- 1Department of Second Institute of Clinical Medicine, Guangzhou University of Traditional Chinese Medicine, Guangzhou, China
- 2Department of Anesthesiology, Guangdong Provincial Hospital of Traditional Chinese Medicine/The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, China
- 3Department of Neurology, Guangdong Provincial Hospital of Traditional Chinese Medicine/The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, China
- 4Department of Anatomy, Sun Yat-Sen School of Medicine, Sun Yat-Sen University, Shenzhen, China
- 5State Key Laboratory of Dampness Syndrome of Chinese Medicine, The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, China
- 6Guangdong Provincial Key Laboratory of Research on Emergency in TCM, Guangzhou, China
Ischemic stroke (IS) is a major neurological disease with high fatality and residual disability burdens. Long noncoding RNAs (lncRNAs) have been found to play an important role in IS. However, the roles and significance of most lncRNAs in IS are still unknown. This study was performed to identify differentially expressed (DE) lncRNAs using a lncRNA microarray in whole blood samples of patients suffering from acute cerebral ischemia. Bioinformatics analyses, including GO, KEGG pathway enrichment analysis, and proximity to putative stroke risk location analysis were performed. The novel lncRNA, ENST00000530525, significantly decreased after IS. Furthermore, we evaluated lncRNA ENST00000530525 expression in cultured hCMEC/D3 cells under oxygen-glucose deprivation/reoxygenation (OGD/R) conditions using fluorescent in situ hybridization (FISH) and quantitative real-time polymerase chain reaction (RT–qPCR) analysis. To investigate the function of lncRNA ENST00000530525, its over-expression (OE) and negative control (NC) plasmids were transfected into hCMEC/D3 cells, and cell viability was detected by a cell counting kit-8 (CCK-8) assay after OGD/R. LncRNA ENST00000530525 and ANO1 expression were investigated using RT–qPCR and immunofluorescence. For blood–brain barrier (BBB) permeability, FITC-dextran transendothelial permeability assay and tight junction (TJ) protein immunofluorescence assays were performed. There were 3352 DE lncRNAs in the blood samples of acute IS patients. The validation results were consistent with the gene chip data. The GO and KEGG results showed that these lncRNAs were mainly related to oxygen and glucose metabolism, leukocyte transendothelial migration, mitophagy and cellular senescence. Among these, lncRNA ENST00000530525 was the most highly downregulated lncRNA and it was mapped within the IS-associated gene anoctamin-1 (ANO1). We further found that lncRNA ENST00000530525 was downregulated in hCMEC/D3 cells under 4 h OGD and 20 h reoxygenation (OGD4/R20) conditions. Upregulating lncRNA ENST00000530525 by plasmid transfection decreased cell viability while increasing ANO1 expression and it contributed to BBB injury in hCMEC/D3 cells after OGD4/R20. The lncRNA ENST00000530525 might play deleterious roles in post-stroke pathogenesis. These results show that some DE lncRNAs in humans participate through characteristic roles in post-stroke pathogenesis; thus, the roles and significance of some novel lncRNAs in IS warrant further study.
Introduction
Stroke is a high risk factor for death and disability worldwide. IS is the most prevalent, accounting for up to 80% of all stroke cases (Wang et al., 2017). Hypertension, diabetes, dyslipidemia, atrial fibrillation, and obesity are prevalent among Chinese individuals aged 40 years and older (Li et al., 2017). This makes stroke a very important health issue.
Cerebral blood flow interruption initiates a cascade of complex and poorly understood pathophysiologic processes, including oxidative stress, inflammation and apoptosis (Schaukowitch and Kim, 2014; Ren and Yang, 2018). Therefore, many scientists have turned to epigenetics to understand the molecular mechanisms, regulation of the complex stroke-induced pathophysiological processes as well as the elucidation of new therapeutic targets and the design of new treatment modalities (Dharap et al., 2012; Liu et al., 2018)). The most promising theme thus far is the alteration of the expression of some noncoding RNAs that could participate in post-stroke processes, both neuroprotective and neurodegenerative.
Long noncoding RNAs (lncRNAs) are more than 200 nucleotides long and do not encode any proteins. Previous studies showed lncRNAs could modulate cell survival, inflammation, and angiogenesis (Bao et al., 2018). They appear to perform these varied functions by interacting with multiple elements involved in the central dogma of biology, such as other RNAs, DNA, and proteins (Akella et al., 2019). Recently, a number of abnormally expressed lncRNAs have been identified in IS though microarrays, high-throughput deep sequencing, and RNA-seq (Bao et al., 2018). However, the importance and significance of most lncRNAs remain poorly understood and warrant further study in IS.
Therefore, this research was performed to evaluate lncRNA expression in patients’ whole blood samples after acute cerebral ischemia. Bioinformatics analyses were also used for the dysregulated genes to reveal the predicted connections and functions of the DE lncRNAs. Furthermore, we found that lncRNA ENST00000530525 is highly decreased in stroke patients relative to controls, and the high conservation of lncRNA ENST00000530525 suggests that it may be important in stroke pathology.
LncRNA ENST00000530525 is located intragenically with its neighbor gene ANO1. However, the role of lncRNA ENST00000530525 and the regulatory mechanism linking lncRNAs and ANO1 signaling in stroke remain enigmatic. ANO1 (also known as transmembrane protein 16A, TMEM16A) widely expresses in eukaryotes and is a molecular indicator of calcium-activated chloride channels. Recent studies have shown that the upregulation of ANO1 is related to the development of many diseases, such as IS (Wang et al., 2012; Wu et al., 2014; Deng et al., 2016). Recently, it was found that ANO1 is strongly expressed in cerebrovascular endothelial cells, IS-induced BBB injury is accompanied by upregulation of ANO1. Inhibiting ANO1 significantly rescued OGD/R-induced downregulation of TJ proteins and upregulation of BBB permeability in vitro (Liu et al., 2019). And the present studies showed that ANO1 affects the functions of brain capillary endothelial cells, which are involved in BBB functions (Suzuki et al., 2020; Liu et al., 2021; Ma et al., 2021; Suzuki et al., 2021).
As we know, BBB is part of the neurovascular unit that plays a vital role in regulating blood-to-brain flux of endogenous and exogenous xenobiotics and associated metabolites. Because of the TJ protein between the endothelial cells of the BBB, blood-borne substances and cells have restricted access to the brain. Under IS condition, the BBB can be destroyed and loss of the TJ integrity, followed by the extravasation of blood components into the brain and compromise of normal neuronal function, future research on the BBB is likely to reveal promising potential therapeutic targets for protecting the BBB and improving patient outcome after IS (Jiang et al., 2018).
Based on the above, the underlying mechanisms and targets of lncRNA ENST00000530525 in hCMEC/D3 cells under OGD/R injury were investigated, which may provide a novel idea for us to better understand the function and significance of lncRNAs in the pathophysiological mechanism of IS. Further studies of these lncRNAs in biological systems after IS may provide opportunities to identify biomarkers and new therapeutic targets.
Materials and Methods
Study Subjects
Ten patients with a diagnosis of IS who were seen at Guangdong Province Traditional Chinese Medical Hospital from September 2016 to November 2017 along with 20 control subjects were recruited to join the study. The protocol was approved by the ethics committee of Guangdong Provincial Hospital of Chinese Medicine (B2016-149-01). After explaining study details, patients and their immediate caregivers or significant others provided written informed consent to participate. There was no incentive to participate.
Patients’ histories, examinations, and evaluations were collected after qualified neurologists diagnosed them as acute IS. Standard stroke assessments performed at our hospital included computed tomographic (CT), electrocardiography (ECG), magnetic resonance imaging (MRI), carotid Doppler, Holter monitor and computed tomographic angiography or magnetic resonance arteriogram of extracranial and intracranial vessels according to the clients’ clinical data. Acute IS patients who presented with a rare pathogenesis other than atherosclerosis and cardiogenic stroke, psychopathology, pregnancy and lactation or those patients who were in other clinical trials when they suffered a stroke were not eligible to participate in this study. The age and sex of the 20 control subjects were matched to those of the stroke patients; the control patients were without a history of stroke and did not have severe heart disease, cardiac insufficiency, hepatitis, renal insufficiency, respiratory failure, malignant tumor or gastrointestinal bleeding.
Derivation and Validation Group
Ten IS patients and 20 non-stroke subjects were recruited to detect DE lncRNAs and randomly distributed them to the derivation group or the validation group. All patients’ race, sex, age, and vascular risk factors (diabetes mellitus, hypertension, and hyperlipidemia) were matching. Exclusion criteria included prior stroke; treatment with thrombolysis or anticoagulant before sample collection; hemorrhagic infarction; infection before or after stroke; subarachnoid hemorrhage; recreational drug abuse; dialysis; cancer; and immunosuppressive therapy for blood abnormalities or steroids. The validation set was used to prove the microarray analysis results. The primer sequences used are provided in Supplementary Table S1.
Blood Collection and RNA Isolation
Venipuncture was performed once on all study participants, and their whole blood was drawn into EDTA tubes (Jingxin Biotechnology Co., Ltd., China). For stroke patients, blood collection time varied from a few hours to 2 weeks after symptom onset. For the control samples, the time of extraction was not applicable. And then the tubes was rapidly frozen with liquid nitrogen and put at -80°C until use. RNA isolation was performed by using TRIzol (Invitrogen, United States), which was added to every cryopreserved serum supernatant sample according to the protocol.
