- Department of Medicine, Institute for Virology and Immunobiology, University of Würzburg, Würzburg, Germany
Most circulating human gamma delta T cells are Vγ9Vδ2 T cells. Their hallmark is the expression of T cell antigen receptors (TCR) whose γ-chains show a Vγ9-JP (Vγ2-Jγ1.2) rearrangement and are paired with Vδ2-containing δ-chains, a dominant TCR configuration, which until recently seemed to occur in primates only. Vγ9Vδ2 T cells respond to phosphoantigens (PAg) such as (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate (HMBPP), which is produced by many pathogens and isopentenyl pyrophosphate (IPP), which accumulates in certain tumors or cells treated with aminobisphosphonates such as zoledronate. A prerequisite for PAg-induced activation is the contact of Vγ9Vδ2 T cells with cells expressing butyrophilin-3 A1 (BTN3A1). We will first critically review models of how BTN3 might act in PAg-mediated Vγ9Vδ2 T cell activation and then address putative co-evolution of Vγ9, Vδ2, and BTN3 genes. In those rodent and lagomorphs used as animal models, all three genes are lost but a data-base analysis showed that they emerged together with placental mammals. A strong concomitant conservation of functional Vγ9, Vδ2, and BTN3 genes in other species suggests co-evolution of these three genes. A detailed analysis was performed for the new world camelid alpaca (Vicugna pacos). It provides an excellent candidate for a non-primate species with presumably functional Vγ9Vδ2 T cells since TCR rearrangements share features characteristic for PAg-reactive primate Vγ9Vδ2 TCR and proposed PAg-binding sites of BTN3A1 have been conserved. Finally, we analyze the possible functional relationship between the butyrophilin-family member Skint1 and the γδ TCR-V genes used by murine dendritic epithelial T cells (DETC). Among placental mammals, we identify five rodents, the cow, a bat, and the cape golden mole as the only species concomitantly possessing potentially functional homologs of murine Vγ3, Vδ4 genes, and Skint1 gene and suggest to search for DETC like cells in these species.
αβ T Cells and γδ T Cells
Jawed vertebrates (Gnathostomata) possess lymphocytes expressing Ig-domain containing antigen-receptors, whose highly diverse antigen-binding sites are generated by RAG-dependent somatic recombination of genes encoding for an antigen-binding variable-domain. Such receptors are broadly classified into three types. αβ and γδ T cell antigen receptors (TCR), which are encoded by αβ and γδ genes, respectively, and the B cell antigen receptors, which are encoded by immunoglobulin heavy and light chain genes (1–3). The exact sequence of emergence of the antigen-receptor genes is controversial. One view is that the γδ TCR encoding genes originally constituted a receptor for soluble antigens, which is primordial to the MHC-restricted αβ TCR, BCR, and antibodies (2, 4). Alternatively, αβ and γδ TCR-genes may have emerged from a common ancestor (5). An interesting case of convergent evolution has occurred in jawless vertebrates (Agnatha). Their lymphocytes express antigen-receptors completely distinct from those of Gnathostomata both in terms of molecular composition (leucine rich repeats instead of Ig domains) and genetic basis of diversity generation [Cytidine deaminase (AID) dependent gene conversion]. Three distinct lineages of lymphocytes expressing distinct antigen receptors variable leukocyte receptors (VLR) (6, 7) have been identified: T-like lymphocytes maturing in the thymus express VLRA and VLRC while B-like lymphocytes produce VLRB as soluble antigen-receptors (8).
Among T cells, those which confer adaptive immunity are MHC-restricted T cells. They express αβ TCR, which bind to complexes of polymorphic MHC molecules and peptide antigens. Their diversity is generated by RAG-mediated recombination of V(D)J genes of both chains. The diversity of their third complementarity-determining regions (CDR3) which are encoded by the VαJα and VβDβJβ transition is further increased by joining flexibility and insertion of P and N-nucleotides. The final composition of TCR specificities (T cell repertoire) is shaped by intrathymic positive and negative selection guided by anatomically controlled presentation of peptide-MHC complexes and avidity of binding of the emerging TCR to those complexes (9). A highly conserved (2, 3), although not absolute feature in all vertebrate species (10), is the division of mature T cells as in MHC-class I restricted CD8 T cells, which exert killer function and MHC-class II restricted CD4 T cells, which regulate immune functions (2, 3). In spite of a likely co-evolution of the peptide presenting MHC molecules with Vα and Vβ TCR genes, it is not possible to predict MHC-class restriction or antigen-specificity of a given T cell from usage of certain TCR genes (11–13).
Alongside “conventional” MHC-restricted αβ T cells, several T cell populations exist with specificities for ligands other than MHC-peptide complexes. These “non-conventional” T cells can express αβ or γδ TCR. The generation of their TCR diversity follows the same genetic mechanisms as for MHC-restricted αβ TCR but in contrast to MHC-restricted T cells TCR-gene usage can be predictive for ontogeny, homing, and effector functions and is used to define distinct T cell subpopulations (12, 14–17). The best understood population of non-conventional T cells are the CD1d-restricted invariant NKT cells (17, 18). Their TCRα chains are invariant with a characteristic VαJα rearrangement and pair with β-chains of restricted Vβ gene usage. The iNKT TCRs bind in a highly conserved manner to complexes of the non-polymorphic MHC-class I like CD1d molecule and microbial or host cell glycolipids. Thus, functionally they resemble pattern recognition receptors of innate immune cells, which do not discriminate between highly variable antigens but recognize molecular patterns (17, 18). In contrast to MHC-restricted αβ T cells, which at least in mammals differentiate into CD8 killer T cells and CD4 helper and regulatory T cells, subpopulations of non-conventional T cells vary strongly between phylogenetic groups, e.g., many mammals lack iNKT cells as well as the restricting CD1d molecule (19, 20) but even closely related species such as mouse and rat differ dramatically in terms of iNKT-cell frequency although the respective genes for iNKT TCR and CD1d are highly conserved (21).
γδ T cells differ strongly between taxa. This difference can be rather global as in case of γδ T cells whose frequencies among blood T cells vary between 1 and 5% in man and mouse to 50% in ruminants (22). The presence or absence of entire populations defined by their TCR gene usage is even more striking (14, 23–25). Two such cases are addressed in this review: at first the human Vγ9Vδ2 T cells, which have so far only been found in higher primates (26). For their function, associated with various microbial and host metabolites, the molecule butyrophilin-3A1 is mandatory and this article will address the putative function of BTN3A1 and current evidence for a co-evolution of Vγ9Vδ2 TCR and BTN3A1. Secondly, we provide a first analysis of phylogeny of Vγ and Vδ genes constituting the TCR of murine dendritic epidermal T cell receptor (DETC) (27–30) and Skint1, a member of the butyrophilin family, which is critical during the ontogeny and function of these cells (31).
Vγ9Vδ2 T Cells: TCR and Phosphoantigen Reactivity
The vast majority of human blood γδ T cells respond to so called “Phosphoantigens” (PAg) (32–34). Their TCR share a characteristic Vγ9JP (alternatively designated as Vγ2Jγ1.2) rearrangement (35–38) and Vδ2-containing chains (35) bearing one of the hydrophobic amino acids (38, 39): Leucine (L), Isoleucine (I), Valine (V), or Glycine (G) at position 97. Unless explicitly mentioned PAg-reactive T cells and Vγ9Vδ2 T cells will be used as synonyms in this article.
Freshly isolated Vγ9Vδ2 T cells share functional features with TH1 cells, CD8 T cells, and NK cells (40) but upon culture they can differentiate to TH17 like (41), and to professional antigen-presenting cells (42). Apart from killing or cytokine release, they promote and regulate immune responses by crosstalk with dendritic cells (43), NK cells (44), and monocytes (45). Numerous preclinical and clinical studies demonstrated their therapeutic potential for treatment of tumors (46, 47) and infection (48, 49). The antigen-dependent activation of Vγ9Vδ2 T cells is strongly modulated by additional receptors (50) including inhibitory and activating NK-cell receptors (51, 52). In case of NKG2D even a direct triggering of some effector functions is possible (53).
PAg are products of isoprenoid synthesis, which specifically activate Vγ9Vδ2 T cells. The building blocks of isoprenoid synthesis are isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate. Both are weak PAg (54, 55) and 1000- to 10000-fold less potent than the strongest naturally occurring PAg (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate (HMBPP) (26, 55, 56). HMBPP is the immediate precursor in the synthesis of IPP by the non-mevalonate pathway also known as DOXP, MEP, or Rohmer pathway. The non-mevalonate-pathway is restricted to eubacteria, cyanobacteria, plants, and apicomplexan protozoa (26, 56), which may have adopted this pathway by endosymbiosis of precursors of chloroplasts and apicoplast, respectively (57, 58). HMBPP is the driving force of a massive Vγ9Vδ2 T cell expansion in infections with HMBPP producing parasites or bacteria, which can lead to an increase of Vγ9Vδ2 T cells from 1 to 5% of blood T cells to more than 50% (26, 49). In mammals and most other animals, IPP is synthesized via the mevalonate pathway whose manipulation can render human cells into Vγ9Vδ2 T cell activators. Cells pulsed with aminobisphosphonates (e.g., zoledronate or pamidronate) become potent activators of primary Vγ9Vδ2 T cells (59, 60) and of Vγ9Vδ2 TCR transductants (61) most likely in consequence of IPP accumulation after inhibition of the IPP metabolizing farnesyl diphosphate synthase (FPPS) (59, 62). The same effects are seen by inhibiting expression of FPPS (63, 64) and of isopentenyl diphosphate isomerase (64). Other modes of activation by IPP accumulation are activation (65) or over-expression of HMG-CoA reductase (66), which is the rate-controlling enzyme upstream of IPP synthesis, and finally inhibition of FPPS by alkylamines (67, 68).
Altogether the Vγ9Vδ2 TCR acts as a kind of pattern recognition receptor, which senses microbial infections as well as metabolic changes found in transformed, infected, or drug treated host cells (69). This reactivity can be harnessed clinically since remission of certain tumor entities after Vγ9Vδ2 T cell activation was observed in clinical trials (46, 47, 59, 60) and it may even contribute to the beneficial effects of zoledronate seen upon treatment of premenopausal women with early-stage breast cancer (70).
Some tumors such as the human B cell lymphoma Daudi (71, 72) spontaneously activate Vγ9Vδ2 T cells. This activation depends on intracellular accumulation of IPP and can be abolished by statins, which inhibit the HMG-CoA reductase and consequently also IPP synthesis (66). In contrast, still unclear despite intensive investigation is the contribution and the mechanistic basis of Vγ9Vδ2 T cell activation by the pro-apoptotic IPP metabolite ApppI (triphosphoric acid 1-adenosin-50-yl ester 3-(3-methylbut-3-enyl) ester), which is synthesized from IPP and AMP by aminoacyl-tRNA-synthetases (73, 74). Also not clear is how PAg-action is associated with the reported binding of the Vγ9Vδ2 TCR G115 to ectopically expressed F1-ATPase (74, 75).
