- 1Department of Surgery, Duke University, Durham, NC, United States
- 2Duke Human Vaccine Institute, Durham, NC, United States
- 3Center for Human Systems Immunology, Durham, NC, United States
- 4Department of Biostatistics and Bioinformatics, Duke University, Durham, NC, United States
- 5Department of Medicine, Duke University, Durham, NC, United States
- 6New Iberia Research Center, University of Louisiana at Lafayette, New Iberia, LA, United States
- 7U.S. Military HIV Research Program, Walter Reed Army Institute of Research, Silver Spring, MD, United States
- 8Department of Pediatrics, Duke University, Durham, NC, United States
- 9Department of Integrative Immunobiology, Duke University, Durham, NC, United States
- 10Department of Molecular Genetics and Microbiology, Duke University, Durham, NC, United States
- 11Center for Virology and Vaccine Research, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA, United States
Rhesus macaques (RMs) are a common pre-clinical model used to test HIV vaccine efficacy and passive immunization strategies. Yet, it remains unclear to what extent the Fc-Fc receptor (FcR) interactions impacting antiviral activities of antibodies in RMs recapitulate those in humans. Here, we evaluated the FcR-related functionality of natural killer cells (NKs) from peripheral blood of uninfected humans and RMs to identify intra- and inter-species variation. NKs were screened for FcγRIIIa (human) and FcγRIII (RM) genotypes (FcγRIII(a)), receptor signaling, and antibody-dependent cellular cytotoxicity (ADCC), the latter mediated by a cocktail of monoclonal IgG1 antibodies with human or RM Fc. FcγRIII(a) genetic polymorphisms alone did not explain differences in NK effector functionality in either species cohort. Using the same parameters, hierarchical clustering separated each species into two clusters. Importantly, in principal components analyses, ADCC magnitude, NK contribution to ADCC, FcγRIII(a) cell-surface expression, and frequency of phosphorylated CD3ζ NK cells all contributed similarly to the first principal component within each species, demonstrating the importance of measuring multiple facets of NK cell function. Although ADCC potency was similar between species, we detected significant differences in frequencies of NK cells and pCD3ζ+ cells, level of cell-surface FcγRIII(a) expression, and NK-mediated ADCC (P<0.001), indicating that a combination of Fc-FcR parameters contribute to overall inter-species functional differences. These data strongly support the importance of multi-parameter analyses of Fc-FcR NK-mediated functions when evaluating efficacy of passive and active immunizations in pre- and clinical trials and identifying correlates of protection. The results also suggest that pre-screening animals for multiple FcR-mediated NK function would ensure even distribution of animals among treatment groups in future preclinical trials.
1 Introduction
Natural killer (NK) cells are critical for protection against, control, and clearance of human immunodeficiency virus type-1 (HIV-1) and simian immunodeficiency virus (SIV) infections (1–4). One of the important NK cell functions is antibody-mediated recognition and elimination of infected cells, known as antibody-dependent cellular cytotoxicity (ADCC). ADCC has been reported to play a role in lowering the risk of and protecting from HIV-1 infection in preclinical (5–7) and clinical studies (8–10) through recruitment of NK cells via the low affinity receptor that binds to the Fc portion of IgG, FcγRIIIa (CD16) or RM FcγRIII, the homologue for human FcγRIIIa. Throughout, we will use the nomenclature FcγRIII(a) when referring to both human FcγRIIIa and RM FcγRIII. The analysis of the FcγRIIIa allelic polymorphism in humans has revealed that a single nucleotide T to G substitution at position 559 in the second extracellular domain of FcγRIIIa results in an amino acid change from phenylalanine (F) at amino acid position 158 to valine (V) (11). The V/V, V/F, and F/F genotypes have been found to display different affinities for the IgG subclasses (12–15), with V/V resulting in the highest affinity for IgG1 and IgG3 (12). It has further been reported how these differences in FcγRIIIa polymorphisms influence predisposition to autoimmunity, impact cancer immunotherapy and infectious disease outcomes (13, 16–20). For instance, patients homozygous for the FcγRIIIa 158V respond better to treatment with low concentrations of anti-CD20 monoclonal Ab (mAb) rituximab in B cell malignancies (21), however, the difference in in vitro binding between different FcγRIIIa polymorphisms is lost with high concentrations of rituximab (22). Conversely, HIV-1-protein vaccinees homozygous for the FcγRIIIa V allele were more susceptible to HIV-1 infection than vaccine recipients with at least one F allele (23). It is important to note that FcγRIIIa genetic polymorphisms have not demonstrated reproducible association with HIV-1 acquisition, progression or vaccine efficacy, suggesting that further investigation of the impact of genetic variations on HIV-1 disease pathogenesis is needed (24).
NK cell functionality typically requires a net triggering of activating over inhibitory signals on the cell surface (25). Among the activation signals, triggering through FcγRIIIa can initiate the most robust signals for NK cell activation and even act as a mono-signal (26). NK cells do not, however, uniformly express FcγRIIIa and its various signaling adaptors. In direct relation to ADCC, previous research conducted by Shah et al. indicated that a subset of adaptive NK cells expresses FcγRIIIa but forgo the typical FcRγ/SYK pathway in favor of the CD3Ç heterodimer or homodimer and ZAP70, resulting in more potent antiviral function (27). Of note, this subset of NK cells, termed FcγRΔg NK cells, have been shown to exhibit enhanced ADCC against HIV/SIV, influenza, and HSV-1 antigens (27–30). Based on these data, we decided to focus our attention on the phosphorylation of CD3ζ as indicator of the activation pathways in NK cells.
Rhesus macaques (RMs) have been a fundamental animal model for the preclinical evaluation of HIV-1 candidate vaccines (31–34), but we are still missing data that can address the degree of similarities between humans and RMs related to NK cell-mediated immunity. Despite extensive testing of HIV-1 vaccine regimens in RMs, we have limited knowledge on the similarities that may exist in recapitulating human antibody Fc-FcR interactions in the RM model. Macaques express FcγRs that are significantly different in sequence, structure or expression profiles from humans (35–40). Although less investigated than human Fc receptors (FcRs), several studies have described genetic polymorphisms of rhesus macaque FcRs and have identified several variants not observed in humans (36, 41–43). However, the functional implications of this genetic variation remain incompletely defined. In fact, no evidence has been reported that genetic polymorphisms of RM FcγRIII may play a role in the protection from SIV/SHIV/HIV-1 infection observed in the pre-clinical studies conducted in RMs. According to our current knowledge, we hypothesize that these interspecies variations may result in substantial Fc-mediated functional differences and, therefore, need to be evaluated. Therefore, in this study we sought to evaluate the diversity of cellular biology factors that may predict Fc-FcR interactions and impact antibody dependent effector functions in vivo. We included FcγRIII(a) genotype as well as phenotypic and functional parameters of NK cell signaling significantly related to ADCC responses (27). We observed that the overall magnitude of ADCC had similar ranges in the two species. However, the frequencies of NK cells and phosphorylated CD3ζ (pCD3ζ+) cells, level of cell-surface FcγRIII(a) expression, and NK cell-mediated ADCC were significantly different between the species. In addition, total ADCC, pCD3ζ+ cells, level of cell-surface FcγRIII(a) expression, and NK cell-mediated ADCC contributed equally to intra-species variation in both humans and RMs. This study indicates that FcR-mediated NK cell functional outcomes are complex and should be analyzed in a multi-parametric approach. This approach will provide a better understanding of correlates of protection in clinical vaccine trials and allow for a proper randomization of animals in pre-clinical protection trials based on FcR-mediated NK functions.
2 Results
2.1 Comparison of FcR-related NK cell functional measurements between humans and rhesus macaques
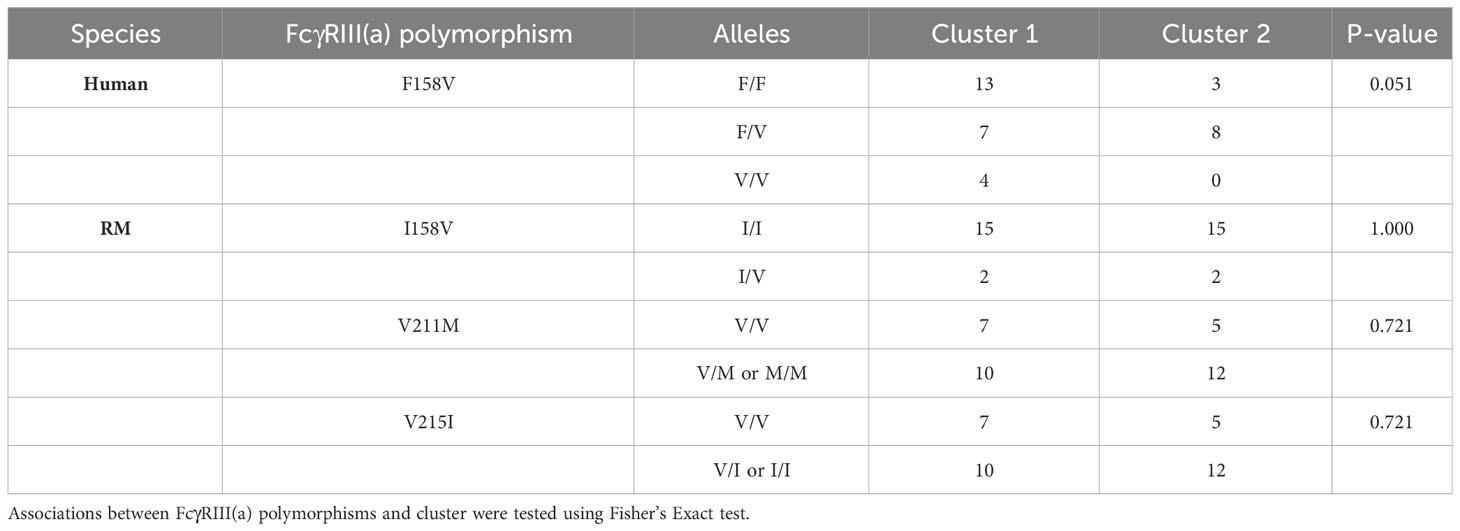
Peripheral blood mononuclear cells (PBMCs) from each human and rhesus macaque (RM) were analyzed for their effector functionality (Figure 1). A flow-based GranToxiLux (GTL) assay (44) was used to determine the ADCC magnitude of human or RM PBMCs. Target cells were coated with the recombinant SHIV1157 QNE Y173H gp120 protein for the GTL assay. To test ADCC potency, we used a combination of four HIV-specific human or RM IgG1 mAbs each at 1µg/ml (C1C2: JR4 (45, 46) and DH677.3 (45, 46); V1V2: DH827 (45, 47) and DH614.2 (45), which previously demonstrated binding to non-overlapping epitopes on gp120, and mediated ADCC against SHIV1157 QNE Y173H gp120-coated cells and SHIV1157 QNE Y173H-infected cells individually and in corresponding human and rhesus combinations (45, 48). We observed similar total ADCC activity across species (P > 0.05 by two-sided Mann-Whitney U test, Figure 1A). To assess the contribution of NK cells and monocytes to the total observed ADCC activity (% NK ADCC), we performed area scaling analysis (ASA, Figure S1A) (49). The contribution of NK cells to the total ADCC was significantly higher in humans compared to RMs (P < 0.001 by two-sided Mann-Whitney U test, Figure 1B) whereas NK cells contribution to the total ADCC was similar or lower than the monocyte contribution in RMs. The frequency of NK cells in PBMC samples was also significantly higher in humans compared to RMs (P < 0.001 by two-sided Mann-Whitney U test, Figures 1C, S1B, C). However, the frequency of CD16+ NK cells was similar across species (Figure 1D). We also analyzed the median fluorescent intensity (MFI) of cell-surface expressed FcγRIII(a) (CD16) within PBMCs, which is essential for ADCC by NK cells and monocytes (50, 51). Intriguingly, the level of cell-surface CD16 expression was significantly lower in humans (P < 0.001 by two-sided Mann-Whitney U test, Figure 1E). FcγRIII(a) expressed on NK cells can transduce signals through FcRγ and CD3ζ chains, where cells signaling through the alternative CD3ζ chain typically demonstrate enhanced ADCC (28, 52). Thus, we also measured the frequency of NK cells signaling through CD3ζ (phosphorylated CD3ζ; hereafter pCD3ζ) in PBMCs following FcγRIII(a) cross-linking (Figure S1D). The frequency of pCD3ζ+ NK cells was significantly higher in humans (P < 0.001 by two-sided Mann-Whitney U test, Figure 1F). Together, these data demonstrate variation in frequency and function of NK cells between the two species.
