- 1Department of Genetics, Development and Cell Biology, Iowa State University, Ames, IA, United States
- 2Department of Agricultural and Environmental Sciences, Tuskegee University, Tuskegee, AL, United States
- 3Crops Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China
Peanut (Arachis hypogaea L.) seed is a rich source of edible oil, comprised primarily of monounsaturated oleic acid and polyunsaturated linoleic acid, accounting for 80% of its fatty acid repertoire. The conversion of oleic acid to linoleic acid, catalyzed by Fatty Acid Desaturase 2 (FAD2) enzymes, is an important regulatory point linked to improved abiotic stress responses while the ratio of these components is a significant determinant of commercial oil quality. Specifically, oleic acid has better oxidative stability leading to longer shelf life and better taste qualities while also providing nutritional based health benefits. Naturally occurring FAD2 gene knockouts that lead to high oleic acid levels improve oil quality at the potential expense of plant health though. We undertook a CRISPR/Cas9 based site-specific genome modification approach designed to downregulate the expression of two homeologous FAD2 genes in seed while maintaining regulation in other plant tissues. Two cis-regulatory elements the RY repeat motif and 2S seed protein motif in the 5′UTR and associated intron of FAD2 genes are potentially important for regulating seed-specific gene expression. Using hairy root and stable germ line transformation, differential editing efficiencies were observed at both CREs when targeted by single gRNAs using two different gRNA scaffolds. The editing efficiencies also differed when two gRNAs were expressed simultaneously. Additionally, stably transformed seed exhibited an increase in oleic acid levels relative to wild type. Taken together, the results demonstrate the immense potential of CRISPR/Cas9 based approaches to achieve high frequency targeted edits in regulatory sequences for the generation of novel transcriptional alleles, which may lead to fine tuning of gene expression and functional genomic studies in peanut.
Introduction
Peanut (Arachis hypogaea L.) or groundnut is an important legume crop due to the seed being a rich source of edible oil, protein and fiber. The oil content is about 50%, of which oleic and linoleic acids account for 80% of the total fatty acid with the ratio of these components largely determining oil quality. Therefore, the conversion of monounsaturated oleic acid to polyunsaturated linoleic acid by the Fatty Acid Desaturase 2 (FAD2) enzyme is a key regulatory point. Peanut oil is naturally high in linoleic acid, making it prone to oxidation, which reduces oil stability leading to rancidity and poor flavor characteristics. However, linoleic acid is important for healthy plant growth and it is also an essential fatty acid for humans (Guan et al., 2012; Marangoni et al., 2020). High oleic acid is beneficial for improved shelf life through oxidative stability, improved flavor characteristics and consumption may lead to better cardiovascular health in humans (O’Byrne et al., 1997; Yu et al., 2008; Lim et al., 2017). The natural FAD2 knockout mutant line, F435, is devoid of FAD2 enzyme activity in seed and accumulates 80% oleic acid as observed by Norden et al., 1987. On the surface, these mutations may seem optimal for oil quality, however, introgression of these mutations into other peanut varieties is time consuming and knockout mutations may leave plants vulnerable during periods of stress (Zhang et al., 2012). Therefore, an ideal strategy to engineer oil composition without compromising plant health may involve downregulation of FAD2 gene expression in seed while minimizing pleiotropic effects in other vegetative tissues through promoter modification.
The FAD2 gene family has been extensively characterized from many plant species including peanut (Janila et al., 2016; Dar et al., 2017). Cultivated peanut is an allotetraploid (AABB, 2n = 4x = 40) and the genome contains two functional homeologous genes, AhFAD2A and AhFAD2B, derived from the diploid ancestors Arachis duranensis and Arachis ipaensis, respectively. FAD2 is the key enzyme that is responsible for biosynthesis of polyunsaturated acid in non-photosynthetic tissues, such as roots and developing seeds in oilseed plants (Miquel and Browse, 1992), while some FAD2 genes are targeted to stems and leaves (Cao et al., 2013; Lee et al., 2020). FAD2 genes typically have a conserved single large intron in the 5′-untranslated region (UTR), which has been suggested to have enhancer activity in mediating transcriptional regulation. This has been shown in sesame (Kim et al., 2006), and Brassica napus (Xiao et al., 2014) by deletion mapping approaches and Arabidopsis and olives by gene association studies (Menard et al., 2017; Salimonti et al., 2020).
Seed development, including embryogenesis and seed maturation, is regulated by a combination of hormonal, genetic and metabolic controls (Gutierrez et al., 2007). Differing clusters of cis-regulatory elements (CREs) in the promoters of seed associated proteins are bound by various combinations of transcription factors (TFs) including LEAFY COTYLEDON1 (LEC1), ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEC2 (Parcy et al., 1997; Boulard et al., 2017; Jo et al., 2019; Tang et al., 2021). These interactions lead to formation of tightly controlled regulatory pathways in seed and other tissues making CREs potentially useful targets for altered gene expression. Among the CREs, the RY repeat element (Bäumlein et al., 1992; Fujiwara and Beachy, 1994) and 2S seed protein motif (Stålberg et al., 1996) are crucial for regulatory activity in a number of seed-specific promoters (Reidt et al., 2000; Chatthai et al., 2004).
RNA programmable CRISPR/Cas nucleases trigger sequence-specific double stranded breaks (DSB)s that are subject to error prone non-homologous end joining (NHEJ) repair or template dependent, homology-directed repair (HDR) pathways. NHEJ, the most frequently used repair mechanism in plants, often leading to insertions or deletions (indels) at the target site. This technology is used to generate gene knockouts when coding regions are targeted, therefore, this approach has been widely used for plant genome editing applications (Manghwar et al., 2019).
Our overarching goal was to utilize CRISPR/Cas9-mediated gene editing to modify cis-regulatory elements in the 5′ UTR and intron of FAD2 genes for functional characterization of these elements and to potentially generate seeds with increased oleic acid content without affecting the fatty acid composition in other plant tissues. We designed guide RNAs (gRNAs) specific to the RY repeat element and 2S seed protein motif of peanut FAD2 genes then transformed peanut tissues to explore the potential of modifying transcriptional regulation of FAD2 gene expression in seeds. Our findings demonstrated high frequency modification of FAD2 RY and 2S motifs by CRISPR/Cas9-mediated gene editing and alteration of the oil profile in seed.
Materials and Methods
FAD2 Gene Sequence Confirmation and Analysis
The AhFAD2A and AhFAD2B genes from genotype GT-C20 were characterized by PCR and sequence analysis to serve as a reference. The gene models for selected isoforms, FAD2A (AH09G29670) and FAD2B (AH19G38370) (Figure 1) and expression data were obtained from the Peanut Genome Resource (http://peanutgr.fafu.edu.cn/index.php; Sinha et al., 2020). The cis-elements that are present in the region upstream of the coding sequence were predicted using PLACE (https://www.dna.affrc.go.jp/PLACE/?action=newplace; Higo et al., 1999) and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al., 2002).
