- 1Department of Agricultural and Environmental Biology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
- 2Faculty of Bioscience and Applied Chemistry, Hosei University, Tokyo, Japan
Phytoplasmas are obligate intracellular plant pathogenic bacteria that can induce phyllody, which is a type of abnormal floral organ development. Phytoplasmas possess phyllogens, which are effector proteins that cause phyllody in plants. Phylogenetic comparisons of phyllogen and 16S rRNA genes have suggested that phyllogen genes undergo horizontal transfer between phytoplasma species and strains. However, the mechanisms and evolutionary implications of this horizontal gene transfer are unclear. Here, we analyzed synteny in phyllogen flanking genomic regions from 17 phytoplasma strains that were related to six ‘Candidatus’ species, including three strains newly sequenced in this study. Many of the phyllogens were flanked by multicopy genes within potential mobile units (PMUs), which are putative transposable elements found in phytoplasmas. The multicopy genes exhibited two distinct patterns of synteny that correlated with the linked phyllogens. The low level of sequence identities and partial truncations found among these phyllogen flanking genes indicate that the PMU sequences are deteriorating, whereas the highly conserved sequences and functions (e.g., inducing phyllody) of the phyllogens suggest that the latter are important for phytoplasma fitness. Furthermore, although their phyllogens were similar, PMUs in strains related to ‘Ca. P. asteris’ were often located in different regions of the genome. These findings strongly indicate that PMUs drive the horizontal transfer of phyllogens among phytoplasma species and strains. These insights improve our understanding of how symptom-determinant genes have been shared among phytoplasmas.
Introduction
Gene acquisition by horizontal gene transfer (HGT) plays a crucial role in the adaptive evolution of most organisms (Keeling and Palmer, 2008; Soucy et al., 2015). In bacteria, HGT is generally mediated by transposable elements (e.g., phages, plasmids, and transposons) that can travel between host bacteria, as well as by homologous recombination between genomes (Soucy et al., 2015; Drew et al., 2021). Horizontal transfer of single genes or genomic islands can influence bacterial phenotypes and pathogenicity (Melnyk et al., 2019; Gluck-Thaler et al., 2020; Drew et al., 2021). In the genus Rhizobium, there is a plasmid carrying genes that can influence root nodulation transfer among different species and determines the host range for root nodule induction (Hooykaas et al., 1982). In the family Xanthomonadaceae, colonization of plant vascular tissues prior to systemic infection was associated with horizontal transfer of a single gene, cbsA (Gluck-Thaler et al., 2020). The mode of HGT-mediated acquisition of pathogenic genes may have profound implications for pathogenic microbial evolution. These HGTs are mainly inferred using parametric or phylogenetic methods (Ravenhall et al., 2015).
Phytoplasmas [‘Candidatus (Ca.) Phytoplasma’ spp.] are obligate intracellular plant pathogenic bacteria in the class Mollicutes. Phytoplasmas are transmitted by phloem-feeding insects of the order Hemiptera and can infect more than 1,000 plant species (Marcone, 2014). Although phytoplasma genomes are small and lack many metabolic genes (Oshima et al., 2004), multicopy genes account for 18%–28% of all phytoplasma genes (Oshima et al., 2004; Bai et al., 2006; Kube et al., 2008; Tran-Nguyen et al., 2008). Most of these multicopy genes occur in gene clusters called potential mobile units (PMUs; Bai et al., 2006; Arashida et al., 2008a; Wei et al., 2008). PMUs are often associated with tra5, which is a putative transposable gene belonging to the IS3 family, among other genes such as fliA, ssb, dam, himA, hflB, smc, tmk, dnaB, and dnaG (Arashida et al., 2008a). Some PMUs have been reported to transfer in phytoplasma genomes and to exist as probably transposable circular extrachromosomal elements (Arashida et al., 2008a; Toruño et al., 2010; Ku et al., 2013). In addition, PMUs can reportedly undergo HGT between phytoplasma genomes, which mediates the acquisition of novel genes by phytoplasmas (Chung et al., 2013; Ku et al., 2013; Music et al., 2019). Although many PMUs have lost their capacity for HGT due to the deletion of genes such as tra5, they are still described as PMUs in many studies, including this one.
Phytoplasma diseases are associated with unique symptoms such as dwarfing, witches’ broom, yellowing, and phyllody (Namba et al., 1993). These symptoms are mainly induced by effector proteins secreted by the phytoplasmas (Hoshi et al., 2009; MacLean et al., 2011; Sugio et al., 2011; Minato et al., 2014; Huang et al., 2021). Phytoplasmas have effectors that cause different disease symptoms. Many of the secreted effector proteins are encoded by PMUs, which phytoplasmas may have acquired by HGT (Sugio and Hogenhout, 2012; Ku et al., 2013). However, to understand the genetics of phytoplasma pathogenicity, it is necessary to confirm that the acquisition of these effectors is associated with HGT of PMUs.
Phyllogens are effector family proteins unique to phytoplasmas that induce phyllody in various eudicots (MacLean et al., 2011; Maejima et al., 2014; Yang et al., 2015; Kitazawa et al., 2017; Iwabuchi et al., 2020). The conserved molecular mechanisms responsible for inducing phyllody have been elucidated (MacLean et al., 2014; Maejima et al., 2014; Kitazawa et al., 2022). Phyllogen genes (phyllogens) can be phylogenetically divided into four groups (i.e., phyl-A, -B, -C, and -D) and the evolutionary history of these phyllogens differs from that of phytoplasmas, as confirmed by analyses of phytoplasma 16S rRNA gene sequences (Iwabuchi et al., 2020; Kalla et al., 2021). Indeed, different groups of phyllogens have been found in the same ‘Ca. Phytoplasma’ sp. These findings suggest that phyllogens have undergone HGT among various phytoplasma species, thereby enabling these phytoplasmas to induce phyllody (Iwabuchi et al., 2020).
However, gene sequence information is insufficient to establish the evolutionary history of all genes and reveal the mechanism of HGT; therefore, a better understanding of the structure of the gene flanking regions may be necessary (Melnyk et al., 2019; Gluck-Thaler et al., 2020). Phyllogens are often found within PMUs (Jomantiene et al., 2007; Sugio and Hogenhout, 2012; Maejima et al., 2014; Wang et al., 2018; Huang et al., 2022); therefore, HGT of phyllogens may be mediated by PMUs (Iwabuchi et al., 2020). However, phyllogen flanking PMUs have only been analyzed in a few phytoplasma strains (Sugio and Hogenhout, 2012; Wang et al., 2018; Huang et al., 2022), and most phyllogen flanking regions have not been studied in detail.
In this study, we analyzed the sequences of phyllogen flanking regions from various phytoplasma species and strains, and evaluated similarities to infer the mechanisms and evolutionary history of phyllogen acquisition. Sequencing and comparison of phyllogen flanking regions revealed synteny among PMU-associated genes for each group of phyllogens, indicating that horizontal transfer of PMUs was associated with phyllogen acquisition. Furthermore, HGT events were followed by hypermutation, pseudogenesis, and deletion of PMU-associated genes, whereas the sequences of phyllogens were highly conserved.
Materials and methods
Phytoplasma DNA
DNA samples from ‘Ca. P. asteris’ HP, RhY, and PaWB-Japan strains, and from ‘Ca. P. phoenicium’ PEY were extracted from infected Hydrangea spp. (Takinami et al., 2013), Rhus javanica (Takinami et al., 2013), Paulownia tomentosa (Kakizawa et al., 2006), and periwinkle (Maejima et al., 2014), respectively.
Identification of genomic sequences flanking phyllogens
The phytoplasma species and strains used in this study are listed in Supplementary Table S1. Contigs of phyllogen sequences were created from draft genomes of the HP, RhY, and PaWB-Japan strains sequenced in this study (see Supplementary Methods), as well as from phytoplasma genomic sequences deposited in GenBank (Supplementary Table S1); we performed tBLASTn searches using amino acid sequences from phyllogen homologs (Iwabuchi et al., 2020). To determine upstream sequences for the RhY, PaWB-Japan, and PEY strain contigs, we performed polymerase chain reaction (PCR) using primer pairs that targeted each contig and conserved regions of phytoplasma fliA, which is frequently found in the first or second open reading frame (ORF) of PMUs (Arashida et al., 2008a; Wang et al., 2018; Supplementary Table S2). The flanking regions of the RhY and PaWB-Japan strain contigs were determined by PCR using primer pairs designed to match the corresponding regions of the HP and PaWB-China genomes, respectively (Supplementary Table S2). The downstream region for the PEY strain was determined by PCR using primer pairs that targeted the phyllogen sequence and dnaG sequence in the AY-WB strain PMU (Supplementary Table S2). PCR was performed using LA Taq (TaKaRa Bio, Inc., Shiga, Japan) and 0.3 µM of each primer, in accordance with the manufacturer’s instructions. Amplified fragments (>1.5 kbp) were purified and sequenced using PCR primers, followed by primer walking. Amplified fragments (<1.5 kbp) were purified, cloned into a pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA, United States), and sequenced by Sanger sequencing. DNA samples from phytoplasma strains containing phyl-A group phyllogens (Supplementary Table S1) were subjected to PCR amplification of the region between hflB-like genes with similarity to hflB and a region downstream of phyllogens using primers described previously (Jomantiene et al., 2007; Supplementary Table S2). The phyllogen flanking sequences were added to the draft genomes and deposited in DDBJ under the accession numbers listed in Supplementary Table S1.