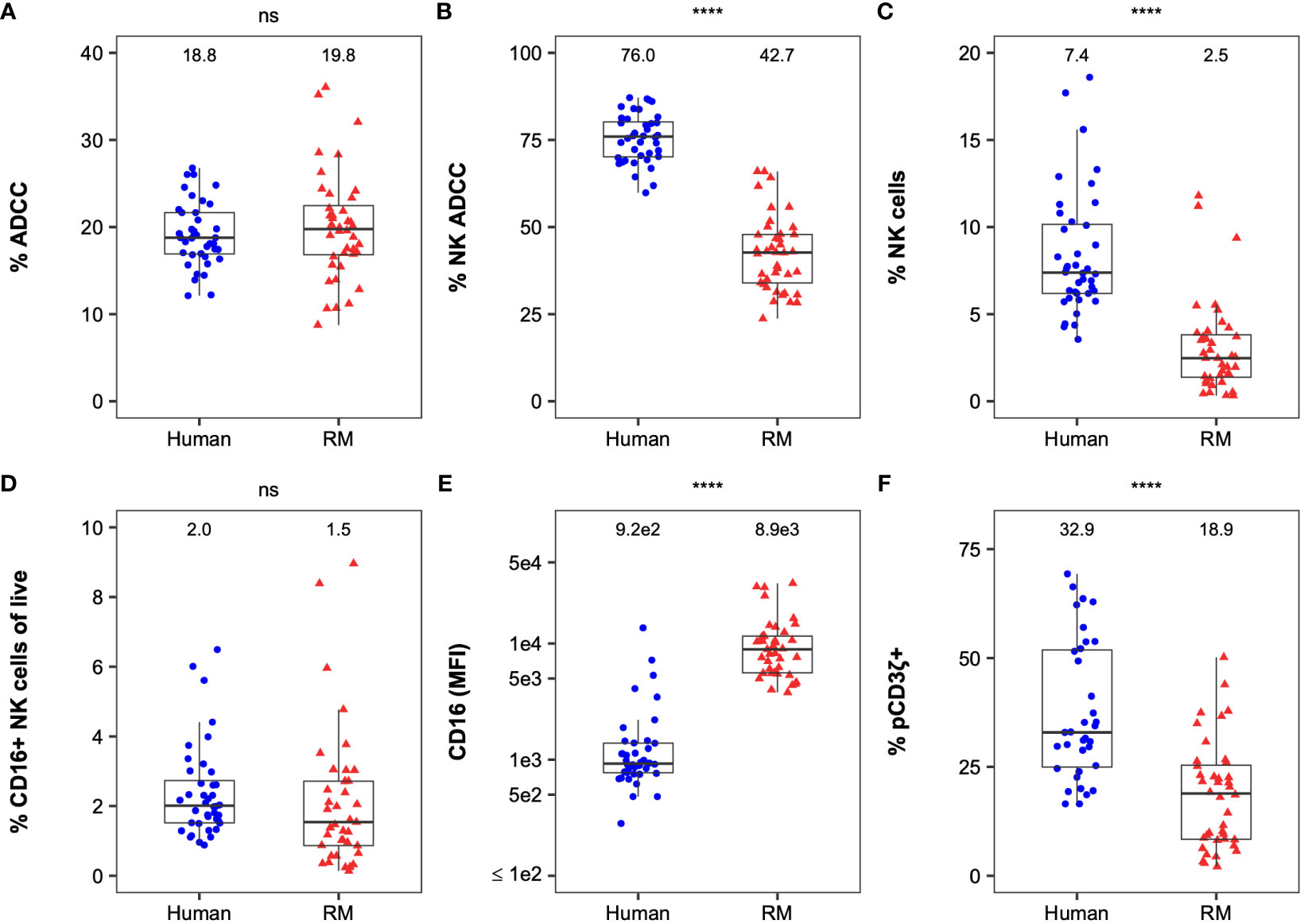
Figure 1 Comparison of FcR-related NK cell functional measurements between humans and RMs. Distribution of ADCC (A), % NK-mediated ADCC (B), % NK cells (C), % CD16+ NK cellsof live cells (D), CD16 expression, (E), and %pCD3ζ+ (F) by species. Boxplots extend from 25th percentile to 75th percentile with a horizontal line at the median. Whiskers extend from the largest value within 25th percentile + 1.5×IQR to the smallest value within 75th percentile - 1.5×IQR, where the interquartile range (IQR) equals 75th percentile - 25th percentile. Medians are shown above the data. Each point represents an individual human or animal. N=40 humans (with the exception of %pCD3ζ+, for which N=35 humans) and N=40 RMs (with the exception of CD16, for which N=39 RMs). The statistical significance of species differences is reported as **** for P < 0.001, and “ns” for non-significant (P > 0.05) on the top of each panel and were calculated using two-sided Mann-Whitney U tests.
2.2 FcγRIII(a) genetic diversity as a predictor of FcR-related NK cell functions
Human PBMCs were genotyped for known polymorphisms of FcγRIIIa at amino acid position 158 (Phe or Val, F/V) in the membrane-proximal, IgG-binding domain, which demonstrated different affinity for antibody Fc with subsequent variation in ADCC potency (14). Among 40 human samples, we identified 17 samples with F/F genotype, 18 with F/V genotype and 5 with V/V genotype (Table S1). For RM cells, we were able to sequence FCGR3 for 34 of the 40 animals (Table S2). Among those 34 RMs, 11 animals had no mutations relative to the reference sequence (Mnul_10, NCBI Genome ID: 215). One animal had only an I158V mutation, which is within the extracellular region of the receptor that forms the binding interface with IgG Fc (Tolbert et al., 2022, in press). 19 animals had both V211M and V215I mutations, which are located within the cytoplasmic tail. In addition, 3 animals had I158V, V211M, and V215I mutations (Table S2).
We then compared NK cell effector functions based on FcγRIII(a) polymorphisms to determine whether genetic diversity was predictive of functional diversity (Figure 2). The median ADCC magnitude was similar between genotypes in humans and RMs (Figures 2A, B). The contribution of NK cells and monocytes to the total observed ADCC activity was also similar among all genotypes in humans and RMs (Figures 2C, D). We observed similarity in cell-surface expression of FcγRIII(a) within PBMCs (Figures 2E, F) and the frequency of NK cells signaling through the CD3ζ chain (Figures 2G, H). Importantly, none of these measurements had a statistically significant association with FcγRIII(a) polymorphism(s) (all P > 0.05 by Kruskal-Wallis tests in humans and Mann-Whitney U tests in RMs, with comparisons of polymorphisms at each amino acid position being analyzed independently), suggesting that genetic diversity of FcγRIII(a) is insufficient to explain heterogeneity in NK cell function in either humans or macaques.
