- 1NHC Key Laboratory for Birth Defect for Research and Prevention, Hunan Provincial Maternal and Child Health Care Hospital, Changsha, China
- 2Department of Women Health Care, Hunan Provincial Maternal and Child Healthcare Hospital, Changsha, China
- 3Department of Maternal and Child Health, Xiangya School of Public Health, Central South University, Changsha, China
Insulin resistance plays an important role in the pathogenesis of polycystic ovarian syndrome (PCOS). Calpain10 (CAPN10) gene was the first identified susceptibility gene for type 2 diabetes mellitus and closely related to insulin sensitivity. A lot of research attention has been attracted on the relationship between CAPN10 polymorphisms and PCOS risk, but they didn’t reach a consistent conclusion. We therefore performed this systematic review and meta-analysis to assess the association of CAPN10 common variants with PCOS susceptibility. A total of 21 studies were eligible for inclusion. Meta-analyses were done for 5 variants that had at least two data sources: UCSNP-19, -43, −44, −56 and −63. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated under five genetic models. Subgroup analyses by ethnicity, PCOS diagnostic criteria, and source of controls were conducted. Moreover, false-positive report probability (FPRP) test and trial sequential analysis (TSA) were performed to assess the significant associations. The results showed a possible negative association between UCSNP-19 and PCOS risk (ins/ins vs. del/del + del/ins: OR = 0.84, 95% CI: 0.72–0.98). In subgroup analyses, FPRP test indicated that noteworthy associations were observed in mixed ethnicities for UCSNP-43 (A vs. G: OR = 1.81, 95% CI: 1.17-2.79; AA + AG vs. GG: OR = 2.14, 95% CI: 1.20-3.80) and in Asians for UCSNP-44 (CC vs. TT: OR = 2.07, 95% CI: 1.21-3.51; CC vs. CT + TT: OR = 2.19, 95% CI: 1.31-3.69), but TSA plots showed that the accumulated sample sizes of these associations were insufficient to draw firm conclusions. In summary, our study suggested that UCSNP-19, UCSNP-43, and UCSNP-44 in CAPN10 gene may be involved in PCOS susceptibility. These findings warrant further studies.
1 Introduction
Polycystic ovarian syndrome (PCOS) is the most common reproductive endocrine disorder affecting 5%-20% of women at childbearing age worldwide, depending on the diagnostic criteria (Azziz et al., 2016; Skiba et al., 2018). The condition is characterized by hyperandrogenism, ovulatory dysfunction and polycystic ovarian morphology, and is commonly accompanied by metabolic abnormalities such as insulin resistance, hyperinsulinemia and dyslipidemia (Fauser et al., 2012). PCOS is the primary cause of anovulatory subfertility and increases the risk of pregnancy complications, resulting in major health and economic costs (Roos et al., 2011; Liu et al., 2021). In addition, women with PCOS have increased lifetime risks for type 2 diabetes mellitus (T2DM), cardiovascular events, psychiatric disorders and gynecological cancers (Anagnostis et al., 2018; Cooney and Dokras, 2018). To data, the exact etiology and pathogenesis of PCOS remains unclear. It is believed to result from complex interactions between genetic, behavioral and environmental factors (Azziz et al., 2016). Multiple genes involved in steroid biosynthesis and metabolism, sex hormone regulation, insulin sensitivity and inflammation may be associated with susceptibility to PCOS (Hiam et al., 2019).
Calpain10 (CAPN10) gene is located on the human chromosome 2q37.3 and consists of 15 exons and 14 introns spanning 31 kb. It is the first susceptibility gene for T2DM identified through a genome-wide scan followed by positional cloning (Hanis et al., 1996; Horikawa et al., 2000). CAPN10 was ubiquitously expressed in most tissues and cell types, and played an important role in insulin secretion and insulin sensitivity (Rasmussen et al., 2002; Sáez et al., 2008). Researchers hypothesized that CAPN10 gene polymorphisms may contribute to the development of PCOS because that insulin resistance was considered as the common pathologic basis of T2DM and PCOS (Diamanti-Kandarakis and Dunaif, 2012).
Several common polymorphisms in CAPN10 gene have been studied concerning their potential effect on PCOS susceptibility: UCSNP-19, -22, -43, -44, -56, -58, -63, and -110. Most studies were focusing on UCSNP-19, -43, -44, and -63. However, the results remain controversial. There have been two meta-analyses that examined the association between CAPN10 gene polymorphisms and PCOS risk in 2012 and 2013, respectively (Huang et al., 2012; Shen et al., 2013). The meta-analysis by Huang et al. (Huang et al., 2012) identified significant associations between UCSNP-19, and -63 and PCOS risk among UCSNP-19, -43, -44 and -63. The latest meta-analysis in 2013 indicated that UCSNP-19, -44 and -63 polymorphisms may be risk factors for PCOS, especially among Asians (Shen et al., 2013). However, the above studies had some obvious shortcomings about study selection, data extraction and statistical analyses (Raihan et al., 2019); besides, adjusted alpha for multiple tests and required information size were not evaluated. Furthermore, several new studies from various countries were conducted to analyze this topic over the past decade. Combining them in updated meta-analysis could increase the overall statistical power to detect an effect.
Therefore, we carried out this systematic review and meta-analysis with trial sequential analysis to pool current evidence together for a more accurate evaluation of the association between CAPN10 gene polymorphisms and PCOS risk.
2 Methods
This systematic review and meta-analysis was designed and reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (Supplementary Table S1) (Page et al., 2021).
2.1 Search strategy
A comprehensive literature search was conducted through the following electronic databases: The Cochrane Library, Web of Science, PubMed, Embase, Chinese Biomedical Database (CBM), China National Knowledge Infrastructure (CNKI), and Wanfang Database. Keyword search was performed using the following terms: (PCOS OR “polycystic ovary syndrome” OR “polycystic ovarian syndrome”) AND (“calpain 10” OR calpain-10 OR calpain10 OR “CAPN 10” OR CAPN-10 OR CAPN10). The full details of search strategy were listed in Supplementary Appendix SA1. There were no restrictions on either date or language. The search was completed on 4 May 2022.
2.2 Inclusion and exclusion criteria
Studies selected in our systematic review had to meet all the following criteria: (1) case-control studies based on unrelated subjects; (2) the diagnosis of PCOS was clear; (3) human studies; (4) the study assessed the association between CAPN10 polymorphisms and PCOS susceptibility. The exclusion criteria included: (1) case-only studies; (2) family-based studies; (3) comments or review articles; (4) duplicate or overlapping data.
All records obtained through the database search were collected in the EndNote software. After removal of duplicates, all the titles and abstracts were screened to exclude the irrelevant studies and then the full texts of the remaining records were reviewed for eligibility independently by two researchers (YML and TH). Reference lists of the eligible studies were also reviewed.
2.3 Data extraction
The following items was extracted from each eligible study: the first author, year of publication, country, ethnicity, sample size, age of cases and controls, diagnostic criteria used for PCOS, source of control subjects, genotyping methods, and allele and genotype frequencies in cases and controls.
2.4 Study quality assessment
The quality of included studies was assessed using the Newcastle–Ottawa Scale (NOS) (Stang, 2010). The NOS criteria use a “star” rating system to judge study methodological quality based on three perspectives: selection, comparability, and exposure. Scores were ranged from 0 stars (worst) to 9 stars (best), with equal or higher than 7 indicating that the methodological quality was generally good.
Two researchers (YML and TH) independently performed study screening, data extraction and quality assessment based on the specified selection criteria. All disagreements were resolved by consensus after discussion with the other authors.
2.5 Quantitative data synthesis
Meta-analysis was performed for genetic variants with at least two available datasets using the STATA version 14.0 software (Stata Corporation, College Station, TX, United States). Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated under five genetic models: allelic model (mutant [M] allele vs. wild [W] allele), heterozygous model (MW vs. WW), homozygous model (MM vs. WW), recessive model (MM vs. WW + MW), and dominant model (MW + MM vs. WW) (Zintzaras and Lau, 2008).
The Cochrane’s Q test and I2 index were used to assess the heterogeneity across the eligible studies (Whitehead and Whitehead, 1991; Higgins and Thompson, 2002). p value for Q test < 0.1 or I2 index > 50% indicated significant heterogeneity, and random effect model (DerSimonian–Laird method) was conducted in meta-analysis. Otherwise, the fixed effect model (Mantel–Haenszel method) was applied.
