- 1Department of Gastroenterology, The Sixth Affiliated Hospital, School of Medicine, South China University of Technology, Foshan, Guangdong, China
- 2Department of Gastroenterology, Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences), Southern Medical University, Guangzhou, China
- 3Department of Cardiology, Guangdong Provincial Hospital of Chinese Medicine, The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, China
Background: Recently, observational studies have reported that gastroesophageal reflux disease (GERD) is commonly associated with irritable bowel syndrome (IBS), but the causal relationship is unclear.
Methods: We conducted a two-sample Mendelian randomization study using summary data from genome-wide association studies (GWASs) to explore a causal relationship between GERD (N cases = 129,080) and IBS (N cases = 4,605) of European ancestry. Furthermore, the inverse-variance weighted (IVW) method and a series of sensitivity analyses were used to assess the accuracy and confidence of our results.
Results: We found a significant association of GERD with IBS (NSNP = 74; OR: 1.375; 95% CI: 1.164–1.624; p < 0.001). Reverse MR analysis showed no evidence of a causal association for IBS with GERD (NSNP = 6; OR: 0.996; 95% CI: 0.960–1.034; p = 0.845).
Conclusion: This study provides evidence that the presence of GERD increases the risk of developing IBS, and it is observed from the reverse MR results that IBS did not increase the risk of GERD.
1 Introduction
Gastroesophageal reflux disease (GERD) is one of the most common diseases of the digestive tract and is characterized by heartburn and reflux symptoms caused by reflux of stomach contents. It is estimated that approximately 20% of adults in the West suffer from GERD (Maret-Ouda et al., 2020; Mehta et al., 2021). Irritable bowel syndrome (IBS) is a chronic intestinal dysfunction characterized by altered intestinal function (frequency and/or consistency) and abdominal pain associated with intestinal function (Ford et al., 2017; Camilleri, 2021), with a prevalence of approximately 7%–16% of population in the United States suffering from IBS(Sperber et al., 2021). Both GERD and IBS have a significant financial impact on healthcare systems and have a negative impact on the quality of life (Black and Ford, 2020; Katzka and Kahrilas, 2020; Sperber et al., 2021).
Evidence from observational studies indicates that there is a degree of overlap between GERD and IBS patients (Monnikes et al., 2011; Plaidum et al., 2022), which indicates that GERD and IBS may share a common pathophysiological phenotype on the basis of pathogenic mechanisms (Gasiorowska et al., 2009). Furthermore, several systematic review studies have shown that patients diagnosed with GERD are at a significantly increased risk of being subsequently diagnosed with IBS, according to Lovell’s research, and the pooled OR for GERD in patients with IBS compared with patients without IBS was 4.17 (95% CI, 2.85–6.09). Similarly, patients with a GERD diagnosis were also at a significantly increased risk of IBS (El-Serag et al., 2009; Lovell and Ford, 2012). A randomized clinical trial study showed that in patients with overlapping GERD and IBS, there was a significant improvement in pre-existing IBS symptoms after proton pump inhibitor (PPI) treatment for GERD (Monnikes et al., 2012). However, single cross-sectional designs are frequently used in these observational studies, and there is uncertainty in the results because they are susceptible to potential bias from reverse causality and confounding factors. So far, few studies have investigated the causal relationship between GERD and IBS.
Mendelian randomization (MR) is a genetic instrumental variable approach that employs a single-nucleotide polymorphism (SNP) as an instrument variable (IV) to infer a causal association between two traits, and it has the advantage of reducing bias owing to reverse causality and confounding factors (Ziegler et al., 2015; Davies et al., 2018; Lord et al., 2021). The two-sample MR analysis is a statistical method based on the natural random allocation of genetic variations in evaluating a causal relationship within the summary statistics of GWASs. Therefore, we used a two-sample MR method to comprehensively assess the causal relationship between GERD and IBS.
