- 1Department of Agricultural, Environmental and Food Sciences, University of Molise, Campobasso, Italy
- 2Department of Agricultural Science, University Federico II, Portici, Napoli, Italy
- 3Institute of Food Science, National Council of Research (ISA-CNR), Avellino, Italy
Introduction: The genus Bifidobacterium is a key component of the honey bee gut microbiota, playing a fundamental role in maintaining host health and colony well-being. Alongside other core genera such as Bombilactobacillus, Gilliamella, Lactobacillus, and Snodgrassella, Bifidobacterium contributes to essential functions including nutrient digestion, immune modulation, and protection against pathogens. Among threats to honey bee health, Chalkbrood disease, caused by fungus Ascosphaera apis, remains a major concern due to detrimental effects on colony strength and honey yield.
Materials and methods: We characterized enzymatic activity and carbohydrate assimilation of nine Bifidobacterium strains isolated from the honey bee intestinal tract. In parallel, we assessed antifungal potential against A. apis strains, focusing on volatile organic compounds (VOCs).
Results and discussion: Notably, Bifidobacterium asteroides 3CP-2B exhibited enzymatic capabilities supporting digestive functions and metabolism of sugars potentially harmful to honey bees. This strain showed marked antifungal activity against A. apis, mediated by volatile and non-volatile bioactive metabolites. Among VOCs identified, propanoic acid, ethanol, acetic acid, ethyl propionate, and 1-propanol were the most prominent compounds associated with the antifungal effect.
1 Introduction
In recent years, honey bees (Apis mellifera L.) have emerged as an important model system for understanding the functional roles of bacteria within the gut microbiome (1, 2). However, it remains unclear how specific members of the gut microbiota influence bee health and physiological state (3). Honey bee gut is primarily dominated by nine bacterial taxa, which together comprise more than 95% of the total gut microbial community. Among these, five phylogenetic lineages are consistently present in every individual and are defined as the core members of the honey bee gut microbiota. These core lineages represent genus-level taxa from distinct bacterial classes: Gilliamella (Gammaproteobacteria), Snodgrassella (Betaproteobacteria), Lactobacillus Firm-4 (including Bombilactobacillus), Lactobacillus Firm-5 (including Apilactobacillus), and Bifidobacterium (Actinobacteria).
This characteristic taxonomic composition of the microbiota, comprising largely species exclusive to social honey bees, along with their essential biochemical contributions to the host, suggests a highly specialized and co-evolved relationship between microbes and honey bees (4, 5). A gut microbiota with a balanced composition plays a crucial role in defending against pathogens and parasites, detoxifying foodborne toxins, and regulating the immunity, metabolism, behavior, and development of honey bees. Conversely, dysbiosis of this community can lead to altered gene expression related to these key functions, potentially compromising overall health and well-being (3).
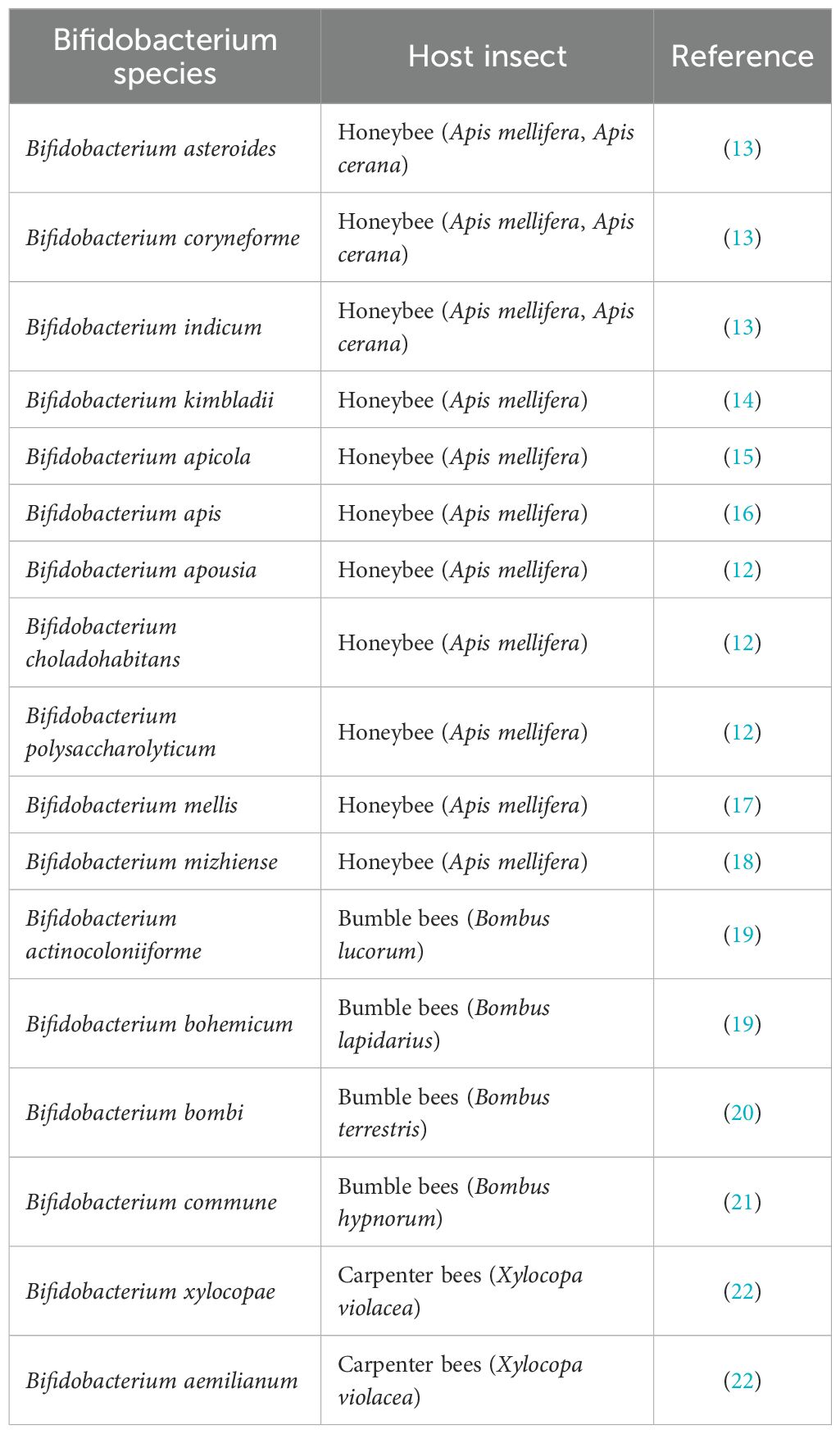
The genus Bifidobacterium encompasses Gram-positive bacteria belonging to the family Bifidobacteriaceae within the phylum Actinomycetota (6). Bifidobacterium spp. are symbiotic microorganisms that contribute to gastrointestinal homeostasis in humans, animals, and insects; in honey bees, they colonize the gut throughout development, with maximal abundance in the adult hindgut (3, 7–10). Although typically less abundant than other core gut taxa, they play a critical role in host metabolism, immune regulation, disease resilience, and adaptation to environmental stressors (11, 12). To date, multiple Bifidobacterium species have been identified and characterized from the gut microbiota of various honey bee species within the family Apidae (Table 1). Recently, Bifidobacterium favimelis, a novel species isolated from black comb honey of A. mellifera, was identified by Li et al. (23). The presence and divergence of Bifidobacterium strains in honey bees is attributed to a long-term coevolutionary process, reflecting their adaptation to various microenvironments within the bee gut and hive, as well as to hive-mediated vertical transmission across generations (24–27). Populations of bifidobacteria in honey bees have been observed to remain relatively stable over time, suggesting that these microorganisms play a consistent and essential role in host physiology (28, 29). Strains of Bifidobacterium inhabiting the honey bee gut are of particular interest due to their potential probiotic properties. For example, B. asteroides has been shown to stimulate the production of host-derived hormones, such as prostaglandins and juvenile hormone derivatives, which are known to influence honey bee development (30). Comprehensive genomic analyses have revealed that Bifidobacterium species harbor a substantial repertoire of genes involved in carbohydrate metabolism, underscoring their functional role in insect physiology (4, 12, 31). Recent studies on pollinator gut microbiota have further elucidated the involvement of bifidobacteria in maintaining immune function, enhancing disease tolerance, and improving resistance to environmental stressors (11). The genus Bifidobacterium supports honey bee health through polysaccharide degradation and immune modulation; however, its abundance and overall gut microbiota stability are influenced by factors such as diet, seasonal changes, caste roles, geography, and exposure to xenobiotics like herbicides and antibiotics, which can disrupt microbial balance and lead to dysbiosis, impairing metabolism and vitamin biosynthesis (14, 29, 31–36). Moreover, a disrupted gut microbiota may increase honey bee susceptibility to parasitic infections, including those caused by Nosema spp. and Ascosphaera apis (3, 36–40).
Chalkbrood, caused by the fungus A. apis, is a widespread fungal disease that primarily affects developing honey bee brood, especially in A. mellifera colonies, although it can also impact various other bee taxa (41–44). Recent evidence indicates an increasing global incidence of chalkbrood, which is contributing to honey bee population declines and significant reductions in colony productivity (45–48); moreover, it has been shown that chalkbrood infection alters the honey bee gut bacteriome and increases the host’s vulnerability to other pests and pathogens (49–52). A. apis is generally regarded as an opportunistic pathogen that is efficiently dispersed and highly prevalent; however, its mere presence in the hive does not necessarily lead to disease manifestation. Rather, one or more predisposing factors must coincide for a clinical outbreak to occur. These include environmental stressors such as damp and cold weather, colony health status, genetic susceptibility, and developmental stress within the brood (53, 54). Infection is initiated when larvae orally ingest fungal ascospores, which subsequently germinate in the posterior midgut. The resulting hyphae invade the epithelial cells and basement membrane, ultimately leading to larval death. Fungal development continues in a necrotrophic phase even after the host’s demise (55).
Over the years, various chemotherapeutic agents have been investigated for their efficacy against A. apis (53, 56), but, none have proven effective in preventing chalkbrood, despite their antifungal activity. Moreover, the presence of antifungal residues in honey represents a potential health hazard for consumers (57). Consequently, there is a growing demand for eco-friendly, and sustainable alternatives for disease control (58–63). In this context, the use of microbial resources as biocontrol agents against honey bee pathogens, including A. apis, offers promising opportunities (64–66). Several studies have demonstrated that Apilactobacillus kunkeei and Lactiplantibacillus plantarum, isolated from the honey bee gut, can inhibit A. apis mycelial growth in vitro, suggesting their potential as prophylactic agents to restore and maintain gut microbial balance (56, 67). Similarly, other beneficial microbes have shown effectiveness in the biocontrol of chalkbrood (68, 69). Notably, Daisley et al. (70) demonstrated that hive treatments with a probiotic formulation containing L. plantarum, Lacticaseibacillus rhamnosus, and A. kunkeei exerted strong antifungal effects against A. apis while also promoting the recovery of symbiotic gut communities. These microbiome shifts were positively correlated with enhanced brood production and colony development (70).
To date, there is limited research on the use of B. asteroides as an anti-fungal or probiotic agent in beekeeping (71). In a study by Alberoni et al. (28), symbiotic species including B. asteroides, B. coryneforme, and B. indicum were shown to enhance colony productivity when administered in sugar syrup as a dietary probiotic. Additionally, Bifidobacterium spp. supplementation led to reduced Nosema infection rates in honey bee colonies (11). More recently, Dengiz et al. (72) reported significant antimicrobial activity by B. asteroides, B. choladohabitans, and B. polysaccharolyticum against key bee pathogens such as Paenibacillus larvae, Melissococcus plutonius, and Serratia marcescens. Conversely, a Bifidobacterium bifidum strain, isolated from human feces, did not exhibit inhibitory effect on A. apis mycelial growth (73). This finding supports the growing consensus that exogenous probiotics, not derived from the honey bee microbiota, may lack beneficial effects, or even pose risks, to health (3, 74–77). Therefore, the identification and characterization of autochthonous Bifidobacterium strains with honey bee-specific probiotic properties is essential for developing effective, safe, and sustainable tools for disease prevention in apiculture.
In the present study, a preliminary characterization of Bifidobacterium strains isolated from the gastrointestinal tract of A. mellifera (collected from apiaries in central-southern Italy) was conducted, including analyses of enzymatic activity and carbohydrate assimilation profiles. Furthermore, the antifungal activity of these strains against multiple A. apis isolates was evaluated, with particular focus on the production of volatile organic compounds (VOCs). This is the first report describing the potential of B. asteroides as a biocontrol agent against chalkbrood disease.
2 Materials and methods
2.1 Fungal cultures
Table 2 provides a detailed overview of the A. apis strains employed in the present study.
