs CT: 4.6 ± 1.8 vs 1.8 ± 0.2; CC vs CT: 2.0 ± 0.4 vs 1.8 ± 0.2, results are expressed as mean ± standard error mean). We also analysed historical RNA-seq data (Li et al., 2017) obtained from AS case PBMCs measuring RUNX3 mRNA expression, stratified on rs6600247: there was no apparent influence from this SNP on RUNX3 expression (Figure 4C).
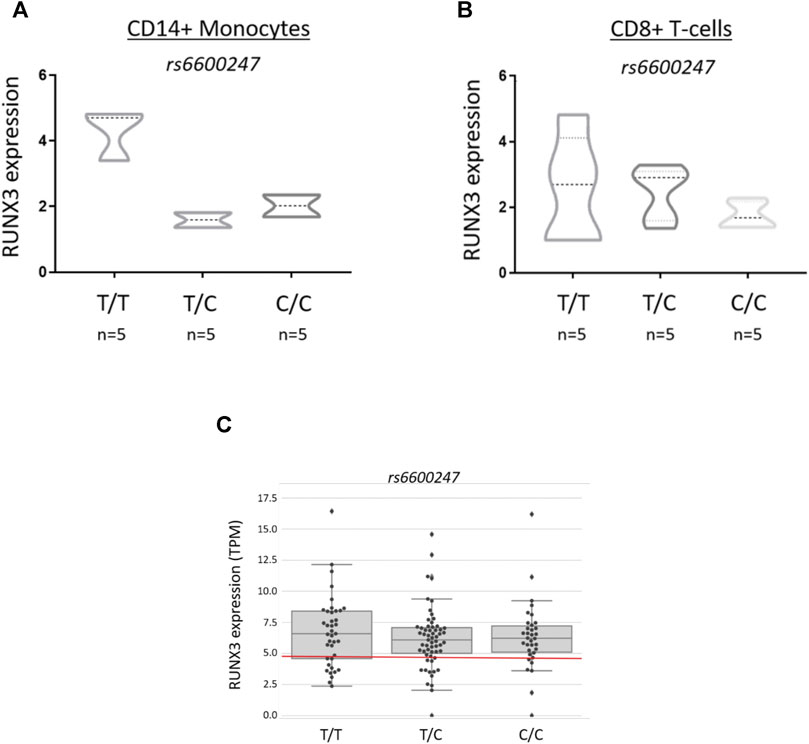
FIGURE 4. rs6600247 genotype shows no regulatory effect on RUNX3 expression RUNX3 expression levels measured by qRT-PCR in freshly isolated (A) CD14+ monocytes and (B) CD8+ T-cells from 15 AS patients stratified according to rs6600247 genotype. Statistical analysis performed with Welch’s two-sample t test; (C) Expression of RUNX3 in an historical RNA-seq dataset (Li et al., 2017) obtained from PBMCs, stratified on rs6600247.
Discussion
In this study, we have demonstrated that the lead AS-associated SNP in the RUNX3 region, rs6600247, affects the binding of c-MYC to the region of DNA 13 kb upstream of the RUNX3 promoter, which lies in a region of open chromatin in CD14+ monocytes. Further, this region showed enrichment for H3K4Me1 modification in the absence of H3K4me3 or H3K27ac enrichment, suggesting a weak or poised enhancer (Gasperini et al., 2020). Although GWAS have identified hundreds of genetic variants associated with AS (Cortes et al., 2013), only a very small portion of these have been investigated to define causal variants. Cell type and stimulation conditions must be taken carefully in consideration in identifying causal SNPs, as both impact on chromatin interaction and gene regulation (Shi et al., 2021).
Recent findings have shown that RUNX3 is highly expressed in monocytes where it has a role in transcriptional repression, metabolic regulation, and in tuning the function of CD14+ monocytes. (Puig-Kröger et al., 2010; Estecha et al., 2012). Expression of RUNX3 has been found also in CD11c + mature intestinal macrophages, suggesting a role for this TF in macrophage maturation (Corbin et al., 2020). Further, both RUNX3 and another TF, ID2 (Inhibitor of DNA binding 2), are required for the differentiation of epidermal Langerhans cells from monocytes (Fainaru et al., 2004).
