- 1Department of Pathology, Immunology and Laboratory Medicine, College of Medicine, University of Florida, Gainesville, FL, USA
- 2Florida Center for AIDS Research, University of Florida, Gainesville, FL, USA
- 3Department of Biostatistics, College of Public Health, University of Florida, Gainesville, FL, USA
- 4Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, FL, USA
- 5Division of Infectious Diseases, College of Medicine, University of South Florida, St. Petersburg, FL, USA
- 6Department of Pediatrics, Division of Allergy, Immunology, and Rheumatology, College of Medicine, University of South Florida, St. Petersburg, FL, USA
Background: HIV-1 infection or systemic lupus erythematosus (SLE) disrupt B cell homeostasis, reduce memory B cells, and impair function of IgG and IgM antibodies.
Objective: To determine how disturbances in B cell populations producing polyclonal antibodies relate to the IgM repertoire, the IgM transcriptome in health and disease was explored at the complementarity determining region 3 (CDRH3) sequence level.
Methods: 454-deep pyrosequencing in combination with a novel analysis pipeline was applied to define populations of IGHM CDRH3 sequences based on absence or presence of somatic hypermutations (SHM) in peripheral blood B cells.
Results: HIV or SLE subjects have reduced biodiversity within their IGHM transcriptome compared to healthy subjects, mainly due to a significant decrease in the number of unique combinations of alleles, although recombination machinery was intact. While major differences between sequences without or with SHM occurred among all groups, IGHD and IGHJ allele use, CDRH3 length distribution, or generation of SHM were similar among study cohorts. Antiretroviral therapy failed to normalize IGHM biodiversity in HIV-infected individuals. All subjects had a low frequency of allelic combinations within the IGHM repertoire similar to known broadly neutralizing HIV-1 antibodies.
Conclusion: Polyclonal expansion would decrease overall IgM biodiversity independent of other mechanisms for development of the B cell repertoire. Applying deep sequencing as a strategy to follow development of the IgM repertoire in health and disease provides a novel molecular assessment of multiple points along the B cell differentiation pathway that is highly sensitive for detecting perturbations within the repertoire at the population level.
Introduction
HIV-1 infection and systemic lupus erythematosus (SLE) each results in defective B cell activation and differentiation (1, 2). Both conditions have decreased proportions of CD27+ class switched memory B cells, increased B cell apoptosis, and increased expression of B cell activation markers (3–6). In HIV infection, disruption of B cell homeostasis and loss of normal B cell architecture within lymphoid tissues occur early in disease and persist even with control of viral replication following antiretroviral therapy (ART) (7, 8). Individuals with SLE display defects in B cell checkpoints in both early and late stage development contributing to impaired tolerance and autoantibody production (9).
The dynamics between defects in B cell function and underlying molecular perturbations in the B cell repertoire during the course of infection or autoimmunity have not been extensively evaluated, reflecting in part the challenge of generating sufficiently robust data sets by conventional clonal sequencing. Massively parallel deep sequencing has revolutionized the capacity to evaluate the depth and breadth of the immunoglobulin (Ig) repertoire (10). Application of deep sequencing to probe the Ig heavy chain variable region (IGH) repertoire along the B cell developmental pathway may pin point HIV-mediated defects over the antibody maturation, and uncover evidence for elusive broadly neutralizing HIV-specific antibodies (bn-HIV-Ab) critical to development of an effective HIV vaccine. This study focused on IGHM repertoire at early B cell developmental stage before isotype switch. IgM is the initial antibody generated when encountering antigen. Activated IgM B cells confer substantial response in acute HIV-infection (11–13). IgM antibodies demonstrated broad spectrum and high affinity to HIV-1 envelope glycoproteins, neutralized HIV-1 more potently than IgG due to pentameric binding nature, and prevented viral entry across the mucous membrane (14, 15).
The limited repertoire of bn-HIV-Ab share biochemical, structural, and functional features resembling natural polyreactive autoantibodies and are produced by individuals with autoimmunity, particularly SLE, and a significant proportion of HIV-infected individuals (16–25). Polyreactive autoantibodies and bn-HIV-Ab are frequently IgG (26–28), or can be IgA (29–32). More than a third of SLE individuals have IgM antibodies reactive with HIV-1 gp41-derived peptides (33), a CD4-reactive IgM Fab clone isolated from a healthy individual inhibited HIV-1 replication (34), and IgM autoantibodies blocked HIV-1 entry (35).
While polyreactive antibodies are well-characterized at the biochemical level, molecular assessment of the Ig repertoire among populations of B cells that produce polyreactive antibodies is limited. Our current in-depth analysis based on deep sequencing of the IgM transcriptome was designed to examine the molecular repertoire of IGH complementarity determining region 3 (CDRH3) in IgM among individuals with SLE or HIV-1 infection and relate the findings to a group of healthy individuals by analysis of biodiversity. Biodiversity is used in population genetics to present a unified view of variation of life forms within habitats based on the premise that genomes are taxonomically similar, randomly distributed, and sufficiently large (36, 37). Assessments of biodiversity from deep sequencing data provide unprecedented views of the richness of immune loci in primates, zebra fish, and humans (38–41). The goal for our study was to determine how disturbances in B cell populations producing polyclonal antibodies relate to biodiversity of the IgM repertoire by examining key components, including allele usage, V-D-J recombination and junctional diversity, and extent of somatic hypermutation (SHM), which collectively contribute to differences in Ig biodiversity between health and disease.
Materials and Methods
Study Cohort
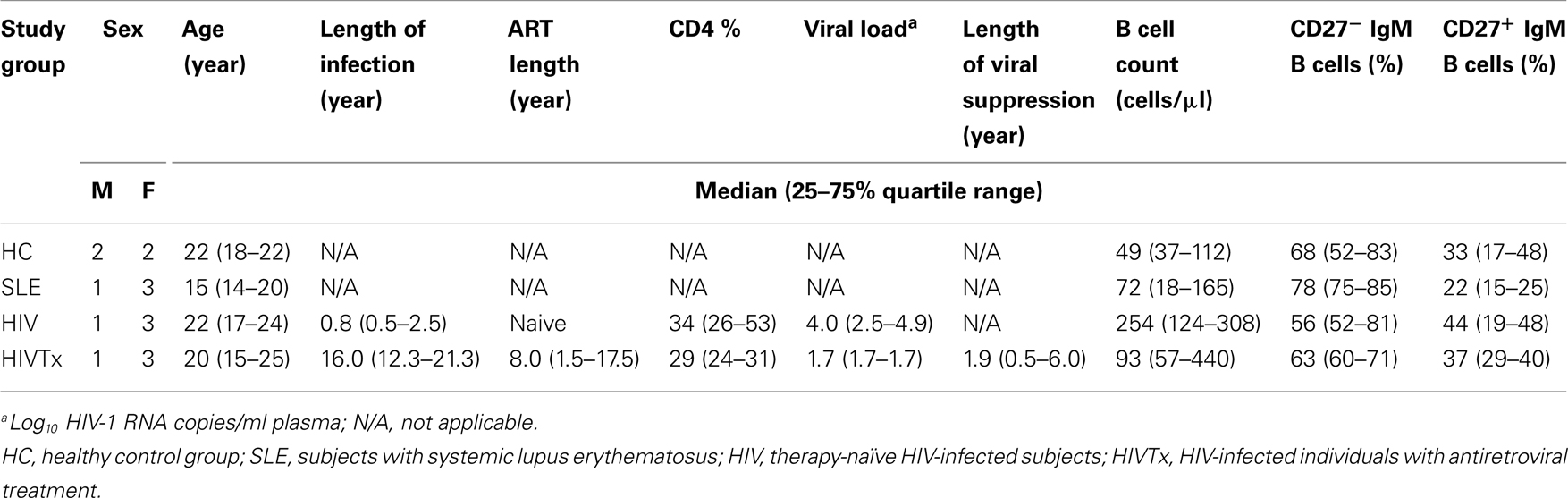
Sixteen individuals, enrolled in a protocol approved by the Institutional Review Boards of the University of Florida and the University of South Florida, included four groups (n = 4 per group): healthy controls (HC), subjects with SLE, and HIV-1 infected individuals either therapy-naïve (HIV) or receiving combination antiretroviral therapy (cART) (HIVTx) (Table 1). Groups within the cohort were age-balanced with median (25–75% quartile range) age of 20 (16–22) years. SLE subjects were untreated and diagnosed by clinical and laboratory criteria defined by the American College of Rheumatology (42). Therapy-naïve subjects were HIV-1 infected by sexual transmission for at least 6 months (Table 1). HIVTx individuals were infected either through maternal to child transmission or by contaminated blood products for 16.0 (12.3–21.3) years, treated for 8.0 (1.5–17.5) years, and achieved viral suppression (log10 HIV-1 RNA copies/ml < 1.7), and CD4 > 25% for 1.9 (0.5–6.0) years at the time of study (Table 1). No subject received any vaccination 30 days prior to study entry, and/or had acute infection to reduce the chance of plasma cell circulating in peripheral blood. Informed consent was obtained from all subjects.
B Cell Profiles by Multiparameter Flow Cytometry
Immunofluorescence staining was performed using the whole blood lysis method (43) and analysis by LSR2 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) with Diva software (BD Biosciences, San Jose, CA, USA). Monoclonal antibody panel included pan B cell marker PE Cy7-conjugated anti-CD19 (BD Biosciences), memory B cell marker Qdot 655-conjugated anti-CD27 (Invitrogen, Carlsbad, CA, USA), and surface IgM with APC-conjugated anti-IgM (BD Biosciences). B cell percentages ranged from 6.6 to 16.1% of peripheral blood mononuclear cells (PBMC) across the groups, CD19+ IgM B cells ranged from 70 to 90% of total B cells (Table 1).
Generation of IGHM CDRH3 Amplicon Libraries
Analysis of the IGHM CDRH3 transcriptome was performed using total peripheral blood mRNA without isolation of IgM B cells. Specificity for the IgM transcriptome was achieved by reverse amplification primers Cμ15, Cμ5, and Cμ2 homologous to the IgM constant region (44) (Figure 1A). An amplicon library was constructed for each subject by RT-PCR using SuperScript™ One-Step RT-PCR with Platinum Taq (Invitrogen) and GoTaq colorless Master Mix (Promega, Madison, WI, USA) from ∼200 ng mRNA (equivalent to about 100,000 B cells or 80,000 IgM B cells) extracted from total PBMC by MicroPoly[A]Purist™ kit (Ambion, Austin, TX, USA).