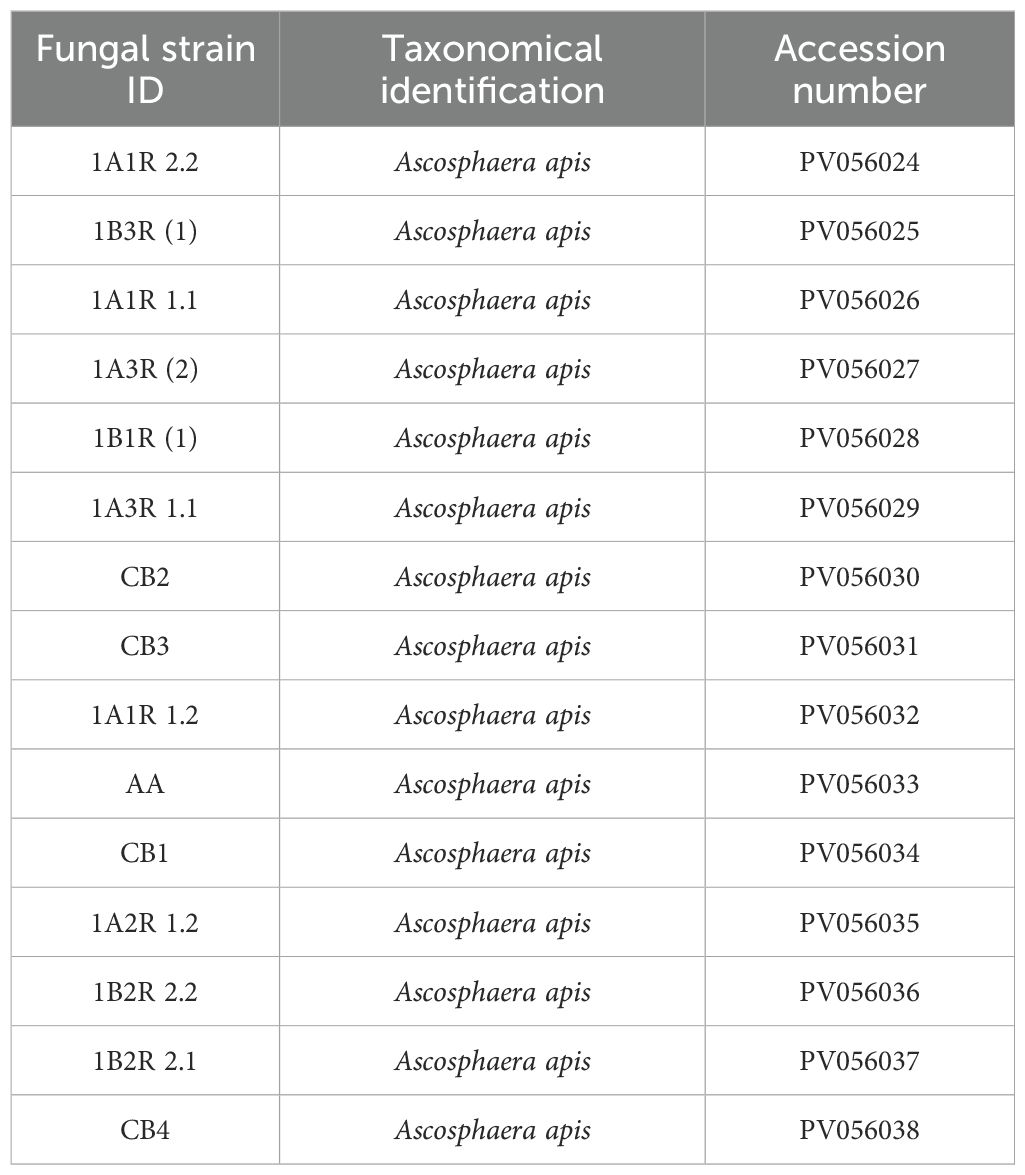
Table 2. Catalogue of A. apis strains utilized in this study, accompanied by their GenBank accession numbers from the National Center for Biotechnology Information (NCBI).
2.2 Isolation of bifidobacteria
Worker bees (A. mellifera subsp. mellifera) were collected from managed apiaries in the Molise and Campania regions (central-southern Italy). After euthanization by rapid cooling on ice, bees were transported under refrigeration to the laboratory on the same day and stored at −80 °C until DNA extraction. Dissection of the intestinal tract was performed under sterile conditions using stainless-steel scissors. Entire guts were placed in sterile glass Petri dishes with physiological saline solution (0.9% NaCl) and homogenized. Serial dilutions of the homogenates were plated on Bifidobacterium Selective Medium agar (BSM; Sigma-Aldrich) and incubated at 37 °C for 72 h under anaerobic conditions (Anaerogen system, Oxoid, Milan, Italy). Colonies showing Gram-positive staining with characteristic bifurcated (Y- or V-shaped), club-shaped, or spatula morphologies were presumptively identified as Bifidobacterium.
2.3 Genotypic characterization
Genomic DNA (gDNA) was extracted using the Bacterial Genomic DNA Isolation Kit (Norgen Biotek, Thorold, ON, Canada) according to the manufacturer’s instructions. The 16S rRNA gene was amplified by PCR using universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′). Each 20 µL reaction included 1× Master Mix (Norgen Biotek), 2.5 µM of each primer, and 10 ng of template DNA. Negative controls using Milli-Q water were included. PCR was performed using a Mastercycler Nexus (Eppendorf, Hamburg, Germany) with cycling parameters: 95 °C for 5 min; 35 cycles of 95 °C for 30 s, primer-specific annealing temperature for 1 min, 72 °C for 1.5 min; followed by a final extension at 72 °C for 5 min. Products were resolved on a 1% (w/v) agarose gel in 1× TAE buffer, visualized under UV light (Bio-Rad, Hercules, CA, USA), and compared against a 1 kb DNA ladder (Norgen Biotek). Amplicons were purified using the QIAquick PCR Purification Kit (QIAGEN GmbH, Hilden, Germany) and sequenced (Eurofins MWG Biotech, Ebersberg, Germany). Sequences were analyzed using BLAST (78) against the NCBI nucleotide database (NCBI 79). Strains with ≥98% identity were assigned species-level taxonomy (80).
2.4 Biochemical characterization
2.4.1 Carbohydrate assimilation patterns
Carbohydrate utilization was tested using Fermentation Broth Base (FBB; Biolife, Milan, Italy) supplemented with bromocresol purple as a pH indicator. Prior to testing, bacterial strains were cultured in BSM broth at 37 °C for 48 hours under anaerobic conditions. Cultures were centrifuged, and the cell pellet washed with 0.9% NaCl solution to remove the residual medium. The pellet was then resuspended in saline to reach a standard turbidity of 0.5 McFarland (approx. 1.5 × 108 CFU/mL) (81), and used as inoculum. Thirteen carbohydrates were tested: D-arabinose, fructose, galactose, glucose, lactose, maltose, mannose, melezitose, melibiose, raffinose, rhamnose, sucrose, and xylose. For each assay, 4.5 mL of FBB were mixed with 500 μL of carbohydrate solution and 100 μL of bacterial suspension. Negative controls were prepared identically but without inoculum. Sugar solutions were sterilized using 0.22 μm syringe filters. Assays were incubated at 37 °C for 48 hours under anaerobic conditions. The change of color from purple to yellow is due to the production of acids during fermentation. All tests were performed in triplicate for each strain–carbohydrate combination.
2.4.2 Enzymatic profile
Enzymatic activity was evaluated using the API ZYM system (BioMérieux, Lyon, France). The cell pellet (CP), prepared as above, was resuspended in 0.9% NaCl solution to achieve a turbidity of 5 McFarland. Wells of the API ZYM strip were inoculated with 65 µL of this suspension and incubated at 37 °C. After 4 hours, enzymatic activity was assessed based on color change, according to the manufacturer’s guidelines.
2.4.3 Biogenic amine production
Biogenic amine production by Bifidobacterium strains was qualitatively assessed using method from Torracca et al. (82), with slight modifications. The Bifidobacterium strains were grown in BSM broth at 37 °C for 48 h anaerobically. Subsequently, the cultures, in a volume of 50 µL, were inoculated using a spot inoculation method onto solid media formulated with the following components: 0.5% tryp-tone, 0.5% yeast extract, 0.5% meat extract, 0.25% NaCl, 0.05% glucose, 0.1% Tween 80, 0.02% MgSO4, 0.005% MnSO4, 0.004% FeSO4, 0.2% ammonium citrate, 0.001% thia-mine, 0.2% K2HPO4, 0.01% CaCO3, 0.005% pyridoxal-5-phosphate, and 1.5% agar. The medium was supplemented with 1% of each amino acid precursor: L-histidine, L-tyrosine, L-lysine monohydrate, and L-ornithine monohydrochloride. Bromocresol purple (0.006%) was incorporated as a pH indicator, and the medium pH was adjusted to 5.3 prior to inoculation. Petri dishes were incubated at 37 °C for 72 hours under anaerobic conditions. The decarboxylation of the amino acids to the corresponding biogenic amines results in an increase in pH, detected by the culture medium color change. A purple coloration indicated the production of histamine, cadaverine, or putrescine, while medium de-colorization suggested tyramine production. Negative controls lacking amino acid precursors were included to confirm the specificity of the reactions. All tests were conducted in triplicate and all reagents were purchased from Merck KGaA (Darmstadt, Germany).
2.5 Antifungal activity assessment
2.5.1 Preliminary evaluation of antifungal activity
Antifungal activity of Bifidobacterium strains was assessed using a method adapted from Iorizzo et al. (69). Three matrices were tested: broth culture (BC), cell-free supernatant (CFS), and CP. Bifidobacteria were grown in BSM broth at 37 °C for 48 hours under anaerobic conditions to a final cell density of 108 CFU/mL. BC was collected without further treatment. For CFS, 5 mL of bacterial culture was centrifuged at 8000 rpm for 15 min at 4 °C, and the supernatant filtered through a 0.22 μm cellulose acetate membrane. CP was prepared by washing and resuspending the pellet in 5 mL of sterile distilled water. Antifungal assays were conducted by transferring a 6 mm mycelial disc of A. apis, pre-cultured on Sabouraud Dextrose Agar (SDA) at 30 °C for 3 days, to the center of 90 mm Petri dishes containing fresh SDA. In different plates containing the pathogenic fungus, 5 mL of each matrix (BC, CFS, or CP) was added alternately. An SDA plate containing only the pathogenic fungus was used as a control. All plates were incubated aerobically at 30 °C, and each experimental condition was tested in triplicate. Following six days of incubation, the radial growth of A. apis mycelium was measured using a digital caliper. The percentage of mycelium radial growth inhibition (% I) was calculated according to the formula: % I = [(C – T)/C] × 100 (83), where C represent the radial growth in the control, and T represents the radial growth in the presence of different matrices obtained from the Bifidobacterium cultures.
2.5.2 Antifungal activity of the VOCs produced by Bifidobacterium
The antifungal activity of VOCs produced by Bifidobacterium strains was evaluated using a modified double-dish system (DDS) based on Ruiz-Moyano et al. (84). A 100 μL aliquot of a 48-hour culture (108 CFU/mL) was spread on BSM agar in 90 mm Petri dishes. Simultaneously, a 6 mm disc of A. apis mycelium (grown on SDA) was placed in the center of a separate plate. Lids of both plates were removed and the dishes sealed together in an inverted DDS configuration using Parafilm (Pechiney Plastic Packaging Co., Milwaukee, WI, USA), with the fungal plate on the bottom. This setup allowed VOCs to diffuse freely. Control DDS setups (no bacteria) were included. After 6 days at 30 °C, radial growth inhibition was measured as previously described. All experiments were performed in triplicate.
2.6 Volatile organic compounds profiling
2.6.1 VOCs extraction
VOCs were extracted using headspace solid-phase microextraction (HS-SPME) (Figure 1). B. asteroids 3CP-2B was cultured for 72 h at 30°C in BSM medium (15 mL) directly in 30 mL screw capped SPME vials (Agilent Technologies, Santa Clara, CA, USA). The vials were sealed with a magnetic screw capped PTFE/silicone liner septum. After 72 hours, the vial was equilibrated to 40 °C for 15 minutes to allow equilibration of the headspace. A 2 cm DVB/CAR/PDMS fiber (50/30 µm; Supelco, Bellefonte, PA, USA) was then inserted into the vial headspace and exposed at 40 °C for 30 minutes to adsorb volatile compounds. Following adsorption, the fiber was immediately transferred to the GC injector port, where desorption was performed at 240 °C for 10 minutes in splitless mode. To distinguish bacterial VOCs from background volatiles, control vials containing non-inoculated media were processed in parallel. Blank runs were also conducted between samples to confirm the absence of carryover or contamination throughout the extraction and analytical procedures. The experiment was done in triplicate.