BTN3 is Mandatory for PAg-Mediated Activation of Vγ9Vδ2 T Cells
PAg act not as soluble molecules but need to be “presented” by cells of human or primate origin (76). This can be seen as evidence for species-specific molecules, which could be PAg-presenting molecules and/or for molecules with special co-stimulatory or cell–cell interaction mediating properties (77, 78). Attempts to identify PAg-binding cell surface molecules by biochemical means, e.g., with the help of photo-activated PAg analogs have failed so far (79, 80) although direct binding of tetramers of rhesus monkey Vγ9Vδ2 TCR to HMBPP pulsed primate dendritic cells was reported (81).
The major breakthrough in defining species-specific molecular requirements for Vγ9Vδ2 T cell activation by PAg was the identification of the pivotal role of Butyrophilin 3 (BTN3) in modulating PAg induced responses (82). BTN3 – also named CD277 – is a membrane protein, which belongs to the butyrophilin family and shares similarity with the B7 family (83). In human beings, three isoforms of BTN3 named BTN3A1, BTN3A2, and BTN3A3 exist whose genes localize to the telomeric end of the MHC complex on the short arm of human chromosome 6 (84). The extracellular domain of BTN3 (BTN3-ED) consists of an N-terminal IgV-like domain (V-domain) and a membrane proximal IgC-like domain (C-domain). The BTN3-ED of the three isoforms is extremely similar and cannot be discriminated by the available antibodies (82, 83). The intracellular domain (ID) of BTN3A1 and BTN3A3 belongs to the B30.2 family (85, 86) and is often found in proteins involved in innate immunity. This includes even molecules encoded by genes found in the hypothetical “proto MHC” of placozoa, which form the most basal branch of Metazoa (87). The BTN3A2 molecule has a truncated intracellular B30.2-negative domain. So far the binding of BTN3 to Vγ9Vδ2 TCR is controversial and a natural ligand or counter-receptor of the BTN3-extracellular domain has not been found yet. Moreover, CD277 antibodies modulate not only responses of Vγ9Vδ2 T cells but also those of αβ T cells and NK cells (88, 89).
The first evidence for a critical role of BTN3 in Vγ9Vδ2 T cell activation came from the agonistic action of the CD277-specific monoclonal antibody (mAb) 20.1 on Vγ9Vδ2 T cells and other CD277-specific antibodies, such as mAb 103.2, being antagonists for PAg-mediated activation (82). Co-cultures of Vγ9Vδ2 T cells or human Vγ9Vδ2 TCR transductants with mAb-pulsed BTN3 positive cells suggested that mAbs work by binding to stimulatory or target cells and not to the human γδ T cells. This was formally proven by demonstrating that mAb 20.1 activates BTN3-negative murine Vγ9Vδ2 TCR transductants in co-cultures with human Raji cells (82). These reporter cells were either mouse hybridoma BW58 (or C58 α-β-) or the rat/mouse T cell hybridoma 53/4-transduced with a Vγ9Vδ2 TCR (TCR-MOP) (82, 90, 91). Interestingly, the PAg-induced IL-2 production by both cell types depends strictly on provision of strong co-stimulatory signal. This can be achieved via ligation of a chimeric rat/mouse CD28 overexpressed on the surface of the reporter cell by CD80 or CD86 on the stimulatory human cell, which can be of natural origin or introduced by gene-transduction (91).
Activation by mAb 20.1 of TCR-MOP transductants but also of primary responder cells is usually less efficient than PAg-induced activation. Nevertheless, it has been shown for primary γδ T cells that PAg- and mAb 20.1-induced activation are quite similar with respect to TCR-mediated signals and phenotypic changes of the cells (92). Furthermore, activation by mAb 20.1 is resistant to statins and not accompanied by accumulation of IPP (80, 82).
Models of PAg-Action: Allosteric Change vs. Antigen Presentation
At present, two major hypotheses to explain PAg-mediated activation of Vγ9Vδ2 T cells compete with each other. One suggests that PAg-binding to the ID of BTN3A1 provokes changes in the BTN3A1-ED either directly or with help of molecules recruited by the ID. These events are accompanied by a reduced mobility of cell surface BTN3A1 and are mandatory for binding of the Vγ9Vδ2 TCR to BTN3A1 or associated molecule(s) alone or in complex with BTN3. We will refer to this concept as the “allosteric model” (82, 93, 94). The other concept suggests a direct binding of BTN3A1-PAg complexes to the Vγ9Vδ2 TCR and describes BTN3A1 as an antigen-presenting molecule. This will be referred to as the “antigen-presentation model” (95).
The Allosteric Model
The work of Harly et al. (82) describes not only the general importance of BTN3 for the PAg response but also that BTN3 isoforms differently support PAg-dependent Vγ9Vδ2 T cell activation while no such differences are found for mAb 20.1-induced activation. Evidence was obtained by comparing BTN3 isoform specific knockdown cells and transductants for their capacity to induce PAg-dependent stimulation of primary Vγ9Vδ2 T cells.
The same pattern was found for the reduction in BTN3-cell surface motility. In photobleaching experiments cells expressing BTN3A1 or BTN3A2 constructs were compared. Aminobisphophonate (pamidronate) pulsed BTN3A1 but not BTN3A2 transductants showed a clear mevastatin-sensitive decrease in cell surface mobility suggesting a PAg-induced BTN3 isoform related effect. In contrast, for mAb 20.1 no differences between the isoforms were revealed although it decreased motility even stronger than PAg. Further important insights came from structural and functional characterization of the BTN3-ED (93). In this study, soluble and crystallized BTN3-ED revealed no major difference between the three BTN3 isoforms and could consequently not help to explain their functional difference. More interestingly, these structural analyses revealed two types of BTN3-homodimers. One had a symmetric paralleled structure where both chains were fixed by interaction of their C-domains. The other one had an asymmetric head to tail conformation with contacts between V- and C-domain, respectively. Importantly, the agonistic but not the antagonistic antibody favors formation of the symmetric dimer. In addition, co-crystals with single chain (sc)20.1 revealed a conformational shift of the BTN3 dimer while sc103.2 has no such effect. These results together with other data lead to model where BTN3 and mAb 20.1 binds with a 2:1 stoichiometry and formed a BTN3 lattice at the cells surface. In contrast, intact mAb 103.2 binds with 1:1 stoichiometry and is expected to inhibit formation of such lattices while no inhibitory effects were seen with sc103.2.
Important clues how IDs might control PAg-dependent stimulation comes from recent crystallographic and functional studies (94). This study first identified by deletion and amino acid exchange mutants a region in the B30.2 domain as critical for PAg-action. Crystallographic analysis identified then a positively charged pocket in this region (the presumed contacts with the PAg are marked in Figure 1B), which could accommodate a PAg. Isothermal calorimetry demonstrated PAg-binding to recombinant BTN3A1-ID, which was considerably stronger for HMBPP than for IPP and was extinguished by the same mutants, which abolished Vγ9Vδ2 TCR activation by respective BTN3 transductants. The importance of the pocket is further corroborated by a single aa mutant in BTN3A1 (H351R), which destroyed the PAg-binding to BTN3A1-ID, the PAg-dependent activation of Vγ9Vδ2 T cells and the zoledronate-induced reduction of cell surface motility. Altogether, evidence is provided that changes induced by PAg-binding to BTN3A1-ID correlate with changes seen in BTN3A1 cell surface motility and PAg-induced Vγ9Vδ2 T cell activation (82, 93, 94). For illustrating the key points of this hypothesis a simplified version of the “allosteric model” is depicted in Figure 2A.
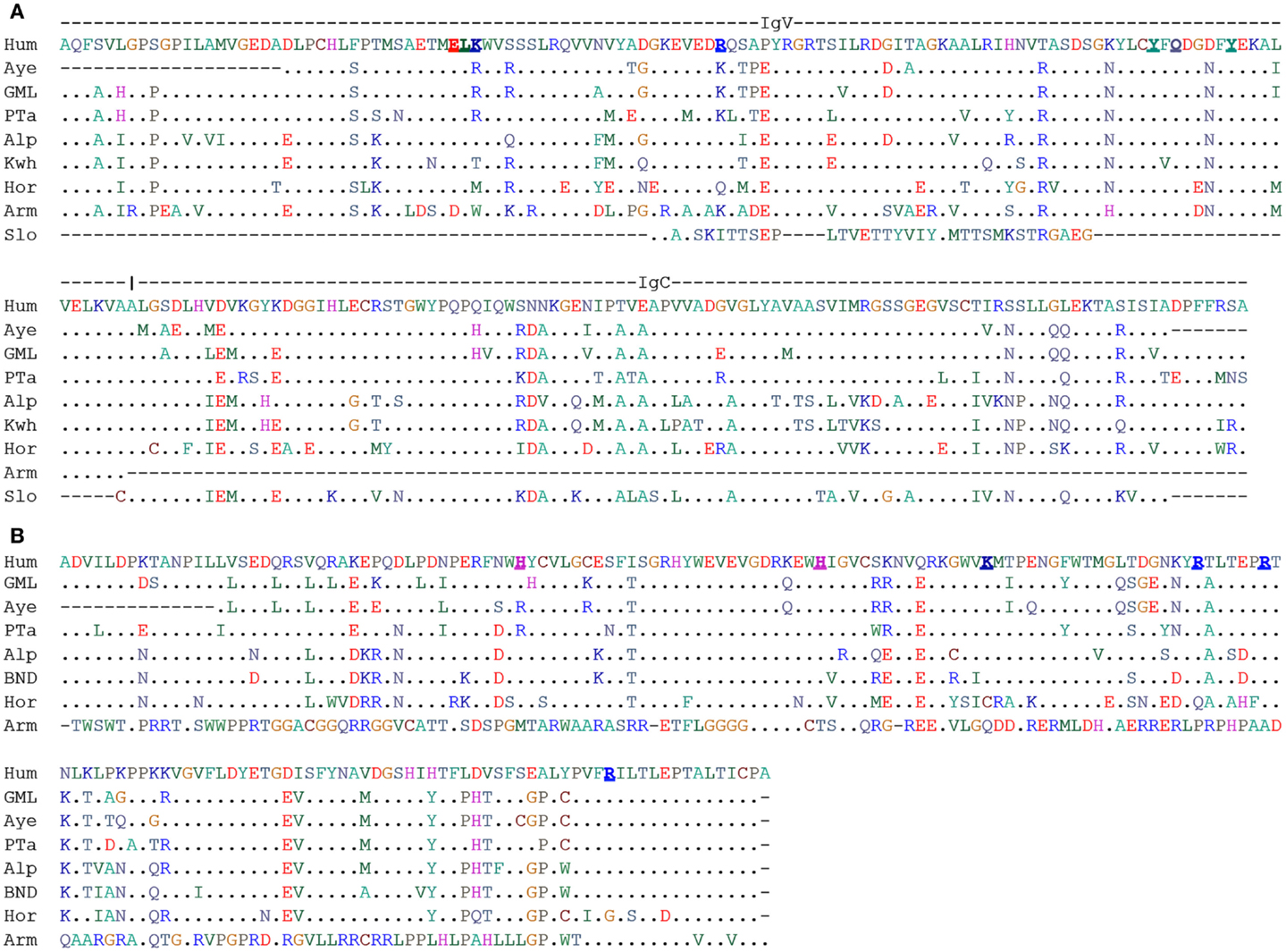
Figure 1. Alignment of human BTN3A1 (V-C) and B30.2 domain. (A) ClustalW2 amino acid alignment of V-C domains of human BTN3A1 extracellular domain with respective homologous sequence identified from WGS database at NCBI. Underlined bold amino acids of human BTN3A1-ED were predicted to interact with PAg (95). (B) ClustalW2 amino acid alignment of intracellular B30.2 domains of human BTN3A1 with respective homologous sequence. Underlined bold amino acids were of human BTN3A1-ID were predicted to interact with PAg (94). Species were abbreviated as Hum, Human; Aye, Aye-aye; GML, Gray mouse lemur; PTa, Philippine tarsier; Alp, Alpaca; Kwh, Killer whales; Hor, Horse; Arm, Armadillo; Slo, Sloth.
