- 1Department of Biostatistics, State Key Laboratory of Organ Failure Research, Ministry of Education, and Guangdong Provincial Key Laboratory of Tropical Disease Research, School of Public Health, Southern Medical University, Guangzhou, China
- 2Guangdong-Hong Hong-Macao Joint Laboratory for Contaminants Exposure and Health, Guangzhou, China
Behavioral disinhibition is one of the important characteristics of many mental diseases. It has been reported in literature that serious behavioral disinhibition will affect people’s health and greatly reduce people’s quality of life. Meanwhile, behavioral disinhibition can easily lead to illegal drug abuse and violent crimes, etc., which will bring great harm to the society. At present, large-scale genome-wide association analysis has identified many loci associated with behavioral disinhibition. However, these studies have not incorporated the parent-of-origin effects (POE) into analysis, which may ignore or underestimate the genetic effects of loci on behavioral disinhibition. Therefore, in this article, we analyzed the five phenotypes related to behavioral disinhibition in the Minnesota Center for Twin and Family Research data (nicotine, alcohol consumption, alcohol dependence, illicit drugs, and non-substance use related behavioral disinhibition), to further explore the POE of variants on behavioral disinhibition. We applied a linear mixed model to test for the POE at a genome-wide scale on five transformed phenotypes, and found nine SNPs with statistically significant POE at the significance level of 5 × 10−8. Among them, SNPs rs4141854, rs9394515, and rs4711553 have been reported to be associated with two neurological disorders (restless legs syndrome and Tourette’s syndrome) which are related to behavioral disinhibition; SNPs rs12960235 and rs715351 have been found to be associated with head and neck squamous cell carcinoma, skin cancer and type I diabetes, while both SNPs have not been identified to be related to behavioral disinhibition in literature; SNPs rs704833, rs6837925, rs1863548, and rs11067062 are novel loci identified in this article, and their function annotations have not been reported in literature. Follow-up study in molecular genetics is needed to verify whether they are surely related to behavioral disinhibition.
Introduction
Behavioral disinhibition refers to the problematic and uncontrolled performance of impulsive behavior, and it has been observed to be an important characteristic of several neurodevelopmental and psychiatric diseases (Sharma et al., 2014). Its core feature is the inability to regulate immediate response inclinations at the expense of long-term gains or losses. There have been substantial studies consistently showing that the individuals with a high degree of behavioral disinhibition are at elevated risk for developing a broad array of behavioral disorders including substance use disorders (McGue et al., 2013). Meanwhile, behavioral disinhibition is a behavioral trait which is hypothesized to represent a general vulnerability in the development of substance use disorders, and the ability to manage immediate impulsive responses is reduced (Vrieze et al., 2013). Studies have shown that those with greater levels of disinhibition are thought to act more impulsively and to be more inclined to seek excitement, without considering the long-term consequences of their behaviors. People with a high degree of behavioral disinhibition are also more likely to use substances and have a more difficult time quitting (Iacono et al., 2008). Severe behavioral disinhibition is more generally related to increased negative health outcomes and increased mortality during the lifespan (Amirian et al., 2010; Kubzansky et al., 2011).
With the completion of the Human Genome Project, large-scale genetic data had been generated, and the research on behavioral disinhibition has also gone further to the genetic level. Some indicators were first used to quantitatively describe the degree of behavioral disinhibition, and then the relationship between single nucleotide polymorphisms (SNPs) and these quantitative indicators related to behavioral disinhibition were explored, aiming to capture the heritable variations which affect some behaviors including dangerous behaviors and impulsive behaviors, and finally they were utilized for discovering genetic loci which are likely to cause behavioral disinhibition disorders. The twin studies found that most of the genetic influences on individual-level behavioral disinhibition disorders can be attributed to genetic influences at the level of general factors, and the heritability of the behavioral disinhibition is estimated to be between 60% and 80% (Young et al., 2000; Krueger et al., 2002; Kendler et al., 2003). Some studies have pointed out that certain genes have common effects on multiple indicators of behavioral inhibition. For example, genetic variations affecting the γ-aminobutyric acid are related to the general addiction process (Krystal et al., 2006). Agrawal et al. (2006) have shown that the SNPs in the GABRA2 receptor gene are associated with severe addiction. Corley et al. (2008) also confirmed that the GABRA2 receptor gene is significantly associated with a variety of drug abuse and antisocial behaviors. Similarly, the mutations in the cholinergic muscarinic receptor 2 gene are associated with the risk for smoking (Mobascher et al., 2010). Vrieze et al. (2013) used the genome-wide complex trait analysis to estimate the aggregate genetic effect between 515,384 SNPs and behavioral disinhibition, and the estimated aggregated SNPs can explain 10%∼30% of the variances in the corresponding traits. McGue et al. (2013) analyzed five quantitative indicators related to behavioral disinhibition, i.e., “nicotine,” “alcohol consumption,” “alcohol dependence,” “illicit drugs,” and “non-substance use related behavioral disinhibition” (BD), and found that SNP rs1868152 is statistically significantly related to illegal drug abuse. At the same time, by adjusting the threshold of the p-values of genome-wide association study (GWAS), 13 candidate SNPs which may be related to behavioral disinhibition were found. Derringer et al. (2015) used 1,901 adolescents for the GWAS of behavioral disinhibition. No single SNP was found to be significantly associated with behavioral disinhibition. However, in the subgroup analysis, it was estimated that 49.3% of the variance in behavioral disinhibition within the Caucasian sub-sample could be explained by the genetic variations, and seven genes were identified to be significantly associated with behavioral disinhibition.
However, GWAS usually regards the alleles inherited from the mother and the father as equivalent, and generally does not consider parent-of-origin effects (POE). POE is an important epigenetic phenomenon, which means that the parental source of the chromosome where the gene is located determines whether the gene is expressed. One of the important mechanisms of parental effects is genomic imprinting. Genes with imprinting effects have parental specificity in their expression. For example, paternal imprinting means that the gene inherited from the father is not expressed and only the gene inherited from the mother is expressed, which is also known as maternally derived effect. On the contrary, maternal imprinting indicates that the gene has a paternally derived effect. Different imprinting effects may lead to different diseases. For example, Prader-Willi syndrome and Angelman syndrome are both caused by the loss of the functional alleles of the genes within the imprinted region of Chromosome 15q11-13. Among them, inheriting a loss of function mutation for the SNRPN gene from the father can cause Prader-Willi syndrome, while inheriting a loss of function mutation for the UBE3A gene from the mother results in Angelman syndrome (Falls et al., 1999; Peters, 2014). Morison et al. (2001) built a database of imprinted genes (http://igc.otago.ac.nz/), and so far there are 355 records related to humans. Similarly, in the geneimprint and Otago imprint databases (https://www.geneimprint.com/), more than 150 imprinted genes have been described in humans, but there may be more imprinted genes which have not been verified (Benonisdottir et al., 2016). Many imprinted genes are essential to the normal growth, neurodevelopment, metabolism of the fetus and adult behavior (Smith et al., 2006). Imprinted mutants can affect the growth and development of individuals and cause various diseases, such as Prader-Willi syndrome, Angelman syndrome, Turner’s syndrome (Skuse et al., 1997), etc.