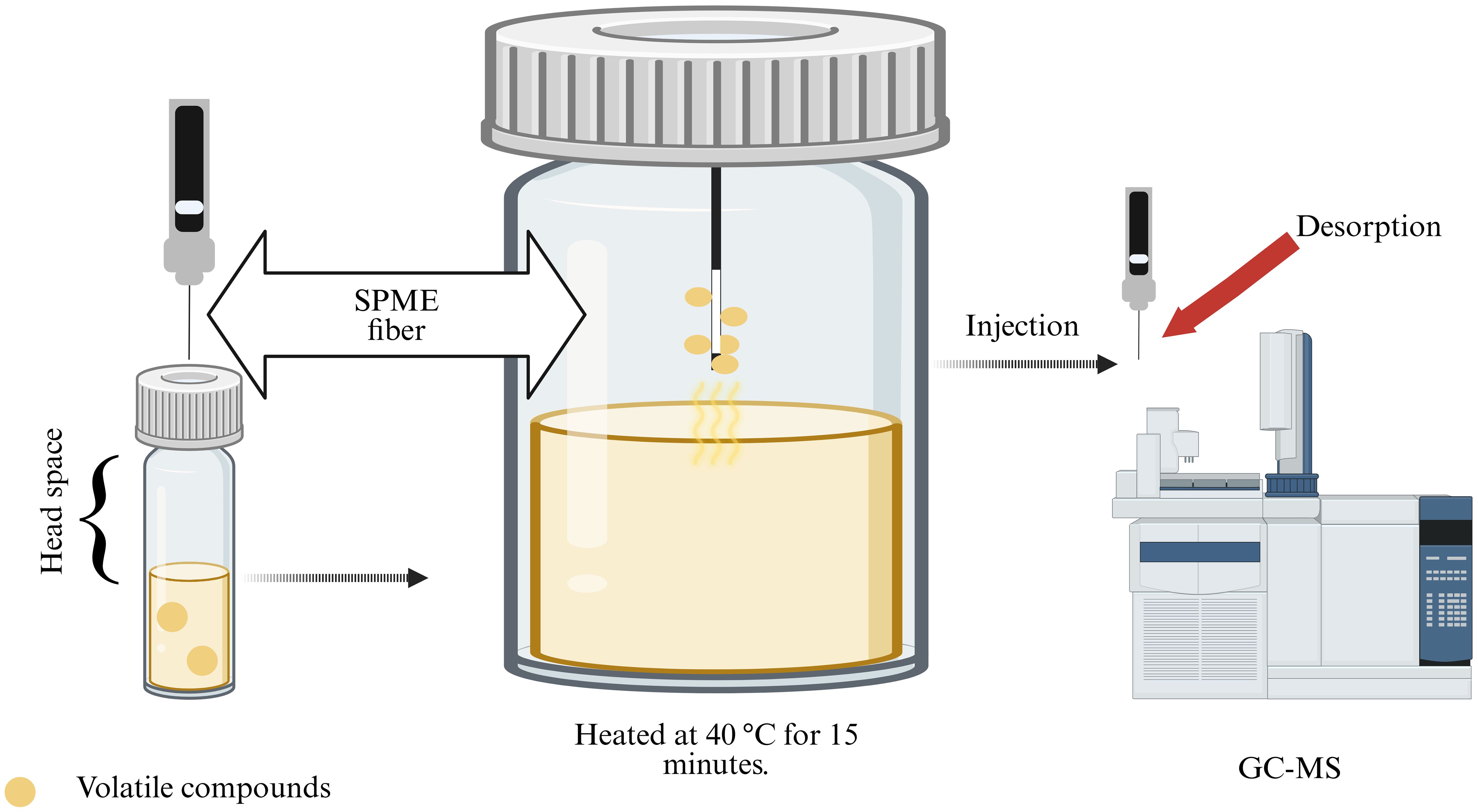
Figure 1. Experimental workflow used to evaluate the production of volatile organic compounds (VOCs) by B asteroides 3CP-2B. VOCs were subsequently captured using a solid-phase microextraction (SPME) fiber and were analyzed using gas chromatography coupled with mass spectrometry (GC-MS) (Created with BioRender.com).
2.6.2 Gas chromatography–mass spectrometry analysis
Analysis of volatile organic compounds was conducted using a GC–MS system consisting of an Agilent 7890A gas chromatograph coupled with a 5975A mass selective detector (Agilent Technologies, Santa Clara, CA, USA). Chromatographic separation was achieved using a polar HP-Innowax capillary column (30 m × 0.25 mm i.d., 0.50 μm film thickness; Agilent Technologies). The method was adapted from Serradilla et al. (85), with the oven temperature program set as follows: initial hold at 40 °C for 3 minutes; ramp to 150 °C at 4 °C/min, held for 1 minute; then increased to 220 °C at 3 °C/min with a final hold for 2 minutes. Helium was employed as the carrier gas at a constant flow rate of 1.0 mL/min. The injector was operated in splitless mode, and the desorbed VOCs were introduced directly into the ion source. Electron impact (EI) ionization was performed at 70 eV. The ion source and quadrupole temperatures were set to 230 °C and 150 °C, respectively. Mass spectra were acquired in full-scan mode over a range of m/z 30–300. Compound identification was performed by comparing the obtained mass spectra and retention indices (linear retention indices, LRI) with entries in the NIST05 and Wiley07 spectral libraries. When available, identification was further confirmed using authentic standards. Semi-quantitative analysis of each VOC was expressed as the relative peak area (RPA%), calculated as the ratio of the individual compound’s area to the total area of all detected VOCs in the total ion chromatogram (TIC).
2.7 Statistical analysis
Statistical analyses were conducted in Rstudio (R version 4.3.0). Results from triplicate experiments were expressed as mean ± standard deviation (SD). One-way ANOVA followed by Tukey’s post hoc test was used to determine significant differences (p < 0.05).
3 Results
3.1 Taxonomical identification
The nine isolates have been identified as members of the species B. asteroides, B. apousia, B. mizhiense and B. choladohabitans as reported in Table 3.
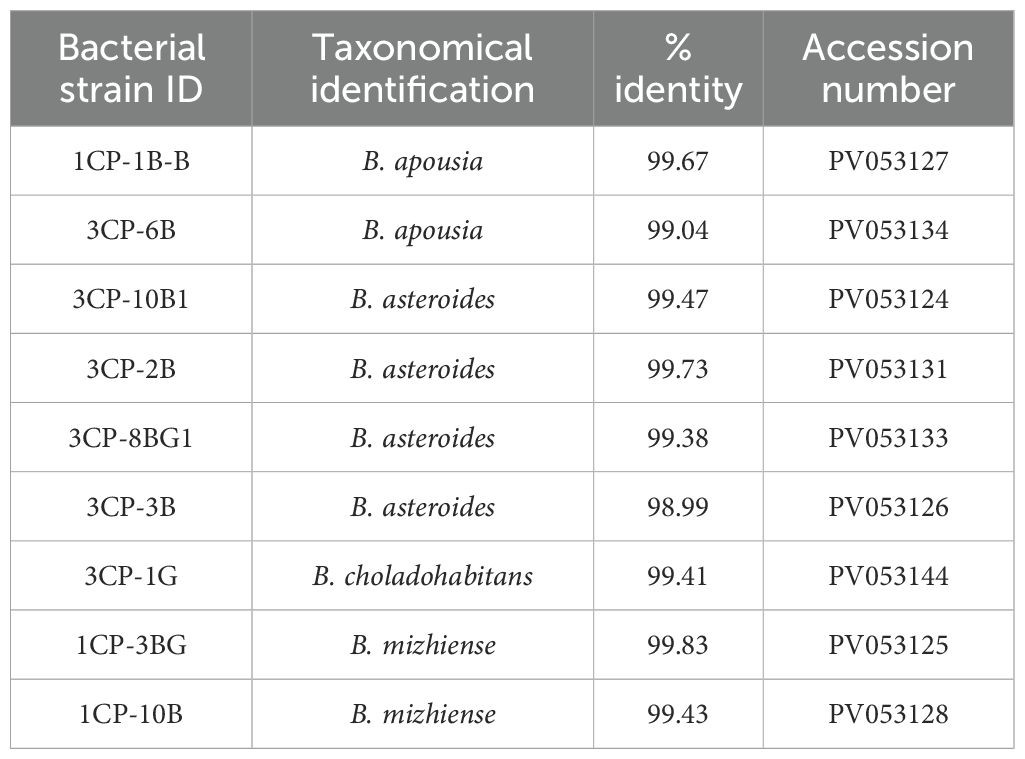
Table 3. List of Bifidobacterium strains isolated in this study with their taxonomic assignment and NCBI GenBank accession number.
3.2 Biochemical characterization
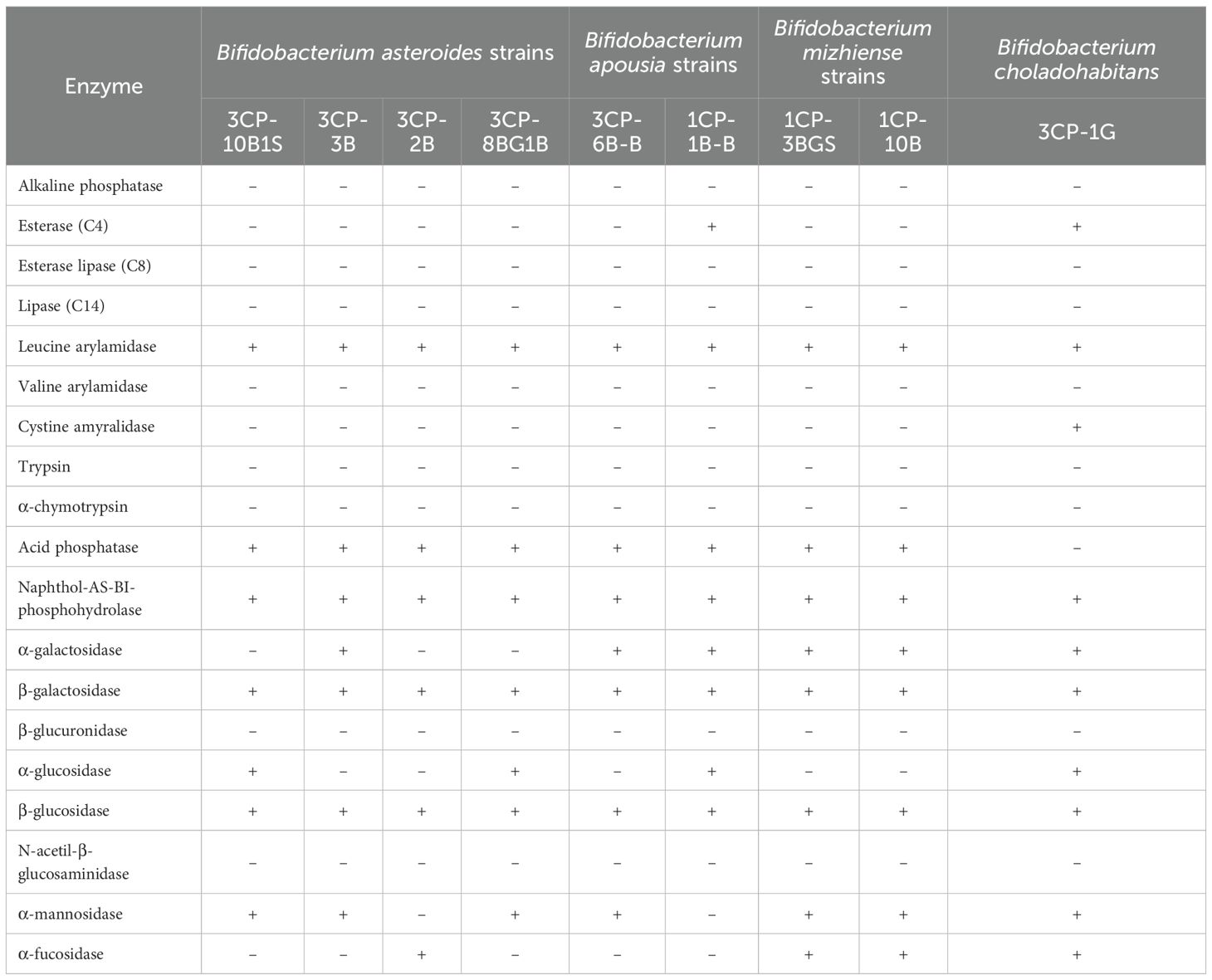
The enzymatic activities of the bacterial isolates were comprehensively assessed using the qualitative API ZYM kit, with results summarized in Table 4. The enzymatic profiles of the Bifidobacterium isolates, including B. asteroides, B. apousia, B. mizhiense, and B. choladohabitans, revealed activities for several enzymes, notably leucine arylamidase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, and β-glucosidase. Enzymatic activities varied within the B. asteroides species. For example, B. asteroides 3CP-3B exhibited α-galactosidase activity, which was absent in other strains of the same species. Additionally, strains 3CP-10B1S and 3CP-8BG1B were the only ones to display α-glucosidase activity. Strains 3CP-10B1, 3CP-3B, and 3CP-8BG1B also showed α-mannosidase activity, while strain 3CP-2B was unique in exhibiting α-fucosidase activity. Among B. apousia strains, 1CP-1B-B was distinct in showing both esterase and α-glucosidase activities. In contrast, B. apousia 3CP-6B-B was the only strain within its species to demonstrate α-mannosidase activity. No enzymatic differences were observed between the two B. mizhiense strains.
