- 1Department of Medical Genetics, Changzhou Maternal and Child Healthcare Hospital, Changzhou Medical Center of Nanjing Medical University, Changzhou, Jiangsu, China
- 2The Affiliated Suqian First People’s Hospital of Nanjing Medical University, Suqian First People's Hospital, Suqian, Jiangsu, China
Objective: To assess the detection rate of exome sequencing (ES) in fetuses diagnosed as skeletal abnormalities (SKA) with normal karyotype or chromosomal microarray analysis (CMA) results.
Methods: We conducted electronic searches in four databases, focusing on studies involving ES in fetuses with SKA. Additional detection rate of ES compared to karyotype/CMA was calculated, followed by a meta-analysis. Subgroup analyses explored the influence of fetal phenotype on diagnostic outcomes.
Results: From 2,393 studies, 21 reports covering 476 fetuses were analyzed. Key findings include: (1) an additional detection rate of ES of 63.2% (Risk Difference (RD), 0.68 [95% CI, 0.60–0.76], p < 0.00001); (2) identification of 76 genes across 304 types of variants, with FGFR3, COL1A1, COL1A2, and COL2A1 being prevalent; (3) lower detection rates in fetuses with isolated short long bones compared to non-isolated conditions, though not significantly different (p = 0.35); (4) higher detection rates in subgroups with abnormal ossification, small chest, suspected long bone fractures or angulations, and skull abnormalities.
Conclusion: The meta-analysis indicates that genetic variation significantly contributes to fetal SKA, primarily due to single-gene variants. Consequently, ES should be used in the prenatal diagnosis of SKA fetuses in clinical practice.
1 Introduction
Fetal skeletal abnormalities (SKA) is a prevalent structural malformation, occurring in approximately 5 per 1,000 fetuses (Schramm and Mommsen, 2018). This congenital disease impacts the composition and structure of bone and cartilage tissue. Clinical manifestations of SKA include various abnormalities in skeletal tissue growth and development, such as short stature, joint malposition, cranial and limb deformities, abnormal spinal curvature, and changes in bone mineral density. Additionally, this condition may co-occur with malformations in other systems and organs (Schramm and Mommsen, 2018; Krakow, 2015; Milks et al., 2017). In severe cases, abnormal skeletal development in fetuses can lead to fetal death. Survivors may face disabilities due to skeletal malformations, and in some instances, various degrees of intellectual disability, severely affecting patient quality of life (Schramm and Mommsen, 2018). Current research indicates that genetic factors, such as chromosomal number and structural abnormalities, chromosomal copy number variations, and single-gene variants, are considered to be the main factors leading to fetal skeletal abnormalities (Krakow, 2015; Kucinska-Chahwan et al., 2022). The impact of SKA on fetal health is significant, being a major cause of birth defects. Therefore, early detection through prenatal screening and diagnosis is crucial for timely clinical intervention. Currently, ultrasound examination is the most effective prenatal diagnostic tool for SKA. However, its efficacy is influenced by several factors, including gestational age, fetal position, maternal abdominal wall conditions, the types of SKA, amniotic fluid volume, and variations in sonographer techniques and experience. These factors contribute to the limitations of ultrasound in the detection of SKA fetuses (Best et al., 2018). In clinical practice, genetic testing is often conducted on fetuses diagnosed with SKA via ultrasound. Traditional genetic diagnosis methods include karyotyping and chromosomal microarray analysis (CMA) of amniotic fluid exfoliated cells (Wapner et al., 2012; Callaway et al., 2013). Most SKA fetuses are monogenic genetic diseases (Best et al., 2018), so the current first-line prenatal diagnosis techniques, karyotyping and CMA, are not fully suitable for SKA fetuses. Consequently, prenatal screening and diagnosis that rely solely on ultrasound combined with chromosome analysis may not be entirely sufficient for diagnosing the genetic etiology of fetal SKA.
Exome Sequencing (ES) encompasses 1%–2% of the genome, yet includes about 85% of known disease-causing genetic variants. Two prospective studies have shown disease detection rates of 24% and 15.4% in SKA fetuses when assessed on a large scale using ES (Tournis and Dede, 2018; Lord et al., 2019). Additionally, a meta-analysis of 66 studies and 72 reports underscored the significant value of ES in prenatal detection. This study found that for fetuses with structural abnormalities, the detection rate using ES was 31% higher compared to CMA or karyotyping. Notably, the detection rate for SKA was the highest at 53% (Mellis et al., 2022).
However, due to the rarity of SKA fetuses, most studies evaluating the additional detection rate of prenatal ES in these cases have been limited to small sample sizes. Moreover, the complexity of SKA types contributes to a lack of data accumulation and evidence guiding the selection of prenatal diagnosis techniques and genetic counselling. In our study, we merged various studies on the application of ES in SKA fetuses with normal karyotypes or CMA results to form a larger cohort. We then conducted a meta-analysis to explore the additional detection value of ES in SKA fetuses with normal karyotypes or CMA.
2 Materials and methods
2.1 Protocol
We devised a systematic review protocol in line with PRISMA guidance (Moher et al., 2009; Page et al., 2021). Study authors agreed the protocol prior to conducting the searches. Any required small amendments were made with the consensus of all authors.
2.2 Data sources and search strategies
2.2.1 Literature source
Searches were conducted in PubMed, EMBASE, Web of Science, and the Cochrane Library for articles published up to May 2023.
2.2.2 Search strategy
This involved using a combination of subject terms and keywords. 1) For English subject terms: ‘skeletal dysplasia’ was represented by ‘skeletal abnormalities’, ‘SD’, ‘SKA’, ‘SDs’, and ‘skeletal dysplasia’s’. Similarly, ‘whole exome sequencing’ and ‘WES‘ were used to refer to ‘exome sequencing’. 2) Additionally, references from identified literature were reviewed to locate other relevant studies. 3) Finally, two researchers independently conducted and evaluated the literature search.