Comparative analysis of phyllogen flanking regions
The genomic regions that flanked the phyllogens were annotated for protein coding genes using MetaGeneAnnotator (Noguchi et al., 2008) in DFAST (Tanizawa et al., 2018) and analyzed using BLASTp searches restricted to sequences >8,000 bp (Altschul et al., 1990; Camacho et al., 2009). Homologous genes were identified when the E-value, identity, and query-cover between two genes were < 1e−6, >30%, and >75%, respectively. To identify genomic locations of phyllogen flanking regions in strains related to ‘Ca. P. asteris,’ genomic regions were subjected to comparative analyses with other genomes of strains related to ‘Ca. P. asteris’ using BLASTp. The results of these comparative analyses were displayed using Clinker ver. 0.0.21 software to enable gene cluster comparisons (Gilchrist and Chooi., 2021). Pairwise sequence identities were calculated using the Sequence Demarcation Tool (SDT; ver. 1.2) (Muhire et al., 2014). Pseudogenes split into multiple ORFs due to premature stop codons were identified by combining the ORFs.
Alignment and phylogenetic analyses
The nucleotide sequences of phyllogen, fliA, himA, and hflB were picked up from the list in Supplementary Table S3 and were aligned using the MUSCLE algorithm in MEGA X software (Kumar et al., 2018). The neighbor-joining (NJ) method (Saitou and Nei, 1987) was used to create phylogenetic trees of these genes using MEGA X software (Kumar et al., 2018). The whole/draft genome-based phylogenetic analysis of phytoplasma strains related to ‘Ca. P. asteris’, except for NJAY strain (GenBank accession number MAPF00000000; completeness of only 91%) and TW strain (GenBank accession number QGKT00000000; draft genome reportedly contaminated with two different strains; Cho et al., 2020), was performed as follows. Single-copy genes shared by the ‘Ca. P. asteris’-related phytoplasma genomes (Supplementary Table S3) were identified using SonicParanoid software ver. 1.3.5 (Cosentino and Iwasaki, 2019). The identified homologs were aligned as described above. Aligned sequences were combined for each strain in the same order, and were subjected to phylogenetic analyses using the NJ method as described above.
In planta expression of phyllogen
A modified tobacco rattle virus (TRV)-based gene expression vector system (Iwabuchi et al., 2019) was used to express phyllogen in planta. In brief, the PHYLRP166 and PHYLNCHU2019 sequences were optimized for plant codons and synthesized by ThermoFisher Scientific (Waltham, MA, United States; Supplementary Table S4). The fragments were cloned into pTRV2-2A-sGFP with the primers shown in Supplementary Table S2 by replacing the sGFP region using the NEBuilder HiFi DNA Assembly Cloning kit (New England Biolabs, Ipswich, MA, United States). PHYLHP and PHYLOYE33 were cloned with the primers shown in Supplementary Table S2 by adding a single amino acid mutation into pTRV2-cloned PHYLJWB (Iwabuchi et al., 2020) and PHYLOY (Iwabuchi et al., 2019), respectively, using the GeneArt site-directed mutagenesis system (Invitrogen). PHYLPEY was cloned with the primers shown in Supplementary Table S2 by incorporating a two-amino acid mutation into pTRV2-cloned PHYLRP166, amplifying the insert by PCR, and amplifying the rest of the plasmid by inverse PCR. The amplified fragments were ligated using the NEBuilder HiFi DNA Assembly Cloning kit (New England Biolabs). Arabidopsis thaliana was maintained as described previously (Iwabuchi et al., 2020). Agrobacterium tumefaciens EHA105 cells containing pTRV1 and pTRV2-empty, pTRV2-PHYLOY, pTRV2-PHYLRP166, pTRV2-PHYLPEY, pTRV2-PHYLNCHU2019, pTRV2-PHYLHP, pTRV2-PHYLJWB, or pTRV2-PHYLOYE33 were adjusted to an OD600 of 0.1, mixed at a ratio of 1:1, and co-infiltrated into 2- and 3-week-old A. thaliana as described previously (Takahashi et al., 2006).
Results
The phylogeny of phyllogens was consistent with the synteny of the PMUs
To understand the mechanisms involved in the HGT of phyllogens across phytoplasmas, we compared phyllogen flanking regions among the genomes of 17 phytoplasma strains related to six species (Figure 1; Supplementary Table S1). Strains harboring the phyl-B group phyllogens were not analyzed because the corresponding genomes have not been characterized. Prior to this analysis, we generated draft genome data for the ‘Ca. P. asteris’ HP, RhY, and PaWB-Japan strains (Supplementary Figure S1; Supplementary Table S5). The draft genome data were used to generate full-length phyllogen sequences for the RhY and PaWB-Japan strains, as well as sequences for phyllogen flanking regions from the HP, RhY, and PaWB-Japan strains. The detailed results about genome sequencing were described in Supplementary Results. The phyllogen flanking regions for the ‘Ca. P. phoenicium’ PEY strains were also determined using PCR. Phylogenetic analyses of phyllogens showed that the genes of the 17 strains could be separated into three clades (Figure 1A) corresponding to the phyl-A (6 strains), phyl-C (1 strains), and phyl-D groups (10 strains) described by a previous study (Iwabuchi et al., 2020); the RhY and PaWB-Japan phyllogens belong to the phyl-D group. In most phyllogen flanking regions (all phyl-A and phyl-C, and seven phyl-D group phyllogens), there were several PMU-associated genes (fliA, ssb, dam, himA, hflB, smc, tmk, dnaB, dnaG, and tra5; Figure 1B; Arashida et al., 2008a). This observation is consistent with previous reports of phytoplasma strains with phyllogens within PMUs (Sugio and Hogenhout, 2012; Maejima et al., 2014).
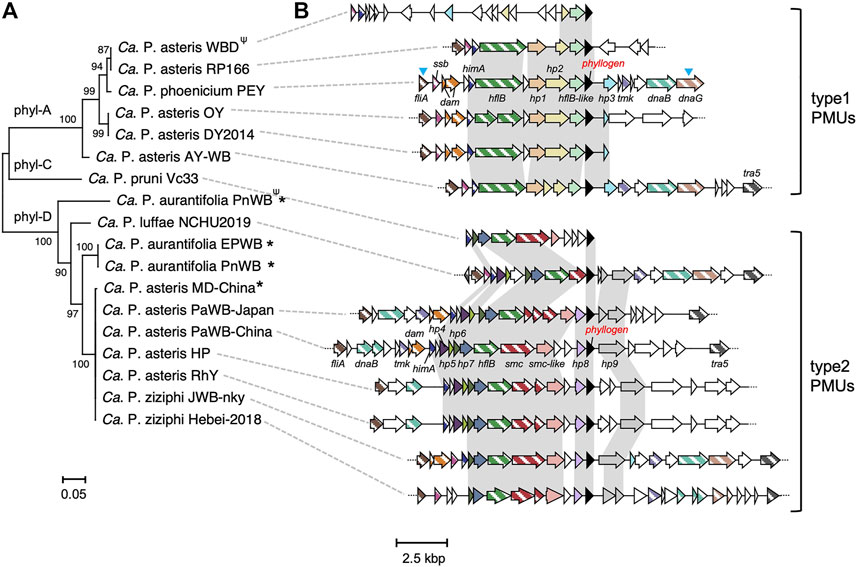
FIGURE 1. Comparative analysis of phyllogen flanking regions. (A) Neighbor-joining phylogenetic tree for phyllogens with known flanking regions of >8,000 bp, analyzed with the complete deletion option. The MUSCLE multiple alignment algorithm (Kumar et al., 2018) was used to align the phyllogen nucleotide sequences. Numbers at the nodes represent bootstrap values for 1,000 replicates (only values >70% are shown). Phylogenetic groups (Iwabuchi et al., 2020) are indicated on the tree branches. The scale bar indicates the number of nucleotide substitutions per site. For phyllogens truncated at the C-terminus due to premature stop codons (indicated by Ψ), the nucleotide regions after the stop codons were also included in the MUSCLE alignment (Kumar et al., 2018). Phytoplasmas marked with asterisks had phyllogens that were not located within PMUs. The ‘Candidatus P. ziziphi’ JWB-nky and ‘Ca. P. luffae’ NCHU2019 strains had several phyllogens with identical sequences; one gene is shown. Full strain names and GenBank accession numbers are listed in Supplementary Table S1. (B) Genomic structure of the phyllogen flanking regions. Homologous genes present in at least five strains are shown in the same color. Phyllogens are shown in black. Diagonal stripe patterns indicate multicopy genes associated with PMUs (Arashida et al., 2008a). The different types of PMU described in this study are indicated on the right. Syntenies unique to the type 1 or type 2 PMUs are highlighted against a light gray background. Open reading frames (ORFs) marked with blue arrowheads have not been completely sequenced. Broken lines at the ends of genomic structures indicate that these regions have been sequenced but are not shown in the figure. The gene names correspond to locus tags in draft genomes (Supplementary Table S8).
