- 1School of Medicine, Department of Pathology, Stanford University, Stanford, CA, United States
- 2Department of Urology, Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh, India
Major fraction of the human genome is transcribed in to the RNA but is not translated in to any specific functional protein. These transcribed but not translated RNA molecules are called as non-coding RNA (ncRNA). There are thousands of different non-coding RNAs present inside the cells, each regulating different cellular pathway/pathways. Over the last few decades non-coding RNAs have been found to be involved in various diseases including cancer. Non-coding RNAs are reported to function both as tumor enhancer and/or tumor suppressor in almost each type of cancer. Urothelial carcinoma of the urinary bladder is the second most common urogenital malignancy in the world. Over the last few decades, non-coding RNAs were demonstrated to be linked with bladder cancer progression by modulating different signalling pathways and cellular processes such as autophagy, metastasis, drug resistance and tumor proliferation. Due to the heterogeneity of bladder cancer cells more in-depth molecular characterization is needed to identify new diagnostic and treatment options. This review emphasizes the current findings on non-coding RNAs and their relationship with various oncological processes such as autophagy, and their applicability to the pathophysiology of bladder cancer. This may offer an understanding of evolving non-coding RNA-targeted diagnostic tools and new therapeutic approaches for bladder cancer management in the future.
Introduction
Urothelial carcinoma of the urinary bladder is the second most commonly diagnosed urogenital cancer (Saginala et al., 2020; Compérat et al., 2022). On initial presentation, 70–75% of patients with bladder tumors are categorized as non-muscle invasive bladder cancer (NMIBC). The initial approach for the management of NMIBC is cystoscopy followed by transurethral resection of the tumor. Approximately 47% of patients have disease recurrence within 5 years of diagnosis of NMIBC and 9% of patients may progress to muscle-invasive disease. At present, bladder cancer management is limited to surgery, chemotherapy, radiotherapy, and immunotherapy. Personalized medicine may offer new windows for the management of bladder cancer (Lemke and Shah, 2018; Pardo et al., 2020). The next-generation sequencing (NGS) technology has discovered extensive tumor heterogeneity in different cancer types. Furthermore, RNA sequencing revealed upregulation of non-coding RNA (ncRNA) specifically microRNA, and long non-coding RNA (lncRNA) in bladder cancer (Cong et al., 2019). The human genome is 98% transcribed, however, most of them do not encode proteins, and are reported as ncRNAs (Pop-Bica et al., 2017; Cong et al., 2019). ncRNAs have been demonstrated to regulate gene expression as well as transcription, splicing, and protein function (Pop-Bica et al., 2017; Liu et al., 2022a). Recently, lncRNAs have been shown to regulate autophagy (Ghafouri-Fard et al., 2022). Autophagy is a conserved catabolic process in which intracellular components are engulfed and degraded, serving various functions depending upon the cellular conditions (Onorati et al., 2018; Ojha et al., 2019; Ishaq et al., 2020). Under normal conditions, it protects the cells from starvation by recycling damaged proteins and organelles. On the other hand, defective autophagy in a cell predisposes it to cancer (Ojha et al., 2015). Autophagy has been shown to help in tumor survival and progression by reducing ROS levels and providing nutrients (Ojha et al., 2014; Ishaq et al., 2016; Ojha et al., 2019). However, the role of autophagy in cancer is very complex and has been reviewed in detail somewhere else (Poillet-Perez et al., 2015; Amaravadi et al., 2019). Role of lnc-RNA in tumor progression via autophagy modulation is actively being investigated. In a recent study by Tan et al. showed that a set of seven autophagy related lnc-RNAs may predict the disease prognosis in MIBC patients (Tan et al., 2022). Similarly, Lai et al. set of 15 lnc-RNA related with autophagy were found to predict the prognosis of bladder cancer. Using the bioinformatics, these authors found that the 15-autophagy related lnc-RNA may regulate bladder cancer progression by modulating cell cycle, cell adhesion, DNA replication and WNT signalling pathways (Lai et al., 2020).
In this review, we will discuss the applicability of ncRNA in the progression and development of bladder cancer. We review the regulation of autophagy by ncRNA, especially miRNA and lncRNA, and the contribution of the ncRNA-autophagy axis in inducing tumor growth and/or tumor suppression in bladder cancer. Also, we will focus on the potential of exploring ncRNA expression profiles for the diagnosis and treatment of bladder cancer.
Bladder cancer and non-coding RNAs (ncRNAs)
Bladder cancer is one of the most commonly diagnosed malignancies of the urinary tract and is characterized by low sensitivity to chemotherapy and a high recurrence rate (Saginala et al., 2020; Compérat et al., 2022). Low-grade bladder cancer has a slow progression rate and hardly presents a threat to patients. On the other hand, high-grade bladder cancer has malignant potential with a greater mortality rate (Berdik, 2017; Daneshmand, 2020). Chemical agents, physical stimuli, and some pathogens are among the common factors responsible for the conversion of normal urothelial cells to the malignant urothelium. 95% of the primary urothelial tumors are confined within the bladder (Siracusano et al., 2020). Patients with this disease need lifelong surveillance because of the high recurrence rate, particularly if the tumor is within the bladder. Therefore, bladder cancer is the most expensive cancer to manage and because of this financial burden, bladder cancer is currently a very important focus of research (Audenet et al., 2018). Almost 60–65% of all bladder cancer show loss of heterozygosity on chromosome 9 and approximately 40% of bladder cancer show loss of heterozygosity on chromosome 17. Loss of heterozygosity on chromosome 9 is believed to be among the initial events in bladder carcinogenesis (Anderson, 2018). In contrast, loss of heterozygosity on chromosome 17 occurs late during cancer development and is related to the aggressiveness of cancer. Gene encoding p53 (TP53) is located on chromosome number 17 and most of the bladder cancer show loss of one allele of 17p resulting in loss of tumor suppressor functions of p53 (Li et al., 2021a). Mutations in the retinoblastoma (tumor suppressor) gene have been also observed in muscle-invasive, high grade and also in superficial bladder cancers (Li et al., 2021a). Bladder cancer cells have a higher expression of anti-apoptotic proteins Bcl-2 and Bcl-xL and the increased expression of these anti-apoptotic proteins correlates with poor prognosis in bladder cancer patients (Real and Malats, 2007). Bcl-2 overexpression in bladder cancer has been also reported to play an important role in cisplatin resistance (Cho et al., 2006). The very first study by Zhong et al. has shown atypical/aberrant expression of ncRNAs in bladder cancer might be exploited as bladder cancer biomarkers (Zhong et al., 2019).
High-throughput next-generation sequencing has shown that the human genome is approximately 90% transcribed. However, the transcribed genome can encode only 20,000 proteins (Reuter et al., 2015). A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. Emerging evidence suggests that ncRNAs are fundamental players in the regulation of gene expression and play a key role in various pathological conditions (Churko et al., 2013). Additionally, ncRNAs have been shown to mediate mRNA degradation, remodeling of chromatin structure, and also in cis and trans gene regulation (Fernandes et al., 2019; Ning et al., 2019). There are mainly two types of ncRNA characterized by their length: small ncRNAs and long ncRNAs (lncRNAs). Small ncRNAs include ribosomal RNA (rRNA), transfer RNA (tRNA), microRNA (miRNA), circular RNA, and piwi-interacting RNA (piRNA) (Zhang et al., 2019a; Papaioannou et al., 2021). lncRNAs include intergenic lncRNAs, intronic ncRNAs, and sense and antisense lncRNAs. Each type of lncRNAs showing different genomic positions in relation to genes and exons. lncRNAs regulate protein translation and post-transcriptional levels (Statello et al., 2021). Circular RNAs are different from linear mRNAs, however exact roles and functions are not fully understood yet (Cech and Steitz, 2014). In the following sections, we will be focusing on the role of miRNA and lncRNAs in different oncological processes in tumor bladder cancer (Figures 1A, B).
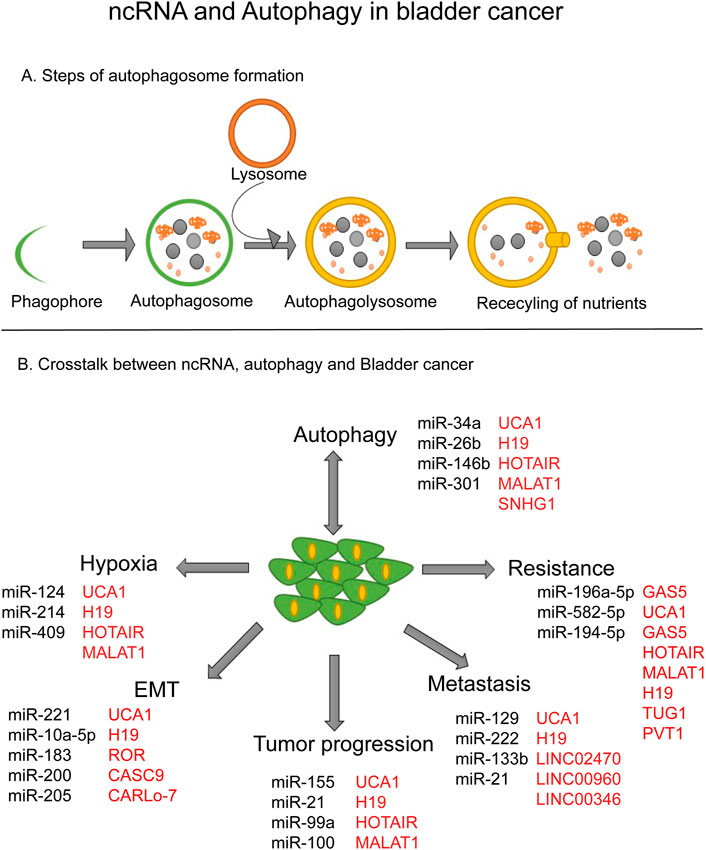
FIGURE 1. ncRNA and Autophagy in bladder cancer. (A) Steps of autophagosome formation. (B) Crosstalk between ncRNA, autophagy and bladder cancer.
miRNA and bladder cancer
miRNAs are single-stranded ncRNAs with a length of nearly 22 nucleotides (O’Brien et al., 2018). miRNAs are responsible for regulating gene expression at the post-transcriptional level, thereby modulating cellular processes like growth, proliferation, differentiation, and apoptosis (Towler et al., 2015). The miRNAs are produced from the conversion of pri-miRNA to pre-miRNA by a protein complex consisting of Rosha and the double-stranded RNA binding protein DiGeorge Syndrome Critical Region 8 (DGCR8). Pre-miRNAs are around 65 nucleotide-long RNA intermediates in the nucleus and are transported to the cytoplasm by Exportin-5. After the transportation, the pre-miRNA is cleaved by DICER and integrated into an RNA-induced silencing complex (RISC). When miRNA is complementary to the 3′UTR region of target mRNA, RISC will inhibit the protein translation and mRNA will be degraded by endonucleases (Michlewski and Cáceres, 2019; Saliminejad et al., 2019; Rani and Sengar, 2022). The biology of miRNA processing and mechanism of action have been explored well and is reviewed by Statello et al. (Statello et al., 2021). Here we will be mainly focusing on the role of miRNAs in various hallmarks of bladder cancer (Figure 2).
Tumor progression
MiR-616 is located on chromosome 12q13.3 and has been shown to be upregulated in many cancers (Zhao et al., 2022). Upregulated expression of miR-616 was shown to increase bladder cancer cell progression by targeting SOX7 (SRY-related HMG-box). SOX7 is a transcription factor belonging to the SRY-related high mobility group (HMG) box family and is known to function as a tumor suppressor. miR-616 downregulates the SOX7 expression by targeting its mRNA for degradation. Inhibition of miR-616 decreased bladder cancer cell proliferation abilities, induced cell cycle arrest in the G2 phase, and enhanced apoptosis (Zhao et al., 2022). MiR-495 is encoded by a gene located on chromosome 14 (q32.31). miR-495 has been linked to various tumors, however, its tumorigenic role is controversial. In bladder cancer, overexpression of miR-495 increased bladder cancer proliferation (Tan et al., 2017). MiR-657 expression was found higher in bladder cancer tissue samples compared to the normal bladder. Overexpression of miR-675 has been reported to promote bladder cancer progression by promoting cell cycle progression. Inhibition of miR-675 decreased p53 activation and increased Bax/Bcl2 ratio and thereby inducing apoptosis in bladder cancer cells (Liu et al., 2016). A high expression of miR-294 was demonstrated in bladder cancer cell lines. Mechanistically, authors have shown that miR-294 upregulates NRAS expression to promote bladder cancer progression. Additionally, overexpression of miR-294 increased the phosphorylation of PI3K, AKT, JNK, and STAT1 (Li et al., 2017a). An upregulated expression of MiR-21 was demonstrated in bladder cancer tissue. MiR-21 overexpression has been shown to increase Bcl2 expression and enhance Akt phosphorylation to promote cell proliferation in bladder cancer cells (Lin et al., 2020).
