- Invitae, San Francisco, CA, United States
Although multiple factors can influence the uptake of cascade genetic testing, the impact of proband indication has not been studied. We performed a retrospective, cross-sectional study comparing cascade genetic testing rates among relatives of probands who received either diagnostic germline testing or non-indication-based proactive screening via next-generation sequencing (NGS)-based multigene panels for hereditary cancer syndromes (HCS) and/or familial hypercholesterolemia (FH). The proportion of probands with a medically actionable (positive) finding were calculated based on genes associated with Centers for Disease Control and Prevention (CDC) Tier 1 conditions, HCS genes, and FH genes. Among probands with a positive finding, cascade testing rates and influencing factors were assessed. A total of 270,715 probands were eligible for inclusion in the study (diagnostic n = 254,281,93.9%; proactive n = 16,434, 6.1%). A positive result in a gene associated with a CDC Tier 1 condition was identified in 10,520 diagnostic probands (4.1%) and 337 proactive probands (2.1%), leading to cascade testing among families of 3,305 diagnostic probands (31.4%) and 36 proactive probands (10.7%) (p < 0.0001). A positive result in an HCS gene was returned to 23,272 diagnostic probands (9.4%) and 970 proactive probands (6.1%), leading to cascade testing among families of 6,611 diagnostic probands (28.4%) and 89 proactive probands (9.2%) (p < 0.0001). Cascade testing due to a positive result in an HCS gene was more commonly pursued when the diagnostic proband was White, had a finding in a gene associated with a CDC Tier 1 condition, or had a personal history of cancer, or when the proactive proband was female. A positive result in an FH gene was returned to 1,647 diagnostic probands (25.3%) and 67 proactive probands (0.62%), leading to cascade testing among families of 360 diagnostic probands (21.9%) and 4 proactive probands (6.0%) (p < 0.01). Consistently higher rates of cascade testing among families of diagnostic probands may be due to a perceived urgency because of personal or family history of disease. Due to the proven clinical benefit of cascade testing, further research on obstacles to systematic implementation and uptake of testing for relatives of any proband with a medically actionable variant is warranted.
Introduction
Cascade testing is the process of providing genetic counseling and testing to at-risk blood relatives following the detection of a pathogenic variant in a disease-causing gene in a family member (i.e., the proband). Confirming the presence (or absence) of a pathogenic variant in at-risk relatives can inform clinical management, including both preventative measures for unaffected relatives and potential changes in treatment for affected relatives. For example, given a proband with diagnosed breast cancer and a pathogenic variant in BRCA1, an unaffected relative who is confirmed to have the same genetic variant may increase mammography screenings or opt for risk-reducing surgery (Daly et al., 2021). The same unaffected relative, if confirmed negative for the pathogenic BRCA1 variant, could likely forgo escalated screenings or other preventive interventions. A recent study estimated that it would take 9.9 years to detect all carriers of a pathogenic variant in one of 18 genes associated with a hereditary cancer syndrome (HCS) in the United States (including BRCA1) if cascade testing were used, compared with 59.5 years if it were not (Offit et al., 2020). Further, cascade testing has the ability to inform reproductive health decisions, especially in relatives who have been identified as carriers of an autosomal recessive disease, and has been demonstrated to be a cost-effective approach for identifying at-risk individuals across many disease types, especially in young, unaffected relatives (Marks et al., 2002; Wonderling et al., 2004; Ademi et al., 2014; Grosse, 2015; Kerr et al., 2017; O’Brien et al., 2021). As a result, cascade testing has immense potential for improving the efficiency of healthcare resource utilization by reducing the burden of care for individuals and families as well as health systems.
Most studies of cascade testing have focused on the genes associated with Tier 1 conditions as established by the Centers for Disease Control and Prevention (CDC), which include hereditary breast and ovarian cancer (HBOC), Lynch syndrome, and familial hypercholesterolemia (FH) (Centers for Disease Control and Prevention, 2021). Evidence demonstrating the utility of cascade testing has led to recommendations and guidelines from professional societies and from the CDC that encourage extending testing to at-risk relatives (Nordestgaard et al., 2013; Hampel et al., 2015; Randall et al., 2017; Committee on Gynecologic Practice, 2018; Sturm et al., 2018; Daly et al., 2021).
Despite mounting evidence on the utility of cascade testing, uptake rates among at-risk relatives remain low overall, though vary across clinical settings (Cernat et al., 2021). Most studies focusing on genes associated with HCS report cascade testing uptake rates between 30 and 60% (Fehniger et al., 2013; Menko et al., 2019; Lee et al., 2021). Uptake rates have been much lower (4–12%) among families with FH in the United States (Ahmad et al., 2016; Gidding et al., 2020; Ajufo et al., 2021), but much higher (30% up to 90%) among families with FH in other Western countries (Marks et al., 2006; Ahmad et al., 2016; van den Heuvel et al., 2020). Limited data are available on cascade testing uptake for proactive or non-indication-based genetic screening, though results from the Electronic Medical Records and Genomics (eMERGE) phase III study demonstrated that only about one-third of probands who received non-indication-based screening reported sharing their test results with their relatives (Wynn et al., 2021). In the present study, we assessed differences in uptake of cascade testing between relatives of probands who received indication-based diagnostic genetic testing and relatives of probands who received proactive, non-indication-based screening for genes associated with HCS or FH.
Materials and Methods
Study Population and Design
Two retrospective cohorts of unrelated probands unselected for sex, self-reported ancestry, or age were compiled with individuals who underwent diagnostic germline genetic testing or proactive screening at Invitae from January 2017 through March 2021.
The diagnostic proband cohort included individuals who had clinician-ordered, indication-based testing via the Invitae Common Hereditary Cancers Panel (up to 47 genes) or the Invitae Familial Hypercholesterolemia Panel (up to 4 genes). Specific clinical criteria that led to clinician-ordered testing (e.g., the individual met guidelines from professional societies for testing) were unknown and thus individuals were unselected for test indication (i.e., personally affected versus family history).
The proactive proband cohort included individuals who were referred by clinicians for screening via the Invitae Cancer Screen (up to 61 genes), the Invitae Cardio Screen (up to 77 genes), or the Invitae Genetic Health Screen (up to 147 genes). Genes for inclusion in these panels were selected based on published guidance from the American College of Medical Genetics and Genomics (ACMG) and ClinGen Working groups, in addition to clinical studies establishing personal risk for monogenic disorders (Foreman et al., 2013; Green et al., 2013; Dewey et al., 2016; Webber et al., 2018). Probands undergoing non-indication-based screening have been described previously (Haverfield et al., 2021). In brief, all probands were included in the analysis, regardless of a personal or family history of cancer or cardiovascular disease. Individuals were excluded only if a familial variant associated with a condition on the screening panel had been previously identified.
In both cohorts, if a proband harbored at least one clinically significant variant (including carrier status), then the proband’s relatives were eligible for cascade testing for the identified variant(s). A clinically significant variant was defined as a pathogenic/likely pathogenic (P/LP) variant, a pathogenic-low penetrance (P[LP]) variant, or an increased risk allele (IRA). P(LP) variants are less penetrant compared to other P/LP variants in the same gene and may result in a less obvious Mendelian pattern of inheritance (e.g., HFE p.Cys282Tyr or p.His63Asp). IRAs are variants in genes that increase the risk for a condition and have stringent criteria (Ioannidis et al., 2008), but are not associated with a Mendelian inheritance pattern (e.g., APC p.Ile1307Lys). Testing was offered at no charge to the relatives for up to 90 days following the proband’s test report date, though the cascade testing window was extended to 150 days after March 30, 2020, due to the COVID-19 pandemic. All blood relatives were eligible for cascade testing, and those who received testing from January 2017 through August 2021 were included in the analysis as long as they were tested for at least one gene in which the proband had a clinically significant variant. Relatives who were tested for the purposes of reclassifying variants of uncertain significance (VUS) in probands within the diagnostic cohort were excluded from the analysis.
Review and analysis of de-identified and aggregated data were approved for waiver of authorization by the WCG Institutional Review Board (study number 1167406).
Genetic Testing
Requested genes were sequenced via a short-read next-generation sequencing (NGS) assay that used genomic DNA extracted from blood or saliva samples as reported previously (Lincoln et al., 2015; Haverfield et al., 2021). A bioinformatics pipeline aligned sequencing reads and utilized community standard and custom algorithms to identify single nucleotide variants (SNVs), small and large insertions or deletions (indels), structural variants, and exon-level copy-number variants (CNVs) (Lincoln et al., 2015, 2021; Truty et al., 2019).
Detected variants were analyzed and interpreted using Sherloc (Nykamp et al., 2017), a points-based framework that incorporates the joint consensus guidelines from the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (Richards et al., 2015). Based on the evidence, variants were classified as benign or likely benign (B/LB), VUS, P/LP, IRA, or P(LP). Clinically significant P/LP, IRA, and P(LP) variants that did not meet stringent NGS quality metrics were confirmed by an orthogonal assay prior to reporting (Lincoln et al., 2019). For individuals who underwent diagnostic testing, variants classified as P/LP, IRA, P(LP), and VUS were reported. For individuals who underwent proactive screening, only P/LP, IRA, and P(LP) findings were reported, as VUS are not reported as part of proactive screening (Haverfield et al., 2021). All results were returned to the ordering healthcare provider, who then oversaw results disclosure to the individual who underwent diagnostic testing or proactive screening.
Individuals were considered to have “positive” findings with medically actionable results if one clinically significant variant was found in a gene associated with an autosomal dominant disorder or two clinically significant variants were found in a gene associated with an autosomal recessive disorder. In addition, male individuals with one clinically significant variant in any gene associated with an X-linked disorder were considered to have positive findings. Female individuals with one clinically significant variant in a gene associated with an X-linked dominant disorder or two clinically significant variants in a gene associated with an X-linked recessive disorder were considered to have positive findings. A carrier finding was classified as one clinically significant variant in a gene associated with an autosomal recessive disorder in any individual or one clinically significant variant in a gene associated with an X-linked recessive disorder in female individuals. Though all results were disclosed to the ordering clinician first, individuals, regardless of result (e.g., no clinically significant result, medically actionable result), could seek post-test genetic counseling through Invitae, though this was not required.