2.3 Eligibility criteria
2.3.1 Inclusion criteria
1) Ultrasound suggested fetal skeletal abnormalities, including SKA fetuses, SKA fetuses with other skeletal or non-skeletal abnormalities. 2) Prospective or retrospective cohort studies. 3) Prenatal ES including Whole Exome Sequencing (WES), Targeted Exome Sequencing (TES), or SKA panel, for diagnosing SKA fetuses. This encompasses studies based on prenatal phenotypes with ES testing completed post-delivery. 4) Cases classified as pathogenic or likely pathogenic variats according to the ACMG (American College of Medical Genetics and Genomics) guidelines and identified as the cause of the fetal ultrasound phenotype, were included in the study. However, cases with only one pathogenic or likely pathogenic variant of an autosomal recessive inheritance, without a second variant, as well as those with Variants of Uncertain Significance (VOUS), were not considered diagnostic cases. Cases where karyotype or CMA results were negative or no diagnostic, and where numerical, structural, and copy number abnormalities (CNVs) of chromosomes were ruled out. 5) Studies that described specific prenatal ultrasound phenotypes. 6) Studies that provide raw data. 7) Full-text reports available in the English language.
2.3.2 Exclusion criteria
1) Literature types such as reviews, case reports, reader letters, animal studies, expert opinions, conference papers, etc.. 2) Studies that did not use Exome Sequencing. 3) Studies lacking a specific subgroup for SKA. 4) Studies where data could not be extracted.
2.4 Study selection
Following the removal of duplicate content, two investigators independently examined titles and abstracts. For abstracts deemed potentially relevant, full texts were scrutinized based on predetermined inclusion and exclusion criteria. Any disagreements between the investigators were resolved through discussion. To prevent data duplication, only studies with the largest or most recent sample sizes were included.
2.5 Data extraction
Data were independently extracted by two investigators, including study setting, sample size, ES method, number of fetuses diagnosed, pregnancy outcomes, and others. If these details were not specified in the literature, the authors were contacted via email for clarification.
2.6 Quality assessment of included studies
Two researchers independently evaluated the quality of the included studies. For this evaluation, the criteria recommended by the Agency for Healthcare Research and Quality (AHRQ) were utilized.
2.7 Data statistics
The incremental diagnostic value of prenatal ES over CMA or karyotype analysis was determined using the 95% confidence interval (CI) from each study. Risk differences were calculated using a random effects model. Statistical analysis was conducted using RevMan 5.3. A statistical difference was considered significant when P < 0.05.
2.8 Heterogeneity test