Although GWAS studies have demonstrated that genetic variations could explain part of the variances of the traits related to behavioral disinhibition, a GWAS of adolescent behavioral disinhibition (Derringer et al., 2015) used the restricted maximum likelihood method to estimate the proportions of the variances that can be explained by SNPs in the phenotypes, and the results showed that there is still part of genetically related variance in the residuals of the model and current GWAS results may underestimate the effect of SNPs on behavioral disinhibition. Other more complicated genetic models need to be considered to explore the realistic effect of SNPs on behavioral disinhibition. The earliest and strongest evidence for the POE on behavioral disinhibition comes from studies of alcohol dependence. Almasy and Borecki (1999) summarized the analyses in Genetic Analysis Workshop 11 based on the data from the Collaborative Study on the Genetics of Alcoholism. They discovered that more significant loci could be found in the samples split by parental origin than in the total sample, and more effects could be seen for paternal transmission than for maternal transmission. Likewise, the findings of Macciardi et al. (1999) showed that the mode of inheritance for alcoholism is probably more complex than traditional Mendelian disorders, and there are significant differences in alcoholism by gender, parent-of-origin effects and other epidemiological factors. Strauch et al. (2005) used 93 Caucasian pedigrees of the Collaborative Study on the Genetics of Alcoholism dataset and found that some loci on Chromosomes 1, 2, 10, 12, 13, 15 and 21 have paternal imprinting on alcohol dependence, and a tendency to maternal imprinting was observed at two loci on Chromosome 7. Moreover, a recent GWAS of the addiction explained that some genes, which were identified as risk factors of smoking, show an unbalanced expression of alleles biased towards paternal alleles, which may be caused by paternally derived effect (Kozlova et al., 2021). On the other hand, more and more studies have claimed that some genes with imprinting effects have a significant impact on psychiatric disorders, such as schizophrenia and bipolar disorder. For example, Ludwig et al. (2009) determined the role of the gene LRRTM1 in the development of schizophrenia/schizoaffective disorder. In their study of 180 parent-offspring trios with schizophrenia, they found that LRRTM1 has maternal imprinting in schizophrenia. Similarly, some genes that have been shown to be imprinted, such as ZDBF2, PPP1R9A and DLGAP2 (Nakabayashi et al., 2004; Luedi et al., 2007), were also found to be genetic causes of schizophrenia or bipolar disorder (Li et al., 2014; Konopaske et al., 2015). Behavioral disinhibition, as an important feature of several neurodevelopmental and psychiatric disorders, may also be influenced by certain imprinted genes. However, there has been currently no POE on behavioral disinhibition systematically discussed in literature.
Therefore, in this article, we explored the POE for behavioral disinhibition based on the data of the Minnesota Center for Twin and Family Research (MCTFR) Genome-Wide Association Study of Behavioral Disinhibition. We applied a linear mixed model to examine the associations of maternally and paternally derived minor alleles with five quantitative indicators of behavioral disinhibition at a genome-wide scale so that the parental sources of genetic variants can be included in the analysis to further reveal the genetic factors which may be missed or underestimated in traditional GWAS. The purpose of this article is to propose new insights into the underlying genetic etiology of behavioral disinhibition, and to identify some novel candidate SNPs for future related researches on behavioral disinhibition in molecular genetics.
Materials and Methods
Samples
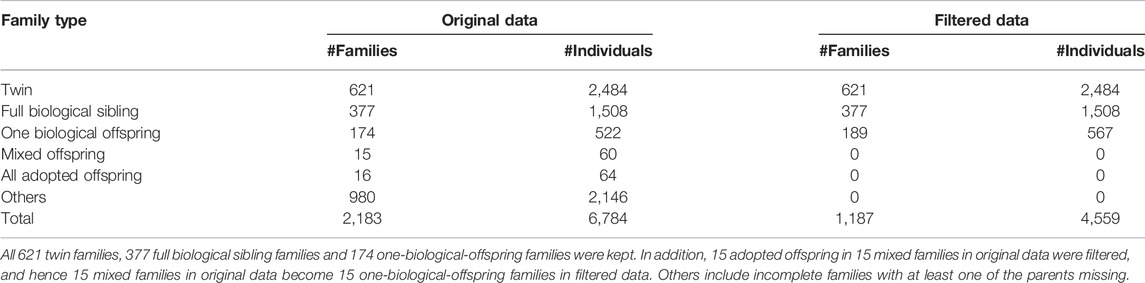
We used the dataset from the MCTFR Genome-Wide Association Study of Behavioral Disinhibition, which is made available from the database of Genotypes and Phenotypes (dbGaP) with accession number 86747-6 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000620.v1.p1). It is a large, ongoing and family-based epidemiological study on substance abuse and related psychopathology, consisting of three cohorts: 1) 17-year-old twins born in Minnesota from 1972 to 1979 and their parents, 2) 11-year-old twins born in Minnesota from 1977 to 1984 and their parents (later, more 11-year-old twins born from 1988 to 1994 and their parents were added), and 3) 15-year-old full biological siblings, adopted siblings, and mixed siblings (one being biologically related to the parents and the other being an adopted child) born in Minnesota from 1978 to 1988 and their parents (Iacono et al., 2006). McGue et al. (2013) conducted a genome-wide association study of behavioral disinhibition using 2,300 Caucasian families and 7,188 individuals for all the three cohorts in MCTFR. As one of the principal investigators of MCTFR, McGue M. uploaded the data of 2,183 families and 6,784 individuals to the dbGaP database for public research. Since the detection of the POE only needs the information of the parents and their biological children in each family, we deleted the adopted children in the families and independent individuals from the original data. Finally, we obtained 1,187 families (totally 4,559 individuals), including 621 twin families, 377 full biological sibling families and 189 one-biological-offspring families for this data analysis. See Table 1 for the details.
Genotypes
There are 527,829 SNPs in the MCTFR dataset. Among them, 515,385 are autosomal SNPs. We used the following quality control criteria to filter the SNPs (McGue et al., 2013): 1) genotype call rate < 99%, 2) minor allele frequency < 1%, and 3) individual call rate < 99%. Since all the filtered data are families, the assumption of Hardy-Weinberg equilibrium is not required. After the quality control, 510,278 autosomal SNPs were included in this data analysis.
Clinical Phenotypes
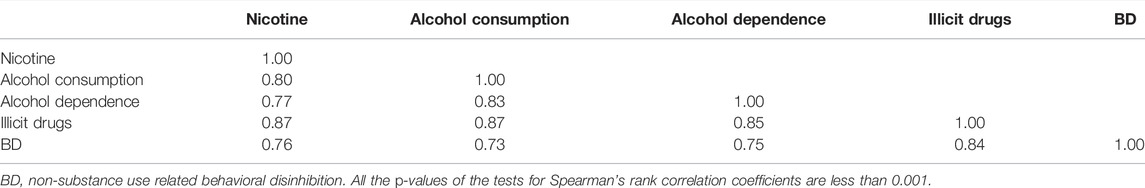
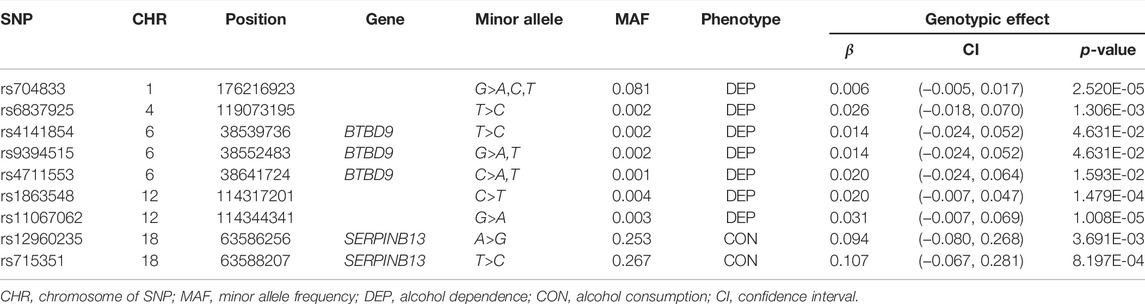
This dataset includes five composite quantitative clinical phenotypes, which are derived using the hierarchical factor analysis method described by Hicks et al. (2011): 1) nicotine (composite score of measures of quantity and frequency of nicotine use and symptoms of nicotine dependence), 2) alcohol consumption (composite score of measures of alcohol use frequency and quantity), 3) alcohol dependence (composite score of diagnostic and statistical manual of mental disorders symptoms of alcohol dependence/abuse and non-diagnostic alcohol-related problems), 4) illicit drugs (composite score of frequency of use of 11 different drug classes and diagnostic and statistical manual of mental disorders symptoms of drug dependence), and 5) BD (composite score of measures of non-substance use related behavioral disinhibition including symptoms of conduct disorder and aggression). Before the analysis, we should firstly carry out the normality tests and the correlation tests for the five phenotypes. We drew the Q-Q plots and performed the Spearman’s rank correlation tests between any two of the five phenotypes based on 1,187 offspring, where only one offspring was randomly selected in each family to remove the correlation between offspring. As shown in Figure 1, we found that none of the five clinical phenotypes follows the normal distribution. There is a significant and high correlation between any two of the phenotypes (ranging from 0.73 to 0.87). See Table 2 for the details. Therefore, to avoid increasing false positive results in the later POE tests at a genome-wide scale, we used the rank-based inverse normal transformation, which was proposed by McCaw et al. (2020), to transform the phenotype data. Then, we conducted the POE tests based on the transformed phenotypes.