Analysis
Medically Actionable (Positive) and Clinically Significant (Carrier) Findings in Probands
The proportion of probands with positive and carrier findings were calculated for the diagnostic and proactive cohorts. The three primary comparisons were based on genes that were analyzed in both cohorts (i.e., CDC Tier 1 conditions, HCS, and FH genes). Demographics of each of these groups were also summarized.
Eleven genes associated with CDC Tier 1 conditions (APOB, BRCA1, BRCA2, EPCAM, LDLR, LDLRAP1, MLH1, MSH2, MSH6, PCSK9, and PMS2) were analyzed in all probands (regardless of panel type). Forty-five HCS genes available to both cohorts were analyzed among patients who underwent diagnostic testing or proactive screening for HCS genes (Supplementary Table S1). Similarly, four FH genes available to both cohorts were analyzed among individuals who underwent testing or screening for FH. Proactive probands who received screening through the Invitae Genetic Health Screen were included in both the HCS and FH cohorts, as this panel included genes across both clinical areas. Diagnostic probands who had both the Invitae Common Hereditary Cancers Panel and the Invitae Familial Hypercholesterolemia Panel ordered were also included in both cohorts.
Additional genes were also analyzed if ordered for probands in either cohort. Diagnostic probands who had the Invitae Common Hereditary Cancers Panel had CTNNA1 and RAD50 analyzed. Proactive probands who had the Invitae Cardio Screen or the Genetic Health Screen had up to an additional 72 genes associated with other cardiology-related conditions or up to 16 genes associated with other HCS analyzed. The Invitae Genetic Health Screen also included 10 genes associated with other hereditary diseases (e.g., hereditary hemochromatosis [HAMP, HFE, HJV, SLC40A1 and TFR2] and malignant hyperthermia susceptibility [CACNA1S and RYR1]) that were analyzed in proactive probands only.
Cascade Testing
Among probands with a medically actionable or clinically significant finding, the proportion who had at least one relative undergo cascade testing through Invitae was calculated (i.e., cascade testing rate). Cascade testing uptake rates were compared between the diagnostic and proactive cohorts by calculating the difference in proportion for two independent samples. In addition, the number of relatives tested per proband was analyzed.
Among relatives, demographic characteristics were calculated and stratified according to the proband’s result type (e.g., medically actionable result in a shared HCS gene). Concordance of findings between the relative and proband was assessed.
Demographic and Clinical Factors Associated With Cascade Testing Utilization
We also assessed whether any demographic or clinical characteristics of probands influenced the rate of cascade testing among relatives. In both diagnostic and proactive cohorts, probands with medically actionable findings and with relatives who had undergone cascade testing were compared with probands who had a medically actionable finding but did not have relatives who had undergone cascade testing. These two groups were compared based on the following factors: age at time of testing, sex, self-reported ethincity, and whether the proband had a post-test genetic counseling session provided through Invitae. Two additional comparisons were made for probands who underwent diagnostic testing or proactive screening for HCS: whether the gene was associated with a CDC Tier 1 condition (diagnostic and proactive cohorts) and reported personal history of cancer (diagnostic cohort only). Differences in categorical data were assessed by comparing proportions for two independent samples; differences in age were assessed using 2-sample, 2-tailed t-tests. No comparisons were made for the proactive FH cohort due to small sample sizes.
Results
Proband Characteristics
A total of 270,715 probands were eligible for inclusion in the study: 254,281 (93.9%) who received indication-based diagnostic testing and 16,434 (6.1%) who received non-indication-based proactive screening (Supplementary Table S2). Diagnostic testing or proactive screening for HCS genes was completed for 247,875 diagnostic probands and 15,984 proactive probands. Diagnostic testing or proactive screening for FH was completed for 6,503 diagnostic probands and 10,776 proactive probands. Of note, 97 diagnostic probands (0.04%) and 10,326 proactive probands (62.8%) had both HCS and FH genes analyzed.
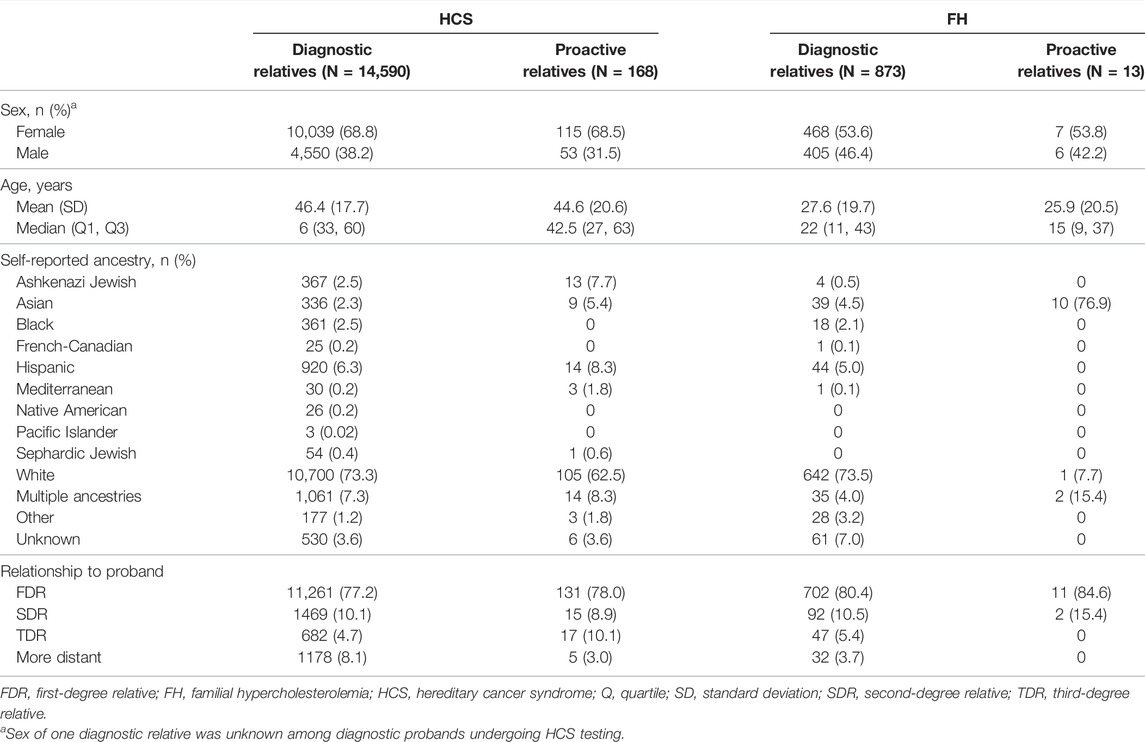
Demographic information for both cohorts based on clinical area is reported in Table 1. Diagnostic probands undergoing genetic testing for HCS were mostly female (87.5%) with a mean age of 55.5 ± 14.5 years. Proactive probands with HCS genes included in the genetic screen were also mostly female (58.0%), with a mean age of 48.4 ± 13.2 years. In both diagnostic and proactive cohorts undergoing testing or screening for FH, approximately half of the probands were female (56.5 and 49.1%, respectively), and the mean ages were 45.0 ± 20.4 years and 48.1 ± 13.0 years, respectively.
TABLE 1. Demographic information of probands by clinical areaa.
Positive Findings and Cascade Testing Rates in Genes Associated With CDC Tier 1 Conditions
A positive result in a gene associated with a CDC Tier 1 condition was identified in 10,520 (4.1%) and 337 (2.1%) of the diagnostic and proactive probands, respectively (Figure 1A). The proportion of patients with positive findings varied by gene (Figure 1B). Significantly more diagnostic probands than proactive probands with a positive finding in a gene associated with a CDC Tier 1 condition had at least one relative pursue cascade testing (diagnostic n = 3,305, 31.4%; proactive n = 36, 10.7%; p = 4.76×10–16; Figure 1A). Compared to proactive probands, a higher proportion of diagnostic probands with a medically actionable finding in each gene had at least one relative pursue cascade testing, ranging from 9.1 to 48.3% (vs. 2.4–28.6% among proactive probands (Figure 1B).
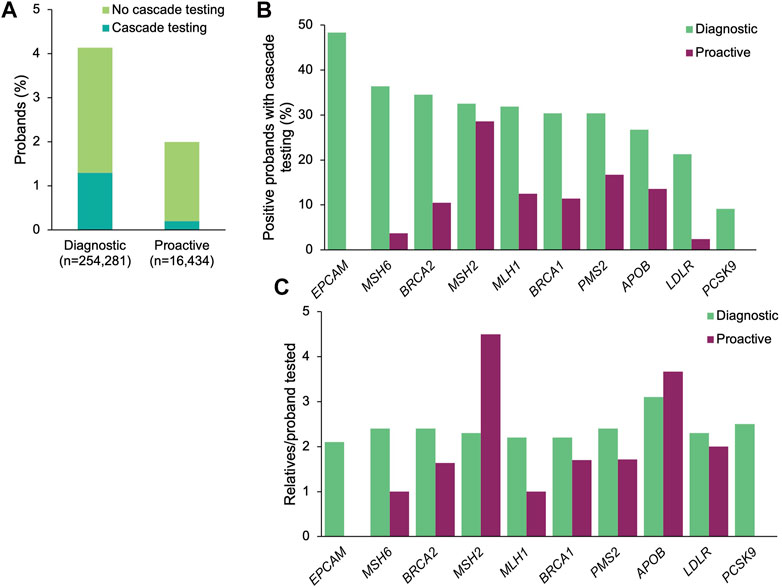
FIGURE 1. Yield of medically actionable findings in probands among the 11 genes associated with a CDC Tier 1 condition and rates of cascade testing. (A) Proportion of diagnostic and proactive probands with a positive result in a gene associated with a CDC Tier 1 condition, stratified by whether cascade testing was pursued. The denominator was the total number of probands that underwent diagnostic testing (n = 254,281) or proactive screening (n = 16,434). (B) Proportion of probands with a medically actionable result in a gene associated with a CDC Tier 1 condition with at least one relative who pursued cascade testing. The denominator was the number of probands with a medically actionable result in each gene associated with a CDC Tier 1 condition for each cohort. Probands with a positive finding in more than one gene associated with a CDC Tier 1 condition were included in calculations for each gene. (C) Mean number of relatives who pursued cascade testing per proband with a positive result in a gene associated with a CDC Tier 1 condition. If cascade testing was pursued for positive findings in more than one gene detected in the proband, the relatives and probands were included in the calculations for each gene. CDC, Centers for Disease Control and Prevention.