The interaction between RUNX3 and c-MYC has previously been investigated in T-cell lymphoma (Selvarajan et al., 2017). Double immunofluorescence revealed co-localization of both proteins in the tumour nuclei. In addition, several binding-sites for c-MYC were identified in the RUNX3 enhancer region. Additional evidence for this interaction stems from colorectal cancer studies where upregulation of RUNX3 by Bone Morphogenetic Protein (BMP) reduces c-MYC expression, thereby exerting c-MYC tumour-suppressor activity (Lee et al., 2010). Recently, it has been demonstrated that two super-enhancers located at 59 and 70 kb upstream of the RUNX3 transcription start site are required for both RUNX3 and MYC expression and function (Hosoi et al., 2021). Other studies also indicate a key role of c-MYC in monocyte/macrophage activation, as it is involved in the regulation of different alternative activation genes (Pello et al., 2012).
Our EMSA/WEMSA experiments confirmed c-MYC binding at the rs6600247 locus, with the AS-risk allele disrupting the binding motif and consequently reducing formation of the c-MYC-DNA complex. Altogether, these observations are consistent with the hypothesis that c-MYC can bind the RUNX3 promoter and/or regulatory elements upstream of the promoter thereby potentially playing a role in the regulation of RUNX3. The processes involved in transcriptional regulation are complex and this finding does not exclude the possibility of other TFs being involved.
The genome is organized in a very dynamic way and TFs mediate chromosome loops to bring enhancers and promoters together (de Wit and de Laat, 2012; Palstra and Grosveld, 2012). 3C and related techniques are the classic approach to demonstrating interactions between target genes and enhancers or enhancer-like regions. Here we demonstrate the presence of chromatin loop between a SNP overlapping a regulatory region and the distal promoter of RUNX3 using 3C followed by qPCR. This method has been used extensively to demonstrate interactions between various regulatory regions in different cell types and it allows one to quantitate the interaction frequency (Sati and Cavalli, 2017; McCord et al., 2020).
We accept that recent findings highlight the fact that contact frequencies from 3C assays sometimes do not correspond to 3D proximity (Williamson et al., 2014), but taken together with the functional data presented here and other recently published findings (Vecellio et al., 2016; Vecellio et al., 2018; Vecellio et al., 2021) we are confident in our results. However, we are aware that other higher throughput techniques have been developed (eg. 4C, 5C, Hi-C) that might give a more general overview of the regulation of the RUNX3 locus and the genetic interactions of the SNPs in this region. These will be incorporated into our ongoing genome-wide studies of chromatin interactions and the regulatory effects of AS-associated genetic variants.
Here, we have demonstrated physical interactions between the distal promoter of RUNX3 and rs4648889 SNP, which we have previously functionally characterized (Vecellio et al., 2016; Vecellio et al., 2021). Conversely, there was a very low interaction frequency with rs6600247 that suggests no functional role for this SNP in chromosome looping in the particular context of CD8+ T-cells or monocytes. As previously shown, in a ∼15 kb linkage disequilibrium (LD) block upstream the promoter of RUNX3, there are 22 RUNX3 SNPs that are strongly associated with AS (p ≤ 10–14) (Vecellio et al., 2016). The SNP analysed in this work, rs6600247 (p = 1.3 × 10–14), is in complete LD with rs4648889 (∼2 kb upstream of the RUNX3 promoter). Conditional analysis established the primacy of the rs4648889 association with AS at RUNX3 (Vecellio et al., 2016), while not excluding additional functional roles for other SNPs in LD with it. The functional experiments described here and in previous publications (Vecellio et al., 2016; Vecellio et al., 2018; Vecellio et al., 2021) represent an approach to identifying more precisely which SNPs in this LD block actually have a functional impact on the RUNX3 regulatory element and its role in the pathogenesis of AS.