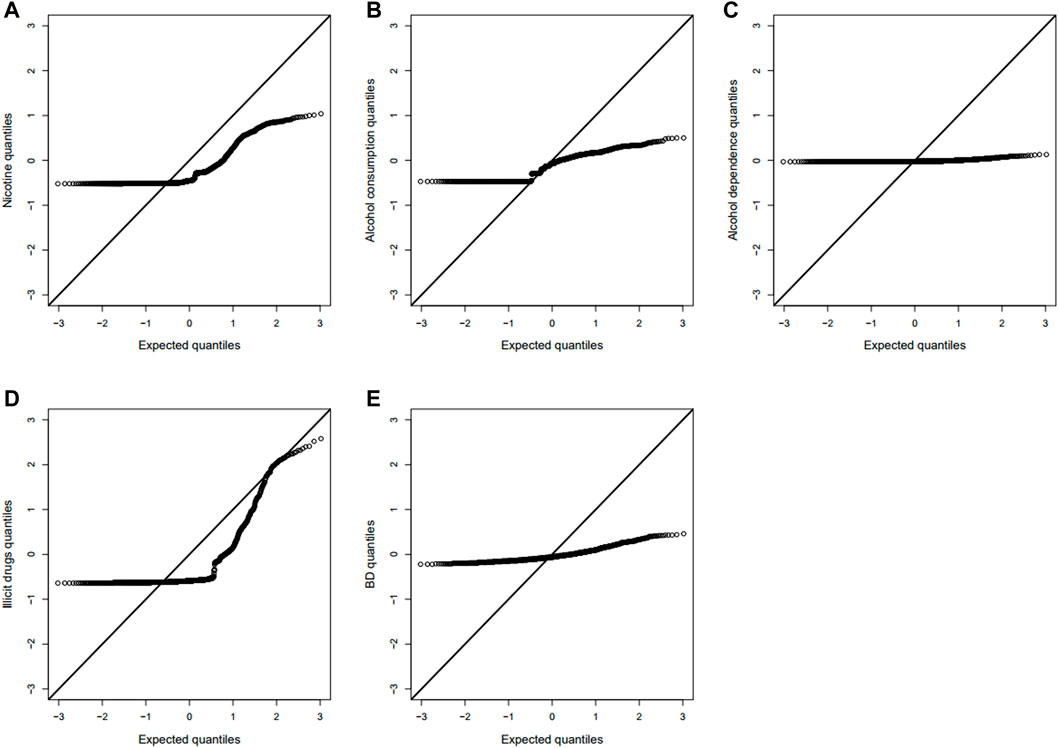
FIGURE 1. Q-Q plots of five clinical phenotypes (N = 1,187). (A) Nicotine, (B) alcohol consumption, (C) alcohol dependence, (D) illicit drugs, and (E) BD, non-substance use related behavioral disinhibition.
Covariates
The models for testing the POE included the covariates age, sex and birth year. Note that the covariate “generation” of the individuals was also collected in the MCTFR dataset. However, the parents in each family were just used to determine the parental origin of minor alleles of their offspring in the POE tests, and then we did not include the covariate “generation” in the models.
Parent-of-Origin Effects Tests
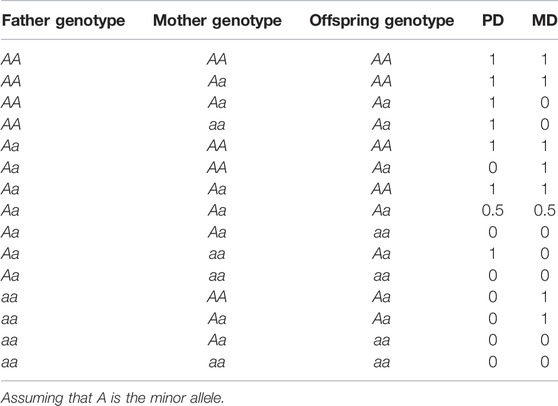
Each of 510,278 autosomal SNPs was used to test for the POE on each of five transformed clinical phenotypes based on 1,187 families (totally 4,559 individuals). To evaluate the individual contribution of maternal and paternal genetic variations to the phenotypes of the offspring, we used the genotype data from families to determine the parental origin of minor alleles in the offspring. For each given SNP locus, we first needed to construct two indicator variables, the paternally derived minor allele indicator variable (PD) and the maternally derived minor allele indicator variable (MD), which are respectively the number of minor alleles inherited from the father and that from the mother, and take the value of either 0 or 1. When the father, the mother, and the offspring are all heterozygous, we could not identify from whom the minor alleles in the offspring were inherited. So, we referred to the methods proposed by Hochner et al. (2015) and set both PD and MD to be 0.5 (Granot-Hershkovitz et al., 2020). Detailed assignments of the values of the two indicator variables are shown in Table 3. Considering the genetic relationship between the offspring in the families, we used a linear mixed model to test for the POE between each SNP and a phenotype. The model is as follows (Granot-Hershkovitz et al., 2020)
where
For comparison, for each candidate SNP, we established another linear mixed model to test for the association between the genotype and the phenotype of the offspring, which does not consider the POE. The model is as follows
where
Implementation in R
All the above analyses were implemented in the R software (version 4.1.1, http://r-project.org). The linear mixed model was fitted using the “lmekin” function in the R package “coxme.”