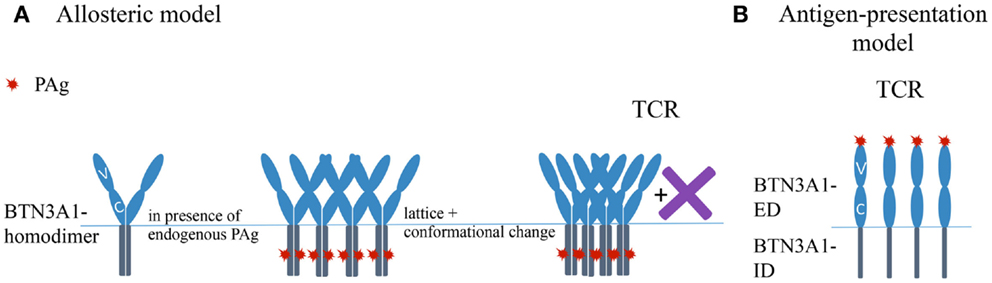
Figure 2. Simplified sketches depicting the “allosteric model” and the “antigen-presenting model.” Note that both models can be partially combined. For reasons of simplicity involvement of hypothetical additional molecule(s) (X) is only depicted were being absolutely necessary. (A) Sketch representing the allosteric model describing possible events involved in PAg mediated activation of Vγ9Vδ2 TCR. X – represents unidentified molecules, which might involve in mechanism. (B) Schematic representation of antigen-presentation model. TCR, T cell receptor; PAg, phosphoantigen.
The Antigen-Presenting Model
Vavassori et al. used a different approach to identify the species-specific factor mandatory for PAg-mediated activation and describe BTN3A1 as an antigen-presenting molecule (95). Their approach takes advantage of the fact that mouse lacks a BTN3 ortholog. At first, murine reporter cells expressing a Vγ9Vδ2 TCR were used to screen mouse-human hybrid cell lines for their capacity to mediate PAg-dependent stimulation with the aim to map this trait to a part of the human genome. By analysis of several of such mouse-hybrid cell lines the telomeric 3–27 Mb region of the human chromosome 6p was found to be mandatory for PAg-presentation. This region comprises the entire MHC as well as the BTN3A1 and BTN3A2 but not the BTN3A3 gene. Thus, genomic localization of the mandatory gene(s) is fully consistent with previously published data that BTN3A1 is mandatory for PAg-mediated activation. The genetic evidence for BTN3A1 as candidate for the molecule involved was further confirmed by knock down and over-expression experiments.
Interestingly, the reporter cells used in this study were not Vγ9Vδ2 TCR-transduced murine hybridoma cells as described above but Vγ9Vδ2 T lymphocytes generated from RAG knock-out mice transgenic for the Vγ9Vδ2 TCR B2G9, which were matured in vivo by administration of anti-CD3 mAb (95, 96). An important difference between data obtained with primary murine reporter cells expressing the Vγ9Vδ2 TCR B2G9 and Vγ9Vδ2 TCR-MOP transduced reporter cells is that the agonistic mAb 20.1 was not stimulatory but inhibitory for the transgenic mouse cells. First results of our group obtained with TCR transductants suggest that this difference reflects variation of the TCR clonotypes, which stands against the idea of mAb 20.1 being a general activator of Vγ9Vδ2 T cells. Nevertheless, to our knowledge, there is no published data on determination of frequencies of mAb 20.1 vs. PAg-reactive cells or direct comparison of sensitivity of different TCR clonotypes for either stimulus supporting this notion. If TCR clonotypes do indeed differ in their sensitivity to both types of stimuli, it would affect models on PAg or mAb 20.1 action. Our interpretation of the presumed clonal differences would rely on substrate competition and inherent qualities of different TCR clonotypes. In the former case, we hypothesize that upon treatment of cells with PAg or mAb 20.1 BTN3A1 adopts a new conformation, which somehow allows binding of Vγ9Vδ2 TCR to BTN3-ED-PAg or mAb complex or to BTN3-ED-associated cell surface molecules(s). This conformation could differ to some extent after exposure of the cell to PAg or mAb 20.1 whereby mAb 20.1 might inhibit conversion into the PAg induced conformation. As a result, some TCR clonotypes cannot bind to the mAb 20.1-induced conformation. Indeed, one could imagine that mAb 20.1-binding “freezes” BTN3-ED in a conformation (93), which is distinct from the PAg-induced one (93, 95). Considering inherent qualities of TCR clonotypes as the basis for their differential capacity in recognizing BTN3A1-ED-PAg complex or BTN3-mAb complex, we propose or speculate that some Vγ9Vδ2 TCR, e.g., TCR B2G9 preferentially bind to a complex of PAg bound to the BTN3A1-ED, whereas others would preferentially bind to the conformationally changed BTN3A1 whose ED does not need to be in complex with the PAg. Consistent with this model would be that the area covered by the mAb 20.1 is rather near to the hypothetical PAg-binding site discussed in the next paragraph. Consequently for some TCR mAb 20.1 would compete with binding of the Vγ9Vδ2 TCR to a BTN3A1-PAg complex while for others mAb 20.1 would still be stimulatory.
De Libero and coworkers (95) provide also a wealth of data in favor of a direct binding of PAg to BTN3A1-ED and of binding of BTN3A1-PAg complexes to the Vγ9Vδ2 TCR: (i) IPP and HMBPP induce a substantial IFNγ secretion by the murine reporter cells cultured in BTN3A1-V domain coated culture plates. (ii) Mass spectrometry data of BTN3A1-V incubated with IPP is consistent with a BTN3A1-IPP complex of 1:1 stoichiometry. (iii) Plasmon resonance analysis of PAg binding to BTN3A1-V domain allowed calculation of Kd values. These are considerably lower than that of MHC-peptide complexes: 66.9 × 10−6 M for binding of BTN3A1-V to IPP and 3.06 × 10−6 M for binding of BTN3A1-V to HMBPP. (iv) Crystal structures of complexes generated from BTN3A1-V and IPP or HMBPP, respectively, identify a shallow Ag-binding groove. The amino acids proposed to interact with HMBPP or IPP respectively are marked in Figure 1A. (v) Plasmon resonance analysis revealed a low affinity binding of recombinant Vγ9Vδ2 TCR multimers (dextramers) (TCR G2B9) to immobilized eukaryotic recombinant BTN3A1-ED (vi) Surface enhanced Raman scattering demonstrates binding of monomeric TCR to BTN3A1 with a Kd of 34 × 10−5 M in the presence and 93 × 10−5 M in the absence of IPP. All these data can be interpreted as evidence that BTN3A1 serves an antigen-presenting molecule (95). A simplified version of this hypothesis is depicted in Figure 2B.
The results and conclusions (95) on BTN3-binding to PAg and Vγ9Vδ2 TCR summarized in the previous paragraph are heavily disputed by proponents of the “allosteric model” (94). In their recent publication, the groups of Adams and Scotet/Bonneville, respectively, give detailed account on failed attempts to detect PAg-binding to the BTN3-ED and of Vγ9Vδ2 TCR (TCR-G115) dextramers to immobilized BTN3-ED. Furthermore, their paper opposes the interpretation of the BTN3-V domain crystallized in presence of PAg as BTN3-PAg complex (94). Based on their analyses of the B30.2 domain and their own negative data on PAg- and TCR-binding, BTN3A1 is refuted as an “antigen-presenting molecule.” Instead, they propose in the discussion section: “a model where PAg binding to the BTN3A1 ID results in recruitment of additional primate-specific factors and/or rearrangement of the BTN3A1 extracellular domain that generates a stimulatory signal directly detected by the Vγ9Vδ2 TCR. This is similar to “Model 2” proposed by Morita and colleagues” (94). The paper of the Morita group (80) mentioned here describes their failed attempts to show binding to BTN3-ED by the use of photo-activatable HMBPP and develops models to explain lacking PAg-binding to BTN3-ED despite its crucial role for the activation of Vγ9Vδ2 T cells.
Some of the data and interpretations reported by the proponents of either model can be reconciled and others not. For example, both agree on the importance of the ID for BTN3A1 mediated PAg activation, and in principle the data on PAg-binding to the BTN3-ID could be adapted in a model with BTN3A1 as an antigen-presenting molecule in following ways: (1) PAg-binding to the BTN3A1-ID may result in intracellular trafficking, e.g., recruitment of a molecule which enables loading of PAg to the BTN3A1-ED and/or involved in proper cell surface distribution of BTN3A1 with the PAg loaded BTN3A1-ED. (2) Another possibility could be that PAg-binding to the BTN3A1-ID induces a conformational change facilitating formation of the BTN-ED-PAg complex or stabilization of such a complex. Concerning the dissent on crystallographic and binding data it would be most desirable – and important for the research community – if controversies on the validity of experimental procedures and their interpretation were solved, e.g., by collaborative attempts of both parties to reproduce data with exchanged reagents, e.g., the different BTN3 preparations and the different TCR clonotypes (TCR-G115 vs. TCR-G2B9). In addition, completely different experimental approaches such as the identification of cofactors controlling PAg-mediated activation by genetic methods might lead to new insights and perspectives and help to solve this controversy.
Evidence for Chr 6 Encoded Gene(s), Which in Addition to BTN3A1 are Mandatory for PAg-Mediated Activation
A first hint that BTN3A1 alone is not the only molecule expressed in primates required for PAg mediated activation comes from experiments where BTN3A1-transduced murine cells pulsed with zoledronate failed to activate human Vγ9Vδ2 T cells (94). Nevertheless, these negative results need to be met with caution since in this xenogeneic setting co-stimulatory receptors and adhesion molecules required for Vγ9Vδ2 TCR-mediated activation might miss their partners on the murine cell and activation may be incomplete.