Heterogeneity within the studies was evaluated using the I2 statistic in the forest plot. An I2 = 0 indicated no heterogeneity. Conversely, a larger I2 statistic reflected greater heterogeneity. Typically, an I2 > 50% suggested significant heterogeneity. The threshold for significance in this study was set at 0.1. If P > 0.1 and I2 was <50%, it was interpreted as a lack of heterogeneity among the studies. If these criteria were not met, heterogeneity among the studies was presumed.
2.9 Assessment of literature publication bias
The analysis results were represented by a Funnel plot, a scatter plot with the size of the effect on the abscissa (horizontal axis) and the sample size on the ordinate (vertical axis). In the absence of publication bias among the included studies, this plot typically exhibits a symmetrical funnel shape.
3 Results
3.1 Data and materials
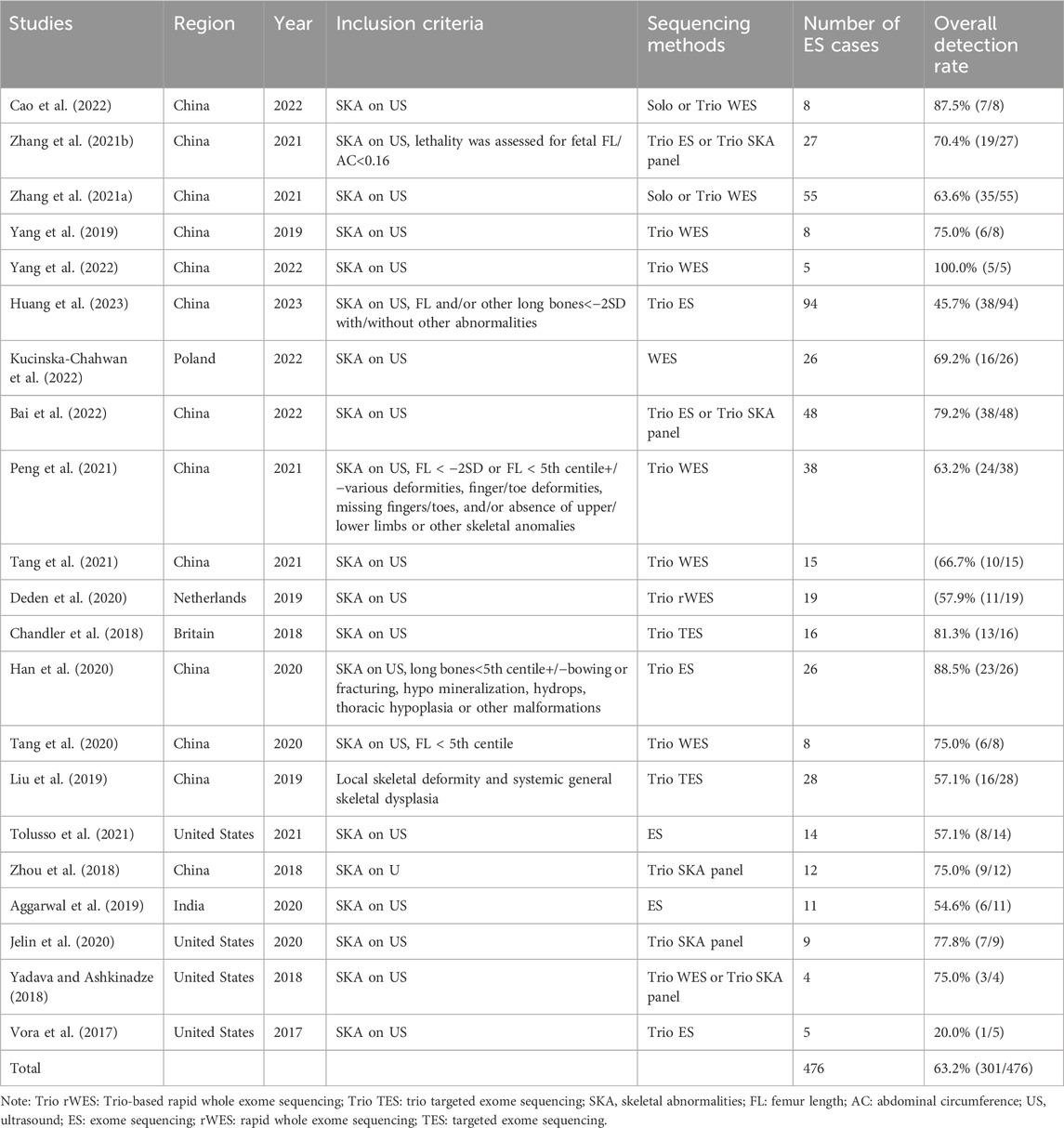
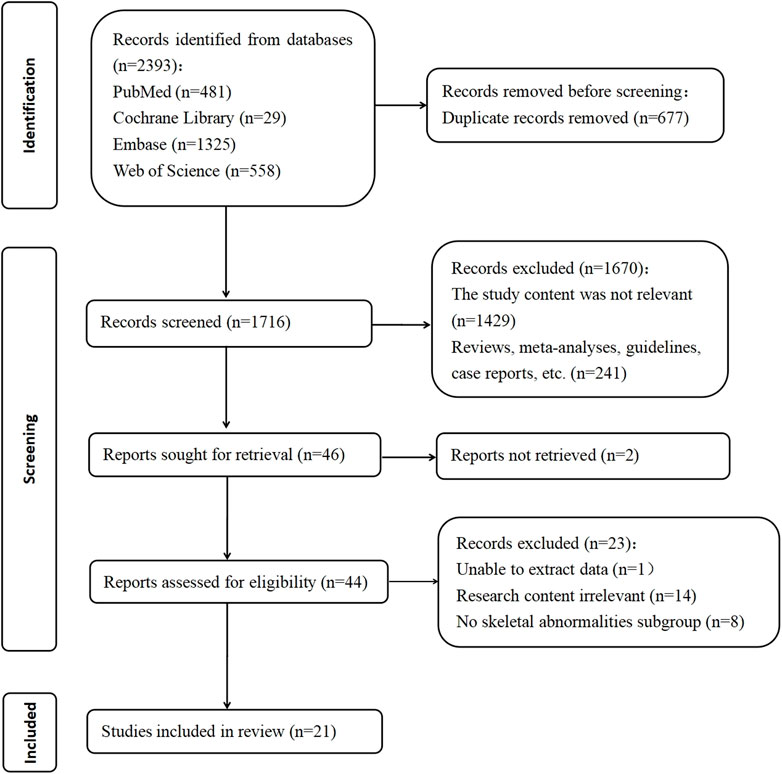
A total of 2,393 manuscripts were retrieved using the specified database. Following the initial exclusion based on title and abstract reading, 46 pieces remained. After a full-text review, 25 pieces were further excluded due to reasons such as the inability to extract relevant data, irrelevance to the research topic, or the absence of the full text. Ultimately, 21 manuscripts that met the inclusion criteria were analyzed, and are listed in Table 1. The process of literature selection is depicted in Figure 1. The quality of these 21 articles was independently assessed based on the 11 items recommended by AHRQ (Supplementary Table 1).
3.2 Study characteristics
Among the 21 studies included in this analysis, 13 (encompassing 372 patients) were conducted in China. The meta-analysis included a total of 476 eligible fetuses, with a mean gestational age of 24 weeks, ranging from 12 to 40+2 weeks. At the time of drafting, 257 pregnancies had been terminated, while there were 51 live births, 5 stillbirths, 12 neonatal deaths, and 3 ongoing pregnancies. The pregnancy outcomes for the remaining 148 patients were not described. Detailed information on phenotype-genotype correlations and molecular diagnoses can be found in the Supplementary Material under ‘Genes’.
3.3 Overall additional monogenic disorder detection rate of ES
Excluding chromosomal abnormalities, the overall rate of abnormal ES testing in fetuses with SKA was 63.2% (Table 1). This result shows a RD of 0.68 with a 95% CI of 0.60–0.76, and p < 0.00001 (Supplementary Figure S1).
Of the 301 cases with positive ES results, 235 cases (78.1%) were autosomal dominant (199 cases de-novo vs. 35 cases inherited), while 66 cases (21.9%) were autosomal recessive (11 cases homozygous vs. 45 cases compound heterozygous) (7 cases de-novo vs. 49 cases inherited). A further 11 cases either did not perform parental testing, or the results of parental analysis were not mentioned in the studies. In these 301 cases, a total of 76 genes across 304 types of variants were detected. The ten most common genes identified were FGFR3, COL1A1, COL1A2, COL2A1, DYNC2H1, ALPL, FLNB, EBP, PPIB, and IFITM5. Their respective frequencies were 31.3%, 14.5%, 10.2%, 8.9%, 3.6%, 2.3%, 1.3%, 1.3%, 1.3%, and 1.0% (Supplementary Figure S2).