A total of 7,750 relatives of diagnostic probands (2.3 relatives/proband) and 71 relatives of proactive probands (2.0 relatives/proband) underwent cascade testing. The majority of relatives in both cohorts were first-degree relatives (diagnostic 76.1%, n = 5,896; proactive 73.2%, n = 52), with the remaining being second-degree (10.8%, n = 838; 12.7%, n = 9), third-degree (5.5%, n = 423; 12.7%, n = 9), and more distant relatives (7.7%, n = 593; 1.4%, n = 1). Genes with the most relatives per family tested were APOB (3.1 relatives/proband) and PCSK9 (2.5 relatives/proband) in the diagnostic cohort and in MSH2 (4.5 relatives/proband) and APOB (3.7 relatives/proband) in the proactive cohort (Figure 1C).
HCS Panels: Proband Results and Cascade Testing Outcomes
Among the 45 shared HCS genes, a positive result was returned to 23,272 (9.4%) of the diagnostic probands and 970 (6.1%) of the proactive probands (Figure 2A). The most common positive findings among diagnostic probands were in CHEK2 (18.6% of positive findings), BRCA2 (15.3%), BRCA1 (11.7%), ATM (9.7%), and APC (7.3%) (Figure 2B). The most common positive findings among proactive probands were in CHEK2 (24.0%), APC (12.6%), BRCA2 (10.8%), ATM (10.6%), and BRCA1 (9.1%). The frequencies of positive findings across all HCS genes are listed in Supplementary Table S3.
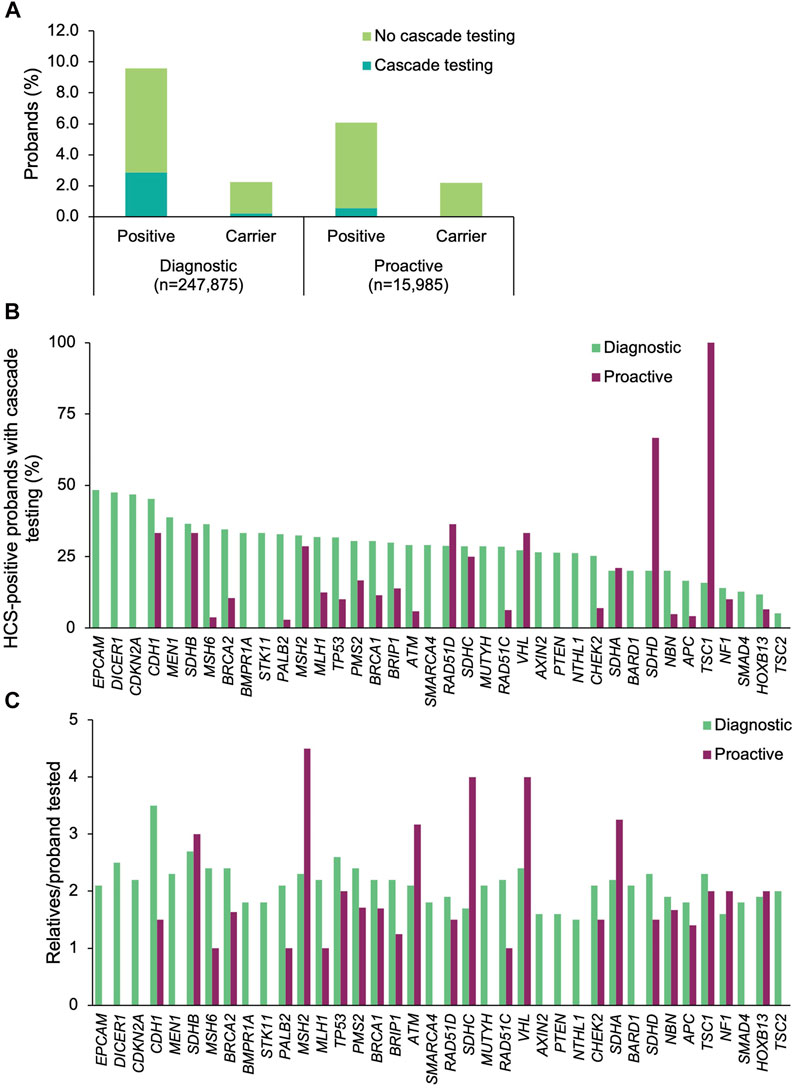
FIGURE 2. Yield of medically actionable (positive) and clinically significant (carrier) findings in probands who underwent diagnostic testing or proactive screening for HCS genes and rates of cascade testing. (A) Proportion of diagnostic and proactive probands with a positive result in an HCS gene, stratified by whether cascade testing was pursued. The denominator was the total number of probands who underwent diagnostic testing (n = 247,875) or proactive screening (n = 15,985) for HCS genes. (B) Proportion of probands with a medically actionable result in each HCS gene common to both diagnostic and proactive panels of interest who had at least one relative undergo cascade testing. The denominator was the number of probands with a positive result in each HCS gene for each cohort. Probands with a positive finding in more than one HCS gene were included in calculations for each gene. Thirty-eight of the 45 shared HCS genes are shown. Data for the remaining seven genes can be found in Supplementary Table S3. (C) Mean number of relatives who pursued cascade testing per proband with a positive result in an HCS gene. If cascade testing was pursued for positive findings in more than one gene detected in the proband, the relatives and probands were included in the calculations for each gene. Data are shown for 38 genes; data for the remaining seven genes can be found in Supplementary Table S3. HCS, hereditary cancer syndrome.
Cascade testing was pursued significantly more often when a positive finding in an HCS gene was returned for diagnostic probands than when it was returned for proactive probands (diagnostic n = 6,611, 28.4%; proactive n = 89, 9.2%; p = 1.01×10–43) (Figure 2A). In general, diagnostic probands were more likely to have at least one relative pursue cascade testing across all HCS genes compared to proactive probands (Figure 2B, Supplementary Table S3). However, cascade testing rates were similar for MSH2, SDHA, RAD51D, NF1, CDH1, SDHB, SDHC, and VHL. A higher proportion of proactive probands with a medically actionable finding in SDHD and TSC1 had at least one relative undergo cascade testing compared to diagnostic probands, but this difference is likely due to the absolute number of probands in each group that had a medically actionable finding in those genes.
A total of 14,590 relatives of diagnostic probands (2.0 relatives/proband) and 168 relatives of proactive probands (1.9 relatives/proband) were tested. Multigene panel testing was ordered for a minority of relatives (diagnostic n = 3,731, 25.6%; proactive n = 29, 17.3%), with the remainder having testing limited to genes with clinically significant and/or medically actionable findings in the proband. Most were first-degree relatives (diagnostic 77.2%, n = 11,261; proactive 78.0%, n = 131), with the remaining being second-degree (10.1%, n = 1469; 8.3%, n = 14), third-degree (4.7%, n = 682; 10.1%, n = 17), or more distant relatives (8.1%, n = 1,178; 3.0%, n = 5). The number of relatives per proband that underwent cascade testing was highest for CDH1 (3.5 relatives/proband), SDHB (2.7 relatives/proband), and TP53 (2.6 relatives/proband) in the diagnostic cohort and for MSH2 (4.5 relatives/proband), SDHC (4.0 relatives/proband), and VHL (4 relatives/proband) in the proactive cohort (Figure 2C).
Relatives in both cohorts were mostly female (diagnostic 68.8%, proactive 68.5%) and self-reported White (diagnostic 73.3%, proactive 62.5%), with a similar mean age at testing (diagnostic 46.4 ± 17.7 years, proactive 44.6 ± 20.6 years) (Table 2). A total of 6,422 (44.0%) and 69 (41.1%) of the relatives of diagnostic and proactive probands, respectively, had at least one clinically significant finding that was consistent with the positive finding in the proband. Additional findings were found in 282 relatives of diagnostic probands, 262 of whom had multigene panel testing. In total, 205 relatives had a finding in another gene on the Common Hereditary Cancers Panel, 45 had a different clinically significant finding in the same gene as the proband’s clinically significant finding, and 32 had a clinically significant finding in a gene that was not analyzed in the proband. One (3.4%) relative of a proactive proband who pursued testing as a result of a positive finding in a shared HCS gene had a positive finding in another gene.
To understand which factors may increase the likelihood of cascade testing, the differences in cascade testing uptake rates among probands with a positive result were compared based on demographics (Supplementary Table S4), whether genetic counseling services through Invitae were utilized, whether the finding was in a gene associated with a CDC Tier 1 condition, and whether a personal history of cancer was reported (diagnostic probands only). No differences in genetic counseling utilization were observed for either cohort (Figure 3A). Cascade testing was more commonly pursued among proactive probands who were female (11.1 vs. 6.8%, p = 0.019), (Figure 3B). It was also pursued more frequently among diagnostic probands who were White (32.3 vs. 21.6%, p = 2.49×10–69) (Figure 3C), had a gene finding associated with a CDC Tier 1 condition (33.5 vs. 25.3%, p = 5.91×10–41) (Figure 3D), or had a personal history of cancer (32.7 vs. 18.6%, p = 4.52×10–108) (Figure 3E).