2 Methods
2.1 Study design
We performed a two-sample MR analysis to investigate the causal effects of GERD and IBS, and the flow diagram of the study design is shown in Supplementary Figure S1. The forward MR analysis considered GERD as the exposure and IBS as the outcome, while the reverse MR analysis considered IBS as the exposure and GERD as the outcome. Because our research relied on summary statistics from previously published studies, no additional ethical approval or informed permission was required.
2.2 Data sources
In this research, the summary statistics of GERD included 129,080 cases and 473,524 controls from a large-scale GWAS meta-analysis (Ong et al., 2022). IBS data were obtained from the FinnGen database, which included a total of 4,605 cases and 182,423 controls (Kurki et al., 2023) (Supplementary Table S1). All the summary statistics for GERD data and IBS data can be downloaded from the Integrative Epidemiology Unit (IEU) Open GWAS database (https://gwas.mrcieu.ac.uk/). All participants in the original study were from European populations.
2.3 Instrument selection
Similar to the majority of MR studies, SNPs associated with GERD were selected at a threshold of p values <5 × 10−8. Because of the limited number of SNPs meeting genome-wide significance in European populations, we used SNPs with a wide threshold (p < 5 × 10−6) as potential IVs for IBS in European populations for reverse casual MR analysis. To minimize the effect of linkage disequilibrium (LD), SNPs apply strict selection criteria, such as R2 = 0.001 with a genetic window of 10,000 kb in the European 1000 Genome reference panel. F statistics for each SNP was calculated with the formula F = β2/SE2, in which F value < 10 was excluded (Burgess et al., 2011; Burgess and Thompson, 2011).
2.4 MR analysis
To investigate the causal relationship between GERD and IBS, we carried out a two-sample MR analysis employing the inverse-variance weighted (IVW) method as the primary method, and p < 0.05 was defined as statistically significant (Burgess et al., 2013). Complementary analyses were conducted employing weighted median, MR-Egger regression, and weighted-mode methods, which provided more credible evidence of causality (Bowden et al., 2015; Bowden et al., 2016). The Wald ratio approach was used to calculate the effect estimate of the association between the selected exposure and the outcome of each SNP. All results were presented as the odds ratio (OR) and 95% confidence interval (95% CI).
2.5 Sensitivity analyses
We conducted sensitivity analyses to calculate the heterogeneity and pleiotropy in the study, as well as any potential genetic outliers, in order to provide a more credible assessment. The heterogeneity was assessed using MR-Egger and IVW; p ≥ 0.05 implies that there is no heterogeneity in the causal relationship. MR-PRESSO outlier analysis and MR-Egger intercept estimate were used to evaluate the horizontal pleiotropy (Verbanck et al., 2018). The leave-one-out analysis removed each SNP from the investigation, and a two-sample MR analysis was conducted using the remaining SNPs as IVs to determine if the whole causal relationship was triggered by a single SNP (Burgess and Thompson, 2017).
2.6 Statistical analyses
R software (version 4.3.0) was employed for all statistical analyses. The R-based package “Two Sample MR” was used to perform the MR analysis (Hemani et al., 2018). p values were normally two-sided, and p < 0.05 was defined as statistically significant.
3 Results
3.1 Genetic instrumental variables
After removing linkage disequilibrium and palindromes, we obtained 74 independent SNPs for GERD and six independent SNPs for IBS. Moreover, the F-statistic of 74 GERD-associated SNPs (the median F-statistic was 35.155, ranging from 29.749 to 96.029) and six IBS-associated SNPs (the median F-statistic was 21.259, ranging from 21.231 to 23.514) was >10 (Supplementary Table S2), which suggests that the causality results obtained in our study can be interpreted without regard to weak instrumental variables. The comprehensive information of the selected SNPs is shown in forest plots (Supplementary Figure S1A, B) and listed in Supplementary Table S3.