3.4 Subgroup analysis of monogenic disorder detection rates
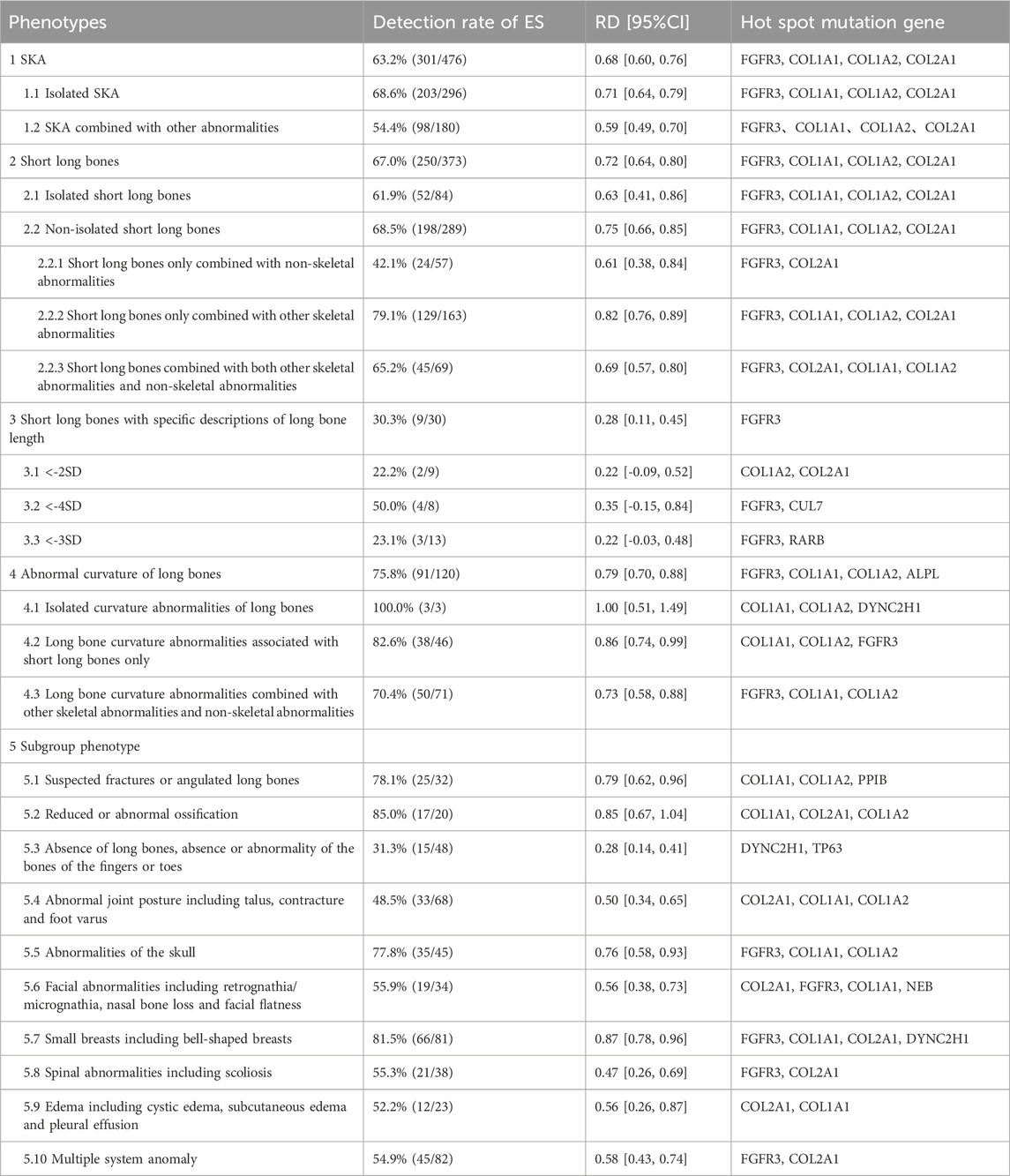
To delve deeper into the individual sonographic features of SKA and their respective clinical implications, a subgroup analysis was conducted. This analysis was based on the described sonographic features, with subgroup categorization partially referencing the groupings from the study by Tse et al. (Tse et al., 2023) and the detailed subgroup and results are presented in Table 2 and Supplementary Table 2.
Among the 476 SKA fetuses included in the study, the detection rate of isolated SKA fetuses was 68.6% (203/296), and the detection rate of non-isolated SKA fetuses was 54.4% (98/180), but the two groups did not achieve statistical significance (p = 0.07). Our study revealed that the detection rate of fetuses with isolated short long bones (52/84, 61.9%) was lower compared to those with non-isolated short long bones (198/289, 68.5%). However, this difference was not statistically significant (p = 0.35). Among the 84 fetuses with isolated short long bones, 30 cases had a detailed description of the length of the long bones shortening. Among these, the femur length of 9 cases was less than −2 standard deviations (SD) of the fetus of the same gestational age, 13 were less than –3SD, and eight were less than –4SD. The detection rates of ES in these subgroups were 22.2%, 23.1%, and 50.0%, respectively. When comparing fetuses with short bones less than –2SD to those with short bones below –4SD, the latter were more likely to be detected genetic abnormality with ES (22.2% for –2SD versus 50.0% for –4SD), but this difference did not reach statistical significance (p = 0.66). In the 9 fetuses with long bones less than –2SD, two cases were still diagnosed by ES. The detection rate of short long bone only combined with other skeletal abnormalities (129/163, 79.1%) was significantly higher than that of short long bone combined with both other skeletal abnormalities and non-skeletal abnormalities (65.2%, 45/69) and p value was 0.04.
Among the 120 fetuses with abnormal curvature of long bones, 111 cases also presented with varying degrees of short long bones. In the three cases with isolated long bone curvature abnormalities, all underwent ES tests which were positive. Two of these cases had only a slight curvature of the long bones, with variant genes identified as COL1A1, COL1A2, and DYNC2H1, respectively. In the 46 cases where abnormal curvature of long bones was combined with short long bones, the detection rate for ES abnormalities was as high as 82.6% (38/46).
Subgroups with higher additional ES detection rates included abnormal ossification (85.0%), small thorax (81.5%), suspected fractures or angulations of long bones (78.1%), and skull abnormalities (77.8%). Conversely, fetuses with absent long bones and abnormal joint posture exhibited relatively lower extra detection rates of 31.3% and 48.5%, respectively, when undergoing ES tests.
The most frequently mutated genes in the long bone loss subgroup were DYNC2H1 and TP63. In other subgroups, the most common variants were found in FGFR3, COL1A1, COL1A2, and COL2A1.