Clearly the presence of an enhancer-promoter loop alone does not ensure activation of a target gene but it provides a platform where transcription factors can bind and regulate gene/s (Espinola et al., 2021; Ing-Simmons et al., 2021). Here, we have confirmed that the genomic regulatory element upstream of the RUNX3 promoter has potentially important cell-type-specific functional effects. We show that the rs6600247 AS-risk allele affects c-MYC binding in monocytes, suggesting that c-MYC/RUNX3 modulated pathways could have a role in the pathophysiology of AS. Nevertheless, the region encompassing rs6600247 has no significant physical interaction with the distal RUNX3 promoter, thereby confirming that rs4648889 appears to be the cardinal genetic variant associated with AS at the RUNX3 locus.
Further studies are required to identify additional higher order chromatin interactions at this locus. For example, HiChIP has been used to delineate promoter-enhancer interactions in keratinocytes and CD8+ T-cell lines exploring psoriasis and psoriatic arthritis disease-associated SNPs and similar methods could be explored in AS (Shi et al., 2020a; Shi et al., 2020b). It is also critical that cell-type and -context specificity are crucial for TF binding and activity, and can also influence chromatin looping data (Nancy et al., 2021). While we have presented here and elsewhere evidence for the involvement of CD8+ T-cells and monocytes in the pathogenesis of AS other cell types must also be considered. These include numerous types found in bone and cartilage, various other components of the immune system and also cells in the gut where chronic low-grade inflammation is a feature of AS in around two-thirds of cases (Ciccia et al., 2016; Shao et al., 2021). In the future targeted RUNX3 enhancer element genomic editing strategies could be used to elucidate their effects on RUNX3 (and other gene) expression and downstream cellular signaling.
In conclusion, this work provides new insights into the complex transcriptional regulation of RUNX3 and the role that AS-associated SNPs may play in this process. We highlight the importance of functional studies in determining which disease associated SNPs are primarily involved in the pathogenesis of such diseases and the importance of interrogating their role in the appropriate cellular context.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by COREC 06/Q1606/139 and OXREC B 07/Q1605/35. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CC, MV, JK and BW conceived and designed the experiments. MV and CC performed the experiments. MV, CC and CD analysed the data. MV, CC, JK and BW drafted the manuscript, and all the authors revised the final version prior submission.
Funding
MV work was supported by Versus Arthritis grant 21428. CC work was supported by Versus Arthritis grant 22053. CD work was supported by Versus Arthritis grant 22198. JK work was supported by the NIHR Oxford Biomedical Research Centre. BW work was supported by the NIHR Oxford Biomedical Research Centre and by the NIHR Oxford Musculoskeletal Biomedical Research Unit. The study received support from the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC) (PB). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.741867/full#supplementary-material