Array Hybridization
Arraystar Human lncRNA V4.0 chip (Arraystar, United States) was used, which can test 20,730 protein-coding and 40,173 lncRNA transcripts. Sample marking and chip hybridization were based on the recommended experimental procedure of Agilent One-Color Microarray-Based Gene Expression Analysis (Agilent Technology, United States). Briefly, a mRNA-ONLY™ Eukaryotic mRNA Isolation Kit (Epicenter, United States) was used to remove the rRNA from mRNA, which was then amplified and transcribed into cRNA with fluorescence by random priming. The marked cRNAs was purified by the RNeasy Mini Kit (Qiagen 74104, Germany). The RNA was quantified by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Fisher, United States), and an Agilent 2100 Bioanalyzer (Agilent, United States) was used to assess the quality of the samples. The GenePix 4000B chip scanner (Agilent, United States) was used to wash and process arrays. DAT file formats were used to save raw values. The scanned results were transformed into digital data and then saved.
Differential Expression Analysis
To identify DE lncRNAs, cuffdiff was used for the differential expression analysis. Only probes with a fold-change higher than 2 and p < 0.05 were selected as significantly DE lncRNAs or mRNAs. To obtain the overall characteristics of the lncRNA and mRNA expression profiles, we used the R package for hierarchical clustering analysis of normalized values of all DE lncRNAs to generate heatmaps. Bioinformatics analyses, including the Gene Ontology project (GO, http://geneontology.org/), the database Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (http://www.kegg.jp/kegg/), and network analysis, were also performed for the identified DE genes. Through a literature review, NONCODE, UCSC Genome Browser, and OMIM®—Online Mendelian Inheritance in Man® databases, we further explained the biology of the differential lncRNAs.
Data Normalization
The original data were exported by GeneSpring GX v12.1 software (Agilent, United States). The data were normalized by preprocess Core in the R package, and log2 transformation was performed to obtain the final normalized data. The standardized data were further analyzed by screening high-quality probes marked as detected in at least three samples out of six. Hierarchical clustering was conducted using R script. Finally, the lncRNAs expressed at significantly different levels between the two groups of samples were screened by volcano plots.
hCMEC/D3 Cell Culture
HCMEC/D3 cells (FuHeng Biology, FH1110, Shanghai, China) were cultured in endothelial cell medium (ECM,ScienCell, Carlsbad, United States) and added fetal bovine serum (FBS, Gibco, Australia) into ECM to making its concentration to 10%, and then incubated the cells in humidified atmosphere at 37°C with 5% CO2. All experiments with hCMEC/D3 cells were used below 15 passages.
OGD/R Condition
To mimic acute IS in vitro, hCMEC/D3 cells were subjected to OGD/R. The establishment of OGD model in this paper refers to the literatures (Fan et al., 2021; Xu et al., 2021). When the cells became confluent, glucose-free DMEM (Cienry, Zhejiang, China) was used to replace the culture medium, and the cells were put into an anaerobic incubator (94% N2, 5% CO2, 1% O2) for 2, 4, 6, and 8 h. After incubation, the cells were returned to regular conditions and normal medium for reoxygenation. The total time of oxygen glucose deprivation and reoxygenation was 24 h.
CCK-8 Assay
The cell counting kit-8 assay (Dojindo, Tokyo, Honshu, Japan) was used to detect cell viability. All of the procedures followed the manufacturer’s instructions. Cell suspensions (100 μl, 1 × 104 cells/mL) were added to a 96-well plate. After incubating with NC or OE (MOI = 50 nM) in 96-well microtiter plates, CCK-8 solution (10 μl) was put into 96-well plate after OGD for 4 h and reoxygenation for 20 h. Finally, absorbance at 450 nm of the wells was measured by the microplate reader (SYNERGY H1, BioTek), and five replicate wells were assessed in each experiment.
RNA FISH
Subcellular localization of the lncRNA ENST00000530525 was detected by the FISH Kit (RiboBio, China). The Cy3-labeled lncRNA ENST00000530525 probe and the control probe 18S were all designed by RiboBio (Guangzhou, China). Briefly, 4% paraformaldehyde was used to fix hCMEC/D3 cells for 15 min at room temperature and washed with PBS three times for 5 min each time. The cells were incubated in proteinase K for 5 min at 4°C and washed as before. Then, prehybridization buffer was added to the cells for incubation at 37°C and then they were incubated in hybridization buffer with specific probes for lncRNA ENST00000530525 and 18S at 37°C overnight. After extensive washing with SSC, the cell nucleus was stained by DAPI for 5 min. The images were took by the fluorescence microscope (Nikon Ti2-E, Japan).
Plasmid Transfection
RiboBio company (Guangzhou, China) provided the lncRNA ENST00000530525 (OE) overexpression plasmid, negative control plasmid (NC) and transfection kit (riboFECT™ CP, C10511-05). HCMEC/D3 cells were cultured and incubated for 3 days with OE or NC (MOI = 50 nM), which was then replaced by glucose-free DMEM for OGD 4 h. RT–qPCR was used to detect the transfection efficiency of lncRNA ENST00000530525.
FITC-Dextran Transendothelial Permeability Assay
In brief, cell suspension was added into polycarbonate 12-well Transwell inserts with a 0.4 μm pore size (Corning, United States) at a density of 4 × 104 cells/cm2. When cells grown to approximately 70% confluence, they were transfected with plasmid. Then FITC-labeled dextran (0.1 mg/ml, MW, 70000, Sigma-Aldrich) was added to the upper chamber and 500 μL PBS was added to the lower cavity after OGD/R. Then we put them in a cell incubator for 5 min. The microplate reader was used to analyze the supernatant from the lower chamber (excitation at 490 nm, emission at 520 nm).
Quantitative Real-Time PCR for lncRNA and mRNA
The protocol of RT-qPCR are as follows. We used TRIzol (Thermo Fisher Scientific, Waltham, MA, United States) to separate total RNA from hCMEC/D3 cells. For reverse transcriptase, we used cDNA synthesis kits (Takara, Dalian, China) to synthesize cDNA. SYBR®Premix Ex Taq™II (Takara, Dalian, China) was used for amplification. The internal reference is human β-actin gene. Comparative quantification was performed by using the 2−ΔΔCt method. The primer sequences used are shown in Supplementary Table S1.
Immunofluorescence
Cells were cultured on 24-well plates for immunofluorescence staining at a seeding density of 7 × 104 cells per well (Shin et al., 2016). For immunofluorescent staining, 4% paraformaldehyde was used to fix the cell at 4°C overnight. After blocking with 5% BSA, the first antibodies: rabbit monoclonal anti-ANO1 (1:200), rabbit monoclonal anti-ZO1 (1:200), rabbit anti-occludin (1:100), and rabbit anti-claudin-5 (1:200) were added to the cells at 4°C overnight. Then the cells were incubated with the corresponding secondary antibody in a dark chamber at 37°C for 1 h. The antibodies and secondary antibodies were obtained from Abcam. After stained with DAPI (1:500, Beyotime), the fluorescence microscope (Nikon TI2-E) was used to take the images.
Transfection and Luciferase Assays
293T cells were inoculated in 24-well plates and cultured until the degree of cell fusion reached over 60%. Plasmid transfections for luciferase assays were performed with 1 µg plasmid and 2 µL X-tremegene HP reagent (ROCHE, Switzerland) as described by the manufacturer. The expression of fluorescent-labeled genes on the plasmid was observed 24–48 h after transfection to determine the transfection efficiency. And then 48 h post transfection, luminescence was detected using the Dual-Luciferase Reporter Assay System as described by the manufacturer (Promega, United States). Data were normalized to the Renilla luminescence and presented relative to control transfected group.
Statistical Analysis
All quantitative experiments were performed at least three times, and the data are showed as the mean ± SD. Statistical tests were performed using SPSS 24.0 statistical software and Graphed Prism 6 was used for graphic representations. The comparisons of patient characteristics between two groups were analyzed by the chi-square test (sex, hypertension history, diabetes history, smoking history, cardiovascular history, and hyperlipidemia) and unpaired two-tailed Student’s t-test (age). One-way ANOVA was used to analyze the comparisons among multiple groups. p < 0.05 was considered statistically significant.
Results
Characteristics
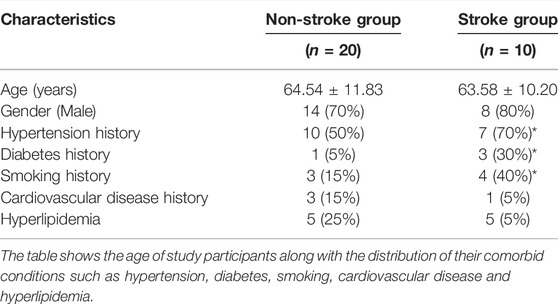
The characteristics of 10 stroke patients and 20 non-stroke cases are shown in Table 1. There were significantly more stroke patients with a positive history of hypertension, diabetes, and smoking.