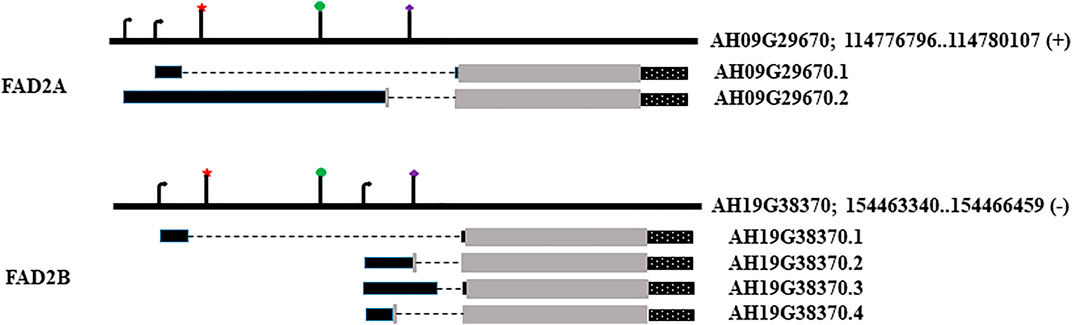
FIGURE 1. Genomic loci, transcript models and location of regulatory elements. Star denotes RY element; circle marks Initiator (INR) motif; diamond marks the 2S seed protein element. The arrows denote transcription start sites. Black rectangles denote 5′UTR, grey for coding sequence and dotted rectangles stand for 3′UTR, broken line represents the intron in the 5′UTR.
Construction of Initial Vectors
A dicot codon optimized Cas9 gene was excised from the vector pDW3602 as a BamHI/SpeI fragment and ligated into pDW3868 using the same enzyme sites to generate pDW3872. This construct has an Arabidopsis U6 promoter from pTF101-AtCa9-GmRCA#1, dual BsaI restriction sites for targeting oligonucleotide pair insertion, an extended gRNA scaffold (Dang. et al., 2015), modified Arabidopsis UBI promoter, Cas9 gene, a CaMV 35s terminator, and a CaMV 35s promoter expressing the Bar gene as a selectable marker. The extended gRNA scaffold was used to test for enhanced targeting efficiency compared to a standard gRNA scaffold. The vector pDW3877 has the same components as pDW3872, except a standard gRNA scaffold replaces the extended gRNA scaffold. The constructs are depicted in Figure 2.
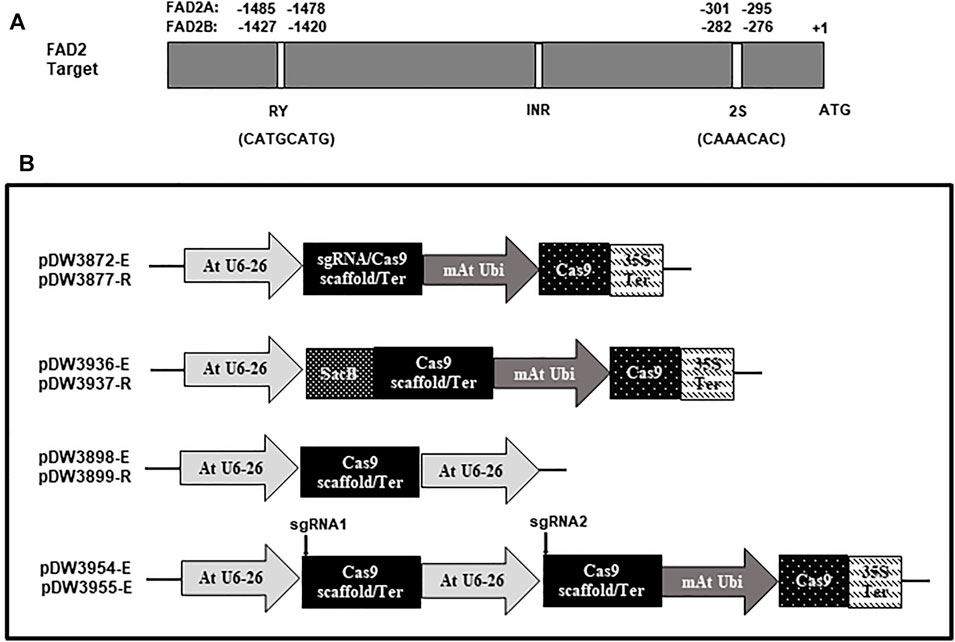
FIGURE 2. Schematic representation of the FAD2 targets and the vectors used in the study. (A). Targets location and sequence in the promoter of FAD2A and FAD2B; (B). Vectors with different scaffolds [Cas9-extended scaffold (-E) and Cas9-regular scaffold (-R)].
Final Vector Construction
gRNA targeting oligonucleotides specific to the predicted RY and 2S seed protein motif CREs were designed using the Cas-Designer tool (Park et al., 2015). The oligonucleotide pairs were synthesized at the Iowa State University DNA facility and were annealed after phosphorylation to generate sticky ends that correspond to the overhangs generated by BsaI restriction digestion of the expression vectors pDW3872 or pDW3877. The sequences of the gRNAs are provided in Table 1. The ligated oligo and vector combinations were transformed into competent DH10B E coli cells, which were selected on LB plates supplemented with 30 μg/ml kanamycin and grown at 37°C overnight. DNA from individual clones was purified using the IBI plasmid purification kit then clones were verified by DNA sequencing.
The cloning strategy for dual gRNA constructs was based on a one step Golden Gate ligation process. For this, a sacB gene was cloned into the BsaI sites of pDW3872 and pDW3877 to confer sucrose sensitivity, generating pDW3936 and pDW3937, respectively (Figure 2) Supplementary plasmids containing a BsaI site, extended or standard gRNA scaffold, an Arabidopsis U6 promoter and a second BsaI site, in that order, were created and designated pDW3898 and pDW3899, respectively (Figure 2). Note that the BsaI sites in the supplementary plasmids was positioned so the Type IIs enzyme will generate unique overhangs that cannot anneal to the Cas9 vector without an intervening oligo pair. Briefly, 625 ng of each plasmid was digested with 2 µL of Promega T4 ligase buffer and 1 μL of BsaI enzyme in a total volume of 18 μL at 37°C for 3 h 600 pmol of each oligo was phosphorylated in a total volume of 30 µL then appropriate pairs were annealed in a total of 60 µL. Finally, the 18 µL plasmid digest, 1 µL of each oligo pair, 1 µL of 10 mM ATP and 1 µL Promega T4 ligase were mixed for a total ligation volume of 22 μLs. The ligations were subjected to one cycle 16°C for 20 min then 49 cycles of 37°C for 5 min and 16°C for 5 min followed by one cycle at 65C for 20 min, in a PCR machine, to denature the enzymes. 2.5 µL of each ligation was transformed as described above. Successful dual oligo ligations were identified by positive/negative selection on LB medium supplemented with 30 μg/ml kanamycin and 5% sucrose. Sequence confirmed single and dual gRNA constructs were mobilized into Agrobacterium rhizogenes strain K599 by electroporation and selected using LB medium supplemented with 50 μg/ml kanamycin and grown at 28°C for 2 days.
