- 1Division of Human Genetics, Department of Pathology and Institute of Infectious Disease and Molecular Medicine (IIDM), Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
- 2SAMRC/UCT Platform for Pharmacogenomics Research and Translation, South African Medical Research Council, Cape Town, South Africa
- 3Department of Pharmaceutical Technology, School of Allied Health Sciences, Harare Institute of Technology, Harare, Zimbabwe
- 4Division of Lipidology and Cape Heart Institute, Department of Medicine, Groote Schuur Hospital and Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
- 5Division of Clinical Pharmacology, Department of Medicine, Groote Schuur Hospital and Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
- 6Department of Applied Sciences, Faculty of Health and Life Sciences, Northumbria University, Newcastle, United Kingdom
- 7Division of Nephrology and Hypertension, Department of Medicine, Groote Schuur Hospital and Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
Introduction: Genetic variation in genes coding for enzymes metabolising antihypertensive drugs, may affect the efficacy of angiotensin converting enzyme (ACE) inhibitors such as enalapril, potentially leading to resistant hypertension (RHTN). We set out to evaluate the contribution of genetic variation in CES1 and NOS3 genes on susceptibility to RHTN, as well as estimate the frequencies of CES1 copy number variation (CNV) in African and Mixed Ancestry (MA) populations of South Africa.
Methods: Using a retrospective age, sex and ethnicity matched case-control study design, 379 participants with hypertension belonging to the African and MA ethnic groups were recruited. Cases were participants with RHTN (i.e., blood pressure (BP) ≥140/90 mmHg on ≥3 antihypertensive drugs or BP < 140/90 mmHg on >3 antihypertensive drugs, including a diuretic). Cases were matched to controls with similar characteristics (age (±5 years), sex and ethnicity) in a 1:1 ratio. Controls were participants with hypertension that was under control (BP < 140/90 mmHg on ≤3 antihypertensive drugs). Five polymorphisms in CES1 and NOS3 were characterized using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), quantitative PCR and validated using Sanger sequencing. The additive model of inheritance and multivariable logistic regression were used to determine associations between genotypes and RHTN while adjusting for potential confounding variables.
Results and discussion: NOS3 rs3918188A/A (aOR: 0.13; CI: 0.04–0.41; P = 0.0009) genotype and NOS3 rs2070744–rs1798883–rs3918188G–T–A haplotype (OR: 0.54; CI: 0.37–0.78; P = 0.001) appeared to confer protection against RHTN among MA participants only. CES1 rs2244613C>A and CES1 CNV were not significantly associated with RHTN. However, there appeared to be quantitative differences in CES1 CNV profiles across ethnic groups. We speculate that NOS3 rs3918188A allele may affect NOS3 gene expression, potentially leading to increased amounts of the vasodilator, nitric oxide (NO) and favourable outcomes in individuals taking antihypertensives drugs such as enalapril.
Conclusion: NOS3 genetic variation seems important in the susceptibility to RHTN among Africans and requires further studies.
Introduction
Hypertension is a serious public health concern, affecting over 1 billion people globally (WHO, 2021). It is a major contributor to the population attributable risk for stroke, myocardial infarction and heart failure, which are among the leading causes of death worldwide (Olsen et al., 2016; Zhou et al., 2021). Resistant hypertension (RHTN) is defined as uncontrolled blood pressure (BP) (≥140/90 mmHg) in an adherent patient taking three or more antihypertensive drugs including a diuretic, or controlled BP (<140/90 mmHg) in an adherent patient taking four or more antihypertensive drugs (Dudenbostel et al., 2016). RHTN affects up to 21% of hypertensive patients globally (Acelajado et al., 2012; Jones et al., 2017), between 4%–19% of hypertensives in Africa (Nansseu et al., 2016), and nearly 13% of South Africans with hypertension (Moosa et al., 2016).
Angiotensin converting enzyme (ACE) inhibitors are one of the most frequently used class of antihypertensive drugs in patients with hypertension, including those with RHTN (Jones et al., 2020; Unger et al., 2020). Enalapril is the preferred ACE inhibitor in the Essential Drug List (EDL) of South Africa and hence the second most prescribed antihypertensive drug in patients attending a tertiary-level Hypertension Clinic at Groote Schuur Hospital, in South Africa (Soko et al., 2023). Response to enalapril has been reported to vary widely among individuals and its use is associated with several potential adverse reactions, such as angioedema, dry-cough, hypotension and hyperkalaemia (Moholisa et al., 2013; Katsukunya et al., 2023). Several enzymes are involved in the pharmacokinetics and pharmacodynamics of enalapril including carboxylesterase-1 (CES1) and endothelial nitric oxide synthase (NOS3/eNOS) respectively. As such, genetic variation in CES1 and NOS3 genes has been reported to influence treatment outcomes in individuals taking enalapril (Oliveira-Paula et al., 2019) and may potentially contribute to susceptibility to RHTN.
CES1 gene is located on chromosome 16q12.2, has 14 exons and is ∼33 kb long and encodes for the CES1 enzyme, a serine esterase, responsible for the conversion of enalapril to enalaprilat. Enalaprilat is up to 20 times more potent in lowering BP than enalapril, thus, this conversion is essential for enalapril efficacy (Davis et al., 2007). CES1 is highly polymorphic, has a complicated structure and is subject to segmental duplications resulting in copy number variations (Yoshimura et al., 2008). Polymorphisms such as CES1 rs2244613C>A and copy number variation (CNV) have been shown to affect the pharmacokinetics of ACE inhibitors, including enalapril. However, studies report inconsistent effects (Ikonnikova et al., 2022a; Wang et al., 2016; Her et al., 2021; Stage et al., 2017a). Recently, CES1 rs2244613C allele carriers were reported to present with reduced peak and trough concentrations of both enalapril and enalaprilat compared to rs2244613A/A genotype carriers in European patients with hypertension (Ikonnikova et al., 2022b).
The NOS3 gene, located on chromosome 7q36.1, has 28 exons and encodes for endothelial nitric oxide synthase (eNOS/NOS3), a 1203 amino acid long protein. According to Oliviera-Paula and colleagues (Oliveira-Paula et al., 2017), ACE-inhibition is not the only mechanism of action of ACE-inhibitors and upregulation of NOS3 activity, stimulating the production of increased nitric oxide (NO), a vasodilator (Oliveira-Paula et al., 2016a) also contributes to BP lowering. Several polymorphisms in NOS3 have been reported in connection with hypertension and antihypertensive drug response. For example, NOS3 rs2070744C>T and rs1799983T>G have been shown to affect susceptibility to essential hypertension (Gamil et al., 2017; Jemaa et al., 2007; Amrani-Midoun et al., 2019; Nassereddine et al., 2018) and response to enalapril (Masilela et al., 2021) whereas NOS3 rs3918188C>A has been reported to be associated with enalapril response among Brazilians, with NOS3 rs3918188A allele carriers showing lower decreases in BP (Oliveira-Paula et al., 2016a).
In Africans, there are currently few studies that have evaluated the impact of genetic variation in CES1 and NOS3 on enalapril response or the likelihood of being diagnosed with RHTN. Given the crucial role of these enzymes in metabolism or response to enalapril and in BP regulating pathways, we hypothesize that they could be of pharmacogenomic importance in African populations, conferring susceptibility to phenotypes associated with poor drug response such as RHTN (Dudenbostel et al., 2016; El Rouby and Cooper-DeHoff, 2015). Thus, our study aimed to evaluate the role of genetic variation in CES1 and NOS3 in susceptibility to RHTN as well as estimate the baseline frequency of CES1 CNV among African and Mixed Ancestry (MA) (i.e., admixed populations of Southern Africa resulting from intermarriages between African, European, San or Asian populations) populations of South Africa. Understanding the pharmacogenetic variants associated with susceptibility to RHTN has the potential to enhance treatment and control of hypertension among African populations.