References
Allen, E. K., Randolph, A. G., Bhangale, T., Dogra, P., Ohlson, M., Oshansky, C. M., et al. (2017). SNP-mediated Disruption of CTCF Binding at the IFITM3 Promoter Is Associated with Risk of Severe Influenza in Humans. Nat. Med. 23 (8), 975–983. Epub 2017/07/18. doi:10.1038/nm.4370
Behr, F. M., Chuwonpad, A., Stark, R., and van Gisbergen, K. P. J. M. (2018). Armed and Ready: Transcriptional Regulation of Tissue-Resident Memory CD8 T Cells. Front. Immunol. 9, 1770. Epub 2018/08/23. doi:10.3389/fimmu.2018.01770
Brewerton, D. A., Hart, F. D., Nicholls, A., Caffrey, M., James, D. C. O., and Sturrock, R. D. (1973). Ankylosing Spondylitis and HL-A 27. The Lancet 301 (7809), 904–907. Epub 1973/04/28. doi:10.1016/s0140-6736(73)91360-3
Bridgewood, C., Sharif, K., Sherlock, J., Watad, A., and McGonagle, D. (2020). Interleukin‐23 Pathway at the Enthesis: The Emerging story of Enthesitis in Spondyloarthropathy. Immunol. Rev. 294 (1), 27–47. Epub 2020/01/21. doi:10.1111/imr.12840
Brown, M. A., Kennedy, L. G., MacGregor, A. J., Darke, C., Duncan, E., Shatford, J. L., et al. (1997). Susceptibility to Ankylosing Spondylitis in Twins the Role of Genes, HLA, and the Environment. Arthritis Rheum. 40 (10), 1823–1828. Epub 1997/10/23. doi:10.1002/art.1780401015
Burton, P. R., Clayton, D. G., Burton, P. R., Clayton, D. G., Cardon, L. R., Craddock, N., et al. (2007). Association Scan of 14,500 Nonsynonymous SNPs in Four Diseases Identifies Autoimmunity Variants. Nat. Genet. 39 (11), 1329–1337. Epub 2007/10/24. doi:10.1038/ng.2007.17
Ciccia, F., Rizzo, A., and Triolo, G. (2016). Subclinical Gut Inflammation in Ankylosing Spondylitis. Curr. Opin. Rheumatol. 28 (1), 89–96. Epub 2015/11/26. doi:10.1097/bor.0000000000000239
Corbin, A. L., Gomez-Vazquez, M., Berthold, D. L., Attar, M., Arnold, I. C., Powrie, F. M., et al. (2020). IRF5 Guides Monocytes toward an Inflammatory CD11c+ Macrophage Phenotype and Promotes Intestinal Inflammation. Sci. Immunol. 5 (47). doi:10.1126/sciimmunol.aax6085
Cortes, A., Cortes, A., Hadler, J., Pointon, J. P., Robinson, P. C., Karaderi, T., et al. (2013). Identification of Multiple Risk Variants for Ankylosing Spondylitis through High-Density Genotyping of Immune-Related Loci. Nat. Genet. 45 (7), 730–738. Epub 2013/06/12. doi:10.1038/ng.2667
Cortes, A., Pulit, S. L., Leo, P. J., Pointon, J. J., Robinson, P. C., Weisman, M. H., et al. (2015). Major Histocompatibility Complex Associations of Ankylosing Spondylitis Are Complex and Involve Further Epistasis with ERAP1. Nat. Commun. 6, 7146. Epub 2015/05/23. doi:10.1038/ncomms8146
de Wit, E., and de Laat, W. (2012). A Decade of 3C Technologies: Insights into Nuclear Organization. Genes Dev. 26 (1), 11–24. Epub 2012/01/05. doi:10.1101/gad.179804.111
Ebihara, T., Song, C., Ryu, S. H., Plougastel-Douglas, B., Yang, L., Levanon, D., et al. (2015). Runx3 Specifies Lineage Commitment of Innate Lymphoid Cells. Nat. Immunol. 16 (11), 1124–1133. Epub 2015/09/29. doi:10.1038/ni.3272