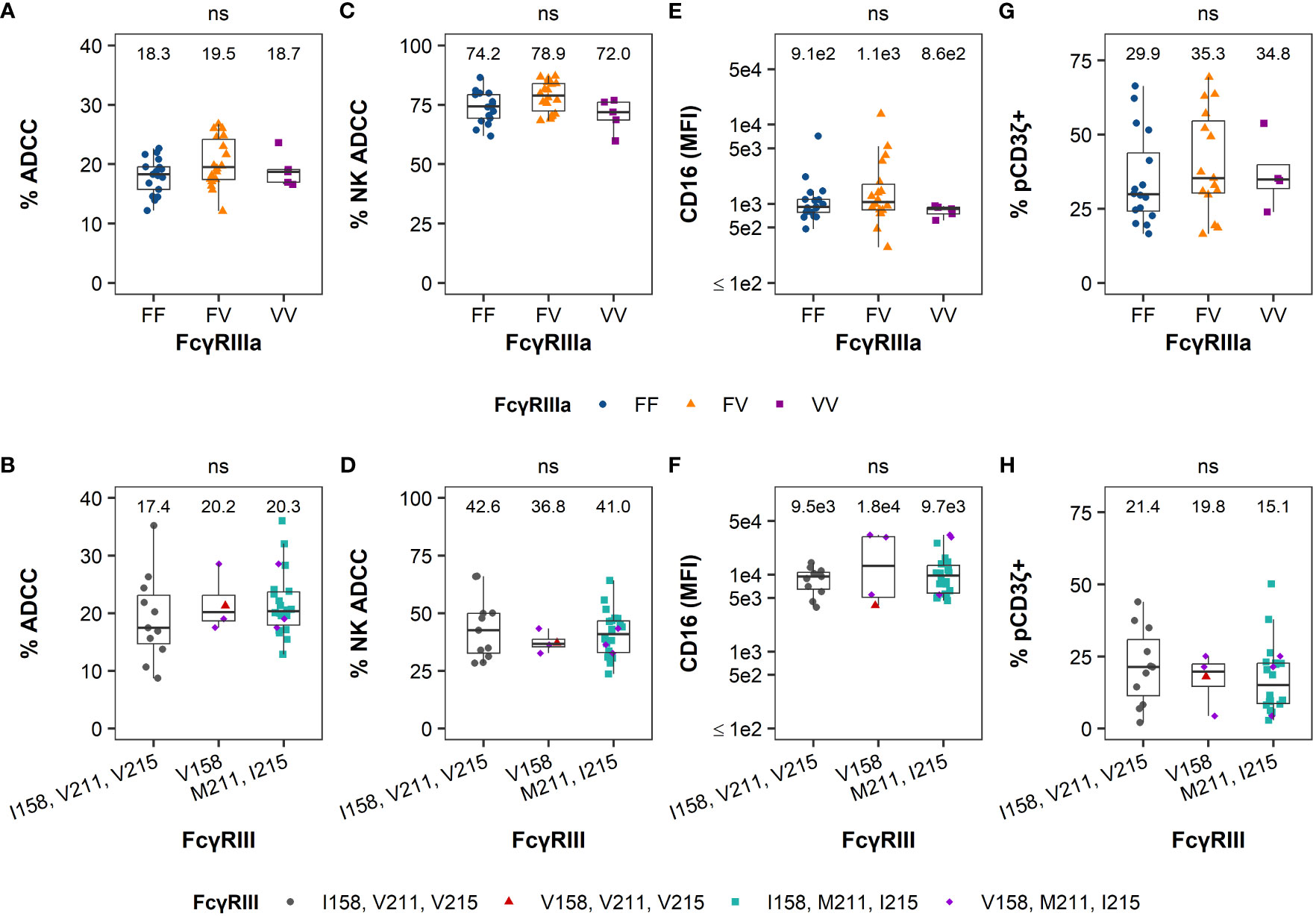
Figure 2 FcγRIII(a) genetic diversity is not a good predictor of FcR-related NK cell function. The distribution of ADCC potency (%ADCC), % NK-mediated ADCC, cell-surface FcγRIII(a) expression (CD16 MFI), and %pCD3ζ+ based on the human 158F/V polymorphism in humans are reported in the panels on the top row from the left to the right, respectively (A, C, E, G). The distribution of the results for the same parameters based on the 158I/V, 211V/M, and 215V/I polymorphisms in RMs are reported in the panels on the bottom row in the same order (B, D, F, H). The values for each parameter are reported on the y-axis and the polymorphisms on the x-axis. For simplicity and due to the low frequency of the I158V mutation in this cohort, all animals with the V158 allele are visualized together and all animals with M211 and I215 alleles are visualized together (with RMs with V158, M211, and I215 polymorphisms being represented twice in each of panels (B, D, F, H) rather than separating all 4 distinct haplotypes. Boxplots extend from 25th percentile to 75th percentile with a horizontal line at the median. Whiskers extend from the largest value within 25th percentile + 1.5×IQR to the smallest value within 75th percentile – 1.5×IQR, where the interquartile range (IQR) equals 75th percentile – 25th percentile. Medians are shown above each boxplot. Each point represents an individual human or animal; the color and shape represent the FcγRIIIa polymorphisms as indicated in the legend at the bottom of the panels). N=40 humans (with the exception of %pCD3ζ+, for which N=35 humans) and N=34 RMs. As indicated by “ns” at the top of each panel, no statistically significant differences in FcR-related NK functions were observed based on FcγRIII(a) polymorphisms among humans or RMs (all P > 0.05 using Kruskal-Wallis tests in (A, C, E, G) and Mann-Whitney U tests in (B, D, F, H). In RM, comparisons of polymorphisms at each amino acid position were analyzed independently.
2.3 Functional data reveal clusters with distinct FcR-related NK functional profiles within each species
We observed heterogeneity among humans and RMs in each of the 4 functional measurements (Figure 1), yet this variability cannot be explained by FcγRIII(a) polymorphisms (Figure 2). Hierarchical clustering based on ADCC magnitude, % NK-mediated ADCC, cell-surface FcγRIII(a) expression (CD16 MFI), and % pCD3ζ+ identified 2 distinct groups within each species cohorts, which are separated on a principal component analysis (PCA) biplot for both humans (Figure 3A) and macaques (Figure 3B). In humans, F/F and V/V genotypes were overrepresented in cluster 1, while the F/V genotype was evenly balanced across clusters. However, the association between polymorphism and cluster was not statistically significant (P = 0.051 by Fisher’s Exact test, Table 1). In RMs, all polymorphisms were represented in each cluster and there were no significant associations between polymorphisms and clusters (P > 0.05 by Fisher’s Exact test, Table 1). In humans, the clusters separated well by principal component 1 (PC1), which explained 46.0% of the total variation within the cohort. All four parameters contributed similarly to PC1 (Figure 3C). The largest contributors to PC2, which explained an additional 26.7% of the total variation, were % NK-mediated ADCC and % pCD3ζ+ NK cells. In RMs, PC1 explained 46.0% of the total variation within the cohort and the magnitude of contributions to PC1 were similar for all four parameters, consistent with results in the human cohort, but the loading value for % NK ADCC was negative while loadings for the other 3 parameters were positive (Figure 3D). The largest contributor to PC2, which explained an additional 20.6% of the total variation, was the total ADCC potency.
Figure 3 FcR-related NK cell functional measurements can separate humans and RMs into distinct clusters. Principal Component Analyses (PCA) are based on ADCC magnitude, % NK-mediated ADCC, CD16 MFI, or %pCD3ζ+ values. PCA biplot are shown by groups and identified using hierarchical clustering in humans (A) and RMs (B), with ellipses showing 68% confidence area for each cluster. Each point represents an individual human or animal; clusters are color coded as indicated in the legend. Variable loadings for the first two principal components, PC1 and PC2, in humans (C) and RMs (D). N=35 humans and N=39 RMs.
The functional profiles of each cluster were further characterized by examining the distribution of the variables by cluster (Figure 4). For human samples, we observed significant differences in ADCC potency (P = 0.002, Figure 4A), % NK ADCC (P < 0.001, Figure 4B), and cell-surface FcγRIIIa expression CD16 (MFI; P < 0.001, Figure 3C) between clusters by Mann-Whitney U tests, with medians for all parameters being higher in cluster 2. Median % pCD3ζ+ was also higher in cluster 2, however there was no statistically significant difference between clusters (P > 0.05 by Mann-Whitney U test, Figure 4D).
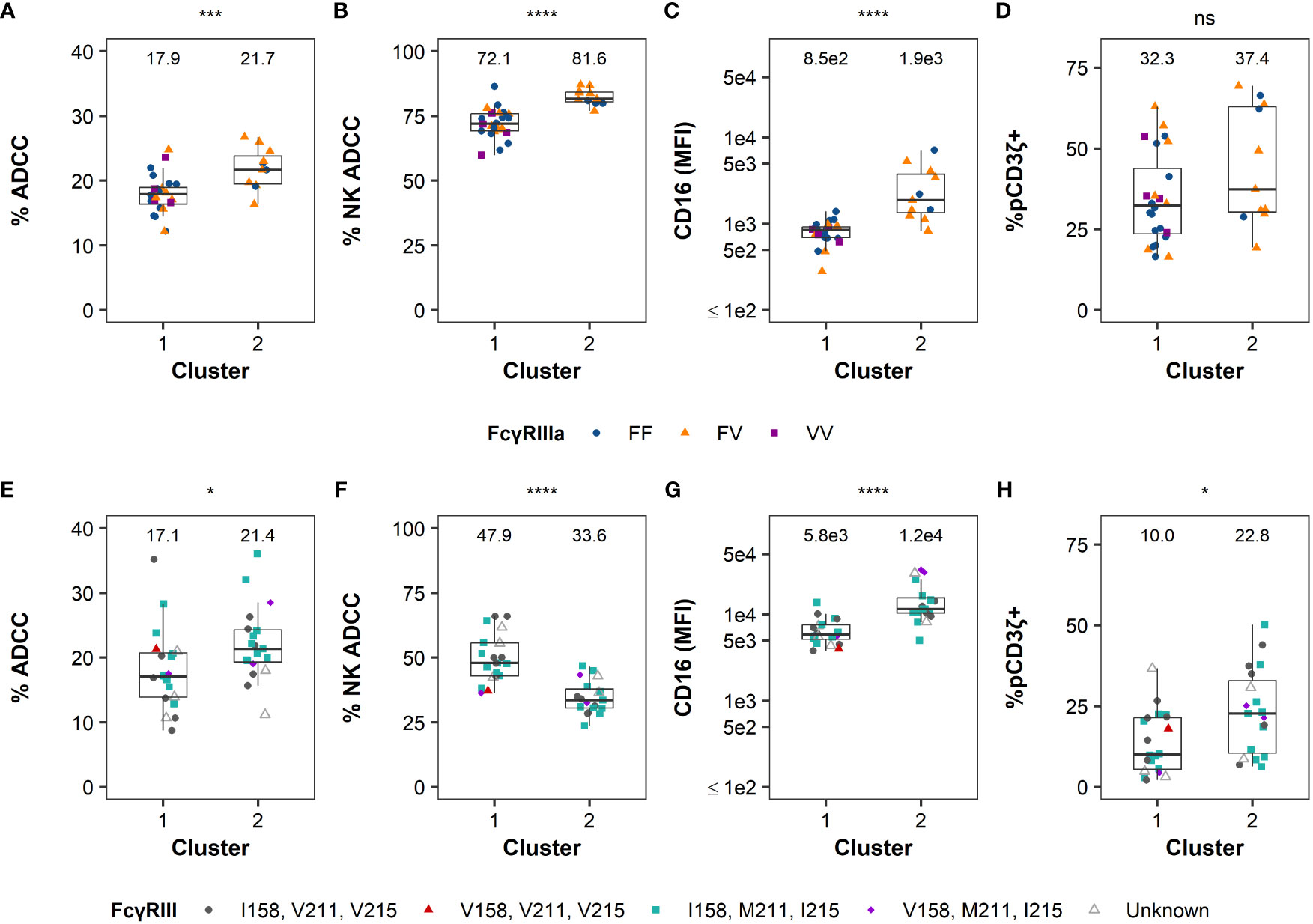
Figure 4 FcR-related NK cell functional measurements by cluster. Distribution of ADCC magnitude, % NK-mediated ADCC, CD16 expression, and %pCD3ζ+ by groups identified using hierarchical clustering are reported in the panels left to right, respectively, in humans (A–D) or RMs (E–H). Boxplots extend from 25th percentile to 75th percentile with a horizontal line at the median. Whiskers extend from the largest value within 25th percentile + 1.5×IQR to the smallest value within 75th percentile - 1.5×IQR, where the interquartile range (IQR) equals 75th percentile - 25th percentile. Data from each individual human and animal shown as a point, with color and shape by FcγRIII(a) polymorphism(s). RMs labeled as unknown if haplotype was not determined. Medians are shown above the data. N=35 humans and N=39 RMs. The statistical significance of the differences is indicated on the top of each panel as * for P < 0.05, ** for P < 0.01, *** for P < 0.005, **** for P < 0.001, and “ns” for non-significant (P > 0.05), and calculated by two-sided Mann-Whitney U tests.