Materials and methods
Study design and participants
This was a retrospective, matched case-control study, enrolling participants attending a tertiary-level Hypertension Clinic at Groote Schuur Hospital in Cape Town, South Africa (33.9413° S, 18.4622° E). This clinic provides specialised care for patients referred for the management of hypertension, including drug response phenotypes such as RHTN. Participants were classified as either poor responders (cases) or good responders (controls) to antihypertensive therapy, based on them having either RHTN or non-RHTN respectively, and matched according to age, sex, and ethnicity in a 1:1 ratio. RHTN was defined as (i) BP > 140/90 mmHg on 3 or more antihypertensive drugs including a diuretic in optimal doses and (ii) BP < 140/90 mmHg on 4 or more antihypertensive drugs in an adherent patient. Where non-adherence was suspected, it was ruled out by monitoring of amlodipine levels as described by with Jones and colleagues (Jones et al., 2017). Non-RHTN was defined as BP < 140/90 mmHg or BP > 140/90 mmHg on less than 3 antihypertensive drugs with no additional clinical indications of true resistant hypertension (Dudenbostel et al., 2016). BP and the number of antihypertensive drugs that the patients were taking were evaluated using information from the most recent visit at the time of recruitment. To ensure accurate BP measurements, an average of 6 BP readings were recorded. If patients had borderline BP (=140/90 ± 5 mmHg) at the most recent visit, an average of BP measurements from prior visits were taken into account, when we were classifying these patients into cases (RHTN) or controls (non-RHTN). The inclusion criteria were participants (i) with a confirmed diagnosis of primary or essential hypertension (defined as hypertension without any known underlying cause), (ii) of African heritage (defined as individuals of African or MA descent), (iii) on at least one antihypertensive drug for at least a year prior, and (iv) > 18 years at the time of recruitment. The exclusion criteria were participants (i) with a confirmed diagnosis of secondary hypertension (defined as hypertension with a known underlying cause), (ii) who were pregnant at the time of recruitment, (iii) with confirmed white coat hypertension or non-adherence, and (iv) not on treatment with any antihypertensive drugs.
Sample size calculation
The sample size was calculated according to Naing et al. (2022), ensuring over 80% power. Briefly, the prevalence of RHTN was taken to be 12.6% as reported by Moosa et al. (2016) and an alpha level of 0.05 was used. This resulted in a required sample size of at least 163 participants with RHTN. Since this was a case-control study by design, 163 participants with non-RHTN would be required to serve as the control group. This resulted in a total sample size of at least 326 participants. However, we incorporated an allowance of at least 15% more participants in each group, to account for potential sample quality issues during genetic characterization. Therefore, our realized sample size was brought to a total of 389 participants, including 190 with RHTN (cases) and 189 with non-RHTN (controls). Although our study was sufficiently powered overall, our cohort was not homogenous as it was made up of African (N = 110) and MA (N = 279) participants. The African group in our study is made up of native Africans predominantly of Xhosa origin, while the MA group is made up of individuals resulting from intermarriages between native Africans and/or European, San or Asian populations (i.e., Cape Coloureds). Thus, to resolve the genetics of the two distinct ethnic groups, subgroup analyses according to ethnicity were performed. However, we acknowledge that the number of African participants, could have limited the power to detect statistically significant associations in this group.
DNA sample preparation
DNA was extracted from 2 mL of venous blood provided by consenting participants using the Chemagic 360® automated nucleic acid extraction system (Chemagen Technologies GMBH/Perkin Elmer Incorporated, United States) according to the manufacturer’s protocol. The extracted genomic DNA was evaluated for quality and quantity using electrophoresis on a 1% (w/v) agarose gel and spectrophotometry on a NanoDrop c1000 UV spectrophotometer (Thermofisher Scientific Corporation, Massachusetts, United States) respectively. DNA of good quality and quantity (at least 50 ng/μL) was used for genetic characterization.
Selection of candidate pharmacogenomic markers in CES1 and NOS3 for genetic characterization
The selection of SNPs in CES1 and NOS3 was guided by querying several pharmacogenomic databases such as the Clinical Pharmacogenetics Implementation Consortium (CPIC) database, Food and Drug Administration (FDA) Table of Pharmacogenomic Biomarkers in Drug Labelling, Pharmacogenomics Knowledge Base (PharmGKB) database and surveying available literature. Variants were prioritised according to (i) annotation in any of the pharmacogenomic databases, (ii) predicted or known functional effect, (iii) minor allele frequency (MAF) > 0.05 in African populations and (iv) published evidence of potential clinical relevance. Furthermore, variants not previously reported in Africans but were reported to influence response/efficacy to enalapril or any other ACE inhibitor were also prioritised, to pronounce on their role in Africans as well. This strategy yielded the following variants for analysis: NOS3 rs1799983G>T (priority 1), rs2070744C>T (priority 1), rs3918188C>A (priority 2), CES1 rs2244613G>T (priority 2) and CES1 CNV (priority 3) (Table 1).
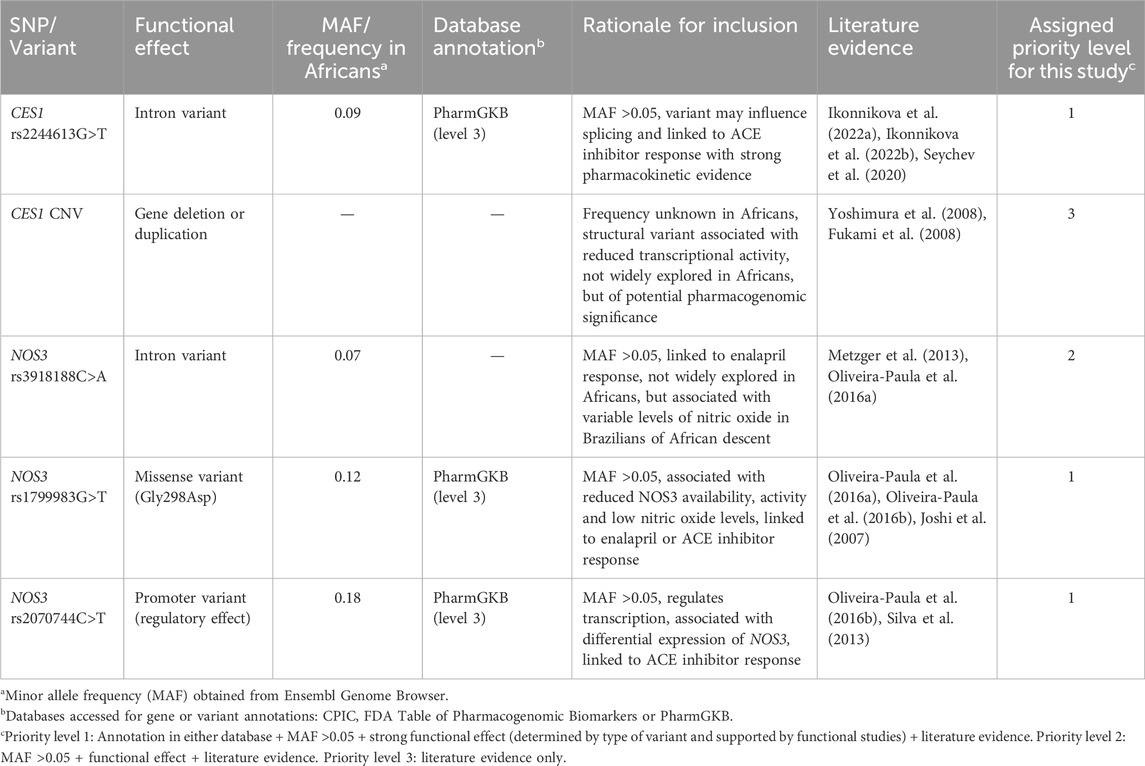
Table 1. Rationale for selection of potential candidate pharmacogenomic biomarkers in CES1 and NOS3 for analysis.
Genetic characterization
Genetic characterization was achieved through polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP), quantitative PCR and validated through Sanger sequencing. Forward and reverse primers flanking regions containing NOS3 rs1799983G>T, rs2070744C>T, rs3918188C>A and CES1 rs2244613G>T SNPs were designed using the NCBI-Primer Blast tool [https://www.ncbi.nlm.nih.gov/tools/primer-blast/(last accessed on 10 February 2025)], Integrated DNA Technologies (IDT) Oligo Analyzer tool [http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/ (last accessed on 10 February 2025)] and synthesized by Inqaba Biotechnical Industries (Muckleneuk, Pretoria, South Africa). Forward and reverse primer sequences, annealing temperatures and PCR product sizes are shown in Supplementary Table S1.
All PCRs were performed in a 25 µL reaction volume containing 100 ng genomic DNA; 1X Green GoTaq Flexi® Reaction Buffer (Promega Corporation, Madison, WI, United States); 0.4 µM of deoxynucleotide triphosphates (dNTPs) (Promega Corporation, Madison, WI, United States); 1.5 mM magnesium chloride (Promega Corporation, Madison, WI, United States); 0.2 µM each of forward and reverse primers (Inqaba Biotechnical Industries (Pty) Ltd., South Africa), 1 U of GoTaq Flexi® DNA Polymerase (Promega Corporation, Madison, WI, United States) and nuclease-free water. Amplification was done using a SimpliAmp™ Thermal Cycler (Applied Biosystems™, Thermofisher Scientific, Massachusetts, United States). The reaction conditions were an initial denaturation at 94°C for 3 min; followed by 35 cycles of further denaturation at 94°C for 30 s; annealing at temperatures specific for each SNP (Supplementary Table S1) for 30 s; initial extension at 72°C for 30 s; and final extension at 72°C for 10 min.