Tumor metastasis
miR-516a is located on chromosome 19q13 and has been shown to be involved in different oncogenic processes. miR-516 expression directly correlated with the invasive properties of bladder cancer cells. miR-516a promotes metastasis by regulating the expression of Matrix metallopeptidase 9 (MMP9). miR-516a decreases Surfeit locus protein 1 (SUMRF1) expression thereby stabilizing and inhibiting the degradation of MMP9 by the proteasomal pathway (Chang et al., 2020). Also, another study by Zhu et al. has shown that miR-146b is upregulated in bladder cancer tissues. Silencing of miR-146b inhibited bladder cancer cell invasion by reducing MMP2 level expression by inhibiting its transcription factor ETS2. Mechanistically, authors have demonstrated that miR-146b inhibition stabilizes ARE/poly(U)-binding/degradation factor 1 mRNA expression by directly binding to its mRNA at 3′ UTR (Zhu et al., 2019). Overexpression of miR-200c in bladder cancer cell line UMUC3 decreases their invasive properties. The high expression of miR-200c resulted in increased expression of E-cadherin and decreased expression of Zinc finger E-box-binding homeobox 1 and 2 (ZEB1) and ZEB2 at both mRNA and protein levels. These authors showed that ZEB1 and ZEB2 are the direct targets of miR-200 (Liu et al., 2022b). Recently Yang et al., 2021, showed that miR-20a-5p expression is highly upregulated in bladder cancer tissues. Overexpression of miR-20a-5p not only increased bladder cancer cell proliferation but also promoted epithelial-to-mesenchymal transition (EMT). The overexpression of miR-20a-5p inhibited the expression of epithelial markers (E-cadherin) and increased the Vimentin expression (mesenchymal marker) (Yang et al., 2021). This study showed that miR-20a-5p selectively targets Nuclear Receptor Subfamily four Group A Member 3 (NR4A3) and decreases its expression. So, by targeting this nucleus receptor protein NR4A3-miR-20a-5p regulates the migration, invasion, and metastasis of bladder cancer cells (Yang et al., 2021). On the contrary, there are some miRNAs such as miR-433, miR-323a-3p, and miR-613 that have inhibitory effects on EMT. miR-433 expression was shown to be downregulated in bladder cancer tissue samples. Overexpression of miR-433 decreased migratory and EMT properties of bladder cancer cells by modulating the c-MET-Akt pathway. MiR-433 directly binds to c-MET and CAMP responsive element binding protein 1 (CREB1) at 3′-UTR and inhibits its mRNA as well as protein expression (Xu et al., 2016). The miR-323a-3p level was found to be downregulated in bladder cancer tissue samples as well as in cell lines. These authors demonstrated that the two key oncogenes Suppressor of Mothers against Decapentaplegic 3 (SMAD3) and MET are the direct targets of miR-323a-3p. Also, miR-323a-3p upregulation was shown to inhibit EMT progression in bladder cancer cells by targeting the AKT- Glycogen synthase kinase-3β (GSKB) axis (Li et al., 2017b). Just like miR-323a-3p, the expression of miR-613 is downregulated in bladder cancer cell lines and tumor tissues. miR-613 overexpression abrogated EMT via increasing the expression of E-cadherin and decreasing that of Vimentin and Snail. miR-613 was found to exert its inhibitory effects on EMT via blocking the expression of Sphingosine kinase 1 (Sphk1) (Yu et al., 2017a).
Chemoresistance
The recurrence rate in bladder cancer is of common occurrence particularly due to the cancer cells developing resistance towards standard drugs like gemcitabine, cisplatin, etc. (Lemke and Shah, 2018). There are various mechanisms by which cancer cells develop resistance towards any anti-cancer drug, but recently microRNAs have been also added to that list (Taheri et al., 2020). The role of miRNAs in drug resistance was first studied by Fojo et al., who showed that miRNA profile can play a key role in chemosensitivity or chemoresistance (Fojo, 2007). In bladder cancer, the first identified drug-resistance-related miRNAs belonged to the miR-200 family. MiR-200 overexpression in bladder cancer cells increases their sensitivity towards the epidermal growth factor receptor (EGFR) inhibitors via regulating the expression of ERBB receptor feedback inhibitor 1 (ERRF1) (Adam et al., 2009). MiR-21 has been shown to be highly upregulated in bladder cancer cells and contributes to doxorubicin resistance. MiR-21 enhanced resistance towards doxorubicin was mediated by increased expression of anti-apoptotic protein Bcl2 (Tao et al., 2011). miR-203 was associated with cancer progression and poor prognosis of bladder cancer patients who received cisplatin-based adjuvant chemotherapy (Zhang et al., 2015). In contrast to miR-21, miR-203 expression is highly reduced in bladder cancer patients. Overexpression of miR-203 in bladder cancer cells increased their sensitivity to cisplatin by inducing apoptosis. In their study, Zhang et al. showed that two pro-apoptotic proteins, Bcl-w and Survivin are the direct targets of miR-203 (Zhang et al., 2015). MiR-193a-3p has been found to be more highly expressed in resistant cell lines than in sensitivity cell lines. Interestingly, miR-193a-3p was shown to promote multi-drug resistance in bladder cancer cells. The mechanism by which miR-193a-3p enhanced drug resistance was shown to be the inhibition of Serine/arginine-rich splicing factor 2 (SRSF2) and lysyl oxidase-like 4 (LOXL4) expression in bladder cancer cells (Deng et al., 2014). In a very interesting study, miR-218 was reported to enhance the cisplatin sensitivity of bladder cancer cells by targeting Glut1 (glucose transporter isoform 1). Glut1 expression was shown to be significantly decreased in miR-218 overexpressed cells. This is very important; because restricting the availability of glucose during chemotherapy has been shown to enhance drug sensitivity in different cancer models (Li et al., 2017c). MiR-222 increases resistance towards cisplatin by regulating Protein Phosphatase 2 Regulatory Subunit Balpha (PPP2R2A)/Akt/mTOR pathway. PPP2R2A is a regulatory subunit of phosphatase 2A and is known to play a role in cancer progression. One of the direct targets of PPP2R2A is Akt. In fact, the overexpression of miR-222 was shown to increase the Akt phosphorylation and activates its downstream target mTOR (Zeng et al., 2016). miR-27A is downregulated in bladder cancer tissue samples and is involved in enhancing cisplatin resistance by regulating the expression of Solute Carrier Family 7 Member 11 (SLC7A11). SLC7A11 is highly overexpressed in cells that are resistant to cisplatin. SLC7A11 is a component of cysteine/glutamate exchanger and is an important factor regulating glutathione (GSH) production. Overexpression of miR-27A significantly decreases SLC7A11 expression and enhances sensitivity to bladder cancer cells toward cisplatin (Drayton et al., 2014). In a recent study, CD44 has been shown to be targeted by miR-34a in muscle-invasive bladder cancer during cisplatin treatment. An increased expression of CD44 has been shown to effectively reverse the effects of miR-34a on bladder cancer cell proliferation and chemosensitivity of muscle invasive-bladder cancer cells (Li et al., 2022a).
Autophagy
Autophagy is an umbrella process for the degradation of aggregated proteins and organelles including mitochondria, endoplasmic reticulum, ribosomes, and nucleus (Dikic and Elazar, 2018). The contribution of autophagy in cancer development is well established although still controversial (Ojha et al., 2015; Ishaq et al., 2020). The role of autophagy is highly dependent on the cancer type, mutational status, and developmental stage of cancer (Onorati et al., 2018; Amaravadi et al., 2019). The regulatory networks involved in autophagy induction and/or inhibition have been well characterized. MiRNAs are the latest addition to the long list of autophagy regulators (Xu et al., 2012; Shan et al., 2021). Inhibition of autophagy via selective targeting of Beclin1 by miR30a was the first report, highlighting the role of miRNAs in autophagy and cancer (Zhu et al., 2009). The role of miRNAs in autophagy regulation was first discovered in 2009 when Beclin1 (BECN1), an autophagy gene, was shown to be regulated by miR30A (Cai et al., 2021). miR-221 induces cell proliferation of colorectal cancer (CRC) via the downregulated of autophagy by targeted Tumor protein 53-induced nuclear protein 1 (TP53INP1) (Liao et al., 2018a).
Autophagy has been associated with both chemoresistance and chemosensitivity (Li et al., 2019a). MiR-222 has been shown to inhibit cisplatin-induced cell death by preventing autophagy induction. MiR-222 inhibits autophagy induction via the activation of the Akt/mTOR signaling axis. mTOR is one of the key negative regulators of autophagy involved in cancer progression (Tsikrika et al., 2018). Tsikrika et al. have reported that bladder cancer patients with high expression of miR-221 have a higher short-term recurrence rate. Moreover, miR-221 overexpression has also been reported to be an independent prognostic value for bladder cancer patients (Tsikrika et al., 2018). Liu et al. have shown that the downregulation of miR-221 enhances autophagy activation via increasing TP53INP1. In addition, the miR-221/TP53INP1/p-ERK axis has been shown to regulate autophagy in bladder cancer (Liu et al., 2020a). miR-24-3p is also overexpressed in bladder cancer cells and has been shown to promote autophagy by inhibiting domain-containing protein (DEDD). Autophagy induction via miR-24-3p helped bladder cancer cell proliferation by apoptosis inhibition (Yu et al., 2017b). Luo et al. have shown that miR-139-5p inhibited bladder cancer cell proliferation by direct binding Bmi-1. Inhibition of Bmi-1 has been demonstrated to increase ATP reduction and AMPK-activated autophagy (Luo et al., 2017). Wang et al. has reported that sodium butyrate inhibited bladder cancer cell migration and induced AMP-activated protein kinase (AMPK)-mTOR axis-dependent autophagy and ROS-mediated apoptosis via the miR-139-5p/Bmi-1 pathway (Wang et al., 2020). Interestingly, Zhang et al. have shown that miRNA-mediated autophagy downregulation has been shown to inhibit bladder cancer cell progression. miR-154 has been shown to downregulate bladder cancer tissue samples. Overexpression of miR-145 inhibits bladder cancer cell proliferation, invasion, and migration by preventing ATG7 expression (Zhang et al., 2019b).
In, multiple myeloma (MM), miR-126 mediates the induction of autophagic flux and HIF1α stabilization (Tomasetti et al., 2016). In bladder cancer, miR-221 expression was shown to be positively regulated by TGFβ1. Inhibition of miR-221 rescued TGFβ1-induced EMT by increasing E-cadherin and decreasing vimentin, Fibronectin, and N-cadherin. Moreover, miR-221/TP53INP1/p-ERK axis expression was shown to induce autophagy and reported to be positively correlated with the malignant property of bladder cancer cells (Shen et al., 2021). The level of miR-133b was found to be reduced in bladder cancer patient tissues and in exosomes from the serum of bladder cancer patients. Exosomal miR-133b leads to the inhibition of cancer cell viability by upregulating dual-specificity protein phosphatase 1 (DUSP1) and an escalation of apoptotic cell death in bladder cancer cells (Cai et al., 2020). Small nucleolar RNA host gene 1 (SNHG1) is another lncRNA (lncRNA), which negatively regulates tumor suppressor genes. Increased expression of SNHG1 has been shown to enhance bladder cancer progression and autophagy via miR-493-5p/ATG14/autophagy pathway (Guo et al., 2021). This finding highlights the potential role of SNHG1 as a target for the management of bladder cancer. miR-21 shows a grade-dependent increase in bladder cancer cells. This study has demonstrated that, in 31 patients, miR-21 was significantly up-regulated, and PTEN level was significantly inhibited in bladder tumor tissue compared to the normal bladder mucosa (Yin et al., 2019). Furthermore, patients with recurrence had a significantly higher miR-21 expression as compared to non-recurrent patients. Also, miR-21 overexpression has been reported to decrease autophagy and promote the metastatic properties of bladder cancer cells (Zhang et al., 2020). This study is interesting because we have also shown that autophagy induction is grade dependent in bladder cancer (Ojha et al., 2014). The grade dependency of miR-21 and autophagy may be unrelated but needs to be experimentally verified.