Hairy Root Transformation
Agrobacterium rhizogenes is a plant pathogenic soil bacterium that induces the formation of adventitious roots, called hairy roots, at the site of infection. Hairy roots are unique in that they are predominantly non-chimeric and derive from a single de novo root meristem (Falasca et al., 2000). Stable transformation is laborious, time consuming and largely genotype dependent in peanuts. Therefore, the availability of rapid, reliable and quantifiable gene editing evaluation systems like hairy root assays help to select and advance functional target and gRNA combinations for later stable transformation efforts.
Genotype GT-C20 seeds were kindly provided by Dr. Baozhu Guo in USDA/ARS at Tifton, GA, which has no known natural mutations of the FAD2 genes (Yuan et al., 2019). Peanut seeds were surface sterilized and incubated on germination medium for 1 week with 16-h photoperiod at 28°C. Hairy root transformation was performed on hypocotyl explants as described by Yuan et al. (2019) with slight modifications. The Agrobacterium culture was suspended in the infection medium at an optical density of 0.6 and incubated for 20 min with constant stirring. The explants were co-cultured for 3 days in the dark and transferred to hairy root induction medium after washing with an antibiotic solution containing timentin (300 mg/L). Explants were incubated under fluorescent lights at 28°C with a 16-h photoperiod. After 1.5–2 weeks, transformed roots were harvested then explants were sub-cultured and maintained for 4–6 weeks.
Amplification and Sequencing of Targets
Hairy roots were harvested aseptically into sterile tubes and kept at –20°C for future processing. Root tissues were crushed in dilution buffer provided in the Phire Plant Direct PCR Kit (Thermo Fisher Scientific), and the liquid fraction was used for direct PCR without purification, as per the manufacturer’s instructions. The oligo sequences for PCR fragment generation and sequence analysis are given in Table 2. The amplified products were purified using the IBI PCR product purification kit then subjected to Sanger Sequencing at the Iowa State University DNA facility.
Calyx Tube Injection Transformation
To avoid time-consuming, genotype-dependent and often recalcitrant tissue culture and regeneration, we have developed an Agrobacterium-mediated calyx tube injection method to quickly obtain putatively edited peanut seeds. CRISPR/Cas9 constructs were mobilized into Agrobacterium strain GV3101 using a freeze and thaw methodology (An et al., 1988). Transformed Agrobacterium were grown on LB plates supplemented with 50 μg/ml kanamycin at 28°C and colonies were picked after 2–3 days’ growth, followed by colony PCR to confirm mobilization of the constructs. Transformation confirmed Agrobacterium colonies were grown in 10 ml liquid LB cultures with shaking overnight at 28°C and supplemented with 50 μg/ml kanamycin. Bacterial cells were pelleted using low speed centrifugation (4,000 rpm) and resuspended into 10 ml sterile inoculation media composed of ½ MS supplemented with 25 g/L sucrose and 200 μL/L Silwet L77. The OD600 was adjusted to 0.6 to 0.8 by addition of inoculation media. Besides GT-C20, two additional genotypes (AU18-46 and Exp27-1516) were also used to test the method of calyx tube injection. 0.2 ml of the Agrobacterium suspension carrying a CRISPR/Cas9 vector was injected into individual peanut plant calyx tubes. Inoculated flowers were labelled and derived seeds were harvested for analysis. DNAs were extracted from T1 plants for sequencing FAD2 targets.
Fatty Acid Composition Analysis
The fatty acid content was measured for each of the harvested seeds from T0 and T1 generations using an Agilent 7890A gas chromatograph (GC) with a flame ionization detector (FID). Seeds were individually crushed. Ground seed material was extracted in 4.0 ml heptane (Fisher Scientific) and converted to fatty acid methyl easters (FAMEs) with 500 µL 0.5 N sodium methoxide (NaOCH3) in methanol. A fatty acid methyl ester (FAME) standard mix RM-3 plus four additional FAMES (Sigma) were mixed and used to establish peak retention times. Fatty acid composition was determined by identifying and calculating relative peak areas (Wang et al., 2009).
Results
The FAD2A (AH09G29670) and FAD2B (AH19G38370) gene models used in this study have a 5′ UTR with a single intron, coding sequence, and a 3′ UTR as shown in Figure 2. FAD2A has two transcript models and FAD2B has four transcript models with altered 5′ UTRs, and two transcriptional start sites predicted for each gene. We considered the transcript model AH09G29670.1 for FAD2A and AH19G38370.1 for FAD2B as a reference for this investigation. Mining of transcriptomic data (http://peanutgr.fafu.edu.cn/index.php) revealed co-expression of both FAD2 homeologous genes in different tissues including seed, leaf, root tip, root nodule, stem tip, inflorescence etc. (Sinha et al., 2020). FAD2 gene expression was significantly upregulated in peanut seeds, especially during advanced embryo development stages, specifically FAD2B had a relatively high expression in root nodules and seed tissues compared to FAD2A (Sinha et al., 2020).
In this study, several conserved cis-elements important for tissue specific responsiveness to hormonal and environmental cues were observed in the AhFAD2A and AhFAD2B upstream 5′UTRs (Supplementary File S1). Among them, the cis-regulatory RY element (CATGCATG) and 2S seed protein motifs (CAAACAC), which are implicated in seed specific gene expression. The initiator elements (INR) with sequences ‘TTCATTCT’ and ‘TTCATTTT’ responsible for basal transcription and light responsiveness were found. The cis-regulatory elements involved in stress response included Anaerobiosis Responsive Element (ARE), Wounding-Responsive Element (WUN motif), W box, G box, Anaeroconsensus among others were also identified. Additionally, the conserved elements for hormonal regulation included Abscisic acid Responsive Element (ABRE), Auxin Responsive Element (AuxRE), GA-responsive elements (GARE motif) etc. were predicted (Supplementary File S1). We chose to focus on the seed specific RY and 2S motifs for this study.
Complete single gRNA CRISPR/Cas9 constructs with a standard or extended gRNA scaffold were developed for the RY and 2S target sites then an Agrobacterium-mediated hairy root assay was employed to efficiently test the assembled constructs because traditional peanut transformation and regeneration methods are technically challenging and time consuming. Site-specific mutation frequencies and the type of DNA modifications were analyzed and recorded for each construct through PCR amplification of the target area followed by purification and Sanger sequencing.