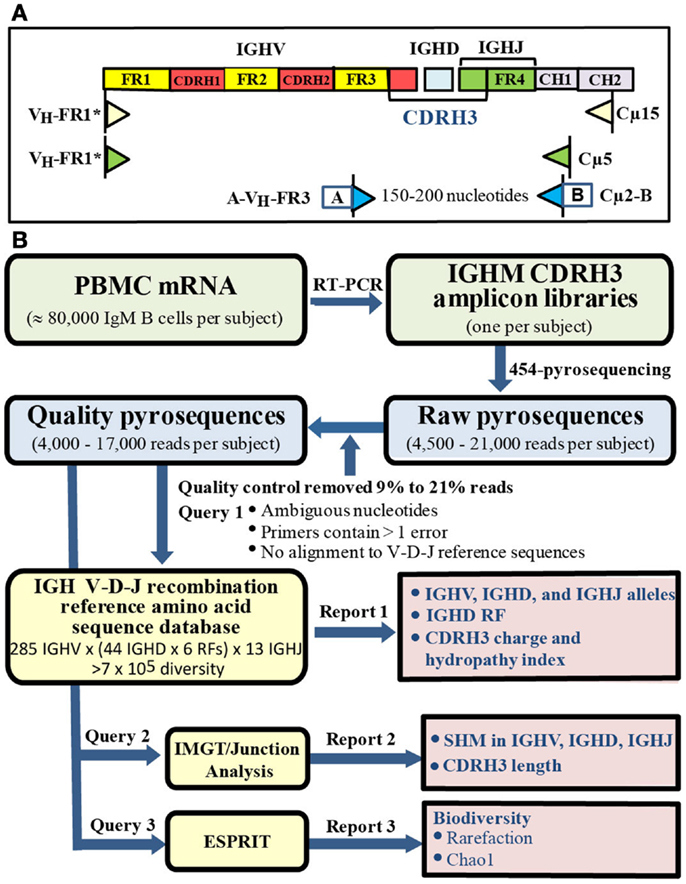
Figure 1. Amplicon libraries and bioinformatic pipeline. (A) Library construction. IgM specificity was determined by reverse primers sequences specific for IgM constant region without IgM B cell purification. (B) Bioinformatic pipeline. Quality sequences (blue box) were analyzed by querying: (1) customized IGH V-D-J recombination reference sequence database, (2) IMGT/Junction Analysis, (3) ESPRIT (yellow boxes) to generate three reports (pink boxes).
Forward inner primer pan VH-FR3 is located in framework 3 and does not overlap CDRH3 (44). Using the software Oligo (Molecular Biology Insights, Inc., Cascade, CO, USA), pan VH-FR3 primer was predicted to bind with similar capacity to a unique sequence in each IGHV family when evaluated against IGHV family specific reference sequences. In contrast, the pan VH-FR1 primer (44) displayed different binding capacity to different IGHV families. Specifically, pan VH-FR1 primer bound preferentially to IGHV3; displayed low binding capacity with IGHV4, IGHV5, and IGHV7; failed to bind to IGHV2 or IGHV6; and bound to false priming sites in IGHV1, IGHV4, and IGHV5. To overcome the different binding capacity of the published primer sequence, we designed a modified panVH-FR1* primer (CAGGTGCAGCTGGAGCAGTCTGG_) that was one nucleotide shorter and substituted A and C for T and G in the published primer sequence, respectively. When evaluated by Oligo, the modified panVH-FR1* primer displayed similar binding capacity to each IGHV family reference sequence and no binding to false priming sites. Forward and reverse primers, pan VH-FR3, and Cμ2, in the final amplification were conjugated at 5′ ends with A (GCCTCCCTCGCGCCATCAG) or B (GCCTTGCCAGCCCGCTCAG) adaptor, respectively, to generate amplicons ranging from 150 to 200 nucleotides. Gel-purified amplicons were submitted to the Interdisciplinary Center for Biotechnology Research (University of Florida) for 454-pyrosequencing using a Genome Sequencer FLX (454 Life Sciences) according to the manufacturer’s protocol.
Bioinformatics Pipeline
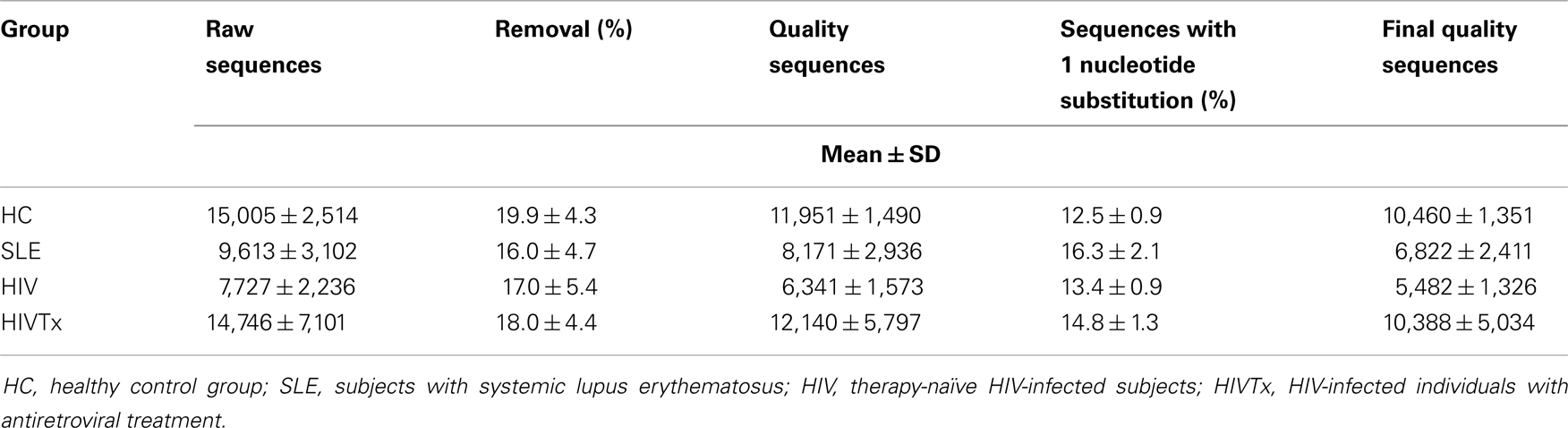
A bioinformatics pipeline was developed to facilitate analysis of large numbers of relatively short IGHM CDRH3 sequences that could not be processed by conventional IMGT/V-QUEST analysis (Figure 1B) (45). Raw reads ranged from 4,500 to 21,000 pyrosequences per subject. A quality control step filtered 9–21% low quality reads with ambiguous nucleotides, more than one error in either primer tag, or failure to align to reference sequences in the germ line IGH V-D-J recombination amino acid reference sequence database (see below), leaving 4,000–17,000 quality sequences per sample with no significant difference in number of sequences among the groups (Table 2).
To overcome the limitation of IMGT/V-QUEST for high throughput classification of relatively short sequences, a novel custom reference sequence database containing over 700,000 germ line IGH V-D-J amino acid (aa) sequences was established by generating in silico all possible combinations of germ line IGHV (285 unique aa sequences), IGHD [44 unique nucleotide sequences translated in 6 reading frames (RF) producing 222 aa sequences], and IGHJ (13 unique aa sequences) alleles downloaded from IMGT (45). Each reference sequence was annotated for IGHV, IGHD, and IGHJ allele use, IGHD RF in the header with C104 and W118, the 5′ and 3′ boarders of CDRH3, marked by aligning to germline IGHV and germline IGHJ reference sequences using FASTA (http://fasta.bioch.virginia.edu/fasta_www2/fasta_down.shtml) (46). After removing adapter sequences, query sequences were then aligned to the reference sequences using FASTY. Use of IGHV, IGHD, and IGHJ alleles, and IGHD RF of each query 454-sequence was extrapolated from the header of the best-matched reference sequence, the C104 and W118 positions of each query sequence were identified, the part of the query sequence matched to the C104-W118 region of the reference sequence was extracted from the entire sequence, and CDRH3 charge and hydropathy index calculated using an in-house code (47, 48). IMGT/Junction Analysis was then performed allowing 3D-GENEs (49). Using this strategy, the frequency distribution of IGHD and IGHJ alleles, with IGHD2, IGHD3 and IGHD6, and IGHJ4 as the dominant genes, was similar to previous reports (50, 51). However, due to the relatively short IGHV sequence, we observed some ambiguity in IGHV4 allele assignment; consequently, no comparisons of IGHV alleles without or with SHM were made.
The total number of mutations including silent- and non-silent-mutations in CDRH3 between C104 and W118 (40) was calculated to summarize extent of SHM as: (ΣSHM within CDRH3 in each SHM+ sequence ÷ ΣCDRH3 nucleotide length in each SHM+ sequence) × 100 nucleotides. The effect of N and P nucleotides to the SHM was similar among the study groups because the number of N and P nucleotides was minimized when the setting for D-GENES was 3, and the frequency of sequences with one D, D-D fusion and D-D-D fusion was similar among study groups (data not shown).
To avoid potential ambiguity in scoring SHM, an average of 14.3 ± 0.6% of reads among groups with a single nucleotide difference from aligned reference sequences was removed from analysis (Table 2) (40). IGHM sequences with two or more mutations in CDRH3 were classified as SHM+ representing IgM memory B cells, while SHM− sequences represent naïve B cells. After correction of deletion and insertion, the PCR and pyrosequencing-induced rate of misincorporation tested in our control analysis of clones of sequences was 0.18 errors per 200 nucleotides, the longest sequence length, similar to other reports (52–54) and well below the level of SHM identified in the IgM populations.
ESPRIT was applied to study the biodiversity (genetic complexity) of nucleotide sequences as well as V-D-J combinations of the IGHM CDRH3 transcriptome repertoire by clustering the sequences at 0% genetic distance (55). Rarefaction analysis measures increase in biodiversity along the depth of sequencing (number of sequences). The deeper the initial slope is, the higher the biodiversity. Left shift or right shift of the curve indicates an increase or decrease of biodiversity. Chao1 analysis inferred maximum biodiversity within the input templates (55, 56). Biodiversity is influenced by input cell number, the coverage (sequence number/input cell number), and clonality (frequency of clusters with more than 10 repeated sequences). Coverage was ∼10% in each individual to minimize the influence of preferential amplification of replicate templates. Biodiversity was weighted by the absolute number of input IgM B cells (calculated from the absolute lymphocyte count multiplied by the percentage of CD19+ IgM B cells) to make the data comparable among study groups. To avoid a potential bias from sequence number, clonality was averaged on the same number of sequences randomly drawn 1,000 times from each sequence pool.
To rule out an influence by differences in sequence numbers among individuals, a bootstrap resampling method was used to generate 1,000 rarefaction curves and Chao1 values from random data sets with the same number of sequences for each individual. Calculated and estimated biodiversity derived from averages of 1,000 rarefaction curves produced similar findings (result not shown).
Statistical Analysis
Comparisons among study groups were performed by ANOVA. Kolmogorov–Smirnov test was used to test Gaussian distribution. Overall difference of distribution of CDRH3 length frequencies, and charge and hydropathy along CDRH3 lengths among study groups were evaluated as described (57). Comparison of frequency of sequences in each gene allele, or each CDRH3 length between SHM+ and SHM− sequences was performed by paired t-test. Pearson correlation evaluated the relationship between frequency of SHM+ sequences and frequency of peripheral CD27+ IgM B cells. A primary purpose of this pilot study with relatively small sample size was to detect meaningful trends, thus parametric methods instead of conservative non-parametric methods were performed. P values were unadjusted for multiple tests to increase sensitivity to identify differences. Statistical analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC, USA) with p < 0.05 (two sided) defined as significant.