Tumor suppression
miR-381 was shown to be downregulated in bladder cancer. Increased expression of miR-381 was demonstrated to inhibit the proliferation and tumor formation capacity of bladder cancer cell lines T24 and RT4 cells. In addition, miR-381 was found to bind B cell-specific Moloney murine leukemia virus integration site 1 (BMI1). The tumor-suppressing properties of miR-381was shown to be blocked by overexpression of BMI. Overexpression of miR-381 decreased RhoA phosphorylation and Rho-associated protein kinase (ROCK2) activation (Chen et al., 2021a). miRNA-145 was significantly downregulated in patients with bladder cancer. miRNA-145 markedly inhibited the ability of bladder cancer cells to migrate and invade. Furthermore, N-cadherin was identified as a target of miRNA-145 in bladder cancer cells. MMP9, acting downstream of N-cadherin, was downregulated in bladder cancer cells by miRNA-145 (Zhang et al., 2018). The expression of miR-146a-3p was found to be downregulated in bladder cancer tissue samples. MiR-146a-3p overexpression has been shown to downregulate the metastatic potential of bladder cancer cells. MiR-146a-3p directly binds to 3′UTR of an oncogene Pituitary tumor-transforming gene 1 (PTTG1) (Xiang et al., 2017). A decreased expression of miR-539 was reported in bladder cancer. Overexpression of miR-539 inhibits bladder cancer cell proliferation. This study showed that IGF1R is a direct target of miR-539. The mimic of miR-539 decreased IGF1R expression via binding to its 3′UTR region. In addition, authors have shown that silencing of miR-539 attenuated the phosphorylation of AKT and ERK. This leads to the inhibition of bladder cancer growth and invasion by the AKT-ERK-IGF1R axis (Liao et al., 2018b).
Long non-coding RNA and bladder cancer
Long non-coding RNAs (lncRNAs) are RNA molecules of more than 200 nucleotides that are not translated. LncRNAs are transcribed by RNA Polymerase-II and are mainly localized to the nucleus. Although lncRNAs are abundant in the nucleus, they are less stable than their cytoplasm-localized counterparts. LncRNAs are stabilized either by polyadenylation or by the formation of secondary structure triple helices at their 3′ end. These 3′end sequences help in the nuclear export of lncRNAs (Nojima and Proudfoot, 2022). LncRNA expression is usually low but shows stronger tissue-specific expression, indicating lncRNAs play an integral role in tissue-specific processes (Aznaourova et al., 2020). Various studies have reported the role of lncRNAs in carcinogenesis as well as other pathologies (Yao et al., 2019a). The role of lncRNA in bladder cancer progression, proliferation, and metastasis has been shown in several studies (Zhang et al., 2021a; Chen et al., 2021b; Li et al., 2021b). In the following section, we will discuss the role of lncRNAs in different oncogenic processes in bladder cancer (Figure 3).
Tumor progression
Urothelial cancer associated 1 (UCA1) is one of the lncRNA associated with bladder cancer progression. UCA1 is located on chromosome 19p13.12 (Mirzaei et al., 2022). The ectopic expression of UCA1 influences bladder tumor progression, revealing UCA1 as an oncogenic player in bladder carcinogenesis (Ding et al., 2021). The role of UCA1 in bladder cancer tumorigenesis has been reported in various studies (Ghafouri-Fard et al., 2022). UCA1 has been demonstrated to increase EMT by zinc finger E-box binding homeobox 1 and 2 (ZEB1 and ZEB2) upregulation in bladder cancer cells (Xue et al., 2016). UCA1 is known to be transported via exosomes and leads to bladder cancer progression under hypoxic conditions (Xue et al., 2017). In a contrast study, Lebrun et al. showed that in a particular type of bladder cancer UCA1 expression is decreased (Lebrun et al., 2018).
Homeobox transcript antisense RNA (HOTAIR) genes reside in the HOXC cluster, located on the human chromosome 12q13.1 (Tang and Hann, 2018). HOX cluster is an important key factor in embryonic development. Suppression of the HOX genes has been demonstrated in tumor progression (Gupta et al., 2010). HOTAIR inhibited p53 expression and the phosphatase and tensin homolog (PTEN) expression, thereby aggravating cancer progression (Zhang et al., 2019c). HOTAIR expression also significantly correlates with the activation of the Wnt/β-catenin pathway (Zhang et al., 2021b). H19 is one of the first discovered ncRNAs, which is located at chromosome 11p15.5. H19 is highly expressed during human embryonic development but is suppressed in adults (Wu et al., 2021). However, H19 has been shown to reactivate during tumorigenesis and play a crucial role in various cancer including bladder cancer (Raveh et al., 2015; Ghafouri-Fard et al., 2020). H19 has been shown to enhance bladder cancer cell proliferation via the upregulation of an inhibitor of DNA binding 2 (ID2) (Luo et al., 2013a). The ID2 upregulation attenuates retinoblastoma protein (Rb) effects on E2F1 expression, thereby promoting bladder cancer progression (Mao et al., 2021). Similarly, H19 has been shown to induce upregulation of p53 protein thereby leading to bladder cancer progression (Atala, 2013). Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been first identified as the prognostic marker in lung cancer and has been reported to have a positive association with many other cancers including prostate cancer and hepatic cancer (Amodio et al., 2018; Sun and Ma, 2019). The overexpression of MALAT1 enhances bladder cancer progression and migration (Xie et al., 2017a; Li et al., 2017d; Liang et al., 2021). Additionally, MALAT1 has been shown to inhibit apoptotic cell death and thereby promote bladder cancer progression.
Tumor Metastasis
UCA1 by regulating the high mobility group box 1 (HMGB1) pathway enhances the properties of bladder cancer invasion and metastasis. HMGB1, a member of the high mobility group box subfamily, has been demonstrated to be associated with various cancer (Wu and Zhou, 2018). HMGB1 acts as an EMT inducer in many human cancer cells (Dong et al., 2022) and is significantly upregulated higher in bladder cancer cells than in normal urothelial cells (Hajjari and Salavaty, 2015). Several studies have reported that HOTAIR is the main player in various malignancies including bladder cancer, colon cancer, lung cancer, and breast cancer (Chi et al., 2019). Liu et al. showed that silencing of HOTAIR inhibits the invasive properties of bladder cancer by downregulating epithelial-to-mesenchymal (EMT), suggesting HOTAIR’s role in regulating metastasis (Liu et al., 2015). In addition, the inhibition of HOTAIR has been demonstrated to regulate Notch1-mediated EMT pathways in bladder cancer and thereby promoting metastatic properties of bladder cancer (Berrondo et al., 2016).
In addition, H19 upregulated expression has been demonstrated to play a vital role in bladder cancer metastasis. Authors have shown that H19 overexpression increased the migratory properties of bladder cancer cells by interaction with EZH2 which leads to activation of Wnt/β-catenin signaling and therefore led to EMT induction (Atala, 2013; Luo et al., 2013b). Zhu et al. has also shown that H19 overexpression leads to enhancing the metastatic properties via EMT pathways (Zhu et al., 2018). The expression of MALAT1 was shown to be higher in invasive and metastatic bladder cancer compared to normal tissue. Additionally, MALAT1 expression was higher in high-grade patients compared to low-grade bladder cancer patients (Li et al., 2017d). Silencing of MALAT1 has an inhibitory effect on the metastatic properties of bladder cancer cells. MALAT1 downregulates ZEB1, ZEB2, and Wnt signaling proteins, while it upregulates E-cadherin (Ying et al., 2012; Fan et al., 2014a). These results indicate that MALAT1 has a key role in initiating metastatic properties in bladder cancer. Recently, LINC02470 was reported to enhance bladder cancer cell viability, migration, and invasion. LIN02470 was demonstrated to activate the SAMD3-TGF β-mediated EMT process in bladder cancer (Huang et al., 2022). Similarly, Haung et al. has shown that two exosome-derived lncRNAs; LINC00960 and LINC02470 increased the malignant properties of bladder cancer cells by upregulating EMT, β-catenin, NOTCH, and SMAD signaling in high-grade bladder cancer (Huang et al., 2020).
Chemoresistance
There are only a few reports where lncRNAs have been shown to facilitate drug resistance in bladder cancer (Liu et al., 2020b). The overexpression of UCA1 was shown to abrogate apoptotic cell death and facilitate acquired resistance during anti-cancer therapy in bladder cancer cells (Wang et al., 2017). This study has demonstrated that increased overexpression of UCA1 decreased the sensitivity of tamoxifen and silencing of UCA1 mediates/enhances drug sensitivity by induction of apoptosis and cell cycle arrest during tamoxifen treatment (Liu et al., 2019). Mechanistically, UCA1 was shown to physically interact with EZH2, which blocked the p21 expression through histone methylation, and parallelly UCA1 expression mediates phosphorylation of CAMP responsive element binding protein (CREB) and PI3K/AKT and thereby facilitates tamoxifen-mediated resistance (Fan et al., 2014b). In addition, UCA1 has also been shown to mediate resistance during cisplatin treatment by Cytochrome P450 Family 1 Subfamily B Member 1 (CYP1B1)-mediated apoptosis via miR-513a-3 upregulation (Li et al., 2022b). UCA1 expression was shown to be positively related to CYP1B1 expression. UCA1 binds with miR-513a-3p to induce CYP1B1 expression. UCA1 silencing promotes chemosensitivity and enhances apoptosis during cisplatin treatment, suggesting the UCA1/miR-513a-3p/CYP1B1 axis plays a key role in mediating chemoresistance (Cheng et al., 2021). The role of UCA1 in colorectal cancer cells (CRC) has also been studied. A study by Liu et al. demonstrated that the UCA1-Wnt/β-catenin axis in CRC plays an imperative role in regulating metastasis and autophagy in vivo (Liu et al., 2019).
Autophagy
Stress is a common feature of tumors, which include hypoxia, and deficiency of nutrient growth factors due to insufficient vasculature (Hanahan, 2022; Ojha et al., 2022). Autophagy is one of the main cellular processes which is induced in many tumors under various stress conditions like hypoxia and starvation (Mulcahy Levy and Thorburn, 2020). For the first time, our group (Ojha et al.) showed that autophagic flux increases in a grade-dependent manner in bladder cancer. AMPK is the key factor for regulating autophagy in bladder cancer cells and functionally autophagy plays a cytoprotective role in these cells (Ojha et al., 2014). Moreover, inhibition of autophagy by both pharmacological and siRNA enhanced the chemotherapeutic effects in bladder cancer cells (Ojha et al., 2016). Gambogic acid, a potent anticancer agent, also has been shown to induce autophagic flux by reactive oxygen species (ROS) mediated JNK activation in the bladder cancer cells (Ishaq et al., 2014).
Autophagy has been demonstrated to be regulated by several mechanisms in cancer cells. Among the known mechanisms, the recently validated mechanism is ncRNAs (Yang et al., 2017). Several miRNAs and lncRNAs have been described to regulate autophagy via distinct mechanisms (Yao et al., 2019b). Various genetic and biochemical studies have illustrated that the inhibition of MEG3 expression enhances autophagy and blocks apoptosis (Xiu et al., 2017). A study by Ying et al. has shown that MEG3 expression was markedly reduced in bladder cancer compared with normal bladder tissues, however, an increased autophagy level was significantly found in tumor tissue compared to normal bladder tissue. Inhibition of MEG3 expression blocked apoptosis and increased cell proliferation while autophagy inhibition augmented MEG3-silencing-mediated apoptosis (Ying et al., 2013). These findings accentuate the significance of MEG3 in tumor suppression and the potential use of targeting MEG3 in the management of bladder cancer. A study by Xiu et al., 2017 has demonstrated that upregulated expression of MEG3 inhibited tumorigenesis in vivo by upregulating ATG3 expression (Xiu et al., 2017). In addition, MEG3 has been shown to protect ATG3 mRNA from degradation during actinomycin D treatment. P53 activation has been demonstrated to regulate MEG3 expression to protect against tumor proliferation (Xiu et al., 2017). MEG3 overexpression has been shown to downregulate miR-96 while upregulating α-tropomyosin 1 (TPM1), which reduced bladder cancer cell viability by increasing apoptotic cell death (Liu et al., 2018).