Differential lncRNA Expression and GO and KEGG Analyses Among IS Patients versus Non-stroke Patients
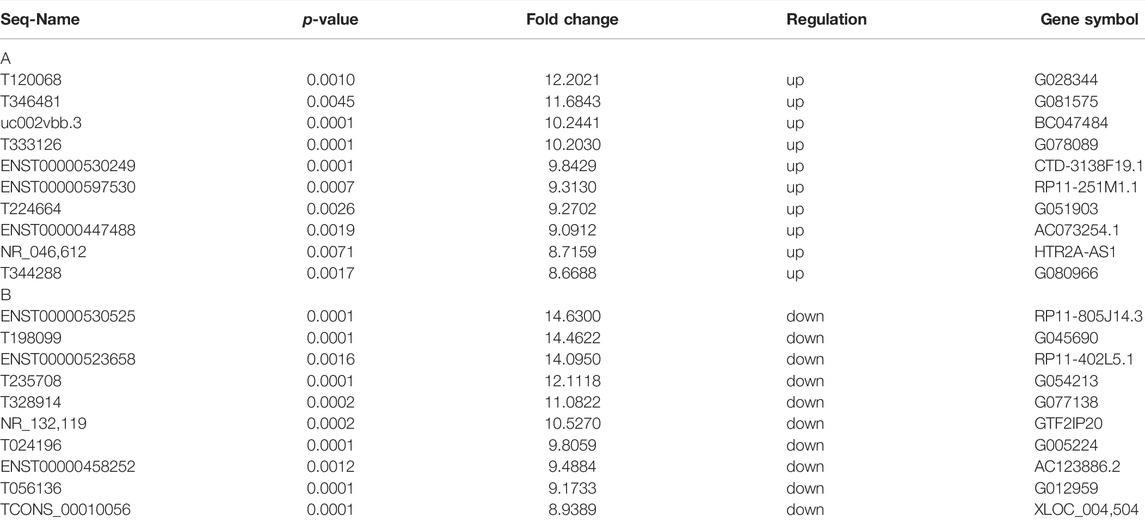
We used the Arraystar Human lncRNA V4.0 chip to analyze the DE lncRNAs between two groups. Unsupervised hierarchical clustering analysis generated from the expression profile of the lncRNAs showed the different expression levels between non-stroke and stroke patients (Figure 1A). Volcano plots were used to show the statistically significant differences expression of lncRNAs. There were 3352 DE lncRNAs, with 1318 upregulated and 2034 downregulated lncRNAs (Figure 1B). The top 10 most significantly up and downregulated lncRNAs are listed in Table 2. T120068 (fold change 12.2021) was the most highly upregulated lncRNAs. ENST00000530525 (fold change -14.63) was the most highly downregulated lncRNAs.
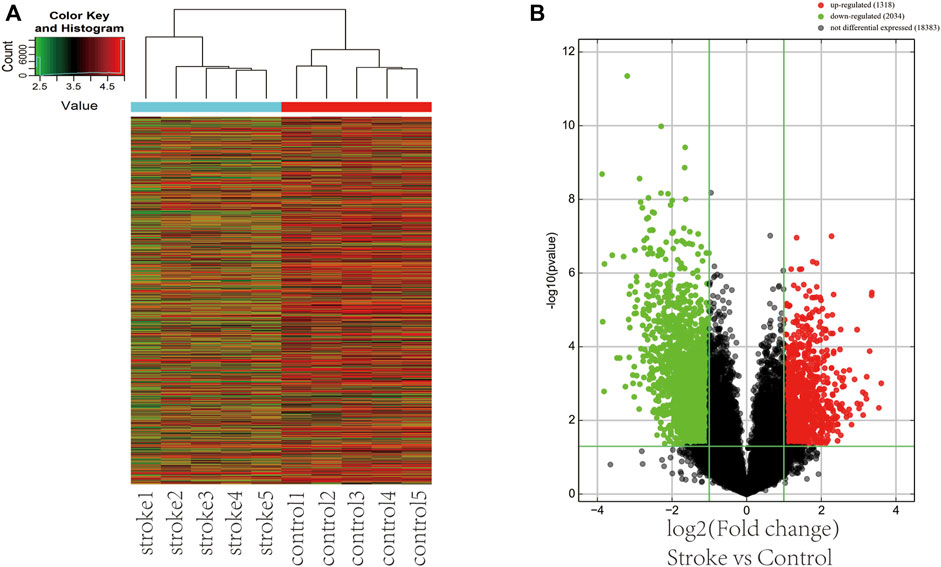
FIGURE 1. The DE lncRNAs among ischemic stroke patients versus non-stroke controls. (A) Hierarchical clustering shows different lncRNA expressions between stroke and nonstroke patients. (B) Volcano plot of the DE lncRNAs in stroke patients and nonstroke controls. Red points in the plot are statistically significant differentially upregulated lncRNAs, while green represents downregulated lncRNAs.
The functions of these lncRNAs were predicted with the co-expressed mRNAs using GO and KEGG pathway annotations. In GO annotation, the upregulated genes were mainly related to ER membrane protein complexes, Toll-like receptor binding, protein localization to the nucleoplasm (Figures 2A–C). The downregulated genes were manly related to RAGE receptor binding, intramolecular oxidoreductase activity, cellular glucuronidation, cellular glucuronation and T cell receptor complexes (Figures 2D–F).
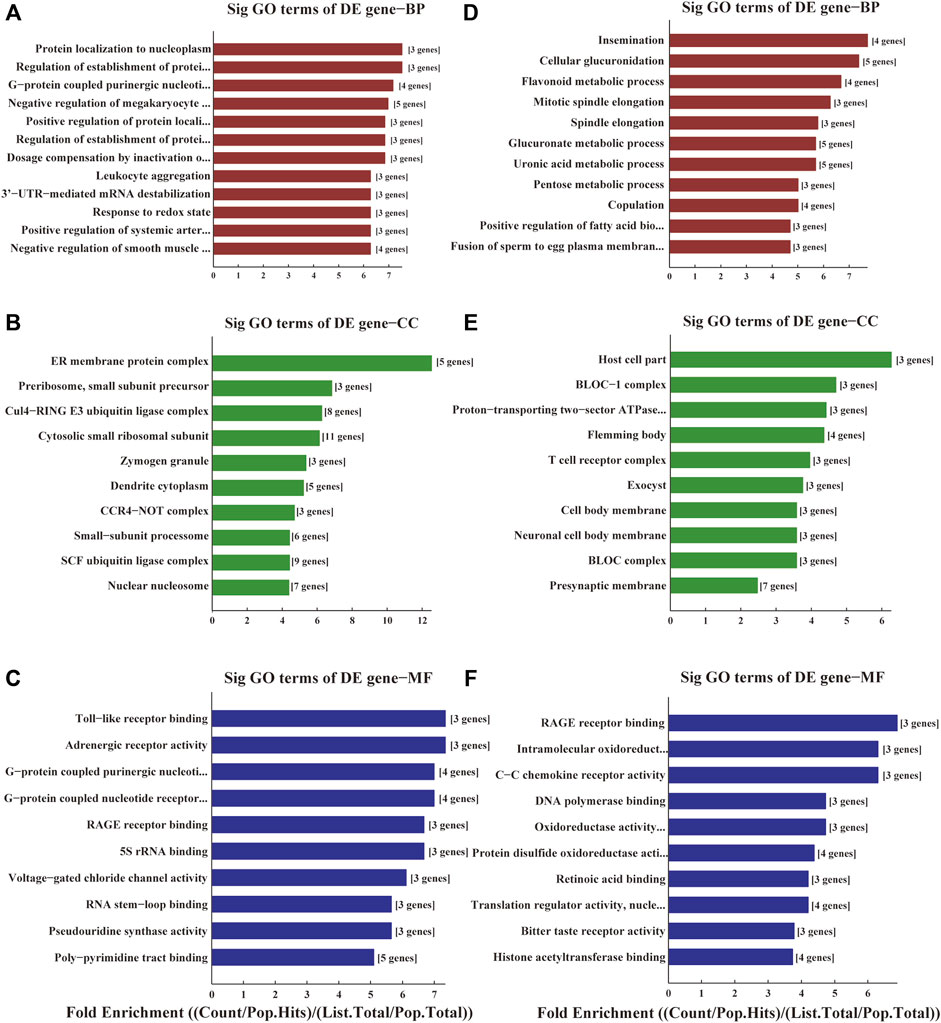
FIGURE 2. GO analyses of the DE genes among ischemic stroke patients versus non-stroke controls. GO terms of Biological Process (BP), Cellular Component (CC) and Molecular Function (MF) for the DE upregulated (A–C) and downregulated genes (D–F).
The results of KEGG enrichment showed that these DE genes were related to pathways in pathophysiological processes underpinning acute cerebral infarction, such as pentose and glucuronate interconversions, regulation of the actin cytoskeleton, RAS signal pathway, focal adhesion, leukocyte transendothelial migration, mitophagy and cellular senescence (Figures 3A,B).
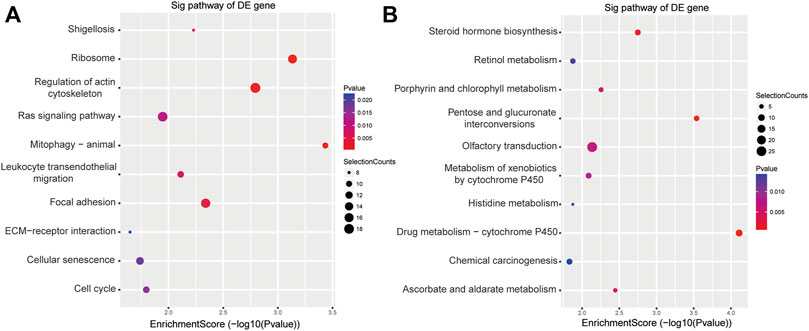
FIGURE 3. KEGG analyses of the DE genes among ischemic stroke patients versus non-stroke controls. KEGG terms for the DE upregulated (A) and downregulated genes (B).