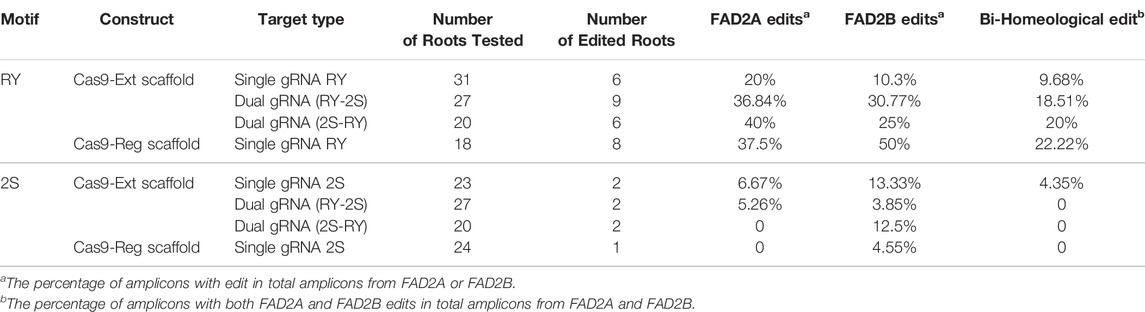
For the single gRNA constructs, a total of 49 hairy roots for RY and 47 hairy roots for 2S were examined. Indels at the distal RY repeat motif ranged from +2 to −33 bp in AhFAD2A and +1 to −7 bp in AhFAD2B and the vast majority of edits were indels of less than 10 bp. The extended scaffold produced mutations in FAD2A at 20%, FAD2B at 10.3% and at both genes 9.68% of the time. The standard scaffold produced mutations in FAD2A at 37.5%, FAD2B at 50% and at both genes 22.22% of the time (Table 3). Most indels caused a disruption of the RY motif (Figure 3). Based on these figures, the standard gRNA scaffold performed better than the extended gRNA scaffold at RY.
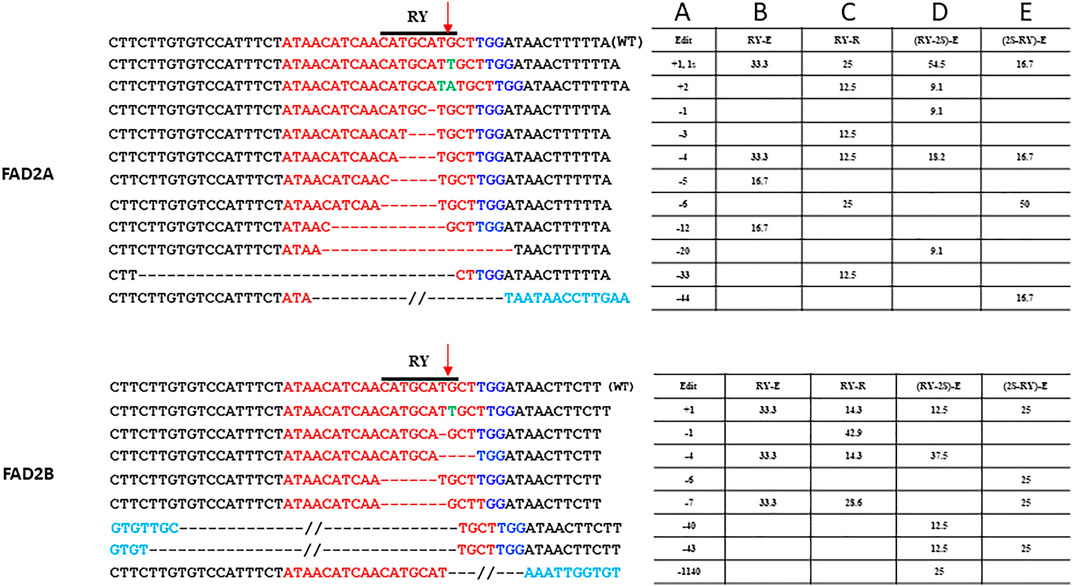
FIGURE 3. Site-specific modifications at RY repeat element in FAD2A and FAD2B Shown are mutations around the RY motif that were detected for the single RY targeted and duel RY and 2S targeted CRISPR/Cas9 constructs using the hairy root system. The target protospacer sequence is in red, the PAM is in dark blue, insertions are in green, bases in light blue are outside of the wild type sequence that is shown, deletions are represented by dashes, a double slash indicates a deletion larger than what is depicted and the arrow indicated the predicted double strand break site (DSB). Numbers in column A indicate indel size, numbers in columns B to E are percentages of total mutations and the E (extended) or R (standard) designation in columns B to E indicate the gRNA scaffold used.
The number of mutations detected at the proximal 2S motif were lower than that observed at the RY motif and the type of edits ranged from indels of −4 bp in FAD2A and +1 to −3 bp for FAD2B with most mutations causing disruption of the 2S motif (Figure 4). The extended scaffold produced mutations in FAD2A at 6.67%, FAD2B at 13.33% and in both genes at 4.35% of the time. The standard scaffold did not produce detectable mutations in FAD2A while 4.55% of mutations were at FAD2B (Table 3) Based on these figures, the extended scaffold performed better than the standard gRNA scaffold at 2S.
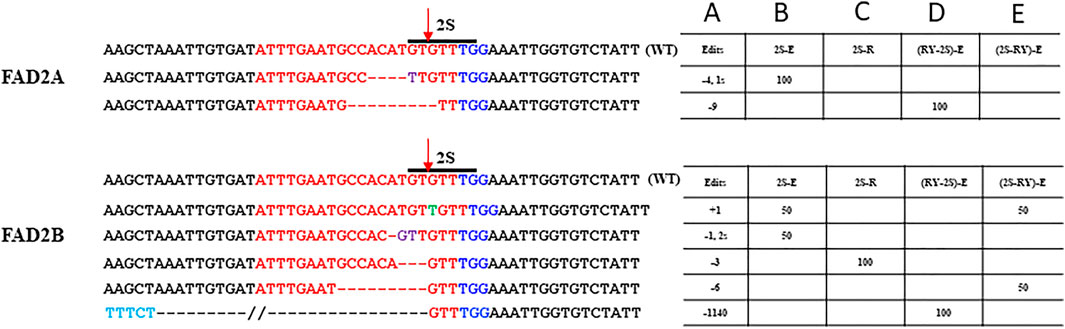
FIGURE 4. Site–specific modifications at 2S Seed protein element. Shown are mutations around the 2S motif that were detected for the single 2S targeted and duel RY and 2S targeted CRISPR/Cas9 constructs used the hairy root assays. The target protospacer sequence is in red, PAM is in blue, substitutions are in purple, insertions are in green, deletions are represented by dashes and the arrow indicated the predicted double strand break site (DSB). Numbers in column A indicate indel size, numbers in columns B to E are percentages of total mutations and the E (extended) or R (standard) designation in columns B to E indicate the gRNA scaffold used.