3.3 Carbohydrate assimilation profiles and biogenic amines production
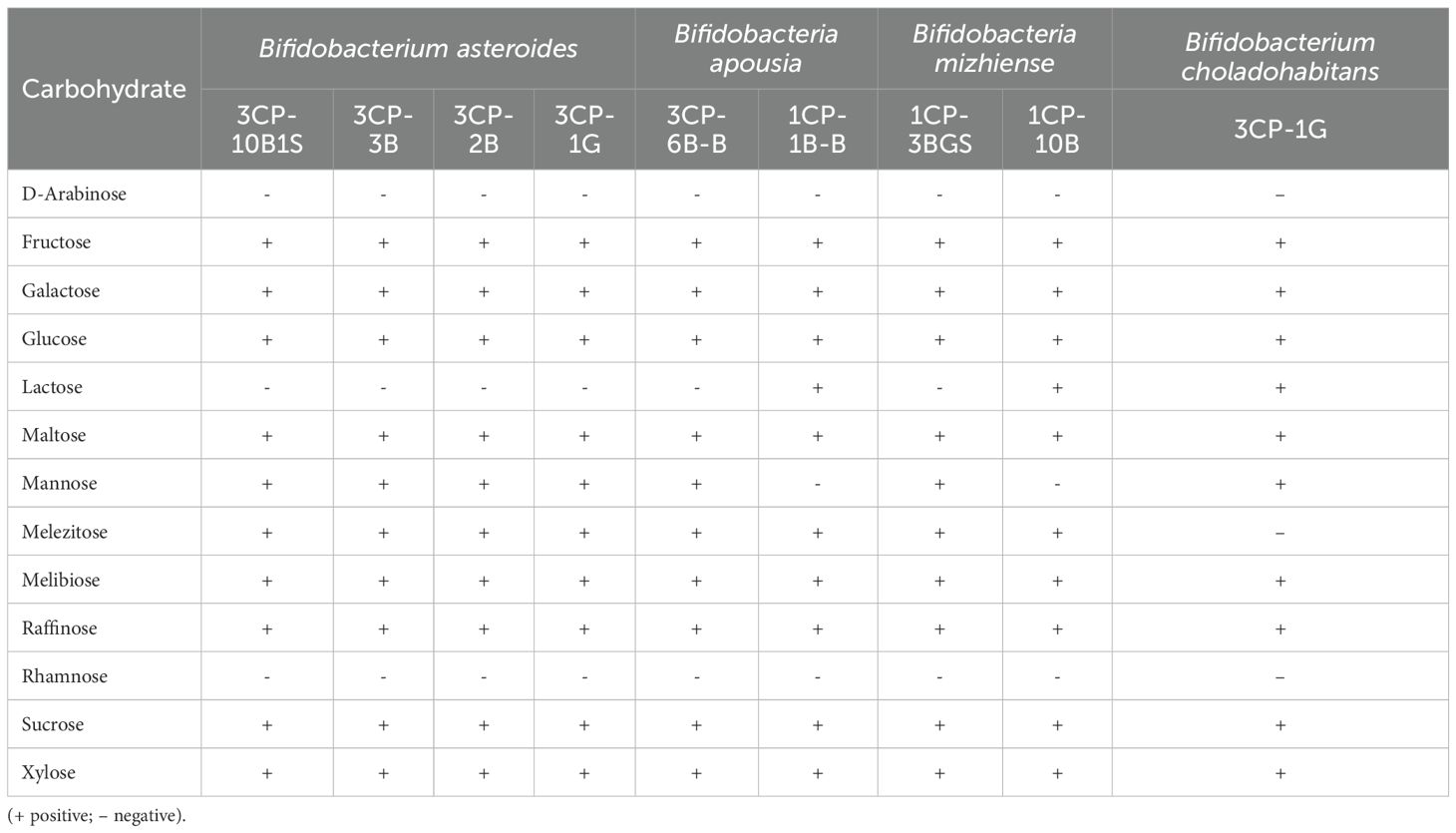
The carbohydrate assimilation abilities of the nine Bifidobacterium strains are detailed in Table 5. All strains were capable of metabolizing fructose, glucose, maltose, melibiose, raffinose, and sucrose. In contrast, none of the strains metabolized D-arabinose or rhamnose. Assimilation of lactose was strain-specific and limited to B. apousia 1CP-1B-B, B. mizhiense 1CP-10B, and B. choladohabitans 3CP-1G. Regarding the production of biogenic amines, none of the strains were able to synthesize these compounds from the tested amino acid precursors.
3.4 Screening of antifungal activity by Bifidobacterium strains
Table 6 presents the antifungal activity of the nine Bifidobacterium isolates against A. apis CB3. In most cases, the use of unprocessed culture broth BC resulted in complete inhibition (100%) of fungal growth. Notable exceptions were isolates 3CP-8BG1B and 3CP-6B-B, which exhibited inhibition rates of 76.7% and 98.9%, respectively. The CFS also showed strong antifungal activity, with inhibition percentages ranging from 65.9% (B. asteroides 3CP-8BG1B) to 100% (B. asteroides 3CP-2B). Regarding the CP fraction, B. mizhiense 1CP-10B exhibited the lowest inhibition (35%), whereas strains 3CP-2B and 1CP-3BGS achieved full inhibition (100%). In terms of VOCs, overall inhibition levels were modest. However, strains 3CP-2B and 3CP-8BG1B showed moderate activity, with inhibition values of 54.2% and 47.2%, respectively.
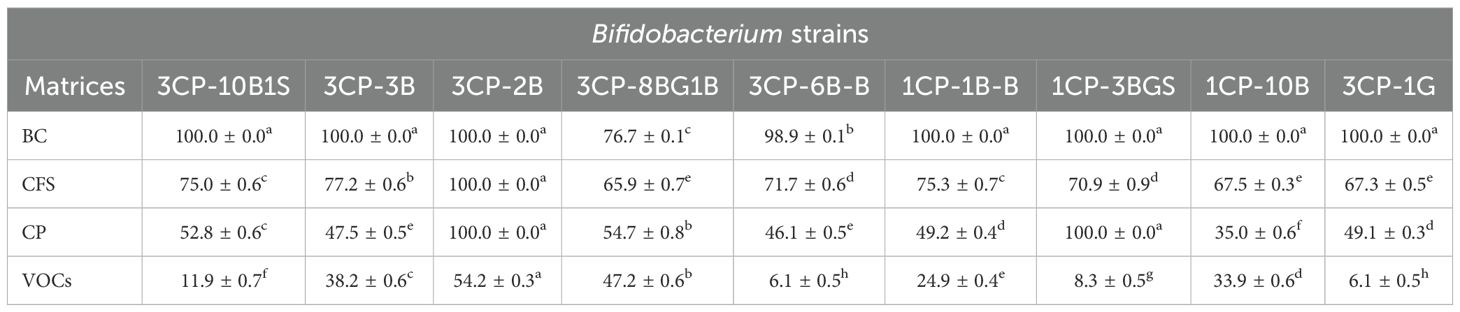
Table 6. Inhibitory effects (%) of Bifidobacterium strains against A. apis CB3 using different matrices: broth culture (BC), cell-free supernatant (CFS), cell pellet (CP), and volatile organic compounds (VOCs).
Subsequently, B. asteroides 3CP-2B, the most effective strain, was selected as the reference bacterium for further antifungal assays against all A. apis strains listed in Table 2.
The inhibitory activity of B. asteroides 3CP-2B against A. apis strains was assessed under four different treatments: BC, CFS, CP, and VOCs (Figure 2). Overall, the BC treatment exhibited the highest and most consistent antifungal efficacy, with most strains achieving complete inhibition. The CFS treatment also resulted in high inhibition levels, ranging from 88.0% (1A1R 1.2) to 100.0% (1A3R 1.1, 1A2R 1.2, 1A3R (2) and CB1). The CP treatment led to moderately reduced inhibition (from 85.6% to 93.3%). VOCs were the least effective treatment, with inhibition ranging widely from 6.2% (CB1) to 72.8% (CB3), and statistically significant differences among nearly all strains.
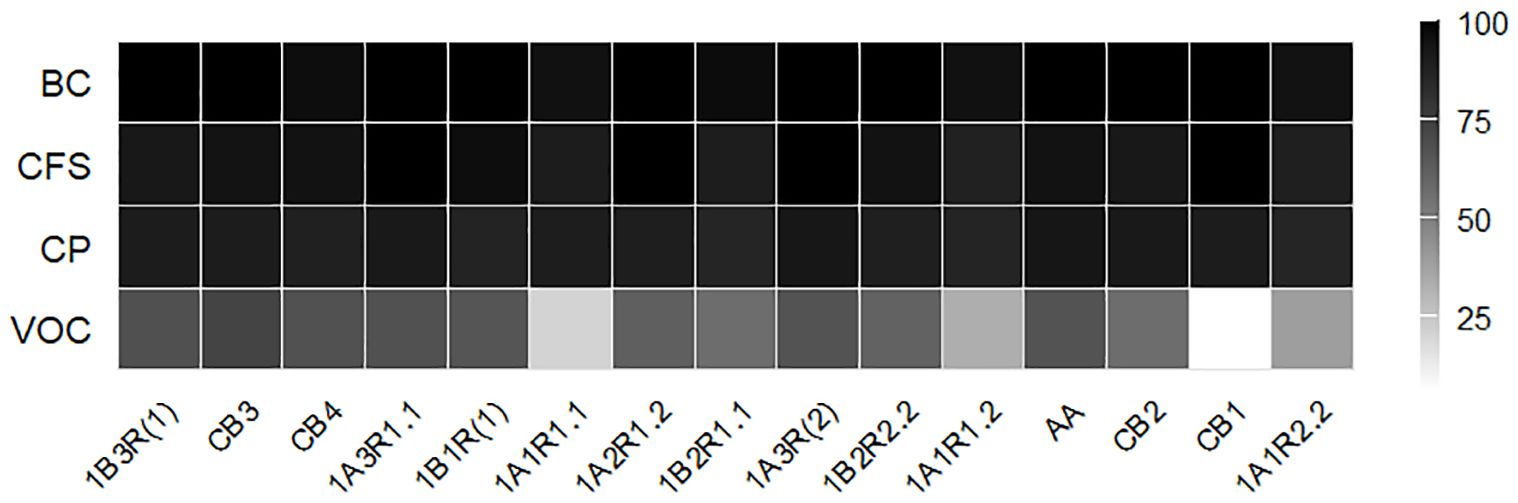
Figure 2. Heatmap showing the inhibition percentages of A apis strains by B asteroides 3CP-2B across different matrices (BC, CFS, CP and VOC).
3.5 Profiling of volatile organic compounds
Supplementary Table 1 lists all 37 VOCs detected by GC–MS analysis, along with their semi-quantitative relative peak area (RPA%) data. Based on peak areas, the major compounds detected were propanoic acid (45.8%), ethanol (25.0%), acetic acid (17.3%), ethyl propionate (3.1%), 1-propanol (2.3%), isoamyl alcohol (1.7%), propyl propionate (0.7%), ethyl acetate (0.4%), butanoic acid (0.3%), benzaldehyde (0.2%), and 2-methyl-propanoic acid (0.2%) (Table 7).
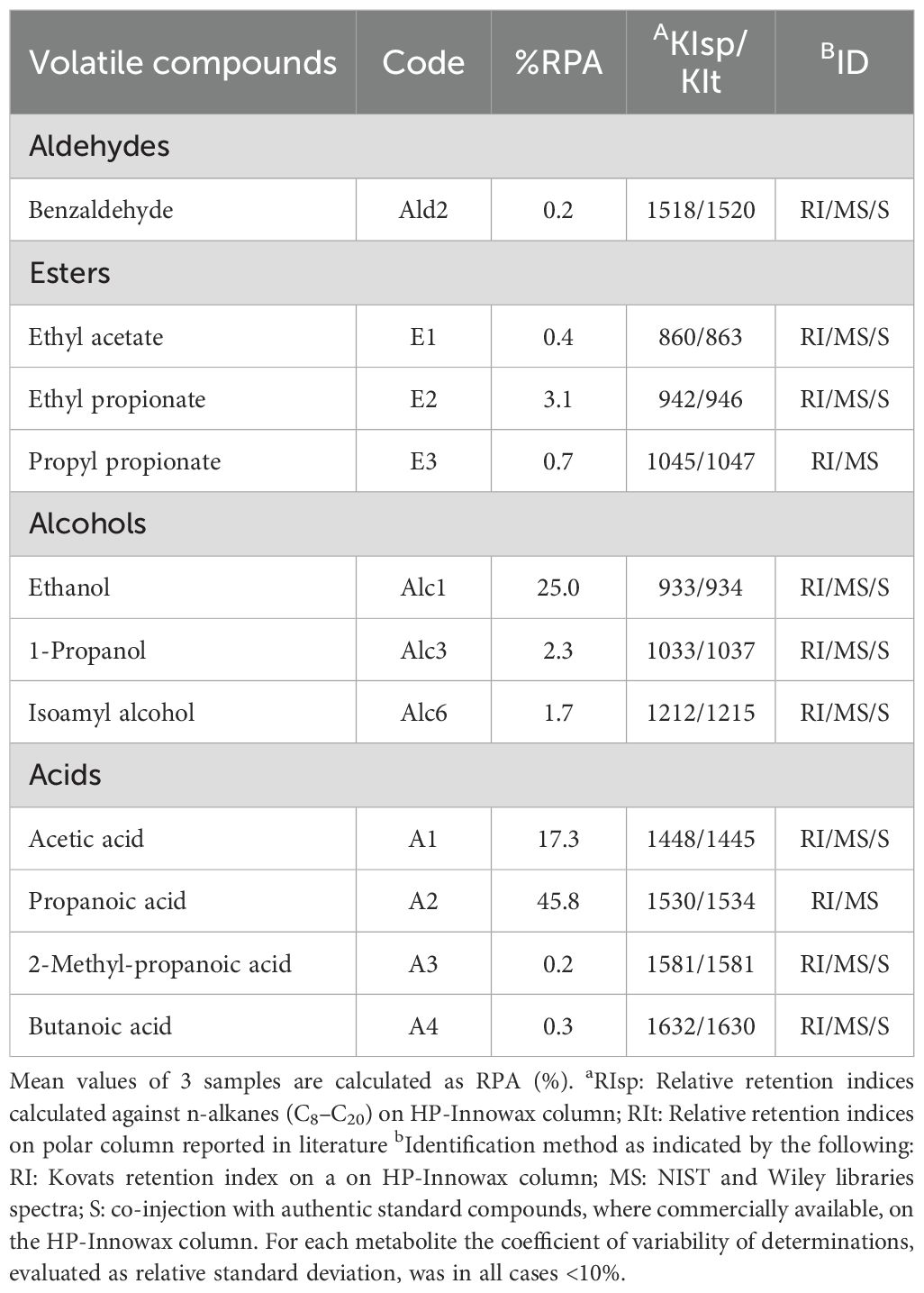
Table 7. Main Volatile Organic Compounds (VOCs) produced by Bifidobacterium asteroides 3CP-2B detected by HS-SPME/GC-MS with corresponding %RPA.