For RM samples, there were significant differences between clusters in all four parameters (by Mann-Whitney U tests) where ADCC potency (P = 0.016, Figure 4E), CD16 MFI (P < 0.001, Figure 4G), and % pCD3ζ (P = 0.013, Figure 4H) were higher in cluster 2, while median % NK ADCC was higher in cluster 1. Altogether, these data indicate that human subjects and RMs can be separated into clusters with distinct functional profiles. Clustering of RMs is independent of FcγRIII genetic diversity, and while there is some evidence that clustering of humans could be influenced by FcγRIIIa polymorphisms, the association was not statistically significant.
Spearman’s rank correlation was used to examine relationships among these functional measurements within humans (Figure S2) and RMs (Figure S3). In humans, no significant correlations were observed to frequency of NK cells (P > 0.05, Figures S2A–D), but positive correlations were observed between total ADCC and CD16 expression (r = 0.465, P = 0.002, Figure S2F), ADCC and the frequency of pCD3ζ+ NK cells (r = 0.425, P = 0.011, Figure S2G), and frequency of NK-cell mediated ADCC and CD16 expression (r = 0.414, P = 0.008, Figure S2H). In RMs, the frequency of NK cells had a negative correlation to total ADCC potency (r = -0.344, P = 0.032, Figure S2A) and a positive correlation to NK-cell mediated ADCC (r = 0.538, P < 0.001, Figure S3B). NK-cell mediated ADCC had negative correlation to the total ADCC potency (r = -0.435, P = 0.005, Figure S3E), CD16 expression (r = -0.383, P = 0.016, Figure S3H), and the frequency of pCD3ζ+ NK cells (r = -0.411, P = 0.009, Figure S3I). CD16 expression had positive correlation to the frequency of pCD3ζ+ NK cells (r = 0.387, P = 0.015, Figure S3J). These weak to moderate correlations, although they reached statistical significance, highlight that no single parameter alone is sufficient to characterize these different aspects of NK cell function.
2.4 Cohort demographics
The human cohort presented here consisted of 24 males and 16 females with CMV status available for 38 individuals (Table S1), while all 40 RM were CMV+ males (Table S2). Based on descriptive statistics we observed little evidence of any NK cell functional differences based on gender or CMV status in this human cohort (Figure S4).
2.5 Randomizing animals for groups with balanced NK cell functions
Based on the heterogeneity observed in FcR-related NK cell functional measurements within these cohorts, we sought to investigate whether we could use screening data to assign treatment groups of RMs in pre-clinical trials to ensure balanced groups. We used covariate constrained randomization to assign animals into the potential treatment groups based on the four functional parameters (Figure 5). We observed that potential treatment groups A and B showed similar % total ADCC (Figure 5A), contribution of NK to the total ADCC (Figure 5B), CD16 expression (Figure 5C), and frequency of pCD3ζ+ NK cells (Figure 5D) with no significant differences (P > 0.05 by Mann-Whitney U test). Together, these data demonstrate that assigning RM to treatment groups using covariate constrained randomization produced balanced groups with similar FcR-related NK cell functional profiles.
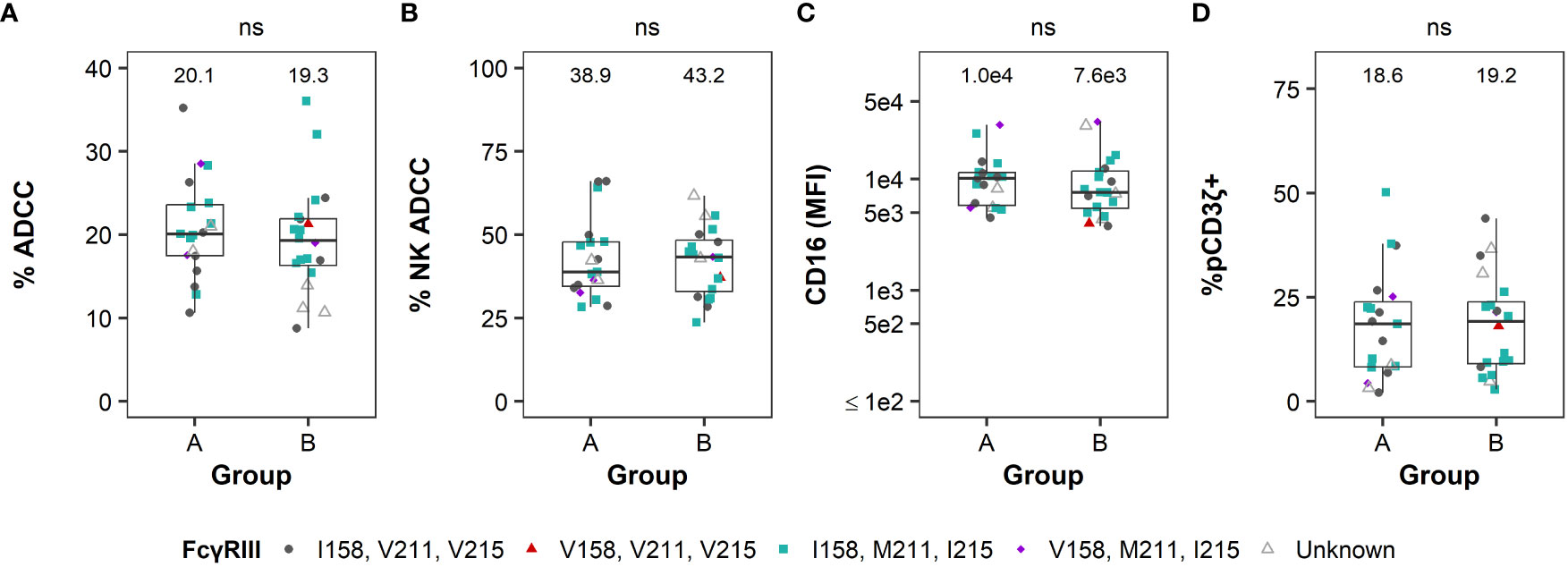
Figure 5 Covariate constrained randomization of RMs produced balanced groups for FcR-related NK cell functional measurements. Distribution of ADCC magnitude (A), % NK-mediated ADCC (B), CD16 MFI (C), and %pCD3ζ+ (D) according to group assignments based on covariate constrained randomization (see Methods for details). Boxplots extend from 25th percentile to 75th percentile with a horizontal line at the median. Whiskers extend from the largest value within 25th percentile + 1.5×IQR to the smallest value within 75th percentile - 1.5×IQR, where the interquartile range (IQR) equals 75th percentile - 25th percentile. Data from each individual animal shown as a point, with color and shape by FcγRIII polymorphisms. RMs labeled as unknown if haplotype was not determined. Medians are shown above the data. N=39. No statistically significant differences in FcR-related NK functions were observed between groups A and B (all P > 0.05 by two-sided Mann-Whitney U tests).
We further sought to investigate whether covariate constrained randomization of animals will introduce bias on non-NK functional parameters, such as antibody-dependent cellular phagocytosis (ADCP). ADCP has been demonstrated to contribute to protection against infection in preclinical (7, 53, 54) and clinical studies (55). We compared phagocytic activity of monocytes, isolated from RM PBMCs, as defined by a phagocytosis score–the mathematical product of the percentage of phagocytic-active cells and the number of phagocytosed particles per cell (56). Although phagocytosis occurs independently of FcγRIII, we analyzed whether FcγRIII polymorphisms or the NK functional clusters identified in Figure 2 were predictive of in vitro phagocytosis. There were no significant differences between FcγRIII polymorphism groups or NK functional clusters for RMs (P > 0.05 by Kruskal-Wallis test, Figures S5A, B). Importantly, covariate constrained randomization of RMs showed similar ADCP score in groups A and B with similar ranges and no significant difference (P > 0.05 by Mann-Whitney U test, Figure S5C). These data demonstrate that assigning RMs into treatment groups using covariate constrained randomization produced groups with similar NK cell functional profiles without introducing bias in non-NK functions, such as ADCP.
3 Discussion
The first clinical trial with administration of monoclonal Ab to treat HIV-1 was introduced in 1994 (57). Today, 36 clinical trials with administration of neutralizing Abs for prevention and treatment of HIV-1 have been launched (58, 59). Both pre-clinical studies (7, 54, 60–65) and clinical trials (55, 66) have demonstrated that Fc-mediated Ab functions contribute to the protection against infection and control of disease progression provided by neutralizing Abs. In addition, the only clinical trial that has shown limited efficacy (RV144) indicated that most of the anti-HIV antibody responses generated by infected individuals did not display neutralizing activities but contributed to protection by mediating ADCC against HIV-infected cells in vaccine recipients with low anti-HIV-1 IgA responses (8, 10). Therefore, these studies suggest the crucial relevance of evaluating Fc-FcR mediated functions as a correlate of risk and/or protection in pre- and clinical trials.
Our study represents the first attempt to address functional, phenotypic, and genetic diversity of effector NK cells among human and RM samples and compare diversity between the two species. Specifically, we analyzed NK cell phenotypic and the extent to which FcγRIII(a) polymorphisms influence the heterogeneity of NK-dependent functional responses. Multivariate analysis results indicated that FCGR3 genotype alone is not sufficient to explain this heterogeneity in human or RM samples. Animals and humans can be clustered into groups with distinct functional profiles; however, clusters were not fully explained by FcγRIII(a) polymorphisms alone. Instead, a combination of four FcR-related NK-cell parameters tested here contributed to the functional differences observed in both human and RM cohorts.
While FcγRIIIa polymorphisms in human NK cells have been linked to treatment efficacies and disease outcomes in many settings (16–20), the effect of rhesus macaque FcγRIII polymorphisms on study outcomes has largely been understudied. Here, we investigated whether genetic polymorphism in FcγRIII had an impact on rhesus macaque FcR-related NK cell functions in vitro. We identified polymorphisms at three sites (I158V, V211M, and V215I) and found that none of these polymorphisms were associated with the identified NK cell functional clusters, nor with any of the individual NK cell functions tested in our study. Previous work demonstrated that rhesus FcγRIII I158 has slightly higher binding affinity for rhesus macaque IgG1 compared to V158 variant (43, 67), but that these differences can vary based on the glycan composition of the FcR and Fc of Ab. Our data is consistent with these results demonstrating that FcγRIII(a) genotype alone cannot predict NK functional differences in either of the species. Our findings are also consistent with recent work demonstrating that polymorphisms at position 158, 211, and 215 of rhesus FcγRIII do not significantly impact ADCC mediated by rhesus IgG1 antibodies (42). However, the abundance of cell-surface FcγRIII can increase ADCC potency of RM NK cells even if NK cells express lower affinity FcγRIII polymorphism (67). Collectively, these observations indicate that rhesus NK cells expressing common allotypes of FcγRIII have similar functionality in vitro and the combination of FcR-related parameters influence functional differences. It is important to note however, that in our study, functional clusters of human NK cells were also not associated with FcγRIIIa SNPs. This in vitro finding is in stark contrast to the numerous in vivo associations linking SNPs and position 158 with human clinical outcomes (16–20). Therefore, although we observed no statistical differences in function and phenotype of rhesus macaque NK cells based on FcγRIII genetic diversity in vitro, it remains possible that these polymorphisms may have in vivo effects that are not yet known.