Heat map analysis of the 10 most common variant genes related to fetal SKA in the meta-analysis results revealed that FGFR3 had the highest frequency in the subgroup of short long bones with other skeletal abnormalities. In isolated short long bones, FGFR3, and in the subgroup of short long bones with other skeletal abnormalities, COL1A1, had the second highest frequency. FGFR3 was also the third most frequent in the small thorax subgroup (Supplementary Figure S3).
3.5 Heterogeneity and publication bias analysis
Given the high level of heterogeneity observed, a random-effects model was employed, although heterogeneity remained relatively substantial. The funnel plot used for the publication bias test displayed significant asymmetry, suggesting the potential presence of publication bias (Supplementary Figure S4).
4 Discussion
4.1 Diagnostic rate
Our review underscores the significance of prenatal ES in fetuses with SKA. The findings reveal that the overall abnormal detection rate of ES compared to karyotyping or CMA was 63.2%. In cases of suspected fetal SKA, the detection rate for CMA testing ranged only between 1.7% and 7.9% (de Wit et al., 2014; Hui et al., 2021). This lower rate is attributed to the fact that SKA is primarily a monogenic disorder, which CMA cannot detect. Pure skeletal dysplasia or classical skeletal dysplasia is a Mendelian monogenic disease (Krakow, 2015), and the detection rate of karyotype or CMA may be lower. Consequently, our research advocates for the routine use of ES in prenatal testing for suspected cases of SKA.
Our study indicated that the detection rate in fetuses with isolated short long bones (61.9%) was slightly lower compared to those with non-isolated short long bones (68.5%), yet this difference was not statistically significant (p = 0.35). This finding aligns with the conclusions of Tse et al.'s study (Tse et al., 2023). We also observed a gradual increase in ES detection rates in fetuses with long bone lengths less than –2SD, –3SD, and –4SD (22.2%, 23.1%, and 50.0%, respectively). In practice, obstetricians frequently encounter pregnancies with fetal short long bones, but few undergo invasive prenatal examinations. However, our data suggest that ES is also crucial for fetuses with mildly shortened long bones.
This study also found that abnormal curvature of long bones often co-occurs with short long bones. In our subgroup analysis, 92.5% (111/120) of cases with abnormal long bone curvature also had varying degrees of short long bones. Among 46 cases with both abnormal curvature and short long bones, the detection rate was high at 82.6%. In three cases of isolated long bone curvature abnormalities, all ES tests were positive, including two with only mild curvature. Thus, fetuses with abnormal curvature of long bones are more likely to carry pathogenic genes related to bone conditions. Clinicians should therefore give considerable attention to such fetal abnormalities in clinical practice.
The detection rate of short long bone only combined with other skeletal abnormalities (129/163, 79.1%) was higher than that of short long bone combined with both other skeletal abnormalities and non-skeletal abnormalities (45/69, 65.2%), and the difference was statistically significant (p = 0.04). Therefore, skeletal system malformations combined with non-skeletal system malformations do not necessarily have higher SKA-related pathogenic genes, because some syndrome diseases may also cause skeletal malformations.
Moreover, the study revealed that specific SKA subgroup characteristics might offer additional detection benefits. The subgroups with abnormal ossification, small chests, suspected fractures or angulations of long bones, and skull abnormalities showed relatively high ES detection rates, being 85.0%, 81.5%, 78.1%, and 77.8%, respectively. Therefore, these features should be meticulously assessed during prenatal ultrasounds, as their identification can inform decisions on whether ES should be pursued.
4.2 Common variant genes
Nosology of genetic skeletal disorders have identified 771 types of hereditary bone diseases, divided into 41 groups involving 552 genes (Unger et al., 2023). Our meta-analysis revealed that FGFR3, COL1A1, COL1A2, and COL2A1 are the most frequently mutated genes in SKA fetuses, accounting for 31.3%, 14.5%, 10.2%, and 8.9% of cases, respectively. In subgroup analyses, DYNC2H1 and TP63 emerged as prominent variant genes in the long bone loss subgroup, while FGFR3, COL1A1, COL1A2, and COL2A1 were prevalent in other subgroups. Thus, a gene panel can be utilized in areas with limited resources when typical sonographic features are identified prenatally.
Our study underscores that pathogenic variants in the Fibroblast Growth Factor Receptor 3 (FGFR3) and collagen genes are the leading genetic causes of SKA. FGFR3, one of four transmembrane tyrosine kinases, serves as a high-affinity receptor for various fibroblast growth factors and plays a crucial role in bone development (Ornitz and Marie, 2015). Pathogenic FGFR3 variants cause achondroplasia, an autosomal dominant skeletal disorder with an incidence of 2–3 cases per 100,000 people (Waller et al., 2008).
Research indicates that 90% of osteogenesis imperfecta (OI) cases result from pathogenic variants in the COL1A1 or COL1A2 genes, which encode the α1 and α2 chains of type I collagen, respectively. These variants affect collagen quantity or structure, with glycine substitutions in the helical domain’s Gly-X-Y triplet being the most common cause of OI (Augusciak-Duma et al., 2018; Zhytnik et al., 2017; Shi et al., 2019; Marini et al., 2017; Petrovski et al., 2019). COL2A1, coding for type II collagen, is involved in the regulation of intramembranous and endochondral osteogenesis. Heterozygous variants in COL2A1 are frequently associated with a range of dwarfism and skeletal dysmorphic disorders (Zhang et al., 2020).
4.3 Impact of prenatal exome sequencing on clinical management decisions
ES will play a crucial role in clinical management and parental decision-making. In our study, pathogenic/likely pathogenic variants were detected in 301 of 476 cases, and 290 cases were tested for trio ES, of which 206 cases (68.4%) were de novo variants and 84 cases (27.9%) were inherited variants from their parents. 21.9% of the cases were autosomal recessive inheritance with a high risk of recurrence, indicating that trio ES is beneficial for data interpretation and genetic counselling. The remaining 11 cases either did not undergo parental testing or did not mention parental analysis results in the study. The cases without trio ES testing cannot provide a reference for clinicians to provide patients with more comprehensive genetic counseling or provide comprehensive guidance for patients in their next pregnancy.