4 Discussion
The isolated microbial cultures were identified as members of the species B. asteroides, B. apousia, B. mizhiense, and B. choladohabitans.
Among these, B. asteroides is of particular interest due to its previously reported oxygen tolerance and its role in carbohydrate metabolism (86). This species metabolizes dietary sugars, including glucose and fructose, and utilizes the malolactic fermentation pathway to convert malic acid into lactate, thereby contributing to the host’s energy metabolism (86). However, functional data on B. apousia and B. mizhiense remain still scarce. Based on the known metabolic capabilities of other Bifidobacterium species, these isolates are hypothesized to play a significant role in sugar degradation, with B. apousia potentially involved in hemicellulose breakdown (12). The balance of the honey bee gut microbiota is crucial for host health, with microbial enzymatic activity directly supporting digestive function (87, 88). Notably, the B. apousia strain 1CP-1B-B and B. choladohabitans 3CP-1G showed esterase activity; a function involved in lipid digestion, and playing a role in detoxification by hydrolyzing or degrading various compounds, including drugs, pesticides, and other xenobiotics (89–92). Honey bees employ a multifaceted detoxification strategy, including enzymatic processes such as those involving cytochrome P450 monooxygenases, glutathione S-transferases, and carboxylesterases. These enzymatic defenses are complemented by behaviors forming a “social detoxification system,” which includes forager discrimination, dilution through pollen mixing, and colony-level food processing via microbial fermentation, reducing the intake of harmful chemicals (93–96, 46). Given the widespread use of insecticides in agriculture, supplementing the honey bee diet with appropriate probiotics, capable of degrading such compounds, may benefit bee health (64, 97, 98).
All tested isolates exhibited leucine arylamidase activity, suggesting a common ability to participate in protein hydrolysis, consistent with other Bifidobacterium species (99). This enzymatic function complements the proteolytic capabilities of other core honey bee gut microbes, such as Snodgrassella alvi and Gilliamella apis (31, 100). Positive activities were also recorded for acid phosphatase, naphthol-AS-BI-phosphohydrolase, β-galactosidase, and β-glucosidase. Among these, β-glucosidase is particularly important for degrading plant-derived polysaccharides such as cellulose and hemicellulose (12, 101), while β-galactosidase catalyzes the hydrolysis of β-D-galactosides, contributing to the digestion of galactose-containing nectar compounds (31). α-Glucosidase activity was detected in B. asteroides 3CP-10B1S and 3CP-8BG1B, B. apousia 1CP-1B-B, and B. choladohabitans 3CP-1G. This enzyme, also secreted by the hypopharyngeal glands of honey bees (52, 102), plays a critical role in maltose hydrolysis and starch degradation (31, 103, 104), thus contributing to the conversion of nectar into honey.
Additional enzymatic functions, including α-galactosidase, α-mannosidase, and α-fucosidase were detected in certain isolates. Although less studied in honey bee-associated Bifidobacterium, these enzymes are involved in degrading complex plant oligosaccharides and polysaccharides. They may contribute to digestion in insects and produce prebiotic compounds that support immune modulation in mammals, including humans (105–107). For example, α-galactosidase breaks down complex carbohydrates such as raffinose and stachyose, important for nutrient absorption in insects (108). α-Fucosidase releases terminal fucose residues, which are key to cell–cell communication and host–microbe interactions in mammals (109, 110). α-Mannosidase hydrolyzes mannose-containing carbohydrates and helps produce prebiotic mannooligosaccharides, which promote the growth of beneficial gut bacteria (111).
Previous studies have suggested that the honey bee gut microbiome may facilitate the metabolism of toxic sugars (112–114). In our study, carbohydrate assimilation profiles revealed both intra- and inter-species variability. All isolates effectively utilized a range of mono- and oligosaccharides commonly found in the honey bee gut, including fructose, glucose, maltose, melezitose, melibiose, raffinose, and sucrose. These results align with earlier findings showing that Bifidobacterium species are well adapted to the bees’ carbohydrate-rich diet (31, 86). Some sugars present in the honey bee diet, such as galactose, mannose, lactose, raffinose, and xylose, can be toxic due to the absence of necessary host enzymes for their degradation (31, 114–116). Gut symbionts enhance the honey bee’s ability to process complex polysaccharides and detoxify harmful sugars, improving dietary efficiency and resistance to diseases (101, 117).
Regarding antifungal activity, our results demonstrated that the different matrices (BC, CFS, CP, and VOCs) derived from the evaluated Bifidobacterium strains were effective (Table 6). Variability in antifungal activity likely reflects differences in the types and quantities of antifungal metabolites produced (118). These metabolites, such as lactic acid, acetic acid, phenyl lactic acid (PLA), short-chain fatty acids (SCFAs), proteins, and others, can disrupt fungal cell membranes, causing damage and inhibiting growth (119; 72, 120–122). The antifungal effects of VOCs are mainly attributed to cell wall and membrane disruption, leakage of intracellular contents, and the induction of oxidative stress (123). Bifidobacteria degrade hexose sugars via the “bifid shunt” pathway, in which fructose-6-phosphoketolase (EC 4.1.2.2) plays a key role and serves as a taxonomic marker for the Bifidobacteriaceae family. This pathway typically yields 3 moles of acetate and 2 moles of lactate per 2 moles of glucose, though other byproducts, such as ethanol, can also be produced (124). Ethanol and acetic acid are known for their antimicrobial properties, including antifungal activity, through mechanisms such as membrane disruption, protein denaturation, and interference with fungal DNA and protein synthesis (125, 126). Similarly, 1-propanol and other alcohols (e.g., isoamyl alcohol, 1-butanol) exhibit antifungal effects likely through membrane disruption, inhibition of spore germination, and interference with transcription and translation processes (127, 128).
In our study, B. asteroides 3CP-2B produced abundant propanoic and butanoic acids, confirming that Bifidobacteria are effective SCFA producers (129, 130). These compounds increase membrane fluidity, causing leakage of intracellular contents and ultimately cell death (131). Propionic acid, in particular, generates reactive oxygen species (ROS), reduces ATP levels, and activates metacaspases, leading to mitochondrial-mediated apoptosis in fungal cells (132). Notably, propionic acid has been identified as a natural constituent of honey, contributing to its flavor and preservation (133).
Esters such as ethyl and propyl propionate have demonstrated antifungal activity (84), while other VOCs, like dimethyl disulfide and limonene, were also detected. Limonene damages fungal hyphae, causing cytoplasmic granulation, membrane detachment, and vacuole formation, ultimately leading to cell death (134, 135). Dimethyl disulfide exhibits antifungal activity by damaging membranes and inhibiting spore germination and hyphal growth (136, 137). B. asteroides 3CP-2B also produces methylpyrazines, aromatic hydrocarbons commonly found in foods and considered safe (138, 139). Pyrazine derivatives have broad biological activity, including antifungal effects (138, 140, 141). Gong et al. (142) demonstrated that methylpyrazine and dimethyl disulfide significantly inhibit fungal growth and spore germination. Transcriptome analysis showed that these VOCs downregulate ribosomal synthesis genes, activate the proteasome system, and suppress genes related to spore development, membrane synthesis, mitochondrial function, and toxin production. Exploring natural antifungal strategies may offer sustainable options for improving bee health. Microbial VOCs can be delivered using formulations designed to overcome their volatility and short lifespan. Recent studies have investigated the use of hydrogels and sprays containing microbial VOCs to control plant pathogenic fungi (123, 143). Similarly, antifungal hydrogel or spray formulations based on symbiotic bacteria like B. asteroides could represent a promising, eco-friendly strategy to manage fungal diseases such as Chalkbrood in honey bee colonies.
5 Conclusions
This study contributes to our understanding of the intricate relationship between honey bees and their gut microbiota. Through a preliminary characterization of Bifidobacterium strains isolated from the honey bee gut, we have demonstrated that certain isolates possess enzymatic activities involved in the detoxification of xenobiotics through hydrolysis or breakdown of harmful compounds. Additionally, several strains exhibited enzymatic capabilities that enhance nutrient bioavailability and facilitate the metabolism of specific sugars, such as mannose, lactose, raffinose, and xylose, that can otherwise be toxic to bees. Notably, B. asteroides 3CP-2B exhibited strong antifungal activity, suggesting its potential application as a probiotic supplement in honey bee diets or as an environmentally friendly biocontrol agent to reduce the incidence of fungal diseases such as chalkbrood. These findings lay a solid foundation for future biocontrol strategies based on honey bee-associated symbionts. However, further studies are essential to evaluate the safety of the VOCs produced by B. asteroides 3CP-2B, particularly their effects on healthy brood development. This will be crucial for developing safe and effective application strategies that do not disrupt the hive’s environmental balance or compromise colony productivity.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PV053127 https://www.ncbi.nlm.nih.gov/genbank/, PV053134 https://www.ncbi.nlm.nih.gov/genbank/, PV053124 https://www.ncbi.nlm.nih.gov/genbank/, PV053131 https://www.ncbi.nlm.nih.gov/genbank/, PV053133 https://www.ncbi.nlm.nih.gov/genbank/, PV053126 https://www.ncbi.nlm.nih.gov/genbank/, PV053144 https://www.ncbi.nlm.nih.gov/genbank/, PV053125 https://www.ncbi.nlm.nih.gov/genbank/, PV053128 https://www.ncbi.nlm.nih.gov/genbank/, From PV056024 to PV056038.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
MI: Resources, Investigation, Funding acquisition, Project administration, Conceptualization, Writing – review & editing, Writing – original draft. SG: Visualization, Validation, Writing – review & editing. BT: Writing – review & editing, Formal Analysis. LD: Formal Analysis, Writing – review & editing. GA: Writing – review & editing, Software, Writing – original draft, Data curation, Formal Analysis. MS: Investigation, Writing – review & editing. FC: Validation, Writing – review & editing, Visualization. RC: Writing – review & editing, Data curation. CM: Writing – review & editing, Formal Analysis. DC: Writing – review & editing, Validation, Visualization. CT: Writing – review & editing, Validation, Visualization, Investigation. AC: Project administration, Conceptualization, Resources, Investigation, Writing – review & editing, Funding acquisition.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by the University of Molise, under the project BIOMOXE, CUP H33C23003280005.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/finsc.2025.1669013/full#supplementary-material
References
1. Engel P, Kwong WK, McFrederick Q, Anderson KE, Barribeau SM, Chandler JA, et al. The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. mBio. (2016) 7:10.1128/mbio.02164–15. doi: 10.1128/mbio.02164-15
2. Romero S, Nastasa A, Chapman A, Kwong WK, and Foster LJ. The honey bee gut microbiota: strategies for study and characterization. Insect Mol Biol. (2019) 28:455–72. doi: 10.1111/imb.12567
3. Motta EV and Moran NA. The honeybee microbiota and its impact on health and disease. Nat Rev Microbiol. (2024) 22:122–37. doi: 10.1038/s41579-023-00990-3
4. Kwong WK and Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol. (2016) 14:374–84. doi: 10.1038/nrmicro.2016.43
5. Liberti J, Kay T, Quinn A, Kesner L, Frank ET, Cabirol A, et al. The gut microbiota affects the social network of honeybees. Nat Ecol Evol. (2022) 6:1471–9. doi: 10.1038/s41559-022-01840-w
6. Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. (2007) 71:495–548. doi: 10.1128/MMBR.00005-07
7. Endo A, Futagawa-Endo Y, and Dicks LMT. Diversity of Lactobacillus and Bifidobacterium in feces of herbivores, omnivores and carnivores. Anaerobe. (2010) 16:590–6. doi: 10.1016/j.anaerobe.2010.10.005
8. Kopečný J, Mrázek J, and Killer J. The presence of bifidobacteria in social insects, fish and reptiles. Folia Microbiol. (2010) 55:336–9. doi: 10.1007/s12223-010-0053-2
9. Bunesova V, Vlkova E, Rada V, Killer J, and Musilova S. Bifidobacteria from the gastrointestinal tract of animals: differences and similarities. Benef Microbes. (2014) 5:377–88. doi: 10.3920/BM2013.0081
10. Alessandri G, van Sinderen D, and Ventura M. The genus bifidobacterium: From genomics to functionality of an important component of the mammalian gut microbiota running title: Bifidobacterial adaptation to and interaction with the host. Comput Struct Biotechnol J. (2021) 19:1472–87. doi: 10.1016/j.csbj.2021.03.006
11. Baffoni L, Gaggìa F, Alberoni D, Cabbri R, Nanetti A, Biavati B, et al. Effect of dietary supplementation of Bifidobacterium and Lactobacillus strains in Apis mellifera L. against Nosema ceranae. Benef Microbes. (2016) 7:45–51. doi: 10.3920/BM2015.0085
12. Chen J, Wang J, and Zheng H. Characterization of Bifidobacterium apousia sp. nov., Bifidobacterium choladohabitans sp. nov., and Bifidobacterium polysaccharolyticum sp. nov., three novel species of the genus Bifidobacterium from honey bee gut. Systematic Appl Microbiol. (2021) 44:126247. doi: 10.1016/j.syapm.2021.126247
13. Scardovi V and Trovatelli LD. New species of bifid bacteria from Apis mellifica L. and Apis indica F. A contribution to the taxonomy and biochemistry of the genus Bifidobacterium. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg. (1969) 123:64–88.