PCR products from amplification of regions flanking the NOS3 rs1799983G>T and CES1 rs2244613G>T SNPs, were digested using BanI and AlwN1 restriction enzymes, respectively (New England BioLabs®, Ipswich, United Kingdom). In each 30 µL digest reaction, 10 µL of PCR product, 1X CutSmart™ Buffer (New England BioLabs®, Ipswich, United Kingdom), 3 U of restriction enzyme specific for each SNP, and nuclease-free water were added. Restriction enzyme digest reactions were incubated and inactivated on a SimpliAmp™ Thermal Cycler (Applied Biosystems™, Thermofisher Scientific, Massachusetts, United States) at temperatures and periods specific for each enzyme according to the manufacture’s protocol [https://nebcloner.neb.com/#!/redigest (last accessed on 10 February 2025)] and resolved using electrophoresis on a 3% (w/v) agarose gel. Restriction enzyme digestion yielded DNA fragments corresponding to the genotype of the samples as shown in Supplementary Table S2.
Commercially available TaqMan™ allelic discrimination and copy number variation (CNV) assays were purchased from Thermofisher Scientific Incorporation (Massachusetts, United States) for genotyping NOS3 rs2070744C>T (C__15903863_10), rs3918188C>A (C__29193459_10) SNPs and for determination of CES1 copy number (Hs00139541_cn). Each TaqMan allelic discrimination assay reaction was set up in a 10 µL reaction volume containing 5 µL of 2X TaqPath™ ProAmp™ Master Mix (Thermofisher Scientific Corporation, Massachusetts, United States), 0.5 µL of 20X TaqMan™ genotyping assay specific for each SNP and 45 ng of genomic DNA. Assays were carried out on a CFX96 Touch Real Time PCR Thermal Cycler (Bio-Rad Laboratories, Inc., California, United States). The reaction conditions were initial denaturation at 95°C for 5 min followed by 50 cycles of further denaturation and annealing at 95°C for 15 s and 60°C for 1 min respectively, then final elongation at 60°C for 30 s.
The TaqMan™ CNV assay targeted intron 11 of the CES1 gene on chromosome 16q12.2 (assay reference genome location: chr.16:55,810,581 on build GRCh38). The reference gene was the gene encoding ribonuclease P located on chromosome 14q11.2 and the VIC dye labelled TaqMan™ CNV RNase P assay (catalogue number: 4,403,326) served as the reference assay (assay reference genome location: chr.14:20,343,370 on build GRCh38). Each reaction was carried out in a total reaction volume of 10 µL containing 5 ng of genomic DNA, 0.5 µL of RNase P assay, 0.5 µL of TaqMan CNV assay (Hs00139541_cn), 5 µL of 2X TaqPath™ ProAmp™ Master Mix and 3 µL of nuclease-free water. The reactions were carried out on the QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems™, Massachusetts, United States) and the reaction conditions were initial denaturation at 95°C for 10 min followed by 40 cycles of further denaturation and annealing at 95°C for 15 s and 60°C for 1 min respectively.
Post-PCR clean-up using exonuclease I (ExoI) and thermosensitive alkaline phosphatase (FastAP™) enzymes from Thermofisher Scientific (Massachusetts, United States) was done as per manufacturer’s protocol prior to Sanger sequencing. Sanger sequencing was done to validate genotyping. Each sequencing reaction was set up in a reaction volume of 10 μL, containing 5 µL of PCR product, 2 µL of BigDye™ Terminator v3.1 5X sequencing buffer, 2 µL of BigDye™ Terminator 3.1 Ready Reaction Mix (Thermofisher Scientific, Massachusetts, United States) and 1 µL of sequencing primer (Supplementary Table S1). Sequencing reactions were carried out on a SimpliAmp™ Thermal Cycler (Applied Biosystems™, Massachusetts, United States). The sequencing reaction cycling conditions and post-sequencing clean-up using EDTA/ethanol precipitation were done according to the manufacturer’s protocol (Thermofisher Scientific, Massachusetts, United States). Sequencing reactions were resolved using capillary electrophoresis on a SeqStudio® Genetic Analyzer (Thermofisher Scientific, Massachusetts, United States) and analyzed using DNAStar-SeqMan Pro Sequence Assembly software (DNAStar®, Madison, WI).
Statistical analyses and in silico functional annotation
Statistical analyses were performed using various packages in R statistical software (version 4.4.1, 2024–06–14, Vienna, Austria). Genotype, allele and haplotype frequencies, as well as linkage disequilibrium (LD) parameters were computed using SHEsis online software (Shen et al., 2016). Calculations of CES1 gene copy number was done using CopyCaller software for Windows (Version 2.0, Applied Biosystems, Massachusetts, United States) which assigned a quality metric to the copy numbers generated for each sample, incorporating absolute Z-scores (±1) and confidence scores (0%–100%). Copy number calls with a confidence metric >50% were included in the final analysis. Categorical data were presented as N (%) [where N = the number of individuals in that category and % = the frequency of individuals within that category]. Continuous data were presented as mean ± standard deviation (SD) or median ± interquartile range (IQR) [25th – 75th percentile] depending on the distribution of the data. The Shapiro-Wilks test was used to assess any deviation from normality, and the Hardy-Weinberg Equilibrium (HWE) was determined for each SNP for African and MA population groups using the Chi-square test with one degree of freedom. Continuous data were compared between cases and controls either using the T-test or the Mann-Whitney U-test (Wilcoxon rank-sum test) for normally and non-normally distributed data, respectively. Categorical data were compared between cases and controls using Pearson’s Chi-square test or Fisher’s exact test.
The main outcome of interest was the association of selected polymorphisms with RHTN. Therefore, the additive model of inheritance was assumed to estimate associations with RHTN. To account for the effect of any confounding variables on the association of genetic factors with RHTN, non-genetic predictor variables (clinical or demographic factors) were screened as univariate and those with a P ≤ 0.2 were retained for analysis as covariates (altogether). A multivariable logistic regression model was fitted, to estimate adjusted odds ratios (aORs) and 95% confidence intervals (CIs) for the association between each genetic variable and RHTN using the generalised linear model (glm (.)) function in R studio. The model included age (in years), smoking status (yes/no), plasma aldosterone concentration (pmol/L), diabetes mellitus (yes/no), dyslipidaemia (yes/no), use of diabetic medications (yes/no), lipid-lowering therapy (yes/no), CES1 CNV, CES1 rs2244613G>T, NOS3 rs1799983G>T, NOS3 rs2070744C>T and NOS3 rs3918188C>A. Adjusted P-values reflected these covariate adjustments, aORs described the strength of associations and 95% confidence intervals (CI) were used as precision estimates. Statistical significance was set at P < 0.05. Bonferroni correction was applied to correct for multiple comparison, although its use is contested. The Bonferroni correction was applied for the 4 SNPs with significant P < 0.0125 (0.05/4).
Finally, intronic SNPs found to be significantly associated with RHTN were further queried in HaploReg v4.2 (Broad Institute, Massachusetts, United States) developed by ENCODE (Ward and Kellis, 2012) to further assess their mechanistic role including potential regulatory function and any other predicted SNPs appearing in high LD in Africans.
Results
Study participant characteristics
A total of 379 participants (29% African and 71% MA populations) were enrolled, and their demographic and clinical characteristics are presented in Table 2. Among the participants, sex and age were similar between cases and controls, in the African and MA groups. Compared to the control group, patients with RHTN (or cases) were more likely to have been active or past smokers (MA group only), on more antihypertensive drugs (i.e., amlodipine, hydrochlorothiazide, atenolol, enalapril, spironolactone or losartan); had left ventricular hypertrophy (LVH) and higher aldosterone levels. Furthermore, there were more cases diagnosed with chronic kidney disease (CKD) (P < 0.001), stroke (P = 0.04) and on statin or other lipid lowering therapy (MA group only, P = 0.001) than controls (Table 2).