Egawa, T., Tillman, R. E., Naoe, Y., Taniuchi, I., and Littman, D. R. (2007). The Role of the Runx Transcription Factors in Thymocyte Differentiation and in Homeostasis of Naive T Cells. J. Exp. Med. 204 (8), 1945–1957. Epub 2007/07/25. doi:10.1084/jem.20070133
Ellinghaus, D., Jostins, L., Jostins, L., Spain, S. L., Cortes, A., Bethune, J., et al. (2016). Analysis of Five Chronic Inflammatory Diseases Identifies 27 New Associations and Highlights Disease-specific Patterns at Shared Loci. Nat. Genet. 48 (5), 510–518. Epub 2016/03/15. doi:10.1038/ng.3528
Espinola, S. M., Götz, M., Bellec, M., Messina, O., Fiche, J.-B., Houbron, C., et al. (2021). Cis-regulatory Chromatin Loops Arise before TADs and Gene Activation, and Are Independent of Cell Fate during Early Drosophila Development. Nat. Genet. 53 (4), 477–486. Epub 2021/04/03. doi:10.1038/s41588-021-00816-z
Estecha, A., Aguilera-Montilla, N., Sánchez-Mateos, P., and Puig-Kröger, A. (2012). RUNX3 Regulates Intercellular Adhesion Molecule 3 (ICAM-3) Expression during Macrophage Differentiation and Monocyte Extravasation. PloS one 7 (3), e33313. Epub 2012/04/06. doi:10.1371/journal.pone.0033313
Evans, D. M., Spencer, C. C., Pointon, J. J., Su, Z., Harvey, D., Kochan, G., et al. (2011). Interaction between ERAP1 and HLA-B27 in Ankylosing Spondylitis Implicates Peptide Handling in the Mechanism for HLA-B27 in Disease Susceptibility. Nat. Genet. 43 (8), 761–767. Epub 2011/07/12. doi:10.1038/ng.873
Fainaru, O., Woolf, E., Lotem, J., Yarmus, M., Brenner, O., Goldenberg, D., et al. (2004). Runx3 Regulates Mouse TGF-β-Mediated Dendritic Cell Function and its Absence Results in Airway Inflammation. Embo J. 23 (4), 969–979. Epub 2004/02/07. doi:10.1038/sj.emboj.7600085
Fishilevich, S., Nudel, R., Rappaport, N., Hadar, R., Plaschkes, I., Iny Stein, T., et al. (2017). GeneHancer: Genome-wide Integration of Enhancers and Target Genes in GeneCards. Database : J. Biol. databases curation 2017, bax028. doi:10.1093/database/bax028
Gasperini, M., Tome, J. M., and Shendure, J. (2020). Towards a Comprehensive Catalogue of Validated and Target-Linked Human Enhancers. Nat. Rev. Genet. 21 (5), 292–310. Epub 2020/01/29. doi:10.1038/s41576-019-0209-0
Hosoi, H., Niibori-Nambu, A., Nah, G. S. S., Bahirvani, A. G., Mok, M. M. H., Sanda, T., et al. (2021). Super-enhancers for RUNX3 Are Required for Cell Proliferation in EBV-Infected B Cell Lines. Gene 774, 145421. Epub 2021/01/15. doi:10.1016/j.gene.2021.145421
Ing-Simmons, E., Vaid, R., Bing, X. Y., Levine, M., Mannervik, M., and Vaquerizas, J. M. (2021). Independence of Chromatin Conformation and Gene Regulation during Drosophila Dorsoventral Patterning. Nat. Genet. 53 (4), 487–499. Epub 2021/04/03. doi:10.1038/s41588-021-00799-x
Lee, C. W. L., Ito, K., and Ito, Y. (2010). Role of RUNX3 in Bone Morphogenetic Protein Signaling in Colorectal Cancer. Cancer Res. 70 (10), 4243–4252. Epub 2010/05/06. doi:10.1158/0008-5472.can-09-3805
Li, Z., Haynes, K., Pennisi, D. J., Anderson, L. K., Song, X., Thomas, G. P., et al. (2017). Epigenetic and Gene Expression Analysis of Ankylosing Spondylitis-Associated Loci Implicate Immune Cells and the Gut in the Disease Pathogenesis. Genes Immun. 18 (3), 135–143. Epub 2017/06/18. doi:10.1038/gene.2017.11