Validation of the DE lncRNAs
We selected nine DE lncRNAs at random and used RT–qPCR analysis to prove our gene chip performance (Figure 4). There were seven upregulated (ENST00000452599, ENST00000527450, ENST00000608826, T013651, T029143, T131416, and T294865) and two downregulated lncRNA transcripts (ENST00000530525 and T058035). The RT–qPCR results were consistent with the gene chip data in the two groups.
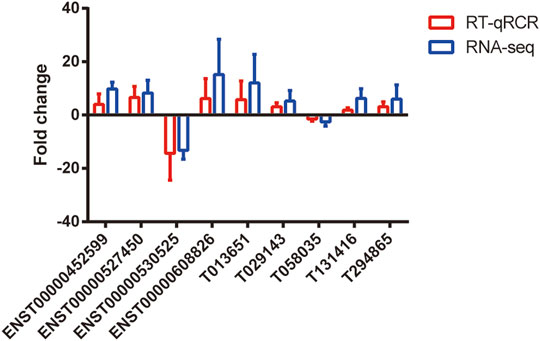
FIGURE 4. Validation of differential expression of lncRNAs. RT-qPCR was used to verify the expression levels of these selected lncRNAs (p < 0.05). The blue points represent RT-qPCR, while the red points represent the gene chip of RNA-seq. n = 5.
Biological Interpretation-Proximity to Putative Stroke Genes
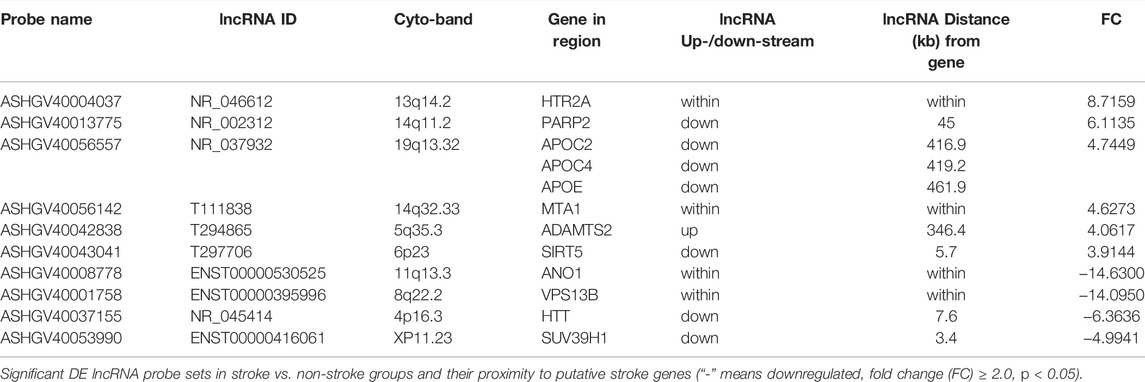
Among the upregulated lncRNAs, ASHGV40004037 was annotated to NR_046612 that mapped to the putative ischemic risk locus HTR2A at cytoband 13q14.2. Similarly, ASHGV40013775 was annotated to NR_002312, which has genomic coordinates 45 kb downstream of the PARP2 cerebral ischemic injury-associated gene at 14q11.2. ASHGV40056557 is expressed as NR_037932, which maps to the stroke-associated genes apolipoprotein C and E within 420 kb, while T111838 (ASHGV40056142) maps at 14q32.33 within metastasis-associated protein 1 (MTA1), a transcriptional regulator-associated gene modifing master chromatin. Additionally, T294865 (ASHGV40042838) mapped within 350 kb of another gene ADAMTS2 associating with stroke at 5q35.3, while T297706 (ASHGV40043041) mapped to Sirtuin 5 (SIRT5) within 6 kb at 6p23. Among the downregulated lncRNAs, the annotation of ASHGV40008778 identified the validated lncRNA ENST00000530525, which is within anoctamin-1 (ANO1), a chloride ion channel gene related to cerebrovascular remodeling. ENST00000395996 (ASHGV40001758) is within VPS13B and is associated with cerebral ischemic injury. Furthermore, NR_045414 (ASHGV40037155) at 4p16.3 mapped within 8 kb of HTT and is associated with post-stroke depression (PSD), while ENST00000416061 (ASHGV40053990)’s genomic coordinates were 3.4 kb downstream of SUV39H1, a histone methyltransferase related to stroke (Table 3).
LncRNA ENST00000530525 was Located Intragenically with ANO1
To futher detect the expression trend of lncRNA ENST00000530525 in stroke patients, we expanded the samples and randomly chose 40 blood samples from stroke patients and 20 blood samples from controls. LncRNA ENST00000530525 expression was significantly decreased in IS group compared with the control group (Supplementary Figure S1). To further research the function of lncRNA ENST00000530525 in stroke pathology, the noncoding nature of lncRNA ENST00000530525 was first confirmed by coding-potential analysis (Supplementary Figure S2) (Qu et al., 2016). Then, we checked Ensembl (Abdillahi et al., 2012), an online database used to predict gene location (Yates et al., 2020), and found that the lncRNA ENST00000530525 is located intragenically with ANO1 (Supplementary Figure S3A). ANO1 is strongly expressed in cerebrovascular endothelial cells (Liu et al., 2019), and study showed that ANO1 contributed to the proliferation and migration of brain capillary endothelial cells, which are involved in BBB functions (Suzuki et al., 2020). We also checked STRING (Lo et al., 2007), an online database, to summarize the network of predicted associations for this protein (Szklarczyk et al., 2015). ANO1 is known to have strong links to epithelial ion channels, such as cystic fibrosis transmembrane conductance regulator (CFTR) and the Bestrophin family (Supplementary Figure S3B). Thus, we further speculated that lncRNA ENST00000530525 and ANO1 are involved in the pathology of stroke. Because ANO1 is strongly expressed in cerebrovascular endothelial cells, in the following study, the cells undergo OGD/R was chosen to mimic IS.
LncRNA ENST00000530525 was DownRegulated in hCMEC/D3 Cells After OGD/R
To investigate lncRNA ENST00000530525 expression in hCMEC/D3 cells after OGD/R injury, CCK-8 assays were used to test cell vitality. The results indicated that the longer the OGD time was, the worse the cell vitality (Figure 5A). The RT–qPCR results showed lncRNA ENST00000530525 expression was significantly downregulated in hCMEC/D3 cells after OGD/R compared with control cells, which is consistent with the results of ChIP sequencing (Figure 5B). The expression of lncRNA ENST00000530525 was highly downregulated after 4 h OGD and 20 h reoxygenation.
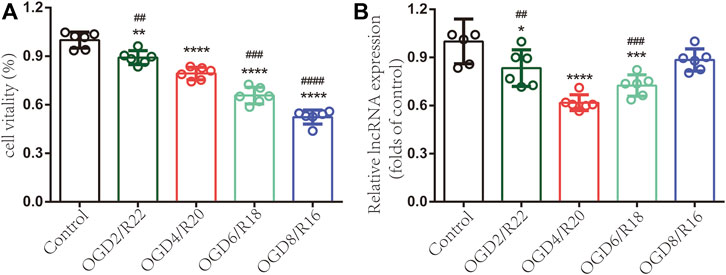
FIGURE 5. LncRNA ENST00000530525 is downregulated in hCMEC/D3 cells under OGD/R condition. Cells were exposed to OGD for 2, 4, 6 or 8 h and reoxygenate for 22, 20, 18 or 16 h. Each bar represents a different OGD duration and reoxygenation time (e.g., OGD2/R22 represents 2 h of OGD and 22 h of reoxygenation). (A) The cell vitality worsened as the OGD time increased (B) LncRNAs highly downregulated under OGD4/R20 conditions. *p < 0.05, **p < 0.01, ****p < 0.0001 (vs. control); ##p < 0.01, ###p < 0.001, ####p < 0.0001 (vs. OGD4/R20), n = 6, one-way ANOVA.
Upregulating lncRNA ENST00000530525 Expression Affected the Viability of hCMEC/D3 Cells After OGD/R
To explore the functions of lncRNA ENST00000530525 in hCMEC/D3 cells after OGD/R, lncRNA ENST00000530525 was upregulated by transfection of an overexpression plasmid. The location of lncRNA ENST00000530525 was tested by FISH. The results showed lncRNA ENST00000530525 was mostly distributed in the cytoplasm, similar to the control group-18S, which means overexpression plasmid transfection would effectively promote the lncRNA’s function (Figure 6A).