14. Modesto M, Scarafile D, Vásquez A, Pukall R, Neumann-Schaal M, Pascarelli S, et al. Phylogenetic characterization of Bifidobacterium kimbladii sp. nov., a novel species from the honey stomach of the honeybee Apis mellifera. Syst Appl Microbiol. (2025) 48:126579. doi: 10.1016/j.syapm.2025.126579
15. Wang T-Y, Wang H, and Gu CT. Bifidobacterium apicola sp. nov., isolated from the gut of honeybee (Apis mellifera). Int J Syst Evol Microbiol. (2024) 74(12). doi: 10.1099/ijsem.0.006599
16. Jiang C-S, Li CY, and Gu CT. Bifidobacterium apis sp. nov., isolated from the gut of honeybee (Apis mellifera). Int J Syst Evol Microbiol. (2024) 74(4). doi: 10.1099/ijsem.0.006358
17. Olofsson TC, Modesto M, Pascarelli S, Scarafile D, Mattarelli P, and Vasquez A. Bifidobacterium mellis sp. nov., isolated from the honey stomach of the honey bee Apis mellifera. Int J Systematic Evolutionary Microbiol. (2023) 73:5766. doi: 10.1099/ijsem.0.005766
18. Li TT, Zhang HX, and Gu CT. Bifidobacterium mizhiense sp. nov., isolated from the gut of honeybee (Apis mellifera). Int J Syst Evol Microbiol. (2022) 72(5). doi: 10.1099/ijsem.0.005390
19. Killer J, Kopečný J, Mrázek J, Koppová I, Havlík J, Benada O, et al. Bifidobacterium actinocoloniiforme sp. nov. and Bifidobacterium bohemicum sp. nov., from the bumblebee digestive tract. Int J Syst Evol Microbiol. (2011) 61:1315–21. doi: 10.1099/ijs.0.022525-0
20. Killer J, Kopecný J, Mrázek J, Rada V, Benada O, Koppová I, et al. Bifidobacterium bombi sp. nov., from the bumblebee digestive tract. Int J Syst Evol Microbiol. (2009) 59:2020–4. doi: 10.1099/ijs.0.002915-0
21. Praet J, Meeus I, Cnockaert M, Aerts M, Smagghe G, and Vandamme P. Bifidobacterium commune sp. nov. isolated from the bumble bee gut. Antonie van Leeuwenhoek. (2015) 107:1307–13. doi: 10.1007/s10482-015-0425-3
22. Alberoni D, Gaggìa F, Baffoni L, Modesto MM, Biavati B, and Di Gioia D. Bifidobacterium xylocopae sp. nov. and Bifidobacterium aemilianum sp. nov., from the carpenter bee (Xylocopa violacea) digestive tract. Syst Appl Microbiol. (2019) 42:205–16. doi: 10.1016/j.syapm.2018.11.005
23. Li Y, Song Q, Yang H, Wei Y, Menghe B, and Liu W. Bifidobacterium favimelis sp. nov., isolated from black comb honey. Int J Syst Evol Microbiol. (2024) 74(11). doi: 10.1099/ijsem.0.006573
24. Mohr KI and Tebbe CC. Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ Microbiol. (2006) 8:258–72. doi: 10.1111/j.1462-2920.2005.00893.x
25. Anderson KE, Johansson A, Sheehan TH, Mott BM, Corby-Harris V, Johnstone L, et al. Draft genome sequences of two Bifidobacterium sp. from the honey bee (Apis mellifera). Gut Pathog. (2013) 5:42. doi: 10.1186/1757-4749-5-42
26. Khan KA, Al-Ghamdi AA, Ghramh HA, Ansari MJ, Ali H, Alamri SA, et al. Structural diversity and functional variability of gut microbial communities associated with honey bees. Microbial Pathogenesis. (2020) 138:103793. doi: 10.1016/j.micpath.2019.103793
27. Lazarova S, Lozanova L, Neov B, Shumkova R, Balkanska R, Palova N, et al. Composition and diversity of bacterial communities associated with honey bee foragers from two contrasting environments. Bull Entomological Res. (2023) 113:693–702. doi: 10.1017/S0007485323000378
28. Alberoni D, Baffoni L, Gaggìa F, Ryan PM, Murphy K, Ross PR, et al. Impact of beneficial bacteria supplementation on the gut microbiota, colony development and productivity of Apis mellifera L. Benef Microbes. (2018) 9(2):269–78. doi: 10.3920/BM2017.0061
29. Bonilla-Rosso G and Engel P. Functional roles and metabolic niches in the honey bee gut microbiota. Curr Opin Microbiol. (2018) 43:69–76. doi: 10.1016/j.mib.2017.12.009
30. Kešnerová L, Mars RAT, Ellegaard KM, Troilo M, Sauer U, and Engel P. Disentangling metabolic functions of bacteria in the honey bee gut. PloS Biol. (2017) 15:e2003467. doi: 10.1371/journal.pbio.2003467
31. Zheng H, Perreau J, Powell JE, Han B, Zhang Z, Kwong WK, et al. Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc Natl Acad Sci. (2019) 116:25909–16. doi: 10.1073/pnas.1916224116
32. Anderson KE and Ricigliano VA. Honey bee gut dysbiosis: a novel context of disease ecology. Curr Opin Insect Sci. (2017) 22:125–32. doi: 10.1016/j.cois.2017.05.020
33. Motta EVS, Raymann K, and Moran NA. Glyphosate perturbs the gut microbiota of honey bees. Proc Natl Acad Sci U.S.A. (2018) 115:10305–10. doi: 10.1073/pnas.1803880115
34. Kešnerová L, Emery O, Troilo M, Liberti J, Erkosar B, and Engel P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. (2020) 14:801–14. doi: 10.1038/s41396-019-0568-8
35. Baffoni L, Alberoni D, Gaggìa F, Braglia C, Stanton C, Ross PR, et al. Honeybee exposure to veterinary drugs: how is the gut microbiota affected? Microbiol Spectr. (2021) 9:e0017621. doi: 10.1128/Spectrum.00176-21
36. Fernandez De Landa G, Alberoni D, Baffoni L, Fernandez De Landa M, Revainera PD, Porrini LP, et al. The gut microbiome of solitary bees is mainly affected by pathogen assemblage and partially by land use. Environ Microbiome. (2023) 18:38. doi: 10.1186/s40793-023-00494-w
37. Hamdi C, Balloi A, Essanaa J, Crotti E, Gonella E, Raddadi N, et al. Gut microbiome dysbiosis and honeybee health. J Appl Entomology. (2011) 135:524–33. doi: 10.1111/j.1439-0418.2010.01609.x
38. Castelli L, Branchiccela B, Garrido M, Invernizzi C, Porrini M, Romero H, et al. Impact of nutritional stress on honeybee gut microbiota, immunity, and nosema ceranae infection. Microb Ecol. (2020) 80:908–19. doi: 10.1007/s00248-020-01538-1
39. Daisley BA, Chmiel JA, Pitek AP, Thompson GJ, and Reid G. Missing microbes in bees: how systematic depletion of key symbionts erodes immunity. Trends Microbiol. (2020) 28:1010–21. doi: 10.1016/j.tim.2020.06.006
40. Borum AE. Microbiota and its importance in honey bees. Bee Stud. (2021) 13:23–30. doi: 10.51458/BSTD.2021.14
41. Maxfield-Taylor SA, Mujic AB, and Rao S. First detection of the larval chalkbrood disease pathogen ascosphaera apis (Ascomycota: eurotiomycetes: ascosphaerales) in adult bumble bees. PloS One. (2015) 10:e0124868. doi: 10.1371/journal.pone.0124868
42. Reynaldi FJ, Lucia M, and Genchi Garcia ML. Ascosphaera apis, the entomopathogenic fungus affecting larvae of native bees (Xylocopa augusti): First report in South America. Rev Iberoam Micol. (2015) 32:261–4. doi: 10.1016/j.riam.2015.01.001
43. Chen D, Guo R, Xiong C, Zheng Y, Hou C, and Fu Z. Morphological and molecular identification of chalkbrood disease pathogen Ascosphaera apis in Apis cerana cerana. J Apicultural Res. (2018) 57:516–21. doi: 10.1080/00218839.2018.1475943
44. Deneke Y. Review on chalkbrood disease of honey bee. Veterinary Med – Open J. (2023) 8:47–55. doi: 10.17140/VMOJ-8-176
45. Tejerina MR, Cabana MJ, and Benitez-Ahrendts MR. Strains of Lactobacillus spp. reduce chalkbrood in Apis mellifera. J Invertebrate Pathol. (2021) 178:107521. doi: 10.1016/j.jip.2020.107521
46. Sevim A, Akpınar R, Karaoğlu Ş.A, Bozdeveci A, and Sevim E. Prevalence and phylogenetic analysis of Ascosphaera apis (Maassen ex Claussen) LS Olive & Spiltoi) isolates from honeybee colonies in Turkey. Biologia. (2022) 77:2689–99. doi: 10.1007/s11756-022-01114-7
47. Das R, Kumar R, Kunal G, Goldar S, Dutta S, and Jha S. Detection of Ascosphaera apis, causing chalkbrood disease in the colonies of European honey bee, Apis mellifera in West Bengal, India. Sociobiology. (2023) 70:e9192–2. doi: 10.13102/sociobiology.v70i4.9129
48. Karthik V, Srinivasan MR, Saminathan VR, Karthikeyan S, and Balasubramani V. Morphological and Molecular Identification and Mating Type Detection of Chalkbrood Fungal Pathogen Ascosphaera apis in Apis mellifera L. @ in Southern India. Indian J Entomology. (2024) 87(2):349–55. doi: 10.55446/IJE.2024.2198
49. Li Z, Su S, Hamilton M, Yan L, and Chen Y. The ability to cause infection in a pathogenic fungus uncovers a new biological feature of honey bee viruses. J Invertebr Pathol. (2014) 120:18–22. doi: 10.1016/j.jip.2014.05.002
50. Cheng X, Zhang L, Luo J, Yang S, Deng Y, Li J, et al. Two pathogenic fungi isolated from chalkbrood samples and honey bee viruses they carried. Front Microbiol. (2022) 13:843842. doi: 10.3389/fmicb.2022.843842
51. Kim DY, Maeng S, Cho S-J, Park HJ, Kim K, Lee JK, et al. The ascosphaera apis infection (Chalkbrood disease) alters the gut bacteriome composition of the honeybee. Pathogens. (2023) 12:734. doi: 10.3390/pathogens12050734
52. Pavlović R, Brodschneider R, Goessler W, Stanisavljević L, Vujčić Z, and Zarić NM. Micronutrient deficiency may be associated with the onset of chalkbrood disease in honey bees. Insects. (2024) 15:269. doi: 10.3390/insects15040269
53. Yoder JA, Jajack AJ, Cornacchione WS, Dunn AL, Cunningham EG, Matchett CL, et al. In vitro evaluation of sugar syrups, antibiotics, and miticides on growth of honey bee pathogen, Ascosphaera apis: Emphasis for chalkbrood prevention is on keeping bees healthy. Apidologie. (2014) 45:568–78. doi: 10.1007/s13592-014-0274-5
54. Castagnino GLB, Mateos A, Meana A, Montejo L, Zamorano Iturralde LV, and Cutuli De Simón MT. Etiology, symptoms and prevention of chalkbrood disease: a literature review. Rev Bras saúde Prod anim. (2020) 21:e210332020. doi: 10.1590/S1519-9940210332020
55. von Knoblauch T, Jensen AB, Mülling CKW, Aupperle-Lellbach H, and Genersch E. Chalkbrood Disease Caused by Ascosphaera apis in Honey Bees (Apis mellifera)—Morphological and Histological Changes in Infected Larvae. Veterinary Sci. (2024) 11:415. doi: 10.3390/vetsci11090415
56. Iorizzo M, Lombardi SJ, Ganassi S, Testa B, Ianiro M, Letizia F, et al. Antagonistic Activity against Ascosphaera apis and Functional Properties of Lactobacillus kunkeei Strains. Antibiotics (Basel). (2020) 9:262. doi: 10.3390/antibiotics9050262
57. Ashraf SA, Mahmood D, Elkhalifa AEO, Siddiqui AJ, Khan MI, Ashfaq F, et al. Exposure to pesticide residues in honey and its potential cancer risk assessment. Food Chem Toxicol. (2023) 180:114014. doi: 10.1016/j.fct.2023.114014
58. Ansari MJ, Al-Ghamdi A, Usmani S, Khan KA, Alqarni AS, Kaur M, et al. In vitro evaluation of the effects of some plant essential oils on Ascosphaera apis, the causative agent of Chalkbrood disease. Saudi J Biol Sci. (2017) 24:1001–6. doi: 10.1016/j.sjbs.2016.04.016