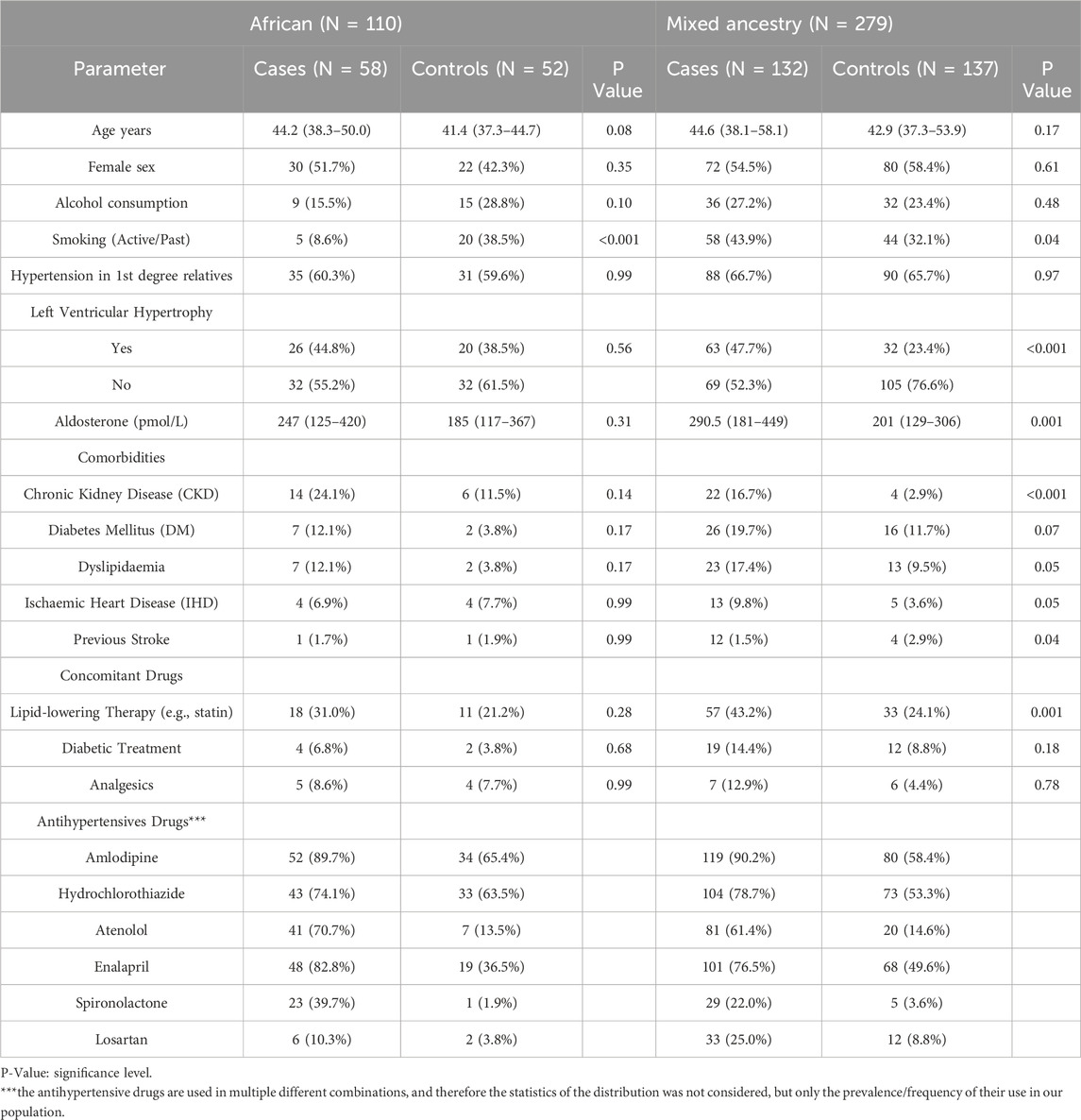
Table 2. Demographic and clinical parameters compared among participants with resistant hypertension (cases) and non-resistant hypertension (controls) for African and Mixed Ancestry groups.
CES1 copy number variation
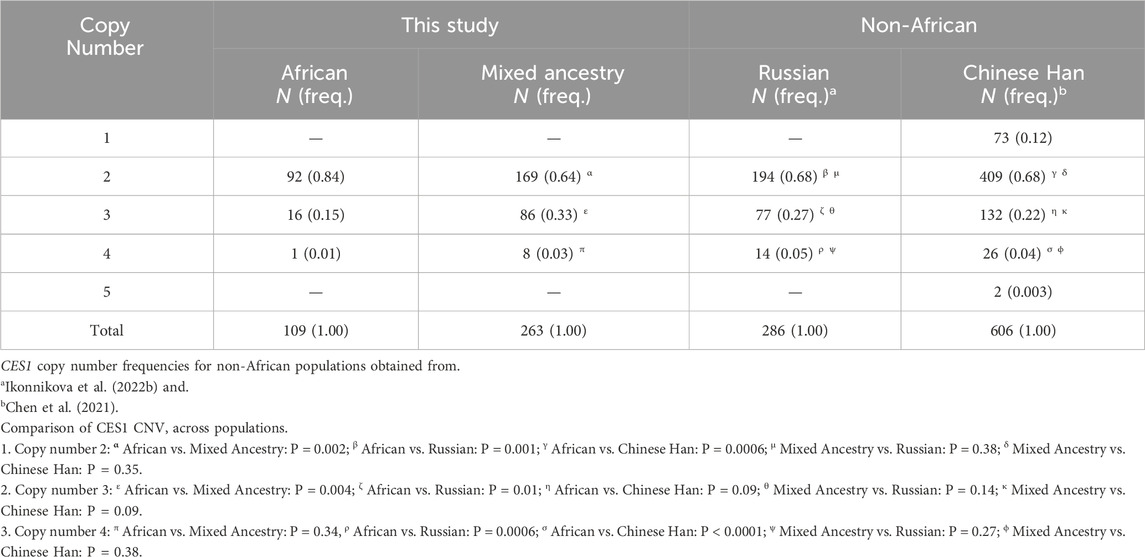
A total of 372 participants included in the final analysis had CES1 copy number calls with confidence scores >50%. The number of copies of the CES1 gene ranged from 2–4 (Table 3). No complete deletion (<2) was observed in our cohort. Copy number 2 was the most frequent in both African (84%) and MA (64%) participants, with a statistically significant difference (P = 0.002) between the two ethnic groups. Copy number 3 was more frequent in MA individuals (33%) compared to Africans (15%) (P = 0.004). No significant difference was observed for copy number 4 (P = 0.34) between African and MA groups. Comparing to non-African populations, there were statistically significant differences (P < 0.05) in frequencies of copy number calls between the African group and the Russian (Ikonnikova et al., 2022b) and Chinese Han (Chen et al., 2021) populations. The copy number calls for the MA group were similar to either the Russian or Chinese Han populations with no statistically significant differences (P > 0.05) observed (Table 3).
No association of CES1 copy number variation with resistant hypertension
The distribution of CES1 copy numbers between cases and controls for the African and MA groups and associations with RHTN after adjusting for potential confounding variables (i.e., age, smoking, aldosterone levels, diabetes mellitus (DM), dyslipidaemia, diabetic treatment and lipid lowering therapy) are shown in Supplementary Table S3. There were no statistically significant differences in the distributions of each copy number between cases and controls (P > 0.05) for both African and MA groups. Similarly, no significant associations with RHTN were observed. CES1 copy numbers were further categorized into copy number neutral (=2) and copy number gain (>2) as previously described by Chen et al. (2021). We observed no significant differences in the distribution of participants who were copy number neutral or had a copy number gain between cases or controls indicating no significant association with RHTN (Supplementary Table S3).
Association of single nucleotide polymorphisms in CES1 and NOS3 with resistant hypertension
Genotype frequency distributions between cases and controls, and associations with RHTN for the CES1 rs2244613G>T, NOS3rs1799983G>T, rs2070744C>T and rs3918188C>A SNPs in the African and MA groups after adjusting for potential confounding variables are shown in Table 4. NOS3 rs3918188A/A genotype carriers were significantly more frequent among the controls compared to cases in the MA group and associated with reduced risk of RHTN [17% vs. 6% respectively (P = 0.0009; aOR: 0.13; CI: 0.04–0.41)]. No statistically significant differences in the distribution of variant alleles (Supplementary Table S4) and genotypes (Table 4) between cases and controls for NOS3 rs1799983G>T, rs2070744C>T and CES1 rs2244613G>T for both African and MA groups were observed.