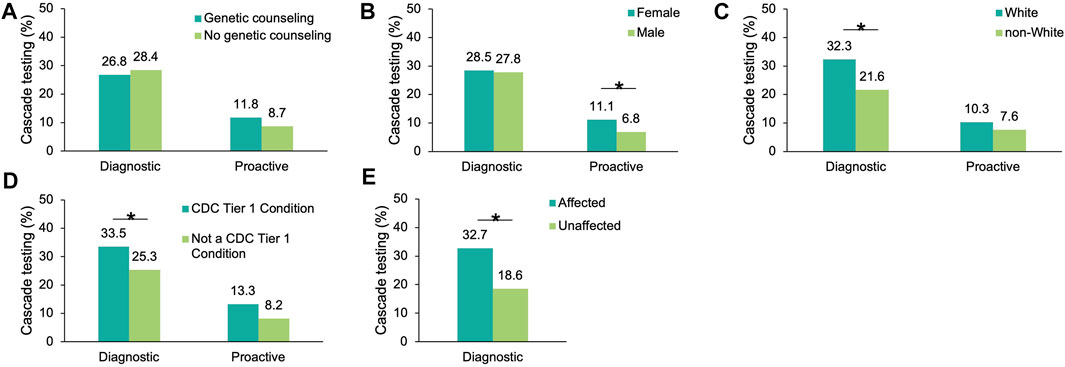
FIGURE 3. Factors influencing cascade testing among probands with a positive result in an HCS gene. Cascade testing rates were calculated for probands with a positive finding in an HCS gene and stratified by proband characteristic: (A) Whether the proband had genetic counseling through Invitae, (B) Sex (C) Self-reported ancestry, (D) Whether the positive finding in the proband was in a gene associated with a CDC Tier 1 condition, and (E) Whether diagnostic probands reported a family or personal history of cancer (this information was not available for proactive probands). HCS, hereditary cancer syndrome.
A small proportion of diagnostic (n = 5,559, 2.2%) and proactive (n = 350, 2.2%) probands had carrier results returned (in the genes included in this analysis) (Figure 2A). At least one relative of 9.4% (n = 524) and 1.7% (n = 6) of the diagnostic and proactive probands, respectively, had cascade testing performed as a result of carrier findings.
FH Panels: Proband Results and Cascade Testing Outcomes
A positive result in at least one of the four FH genes was returned to 1,647 (25.3%) of the diagnostic probands and 67 (0.62%) of the proactive probands (Figure 4A). The most common positive findings among diagnostic probands were in LDLR (86.4%), APOB (12.3%), PCSK9 (1.3%), and LDLRAP1 (0.3%). The most common positive findings among proactive probands were in LDLR (62.7%), APOB (32.8%), PCSK9 (4.5%). No proactive probands had a positive finding in LDLRAP1, though four probands were carriers (see below).
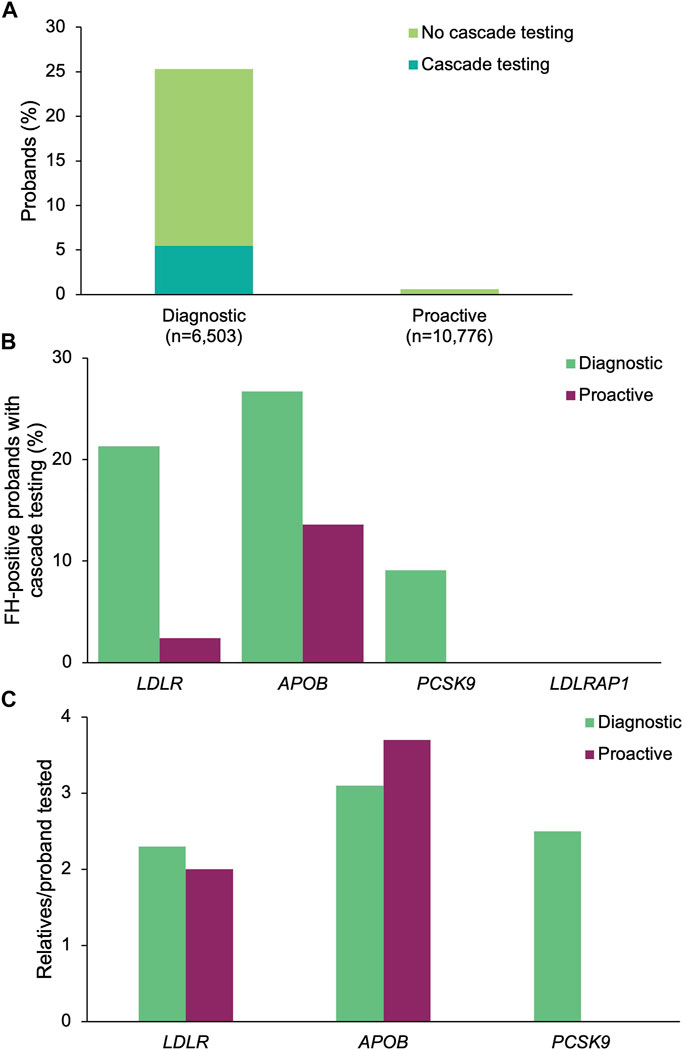
FIGURE 4. Yield of medically actionable (positive) findings in probands who underwent diagnostic testing or proactive screening for FH genes and rates of cascade testing. (A) Proportion of diagnostic and proactive probands with a positive result in an FH gene, stratified by whether cascade testing was pursued. The denominator was the total number of probands who underwent diagnostic testing (n = 6,503) or proactive screening (n = 10,776) for FH genes. (B) Proportion of probands with a positive result in each FH gene common to both diagnostic and proactive panels of interest who had at least one relative pursue cascade testing. The denominator was the number of probands with a positive result in each FH gene for each cohort. Probands with a positive finding in more than one FH gene were included in calculations for each gene. (C) Mean number of relatives who pursued cascade testing per proband with a positive result in an FH gene. If cascade testing was pursued for positive findings in more than one gene detected in the proband, the relatives and probands were included in the calculations for each gene. FH, familial hypercholesterolemia.
A positive finding in an FH gene in 360 (21.9%) of the diagnostic probands and 4 (6.0%) of the proactive probands led to cascade testing in at least one relative (p = 0.00183) (Figure 4A). Cascade testing was pursued in relatives of diagnostic probands with positive findings in LDLR (n = 304, 21.3%), APOB (n = 54, 26.7%), and PCSK9 (n = 2, 9.1%) (Figure 4B). Proactive probands with positive findings in APOB (n = 3, 13.6%) and LDLR (n = 1, 2.4%) led to cascade testing. A total of 873 relatives of diagnostic probands (2.4 relatives/proband) and 13 relatives of proactive probands (3.3 relatives/proband) were tested, of whom 37 (4.2%) diagnostic relatives and 2 (15.4%) proactive relatives had multigene panels ordered. The remainder had testing limited to genes with clinically significant and/or medically actionable findings in the proband. Relatives who underwent cascade testing were mostly first-degree relatives (diagnostic 80.4%, n = 702; proactive 84.6%, n = 11), with the remaining reported to be second-degree (10.5%, n = 92; 15.4%, n = 2), third-degree (5.4%, n = 47; n = 0), or more distant related (3.7%, n = 32; n = 0). The number of relatives per proband who underwent cascade testing was highest for APOB (3.1 relatives/proband), PCSK9 (2.5 relatives/proband), and LDLR (2.3 relatives/proband) in the diagnostic cohort and for APOB (3.7 relatives/proband) and LDLR (2.0 relatives/proband) in the proactive cohort (Figure 4C). Demographic characteristics of relatives in both cohorts were similar (Table 2). A total of 496 (56.8%) and 4 (30.8%) relatives of diagnostic or proactive probands, respectively, had a positive finding, all of which were consistent with the positive finding in the proband. No relatives of diagnostic or proactive probands who pursued testing as a result of a medically actionable finding in a shared FH gene had a positive finding in another gene. Two relatives of diagnostic probands were carriers for ABCG8 and one relative of a proactive proband who had a positive finding in APOB was also identified as a carrier for RYR1. Two (5.4%) diagnostic relatives and zero proactive relatives who had multigene panel testing had a clinically significant finding returned outside of the proband’s diagnostic testing or proactive screening results.
To understand which factors may increase the likelihood of cascade testing, the differences in cascade testing uptake rates among probands with a positive result were compared based on demographics (Supplementary Table S5) and whether post-test genetic counseling services through Invitae were utilized. Cascade testing was more commonly pursued in proactive probands who had genetic counseling (11.8 vs. 8.7%, Figure 5A), were male (7.7 vs. 3.6%, Figure 5B), or non-White (10.1 vs. 8.0%, Figure 5C). Results in the proactive cohort should be interpreted with caution as the sample size of proactive probands with cascade testing was small (n = 4). Cascade testing was pursued more frequently among diagnostic probands who were younger at time of testing (29.4 ± 19.8 years vs. 38.6 ± 22.7 years, p = 1.28 × 10–13) or self-reported White (28.3 vs. 15.2%, p = 1.36 × 10–10) (Figure 5C).
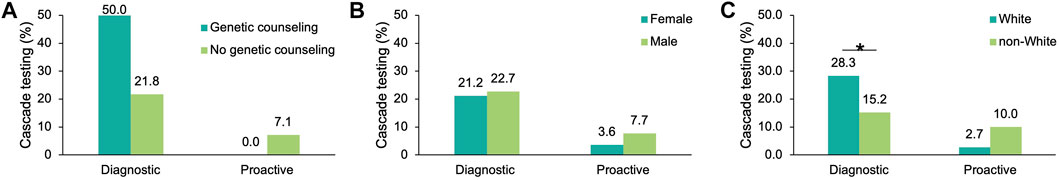
FIGURE 5. Factors influencing cascade testing rates among probands with a positive result in an FH gene. Cascade testing rates were calculated for probands with a positive finding in an FH gene and stratified by characteristic: (A) Whether the proband had genetic counseling through Invitae, (B) Sex, and (C) Self-reported ancestry. FH, familial hypercholesterolemia.