To solve this problem, murine reporter cells provide a valuable tool. We tested mouse-human hybridomas generated in our laboratory for their capacity to activate reporter cells (TCR-MOP transductants) (97) similarly, as it was done in (95). Consistent with the localization of BTN3A1 on chromosome 6 (Chr. 6) only hybridoma carrying the human Chr. 6 were able to induce PAg-dependent Vγ9Vδ2 TCR responses. To test whether BTN3A1 alone or BTN3A1 and other genes on Chr. 6 allow the PAg-mediated stimulation and whether the same accounts for mAb 20.1-induced activation, different types of Chinese hamster ovary (CHO) cells were tested. We compared CHO cells and CHO cells containing human Chr. 6 and the BTN3A1 transductants of either cell type. In a nutshell, Chr. 6 was found to be sufficient and mandatory to induce activation in the presence of HMBPP and zoledronate while BTN3A1 alone allowed mAb 20.1-induced activation even in the absence of Chr. 6 (97).
These data as well inhibition studies with BTN3-specific antibodies are in full agreement with the pivotal role of BTN3A1 in PAg-mediated activation but imply also the existence of one or more human gene(s) controlling PAg-action, which are missing in rodent cells. Such gene(s) could be involved in PAg-loading onto BTN3A1 analogous to the TAP being mandatory for MHC-class I mediated peptide-presentation. Other possibilities would be, e.g., control of BTN3A1 related signaling pathways steering cellular distribution of BTN3A1 or a factor X directly associated with BTN3A1. In any case identification of these molecules in primates will be essential for the design of rodent models with functional Vγ9Vδ2 T cells, which can be expected to provide fresh insights of Vγ9Vδ2 T cell physiology and facilitate preclinical research on Vγ9Vδ2 T cells and Vγ9Vδ2 T cell-activating drugs (97). The search for such molecule(s) could be facilitated by the murine reporter cells (95, 97), which allow functional screens for the missing genes, e.g., of rodent-human radiation hybrids containing fragments of human Chr. 6 or human expression libraries.
An important implication of the finding that mAb 20.1 permits Vγ9Vδ2 TCR-mediated activation by BTN3A1 expressing rodent cells is that species (human) specific gene(s) can be dispensable for PAg-independent Vγ9Vδ2 TCR-mediated activation. We suggest that mAb 20.1 induces a conformational change of BTN3A1, which allows the Vγ9Vδ2 TCR to directly interact with BTN3A1 or a highly conserved molecule, which upon mAb 20.1 incubation interacts with BTN3A1 and is then recognized by the TCR.
We tested also CD69 up-regulation of human Vγ9Vδ2 T cells with PBMC co-cultured with different CHO cell variants pulsed with zoledronate. Only co-cultures with Chr. 6 consomic CHO cells induced a Vγ9Vδ2 T cell specific activation. Interestingly, we failed to detect CD107a induction suggesting that in this setting Vγ9Vδ2 TCR-mediated activation is incomplete (97). This is of interest since Sandstrom et al. (94) who tested another xenogeneic setting namely co-culture of primary Vγ9Vδ2 T cells with zoledronate or mAb 20.1 pulsed BTN3A1-transduced murine cells used CD107a expression as read out and might have missed a partial activation. Not excluded can be that BTN3A1-transduced CHO cells, which are of hamster origin might express species- or cells-specific factors that are missing in murine BTN3A1 transductants (94) and allowed activation of our reporter cells (TCR-MOP transductant).
Vγ9Vδ2 TCR: The Neglected Interaction with Protein Antigens
The identification of PAg as activator for the vast majority of Vγ9Vδ2 T cells does not exclude other physiological modes of TCR-triggered Vγ9Vδ2 T cell activation. In a series of studies He and colleagues investigated tumor infiltrating γδ T cells, i.e., especially malignancies of ovarial origin, and identified specific ligands whose recognition depended largely on the CDR3δ and CDR3δ flanking regions of the TCR (98–103). This was demonstrated with help of TCR transductants but also with Ig constructs with implanted CDR3δ, which bind to tumors in vitro and trigger tumor elimination in xenografted mice (103). Among the proposed antigens were MutS homolog 2, hsp 60 (99), and the NKG2D ligand ULBP-4 (100). In one case tumor-specific and PAg-mediated activation were compared and mutagenesis of amino acid 97 in the CDR3δ abolished PAg but not an anti-tumor response (102). This CDR3δ-controlled recognition is reminiscent of that of T22 molecules by murine γδ TCR (12, 104) and could also be important for other human γδ T cells. In any case, apart from its therapeutic potential the possibility of PAg-independent tumor recognition by Vγ9Vδ2 T cells demonstrates a function of Vγ9Vδ2 T cells beyond that of a sensor of metabolic aberrations or of microbial metabolites (69). Indeed, it is rather likely that such (oligo)clonal responses might have been missed in the analysis of PAg-responding cells given the presumed low frequency of such protein antigen-specific Vγ9Vδ2 TCR in comparison to PAg-reactive cells. Furthermore, these oligoclonal PAg-independent responses might contribute to the changes in the Vγ9Vδ2 T cell repertoire seen during infections with HIV or with Mycobacterium tuberculosis (49, 105).
Vγ9Vδ2 TCR and BTN3 in Primates
Monkeys including hominids possess Vγ9Vδ2 TCR and functional BTN3A1 genes (80, 93, 106, 107). PAg reactivity of Vγ9Vδ2 T cells has been directly demonstrated for simian species (simiiformes), which include the new world monkey Nancy Ma’s night monkey (Aotus nancymaae) (108) and the common marmorset (Callithrix jacchus) (109) and the old world monkeys rhesus macaque (Macaca mulatta) (110) and cynomologous macaque (Macaca fascicularis) (111), which bear a hydrophobic amino acid at position 97 of delta chain and Vγ9JP rearrangements. Human and most primate species have a K(Lysine)KIK motif in JP, which is sometimes changed to R(Arginine)KIK without apparent consequences for PAg reactivity (107, 110).
An update in September 2014 of our previous searches (107) identified translatable Vγ9, Vδ2, and BTN3A1 sequences in lower primates for the representative of non-simiiforme haplorrhini, the philippine tarsier (Carlito syrichta). The same was found for aye aye (Daubentonia madagascariensis) and the gray mouse lemur (Microcebus murinus) representing the two lemur clades of the strepsirrhini suborder. The BTN3A1 sequences were largely conserved in the proposed PAg-binding motif of the BTN3-ED and in the BTN3-ID. The BTN3-ID of aye aye and tarsier showed the H351R substitution, which abrogates PAg-binding of BTN3A3 (Figure 1). Therefore, it would be interesting to learn whether this also leads to loss of PAg-binding or is compensated by other sequence changes. If PAg-binding of the BTN3-ID was indeed lost and if PAg involvement is needed to maintain the typical Vγ9Vδ2 TCR repertoire then one would expect consequential effects on this repertoire, e.g., less or no Vγ9JP rearrangements.
Vγ9, Vδ2, and BTN3 Co-Emerged and may have Co-Evolved in Placental Mammals
Studies on rodent genomes and on TCR expressed in farm animal species (cow, pig, horse) provided no evidence for Vγ9 or Vδ2 homologs, and it was generally assumed that Vγ9Vδ2 T cells might be restricted to (higher) primates. Taking advantage of increasing number of public data genome data bases, especially the 29 mammals project, which covered all mammalian orders (112), we blasted against genomes of Eutherian mammalian species in search for genes homologous to human Vγ9, Vδ2, and BTN3-ED. We asked for at least 80% sequence coverage and 70% nucleotide identity to human genes in order to identify new species as potential Vγ9Vδ2 T cell carriers. Homology was confirmed by reverse blasting and different types of phylogenetic trees were generated. Most of these results have been published recently (107) but we take the opportunity of this paper to present changes resulting from recent updates in the database and from inclusion of the BTN3-ID in our analysis (Figure 1).
Truly surprising results were obtained from the analysis of non-primate species. The timing of the origin of placental mammals is subject of a protracted debate and has been estimated to occur between 165 and 65 million years ago (113–115). Nevertheless, there is consensus that Xenathra – represented in the databases by the nine-banded armadillo (Dasypus novemcinctus) and the two fingered sloth (Choloepus hoffmanni) belong to a clade distinct from Boreoeutheria, which represents the other placental mammals. Thus, detection of a gene in a species of Xenathra and Boreoeutheria proves its presence in a common placental predecessor. Exactly, this is seen for Vγ9, Vδ2, and BTN3-ED since they are found in sloth and armadillo. While TCR-V genes were rather conserved (107), Figure 1 shows that BTN3-ED of sloth lacks a major part of the V domain while that of armadillo lacks parts of the C domain. A BTN3-ID like domain could not be identified for sloth and the B30.2 domain of armadillo was identified as a homolog of BTN2-ID. With the current knowledge on BTN3-structure-function relationship and PAg-mediated Vγ9Vδ2 T cell activation, it seems unlikely that these molecules could function as proposed for human BTN3. Consequently, we would not expect maintenance of typical Vγ9Vδ2 T cells (e.g., dominance of the characteristic Vγ9JP rearrangements) but if such cells were found, then they should be tested for PAg-reactivity or -binding to other ligands. Testing is especially relevant for armadillo, since it is a natural host Mycobacterium leprae, an important human pathogen and potential target for Vγ9Vδ2 T cells (116).
Many of the tested genomes had lost all three genes. This was true for lagomorphs and most rodents and explains why PAg-reactive cells have never been observed in the classical small animal models (rat, mouse, guinea pig, and rabbit). A notable exception could be the 13-lined squirrel (Spermophilus tridecemlineatus), which conserved a translatable Vγ9 and a presumably functional BTN3 (Figure 1) but lacks a Vδ2. Therefore, it will be of special interest to learn whether this Vδ2 is truly missing or whether the searched database is incomplete. Species of other orders such as those representing Bovidae (cow, sheep, goat, tibetan antelope) kept all three genes but at least one of them was either not translatable (e.g., BTN3-ID of tibetan antelope) or otherwise non-functional as in the case of the horse (Perissodactyla) whose Vγ9 and Vδ2 genes lacked one of the cysteine required for Ig domain disulfide bond (107).
It is very striking that all species (11 species representing 9 families representing 3 of the 4 mammalian superorders) with translatable Vγ9 and Vδ2 TCR gene possess translatable BTN3-ED. The inverse correlation is not as strict, since BTN3A1 of horse (Equidae), white rhinoceros (Rhinocerotidae), several bats (Vespertilionidae), and the thirteen-lined squirrel (Sciuridae) are translatable, although the species lack either functional Vγ9 and/or Vδ2 genes. However, inspection of BTN3-ID (Figure 1) of horse, rhinoceros and bats shows considerable differences in the proposed PAg-binding site suggesting loss of PAg-related function. These species show non-conservative substitutions in the proposed extracellular and intracellular PAg-binding sites while the other species with a complete and translatable BTN3-ED showed no or only conservative changes (K to R and vice versa). Altogether, the data suggest an interdependence in conservation of function between Vγ9, Vδ2, and BTN3 genes and indicate molecular co-evolution of Vγ9Vδ2 TCR and BTN3 (117).