Results
Somatic Hypermutations
IGHM CDRH3 sequences were initially classified by absence or presence of SHM as a molecular means to distinguish naïve B cells from IgM memory B cells. Frequency of sequences with SHM in HIV or HIVTx groups was similar to HC group, but significantly reduced in individuals with SLE (p = 0.02) (Figure 2). In contrast, extent of SHM (mutations/100 nucleotides) in IGHM CDRH3 among groups was similar. While the proportion of B cells expressing CD27+ IgM memory phenotype was similar among groups, percentage of phenotypically mature CD27+ IgM B cells was significantly less than the frequency of IgM sequences with SHM within each group (Figure 2). No correlation occurred between the proportion of CD27+ B cells and SHM (r2 = 0.06, p = 0.35). Subsequent analysis of the IgM transcriptome was based on molecular assessments of sequences without or with SHM in IGHM CDRH3.
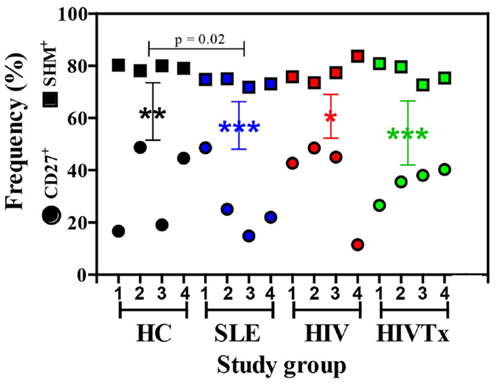
Figure 2. Frequency of SHM+ IgM sequences exceeds the frequency of CD27+ IgM B cells in peripheral blood. Frequency of SHM+ IgM sequences in HIV and HIVTx (treated) groups was similar to HC (healthy controls), but significantly reduced in individuals with SLE (systemic lupus erythematosus) (p = 0.02). Frequency of SHM+ IgM sequences was significantly greater than the proportion of CD27+ IgM B cells in each group (HC, **p = 0.001; SLE, ***p < 0.0001; HIV, *p = 0.004; HIVTx, ***p < 0.0001). Reduced frequency of CD27+ IgM B cells in SLE individuals compared to HC failed to reach statistical significance. Frequency of SHM+ sequence did not correlate with percent of CD27+ IgM B cells (r2 = 0.06, p = 0.35). Symbols: square, IGHM CDRH3 sequences with SHM; circle, CD27+ IgM B cells.
Biodiversity of IGHM CDRH3 Transcriptome Repertoire in Health and Disease
Deep sequencing data sets support application of novel rarefaction analysis to Ig biodiversity that cannot be inferred from analysis of limited numbers of sequences. Rarefaction curves provide comparison of biodiversity along with the depth of sequencing among study groups; deeper slopes indicate greater biodiversity. IGHM CDRH3 among healthy young adults displayed a range of biodiversity that overall was greater than biodiversity in SLE or HIV-1 infection (Figures 3A,B). Differences in biodiversity between health and disease were apparent in populations of IgM sequences without SHM, and to a greater extent among sequences with SHM due to contribution of SHM. At the depth of sequencing, none of the rarefaction curves reached saturation, indicating a preponderance of unique sequences and an extent of IGHM CDRH3 diversity that exceeded the depth of sequencing.
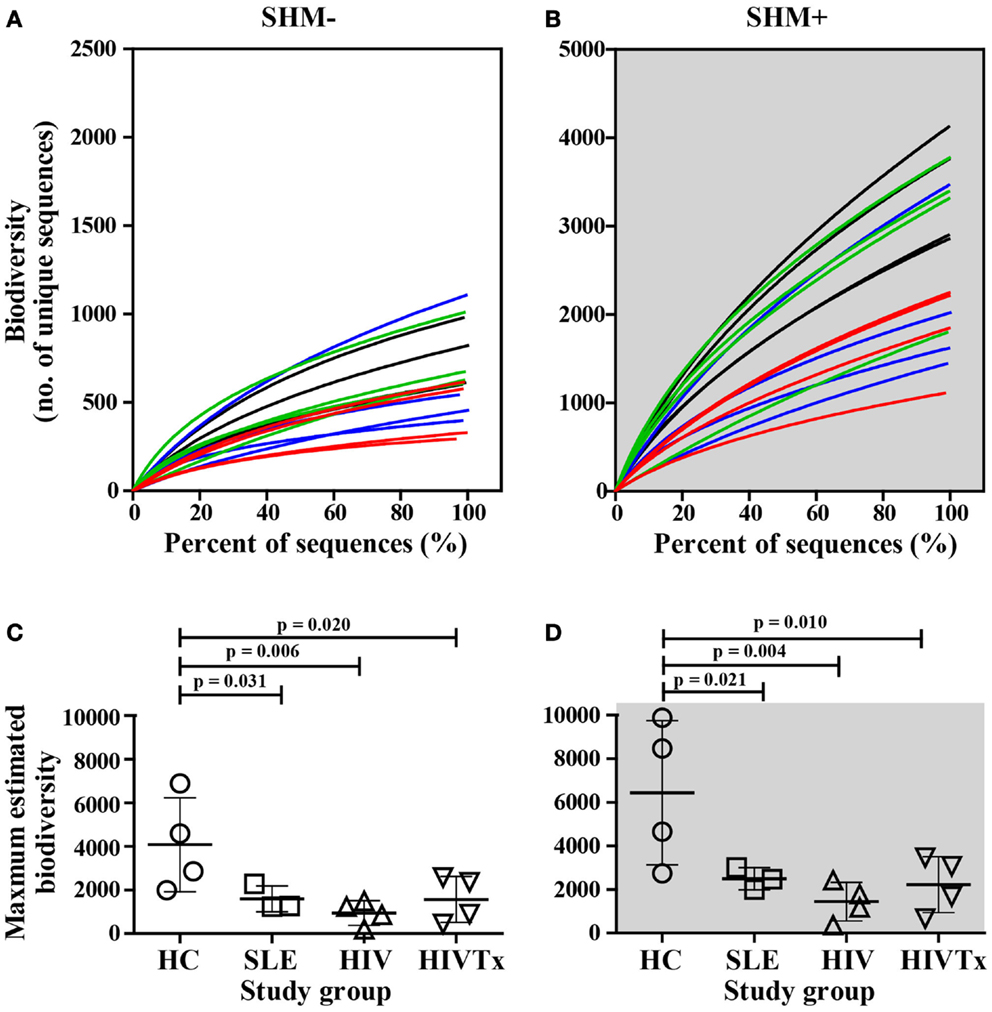
Figure 3. Greater biodiversity among populations of IGHM CDRH3 sequences in health control group than in groups with SLE or HIV-1 infection in sequences without or with SHM. Biodiversity in IGHM CDRH3 sequences without (A) or with (B) SHM in young adults with SLE (blue lines) and among HIV-infected individuals (red lines, untreated; green lines, treated) in comparison to healthy counterparts (black lines). Maximum IGHM CDRH3 biodiversity in input IgM B cells without (C) or with (D) was significantly lower in SLE (SHM−, p = 0.031; SHM+, p = 0.021), HIV (SHM−, p = 0.006; SHM+, p = 0.004), and HIVTx (SHM−, p = 0.020; SHM+, p = 0.010) in comparison with HC. ART failed to normalize IGHM CDRH3 biodiversity in SHM− or SHM+ IGHV CDRH3 repertoire in HIV-infected subjects (HIVTx vs. HIV, p > 0.05).
When maximum IGHM CDRH3 biodiversity was inferred, the SHM-negative repertoire within healthy individuals displayed significantly greater maximum biodiversity than SLE (p = 0.031), HIV (p = 0.006), or HIVTx (p = 0.020) (Figure 3C). Likewise, the repertoire with SHM among healthy individuals also displayed significantly greater biodiversity than SLE (p = 0.021), HIV (p = 0.004), or HIVTx (p = 0.010) (Figure 3D). ART failed to normalize biodiversity in IGHM CDRH3 repertoire either without or with SHM in HIV-infected subjects (Figures 3C,D).
Multiple factors contribute to biodiversity. SHM is one factor, but extent of SHM was similar among study groups. Likewise, extent and distribution of clonality were similar among sequences without or with SHM in each study group with a frequency of ∼99% for unique clusters about 0.2% of clusters with more than 10 repeated sequences. Consequently, to identify molecular differences that might contribute to reduced biodiversity without or with SHM among SLE or HIV-infected groups, CDRH3 length variation, charge, and hydropathy distribution, as well as IGHD and IGHJ allelic use, and diversity of allele combinations were investigated.
CDRH3 Length Variation
In all groups, CDRH3 length variation displayed Gaussian distributions (Figure 4). CDRH3 regions without SHM ranged from 4 to 31 amino acid residues with a peak frequency of 15 amino acids (Figure 4A). SHM+ CDRH3 regions ranged from 5 to 34 amino acids, with lengths of 14 amino acids occurring most frequently (Figure 4B). Overall length distribution differed significantly between sequences without or with SHM (p = 0.004) (Figure 4C). In general, CDRH3 lengths of 10–17 amino acids occurred with greater frequency among sequences with SHM than in sequences without SHM (p < 0.0001). The frequency of long CDRH3 regions (27–34 amino acids) ranged from 0.03 (± 0.06%) to 0.13% (± 0.12%) in sequences without SHM and 0.13 (± 0.04%) to 0.16% (± 0.10%) in sequences with SHM with no significant differences among study groups (Figure 5). Charge and hydropathy distribution across CDRH3 amino acid length range were similar among all groups indicating that junctional modifications in sequences without or with SHM were unchanged (Figure 6).
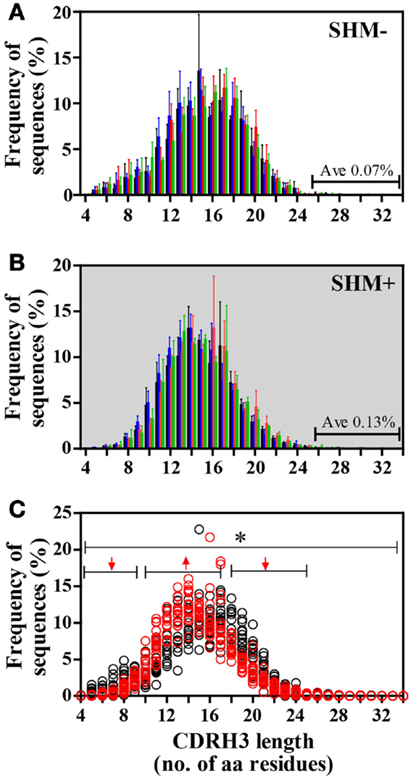
Figure 4. Frequency distribution of IGHM CDRH3 amino acid lengths differed between IgM B cells without or with SHM. (A) Lengths of SHM− CDRH3 regions displayed a Gaussian distribution ranging from 4 to 31 amino acid residues; lengths of 15 amino acids occurred most frequently. (B) Lengths of SHM+ CDRH3 regions showed Gaussian distribution ranging from 5 to 34 amino acids with lengths of 14 amino acids used most often. (C) Overall length distribution between SHM− and SHM+ sequences differed [*p = 0.004]. Among SHM+ sequences, shorter lengths [4–9 amino acids] and longer lengths [18–25 amino acids] were expressed significantly less [↓, p < 0.0001], while lengths of 10–17 amino acids were used more frequently in comparison to SHM− sequences [↑, p < 0.0001]. Bars: HC, black; SLE, blue; HIV-infected individuals: red, untreated; green, treated. Circles: SHM−, open black; SHM+, open red.