3.2 Causal effect of GERD on the risk of IBS
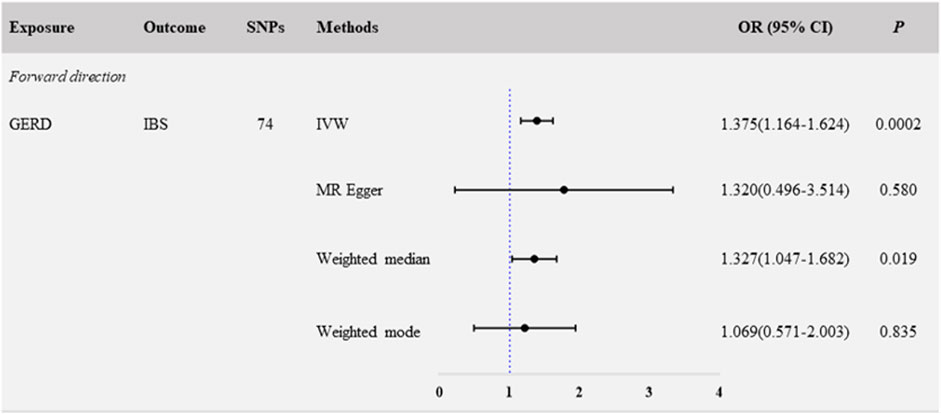
In the primary analyses using IVW, the results showed that genetically predicted GERD was positively related to IBS (NSNP = 74; OR: 1.375; 95% CI: 1.164–1.624; p < 0.001) (Figure 1; Figure 3A). Consistent causal effect estimates of GERD on the risk of IBS were observed using the weighted-median regression method (NSNP = 74; OR: 1.327; 95% CI: 1.047–1.682; p = 0.019) (Figure 1; Figure 3A).
We next performed a sensitivity analysis to assess the robustness of the causal relationship between GERD and IBS. The Egger-intercept of the MR-Egger analysis result (Table 1, intercept = 0.001, p = 0.934) and the MR PRESSO result (Table 1, global test p = 0.806) showed no evidence of potential directional pleiotropy. In addition, combining of Cochran’s Q p-value in IVW (Table 1, Q = 62.633, p = 0.801) and MR-Egger (Table 1, Q = 62.626, p = 0.777) methods with the funnel plot (Supplementary Figure S2A) indicated that the causal relationship between GERD and IBS was without heterogeneity. We also systematically excluded individual SNPs and repeated the MR analysis sequentially; no potential driving SNPs for the evaluated causal relationship between GERD and IBS were observed in the leave-one-out analysis (Supplementary Figure S3A). These findings showed that GERD in patients is a causal risk factor for the development of IBS.

Table 1. Pleiotropy and heterogeneity analysis between gastroesophageal reflux disease and irritable bowel syndrome.
3.3 No causal effect of IBS on GERD
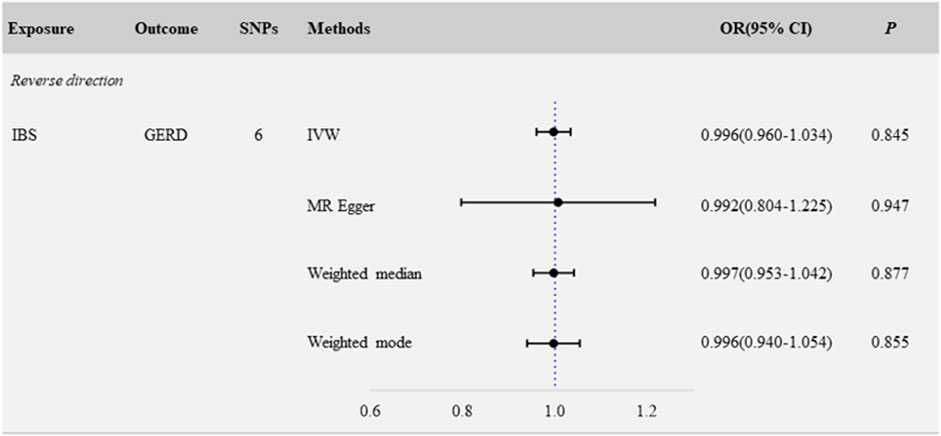
We evaluated the causal associations of IBS on GERD by IVW, and the result revealed that IBS was not causally relevant to critical GERD (NSNP = 6; OR: 0.996; 95% CI: 0.960–1.034; p = 0.845) (Figure 2, Figure 3B). No significant association was found in the other methods, including MR Egger (NSNP = 6; OR: 0.992; 95% CI: 0.804–1.225; p = 0.947), weighted-median (NSNP = 6; OR: 0.997; 95% CI: 0.953–1.042; p = 0.877), and weighted-mode (NSNP = 6; OR: 0.996; 95% CI: 0.940–1.054; p = 0.855) (Figure 2, Figure 3B, Supplementary Figure S2B).