We observed that on average human PBMCs have higher frequencies of NK cells compared to RM PBMCs, whereas cell-surface FcγRIIIa expression (CD16) on human NK cells was significantly lower than CD16 on RM NK cells. Despite the differences in CD16 MFI between humans and RMs, CD16 expression was neither significantly different among SNPs in humans or RMs, nor did it contribute to functional clustering in humans or RMs. Interestingly, the contribution of NK to ADCC was higher in human samples compared to RMs. These dissimilarities are related not only to the cell-surface expression of FcγRIII(a) but also to the difference in binding affinity of human Ab Fc to FcγRIIIa with different polymorphisms vs RM counterparts (43, 67, 68). Here, for simplicity, we generated all mAbs with either human IgG1 or RM IgG1 Fc. Human FcγRIIIa has the highest affinity to IgG1 compared to other isotypes. Human IgG2 binds weakly to FcγRIIIa (15) and is not effective at recruiting NK cells for ADCC (69). Human IgG3 isotype Abs have longer hinge region, better flexibility, and mostly contribute to neutralization and ADCP responses (70). NHP IgG3 Abs have shorter hinge region and have similar binding affinity to FcγRIII and comparable functionality to human IgG1, NHP IgG1 and IgG2 (40, 71). Since the focus of this study is the ADCC-mediated activity of NK cells through Ab Fc-NK FcγRIIIa interaction and did not include evaluation of binding affinities between Fcs and FcRs with different polymorphisms, we chose to produce mAbs with IgG1 Fc for the most optimal ADCC activity.
CD16 does still require physically associated transmembrane adaptors for subsequent signal transduction and typically associates with FcγR and its adaptor, SYK (72). Interestingly, in primates (including humans), CD16 can also utilize CD3Ç in lieu of FcRγ chain, which presents as a disulfide-linked homodimer or as a heterodimer with FcRγ (72). Upon experimental cross-linking of CD16 or antibody ligation, immunoreceptor tyrosine-based activation motifs (ITAMs) on these adaptors are phosphorylated and act as docking sites for the cytoplasmic kinases SYK and ZAP70, which in turn activate downstream signaling pathways inducing ADCC (72). Importantly, NK cell signaling through CD3Ç results in higher ADCC functionality compared to canonical γ-chain (27, 73), these observations drove our decision to focus on this signaling pathway to understand the diversity of Fc-FcR interaction in human and RMs. In our study, we observed that subsets of both human and RM NK cells use CD3ζ/Zap70 signaling, but a higher frequency of human NK cells signal through CD3ζ compared to RMs; however, in both species the level of signaling neither correlates nor cluster with the FcγRIII(a) polymorphisms.
We decided to use covariate constrained randomization to separate animals into two groups equally balanced for Fc-mediated NK functions by using the combination of four FcγRIII-related phenotypic and functional parameters, such as total ADCC, CD16 MFI, %pCD3ζ and %NK ADCC. It should be noted, randomization of animals into two groups based on Fc-FcR related parameters did not introduce bias in non-NK cell functions. While animals with protective alleles (Mamu-B*17+ and B*08+) are excluded from pre-clinical trials (74), we do not suggest the exclusion of animals based on polymorphisms, but propose that screening animals for FcR-mediated NK cell functions will be useful prior to initiating pre-clinical studies, which often have small sample sizes, to ensure even balance in these measures among study arms. Here we used only males rhesus macaques, however, both sexes should be considered to ensure there are no additional gender-related variability that may contribute to NK functionality. Prospective interrogation of RM FcR-related functional parameters will ensure correct randomization of Rhesus macaques in pre-clinical studies intended to evaluate correlates of risk and/or protection, while retrospective multivariate analysis of human Fc-mediated NK functions in autologous systems utilizing Abs and NK cells from the same participants may lead to a better understanding of correlates of risk and/or protection in vaccine and passive infusion efficacy clinical trial.
We have not yet been able to analyze samples collected from humans and RMs who were vaccinated or used in pre-clinical efficacy studies, respectively, but this study indicates that FcR-mediated NK cell functional outcomes are complex and should be analyzed in a multi-parametric approach. In this study we focused primarily on F158V polymorphism of human NK cells which is known to affect affinity for the IgG subclasses and directly impact NK-mediated ADCC. Additional genetic variations should be explored as potential contributors to NK functionality. Additionally, we did not evaluate copy number of FCGR3A alleles that also were reported to affiliate with FcγRIIIa expression on NK cells (75). Future studies including NK functionality should consider including these parameters.
In conclusion, our studies strongly demonstrated that Fc-FcR interaction in pre- and clinical studies cannot be evaluated by simple analysis of the genetic polymorphisms but need multiparameter in-depth functional analyses of FcR-related NK functions in order to link efficacies of passive and active immunization, correlates of protection in pre- and clinical trials, and to ensure even distribution of treatment groups.
4 Materials and methods
4.1 Ethics statement
The studies were reviewed and approved by the Duke University Medical Center Institutional Review Board, and all participants provided written informed consent. All research was performed in accordance with the Duke University School of Medicine guidelines and regulations. Indian origin rhesus macaques (Macaca mulatta) were used in this study. The animals were housed at the New Iberia Research Center, animal experiments were approved by the New Iberia Primate Research Center IACUC (Protocol 2021-010-8823) and conducted in compliance with the principles described in the Guide for the Care and Use of Laboratory Animals (76). Animals with B*17+ and B*08+ alleles were excluded from this cohort due to the protective effect of these alleles (74). Human samples were collected at and experiments with human samples were approved by the Duke IRB (Pro00000873).
4.2 Human Fc Receptor sequence analysis
FcγRIIIa is encoded by the gene FCGR3A. Genotyping of the donors for SNP rs396991 was determined by TaqMan SNP Genotyping Pre-Validated Assay (Applied Biosystems, Foster City, CA) and an ABI7900 Sequence Detection System (ABI). The genotyping assay was performed according to manufacturer’s guidelines using 10 ng of DNA. Standard TaqMan thermocycling conditions were 10 min at 95°C, then 40 cycles of 15 s at 92°C and 1 min at 60°C. The endpoint fluorescence was read on an ABI 7900 Sequence Analyzer. The allele call was made by using the ABI proprietary software SDS version 2.4.
4.3 Rhesus macaque Fc receptor sequence analysis
FCGR3 sequencing was performed using long-read RNA sequencing as previously described (PMID: 34093580). Briefly, RNA was isolated from RM PBMC samples using the AllPrep DNA/RNA isolation kit (Qiagen, Germantown, MD), reverse transcribed using the Qiagen QuantiTect Reverse Transcription Kit (Qiagen), and Fc receptor cDNA was amplified using gene-specific primers designed with PacBio barcodes (Pacific Biosciences, Menlo Park, CA). PCR products were purified and PacBio SMRTbell library preparation was performed in accordance with manufacturer’s recommendations (Pacific Biosciences). Sequencing was performed on a PacBio Sequel II instrument using 2.1 or 3.0 chemistry (Pacific Biosciences). Datasets were loaded into PacBio SMRT Link 7.0.1 software package for demultiplexing of subreads and generating circular consensus sequences (CCS) and data analysis was conducted using a pipeline similar to that previously described (77). FCGR3 sequences were aligned to Mmul_10 reference sequence (NCBI Genome ID: 215) using a long-read sequence alignment tool (78) and variants were identified using the GATK Haplotype Caller (79), and annotated with ANNOVAR (80).
4.4 Monoclonal antibody production
Human IgG1 and RM IgG1 mAbs were produced by transient transfection of heavy and light chain plasmids into Expi293-F cells with Expifectamine (Thermo Fisher Scientific, Waltham, MA) and purified from cell culture supernatants by protein A resin columns as previously described (81–83).
4.5 HIV-1 Env gp120-specific ADCC
The ADCC-GranToxiLux assay was used to measure the ability of immune effector cells present in rhesus macaque (RM) PBMC to mediate Env gp120-specific ADCC when directed by HIV-specific RM IgG1 monoclonal antibodies. The assay was performed as previously described (44, 49). Target cells were a clonal isolate of the CEM.NKRCCR5 CD4+ T cell line (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: from Dr. Alexandra Trkola (84)) coated with SHIV1157 QNE Y173H gp120 protein (7). Human or RM PBMC effector cells were used at an effector cell to target cell ratio of 30:1 or 60:1, respectively. The antibodies used were a combination of the C1C2 cluster A-region specific antibodies JR4 (85) and DH677.3 (46), and V1V2-specific antibodies DH614.2 (86) and DH827 (47). JR4 and DH614.2 were originally isolated from RM after SHIV-infection or HIV-vaccination, respectively. DH677.3 and DH827 were originally isolated from human volunteers in the RV306 clinical trial. All antibodies were recombinantly produced as human and rhesus IgG1 (82). The combination of four human or four rhesus mAb were tested at a concentration of 1 μg/ml each. The influenza specific human or rhesusized CH65-IgG1 (anti-HA) antibodies (87) were used at the equivalent concentration of 4 μg/mL as negative controls. Data were reported as the maximum proportion of cells positive for proteolytically active granzyme B (GzB) out of the total viable target cell population (maximum %GzB activity) after subtracting the background activity observed in wells containing effector and target cells in the absence of antibodies. Area scaling analysis (ASA) of the GzB+ cells was used to evaluate antibody-dependent recruitment of NK cells (% NK ADCC) and monocytes as previously described (49).