Dual gRNA editing constructs were generated that target both RY and 2S motifs using the validated targeting oligo sequences used in the single gRNA constructs. These constructs used the extended scaffold because they exhibited improved efficiency overall and were meant to test the efficiency of editing both targets simultaneously, targeting both genes simultaneously the possibility of generating large deletions between RY and 2S. To test for position effect in the gRNA order, two constructs were generated: one with a RY gRNA in the first position and 2S in the second position while the reverse order was used for the second construct.
Analysis of transformed hairy roots showed that overall editing efficiency at RY was greater than at 2S regardless of gRNA order; a result similar to that observed with the single gRNA study. This remained true for edits in both the FAD2A and FAD2B genes. Most indels at RY were less than 7 bp (80–90% in FAD2A and 50–75% in FAD2B) and a few (16–25%) deletions were greater than 40 bp (43 bp, 44 bp and 46 bp specifically) (Figure 3). Approximately 18–20% of the lines had RY edits in both FAD2A and FAD2B using either gRNA order (Table 3). Also noted was a single root containing a large deletion of 1,140 bp between the RY and 2S sites in FAD2B (Figure 3).
Using the dual editing constructs, it was noted that editing at the 2S motif was much less effective with only one edit of -9 bp in FAD2A when the RY gRNA was in the first position and no edits were observed when 2S was in the first position for FAD2A. The opposite was true for FAD2B where no edits were observed when the RY gRNA was in the first position, but a +1 edit and a −6 bp edit were observed when the 2S gRNA was in the first position (Figure 4). Additionally, the dual gRNA constructs produced no simultaneous 2S mutations in both FAD2A and FAD2B (Table 3).
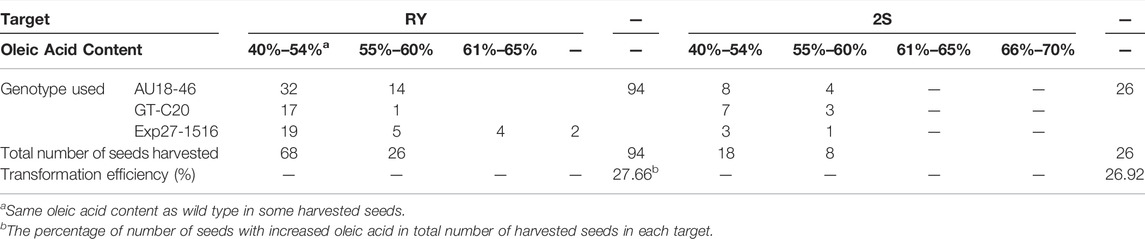
To access the consequences of FAD2 gene editing on oil quality, Agrobacterium-mediated transformation through calyx tube injection was conducted. Single gRNA constructs with the extended scaffold targeting either RY or 2S were used to compare the mutation effect on oil quality at each motif. Approximate 650 calyx tubes were injected with the RY construct strain and 200 of calyx tubes were injected with the 2S construct strain. Following injection, a total of 120 potentially edited seeds were harvested. Among these, 26 seeds had an increased oleic acid content ranging from 55 to 70% when the RY motif was targeted and 7 seeds had an increased oleic acid content ranging from 55 to 60% when the 2S motif was targeted in compared to untransformed seeds with oleic acid concentration of 40–54% (Table 4). The changes of fatty acid in T0 seeds were listed in the Supplementary File S2. Although the apparent transformation efficiency was similar (27.66 RY vs 26.92% 2S) for both constructs, the resulting oleic acid content in the edited RY motif was higher than the edited 2S motif. It should be noted that the oleic acid content of two seeds for the RY motif was 66–70%, while the majority of presumed edits had a lower oleic acid content. Regardless, apparent mutations increased oleic acid concentration in these seed whether the RY or 2S motifs were targeted showing efficacy of this approach. However, amplifications of T1 plant DNAs showed no mutations in the targets from either FAD2A or FAD2B, and all seeds harvested from T1 plants derived from these T0 edited seeds showed oleic acid content less than 55%. It might indicate that the CRISPR components were delivered into somatic cells rather than germ cells in T0 seeds using the calyx tube injection method.
Discussion
Gene Editing has proven to be a powerful tool for generating gene knockouts for the study of gene function. Although less well studied, promoter editing has been successfully employed to engineer desirable traits such as disease resistance in rice (Oliva et al., 2019) and yield traits in tomato (Rodríguez-Leal et al., 2017). As demonstrated in this study and others, this approach has the potential to generate a wide spectrum of novel transcriptional alleles for genetic fine-tuning and optimized gene expression in crop plants.
The RY repeat motif is implicated in quantitative expression in seeds in diverse species (Baumlein et al., 1992; Lelievre et al., 1992; Dickinson et al., 1998) and is known to interact with B3 domain transcription factors involved in Abscisic acid (ABA) mediated transcription, embryogenesis and seed development (Reidt et al., 2000). Promoter studies by Ellerström et al. (1996) demonstrated that alteration of RY motifs in the Brassica napus nap A promoter resulted in changes in tissue specific gene expression. In addition, the 2S seed protein motif is conserved in the promoters of several seed storage protein genes and is implicated in higher activity of the nap A promoter (Stålberg et al., 1996). Deletion analysis specifically showed the 2S motif was important in regulation of seed protein accumulation while alterations to the promoter changed gene expression patterns (Stålberg et al., 1993). In this study, we investigated the feasibility and efficiency of simultaneously editing regulatory elements in two related peanut genes. Specifically, the RY and 2S motifs in the FAD2 genes were altered and an expected increase in oleic acid content of seed was detected suggesting reduced FAD2 gene expression as a consequence of targeted promoter alteration.
Initial testing of gene editing technology in peanut focused on determining the efficacy of CRISPR/Cas9 technology when single gRNAs were employed. Previously tested vector components were ligated together to create a basic vector platform that contained either a standard or an extended gRNA scaffold then targeting oligos for the RY or 2S motifs were added. Testing of constructs in a hairy root assay demonstrated the following: 1) editing efficiency at the distal RY motif was higher than at the proximal 2S motif, 2) the standard gRNA scaffold was more effective at RY than the extended gRNA scaffold, 3) the extended gRNA scaffold was more effective at 2S than the standard scaffold, 4) FAD2B was targeted more efficiently than FAD2A, and 5) the predominance of edits involving deletions less than 10 bp with a single gRNA target was consistent with earlier studies (Gisler et al., 2019). Taken together, these data demonstrate the efficacy of CRISPR/Cas9 mediated editing of peanut FAD2 promoter sequences and suggest that some positions within a promoter may be more accessible than others, use of different gRNA scaffolds may give some flexibility in target efficiency and there may be some difference in homeolog accessibility when targeting gene families.