The overexpression of H19 was found to inhibit GTP-binding protein Di-Ras3 (DIRAS3) expression and induce phosphorylation of mTOR, which leads to the inhibition of autophagy in cardiomyocytes (Zhuo et al., 2017). Treatment of bladder cancer cells with exosomes isolated from tumor-associated macrophages enhances H19 expression and autophagic response. Inhibition of H19 in TAMs-exosomes blocked autophagy flux (Guo et al., 2022). This study suggests that H19 plays a role in autophagy induction in bladder cancer cells and targeting TAMs-Exosomes-H19 is an encouraging therapeutic approach for the management of bladder cancer. Overexpression of GAS5 induced chemoresistance to cisplatin, which was not rescued by 3-MA-mediated inhibition of autophagy, signifying that GAS5 promotes chemosensitivity in an autophagy-independent manner (Xu et al., 2020). Silencing of GAS5 decreased cancer cell viability and reduced autophagy via regulating miR-23a expression (Li et al., 2018). The results suggested that the GAS5-miR-23a complex might be involved in the regulation of autophagy. Collectively, these results indicated that GAS5 participates in carcinogenesis by stimulating the autophagy response. To the best of our knowledge of GAS5 and autophagy in bladder cancer is not studied yet. HOTAIR was shown to increase autophagy and promote imatinib sensitivity of gastrointestinal stromal tumors (GIST). This study has shown that miR-130a and HOTAIR have downstream target autophagy-related protein 2 homolog B (ATG2B). Downregulation of ATG2B blocks the effect of HOTAIR on imatinib sensitivity in GIST (Zhang et al., 2021b). In addition, inhibition of HOTAIR has been demonstrated to enhance the sensitivity to radiotherapy by inhibiting autophagy through the downregulation of the Wnt signaling pathway in cervical cancer (Trujano-Camacho et al., 2021). To the best of our knowledge, there are no reports about the role of HOTAIR in regulating autophagy in Bladder cancer.
Tumor suppressor
Growth arrest-specific 5 (GAS5) was initially shown to regulate cell proliferation and cell cycle during embryogenesis and tumor progression (Yu and Hann, 2019; Xu et al., 2020). However, several studies have demonstrated that the downregulation of GAS5 is potentiating tumor progression in various cancers including bladder cancer (Cao et al., 2016). Similarly, GAS5 has been demonstrated as a tumor suppressor role in bladder cancer via inhibiting EZH2 expression and augmenting apoptosis (Kaur et al., 2022). GAS5 has been also shown to inhibit Calcium-Activated Chloride Channel 1 (CCLA1) expression and thus contributes to the suppression of cancer growth (Jia et al., 2015). Additionally, genetic studies have demonstrated that GAS5 suppresses cell division protein kinase 6 (CDK6), thereby inhibiting bladder cancer progression (Wang et al., 2012). Furthermore, GAS5 has been reported to promote apoptosis by suppressing EZH2 transcription via the recruitment of transcription factor E2F4 to EZH2 promoter in bladder cancer cells (Cao et al., 2016). Altogether, these studies suggest that GAS5 could use for bladder cancer patient’s treatment. However, GAS5 silencing was reported to reduce apoptosis via glucocorticoid receptors during nutrient deprivation (Kino et al., 2010).
Maternally expressed gene 3 (MEG3) is highly expressed in human tissue and has been demonstrated to play as a tumor suppressor (He et al., 2017). MEG3 expression has been shown to inhibit bladder cancer progression (He et al., 2017). Ying et al. have reported that MEG3 expression was downregulated in bladder cancer compared with normal tissue (Liu et al., 2018). Further genetic studies demonstrated the silencing of MEG3 increases autophagy and abrogated apoptosis in vitro (Ying et al., 2013). These studies underscore the significance of MEG3 in tumor suppression in bladder cancer treatment.
Clinical relevance of non-coding RNA
The clinical use of ncRNAs has recently gained momentum because of some advantages over the currently used drugs. First, ncRNAs are synthesized by the cells themselves, so will not be treated as alien molecules. Second, ncRNAs usually target a set of different mRNAs, which encode proteins regulating a specific process. However, so far only one ncRNA, Prostate Cancer Associated 3 (PCA3) is used in clinical settings as a biomarker for prostate cancer. Various ncRNAs are under clinical trials for different pathologies including cancer. However, the effective use of ncRNAs in therapeutics is limited because of issues like; delivery to target regions, bioavailability, and specificity (Wang et al., 2019; Winkle et al., 2021). In the following section we will be highlighting the clinical relevance of ncRNAs in bladder cancer.
Clinical utility of miRNA: Diagnosis and prognosis
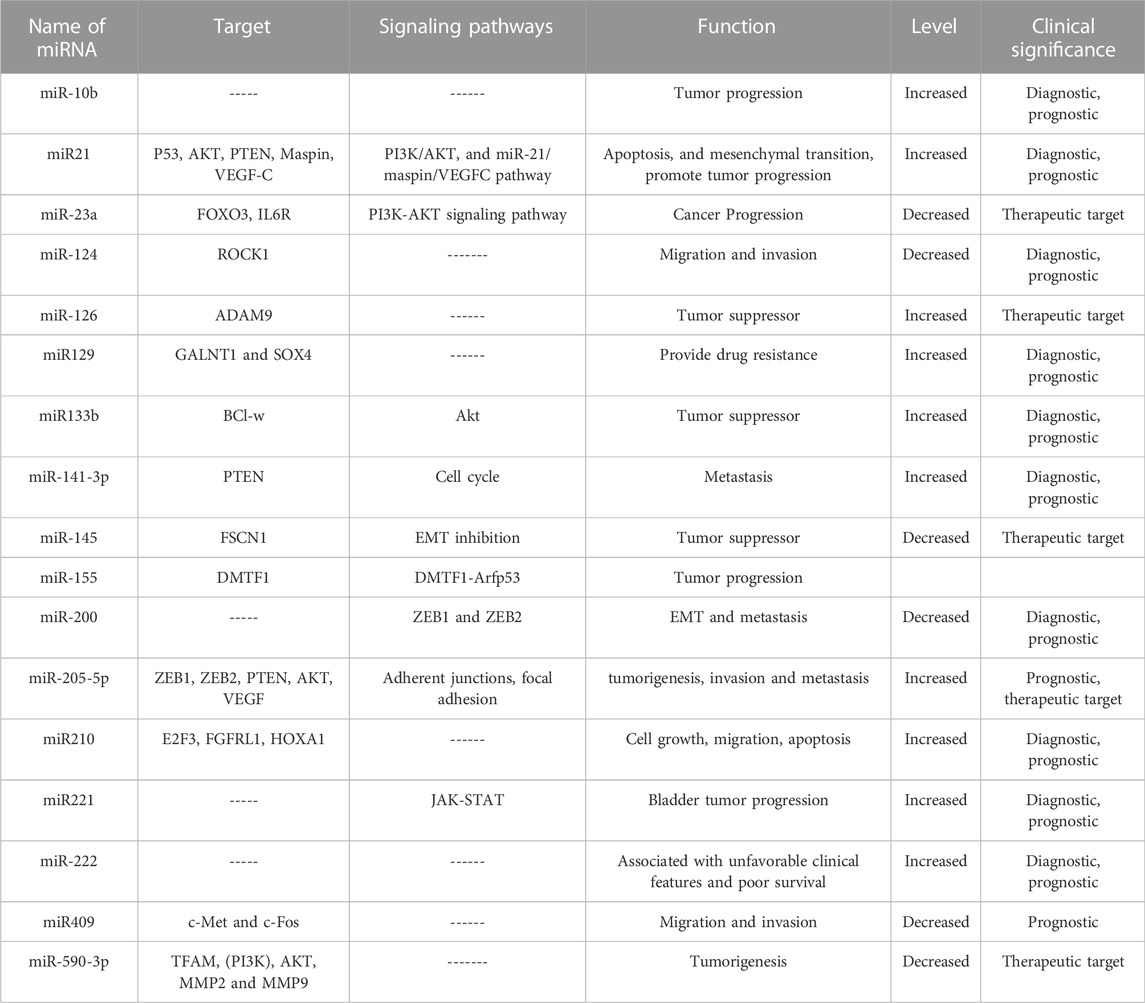
Various biomarker studies have demonstrated the potential of miRNAs in bladder cancer diagnosis, for example high expression of the miR-200 family correlated with better overall and recurrence-free survival (Table 1). Mei et al. concluded that high expression of the miR-200 family is strongly associated with better prognosis in bladder cancer patients and may significantly improve bladder cancer management (Mei et al., 2020). Similarly, higher expressions of hsa-miR-663a and hsa-miR-3648, and lower expression of hsa-miR-185-5p, hsa-miR-30c-5p, hsa-miR-1270, hsa-miR-200c-3p, and hsa-miR-29c-5p, significantly correlated with shorter overall survival of bladder cancer patients (Homami and Ghazi, 2016; Lin et al., 2019). A combination of 7 miRNAs (7-miRNA panel: miR-6087, miR-6724-5p, miR-3960, miR-1343-5p, miR-1185-1-3p, miR-6831-5p and miR-4695-5p) was shown to accurately discriminate bladder cancer from non-cancer and other types of tumors with the high specificity (Usuba et al., 2019). Another study showed that the combination of four miRNAs; miR-181b-5p, miR-183-5p, miR-199-5p and miR-221-3p can be used as a stable biomarker for bladder cancer diagnosis (Li et al., 2022c).
Recent studies showed circulating miRNA by liquid biopsy could be the potential biomarker for bladder cancer. Mitesh et al. found miR-7-5p, miR-22-3p, miR-29a-3p, miR-126-5p, miR-200a-3p, miR-375, and miR-423-5p in urine could serve as non-invasive biomarkers for bladder cancer (Mitash et al., 2017). Lian et al. found miR-148b-3p, miR-3187-3p, miR-15b-5p, miR-27a-3p, and miR-30a-5p in serum samples could be the potential biomarkers for the prognosis of bladder cancer (Lian et al., 2018). A study by Xie et al. identified seven miRNAs were up-regulated in bladder serum cancer samples compared to control samples, including hsa-miR-185-5p, hsa-miR-663a, hsa-miR-30c-5p, hsa-miR-3648, hsa-miR-1270, hsa-miR-200c-3p, and hsa-miR-29c-5p (Xie et al., 2017b; Li et al., 2022c). The dysregulation of these miRNAs correlated with the advanced stage and overall survival in bladder cancer patients.