Results
To determine the parental origin of the minor alleles in offspring, we only selected those complete families for analysis. Therefore, we used 1,187 families (4,559 individuals) in the filtered data of Table 1, of which 2,374 were parents and 2,185 were offspring. The mean and the standard deviation (SD) of the age of the fathers were 44.6
We tested the POE between 510,278 autosomal SNPs and five phenotypes by model 1 for 2,185 offspring. The significance levels for testing
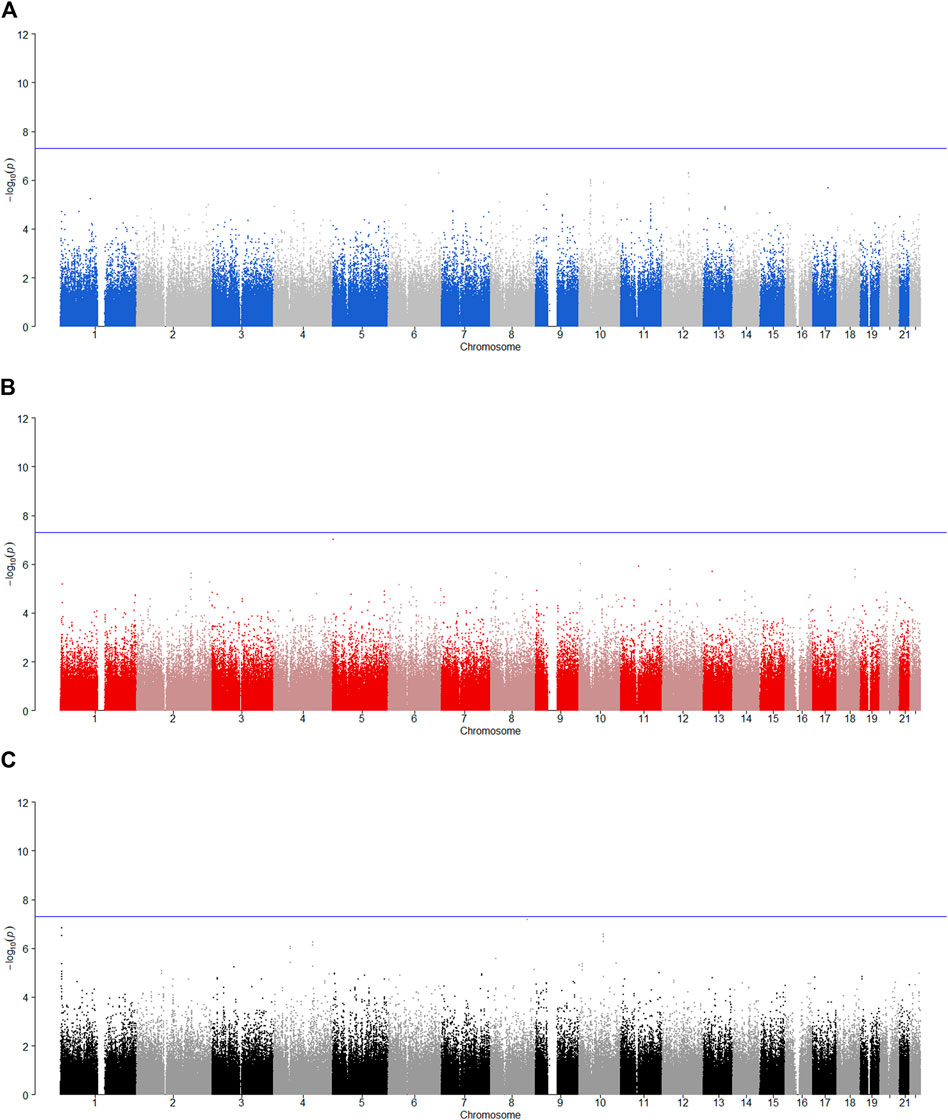
FIGURE 2. Manhattan plot for POE and genotypic effect tests of nicotine (N = 2,185). The blue line represents the significance level
FIGURE 3. Manhattan plot for POE and genotypic effect tests of alcohol consumption (N = 2,185). The blue line represents the significance level
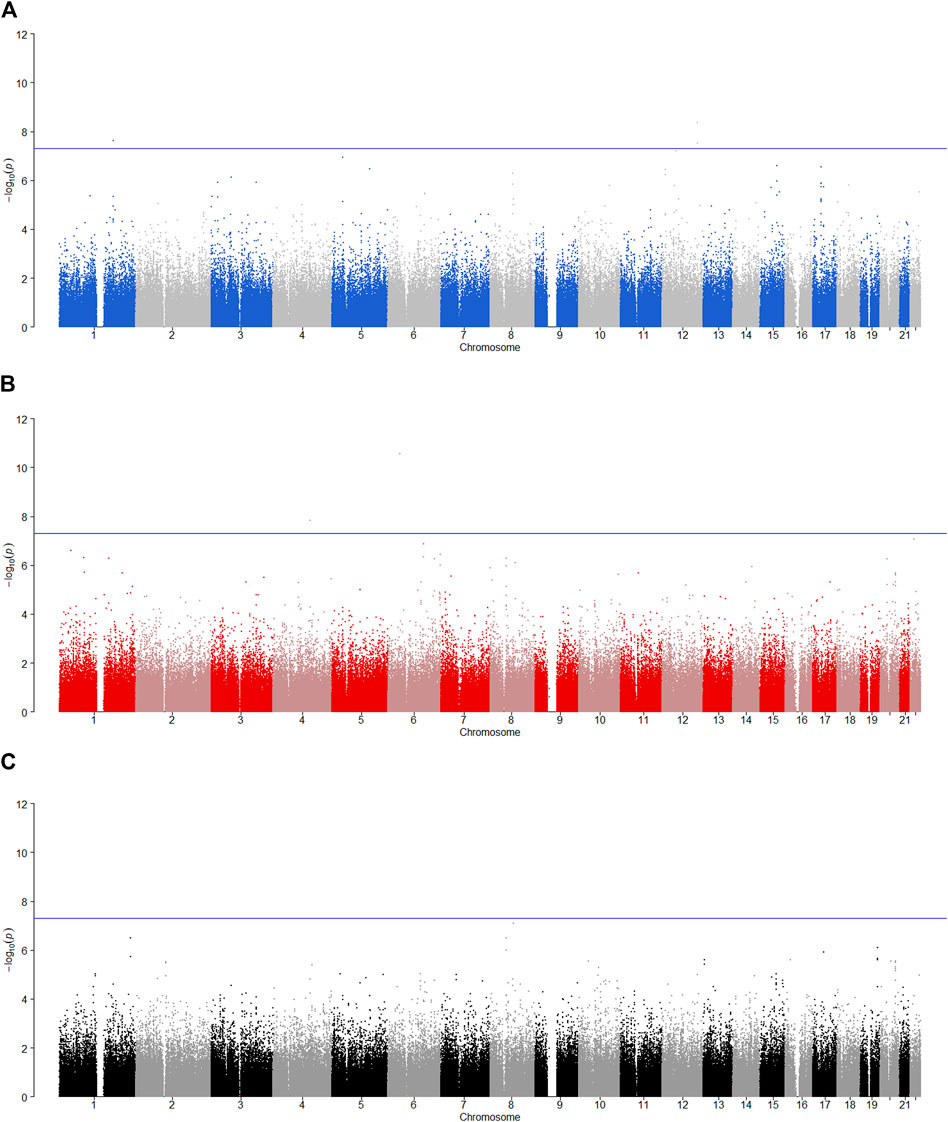
FIGURE 4. Manhattan plot for POE and genotypic effect tests of alcohol dependence (N = 2,185). The blue line represents the significance level
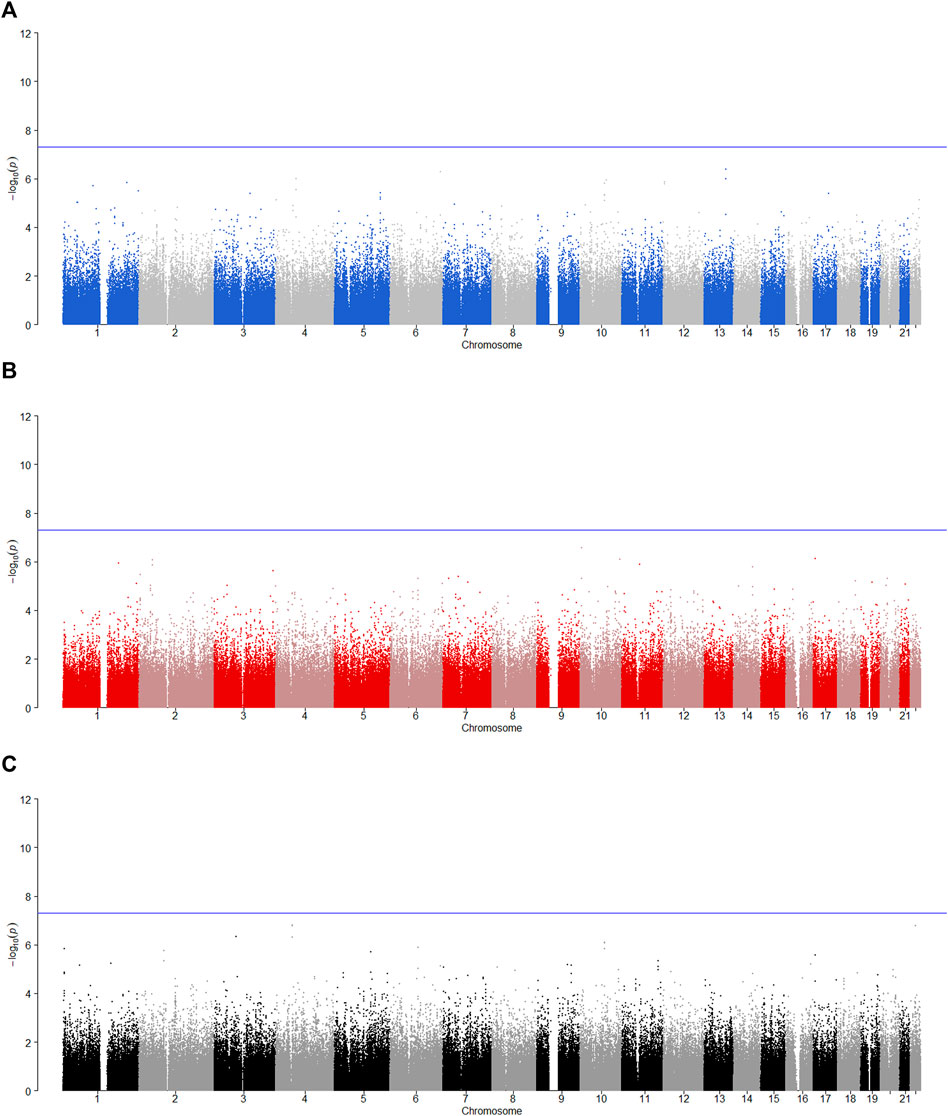
FIGURE 5. Manhattan plot for POE and genotypic effect tests of illicit drugs (N = 2,185). The blue line represents the significance level
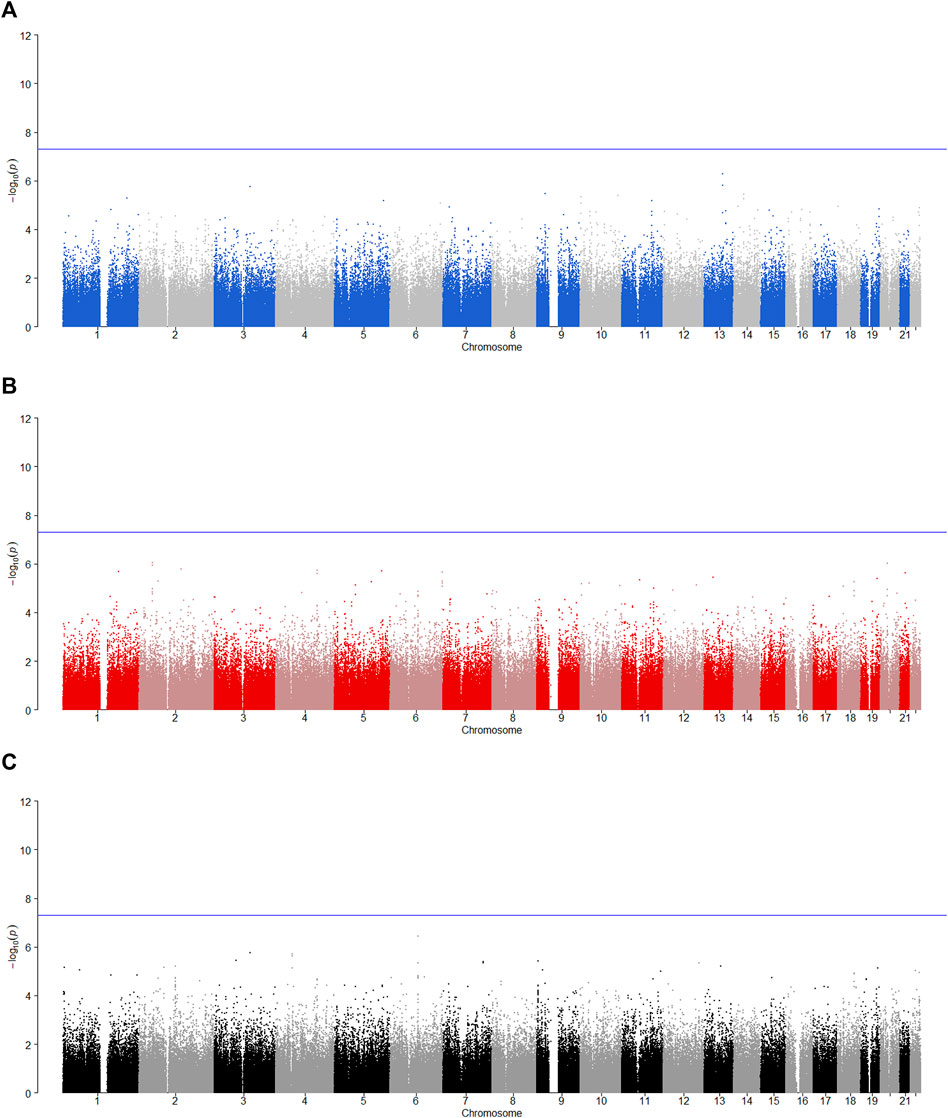
FIGURE 6. Manhattan plot for POE and genotypic effect tests of non-substance use related behavioral disinhibition (N = 2,185). The blue line represents the significance level
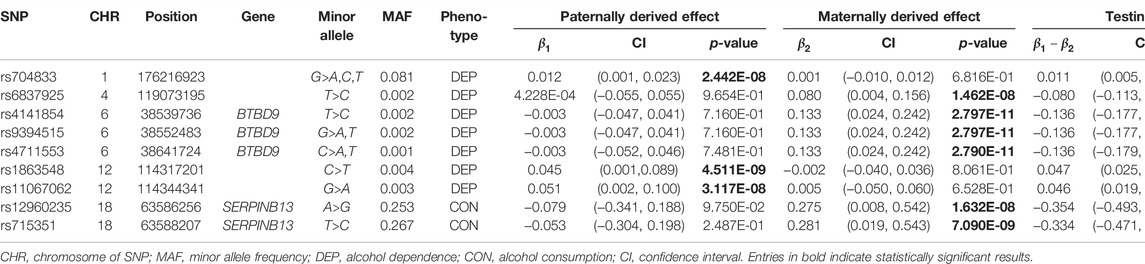
SNP rs704833 is located on Chromosome 1 and the results demonstrate that there is a significant POE at rs704833 on alcohol dependence. The minor allele inherited from the father has the significant association with alcohol dependence (
For comparison, Table 5 displays the results of genotypic effect tests for the above-mentioned 9 SNPs with the POE based on model 2. It is shown in Table 5 that model 2 could not identify the association between the SNPs and the phenotypes at the significance level of
Discussion
Behavioral disinhibition could easily cause great harm to human health and social stability. At present, the research on the influencing factors of behavioral disinhibition has gone further to the genetic level. However, the existing studies have not considered the POE in the analysis, which would miss or underestimate the genetic influence of certain loci on behavioral disinhibition. Therefore, in this study, we explored the POE of genetic loci on behavioral disinhibition based on the MCTFR data. Specifically, we first filtered the MCTFR data and obtained nuclear family data we needed, and then performed the normality tests of the five phenotypes closely related to behavioral disinhibition in the MCTFR data. Note that all the five phenotypes do not follow normal distributions. As such, we used the rank-based inverse normal transformation on the phenotypes to avoid increasing false positive results. Then, the method proposed in Hochner et al. (2015) was utilized to carry out the POE tests for all 510,278 autosomal SNPs at a genome-wide level, and finally nine SNPs were identified to have the significant POE on behavioral disinhibition.