Subgroup analyses were performed by ethnicity, diagnostic criteria, and source of controls to determine the source of heterogeneity. Funnel plots and Egger’s linear regression test were used to assess potential publication bias of included studies (Egger et al., 1997). Sensitivity analyses were conducted by excluding studies whose genotype frequencies in controls exhibited significant deviation from the Hardy–Weinberg equilibrium (HWE). Besides, leave-one-out sensitivity analyses were also performed to evaluate the influence of each study on the overall estimate.
The associations were considered statistically significant if the reported p value was <0.05. The false positive report probability (FPRP) values at different prior probability levels for all significant associations were calculated by the Excel spreadsheet which was offered on Wacholder’s website (Wacholder et al., 2004). An FPRP value < 0.2 indicated a noteworthy association.
2.6 Trial sequential analysis (TSA)
TSA was conducted to calculate the required information size (RIS), and to assess if quantitative findings were robust (Wetterslev et al., 2008). We estimated the RIS based on a type I error of 5%, power of 80% and relative risk reduction assumption of 20% (Møller et al., 2008). If a trial sequential monitoring boundary is crossed before the RIS is reached in a cumulative meta-analysis, firm evidence may have been established and further studies were superfluous; if the boundaries are not surpassed, it is most probably necessary to conduct more studies in order to detect or reject a certain association (Møller et al., 2008). TSA was performed using TSA 0.9.5.10 Beta software (Copenhagen Trial Unit, Denmark).
3 Results
3.1 Study selection
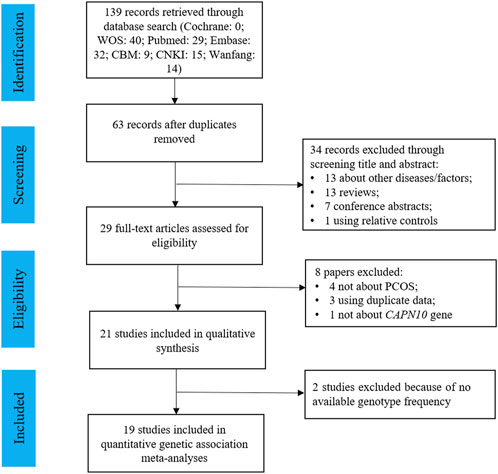
Figure 1 presented the study selection process for this review. Initially, a total of 139 articles were identified by searching various databases, and 63 non-duplicated articles were included in the title and abstract screening. 34 articles were excluded for not about PCOS (n = 7), not about CAPN10 polymorphisms (n = 6), reviews (n = 13), conference abstracts (n = 7), or family-based studies (n = 1), and the remaining 29 papers were included in the full-text review. Finally, 21 studies were eligible for the systematic review, and 19 of them had sufficient data for meta-analysis for 5 common variants: UCSNP-19, -43, -44, -56 and -63.
3.2 Characteristics of eligible studies
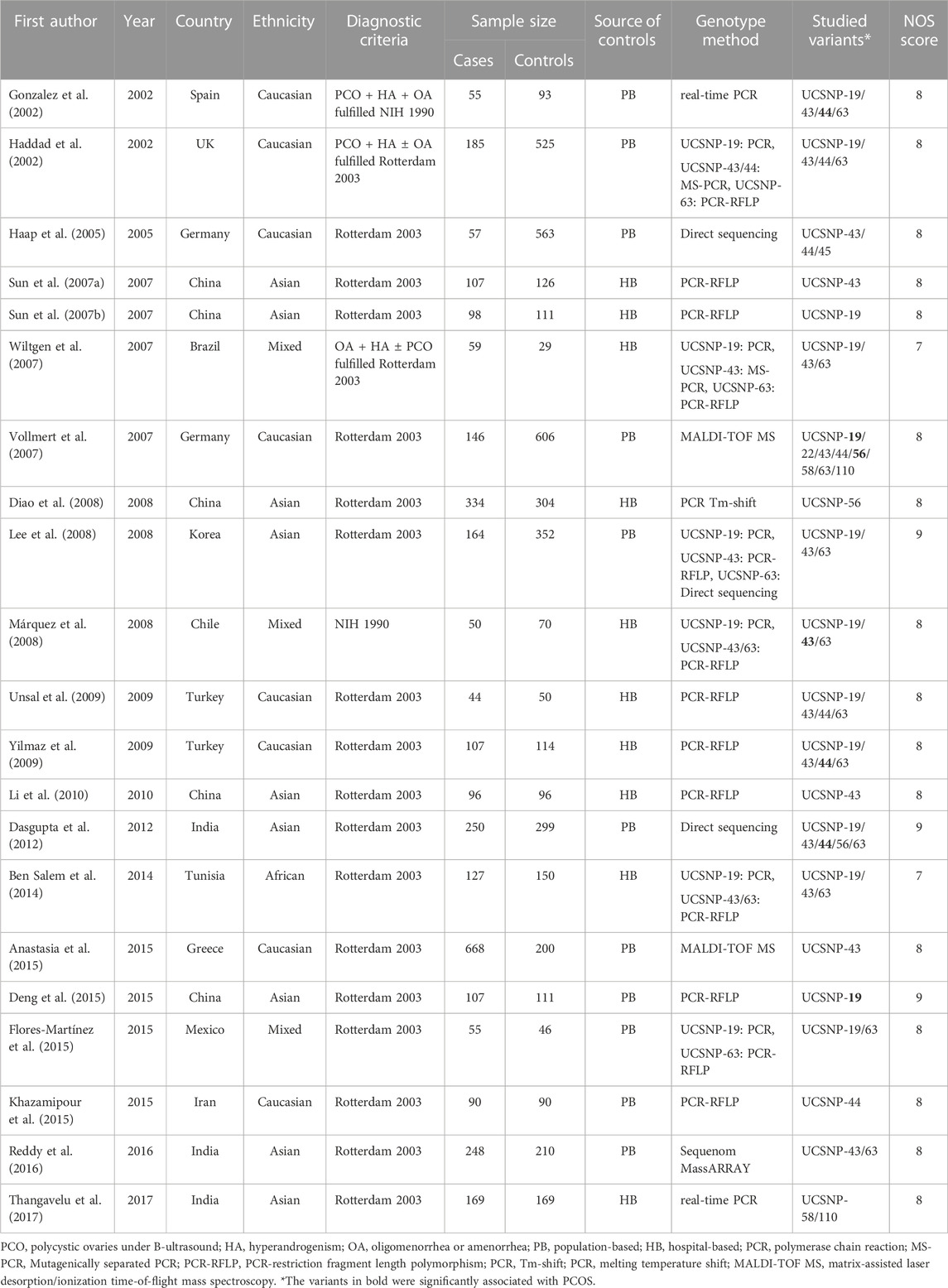
Table 1 summarized the main characteristics of the 21 studies included in the systematic review. All the studies were published between 2002 and 2017. Among them, 9 studies were conducted in Asians, 8 studies in Caucasians, 3 studies in mixed population, and 1 in Africans. 11 studies used population-based controls, and 10 studies used hospital-based controls. The majority of studies adopted the revised Rotterdam criteria for definition of PCOS (n = 19), and the remaining 2 studies adopted the National Institutes of Health (NIH) criteria. All the included studies had good quality with the NOS score ranging from 7 to 9.
Nine common variants in CAPN10 gene were investigated, including UCSNP-19 (rs3842570 Del>Ins), UCSNP-22 (rs2953152 G>A), UCSNP-43 (rs3792267 G>A), UCSNP-44 (rs2975760 T>C), UCSNP-45 (rs2241766 A>C), UCSNP-56 (rs2975762 G>A), UCSNP-58 (rs2975766 A>G), UCSNP-63 (rs5030952 C>T) and UCSNP-110 (rs7607759 A>G). UCSNP-43 was the focus of most studies (n = 15), followed by UCSNP-19 (n = 13), UCSNP-63 (n = 12) and UCSNP-44 (n = 8). There were 3, 2 and 2 studies for UCSNP-56, UCSNP-58 and UCSNP-110, and one study for each of UCSNP-22 and UCSNP-45.