4.6 NK cell frequency and expression of cell-surface CD16
Flow cytometry was used to assess the frequency of NK cells in human and RM PBMC, and to measure the levels of cell surface CD16. Cryopreserved PBMC were thawed and incubated overnight (18 hours) in RPMI1640 medium supplemented with 10% FBS at 37°C, 5% CO2. The cells were then washed with PBS and stained with a viability marker (Fixable Aqua Dead Cell Stain Kit, Thermo Fisher Scientific, San Diego, CA) prior to surface staining with fluorescently conjugated monoclonal antibodies using standard techniques. The panels of fluorescently conjugated antibodies for NK cell phenotyping were modified from what previously published (88, 89). Rhesus PBMCs were phenotyped with: CD3 (Pacific Blue, clone SP34.2, BD Biosciences), CD20 (Alexa Fluor 700, clone 2H7, BD Biosciences), CD8 (APC-H7, clone SK1, BD Biosciences), NKG2A/C (CD159a/c; PE, clone Z199, Beckman-Coulter), CD14 (BV650, clone M5E2, BD Biosciences), CD16 (PE-CF594, clone 3G8, BD Biosciences). Human PBMCs were phenotyped with: CD3 (PE-CF594, clone S4.1/7D6, ThermoFisher), CD4 (PerCPCy5.5, clone OKT4, eBioscience), CD8 (BV650, clone RPA-T8, Biolegend), NKG2A/C (CD159a/c; PE, clone Z199, Beckman-Coulter), CD14 (PE-Cy5, clone Tuk4, Life Technologies), CD19 (PE-Cy5, clone SJ25-C, Invitrogen), CD16 (APC-Cy7, clone 3G8, Beckman Coulter), CD56 (PE-Cy7, clone NCAM16.2, BD Biosciences).
Data analyses were performed using FlowJo software (v10.5.3). RM NK cells were defined as live CD3- CD14- CD20- CD8+ NKG2A/C+, as previously described (90). CD16 expression for human and RM cells is represented by APC-Cy7 or PE-CF594, respectively, median fluorescence intensity (MFI) of the CD16+ NK cell population. All samples were analyzed on a BD LSRFortessa flow cytometer (BD Biosciences) that has been optimized and rigorously maintained under quality control procedures described by Perfetto and colleagues (91).
4.7 Phospho-flow
Phosphorylation of CD3ζ chain (pCD3ζ) was determined as previously described (27). BD Perm buffer III (BD Biosciences, La Jolla, CA) was used for phospho-flow analyses according to the manufacturers’ protocols. Briefly, cells were opsonized with anti-human CD16 (BD Biosciences, clone 3G8, La Jolla, CA) followed by cross-linking with secondary goat-anti-mouse F(ab)’2, (Jackson ImmunoResearch Laboratories, West Grove, PA). Reactions were immediately stopped after 2.5 minutes (experimentally determined, data not shown) with equal volumes of Fix Buffer I (BD Biosciences, La Jolla, CA). After washing, unused/unoccupied sites of the secondary antibody were blocked with normal mouse serum (ThermoFisher) for 10 minutes at room temperature. Fixation was followed by surface stain, permeabilization with Perm Buffer III for 30 minutes on ice, followed by intracellular staining at room temperature according to the manufacturer’s protocol.
4.8 Antibody dependent cellular phagocytosis
ADCP was performed as previously described (56, 92). Briefly, quantification of ADCP was performed by covalently binding SHIV1157 QNE Y173H gp120 protein (7) to fluorescent beads and forming immune complexes by incubating in the presence of combination of four rhesus mAbs each at 25 µg/mL. Immune complexes were then incubated in the presence of rhesus monocytes isolated (Miltenyi Biotec catalog #130-091-097) from cryo-preserved rhesus PBMCs rested overnight (18 hours) in RPMI1640 medium supplemented with 10% FBS at 37°C, 5% CO2 and then washed with PBS. Fluorescence of the cells was detected using flow cytometry (BD Fortessa cytometer). The magnitude of the ADCP immune response was calculated as an ADCP score by multiplying the mean fluorescence intensity (MFI) and frequency of phagocytosis-positive cells and dividing by the MFI and frequency of the bead-positive cells in an antibody-negative (PBS) control well. The influenza specific rhesusized CH65-IgG1 (anti-HA) antibody (87) at the equivalent concentration of 100 μg/mL was used as a negative control. Each sample was tested once, and ADCP score is the mean score from two biological replicates.
4.9 Statistical analysis
Statistical analyses were performed using R statistical software (93). Clusters were identified using Ward’s hierarchical clustering based on Euclidian distance. Clustering was performed using ADCC, % NK ADCC, % pCD3ζ+, and CD16 expression data with and without the available demographic variables gender, age, race, weight and CMV status. However, these demographic variables did not impact the clustering results in humans or RMs. Principal components analysis was performed using the same 4 functional measurements that went into the cluster analysis and visualized using the ‘factoextra’ package (94). Five humans were missing % pCD3ζ+ values and one RM was missing a CD16 expression value; these subjects were excluded from clustering and PCA. Prior to clustering and PCA, CD16 expression values were log-transformed, and then ADCC, % NK ADCC, % pCD3ζ+, and CD16 were each scaled to have mean 0 and standard deviation 1 within each species cohort. To obtain the groups shown in Figure 5, we used covariate constrained randomization to assign the potential treatment arms based on the four variables discussed. The randomization was done using the R package cvcrand (95), examining 50,000 potential randomization schemes with the potential assignment sampled from the lowest 10% of the resulting inverse variance weighted balance scores (96).
Kruskal-Wallis tests were used when there were more than 2 groups for comparison (Figure 4). Two-sided Mann-Whitney U tests were used for pairwise comparisons (Figures 1, 4, 5). Two-sided Fisher’s exact tests were used to test for association of categorical variables (Table 1). All analyses were performed separately by species, except for the direct comparison of humans and macaques shown in Figure 1. All comparisons were considered significant at the 0.05 level. Due to the exploratory nature of these analyses, no corrections were made for multiple testing.
Data availability statement
The RM sequencing data presented in the study are deposited in The National Center for Biotechnology Information (NCBI) repository, under the umbrella BioProject PRJNA1030012. RM demographic and functional data are available for download at doi.org/10.5281/zenodo.8436837. Human data is available upon request.
Ethics statement
The studies involving humans were approved by Duke University Medical Center Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by Duke University Medical Center Institutional Review Board. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MT: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. RS: Conceptualization, Formal Analysis, Writing – original draft, Writing – review & editing. BH: Methodology, Writing – review & editing. JN: Methodology, Writing – review & editing. DG: Methodology, Writing – review & editing. CC: Formal Analysis, Supervision, Writing – review & editing. RB: Formal Analysis, Methodology, Writing – review & editing. WB: Methodology, Writing – review & editing. SJ: Formal Analysis, Writing – review & editing. SA: Methodology, Writing – review & editing. KW: Methodology, Writing – review & editing. MH: Methodology, Writing – review & editing. DE: Methodology, Writing – review & editing. HC: Methodology, Writing – review & editing. TH: Methodology, Writing – review & editing. TG: Methodology, Writing – review & editing. CJ: Methodology, Writing – review & editing. NA: Methodology. FV: Resources, Writing – review & editing. RT: Methodology, Writing – review & editing. TD: Writing – review & editing, Resources. MM: Methodology, Supervision, Writing – review & editing. GT: Funding acquisition, Resources, Supervision, Writing – review & editing. RR: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft. JP: Conceptualization, Funding acquisition, Resources, Writing – original draft. GF: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. National Institutes of Health grant P01 AI120756 (GDT), R01AI143457, Duke Center for AIDS Research (CFAR) AI064518. Biostatistics, Epidemiology and Research Design (BERD) Methods Core funded through grant UL1TR002553 from the National Center for Advancing Translational Sciences (NCATS), a component of the NIH.
Acknowledgments
The authors thank Wes Rountree and Yunfei Wang for statistical expertise and Melissa Kerkau, Val Bekker, and Sarah Mudrak for program management.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1260377/full#supplementary-material
References
1. Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet (2007) 39(6):733–40. doi: 10.1038/ng2035
2. Garrido C, Abad-Fernandez M, Tuyishime M, Pollara JJ, Ferrari G, Soriano-Sarabia N, et al. Interleukin-15-stimulated natural killer cells clear HIV-1-infected cells following latency reversal ex vivo. J Virol (2018) 92(12). doi: 10.1128/JVI.00235-18
3. Alter G, Altfeld M. NK cells in HIV-1 infection: evidence for their role in the control of HIV-1 infection. J Intern Med (2009) 265(1):29–42. doi: 10.1111/j.1365-2796.2008.02045.x
4. Quillay H, El Costa H, Duriez M, Marlin R, Cannou C, Madec Y, et al. NK cells control HIV-1 infection of macrophages through soluble factors and cellular contacts in the human decidua. Retrovirology (2016) 13(1):39. doi: 10.1186/s12977-016-0271-z
5. Pegu P, Vaccari M, Gordon S, Keele BF, Doster M, Guan Y, et al. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. J Virol (2013) 87(3):1708–19. doi: 10.1128/JVI.02544-12
6. Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature (2007) 449(7158):101–4. doi: 10.1038/nature06106
7. Bradley T, Pollara J, Santra S, Vandergrift N, Pittala S, Bailey-Kellogg C, et al. Pentavalent HIV-1 vaccine protects against simian-human immunodeficiency virus challenge. Nat Commun (2017) 8:15711. doi: 10.1038/ncomms15711
8. Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med (2012) 366(14):1275–86. doi: 10.1056/NEJMoa1113425
9. Yates NL, Liao HX, Fong Y, deCamp A, Vandergrift NA, Williams WT, et al. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med (2014) 6(228):228ra39. doi: 10.1126/scitranslmed.3007730
10. Pollara J, Bonsignori M, Moody MA, Liu P, Alam SM, Hwang KK, et al. HIV-1 vaccine-induced C1 and V2 Env-specific antibodies synergize for increased antiviral activities. J Virol (2014) 88(14):7715–26. doi: 10.1128/JVI.00156-14