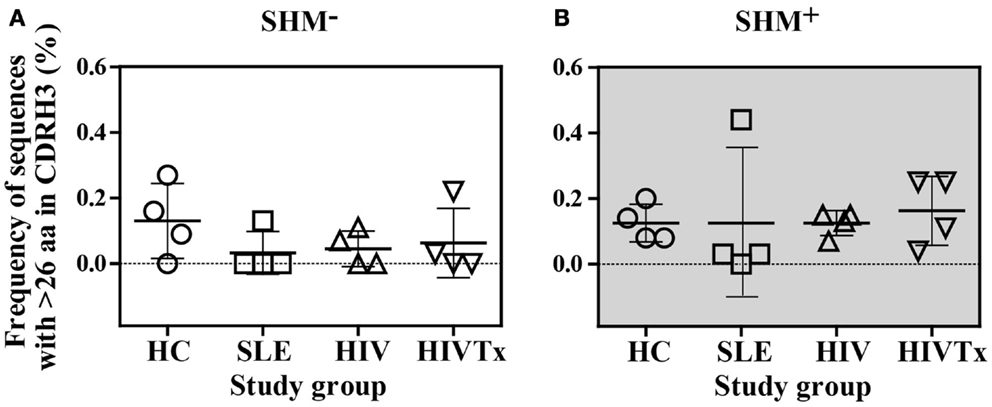
Figure 5. Similar frequency of sequences with CDRH3 lengths longer than 26 amino acids among study groups. (A) Among SHM− sequences, frequency of sequences with greater than 26 amino acids CDR3 length ranged from 0.03 [± 0.06%] to 0.13% [± 0.12%], which was similar among the four study groups. (B). Among SHM+ sequences, frequency of sequences with greater than 26 amino acids CDR3 length ranged from 0.13 [± 0.04%] to 0.16% [± 0.10%], which was similar among the four study groups.
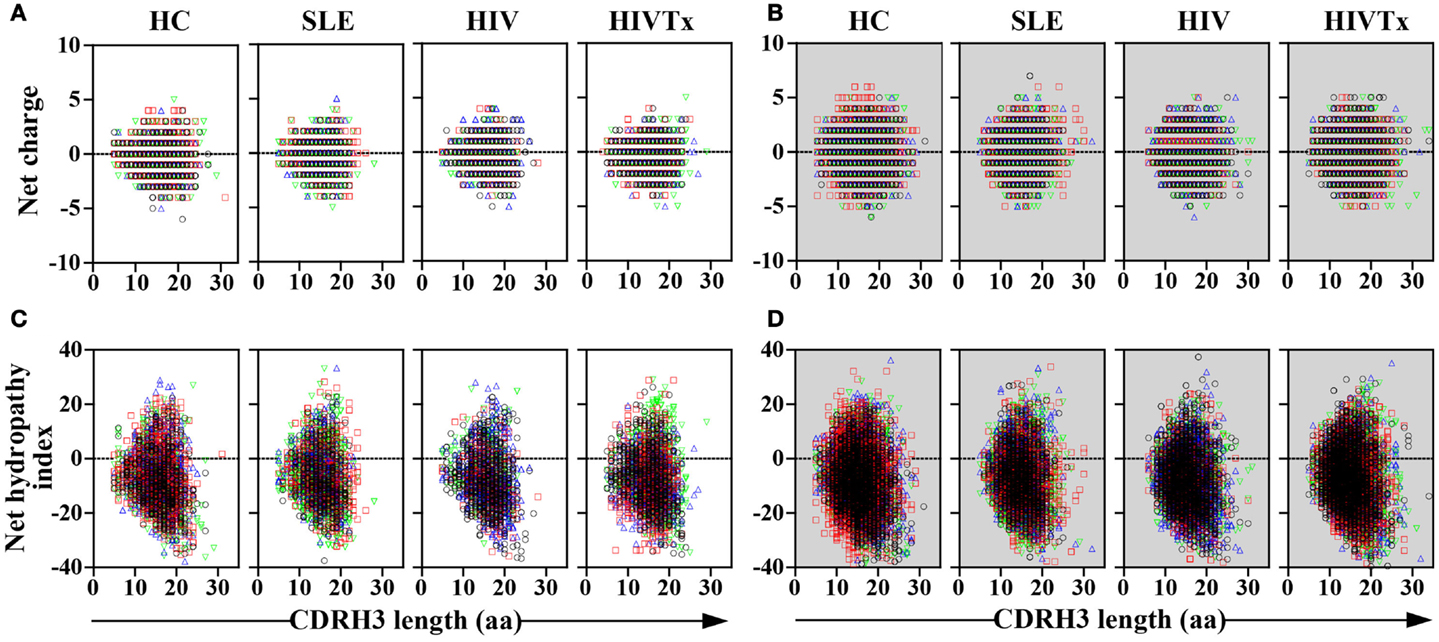
Figure 6. Similar distribution of charge and hydropathy across IGHM CDRH3 length among study groups. Distribution of charge and hydropathy across IGHM CDRH3 amino acid length range were similar among the four study groups in sequences without [charge, (A); hydropathy index, (C)] or with SHM [charge, (B); hydropathy index, (D)].
Allelic Frequency of IGHD and IGHJ Genes
Nearly 90% of the 44 IGHD alleles were detected across the study groups (Figures 7A,B). Sequences with SHM showed significant increases in use of multiple IGHD alleles compared with sequences lacking SHM. Similarly, over 75% of 13 IGHJ alleles, predominantly in IGHJ3 and IGHJ4 families, were identified independent of SHM (Figures 7C,D). A significant increase in three IGHJ alleles was detected in sequences with SHM (Figure 7D). While all study groups had similar IGHD and IGHJ allele usage, the major difference in allele usage was between sequences with or without SHM.
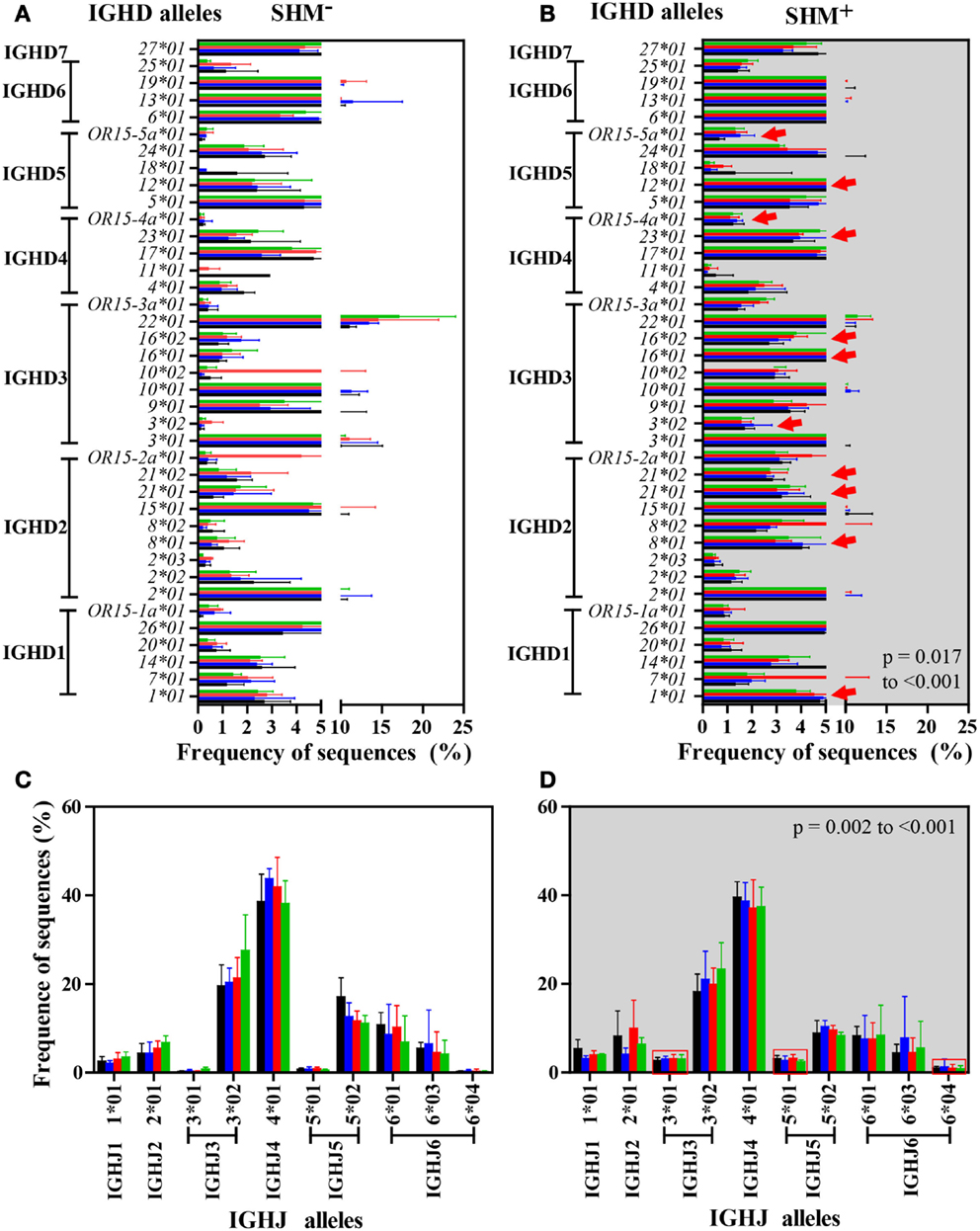
Figure 7. IGHM IGHD and IGHJ alleles were expressed similarly among study groups but differently between SHM− and SHM+ IgM B cells. Number and frequency of IGHD and IGHJ alleles were similar among the four study groups in IGHM CDRH3 sequences without [IGHD, (A); IGHJ, (C)] or with SHM [IGHD, (B); IGHJ, (D)]. A significant difference in usage of 11 IGHD alleles, including IGHD1-1*01, IGHD2-8*01, IGHD2-21*01, IGHD2-21*02, IGHD3-3*02, IGHD3-16*01, IGHD3-16*02, IGHD4-23*01, IGHD4/OR15-4a*01, IGHD5-12*01, and IGHD5/OR15-5a*01 (red arrows), and 3 IGHJ alleles, including IGHJ3*01, IGHJ5*01, and IGHJ6*04 (squared by red), was observed across the groups in SHM+ sequences in comparison to SHM− sequences (p < 0.001 respectively). Bars (mean/SD): HC, black; SLE, blue; HIV-infected individuals: untreated, red; treated, green.