As shown in Supplementary Table S2, more than two available sets of allele or genotype frequency data were extracted from these studies about UCSNP-19, -43, -44, -56, and -63. The genotype distribution in controls was consistent with HWE for UCSNP-19, and -56. However, significant deviation from HWE was detected in genotype frequencies of controls in one study for UCSNP-43 (Dasgupta et al., 2012), in two studies for UCSNP-44 (Yilmaz et al., 2009; Dasgupta et al., 2012) and in three studies for UCSNP-63 (Lee et al., 2008; Yilmaz et al., 2009; Dasgupta et al., 2012).
3.3 Association between UCSNP-19 and PCOS
12 studies consisting of 1299 PCOS patients and 1948 controls were included in the meta-analysis for UCSNP-19. The pooled estimate of recessive model revealed a significant association (ins/ins vs. del/del + del/ins: OR = 0.84, 95% CI: 0.72–0.98) (Table 2; Figure 2). In the subgroup analyses, only the recessive model in studies with population-based controls showed a significant association (ins/ins vs. del/del + del/ins: OR = 0.81, 95% CI: 0.68–0.98) (Table 3; Figure 2). Further, the significant association between UCSNP-19 and PCOS remained noteworthy at the prior probability of 0.1 and FPRP threshold of 0.2 (Supplementary Table S3).
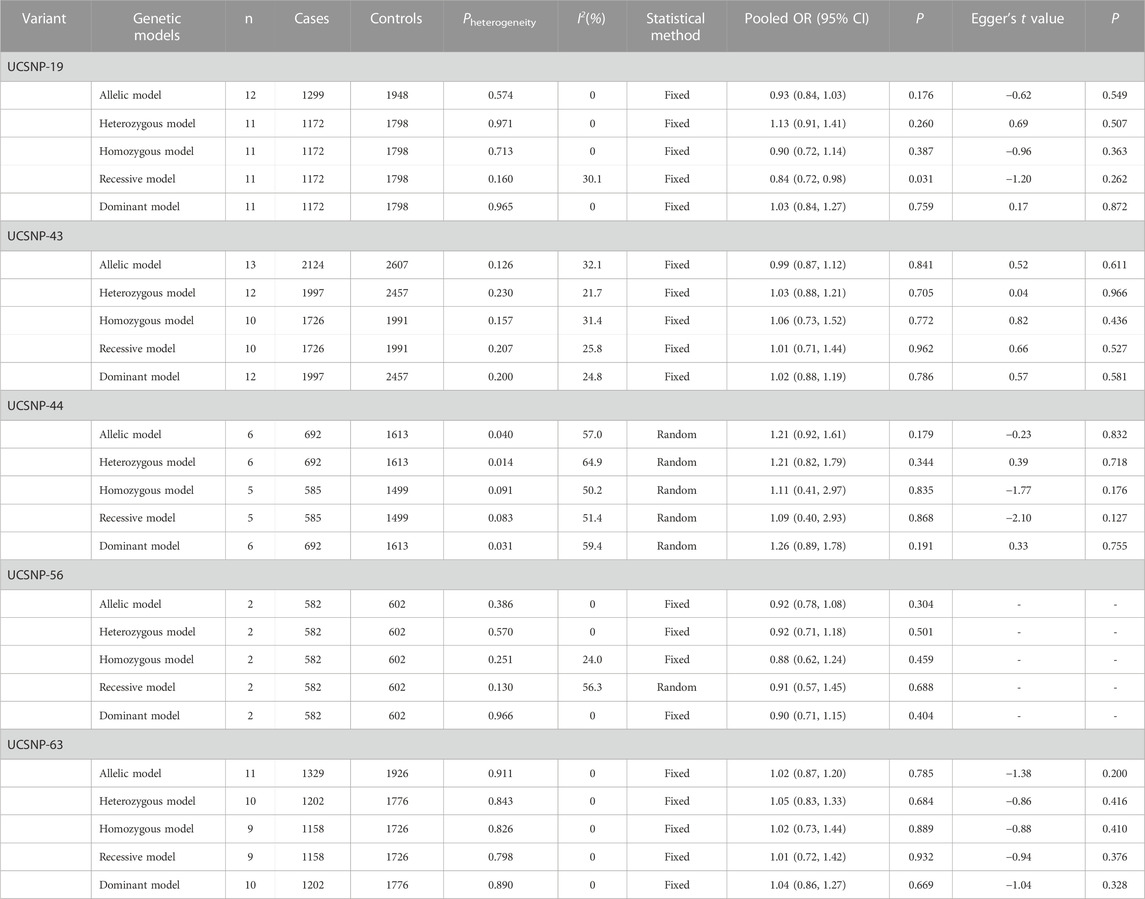
TABLE 2. Overall meta-analysis results for the association between CAPN10 polymorphisms and PCOS risk.
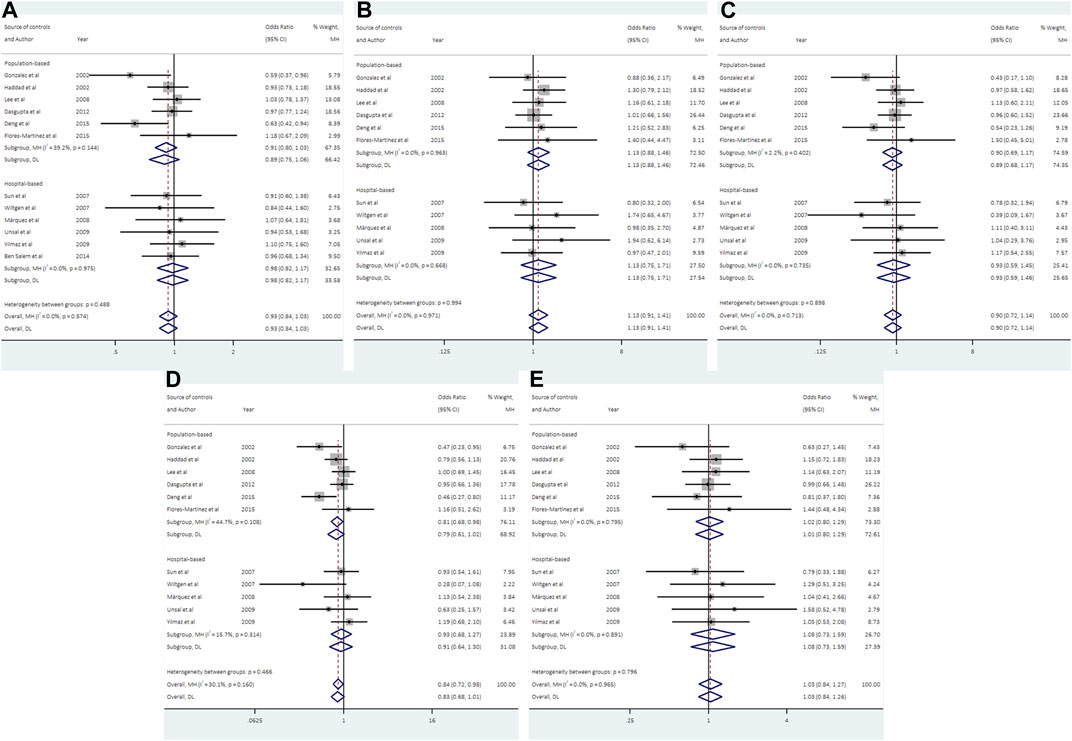
FIGURE 2. Forest plots of association between UCSNP-19 and PCOS susceptibility by source of controls [(A): allelic model, (B) heterozygous model, (C) homozygous model, (D) recessive model, (E) dominant model].