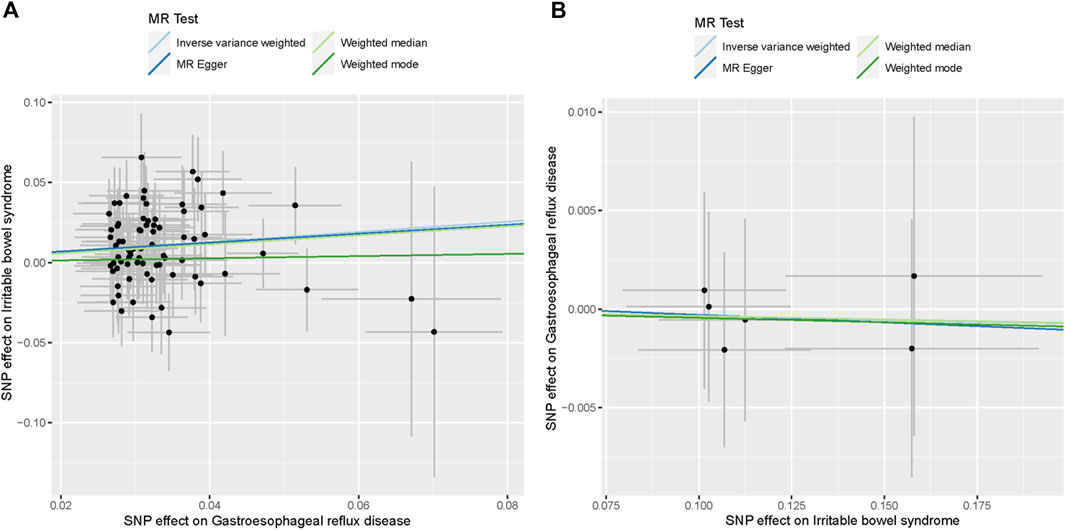
Figure 3. Scatter plots of the causal association between GERD and IBS. (A) Scatter plot of the causal association between GERD and IBS. (B) Scatter plot of the causal association between IBS and GERD.
Sensitivity analyses were performed to evaluate the robustness of the causal association between IBS and GERD. The Egger-intercept of the MR-Egger analysis result (Table 1, intercept = 0.0005, p = 0.972) and the MR PRESSO result (Table 1, global test p = 0.993) showed no evidence of potential directional pleiotropy. Assessment of Cochran’s Q p-value in IVW (Table 1, Q = 0.322, p = 0.997) and MR-Egger (Table 1, Q = 0.321, p = 0.988) methods indicated no heterogeneity. Furthermore, when any single SNP was removed, the observed associations were not altered notably in the leave-one-out analysis (Supplementary Figure S3B).
4 Discussion
In general, our findings provide relatively strong evidence that GERD patients might contribute to the development and advancement of IBS when compared to healthy individuals. However, the reverse MR analysis showed no evidence for the causal relationship of IBS on GERD. Furthermore, we performed a sensitivity analysis on the findings mentioned above, and the results of the causal relationship were consistent and robust, and vice versa. Therefore, our findings indicate a complex genetic interaction between GERD and IBS and suggest that IBS could be caused by specific pathogenic mechanisms observed in GERD.