11. Ravetch JV, PeRussia B. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J Exp Med (1989) 170(2):481–97. doi: 10.1084/jem.170.2.481
12. van Sorge NM, van der Pol WL, van de Winkel JG. FcgammaR polymorphisms: Implications for function, disease susceptibility and immunotherapy. Tissue Antigens (2003) 61(3):189–202. doi: 10.1034/j.1399-0039.2003.00037.x
13. Wu J, Edberg JC, Redecha PB, Bansal V, Guyre PM, Coleman K, et al. A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest (1997) 100(5):1059–70. doi: 10.1172/JCI119616
14. Koene HR, Kleijer M, Algra J, Roos D, von dem Borne AE, de Haas M. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood (1997) 90(3):1109–14. doi: 10.1182/blood.V90.3.1109
15. Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood (2009) 113(16):3716–25. doi: 10.1182/blood-2008-09-179754
16. Arriga R, Caratelli S, Lanzilli G, Ottaviani A, Cenciarelli C, Sconocchia T, et al. CD16-158-valine chimeric receptor T cells overcome the resistance of KRAS-mutated colorectal carcinoma cells to cetuximab. Int J cancer (2020) 146(9):2531–8. doi: 10.1002/ijc.32618
17. Veeramani S, Wang SY, Dahle C, Blackwell S, Jacobus L, Knutson T, et al. Rituximab infusion induces NK activation in lymphoma patients with the high-affinity CD16 polymorphism. Blood (2011) 118(12):3347–9. doi: 10.1182/blood-2011-05-351411
18. Rataj F, Jacobi SJ, Stoiber S, Asang F, Ogonek J, Tokarew N, et al. High-affinity CD16-polymorphism and Fc-engineered antibodies enable activity of CD16-chimeric antigen receptor-modified T cells for cancer therapy. Br J Cancer (2019) 120(1):79–87. doi: 10.1038/s41416-018-0341-1
19. Arnold M-L, Heinemann FM, Oesterreich S, Wilde B, Gäckler A, Goldblatt D, et al. Correlation of fc receptor polymorphisms with pneumococcal antibodies in vaccinated kidney transplant recipients. Vaccines (2022) 10(5):725. doi: 10.3390/vaccines10050725
20. Das LK, Ide K, Tanaka A, Morimoto H, Shimizu S, Tanimine N, et al. Fc-gamma receptor 3A polymorphism predicts the incidence of urinary tract infection in kidney-transplant recipients. Hum Immunol (2017) 78(4):357–62. doi: 10.1016/j.humimm.2017.03.006
21. Dall'Ozzo S, Tartas S, Paintaud G, Cartron G, Colombat P, Bardos P, et al. Rituximab-dependent cytotoxicity by natural killer cells: influence of FCGR3A polymorphism on the concentration-effect relationship. Cancer Res (2004) 64(13):4664–9. doi: 10.1158/0008-5472.CAN-03-2862
22. Congy-Jolivet N, Bolzec A, Ternant D, Ohresser M, Watier H, Thibault G. Fc gamma RIIIa expression is not increased on natural killer cells expressing the Fc gamma RIIIa-158V allotype. Cancer Res (2008) 68(4):976–80. doi: 10.1158/0008-5472.CAN-07-6523
23. Forthal DN, Gabriel EE, Wang A, Landucci G, Phan TB. Association of Fcγ receptor IIIa genotype with the rate of HIV infection after gp120 vaccination. Blood (2012) 120(14):2836–42. doi: 10.1182/blood-2012-05-431361
24. Geraghty DE, Thorball CW, Fellay J, Thomas R. Effect of fc receptor genetic diversity on HIV-1 disease pathogenesis. Front Immunol (2019) 10:970. doi: 10.3389/fimmu.2019.00970
25. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol (2008) 9(5):495–502. doi: 10.1038/ni1581
26. Dixon KJ, Wu J, Walcheck B. Engineering anti-tumor monoclonal antibodies and fc receptors to enhance ADCC by human NK cells. Cancers (Basel) (2021) 13(2):312. doi: 10.3390/cancers13020312
27. Shah SV, Manickam C, Ram DR, Kroll K, Itell H, Permar SR, et al. CMV primes functional alternative signaling in adaptive Δg NK cells but is subverted by lentivirus infection in rhesus macaques. Cell Rep (2018) 25(10):2766–74.e3. doi: 10.1016/j.celrep.2018.11.020
28. Zhou J, Amran FS, Kramski M, Angelovich TA, Elliott J, Hearps AC, et al. An NK cell population lacking fcRγ Is expanded in chronically infected HIV patients. J Immunol (Baltimore Md 1950) (2015) 194(10):4688–97. doi: 10.4049/jimmunol.1402448
29. Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity (2015) 42(3):431–42. doi: 10.1016/j.immuni.2015.02.013
30. Zhang T, Scott JM, Hwang I, Kim S. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRγ deficiency. J Immunol (2013) 190(4):1402–6. doi: 10.4049/jimmunol.1203034
31. Staprans SI, Feinberg MB, Shiver JW, Casimiro DR. Role of nonhuman primates in the evaluation of candidate AIDS vaccines: an industry perspective. Curr Opin HIV AIDS (2010) 5(5):377–85. doi: 10.1097/COH.0b013e32833d2e19
32. Morgan C, Marthas M, Miller C, Duerr A, Cheng-Mayer C, Desrosiers R, et al. The use of nonhuman primate models in HIV vaccine development. PloS Med (2008) 5(8):e173. doi: 10.1371/journal.pmed.0050173
33. Hessell AJ, Malherbe DC, Haigwood NL. Passive and active antibody studies in primates to inform HIV vaccines. Expert Rev Vaccines (2018) 17(2):127–44. doi: 10.1080/14760584.2018.1425619
34. Terrade G, Huot N, Petitdemange C, Lazzerini M, Orta Resendiz A, Jacquelin B, et al. Interests of the non-human primate models for HIV cure research. Vaccines (Basel) (2021) 9(9):958. doi: 10.3390/vaccines9090958
35. Warncke M, Calzascia T, Coulot M, Balke N, Touil R, Kolbinger F, et al. Different adaptations of IgG effector function in human and nonhuman primates and implications for therapeutic antibody treatment. J Immunol (2012) 188(9):4405–11. doi: 10.4049/jimmunol.1200090
36. Rogers KA, Scinicariello F, Attanasio R. IgG Fc receptor III homologues in nonhuman primate species: genetic characterization and ligand interactions. J Immunol (2006) 177(6):3848–56. doi: 10.4049/jimmunol.177.6.3848
37. Choi EI, Wang R, Peterson L, Letvin NL, Reimann KA. Use of an anti-CD16 antibody for in vivo depletion of natural killer cells in rhesus macaques. Immunology (2008) 124(2):215–22. doi: 10.1111/j.1365-2567.2007.02757.x
38. Hong HS, Rajakumar PA, Billingsley JM, Reeves RK, Johnson RP. No monkey business: why studying NK cells in non-human primates pays off. Front Immunol (2013) 4:32. doi: 10.3389/fimmu.2013.00032
39. Tolbert WD, Subedi GP, Gohain N, Lewis GK, Patel KR, Barb AW, et al. From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses. MAbs (2019) 11(4):709–24. doi: 10.1080/19420862.2019.1589852
40. Scinicariello F, Engleman CN, Jayashankar L, McClure HM, Attanasio R. Rhesus macaque antibody molecules: sequences and heterogeneity of alpha and gamma constant regions. Immunology (2004) 111(1):66–74. doi: 10.1111/j.1365-2567.2004.01767.x
41. Nguyen DC, Scinicariello F, Attanasio R. Characterization and allelic polymorphisms of rhesus macaque (Macaca mulatta) IgG Fc receptor genes. Immunogenetics (2011) 63(6):351–62. doi: 10.1007/s00251-011-0514-z
42. Grunst MW, Grandea AG 3rd, Janaka SK, Hammad I, Grimes P, Karl JA, et al. Functional interactions of common allotypes of rhesus macaque fcγR2A and fcγR3A with human and macaque igG subclasses. J Immunol (2020) 205(12):3319–32. doi: 10.4049/jimmunol.2000501
43. Chan YN, Boesch AW, Osei-Owusu NY, Emileh A, Crowley AR, Cocklin SL, et al. IgG binding characteristics of rhesus macaque fcγR. J Immunol (Baltimore Md 1950) (2016) 197(7):2936–47. doi: 10.4049/jimmunol.1502252
44. Pollara J, Hart L, Brewer F, Pickeral J, Packard BZ, Hoxie JA, et al. High-throughput quantitative analysis of HIV-1 and SIV-specific ADCC-mediating antibody responses. Cytometry A (2011) 79(8):603–12. doi: 10.1002/cyto.a.21084
45. Tolbert WD, Nguyen DN, Tuyishime M, Crowley AR, Chen Y, Jha S, et al. Structure and fc-effector function of rhesusized variants of human anti-HIV-1 igG1s. Front Immunol (2021) 12:787603. doi: 10.3389/fimmu.2021.787603
46. Easterhoff D, Pollara J, Luo K, Tolbert WD, Young B, Mielke D, et al. Boosting with AIDSVAX B/E enhances env constant region 1 and 2 antibody-dependent cellular cytotoxicity breadth and potency. J Virol (2020) 94(4). doi: 10.1128/JVI.01120-19
47. Easterhoff D, Pollara J, Luo K, Janus B, Gohain N, Williams LD, et al. HIV vaccine delayed boosting increases Env variable region 2-specific antibody effector functions. JCI Insight (2020) 5(2). doi: 10.1172/jci.insight.131437
48. Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, et al. Molecularly cloned SHIV-1157ipd3N4: a highly replication- competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J Virol (2006) 80(17):8729–38. doi: 10.1128/JVI.00558-06
49. Pollara J, Orlandi C, Beck C, Edwards RW, Hu Y, Liu S, et al. Application of area scaling analysis to identify natural killer cell and monocyte involvement in the GranToxiLux antibody dependent cell-mediated cytotoxicity assay. Cytometry A (2018) 93(4):436–47. doi: 10.1002/cyto.a.23348
50. Caligiuri MA. Human natural killer cells. Blood (2008) 112(3):461–9. doi: 10.1182/blood-2007-09-077438
51. Yeap WH, Wong KL, Shimasaki N, Teo EC, Quek JK, Yong HX, et al. CD16 is indispensable for antibody-dependent cellular cytotoxicity by human monocytes. Sci Rep (2016) 6:34310. doi: 10.1038/srep34310
52. Vivier E, Ackerly M, Rochet N, Anderson P. Structure and function of the CD16:zeta:gamma complex expressed on human natural-killer cells. Int J Cancer Suppl (1992) 7:11–4.
53. Ackerman ME, Das J, Pittala S, Broge T, Linde C, Suscovich TJ, et al. Route of immunization defines multiple mechanisms of vaccine-mediated protection against SIV. Nat Med (2018) 24(10):1590–8. doi: 10.1038/s41591-018-0161-0
54. Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, et al. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science (2015) 349(6245):320–4. doi: 10.1126/science.aab3886
55. Neidich SD, Fong Y, Li SS, Geraghty DE, Williamson BD, Young WC, et al. Antibody Fc effector functions and IgG3 associate with decreased HIV-1 risk. J Clin Invest (2019) 129(11):4838–49. doi: 10.1172/JCI126391
56. Ackerman ME, Moldt B, Wyatt RT, Dugast AS, McAndrew E, Tsoukas S, et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J Immunol Methods (2011) 366(1-2):8–19. doi: 10.1016/j.jim.2010.12.016
57. Günthard HF, Gowland PL, Schüpbach J, Fung MS, Böni J, Liou RS, et al. A phase I/IIA clinical study with a chimeric mouse-human monoclonal antibody to the V3 loop of human immunodeficiency virus type 1 gp120. J Infect Dis (1994) 170(6):1384–93. doi: 10.1093/infdis/170.6.1384
58. Mahomed S, Garrett N, Baxter C, Abdool Karim Q, Abdool Karim SS. Clinical trials of broadly neutralizing monoclonal antibodies for human immunodeficiency virus prevention: A review. J Infect Diseases (2020) 223(3):370–80. doi: 10.1093/infdis/jiaa377
59. ClinicalTrials.gov 2022 (2022). Available at: https://clinicaltrials.gov/ct2/results?term=bNAb%2C+antibody&cond=HIV&Search=Apply&age_v=&gndr=&type=&rslt=.