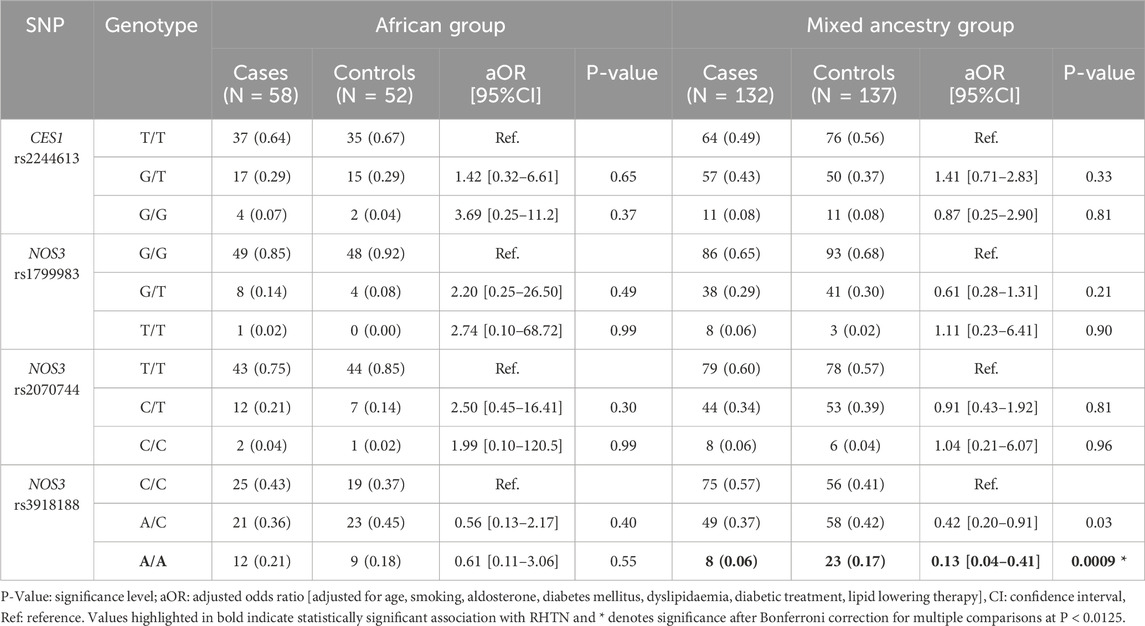
Table 4. Comparison of genotype frequency distributions between resistant hypertension (cases) and non-resistant hypertension (controls) participants among African and Mixed Ancestry groups and associations with resistant hypertension.
No association of polymorphisms in CES1 with resistant hypertension in individuals taking enalapril only
CES1 is directly involved in the metabolism of enalapril. Thus, to further resolve the role of CES1 in RHTN, an additional analysis step was done for African (i.e., cases: N = 48; controls: N = 19) and MA (i.e., cases: N = 101; controls: N = 68) participants that were on enalapril for associations of CES1 polymorphisms with RHTN. Allele and genotype frequencies are shown in Supplementary Tables S4, S5 respectively. It appears that there were no statistically significant differences in the distribution of CES1 rs2244613G>T genotypes and CES1 copy number calls (P > 0.05) between cases and controls on enalapril in both African and MA groups, even after adjusting for confounding variables. In addition, no significant associations with RHTN were observed (Supplementary Table S5).
Linkage disequilibrium analyses and association of NOS3 haplotypes/diplotypes with resistant hypertension
Pairwise linkage disequilibrium (LD) values for NOS3 rs1799983G>T, rs2070744C>T and rs3918188C>A are shown in Figure 1. There is stronger allelic association between NOS3 rs3918188C>A and rs1799983G>T (African: D’ = 0.999; MA: D’ = 0.823); and moderate allelic association between NOS3 rs3918188C>A and rs2070744C>T (African: D’ = 0.622; MA: D’ = 0.500). The frequencies of possible haplotypes constructed from NOS3 rs1799983G>T, rs2070744C>T and rs3918188C>A SNPs are also shown in Table 5. It seems that NOS3 rs1799983 – rs2070744 – rs3918188 G–T–A haplotype carriers were significantly (P = 0.001) more frequent in controls (36%) than cases (24%) and were significantly associated with reduced risk of RHTN (OR: 0.54; CI: 0.37–0.78) for MA participants only. Humans are diploid and typically inherit two haplotypes-one from either parent. Therefore, to capture the full genetic variation across both copies of the NOS3 chromosome, the frequencies of the possible haplotypes in the diploid state were determined, and associations with RHTN after adjusting for confounding variables are shown in Supplementary Table S6. It appears that participants carrying two copies of the NOS3 rs1799983 – rs2070744 – rs3918188 G–T–A haplotype or G–T–A diplotype (G–T–A/G–T–A) were significantly (P = 0.008) more frequent in the controls (15%) than cases (6%) and associated with significantly reduced risk of RHTN in the MA group only (aOR: 0.23, CI: 0.07–0.64).

Figure 1. Linkage disequilibrium (LD) plots (D’) for African and Mixed Ancestry participants for NOS3 rs1799983G>T, rs2070744C>T and rs3918188C>A SNPs.
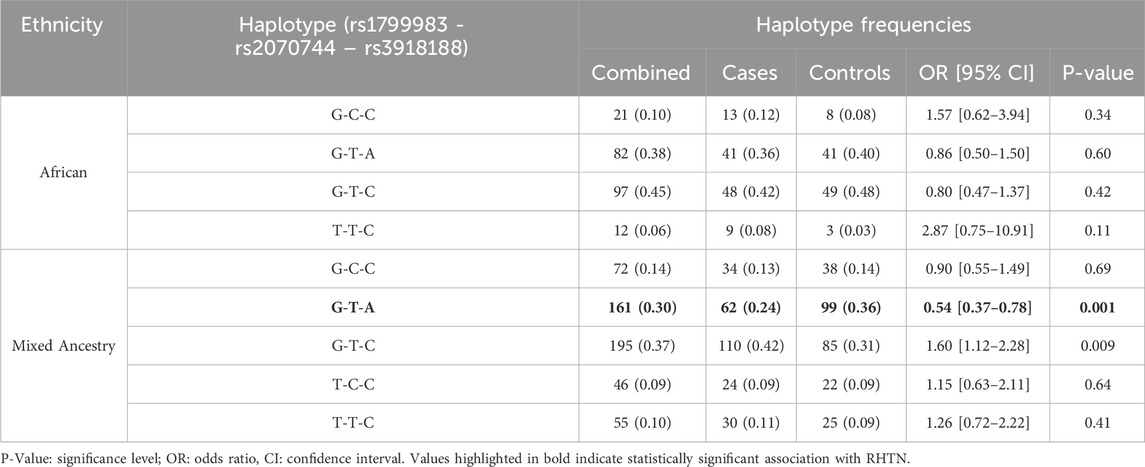
Table 5. Haplotype frequency distributions between participants with resistant hypertension (cases) and non-resistant hypertension (controls) and associations with resistant hypertension for African and Mixed Ancestry groups.
In-silico functional annotation of NOS3 rs3918188C>A
A summary of a HaploReg query for NOS3 rs3918188C>A is presented in Supplementary Table S7. It appears that rs3918188C>A is in strong LD in Africans, with additional SNPs: rs3918181G>A, rs3918182G>A and rs3918184C>T and it coincides with chromatin features of active regulatory elements. The loci have a promoter-like histone signature in liver (H3K4me3) and enhancer-like marks (H3K27ac) in vascular tissue in addition to a DNAse hypersensitive site. This is consistent with regulatory potential. NOS3 rs3918188C>A is also an expression quantitative trait locus (eQTL) for nearby genes including KCNH2 important in BP regulation and antihypertensive drug response.
Discussion and conclusion
We set out to investigate whether genomic variation in CES1 and NOS3 contributes to susceptibility to RHTN. Our findings show that NOS3 rs3918188A allele and NOS3 rs1799983 – rs2070744 – rs3918188 G–T–A haplotype are associated with reduced risk for RHTN in South Africans, especially among the MA ethnic group, suggesting potential implications for drug response. However, the genetic variants studied in CES1 showed no significant association with susceptibility to RHTN.