Except for those from the WBD strain, phyllogen flanking PMUs frequently contained genes (e.g., fliA, himA, and hflB) in their upstream regions. These PMUs could be categorized as type 1 or 2 based on some characteristic genes and synteny (Figure 1B). PMU typing was already conducted based on the order of tmk and dnaB in a previous paper (Huang et al., 2022). Since these genes were not all located in the phyllogen flanking PMUs, we classified them based on other genes as described below.
The type 1 PMUs were found in six strains (‘Ca. P. asteris’ AY-WB, DY 2014, OY, RP166, WBD, and ‘Ca. P. phoenicium’ PEY), all of which contained phyl-A group phyllogens. These type 1 PMUs also contained hypothetical protein 1 (hp1), hp2, and an hflB-like gene in their upstream regions (Figure 1B). Additionally, the type 1 PMUs exhibited conserved synteny of fliA, ssb, himA, hflB, hp1, hp2, hflB-like, phyllogen, and hp3, except for the PMUs of the WBD strain. In the WBD strain, only the synteny of hp2, hflB-like, and the phyllogens was conserved. In the ‘Ca. P. pruni’ CP and ‘Ca. P. trifolii’ CPS strains, as well as in some strains related to ‘Ca. P. asteris,’ only the region between the hflB-like genes and the intergenic regions downstream of the phyllogens were characterized; these regions were conserved in all of these strains, and in the OY and WBD strains (Supplementary Figure S2). This result suggests that the type 1 PMUs that harbor phyl-A group phyllogens are conserved in ‘Ca. P. pruni,’ ‘Ca. P. trifolii,’ ‘Ca. P. asteris,’ and ‘Ca. P. phoenicium’.
Type 2 PMUs were found in seven strains that have phyl-D group phyllogens (‘Ca. P. asteris’ HP, RhY, PaWB-Japan, PaWB-China, ‘Ca. P. luffae’ NCHU2019, ‘Ca. P. ziziphi’ JWB-nky, and Hebei-2018). Type 2 PMUs were also found in the ‘Ca. P. pruni’ Vc33 strain, which has a phyl-C group phyllogen. The type 2 PMUs contained hp4, hp5, hp6, hp7, smc, and smc-like genes with similarity to smc and hp8 genes in the upstream region and hp9 genes in the downstream region. In the PaWB-Japan, PaWB-China, HP, and RhY strains, the type 2 PMUs exhibited conserved synteny of fliA, himA, hp4, hp5, hp6, hp7, hflB, smc, smc-like genes, hp8, phyllogen, and hp9. The JWB-nky and Hebei-2018 strains had another phyllogen, and its flanking region also contained genes that were characteristic of type 2 PMUs, although synteny was only partly conserved (Supplementary Figure S3A). The NCHU2019 strain had three type 2 PMUs with the same gene synteny (Supplementary Figure S3B). These results suggest that type 2 PMUs harboring phyl-D or phyl-C group phyllogens are conserved in four different species (‘Ca. P. asteris,’ ‘Ca. P. luffae,’ ‘Ca. P. ziziphi,’ and ‘Ca. P. pruni’). Additionally, phylogenetic trees based on genes shared by type 1 and 2 PMUs (i.e., fliA, himA, and hflB) indicate that the evolutionary history of these genes differs from that of phytoplasmas. In particular, the hflB tree formed two clades, with type 1 and 2 PMU strains (Supplementary Figure S4). Thus, our data indicate that the phylogeny of phyllogens is consistent with PMU type (Figure 1).
On the other hand, several phyllogens were not located within PMUs (indicated by asterisks; Figure 1). No PMU-associated genes were found in the 20 kbp upstream or downstream of the phyl-D group phyllogen in ‘Ca. P. aurantifolia’ PnWB, or in the ‘Ca. P. asteris’ MD-China strains (except for a dam gene in the MD-China strain; Supplementary Figure S6). Additionally, the phyl-D group phyllogen in the ‘Ca. P. aurantifolia’ EPWB strain was located near the PMU-associated genes, but outside of PMU regions (Supplementary Figure S5).
Phyllogen genes were highly conserved but phyllogen flanking PMU genes were not
Some PMUs contain fewer genes than PMUs reported to exist as probably transposable circular extrachromosomal elements such as AY-WB PMU1 (Toruño et al., 2010) and some PMU-associated genes are truncated (Bai et al., 2006; Ku et al., 2013). To understand the evolution of PMUs harboring phyllogens, we analyzed the conservation of genes in type 1 and 2 PMUs using pairwise amino acid sequence comparisons. In the type 1 PMUs, phyllogens were highly conserved (87%–100%) between ‘Ca. P. asteris’ and ‘Ca. P. phoenicium.’ However, genes in the phyllogen flanking region (fliA, ssb, himA, hflB, hp1, hp2, and hflB-like) exhibited low sequence identities (>57%), except in the closely related OY and DY2014 strains (Supplementary Table S6; Figure 2A). In the type 2 PMUs, phyllogens were also highly conserved (91%–100%) among ‘Ca. P. asteris’, ‘Ca. P. luffae,’ and ‘Ca. P. ziziphi,’ except for the Vc33 strain phyllogen that belongs to a different phyl-C group. However, the genes in the phyllogen flanking regions (fliA, himA, hp6, hp7, hflB, smc, smc-like, and hp9) exhibited low sequence identities (>32%), except for the closely related HP and RhY strains, and JWB-nky and Hebei-2018 strains (Supplementary Table S7; Figure 2B). Additionally, several PMU-associated genes, including hflB in the OY strain and smc in the HP, RhY, and PaWB-Japan strains, were truncated by premature stop codons due to frameshifts or single-nucleotide polymorphisms (Figure 1B). Moreover, the RP166, OY, PaWB-Japan, PaWB-China, RhY, and HP strains lacked PMU-associated genes downstream of phyllogens (e.g., tmk, dnaB, dnaG, and tra5; Figure 1B). These results indicate that although phyllogens are highly conserved in both types of PMU, phyllogen flanking genes have accumulated mutations, undergone pseudogenesis, and sometimes been lost altogether.
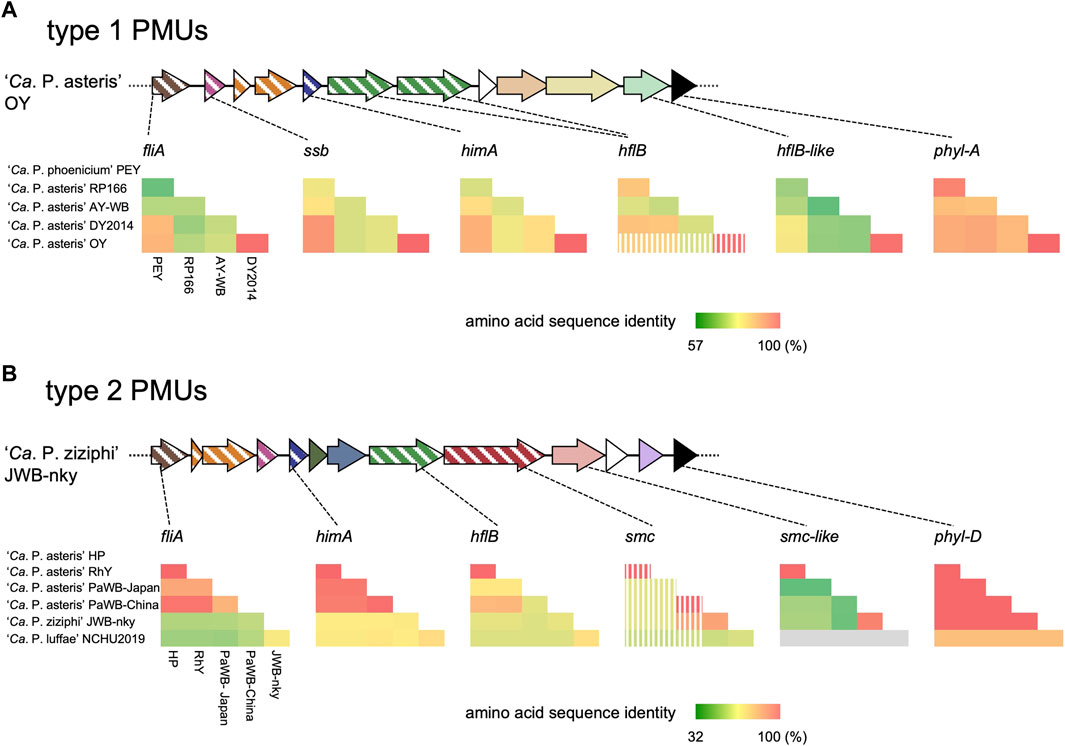
FIGURE 2. Pairwise sequence comparisons of ORFs flanking phyl-A (A) and phyl-D (B) group phyllogens. Genomic structures are represented as in Figure 1B. Pairwise amino acid sequence identities between phytoplasma species/strains are represented by heatmaps. Strain names are shown on the left and below the heatmaps; full names are listed in Supplementary Table S1. Stripes on the heatmap indicate that truncated genes were used in the analysis. The absence of an ORF from the corresponding genomic region of the NCHU2019 strain is indicated in gray. Results are shown for one of two phyllogens for ‘Ca. P. ziziphi’ and one of three phyllogens for ‘Ca. P. luffae.’