Previous studies have extensively reported the frequent co-occurrence of GERD and IBS (de Bortoli et al., 2013; Lacy et al., 2019; Lei et al., 2019; Nwokediuko et al., 2020; Patcharatrakul et al., 2021). A systematic review and meta-analysis demonstrated a high degree of overlap between GERD and IBS (Eusebi et al., 2018). Specifically, a single-center cross-sectional study using data from 168 IBS patients aged 18–60 in southern Iran found that IBS was associated with a higher risk of GERD (Gholamnezhad et al., 2023). Similarly, in a comparative study in Taiwan including 273 GERD patients, it was discovered that GERD was associated with an increased risk of IBS (Lei et al., 2019). Furthermore, a recent MR study has observed a causal association between GERD and IBS, which supports our findings to a certain extent (Li et al., 2023). However, in their study, patients with IBS had an increased risk of developing GERD, which is inconsistent with our findings. In our study, the risk of IBS was increased in patients with GERD, but in patients with existing IBS, no increased risk of GERD was observed. In contrast to previous studies, we have evaluated the causal relationship between GERD and IBS by leveraging the latest GWAS summary data, and there was no sample overlap between exposures and outcomes, which enhances evidence to assessment of causality between IBS and GERD disorders, and our study also conducted reverse validation to assist in clarifying the direction of the relationship. In addition, we validate our results by employing a variety of methods and a series of sensitivity analyses to make the results more reliable.
In patients with IBS, the frequent coexistence of features, or comorbidity, with GERD may underlie a common genetic cause and/or pathogenic mechanism between the two disorders. It is well known that although IBS is considered to have no pathological features, its pathogenesis is clearly related to motor (transport) disorders and local visceral hypersensitivity (Holtmann et al., 2016; Simren et al., 2019). It has been postulated that motor (transport) disorders and local visceral hypersensitivity are associated with GERD patients (Yoshida et al., 2013; Ustaoglu and Woodland, 2023). In addition, prior studies reported that GERD and IBS have found genetic overlaps with the established risk factors with depression (Wu et al., 2021). Diet plays an important role. Several randomized controlled studies on both GERD and IBS have reported the benefits of a low-FODMAP diet on gastrointestinal symptoms and the quality of life in patients with IBS (McIntosh et al., 2017; Wilson et al., 2020). A recent study has demonstrated that the symptoms of GERD and gastric reflux are aggravated by a high-FODMAP diet (Plaidum et al., 2022). It is critical that we investigate the mechanisms of GERD and IBS because we do not yet understand the inherited genetic makeup or common etiology of these conditions. Our findings clarify the causal relationship between GERD and IBS, which should encourage clinicians to pay more attention to the clinical manifestations of IBS in patients with GERD and provide clinical strategies for early detection and treatment of IBS.
The main advantage of this study is that we employed a bidirectional Mendelian randomization analysis method for the first time and confirmed the positive causal relationship of GERD and IBS. Compared with previous observational studies, the application of Mendelian randomization assists in reducing reverse causation bias and residual confounding encountered in observational studies. In addition, we found no heterogeneity and multiple effects influencing the outcomes between GERD and IBS, which properly avoided the interference of negative results and supports the accuracy of our findings. However, there are some limitations to our study. First, only people with European ancestry were included in our study, and our findings may not be generalized to other races and ethnicities. Second, since sex-stratified GWAS data were not available, we were unable to perform a deeper analysis to investigate the specific subpopulation, such as sex affection between two diseases. The availability of high-quality GWASs focused on the specific sex subgroup of GERD or IBS in the future would be valuable. In addition, although the results of all methods are robust, a bias could still exist due to a pool of genetic instruments, a small sample size, or probable sample overlap between exposure and the outcome.