The functional impact of the NOS3 rs3918188C>A polymorphism remains uncertain, as it is intronic. However, there are studies, particularly in Brazilian populations of African descent (Metzger et al., 2013), linking NOS3 tagSNPs including rs3918188C>A to variable levels of circulating NO. Therefore, it is likely that rs3918188C>A potentially affects gene expression (Vaz-Drago et al., 2017), leading to changes in NO levels. In this case, increased NO levels. This is supported by HaploReg annotations (Supplementary Table S7), which place rs3918188C>A—and its African-LD proxies (rs3918181G>A, rs3918182G>A, rs3918184C>T)—in chromatin contexts characteristic of active regulatory elements, with promoter-associated H3K4me3 in liver and enhancer-associated H3K27ac plus DNAse hypersensitivity in vascular tissue, suggesting allele-dependent transcriptional modulation. NOS3 rs3918188C>A is also an eQTL for the vascular potassium-channel gene, KCNH2, and it is likely that it influences vascular excitability, providing a mechanistic basis for inter-individual differences in baseline vascular tone. Since ACE inhibitors such as enalapril, exert part of their antihypertensive effect through NO-mediated vasodilation, variation at this locus could also underlie the differential susceptibility to RHTN or drug response observed. Functional validation—ideally via allele-specific reporter assays in endothelial and hepatic cells—and fine-mapping of the LD block may be essential next steps to confirm causality.
Other SNPs, such as NOS3 rs3918226C>T, rs743506G>A, rs1799983G>T and rs2070744C>T, have also been found to be in high LD with NOS3 rs3918188C>A (Oliveira-Paula et al., 2016a). Our study also found NOS3 rs1799983G>T and rs2070744C>T, to be in LD with rs3918188C>A (Figure 1), and the combination of NOS3 rs1799983G>T - rs2070744C>T - rs3918188C>A; to form a G-T-A haplotype or diplotype, to be associated with reduced risk for RHTN. This shows that the reduced risk for RHTN conferred by the NOS3 rs3918188A allele might also be as a result of allelic associations with rs1799983G>T and rs2070744C>T. The functional consequences of the NOS3 rs1799983G>T and rs2070744C>T SNPs have been well defined in literature. For example, NOS3 rs1799983T allele has been shown to be associated with NOS3 dysregulation and diminished NO bioavailability compared to NOS3 rs1799983G allele (Joshi et al., 2007). NOS3 rs2070744C allele has been shown to reduce NOS3 promoter activity, leading to reduced NO levels compared to rs2070744T allele (Nakayama et al., 1999; Senthil et al., 2005; Nagassaki et al., 2005). Among Africans, NOS3 rs1799983T and rs2070744C have both been shown to be associated with essential hypertension (Algerian, Moroccan and Sudanese populations, although not typical of people from Sub-Saharan Africa) (Gamil et al., 2017; Jemaa et al., 2007; Amrani et al., 2015). NOS3 rs2070744T/T has been shown to be associated with favourable BP response to enalapril among South Africans (Swati, Ndebele, Xhosa, and Zulu ethnic groups) (Masilela et al., 2021). It appears that the NOS3 rs1799983G and rs2070744T alleles are protective against high BP and are determinants of good BP response, respectively. Therefore, it is possible that the protective effect of the NOS3 rs3918188A allele against RHTN observed may also be due to the additive effects of these polymorphisms.
Polymorphisms in CES1 gene are well-documented to affect the response to ACE inhibitors. However, CES1 rs2244613C>A and CNV did not show an association with RHTN despite evidence of associations with enalapril response in previous studies (Ikonnikova et al., 2022b). Nonetheless, we present, for the first time, the relative frequencies of CES1 CNV in Africans. The CNV assay detected the presence of a CES1 subtype, CES1A2, which has been reported to be associated with reduced transcriptional activity (Fukami et al., 2008) although findings are inconsistent (Yoshimura et al., 2008). We observed CNV patterns not previously documented in African populations, with an overall profile comparable to non-African cohorts (Table 3); the only notable difference was the absence of individuals with copy number calls of 1 and >4, which have been reported in a Chinese Han population (Chen et al., 2021). It remains possible that more diverse African groups may have a greater range of CES1 CNV, as we only investigated South African participants of predominantly Xhosa and MA origin.
CES1 is one of the most abundant drug-metabolising enzymes in the liver (Her and Zhu, 2020; Boberg et al., 2017), whose expression levels have been reported to surpass the CYP3A4 enzyme (Boberg et al., 2017). In addition to ACE inhibitors, CES1 is involved in the metabolism of several clinically important drugs, including clopidogrel, methylphenidate, dabigatran, and simvastatin, which are frequently prescribed among Africans (WHO, 2025). CNVs at the CES1 locus—arising mainly from non-allelic homologous recombination events between CES1 and its nearby pseudogenes (CES1P1 and CES1A2)—can alter gene dosage and potentially affect enzyme expression or activity (Rasmussen et al., 2018). The population-specific patterns observed (Table 3) likely reflect any of recombination, genetic drift, founder effects, and selection pressures from local environmental exposures (Rasmussen et al., 2018; Stage et al., 2017b). While we did not observe significant associations in this study, the pharmacogenomic relevance of CES1 CNVs warrants further investigation. Future studies should assess allele-specific expression and protein levels by copy number, perform in vitro enzyme activity assays, and explore pharmacokinetic correlations to clarify the impact of CES1 CNVs on drug response in African populations, as well as leverage other emerging functional genomics techniques (Liu et al., 2023; Zhang et al., 2024; Baysoy et al., 2024).
Our study has some limitations, the most significant being the small sample size. Although the sample size was calculated, the number of individuals belonging to the African group was relatively small in our analyses. This can be attributed to the catchment area of our recruitment site, Groote Schuur Hospital, whose surrounding areas are densely populated by individuals of MA compared to African and our study mirrors this (Statistics South Africa, 2025). Furthermore, there is evidence of differences in heath seeking behaviour among different population groups, and differences in the use of alternative complementary medicines to manage hypertension which is reportedly higher among African population groups compared to the MA (Gohar et al., 2008; Simpson, 1998). These scenarios may have limited our ability to draw definitive conclusions for the African group as access to more participants in this group was limited. Further studies with larger samples of African individuals are needed.
In conclusion, our study sheds light on the association between genetic variation and RHTN in an African population. We identified associations between NOS3 genetic variation and reduced risk for RHTN, among the MA group. In this study, we take the opportunity to give probably the first report on CES1 CNV distribution in an African population. Moving forward, larger prospective cohort studies involving gene expression analyses or biochemical measurements of plasma nitric oxide levels to elucidate on the functional effect of NOS3 polymorphisms are essential, especially in African populations. In addition, non-targeted and hypothesis free approaches [i.e., genome wide association analyses (GWAS)] that would capture more variation are needed. Such efforts will not only enhance our understanding of genetic factors underlying RHTN but also pave the way for personalized treatment strategies tailored to African hypertensive patients (Katsukunya et al., 2023).
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: 10.5281/zenodo.15525258.
Ethics statement
The studies involving humans were approved by the Human Research Ethics Committee (HREC), University of Cape Town. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
JK: Data curation, Formal Analysis, Investigation, Methodology, Project administration, Writing – original draft, Writing – review and editing, Software, Validation. RN: Data curation, Investigation, Writing – review and editing, Formal Analysis. NS: Conceptualization, Data curation, Formal Analysis, Methodology, Project administration, Supervision, Validation, Writing – review and editing. DB: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Validation, Writing – review and editing. PS: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Writing – review and editing. EC: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – review and editing. BR: Investigation, Methodology, Project administration, Supervision, Writing – review and editing, Resources. EJ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Supervision, Writing – review and editing. CD: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Supervision, Writing – review and editing, Funding acquisition, Project administration, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research reported in this publication was supported by the South African Medical Research Council (SAMRC) with funds received from the South African Department of Science and Innovation. The Platform for Pharmacogenomics Research and Translation Unit is funded by the SAMRC through an extramural grant.