The phylogenetic relationships among ‘Ca. P. asteris’ strains containing phyllogens were complex
Next, we focused on relationships among phyllogens in ‘Ca. P. asteris’ because sequence information and genetic diversity were richer in ‘Ca. P. asteris’ than in other phytoplasma species (Iwabuchi et al., 2020). Although many strains related to ‘Ca. P. asteris’ had phyl-A group phyllogens, several strains had phyl-D group phyllogens.
First, intraspecies evolutionary relationships among strains with phyllogens were assessed by analyzing single-copy genes present in all whole/draft genome sequences. In total, 16 strains related to ‘Ca. P. asteris’ were analyzed, with a ‘Ca. P. meliae’ strain used as an outgroup. A WBD strain, which had recently been proposed for reclassification to ‘Ca. P. tritici’ (Zhao et al., 2021), was also used in this analysis. The resulting phylogenetic tree showed that strains related to ‘Ca. P. asteris’ formed two clades (Figure 3A). The minor clade comprised the AY-WB and WBD strains, each of which contained phyl-A group phyllogens. The major clade comprised four subclades and the TBZ1 strain. Two subclades comprised the strains with phyl-D group phyllogens; one of these subclades comprised the HP, RhY, and MD-China strains, and the other comprised the PaWB-Japan and PaWB-China strains. No phyllogens were found in the draft genome of the SW86 strain, which belonged to the former subclade. Another subclade comprised the strains containing phyl-A group phyllogens (i.e., the CYP, RP166, DY 2014, and OY strains). The final subclade comprised strains with complete (De Villa and M3 strains) or draft (LD1 strain) genomes that lacked phyllogens. The TBZ1 strain that contained a phyl-A group phyllogen did not belong to any of these subclades. These results indicate that the strains with phyl-D group phyllogens formed two subclades in ‘Ca. P. asteris’, while the strains with phyl-A group phyllogens formed one clade and one subclade. They also indicate that although strains related to ‘Ca. P. asteris’ retained the same group of phyllogens, at least at the subclade level, the overall relationships were complex.
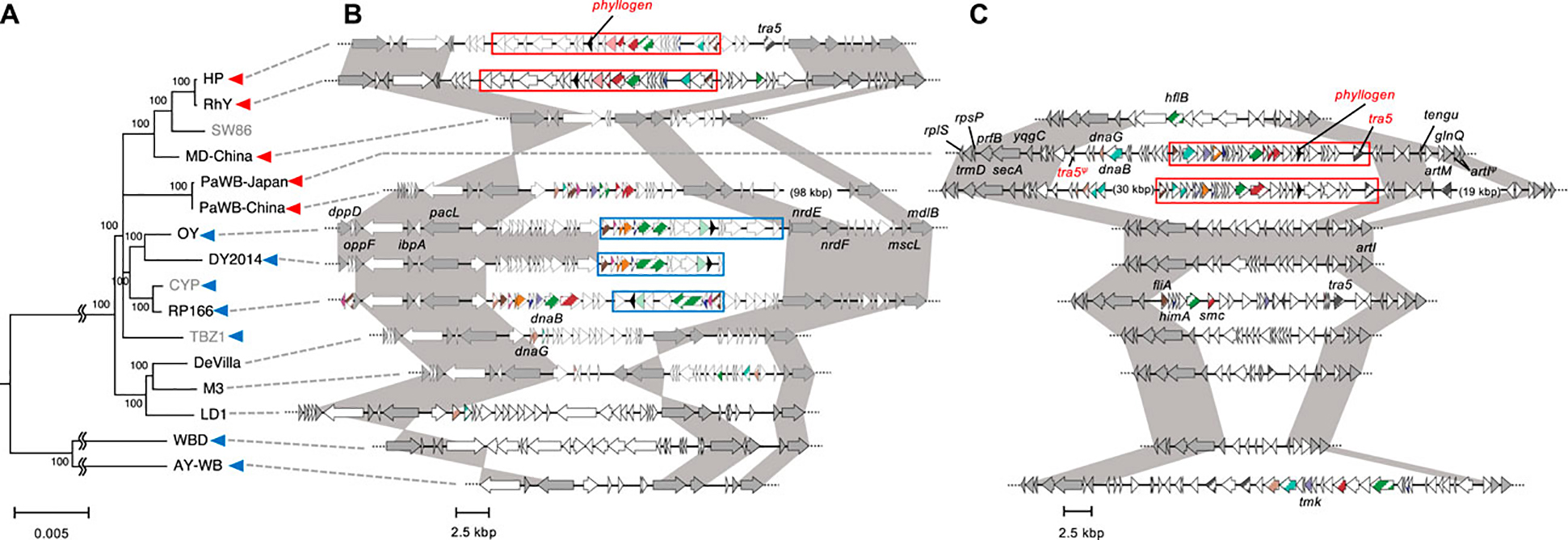
FIGURE 3. Genomic structure of regions flanking PMUs harboring phyllogens. (A) Whole/draft genome-based phylogenetic tree of ‘Ca. P. asteris’ with the distribution of phyllogens. The tree was constructed using the neighbor-joining method and includes single-copy genes shared by all the strains related to ‘Ca. P. asteris’ listed in Supplementary Table S3. The ‘Ca. P. meliae’ ChTYXIII strain was used as an outgroup. Numbers at the nodes represent bootstrap values for 1,000 replicates (only values >70% are shown). The scale bar indicates the number of nucleotide substitutions per site. The full names and GenBank accession numbers of the strains used in the analysis are listed in Supplementary Table S3. The conserved regions shown in (B) and (C) were not linked in the strains colored gray. Blue and red arrowheads indicate the presence of phyl-A and phyl-D groups in the genome, respectively. (B, C) Genomic structure of the regions surrounding PMUs harboring phyllogens from HP, RhY, OY, DY 2014, and RP166 strains (B), and PaWB-Japan and PaWB-China strains (C), compared with other ‘Ca. P. asteris’ genomes. ORF structures are represented as in Figure 1B. Dark gray ORFs indicate single-copy ORFs annotated by MetaGeneAnnotator (Noguchi et al., 2008). Phyllogens are shown in black. Diagonal stripe patterns indicate multicopy genes associated with PMUs (Arashida et al., 2008a). ORFs that are not associated with PMUs are shown in white. Truncated ORFs are marked by Ψ. Conserved regions in strains related to ‘Ca. P. asteris’ are shown in dark gray. Type 1 and 2 PMUs that flank phyllogens are shown in Figure 1 enclosed by blue and red borders, respectively; the corresponding synteny is highlighted in Figure 1. Broken lines at the ends of genomic structures indicate that these regions have been sequenced but are not shown in the figure.
PMUs were inserted into different genomic regions in different ‘Ca. P. asteris’ subgroups
The positions of the mobile elements within genomes may provide important evolutionary information. Therefore, we analyzed the genomic locations of PMUs in strains related to ‘Ca. P. asteris,’ based on their flanking genomic regions. First, we analyzed the genomic locations of the type 1 PMUs. The PMUs and surrounding genomic regions in the OY, DY 2014, RP166, and AY-WB strains are shown in Figure 3B; Supplementary Figure S6B. The type 1 PMUs of the OY, DY 2014, and RP166 strains, which belong to the same subclade as ‘Ca. P. asteris’ (Figure 3A), are flanked by several single-copy genes. These genes include dppD, oppF, ibpA, and pacL in the upstream region, and nrdE, nrdF, mscL, and mdlB in the downstream region (Figure 3B). The syntenies of these single-copy genes were also conserved in the other ‘Ca. P. asteris’ genomes, but there were no phyllogens in the corresponding regions, except in the HP and RhY strains. However, type 2 PMUs harboring phyllogens were found in these corresponding genomic regions. The type 1 PMU in the AY-WB strain was in a different genomic region, and was flanked by several single-copy genes including pdhC, acoL, tatD, and plsX in the upstream region and rpsD, mgtA, degV, and tsaD in the downstream region (Supplementary Figure S6B). The syntenies of these genes were conserved in other ‘Ca. P. asteris’ genomes, although no type 1 PMUs or phyllogens were found. These results indicate that the type 1 PMUs were inserted into different regions in the AY-WB and OY/DY2014/RP166 strains. Next, we analyzed the genomic locations of the type 2 PMUs. The PMUs and surrounding genomic regions in the PaWB-Japan and PaWB-China strains, which belong to the same subgroup, are shown in Figure 3C. These PMUs were delimited by a complete and truncated tra5 gene downstream and upstream of the phyllogens, respectively (Bai et al., 2006). Several single-copy genes were also found near the PMUs, but these genes were different from those surrounding the type 1 PMUs; they included rpsP, prfB, secA, and ypgC in the upstream region and tengu, glnQ, and artl in the downstream region (Figure 3C). Although the syntenies of these single-copy genes were also conserved in other ‘Ca. P. asteris’ genomes, there were no type 2 PMUs or phyllogens. The genomic region between the ypgC and tengu genes in the OY, DY 2014, De Villa, M3, and WBD strains was <13 kbp, which was much shorter than in the PaWB-Japan (35 kbp) and PaWB-China (67 kbp) strains. Several PMU-associated genes were found in this genomic region in the RP166 and AY-WB strains; however, the syntenies of these genes differed from those of the type 2 PMUs (Figure 3C). On the other hand, the type 2 PMUs in the HP and RhY strains were flanked by several single-copy genes different from those present in the PaWB strains (Figure 3B). The phyl-D group phyllogen present in the MD-China strain was also flanked by single-copy genes different from the genes flanking other phyllogens (Supplementary Figure S6). These results indicate that type 2 PMUs were inserted into different regions in the PaWB-Japan/PaWB-China, MD-China, and HP/RhY strains.