To further explore the limits of promoter sequence editing, dual gRNA constructs, using the extended gRNA scaffold, were generated to target RY and 2S motifs simultaneously. Two versions of the vector were constructed with RY and 2S targeting oligos in the first and second positions respectively or in the reverse order. The results demonstrated differential editing rates at the target sites with RY being targeted at a higher rate than 2S. Again, FAD2A was targeted more frequently than FAD2B for the RY motif and FAD2B was targeted more frequently than FAD2A for the 2S motif. This could be related to the sequence context, local DNA methylation levels, or the chromatin configuration modulating accessibility as suggested by Liu et al., 2019. It should also be noted that this observation was independent of which targeting oligo was in the first position or second position.
Most indels created by the dual gRNA vectors were less than 12 bases while a few were larger. A few longer deletions (>40 bp) at a single site seems to suggest a plausible interaction effect of editing at closely linked sites possibly affecting the local chromatin context. This type of local opening up of chromatin can possibly modulate the timing and activity of the individual gRNAs. This seems analogous to the strategy involving coupling of proximal dead sgRNA (dsgRNA) to the functional sgRNA target, to open up the closed chromatin in rice cells and thereby enhancing editing efficiency (Liu et al., 2019). It should also be noted that one instance of a large deletion (1,140 bp) was detected, which demonstrates that it is possible to generate deletions between to gRNA targets albeit at a low frequency. Taken together this dual gRNA construct data demonstrates that multiple targets can be altered in a gene family in peanut, generating many different types of indels that may be useful for promoter modulation.
With the efficacy of single and dual gRNA constructs demonstrated in peanut, a test of practical application would demonstrate the usefulness of promoter modification. Fatty acids are essential components of plant cells and cell membranes, playing an important role in regulating different abiotic stress responses by modulating cell membrane properties (Iba, 2002; Kachroo and Kachroo, 2009). Thus, the desaturation of fatty acids and the number and position of double bonds in fatty acid chains influences the physical and physiological properties of membranes (Shanklin and Cahoon, 1998; Wu et al., 2009), which impacts proper plant growth and development (Traore and He, 2021). Therefore, general FAD2 loss of function mutations would be predicted to negatively impact cell membranes, leading to poor plant growth and development under abiotic stress conditions. This suggests that existing high oleic acid varieties resulting from mutations in the coding sequences of both FAD2A and FAD2B, while commercially useful, may be agriculturally detrimental (Lopez et al., 2000). CRISPR/Cas9 gene editing in peanut may afford a more nuance approach whereby the FAD2 gene activity in seed could be reduced while maintaining gene activity in other plant tissues.
A calyx tube transformation method was employed using the previously demonstrated RY and 2S single gRNA constructs to test if RY or 2S modification would impact oleic acid content in seed. A total of 120 seeds were generated through this method—94 for the RY construct and 26 for the 2S construct. The transformation efficiency for both constructs was relatively high at 27.66% for the RY construct and 26.92% for the 2S construct based on elevated oleic acid content relative to wild type. When tested by gas chromatograph method, the majority of the seeds had an oleic acid content ranging from 55% to 65%. While none of the 2S seeds tested above 61% oleic acid content, four of the RY edited seed were between 61 and 65% and two seeds were in the range of 66%–70% oleic acid. Taken together, this data suggests that editing of the RY or 2S motifs in the peanut FAD2 genes can impact oleic acid concentration in seed. Because the increased oleic acid content in T0 seeds might be derived from somatic cell mutations, a further experiment will be needed to optimize the parameter for inoculation media, the suitable time and the location of the calyx tube for injection to improve the delivery of construct into reproductive cells with the calyx tube injection method.
Using gene editing to target cis-regulatory elements of the FAD2 gene promoters provides a promising approach to manipulate FAD2 gene expression in seeds and potentially minimize undesirable pleiotropic effects on other plant tissues while improving the fatty acid profile of seed. The use of CRISPR/Cas9 based promoter or enhancer editing may lead to the development of “cisgenic” plants with optimized FAD2 gene expression, which may serve as ideal breeding materials for trait introgression, without introducing potentially deleterious alleles and linkage drag, thereby accelerating the pace of cultivar development.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AN, DW, MS and GH conceived and designed the study and developed the manuscript. AN, XC and ST conducted experiment and data analysis. All authors proofread and approved the final version.
Funding
The authors are grateful for financial support from USDA/NIFA (Grant No: 2018-67014-27572 and 2018-38821-27758).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Brandon Tonnis and Ming Li Wang from USDA-ARS Plant Genetic Resources Conservation Unit at Griffin, GA, for the measurement of fatty acid composition.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.849961/full#supplementary-material
References
An, G., Ebert, P. R., Mitra, A., and Ha, S. B. (1988). “Binary Vectors,” in Plant Molecular Biology Manual. Editors S. B. R. A. GelvinSchilperoort, and D. P. S. Verma (Dordrecht, Netherlands: Kluwer Academic), A3, 1–19. doi:10.1007/978-94-017-5294-7_3
Bäumlein, H., Nagy, I., Villarroel, R., Inzé, D., and Wobus, U. (1992). Cis-analysis of a Seed Protein Gene Promoter: the Conservative RY Repeat CATGCATG within the Legumin Box Is Essential for Tissue-specific Expression of a Legumin Gene. Plant J. Cel Mol. Biol. 2, 233
Boulard, C., Fatihi, A., Lepiniec, L., and Dubreucq, B. (2017). Regulation and Evolution of the Interaction of the Seed B3 Transcription Factors With NF-Y Subunits. Biochim. Biophys. Acta. Gene Regul. Mech. 1860 (10), 1069–1078.