McCord, R. P., Kaplan, N., and Giorgetti, L. (2020). Chromosome Conformation Capture and beyond: Toward an Integrative View of Chromosome Structure and Function. Mol. Cel. 77 (4), 688–708. Epub 2020/02/01. doi:10.1016/j.molcel.2019.12.021
Miele, A., Gheldof, N., Tabuchi, T. M., Dostie, J., and Dekker, J. (2006). Mapping Chromatin Interactions by Chromosome Conformation Capture. Current. Protocols. in. Mol. Biol. 21, 21.11 doi:10.1002/0471142727.mb2111s74
Nancy, Z., Yan, L., Hui, S., Paul, B., and Liye, C. (2021). From the Genetics of Ankylosing Spondylitis to New Biology and Drug Target Discovery. Front. Immunol. 12, 624632. Epub 2021/03/09. doi:10.3389/fimmu.2021.624632
Palstra, R.-J., and Grosveld, F. (2012). Transcription Factor Binding at Enhancers: Shaping a Genomic Regulatory Landscape in Flux. Front. Gene 3, 195. Epub 2012/10/13. doi:10.3389/fgene.2012.00195
Pello, O. M., De Pizzol, M., Mirolo, M., Soucek, L., Zammataro, L., Amabile, A., et al. (2012). Role of C-MYC in Alternative Activation of Human Macrophages and Tumor-Associated Macrophage Biology. Blood 119 (2), 411–421. Epub 2011/11/10. doi:10.1182/blood-2011-02-339911
Puig-Kröger, A., Aguilera-Montilla, N., Martínez-Nuñez, R., Domínguez-Soto, A., Sánchez-Cabo, F., Martín-Gayo, E., et al. (2010). The Novel RUNX3/p33 Isoform Is Induced upon Monocyte-Derived Dendritic Cell Maturation and Downregulates IL-8 Expression. Immunobiology 215 (9-10), 812–820. Epub 2010/07/10. doi:10.1016/j.imbio.2010.05.018
Reveille, J. D., Reveille, J. D., Sims, A. M., Danoy, P., Evans, D. M., Leo, P., et al. (2010). Genome-wide Association Study of Ankylosing Spondylitis Identifies Non-MHC Susceptibility Loci. Nat. Genet. 42 (2), 123–127. Epub 2010/01/12. doi:10.1038/ng.513
Rizzo, A., Ferrante, A., Guggino, G., and Ciccia, F. (2017). Gut Inflammation in Spondyloarthritis. Best Pract. Res. Clin. Rheumatol. 31 (6), 863–876. Epub 2018/12/05. doi:10.1016/j.berh.2018.08.012
Robinson, P. C., Leo, P. J., Leo, P. J., Pointon, J. J., Harris, J., Cremin, K., et al. (2016). The Genetic Associations of Acute Anterior Uveitis and Their Overlap with the Genetics of Ankylosing Spondylitis. Genes Immun. 17 (1), 46–51. Epub 2015/11/27. doi:10.1038/gene.2015.49
Sati, S., and Cavalli, G. (2017). Chromosome Conformation Capture Technologies and Their Impact in Understanding Genome Function. Chromosoma 126 (1), 33–44. Epub 2016/05/01. doi:10.1007/s00412-016-0593-6
Schlosstein, L., Terasaki, P. I., Bluestone, R., and Pearson, C. M. (1973). High Association of an HL-A Antigen, W27, with Ankylosing Spondylitis. N. Engl. J. Med. 288 (14), 704–706. Epub 1973/04/05. doi:10.1056/nejm197304052881403
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 9 (7), 671–675. Epub 2012/08/30. doi:10.1038/nmeth.2089
Selvarajan, V., Osato, M., Nah, G. S. S., Yan, J., Chung, T.-H., Voon, D. C.-C., et al. (2017). RUNX3 Is Oncogenic in Natural killer/T-Cell Lymphoma and Is Transcriptionally Regulated by MYC. Leukemia 31 (10), 2219–2227. Epub 2017/01/26. doi:10.1038/leu.2017.40
Shao, F., Liu, Q., Zhu, Y., Fan, Z., Chen, W., Liu, S., et al. (2021). Targeting Chondrocytes for Arresting Bony Fusion in Ankylosing Spondylitis. Nat. Commun. 12 (1), 6540. Epub 2021/11/13. doi:10.1038/s41467-021-26750-6