A significant challenge for obstetricians lies in counselling couples where no genetic cause is identified. The most complex cases are not those with clear, fatal features of SKA, but rather those with mild to moderate features and a normal CMA. In these instances, ES can provide additional insights into the etiology, assisting physicians in offering more informed genetic counselling.
4.4 Strengths and limitations
This systematic review and meta-analysis represents a detailed and comprehensive examination of fetal SKA, incorporating 21 studies from four databases. Given the rarity of SKA, most studies have small sample sizes. This review systematically amalgamates these studies to derive an overall diagnostic yield, applying stringent criteria to all studies and excluding all VOUS.
The primary limitation of our review is the high heterogeneity among the included studies, which impacts the accuracy of our comparisons. Even when analyzing the effect of case selection criteria or fetal phenotype groups on ES diagnostic yield, heterogeneity persisted within and across many subgroups. This suggests that these factors do not fully account for the observed heterogeneity.
Funnel plot asymmetry in our review indicates potential publication bias, which might reflect a correlation between small sample sizes and elevated diagnosis rates. The studies in this review were predominantly selected for small cohorts with a genetic inclination towards a monogenic cause, often identified after expert genetic evaluation. This highly selective approach towards monogenic diseases could lead to a higher diagnostic yield. Furthermore, with 13 of the 21 studies conducted in China, the applicability of our findings may be influenced by specific demographic data.
Cases that had undergone whole genome sequencing (WGS) were considered in our screening of the literature, but because there are relatively few published studies of WGS testing in prenatal diagnosis of SKA fetuses, none were eligible for inclusion. In the future, with the wide application of WGS in prenatal diagnosis of SKA fetuses, more genetic causes may be found, and the detection rate of monogenic diseases in SKA fetuses will also increase. Most studies have evaluated the diagnostic value of prenatal ES for SKA fetuses only with a small sample size, and the low number of cases would make it difficult to find valuable information in our analysis. So in order to achieve a larger study cohort, we included different ES methods for detecting SKA fetuses, including gene panel testing, which may result in a slightly lower diagnostic yield. These are indeed a limitation of this study.
The final conclusions of this meta-analysis are indeed less valuable than we expected. At the beginning, we sought to find the difference in the detection rate of isolated and non-isolated short long bone cases and the correlation between the degree of shortening in isolated short long bone cases and the detection rate, but unfortunately, we did not find meaningful results. This is also a weakness of the analysis.
5 Conclusion
This meta-analysis concludes that genetic variation plays a significant role in the causation of fetal SKA, with the majority of cases attributed to single-gene variants. Consequently, it is essential to advocate for the prenatal diagnosis of SKA using ES in clinical settings. Trio ES should be performed first, especially in fetuses with pure skeletal dysplasia or classical skeletal dysplasia. In the future, with the wide application of WGS in the prenatal diagnosis of SKA fetuses, more genetic causes of SKA may be found, and the detection rate of SKA fetuses will also be improved.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
MJ: Writing – original draft, Writing – review and editing. BZ: Data curation, Investigation, Writing – review and editing. JW: Formal Analysis, Writing – review and editing. CW: Investigation, Writing – review and editing. XM: Writing – original draft. BY: Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by a project supported by Changzhou Key Laboratory of Medical Genetics (CM20223012), Project funding for the training of high level health professionals in Changzhou (2022CZZY007), the Changzhou Science and Technology Support Project (Social Development: CE20225066), Key Laboratory of Prenatal Diagnosis Center of Suqian Supported by Suqian Sci&Tech Program (M202303), Nanjing Medical University Changzhou Medical Center Project (CMCC202421) and Suqian Sci&Tech Program (Z2022082).
Acknowledgments
We thank all of the project participants for their contributions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1502538/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Forest plot of total detection rate.
SUPPLEMENTARY FIGURE S2 | The proportion of various genes. In these 301 cases, a total of 76 genes across 304 types of variants were detected. The ten most common genes identified were FGFR3, COL1A1, COL1A2, COL2A1, DYNC2H1, ALPL, FLNB, EBP, PPIB, and IFITM5. Their respective frequencies were 31.3%, 14.5%, 10.2%, 8.9%, 3.6%, 2.3%, 1.3%, 1.3%, 1.3%, and 1.0%.
SUPPLEMENTARY FIGURE S3 | Subgroup distribution of common variant genes.
SUPPLEMENTARY FIGURE S4 | Funnel plot of total detection rate.