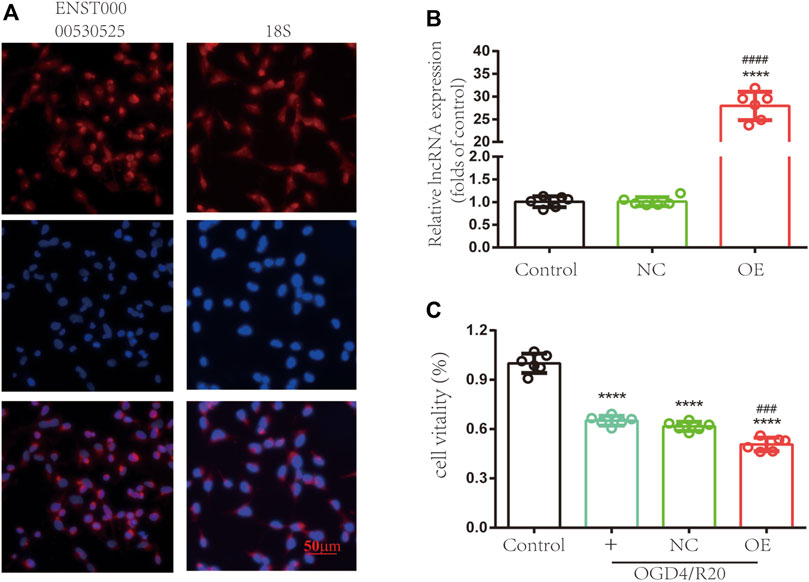
FIGURE 6. Upregulating the expression of lncRNA ENST00000530525 affects the vitality of hCMEC/D3 cells. (A) FISH showed that ENST00000530525 was mainly distributed in the cytoplasm, similar to the control group 18S. Scale bar: 50 μm. (B) The transduction efficiency detected by RT–qPCR. (C) The CCK-8 assay revealed cell vitality. NC: negative control; OE: over expression; OGD4/R20: 4 h of OGD and 20 h of reoxygenation. ****p < 0.0001 (vs. control); ###p < 0.001, ####p < 0.0001 (vs. NC), n = 6, one-way ANOVA.
The RT–qPCR results uncovered lncRNA ENST00000530525 expression in the OE group was effectively upregulated by 28-fold compared to it in the NC group after 72 h of transfection in hCMEC/D3 cells, while there was no difference between the control and NC-treated groups (Figure 6B). This result suggested that the transient plasmid transfection of lncRNA into cultured hCMEC/D3 cells could selectively upregulate lncRNA ENST00000530525 expression.
We investigated whether overexpression of lncRNA ENST00000530525 can influence the viability of hCMEC/D3 cells after OGD/R. The results revealed the cell viability was 1.00 ± 0.05 in untreated cells as a control, 0.65 ± 0.03 in the normal control group, 0.62 ± 0.03 in the NC-treated group, and 0.51 ± 0.04 in the OE-treated group. Statistical analysis showed the cell viability of the OE-treated group was decreased compared to that of the NC-treated group after OGD/R injury (Figure 6C), which indicated that overexpression of lncRNA ENST00000530525 in cultured hCMEC/D3 cells could lead to more severe OGD/R ischemia injury.
Upregulating lncRNA ENST00000530525 Increased the Expression of ANO1
To test whether upregulation of lncRNA ENST00000530525 affects the expression of ANO1, RT–qPCR and immunofluorescence were used. RT–qPCR results showed the expression of ANO1 in the OGD4/R20 group was upregulated. Meanwhile, the ANO1 expression in the OE group was effectively upregulated by 4.27-fold compared to that in the NC-treated cells after 72 h of transfection in hCMEC/D3 cells, but there was no significant difference between OGD4/R20 group and NC group (Figure 7A).
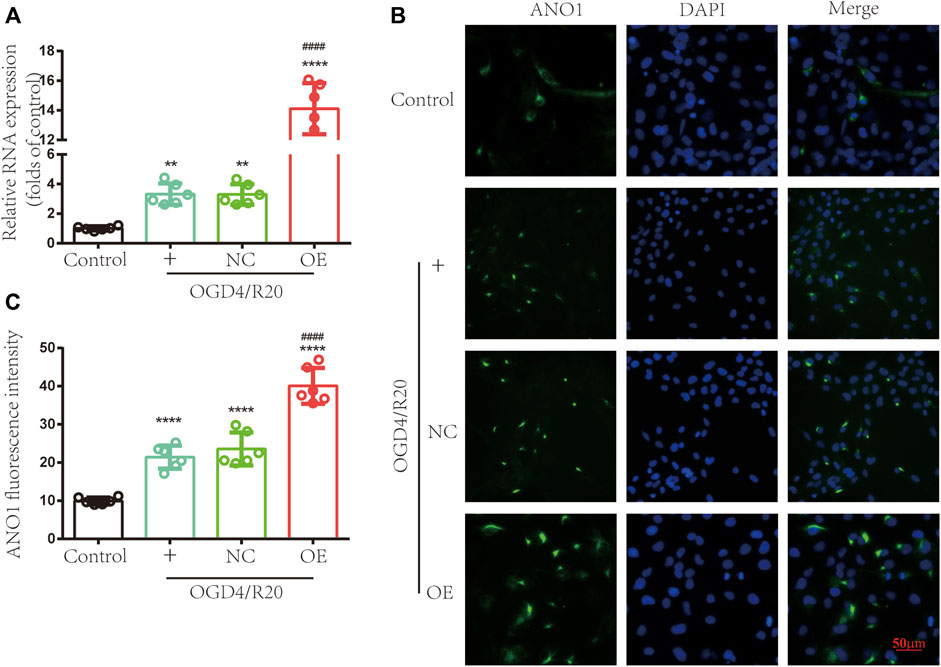
FIGURE 7. Upregulating lncRNA ENST00000530525 increased the expression of ANO1. The effects of lncRNA ENST00000530525 upregulation by plasmid on the gene and protein expression of ANO1 in hCMEC/D3 cells after OGD4/R20 conditions. (A) The RT–qPCR results show that ANO1 increased after OGD4/R20, which increased more significantly when lncRNA ENST00000530525 was upregulated by the plasmid. (B,C) The immunofluorescence results show that transfecting a plasmid into hCMEC/D3 cells resulted in an increase in ANO1 protein expression. Scale bar: 50 μm. NC: negative control; OE: over expression; OGD4/R20: 4 h of OGD and 20 h of reoxygenation. **p < 0.01, ****p < 0.0001 (vs. Control); ####p < 0.0001 (vs. NC), n = 6, one-way ANOVA.
Immunofluorescence showed that the protein expression of ANO1 increased after OGD4/R20. Meanwhile, when lncRNA ENST00000530525 was upregulated by transfection of the overexpression plasmid, ANO1 protein expression in the OE group increased almost 2-fold compared to that in the NC group, while there was no significant difference between OGD4/R20 group and NC group (Figures 7B,C). These results demonstrated that altered expression of lncRNA ENST00000530525 could influence the expression of ANO1 in hCMEC/D3 cells after OGD4/R20.
Targeted Increases in lncRNA ENST00000530525 Deteriorated BBB Disruption After OGD4/R20
FITC-dextran permeability was used to evaluate BBB permeability. The results showed in the OGD4/R20 group FITC-dextran permeability was upregulated compared to the control group. However, FITC-dextran permeability in the OE group was effectively upregulated by 1.41-fold compared to that in the NC-treated cells 72 h after plasmid transfection, while there was no difference between the OGD4/R20 group and NC group (Figure 8A).
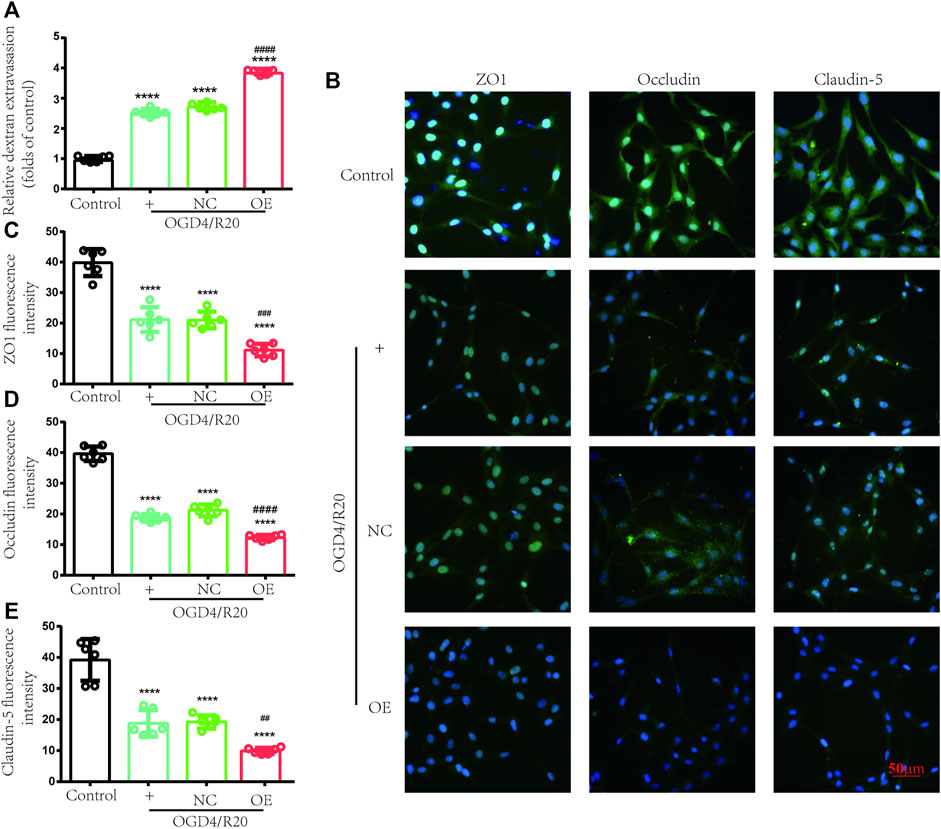
FIGURE 8. Increasing lncRNA ENST00000530525 deteriorated transendothelial permeability under OGD/R condition. (A) FITC-dextran leakage was used to quantificat transendothelial permeability. (B–E) Representative immunofluorescence indensity of ZO-1, occludin and claudin-5. Scale bar: 50 μm. NC: negative control; OE: over expression; OGD4/R20: 4 h of OGD and 20 h of reoxygenation. ****p < 0.0001 (vs. Control); #p < 0.001, ##p < 0.01; ###p < 0.001, ####p < 0.0001 (vs. NC), n = 6, one-way ANOVA.