Alpaca (Vicugna pacos) as Model to Study Vγ9Vδ2 T Cells in a Non-Primate Species
Of the nine species originally found to possess translatable Vγ9, Vδ2, and BTN3-ED genes (107), alpaca was the only one accessible for further analysis. Vγ9-Cγ PCR products from PBMC cDNA were cloned and although different Jγ segments were identified, 90% of the clones showed a JP rearrangement. The primate KKIK or RKIK motif is largely conserved in the three alpaca JP, which have a KTIK or RTIK motif. In contrast to human Vγ9Vδ2 T cells, the Vδ2 gene rearranged (25 out of 25 clones) always with a single Jδ gene, which is highly homologous to human Jδ4 (TRDJ*04.) The amino acids L, I, V, and G at position 97, which are typical for PAg reactive human Vγ9Vδ2 TCR were found in 8 out of 17 clones bearing alpaca Vδ2 chains. Similar to human, alpaca Vδ2 chains also show high diversity in CDR3 lengths ranging from 11 to 18 amino acids (38, 107), while CDR3γ length is rather restricted (107). Transduction of 58α-β-cells (BW58) with full length alpaca Vγ9 and Vδ2 TCR chain genes led to their cell surface expression as assessed by surface staining for murine CD3 and detection of IL-2 upon culture with immobilized anti-CD3 antibodies. Due to lack of appropriate reagents, the formal proof of cells concomitantly expressing Vγ9 and Vδ2 TCR chains is still missing.
Nevertheless, at least with respect to BTN3A1 and the TCR genes, there is currently no obvious reason to assume that PAg could not activate alpaca Vγ9Vδ2 T cells. Indeed, the BTN3A1 has been cloned and expressed and showed complete identity with human BTN3 in the published intracellular- and extracellular PAg-binding sites (118). The functionality with respect to PAg-responses will be tested with γδ TCR transductants and camelid stimulator cells but also with alpaca PBMC.
If PAg-reactivity can be confirmed, alpaca will provide an outgroup allowing identification of common denominators of PAg-reactivity of Vγ9Vδ2 T cells, and it will allow analysis of conservation of the molecular mechanisms of PAg-dependent stimulation or presentation, respectively. If PAg-reactivity is missing, then interesting questions arise. Are there alternative modes of Vγ9Vδ2 T cell activation and are they also BTN3A1-dependent? If there is a BTN3-dependent Vγ9Vδ2 T cell activation, new questions arise such as, did other molecules take over the role of PAg and can introduction of human genes (favorably encoded on chromosome 6) generate a BTN3A1-dependent PAg-response similar to that seen in humans?
Identifying Candidate Species for Dendritic Epidermal T Cells by Investigating γδ TCR Genes and Skint1 Homolog in Mammals
At the end of this review, we want to discuss a possible co-evolution of γδ TCR genes and their relation to another member of the butyrophilin family. Dendritic epidermal T cells (DETC) present an extreme case of a highly specialized γδ T cell population, which so far has only been found in mice and rats (28, 119, 120). As the name indicates these T cells have dendritic shape and reside in the epidermis. They appear as the earliest T cells during development in the fetal thymus and subsequently migrate to skin. In the skin, they fulfill TCR-dependent and TCR-independent functions in body barrier surveillance including control of tumor development, skin repair, and allergy control. A hallmark of these cells is expression of a single TCR, which with unique rearrangements containing Vγ3 and Vδ4 (IMGT-nomenclature; other common nomenclatures are Vγ3Vδ1 and Vγ5Vδ1). Canonical DETC can be replaced by cells with a polyclonal TCR repertoire but these do not fulfill all of their functions (15, 25, 30). Mandatory for the development of canonical DETC is the molecule Skint1, which is a member of the butyrophilin family. It consists of a V-C domains containing extracellular domain and a three times transmembrane-spanning domain (31, 121, 122). Its role in DETC development was discovered by analysis of FVB/N mice from Taconics laboratories. These mice showed changes in the DETC TCR repertoire, which were correlated with a skin phenotype (spontaneous ear inflammation and exaggerated irritant contact dermatitis response to tetradecanoyl-phorbol acetate). The genetic basis of this phenotype is a termination mutation in codon 324 of Skint1 immediately upstream of the third transmembrane domain of the molecule (31, 121). In a recent study on structure-function relationship of Skint1, it was found that a tightly regulated cell surface expression on medullary thymic epithelial cells was mandatory for efficient DETC development. Furthermore, mutagenesis and domain exchange proved that each Skint1 domain is non-redundant, including a unique decamer specifying V-domain processing (122) (marked in Figure 3).
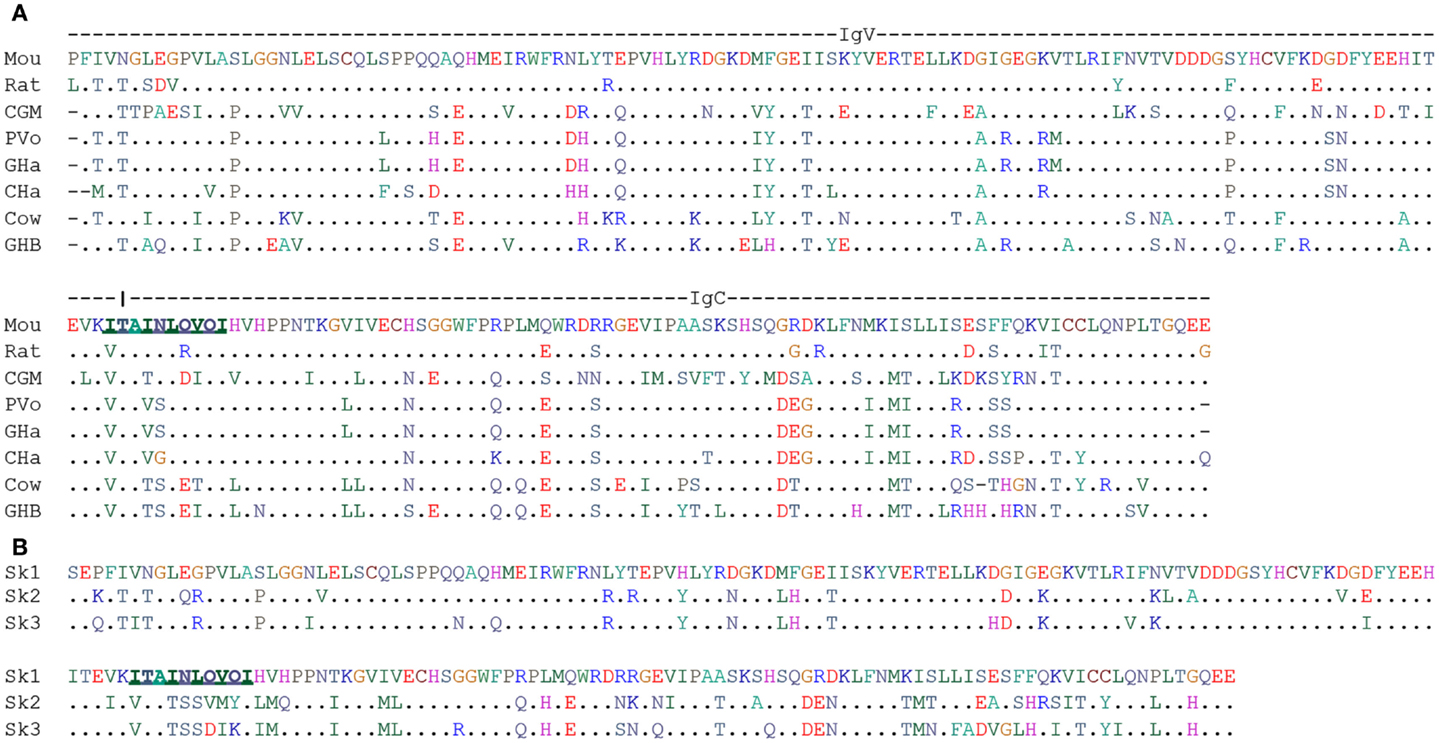
Figure 3. Alignment of human BTN3A1 (V-C) and B30.2 domain. (A) ClustalW2 amino acid alignment of V-C domains of mouse Skint1 extracellular domain with respective homologous sequence identified from WGS database at NCBI. Underlined bold 10 amino acids stretch is predicted to confer functionality for mouse Skint1 (122). (B) ClustalW2 amino acid alignment of V-C domains of mouse Skint1 extracellular domain with its paralogs. Underlined bold 10 amino acids stretch is predicted to confer functionality for mouse Skint1 (122). Species were abbreviated as Mou, Mouse; CGM, Cape golden mole; PVo, Prairie vole; GHa, Golden hamster; CHa, Chinese hamster; GHB, Greater horseshoe bat.
Prompted by the striking concomitant conservation of TCR-V genes and BTN3A1 and the success with identification of alpaca as a candidate for a Vγ9Vδ2 T cell positive species, we decided to search for homologs of DETC TCR-V genes Vγ3 and Vδ4 and of Skint1 as a gene controlling DETC development using the same methods as for BTN3 and Vγ9Vγ2 TCR genes (107). Homologs for at least one of the three genes were identified in 69 species, all of them were Eutheria. No hits were found for Xenathra but the cape golden mole was identified as an afrotherian species being “triple positive.” In case of the TCR-V genes, the assignment was always clear. In case of Skint1, forward blasting of the V-C domains let to identification of genes as Skint1 homolog, which in reverse blast turned out to be Skint2 or Skint3 homologs. We marked this in our table (Table S1 in Supplementary Material) but the limited knowledge about Skint structure and mode of action does not allow to make any predictions to which extent the Skint homologs in these species are redundant – which at least in mouse they are not (122). Also statements on translatability have to be taken with some caution since transmembrane encoding exons were not included in the search.
Nevertheless, the retrieved data (Table S1 in Supplementary Material and Figure 3) could help to identify non-murine species as candidates for a search for rearranged DETC TCR genes and functional Skint: prime candidates are rodent species, which do not belong to the family of murideae but are still phylogenetically not too distant. These would be the two hamster species. Also interesting but unfortunately not yet sequenced at genomic level is the cotton rat (Sigmodon hispidus) and therefore not to be found in the Table S1 in Supplementary Material and Figure 3. This “new world mouse” species is a well-established animal model for a number of infectious diseases (123). Triple positive members were also found in other mammalian superorders than the rodent-containing Euarchontoglires. These are Afrotheria with the golden cape mole (Chrysochloris asiatica) and Laurasiatheria species with the greater horseshoe bat (Rhinolopus ferrumequinum) and the cow (Bos tauris). Nearly, all bats (Chiroptera) carry translatable Vγ3, Vδ4, and either Skint1 or Skint2. Finally, the cow was also triple positive and was the only Artiodactyla species with a translatable Skint. Given the limits of our data-base search, however, at first it needs to be tested whether the latter is still functional. If expression of a Skint1 can be confirmed, it will be of interest to test whether a cow DETC population might be hidden among other populations such as the circulating polyclonal dermal γδ T cells or the small epidermal γδ T cell population (124).