Among the nine identified SNPs, seven SNPs have statistically significant POE on alcohol dependence and two SNPs have statistically significant POE on alcohol consumption. SNPs rs4141854, rs9394515, and rs4711553 on Chromosome 6, which have statistically significant POE on alcohol dependence, were located in the same gene, BTBD9. The BTBD9 gene is located on the short arm of Chromosome 6 and is highly expressed throughout the human brain (Freeman et al., 2012), and its mutation has been shown to be associated with restless legs syndrome and Tourette’s syndrome (Rivière et al., 2009; Schormair et al., 2017; Lyu et al., 2019). Restless legs syndrome is a pervasive chronic neurological disorder that causes nerve impulses to stimulate leg muscle twitching uncontrollably at night and at rest (Schormair et al., 2017). Tourette’s syndrome is similar to restless legs syndrome, except that nerve impulses are transferred from the legs to the face (Rivière et al., 2009). Studies have shown that due to the uncontrollable nerve impulses of these two diseases at night, two prominent symptoms of the diseases are the insomnia and the severely reduced quality of life, and the latter leads to a significant increase in the risk of depression, anxiety and alcoholism (Allen et al., 2011; Blaty and DelRosso, 2022). Studies have also shown that insomnia and alcohol dependence are significantly positively correlated (Chakravorty et al., 2016). Regarding the POE of Tourette’s syndrome, some studies have pointed out that the age of the onset of the maternally derived offspring is earlier than that of the paternally derived offspring significantly, which suggests that the maternally derived effect of the BTBD9 gene on Tourette’s syndrome could be explained by meiotic events or even intrauterine environmental influences (Eapen et al., 1997), and this supports our findings. The remaining four SNPs rs704833, rs6837925, rs1863548, and rs11067062 having statistically significant POE on alcohol dependence do not belong to any gene, and there has been no study to explain the functions of these four SNPs. These novel SNPs we have discovered may be associated with behavioral disinhibition, but this still needs to be confirmed by subsequent molecular genetics.
SNPs rs12960235 and rs715351 on Chromosome 18 have statistically significant POE on alcohol consumption and belong to the SERPINB13 gene. The SERPINB13 gene is a protein-coding gene, and the protein function annotations associated with this gene include serine-type endopeptidase inhibitor activity and cysteine-type endopeptidase inhibitor activity. The SERPINB13-related diseases include head and neck squamous cell carcinoma (de Koning et al., 2009), skin cancer (Moussali et al., 2005) and type I diabetes (Kryvalap et al., 2021). However, this gene has not been found to be associated with alcohol consumption in literature, and only studies have shown that alcoholism is a risk factor for diabetes (Tetzschner et al., 2018). However, we think that this is a normal phenomenon, because few people have considered the POE in the previous association studies, which may reduce the test power of the GWAS and make some positive SNPs not detected. On the other hand, McGue et al. (2013) conducted a GWAS on the basis of the MCTFR data and found a significant SNP rs1868152 (p-value =
We searched the imprinted-gene database (http://igc.otago.ac.nz/) constructed by Morison et al. (2001) and the geneimprint and Otago imprint databases (https://www.geneimprint.com/), while the 9 SNPs with the POE identified in our study are not included in these databases. Strauch et al. (2005) conducted a parametric single-marker linkage analysis and the POE test on alcohol dependence and alcohol consumption data of 93 Caucasian pedigrees composed of 919 people. The results showed that paternally derived effects were found on Chromosome 7, and maternally derived effects were found on Chromosomes 1, 2, 10, 13, 15 and 21. Another study based on alcoholism data (Liu et al., 2005) found the evidence of paternally derived effects on Chromosome 7 and maternally derived effects on Chromosomes 10 and 12. In our study, we found the paternally derived effects on Chromosomes 1 and 12, and the maternally derived effects on Chromosomes 4, 6, and 18. This may suggest that there is the POE on behavioral disinhibition, especially alcohol dependence and alcohol consumption.
We used two models to conduct the association analysis in Tables 4, 5, respectively with the POE and without the POE, to compare the performances of these two models at the nine SNPs. It can be seen from Table 5 that model 2 not considering the POE does not find any significant SNPs among the nine SNPs. This suggests that if the POE is present but is not considered in the association analysis, the genetic factors of the phenotypes may be ignored or underestimated. Granot-Hershkovitz et al. (2020) and Nudel et al. (2014) have also drawn similar conclusions in their studies. On the other hand, there are several limitations in our study. First, the MCTFR dataset only contains a few covariates (age, sex, birth year and generation). Although we adjusted them (except for the covariate generation) in the model, we are not sure if we have considered most of the variances of environmental factors. Second, the POE is closely related to methylation while we did not include any methylation data in the analysis. To obtain more significant results, methylation data should be incorporated into the detection of the POE in future. In addition, there has been a novel method available for inferring the POE in literature (Hofmeister et al., 2021), which does not require parental genomes nor prior knowledge of genealogy. As such, it is suitable for many data types, besides nuclear families with both parents. We will use this method to reanalyze the MCTFR data for the POE on behavioral disinhibition in future. Finally, this study has the following contributions. 1) Facing the challenge that the five behavioral inhibition phenotypes do not follow normal distributions, we used the rank-based inverse normal transformation method on the five phenotypes to avoid increasing false positive results. 2) Verify the conclusion proposed by Granot-Hershkovitz et al. (2020) that taking account of the POE in GWAS can enhance the ability to detect genetic association and population characteristics. 3) Provide novel evidence for the presence of the POE on behavioral disinhibition.
Data Availablity Statement
The Minnesota Center for Twin and Family Research data used for this study can be found on the database of Genotypes and Phenotypes with accession number 86747-6 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000620.v1.p1.