A small proportion of diagnostic (n = 3, 0.05%) and proactive (n = 4, 0.04%) probands had carrier results returned. No relatives pursued cascade testing as a result of these carrier findings in either cohort.
Findings in Additional Genes Unique to the Diagnostic and Proactive Panels
In addition to the 49 genes that were available on both diagnostic and proactive gene panels of interest, an additional two genes (RAD50 and CTNNA1) were available only on diagnostic panels. A small number (626, 0.3%) of diagnostic probands who had testing via the Invitae Common Hereditary Cancers Panel had a positive result in RAD50 (no probands had a positive result in CTNNA1), 108 (17.3%) of whom had 205 relatives (1.9 relatives/proband) pursue cascade testing.
An additional 98 genes were available only on the proactive panels, including genes associated with HCS (n = 16), other non-FH cardiology conditions (n = 73), or other conditions (n = 10) (Supplementary Table S1). The proportion of positive results in one of these genes ranged from 0.6% in HCS genes to 9.2% in cardiology genes (Supplementary Figure S1). Cascade testing was most commonly pursued for positive findings in an HCS gene (HCS 10.8%, cardiology 2.4%, other clinical areas 1.6%).
Discussion
In addition to the potential utility of genetic testing results to inform an individual’s clinical care and outcomes, a positive result has implications for that individual’s family. Studies assessing the uptake of cascade testing among relatives in a diagnostic setting have consistently demonstrated that rates are generally low, though they vary based on clinical area and focus only on just a few genes (Fehniger et al., 2013; Menko et al., 2019; Ajufo et al., 2021; Lee et al., 2021). As population-based and proactive screening methods begin to become more widespread, it is critical to understand how these testing approaches may impact at-risk relatives. Currently, the utilization of cascade testing in a non-indication-based, proactive setting is less well understood and uptake rates have not yet been reported. This study compared findings between two cohorts that differed in how NGS was pursued: indication-based diagnostic testing versus non-indication-based proactive screening. The findings reported allow not only for insights into differences between diagnostic and proactive results, but also more generally to ordering patterns for diagnostic testing or proactive screening for HCS and FH. Interestingly, we also gain tangential and preliminary insights into the potential benefits of multigene panel testing in at-risk relatives. This study demonstrates that there is an even larger gap in the uptake of cascade testing in a proactive versus diagnostic setting and highlights the need for further research to understand both the reasons for underutilization of cascade testing and the approaches that could lead to increased uptake rates.
In this study, we find that cascade testing rates were significantly higher among diagnostic probands compared to proactive probands across all comparisons, including testing or screening for any CDC Tier 1 condition, for HCS, and for FH. The findings from this study are the first to begin to investigate which factors may be associated with cascade testing utilization in a proactive setting. Proband characteristics shown to be associated with cascade testing in a diagnostic setting were consistent with our cohort, including self-reported ancestry, sex, and a personal history of disease (Dugan et al., 2003; Hamilton et al., 2005; Gaff et al., 2007; Sharaf et al., 2013; Roberts et al., 2018; Caswell-Jin et al., 2019; Menko et al., 2019; Braley et al., 2021). The only factor that resulted in a significant difference in cascade testing rates in the proactive cohort was sex, with rates higher among female probands. These preliminary findings demonstrate that there may be different factors that influence the utilization of cascade testing depending on the method of testing in the index case. Further prospective studies exploring a wider variety of proband and relative characteristics in relation to cascade testing rates will be critical to developing tools for encouraging and facilitating cascade testing that are tailored to various testing methods (i.e., diagnostic versus proactive).
Two large hurdles must be overcome in order for cascade testing to be pursued; first, the proband must share results with at-risk relatives and second, the relative must make the choice to seek genetic testing. It has been established that results sharing is poor regardless of whether diagnostic testing or proactive screening is ordered (Dugan et al., 2003; Hamilton et al., 2005; Gaff et al., 2007; Elrick et al., 2017; Wurtmann et al., 2018). Reasons for a lack of cascade testing utilization is limited to a diagnostic setting, with no research yet focusing on potential barriers for proband testing in a proactive setting. However, it is likely some of the reasons are similar for both approaches. Diagnostic probands have cited a perceived lack of clinician support and familial relationships as barriers (Dugan et al., 2003; Chivers Seymour et al., 2010; Muir et al., 2012; Hardcastle et al., 2015; Pollard et al., 2020; Srinivasan et al., 2020). Among the limited pool of at-risk relatives who do have results shared with them, only a small proportion end up seeking cascade testing. Previous research has shown that relatives of diagnostic probands do not seek testing because of several perceived hurdles, including cost, the need to make an appointment with a clinician, and concerns about insurance or employment discrimination, even with current legislation barring such discrimination (EEOC, 2008; Hendricks-Sturrup et al., 2019; Srinivasan et al., 2020).
Approaches to encouraging and facilitating cascade testing have been largely limited to a diagnostic setting. However, ongoing studies, such as the IMPACT-FH (Identification Methods, Patient Activation, and Cascade Testing for FH) study (Campbell-Salome et al., 2021), are exploring strategies that increase the uptake of cascade testing in population-based screening programs. However, learnings from the diagnostic setting may provide some insights, including the availability of clinician-drafted letters (Newson and Humphries, 2005; Suthers et al., 2006; Hadfield et al., 2009; Dilzell et al., 2014; Petersen et al., 2019; Kurian and Katz, 2020; Neuner et al., 2020), access to support from foundations focused on a single condition or clinical area (Bell et al., 2015; Wald et al., 2016; McGowan et al., 2021), and access to educational materials that are easily shared outside of a clinical setting (Kardashian et al., 2012; Petersen et al., 2019; Bowen et al., 2020; Jujjavarapu et al., 2021; Nazareth et al., 2021; Nitecki et al., 2021; Snir et al., 2021). When considering approaches to encouraging at-risk relatives to ultimately seek cascade testing, programs have been designed to offer cascade testing at reduced rates or at no-charge for relatives (Aktan-Collan et al., 2007; Caswell-Jin et al., 2019; Courtney et al., 2019; Invitae, 2021). Preliminary findings in a recent study demonstrated that chatbots are an effective means to facilitating cascade testing (data in press Schmidlen et al., 2022). Regardless of the approach, it is critical that methods used to encourage both results sharing and subsequent cascade testing are accessible to diverse populations (Milo Rasouly et al., 2021).
The sum of these observations demonstrates that there is not likely a one size fits all approach to encouraging cascade testing, and that having several avenues available for both facilitating results sharing and streamlining testing processes will maximize the success of cascade testing initiatives. Especially for probands who are identified in non-indication-based settings, additional efforts to educate probands, as well as tools to help them share information with relatives, will be essential as genetic screening in healthy individuals becomes more widespread. For example, novel approaches utilizing chatbots may not only improve communication with probands but also facilitate results sharing and subsequently help connect relatives to a clinician for cascade testing. This is especially important as genetic counseling may not be sought prior to or after screening in the proband.
In addition to insights into differences between diagnostic and proactive cohorts, ordering behaviors among probands seeking testing or screening for HCS and FH were very different. Strikingly, the absolute number of probands who were tested for HCS and FH panels was very different for both the diagnostic and proactive cohorts. It is possible that this is due to the increased awareness of and testing for hereditary breast and ovarian cancer and Lynch syndrome compared to that of FH. While we observe higher cascade testing uptake for HCS, the number of relatives tested per proband is higher for FH compared to HCS (∼3 relatives/proband vs. 2 relatives/proband). We suspect that this is because genetics specialists are providing care to probands being tested for FH in collaboration with the treating clinician (Ingles et al., 2020; Musunuru et al., 2020), while oncologists may have an increased experience and comfort with ordering genetic testing themselves (Hamilton et al., 2021). So while more probands could be referred for HCS testing by a non-genetics specialist, probands tested for FH may more likely be receiving counseling from genetic counselors and as a result, may have higher numbers of relatives tested once results are shared.
Finally, though limited to a minority of relatives, anywhere from ∼5 to ∼25% of relatives have additional genes (often multigene panel testing) ordered. Among relatives who had probands undergo diagnostic testing or proactive screening for HCS, 7.0% of relatives of diagnostic probands and 3.4% of relatives of proactive probands had a clinically significant finding outside of the proband’s findings. This finding demonstrates that panel testing does in fact identify additional risks that would have otherwise been missed had gene-specific testing been ordered. Reasons for missing these clinically significant findings could be a result of, among others, the proband not being tested for that gene or that the relative has a family history associated with another relative unrelated to the proband. Relatives who received a negative result following targeted testing based on proband results could have a false sense of reassurance without understanding that they could have medically actionable variants in other genes. This may be the case even though clinicians take into account an individual’s full family history and genetic counseling based on a negative result centers around residual risk. While there may be higher costs related to testing for additional genes, these results underscore the possible benefits of considering broader testing for relatives seeking cascade testing.