To correlate the inserted type 2 PMUs with other genomes at the nucleotide level, the upstream and downstream sequences of the PMUs in the PaWB strains were aligned with the corresponding genomic regions in the OY, De Villa, and DY2014 strains. The alignments identified regions of approximately 350 bp at both ends of the insertions that contained inverted repeat sequences, which is characteristic of transposon insertion (Szeverényi et al., 2003), and these inverted repeats were conserved between the PaWB strains (Figure 4; Supplementary Figure S7). Although these inverted repeat sequences were not identical in the other three strains, there were nearby sequences with strong similarity among these genomes (Figure 4), suggesting that PMU boundaries were located near the inverted repeats. These results indicate that a type 2 PMU was inserted at this position in the common ancestor of the PaWB strains.
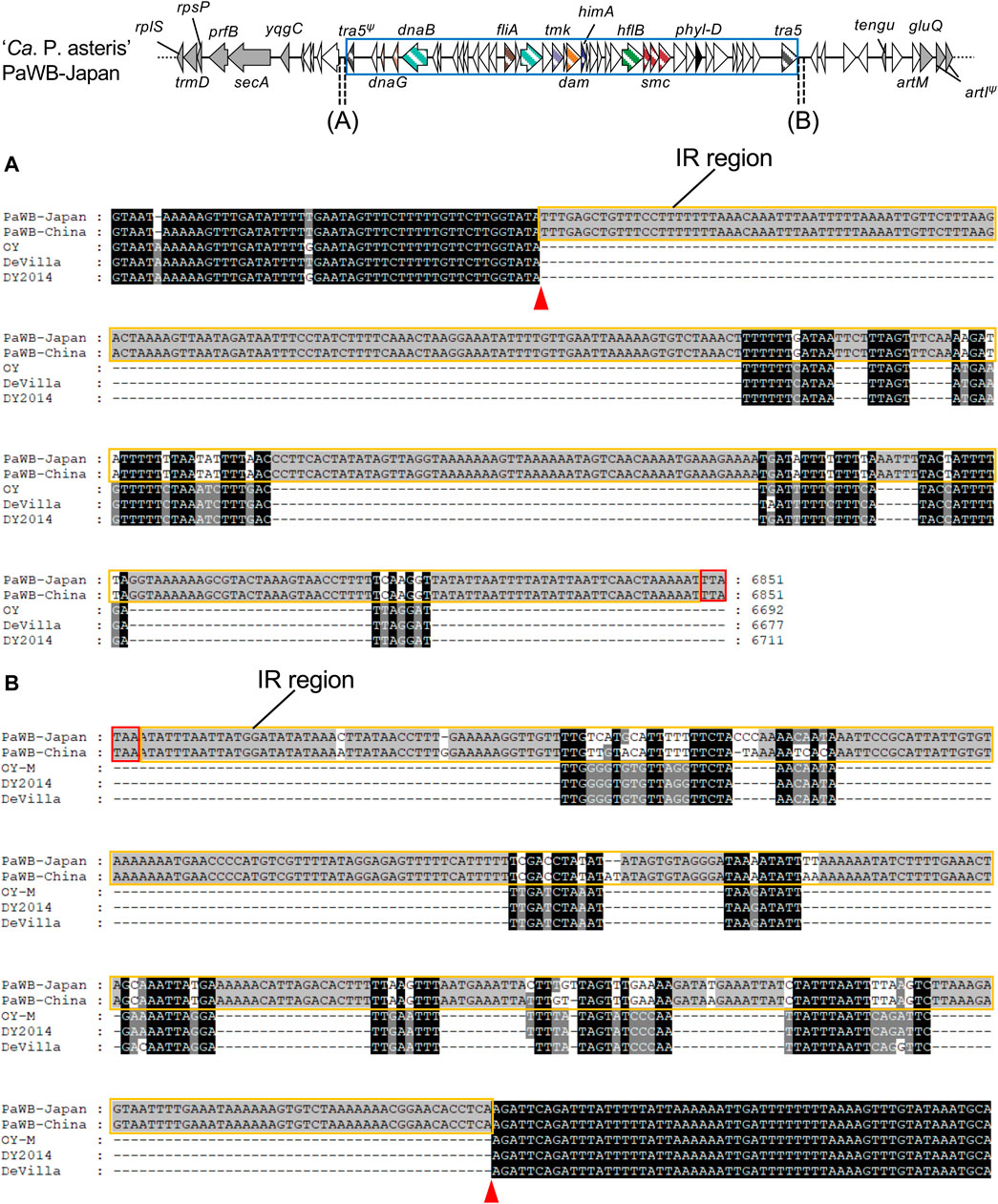
FIGURE 4. Nucleotide sequence alignment of the downstream sequences, beginning at stop codons of truncated tra5 in upstream regions (A) or intact tra5 in downstream regions (B) of phyllogens. Stop codons in upstream regions of truncated tra5 genes were identified by comparing nucleotide sequences with the intact tra5 gene. Tra5 stop codons are enclosed by red borders. Inverted repeat regions are enclosed by orange borders. Putative PMU boundaries are marked with red arrowheads. Genomic structures of the phyllogen flanking regions in the PaWB-Japan strain are represented as in Figure 3. PMUs are enclosed by blue borders.
Functions of PMU-associated phyllogens were conserved
Comparative analyses revealed conservation of amino acid sequences of the PMU-associated phyllogens. Thus, we examined functional conservation of the phyllogens by testing the capacity of various phyllogens to induce phyllody. We tested the phyl-A group phyllogens of RP166 (PHYLRP166) and the PEY strains (PHYLPEY), as well as the phyl-D group phyllogens of NCHU2019 (PHYLNCHU2019) and the HP strains (PHYLHP). We used the PHYLOY phyllogen of the OY strain (phyl-A group) and the PHYLJWB phyllogen of the JWB strain (phyl-D group) as controls known to induce phyllody (Iwabuchi et al., 2020). Each phyllogen was expressed in A. thaliana using a TRV-based gene expression vector system. All of the tested phyllogens converted sepals, petals, and stamens into leaf-like structures with stellate trichomes, changed pistils into secondary vegetative shoot-like structures, and enlarged flowers (Figure 5A), as did the PHYLJWB phyllogen control. These results showed that the functions of the type 1 and 2 PMU-associated phyllogens were as highly conserved as their sequences.
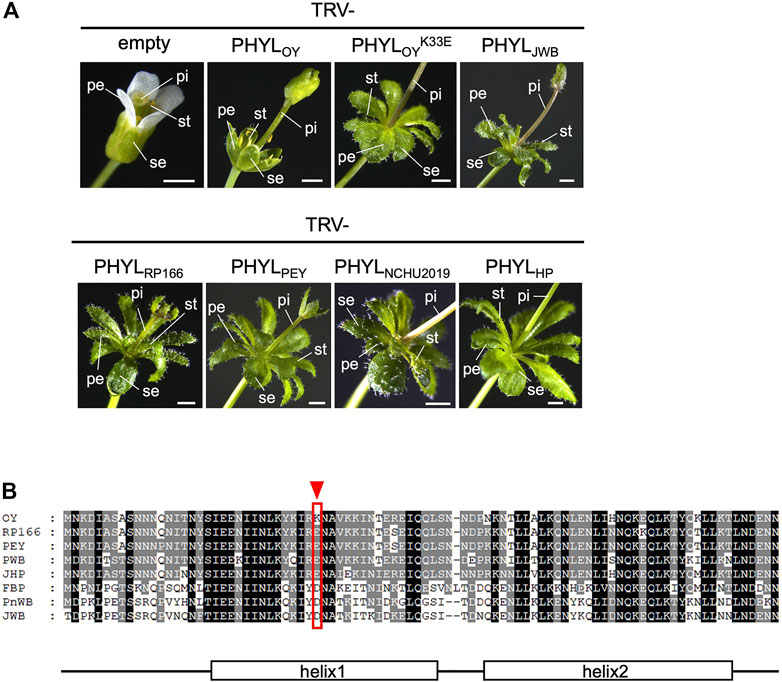
FIGURE 5. (A) Functional analysis of phyllogens associated with PMUs and PHYLOYK33E. Arabidopsis thaliana plants were infected with the tobacco rattle virus (TRV) vectors carrying phyllogens from either the phyl-A or phyl-D group. The following floral organs are shown: sepals (se), petals (pe), stamens (st), and pistils (pi). White scale bar: 1 mm. (B) Amino acid alignment of the secreted regions of PHYLOY and the phyl-A or phyl-D group phyllogens, which induce severe phyllody. The red arrowhead and border indicate a unique polymorphism at position 33 of PHYLOY. Consensus secondary structure elements of phyllogens are shown (Iwabuchi et al., 2019).