Cao, S., Zhou, X.-R., Wood, C. C., Green, A. G., Singh, S. P., Liu, L., et al. (2013). A Large and Functionally Diverse Family of Fad2 Genes in Safflower (Carthamus tinctoriusL.). BMC Plant Biol. 13, 5. doi:10.1186/1471-2229-13-5
Chatthai, M., Forward, B. S., Yevtushenko, D., Stefanov, I., Osuska, L., Osusky, M., et al. (2004). 2S Storage Protein Gene of Douglas-fir: Characterization and Activity of Promoter in Transgenic Tobacco Seeds. Plant Physiol. Biochem. 42, 417–423. doi:10.1016/j.plaphy.2004.04.008
Dang, Y., Jia, G., Choi, J., Ma, H., Anaya, E., Ye, C., et al. (2015). Optimizing sgRNA Structure to Improve CRISPR-Cas9 Knockout Efficiency. Genome Biol. 16, 280. doi:10.1186/s13059-015-0846-3
Dar, A. A., Choudhury, A. R., Kancharla, P. K., and Arumugam, N. (2017). The FAD2 Gene in Plants: Occurrence, Regulation, and Role. Front. Plant Sci. 8, 1789. doi:10.3389/fpls.2017.01789
Dickinson, C. D., Evans, R. P., and Nielsen, N. C. (1988). RY Repeats Are Conserved in the 5′-flanking Regions of Legume Seed-Protein Genes. Nucl. Acids Res. 16, 371. doi:10.1093/nar/16.1.371
Ellerström, M., Stålbery, K., Ezcurra, I., and Rask, L. (1996). Functional Dissection of a Napin Gene Promoter: Identification of Promoter Elements Required for Embryo and Endosperm Specific Transcription. Plant Mol. Biol. 32, 1019
Falasca, G., Reverberi, M., Lauri, P., Caboni, E., De Stradis, A., and Altamura, M. M. (2000). How Agrobacterium Rhizogenes Triggers de novo Root Formation in a Recalcitrant Woody Plant: An Integrated Histological, Ultrastructural and Molecular Analysis. The New Phytologist. 145 (1), 77–93.
Fujiwara, T., and Beachy, R. N. (1994). Tissue-specific and Temporal Regulation of a ?-conglycinin Gene: Roles of the RY Repeat and Other Cis-Acting Elements. Plant Mol. Biol. 24, 261–272. doi:10.1007/bf00020166
Gisler, S., Gonçalves, J. P., Akhtar, W., de Jong, J., Pindyurin, A. V., Wessels, L. F. A., et al. (2019). Multiplexed Cas9 Targeting Reveals Genomic Location Effects and gRNA-Based Staggered Breaks Influencing Mutation Efficiency. Nat. Commun. 10, 1598. doi:10.1038/s41467-019-09551-w
Guan, L.-L., Wang, Y.-B., Shen, H., Hou, K., Xu, Y.-W., and Wu, W. (2012). Molecular Cloning and Expression Analysis of Genes Encoding Two Microsomal Oleate Desaturases (FAD2) from Safflower (Carthamus tinctorius L.). Plant Mol. Biol. Rep. 30, 139–148. doi:10.1007/s11105-011-0322-5
Gutierrez, L., Van Wuytswinkel, O., Castelain, M., and Bellini, C. (2007). Combined Networks Regulating Seed Maturation. Trends Plant Sci. 12, 294–300. doi:10.1016/j.tplants.2007.06.003
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant Cis-Acting Regulatory DNA Elements (PLACE) Database: 1999. Nucleic Acids Res. 27 (1), 297–300. doi:10.1093/nar/27.1.297
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol. 31 (9), 827–832. doi:10.1038/nbt.2647
Iba, K. (2002). Acclimative Response to Temperature Stress in Higher Plants: Approaches of Gene Engineering for Temperature Tolerance. Annu. Rev. Plant Biol. 53, 225–245. doi:10.1146/annurev.arplant.53.100201.160729
Janila, P., Pandey, M. K., Shasidhar, Y., Variath, M. T., Sriswathi, M., Khera, P., et al. (2016). Molecular Breeding for Introgression of Fatty Acid Desaturase Mutant Alleles ( ahFAD2A and ahFAD2B ) Enhances Oil Quality in High and Low Oil Containing Peanut Genotypes. Plant Sci. 242, 203–213. doi:10.1016/j.plantsci.2015.08.013
Jo, L., Pelletier, J. M., and Harada, J. J. (2019). Central Role of the LEAFY COTYLEDON1 Transcription Factor in Seed Development. J. Integr. Plant Biol. 61 (5), 564–580.
Kachroo, A., and Kachroo, P. (2009). Fatty Acid-Derived Signals in Plant Defense. Annu. Rev. Phytopathol. 47, 153–176. doi:10.1146/annurev-phyto-080508-081820
Kim, M. J., Kim, H., Shin, J. S., Chung, C.-H., Ohlrogge, J. B., and Suh, M. C. (2006). Seed-specific Expression of Sesame Microsomal Oleic Acid Desaturase Is Controlled by Combinatorial Properties between Negative Cis-Regulatory Elements in the SeFAD2 Promoter and Enhancers in the 5′-UTR Intron. Mol. Genet. Genomics 276 (4), 351–368. doi:10.1007/s00438-006-0148-2
Lee, M. W., Padilla, C. S., Gupta, C., Galla, A., Pereira, A., Li, J., et al. (2020). The FATTY ACID DESATURASE2 Family in Tomato Contributes to Primary Metabolism and Stress Responses. Plant Physiol. 182, 1083–1099. doi:10.1104/pp.19.00487
Lelievre, J.-M., Oliveira, L. O., and Nielsen, N. C. (1992). 5′CATGCAT-3′ Elements Modulate the Expression of Glycinin Genes. Plant Physiol. 98, 387–391. doi:10.1104/pp.98.1.387
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Lim, D. K., Long, N. P., Mo, C., Dong, Z., Cui, L., Kim, G., et al. (2017). Combination of Mass Spectrometry-based Targeted Lipidomics and Supervised Machine Learning Algorithms in Detecting Adulterated Admixtures of White Rice. Food Res. Int. 100, 814–821.
Liu, G., Yin, K., Zhang, Q., Gao, C., and Qiu, J.-L. (2019). Modulating Chromatin Accessibility by Transactivation and Targeting Proximal dsgRNAs Enhances Cas9 Editing Efficiency In Vivo. Genome Biol. 20, 145. doi:10.1186/s13059-019-1762-8
López, Y., Nadaf, H. L., Smith, O. D., Connell, J. P., Reddy, A. S., and Fritz, A. K. (2000). Isolation and Characterization of the Δ12-fatty Acid Desaturase in Peanut (Arachis hypogaea L.) and Search for Polymorphisms for the High Oleate Trait in Spanish Market-type Lines. Theor. Appl. Genet. 101, 1131–1138. doi:10.1007/s001220051589
Manghwar, H., Lindsey, K., Zhang, X., and Jin, S. (2019). CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 24, 1102–1125. doi:10.1016/j.tplants.2019.09.006
Marangoni, F., Agostoni, C., Borghi, C., Catapano, A. L., Cena, H., Ghiselli, A., et al. (2020). Dietary Linoleic Acid and Human Health: Focus on Cardiovascular and Cardiometabolic Effects. Atherosclerosis 292, 90–98.