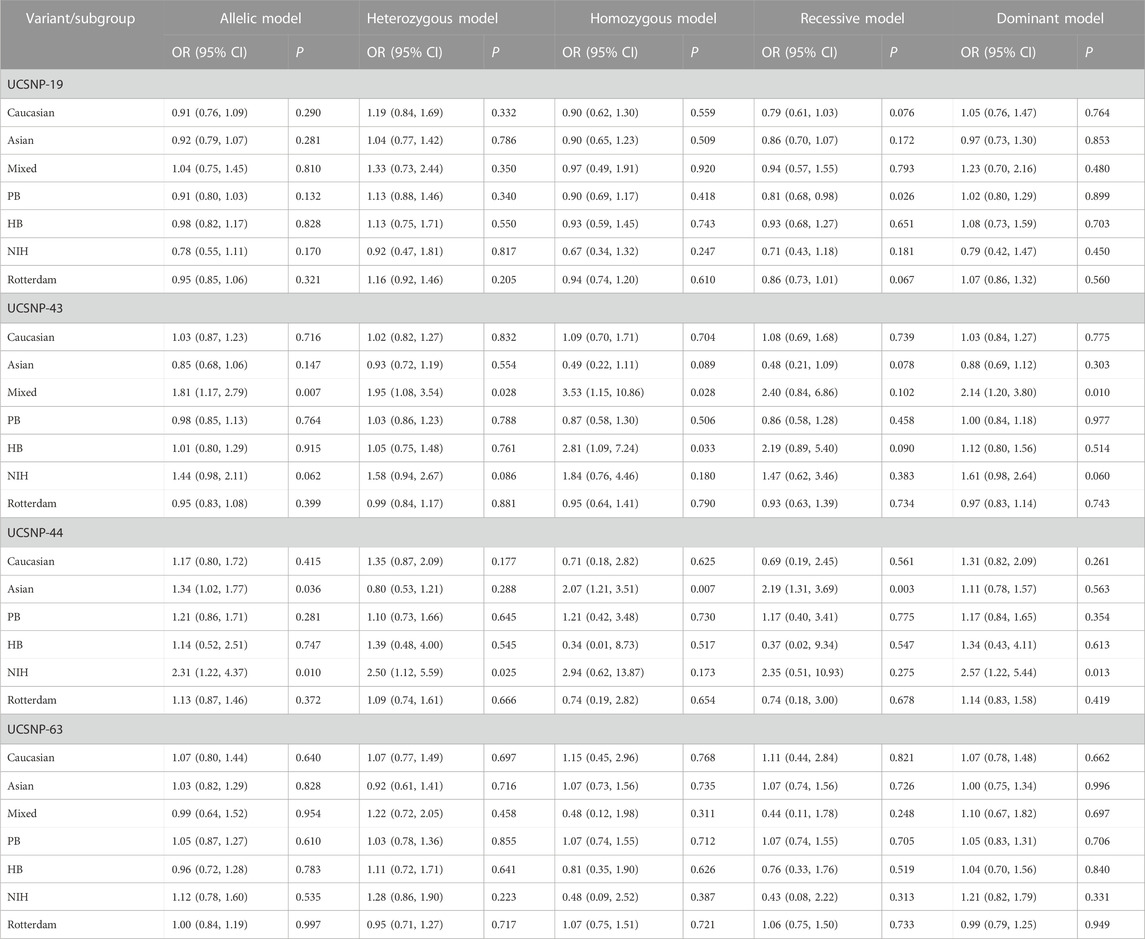
TABLE 3. Stratified meta-analysis results for the association between CAPN10 polymorphisms and PCOS risk.
3.4 Association between UCSNP-43 and PCOS
With regard to UCSNP-43, 13 studies involving 2124 cases and 2607 controls met the eligibility criteria for meta-analysis. No significant associations between UCSNP-43 and PCOS susceptibility were observed under all genetic models (Table 2; Figure 3). In the subgroup analyses by ethnicity, significant associations were observed in allelic model (A vs. G: OR = 1.81, 95% CI: 1.17-2.79), heterozygous model (AG vs. GG: OR = 1.95, 95% CI: 1.08-3.54), homozygous model (AA vs. GG: OR = 3.53, 95% CI: 1.15-10.86) and dominant model (AA + AG vs. GG: OR = 2.14, 95% CI: 1.20-3.80) in mixed ethnicities (Table 3; Figure 3). When stratified by source of controls, the significant association of homozygous model (AA vs. GG: OR = 2.81, 95% CI: 1.09-7.24) was found in the subgroup of hospital-based controls (Table 3). But FPRP test indicated that the associations were noteworthy in allelic model and dominant model among mixed ethnicities (Supplementary Table S2).
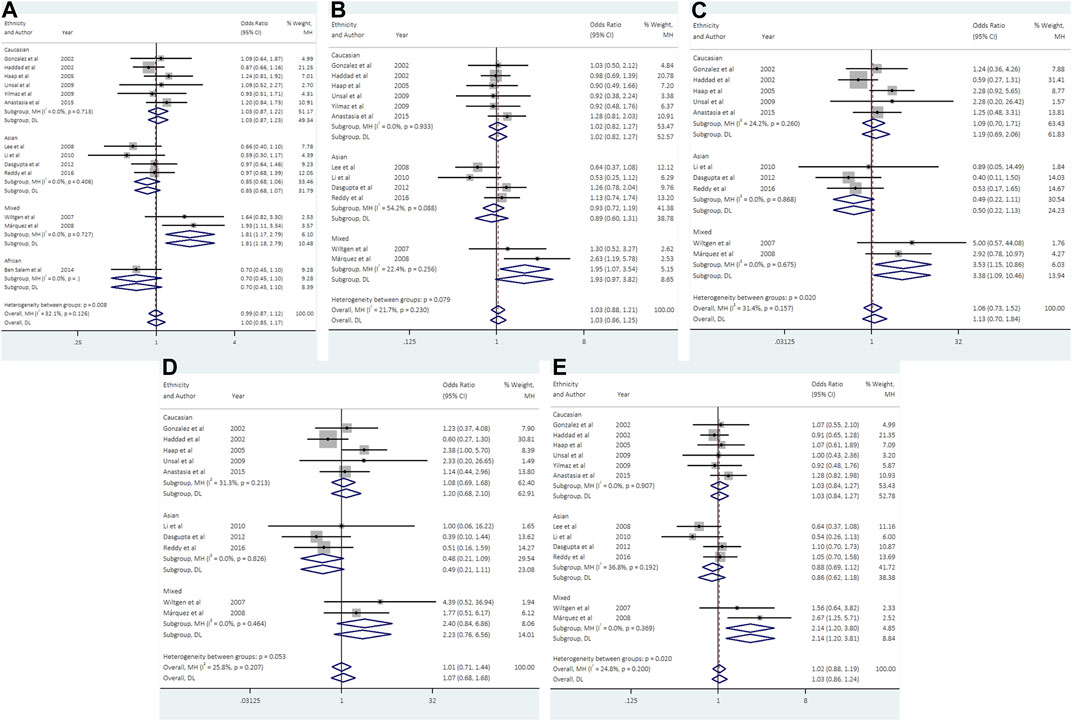
FIGURE 3. Forest plots of association between UCSNP-43 and PCOS susceptibility by ethnicity [(A): allelic model, (B) heterozygous model, (C) homozygous model, (D) recessive model, (E) dominant model].
3.5 Association between UCSNP-44 and PCOS
Data from 6 studies (692 cases and 1613 controls) showed that UCSNP-44 was not significantly associated with PCOS risk in all genetic models (Table 2; Figure 4). In the subgroup analyses by ethnicity, significant associations were observed in allelic model (C vs. T: OR = 1.34, 95% CI: 1.02-1.77), homozygous model (CC vs. TT: OR = 2.07, 95% CI: 1.21-3.51) and recessive model (CC vs. CT + TT: OR = 2.19, 95% CI: 1.31-3.69) in Asians (Table 3; Figure 4). When stratified according to PCOS diagnostic criteria, we observed significant associations of allelic model (C vs. T: OR = 2.31, 95% CI: 1.22-4.37), heterozygous model (CT vs. TT: OR = 2.50, 95% CI: 1.12-5.59) and dominant model (CC + CT vs. TT: OR = 2.57, 95% CI: 1.22-5.44) in NIH subgroup (Table 3). The significant associations remained noteworthy in homozygous model and recessive model among Asians in the FPRP tests (Supplementary Table S2).