Interestingly, in the PHYLOY expression test, the stamens of most flowers were not converted into leaf-like structures, the other floral organs had fewer trichomes, and the flowers did not enlarge (Figure 5A). This suggested that PHYLOY exhibited a reduced capacity to induce phyllody. Sequence comparisons revealed that PHYLOY had a unique polymorphism at position 33 (lysine; Figure 5B). A PHYLOY mutant with a reciprocal substitution at this position (glutamate; PHYLOYK33E) induced phyllody to the same extent as the other homologs (Figure 5A). Therefore, this amino acid is important for strong induction of phyllody.
Discussion
Interspecies acquisition of phyllogens via horizontal transfer of PMUs
The apparent differences between the evolutionary histories of phyllogens and 16S rRNA genes in phytoplasmas suggest that phyllogens may have undergone HGT (Iwabuchi et al., 2020; Kalla et al., 2021). However, the mechanisms involved were previously unclear due to the lack of phyllogen flanking-region sequence data. In this study, we determined draft genomes for three strains and analyzed phyllogen flanking regions from 17 phytoplasma strains related to six different species. We found that most phyllogens were associated with PMUs (Figure 1). Furthermore, phyllogen groupings correlated closely with the types of PMU (Figure 1). These results indicate that PMUs drive the horizontal transfer of distinct groups of phyllogens between phytoplasma species and strains. This study demonstrates that the symptom-determinant effectors of phytoplasmas are evolutionarily correlated with the types of PMU rather than the phytoplasma genomes.
In addition to phyllogens, three other effectors (TENGU, SAP11, and SAP05) have been linked experimentally with phytoplasma virulence (Hoshi et al., 2009; Sugawara et al., 2013; Minato et al., 2014; Cho et al., 2019; Huang et al., 2021). Among these, the SAP11 and SAP05 genes are also located near PMUs; they are conserved in various species (Sugio and Hogenhout., 2012; Cho et al., 2019; Huang et al., 2022) and have phylogenetic trees that differ from those of phytoplasmas (Chang et al., 2018; Huang et al., 2022). These observations suggest that PMU-mediated HGT may play a major role in the transfer of virulence genes across phytoplasma species.
Many studies have reported HGT of bacterial virulence genes based on genomic analyses of individual species (Ma et al., 2006; McCann and Guttman, 2008; Drew et al., 2021), but few have shown how HGT can occur via transposable elements (such as transposons) by comparing the genomes of different bacterial species. Thus, this study provides new insight into the mechanisms of HGT of virulence genes.
Phyllogen sequences and functions are conserved but PMU sequences may deteriorate
Genetic elements may become immobilized due to inactivation or deletion of genes necessary for their transfer (Dobrindt et al., 2004). Degeneration of PMUs has also been reported in several phytoplasma strains, including AY-WB (Bai et al., 2006; Ku et al., 2013). In this study, some PMUs harboring phyllogens lacked tra5 (Figure 1B). In addition, some PMU genes had accumulated mutations, were truncated, or had been lost altogether (Figures 1B, 2), including genes with putative roles in DNA recombination, replication, and transposition (Arashida et al., 2008a; Ku et al., 2013). Therefore, most of these PMUs may begin to lose their capacity for transposition after the phyllogens have been acquired. However, the type 2 PMUs of the PaWB-Japan and PaWB-China strains retained their tra5 genes and inverted repeat-like sequences at both ends (Figures 3, 4), suggesting that they had also retained their capacity for transposition.
Despite the deterioration of PMU-associated genes, phyllogens from different species had highly conserved sequences, especially phyllogens from the phyl-A and -D groups (Figure 2). Furthermore, phyllogen functions were conserved (Figure 5). Several phyllogens were not located within PMUs (Figure 1A; Chung et al., 2013; Luo et al., 2022) and their flanking regions exhibited considerable variation (data not shown), suggesting that after phyllogen transfer, PMU sequences may deteriorate completely. Therefore, phyllogens may become fixed in many different phytoplasma genomes after their acquisition, suggesting that they are crucial for their hosts. Previous studies have shown that phyllody symptoms facilitate phytoplasma accumulation/localization within host plants (Arashida et al., 2008b; Su et al., 2011), and phyllogens can attract insect vectors (MacLean et al., 2014). Although further studies are needed to investigate the roles of phyllody symptoms in host adaptability, the fact that phyllogens have been strongly conserved throughout evolution suggests that they are critical for phytoplasma survival.
Evolutionary history of phyllogen acquisition in ‘Ca. P. asteris’
To date, three different groups of phyllogens have been identified in strains related to ‘Ca. P. asteris’ (phyl-A, -B, and -D; Iwabuchi et al., 2020). In this study, we found that the acquisition of these phyllogens was a complex process, at least for phyl-A and phyl-D. In ‘Ca. P. asteris,’ we identified one subclade comprising strains that retained phyl-A phyllogens and two subclades comprising strains that retained phyl-D phyllogens (Figure 3A). In the former subclade, the PMUs retaining phyl-A were found in the same region of the genomes (Figure 3), suggesting that these PMUs were acquired by their common ancestor. The same region also surrounded PMUs of the HP and RhY strains (Figure 3). Therefore, this region may be a hot spot for genome rearrangement in phytoplasmas, as described by a previous study (Arashida et al., 2008b). PMUs retaining phyl-D were found in different genomic regions in the PaWB-Japan/PaWB-China, MD-China, and HP/RhY strains (Figure 3; Supplementary Figure S6). This observation suggests that PMUs harboring phyllogens either moved frequently within the genome or were acquired separately. The PaWB strains had complete PMUs retaining phyl-D, suggesting that these were acquired more recently than the PMUs in the HP and RhY strains. Therefore, the latter hypothesis seems more plausible. We could not determine the acquisition order of the phyl-A and phyl-D group phyllogens in ‘Ca. P. asteris’ due to insufficient genomic data. Therefore, further studies of phyllogen flanking regions and their genomic positions are needed.
HGT may occur between organisms that share an ecological niche, irrespective of their phylogenetic relationships (Polz et al., 2013). Phylogenetic analyses have shown that all of the phyl-D group phyllogens of ‘Ca. P. asteris’ are most closely related to phyllogens of ‘Ca. P. ziziphi’ (Iwabuchi et al., 2020; Figure 1A). Furthermore, several lines of evidence indicate that strains related to ‘Ca. P. asteris’ and ‘Ca. P. ziziphi’ share common ecological niches. First, the distribution of ‘Ca. P. ziziphi’ is limited to eastern and southern Asia (Jung et al., 2003; Rao et al., 2017), as is the distribution of strains related to ‘Ca. P. asteris’ that retain phyl-D (Kakizawa et al., 2006; Takinami et al., 2013; Luo et al., 2022). Second, co-infection of ‘Ca. P. asteris’ and ‘Ca. P. ziziphi’ has been reported in jujube plants (Sun et al., 2013). Third, Hishimonus sellatus, a vector of ‘Ca. P. ziziphi,’ can transmit two strains related to ‘Ca. P. asteris’ that retain phyl-D (RhY and MD-China; Tanaka et al., 2000; Kusunoki et al., 2002). Therefore, type 2 PMUs in ‘Ca. P. asteris’ may be acquired from ‘Ca. P. ziziphi’ in the same ecological niche. Further accumulation of information on the synteny of PMUs will reveal how PMUs and symptom-determinant genes located in them are shared among phytoplasmas in the same niche.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, BSDA00000000; https://www.ddbj.nig.ac.jp/, BSCX00000000; https://www.ddbj.nig.ac.jp/, BSCY00000000; https://www.ddbj.nig.ac.jp/, LC740440; https://www.ddbj.nig.ac.jp/, LC740441; https://www.ddbj.nig.ac.jp/, LC740442; https://www.ddbj.nig.ac.jp/, LC740443; https://www.ddbj.nig.ac.jp/, LC740444; https://www.ddbj.nig.ac.jp/, LC740445; https://www.ddbj.nig.ac.jp/, LC740446; https://www.ddbj.nig.ac.jp/, LC740447; https://www.ddbj.nig.ac.jp/, LC740448; https://www.ddbj.nig.ac.jp/, LC740449; https://www.ddbj.nig.ac.jp/, LC740450; https://www.ddbj.nig.ac.jp/, LC740451; https://www.ddbj.nig.ac.jp/, LC740452; https://www.ddbj.nig.ac.jp/, LC740453.
Author contributions
RT, NI, and KM designed the research. YK, TN, KO, SN, and YY supervised the experiments. RT performed most experiments. MS provided technical assistance in the experiments. RT wrote the initial draft of the article. MS complemented the writing. NI, YK, TN, KM, KO, SN, and YY critically revised and edited the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) through Grants-in-Aid for Scientific Research (nos. 19K15840, 20H02991, 20J23168, 20K22562, 20KK0132, 21H04722, 21K14847, 21K14853, and 21K19239).