Similar to other retrospective cohort studies, this study was limited in the data available for analysis. Our analysis compared two cohorts based on how probands were referred: indication-based diagnostic testing or non-indication-based proactive screening. As a commercial testing laboratory, orders are received from clinicians requesting diagnostic testing as well as individuals seeking proactive screening. The majority of individuals in these cohorts indicated a self-reported White ancestry, which may have biased the results. The socioeconomic factors demonstrated to impact the utilization of genetic testing were not controlled for in this study (Gómez-Trillos et al., 2020; McKinney et al., 2020; Giri et al., 2021). While not a focus in this study, Invitae has sponsored testing programs that eliminate potential financial barriers to diagnostic genetic testing for a number of clinical indications, in addition to research initiatives to help facilitate population screening and cascade testing across more diverse groups (staff reporter, 2021). Novel approaches to improving genetics literacy and awareness across diverse populations have proven to be successful (Milo Rasouly et al., 2021). Among diagnostic probands, the specific reason for testing could not be determined because the test requisition form did not require disclosure of whether the individual for whom testing was ordered had a personal or family history of an HCS or FH. Thus, whether a proband had a personal or family history was unknown for many individuals and was not uniform when shared. However, for a number of hereditary cancer conditions and familial cardiac conditions, current guidelines recommend that family history alone, when meeting certain requirements, is a standalone indication for diagnostic genetic testing in an otherwise unaffected individual (e.g., family history of breast cancer in multiple first degree relatives) (Daly et al., 2021). For these logistic and clinical reasons, we could only assume that diagnostic testing was warranted based on the ordering clinician’s evaluation of the individual. Another limitation is that, as the testing laboratory, the total number of relatives that were offered cascade testing could not be determined. As such, the cascade testing uptake rate is based purely on those individuals tested through Invitae with reported relationships disclosed at the time of test requisition. The number of probands with clinically significant results who shared their results with relatives and the number of relatives who ultimately sought testing could not be determined. However, as reported from other studies, it is clear that results sharing and subsequent testing rates are generally low. Further, it is unknown how many probands sought genetic counseling outside of Invitae. It is expected that most diagnostic probands, but far fewer proactive probands, had received counseling through a clinician or an adjacent clinical service (such as a medical geneticist or genetic counselor). However, this information was not well documented, so assessing the rate of cascade testing based on genetic counseling through Invitae may be an underestimate. Although limited, this study helps to establish preliminary findings that can help to guide future prospective studies.
The results of this study have demonstrated that cascade testing uptake is significantly lower among probands who seek testing in a non-indication-based, proactive setting than among those who are referred for indication-based testing. The barriers and facilitators of cascade testing seem to be similar between the two cohorts, suggesting that approaches that promote family testing in a diagnostic setting could be similarly applied to proactive settings. However, the tools and methods may need to be tailored to these different settings in order to increase cascade testing rates. Such an investigation is underway in individuals undergoing testing for FH as part of a population-based genomic research study (Campbell-Salome et al., 2021). The findings from the present study establish a baseline for future prospective studies designed to understand the reasons for results sharing (or not) among probands and the subsequent influences that encourage relatives to engage with cascade testing.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by WCG Institutional Review Board. Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author Contributions
TS, EE, RN, and EH contributed to the conception of the study. TS, KH, SB, and EH contributed to the methodology of the study. KH generated the study databases, ran formal statistical analyses, and validated the results. TS, SB, KH, and EH curated the databases. SB prepared the first draft of the manuscript and generated figures. EH provided supervision over manuscript development. TS oversaw project administration. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was funded by Invitae.
Conflict of Interest
TS, SB, KH, EE, RN, and EH are employed by and shareholders in the company Invitae.
The reviewer [PH] declared a past co-authorship with the authors [KH, EE, RN, and EH] to the handling editor.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.867226/full#supplementary-material
References
Ademi, Z., Watts, G. F., Pang, J., Sijbrands, E. J. G., van Bockxmeer, F. M., O'Leary, P., et al. (2014). Cascade Screening Based on Genetic Testing Is Cost-Effective: Evidence for the Implementation of Models of Care for Familial Hypercholesterolemia. J. of Clin. Lipidol. 8, 390–400. doi:10.1016/j.jacl.2014.05.008
Ahmad, Z. S., Andersen, R. L., Andersen, L. H., O'Brien, E. C., Kindt, I., Shrader, P., et al. (2016). US Physician Practices for Diagnosing Familial Hypercholesterolemia: Data from the CASCADE-FH Registry. J. of Clin. Lipidol. 10, 1223–1229. doi:10.1016/j.jacl.2016.07.011
Ajufo, E., deGoma, E. M., Raper, A., Yu, K. D., Cuchel, M., and Rader, D. J. (2021). A Randomized Controlled Trial of Genetic Testing and Cascade Screening in Familial Hypercholesterolemia. Genet. in Med. 23, 1697–1704. doi:10.1038/s41436-021-01192-z
Aktan-Collan, K., Haukkala, A., Pylvanainen, K., Jarvinen, H. J., Aaltonen, L. A., Peltomaki, P., et al. (2007). Direct Contact in Inviting High-Risk Members of Hereditary Colon Cancer Families to Genetic Counselling and DNA Testing. J. of Med. Genet. 44, 732–738. doi:10.1136/jmg.2007.051581
Bell, D. A., Pang, J., Burrows, S., Bates, T. R., van Bockxmeer, F. M., Hooper, A. J., et al. (2015). Effectiveness of Genetic Cascade Screening for Familial Hypercholesterolaemia Using a Centrally Co-Ordinated Clinical Service: An Australian Experience. Atherosclerosis 239, 93–100.
Bowen, D. J., Hyams, T., Laurino, M., Woolley, T., Cohen, S., Leppig, K. A., et al. (2020). Development of FamilyTalk: An Intervention to Support Communication and Educate Families About Colorectal Cancer Risk. J. Cancer Educ. 35, 470–478.
Braley, E. F., Bedard, A. C., Nuk, J., Hong, Q., Bedard, J. E. J., Sun, S., et al. (2021). Patient Ethnicity and Cascade Genetic Testing: a Descriptive Study of a Publicly Funded Hereditary Cancer Program. Fam. Cancer, 1–6. doi:10.1007/s10689-021-00270-0
Campbell-Salome, G., Jones, L. K., Masnick, M. F., Walton, N. A., Ahmed, C. D., Buchanan, A. H., et al. (2021). Developing and Optimizing Innovative Tools to Address Familial Hypercholesterolemia Underdiagnosis. Circulation Genomic and Precis. Med. 14, e003120. doi:10.1161/circgen.120.003120
Caswell-Jin, J. L., Zimmer, A. D., Stedden, W., Kingham, K. E., Zhou, A. Y., and Kurian, A. W. (2019). Cascade Genetic Testing of Relatives for Hereditary Cancer Risk: Results of an Online Initiative. J. Natl. Cancer Inst. 111, 95–98. doi:10.1093/jnci/djy147
Centers for Disease Control and Prevention (2021). Tier 1 Genomics Applications and Their Importance to Public Health. Available at: https://www.cdc.gov/genomics/implementation/toolkit/tier1.htm [Accessed December 8, 2021].
Cernat, A., Hayeems, R. Z., and Ungar, W. J. (2021). Cascade Health Service Use in Family Members Following Genetic Testing in Children: a Scoping Literature Review. Eur. J. Hum. Genet. 29, 1601–1610. doi:10.1038/s41431-021-00952-4
Chivers Seymour, K., Addington-Hall, J., Lucassen, A. M., and Foster, C. L. (2010). What Facilitates or Impedes Family Communication Following Genetic Testing for Cancer Risk? A Systematic Review and Meta-Synthesis of Primary Qualitative Research. J. Genet. Couns. 19, 330–342. doi:10.1007/s10897-010-9296-y
Committee on Gynecologic Practice (2018). ACOG Committee Opinion No. 727 Summary: Cascade Testing: Testing Women for Known Hereditary Genetic Mutations Associated with Cancer. Obstet. Gynecol. 131, 194–195. doi:10.1097/AOG.0000000000002451
Courtney, E., Chok, A. K.-L., Ting Ang, Z. L., Shaw, T., Li, S.-T., Yuen, J., et al. (2019). Impact of Free Cancer Predisposition Cascade Genetic Testing on Uptake in Singapore. npj Genom. Med. 4, 22. doi:10.1038/s41525-019-0096-5
Daly, M. B., Pal, T., Berry, M. P., Buys, S. S., Dickson, P., Domchek, S. M., et al. (2021). Genetic/familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 19, 77–102. doi:10.6004/jnccn.2021.0001
Dewey, F. E., Murray, M. F., Overton, J. D., Habegger, L., Leader, J. B., Fetterolf, S. N., et al. (2016). Distribution and Clinical Impact of Functional Variants in 50,726 Whole-Exome Sequences from the DiscovEHR Study. Science 354, aaf6814. doi:10.1126/science.aaf6814
Dilzell, K., Kingham, K., Ormond, K., and Ladabaum, U. (2014). Evaluating the Utilization of Educational Materials in Communicating about Lynch Syndrome to At-Risk Relatives. Fam. Cancer 13, 381–389. doi:10.1007/s10689-014-9720-9
Dugan, R. B., Wiesner, G. L., Juengst, E. T., O'Riordan, M., Matthews, A. L., and Robin, N. H. (2003). Duty to Warn At-Risk Relatives for Genetic Disease: Genetic Counselors' Clinical Experience. Am. J. Med. Genet. 119C, 27–34. doi:10.1002/ajmg.c.10005
EEOC(2008). The Genetic Information Nondiscrimination Act of 2008 Available at: https://www.eeoc.gov/statutes/genetic-information-nondiscrimination-act-2008 [Accessed December 13, 2021].