Acknowledgments
Thank you to the study participants, members of the Pharmacogenomics and Drug Metabolism Research group (both current and former) and to the South African Medical Research Council (SAMRC), the National Research Foundation (NRF), the University of Cape Town for funding assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2025.1608423/full#supplementary-material
References
Acelajado, M. C., Pisoni, R., Dudenbostel, T., Dell'Italia, L. J., Cartmill, F., Zhang, B., et al. (2012). Refractory hypertension: definition, prevalence, and patient characteristics. J. Clin. Hypertens. (Greenwich) 14 (1), 7–12. doi:10.1111/j.1751-7176.2011.00556.x
Amrani, A., Baba Hamed, M. B., and Mesli Talebbendiab, F. (2015). Association study between some renin-angiotensin system gene variants and essential hypertension in a sample of Algerian population: case control study. Ann. Biol. Clin. Paris. 73 (5), 557–563. doi:10.1684/abc.2015.1069
Amrani-Midoun, A., Kiando, S. R., Treard, C., Jeunemaitre, X., and Bouatia-Naji, N. (2019). Genetic association study between T-786C NOS3 polymorphism and essential hypertension in an Algerian population of the Oran city. Diabetes Metab. Syndr. 13 (2), 1317–1320. doi:10.1016/j.dsx.2019.02.024
Baysoy, A., Tian, X., Zhang, F., Renauer, P., Bai, Z., Shi, H., et al. (2024). Spatially resolved in vivo CRISPR screen sequencing via perturb-DBiT. bioRxiv. doi:10.1101/2024.11.18.624106
Boberg, M., Vrana, M., Mehrotra, A., Pearce, R. E., Gaedigk, A., Bhatt, D. K., et al. (2017). Age-dependent absolute abundance of hepatic carboxylesterases (CES1 and CES2) by LC-MS/MS proteomics: application to PBPK modeling of oseltamivir in vivo pharmacokinetics in infants. Drug Metab. Dispos. 45 (2), 216–223. doi:10.1124/dmd.116.072652
Chen, B. B., Yan, J. H., Zheng, J., Peng, H. W., Cai, X. L., Pan, X. T., et al. (2021). Copy number variation in the CES1 gene and the risk of non-alcoholic fatty liver in a Chinese Han population. Sci. Rep. 11 (1), 13984. doi:10.1038/s41598-021-93549-2
Davis, S. (2007). “Enalapril,” in xPharm: the comprehensive pharmacology reference. Editors S. J. Enna, and D. B. Bylund (New York: Elsevier), 1–6.
Dudenbostel, T., Siddiqui, M., Oparil, S., and Calhoun, D. A. (2016). Refractory hypertension: a novel phenotype of antihypertensive treatment failure. Hypertension 67 (6), 1085–1092. doi:10.1161/HYPERTENSIONAHA.116.06587
El Rouby, N., and Cooper-DeHoff, R. M. (2015). Genetics of resistant hypertension: a novel pharmacogenomics phenotype. Curr. Hypertens. Rep. 17 (9), 583. doi:10.1007/s11906-015-0583-8
Fukami, T., Nakajima, M., Maruichi, T., Takahashi, S., Takamiya, M., Aoki, Y., et al. (2008). Structure and characterization of human carboxylesterase 1A1, 1A2, and 1A3 genes. Pharmacogenet Genomics. 18 (10), 911–920. doi:10.1097/FPC.0b013e32830b0c5e
Gamil, S., Erdmann, J., Abdalrahman, I. B., and Mohamed, A. O. (2017). Association of NOS3 gene polymorphisms with essential hypertension in Sudanese patients: a case control study. BMC Med. Genet. 18 (1), 128. doi:10.1186/s12881-017-0491-7
Gohar, F., Greenfield, S. M., Beevers, D. G., Lip, G. Y., and Jolly, K. (2008). Self-care and adherence to medication: a survey in the hypertension outpatient clinic. BMC complementary Altern. Med. 8, 4–9. doi:10.1186/1472-6882-8-4
Her, L., and Zhu, H.-J. (2020). Carboxylesterase 1 and precision pharmacotherapy: pharmacogenetics and nongenetic regulators. Drug Metabolism Dispos. 48 (3), 230–244. doi:10.1124/dmd.119.089680
Her, L. H., Wang, X., Shi, J., Choi, H. J., Jung, S. M., Smith, L. S., et al. (2021). Effect of CES1 genetic variation on enalapril steady-state pharmacokinetics and pharmacodynamics in healthy subjects. Br. J. Clin. Pharmacol. 87 (12), 4691–4700. doi:10.1111/bcp.14888
Ikonnikova, A., Kazakov, R., Rodina, T., Dmitriev, A., Melnikov, E., Zasedatelev, A., et al. (2022b). The influence of structural variants of the CES1 gene on the pharmacokinetics of enalapril, presumably due to linkage disequilibrium with the intronic rs2244613. Genes (Basel) 13 (12), 2225. doi:10.3390/genes13122225
Ikonnikova, A., Rodina, T., Dmitriev, A., Melnikov, E., Kazakov, R., and Nasedkina, T. (2022a). The influence of the CES1 genotype on the pharmacokinetics of enalapril in patients with arterial hypertension. J. Pers. Med. 12 (4), 580. doi:10.3390/jpm12040580
Jemaa, R., Kallel, A., Ben Ali, S., Omar, S., Chabrak, S., Elasmi, M., et al. (2007). Association of a 27-bp repeat polymorphism in intron 4 of endothelial constitutive nitric oxide synthase gene with myocardial infarction in Tunisian patients. Clin. Chem. Lab. Med. 45 (11), 1476–1480. doi:10.1515/CCLM.2007.312
Jones, E. S., Damasceno, A., Ogola, E. N., Ojji, D. B., Dzudie, A., and Rayner, B. L. (2020). PASCAR commentary on the International Society of Hypertension global guidelines 2020: relevance to sub-Saharan Africa. Cardiovasc J. Afr. 31 (6), 325–329. doi:10.5830/CVJA-2020-055
Jones, E. S. W., Lesosky, M., Blockman, M., Castel, S., Decloedt, E. H., Schwager, S. L. U., et al. (2017). Therapeutic drug monitoring of amlodipine and the Z-FHL/HHL ratio: adherence tools in patients referred for apparent treatment-resistant hypertension. S Afr. Med. J. 107 (10), 887–891. doi:10.7196/SAMJ.2017.v107i10.12268
Joshi, M. S., Mineo, C., Shaul, P. W., and Bauer, J. A. (2007). Biochemical consequences of the NOS3 Glu298Asp variation in human endothelium: altered caveolar localization and impaired response to shear. FASEB J. 21 (11), 2655–2663. doi:10.1096/fj.06-7088com
Katsukunya, J. N., Soko, N. D., Naidoo, J., Rayner, B., Blom, D., Sinxadi, P., et al. (2023). Pharmacogenomics of hypertension in Africa: paving the way for a pharmacogenetic-based approach for the treatment of hypertension in Africans. Int. J. Hypertens. 2023, 9919677. doi:10.1155/2023/9919677
Liu, Y., DiStasio, M., Su, G., Asashima, H., Enninful, A., Qin, X., et al. (2023). High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 41 (10), 1405–1409. doi:10.1038/s41587-023-01676-0
Masilela, C., Pearce, B., Ongole, J. J., Adeniyi, O. V., Johnson, R., and Benjeddou, M. (2021). Cross-sectional study of the association of 5 single nucleotide polymorphisms with enalapril treatment response among South African adults with hypertension. Med. Baltim. 100 (46), e27836. doi:10.1097/MD.0000000000027836
Metzger, I. F., Luizon, M. R., Lacchini, R., Ishizawa, M. H., and Tanus-Santos, J. E. (2013). Effects of endothelial nitric oxide synthase tagSNPs haplotypes on nitrite levels in black subjects. Nitric Oxide 28, 33–38. doi:10.1016/j.niox.2012.10.002
Moholisa, R. R., Rayner, B. R., Patricia Owen, E., Schwager, S. L., Stark, J. S., Badri, M., et al. (2013). Association of B2 receptor polymorphisms and ACE activity with ACE inhibitor-induced angioedema in black and mixed-race South Africans. J. Clin. Hypertens. (Greenwich) 15 (6), 413–419. doi:10.1111/jch.12104
Moosa, M. S., Kuttschreuter, L. S., and Rayner, B. L. (2016). Evaluation and management of patients referred to a tertiary-level hypertension clinic in Cape Town, South Africa. S Afr. Med. J. 106 (8), 797–800. doi:10.7196/SAMJ.2016.v106i8.9610
Nagassaki, S., Metzger, I. F., Souza-Costa, D. C., Marroni, A. S., Uzuelli, J. A., and Tanus-Santos, J. E. (2005). eNOS genotype is without effect on circulating nitrite/nitrate level in healthy male population. Thromb. Res. 115 (5), 375–379. doi:10.1016/j.thromres.2004.09.003