Endothelial cells, which were connected by TJ proteins, are a major component of BBB. TJ proteins (ZO-1, occludin, and claudin-5) were evaluated by immunofluorescence to assess the state of BBB. After OGD4/R20, the fluorescence intensity of these proteins significantly decreased compared to the control group. Meanwhile, lncRNA ENST00000530525 overexpression futher decreased the fluorescence intensity of these proteins compared to the NC group (Figures 8B–E). These results also showed that lncRNA ENST00000530525 overexpression downregulated the expression of ZO-1, occludin and claudin-5 under OGD4/R20 condition.
LncRNA ENST00000530525 Not Directly Bind to 3′UTR of ANO1
In order to investigate whether lncRNA ENST00000530525 directly bind to 3′UTR of ANO1 and affect its expression, a double luciferase assay was used. Luc-ANO1-NC, Luc-ANO1-mimic, lncRNA-NC, and lncRNA-OE plasmids were constructed and transfected into 293T cells. After 48 h transfection, firefly luminescence and renilla luminescence were detected. However, there was no significant difference between Luc-ANO1-mimic group and Luc-ANO1-NC group when lncRNA ENST00000530525 was over-expressed (p < 0.05 but the difference was less than 20%), which indicated that lncRNA could not directly bind to the target gene ANO1 and affect its expression (Supplementary Figure S4).
Discussion
In our study, we uncovered lncRNA ENST00000530525 was downregulated after OGD4/R20, but ANO1 was upregulated. After IS, ANO1 was showed to be upregulated and to destroy the BBB integrity by regulating the NF-kB signaling pathway (Liu et al., 2019). This study was consistent with these researches. Notably, when lncRNA ENST00000530525 was upregulated by plasmid transfection, the RNA and protein expression of ANO1 was also increased, while the vitality of hCMEC/D3 cells and the BBB integrity were inhibited. This indicated that lncRNA ENST00000530525 might affect its neighbor gene ANO1 to play an important role in BBB integrity (Figure 9).
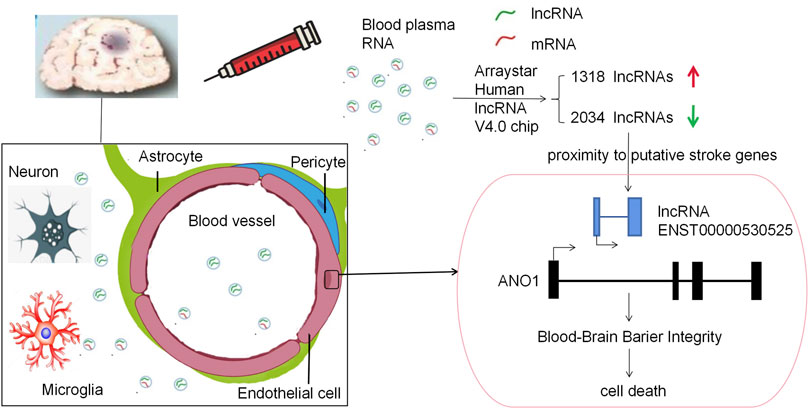
FIGURE 9. Schematic diagram of this paper. This study used a RNA-chip sequencing in whole blood samples of patients suffering from acute cerebral ischemia to identify DE lncRNAs. The Arraystar Human lncRNA V4.0 chip results showed there were 3352 lncRNA transcripts DE, with 1318 upregulated and 2034 downregulated lncRNAs. LncRNA ENST00000530525 was the highest downregulated lncRNA, while ANO1 was in proximity to the putative stroke gene. Upregulating lncRNA ENST00000530525 improved the expression of ANO1, which further destroyed the BBB integrity, in the hCMEC/D3 cells after OGD4/R20.
Stroke is a high risk factor for death and disability worldwide. However, the narrow time window for treatment of its standard therapy and the difficulty in making a quick diagnosis in most low-resource medical facilities (no brain imaging) indicates a need to understand the molecular regulation of stroke and the identification of its biomarkers. Such knowledge would enable the identification of biomarkers among the DE proteins and mRNAs as well as to provide new opinions to the mechanisms of stroke pathogenesis in patients (ENCODE Project Consortium, 2012; Falcione et al., 2020). Consequently, this study identified several stroke-induced changes in the expression of some circulating lncRNAs and their probable target genes.
Among the upregulated lncRNAs, NR_002312 at 14q11.2 is 45 kb away from poly (ADP-ribose) polymerase-2 (PARP2). PARP2 is responsive to DNA damage and drives cell death pathways in IS (Kofler et al., 2006). Additionally, lncRNA NR_037,932 (upregulated) was mapped to APOC2 and APOC4 within 420 kb and APOE within 462 kb at 19q13.32. APOE is central to the transport and metabolism of lipids and plays an important role in both cerebral IS and coronary heart disease (Satizabal et al., 2018). APOC4 correlates with stroke recovery, while APOC2 is involved in the pathophysiology of post-stroke depression (Zhan et al., 2014; Plubell et al., 2020). These associations may be key to the functions of lncRNAs in the evolution of stroke and can serve as potential biomarkers if pursued further. Another significant DE lncRNA, T297706, is within 6 kb of SIRT5 at 6p23, which increases blood–brain barrier permeability by degrading occludins (Diaz-Cañestro et al., 2018) and it promotes arterial thrombosis via endothelial PAI-1 expression (Liberale et al., 2020).
Among the downregulated lncRNAs, ENST00000395996 is within vacuolar protein sorting 13 homolog B (VPS13B). VPS13B participates in the pathoevolution of atherosclerosis-induced IS (Ruan et al., 2020). The HTT gene is less than 8 kb away from the downregulated lncRNA NR_045414, while the upregulated lncRNA is within the HTR2A gene. Both HTT and HTR2A are associated with post-stroke disease (Kim et al., 2012). The highly downregulated lncRNA ENST00000530525 is within the ANO1 gene. It is plausible that the lncRNA ENST00000530525 may affect the expression of the ANO1 gene after IS due to proximity. Our guess was identified by plasmid transfection assays, which mean upregulation of the lncRNA ENST00000530525 could influence the expression of ANO1. But the double luciferase assay result showed, the lncRNA ENST00000530525 could not directly bind to and regulate its neighbor gene ANO1.
Previous studies in rodents revealed the mechanisms of action of some of the dysregulated lncRNAs. There are four archetypes of molecular functions that lncRNAs execute: decoys, guides, signals, and scaffolds (Wang and Chang, 2011). For instance, cis-regulation, which means lncRNA can somehow bind to adjacent DNA in chromosomes and regulate its expression, is one kind of signal and plays a crucial role in stroke (Yan et al., 2017). LncRNA Peril acts in this manner to positively regulate the expression of two genes 1.5 million base pairs (Mb) away from its transcription location (Groff et al., 2018). Accordingly, it is possible that other cis-lncRNAs may regulate cerebral ischemic stroke genes in this way, as several significantly DE lncRNAs in our study mapped within or to the neighboring genes associated with IS (Table 3).
Recent studies have provided intriguing evidence for decoy lncRNAs. LncRNA U90926 binds directly to malate dehydrogenase 2 (MDH2), competitively inhibiting the binding of MDH2 to the untranslated region (UTR) of CXCL2, thereby protecting the mRNA of CXCL2 from MDH2-mediated attenuation (Chen et al., 2021). As for signal, the lncRNAs in this archetype can act as markers of functionally significant biological events. For example, HOTTIP, a lncRNA found at the distal end of the human HOXA cluster, directly binds the adaptor protein WDR5 and targets WDR5/MLL complexes across HOXA, driving histone H3 lysine 4 trimethylation and gene transcription (Wang et al., 2011). Another archetype of lncRNA is the guide—RNA binds proteins (usually transcription factors), then directs the protein complex to specific targets. Engreitz found that 5 of 12 lncRNA loci regulate their neighboring gene transcription. In most of these lncRNA, local effects are mediated by enhancer-like functions of DNA elements, while in one locus perhaps by recruit more transcription-associated factors (Engreitz et al., 2016). In conclusion, the lncRNA ENST00000530525 may recruit transcription factors or enhancers to promote ANO1 gene transcription. In this study, the mechanism by which lncRNA ENST00000530525 affects its target gene ANO1 requires further research.
There are still some problems being worthy of consideration and in-depth exploration in this study. The basic disease of stroke patients is complex and diverse in clinical, the incidence of ischemia stroke is common in the middle-aged and elderly who have high-risk factors (such as Hypertension, diabetes, and hyperlipidemia). The race, sex, age, and vascular risk factors (diabetes mellitus, hypertension, and hyperlipidemia) were matching of all patients enrolled in this study and there were significantly more stroke patients with a positive history of hypertension, diabetes, and smoking in Table 1, the expression of the novel lncRNA ENST00000530525 in stroke patients without risk factors has not been verified due to the limitation of sample, However, we are randomly sampling 40 blood samples from stroke patients and 20 blood samples from controls to detect the expression trend of lncRNA ENST00000530525, which is downregulated in stroke group compared with the control group (See Supplementary Figure S1). Moreover, the gene chip results were gotten from patients’ blood, but our further validation used hCMEC/D3 cells. It is not clear how the reduced circulating levels of lncRNA in patients’ blood is linked to the downregulation of its expression in the hCMEC/D3 cells, and the relationship between them needs to be further studied. In addition, there are varying times of BBB permeability detection according to the literatures. In Cowan’s study, FITC-dextran was added to the inserts, which were then transferred every 5 min over 30 min to a series of collecting wells to measure the BBB permeability (Cowan and Easton, 2010). Gerhartl used FITC-dextran to assess BBB permeability after transferring 30 min (Gerhartl et al., 2020). While in Liu’s study, the BBB permeability test is performed for 20 min (Liu et al., 2019). There were still other researchers added FITC-Dextran the upper chamber and then incubated for 1 h to test (Ni et al., 2017; Bergman et al., 2021). BBB permeability test just performed for 5 min in our study, the test time may need further verification. In this study, if the expression of ANO1 and cell viability under upregulating and downregulating lncRNA conditions by transfection, and also ANO1 expression in lncRNA overexpressed cells under normal conditions (not just OGD/R pathological condition) were investigated, the conclusion would be more convincing.