Clinical utility of long non-coding RNA: Diagnosis and prognosis
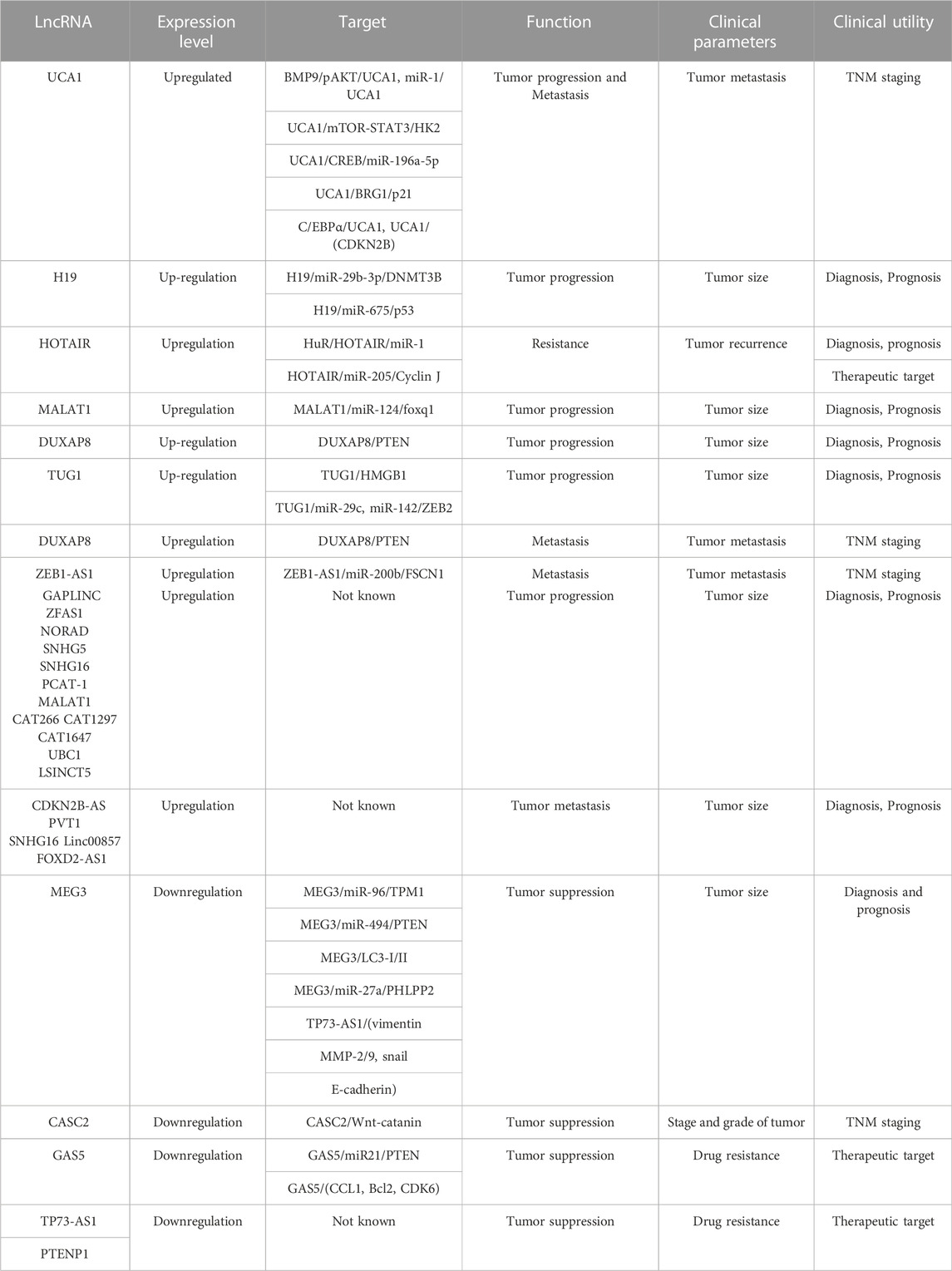
Just like miRNAs, lncRNAs have been explored as possible diagnostic biomarkers for tumor bladder cancer (Table 2). UCA1 is among the well-studied and good candidates for bladder cancer biomarkers. UCA1 mRNA expression shows a significant association with the stage and grade of bladder cancer (Lebrun et al., 2018). Yu et al. have shown increased expression of UCA1 in urine samples of high-grade bladder tumor patients (Yu et al., 2020). UCA1 has been shown to have high sensitivity for the T2-T4 stage of bladder cancer, suggesting that UCA1 may be considered a marker for the better prognosis of bladder cancer (Avgeris et al., 2019). Expression of lncRNAs H19 and HOTAIR are significantly increased in bladder cancer tissues compared to normal bladder tissues (Wang and Yin, 2017). High expression of HOTAIR and GAS5 was shown to be associated with poor survival in bladder cancer (Li et al., 2019b). A study by Lodewijk et al. has demonstrated that UCA1, HOTAIR, and MALAT1, can be used as urinary biomarkers for bladder cancer patients (Lodewijk et al., 2018). Another study showed that overexpression of HOTAIR, MALAT1 and LINC00477 was found in urine exosomes of high-grade muscle-invasive bladder cancer patients. Irregular expression of HOTAIR, UBC1, H19, and GAS5 has been reported to be related to overall survival in bladder cancer (Berrondo et al., 2016) however, more confirmative studies are needed to make a definite conclusion. HOTAIR expression was shown to be associated with prognosis, and poor disease-free survival in bladder cancer (Heubach et al., 2015). Another study by Yan et al. demonstrated a correlation between HOTAIR high expression with histological grade and recurrence rate (Zhang et al., 2022). Additionally, HOTAIR was also shown to be enriched in urinary exosomes of high-grade muscle-invasive bladder cancer (Berrondo et al., 2016). Another lncRNA GAS5 was found to be decreased in bladder cancer. Downregulated expression of GAS5 was shown to be related to poor disease-free survival (Cao et al., 2016). H19 expression was also found to correlate with overall survival (Luo et al., 2013b). MALAT1 overexpression was reported to have a significant association with the grade and metastasis in bladder cancer (Amodio et al., 2018), indicating that MALAT1 may be used as a prognostic biomarker in bladder cancer.
Future perspectives
With advancing research, it has been well established that ncRNAs are linked to tumor progression, metastasis, and chemoresistance (Cho et al., 2006; Real and Malats, 2007). Several clinical studies have indicated that upregulated or downregulated expression of ncRNAs is associated with the grade, stage, and metastatic potential of cancer cells. Therefore, ncRNAs can be utilized for the early detection and better management of bladder cancer (Real and Malats, 2007). However, the functional role of ncRNAs is still perplexing due to the intricacy and multiplicity of their expression profiles and targeting proteins. Both miRNA and lncRNAs interact with mRNAs which affect different cellular processes like immune response, proliferation, and autophagy (Ning et al., 2019). The effects of ncRNAs as autophagy inducers or inhibitors have been studied; however, there is a prerequisite for more experimental studies in preclinical and clinical settings. It is worth noting that autophagy plays a dual role in cancer, acting not only as a tumor suppressor but also tumor enhancer process. Therefore, the relationship between autophagy and ncRNAs may not be simple. The effect of autophagy inhibitors in combination with targeted knockdown or overexpression of specific ncRNAs on bladder cancer progression needs to be explored. Blocking autophagy has been shown to sensitize the bladder cancer cells to gemcitabine and cisplatin. In the future, ncRNAs which are strong inhibitors of autophagy can be used in combination with these standard drugs to increase their efficacy in bladder cancer patients. Since autophagy is a key process in developing chemoresistance, targeted inhibition of autophagy by ncRNAs may sensitize these cells toward the standard chemotherapeutic drugs. The diagnostic, therapeutic, and prognostic value of the ncRNA and autophagy in urologic cancers needs to be further investigated for the better treatment and management of bladder cancer. In addition to specific proteins and the corresponding signaling pathways, some other factors, such as tumor microenvironment and tumor immunity also play roles in the development of urologic cancers. The role of ncRNAs in these processes will be really interesting to study. The molecular mechanism of the ncRNA-autophagy-immunity axis and its role in urologic tumorigenesis must be further clarified and explored. The answer to these questions may be helpful for finding new diagnostic biomarkers and innovative therapeutic alternatives in urologic malignancies.
Conclusion
Various studies have clearly established the role of ncRNAs in tumor progression, mediating metastatic potential and drug resistance in bladder cancer. Also, the role of ncRNAs as diagnostic and prognostic markers for bladder cancer has been demonstrated by several studies. Once the role of ncRNAs is well explored, the therapeutic strategies can be precisely tailored for the effective treatment of bladder cancer. For example, by targeting a specific ncRNA the sensitivity towards routine drugs can be significantly increased. In addition, the combinatorial intervention of targeting both ncRNAs and autophagy might be a favorable therapeutic strategy for bladder cancer. With the help of new cutting-edge technologies, the role of ncRNA will be crucial for the development of precision medicine.
Author contributions
IT wrote the first draft of manuscript. RO and AS performed the literature search. All authors reviewed the final draft and approved the final draft of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adam, L., Zhong, M., Choi, W., Qi, W., Nicoloso, M., Arora, A., et al. (2009). miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 15 (16), 5060–5072. doi:10.1158/1078-0432.CCR-08-2245
Amaravadi, R. K., Kimmelman, A. C., and Debnath, J. (2019). Targeting autophagy in cancer: Recent advances and future directions. Cancer Discov. 9 (9), 1167–1181. doi:10.1158/2159-8290.CD-19-0292
Amodio, N., Raimondi, L., Juli, G., Stamato, M. A., Caracciolo, D., Tagliaferri, P., et al. (2018). MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 11 (1), 63. doi:10.1186/s13045-018-0606-4
Anderson, B. (2018). Bladder cancer: Overview and disease management. Part 1: Non-muscle-invasive bladder cancer. Br. J. Nurs. 27 (9), S27–S37. doi:10.12968/bjon.2018.27.9.S27
Atala, A. (2013). Re: Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. J. Urol. 190 (6), 2306. doi:10.1016/j.juro.2013.08.057
Audenet, F., Attalla, K., and Sfakianos, J. P. (2018). The evolution of bladder cancer genomics: What have we learned and how can we use it? Urol. Oncol. 36 (7), 313–320. doi:10.1016/j.urolonc.2018.02.017
Avgeris, M., Tsilimantou, A., Levis, P. K., Rampias, T., Papadimitriou, M. A., Panoutsopoulou, K., et al. (2019). Unraveling UCA1 lncRNA prognostic utility in urothelial bladder cancer. Carcinogenesis 40 (8), 965–974. doi:10.1093/carcin/bgz045
Aznaourova, M., Schmerer, N., Schmeck, B., and Schulte, L. N. (2020). Disease-causing mutations and rearrangements in long non-coding RNA gene loci. Front. Genet. 11, 527484. doi:10.3389/fgene.2020.527484
Berrondo, C., Flax, J., Kucherov, V., Siebert, A., Osinski, T., Rosenberg, A., et al. (2016). Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS One 11 (1), e0147236. doi:10.1371/journal.pone.0147236
Cai, X., Qu, L., Yang, J., Xu, J., Sun, L., Wei, X., et al. (2020). Exosome-transmitted microRNA-133b inhibited bladder cancer proliferation by upregulating dual-specificity protein phosphatase 1. Cancer Med. 9 (16), 6009–6019. doi:10.1002/cam4.3263
Cai, Y., An, B., Yao, D., Zhou, H., and Zhu, J. (2021). MicroRNA miR-30a inhibits cisplatin resistance in ovarian cancer cells through autophagy. Bioengineered 12 (2), 10713–10722. doi:10.1080/21655979.2021.2001989
Cao, Q., Wang, N., Qi, J., Gu, Z., and Shen, H. (2016). Long non-coding RNA-GAS5 acts as a tumor suppressor in bladder transitional cell carcinoma via regulation of chemokine (C-C motif) ligand 1 expression. Mol. Med. Rep. 13 (1), 27–34. doi:10.3892/mmr.2015.4503
Cech, T. R., and Steitz, J. A. (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157 (1), 77–94. doi:10.1016/j.cell.2014.03.008
Chang, Y., Jin, H., Li, H., Ma, J., Zheng, Z., Sun, B., et al. (2020). MiRNA-516a promotes bladder cancer metastasis by inhibiting MMP9 protein degradation via the AKT/FOXO3A/SMURF1 axis. Clin. Transl. Med. 10 (8), e263. doi:10.1002/ctm2.263
Chen, D., Cheng, L., Cao, H., and Liu, W. (2021). Role of microRNA-381 in bladder cancer growth and metastasis with the involvement of BMI1 and the Rho/ROCK axis. BMC Urol. 21 (1), 5. doi:10.1186/s12894-020-00775-3
Chen, Y., Li, Z., Chen, X., and Zhang, S. (2021). Long non-coding RNAs: From disease code to drug role. Acta Pharm. Sin. B 11 (2), 340–354. doi:10.1016/j.apsb.2020.10.001
Cheng, H., Sharen, G., Wang, Z., and Zhou, J. (2021). LncRNA UCA1 enhances cisplatin resistance by regulating CYP1B1-mediated apoptosis via miR-513a-3p in human gastric cancer. Cancer Manag. Res. 13, 367–377. doi:10.2147/CMAR.S277399
Chi, Y., Wang, D., Wang, J., Yu, W., and Yang, J. (2019). Long non-coding RNA in the pathogenesis of cancers. Cells 8 (9), 1015. doi:10.3390/cells8091015
Cho, H. J., Kim, J. K., Kim, K. D., Yoon, H. K., Cho, M. Y., Park, Y. P., et al. (2006). Upregulation of Bcl-2 is associated with cisplatin-resistance via inhibition of Bax translocation in human bladder cancer cells. Cancer Lett. 237 (1), 56–66. doi:10.1016/j.canlet.2005.05.039
Churko, J. M., Mantalas, G. L., Snyder, M. P., and Wu, J. C. (2013). Overview of high throughput sequencing technologies to elucidate molecular pathways in cardiovascular diseases. Circ. Res. 112 (12), 1613–1623. doi:10.1161/CIRCRESAHA.113.300939