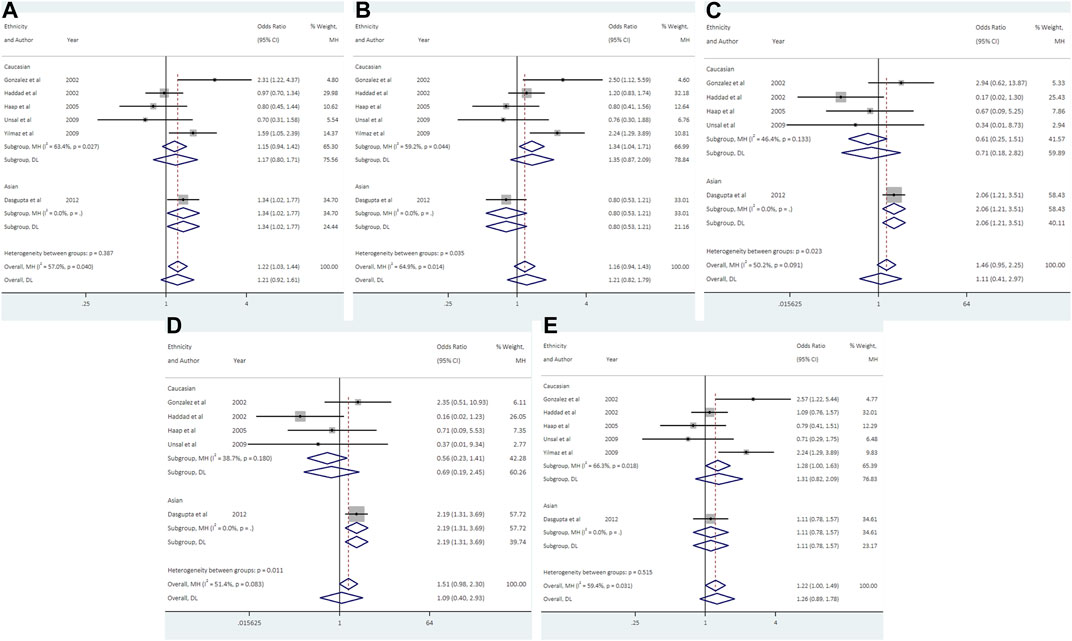
FIGURE 4. Forest plots of association between UCSNP-44 and PCOS susceptibility by ethnicity [(A): allelic model, (B) heterozygous model, (C) homozygous model, (D) recessive model, (E) dominant model].
3.6 Association between UCSNP-56, UCSNP-63 and PCOS
For UCSNP-56, 2 studies consisting of 582 cases and 602 controls were included in the meta-analysis. No significant associations were observed under all genetic models (Table 2; Supplementary Figure S1). The subgroup analyses were not performed because of the limited studies included.
For UCSNP-63, 11 studies involving 1329 cases and 1926 controls were eligible for meta-analysis. No significant associations were observed under all genetic models in overall and subgroup analyses (Tables 2, 3; Supplementary Figure S2).
3.7 Publication bias
The results of Egger’s test still did not suggest any evidence of publication bias in the analysis (Table 2). Besides, the shape of the funnel plots did not reveal any evidence of obvious asymmetry in all genetic models for these five SNPs (Supplementary Figures S3–S7).
3.8 Sensitivity analyses
Sensitivity analyses were carried out by excluding studies not in HWE for UCSNP-43, UCSNP-44, and UCSNP-63, and all the results remained practically unchanged (Supplementary Table S3). Sensitivity analyses were also performed to evaluate the influence of a specific study on the overall estimate. We observed that omission of any single study had little effect on the combined results for UCSNP-43 and UCSNP-63. For UCSNP-19, the pooled ORs were insignificant under the recessive model when omitting each of the following studies: Gonzalez et al., 2002, Haddad et al., 2002, Wiltgen et al., 2007, and Deng et al., 2015. Conversely, the pooled ORs of homozygous model and recessive model for UCSNP-44 become statistically significant after exclusion of the study by Haddad et al., 2002 (Supplementary Table S4).
3.9 Trial sequential analysis
The TSA showed that cumulative Z curve reached the RIS and crossed the conventional boundary, but it did not cross the trial sequential monitoring boundary, suggesting that the correlation within UCSNP-19 polymorphism and PCOS susceptibility may be invalid and more studies are needed for stable conclusion (Figure 5). For UCSNP-43 and UCSNP-44, the cumulative Z curves crossed the conventional boundary, but it did not cross the trial sequential monitoring boundary and reach the RIS, which suggested that more studies were needed to confirm these findings in subgroup analyses (Supplementary Figures S8–S11).
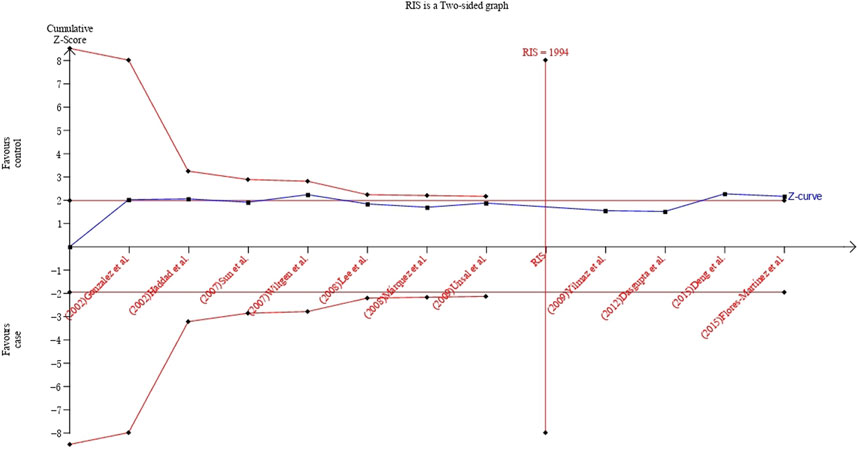
FIGURE 5. TSA plot of the recessive model for the association between UCSNP-19 and PCOS susceptibility.
4 Discussion
The identification of genetic variants associated with PCOS and related traits has the potential to inform our understanding for pathogenesis of PCOS. The current study provided a systematic assessment of the association between CAPN10 polymorphisms and susceptibility to PCOS. The results showed a possible negative association between UCSNP-19 and PCOS susceptibility. UCSNP-43 and UCSNP-44 may be associated with increased risk of PCOS among mixed ethnicities and Asians, respectively. However, no significant association was observed between UCSNP-22, -45, -56, -58, -63 and -110 polymorphisms and PCOS risk.
Our finding about the significant association of UCSNP-19 with PCOS was consistent with the previous meta-analyses by Huang et al. (2012) and Shen et al. (2013). UCSNP-19, localized in intron 6 of CAPN10 gene, is a 2-allele polymorphism consisting of 2 or 3 copies of 32-bp sequence. The association may be attributed to the effect of UCSNP-19 polymorphism or the possible haplotypes of CAPN10 gene. Although UCSNP-19 did not alter the amino acid sequence, the variable number of tandem repeat polymorphism may be involved in the transcription modification and consequent protein expression (Nakamura et al., 1998; Vollmert et al., 2007). The haplotype combination defined by UCSNP-43, -19, and -63 alleles was reported to confer increased risk of PCOS or T2DM or insulin resistance in different studies (Horikawa et al., 2000; Ehrmann et al., 2002; Sáez et al., 2008; Lee et al., 2009). Besides, UCSNP-19 and UCSNP-56 were found to be in strong linkage disequilibrium and showed significant association with PCOS in German women (Vollmert et al., 2007).
In subgroup analyses, ethnic-specific genetic associations of UCSNP-43 and -44 with PCOS were observed in our study. The role of these two SNPs in PCOS susceptibility have been attracted much attention due to the strong relevance between two SNPs and T2DM. The UCSNP-43 in intron 3 was the variant most strongly linked with T2DM among Mexican Americans (Horikawa et al., 2000). Afterwards, the UCSNP-43, either individually or in combination with UCSNP-19 and -63, was found to contribute to PCOS susceptibility in specific population (Ehrmann et al., 2002; Márquez et al., 2008; Lee et al., 2009). UCSNP-44, located in a transcription enhancer element next to UCSNP-43, was shown to be associated with T2DM in Mexican Americans and Europeans (Evans et al., 2001) and related to PCOS in several studies (Gonzalez et al., 2002; Yilmaz et al., 2009; Dasgupta et al., 2012). Moreover, experimental studies have suggested that the polymorphisms defined by UCSNP-43 and UCSNP-44 in intron 3 appear to regulate the expression of CAPN10 gene (Horikawa et al., 2000; Evans et al., 2001). Ethnicity is considered as a multidimensional construct that may vary greatly in genetic background, socioeconomic status, and lifestyle (Beatty Moody et al., 2021). These factors might have complex influence on effect allele frequencies of susceptibility variants, gene-environment interaction, and the incidence of PCOS (Kim and Choi, 2019; Khil et al., 2022). But it is worth noting that some ethnic subgroups in our study contained few studies without sufficient statistical power, and more studies in various populations are needed to confirm the results.