Due to the relatively recent discoveries, the expression and functions of the majority of lncRNAs in the post-stroke brain are largely unknown. However, numerous lncRNAs are emerging as important regulators of transcription and translation. These findings of dysregulated lncRNAs may help to guide evidence-based preventive measures and the search for a cure. Further studies will be needed to establish whether these remaining lncRNAs can modulate theis neighboring and stroke-associated genes in vivo or in vitro.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GEO—GSE198710
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of Guangdong Provincial Hospital of Chinese Medicine Guangdong Provincial Hospital of Chinese Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
WJ performed the experiments, data statistics, and was a major contributor in writing the manuscript. JL and YC participated in drafting and revised the manuscript. WL analyzed and interpreted the patient data regarding the stroke patients. MC, XX, MD, and WS carried out experiments and analyzed the data. JS, LZ, YH, and XC supervised the design of the study and reviewed this manuscript. All authors reviewed and approved the final version of this paper. All authors read and approved the final manuscript.
Funding
This study was financially supported by the State key R & D Program (No. 2019YFC1708601), the National Natural Science Foundation of China, Nos. 81303115, 81774042, 81771353, and 81904104; the Pearl River S & T Nova Program of Guangzhou, No. 201806010025; the Youth Pilot Project of Chinese Society of Traditional Chinese Medicine, No. CACM-2018-QNRC2-C09; the Science and Technology Program of Guangzhou City of China, Nos. 201604020003 and 202102010268; Guangdong Provincial Key Laboratory of Research on Emergency in TCM, No. 2017B030314176; 2018-75 and 2019-140; the Administration of Traditional Chinese Medicine of Guangdong Province, No. 20211203; Postdoctoral Science Foundation of China, No. 2021M690759. Guangzhou Science and Technology Project (Nos. 202102010268 and 202102010339). The Specialty Program of Guangdong Province Hospital of Traditional Chinese Medicine of China, Nos. YN2016MJ07, 2015KT1294, YN2015MS02, and YN2018ZD07.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank AJE (https://www.aje.cn/) for editing this manuscript. The content of this manuscript has previously appeared online in a preprint. Available at: https://www.researchsquare.com/article/rs-968571/v1 (Accessed 10 January 2022).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.873230/full#supplementary-material
Abbreviations
ANO1, Anoctamin-1; BBB, Blood-brain barrier; CCK-8, cell counting kit-8; CT, Computed tomographic; CFTR, Cystic fibrosis transmembrane conductance regulator; CSF, Codon substitution frequency; DE, Differential expressed; ECG, Electrocardiograph; FE, Feature Extraction; FISH, Fluorescent in situ hybridization; IS, Ischemic stroke; lncRNAs, Long noncoding RNAs; NC, Negative control; OE, Over-expression; ORF, Open reading frame; OGD/R, Oxygen-glucose deprivation/reoxygenation; RT-qPCR, Quantitative real-time polymerase chain reaction; TMEM16A, Transmembrane protein 16A; TJ, Tight junction.
References
Abdillahi, M., Ananthakrishnan, R., Vedantham, S., Shang, L., Zhu, Z., Rosario, R., et al. (2012). Aldose Reductase Modulates Cardiac Glycogen Synthase Kinase-3β Phosphorylation during Ischemia-Reperfusion. Am. J. Physiology-Heart Circulatory Physiology 303 (3), H297–H308. doi:10.1152/ajpheart.00999.2011
Akella, A., Bhattarai, S., and Dharap, A. (2019). Long Noncoding RNAs in the Pathophysiology of Ischemic Stroke. Neuromol Med. 21 (4), 474–483. doi:10.1007/s12017-019-08542-w
Bao, M.-H., Szeto, V., Yang, B. B., Zhu, S.-z., Sun, H.-S., and Feng, Z.-P. (2018). Long Non-coding RNAs in Ischemic Stroke. Cell Death Dis. 9 (3), 281. doi:10.1038/s41419-018-0282-x
Bergman, L., Acurio, J., Leon, J., Gatu, E., Friis, T., Nelander, M., et al. (2021). Preeclampsia and Increased Permeability over the Blood-Brain Barrier: A Role of Vascular Endothelial Growth Receptor 2. Am. J. Hypertens. 34 (1), 73–81. doi:10.1093/ajh/hpaa142
Chen, J., Jin, J., Zhang, X., Yu, H., Zhu, X., Yu, L., et al. (2021). Microglial Lnc-U90926 Facilitates Neutrophil Infiltration in Ischemic Stroke via MDH2/CXCL2 axis. Mol. Ther. 29 (9), 2873–2885. doi:10.1016/j.ymthe.2021.04.025
Cowan, K. M., and Easton, A. S. (2010). Neutrophils Block Permeability Increases Induced by Oxygen Glucose Deprivation in a Culture Model of the Human Blood-Brain Barrier. Brain Res. 1332, 20–31. doi:10.1016/j.brainres.2010.03.066
Deng, L., Yang, J., Chen, H., Ma, B., Pan, K., Su, C., et al. (2016). Knockdown of TMEM16A Suppressed MAPK and Inhibited Cell Proliferation and Migration in Hepatocellular Carcinoma. Onco Targets Ther. 9, 325–333. doi:10.2147/OTT.S95985
Dharap, A., Nakka, V. P., and Vemuganti, R. (2012). Effect of Focal Ischemia on Long Noncoding RNAs. Stroke 43 (10), 2800–2802. doi:10.1161/strokeaha.112.669465
Diaz-Cañestro, C., Merlini, M., Bonetti, N. R., Liberale, L., Wüst, P., Briand-Schumacher, S., et al. (2018). Sirtuin 5 as a Novel Target to Blunt Blood-Brain Barrier Damage Induced by Cerebral Ischemia/reperfusion Injury. Int. J. Cardiol. 260, 148–155. doi:10.1016/j.ijcard.2017.12.060
Engreitz, J. M., Haines, J. E., Perez, E. M., Munson, G., Chen, J., Kane, M., et al. (2016). Local Regulation of Gene Expression by lncRNA Promoters, Transcription and Splicing. Nature 539 (7629), 452–455. doi:10.1038/nature20149
Falcione, S., Kamtchum-Tatuene, J., Sykes, G., and Jickling, G. C. (2020). RNA Expression Studies in Stroke: what Can They Tell Us about Stroke Mechanism? Curr. Opin. neurology 33 (1), 24–29. doi:10.1097/wco.0000000000000786
Fan, X.-D., Yao, M.-J., Yang, B., Han, X., Zhang, Y.-H., Wang, G.-R., et al. (2021). Chinese Herbal Preparation SaiLuoTong Alleviates Brain Ischemia via Nrf2 Antioxidation Pathway-dependent Cerebral Microvascular Protection. Front. Pharmacol. 12, 748568. doi:10.3389/fphar.2021.748568
Gerhartl, A., Pracser, N., Vladetic, A., Hendrikx, S., Friedl, H.-P., and Neuhaus, W. (2020). The Pivotal Role of Micro-environmental Cells in a Human Blood-Brain Barrier In Vitro Model of Cerebral Ischemia: Functional and Transcriptomic Analysis. Fluids Barriers CNS 17 (1), 19. doi:10.1186/s12987-020-00179-3
Groff, A. F., Barutcu, A. R., Lewandowski, J. P., and Rinn, J. L. (2018). Enhancers in the Peril lincRNA Locus Regulate Distant but Not Local Genes. Genome Biol. 19 (1), 219. doi:10.1186/s13059-018-1589-8
ENCODE Project Consortium (2012). An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 489(7414), 57–74. doi:10.1038/nature11247
Jiang, X., Andjelkovic, A. V., Zhu, L., Yang, T., Bennett, M. V. L., Chen, J., et al. (2018). Blood-brain Barrier Dysfunction and Recovery after Ischemic Stroke. Prog. Neurobiol. 163-164, 144–171. doi:10.1016/j.pneurobio.2017.10.001
Kim, J.-M., Stewart, R., Bae, K.-Y., Kim, S.-W., Kang, H.-J., Shin, I.-S., et al. (2012). Serotonergic and BDNF Genes and Risk of Depression after Stroke. J. Affect. Disord. 136 (3), 833–840. doi:10.1016/j.jad.2011.09.029