Nevertheless, despite all possible pitfalls in database analysis, we are confident that comparative analysis of TCR genes and genes of putative TCR ligands or of molecules controlling development and function of non-conventional T cells will allow to identify genetic or functional homologs to human non-conventional T cells. The identification of such homologous populations in phylogenetic distant species or species with different life style could help to identify common themes on preservation and flexibility of genes and of functions of such cells. Going back to human beings (or mice) and seeing old acquaintances in a new light may help for a better understanding of the human system and identification of targets of genetic and immune-intervention.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Thomas Göbel, Walter Dressel, Niklas Beyersdorf, and specially Thomas Hünig for comments on the manuscript. This work was supported by the Wilhelm Sander-Stiftung 2013.709.1 and the Deutsche Forschungsgemeinschaft DFG HE 2346/7-1. This publication was funded by the German Research Foundation (DFG) and the University of Wuerzburg in the funding programme Open Access Publishing.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fimmu.2014.00648/abstract
Abbreviations
aa, amino acids; BTN, butyrophilin; BTN3A1-ED, butyrophilin-3 A1 extracellular domain; BTN3A1-ID, butyrophilin-3 A1 intracellular domain; Chr. 6, chromosome 6; DETC, dendritic epidermal T cells; DOXP, 1-deoxy-d-xylulose 5-phosphate pathway; FPPS, farnesyl diphosphate synthase; HMBPP, (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate; IPP, isopentenyl pyrophosphate; mAb, monoclononal antibody; MEP, 2-C-methyl-d-erythritol 4-phosphate pathway; PAg, phosphoantigen; VLR, variable leukocyte receptors.
References
1. Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, Litman GW. Alpha, beta, gamma, and delta T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity (1997) 6(1):1–11. doi:10.1016/S1074-7613(00)80237-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Litman GW, Rast JP, Fugmann SD. The origins of vertebrate adaptive immunity. Nat Rev Immunol (2010) 10(8):543–53. doi:10.1038/nri2807
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet (2010) 11(1):47–59. doi:10.1038/nrg2703
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Richards MH, Nelson JL. The evolution of vertebrate antigen receptors: a phylogenetic approach. Mol Biol Evol (2000) 17(1):146–55. doi:10.1093/oxfordjournals.molbev.a026227
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Glusman G, Rowen L, Lee I, Boysen C, Roach JC, Smit AF, et al. Comparative genomics of the human and mouse T cell receptor loci. Immunity (2001) 15(3):337–49. doi:10.1016/S1074-7613(01)00200-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol (2006) 24:497–518. doi:10.1146/annurev.immunol.24.021605.090542
7. Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD. VLR-based adaptive immunity. Annu Rev Immunol (2012) 30:203–20. doi:10.1146/annurev-immunol-020711-075038
8. Hirano M, Guo P, McCurley N, Schorpp M, Das S, Boehm T, et al. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature (2013) 501(7467):435–8. doi:10.1038/nature12467
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol (2014) 14(6):377–91. doi:10.1038/nri3667
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, Gregers TF, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature (2011) 477(7363):207–10. doi:10.1038/nature10342
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Yin L, Scott-Browne J, Kappler JW, Gapin L, Marrack P. T cells and their eons-old obsession with MHC. Immunol Rev (2012) 250(1):49–60. doi:10.1111/imr.12004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol (2013) 31:529–61. doi:10.1146/annurev-immunol-032712-095912
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Eckle SB, Turner SJ, Rossjohn J, McCluskey J. Predisposed alphabeta T cell antigen receptor recognition of MHC and MHC-I like molecules? Curr Opin Immunol (2013) 25(5):653–9. doi:10.1016/j.coi.2013.07.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Bonneville M, O’Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol (2010) 10(7):467–78. doi:10.1038/nri2781
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Vantourout P, Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol (2013) 13(2):88–100. doi:10.1038/nri3384
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Le Bourhis L, Mburu YK, Lantz O. MAIT cells, surveyors of a new class of antigen: development and functions. Curr Opin Immunol (2013) 25(2):174–80. doi:10.1016/j.coi.2013.01.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol (2012) 12(12):845–57. doi:10.1038/nri3328
18. Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol (2007) 25:297–336. doi:10.1146/annurev.immunol.25.022106.141711
19. Dascher CC. Evolutionary biology of CD1. Curr Top Microbiol Immunol (2007) 314:3–26. doi:10.1007/978-3-540-69511-0_1
20. Looringh van Beeck FA, Reinink P, Hermsen R, Zajonc DM, Laven MJ, Fun A, et al. Functional CD1d and/or NKT cell invariant chain transcript in horse, pig, African elephant and guinea pig, but not in ruminants. Mol Immunol (2009) 46(7):1424–31. doi:10.1016/j.molimm.2008.12.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Monzon-Casanova E, Paletta D, Starick L, Muller I, Sant’Angelo DB, Pyz E, et al. Direct identification of rat iNKT cells reveals remarkable similarities to human iNKT cells and a profound deficiency in LEW rats. Eur J Immunol (2013) 43(2):404–15. doi:10.1002/eji.201242565
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Hein WR, Mackay CR. Prominence of gamma delta T cells in the ruminant immune system. Immunol Today (1991) 12(1):30–4. doi:10.1016/0167-5699(91)90109-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Hein WR. Structural and functional evolution of the extracellular regions of T cell receptors. Semin Immunol (1994) 6(6):361–72. doi:10.1006/smim.1994.1046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. Clark SP, Arden B, Kabelitz D, Mak TW. Comparison of human and mouse T-cell receptor variable gene segment subfamilies. Immunogenetics (1995) 42(6):531–40. doi:10.1007/BF00172178
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Hayday AC. [Gamma][delta] cells: a right time and a right place for a conserved third way of protection. Annu Rev Immunol (2000) 18:975–1026. doi:10.1146/annurev.immunol.18.1.975
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Morita CT, Jin C, Sarikonda G, Wang H. Nonpeptide antigens, presentation mechanisms, and immunological memory of human Vgamma2Vdelta2 T cells: discriminating friend from foe through the recognition of prenyl pyrophosphate antigens. Immunol Rev (2007) 215:59–76. doi:10.1111/j.1600-065X.2006.00479.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Raulet DH. The structure, function, and molecular genetics of the gamma/delta T cell receptor. Annu Rev Immunol (1989) 7:175–207. doi:10.1146/annurev.iy.07.040189.001135
28. Allison JP, Havran WL. The immunobiology of T cells with invariant gamma delta antigen receptors. Annu Rev Immunol (1991) 9:679–705. doi:10.1146/annurev.iy.09.040191.003335
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Tigelaar RE, Lewis JM, Bergstresser PR. TCR gamma/delta+ dendritic epidermal T cells as constituents of skin-associated lymphoid tissue. J Invest Dermatol (1990) 94(6 Suppl):58S–63S. doi:10.1111/1523-1747.ep12875138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Macleod AS, Havran WL. Functions of skin-resident gammadelta T cells. Cell Mol Life Sci (2011) 68(14):2399–408. doi:10.1007/s00018-011-0702-x
31. Boyden LM, Lewis JM, Barbee SD, Bas A, Girardi M, Hayday AC, et al. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet (2008) 40(5):656–62. doi:10.1038/ng.108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Pfeffer K, Schoel B, Gulle H, Kaufmann SH, Wagner H. Primary responses of human T cells to mycobacteria: a frequent set of gamma/delta T cells are stimulated by protease-resistant ligands. Eur J Immunol (1990) 20(5):1175–9. doi:10.1002/eji.1830200534
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. De Libero G, Casorati G, Giachino C, Carbonara C, Migone N, Matzinger P, et al. Selection by two powerful antigens may account for the presence of the major population of human peripheral gamma/delta T cells. J Exp Med (1991) 173(6):1311–22. doi:10.1084/jem.173.6.1311
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Kabelitz D, Bender A, Prospero T, Wesselborg S, Janssen O, Pechhold K. The primary response of human gamma/delta + T cells to Mycobacterium tuberculosis is restricted to V gamma 9-bearing cells. J Exp Med (1991) 173(6):1331–8. doi:10.1084/jem.173.6.1331
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. Casorati G, De Libero G, Lanzavecchia A, Migone N. Molecular analysis of human gamma/delta+ clones from thymus and peripheral blood. J Exp Med (1989) 170(5):1521–35. doi:10.1084/jem.170.5.1521
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Bukowski JF, Morita CT, Band H, Brenner MB. Crucial role of TCR gamma chain junctional region in prenyl pyrophosphate antigen recognition by gamma delta T cells. J Immunol (1998) 161(1):286–93.
