- 1Molecular Parasitology and Systems Biology Lab, Department of Biological Sciences, Birla Institute of Technology and Science (BITS), Pilani, Rajasthan, India
- 2Department of Medicine, S.P. Medical College, Bikaner, Rajasthan, India
- 3Genotypic Technology Pvt. Ltd., Bangalore, Karnataka, India
Background: Copy number variations (CNVs) in the Plasmodium vivax genome can influence key parasite traits such as erythrocyte invasion, immune evasion, drug resistance, and survival in the human host. Their potential role in severe manifestations of P. vivax malaria, such as cerebral malaria (CM) remains underexplored. In regions like India, where P. vivax is endemic, understanding genomic factors that contribute to disease severity is crucial. Given the limited understanding of genomic factors contributing to disease severity in P. vivax, this study aims to investigate genome-wide CNVs in clinical isolates from patients with cerebral and uncomplicated malaria.
Methods: We employed a high-resolution, custom-designed 2 × 400K tiling microarray for array-based comparative genomic hybridization (aCGH), using probes with an average spacing of 56 base pairs covering the entire P. vivax genome. Genomic DNA from cerebral malaria isolates was differentially labeled and hybridized against reference DNA from uncomplicated malaria isolates. CNVs were inferred based on fluorescence intensity ratios, indicating chromosomal regions with copy number gains or losses.
Results: Utilizing probes based on the P. vivax Sal-1 reference genome, we detected significant CNVs across all 14 chromosomes, affecting 2,138 genes. CNVs ranged from 100 bp to approximately 1,429 kb in cerebral malaria isolates compared to uncomplicated cases. Altered regions having gains or losses included genes encoding surface antigens such as 6-cysteine proteins, tryptophan-rich antigens (TRAGs), serine-repeat antigen (SERA), apical membrane antigen (AMA), as well as drug resistance markers. The most extensive CNV spanned ~1,450 kb on chromosome 12. CNVs were also observed in intergenic regions, suggesting potential regulatory impacts.
Discussion: This study identifies CNVs in the genome of P. vivax isolates from cerebral malaria cases, in genes involved in immune evasion, drug resistance, and host-pathogen interactions. Although the precise impact of these CNVs on disease severity remains unclear, the findings highlight genetic differences between isolates from severe and uncomplicated malaria cases, including variations in intergenic regions. These findings emphasize the need to further investigate CNVs that may contribute to P. vivax pathogenesis and resistance. A deeper understanding of these variations could aid in identifying biomarkers for severe disease and support the development of more effective malaria control and treatment strategies.
1 Introduction
Plasmodium vivax is the most widespread parasite extending into temperate climates among the 5 Plasmodium species known to cause malaria in humans. Earlier, P. vivax was considered to be benign, but in the last few years there has been a change in this trend as life-threatening symptoms similar to those of P. falciparum cases such as hepatic dysfunction, cerebral malaria, severe anemia, acute respiratory distress syndrome (ARDS), and acute kidney injury have been observed in P. vivax cases as well (Kochar et al., 2005, 2007, 2009; Nautiyal et al., 2005; Sarkar and Bhattacharya, 2008; Alexandre et al., 2010; Sarkar et al., 2010; Douglas et al., 2012; Kute et al., 2012; Naing et al., 2014; Phyo et al., 2022; White, 2022).
The genome of an organism exhibits a spectrum of variations, encompassing significant structural changes and extending to single nucleotide polymorphisms. Structural variations comprise chromosomal size polymorphisms, gene copy number variations (CNVs), inversions, and translocations. Selection pressures, genetic drift, and migration collectively influence the diverse genomic variations present in a population (Mackinnon and Marsh, 2010; Chang et al., 2013; Kennedy and Dwyer, 2018). Recent interest in CNVs in malaria parasites has been driven by increasing evidence of the CNVs role in adaptation, evolution, and disease in a number of organisms (Anderson et al., 2009; Henrichsen et al., 2009; Wellcome Trust Case Control Consortium et al., 2010; Chaignat et al., 2011; Angstadt et al., 2013; Kirov et al., 2014; Pös et al., 2021). CNVs include both deletions and amplifications of a single gene or a cluster of adjacent genes. Furthermore, CNVs are believed to impact gene expression levels directly by changing gene dosage and indirectly by altering the chromatin environment in the proximity of the CNV (Hastings et al., 2009). Therefore, these CNVs may influence parasite phenotypes such as drug resistance, erythrocyte invasion, and transmissibility.