5 Conclusion
Taken together, our bidirectional MR study demonstrated that patients with GERD were more likely to develop IBS, but the patients with IBS were less likely to develop GERD. To better prevent IBS, it is recommended that clinicians pay more attention to whether individuals with GERD have clinical manifestations of IBS. Moreover, it is necessary to conduct additional research on the pathophysiological mechanisms underlying the interaction between GERD and IBS.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the studies involving humans because our research relied on summary statistics from previously published studies, and no additional ethical approval or informed permission was required. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements because our research relied on summary statistics from previously published studies, and no additional ethical approval or informed permission was required.
Author contributions
HW: validation and writing–original draft. JL: validation and writing–original draft. FL: data curation, investigation, and writing–review and editing. WL: funding acquisition, supervision, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work is supported by the Health Commission of Guangdong Province (Grant No. B2021311).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1328327/full#supplementary-material
References
Black, C. J., and Ford, A. C. (2020). Global burden of irritable bowel syndrome: trends, predictions and risk factors. Nat. Rev. Gastroenterol. Hepatol. 17, 473–486. doi:10.1038/s41575-020-0286-8
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. doi:10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314. doi:10.1002/gepi.21965
Burgess, S., Butterworth, A., and Thompson, S. G. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665. doi:10.1002/gepi.21758
Burgess, S., and Thompson, S. G. (2011). Bias in causal estimates from Mendelian randomization studies with weak instruments. Stat. Med. 30, 1312–1323. doi:10.1002/sim.4197
Burgess, S., and Thompson, S. G.CRP CHD Genetics Collaboration (2011). Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 40, 755–764. doi:10.1093/ije/dyr036
Burgess, S., and Thompson, S. G. (2017). Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389. doi:10.1007/s10654-017-0255-x
Camilleri, M. (2021). Diagnosis and treatment of irritable bowel syndrome: a review. JAMA 325, 865–877. doi:10.1001/jama.2020.22532
Davies, N. M., Holmes, M. V., and Davey Smith, G. (2018). Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362, k601. doi:10.1136/bmj.k601
de Bortoli, N., Martinucci, I., Bellini, M., Savarino, E., Savarino, V., Blandizzi, C., et al. (2013). Overlap of functional heartburn and gastroesophageal reflux disease with irritable bowel syndrome. World J. Gastroenterol. 19, 5787–5797. doi:10.3748/wjg.v19.i35.5787
El-Serag, H., Hill, C., and Jones, R. (2009). Systematic review: the epidemiology of gastro-oesophageal reflux disease in primary care, using the UK General Practice Research Database. Aliment. Pharmacol. Ther. 29, 470–480. doi:10.1111/j.1365-2036.2008.03901.x
Eusebi, L. H., Ratnakumaran, R., Bazzoli, F., and Ford, A. C. (2018). Prevalence of dyspepsia in individuals with gastroesophageal reflux-type symptoms in the community: a systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 16, 39–48. doi:10.1016/j.cgh.2017.07.041
Ford, A. C., Lacy, B. E., and Talley, N. J. (2017). Irritable bowel syndrome. N. Engl. J. Med. 376, 2566–2578. doi:10.1056/NEJMra1607547
Gasiorowska, A., Poh, C. H., and Fass, R. (2009). Gastroesophageal reflux disease (GERD) and irritable bowel syndrome (IBS)--is it one disease or an overlap of two disorders? Dig. Dis. Sci. 54, 1829–1834. doi:10.1007/s10620-008-0594-2