60. Gómez-Román VR, Florese RH, Peng B, Montefiori DC, Kalyanaraman VS, Venzon D, et al. An adenovirus-based HIV subtype B prime/boost vaccine regimen elicits antibodies mediating broad antibody-dependent cellular cytotoxicity against non-subtype B HIV strains. J Acquir Immune Defic Syndr (2006) 43(3):270–7. doi: 10.1097/01.qai.0000230318.40170.60
61. Felber BK, Lu Z, Hu X, Valentin A, Rosati M, Remmel CAL, et al. Co-immunization of DNA and protein in the same anatomical sites induces superior protective immune responses against SHIV challenge. Cell Rep (2020) 31(6):107624. doi: 10.1016/j.celrep.2020.107624
62. Wang P, Gajjar MR, Yu J, Padte NN, Gettie A, Blanchard JL, et al. Quantifying the contribution of Fc-mediated effector functions to the antiviral activity of anti-HIV-1 IgG1 antibodies in vivo. Proc Natl Acad Sci USA (2020) 117(30):18002–9. doi: 10.1073/pnas.2008190117
63. Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell (2014) 158(6):1243–53. doi: 10.1016/j.cell.2014.08.023
64. Lu CL, Murakowski DK, Bournazos S, Schoofs T, Sarkar D, Halper-Stromberg A, et al. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science (2016) 352(6288):1001–4. doi: 10.1126/science.aaf1279
65. Parsons MS, Chung AW, Kent SJ. Importance of Fc-mediated functions of anti-HIV-1 broadly neutralizing antibodies. Retrovirology (2018) 15(1):58. doi: 10.1186/s12977-018-0438-x
66. Ackerman ME, Mikhailova A, Brown EP, Dowell KG, Walker BD, Bailey-Kellogg C, et al. Polyfunctional HIV-specific antibody responses are associated with spontaneous HIV control. PloS Pathog (2016) 12(1):e1005315. doi: 10.1371/journal.ppat.1005315
67. Tolbert WD, Gohain N, Kremer PG, Hederman AP, Nguyen DN, Van V, et al. Decoding human-macaque interspecies differences in Fc-effector functions: The structural basis for CD16-dependent effector function in Rhesus macaques. Front Immunol (2022) 13:960411. doi: 10.3389/fimmu.2022.960411
68. Crowley AR, Richardson SI, Tuyishime M, Jennewein M, Bailey MJ, Lee J, et al. Functional consequences of allotypic polymorphisms in human immunoglobulin G subclasses. Immunogenetics (2022) 75(1):1–16. doi: 10.1007/s00251-022-01272-7
69. Schneider-Merck T, Lammerts van Bueren JJ, Berger S, Rossen K, van Berkel PH, Derer S, et al. Human IgG2 antibodies against epidermal growth factor receptor effectively trigger antibody-dependent cellular cytotoxicity but, in contrast to IgG1, only by cells of myeloid lineage. J Immunol (2010) 184(1):512–20. doi: 10.4049/jimmunol.0900847
70. Scharf O, Golding H, King LR, Eller N, Frazier D, Golding B, et al. Immunoglobulin G3 from polyclonal human immunodeficiency virus (HIV) immune globulin is more potent than other subclasses in neutralizing HIV type 1. J Virol (2001) 75(14):6558–65. doi: 10.1128/JVI.75.14.6558-6565.2001
71. Boesch AW, Osei-Owusu NY, Crowley AR, Chu TH, Chan YN, Weiner JA, et al. Biophysical and functional characterization of rhesus macaque igG subclasses. Front Immunol (2016) 7:589. doi: 10.3389/fimmu.2016.00589
72. Sugawara S, Manickam C, Reeves RK. TRIGGERED: could refocused cell signaling be key to natural killer cell-based HIV immunotherapeutics? Aids (2021) 35(2):165–76. doi: 10.1097/QAD.0000000000002743
73. Hart GT, Tran TM, Theorell J, Schlums H, Arora G, Rajagopalan S, et al. Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J Exp Med (2019) 216(6):1280–90. doi: 10.1084/jem.20181681
74. Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature (2011) 473(7348):523–7. doi: 10.1038/nature10003
75. Breunis WB, van Mirre E, Geissler J, Laddach N, Wolbink G, van der Schoot E, et al. Copy number variation at the FCGR locus includes FCGR3A, FCGR2C and FCGR3B but not FCGR2A and FCGR2B. Hum Mutat (2009) 30(5):E640–50. doi: 10.1002/humu.20997
76. Council NR. Guide for the care and use of laboratory animals. (2010) (Washington DC, USA: The National Acamedies Press).
77. Wenger AM, Peluso P, Rowell WJ, Chang PC, Hall RJ, Concepcion GT, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol (2019) 37(10):1155–62. doi: 10.1038/s41587-019-0217-9
78. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics (2018) 34(18):3094–100. doi: 10.1093/bioinformatics/bty191
79. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet (2011) 43(5):491–8. doi: 10.1038/ng.806
80. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res (2010) 38(16):e164. doi: 10.1093/nar/gkq603
81. Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods (2009) 158(1-2):171–9. doi: 10.1016/j.jviromet.2009.02.014
82. Saunders KO, Pegu A, Georgiev IS, Zeng M, Joyce MG, Yang ZY, et al. Sustained delivery of a broadly neutralizing antibody in nonhuman primates confers long-term protection against simian/human immunodeficiency virus infection. J Virol (2015) 89(11):5895–903. doi: 10.1128/JVI.00210-15
83. Pollara J, Tay MZ, Edwards RW, Goodman D, Crowley AR, Edwards RJ, et al. Functional homology for antibody-dependent phagocytosis across humans and rhesus macaques. Front Immunol (2021) 12:678511. doi: 10.3389/fimmu.2021.678511
84. Trkola A, Matthews J, Gordon C, Ketas T, Moore JP. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J Virol (1999) 73(11):8966–74. doi: 10.1128/JVI.73.11.8966-8974.1999
85. Gohain N, Tolbert WD, Acharya P, Yu L, Liu T, Zhao P, et al. Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1. J Virol (2015) 89(17):8840–54. doi: 10.1128/JVI.01232-15
86. Luo K, Liao HX, Zhang R, Easterhoff D, Wiehe K, Gurley TC, et al. Tissue memory B cell repertoire analysis after ALVAC/AIDSVAX B/E gp120 immunization of rhesus macaques. JCI Insight (2016) 1(20):e88522. doi: 10.1172/jci.insight.88522
87. Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA (2011) 108(34):14216–21. doi: 10.1073/pnas.1111497108
88. Pollara J, Edwards RW, Jha S, Lam CK, Liu L, Diedrich G, et al. Redirection of cord blood T cells and natural killer cells for elimination of autologous HIV-1-infected target cells using bispecific DART® Molecules. Front Immunol (2020) 11:713. doi: 10.3389/fimmu.2020.00713
89. Fisher L, Zinter M, Stanfield-Oakley S, Carpp LN, Edwards RW, Denny T, et al. Vaccine-induced antibodies mediate higher antibody-dependent cellular cytotoxicity after interleukin-15 pretreatment of natural killer effector cells. Front Immunol (2019) 10:2741. doi: 10.3389/fimmu.2019.02741
90. Reeves RK, Li H, Jost S, Blass E, Li H, Schafer JL, et al. Antigen-specific NK cell memory in rhesus macaques. Nat Immunol (2015) 16(9):927–32. doi: 10.1038/ni.3227
91. Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay PK, Roederer M. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat Protoc (2012) 7(12):2067–79. doi: 10.1038/nprot.2012.126
92. Tay MZ, Liu P, Williams LD, McRaven MD, Sawant S, Gurley TC, et al. Antibody-mediated internalization of infectious HIV-1 virions differs among antibody isotypes and subclasses. PloS Pathog (2016) 12(8):e1005817. doi: 10.1371/journal.ppat.1005817
93. R Core Team. R: A language and environment for statistical computing. (2021) (Vienna, Austria: R Foundation for Statistical Comput).
94. Kassambara A, Mundt F. factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R package version 1.0.7 (2020).
95. Yu H, Li F, Gallis JA, Turner EL. cvcrand: Efficient Design and Analysis of Cluster Randomized Trials. R package version 0.1.0 (2020).
Keywords: NK cells, ADCC, Fc gamma receptor, antibody, FcR-mediated effector functions, rhesus macaques, principal component analysis
Citation: Tuyishime M, Spreng RL, Hueber B, Nohara J, Goodman D, Chan C, Barfield R, Beck WE, Jha S, Asdell S, Wiehe K, He MM, Easterhoff D, Conley HE, Hoxie T, Gurley T, Jones C, Adhikary ND, Villinger F, Thomas R, Denny TN, Moody MA, Tomaras GD, Pollara J, Reeves RK and Ferrari G (2023) Multivariate analysis of FcR-mediated NK cell functions identifies unique clustering among humans and rhesus macaques. Front. Immunol. 14:1260377. doi: 10.3389/fimmu.2023.1260377
Received: 17 July 2023; Accepted: 04 October 2023;
Published: 06 December 2023.
Edited by:
Ricciarda Galandrini, Sapienza University of Rome, ItalyReviewed by:
Charles Daniel Murin, The Scripps Research Institute, United StatesSilke Paust, Jackson Laboratory for Genomic Medicine, United States
Caroline T. Tiemessen, National Institute of Communicable Diseases (NICD), South Africa
Copyright © 2023 Tuyishime, Spreng, Hueber, Nohara, Goodman, Chan, Barfield, Beck, Jha, Asdell, Wiehe, He, Easterhoff, Conley, Hoxie, Gurley, Jones, Adhikary, Villinger, Thomas, Denny, Moody, Tomaras, Pollara, Reeves and Ferrari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marina Tuyishime, marina.tuyishime@duke.edu
†Present addresses: Stephanie Asdell, University of Chicago Medical School, Chicago, IL, United States
David Easterhoff, Moderna Therapeutics, Cambridge, MA, United States
‡These authors have contributed equally to this work and share first authorship
§These authors have contributed equally to this work and share senior authorship
 Marina Tuyishime
Marina Tuyishime Rachel L. Spreng
Rachel L. Spreng Brady Hueber3
Brady Hueber3 Derrick Goodman
Derrick Goodman Cliburn Chan
Cliburn Chan Richard Barfield
Richard Barfield Kevin Wiehe
Kevin Wiehe Haleigh E. Conley
Haleigh E. Conley Taylor Hoxie
Taylor Hoxie Rasmi Thomas
Rasmi Thomas Thomas N. Denny
Thomas N. Denny Michael Anthony Moody
Michael Anthony Moody R. Keith Reeves
R. Keith Reeves Guido Ferrari
Guido Ferrari