However, the present study did not show significant association between UCSNP-63 and PCOS susceptibility overall or within any subgroup, which was somewhat different from the previous meta-analyses (Huang et al., 2012; Shen et al., 2013). The discrepancies may be explained by the improper data extraction in the previous studies. The dataset erroneously derived from the haplotype data of a Korean study was found to be the outlier of forest plots for UCSNP-63, which were also proved by the leave-one-out sensitivity analyses (Huang et al., 2012; Shen et al., 2013). In fact, the association between UCSNP-63 and PCOS had not been observed in the individual studies. Our study further confirmed that the single variant UCSNP-63 probably has no effect on PCOS in the pooled sample.
Although the mechanisms underlying PCOS remain elusive, recent studies have suggested that hyperandrogenism and insulin resistance were the core etiology and primary endocrine characteristics of this condition (Diamanti-Kandarakis and Dunaif, 2012; Wang et al., 2019). Insulin resistance was found in approximately three-quarters of affected women, and played a crucial role in the pathophysiology of PCOS (Tosi et al., 2017; Ding et al., 2021). Hyperinsulinemia from insulin resistance contributes to hyperandrogenism via stimulation of ovarian androgen secretion and inhibition of hepatic sex hormone-binding globulin synthesis, leading to follicle arrest and anovulation (Diamanti-Kandarakis and Dunaif, 2012). In turn, androgen excess could impair insulin action in skeletal muscle and adipose tissue of women with PCOS, potentially setting up a vicious cycle (Corbould, 2008). As in T2DM, intrinsic defects in insulin secretion and action are critical for PCOS development (Diamanti-Kandarakis and Papavassiliou, 2006; Tripathy and Chavez, 2010; Shaaban et al., 2021). CAPN10 gene is the first putative diabetes gene and encodes the cysteine protease calpain10 that involved in pro-insulin processing and insulin secretion and action (Sreenan et al., 2001). Multiple polymorphisms in CAPN10 gene individually or in combination were demonstrated to regulate gene expression and be associated with insulin resistance and PCOS (Horikawa et al., 2000; Shaaban et al., 2021). Accordingly, CAPN10 may be an important susceptibility gene for PCOS.
4.1 Strengths and limitations
This systematic review had several strengths. First, our study has been well-designed with explicit criteria and methods for study selection, data extraction and data analysis compared with the previous meta-analyses (Huang et al., 2012; Shen et al., 2013), which make it advantageous to provide powerful and valid results. Second, FPRP test and TSA were used to justify the results and enhance the reliability of the results. However, our findings should be interpreted in the light of the following limitations. First of all, only published literatures with sufficient data were included in the meta-analysis. Therefore, publication bias may have occurred, even though the results of Egger’s linear regression tests did not show it. Second, the meta-analyses for some variants (e.g., UCSNP-56, UCSNP-44) with few studies and limited sample size may not have enough statistical power to explore the real association. Further large-scale validation studies are needed for these polymorphisms. Third, significant changes were observed in the leave-one-out sensitivity analyses of the recessive model for UCSNP-19 as well as the homozygous and recessive model for UCSNP-44, which had some impact on the robustness of the conclusions. Finally, PCOS is thought to be a complex disease resulting from interactions between multiple genes and environmental factors, but it is difficult to eliminate the effect of confounding factors in meta-analysis, which may affect the final results.
In conclusion, our study showed a possible negative association between UCSNP-19 and PCOS susceptibility, while UCSNP-43 and UCSNP-44 may be associated with increased risk of PCOS in specific ethnicities. Although the findings reflected the correlation between CAPN10 gene polymorphisms and PCOS, they were not robust enough to withstand statistical interrogation. Further rigorous studies in various populations are needed to draw more definite conclusions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
YML, YLW, and JYL designed the study; YML and TH conducted literature search and data extraction; YML conducted statistical analysis and wrote the manuscript; YXW, JG, and JLZ participated in interpreting the results and revised the draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Hunan Province (Grant Number: 2023JJ40357).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1153960/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Forest plots of association between UCSNP-56 and PCOS susceptibility [(A): allelic model, (B): heterozygous model, (C): homozygous model, (D): recessive model, (E): dominant model].
SUPPLEMENTARY FIGURE S2 | Forest plots of association between UCSNP-63 and PCOS susceptibility [(A): allelic model, (B): heterozygous model, (C): homozygous model, (D): recessive model, (E): dominant model].
SUPPLEMENTARY FIGURE S3 | Funnel plots of the five genetic models for the association between UCSNP-19 and PCOS susceptibility [(A): allelic model, (B): heterozygous model, (C): homozygous model, (D): recessive model, (E): dominant model].
SUPPLEMENTARY FIGURE S4 | Funnel plots of the five genetic models for the association between UCSNP-43 and PCOS susceptibility [(A): allelic model, (B): heterozygous model, (C): homozygous model, (D): recessive model, (E): dominant model].
SUPPLEMENTARY FIGURE S5 | Funnel plots of the five genetic models for the association between UCSNP-44 and PCOS susceptibility [(A): allelic model, (B): heterozygous model, (C): homozygous model, (D): recessive model, (E): dominant model].
SUPPLEMENTARY FIGURE S6 | Funnel plots of the five genetic models for the association between UCSNP-56 and PCOS susceptibility [(A): allelic model, (B): heterozygous model, (C): homozygous model, (D): recessive model, (E): dominant model].
SUPPLEMENTARY FIGURE S7 | Funnel plots of the five genetic models for the association between UCSNP-63 and PCOS susceptibility [(A): allelic model, (B): heterozygous model, (C): homozygous model, (D): recessive model, (E): dominant model].
SUPPLEMENTARY FIGURE S8 | TSA plot of the allelic model for the association between UCSNP-43 and PCOS susceptibility among mixed ethnicities.
SUPPLEMENTARY FIGURE S9 | TSA plot of the dominant model for the association between UCSNP-43 and PCOS susceptibility among mixed ethnicities.
SUPPLEMENTARY FIGURE S10 | TSA plot of the homozygous model for the association between UCSNP-44 and PCOS susceptibility among Asians.
SUPPLEMENTARY FIGURE S11 | TSA plot of the recessive model for the association between UCSNP-44 and PCOS susceptibility among Asians.
References
Anagnostis, P., Tarlatzis, B. C., and Kauffman, R. P. (2018). Polycystic ovarian syndrome (PCOS): long-term metabolic consequences. Metabolism 86, 33–43. doi:10.1016/j.metabol.2017.09.016
Anastasia, K., Koika, V., Roupas, N. D., Armeni, A., Marioli, D., Panidis, D., et al. (2015). Association of Calpain (CAPN) 10 (UCSNP-43, rs3792267) gene polymorphism with elevated serum androgens in young women with the most severe phenotype of polycystic ovary syndrome (PCOS). Gynecol. Endocrinol. 31 (8), 630–634. doi:10.3109/09513590.2015.1032932
Azziz, R., Carmina, E., Chen, Z., Dunaif, A., Laven, J. S., Legro, R. S., et al. (2016). Polycystic ovary syndrome. Nat. Rev. Dis. Prim. 2, 16057. doi:10.1038/nrdp.2016.57
Beatty Moody, D. L., Waldstein, S. R., Leibel, D. K., Hoggard, L. S., Gee, G. C., Ashe, J. J., et al. (2021). Race and other sociodemographic categories are differentially linked to multiple dimensions of interpersonal-level discrimination: implications for intersectional, health research. PLoS One 16 (5), e0251174. doi:10.1371/journal.pone.0251174
Ben Salem, A., Attaoua, R., Mtiraoui, N., Belkahla, S., Ezzidi, I., Ajina, M., et al. (2014). Common polymorphisms of calpain-10 and the risk of polycystic ovary syndrome in Tunisian population: A case-control study. Mol. Biol. Rep. 41 (10), 6569–6574. doi:10.1007/s11033-014-3540-8
Cooney, L. G., and Dokras, A. (2018). Beyond fertility: polycystic ovary syndrome and long-term health. Fertil. Steril. 110 (5), 794–809. doi:10.1016/j.fertnstert.2018.08.021
Corbould, A. (2008). Effects of androgens on insulin action in women: is androgen excess a component of female metabolic syndrome? Diabetes Metab. Res. Rev. 24 (7), 520–532. doi:10.1002/dmrr.872
Dasgupta, S., Sirisha, P. V., Neelaveni, K., Anuradha, K., and Reddy, B. M. (2012). Association of CAPN10 SNPs and haplotypes with polycystic ovary syndrome among South Indian Women. PLoS One 7 (2), e32192. doi:10.1371/journal.pone.0032192
Deng, M., Liu, Y., Zhang, H., Cen, J., and Zhang, Z. (2015). Association of CAPN-10 SNP-19 with polycystic ovary syndrome. Zhejiang Med. J. 37 (11), 912–915. 30. Available at: http://www.zjyxzzs.com/ch/reader/view_abstract.aspx?file_no=2014-787&flag=1.