Gholamnezhad, F., Qeisari, A., and Shahriarirad, R. (2023). Gastroesophageal reflux disease incidence among male patients with irritable bowel syndrome: a single-center cross-sectional study in southern Iran. JGH Open 7, 152–156. doi:10.1002/jgh3.12867
Hemani, G., Zheng, J., Elsworth, B., Wade, K. H., Haberland, V., Baird, D., et al. (2018). The MR-Base platform supports systematic causal inference across the human phenome. Elife 7, e34408. doi:10.7554/eLife.34408
Holtmann, G. J., Ford, A. C., and Talley, N. J. (2016). Pathophysiology of irritable bowel syndrome. Lancet Gastroenterol. Hepatol. 1, 133–146. doi:10.1016/S2468-1253(16)30023-1
Katzka, D. A., and Kahrilas, P. J. (2020). Advances in the diagnosis and management of gastroesophageal reflux disease. BMJ 371, m3786. doi:10.1136/bmj.m3786
Kurki, M. I., Karjalainen, J., Palta, P., Sipila, T. P., Kristiansson, K., Donner, K. M., et al. (2023). FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518. doi:10.1038/s41586-022-05473-8
Lacy, B. E., Harris, L. A., Chang, L., Lucak, S., Gutman, C., Dove, L. S., et al. (2019). Impact of patient and disease characteristics on the efficacy and safety of eluxadoline for IBS-D: a subgroup analysis of phase III trials. Ther. Adv. Gastroenterol. 12, 1756284819841290. doi:10.1177/1756284819841290
Lei, W. Y., Chang, W. C., Wen, S. H., Wong, M. W., Hung, J. S., Yi, C. H., et al. (2019). Impact of concomitant dyspepsia and irritable bowel syndrome on symptom burden in patients with gastroesophageal reflux disease. J. Formos. Med. Assoc. 118, 797–806. doi:10.1016/j.jfma.2018.12.002
Li, C., Chen, Y., Ying, Z., Hu, Y., Kuang, Y., Yang, H., et al. (2023). The causal association of irritable bowel syndrome with multiple disease outcomes: a phenome-wide mendelian randomization study. J. Clin. Med. 3, 1106. doi:10.3390/jcm12031106
Lord, J., Jermy, B., Green, R., Wong, A., Xu, J., Legido-Quigley, C., et al. (2021). Mendelian randomization identifies blood metabolites previously linked to midlife cognition as causal candidates in Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 118, e2009808118. doi:10.1073/pnas.2009808118
Lovell, R. M., and Ford, A. C. (2012). Prevalence of gastro-esophageal reflux-type symptoms in individuals with irritable bowel syndrome in the community: a meta-analysis. Am. J. Gastroenterol. 107, 1793–1801. doi:10.1038/ajg.2012.336
Maret-Ouda, J., Markar, S. R., and Lagergren, J. (2020). Gastroesophageal reflux disease. JAMA 324, 2565. doi:10.1001/jama.2020.21573
McIntosh, K., Reed, D. E., Schneider, T., Dang, F., Keshteli, A. H., De Palma, G., et al. (2017). FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut 66, 1241–1251. doi:10.1136/gutjnl-2015-311339
Mehta, R. S., Staller, K., and Chan, A. T. (2021). Review of gastroesophageal reflux disease. JAMA 325, 1472. doi:10.1001/jama.2021.1438
Monnikes, H., Heading, R. C., Schmitt, H., and Doerfler, H. (2011). Influence of irritable bowel syndrome on treatment outcome in gastroesophageal reflux disease. World J. Gastroenterol. 17, 3235–3241. doi:10.3748/wjg.v17.i27.3235
Monnikes, H., Schwan, T., van Rensburg, C., Straszak, A., Theek, C., Sander, P., et al. (2012). Randomised clinical trial: sustained response to PPI treatment of symptoms resembling functional dyspepsia and irritable bowel syndrome in patients suffering from an overlap with erosive gastro-oesophageal reflux disease. Aliment. Pharmacol. Ther. 35, 1279–1289. doi:10.1111/j.1365-2036.2012.05085.x