Biodiversity of Allele Combinations
Biodiversity resulting from different combinations of alleles comprising the IGHM CDRH3 regions was assessed in health and disease groups. Every sequence was composed of V-D-J gene segments suggesting intact recombination machinery in all individuals independent of health or disease. When comparing numbers of different combinations along the depth of sequencing using rarefaction curves, a left shift was observed in healthy individuals, indicating greater diversification of combinations whether or not sequences contained SHM (Figures 8A,B). The difference between health and disease was more pronounced when maximum biodiversity was inferred by Chao1 algorithm based on both the richness and evenness of different V-D-J combinations (Figures 8C,D). ART failed to restore biodiversity in either group of IgM transcripts. Overall, reduced diversity of V-D-J combinations was a major contributor to the difference in biodiversity within IGHM CDRH3 sequences between groups.
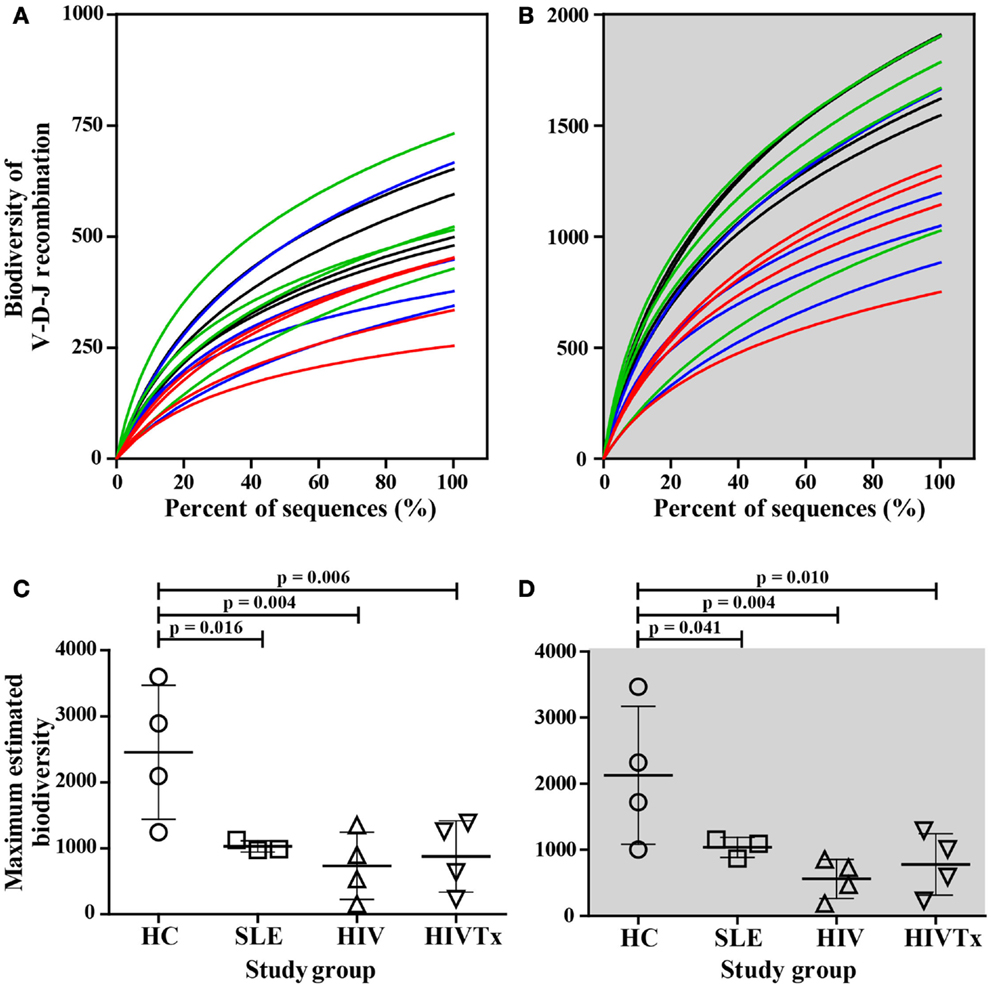
Figure 8. Significant decrease in biodiversity resulting from V-D-J allele recombination in young adults with SLE and HIV-1 infection in comparison to healthy controls. As assessed by rarefaction, biodiversity of V-D-J allele recombination along sequence depths decreased in young adults with SLE (blue lines) and untreated HIV-infected individuals (red lines) in comparison to their healthy counterparts (black lines) in IgM B cells without (A) or with SHM (B). Control of viral replication by ART failed to restore biodiversity in either cell type [green lines in (A,B)]. The maximum biodiversity of V-D-J allele recombination estimated by Chao1 was significantly greater in HC subjects than in individuals with SLE and HIV infection in input IgM B cells without (C) or with SHM (D) (SLE, p = 0.016 or 0.041; HIV, p = 0.004 for SHM+ or SHM+; HIVTx, p = 0.006 or 0.01, respectively).
IgM Sequences Resembling bn-HIV-Abs
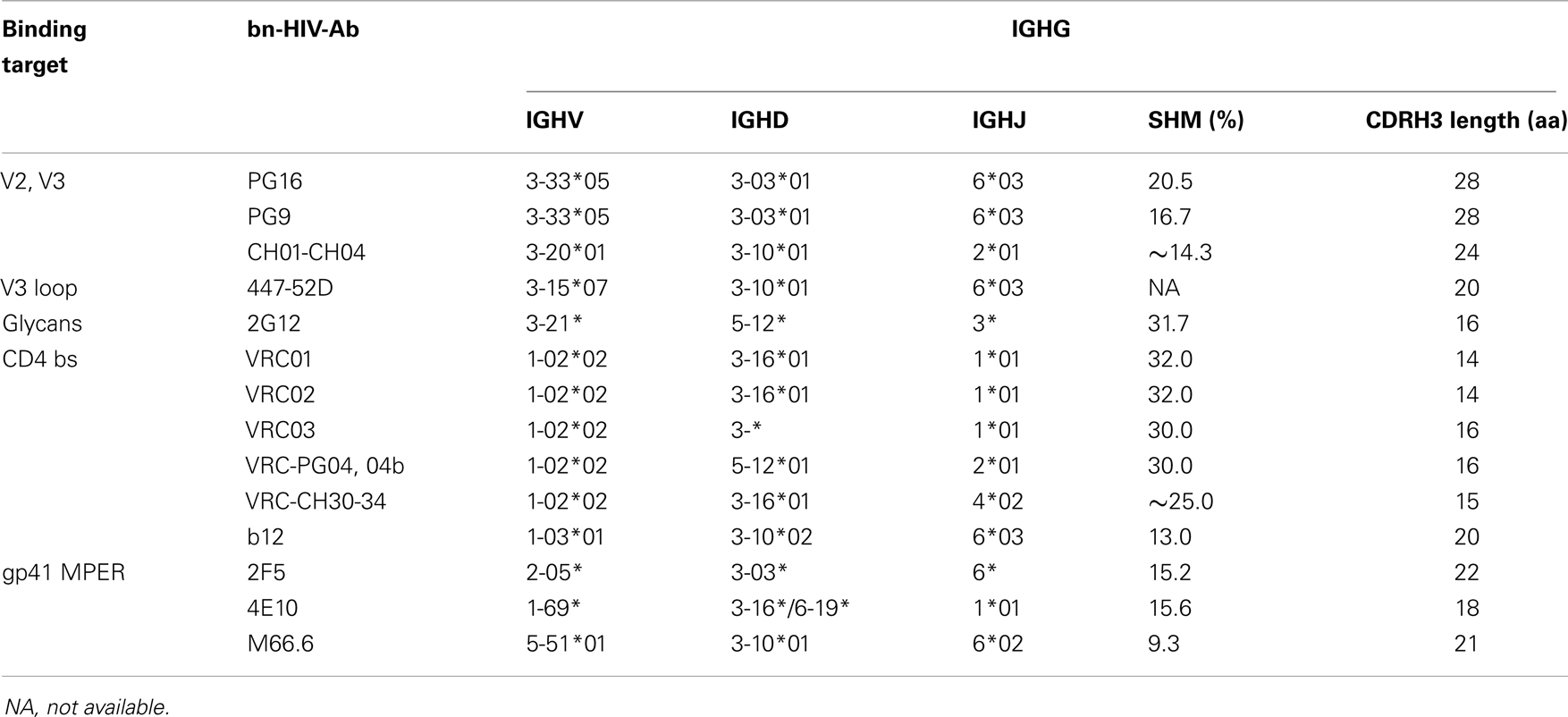
Combinations of alleles within IgM sequences that might be similar to IgG bn-HIV-Abs were evaluated, but undetected among any individuals (Table 3) (23, 26, 27, 58–68). In contrast, combinations of families found in many bn-HIV-Abs were identified among IgM sequences across the four study groups (Table 4). While few, if any, family combinations similar to M66.6/MPER or CD4 binding site antibodies b12 and VRC-PG04 were detected, combinations similar to other bn-HIV-Abs were identified among IgM sequences in a majority of subjects. In general, family combinations were more frequently represented among IgM sequences with SHM, although frequencies greater than 0.2% appeared in sequences with or without SHM. Frequency of family combinations was not a function of the targets in Envelope, as combinations directed toward CD4 binding site, N-linked glycans, V2/V3, or MPER were detected in almost all individuals.