Diamanti-Kandarakis, E., and Dunaif, A. (2012). Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr. Rev. 33 (6), 981–1030. doi:10.1210/er.2011-1034
Diamanti-Kandarakis, E., and Papavassiliou, A. G. (2006). Molecular mechanisms of insulin resistance in polycystic ovary syndrome. Trends Mol. Med. 12 (7), 324–332. doi:10.1016/j.molmed.2006.05.006
Diao, X. H., Shi, Y. H., Gao, Q., Wang, L. C., Tang, R., and Chen, Z. J. (2008). Relationship between single nucleotide polymorphism-56 of calpain-10 gene and glucose and lipid metabolism in polycystic ovary syndrome patients. Zhonghua Fu Chan Ke Za Zhi 43 (2), 106–109. doi:10.3321/j.issn:0529-567X.2008.02.007
Ding, H., Zhang, J., Zhang, F., Zhang, S., Chen, X., Liang, W., et al. (2021). Resistance to the insulin and elevated level of androgen: A major cause of polycystic ovary syndrome. Front. Endocrinol. (Lausanne) 12, 741764. doi:10.3389/fendo.2021.741764
Egger, M., Davey Smith, G., Schneider, M., and Minder, C. (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ 315 (7109), 629–634. doi:10.1136/bmj.315.7109.629
Ehrmann, D. A., Schwarz, P. E., Hara, M., Tang, X., Horikawa, Y., Imperial, J., et al. (2002). Relationship of calpain-10 genotype to phenotypic features of polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 87 (4), 1669–1673. doi:10.1210/jcem.87.4.8385
Evans, J. C., Frayling, T. M., Cassell, P. G., Saker, P. J., Hitman, G. A., Walker, M., et al. (2001). Studies of association between the gene for calpain-10 and type 2 diabetes mellitus in the United Kingdom. Am. J. Hum. Genet. 69 (3), 544–552. doi:10.1086/323315
Fauser, B. C., Tarlatzis, B. C., Rebar, R. W., Legro, R. S., Balen, A. H., Lobo, R., et al. (2012). Consensus on women's health aspects of polycystic ovary syndrome (PCOS): the Amsterdam ESHRE/ASRM-Sponsored 3rd PCOS consensus workshop group. Fertil. Steril. 97 (1), 28–38. doi:10.1016/j.fertnstert.2011.09.024
Flores-Martínez, S. E., Castro-Martínez, A. G., López-Quintero, A., García-Zapién, A. G., Torres-Rodríguez, R. N., and Sánchez-Corona, J. (2015). Association analysis of SNP-63 and indel-19 variant in the calpain-10 gene with polycystic ovary syndrome in women of reproductive age. Cir. Cir. 83 (1), 35–42. doi:10.1016/j.circir.2015.04.021
Gonzalez, A., Abril, E., Roca, A., Aragón, M. J., Figueroa, M. J., Velarde, P., et al. (2002). Comment: CAPN10 alleles are associated with polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 87 (8), 3971–3976. doi:10.1210/jcem.87.8.8793
Haap, M., Machicao, F., Stefan, N., Thamer, C., Tschritter, O., Schnuck, F., et al. (2005). Genetic determinants of insulin action in polycystic ovary syndrome. Exp. Clin. Endocrinol. Diabetes 113 (5), 275–281. doi:10.1055/s-2005-837665
Haddad, L., Evans, J. C., Gharani, N., Robertson, C., Rush, K., Wiltshire, S., et al. (2002). Variation within the type 2 diabetes susceptibility gene calpain-10 and polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 87 (6), 2606–2610. doi:10.1210/jcem.87.6.8608
Hanis, C. L., Boerwinkle, E., Chakraborty, R., Ellsworth, D. L., Concannon, P., Stirling, B., et al. (1996). A genome-wide search for human non-insulin-dependent (type 2) diabetes genes reveals a major susceptibility locus on chromosome 2. Nat. Genet. 13 (2), 161–166. doi:10.1038/ng0696-161
Hiam, D., Moreno-Asso, A., Teede, H. J., Laven, J. S. E., Stepto, N. K., Moran, L. J., et al. (2019). The genetics of polycystic ovary syndrome: an overview of candidate gene systematic reviews and genome-wide association studies. J. Clin. Med. 8 (10), 1606. doi:10.3390/jcm8101606
Higgins, J. P., and Thompson, S. G. (2002). Quantifying heterogeneity in a meta-analysis. Stat. Med. 21 (11), 1539–1558. doi:10.1002/sim.1186
Horikawa, Y., Oda, N., Cox, N. J., Li, X., Orho-Melander, M., Hara, M., et al. (2000). Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 26 (2), 163–175. doi:10.1038/79876
Huang, M., Xiao, J., Zhao, X., Liu, C., and Chen, Q. (2012). Four polymorphisms of the CAPN 10 gene and their relationship to polycystic ovary syndrome susceptibility: A meta-analysis. Clin. Endocrinol. (Oxf) 76 (3), 431–438. doi:10.1111/j.1365-2265.2011.04218.x
Khazamipour, A., Nejatizadeh, A., Kalantar, S. M., Khashavi, Z., and Kalantar, S. M. (2015). Association study of calpain-10 gene polymorphism in patients with polycystic ovarian syndrome. Genet. Third Millenn. 13 (1), 3924–3930. Available at: https://www.researchgate.net/publication/281999427.