37. Miyagawa F, Tanaka Y, Yamashita S, Mikami B, Danno K, Uehara M, et al. Essential contribution of germline-encoded lysine residues in Jgamma1.2 segment to the recognition of nonpeptide antigens by human gammadelta T cells. J Immunol (2001) 167(12):6773–9. doi:10.4049/jimmunol.167.12.6773
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Wang H, Fang Z, Morita CT. Vgamma2Vdelta2 T cell receptor recognition of prenyl pyrophosphates is dependent on all CDRs. J Immunol (2010) 184(11):6209–22. doi:10.4049/jimmunol.1000231
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Nishimura H, Hirokawa M, Fujishima N, Fujishima M, Miura I, Sawada K. Contribution of complementarity-determining region 3 of the T-cell receptor Vdelta2 chain to the recognition of aminobisphosphonates by human gammadelta T-cells. Int J Hematol (2004) 79(4):369–76. doi:10.1532/IJH97.03157
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Pont F, Familiades J, Dejean S, Fruchon S, Cendron D, Poupot M, et al. The gene expression profile of phosphoantigen-specific human gammadelta T lymphocytes is a blend of alphabeta T-cell and NK-cell signatures. Eur J Immunol (2012) 42(1):228–40. doi:10.1002/eji.201141870
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Caccamo N, La Mendola C, Orlando V, Meraviglia S, Todaro M, Stassi G, et al. Differentiation, phenotype, and function of interleukin-17-producing human Vgamma9Vdelta2 T cells. Blood (2011) 118(1):129–38. doi:10.1182/blood-2011-01-331298
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Khan MW, Curbishley SM, Chen HC, Thomas AD, Pircher H, Mavilio D, et al. Expanded human blood-derived gammadelta T cells display potent antigen-presentation functions. Front Immunol (2014) 5:344. doi:10.3389/fimmu.2014.00344
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Scotet E, Nedellec S, Devilder MC, Allain S, Bonneville M. Bridging innate and adaptive immunity through gammadelta T-dendritic cell crosstalk. Front Biosci (2008) 13:6872–85. doi:10.2741/3195
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Nussbaumer O, Gruenbacher G, Gander H, Thurnher M. DC-like cell-dependent activation of human natural killer cells by the bisphosphonate zoledronic acid is regulated by gammadelta T lymphocytes. Blood (2011) 118(10):2743–51. doi:10.1182/blood-2011-01-328526
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Eberl M, Moser B. Monocytes and gammadelta T cells: close encounters in microbial infection. Trends Immunol (2009) 30(12):562–8. doi:10.1016/j.it.2009.09.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Castella B, Vitale C, Coscia M, Massaia M. Vgamma9Vdelta2 T cell-based immunotherapy in hematological malignancies: from bench to bedside. Cell Mol Life Sci (2011) 68(14):2419–32. doi:10.1007/s00018-011-0704-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Braza MS, Klein B. Anti-tumour immunotherapy with Vgamma9Vdelta2 T lymphocytes: from the bench to the bedside. Br J Haematol (2013) 160(2):123–32. doi:10.1111/bjh.12090
48. Chen ZW. Immune biology of Ag-specific gammadelta T cells in infections. Cell Mol Life Sci (2011) 68(14):2409–17. doi:10.1007/s00018-011-0703-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Chen ZW. Multifunctional immune responses of HMBPP-specific Vgamma2Vdelta2 T cells in M. tuberculosis and other infections. Cell Mol Immunol (2013) 10(1):58–64. doi:10.1038/cmi.2012.46
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Ribot JC, deBarros A, Silva-Santos B. Searching for “signal 2”: costimulation requirements of gammadelta T cells. Cell Mol Life Sci (2011) 68(14):2345–55. doi:10.1007/s00018-011-0698-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Moris A, Rothenfusser S, Meuer E, Hangretinger R, Fisch P. Role of gammadelta T cells in tumor immunity and their control by NK receptors. Microbes Infect (1999) 1(3):227–34. doi:10.1016/S1286-4579(99)80038-0
52. De Libero G. Control of gammadelta T cells by NK receptors. Microbes Infect (1999) 1(3):263–7. doi:10.1016/S1286-4579(99)80043-4
53. Rincon-Orozco B, Kunzmann V, Wrobel P, Kabelitz D, Steinle A, Herrmann T. Activation of V gamma 9V delta 2 T cells by NKG2D. J Immunol (2005) 175(4):2144–51. doi:10.4049/jimmunol.175.4.2144
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Tanaka Y, Morita CT, Tanaka Y, Nieves E, Brenner MB, Bloom BR. Natural and synthetic non-peptide antigens recognized by human gamma delta T cells. Nature (1995) 375(6527):155–8. doi:10.1038/375155a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Amslinger S, Hecht S, Rohdich F, Eisenreich W, Adam P, Bacher A, et al. Stimulation of Vgamma9/Vdelta2 T-lymphocyte proliferation by the isoprenoid precursor, (E)-1-hydroxy-2-methyl-but-2-enyl 4-diphosphate. Immunobiology (2007) 212(1):47–55. doi:10.1016/j.imbio.2006.08.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Eberl M, Hintz M, Reichenberg A, Kollas AK, Wiesner J, Jomaa H. Microbial isoprenoid biosynthesis and human gammadelta T cell activation. FEBS Lett (2003) 544(1–3):4–10. doi:10.1016/S0014-5793(03)00483-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Kohler S, Delwiche CF, Denny PW, Tilney LG, Webster P, Wilson RJ, et al. A plastid of probable green algal origin in apicomplexan parasites. Science (1997) 275(5305):1485–9. doi:10.1126/science.275.5305.1485
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Yeh E, DeRisi JL. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol (2011) 9(8):e1001138. doi:10.1371/journal.pbio.1001138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Kunzmann V, Bauer E, Feurle J, Weissinger F, Tony HP, Wilhelm M. Stimulation of gammadelta T cells by aminobisphosphonates and induction of antiplasma cell activity in multiple myeloma. Blood (2000) 96(2):384–92.
60. Wilhelm M, Kunzmann V, Eckstein S, Reimer P, Weissinger F, Ruediger T, et al. Gammadelta T cells for immune therapy of patients with lymphoid malignancies. Blood (2003) 102(1):200–6. doi:10.1182/blood-2002-12-3665
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Das H, Wang L, Kamath A, Bukowski JF. Vgamma2Vdelta2 T-cell receptor-mediated recognition of aminobisphosphonates. Blood (2001) 98(5):1616–8. doi:10.1182/blood.V98.5.1616
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Thompson K, Rogers MJ. Statins prevent bisphosphonate-induced gamma,delta-T-cell proliferation and activation in vitro. J Bone Miner Res (2004) 19(2):278–88. doi:10.1359/JBMR.0301230
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Li J, Herold MJ, Kimmel B, Muller I, Rincon-Orozco B, Kunzmann V, et al. Reduced expression of the mevalonate pathway enzyme farnesyl pyrophosphate synthase unveils recognition of tumor cells by Vgamma9Vdelta2 T cells. J Immunol (2009) 182(12):8118–24. doi:10.4049/jimmunol.0900101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Wang H, Sarikonda G, Puan KJ, Tanaka Y, Feng J, Giner JL, et al. Indirect stimulation of human Vgamma2Vdelta2 T cells through alterations in isoprenoid metabolism. J Immunol (2011) 187(10):5099–113. doi:10.4049/jimmunol.1002697
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Kistowska M, Rossy E, Sansano S, Gober HJ, Landmann R, Mori L, et al. Dysregulation of the host mevalonate pathway during early bacterial infection activates human TCR gamma delta cells. Eur J Immunol (2008) 38(8):2200–9. doi:10.1002/eji.200838366
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Gober HJ, Kistowska M, Angman L, Jeno P, Mori L, De Libero G. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med (2003) 197(2):163–8. doi:10.1084/jem.20021500
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Bukowski JF, Morita CT, Brenner MB. Human gamma delta T cells recognize alkylamines derived from microbes, edible plants, and tea: implications for innate immunity. Immunity (1999) 11(1):57–65. doi:10.1016/S1074-7613(00)80081-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Thompson K, Rojas-Navea J, Rogers MJ. Alkylamines cause Vgamma9Vdelta2 T-cell activation and proliferation by inhibiting the mevalonate pathway. Blood (2006) 107(2):651–4. doi:10.1182/blood-2005-03-1025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. De Libero G. Sentinel function of broadly reactive human gamma delta T cells. Immunol Today (1997) 18(1):22–6. doi:10.1016/S0167-5699(97)80010-2
70. Kunzmann V, Wilhelm M. Adjuvant zoledronic acid for breast cancer: mechanism of action? Lancet Oncol (2011) 12(11):991–2. doi:10.1016/S1470-2045(11)70252-2
71. Fisch P, Malkovsky M, Braakman E, Sturm E, Bolhuis RL, Prieve A, et al. Gamma/delta T cell clones and natural killer cell clones mediate distinct patterns of non-major histocompatibility complex-restricted cytolysis. J Exp Med (1990) 171(5):1567–79. doi:10.1084/jem.171.5.1567
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Davodeau F, Peyrat MA, Hallet MM, Gaschet J, Houde I, Vivien R, et al. Close correlation between Daudi and mycobacterial antigen recognition by human gamma delta T cells and expression of V9JPC1 gamma/V2DJC delta-encoded T cell receptors. J Immunol (1993) 151(3):1214–23.
73. Monkkonen H, Auriola S, Lehenkari P, Kellinsalmi M, Hassinen IE, Vepsalainen J, et al. A new endogenous ATP analog (ApppI) inhibits the mitochondrial adenine nucleotide translocase (ANT) and is responsible for the apoptosis induced by nitrogen-containing bisphosphonates. Br J Pharmacol (2006) 147(4):437–45. doi:10.1038/sj.bjp.0706628
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Champagne E. Gammadelta T cell receptor ligands and modes of antigen recognition. Arch Immunol Ther Exp (Warsz) (2011) 59(2):117–37. doi:10.1007/s00005-011-0118-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Scotet E, Martinez LO, Grant E, Barbaras R, Jeno P, Guiraud M, et al. Tumor recognition following Vgamma9Vdelta2 T cell receptor interactions with a surface F1-ATPase-related structure and apolipoprotein A-I. Immunity (2005) 22(1):71–80. doi:10.1016/j.immuni.2004.11.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Morita CT, Beckman EM, Bukowski JF, Tanaka Y, Band H, Bloom BR, et al. Direct presentation of nonpeptide prenyl pyrophosphate antigens to human gamma delta T cells. Immunity (1995) 3(4):495–507. doi:10.1016/1074-7613(95)90178-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Kato Y, Tanaka Y, Tanaka H, Yamashita S, Minato N. Requirement of species-specific interactions for the activation of human gamma delta T cells by pamidronate. J Immunol (2003) 170(7):3608–13. doi:10.4049/jimmunol.170.7.3608
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Green AE, Lissina A, Hutchinson SL, Hewitt RE, Temple B, James D, et al. Recognition of nonpeptide antigens by human V gamma 9V delta 2 T cells requires contact with cells of human origin. Clin Exp Immunol (2004) 136(3):472–82. doi:10.1111/j.1365-2249.2004.02472.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Sarikonda G, Wang H, Puan KJ, Liu XH, Lee HK, Song Y, et al. Photoaffinity antigens for human gammadelta T cells. J Immunol (2008) 181(11):7738–50. doi:10.4049/jimmunol.181.11.7738
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Wang H, Henry O, Distefano MD, Wang YC, Raikkonen J, Monkkonen J, et al. Butyrophilin 3A1 plays an essential role in prenyl pyrophosphate stimulation of human Vgamma2Vdelta2 T cells. J Immunol (2013) 191(3):1029–42. doi:10.4049/jimmunol.1300658
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Wei H, Huang D, Lai X, Chen M, Zhong W, Wang R, et al. Definition of APC presentation of phosphoantigen (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate to Vgamma2Vdelta 2 TCR. J Immunol (2008) 181(7):4798–806. doi:10.4049/jimmunol.181.7.4798
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Harly C, Guillaume Y, Nedellec S, Peigne CM, Monkkonen H, Monkkonen J, et al. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human gammadelta T-cell subset. Blood (2012) 120(11):2269–79. doi:10.1182/blood-2012-05-430470
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Compte E, Pontarotti P, Collette Y, Lopez M, Olive D. Frontline: characterization of BT3 molecules belonging to the B7 family expressed on immune cells. Eur J Immunol (2004) 34(8):2089–99. doi:10.1002/eji.200425227
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Rhodes DA, Stammers M, Malcherek G, Beck S, Trowsdale J. The cluster of BTN genes in the extended major histocompatibility complex. Genomics (2001) 71(3):351–62. doi:10.1006/geno.2000.6406
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Abeler-Dorner L, Swamy M, Williams G, Hayday AC, Bas A. Butyrophilins: an emerging family of immune regulators. Trends Immunol (2012) 33(1):34–41. doi:10.1016/j.it.2011.09.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Afrache H, Gouret P, Ainouche S, Pontarotti P, Olive D. The butyrophilin (BTN) gene family: from milk fat to the regulation of the immune response. Immunogenetics (2012) 64(11):781–94. doi:10.1007/s00251-012-0619-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Suurvali J, Jouneau L, Thepot D, Grusea S, Pontarotti P, Du Pasquier L, et al. The proto-MHC of placozoans, a region specialized in cellular stress and ubiquitination/proteasome pathways. J Immunol (2014) 193(6):2891–901. doi:10.4049/jimmunol.1401177
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Yamashiro H, Yoshizaki S, Tadaki T, Egawa K, Seo N. Stimulation of human butyrophilin 3 molecules results in negative regulation of cellular immunity. J Leukoc Biol (2010) 88(4):757–67. doi:10.1189/jlb.0309156
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Messal N, Mamessier E, Sylvain A, Celis-Gutierrez J, Thibult ML, Chetaille B, et al. Differential role for CD277 as a co-regulator of the immune signal in T and NK cells. Eur J Immunol (2011) 41(12):3443–54. doi:10.1002/eji.201141404
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Kreiss M, Asmuss A, Krejci K, Lindemann D, Miyoshi-Akiyama T, Uchiyama T, et al. Contrasting contributions of complementarity-determining region 2 and hypervariable region 4 of rat BV8S2+ (Vbeta8.2) TCR to the recognition of myelin basic protein and different types of bacterial superantigens. Int Immunol (2004) 16(5):655–63. doi:10.1093/intimm/dxh068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Li J. Modulating the Expression of Enzymes of Isoprenoid Synthesis: Effects on Vgamma9Vdelta2 T Cell Activation and Tumor Cell Growth [PhD]. Würzburg: Julius Maximilians Universität Würzburg (2010).