Acknowledgments
We are grateful to Drs T. Shiomi (Department of Biological Safety, National Institute for Agro-Environmental Sciences, Ibaraki, Japan), T. Usugi (Plant Protection Division, National Agricultural Research Center, Ibaraki, Japan), N. Nishimura and T. Tsuchizaki (Department of Agribusiness, Koibuchi College of Agriculture and Nutrition, Ibaraki, Japan), and A. Bertaccini (DipSA, University of Bologna, Bologna, Italy) for providing phytoplasma DNA samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1132432/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi:10.1016/S0022-2836(05)80360-2
Arashida, R., Kakizawa, S., Hoshi, A., Ishii, Y., Jung, H. Y., Kagiwada, S., et al. (2008a). Heterogeneic dynamics of the structures of multiple gene clusters in two pathogenetically different lines originating from the same phytoplasma. DNA Cell Biol. 27, 209–217. doi:10.1089/dna.2007.0654
Arashida, R., Kakizawa, S., Ishii, Y., Hoshi, A., Jung, H. Y., Kagiwada, S., et al. (2008b). Cloning and characterization of the antigenic membrane protein (AMP) gene and in situ detection of AMP from malformed flowers infected with Japanese Hydrangea phyllody phytoplasma. Phytopathology 98, 769–775. doi:10.1094/PHYTO-98-7-0769
Bai, X., Zhang, J., Ewig, A., Miller, S. A., Radek, A. J., Shevchenko, D. V., et al. (2006). Living with genome instability: The adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188, 3682–3696. doi:10.1128/JB.188.10.3682-3696.2006
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and applications. BMC Bioinform 10, 421. doi:10.1186/1471-2105-10-421
Chang, S. H., Tan, C. M., Wu, C. T., Lin, T. H., Jiang, S. Y., Liu, R. C., et al. (2018). Alterations of plant architecture and phase transition by the phytoplasma virulence factor SAP11. J. Exp. Bot. 69, 5389–5401. doi:10.1093/jxb/ery318
Cho, S. T., Kung, H. J., Huang, W., Hogenhout, S. A., and Kuo, C. H. (2020). Species boundaries and molecular markers for the classification of 16SrI phytoplasmas inferred by genome analysis. Front. Microbiol. 11, 1531. doi:10.3389/fmicb.2020.01531
Cho, S. T., Lin, P., and Kuo, C. H. (2019). Genomic characterization of the periwinkle leaf yellowing (PLY) phytoplasmas in Taiwan. Front. Microbiol. 10, 2194. doi:10.3389/fmicb.2019.02194
Chung, W. C., Chen, L. L., Lo, W. S., Lin, C. P., and Kuo, C. H. (2013). Comparative analysis of the peanut witches'-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. PLoS One 8, e62770. doi:10.1371/journal.pone.0062770
Cosentino, S., and Iwasaki, W. (2019). SonicParanoid: Fast, accurate and easy orthology inference. Bioinformatics 35, 149–151. doi:10.1093/bioinformatics/bty631
Dobrindt, U., Hochhut, B., Hentschel, U., and Hacker, J. (2004). Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2, 414–424. doi:10.1038/nrmicro884
Drew, G. C., Stevens, E. J., and King, K. C. (2021). Microbial evolution and transitions along the parasite–mutualist continuum. Nat. Rev. Microbiol. 19, 623–638. doi:10.1038/s41579-021-00550-7
Gilchrist, C. L. M., and Chooi, Y. H. (2021). Clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 37, 2473–2475. doi:10.1093/bioinformatics/btab007
Gluck-Thaler, E., Cerutti, A., Perez-Quintero, A. L., Butchacas, J., Roman-Reyna, V., Madhavan, V. N., et al. (2020). Repeated gain and loss of a single gene modulates the evolution of vascular plant pathogen lifestyles. Sci. Adv. 6, eabc4516. doi:10.1126/sciadv.abc4516
Hooykaas, P. J. J., Snijdewint, F. G. M., and Schilperoort, R. A. (1982). Identification of the sym plasmid of Rhizobium leguminosarum strain 1001 and its transfer to and expression in other Rhizobia and Agrobacterium tumefaciens. Plasmid 8, 73–82. doi:10.1016/0147-619x(82)90042-7
Hoshi, A., Oshima, K., Kakizawa, S., Ishii, Y., Ozeki, J., Hashimoto, M., et al. (2009). A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc. Natl. Acad. Sci. U. S. A. 106, 6416–6421. doi:10.1073/pnas.0813038106
Huang, C. T., Cho, S. T., Lin, Y. C., Tan, C. M., Chiu, Y. C., Yang, J. Y., et al. (2022). Comparative genome analysis of ‘Candidatus Phytoplasma luffae’ reveals the influential roles of potential mobile units in phytoplasma evolution. Front. Microbiol. 13, 773608. doi:10.3389/fmicb.2022.773608
Huang, W., MacLean, A. M., Sugio, A., Maqbool, A., Busscher, M., Cho, S. T., et al. (2021). Parasitic modulation of host development by ubiquitin-independent protein degradation. Cell 184, 5201–5214.e12. doi:10.1016/j.cell.2021.08.029
Iwabuchi, N., Kitazawa, Y., Maejima, K., Koinuma, H., Miyazaki, A., Matsumoto, O., et al. (2020). Functional variation in phyllogen, a phyllody-inducing phytoplasma effector family, attributable to a single amino acid polymorphism. Mol. Plant. Pathol. 21, 1322–1336. doi:10.1111/mpp.12981
Iwabuchi, N., Maejima, K., Kitazawa, Y., Miyatake, H., Nishikawa, M., Tokuda, R., et al. (2019). Crystal structure of phyllogen, a phyllody-inducing effector protein of phytoplasma. Biochem. Biophys. .Res. Commun. 513, 952–957. doi:10.1016/j.bbrc.2019.04.060
Jomantiene, R., Zhao, Y., and Davis, R. E. (2007). Sequence-variable mosaics: Composites of recurrent transposition characterizing the genomes of phylogenetically diverse phytoplasmas. DNA Cell. Biol. 26, 557–564. doi:10.1089/dna.2007.0610
Jung, H. Y., Sawayanagi, T., Kakizawa, S., Nishigawa, H., Wei, W., Oshima, K., et al. (2003). ‘Candidatus Phytoplasma ziziphi’, a novel phytoplasma taxon associated with jujube witches’-broom disease. Int. J. Syst. Evol. Microbiol. 53, 1037–1041. doi:10.1099/ijs.0.02393-0
Kakizawa, S., Oshima, K., Jung, H. Y., Suzuki, S., Nishigawa, H., Arashida, R., et al. (2006). Positive selection acting on a surface membrane protein of the plant-pathogenic phytoplasmas. J. Bacteriol. 188, 3424–3428. doi:10.1128/JB.188.9.3424-3428.2006
Kalla, S., Verma, P., and Lakhanpaul, S. (2021). Understanding the evolution of SAP54, a phyllody associated phytoplasma effector gene through phylogenetic network analysis. Phytopathog. Mollicutes. 11, 36–44. doi:10.5958/2249-4677.2021.00005.0
Keeling, P. J., and Palmer, J. D. (2008). Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618. doi:10.1038/nrg2386
Kitazawa, Y., Iwabuchi, N., Himeno, M., Sasano, M., Koinuma, H., Nijo, T., et al. (2017). Phytoplasma-conserved phyllogen proteins induce phyllody across the Plantae by degrading floral MADS domain proteins. J. Exp. Bot. 68, 2799–2811. doi:10.1093/jxb/erx158
Kitazawa, Y., Iwabuchi, N., Maejima, K., Sasano, M., Matsumoto, O., Koinuma, H., et al. (2022). A phytoplasma effector acts as a ubiquitin-like mediator between floral MADS-box proteins and proteasome shuttle proteins. Plant Cell 34, 1709–1723. doi:10.1093/plcell/koac062
Ku, C., Lo, W. S., and Kuo, C. H. (2013). Horizontal transfer of potential mobile units in phytoplasmas. Mob. Genet. Elem. 3, e26145. doi:10.4161/mge.26145
Kube, M., Schneider, B., Kuhl, H., Dandekar, T., Heitmann, K., Migdoll, A. M., et al. (2008). The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus Phytoplasma Mali. BMC Genom 9, 306. doi:10.1186/1471-2164-9-306
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Kusunoki, M., Shiomi, T., Kobayashi, M., Okudaira, T., Ohashi, A., and Nohira, T. (2002). A leafhopper (Hishimonus sellatus) transmits phylogenetically distant phytoplasmas: Rhus yellows and hovenia witches’ broom phytoplasma. J. Gen. Plant. Pathol. 68, 147–154. doi:10.1007/PL00013068
Luo, L., Zhang, X., Meng, F., Wang, Y., Zhou, Y., and Liu, J. (2022). Draft genome sequences resources of mulberry dwarf phytoplasma strain MDGZ-01 associated with mulberry yellow dwarf (MYD) diseases. Plant Dis. 106, 2239–2242. doi:10.1094/PDIS-12-21-2671-A
Ma, W., Dong, F. F. T., Stavrinides, J., and Guttman, D. S. (2006). Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS. Genet. 2, e209–e2142. doi:10.1371/journal.pgen.0020209