Author Contributions
Y-FK, M-KL, and J-YZ contributed to the data analysis, the interpretation of the result of the data analysis and the writing of the manuscript. Y-XY, Z-YY, W-YY, and P-ZZ reviewed the whole paper and revised the article critically. J-YZ helped design the study and directed its implementation. All authors read and approved this version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of China grant nos. 82173619 and 81773544, and the Science and Technology Planning Project of Guangdong Province grant no. 2020B1212030008. The Minnesota Center for Twin and Family Research (MCTFR) was supported by the National Institute on Drug Abuse grant no. U01 DA024417. The sample ascertainment and data collection in MCTFR data were supported by the National Institute on Drug Abuse grant nos. R37 DA05147 and R01 DA13240, the National Institute on Alcohol Abuse and Alcoholism grant nos. R01 AA09367 and R01 AA11886, and the National Institute of Mental Health grant no. R01 MH66140.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agrawal, A., Edenberg, H. J., Foroud, T., Bierut, L. J., Dunne, G., Hinrichs, A. L., et al. (2006). Association of GABRA2 with Drug Dependence in the Collaborative Study of the Genetics of Alcoholism Sample. Behav. Genet. 36, 640–650. doi:10.1007/s10519-006-9069-4
Allen, R. P., Bharmal, M., and Calloway, M. (2011). Prevalence and Disease burden of Primary Restless Legs Syndrome: Results of a General Population Survey in the United States. Mov. Disord. 26, 114–120. doi:10.1002/mds.23430
Almasy, L., and Borecki, I. B. (1999). Exploring Genetic Analysis of Complex Traits through the Paradigm of Alcohol Dependence: Summary of GAW11 Contributions. Genet. Epidemiol. 17 (Suppl. 1), S1–S24. doi:10.1002/gepi.1370170704
Amirian, E., Baxter, J., Grigsby, J., Curran-Everett, D., Hokanson, J. E., and Bryant, L. L. (2010). Executive Function (Capacity for Behavioral Self-Regulation) and Decline Predicted Mortality in a Longitudinal Study in Southern Colorado. J. Clin. Epidemiol. 63, 307–314. doi:10.1016/j.jclinepi.2009.06.004
Benonisdottir, S., Oddsson, A., Helgason, A., Kristjansson, R. P., Sveinbjornsson, G., Oskarsdottir, A., et al. (2016). Epigenetic and Genetic Components of Height Regulation. Nat. Commun. 7, 13490. doi:10.1038/ncomms13490
Blaty, J. L., and DelRosso, L. M. (2022). Tourette Disorder and Sleep. Biomed. J. S2319-4170, 00002–00006. doi:10.1016/j.bj.2022.01.002
Chakravorty, S., Chaudhary, N. S., and Brower, K. J. (2016). Alcohol Dependence and its Relationship with Insomnia and Other Sleep Disorders. Alcohol. Clin. Exp. Res. 40, 2271–2282. doi:10.1111/acer.13217
Corley, R. P., Zeiger, J. S., Crowley, T., Ehringer, M. A., Hewitt, J. K., Hopfer, C. J., et al. (2008). Association of Candidate Genes with Antisocial Drug Dependence in Adolescents. Drug Alcohol Depend. 96, 90–98. doi:10.1016/j.drugalcdep.2008.02.004
de Koning, P. J. A., Bovenschen, N., Leusink, F. K. J., Broekhuizen, R., Quadir, R., van Gemert, J. T. M., et al. (2009). Downregulation of SERPINB13 Expression in Head and Neck Squamous Cell Carcinomas Associates with Poor Clinical Outcome. Int. J. Cancer 125, 1542–1550. doi:10.1002/ijc.24507
Derringer, J., Corley, R. P., Haberstick, B. C., Young, S. E., Demmitt, B. A., Howrigan, D. P., et al. (2015). Genome-Wide Association Study of Behavioral Disinhibition in a Selected Adolescent Sample. Behav. Genet. 45, 375–381. doi:10.1007/s10519-015-9705-y
Eapen, V., O'Neill, J., D Gurling, H. M., and Robertson, M. M. (1997). Sex of Parent Transmission Effect in Tourette's Syndrome: Evidence for Earlier Age at Onset in Maternally Transmitted Cases Suggests a Genomic Imprinting Effect. Neurology 48, 934–936. doi:10.1212/wnl.48.4.934
Falls, J. G., Pulford, D. J., Wylie, A. A., and Jirtle, R. L. (1999). Genomic Imprinting: Implications for Human Disease. Am. J. Pathol. 154, 635–647. doi:10.1016/S0002-9440(10)65309-6
Freeman, A., Pranski, E., Miller, R. D., Radmard, S., Bernhard, D., Jinnah, H. A., et al. (2012). Sleep Fragmentation and Motor Restlessness in a drosophila Model of Restless Legs Syndrome. Curr. Biol. 22, 1142–1148. doi:10.1016/j.cub.2012.04.027
Granot-Hershkovitz, E., Wu, P., Karasik, D., Peter, I., Peloso, G. M., Levy, D., et al. (2020). Searching for Parent-Of-Origin Effects on Cardiometabolic Traits in Imprinted Genomic Regions. Eur. J. Hum. Genet. 28, 646–655. doi:10.1038/s41431-019-0568-1
Hicks, B. M., Schalet, B. D., Malone, S. M., Iacono, W. G., and McGue, M. (2011). Psychometric and Genetic Architecture of Substance Use Disorder and Behavioral Disinhibition Measures for Gene Association Studies. Behav. Genet. 41, 459–475. doi:10.1007/s10519-010-9417-2
Hochner, H., Allard, C., Granot-Hershkovitz, E., Chen, J., Sitlani, C. M., Sazdovska, S., et al. (2015). Parent-of-Origin Effects of the APOB Gene on Adiposity in Young Adults. Plos Genet. 11, e1005573. doi:10.1371/journal.pgen.1005573
Hofmeister, R. J., Rubinacci, S., Ribeiro, D. M., Kutalik, Z., Buil, A., and Delaneau, O. (2021). Parent-of-Origin Effects in the UK Biobank. bioRxiv [Preprint]. Available at: https://www.biorxiv.org/content/10.1101/2021.11.03.467079v1 (Accessed November 06, 2021)
Iacono, W. G., McGue, M., and Krueger, R. F. (2006). Minnesota center for Twin and Family Research. Twin Res. Hum. Genet. 9, 978–984. doi:10.1375/18324270677946264210.1375/twin.9.6.978
Iacono, W. G., Malone, S. M., and McGue, M. (2008). Behavioral Disinhibition and the Development of Early-Onset Addiction: Common and Specific Influences. Annu. Rev. Clin. Psychol. 4, 325–348. doi:10.1146/annurev.clinpsy.4.022007.141157
Kendler, K. S., Prescott, C. A., Myers, J., and Neale, M. C. (2003). The Structure of Genetic and Environmental Risk Factors for Common Psychiatric and Substance Use Disorders in Men and Women. Arch. Gen. Psychiatry 60, 929–937. doi:10.1001/archpsyc.60.9.929
Konopaske, G. T., Subburaju, S., Coyle, J. T., and Benes, F. M. (2015). Altered Prefrontal Cortical MARCKS and PPP1R9A mRNA Expression in Schizophrenia and Bipolar Disorder. Schizophrenia Res. 164, 100–108. doi:10.1016/j.schres.2015.02.005
Kozlova, A., Butler, R. R., Zhang, S., Ujas, T., Zhang, H., Steidl, S., et al. (2021). Sex-specific Nicotine Sensitization and Imprinting of Self-Administration in Rats Inform GWAS Findings on Human Addiction Phenotypes. Neuropsychopharmacol. 46, 1746–1756. doi:10.1038/s41386-021-01027-0
Krueger, R. F., Hicks, B. M., Patrick, C. J., Carlson, S. R., Iacono, W. G., and McGue, M. (2002). Etiologic Connections Among Substance Dependence, Antisocial Behavior and Personality: Modeling the Externalizing Spectrum. J. Abnormal Psychol. 111, 411–424. doi:10.1037/0021-843X.111.3.411
Krystal, J. H., Staley, J., Mason, G., Petrakis, I. L., Kaufman, J., Harris, R. A., et al. (2006). Gamma-aminobutyric Acid Type a Receptors and Alcoholism: Intoxication, Dependence, Vulnerability, and Treatment. Arch. Gen. Psychiatry 63, 957–968. doi:10.1001/archpsyc.63.9.957