Over the years, the natural evolution of malaria parasites has led to a massive amount of genetic diversity, either via CNVs or acquisition of new single nucleotide polymorphisms (SNPs) (Feng et al., 2003; Cheeseman et al., 2009; Costa et al., 2017; Simam et al., 2018; Qidwai, 2020). This allows parasites to acquire a high capacity to adapt to environmental shifts. The malaria parasite employs gene amplifications or duplications and deletions (CNVs) as a general strategy to enhance its survival and spread. CNVs in genes related to antifolate resistance, such as those encoding GTP cyclohydrolase (gch1), have been implicated in the development of resistance to antimalarial drugs in P. falciparum (Heinberg et al., 2013; Srisutham et al., 2022). Studies have identified CNVs in genes associated with multidrug resistance in P. falciparum, including P. falciparum multidrug resistance gene 1 (pfmdr1) and P. falciparum chloroquine resistance transporter (pfcrt), which play important roles in the resistance to multiple antimalarial drugs (Gadalla et al., 2011; Gupta et al., 2018b; Wicht et al., 2020; Gil and Fançony, 2021; Ward et al., 2022). Similarly, amplifications of pvmdr1 and pvcrt have been identified in isolates from Brazil, French Guiana, Thailand, Laos, Myanmar, Cambodia, and India, and are associated with altered susceptibility to multiple antimalarials (Imwong et al., 2008; Suwanarusk et al., 2008; Vargas-Rodríguez et al., 2012; Faway et al., 2016; Costa et al., 2017; Silva et al., 2018; Buyon et al., 2021)vivaxvivax. Field studies have also linked pvcrt overexpression, resulting from increased gene copy number, with chloroquine resistance (Melo et al., 2014; Costa et al., 2017)). Beyond drug resistance, recent genomic studies across Southeast Asia, South America, and Africa have shown that CNVs in P. vivax are frequently concentrated in host–parasite interaction gene families, including the Duffy binding protein (PvDBP), merozoite surface protein 3 (MSP3 family), and reticulocyte binding proteins (PvRBPs). Independent expansions of PvDBP and MSP3.11 were observed in Ethiopian isolates (Lo et al., 2019). Recently, Pvdbp gene amplification has been reported to allow P.vivax to evade host anti-PvDBP humoral immunity (Popovici et al., 2020). Similarly, CNVs in multiple PvRBP and MSP genes have been identified in Ethiopian and Southeast Asian isolates using whole genome sequencing (WGS) (Ford et al., 2020).
Several laboratory-based approaches have been developed for detecting CNVs, including multiplex ligation-dependent probe amplification (MLPA), microarray based comparative genomic hybridization (aCGH) and SNP microarrays, whole genome sequencing and RNA sequencing, fluorescence in situ hybridization (FISH) and PCR based methods (Singh et al., 2021).
Here, we employed an aCGH tiling array approach to identify CNVs in P. vivax field isolates from the Indian subcontinent associated with cerebral malaria (CM) by comparing them to isolates from uncomplicated malaria cases. The CGH array offers several benefits in detecting CNVs. Its enhanced sensitivity allows for the detection of small differences in copy number, as it assesses both copy loss and gain variations in one experiment by comparing the relative hybridization intensity of fluorescently labeled test samples with a single reference DNA sample. Moreover, a dense and uniform CGH array can be swiftly synthesized and tailored to focus on virtually any region of interest, including regions abundant in repeats (Liu et al., 2019).