Nwokediuko, S. C., Adekanle, O., Akere, A., Olokoba, A., Anyanechi, C., Umar, S. M., et al. (2020). Gastroesophageal reflux disease in a typical African population: a symptom-based multicenter study. BMC Gastroenterol. 20, 107. doi:10.1186/s12876-020-01261-8
Ong, J. S., An, J., Han, X., Law, M. H., Nandakumar, P., andMe Research, t., et al. (2022). Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett's oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut 71, 1053–1061. doi:10.1136/gutjnl-2020-323906
Patcharatrakul, T., Linlawan, S., Plaidum, S., and Gonlachanvit, S. (2021). The effect of rice vs. Wheat ingestion on postprandial gastroesophageal reflux (GER) symptoms in patients with overlapping GERD-irritable bowel syndrome (IBS). Foods 11, 26. doi:10.3390/foods11010026
Plaidum, S., Patcharatrakul, T., Promjampa, W., and Gonlachanvit, S. (2022). The effect of fermentable, oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAP) meals on transient lower esophageal relaxations (TLESR) in gastroesophageal reflux disease (GERD) patients with overlapping irritable bowel syndrome (IBS). Nutrients 14, 1755. doi:10.3390/nu14091755
Simren, M., Tornblom, H., Palsson, O. S., Van Oudenhove, L., Whitehead, W. E., and Tack, J. (2019). Cumulative effects of psychologic distress, visceral hypersensitivity, and abnormal transit on patient-reported outcomes in irritable bowel syndrome. Gastroenterology 157, 391–402. doi:10.1053/j.gastro.2019.04.019
Sperber, A. D., Bangdiwala, S. I., Drossman, D. A., Ghoshal, U. C., Simren, M., Tack, J., et al. (2021). Worldwide prevalence and burden of functional gastrointestinal disorders, results of Rome foundation global study. Gastroenterology 160, 99–114 e3. doi:10.1053/j.gastro.2020.04.014
Ustaoglu, A., and Woodland, P. (2023). Sensory phenotype of the oesophageal mucosa in gastro-oesophageal reflux disease. Int. J. Mol. Sci. 24, 2502. doi:10.3390/ijms24032502
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi:10.1038/s41588-018-0099-7
Wilson, B., Rossi, M., Kanno, T., Parkes, G. C., Anderson, S., Mason, A. J., et al. (2020). β-Galactooligosaccharide in conjunction with low FODMAP diet improves irritable bowel syndrome symptoms but reduces fecal bifidobacteria. Am. J. Gastroenterol. 115, 906–915. doi:10.14309/ajg.0000000000000641
Wu, Y., Murray, G. K., Byrne, E. M., Sidorenko, J., Visscher, P. M., and Wray, N. R. (2021). GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other gastrointestinal disorders and depression. Nat. Commun. 12, 1146. doi:10.1038/s41467-021-21280-7
Yoshida, N., Kuroda, M., Suzuki, T., Kamada, K., Uchiyama, K., Handa, O., et al. (2013). Role of nociceptors/neuropeptides in the pathogenesis of visceral hypersensitivity of nonerosive reflux disease. Dig. Dis. Sci. 58, 2237–2243. doi:10.1007/s10620-012-2337-7
Keywords: gastroesophageal reflux disease, irritable bowel syndrome, Mendelian randomization, causal association, clinical guidance
Citation: Wu H, Li J, Li F and Lun W (2024) Causal association of gastroesophageal reflux disease on irritable bowel syndrome: a two-sample Mendelian randomization study. Front. Genet. 15:1328327. doi: 10.3389/fgene.2024.1328327
Received: 26 October 2023; Accepted: 11 March 2024;
Published: 27 March 2024.
Edited by:
Phillip E. Melton, University of Tasmania, AustraliaReviewed by:
Adam de Smith, University of Southern California, United StatesMohsen Norouzinia, Shahid Beheshti University of Medical Sciences, Iran
Copyright © 2024 Wu, Li, Li and Lun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijian Lun, bHdqMTk4NDA3QDE2My5jb20=
†These authors have contributed equally to this work
 Huihuan Wu
Huihuan Wu Jingwei Li2†
Jingwei Li2† Weijian Lun
Weijian Lun