Kofler, J., Otsuka, T., Zhang, Z., Noppens, R., Grafe, M. R., Koh, D. W., et al. (2006). Differential Effect of PARP-2 Deletion on Brain Injury after Focal and Global Cerebral Ischemia. J. Cereb. Blood Flow. Metab. 26 (1), 135–141. doi:10.1038/sj.jcbfm.9600173
Li, Q., Wu, H., Yue, W., Dai, Q., Liang, H., Bian, H., et al. (2017). Prevalence of Stroke and Vascular Risk Factors in China: a Nationwide Community-Based Study. Sci. Rep. 7 (1), 6402. doi:10.1038/s41598-017-06691-1
Liberale, L., Akhmedov, A., Vlachogiannis, N. I., Bonetti, N. R., Nageswaran, V., Miranda, M. X., et al. (2020). Sirtuin 5 Promotes Arterial Thrombosis by Blunting the Fibrinolytic System. Cardiovasc. Res. 117 (10), 2275–2288. doi:10.1093/cvr/cvaa268
Liu, C., Yang, J., Zhang, C., Liu, M., Geng, X., Ji, X., et al. (2018). Analysis of Long Non-coding RNA Expression Profiles Following Focal Cerebral Ischemia in Mice. Neurosci. Lett. 665, 123–129. doi:10.1016/j.neulet.2017.11.058
Liu, P.-y., Zhang, Z., Liu, Y., Tang, X.-l., Shu, S., Bao, X.-y., et al. (2019). TMEM16A Inhibition Preserves Blood-Brain Barrier Integrity after Ischemic Stroke. Front. Cell. Neurosci. 13, 360. doi:10.3389/fncel.2019.00360
Liu, Y., Liu, Z., and Wang, K. (2021). The Ca2+-Activated Chloride Channel ANO1/TMEM16A: An Emerging Therapeutic Target for Epithelium-Originated Diseases? Acta Pharm. Sin. B 11 (6), 1412–1433. doi:10.1016/j.apsb.2020.12.003
Lo, A. C., Cheung, A. K., Hung, V. K., Yeung, C.-M., He, Q.-Y., Chiu, J.-F., et al. (2007). Deletion of Aldose Reductase Leads to Protection against Cerebral Ischemic Injury. J. Cereb. Blood Flow. Metab. 27 (8), 1496–1509. doi:10.1038/sj.jcbfm.9600452
Ma, K., Liu, S., Liang, H., Wang, G., Wang, T., Luo, S., et al. (2021). Ca2+-activated Cl− Channel TMEM16A Inhibition by Cholesterol Promotes Angiogenesis in Endothelial Cells. J. Adv. Res. 29, 23–32. doi:10.1016/j.jare.2020.09.003
Ni, Y., Teng, T., Li, R., Simonyi, A., Sun, G. Y., and Lee, J. C. (2017). TNFα Alters Occludin and Cerebral Endothelial Permeability: Role of p38MAPK. PLoS One 12 (2), e0170346. doi:10.1371/journal.pone.0170346
Plubell, D. L., Fenton, A. M., Rosario, S., Bergstrom, P., Wilmarth, P. A., Clark, W. M., et al. (2020). High-Density Lipoprotein Carries Markers that Track with Recovery from Stroke. Circ. Res. 127 (10), 1274–1287. doi:10.1161/circresaha.120.316526
Qu, L., Ding, J., Chen, C., Wu, Z.-J., Liu, B., Gao, Y., et al. (2016). Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 29 (5), 653–668. doi:10.1016/j.ccell.2016.03.004
Ren, W., and Yang, X. (2018). Pathophysiology of Long Non-coding RNAs in Ischemic Stroke. Front. Mol. Neurosci. 11, 96. doi:10.3389/fnmol.2018.00096
Ruan, W., Wu, J., Su, J., Jiang, Y., Pang, T., and Li, J. (2020). Altered lncRNAs Transcriptomic Profiles in Atherosclerosis-Induced Ischemic Stroke. Cell Mol. Neurobiol. 42 (1), 265–278. doi:10.1007/s10571-020-00918-y
Satizabal, C. L., Samieri, C., Davis-Plourde, K. L., Voetsch, B., Aparicio, H. J., Pase, M. P., et al. (2018). APOE and the Association of Fatty Acids with the Risk of Stroke, Coronary Heart Disease, and Mortality. Stroke 49 (12), 2822–2829. doi:10.1161/strokeaha.118.022132
Schaukowitch, K., and Kim, T.-K. (2014). Emerging Epigenetic Mechanisms of Long Non-coding RNAs. Neuroscience 264, 25–38. doi:10.1016/j.neuroscience.2013.12.009
Shin, J. A., Yoon, J. C., Kim, M., and Park, E.-M. (2016). Activation of Classical Estrogen Receptor Subtypes Reduces Tight Junction Disruption of Brain Endothelial Cells under Ischemia/reperfusion Injury. Free Radic. Biol. Med. 92, 78–89. doi:10.1016/j.freeradbiomed.2016.01.010
Suzuki, T., Suzuki, Y., Asai, K., Imaizumi, Y., and Yamamura, H. (2021). Hypoxia Increases the Proliferation of Brain Capillary Endothelial Cells via Upregulation of TMEM16A Ca2+-Activated Cl− Channels. J. Pharmacol. Sci. 146 (1), 65–69. doi:10.1016/j.jphs.2021.03.002
Suzuki, T., Yasumoto, M., Suzuki, Y., Asai, K., Imaizumi, Y., and Yamamura, H. (2020). TMEM16A Ca2+-Activated Cl- Channel Regulates the Proliferation and Migration of Brain Capillary Endothelial Cells. Mol. Pharmacol. 98 (1), 61–71. doi:10.1124/mol.119.118844
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING V10: Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 43 (Database issue), D447–D452. doi:10.1093/nar/gku1003
Wang, K. C., and Chang, H. Y. (2011). Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 43 (6), 904–914. doi:10.1016/j.molcel.2011.08.018
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A Long Noncoding RNA Maintains Active Chromatin to Coordinate Homeotic Gene Expression. Nature 472 (7341), 120–124. doi:10.1038/nature09819
Wang, M., Yang, H., Zheng, L.-Y., Zhang, Z., Tang, Y.-B., Wang, G.-L., et al. (2012). Downregulation of TMEM16A Calcium-Activated Chloride Channel Contributes to Cerebrovascular Remodeling during Hypertension by Promoting Basilar Smooth Muscle Cell Proliferation. Circulation 125 (5), 697–707. doi:10.1161/circulationaha.111.041806
Wang, W., Jiang, B., Sun, H., Ru, X., Sun, D., Wang, L., et al. (2017). Prevalence, Incidence, and Mortality of Stroke in China. Circulation 135 (8), 759–771. doi:10.1161/circulationaha.116.025250
Wu, M. M., Lou, J., Song, B. L., Gong, Y. F., Li, Y. C., Yu, C. J., et al. (2014). Hypoxia Augments the Calcium‐activated Chloride Current Carried by Anoctamin‐1 in Cardiac Vascular Endothelial Cells of Neonatal Mice. Br. J. Pharmacol. 171 (15), 3680–3692. doi:10.1111/bph.12730
Xu, S. y., Bian, H. j., Shu, S., Xia, S. n., Gu, Y., Zhang, M. j., et al. (2021). AIM2 Deletion Enhances Blood‐brain Barrier Integrity in Experimental Ischemic Stroke. CNS Neurosci. Ther. 27 (10), 1224–1237. doi:10.1111/cns.13699
Yan, P., Luo, S., Lu, J. Y., and Shen, X. (2017). Cis- and Trans-acting lncRNAs in Pluripotency and Reprogramming. Curr. Opin. Genet. Dev. 46, 170–178. doi:10.1016/j.gde.2017.07.009
Yates, A. D., Achuthan, P., Akanni, W., Allen, J., Allen, J., Alvarez-Jarreta, J., et al. (2020). Ensembl 2020. Nucleic acids Res. 48 (D1), D682–8. doi:10.1093/nar/gkz966
Keywords: ischemic stroke, ANO1, lncRNA, ENST00000530525, oxygen-glucose deprivation and reperfusion, human cerebrovascular endothelial cells
Citation: Jiang W, Li J, Cai Y, Liu W, Chen M, Xu X, Deng M, Sun J, Zhou L, Huang Y, Wu S and Cheng X (2022) The Novel lncRNA ENST00000530525 Affects ANO1, Contributing to Blood–Brain Barrier Injury in Cultured hCMEC/D3 Cells Under OGD/R Conditions. Front. Genet. 13:873230. doi: 10.3389/fgene.2022.873230
Received: 10 February 2022; Accepted: 19 May 2022;
Published: 08 June 2022.
Edited by:
William C. Cho, QEH, Hong Kong SAR, ChinaReviewed by:
Malgorzata Burek, Julius Maximilian University of Würzburg, GermanyRavi K. Sajja, Nelson Laboratories, United States
Abraham Jacob Al-Ahmad, Jerry H. Hodge School of Pharmacy, United States
Copyright © 2022 Jiang, Li, Cai, Liu, Chen, Xu, Deng, Sun, Zhou, Huang, Wu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Cheng, Y2hlbmd4aWFvbHVja3lAMTI2LmNvbQ==
 Wen Jiang
Wen Jiang Jie Li
Jie Li Yuefang Cai
Yuefang Cai Wenchen Liu
Wenchen Liu Mei Chen
Mei Chen Xiaoying Xu
Xiaoying Xu Minzhen Deng
Minzhen Deng Jingbo Sun
Jingbo Sun Lihua Zhou
Lihua Zhou Yan Huang
Yan Huang Shuang Wu
Shuang Wu Xiao Cheng
Xiao Cheng