92. Decaup E, Duault C, Bezombes C, Poupot M, Savina A, Olive D, et al. Phosphoantigens and butyrophilin 3A1 induce similar intracellular activation signaling in human TCRVgamma9+ gammadelta T lymphocytes. Immunol Lett (2014) 161(1):133–7. doi:10.1016/j.imlet.2014.05.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Palakodeti A, Sandstrom A, Sundaresan L, Harly C, Nedellec S, Olive D, et al. The molecular basis for modulation of human Vgamma9Vdelta2 T cell responses by CD277/butyrophilin-3 (BTN3A)-specific antibodies. J Biol Chem (2012) 287(39):32780–90. doi:10.1074/jbc.M112.384354
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Sandstrom A, Peigne CM, Leger A, Crooks JE, Konczak F, Gesnel MC, et al. The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vgamma9Vdelta2 T cells. Immunity (2014) 40(4):490–500. doi:10.1016/j.immuni.2014.03.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Vavassori S, Kumar A, Wan GS, Ramanjaneyulu GS, Cavallari M, El Daker S, et al. Butyrophilin 3A1 binds phosphorylated antigens and stimulates human gammadelta T cells. Nat Immunol (2013) 14(9):908–16. doi:10.1038/ni.2665
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Kistowska M. Antigen Recognition and thymic maturation of human TCR Vgamma9-Vdelta2 cells. Basel: Universität Basel (2007).
97. Riano F, Karunakaran MM, Starick L, Li J, Scholz CJ, Kunzmann V, et al. Vgamma9Vdelta2 TCR-activation by phosphorylated antigens requires butyrophilin 3 A1 (BTN3A1) and additional genes on human chromosome 6. Eur J Immunol (2014) 44(9):2571–6. doi:10.1002/eji.201444712
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Xu C, Zhang H, Hu H, He H, Wang Z, Xu Y, et al. Gammadelta T cells recognize tumor cells via CDR3delta region. Mol Immunol (2007) 44(4):302–10. doi:10.1016/j.molimm.2006.03.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Chen H, He X, Wang Z, Wu D, Zhang H, Xu C, et al. Identification of human T cell receptor gammadelta-recognized epitopes/proteins via CDR3delta peptide-based immunobiochemical strategy. J Biol Chem (2008) 283(18):12528–37. doi:10.1074/jbc.M708067200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Kong Y, Cao W, Xi X, Ma C, Cui L, He W. The NKG2D ligand ULBP4 binds to TCRgamma9/delta2 and induces cytotoxicity to tumor cells through both TCRgammadelta and NKG2D. Blood (2009) 114(2):310–7. doi:10.1182/blood-2008-12-196287
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Xi X, Guo Y, Chen H, Xu C, Zhang H, Hu H, et al. Antigen specificity of gammadelta T cells depends primarily on the flanking sequences of CDR3delta. J Biol Chem (2009) 284(40):27449–55. doi:10.1074/jbc.M109.011684
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Xi X, Cui L, He W. The recognition of gammadelta TCR to protein antigen does not depend on the hydrophobic I97 residue of CDR3delta. Int Immunol (2010) 22(4):299–306. doi:10.1093/intimm/dxq011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Zheng J, Guo Y, Ji X, Cui L, He W. A novel antibody-like TCRgammadelta-Ig fusion protein exhibits antitumor activity against human ovarian carcinoma. Cancer Lett (2013) 341(2):150–8. doi:10.1016/j.canlet.2013.07.036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Adams EJ, Chien YH, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science (2005) 308(5719):227–31. doi:10.1126/science.1106885
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Li H, Chaudhry S, Poonia B, Shao Y, Pauza CD. Depletion and dysfunction of Vgamma2Vdelta2 T cells in HIV disease: mechanisms, impacts and therapeutic implications. Cell Mol Immunol (2013) 10(1):42–9. doi:10.1038/cmi.2012.50
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Kazen AR, Adams EJ. Evolution of the V, D, and J gene segments used in the primate gammadelta T-cell receptor reveals a dichotomy of conservation and diversity. Proc Natl Acad Sci U S A (2011) 108(29):E332–40. doi:10.1073/pnas.1105105108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
107. Karunakaran MM, Gobel TW, Starick L, Walter L, Herrmann T. Vgamma9 and Vdelta2 T cell antigen receptor genes and butyrophilin 3 (BTN3) emerged with placental mammals and are concomitantly preserved in selected species like alpaca (Vicugna pacos). Immunogenetics (2014) 66(4):243–54. doi:10.1007/s00251-014-0763-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
108. Daubenberger CA, Salomon M, Vecino W, Hubner B, Troll H, Rodriques R, et al. Functional and structural similarity of V gamma 9V delta 2 T cells in humans and Aotus monkeys, a primate infection model for Plasmodium falciparum malaria. J Immunol (2001) 167(11):6421–30. doi:10.4049/jimmunol.167.11.6421
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
109. Rowland CA, Laws TR, Oyston PC. An assessment of common marmoset (Callithrix jacchus) gamma9(+) T cells and their response to phosphoantigen in vitro. Cell Immunol (2012) 280(2):132–7. doi:10.1016/j.cellimm.2012.12.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
110. Wang H, Lee HK, Bukowski JF, Li H, Mariuzza RA, Chen ZW, et al. Conservation of nonpeptide antigen recognition by rhesus monkey V gamma 2V delta2 T cells. J Immunol (2003) 170(7):3696–706. doi:10.4049/jimmunol.170.7.3696
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Ali Z, Shao L, Halliday L, Reichenberg A, Hintz M, Jomaa H, et al. Prolonged (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate-driven antimicrobial and cytotoxic responses of pulmonary and systemic Vgamma2Vdelta2 T cells in macaques. J Immunol (2007) 179(12):8287–96. doi:10.4049/jimmunol.179.12.8287
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature (2011) 478(7370):476–82. doi:10.1038/nature10530
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, et al. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science (2011) 334(6055):521–4. doi:10.1126/science.1211028
114. O’Leary MA, Bloch JI, Flynn JJ, Gaudin TJ, Giallombardo A, Giannini NP, et al. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science (2013) 339(6120):662–7. doi:10.1126/science.1229237
115. dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PC, Yang Z. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc Biol Sci (2012) 279(1742):3491–500. doi:10.1098/rspb.2012.0683
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Sharma R, Lahiri R, Scollard DM, Pena M, Williams DL, Adams LB, et al. The armadillo: a model for the neuropathy of leprosy and potentially other neurodegenerative diseases. Dis Model Mech (2013) 6(1):19–24. doi:10.1242/dmm.010215
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
117. Lovell SC, Robertson DL. An integrated view of molecular coevolution in protein-protein interactions. Mol Biol Evol (2010) 27(11):2567–75. doi:10.1093/molbev/msq144
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
118. Karunakaran MM. Evolution of Vgamma9Vdelta2 T cells [PhD]. Würzburg: Julius Maximilians Universität Würzburg (2014).
119. Stingl G, Koning F, Yamada H, Yokoyama WM, Tschachler E, Bluestone JA, et al. Thy-1+ dendritic epidermal cells express T3 antigen and the T-cell receptor gamma chain. Proc Natl Acad Sci U S A (1987) 84(13):4586–90. doi:10.1073/pnas.84.13.4586
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
120. Kuhnlein P, Mitnacht R, Torres-Nagel NE, Herrmann T, Elbe A, Hunig T. The canonical T cell receptor of dendritic epidermal gamma delta T cells is highly conserved between rats and mice. Eur J Immunol (1996) 26(12):3092–7. doi:10.1002/eji.1830261240
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
121. Lewis JM, Girardi M, Roberts SJ, Barbee SD, Hayday AC, Tigelaar RE. Selection of the cutaneous intraepithelial gammadelta+ T cell repertoire by a thymic stromal determinant. Nat Immunol (2006) 7(8):843–50. doi:10.1038/ni1363
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
122. Barbee SD, Woodward MJ, Turchinovich G, Mention JJ, Lewis JM, Boyden LM, et al. Skint-1 is a highly specific, unique selecting component for epidermal T cells. Proc Natl Acad Sci U S A (2011) 108(8):3330–5. doi:10.1073/pnas.1010890108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
123. Green MG, Huey D, Niewiesk S. The cotton rat (Sigmodon hispidus) as an animal model for respiratory tract infections with human pathogens. Lab Anim (2013) 42(5):170–6. doi:10.1038/laban.188
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
124. Hein WR, Dudler L. TCR gamma delta+ cells are prominent in normal bovine skin and express a diverse repertoire of antigen receptors. Immunology (1997) 91(1):58–64. doi:10.1046/j.1365-2567.1997.00224.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: γδ T cell, Vγ9Vδ2 T cell, phosphoantigen, BTN3, alpaca, co-evolution, DETC and Skint1
Citation: Karunakaran MM and Herrmann T (2014) The Vγ9Vδ2 T cell antigen receptor and butyrophilin-3 A1: models of interaction, the possibility of co-evolution, and the case of dendritic epidermal T cells. Front. Immunol. 5:648. doi: 10.3389/fimmu.2014.00648
Received: 30 September 2014; Paper pending published: 10 November 2014;
Accepted: 04 December 2014; Published online: 19 December 2014.
Edited by:
Dieter Kabelitz, Christian-Albrechts University Kiel, GermanyReviewed by:
David L. Wiest, Fox Chase Cancer Center, USADieter Kabelitz, Christian-Albrechts University Kiel, Germany
Christelle Harly, National Institutes of Health, USA
Copyright: © 2014 Karunakaran and Herrmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Herrmann, Institut für Virologie und Immunbiologie, Versbacher Strasse 7, Würzburg 97078, Germany e-mail:aGVycm1hbm4tdEB2aW0udW5pLXd1ZXJ6YnVyZy5kZQ==
 Mohindar M. Karunakaran
Mohindar M. Karunakaran Thomas Herrmann
Thomas Herrmann