Elrick, A., Ashida, S., Ivanovich, J., Lyons, S., Biesecker, B. B., Goodman, M. S., et al. (2017). Psychosocial and Clinical Factors Associated with Family Communication of Cancer Genetic Test Results Among Women Diagnosed with Breast Cancer at a Young Age. J. Genet. Couns. 26, 173–181. doi:10.1007/s10897-016-9995-0
Fehniger, J., Lin, F., Beattie, M. S., Joseph, G., and Kaplan, C. (2013). Family Communication of BRCA1/2 Results and Family Uptake of BRCA1/2 Testing in a Diverse Population of BRCA1/2 Carriers. Jrnl of Gene Coun 22, 603–612. doi:10.1007/s10897-013-9592-4
Foreman, A. K. M., Lee, K., and Evans, J. P. (2013). The NCGENES Project: Exploring the New World of Genome Sequencing. North Carolina Med. J. 74, 500–504. doi:10.18043/ncm.74.6.500
Gaff, C. L., Clarke, A. J., Atkinson, P., Sivell, S., Elwyn, G., Iredale, R., et al. (2007). Process and Outcome in Communication of Genetic Information within Families: a Systematic Review. Eur. J. Hum. Genet. 15, 999–1011. doi:10.1038/sj.ejhg.5201883
Gidding, S. S., Sheldon, A., Neben, C. L., Williams, H. E., Law, S., Zhou, A. Y., et al. (2020). Patient Acceptance of Genetic Testing for Familial Hypercholesterolemia in the CASCADE FH Registry. J. of Clin. Lipidol. 14, 218–223. doi:10.1016/j.jacl.2020.02.001
Giri, V. N., Shimada, A., and Leader, A. E. (2021). Predictors of Population Awareness of Cancer Genetic Tests: Implications for Enhancing Equity in Engaging in Cancer Prevention and Precision Medicine. JCO Precis. Oncol. 5, 1699–1708. doi:10.1200/PO.21.00231
Gómez-Trillos, S., Sheppard, V. B., Graves, K. D., Song, M., Anderson, L., Ostrove, N., et al. (2020). Latinas' Knowledge of and Experiences with Genetic Cancer Risk Assessment: Barriers and Facilitators. J. Genet. Couns. 29, 505–517. doi:10.1002/jgc4.1201
Green, R. C., Berg, J. S., Grody, W. W., Kalia, S. S., Korf, B. R., Martin, C. L., et al. (2013). ACMG Recommendations for Reporting of Incidental Findings in Clinical Exome and Genome Sequencing. Genet. in Med. 15, 565–574. doi:10.1038/gim.2013.73
Grosse, S. (2015). When Is Genomic Testing Cost-Effective? Testing for Lynch Syndrome in Patients with Newly-Diagnosed Colorectal Cancer and Their Relatives. Healthcare 3, 860–878. doi:10.3390/healthcare3040860
Hadfield, S. G., Horara, S., Starr, B. J., Yazdgerdi, S., Marks, D., Bhatnagar, D., et al. (2009). Family Tracing to Identify Patients with Familial Hypercholesterolaemia: the Second Audit of the Department of Health Familial Hypercholesterolaemia Cascade Testing Project. Ann. Clin. Biochem. Biochem. 46, 24–32. doi:10.1258/acb.2008.008094
Hamilton, J. G., Symecko, H., Spielman, K., Breen, K., Mueller, R., Catchings, A., et al. (2021). Uptake and Acceptability of a Mainstreaming Model of Hereditary Cancer Multigene Panel Testing Among Patients with Ovarian, Pancreatic, and Prostate Cancer. Genet. in Med. 23, 2105–2113. doi:10.1038/s41436-021-01262-2
Hamilton, R. J., Bowers, B. J., and Williams, J. K. (2005). Disclosing Genetic Test Results to Family Members. J. Nurs. Scholarsh. 37, 18–24. doi:10.1111/j.1547-5069.2005.00007.x
Hampel, H., Bennett, R. L., Buchanan, A., Pearlman, R., and Wiesner, G. L. (2015). Guideline Development Group, American College of Medical Genetics and Genomics Professional Practice and Guidelines Committee and National Society of Genetic Counselors Practice Guidelines CommitteeA Practice Guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: Referral Indications for Cancer Predisposition Assessment. Genet. in Med. 17, 70–87. doi:10.1038/gim.2014.147
Hardcastle, S. J., Legge, E., Laundy, C. S., Egan, S. J., French, R., Watts, G. F., et al. (2015). Patients' Perceptions and Experiences of Familial Hypercholesterolemia, Cascade Genetic Screening and Treatment. Int.J. Behav. Med. 22, 92–100. doi:10.1007/s12529-014-9402-x
Haverfield, E. V., Esplin, E. D., Aguilar, S. J., Hatchell, K. E., Ormond, K. E., Hanson-Kahn, A., et al. (2021). Physician-directed Genetic Screening to Evaluate Personal Risk for Medically Actionable Disorders: a Large Multi-Center Cohort Study. BMC Med. 19, 199. doi:10.1186/s12916-021-01999-2
Hendricks-Sturrup, R. M., Mazor, K. M., Sturm, A. C., and Lu, C. Y. (2019). Barriers and Facilitators to Genetic Testing for Familial Hypercholesterolemia in the United States: A Review. Jpm 9, 32. doi:10.3390/jpm9030032
Ingles, J., Macciocca, I., Morales, A., and Thomson, K. (2020). Genetic Testing in Inherited Heart Diseases. Heart, Lung and Circulation 29, 505–511. doi:10.1016/j.hlc.2019.10.014
Invitae (2021). Flexible Follow-Up Testing. Available at: https://www.invitae.com/en/providers/follow-up-testing [Accessed May 9, 2022].
Ioannidis, J. P., Boffetta, P., Little, J., O'Brien, T. R., Uitterlinden, A. G., Vineis, P., et al. (2008). Assessment of Cumulative Evidence on Genetic Associations: Interim Guidelines. Int. J. of Epidemiol. 37, 120–132. doi:10.1093/ije/dym159
Jujjavarapu, C., Anandasakaran, J., Amendola, L. M., Haas, C., Zampino, E., Henrikson, N. B., et al. (2021). ShareDNA: A Smartphone App to Facilitate Family Communication of Genetic Results. BMC Med. Genomics 14, 10.
Kardashian, A., Fehniger, J., Creasman, J., Cheung, E., and Beattie, M. S. (2012). A Pilot Study of the Sharing Risk Information Tool (ShaRIT) for Families with Hereditary Breast and Ovarian Cancer Syndrome. Hered. Cancer Clin. Pract. 10, 4.
Kerr, M., Pears, R., Miedzybrodzka, Z., Haralambos, K., Cather, M., Watson, M., et al. (2017). Cost Effectiveness of Cascade Testing for Familial Hypercholesterolaemia, Based on Data from Familial Hypercholesterolaemia Services in the UK. Eur. Heart J. 38, 1832–1839. doi:10.1093/eurheartj/ehx111
Kurian, A. W., and Katz, S. J. (2020). Emerging Opportunity of Cascade Genetic Testing for Population-wide Cancer Prevention and Control. Jco 38, 1371–1374. doi:10.1200/jco.20.00140
Lee, D. S. C., Meiser, B., Mariapun, S., Hassan, T., Yip, C. H., Mohd Taib, N. A., et al. (2021). Communication about Positive BRCA1 and BRCA2 Genetic Test Results and Uptake of Testing in Relatives in a Diverse Asian Setting. Jrnl of Gene Coun 30, 720–729. doi:10.1002/jgc4.1360
Lincoln, S. E., Hambuch, T., Zook, J. M., Bristow, S. L., Hatchell, K., Truty, R., et al. (2021). One in Seven Pathogenic Variants Can Be Challenging to Detect by NGS: an Analysis of 450,000 Patients with Implications for Clinical Sensitivity and Genetic Test Implementation. Genet. in Med. 23, 1673–1680. doi:10.1038/s41436-021-01187-w
Lincoln, S. E., Kobayashi, Y., Anderson, M. J., Yang, S., Desmond, A. J., Mills, M. A., et al. (2015). A Systematic Comparison of Traditional and Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Genes in More Than 1000 Patients. The J. of Mol. Diagnostics 17, 533–544. doi:10.1016/j.jmoldx.2015.04.009
Lincoln, S. E., Truty, R., Lin, C.-F., Zook, J. M., Paul, J., Ramey, V. H., et al. (2019). A Rigorous Interlaboratory Examination of the Need to Confirm Next-Generation Sequencing-Detected Variants with an Orthogonal Method in Clinical Genetic Testing. The J. of Mol. Diagnostics 21, 318–329. doi:10.1016/j.jmoldx.2018.10.009
Marks, D., Thorogood, M., Neil, S. M., Humphries, S. E., and Neil, H. A. W. (2006). Cascade Screening for Familial Hypercholesterolaemia: Implications of a Pilot Study for National Screening Programmes. J. Med. Screen. 13, 156–159. doi:10.1258/096914106778440617
Marks, D., Wonderling, D., Thorogood, M., Lambert, H., Humphries, S. E., and Neil, H. A. W. (2002). Cost Effectiveness Analysis of Different Approaches of Screening for Familial Hypercholesterolaemia. BMJ 324, 1303. doi:10.1136/bmj.324.7349.1303
McGowan, M. P., Cuchel, M., Ahmed, C. D., Khera, A., Weintraub, W. S., Wilemon, K. A., et al. (2021). A Proof-of-Concept Study of Cascade Screening for Familial Hypercholesterolemia in the US, Adapted from the Dutch Model. Am. J. Prev. Cardiol. 6, 100170.
McKinney, L. P., Gerbi, G. B., Caplan, L. S., Claridy, M. D., and Rivers, B. M. (2020). Predictors of Genetic Beliefs toward Cancer Risk Perceptions Among Adults in the United States: Implications for Prevention or Early Detection. Jrnl of Gene Coun 29, 494–504. doi:10.1002/jgc4.1228
Menko, F. H., Ter Stege, J. A., van der Kolk, L. E., Jeanson, K. N., Schats, W., Moha, D. A., et al. (2019). The Uptake of Presymptomatic Genetic Testing in Hereditary Breast-Ovarian Cancer and Lynch Syndrome: a Systematic Review of the Literature and Implications for Clinical Practice. Fam. Cancer 18, 127–135. doi:10.1007/s10689-018-0089-z
Milo Rasouly, H., Cuneo, N., Marasa, M., DeMaria, N., Chatterjee, D., Thompson, J. J., et al. (2021). GeneLiFT: A Novel Test to Facilitate Rapid Screening of Genetic Literacy in a Diverse Population Undergoing Genetic Testing. Jrnl of Gene Coun 30, 742–754. doi:10.1002/jgc4.1364
Muir, L. A., George, P. M., and Whitehead, L. (2012). Using the Experiences of People with Familial Hypercholesterolaemia to Help Reduce the Risk of Cardiovascular Disease: a Qualitative Systematic Review. J. Adv. Nurs. 68, 1920–1932. doi:10.1111/j.1365-2648.2012.05957.x
Musunuru, K., Hershberger, R. E., Day, S. M., Klinedinst, N. J., Landstrom, A. P., Parikh, V. N., et al. (2020). Genetic Testing for Inherited Cardiovascular Diseases: A Scientific Statement from the American Heart Association. Circ. Genom Precis. Med. 13, e000067. doi:10.1161/HCG.0000000000000067
Nazareth, S., Hayward, L., Simmons, E., Snir, M., Hatchell, K. E., Rojahn, S., et al. (2021). Hereditary Cancer Risk Using a Genetic Chatbot Before Routine Care Visits. Obstet. Gynecol. 138, 860–870.