Khil, J., Darbinian, J. A., Guo, L., Greenspan, L. C., Ramalingam, N. D., and Lo, J. C. (2022). Ethnic diversity and burden of polycystic ovary syndrome among US adolescent females. J. Pediatr. Endocrinol. Metab. 35 (6), 821–825. doi:10.1515/jpem-2022-0160
Kim, J. J., and Choi, Y. M. (2019). Phenotype and genotype of polycystic ovary syndrome in asia: ethnic differences. J. Obstet. Gynaecol. Res. 45 (12), 2330–2337. doi:10.1111/jog.14132
Lee, H., Pyun, G., Byun, E., Oh, J., Oh, J-Y., Hong, Y., et al. (2008). Caffeic acid phenethyl ester inhibits invasion and expression of matrix metalloproteinase in SK-Hep1 human hepatocellular carcinoma cells by targeting nuclear factor kappa B. J. Korean Endocr. Soc. 23 (5), 319–322. doi:10.1007/s12263-007-0067-9
Lee, J. Y., Lee, W. J., Hur, S. E., Lee, C. M., Sung, Y. A., and Chung, H. W. (2009). 111/121 diplotype of Calpain-10 is associated with the risk of polycystic ovary syndrome in Korean women. Fertil. Steril. 92 (2), 830–833. doi:10.1016/j.fertnstert.2008.06.023
Li, R., Xu, L., and Xiong, D. (2010). Association between calpain-10 gene polymorphisms and susceptibility to PCOS. J. Sichuan Univ. Med. Sci. Ed. 41 (2), 337–339. 47. doi:10.13464/j.scuxbyxb.2010.02.028
Liu, J., Wu, Q., Hao, Y., Jiao, M., Wang, X., Jiang, S., et al. (2021). Measuring the global disease burden of polycystic ovary syndrome in 194 countries: global burden of disease study 2017. Hum. Reprod. 36 (4), 1108–1119. doi:10.1093/humrep/deaa371
Márquez, J. L., Pacheco, A., Valdés, P., and Salazar, L. A. (2008). Association between CAPN10 UCSNP-43 gene polymorphism and polycystic ovary syndrome in Chilean women. Clin. Chim. Acta 398 (1-2), 5–9. doi:10.1016/j.cca.2008.07.028
Møller, C. H., Penninga, L., Wetterslev, J., Steinbrüchel, D. A., and Gluud, C. (2008). Clinical outcomes in randomized trials of off-vs. on-pump coronary artery bypass surgery: systematic review with meta-analyses and trial sequential analyses. Eur. Heart J. 29 (21), 2601–2616. doi:10.1093/eurheartj/ehn335
Nakamura, Y., Koyama, K., and Matsushima, M. (1998). VNTR (variable number of tandem repeat) sequences as transcriptional, translational, or functional regulators. J. Hum. Genet. 43 (3), 149–152. doi:10.1007/s100380050059
Page, M. J., McKenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372, 790–799. doi:10.1016/j.rec.2021.07.010
Raihan, S., Shariff, D., and Bizzari, S. (2019). Comments on the latest meta-analysis of CAPN10 polymorphism associations with polycystic ovary syndrome. Genet. Res. (Camb) 717, 144006. doi:10.1016/j.gene.2019.144006
Rasmussen, S. K., Urhammer, S. A., Berglund, L., Jensen, J. N., Hansen, L., Echwald, S. M., et al. (2002). Variants within the calpain-10 gene on chromosome 2q37 (NIDDM1) and relationships to type 2 diabetes, insulin resistance, and impaired acute insulin secretion among Scandinavian Caucasians. Diabetes 51 (12), 3561–3567. doi:10.2337/diabetes.51.12.3561
Reddy, B. M., Kommoju, U. J., Dasgupta, S., and Rayabarapu, P. (2016). Association of type 2 diabetes mellitus genes in polycystic ovary syndrome aetiology among women from southern India. Indian J. Med. Res. 144 (3), 400–408. doi:10.4103/0971-5916.198678
Roos, N., Kieler, H., Sahlin, L., Ekman-Ordeberg, G., Falconer, H., and Stephansson, O. (2011). Risk of adverse pregnancy outcomes in women with polycystic ovary syndrome: population based cohort study. BMJ 343, d6309. doi:10.1136/bmj.d6309
Sáez, M. E., González-Sánchez, J. L., Ramírez-Lorca, R., Martínez-Larrad, M. T., Zabena, C., González, A., et al. (2008). The CAPN10 gene is associated with insulin resistance phenotypes in the Spanish population. PLoS One 3 (8), e2953. doi:10.1371/journal.pone.0002953
Shaaban, Z. K. A., Amiri-Yekta, A., Nowzari, F., Jafarzadeh Shirazi, M. R., and Tamadon, A. (2021). Pathophysiologic mechanisms of insulin secretion and signaling-related genes in etiology of polycystic ovary syndrome. Genet. Res. (Camb). 2021, 7781823. doi:10.1155/2021/7781823
Shen, W., Li, T., Hu, Y., Liu, H., and Song, M. (2013). Calpain-10 genetic polymorphisms and polycystic ovary syndrome risk: A meta-analysis and meta-regression. Gene 531 (2), 426–434. doi:10.1016/j.gene.2013.08.072
Skiba, M. A., Islam, R. M., Bell, R. J., and Davis, S. R. (2018). Understanding variation in prevalence estimates of polycystic ovary syndrome: A systematic review and meta-analysis. Hum. Reprod. Update 24 (6), 694–709. doi:10.1093/humupd/dmy022
Sreenan, S. K., Zhou, Y. P., Otani, K., Hansen, P. A., Currie, K. P., Pan, C. Y., et al. (2001). Calpains play a role in insulin secretion and action. Diabetes 50 (9), 2013–2020. doi:10.2337/diabetes.50.9.2013
Stang, A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25 (9), 603–605. doi:10.1007/s10654-010-9491-z
Sun, X., Shi, Y., Gao, Q., Wang, L., Yan, J., and Chen, Z. (2007a). Research on association of Calpain10 gene SNP-43 with polycystic ovary syndrome. J. Pract. Obstetrics Gynecol. 23 (6), 345–347. doi:10.3969/j.issn.1003-6946.2007.06.011
Sun, X., Gao, Q., Shi, Y., Yan, J., Zhao, L., Tang, R., et al. (2007b). Research on association of calpain10 gene SNP-19 with polycystic ovary syndrome. Chin. J. Obstetrics Gynecol. 42 (9), 628–630. doi:10.3760/j.issn:0529-567x.2007.09.013
Thangavelu, M., Godla, U. R., Paul Solomon, F. D., and Maddaly, R. (2017). Single-nucleotide polymorphism of INS, INSR, IRS1, IRS2, PPAR-G and CAPN10 genes in the pathogenesis of polycystic ovary syndrome. J. Genet. 96 (1), 87–96. doi:10.1007/s12041-017-0749-z
Tosi, F., Bonora, E., and Moghetti, P. (2017). Insulin resistance in a large cohort of women with polycystic ovary syndrome: A comparison between euglycaemic-hyperinsulinaemic clamp and surrogate indexes. Hum. Reprod. 32 (12), 2515–2521. doi:10.1093/humrep/dex308
Tripathy, D., and Chavez, A. O. (2010). Defects in insulin secretion and action in the pathogenesis of type 2 diabetes mellitus. Curr. Diab Rep. 10 (3), 184–191. doi:10.1007/s11892-010-0115-5
Unsal, T., Konac, E., Yesilkaya, E., Yilmaz, A., Bideci, A., Ilke Onen, H., et al. (2009). Genetic polymorphisms of FSHR, CYP17, CYP1A1, CAPN10, INSR, SERPINE1 genes in adolescent girls with polycystic ovary syndrome. J. Assist. Reprod. Genet. 26 (4), 205–216. doi:10.1007/s10815-009-9308-8
Vollmert, C., Hahn, S., Lamina, C., Huth, C., Kolz, M., Schöpfer-Wendels, A., et al. (2007). Calpain-10 variants and haplotypes are associated with polycystic ovary syndrome in Caucasians. Am. J. Physiol. Endocrinol. Metab. 292 (3), E836–E844. doi:10.1152/ajpendo.00584.2005
Wacholder, S., Chanock, S., Garcia-Closas, M., El Ghormli, L., and Rothman, N. (2004). Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 96 (6), 434–442. doi:10.1093/jnci/djh075
Wang, J., Wu, D., Guo, H., and Li, M. (2019). Hyperandrogenemia and insulin resistance: the chief culprit of polycystic ovary syndrome. Life Sci. 236, 116940. doi:10.1016/j.lfs.2019.116940
Wetterslev, J., Thorlund, K., Brok, J., and Gluud, C. (2008). Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J. Clin. Epidemiol. 61 (1), 64–75. doi:10.1016/j.jclinepi.2007.03.013
Whitehead, A., and Whitehead, J. (1991). A general parametric approach to the meta-analysis of randomized clinical trials. Stat. Med. 10 (11), 1665–1677. doi:10.1002/sim.4780101105
Wiltgen, D., Furtado, L., Kohek, M. B., and Spritzer, P. M. (2007). CAPN10 UCSNP-43, UCSNP-19 and UCSNP-63 polymorphisms and metabolic syndrome in polycystic ovary syndrome. Gynecol. Endocrinol. 23 (3), 173–178. doi:10.1080/09513590701233661
Yilmaz, M., Yurtçu, E., Demirci, H., Ergün, M. A., Ersoy, R., Karakoç, A., et al. (2009). Calpain 10 gene single-nucleotide 44 polymorphism may have an influence on clinical and metabolic features in patients with polycystic ovary syndrome. J. Endocrinol. Invest. 32 (1), 13–17. doi:10.1007/BF03345671
Keywords: Calpain10 gene, polymorphism, polycystic ovarian syndrome, systematic review, meta-analysis, false-positive report probability, trial sequential analysis
Citation: Li Y, Han T, Wang Y, Gao J, Zhang J, Wu Y and Luo J (2023) Association of Calpain10 polymorphisms with polycystic ovarian syndrome susceptibility: a systematic review and meta-analysis with trial sequential analysis. Front. Genet. 14:1153960. doi: 10.3389/fgene.2023.1153960
Received: 30 January 2023; Accepted: 21 August 2023;
Published: 01 September 2023.
Edited by:
Masood Bazrgar, Royan Institute, IranCopyright © 2023 Li, Han, Wang, Gao, Zhang, Wu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinglan Wu, eWluZ2xhbnd1MDkxN0AxMjYuY29t; Jiayou Luo, amlheW91bHVvQDEyNi5jb20=
 Yamei Li
Yamei Li Ting Han2
Ting Han2 Jiayou Luo
Jiayou Luo