Menard, G. N., Moreno, J. M., Bryant, F. M., Munoz-Azcarate, O., Kelly, A. A., Hassani-Pak, K., et al. (2017). Genome Wide Analysis of Fatty Acid Desaturation and its Response to Temperature. Plant Physiol. 173, 1594–1605. doi:10.1104/pp.16.01907
Miquel, M., and Browse, J. (1992). Arabidopsis Mutants Deficient in Polyunsaturated Fatty Acid Synthesis. Biochemical and Genetic Characterization of a Plant Oleoyl-Phosphatidylcholine Desaturase. J. Biol. Chem. 267, 1502–1509. doi:10.1016/s0021-9258(18)45974-1
Norden, A. J., Gorbet, D. W., Knauft, D. A., and Young, C. T. (1987). Variability in Oil Quality Among Peanut Genotypes in the Florida Breeding Program1. Peanut Sci. 14, 7–11. doi:10.3146/i0095-3679-14-1-3
O’Byrne, D. J., Knauft, D. A., and Shireman, R. B. (1997). Low Fat Monounsaturated Rich Diets Containing High-Oleic Peanuts Improve Serum Lipoprotein Profiles. Lipids 32, 678
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum Resistance to Bacterial Blight in rice Using Genome Editing. Nat. Biotechnol. 37 (11), 1344–1350. doi:10.1038/s41587-019-0267-z
Parcy, F., Valon, C., Kohara, A., Miséra, S., and Giraudat, J. (1997). The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 Loci Act in Concert to Control Multiple Aspects of Arabidopsis Seed Development. Plant Cell 9 (8), 1265–1277.
Park, J., Bae, S., and Kim, J. S. (2015). Cas-Designer: A Web-Based Tool for Choice of CRISPR-Cas9 Target Sites. Bioinformatics 31, 4014–4016. doi:10.1093/bioinformatics/btv537
Reidt, W., Wohlfarth, T., Ellerstrom, M., Czihal, A., Tewes, A., Ezcurra, I., et al. (2000). Gene Regulation during Late Embryogenesis: the RY Motif of Maturation-specific Gene Promoters Is a Direct Target of the FUS3 Gene Product. Plant J. 21, 401–408. doi:10.1046/j.1365-313x.2000.00686.x
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 171 (2), 470–e8. doi:10.1016/j.cell.2017.08.030
Salimonti, A., Carbone, F., Romano, E., Pellegrino, M., Benincasa, C., Micali, S., et al. (2020). Association Study of the 5′UTR Intron of the FAD2-2 Gene with Oleic and Linoleic Acid Content in Olea Europaea L. Front. Plant Sci. 11, 66. doi:10.3389/fpls.2020.00066
Shanklin, J., and Cahoon, E. B. (1998). Desaturation and Related Modifications of Fatty Acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 611–641. doi:10.1146/annurev.arplant.49.1.611
Sinha, P., Bajaj, P., Pazhamala, L. T., Nayak, S. N., Pandey, M. K., Chitikineni, A., et al. (2020). Arachis hypogaea Gene Expression Atlas for Fastigiata Subspecies of Cultivated Groundnut to Accelerate Functional and Translational Genomics Applications. Plant Biotechnol. J. 18 (11), 2187–2200. doi:10.1111/pbi.13374
Stålberg, K., Ellerstöm, M., and Ezcurra, I. (1996). Disruption of an Overlapping E-Box/ABRE Motif Abolished High Transcription of the napA Storage-Protein Promoter in Transgenic Brassica Napus Seeds. Planta 199, 515
Stålberg, K., Ellerström, M., Josefsson, L. G., and Rask, L. (1993). Deletion Analysis of a 2S Seed Storage Protein Promoter of Brassica Napus in Transgenic Tobacco. Plant Mol. Biol. 23, 671
Tang, G., Xu, P., Li, P., Zhu, J., Chen, G., Shan, L., et al. (2021). Cloning and Functional Characterization of Seed-specific LEC1A Promoter from Peanut (Arachis hypogaea L.). PLoS ONE 16 (3), e0242949. doi:10.1371/journal.pone.0242949
Traore, S. M., and He, G. (2021). “Soybean as a Model Crop to Study Plant Oil Genes: Mutations in FAD2 Gene Family,” in Book Chapter in: Model Organisms in Plant Genetics. doi:10.5772/intechopen.99752
Wang, M. L., Chen, C. Y., Davis, J., Guo, B., Stalker, H. T., and Pittman, R. N. (2009). Assessment of Oil Content and Fatty Acid Composition Variability in Different Peanut Subspecies and Botanical Varieties. Plant Genet. Res. 8, 71–73. doi:10.1017/s1479262109990177
Wu, Q., Liu, T., Liu, H., and Zheng, G. C. (2009). Unsaturated Fatty Acid: Metabolism, Synthesis and Gene Regulation. Afr. J. Biotechnol. 8 (9), 1782–1785. http://www.academicjournals.org/AJB.
Xiao, G., Zhang, Z. Q., Yin, C. F., Liu, R. Y., Wu, X. M., Tan, T. L., et al. (2014). Characterization of the Promoter and 5′-UTR Intron of Oleic Acid Desaturase (FAD2) Gene in Brassica Napus. Gene 545, 45–55. doi:10.1016/j.gene.2014.05.008
Yu, S., Pan, L., Yang, Q., Min, P., Ren, Z., and Zhang, H. (2008). Comparison of the Δ12 Fatty Acid Desaturase Gene between High-Oleic and normal-oleic Peanut Genotypes. J. Genet. Genomics 35, 679–685. doi:10.1016/s1673-8527(08)60090-9
Yuan, M., Zhu, J., Gong, L., He, L., Lee, C., Han, S., et al. (2019). Mutagenesis of FAD2 Genes in Peanut with CRISPR/Cas9 Based Gene Editing. BMC Biotechnol. 19, 24. doi:10.1186/s12896-019-0516-8
Keywords: regulatory element, gene editing, agrobacterium-mediated transformation, fatty acid, peanut
Citation: Neelakandan AK, Wright DA, Traore SM, Chen X, Spalding MH and He G (2022) CRISPR/Cas9 Based Site-Specific Modification of FAD2 cis-Regulatory Motifs in Peanut (Arachis hypogaea L). Front. Genet. 13:849961. doi: 10.3389/fgene.2022.849961
Received: 06 January 2022; Accepted: 12 April 2022;
Published: 27 April 2022.
Edited by:
Jianping Wang, University of Florida, United StatesReviewed by:
Faiz Ahmad Joyia, University of Agriculture, PakistanHai Zhou, South China Agricultural University, China
Copyright © 2022 Neelakandan, Wright, Traore, Chen, Spalding and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guohao He, Z2hlQHR1c2tlZ2VlLmVkdQ==
 Anjanasree K. Neelakandan
Anjanasree K. Neelakandan David A. Wright1
David A. Wright1 Sy M. Traore
Sy M. Traore Xiangyu Chen
Xiangyu Chen Martin H. Spalding
Martin H. Spalding Guohao He
Guohao He