References
Aggarwal, S., Vineeth, V. S., Das Bhowmik, A., Tandon, A., Kulkarni, A., Narayanan, D. L., et al. (2019). Exome sequencing for perinatal phenotypes: the significance of deep phenotyping. Prenat. Diagn. 40 (2), 260–273. doi:10.1002/pd.5616
Augusciak-Duma, A., Witecka, J., Sieron, A. L., Janeczko, M., Pietrzyk, J. J., Ochman, K., et al. (2018). Mutations in the COL1A1 and COL1A2 genes associated with osteogenesis imperfecta (OI) types I or III. Acta Biochim. Pol. 65 (1), 79–86. doi:10.18388/abp.2017_1612
Bai, Y., Sun, Y., Liu, N., Wang, L., Jiao, Z. H., Hou, Y. Q., et al. (2022). Genetic analysis of 55 cases with fetal skeletal dysplasia. Orphanet J. Rare Dis. 17 (1), 410. doi:10.1186/s13023-022-02559-4
Best, S., Wou, K., Vora, N., Van der Veyver, I. B., Wapner, R., and Chitty, L. S. (2018). Promises, pitfalls and practicalities of prenatal whole exome sequencing. Prenat. Diagn 38 (1), 10–19. doi:10.1002/pd.5102
Callaway, J. L., Shaffer, L. G., Chitty, L. S., Rosenfeld, J. A., and Crolla, J. A. (2013). The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: a review of the literature. Prenat. Diagn 33 (12), 1119–1123. doi:10.1002/pd.4209
Cao, J., Chen, A. E., Tian, L. Y., Yan, L. L., Li, H. B., and Zhou, B. H. (2022). Application of whole exome sequencing in fetal cases with skeletal abnormalities. Heliyon 8 (7), e09819. doi:10.1016/j.heliyon.2022.e09819
Chandler, N., Best, S., Hayward, J., Faravelli, F., Mansour, S., Kivuva, E., et al. (2018). Rapid prenatal diagnosis using targeted exome sequencing: a cohort study to assess feasibility and potential impact on prenatal counseling and pregnancy management. Genet. Med. 20 (11), 1430–1437. doi:10.1038/gim.2018.30
Deden, C., Neveling, K., Zafeiropopoulou, D., Gilissen, C., Pfundt, R., Rinne, T., et al. (2020). Rapid whole exome sequencing in pregnancies to identify the underlying genetic cause in fetuses with congenital anomalies detected by ultrasound imaging. Prenat. Diagn. 40 (8), 972–983. doi:10.1002/pd.5717
de Wit, M. C., Srebniak, M. I., Govaerts, L. C., Van Opstal, D., Galjaard, R. J., and Go, A. T. (2014). Additional value of prenatal genomic array testing in fetuses with isolated structural ultrasound abnormalities and a normal karyotype: a systematic review of the literature. Ultrasound Obstet. Gynecol. 43 (2), 139–146. doi:10.1002/uog.12575
Han, J., Yang, Y. D., He, Y., Liu, W. J., Zhen, L., Pan, M., et al. (2020). Rapid prenatal diagnosis of skeletal dysplasia using medical trio exome sequencing: benefit for prenatal counseling and pregnancy management. Prenat. Diagn. 40 (5), 577–584. doi:10.1002/pd.5653
Huang, Y. L., Liu, C., Ding, H. K., Wang, Y. A., Yu, L. H., Guo, F. F., et al. (2023). Exome sequencing in fetuses with short long bones detected by ultrasonography: a retrospective cohort study. Front. Genet. 14, 1032346. doi:10.3389/fgene.2023.1032346
Hui, A. S., Chau, M. H. K., Chan, Y. M., Cao, Y., Kwan, A. H., Zhu, X., et al. (2021). The role of chromosomal microarray analysis among fetuses with normal karyotype and single system anomaly or nonspecific sonographic findings. Acta Obstet. Gynecol. Scand. 100 (2), 235–243. doi:10.1111/aogs.14003
Jelin, A. C., Blakemore, K., Trebes, S., Sagaser, K., Forster, K. R., Russo, M., et al. (2020). Molecular testing strategies in the evaluation of fetal skeletal dysplasia. J. Maternal-Fetal & Neonatal Med. 35 (14), 2788–2794. doi:10.1080/14767058.2020.1802715
Krakow, D. (2015). Skeletal dysplasias. Clin. Perinatology 42 (2), 301–319. doi:10.1016/j.clp.2015.03.003
Kucinska-Chahwan, A., Roszkowski, T., Nowakowska, B., Geremek, M., Paczkowska, M., Bijok, J., et al. (2022). Extended genetic testing in fetuses with sonographic skeletal system abnormalities. Ultrasound Obstetrics & Gynecol. 59 (5), 660–667. doi:10.1002/uog.23722
Liu, Y., Wang, L., Yang, Y.-K., Liang, Y., Zhang, T.-J., Liang, N., et al. (2019). Prenatal diagnosis of fetal skeletal dysplasia using targeted next-generation sequencing: an analysis of 30 cases. Diagn. Pathol. 14 (1), 76. doi:10.1186/s13000-019-0853-x
Lord, J., McMullan, D. J., Eberhardt, R. Y., Rinck, G., Hamilton, S. J., Quinlan-Jones, E., et al. (2019). Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet 393 (10173), 747–757. doi:10.1016/S0140-6736(18)31940-8
Marini, J. C., Forlino, A., Bachinger, H. P., Bishop, N. J., Byers, P. H., Paepe, A., et al. (2017). Osteogenesis imperfecta. Nat. Rev. Dis. Prim. 3, 17052. doi:10.1038/nrdp.2017.52
Mellis, R., Oprych, K., Scotchman, E., Hill, M., and Chitty, L. S. (2022). Diagnostic yield of exome sequencing for prenatal diagnosis of fetal structural anomalies: a systematic review and meta-analysis. Prenat. Diagn. 42 (6), 662–685. doi:10.1002/pd.6115
Milks, K. S., Hill, L. M., and Hosseinzadeh, K. (2017). Evaluating skeletal dysplasias on prenatal ultrasound: an emphasis on predicting lethality. Pediatr. Radiol. 47 (2), 134–145. doi:10.1007/s00247-016-3725-5
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G.PRISMA Group (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6 (7), e1000097. doi:10.1371/journal.pmed.1000097
Ornitz, D. M., and Marie, P. J. (2015). Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 29 (14), 1463–1486. doi:10.1101/gad.266551.115
Page, M. J., McKenzie, J. E., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. PLOS Med. 18 (3), e1003583. doi:10.1371/journal.pmed.1003583