59. Khan SU, Anjum SI, Ansari MJ, Khan MHU, Kamal S, Rahman K, et al. Antimicrobial potentials of medicinal plant’s extract and their derived silver nanoparticles: A focus on honey bee pathogen. Saudi J Biol Sci. (2019) 26:1815–34. doi: 10.1016/j.sjbs.2018.02.010
60. Pusceddu M, Floris I, Mangia NP, Angioni A, and Satta A. In Vitro Activity of Several Essential Oils Extracted from Aromatic Plants against Ascosphaera apis. Veterinary Sci. (2021) 8:80. doi: 10.3390/vetsci8050080
61. Krutmuang P, Rajula J, Pittarate S, Chatima C, Thungrabeab M, Mekchay S, et al. The inhibitory action of plant extracts on the mycelial growth of Ascosphaera apis, the causative agent of chalkbrood disease in Honey bee. Toxicol Rep. (2022) 9:713–9. doi: 10.1016/j.toxrep.2022.03.036
62. Usta M. Biological control of honey bee diseases and pests. In: Melittology - new advances. IntechOpen (2023). doi: 10.5772/intechopen.1003750
63. Boonmee T, Sinpoo C, Thayatham K, Suanpoot P, Disayathanoowat T, Pettis JS, et al. Atmospheric non-thermal plasma inactivation of Ascosphaera apis, the causative agent of chalkbrood disease in honeybee. Sci Rep. (2024) 14:1831. doi: 10.1038/s41598-024-52221-1
64. Iorizzo M, Letizia F, Ganassi S, Testa B, Petrarca S, Albanese G, et al. Functional properties and antimicrobial activity from lactic acid bacteria as resources to improve the health and welfare of honey bees. Insects. (2022) 13:308. doi: 10.3390/insects13030308
65. Abdi K, Ben Said M, Crotti E, Masmoudi AS, and Cherif A. The promise of probiotics in honeybee health and disease management. Arch Microbiol. (2023) 205:73. doi: 10.1007/s00203-023-03416-z
66. Rodríguez MA, Fernández LA, Díaz ML, Pérez M, Corona M, and Reynaldi FJ. Microbiological and chemical characterization of water kefir: An innovative source of potential probiotics for bee nutrition. Rev Argent Microbiología. (2023) 55:176–80. doi: 10.1016/j.ram.2022.09.003
67. Iorizzo M, Testa B, Ganassi S, Lombardi SJ, Ianiro M, Letizia F, et al. Probiotic properties and potentiality of lactiplantibacillus plantarum strains for the biological control of chalkbrood disease. J Fungi (Basel). (2021) 7:379. doi: 10.3390/jof7050379
68. Tejerina MR, Cabana MJ, Enríquez PA, Benítez-Ahrendts MR, and Fonseca MI. Bacterial strains isolated from stingless bee workers inhibit the growth of apis mellifera pathogens. Curr Microbiol. (2024) 81:106. doi: 10.1007/s00284-024-03618-8
69. Iorizzo M, Coppola F, Pannella G, Ganassi S, Matarazzo C, Albanese G, et al. First Report on Antifungal Activity of Metschnikowia pulcherrima Against Ascosphaera apis, the Causative Agent of Chalkbrood Disease in Honeybee (Apis mellifera L.) Colonies. J Fungi. (2025) 11:336. doi: 10.3390/jof11050336
70. Daisley BA, Pitek AP, Torres C, Lowery R, Adair BA, Al KF, et al. Delivery mechanism can enhance probiotic activity against honey bee pathogens. ISME J. (2023) 17:1382–95. doi: 10.1038/s41396-023-01422-z
71. Pino A, Benkaddour B, Inturri R, Amico P, Vaccaro SC, Russo N, et al. Characterization of bifidobacterium asteroides isolates. Microorganisms. (2022) 10:655. doi: 10.3390/microorganisms10030655
72. Dengiz B, Killer J, Havlík J, Dobeš P, and Hyršl P. Selection of Probiotics for Honey Bees: The In Vitro Inhibition of Paenibacillus larvae, Melissococcus plutonius, and Serratia marcescens Strain Sicaria by Host-Specific Lactobacilli and Bifidobacteria. Microorganisms. (2025) 13:1159. doi: 10.3390/microorganisms13051159
73. Moradi M and Ownagh A. Antifungal Effects of Lactobacillus casei, Lactobacillus acidophilus and Bifidobacterium bifidum on the Ascospharea apis Causative Agent of Honey bee Chalkbrood Disease. J Veterinary Res. (2019) 74:273–82. doi: 10.22059/jvr.2019.217394.2533
74. Ptaszyńska AA, Borsuk G, Zdybicka-Barabas A, Cytryńska M, and Małek W. Are commercial probiotics and prebiotics effective in the treatment and prevention of honeybee nosemosis C? Parasitol Res. (2016) 115:397–406. doi: 10.1007/s00436-015-4761-z
75. Kwong WK, Mancenido AL, and Moran NA. Immune system stimulation by the native gut microbiota of honey bees. R Soc Open Sci. (2017) 4:170003. doi: 10.1098/rsos.170003
76. El Khoury S, Rousseau A, Lecoeur A, Cheaib B, Bouslama S, Mercier P-L, et al. Deleterious Interaction Between Honeybees (Apis mellifera) and its Microsporidian Intracellular Parasite Nosema ceranae Was Mitigated by Administrating Either Endogenous or Allochthonous Gut Microbiota Strains. Front Ecol Evol. (2018) 6:58. doi: 10.3389/fevo.2018.00058
77. Todorov SD, Alves MV, Bueno GCA, Alves VF, and Ivanova IV. Bee-associated beneficial microbes—Importance for bees and for humans. Insects. (2024) 15:430. doi: 10.3390/insects15060430
78. Zhang Z, Schwartz S, Wagner L, and Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. (2000) 7:203–14. doi: 10.1089/10665270050081478
79. Resource Coordinators NCBI. Database resources of the national center for biotechnology information. Nucleic Acids Res. (2018) 46:D8–D13. doi: 10.1093/nar/gkx1095
80. Johnson JS, Spakowicz DJ, Hong B-Y, Petersen LM, Demkowicz P, Chen L, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. (2019) 10:5029. doi: 10.1038/s41467-019-13036-1
81. Lonsway DR. Preparation of routine media and reagents used in antimicrobial susceptibility testing. ClinMicroNow. (2023), 1–25. doi: 10.1002/9781683670438.cmph0082
82. Torracca B, Pedonese F, Turchi B, Fratini F, and Roberta N. Qualitative and quantitative evaluation of biogenic amines in vitro production by bacteria isolated from ewes’ milk cheeses. Eur Food Res Technol. (2018) 244(10):721–8. doi: 10.1007/s00217-017-2992-1
83. Paca C, Matei IA, Diaconeasa Z, Rotaru A, Erler S, and Dezmirean DS. Biologically active extracts from different medicinal plants tested as potential additives against bee pathogens. Antibiotics. (2021) 10:960. doi: 10.3390/antibiotics10080960
84. Ruiz-Moyano S, Hernández A, Galvan AI, Córdoba MG, Casquete R, Serradilla MJ, et al. Selection and application of antifungal VOCs-producing yeasts as biocontrol agents of grey mould in fruits. Food Microbiol. (2020) 92:103556. doi: 10.1016/j.fm.2020.103556
85. Serradilla MJ, Martín A, Hernandez A, López-Corrales M, Lozano M, Córdoba M, et al. Effect of the commercial ripening stage and postharvest storage on microbial and aroma changes of ‘Ambrunés’ Sweet cherries. J Agric Food Chem. (2010) 58:9157–63. doi: 10.1021/jf102004v
86. Bottacini F, Milani C, Turroni F, Sánchez B, Foroni E, Duranti S, et al. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PloS One. (2012) 7:e44229. doi: 10.1371/journal.pone.0044229
87. Powell JE, Martinson VG, Urban-Mead K, and Moran NA. Routes of acquisition of the gut microbiota of the honey bee apis mellifera. Appl Environ Microbiol. (2014) 80:7378–87. doi: 10.1128/AEM.01861-14
88. Raymann K and Moran NA. The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci. (2018) 26:97–104. doi: 10.1016/j.cois.2018.02.012
89. Devonshire AL, Moores GD, and Ffrench-Constant RH. Detection of insecticide resistance by immunological estimation of carboxylesterase activity in Myzus persicae (Sulzer) and cross reaction of the antiserum with Phorodon humuli (Schrank) (Hemiptera: Aphididae). Bull Entomological Res. (1986) 76:97–107. doi: 10.1017/S0007485300015327
90. Kishino S, Takeuchi M, Park S-B, Hirata A, Kitamura N, Kunisawa J, et al. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc Natl Acad Sci U.S.A. (2013) 110:17808–13. doi: 10.1073/pnas.1312937110
91. Milone J, Rinkevich F, Mcafee A, Foster L, and Tarpy D. Differences in larval pesticide tolerance and esterase activity across honey bee (Apis mellifera) stocks. Ecotoxicology Environ Saf. (2020) 206:111213. doi: 10.1016/j.ecoenv.2020.111213
92. Bosch-Serra D, Rodríguez MA, Avilla J, Sarasúa MJ, and Miarnau X. Esterase, glutathione S-transferase and NADPH-cytochrome P450 reductase activity evaluation in(Hemiptera: psyllidae) individual adults. Insects. (2021) 12:329. doi: 10.3390/insects12040329
93. Claudianos C, Ranson H, Johnson RM, Biswas S, Schuler MA, Berenbaum MR, et al. A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. (2006) 15:615–36. doi: 10.1111/j.1365-2583.2006.00672.x
94. Berenbaum MR and Johnson RM. Xenobiotic detoxification pathways in honey bees. Curr Opin Insect Sci. (2015) 10:51–8. doi: 10.1016/j.cois.2015.03.005
95. Rand EE, Smit S, Beukes M, Apostolides Z, Pirk CWW, and Nicolson SW. Detoxification mechanisms of honey bees (Apis mellifera) resulting in tolerance of dietary nicotine. Sci Rep. (2015) 5:11779. doi: 10.1038/srep11779
96. Gong Y and Diao Q. Current knowledge of detoxification mechanisms of xenobiotic in honey bees. Ecotoxicology. (2017) 26:1–12. doi: 10.1007/s10646-016-1742-7
97. Chmiel JA, Daisley BA, Pitek AP, Thompson GJ, and Reid G. Understanding the effects of sublethal pesticide exposure on honey bees: A role for probiotics as mediators of environmental stress. Front Ecol Evol. (2020) 8:22. doi: 10.3389/fevo.2020.00022
98. Shamjana U, Vasu DA, Hembrom PS, Nayak K, and Grace T. The role of insect gut microbiota in host fitness, detoxification and nutrient supplementation. Antonie van Leeuwenhoek. (2024) 117:71. doi: 10.1007/s10482-024-01970-0
99. Cui S, Gu Z, Wang W, Tang X, Zhang Q, Mao B, et al. Characterization of peptides available to different bifidobacteria. LWT. (2022) 169:113958. doi: 10.1016/j.lwt.2022.113958
100. Li Y, Leonard SP, Powell JE, and Moran NA. Species divergence in gut-restricted bacteria of social bees. Proc Natl Acad Sci. (2022) 119:e2115013119. doi: 10.1073/pnas.2115013119
101. Zheng H, Nishida A, Kwong WK, Koch H, Engel P, Steele MI, et al. Metabolism of toxic sugars by strains of the bee gut symbiont gilliamella apicola. mBio. (2016) 7:10.1128/mbio.01326–16. doi: 10.1128/mbio.01326-16
102. Kubota M, Tsuji M, Nishimoto M, Wongchawalit J, Okuyama M, Mori H, et al. Localization of alpha-glucosidases I, II, and III in organs of European honeybees, Apis mellifera L., and the origin of alpha-glucosidase in honey. Biosci Biotechnol Biochem. (2004) 68:2346–52. doi: 10.1271/bbb.68.2346
103. Pokusaeva K, O’Connell-Motherway M, Zomer A, Fitzgerald GF, and van Sinderen D. Characterization of two novel α-glucosidases from bifidobacterium breve UCC2003. Appl Environ Microbiol. (2009) 75:1135–43. doi: 10.1128/AEM.02391-08