Compérat, E., Wasinger, G., Oszwald, A., and Shariat, S. F. (2022). Brief update of the new WHO classification for urothelial carcinoma. Curr. Opin. Urol. 32 (5), 511–516. doi:10.1097/MOU.0000000000001020
Cong, L., Yang, Q., Hu, C., Yu, Q., Hao, S., and Li, D. (2019). Current status of functional studies on circular RNAs in bladder cancer and their potential role as diagnostic and prognostic biomarkers: A review. Med. Sci. Monit. 25, 3425–3434. doi:10.12659/MSM.916697
Daneshmand, S. (2020). Bladder cancer: Advances and innovations. Eur. Urol. Focus 6 (4), 613–614. doi:10.1016/j.euf.2020.02.012
Deng, H., Lv, L., Li, Y., Zhang, C., Meng, F., Pu, Y., et al. (2014). miR-193a-3p regulates the multi-drug resistance of bladder cancer by targeting the LOXL4 gene and the oxidative stress pathway. Mol. Cancer 13, 234. doi:10.1186/1476-4598-13-234
Dikic, I., and Elazar, Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19 (6), 349–364. doi:10.1038/s41580-018-0003-4
Ding, Z., Ying, W., He, Y., Chen, X., Jiao, Y., Wang, J., et al. (2021). lncRNA-UCA1 in the diagnosis of bladder cancer: A meta-analysis. Med. Baltim. 100 (11), e24805. doi:10.1097/MD.0000000000024805
Dong, H., Zhang, L., and Liu, S. (2022). Targeting HMGB1: An available therapeutic strategy for breast cancer therapy. Int. J. Biol. Sci. 18 (8), 3421–3434. doi:10.7150/ijbs.73504
Drayton, R. M., Dudziec, E., Peter, S., Bertz, S., Hartmann, A., Bryant, H. E., et al. (2014). Reduced expression of miRNA-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin. Cancer Res. 20 (7), 1990–2000. doi:10.1158/1078-0432.CCR-13-2805
Fan, Y., Shen, B., Tan, M., Mu, X., Qin, Y., Zhang, F., et al. (2014). Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 281 (7), 1750–1758. doi:10.1111/febs.12737
Fan, Y., Shen, B., Tan, M., Mu, X., Qin, Y., Zhang, F., et al. (2014). TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 20 (6), 1531–1541. doi:10.1158/1078-0432.CCR-13-1455
Fernandes, J. C. R., Acuña, S. M., Aoki, J. I., Floeter-Winter, L. M., and Muxel, S. M. (2019). Long non-coding RNAs in the regulation of gene expression: Physiology and disease. Noncoding. RNA 5 (1), 17. doi:10.3390/ncrna5010017
Fojo, T. (2007). Multiple paths to a drug resistance phenotype: Mutations, translocations, deletions and amplification of coding genes or promoter regions, epigenetic changes and microRNAs. Drug resist. updat. 10 (1–2), 59–67. doi:10.1016/j.drup.2007.02.002
Ghafouri-Fard, S., Esmaeili, M., and Taheri, M. (2020). H19 lncRNA: Roles in tumorigenesis. Biomed. Pharmacother. 123, 109774. doi:10.1016/j.biopha.2019.109774
Ghafouri-Fard, S., Shoorei, H., Mohaqiq, M., Majidpoor, J., Moosavi, M. A., and Taheri, M. (2022). Exploring the role of non-coding RNAs in autophagy. Autophagy 18 (5), 949–970. doi:10.1080/15548627.2021.1883881
Guo, C., Li, X., Xie, J., Liu, D., Geng, J., Ye, L., et al. (2021). Long noncoding RNA SNHG1 activates autophagy and promotes cell invasion in bladder cancer. Front. Oncol. 11, 660551. doi:10.3389/fonc.2021.660551
Guo, Y., Sun, W., Gao, W., Li, L., Liang, Y., Mei, Z., et al. (2022). Long noncoding RNA H19 derived from M2 tumor-associated macrophages promotes bladder cell autophagy via stabilizing ULK1. J. Oncol. 2022, 3465459. doi:10.1155/2022/3465459
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464 (7291), 1071–1076. doi:10.1038/nature08975
Hajjari, M., and Salavaty, A. (2015). HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 12 (1), 1–9. doi:10.7497/j.issn.2095-3941.2015.0006
Hanahan, D. (2022). Hallmarks of cancer: New dimensions. Cancer Discov. 12 (1), 31–46. doi:10.1158/2159-8290.CD-21-1059
He, Y., Luo, Y., Liang, B., Ye, L., Lu, G., and He, W. (2017). Potential applications of MEG3 in cancer diagnosis and prognosis. Oncotarget 8 (42), 73282–73295. doi:10.18632/oncotarget.19931
Heubach, J., Monsior, J., Deenen, R., Niegisch, G., Szarvas, T., Niedworok, C., et al. (2015). The long noncoding RNA HOTAIR has tissue and cell type-dependent effects on HOX gene expression and phenotype of urothelial cancer cells. Mol. Cancer 14, 108. doi:10.1186/s12943-015-0371-8
Homami, A., and Ghazi, F. (2016). MicroRNAs as biomarkers associated with bladder cancer. Med. J. Islam. Repub. Iran. 30, 475.
Huang, C. S., Ho, J. Y., Chiang, J. H., Yu, C. P., and Yu, D. S. (2020). Exosome-derived LINC00960 and LINC02470 promote the epithelial-mesenchymal transition and aggressiveness of bladder cancer cells. Cells 9 (6), 1419. doi:10.3390/cells9061419
Huang, C. S., Tsai, C. H., Yu, C. P., Wu, Y. S., Yee, M. F., Ho, J. Y., et al. (2022). Long noncoding RNA LINC02470 sponges MicroRNA-143-3p and enhances SMAD3-mediated epithelial-to-mesenchymal transition to promote the aggressive properties of bladder cancer. Cancers (Basel) 14 (4), 968. doi:10.3390/cancers14040968
Ishaq, M., Khan, M. A., Sharma, K., Sharma, G., Dutta, R. K., and Majumdar, S. (2014). Gambogic acid induced oxidative stress dependent caspase activation regulates both apoptosis and autophagy by targeting various key molecules (NF-κB, Beclin-1, p62 and NBR1) in human bladder cancer cells. Biochim. Biophys. Acta 1840 (12), 3374–3384. doi:10.1016/j.bbagen.2014.08.019
Ishaq, M., Ojha, R., Sharma, A. P., and Singh, S. K. (2020). Autophagy in cancer: Recent advances and future directions. Semin. Cancer Biol. 66, 171–181. doi:10.1016/j.semcancer.2020.03.010
Ishaq, M., Ojha, R., Sharma, K., Sharma, G., Singh, S. K., and Majumdar, S. (2016). Functional inhibition of Hsp70 by Pifithrin-μ switches Gambogic acid induced caspase dependent cell death to caspase independent cell death in human bladder cancer cells. Biochim. Biophys. Acta 1863 (11), 2560–2573. doi:10.1016/j.bbamcr.2016.07.001
Jia, L., Liu, W., Guan, L., Lu, M., and Wang, K. (2015). Inhibition of calcium-activated Chloride Channel ANO1/tmem16a suppresses tumor growth and invasion in human lung cancer. PLoS One 10 (8), e0136584. doi:10.1371/journal.pone.0136584
Kaur, J., Salehen, N., Norazit, A., Rahman, A. A., Murad, N. A. A., Rahman, N. M. A. N. A., et al. (2022). Tumor suppressive effects of GAS5 in cancer cells. Noncoding. RNA 8 (3), 39. doi:10.3390/ncrna8030039
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., and Chrousos, G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3 (107), ra8. doi:10.1126/scisignal.2000568
Lai, C., Wu, Z., Shi, J., Li, K., Zhu, J., Chen, Z., et al. (2020). Autophagy-related long noncoding RNAs can predict prognosis in patients with bladder cancer. Aging (Albany NY) 12 (21), 21582–21596. doi:10.18632/aging.103947
Lebrun, L., Milowich, D., Le Mercier, M., Allard, J., Van Eycke, Y. R., Roumeguere, T., et al. (2018). UCA1 overexpression is associated with less aggressive subtypes of bladder cancer. Oncol. Rep. 40 (5), 2497–2506. doi:10.3892/or.2018.6697
Lemke, E. A., and Shah, A. Y. (2018). Management of advanced bladder cancer: An update. J. Adv. Pract. Oncol. 9 (4), 410–416.
Li, C., Cui, Y., Liu, L. F., Ren, W. B., Li, Q. Q., Zhou, X., et al. (2017). High expression of long noncoding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin. Genitourin. Cancer 15 (5), 570–576. doi:10.1016/j.clgc.2017.05.001
Li, H., Yu, G., Shi, R., Lang, B., Chen, X., Xia, D., et al. (2022). Correction: Cisplatin-induced epigenetic activation of miR-34a sensitizes bladder cancer cells to chemotherapy. Mol. Cancer 21 (1), 8. doi:10.1186/1476-4598-13-8
Li, H. J., Gong, X., Li, Z. K., Qin, W., He, C. X., Xing, L., et al. (2021). Role of long non-coding RNAs on bladder cancer. Front. Cell Dev. Biol. 9, 672679. doi:10.3389/fcell.2021.672679
Li, J., Li, Z., Bai, X., Chen, X., Wang, M., Wu, Y., et al. (2022). LncRNA UCA1 promotes the progression of AML by upregulating the expression of CXCR4 and CYP1B1 by affecting the stability of METTL14. J. Oncol. 2022, 2756986. doi:10.1155/2022/2756986
Li, J., Xu, X., Meng, S., Liang, Z., Wang, X., Xu, M., et al. (2017). MET/SMAD3/SNAIL circuit mediated by miR-323a-3p is involved in regulating epithelial-mesenchymal transition progression in bladder cancer. Cell Death Dis. 8 (8), e3010. doi:10.1038/cddis.2017.331
Li, L., Huang, C., He, Y., Sang, Z., Liu, G., and Dai, H. (2018). Knockdown of long non-coding RNA GAS5 increases miR-23a by targeting ATG3 involved in autophagy and cell viability. Cell. Physiol. biochem. 48 (4), 1723–1734. doi:10.1159/000492300
Li, P., Yang, X., Cheng, Y., Zhang, X., Yang, C., Deng, X., et al. (2017). MicroRNA-218 increases the sensitivity of bladder cancer to cisplatin by targeting Glut1. Cell. Physiol. biochem. 41 (3), 921–932. doi:10.1159/000460505
Li, X., Chen, W., Li, R., Chen, X., Huang, G., Lu, C., et al. (2022). Bladder cancer diagnosis with a four-miRNA panel in serum. Future Oncol. 18 (29), 3311–3322. doi:10.2217/fon-2022-0448
Li, X., Zhou, Y., Li, Y., Yang, L., Ma, Y., Peng, X., et al. (2019). Autophagy: A novel mechanism of chemoresistance in cancers. Biomed. Pharmacother. 119, 109415. doi:10.1016/j.biopha.2019.109415
Li, Y., Shan, Z., Liu, C., Yang, D., Wu, J., Men, C., et al. (2017). MicroRNA-294 promotes cellular proliferation and motility through the PI3K/AKT and JAK/STAT pathways by upregulation of NRAS in bladder cancer. Biochemistry. 82 (4), 474–482. doi:10.1134/S0006297917040095
Li, Y., Sun, L., Guo, X., Mo, N., Zhang, J., and Li, C. (2021). Frontiers in bladder cancer genomic research. Front. Oncol. 11, 670729. doi:10.3389/fonc.2021.670729
Li, Z., Yu, D., Li, H., Lv, Y., and Li, S. (2019). Long non-coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int. J. Oncol. 54 (3), 1033–1042. doi:10.3892/ijo.2019.4679
Lian, J., Lin, S. H., Ye, Y., Chang, D. W., Huang, M., Dinney, C. P., et al. (2018). Serum microRNAs as predictors of risk for non-muscle invasive bladder cancer. Oncotarget 9 (19), 14895–14908. doi:10.18632/oncotarget.24473
Liang, T., Xu, F., Wan, P., Zhang, L., Huang, S., Yang, N., et al. (2021). Malat-1 expression in bladder carcinoma tissues and its clinical significance. Am. J. Transl. Res. 13 (4), 3555–3560.