Peng, Y., Yang, S. T., Huang, X. L., Pang, J. L., Liu, J., Hu, J. C., et al. (2021). Whole exome sequencing analysis in fetal skeletal dysplasia detected by ultrasonography: an analysis of 38 cases. Front. Genet. 12, 728544. doi:10.3389/fgene.2021.728544
Petrovski, S., Aggarwal, V., Giordano, J. L., Stosic, M., Wou, K., Bier, L., et al. (2019). Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet 393 (10173), 758–767. doi:10.1016/S0140-6736(18)32042-7
Schramm, T., and Mommsen, H. (2018). Fetal skeletal disorders. Ultraschall der Medizin - Eur. J. Ultrasound 39 (06), 610–634. doi:10.1055/a-0660-9417
Shi, J., Ren, M., Jia, J., Tang, M., Guo, Y., Ni, X., et al. (2019). Genotype-phenotype association analysis reveals new pathogenic factors for osteogenesis imperfecta disease. Front. Pharmacol. 10, 1200. doi:10.3389/fphar.2019.01200
Tang, H., Zhang, Q., Xiang, J. J., Yin, L. L., Wang, J., and Wang, T. (2021). Whole exome sequencing aids the diagnosis of fetal skeletal dysplasia. Front. Genet. 12, 599863. doi:10.3389/fgene.2021.599863
Tang, J., Zhou, C. L., Shi, H. H., Mo, Y. Y., Tan, W. L., Sun, T. L., et al. (2020). Prenatal diagnosis of skeletal dysplasias using whole exome sequencing in China. Clin. Chim. Acta 507, 187–193. doi:10.1016/j.cca.2020.04.031
Tolusso, L. K., Hazelton, P., Wong, B., and Swarr, D. T. (2021). Beyond diagnostic yield: prenatal exome sequencing results in maternal, neonatal, and familial clinical management changes. Genet. Med. 23 (5), 909–917. doi:10.1038/s41436-020-01067-9
Tournis, S., and Dede, A. D. (2018). Osteogenesis imperfecta - a clinical update. Metabolism 80, 27–37. doi:10.1016/j.metabol.2017.06.001
Tse, K. Y., Surya, I. U., Irwinda, R., Leung, K. Y., Ting, Y. H., Cao, Y., et al. (2023). Diagnostic yield of exome sequencing in fetuses with sonographic features of skeletal dysplasias but normal karyotype or chromosomal microarray analysis: a systematic review. Genes 14 (6), 1203. doi:10.3390/genes14061203
Unger, S., Ferreira, C. R., Mortier, G. R., Ali, H., Bertola, D. R., Calder, A., et al. (2023). Nosology of genetic skeletal disorders: 2023 revision. Am. J. Med. Genet. Part A 191 (5), 1164–1209. doi:10.1002/ajmg.a.63132
Vora, N. L., Powell, B., Brandt, A., Strande, N., Hardisty, E., Gilmore, K., et al. (2017). Prenatal exome sequencing in anomalous fetuses: new opportunities and challenges. Genet. Med. 19 (11), 1207–1216. doi:10.1038/gim.2017.33
Waller, D. K., Correa, A., Vo, T. M., Wang, Y., Hobbs, C., Langlois, P. H., et al. (2008). The population-based prevalence of achondroplasia and thanatophoric dysplasia in selected regions of the US. Am. J. Med. Genet. Part A 146A (18), 2385–2389. doi:10.1002/ajmg.a.32485
Wapner, R. J., Martin, C. L., Levy, B., Ballif, B. C., Eng, C. M., Zachary, J. M., et al. (2012). Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 367 (23), 2175–2184. doi:10.1056/NEJMoa1203382
Yadava, S. M., and Ashkinadze, E. (2018). Whole exome sequencing for prenatal diagnosis in cases with fetal anomalies: criteria to improve diagnostic yield. J. Genet. Couns. 28 (2), 251–255. doi:10.1002/jgc4.1045
Yang, K., Shen, M., Yan, Y. S., Tan, Y., Zhang, J., Wu, J., et al. (2019). Genetic analysis in fetal skeletal dysplasias by trio whole-exome sequencing. Biomed Res. Int. 2019, 2492590. doi:10.1155/2019/2492590
Yang, Y., Wang, M., and Wang, H. (2022). Prenatal trio-based whole exome sequencing in fetuses with abnormalities of the skeletal system. Mol. Genet. Genomics 297 (4), 1017–1026. doi:10.1007/s00438-022-01899-x
Zhang, B., Zhang, Y., Wu, N., Li, J., Liu, H., and Wang, J. (2020). Integrated analysis of COL2A1 variant data and classification of type II collagenopathies. Clin. Genet. 97 (3), 383–395. doi:10.1111/cge.13680
Zhang, L., Pan, L. J., Teng, Y. L., Liang, D. S., Li, Z., and Wu, L. Q. (2021a). Molecular diagnosis for 55 fetuses with skeletal dysplasias by whole-exome sequencing: a retrospective cohort study. Clin. Genet. 100 (2), 219–226. doi:10.1111/cge.13976
Zhang, X. Y., Ren, Y., Song, R., Wang, L. X., Xu, H., Xie, X. X., et al. (2021b). Combined exome sequencing and deep phenotyping in highly selected fetuses with skeletal dysplasia during the first and second trimesters improves diagnostic yield. Prenat. Diagn. 41 (11), 1401–1413. doi:10.1002/pd.5974
Zhou, X., Chandler, N., Deng, L., Zhou, J., Yuan, M., and Sun, L. (2018). Prenatal diagnosis of skeletal dysplasias using a targeted skeletal gene panel. Prenat. Diagn. 38 (9), 692–699. doi:10.1002/pd.5298
Keywords: exome sequencing, skeletal abnormalities, prenatal diagnosis, karyotyping, chromosomal microarray analysis
Citation: Jiang M, Zhang B, Wang J, Wei C, Mao X and Yu B (2025) Exome sequencing and prenatal skeletal abnormalities: comprehensive review and meta-analysis and way forward. Front. Genet. 16:1502538. doi: 10.3389/fgene.2025.1502538
Received: 27 September 2024; Accepted: 14 May 2025;
Published: 11 June 2025.
Edited by:
Paulo Ricardo Gazzola Zen, Federal University of Health Sciences of Porto Alegre, BrazilReviewed by:
Cristina Skrypnyk, Arabian Gulf University, BahrainLauren Wasson, Harvard Medical School, United States
Copyright © 2025 Jiang, Zhang, Wang, Wei, Mao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Yu, YmlueXVAbmptdS5lZHUuY24=, Xiuzhen Mao, Ynl4aXV6aGVubWFvQDE2My5jb20=
 Mengting Jiang1,2
Mengting Jiang1,2 Bin Zhang
Bin Zhang Jing Wang
Jing Wang Bin Yu
Bin Yu