MacLean, A. M., Orlovskis, Z., Kowitwanich, K., Zdziarska, A. M., Angenent, G. C., Immink, R. G. H., et al. (2014). Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 12, e1001835. doi:10.1371/journal.pbio.1001835
MacLean, A. M., Sugio, A., Makarova, O. V., Findlay, K. C., Grieve, V. M., Toth, R., et al. (2011). Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 157, 831–841. doi:10.1104/pp.111.181586
Maejima, K., Iwai, R., Himeno, M., Komatsu, K., Kitazawa, Y., Fujita, N., et al. (2014). Recognition of floral homeotic MADS-domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody. Plant J. 78, 541–554. doi:10.1111/tpj.12495
Marcone, C. (2014). Molecular biology and pathogenicity of phytoplasmas. Ann. Appl. Biol. 165, 199–221. doi:10.1111/aab.12151
McCann, H. C., and Guttman, D. S. (2008). Evolution of the type III secretion system and its effectors in plant–microbe interactions. New. Phytol. 177, 33–47. doi:10.1111/j.1469-8137.2007.02293.x
Melnyk, R. A., Hossain, S. S., and Haney, C. H. (2019). Convergent gain and loss of genomic islands drive lifestyle changes in plant-associated Pseudomonas. ISME J. 13, 1575–1588. doi:10.1038/s41396-019-0372-5
Minato, N., Himeno, M., Hoshi, A., Maejima, K., Komatsu, K., Takebayashi, Y., et al. (2014). The phytoplasmal virulence factor TENGU causes plant sterility by downregulating of the jasmonic acid and auxin pathways. Sci. Rep. 4, 7399. doi:10.1038/srep07399
Muhire, B. M., Varsani, A., and Martin, D. P. (2014). Sdt: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9, e108277. doi:10.1371/journal.pone.0108277
Music, M. S., Samarzija, I., Hogenhout, S. A., Haryono, M., Cho, S. T., and Kuo, C. H. (2019). The genome of ‘Candidatus Phytoplasma solani’ strain SA-1 is highly dynamic and prone to adopting foreign sequences. Syst. Appl. Microbiol. 42, 117–127. doi:10.1016/j.syapm.2018.10.008
Namba, S., Oyaizu, H., Kato, S., Iwanami, S., and Tsuchizaki, T. (1993). Phylogenetic diversity of phytopathogenic mycoplasmalike organisms. Int. J. Syst. Bacteriol. 43, 461–467. doi:10.1099/00207713-43-3-461
Noguchi, H., Taniguchi, T., and Itoh, T. (2008). MetaGeneAnnotator: Detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 15, 387–396. doi:10.1093/dnares/dsn027
Oshima, K., Kakizawa, S., Nishigawa, H., Jung, H. Y., Wei, W., Suzuki, S., et al. (2004). Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 36, 27–29. doi:10.1038/ng1277
Polz, M. F., Alm, E. J., and Hanage, W. P. (2013). Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 29, 170–175. doi:10.1016/j.tig.2012.12.006
Rao, G. P., Priya, M., Thorat, V., Manimekalai, R., Tiwari, K., and Yadav, A. (2017). A century progress of research on phytoplasma diseases in India. Phytopathog. Mollicutes. 7, 1–38. doi:10.5958/2249-4677.2017.00001.9
Ravenhall, M., Škunca, N., Lassalle, F., and Dessimoz, C. (2015). Inferring horizontal gene transfer. PLoS Comput. Biol. 11, e1004095. doi:10.1371/journal.pcbi.1004095
Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi:10.1093/oxfordjournals.molbev.a040454
Soucy, S., Huang, J., and Gogarten, J. (2015). Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 16, 472–482. doi:10.1038/nrg3962
Su, Y. T., Chen, J. C., and Lin, C. P. (2011). Phytoplasma-induced floral abnormalities in Catharanthus roseus are associated with phytoplasma accumulation and transcript repression of floral organ identity genes. Mol. Plant Microbe Interact. 24, 1502–1512. doi:10.1094/MPMI-06-11-0176
Sugawara, K., Honma, Y., Komatsu, K., Himeno, M., Oshima, K., and Namba, S. (2013). The alteration of plant morphology by small peptides released from the proteolytic processing of the bacterial peptide TENGU. Plant Physiol. 162, 2005–2014. doi:10.1104/pp.113.218586
Sugio, A., and Hogenhout, S. A. (2012). The genome biology of phytoplasma: Modulators of plants and insects. Curr. Opin. Microbiol. 15, 247–254. doi:10.1016/j.mib.2012.04.002
Sugio, A., Kingdom, H. N., MacLean, A. M., Grieve, V. M., and Hogenhout, S. A. (2011). Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 108, E1254–E1263. doi:10.1073/pnas.1105664108
Sun, X. C., Mou, H. Q., Li, T. T., Tian, Q., and Zhao, W. J. (2013). Mixed infection of two groups (16SrI & V) of phytoplasmas in a single jujube tree in China. J. Phytopathol. 161, 661–665. doi:10.1111/jph.12101
Szeverényi, I., Nagy, Z., Farkas, T., Olasz, F., and Kiss, J. (2003). Detection and analysis of transpositionally active head-to-tail dimers in three additional Escherichia coli IS elements. Microbiology 149, 1297–1310. doi:10.1099/mic.0.26121-0
Takahashi, S., Komatsu, K., Kagiwada, S., Ozeki, J., Mori, T., Hirata, H., et al. (2006). The efficiency of interference of potato virus X infection depends on the target gene. Virus Res. 116, 214–217. doi:10.1016/j.virusres.2005.11.002
Takinami, Y., Maejima, K., Takahashi, A., Keima, T., Shiraishi, T., Okano, Y., et al. (2013). First report of 'Candidatus Phytoplasma asteris' infecting hydrangea showing phyllody in Japan. J. Gen. Plant Pathol. 79, 209–213. doi:10.1007/s10327-013-0445-7
Tanaka, M., Osada, S., Matsuda, I., and Marks, R. (2000). Transmission of rhus (Rhus javanica L.) yellows by Hishimonus sellatus and host range of the causal phytoplasma. J. Gen. Plant Pathol. 66, 323–326. doi:10.1046/j.1365-2230.2000.00653.x
Tanizawa, Y., Fujisawa, T., and Nakamura, Y. (2018). Dfast: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34, 1037–1039. doi:10.1093/bioinformatics/btx713
Toruño, T. Y., Musić, M. S., Simi, S., Nicolaisen, M., and Hogenhout, S. A. (2010). Phytoplasma PMU1 exists as linear chromosomal and circular extrachromosomal elements and has enhanced expression in insect vectors compared with plant hosts. Mol. Microbiol. 77, 1406–1415. doi:10.1111/j.1365-2958.2010.07296.x
Tran-Nguyen, L. T., Kube, M., Schneider, B., Reinhardt, R., and Gibb, K. S. (2008). Comparative genome analysis of ‘Candidatus Phytoplasma australiense’ (subgroup tuf-Australia I; rp-A) and ‘Ca. Phytoplasma asteris’ strains OY-M and AY-WB. J. Bacteriol. 190, 3979–3991. doi:10.1128/JB.01301-07
Wang, J., Song, L., Jiao, Q., Yang, S., Gao, R., Lu, X., et al. (2018). Comparative genome analysis of jujube witches’-broom Phytoplasma, an obligate pathogen that causes jujube witches’-broom disease. BMC Genom 19, 689. doi:10.1186/s12864-018-5075-1
Wei, W., Davis, R. E., Jomantiene, R., and Zhao, Y. (2008). Ancient, recurrent phage attacks and recombination shaped dynamic sequence-variable mosaics at the root of phytoplasma genome evolution. Proc. Natl. Acad. Sci. U. S. A. 105, 11827–11832. doi:10.1073/pnas.0805237105
Yang, C. Y., Huang, Y. H., Lin, C. P., Lin, Y. Y., Hsu, H. C., Wang, C. N., et al. (2015). MicroRNA396-targeted SHORT VEGETATIVE PHASE is required to repress flowering and is related to the development of abnormal flower symptoms by the phyllody symptoms1 effector. Plant Physiol. 168, 1702–1716. doi:10.1104/pp.15.00307
Keywords: phyllogen, phytoplasma, effector, horizontal gene transfer, potential mobile unit, pathogenicity, symptom
Citation: Tokuda R, Iwabuchi N, Kitazawa Y, Nijo T, Suzuki M, Maejima K, Oshima K, Namba S and Yamaji Y (2023) Potential mobile units drive the horizontal transfer of phytoplasma effector phyllogen genes. Front. Genet. 14:1132432. doi: 10.3389/fgene.2023.1132432
Received: 27 December 2022; Accepted: 03 April 2023;
Published: 11 May 2023.
Edited by:
Satyanarayana Tulasi, University of Delhi, IndiaReviewed by:
Inderjit Singh Yadav, Punjab Agricultural University, IndiaJacqueline Fletcher, Oklahoma State University, United States
Valeria Trivellone, University of Illinois at Urbana-Champaign, United States
Copyright © 2023 Tokuda, Iwabuchi, Kitazawa, Nijo, Suzuki, Maejima, Oshima, Namba and Yamaji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kensaku Maejima, YW1hZWppbWFAZy5lY2MudS10b2t5by5hYy5qcA==
 Ryosuke Tokuda1
Ryosuke Tokuda1 Nozomu Iwabuchi
Nozomu Iwabuchi Kensaku Maejima
Kensaku Maejima Kenro Oshima
Kenro Oshima Yasuyuki Yamaji
Yasuyuki Yamaji