Neuner, J., Dimmock, D., Kirschner, A. L. P., Beaudry, H., Paradowski, J., and Orlando, L. (2020). Results and Lessons of a Pilot Study of Cascade Screening for Familial Hypercholesterolemia in US Primary Care Practices. J. Gen. Intern. Med. 35, 351–353. doi:10.1007/s11606-019-05485-7
Newson, A. J., and Humphries, S. E. (2005). Cascade Testing in Familial Hypercholesterolaemia: How Should Family Members Be Contacted? Eur. J. Hum. Genet. 13, 401–408. doi:10.1038/sj.ejhg.5201360
Nitecki, R., Moss, H. A., Watson, C. H., Urbauer, D. L., Melamed, A., Lu, K. H., et al. (2021). Facilitated Cascade Testing (FaCT): A Randomized Controlled Trial. Int. J. Gynecol. Cancer 31, 779–783.
Nordestgaard, B. G., Chapman, M. J., Humphries, S. E., Ginsberg, H. N., Masana, L., Descamps, O. S., et al. (2013). Familial Hypercholesterolaemia Is Underdiagnosed and Undertreated in the General Population: Guidance for Clinicians to Prevent Coronary Heart Disease: Consensus Statement of the European Atherosclerosis Society. Eur. Heart J. 34, 3478–3490. doi:10.1093/eurheartj/eht273
Nykamp, K., Anderson, M., Powers, M., Garcia, J., Herrera, B., Ho, Y.-Y., et al. (2017). Sherloc: a Comprehensive Refinement of the ACMG-AMP Variant Classification Criteria. Genet. in Med. 19, 1105–1117. doi:10.1038/gim.2017.37
O'Brien, G., Christensen, K. D., Sullivan, H. K., Stout, N. K., Diller, L., Yeh, J. M., et al. (2021). Estimated Cost-Effectiveness of Genetic Testing in Siblings of Newborns with Cancer Susceptibility Gene Variants. JAMA Netw. Open 4, e2129742. doi:10.1001/jamanetworkopen.2021.29742
Offit, K., Tkachuk, K. A., Stadler, Z. K., Walsh, M. F., Diaz-Zabala, H., Levin, J. D., et al. (2020). Cascading after Peridiagnostic Cancer Genetic Testing: An Alternative to Population-Based Screening. Jco 38, 1398–1408. doi:10.1200/jco.19.02010
Petersen, H. V., Frederiksen, B. L., Lautrup, C. K., Lindberg, L. J., Ladelund, S., and Nilbert, M. (2019). Unsolicited Information Letters to Increase Awareness of Lynch Syndrome and Familial Colorectal Cancer: Reactions and Attitudes. Fam. Cancer 18, 43–51. doi:10.1007/s10689-018-0083-5
Pollard, S., Kalloger, S., Weymann, D., Sun, S., Nuk, J., Schrader, K. A., et al. (2020). Genetic Testing for Hereditary Cancer Syndromes: Patient Recommendations for Improved Risk Communication. Health Expect. 23, 884–892. doi:10.1111/hex.13062
Randall, L. M., Pothuri, B., Swisher, E. M., Diaz, J. P., Buchanan, A., Witkop, C. T., et al. (2017). Multi-disciplinary Summit on Genetics Services for Women with Gynecologic Cancers: A Society of Gynecologic Oncology White Paper. Gynecol. Oncol. 146, 217–224. doi:10.1016/j.ygyno.2017.06.002
Richards, S., Aziz, N., Bale, S., Bick, D., Das, S., Gastier-Foster, J., et al. (2015). Standards and Guidelines for the Interpretation of Sequence Variants: a Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. in Med. 17, 405–424. doi:10.1038/gim.2015.30
Roberts, M. C., Dotson, W. D., DeVore, C. S., Bednar, E. M., Bowen, D. J., Ganiats, T. G., et al. (2018). Delivery of Cascade Screening for Hereditary Conditions: A Scoping Review of the Literature. Health Aff. 37, 801–808. doi:10.1377/hlthaff.2017.1630
Schmidlen, T., Jones, C. L., Campbell-Salome, G., McCormick, C. Z., Vanenkevort, E., and Sturm, A. C. (2022). Use of a Chatbot to Increase Uptake of Cascade Genetic Testing. J. Genet. Couns. Advance Online Publication. doi:10.1002/jgc4.1592
Sharaf, R. N., Myer, P., Stave, C. D., Diamond, L. C., and Ladabaum, U. (2013). Uptake of Genetic Testing by Relatives of Lynch Syndrome Probands: a Systematic Review. Clin. Gastroenterology and Hepatology 11, 1093–1100. doi:10.1016/j.cgh.2013.04.044
Snir, M., Nazareth, S., Simmons, E., Hayward, L., Ashcraft, K., Bristow, S. L., et al. (2021). Democratizing Genomics: Leveraging Software to Make Genetics an Integral Part of Routine Care. Am. J. Med. Genet. C Semin. Med. Genet. 187, 14–27.
Srinivasan, S., Won, N. Y., Dotson, W. D., Wright, S. T., and Roberts, M. C. (2020). Barriers and Facilitators for Cascade Testing in Genetic Conditions: a Systematic Review. Eur. J. Hum. Genet. 28, 1631–1644. doi:10.1038/s41431-020-00725-5
staff reporter (2021). Invitae to Provide Genetic Testing within eMERGE Population Screening Study. Available at: https://www.genomeweb.com/molecular-diagnostics/invitae-provide-genetic- testing-within-emerge-population-screening-study#.YnkzFRPML0r [Accessed May 9, 2022].
Sturm, A. C., Knowles, J. W., Gidding, S. S., Ahmad, Z. S., Ahmed, C. D., Ballantyne, C. M., et al. (2018). Clinical Genetic Testing for Familial Hypercholesterolemia. J. of the Am. Coll. of Cardiol. 72, 662–680. doi:10.1016/j.jacc.2018.05.044
Suthers, G. K., Armstrong, J., McCormack, J., and Trott, D. (2006). Letting the Family Know: Balancing Ethics and Effectiveness when Notifying Relatives about Genetic Testing for a Familial Disorder. J. of Med. Genet. 43, 665–670. doi:10.1136/jmg.2005.039172
Truty, R., Patil, N., Sankar, R., Sullivan, J., Millichap, J., Carvill, G., et al. (2019). Possible Precision Medicine Implications from Genetic Testing Using Combined Detection of Sequence and Intragenic Copy Number Variants in a Large Cohort with Childhood Epilepsy. Epilepsia Open 4, 397–408. doi:10.1002/epi4.12348
van den Heuvel, L. M., van Teijlingen, M. O., van der Roest, W., van Langen, I. M., Smets, E. M. A., van Tintelen, J. P., et al. (2020). Long-Term Follow-Up Study on the Uptake of Genetic Counseling and Predictive DNA Testing in Inherited Cardiac Conditions. Circ Genomic and Precis. Med. 13, 524–530. doi:10.1161/circgen.119.002803
Wald, D. S., Bestwick, J. P., Morris, J. K., Whyte, K., Jenkins, L., and Wald, N. J. (2016). Child-Parent Familial Hypercholesterolemia Screening in Primary Care. N. Engl. J. Med. 375, 1628–1637.
Webber, E. M., Hunter, J. E., Biesecker, L. G., Buchanan, A. H., Clarke, E. V., Currey, E., et al. (2018). Evidence‐based Assessments of Clinical Actionability in the Context of Secondary Findings: Updates from ClinGen's Actionability Working Group. Hum. Mutat. 39, 1677–1685. doi:10.1002/humu.23631
Wonderling, D., Umans-Eckenhausen, M., Marks, D., Defesche, J., Kastelein, J., and Thorogood, M. (2004). Cost-effectiveness Analysis of the Genetic Screening Program for Familial Hypercholesterolemia in The Netherlands. Seminars in Vasc. Med. 4, 97–104. doi:10.1055/s-2004-822992
Wurtmann, E., Steinberger, J., Veach, P. M., Khan, M., and Zierhut, H. (2018). Risk Communication in Families of Children with Familial Hypercholesterolemia: Identifying Motivators and Barriers to Cascade Screening to Improve Diagnosis at a Single Medical Center. J. Genet. Couns. 28, 50–58. doi:10.1007/s10897-018-0290-0
Keywords: cascade testing, genetic testing, diagnostic testing, proactive screening, hereditary cancer syndromes, familial hypercholesterolemia, CDC tier 1 conditions
Citation: Schmidlen TJ, Bristow SL, Hatchell KE, Esplin ED, Nussbaum RL and Haverfield EV (2022) The Impact of Proband Indication for Genetic Testing on the Uptake of Cascade Testing Among Relatives. Front. Genet. 13:867226. doi: 10.3389/fgene.2022.867226
Received: 31 January 2022; Accepted: 18 May 2022;
Published: 16 June 2022.
Edited by:
Muin J. Khoury, Centers for Disease Control and Prevention, United StatesReviewed by:
Maria Katapodi, University of Basel, SwitzerlandIftikhar Kullo, Mayo Clinic, United States
Copyright © 2022 Schmidlen, Bristow, Hatchell, Esplin, Nussbaum and Haverfield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eden V. Haverfield, ZWRlbi5oYXZlcmZpZWxkQGludml0YWUuY29t
 Tara J. Schmidlen
Tara J. Schmidlen Sara L. Bristow
Sara L. Bristow Kathryn E. Hatchell
Kathryn E. Hatchell Edward D. Esplin
Edward D. Esplin Robert L. Nussbaum
Robert L. Nussbaum