Liao, D., Li, T., Ye, C., Zeng, L., Li, H., Pu, X., et al. (2018). miR-221 inhibits autophagy and targets TP53INP1 in colorectal cancer cells. Exp. Ther. Med. 15 (2), 1712–1717. doi:10.3892/etm.2017.5522
Liao, G., Chen, F., Zhong, J., and Jiang, X. (2018). MicroRNA-539 inhibits the proliferation and invasion of bladder cancer cells by regulating IGF-1R. Mol. Med. Rep. 17 (4), 4917–4924. doi:10.3892/mmr.2018.8497
Lin, F., Yin, H. B., Li, X. Y., Zhu, G. M., He, W. Y., and Gou, X. (2020). Bladder cancer cell-secreted exosomal miR-21 activates the PI3K/AKT pathway in macrophages to promote cancer progression. Int. J. Oncol. 56 (1), 151–164. doi:10.3892/ijo.2019.4933
Lin, G. B., Zhang, C. M., Chen, X. Y., Wang, J. W., Chen, S., Tang, S. Y., et al. (2019). Identification of circulating miRNAs as novel prognostic biomarkers for bladder cancer. Math. Biosci. Eng. 17 (1), 834–844. doi:10.3934/mbe.2020044
Liu, C., Chen, Z., Fang, J., Xu, A., Zhang, W., and Wang, Z. (2016). H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol. 37 (1), 263–270. doi:10.1007/s13277-015-3779-2
Liu, C., Ji, L., and Song, X. (2019). Long non coding RNA UCA1 contributes to the autophagy and survival of colorectal cancer cells via sponging miR-185-5p to up-regulate the WISP2/β-catenin pathway. RSC Adv. 9 (25), 14160–14166. doi:10.1039/c8ra10468a
Liu, G., Zhao, X., Zhou, J., Cheng, X., Ye, Z., and Ji, Z. (2018). Long non-coding RNA MEG3 suppresses the development of bladder urothelial carcinoma by regulating miR-96 and TPM1. Cancer Biol. Ther. 19 (11), 1039–1056. doi:10.1080/15384047.2018.1480279
Liu, K., Gao, L., Ma, X., Huang, J. J., Chen, J., Zeng, L., et al. (2020). Long non-coding RNAs regulate drug resistance in cancer. Mol. Cancer 19 (1), 54. doi:10.1186/s12943-020-01162-0
Liu, L., Qiu, M., Tan, G., Liang, Z., Qin, Y., Chen, L., et al. (2022). Correction to: miR-200c inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3. J. Transl. Med. 20 (1), 305. doi:10.1186/s12967-014-0305-z
Liu, S., Chen, X., and Lin, T. (2022). Emerging strategies for the improvement of chemotherapy in bladder cancer: Current knowledge and future perspectives. J. Adv. Res. 39, 187–202. doi:10.1016/j.jare.2021.11.010
Liu, X., Zhou, Z., Wang, Y., Zhu, K., Deng, W., Li, Y., et al. (2020). Downregulation of HMGA1 mediates autophagy and inhibits migration and invasion in bladder cancer via miRNA-221/TP53INP1/p-ERK Axis. Front. Oncol. 10, 589. doi:10.3389/fonc.2020.00589
Liu, Y. W., Sun, M., Xia, R., Zhang, E. B., Liu, X. H., Zhang, Z. H., et al. (2015). LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 6 (7), e1802. doi:10.1038/cddis.2015.150
Lodewijk, I., Dueñas, M., Rubio, C., Munera-Maravilla, E., Segovia, C., Bernardini, A., et al. (2018). Liquid biopsy biomarkers in bladder cancer: A current need for patient diagnosis and monitoring. Int. J. Mol. Sci. 19 (9), 2514. doi:10.3390/ijms19092514
Luo, H., Yang, R., Li, C., Tong, Y., Fan, L., Liu, X., et al. (2017). MicroRNA-139-5p inhibits bladder cancer proliferation and self-renewal by targeting the Bmi1 oncogene. Tumour Biol. 39 (7), 1010428317718414. doi:10.1177/1010428317718414
Luo, M., Li, Z., Wang, W., Zeng, Y., Liu, Z., and Qiu, J. (2013). Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 333 (2), 213–221. doi:10.1016/j.canlet.2013.01.033
Luo, M., Li, Z., Wang, W., Zeng, Y., Liu, Z., and Qiu, J. (2013). Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 280 (7), 1709–1716. doi:10.1111/febs.12185
Mao, W., Wang, K., Sun, S., Wu, J., Chen, M., Geng, J., et al. (2021). ID2 inhibits bladder cancer progression and metastasis via PI3K/AKT signaling pathway. Front. Cell Dev. Biol. 9, 738364. doi:10.3389/fcell.2021.738364
Mei, Y., Zheng, J., Xiang, P., Liu, C., and Fan, Y. (2020). Prognostic value of the miR-200 family in bladder cancer: A systematic review and meta-analysis. Med. Baltim. 99 (47), e22891. doi:10.1097/MD.0000000000022891
Michlewski, G., and Cáceres, J. F. (2019). Post-transcriptional control of miRNA biogenesis. RNA 25 (1), 1–16. doi:10.1261/rna.068692.118
Mirzaei, S., Paskeh, M. D. A., Hashemi, F., Zabolian, A., Hashemi, M., Entezari, M., et al. (2022). Long non-coding RNAs as new players in bladder cancer: Lessons from pre-clinical and clinical studies. Life Sci. 288, 119948. doi:10.1016/j.lfs.2021.119948
Mitash, N., Tiwari, S., Agnihotri, S., and Mandhani, A. (2017). Bladder cancer: Micro RNAs as biomolecules for prognostication and surveillance. Indian J. Urol. 33 (2), 127–133. doi:10.4103/0970-1591.203412
Mulcahy Levy, J. M., and Thorburn, A. (2020). Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 27 (3), 843–857. doi:10.1038/s41418-019-0474-7
Ning, B., Yu, D., and Yu, A. M. (2019). Advances and challenges in studying noncoding RNA regulation of drug metabolism and development of RNA therapeutics. Biochem. Pharmacol. 169, 113638. doi:10.1016/j.bcp.2019.113638
Nojima, T., and Proudfoot, N. J. (2022). Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat. Rev. Mol. Cell Biol. 23 (6), 389–406. doi:10.1038/s41580-021-00447-6
O’Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9, 402. doi:10.3389/fendo.2018.00402
Ojha, R., Ishaq, M., and Singh, S. K. (2015). Caspase-mediated crosstalk between autophagy and apoptosis: Mutual adjustment or matter of dominance. J. Cancer Res. Ther. 11 (3), 514–524. doi:10.4103/0973-1482.163695
Ojha, R., Jha, V., and Singh, S. K. (2016). Gemcitabine and mitomycin induced autophagy regulates cancer stem cell pool in urothelial carcinoma cells. Biochim. Biophys. Acta 1863 (2), 347–359. doi:10.1016/j.bbamcr.2015.12.002
Ojha, R., Leli, N. M., Onorati, A., Piao, S., Verginadis, , Tameire, F., et al. (2019). ER translocation of the MAPK pathway drives therapy resistance in BRAF-mutant melanoma. Cancer Discov. 9 (3), 396–415. doi:10.1158/2159-8290.CD-18-0348
Ojha, R., Singh, S. K., Bhattacharyya, S., Dhanda, R. S., Rakha, A., Mandal, A. K., et al. (2014). Inhibition of grade dependent autophagy in urothelial carcinoma increases cell death under nutritional limiting condition and potentiates the cytotoxicity of chemotherapeutic agent. J. Urol. 191 (6), 1889–1898. doi:10.1016/j.juro.2014.01.006
Ojha, R., Tantray, I., Rimal, S., Mitra, S., Cheshier, S., and Lu, B. (2022). Regulation of reverse electron transfer at mitochondrial complex I by unconventional Notch action in cancer stem cells. Dev. Cell 57 (2), 260–276.e9. doi:10.1016/j.devcel.2021.12.020
Onorati, A. V., Dyczynski, M., Ojha, R., and Amaravadi, R. K. (2018). Targeting autophagy in cancer. Cancer 124 (16), 3307–3318. doi:10.1002/cncr.31335
Papaioannou, E., González-Molina, M. D. P., Prieto-Muñoz, A. M., Gámez-Reche, L., and González-Martín, A. (2021). Regulation of adaptive tumor immunity by non-coding RNAs. Cancers (Basel) 13 (22), 5651. doi:10.3390/cancers13225651
Pardo, J. C., Ruiz de Porras, V., Plaja, A., Carrato, C., Etxaniz, O., Buisan, O., et al. (2020). Moving towards personalized medicine in muscle-invasive bladder cancer: Where are we now and where are we going? Int. J. Mol. Sci. 21 (17), 6271. doi:10.3390/ijms21176271
Poillet-Perez, L., Despouy, G., Delage-Mourroux, R., and Boyer-Guittaut, M. (2015). Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 4, 184–192. doi:10.1016/j.redox.2014.12.003
Pop-Bica, C., Gulei, D., Cojocneanu-Petric, R., Braicu, C., Petrut, B., and Berindan-Neagoe, I. (2017). Understanding the role of non-coding RNAs in bladder cancer: From dark matter to valuable therapeutic targets. Int. J. Mol. Sci. 18 (7), 1514. doi:10.3390/ijms18071514
Rani, V., and Sengar, R. S. (2022). Biogenesis and mechanisms of microRNA-mediated gene regulation. Biotechnol. Bioeng. 119 (3), 685–692. doi:10.1002/bit.28029
Raveh, E., Matouk, I. J., Gilon, M., and Hochberg, A. (2015). The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol. Cancer 14, 184. doi:10.1186/s12943-015-0458-2
Real, F. X., and Malats, N. (2007). Bladder cancer and apoptosis: Matters of life and death. Lancet. Oncol. 8 (2), 91–92. doi:10.1016/S1470-2045(07)70008-6
Reuter, J. A., Spacek, D. V., and Snyder, M. P. (2015). High-throughput sequencing technologies. Mol. Cell 58 (4), 586–597. doi:10.1016/j.molcel.2015.05.004
Saginala, K., Barsouk, A., Aluru, J. S., Rawla, P., Padala, S. A., and Barsouk, A. (2020). Epidemiology of bladder cancer. Med. Sci. 8 (1), 15. doi:10.3390/medsci8010015
Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S., and Ghaffari, S. H. (2019). An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 234 (5), 5451–5465. doi:10.1002/jcp.27486
Shan, C., Chen, X., Cai, H., Hao, X., Li, J., Zhang, Y., et al. (2021). The emerging roles of autophagy-related MicroRNAs in cancer. Int. J. Biol. Sci. 17 (1), 134–150. doi:10.7150/ijbs.50773
Shen, M., Li, X., Qian, B., Wang, Q., Lin, S., Wu, W., et al. (2021). Crucial roles of microRNA-mediated autophagy in urologic malignancies. Int. J. Biol. Sci. 17 (13), 3356–3368. doi:10.7150/ijbs.61175
Siracusano, S., Rizzetto, R., and Porcaro, A. B. (2020). Bladder cancer genomics. Urologia 87 (2), 49–56. doi:10.1177/0391560319899011
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22 (2), 96–118. doi:10.1038/s41580-020-00315-9
Sun, Y., and Ma, L. (2019). New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel) 11 (2), 216. doi:10.3390/cancers11020216
Taheri, M., Shirvani-Farsani, Z., Ghafouri-Fard, S., and Omrani, M. D. (2020). Expression profile of microRNAs in bladder cancer and their application as biomarkers. Biomed. Pharmacother. 131, 110703. doi:10.1016/j.biopha.2020.110703
Tan, M., Mu, X., Liu, Z., Tao, L., Wang, J., Ge, J., et al. (2017). microRNA-495 promotes bladder cancer cell growth and invasion by targeting phosphatase and tensin homolog. Biochem. Biophys. Res. Commun. 483 (2), 867–873. doi:10.1016/j.bbrc.2017.01.019
Tan, W., Yuan, Y., Huang, H., Ma, J., Li, Y., Gou, Y., et al. (2022). Comprehensive analysis of autophagy related long non-coding RNAs in prognosis, immunity, and treatment of muscular invasive bladder cancer. Sci. Rep. 12 (1), 11242. doi:10.1038/s41598-022-13952-1
Tang, Q., and Hann, S. S. (2018). HOTAIR: An oncogenic long non-coding RNA in human cancer. Cell. Physiol. biochem. 47 (3), 893–913. doi:10.1159/000490131
Tao, J., Lu, Q., Wu, D., Li, P., Xu, B., Qing, W., et al. (2011). microRNA-21 modulates cell proliferation and sensitivity to doxorubicin in bladder cancer cells. Oncol. Rep. 25 (6), 1721–1729. doi:10.3892/or.2011.1245
Tomasetti, M., Monaco, F., Manzella, N., Rohlena, J., Rohlenova, K., Staffolani, S., et al. (2016). MicroRNA-126 induces autophagy by altering cell metabolism in malignant mesothelioma. Oncotarget 7 (24), 36338–36352. doi:10.18632/oncotarget.8916
Towler, B. P., Jones, C. I., and Newbury, S. F. (2015). Mechanisms of regulation of mature miRNAs. Biochem. Soc. Trans. 43 (6), 1208–1214. doi:10.1042/BST20150157
Trujano-Camacho, S., Cantú-de León, D., Delgado-Waldo, I., Coronel-Hernández, J., Millan-Catalan, O., Hernández-Sotelo, D., et al. (2021). Inhibition of wnt-β-catenin signaling by ICRT14 drug depends of post-transcriptional regulation by HOTAIR in human cervical cancer HeLa cells. Front. Oncol. 11, 729228. doi:10.3389/fonc.2021.729228
Tsikrika, F. D., Avgeris, M., Levis, P. K., Tokas, T., Stravodimos, K., and Scorilas, A. (2018). miR-221/222 cluster expression improves clinical stratification of non-muscle invasive bladder cancer (TaT1) patients’ risk for short-term relapse and progression. Genes Chromosom. Cancer 57 (3), 150–161. doi:10.1002/gcc.22516
Usuba, W., Urabe, F., Yamamoto, Y., Matsuzaki, J., Sasaki, H., Ichikawa, M., et al. (2019). Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 110 (1), 408–419. doi:10.1111/cas.13856
Wang, F., Wu, H., Fan, M., Yu, R., Zhang, Y., Liu, J., et al. (2020). Sodium butyrate inhibits migration and induces AMPK-mTOR pathway-dependent autophagy and ROS-mediated apoptosis via the miR-139-5p/Bmi-1 axis in human bladder cancer cells. FASEB J. 34 (3), 4266–4282. doi:10.1096/fj.201902626R
Wang, G., Zheng, L., Yu, Z., Liao, G., Lu, L., Xu, R., et al. (2012). Increased cyclin-dependent kinase 6 expression in bladder cancer. Oncol. Lett. 4 (1), 43–46. doi:10.3892/ol.2012.695
Wang, H., Guan, Z., He, K., Qian, J., Cao, J., and Teng, L. (2017). LncRNA UCA1 in anti-cancer drug resistance. Oncotarget 8 (38), 64638–64650. doi:10.18632/oncotarget.18344
Wang, W., and Yin, Z. (2017). Diagnostic value of long non-coding RNA H19, UCA1, and HOTAIR as promising biomarkers in human bladder cancer. Int. J. Clin. Exp. Pathol. 10 (12), 11659–11665.