104. Stanley D, Rejzek M, Naested H, Smedley M, Otero S, Fahy B, et al. The role of alpha-glucosidase in germinating barley grains. Plant Physiol. (2011) 155:932–43. doi: 10.1104/pp.110.168328
105. Guzik J, Nakonieczny M, Tarnawska M, Bereś PK, Drzewiecki S, and Migula P. The Glycolytic Enzymes Activity in the Midgut of Diabrotica virgifera virgifera (Coleoptera: Chrysomelidae) adult and their Seasonal Changes. J Insect Sci. (2015) 15:56. doi: 10.1093/jisesa/iev036
106. Lezyk M, Jers C, Kjaerulff L, Gotfredsen CH, Mikkelsen MD, and Mikkelsen JD. Novel α-L-fucosidases from a soil metagenome for production of fucosylated human milk oligosaccharides. PloS One. (2016) 11:e0147438. doi: 10.1371/journal.pone.0147438
107. Gama M, do VF, Alexandre Y, Do N, Pereira da Silva JM, Castro DP, et al. Digestive α-L-fucosidase activity in Rhodnius prolixus after blood feeding: effect of secretagogue and nutritional stimuli. Front Physiol. (2023) 14:1123414. doi: 10.3389/fphys.2023.1123414
108. Grossmann GA and Terra WR. α-Galactosidases from the larval midgut of Tenebrio molitor (Coleoptera) and Spodoptera frugiperda (Lepidoptera). Comp Biochem Physiol Part B: Biochem Mol Biol. (2001) 128:109–22. doi: 10.1016/S1096-4959(00)00306-7
109. Becker DJ and Lowe JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. (2003) 13:41R–53R. doi: 10.1093/glycob/cwg054
110. Pickard JM and Chervonsky AV. Intestinal fucose as a mediator of host–microbe symbiosis. J Immunol. (2015) 194:5588–93. doi: 10.4049/jimmunol.1500395
111. Hlalukana N, Magengelele M, Malgas S, and Pletschke BI. Enzymatic conversion of mannan-rich plant waste biomass into prebiotic mannooligosaccharides. Foods. (2021) 10:2010. doi: 10.3390/foods10092010
112. Ricigliano VA, Fitz W, Copeland DC, Mott BM, Maes P, Floyd AS, et al. The impact of pollen consumption on honey bee (Apis mellifera) digestive physiology and carbohydrate metabolism. Arch Insect Biochem Physiol. (2017) 96:e21406. doi: 10.1002/arch.21406
113. Iorizzo M, Pannella G, Lombardi SJ, Ganassi S, Testa B, Succi M, et al. Inter- and Intra-Species Diversity of Lactic Acid Bacteria in Apis mellifera ligustica Colonies. Microorganisms. (2020) 8:1578. doi: 10.3390/microorganisms8101578
114. Iorizzo M, Testa B, Lombardi SJ, Ganassi S, Ianiro M, Letizia F, et al. Antimicrobial activity against Paenibacillus larvae and functional properties of Lactiplantibacillus plantarum strains: Potential benefits for honeybee health. Antibiotics. (2020) 9:442. doi: 10.3390/antibiotics9080442
115. Tan K, Guo YH, Nicolson SW, Radloff SE, Song QS, and Hepburn HR. Honeybee (Apis cerana) foraging responses to the toxic honey of Tripterygium hypoglaucum (Celastraceae): changing threshold of nectar acceptability. J Chem Ecol. (2007) 33:2209–17. doi: 10.1007/s10886-007-9384-0
116. Taylor MA, Robertson AW, Biggs PJ, Richards KK, Jones DF, and Parkar SG. The effect of carbohydrate sources: Sucrose, invert sugar and components of mānuka honey, on core bacteria in the digestive tract of adult honey bees (Apis mellifera). PloS One. (2019) 14:e0225845. doi: 10.1371/journal.pone.0225845
117. Lee FJ, Rusch DB, Stewart FJ, Mattila HR, and Newton ILG. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol. (2015) 17:796–815. doi: 10.1111/1462-2920.12526
118. Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sánchez B, and Margolles A. Bifidobacteria and their health-promoting effects. Microbiol Spectr. (2017) 5(3). doi: 10.1128/microbiolspec.BAD-0010-2016
119. Herranz YS, Amore M, and Del CC. Compounds with antifungal activity, which are produced by bifidobacterium spp (2006). Available online at: https://patents.google.com/patent/WO2006070041A2/en (Accessed July 3, 2025).
120. Qiao N, Yu L, Zhang C, Wei C, Zhao J, Zhang H, et al. A comparison of the inhibitory activities of Lactobacillus and Bifidobacterium against Penicillium expansum and an analysis of potential antifungal metabolites. FEMS Microbiol Lett. (2020) 367:fnaa130. doi: 10.1093/femsle/fnaa130
121. Yu L, Qiao N, Wei C, Hu Q, Zhai Q, Yan B, et al. Underlying mechanisms of the antagonistic effects of Bifidobacterium adolescentis CCFM1108 on Penicillium expansum: Based on comparative transcriptome analysis. Food Bioscience. (2022) 47:101693. doi: 10.1016/j.fbio.2022.101693
122. Kim H, Maigoro AY, Lee J-H, Frunze O, and Kwon H-W. The improving effects of probiotic-added pollen substitute diets on the gut microbiota and individual health of honey bee (Apis mellifera L.). Microorganisms. (2024) 12:1567. doi: 10.3390/microorganisms12081567
123. Zhao X, Zhou J, Tian R, and Liu Y. Microbial volatile organic compounds: Antifungal mechanisms, applications, and challenges. Front Microbiol. (2022) 13:922450. doi: 10.3389/fmicb.2022.922450
124. Pokusaeva K, Fitzgerald GF, and van Sinderen D. Carbohydrate metabolism in bifidobacteria. Genes Nutr. (2011) 6:285–306. doi: 10.1007/s12263-010-0206-6
125. Rane HS, Bernardo SM, Walraven CJ, and Lee SA. In Vitro Analyses of Ethanol Activity against Candida albicans Biofilms. Antimicrob Agents Chemother. (2012) 56:4487–9. doi: 10.1128/AAC.00263-12
126. Zinn M-K and Bockmühl D. Did granny know best? Evaluating the antibacterial, antifungal and antiviral efficacy of acetic acid for home care procedures. BMC Microbiol. (2020) 20:265. doi: 10.1186/s12866-020-01948-8
127. Suwannarach N, Kumla J, Bussaban B, Nuangmek W, Matsui K, and Lumyong S. Biofumigation with the endophytic fungus Nodulisporium spp. CMU-UPE34 to control postharvest decay of citrus fruit. Crop Prot. (2013) 45:63–70. doi: 10.1016/j.cropro.2012.11.015
128. Williamson DA, Carter GP, and Howden BP. Current and emerging topical antibacterials and antiseptics: agents, action, and resistance patterns. Clin Microbiol Rev. (2017) 30:827–60. doi: 10.1128/CMR.00112-16
129. Usta-Gorgun B and Yilmaz-Ersan L. Short-chain fatty acids production by Bifidobacterium species in the presence of salep. Electronic J Biotechnol. (2020) 47:29–35. doi: 10.1016/j.ejbt.2020.06.004
130. Fusco W, Lorenzo MB, Cintoni M, Porcari S, Rinninella E, Kaitsas F, et al. Short-chain fatty-acid-producing bacteria: key components of the human gut microbiota. Nutrients. (2023) 15:2211. doi: 10.3390/nu15092211
131. Bhattacharyya A, Sinha M, Singh H, Patel RS, Ghosh S, Sardana K, et al. Mechanistic insight into the antifungal effects of a fatty acid derivative against drug-resistant fungal infections. Front Microbiol. (2020) 11:2116. doi: 10.3389/fmicb.2020.02116
132. Yun J and Lee DG. A novel fungal killing mechanism of propionic acid. FEMS Yeast Res. (2016) 16:fow089. doi: 10.1093/femsyr/fow089
133. Costa Dos Santos A, Carina Biluca F, Brugnerotto P, Valdemiro Gonzaga L, Carolina Oliveira Costa A, and Fett R. Brazilian stingless bee honey: Physicochemical properties and aliphatic organic acids content. Food Res Int. (2022) 158:111516. doi: 10.1016/j.foodres.2022.111516
134. Chee HY, Kim H, and Lee MH. In vitro Antifungal Activity of Limonene against Trichophyton rubrum. Mycobiology. (2009) 37:243–6. doi: 10.4489/MYCO.2009.37.3.243
135. Giorgio A, De Stradis A, Lo Cantore P, and Iacobellis NS. Biocide effects of volatile organic compounds produced by potential biocontrol rhizobacteria on Sclerotinia sclerotiorum. Front Microbiol. (2015) 6:1056. doi: 10.3389/fmicb.2015.01056
136. Tyagi S, Lee K-J, Shukla P, and Chae J-C. Author Correction: Dimethyl disulfide exerts antifungal activity against Sclerotinia minor by damaging its membrane and induces systemic resistance in host plants. Sci Rep. (2020) 10:18183. doi: 10.1038/s41598-020-74901-4
137. Chen X, Liu J, Chen AJ, Wang L, Jiang X, Gong A, et al. Burkholderia ambifaria H8 as an effective biocontrol strain against maize stalk rot via producing volatile dimethyl disulfide. Pest Manage Sci. (2024) 80:4125–36. doi: 10.1002/ps.8119
138. Adams TB, Doull J, Feron VJ, Goodman JI, Marnett LJ, Munro IC, et al. The FEMA GRAS assessment of pyrazine derivatives used as flavor ingredients. Flavor and Extract Manufacturers Association. Food Chem Toxicol. (2002) 40:429–51. doi: 10.1016/s0278-6915(01)00123-5
139. Dolezal M and Zitko J. Pyrazine derivatives: a patent review (June 2012 - present). Expert Opin Ther Pat. (2015) 25:33–47. doi: 10.1517/13543776.2014.982533
140. Kucerova-Chlupacova M, Kunes J, Buchta V, Vejsova M, and Opletalova V. Novel pyrazine analogs of chalcones: synthesis and evaluation of their antifungal and antimycobacterial activity. Molecules. (2015) 20:1104–17. doi: 10.3390/molecules20011104
141. Su F, Su Z, Zhao Q, Zhao Z, Wu Z, Zhao M, et al. Synthesis, thermal property and antifungal evaluation of pyrazine esters. Arabian J Chem. (2022) 15:104351. doi: 10.1016/j.arabjc.2022.104351
142. Gong Y, Liu J-Q, Xu M-J, Zhang C-M, Gao J, Li C-G, et al. Antifungal volatile organic compounds from streptomyces setonii WY228 control black spot disease of sweet potato. Appl Environ Microbiol. (2022) 88:e0:231721. doi: 10.1128/aem.02317-21
Keywords: honey bee, gut microbiota, Bifidobacterium asteroides, chalkbrood disease, Ascosphaera apis
Citation: Iorizzo M, Ganassi S, Testa B, Di Donato LM, Albanese G, Succi M, Coppola F, Cozzolino R, Matarazzo C, Di Criscio D, Tedino C and De Cristofaro A (2025) Ascosphaera apis as a target for the antifungal activity of symbiotic Bifidobacteria in honey bees. Front. Insect Sci. 5:1669013. doi: 10.3389/finsc.2025.1669013
Received: 18 July 2025; Accepted: 17 September 2025;
Published: 01 October 2025.
Edited by:
Maristella Mastore, University of Insubria, ItalyReviewed by:
Walaa Ahmed Elsayeh, Newcastle University, United KingdomChun Tao Gu, Northeast Agricultural University, China
Copyright © 2025 Iorizzo, Ganassi, Testa, Di Donato, Albanese, Succi, Coppola, Cozzolino, Matarazzo, Di Criscio, Tedino and De Cristofaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sonia Ganassi, c29uaWEuZ2FuYXNzaUB1bmltb2wuaXQ=; Gianluca Albanese, Z2lhbmx1Y2EuYWxiYW5lc2VAdW5pbW9sLml0
 Massimo Iorizzo
Massimo Iorizzo Sonia Ganassi
Sonia Ganassi Bruno Testa
Bruno Testa Licia Maria Di Donato
Licia Maria Di Donato Gianluca Albanese
Gianluca Albanese Mariantonietta Succi
Mariantonietta Succi Francesca Coppola
Francesca Coppola Rosaria Cozzolino
Rosaria Cozzolino Cristina Matarazzo
Cristina Matarazzo Dalila Di Criscio
Dalila Di Criscio Cosimo Tedino
Cosimo Tedino Antonio De Cristofaro
Antonio De Cristofaro