Naing, L., Nordin, R. B., Abdul Rahman, H., and Naing, Y. T. (2022). Sample size calculation for prevalence studies using Scalex and ScalaR calculators. BMC Med Res Methodol. 22, (1) 209. doi:10.1186/s12874-022-01694-7
Nakayama, M., Yasue, H., Yoshimura, M., Shimasaki, Y., Kugiyama, K., Ogawa, H., et al. (1999). T-786 C mutation in the 5'-flanking region of the endothelial nitric oxide synthase gene is associated with coronary spasm. Circulation 99 (22), 2864–2870. doi:10.1161/01.cir.99.22.2864
Nansseu, J. R. N., Noubiap, J. J. N., Mengnjo, M. K., Aminde, L. N., Essouma, M., Jingi, A. M., et al. (2016). The highly neglected burden of resistant hypertension in Africa: a systematic review and meta-analysis. BMJ Open 6 (9), e011452. doi:10.1136/bmjopen-2016-011452
Nassereddine, S., Hassani Idrissi, H., Habbal, R., Abouelfath, R., Korch, F., Haraka, M., et al. (2018). The polymorphism G894 T of endothelial nitric oxide synthase (eNOS) gene is associated with susceptibility to essential hypertension (EH) in Morocco. BMC Med. Genet. 19, 127–128. doi:10.1186/s12881-018-0638-1
Oliveira-Paula, G. H., Lacchini, R., and Tanus-Santos, J. E. (2016b). Endothelial nitric oxide synthase: from biochemistry and gene structure to clinical implications of NOS3 polymorphisms. Gene 575 (2 Pt 3), 584–599. doi:10.1016/j.gene.2015.09.061
Oliveira-Paula, G. H., Lacchini, R., and Tanus-Santos, J. E. (2017). Clinical and pharmacogenetic impact of endothelial nitric oxide synthase polymorphisms on cardiovascular diseases. Nitric Oxide 63, 39–51. doi:10.1016/j.niox.2016.08.004
Oliveira-Paula, G. H., Pereira, S. C., Tanus-Santos, J. E., and Lacchini, R. (2019). Pharmacogenomics and hypertension: current insights. Pharmgenomics Pers. Med. 12, 341–359. doi:10.2147/PGPM.S230201
Oliveira-Paula, , Gustavo, H., Lacchini, R., Luizon, M. R., Fontana, V., Silva, P. S., et al. (2016a). Endothelial nitric oxide synthase tagSNPs influence the effects of enalapril in essential hypertension. Nitric Oxide 55, 62–69. doi:10.1016/j.niox.2016.03.006
Olsen, M. H., Angell, S. Y., Asma, S., Boutouyrie, P., Burger, D., Chirinos, J. A., et al. (2016). A call to action and a lifecourse strategy to address the global burden of raised blood pressure on current and future generations: the Lancet Commission on hypertension. Lancet 388 (10060), 2665–2712. doi:10.1016/S0140-6736(16)31134-5
Rasmussen, H. B., Madsen, M. B., and Consortium, I. (2018). Carboxylesterase 1 genes: systematic review and evaluation of existing genotyping procedures. Drug Metabolism Personalized Ther. 33 (1), 3–14. doi:10.1515/dmpt-2017-0023
Senthil, D., Raveendran, M., Shen, Y. H., Utama, B., Dudley, D., Wang, J., et al. (2005). Genotype-dependent expression of endothelial nitric oxide synthase (eNOS) and its regulatory proteins in cultured endothelial cells. DNA Cell Biol. 24 (4), 218–224. doi:10.1089/dna.2005.24.218
Shen, J., Li, Z., Chen, J., Song, Z., Zhou, Z., and Shi, Y. (2016). SHEsisPlus, a toolset for genetic studies on polyploid species. Sci. Rep. 6, 24095. doi:10.1038/srep24095
Silva, P., Fontana, V., Luizon, M. R., Lacchini, R., Silva, W., Biagi, C., et al. (2013). eNOS and BDKRB2 genotypes affect the antihypertensive responses to enalapril. Eur. J. Clin. Pharmacol. 69, 167–177. doi:10.1007/s00228-012-1326-2
Simpson, D. (1998). Buchu—South Africa's amazing herbal remedy. Scott. Med. J. 43 (6), 189–191. doi:10.1177/003693309804300610
Soko, N. D., Muyambo, S., Dandara, M. T. L., Kampira, E., Blom, D., Jones, E. S. W., et al. (2023). Towards evidence-based implementation of pharmacogenomics in southern Africa: comorbidities and polypharmacy profiles across diseases. J. Pers. Med. 13 (8), 1185. doi:10.3390/jpm13081185
Stage, C., Jürgens, G., Guski, L. S., Thomsen, R., Bjerre, D., Ferrero-Miliani, L., et al. (2017a). The pharmacokinetics of enalapril in relation to CES1 genotype in healthy Danish volunteers. Basic Clin. Pharmacol. Toxicol. 121 (6), 487–492. doi:10.1111/bcpt.12835
Stage, C., Jürgens, G., Guski, L. S., Thomsen, R., Bjerre, D., Ferrero-Miliani, L., et al. (2017b). The impact of CES1 genotypes on the pharmacokinetics of methylphenidate in healthy Danish subjects. Br. J. Clin. Pharmacol. 83 (7), 1506–1514. doi:10.1111/bcp.13237
Statistics South Africa (2025). Cape Town. Available online at: https://www.statssa.gov.za/?page_id=4286&id=331:GovernmentofSouthAfrica2025.
Sychev, D., Skripka, A., Ryzhikova, K., Bochkov, P., Shevchenko, R., Krupenin, P., et al. (2020). Effect of CES1 and ABCB1 genotypes on the pharmacokinetics and clinical outcomes of dabigatran etexilate in patients with atrial fibrillation and chronic kidney disease. Drug metabolism personalized Ther. 35 (1). doi:10.1515/dmpt-2019-0029
Unger, T., Borghi, C., Charchar, F., Khan, N. A., Poulter, N. R., Prabhakaran, D., et al. (2020). 2020 international society of hypertension global hypertension practice guidelines. Hypertension 75 (6), 1334–1357. doi:10.1161/HYPERTENSIONAHA.120.15026
Vaz-Drago, R., Custódio, N., and Carmo-Fonseca, M. (2017). Deep intronic mutations and human disease. Hum. Genet. 136 (9), 1093–1111. doi:10.1007/s00439-017-1809-4
Wang, X., Wang, G., Shi, J., Aa, J., Comas, R., Liang, Y., et al. (2016). CES1 genetic variation affects the activation of angiotensin-converting enzyme inhibitors. Pharmacogenomics J. 16 (3), 220–230. doi:10.1038/tpj.2015.42
Ward, L. D., and Kellis, M. (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40 (Database issue), D930–D934. doi:10.1093/nar/gkr917
WHO (2025). WHO Model Lists of Essential Medicines. Available online at: https://www.who.int/groups/expert-committee-on-selection-and-use-of-essential-medicines/essential-medicines-lists.
WHO (2023). Hypertension. Available online at: https://www.who.int/news-room/fact-sheets/detail/hypertension.
Yoshimura, M., Kimura, T., Ishii, M., Ishii, K., Matsuura, T., Geshi, E., et al. (2008). Functional polymorphisms in carboxylesterase1A2 (CES1A2) gene involves specific protein 1 (Sp1) binding sites. Biochem. Biophys. Res. Commun. 369 (3), 939–942. doi:10.1016/j.bbrc.2008.02.120
Zhang, D., Rubio Rodríguez-Kirby, L. A., Lin, Y., Song, M., Wang, L., Wang, L., et al. (2024). Spatial dynamics of mammalian brain development and neuroinflammation by multimodal tri-omics mapping. bioRxiv. doi:10.21203/rs.3.rs-4814866/v1
Keywords: CES1, NOS3, pharmacogenomics, hypertension, Africans, ACE inhibitors, enalapril
Citation: Katsukunya JN, Naicker R, Soko ND, Blom D, Sinxadi P, Chimusa ER, Rayner B, Jones E and Dandara C (2025) NOS3 rs3918188C>A is associated with susceptibility to resistant hypertension while CES1 genetic variation was not associated with resistant hypertension among South Africans. Front. Genet. 16:1608423. doi: 10.3389/fgene.2025.1608423
Received: 08 April 2025; Accepted: 15 May 2025;
Published: 04 June 2025.
Edited by:
José A. G. Agúndez, University of Extremadura, SpainReviewed by:
Fu Gao, Yale University, United StatesXiang Sun, St. Jude Children’s Research Hospital, United States
Shuozhen Bao, Yale University, United States
Yujia Ji, Stony Brook University, United States
Copyright © 2025 Katsukunya, Naicker, Soko, Blom, Sinxadi, Chimusa, Rayner, Jones and Dandara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Collet Dandara, Y29sbGV0LmRhbmRhcmFAdWN0LmFjLnph
 Jonathan N. Katsukunya
Jonathan N. Katsukunya Revina Naicker1,2
Revina Naicker1,2 Dirk Blom
Dirk Blom Phumla Sinxadi
Phumla Sinxadi Emile R. Chimusa
Emile R. Chimusa Collet Dandara
Collet Dandara