Shi, C., Ray-Jones, H., Ding, J., Duffus, K., Fu, Y., Gaddi, V. P., et al. (2021). Chromatin Looping Links Target Genes with Genetic Risk Loci for Dermatological Traits. J. Invest. Dermatol. Epub 2021/02/20. doi:10.1016/j.jid.2021.01.015
Shi, C., Rattray, M., Barton, A., Bowes, J., and Orozco, G. (2020). Using Functional Genomics to advance the Understanding of Psoriatic Arthritis. Rheumatology (Oxford) 59 (11), 3137–3146. Epub 2020/08/12. doi:10.1093/rheumatology/keaa283
Shi, C., Rattray, M., and Orozco, G. (2020). HiChIP-Peaks: a HiChIP Peak Calling Algorithm. Bioinformatics 36 (12), 3625–3631. Epub 2020/03/25. doi:10.1093/bioinformatics/btaa202
Stolwijk, C., van Tubergen, A., Castillo-Ortiz, J. D., and Boonen, A. (2015). Prevalence of Extra-articular Manifestations in Patients with Ankylosing Spondylitis: a Systematic Review and Meta-Analysis. Ann. Rheum. Dis. 74 (1), 65–73. Epub 2013/09/04. doi:10.1136/annrheumdis-2013-203582
Vecellio, M., Chen, L., Cohen, C. J., Cortes, A., Li, Y., Bonham, S., et al. (2021). Functional Genomic Analysis of a RUNX3 Polymorphism Associated with Ankylosing Spondylitis. Arthritis Rheumatol. 73 (6), 980–990. Epub 2020/12/29. doi:10.1002/art.41628
Vecellio, M., Cortes, A., Roberts, A. R., Ellis, J., Cohen, C. J., Knight, J. C., et al. (2018). Evidence for a Second Ankylosing Spondylitis-Associated RUNX3 Regulatory Polymorphism. RMD open 4 (1), e000628. Epub 2018/03/14. doi:10.1136/rmdopen-2017-000628
Vecellio, M., Roberts, A. R., Cohen, C. J., Cortes, A., Knight, J. C., Bowness, P., et al. (2016). The Genetic Association ofRUNX3with Ankylosing Spondylitis Can Be Explained by Allele-specific Effects on IRF4 Recruitment that Alter Gene Expression. Ann. Rheum. Dis. 75 (8), 1534–1540. Epub 2015/10/11. doi:10.1136/annrheumdis-2015-207490
Williamson, I., Berlivet, S., Eskeland, R., Boyle, S., Illingworth, R. S., Paquette, D., et al. (2014). Spatial Genome Organization: Contrasting Views from Chromosome Conformation Capture and Fluorescence In Situ Hybridization. Genes Dev. 28 (24), 2778–2791. Epub 2014/12/17. doi:10.1101/gad.251694.114
Keywords: ankylosing spondylitis, single nucleotide polymorphism (SNP), chromosome conformation capture (3C), RUNX3, c-Myc
Citation: Cohen CJ, Davidson C, Selmi C, Bowness P, Knight JC, Wordsworth BP and Vecellio M (2022) Disruption of c-MYC Binding and Chromosomal Looping Involving Genetic Variants Associated With Ankylosing Spondylitis Upstream of the RUNX3 Promoter. Front. Genet. 12:741867. doi: 10.3389/fgene.2021.741867
Received: 15 July 2021; Accepted: 20 December 2021;
Published: 07 January 2022.
Edited by:
Tommaso Pippucci, Unità Genetica Medica, Policlinico Sant’Orsola-Malpighi, ItalyReviewed by:
Elisa Giorgio, University of Turin, ItalyMalte Spielmann, Max Planck Institute for Molecular Genetics, Germany
Copyright © 2022 Cohen, Davidson, Selmi, Bowness, Knight, Wordsworth and Vecellio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matteo Vecellio, bWF0dGVvLnZlY2VsbGlvQG5kb3Jtcy5veC5hYy51aw==
 Carla J. Cohen
Carla J. Cohen Connor Davidson1,2,3
Connor Davidson1,2,3 Paul Bowness
Paul Bowness Matteo Vecellio
Matteo Vecellio