Kryvalap, Y., Jiang, M. L., Kryvalap, N., Hendrickson, C., and Czyzyk, J. (2021). SerpinB13 Antibodies Promote β Cell Development and Resistance to Type 1 Diabetes. Sci. Transl. Med. 13, eabf1587. doi:10.1126/scitranslmed.abf1587
Kubzansky, L. D., Park, N., Peterson, C., Vokonas, P., and Sparrow, D. (2011). Healthy Psychological Functioning and Incident Coronary Heart Disease: the Importance of Self-Regulation. Arch. Gen. Psychiatry 68, 400–408. doi:10.1001/archgenpsychiatry.2011.23
Li, J.-M., Lu, C.-L., Cheng, M.-C., Luu, S.-U., Hsu, S.-H., Hu, T.-M., et al. (2014). Role of the DLGAP2 Gene Encoding the SAP90/PSD-95-Associated Protein 2 in Schizophrenia. PLoS One 9, e85373. doi:10.1371/journal.pone.0085373
Liu, X.-Q., Greenwood, C. M., Wang, K.-S., and Paterson, A. D. (2005). A Genome Scan for Parent-Of-Origin Linkage Effects in Alcoholism. BMC Genet. 6 (Suppl. 1), S160. doi:10.1186/1471-2156-6-S1-S160
Ludwig, K. U., Mattheisen, M., Mühleisen, T. W., Roeske, D., Schmäl, C., Breuer, R., et al. (2009). Supporting Evidence for LRRTM1 Imprinting Effects in Schizophrenia. Mol. Psychiatry 14, 743–745. doi:10.1038/mp.2009.28
Luedi, P. P., Dietrich, F. S., Weidman, J. R., Bosko, J. M., Jirtle, R. L., and Hartemink, A. J. (2007). Computational and Experimental Identification of Novel Human Imprinted Genes. Genome Res. 17, 1723–1730. doi:10.1101/gr.6584707
Lyu, S., Xing, H., DeAndrade, M. P., Liu, Y., Perez, P. D., Yokoi, F., et al. (2019). The Role of BTBD9 in Striatum and Restless Legs Syndrome. eNeuro 6, ENEURO.0277-19.2019. doi:10.1523/ENEURO.0277-19.2019
Macciardi, F., Morenghi, E., and Morabito, A. (1999). Alcoholism as a Complex Trait: Comparison of Genetic Models and Role of Epidemiological Risk Factors. Genet. Epidemiol. 17 (Suppl. 1), S247–S252. doi:10.1002/gepi.1370170742
McCaw, Z. R., Lane, J. M., Saxena, R., Redline, S., and Lin, X. (2020). Operating Characteristics of the Rank‐based Inverse normal Transformation for Quantitative Trait Analysis in Genome‐wide Association Studies. Biometrics 76, 1262–1272. doi:10.1111/biom.13214
McGue, M., Zhang, Y., Miller, M. B., Basu, S., Vrieze, S., Hicks, B., et al. (2013). A Genome-wide Association Study of Behavioral Disinhibition. Behav. Genet. 43, 363–373. doi:10.1007/s10519-013-9606-x
Mobascher, A., Rujescu, D., Mittelstraß, K., Giegling, I., Lamina, C., Nitz, B., et al. (2010). Association of a Variant in the Muscarinic Acetylcholine Receptor 2 Gene (CHRM2 ) with Nicotine Addiction. Am. J. Med. Genet. 153B, 684–690. doi:10.1002/ajmg.b.31011
Morison, I. M., Paton, C. J., and Cleverley, S. D. (2001). The Imprinted Gene and Parent-Of-Origin Effect Database. Nucleic Acids Res. 29, 275–276. doi:10.1093/nar/29.1.275
Moussali, H., Bylaite, M., Welss, T., Abts, H. F., Ruzicka, T., and Walz, M. (2005). Expression of Hurpin, a Serine Proteinase Inhibitor, in normal and Pathological Skin: Overexpression and Redistribution in Psoriasis and Cutaneous Carcinomas. Exp. Dermatol. 14, 420–428. doi:10.1111/j.0906-6705.2005.00300.x
Mozaffari, S. V., DeCara, J. M., Shah, S. J., Sidore, C., Fiorillo, E., Cucca, F., et al. (2019). Parent-of-origin Effects on Quantitative Phenotypes in a Large Hutterite Pedigree. Commun. Biol. 2, 28. doi:10.1038/s42003-018-0267-4
Nakabayashi, K., Makino, S., Minagawa, S., Smith, A. C., Bamforth, J. S., Stanier, P., et al. (2004). Genomic Imprinting of PPP1R9A Encoding Neurabin I in Skeletal Muscle and Extra-embryonic Tissues. J. Med. Genet. 41, 601–608. doi:10.1136/jmg.2003.014142
Nudel, R., Simpson, N. H., Baird, G., O'Hare, A., Conti‐Ramsden, G., Bolton, P. F., et al. (2014). Genome‐wide Association Analyses of Child Genotype Effects and Parent‐of‐origin Effects in Specific Language Impairment. Genes, Brain Behav. 13, 418–429. doi:10.1111/gbb.12127
Peters, J. (2014). The Role of Genomic Imprinting in Biology and Disease: an Expanding View. Nat. Rev. Genet. 15, 517–530. doi:10.1038/nrg3766
Rivière, J.-B., Xiong, L., Levchenko, A., St-Onge, J., Gaspar, C., Dion, Y., et al. (2009). Association of Intronic Variants of the BTBD9 Gene with Tourette Syndrome. Arch. Neurol. 66, 1267–1272. doi:10.1001/archneurol.2009.213
Schormair, B., Zhao, C., Bell, S., Tilch, E., Salminen, A. V., Pütz, B., et al. (2017). Identification of Novel Risk Loci for Restless Legs Syndrome in Genome-wide Association Studies in Individuals of European Ancestry: a Meta-Analysis. Lancet Neurol. 16, 898–907. doi:10.1016/S1474-4422(17)30327-7
Sharma, L., Markon, K. E., and Clark, L. A. (2014). Toward a Theory of Distinct Types of "impulsive" Behaviors: A Meta-Analysis of Self-Report and Behavioral Measures. Psychol. Bull. 140, 374–408. doi:10.1037/a0034418
Skuse, D. H., James, R. S., Bishop, D. V. M., Coppin, B., Dalton, P., Aamodt-Leeper, G., et al. (1997). Evidence from Turner's Syndrome of an Imprinted X-Linked Locus Affecting Cognitive Function. Nature 387, 705–708. doi:10.1038/42706
Smith, F. M., Garfield, A. S., and Ward, A. (2006). Regulation of Growth and Metabolism by Imprinted Genes. Cytogenet. Genome Res. 113, 279–291. doi:10.1159/000090843
Strauch, K., Fürst, R., Rüschendorf, F., Windemuth, C., Dietter, J., Flaquer, A., et al. (2005). Linkage Analysis of Alcohol Dependence Using MOD Scores. BMC Genet. 6 (Suppl. 1), S162. doi:10.1186/1471-2156-6-S1-S162
Tetzschner, R., Nørgaard, K., and Ranjan, A. (2018). Effects of Alcohol on Plasma Glucose and Prevention of Alcohol-Induced Hypoglycemia in Type 1 Diabetes-A Systematic Review with GRADE. Diabetes Metab. Res. Rev. 34, e2965. doi:10.1002/dmrr.2965
Vrieze, S. I., McGue, M., Miller, M. B., Hicks, B. M., and Iacono, W. G. (2013). Three Mutually Informative Ways to Understand the Genetic Relationships Among Behavioral Disinhibition, Alcohol Use, Drug Use, Nicotine Use/dependence, and Their Co-occurrence: Twin Biometry, GCTA, and Genome-wide Scoring. Behav. Genet. 43, 97–107. doi:10.1007/s10519-013-9584-z
Keywords: behavioral disinhibition, genome-wide association study, parent-of-origin effects, Minnesota Center for Twin and Family Research data, rank-based inverse normal transformation
Citation: Kong Y-F, Li M-K, Yuan Y-X, Yang Z-Y, Yu W-Y, Zhao P-Z and Zhou J-Y (2022) Detection of Parent-of-Origin Effects for the Variants Associated With Behavioral Disinhibition in the MCTFR Data. Front. Genet. 13:831685. doi: 10.3389/fgene.2022.831685
Received: 08 December 2021; Accepted: 28 March 2022;
Published: 26 April 2022.
Edited by:
Julie Demars, Institut National de Recherche Pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceReviewed by:
Bertrand Servin, Institut National de la Recherche Agronomique de Toulouse, FranceJonathan Covault, University of Connecticut Health Center, United States
Copyright © 2022 Kong, Li, Yuan, Yang, Yu, Zhao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji-Yuan Zhou, emhvdWppeXVhbjU0NjBAaG90bWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
 Yi-Fan Kong
Yi-Fan Kong Meng-Kai Li
Meng-Kai Li Yu-Xin Yuan
Yu-Xin Yuan Zi-Ying Yang
Zi-Ying Yang Wen-Yi Yu
Wen-Yi Yu Pei-Zhen Zhao
Pei-Zhen Zhao Ji-Yuan Zhou
Ji-Yuan Zhou