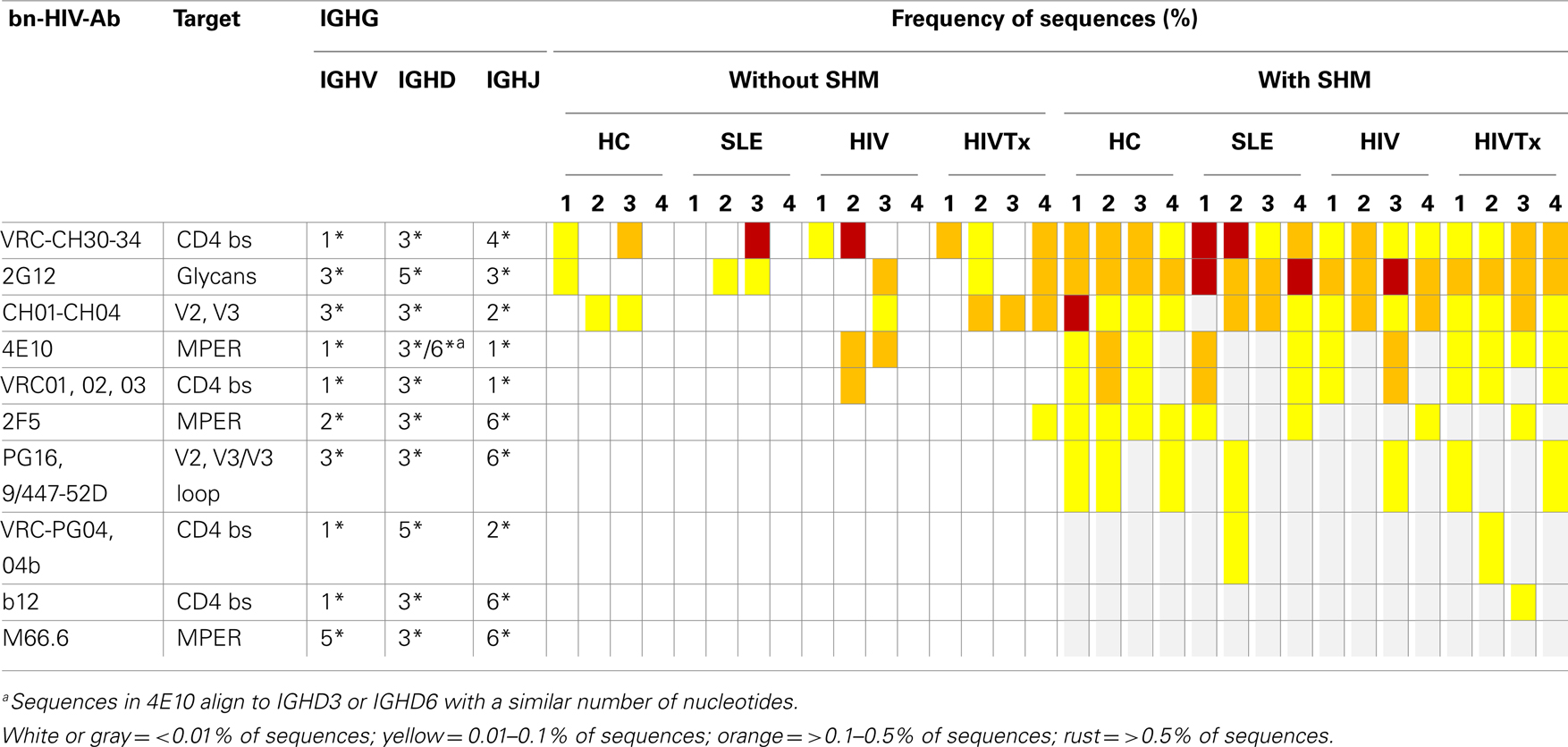
Table 4. IGHV, IGHD, and IGHJ combinations similar to known bn-HIV-Ab in IgM repertoire in peripheral blood.
Discussion
Little is known about how B cell abnormalities in HIV infection or SLE impact IGH repertoire at the molecular level (1–5, 69–72). Applying deep sequencing to IgM transcriptomes in total B cell populations is a strategy to follow development of the IgM repertoire as a novel molecular assessment at multiple points along the B cell differentiation pathway, which is highly sensitive for detecting perturbations within the repertoire in health and disease. Evaluation of CDRH3 regions in IgM focuses assessment on transition of the repertoire from initial V-D-J recombination in the bone marrow, through antigen-induced clonal expansion, SHM, and establishment of IgM memory B cells (73, 74).
A memory of antigen is imprinted irreversibly by SHM in Ig variable region genes, which renders SHM the most accurate marker to distinguished memory from naïve B cells than B cell phenotype defined by surface expression of IgM, IgD, and CD27. While memory CD27+ IgM B cells undergo robust SHM in germinal centers, CD27 is not an immutable indicator of B cell memory (74–77). We observed IGHM sequences with SHM in CDRH3, in accordance with observations suggesting an IgM-expressing memory B cell compartment in the marginal zone (52, 54, 78, 79). The high percentage of IGHM sequences with SHM may be due to a larger IgM memory B cell compartment than previously thought, as well as to increased mRNA levels in IgM B cells with SHM (54), It is unlikely that plasma cell sequences contributed to the results as plasma cell frequency in peripheral blood is minimal (80, 81) and our study design included subjects with no recent immunizations and/or acute infections to minimize the proportion of plasmacytoid B cells undergoing clonal expansion that may skew the IgM sequence repertoire (82).
A small but significant difference was detected between the frequency of SHM+ IgM B cells among subjects with SLE, but not HIV-1 infection, compared to healthy individuals (5, 83, 84). These cells resemble recirculating marginal zone memory B cells that contain somatic mutations and can create considerable IGH diversity during early childhood in the absence of specific antigenic stimulation (74, 85).
Rarefaction/Chao1 analysis provides the capacity to compare IGHM biodiversity between sequences expressed by naïve/transitional B cells and IgM memory B cells defined by SHM in peripheral blood B cell populations. Within individuals, CDRH3 sequence biodiversity resulting from V-D-J recombination generated in the bone marrow was significantly limited relative to sequence biodiversity following antigen activation and development of SHM, consistent with estimates of the frequency of SHM within the B cell repertoire (54). A critical finding is that biodiversity of the IgM repertoire in the absence or presence of SHM distinguishes between health and disease. Study groups were similar ages, an important aspect of the design, as age profoundly impacts IGH diversity, particularly with respect to the extent of SHM (86). Overall, IGH affinity maturation is more accurately assessed by direct molecular analysis through pyrosequencing of the IgM transcriptome than by phenotypic analysis of B cell subsets.
While effective treatment restores many HIV-induced B cell defects, overall B cell function remains impaired (8, 72, 87–89). Control of HIV replication by cART failed to restore IgM sequence diversity, although longitudinal studies are needed to evaluate directly the effects of cART on the B cell repertoire. Abnormalities of IgM B cell populations in HIV-1 infection may persist as the result of chronic inflammation that continues to impair B cell populations even when viral replication is optimally controlled (90). Furthermore, persistent low-level viral replication in the secondary lymphoid tissues where B cells and CD4 T-cells co-localize may induce lymphoid apoptosis and/or clonal expansion, contracting the IgM repertoire within memory B cells (4, 91).
We observed an overall shift of distribution of CDRH3 lengths toward shorter lengths in sequences with SHM, but no shifts in hydropathy or charge distribution across the CDRH3 lengths, as found in assessments using sorted IgM memory B cells (51, 92). Differences may reflect a distinct developmental program for CD27+ IgM marginal zone B cells that develop outside typical T-dependent or T-independent pathways (85). Overall, neither HIV-1 infection nor SLE produces profound alterations in allele usage, CDRH3 length, hydropathy, or charge distribution within the IgM repertoire, indicating that intrinsic mechanisms for generating junctional diversity and SHM are functional.
Decreased biodiversity in IGHM CDRH3 sequences without SHM supports the observation that HIV-1 reduces the breadth of the IgM repertoire early in B cell development (93, 94). In contrast, reduced biodiversity within the SHM+ IGHM repertoire of HIV-infected or SLE subjects might reflect polyclonal expansion, reduced B cell precursors, and/or B cell exhaustion. While pyrosequencing provides an unprecedented view of the human IgM B cell repertoire, depth of sampling of peripheral blood B cell populations was insufficient to detect directly expanded B cell clones, which may be expected from polyclonal expansion. In a study of healthy individuals the estimated IGH pool was greater than 1,000,000 different rearrangements so that even with deep sequencing no single sequence would be represented multiple times (39). Polyclonal expansion of limited number of clones would decrease overall diversity without impacting junctional modification, SHM generation, CDRH3 length distribution, or biochemical characteristics of amino acid residues. The healthy subjects enrolled in our study had no recent illnesses or immunizations that might skew their repertoire (86). In contrast, both the SLE and untreated HIV-infected subjects had active disease, yet CDRH3 length diversity and gene usage was not impacted. Decreased B cell numbers in the peripheral blood are restored by control of viral replication but therapy failed to restore sequence biodiversity in either SHM+ or SHM− populations (88, 89). This assessment of the IGHM sequence repertoire provides a basis for examining late stage B cell development through pyrosequencing of IGH in IgG and IgA repertoires within the same mRNA used for the IgM transcriptome, which will expand molecular understanding of the biodiversity of the peripheral blood B cell repertoire in health and disease.
Autoantibodies can result from defective early B cell tolerance in the bone marrow or arise as naïve B cells encounter antigen, undergo SHM and affinity maturation (6). Our study identified no skewing with the IGHM repertoire based on assessment of CDRH3 length, charge, and hydropathy within sequences that contained or lacked SHM, which is not surprising as assessment of the B cell repertoire using clonal sequencing of V-D-J recombination in SLE subjects also failed to reveal major abnormalities (6, 95). Overall, results provide a framework for in-depth studies of the molecular mechanisms that lead to lower biodiversity in SLE and HIV-infected individuals.
An intense area of study in the development of an effective HIV-1 vaccine is the capacity of the repertoire to generate bn-HIV-Abs. Most characterized bn-HIV-Abs are IgG isotype with extensive affinity maturation and SHM (58, 61, 62, 66, 67, 95). IgA autoantibodies, including anti-CCR5 or anti-gp41, produced by a subset of the HIV-1-exposed seronegative individuals or long-term non-progressors, contribute to effective prevention of viral entry at major mucosal portals (29, 32, 96–98). Both IgG and IgA are derived from IgM. Our assessment of the IgM repertoire revealed in all subjects low frequency B cells with the same combinations of IGHV, IGHD, and IGHJ gene families found in known IgG bn-HIV-Abs, particularly those which bind to CD4 binding site, gp41 MPER, V2/V3, glycans in C2, C3, V4 and C4, and V3 loop (23, 26, 27, 58, 60–67, 99, 100). Clearly the IgM repertoire across individuals normally includes B cells with the potential to express IgG bn-HIV-Abs, holding promise that these antibodies can be induced as part of an overall HIV vaccine strategy. Biochemical characteristics of IgM antibody sequences provide an assessment of functional potential of IgM antibodies. In our study, significant differences in frequency distribution of CDRH3 lengths, or distribution of charge or hydropathy along CDRH3 lengths in IgM repertoire of SLE or HIV-infected individuals were found in comparisons with HC. Results suggest that IgM antibodies with characteristics of IgG bn-HIV-Ab were not produced at levels sufficient to perturb IgM repertoire in SLE or HIV-infected individuals.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors are grateful to the individuals who volunteered for the study and to Gary Litman, Ph.D. and Donna Eason, Ph.D. for review and discussions of the research and the manuscript. This work was funded by NIH grants, including R21 AI078450, R01 AI100147, and T32 AR007603 Postdoctoral Training Award for Immunologic/Genetic Mechanisms in Rheumatic Diseases, awards from University of Florida, including Incentive Award, Thomas H. Maren Junior Investigator Postdoctoral Award, Laura McClamma Fellowship for Research in Pediatric Immune Deficiency, Robert A. Good Endowed Chair in Immunology, and Stephany W. Holloway University Chair for AIDS Research.