Wang, W. T., Han, C., Sun, Y. M., Chen, T. Q., and Chen, Y. Q. (2019). Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 12 (1), 55. doi:10.1186/s13045-019-0748-z
Winkle, M., El-Daly, S. M., Fabbri, M., and Calin, G. A. (2021). Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 20 (8), 629–651. doi:10.1038/s41573-021-00219-z
Wu, B., Zhang, Y., Yu, Y., Zhong, C., Lang, Q., Liang, Z., et al. (2021). Long noncoding RNA H19: A novel therapeutic target emerging in oncology via regulating oncogenic signaling pathways. Front. Cell Dev. Biol. 9, 796740. doi:10.3389/fcell.2021.796740
Wu, H., and Zhou, C. (2018). Long non-coding RNA UCA1 promotes lung cancer cell proliferation and migration via microRNA-193a/HMGB1 axis. Biochem. Biophys. Res. Commun. 496 (2), 738–745. doi:10.1016/j.bbrc.2018.01.097
Xiang, W., Wu, X., Huang, C., Wang, M., Zhao, X., Luo, G., et al. (2017). PTTG1 regulated by miR-146a-3p promotes bladder cancer migration, invasion, metastasis and growth. Oncotarget 8 (1), 664–678. doi:10.18632/oncotarget.13507
Xie, H., Liao, X., Chen, Z., Fang, Y., He, A., Zhong, Y., et al. (2017). LncRNA MALAT1 inhibits apoptosis and promotes invasion by antagonizing miR-125b in bladder cancer cells. J. Cancer 8 (18), 3803–3811. doi:10.7150/jca.21228
Xie, Y., Ma, X., Chen, L., Li, H., Gu, L., Gao, Y., et al. (2017). MicroRNAs with prognostic significance in bladder cancer: A systematic review and meta-analysis. Sci. Rep. 7 (1), 5619. doi:10.1038/s41598-017-05801-3
Xiu, Y. L., Sun, K. X., Chen, X., Chen, S., Zhao, Y., Guo, Q. G., et al. (2017). Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget 8 (19), 31714–31725. doi:10.18632/oncotarget.15955
Xu, J., Wang, Y., Tan, X., and Jing, H. (2012). MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy 8 (6), 873–882. doi:10.4161/auto.19629
Xu, W., Yan, Z., Hu, F., Wei, W., Yang, C., and Sun, Z. (2020). Long non-coding RNA GAS5 accelerates oxidative stress in melanoma cells by rescuing EZH2-mediated CDKN1C downregulation. Cancer Cell Int. 20, 116. doi:10.1186/s12935-020-01167-1
Xu, X., Zhu, Y., Liang, Z., Li, S., Xu, X., Wang, X., et al. (2016). c-Met and CREB1 are involved in miR-433-mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3β/Snail signaling. Cell Death Dis. 7 (2), e2088. doi:10.1038/cddis.2015.274
Xue, M., Chen, W., Xiang, A., Wang, R., Chen, H., Pan, J., et al. (2017). Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol. Cancer 16 (1), 143. doi:10.1186/s12943-017-0714-8
Xue, M., Pang, H., Li, X., Li, H., Pan, J., and Chen, W. (2016). Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 107 (1), 18–27. doi:10.1111/cas.12844
Yang, H., Chen, Z., and Liu, Z. (2021). MiR-20a-5p negatively regulates NR4A3 to promote metastasis in bladder cancer. J. Oncol. 2021, 1377989. doi:10.1155/2021/1377989
Yang, L., Wang, H., Shen, Q., Feng, L., and Jin, H. (2017). Long non-coding RNAs involved in autophagy regulation. Cell Death Dis. 8 (10), e3073. doi:10.1038/cddis.2017.464
Yao, H., Han, B., Zhang, Y., Shen, L., and Huang, R. (2019). Non-coding RNAs and autophagy. Adv. Exp. Med. Biol. 1206, 199–220. doi:10.1007/978-981-15-0602-4_10
Yao, R. W., Wang, Y., and Chen, L. L. (2019). Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21 (5), 542–551. doi:10.1038/s41556-019-0311-8
Yin, X. H., Jin, Y. H., Cao, Y., Wong, Y., Weng, H., Sun, C., et al. (2019). Development of a 21-miRNA signature associated with the prognosis of patients with bladder cancer. Front. Oncol. 9, 729. doi:10.3389/fonc.2019.00729
Ying, L., Chen, Q., Wang, Y., Zhou, Z., Huang, Y., and Qiu, F. (2012). Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol. Biosyst. 8 (9), 2289–2294. doi:10.1039/c2mb25070e
Ying, L., Huang, Y., Chen, H., Wang, Y., Xia, L., Chen, Y., et al. (2013). Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. Biosyst. 9 (3), 407–411. doi:10.1039/c2mb25386k
Yu, G., Jia, Z., and Dou, Z. (2017). miR-24-3p regulates bladder cancer cell proliferation, migration, invasion and autophagy by targeting DEDD. Oncol. Rep. 37 (2), 1123–1131. doi:10.3892/or.2016.5326
Yu, H., Duan, P., Zhu, H., and Rao, D. (2017). miR-613 inhibits bladder cancer proliferation and migration through targeting SphK1. Am. J. Transl. Res. 9 (3), 1213–1221.
Yu, X., Wang, R., Han, C., Wang, Z., and Jin, X. (2020). A panel of urinary long non-coding RNAs differentiate bladder cancer from urocystitis. J. Cancer 11 (4), 781–787. doi:10.7150/jca.37006
Yu, Y., and Hann, S. S. (2019). Novel tumor suppressor lncRNA growth arrest-specific 5 (GAS5) in human cancer. Onco. Targets. Ther. 12, 8421–8436. doi:10.2147/OTT.S221305
Zeng, L. P., Hu, Z. M., Li, K., and Xia, K. (2016). miR-222 attenuates cisplatin-induced cell death by targeting the PPP2R2A/Akt/mTOR Axis in bladder cancer cells. J. Cell. Mol. Med. 20 (3), 559–567. doi:10.1111/jcmm.12760
Zhang, H. H., Huang, Z. X., Zhong, S. Q., Fei, K. L., and Cao, Y. H. (2020). miR-21 inhibits autophagy and promotes malignant development in the bladder cancer T24 cell line. Int. J. Oncol. 56 (4), 986–998. doi:10.3892/ijo.2020.4984
Zhang, J., Chen, K., Tang, Y., Luan, X., Zheng, X., Lu, X., et al. (2021). LncRNA-HOTAIR activates autophagy and promotes the imatinib resistance of gastrointestinal stromal tumor cells through a mechanism involving the miR-130a/ATG2B pathway. Cell Death Dis. 12 (4), 367. doi:10.1038/s41419-021-03650-7
Zhang, J., Mao, S., Wang, L., Zhang, W., Zhang, Z., Guo, Y., et al. (2019). MicroRNA-154 functions as a tumor suppressor in bladder cancer by directly targeting ATG7. Oncol. Rep. 41 (2), 819–828. doi:10.3892/or.2018.6879
Zhang, P., Wu, W., Chen, Q., and Chen, M. (2019). Non-coding RNAs and their integrated networks. J. Integr. Bioinform. 16 (3), 20190027. doi:10.1515/jib-2019-0027
Zhang, W., Wu, Q., Liu, Y., Wang, X., Ma, C., and Zhu, W. (2022). LncRNA HOTAIR promotes chemoresistance by facilitating epithelial to mesenchymal transition through miR-29b/PTEN/PI3K signaling in cervical cancer. Cells Tissues Organs 211 (1), 16–29. doi:10.1159/000519844
Zhang, X., Zhang, Y., Liu, X., Fang, A., Li, P., Li, Z., et al. (2015). MicroRNA-203 is a prognostic indicator in bladder cancer and enhances chemosensitivity to cisplatin via apoptosis by targeting bcl-w and Survivin. PLoS One 10 (11), e0143441. doi:10.1371/journal.pone.0143441
Zhang, X. F., Zhang, X. Q., Chang, Z. X., Wu, C. C., and Guo, H. (2018). microRNA-145 modulates migration and invasion of bladder cancer cells by targeting N-cadherin. Mol. Med. Rep. 17 (6), 8450–8456. doi:10.3892/mmr.2018.8910
Zhang, X. H., Hu, P., Xie, Y. Q., Kang, Y. J., and Li, M. (2019). Long noncoding RNA HOTAIR promotes endometrial carcinoma cell proliferation by binding to PTEN via the activating phosphatidylinositol 3-kinase/akt signaling pathway. Mol. Cell. Biol. 39 (23), 1–18. doi:10.1128/MCB.00251-19
Zhang, Y., Chen, X., Lin, J., and Jin, X. (2021). Biological functions and clinical significance of long noncoding RNAs in bladder cancer. Cell Death Discov. 7 (1), 278. doi:10.1038/s41420-021-00665-z
Zhao, X., Li, D., Zhao, S. T., Zhang, Y., Xu, A., Hu, Y. Y., et al. (2022). MiRNA-616 aggravates the progression of bladder cancer by regulating cell proliferation, migration and apoptosis through downregulating SOX7. Eur. Rev. Med. Pharmacol. Sci. 26 (12), 9304–9312. doi:10.26355/eurrev_201911_19423
Zhong, Y., Zhang, Y., Li, H., and Ma, W. (2019). LncRNAs act as prognostic biomarkers in bladder carcinoma: A meta-analysis. Heliyon 5 (11), e02785. doi:10.1016/j.heliyon.2019.e02785
Zhu, H., Wu, H., Liu, X., Li, B., Chen, Y., Ren, X., et al. (2009). Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 5 (6), 816–823. doi:10.4161/auto.9064
Zhu, J., Xu, C., Ruan, L., Wu, J., Li, Y., and Zhang, X. (2019). MicroRNA-146b overexpression promotes human bladder cancer invasion via enhancing ETS2-mediated mmp2 mRNA transcription. Mol. Ther. Nucleic Acids 16, 531–542. doi:10.1016/j.omtn.2019.04.007
Zhu, Z., Xu, L., Wan, Y., Zhou, J., Fu, D., Chao, H., et al. (2018). Inhibition of E-cadherin expression by lnc-RNA H19 to facilitate bladder cancer metastasis. Cancer Biomark. 22 (2), 275–281. doi:10.3233/CBM-170998
Keywords: non-coding RNA, autophagy, bladder cancer, drug resistance, miRNA and lncRNA
Citation: Tantray I, Ojha R and Sharma AP (2023) Non-coding RNA and autophagy: Finding novel ways to improve the diagnostic management of bladder cancer. Front. Genet. 13:1051762. doi: 10.3389/fgene.2022.1051762
Received: 23 September 2022; Accepted: 07 December 2022;
Published: 04 January 2023.
Edited by:
Zhongbao Zhou, Capital Medical University, ChinaReviewed by:
Sohini Chakraborty, New York University, United StatesXiaomei Yang, Johns Hopkins Medicine, United States
Copyright © 2023 Tantray, Ojha and Sharma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rani Ojha, b2poYS5yYW5pQHBnaW1lci5lZHUuaW4=; Aditya P. Sharma, YWRpdHlhLnAuc2hhcm1hQGdtYWlsLmNvbQ==
 Ishaq Tantray1
Ishaq Tantray1 Rani Ojha
Rani Ojha Aditya P. Sharma
Aditya P. Sharma