References
1. Lipsky PE. Systemic lupus erythematosus: an autoimmune disease of B cell hyperactivity. Nat Immunol (2001) 2:764–6. doi: 10.1038/ni0901-764
2. Moir S, Fauci AS. B cells in HIV infection and disease. Nat Rev Immunol (2009) 9:235–45. doi:10.1038/nri2524
3. Ho J, Moir S, Malaspina A, Howell ML, Wang W, DiPoto AC, et al. Two overrepresented B cell populations in HIV-infected individuals undergo apoptosis by different mechanisms. Proc Natl Acad Sci U S A (2006) 103:19436–41. doi:10.1073/pnas.0609515103
4. Moir S, Ho J, Malaspina A, Wang W, DiPoto AC, O’Shea MA, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med (2008) 205:1797–805. doi:10.1084/jem.20072683
5. Rodriguez-Bayona B, Ramos-Amaya A, Perez-Venegas JJ, Rodriguez C, Brieva JA. Decreased frequency and activated phenotype of blood CD27 IgD IgM B lymphocytes is a permanent abnormality in systemic lupus erythematosus patients. Arthritis Res Ther (2010) 12:R108. doi:10.1186/ar3042
6. Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V, et al. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med (2005) 201:703–11. doi:10.1084/jem.20042251
7. Brenchley JM, Price DA, Douek DC. HIV disease: fallout from a mucosal catastrophe? Nat Immunol (2006) 7:235–9. doi:10.1038/ni1316
8. Jacobson MA, Khayam-Bashi H, Martin JN, Black D, Ng V. Effect of long-term highly active antiretroviral therapy in restoring HIV-induced abnormal B-lymphocyte function. J Acquir Immune Defic Syndr (2002) 31:472–7. doi:10.1097/00126334-200212150-00003
9. Jacobi AM, Diamond B. Balancing diversity and tolerance: lessons from patients with systemic lupus erythematosus. J Exp Med (2005) 202:341–4. doi:10.1084/jem.20050221
10. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature (2005) 437:376–80. doi:10.1038/nature03959
11. Bomsel M, Heyman M, Hocini H, Lagaye S, Belec L, Dupont C, et al. Intracellular neutralization of HIV transcytosis across tight epithelial barriers by anti-HIV envelope protein dIgA or IgM. Immunity (1998) 9:277–87. doi:10.1016/S1074-7613(00)80610-X
12. Sheppard NC, Bates AC, Sattentau QJ. A functional human IgM response to HIV-1 Env after immunization with NYVAC HIV C. AIDS (2007) 21:524–7. doi:10.1097/QAD.0b013e32803277f9
13. Toran JL, Kremer L, Sanchez-Pulido L, de Alboran IM, del RG, Llorente M, et al. Molecular analysis of HIV-1 gp120 antibody response using isotype IgM and IgG phage display libraries from a long-term non-progressor HIV-1-infected individual. Eur J Immunol (1999) 29:2666–75. doi:10.1002/(SICI)1521-4141(199909)29:09<2666::AID-IMMU2666>3.0.CO;2-Q
14. Chen W, Zhu Z, Liao H, Quinnan GV Jr, Broder CC, Haynes BF, et al. Cross-reactive human IgM-derived monoclonal antibodies that bind to HIV-1 envelope glycoproteins. Viruses (2010) 2:547–65. doi:10.3390/v2020547
15. Wolbank S, Kunert R, Stiegler G, Katinger H. Characterization of human class-switched polymeric (immunoglobulin M [IgM] and IgA) anti-human immunodeficiency virus type 1 antibodies 2F5 and 2G12. J Virol (2003) 77:4095–103. doi:10.1128/JVI.77.7.4095-4103.2003
16. Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity (2005) 22:163–73. doi:10.1016/j.immuni.2004.12.011
17. Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science (2005) 308:1906–8. doi:10.1126/science.1111781
18. Kopelman RG, Zolla-Pazner S. Association of human immunodeficiency virus infection and autoimmune phenomena. Am J Med (1988) 84:82–8. doi:10.1016/0002-9343(88)90012-5
19. Massabki PS, Accetturi C, Nishie IA, da Silva NP, Sato EI, Andrade LE. Clinical implications of autoantibodies in HIV infection. AIDS (1997) 11:1845–50. doi:10.1097/00002030-199715000-00009
20. Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol (2004) 78:10724–37. doi:10.1128/JVI.78.19.10724-10737.2004
21. Petrovas C, Vlachoyiannopoulos PG, Kordossis T, Moutsopoulos HM. Anti-phospholipid antibodies in HIV infection and SLE with or without anti-phospholipid syndrome: comparisons of phospholipid specificity, avidity and reactivity with beta2-GPI. J Autoimmun (1999) 13:347–55. doi:10.1006/jaut.1999.0324
22. Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, et al. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science (2001) 293:1155–9. doi:10.1126/science.1061692
23. Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure (2004) 12:193–204. doi:10.1016/j.str.2004.01.003
24. Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science (2003) 301:1374–7. doi:10.1126/science.1086907
25. Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol (2001) 75:10892–905. doi:10.1128/JVI.75.24.12198-12208.2001
26. Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses (1994) 10:359–69. doi:10.1089/aid.1994.10.359
27. Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, et al. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol (1992) 66:7538–42.
28. Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature (2009) 458:636–40. doi:10.1038/nature07930
29. Bomsel M, Pastori C, Tudor D, Alberti C, Garcia S, Ferrari D, et al. Natural mucosal antibodies reactive with first extracellular loop of CCR5 inhibit HIV-1 transport across human epithelial cells. AIDS (2007) 21:13–22. doi:10.1097/QAD.0b013e328011049b
30. Devito C, Broliden K, Kaul R, Svensson L, Johansen K, Kiama P, et al. Mucosal and plasma IgA from HIV-1-exposed uninfected individuals inhibit HIV-1 transcytosis across human epithelial cells. J Immunol (2000) 165:5170–6.
31. Devito C, Hinkula J, Kaul R, Lopalco L, Bwayo JJ, Plummer F, et al. Mucosal and plasma IgA from HIV-exposed seronegative individuals neutralize a primary HIV-1 isolate. AIDS (2000) 14:1917–20. doi:10.1097/00002030-200009080-00006
32. Nguyen M, Pean P, Lopalco L, Nouhin J, Phoung V, Ly N, et al. HIV-specific antibodies but not T-cell responses are associated with protection in seronegative partners of HIV-1-infected individuals in Cambodia. J Acquir Immune Defic Syndr (2006) 42:412–9. doi:10.1097/01.qai.0000222289.97825.35
33. Koshino K, Tokano Y, Hishikawa T, Sekigawa I, Takasaki Y, Hashimoto H. Detection of antibodies to HIV-1 gp41- and HLA class II antigen-derived peptides in SLE patients. Scand J Rheumatol (1995) 24:288–92. doi:10.3109/03009749509095165
34. Hamatake M, Komano J, Urano E, Maeda F, Nagatsuka Y, Takekoshi M. Inhibition of HIV replication by a CD4-reactive Fab of an IgM clone isolated from a healthy HIV-seronegative individual. Eur J Immunol (2010) 40:1504–9. doi:10.1002/eji.200939479
35. Lobo PI, Schlegel KH, Yuan W, Townsend GC, White JA. Inhibition of HIV-1 infectivity through an innate mechanism involving naturally occurring IgM anti-leukocyte autoantibodies. J Immunol (2008) 180:1769–79.
36. Campbell A. Save those molecules: molecular biodiversity and life. J Appl Ecol (2003) 40:193–203. doi:10.1046/j.1365-2664.2003.00803.x
37. Newton AC. Forest Ecology and Preservation: A Handbook of Techniques. New York: Oxford University Press (1999).
38. Bimber BN, Burwitz BJ, O’Connor S, Detmer A, Gostick E, Lank SM, et al. Ultradeep pyrosequencing detects complex patterns of CD8+ T-lymphocyte escape in simian immunodeficiency virus-infected macaques. J Virol (2009) 83:8247–53. doi:10.1128/JVI.00897-09
39. Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med (2009) 1:12ra23. doi:10.1126/scitranslmed.3000540
40. Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U S A (2009) 106:20216–21. doi:10.1073/pnas.0909775106
41. Weinstein JA, Jiang N, White RA III, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science (2009) 324:807–10. doi:10.1126/science.1170020
42. Petri M, Orbai AM, Alarcon GS, Gordon C, Merrill JT, Fortin PR, et al. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum (2012) 64:2677–86. doi:10.1002/art.34473
43. Bossuyt X, Marti GE, Fleisher TA. Comparative analysis of whole blood lysis methods for flow cytometry. Cytometry (1997) 30:124–33. doi:10.1002/(SICI)1097-0320(19970615)30:3<124::AID-CYTO3>3.0.CO;2-L
44. Gokmen E, Raaphorst FM, Boldt DH, Teale JM. Ig heavy chain third complementarity determining regions (H CDR3s) after stem cell transplantation do not resemble the developing human fetal H CDR3s in size distribution and Ig gene utilization. Blood (1998) 92:2802–14.
45. IMGT website. (2012). Available from: http://www.imgt.org
46. Lefranc MP, Duprat E, Kaas Q, Tranne M, Thiriot A, Lefranc G. IMGT unique numbering for MHC groove G-DOMAIN and MHC superfamily (MhcSF) G-LIKE-DOMAIN. Dev Comp Immunol (2005) 29:917–38. doi:10.1016/j.dci.2005.03.003
47. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res (2004) 14:1188–90. doi:10.1101/gr.849004
48. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol (1982) 157:105–32. doi:10.1016/0022-2836(82)90515-0
49. Yousfi MM, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics (2004) 20(Suppl 1):i379–85. doi:10.1093/bioinformatics/bth945
50. Link JM, Schroeder HW Jr. Clues to the etiology of autoimmune diseases through analysis of immunoglobulin genes. Arthritis Res (2002) 4:80–3. doi:10.1186/ar393
51. Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood (2010) 116:1070–8. doi:10.1182/blood-2010-03-275859
52. Dunn-Walters DK, Isaacson PG, Spencer J. Analysis of mutations in immunoglobulin heavy chain variable region genes of microdissected marginal zone (MGZ) B cells suggests that the MGZ of human spleen is a reservoir of memory B cells. J Exp Med (1995) 182:559–66. doi:10.1084/jem.182.2.559
53. Gilles A, Meglecz E, Pech N, Ferreira S, Malausa T, Martin JF. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics (2011) 12:245. doi:10.1186/1471-2164-12-245
54. Klein U, Kuppers R, Rajewsky K. Evidence for a large compartment of IgM-expressing memory B cells in humans. Blood (1997) 89:1288–98.
55. Sun Y, Cai Y, Liu L, Yu F, Farrell ML, McKendree W, et al. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res (2009) 37:e76. doi:10.1093/nar/gkp285
56. Yin L, Liu L, Sun Y, Hou W, Lowe AC, Gardner BP, et al. High-resolution deep sequencing reveals biodiversity, population structure, and persistence of HIV-1 quasispecies within host ecosystems. Retrovirology (2012) 9:108. doi:10.1186/1742-4690-9-108
57. Kou ZC, Puhr JS, Wu SS, Goodenow MM, Sleasman JW. Combination antiretroviral therapy results in a rapid increase in T cell receptor variable region beta repertoire diversity within CD45RA CD8 T cells in human immunodeficiency virus-infected children. J Infect Dis (2003) 187:385–97. doi:10.1086/367674
58. Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol (2011) 85:9998–10009. doi:10.1128/JVI.05045-11
59. Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One (2010) 5:e8805. doi:10.1371/journal.pone.0008805
60. Kunert R, Ruker F, Katinger H. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res Hum Retroviruses (1998) 14:1115–28. doi:10.1089/aid.1998.14.1115
61. Li Y, O’Dell S, Walker LM, Wu X, Guenaga J, Feng Y, et al. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol (2011) 85:8954–67. doi:10.1128/JVI.00754-11
62. Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol (2009) 83:7337–48. doi:10.1128/JVI.00110-09
63. Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, et al. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses (2001) 17:1757–65. doi:10.1089/08892220152741450
64. Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol (1996) 70:1100–8.
65. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science (2009) 326:285–9. doi:10.1126/science.1178746
66. Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (2010) 329:856–61. doi:10.1126/science.1187659
67. Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science (2011) 333:1593–602. doi:10.1126/science.1207532
68. Zhu Z, Qin HR, Chen W, Zhao Q, Shen X, Schutte R, et al. Cross-reactive HIV-1-neutralizing human monoclonal antibodies identified from a patient with 2F5-like antibodies. J Virol (2011) 85:11401–8. doi:10.1128/JVI.05312-11
69. Hart M, Steel A, Clark SA, Moyle G, Nelson M, Henderson DC, et al. Loss of discrete memory B cell subsets is associated with impaired immunization responses in HIV-1 infection and may be a risk factor for invasive pneumococcal disease. J Immunol (2007) 178:8212–20.
70. Kardava L, Moir S, Wang W, Ho J, Buckner CM, Posada JG, et al. Attenuation of HIV-associated human B cell exhaustion by siRNA downregulation of inhibitory receptors. J Clin Invest (2011) 121:2614–24. doi:10.1172/JCI45685
71. Longwe H, Gordon S, Malamba R, French N. Characterising B cell numbers and memory B cells in HIV infected and uninfected Malawian adults. BMC Infect Dis (2010) 10:280. doi:10.1186/1471-2334-10-280
72. Titanji K, De MA, Cagigi A, Thorstensson R, Grutzmeier S, Atlas A, et al. Loss of memory B cells impairs maintenance of long-term serologic memory during HIV-1 infection. Blood (2006) 108:1580–7. doi:10.1182/blood-2005-11-013383
73. Seifert M, Kuppers R. Molecular footprints of a germinal center derivation of human IgM+(IgD+)CD27+ B cells and the dynamics of memory B cell generation. J Exp Med (2009) 206:2659–69. doi:10.1084/jem.20091087
74. Weller S, Braun MC, Tan BK, Rosenwald A, Cordier C, Conley ME, et al. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood (2004) 104:3647–54. doi:10.1182/blood-2004-01-0346
75. Griffin DO, Holodick NE, Rothstein TL. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70−. J Exp Med (2011) 208:67–80. doi:10.1084/jem.20101499
76. Tangye SG, Good KL. Human IgM+CD27+ B cells: memory B cells or “memory” B cells? J Immunol (2007) 179:13–9.
77. Wei C, Anolik J, Cappione A, Zheng B, Pugh-Bernard A, Brooks J, et al. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J Immunol (2007) 178:6624–33.
78. Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med (1998) 188:1679–89. doi:10.1084/jem.188.9.1679
79. Weston-Bell N, Townsend M, Di GG, Forconi F, Sahota SS. Defining origins of malignant B cells: a new circulating normal human IgM(+)D(+) B-cell subset lacking CD27 expression and displaying somatically mutated IGHV genes as a relevant memory population. Leukemia (2009) 23:2075–80. doi:10.1038/leu.2009.178
80. Medina F, Segundo C, Campos-Caro A, Gonzalez-Garcia I, Brieva JA. The heterogeneity shown by human plasma cells from tonsil, blood, and bone marrow reveals graded stages of increasing maturity, but local profiles of adhesion molecule expression. Blood (2002) 99:2154–61. doi:10.1182/blood.V99.6.2154
81. Minges Wols HA. Plasma cells. In: Encyclopedia Life Science. John Wiley & Sons, Ltd. (2006). p. 1–8. doi:10.1038/npg.els.0004030
82. Blanchard-Rohner G, Pulickal AS, Jol-van der Zijde CM, Snape MD, Pollard AJ. Appearance of peripheral blood plasma cells and memory B cells in a primary and secondary immune response in humans. Blood (2009) 114:4998–5002. doi:10.1182/blood-2009-03-211052
83. De Milito A, Morch C, Sonnerborg A, Chiodi F. Loss of memory (CD27) B lymphocytes in HIV-1 infection. AIDS (2001) 15:957–64. doi:10.1097/00002030-200105250-00003
84. Jacobi AM, Reiter K, Mackay M, Aranow C, Hiepe F, Radbruch A, et al. Activated memory B cell subsets correlate with disease activity in systemic lupus erythematosus: delineation by expression of CD27, IgD, and CD95. Arthritis Rheum (2008) 58:1762–73. doi:10.1002/art.23498
85. Weller S, Mamani-Matsuda M, Picard C, Cordier C, Lecoeuche D, Gauthier F, et al. Somatic diversification in the absence of antigen-driven responses is the hallmark of the IgM+ IgD+ CD27+ B cell repertoire in infants. J Exp Med (2008) 205:1331–42. doi:10.1084/jem.20071555
86. Wu YC, Kipling D, Dunn-Walters DK. Age-related changes in human peripheral blood IGH repertoire following vaccination. Front Immunol (2012) 3:193. doi:10.3389/fimmu.2012.00193
87. Chong Y, Ikematsu H, Yamamoto M, Murata M, Yamaji K, Nishimura M, et al. Increased frequency of CD27- (naive) B cells and their phenotypic alteration in HIV type 1-infected patients. AIDS Res Hum Retroviruses (2004) 20:621–9. doi:10.1089/0889222041217455
88. Moir S, Malaspina A, Ho J, Wang W, DiPoto AC, O’Shea MA, et al. Normalization of B cell counts and subpopulations after antiretroviral therapy in chronic HIV disease. J Infect Dis (2008) 197:572–9. doi:10.1086/526789
89. Sleasman JW, Nelson RP, Goodenow MM, Wilfret D, Hutson A, Baseler M, et al. Immunoreconstitution after ritonavir therapy in children with human immunodeficiency virus infection involves multiple lymphocyte lineages. J Pediatr (1999) 134:597–606. doi:10.1016/S0022-3476(99)70247-7
90. Wallet MA, Rodriguez CA, Yin L, Saporta S, Chinratanapisit S, Hou W, et al. Microbial translocation induces persistent macrophage activation unrelated to HIV-1 levels or T-cell activation following therapy. AIDS (2010) 24:1281–90. doi:10.1097/QAD.0b013e328339e228
91. Cohen OJ, Pantaleo G, Lam GK, Fauci AS. Studies on lymphoid tissue from HIV-infected individuals: implications for the design of therapeutic strategies. Springer Semin Immunopathol (1997) 18:305–22. doi:10.1007/BF00813500
92. Wu YC, Kipling D, Dunn-Walters DK. The relationship between CD27 negative and positive B cell populations in human peripheral blood. Front Immunol (2011) 2:81. doi:10.3389/fimmu.2011.00081
93. Carter CC, McNamara LA, Onafuwa-Nuga A, Shackleton M, Riddell J, Bixby D, et al. HIV-1 utilizes the CXCR4 chemokine receptor to infect multipotent hematopoietic stem and progenitor cells. Cell Host Microbe (2011) 9:223–34. doi:10.1016/j.chom.2011.02.005
94. Koka PS, Reddy ST. Cytopenias in HIV infection: mechanisms and alleviation of hematopoietic inhibition. Curr HIV Res (2004) 2:275–82. doi:10.2174/1570162043351282
95. Barbas CF III, Collet TA, Amberg W, Roben P, Binley JM, Hoekstra D, et al. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol (1993) 230:812–23. doi:10.1006/jmbi.1993.1203
96. Barassi C, Lazzarin A, Lopalco L. CCR5-specific mucosal IgA in saliva and genital fluids of HIV-exposed seronegative subjects. Blood (2004) 104:2205–6. doi:10.1182/blood-2004-06-2134
97. Lopalco L. Natural anti-CCR5 antibodies in HIV-infection and -exposure. J Transl Med (2010) 9(Suppl 1):S4. doi:10.1186/1479-5876-9-S1-S4
98. Lopalco L, Barassi C, Paolucci C, Breda D, Brunelli D, Nguyen M, et al. Predictive value of anti-cell and anti-human immunodeficiency virus (HIV) humoral responses in HIV-1-exposed seronegative cohorts of European and Asian origin. J Gen Virol (2005) 86:339–48. doi:10.1099/vir.0.80585-0
99. Kunert R, Wolbank S, Stiegler G, Weik R, Katinger H. Characterization of molecular features, antigen-binding, and in vitro properties of IgG and IgM variants of 4E10, an anti-HIV type 1 neutralizing monoclonal antibody. AIDS Res Hum Retroviruses (2004) 20:755–62. doi:10.1089/0889222041524571
Keywords: IgM antibody transcriptome repertoire, biodiversity, HIV-1, systemic lupus erythematosus, somatic hypermutation, naïve B cells, IgM memory B cells, pyrosequencing
Citation: Yin L, Hou W, Liu L, Cai Y, Wallet MA, Gardner BP, Chang K, Lowe AC, Rodriguez CA, Sriaroon P, Farmerie WG, Sleasman JW and Goodenow MM (2013) IgM repertoire biodiversity is reduced in HIV-1 infection and systemic lupus erythematosus. Front. Immunol. 4:373. doi: 10.3389/fimmu.2013.00373
Received: 14 June 2013; Accepted: 30 October 2013;
Published online: 15 November 2013.
Edited by:
Teunis Geijtenbeek, University of Amsterdam, NetherlandsReviewed by:
Deborah K. Dunn-Walters, King’s College London School of Medicine, UKFrancisco Veas, Institut de recherche pour le Développement, France
Copyright: © 2013 Yin, Hou, Liu, Cai, Wallet, Gardner, Chang, Lowe, Rodriguez, Sriaroon, Farmerie, Sleasman and Goodenow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Yin, Department of Pathology, Immunology and Laboratory Medicine, College of Medicine, University of Florida, 2033 Mowry Road, POB 103633, Gainesville, FL 32610-3633, USA e-mail:eWluQHBhdGhvbG9neS51ZmwuZWR1
†Present address: John William Sleasman, Division of Allergy and Immunology, Duke University School of Medicine, DUMC Box 2644, 203 Research Dr., Room 133B, MSRB 1, Durham, NC 27710, USA
‡John William Sleasman and Maureen Michels Goodenow have contributed equally to this work.
 Li Yin
Li Yin Wei Hou
Wei Hou Li Liu
Li Liu Yunpeng Cai4
Yunpeng Cai4 Mark Andrew Wallet
Mark Andrew Wallet Brent Paul Gardner
Brent Paul Gardner Kaifen Chang
Kaifen Chang Amanda Catherine Lowe
Amanda Catherine Lowe Carina Adriana Rodriguez
Carina Adriana Rodriguez Panida Sriaroon
Panida Sriaroon William George Farmerie
William George Farmerie John William Sleasman
John William Sleasman Maureen Michels Goodenow
Maureen Michels Goodenow