2 Materials and methods
2.1 Collection of patient blood sample
Blood samples (5 ml) were obtained from patients infected with P. vivax who were admitted to S.P. Medical College in Bikaner, India. The collection of patient samples was conducted following informed consent in accordance with hospital guidelines. Approval for sample collection was granted by the hospital ethics committee (No.F.(Acad) SPMC/2003/2395, No.F29(Acad)SPMC/2020/3151). Infection with P. vivax was confirmed by microscopy and rapid diagnostic tests (OptiMal test; Diamed AG, Cressier sur Morat, Switzerland; Falcivax test; Zephyr Biomedical SystemGoa, India) in the hospital. Blood was immediately (within 15 min) subjected to density gradient-based separation (Histopaque 1077, Sigma Aldrich, USA) to isolate peripheral blood mononuclear cells (PBMCs) from the infected blood samples as per the manufacturer’s instructions. Both fractions were then washed twice with phosphate-buffered saline (PBS), lysed with 4 volumes of TRI-reagent, and stored at -80 °C. These samples were maintained within the cold chain during transportation to BITS, where they were processed and evaluated using 18S rRNA gene-based multiplex PCR to ensure the absence of P. falciparum co-infection (Pakalapati et al., 2013). The PCR conditions used for the amplification of the 18S rRNA gene involved initial denaturation at 94 °C for 2 min, followed by denaturation at 94 °C for 1 min 30 s, annealing at 52 °C for 2 min, extension at 72 °C for 3 minutes, after initial denaturation all the reactions were run for 30 cycles. The TRI protocol was used to investigate both RNA and DNA from previous clinical samples (Chomczynski, 1993). The criteria for determining complicated cases were based on the World Health Organization guidelines (World Health Organization, 2010). For CGH array hybridization, isolates from six patients were considered, of which three showed cerebral malarial complications, while the other three were uncomplicated malaria cases (Supplementary Table 1). Cerebral malaria (CM) with a Glasgow coma scale score lesser than 11 was considered a complicated case.
2.2 Array CGH design
A custom P. vivax 2 × 400 K CGH microarray (AMADID: 25335) was designed on an Agilent platform by Genotypic Technology Private Limited, Bangalore, India, using the genomic location and direction of transcriptional regulation data retrieved from NCBI. The 60mer probes were designed based on the optimal GC% and melting temperature using Genotypic_Probe_Parser.pl (Perl program developed by Genotypic Technology). All oligonucleotides were designed and synthesized in situ according to standard algorithms and methodologies used by Agilent Technologies. The array design comprised 418577 probes tilled across the whole genome, with an average probe spacing of 56 bp. This covers the entire genome, irrespective of intergenic or intronic sequences.
2.3 Genomic DNA isolation, preparation, labeling, and hybridization
Genomic DNA preparation, labeling, and hybridization of the CGH array were performed at the Agilent certified microarray facility at Genotypic Technology, Bengaluru, India. Total DNA was extracted from the samples following the manufacturer’s protocol (TRI Reagent, Sigma Aldrich, USA). Subsequently, both DNA samples (PVC and PVU) were run on an agarose gel and confirmed to be intact. The purity and concentration of total DNA were determined using a Nanodrop ND-1000 UV-visible spectrophotometer (Nanodrop Technologies, Rockland, USA). The A260/A280 ratio indicated the absence of proteins and RNA. DNA from the three patient samples showing CM manifestations were pooled (PVC) and used as test, while the same was performed for the uncomplicated samples (PVU) as control. This approach was designed to identify CNVs consistently enriched in cerebral malaria compared to uncomplicated malaria isolates, with pooling providing a representative, group-level overview of robust variations associated with severe disease. The samples for Comparative Genomic DNA Hybridization were labeled using an Agilent Genomic DNA Labeling Kit (Part Number: 5190-0453). Two micrograms of each sample were digested using Alu1 and Rsa1. The restricted DNA was then labeled with Cy3 (PVU) and Cy5 (PVC) dUTP using a random primer labeling method. The labeled DNA was then concentrated, and the quality was assessed for yield and specific activity. The labeled DNA samples were hybridized on a collaboratively designed P. vivax 2x400K CGH microarray (AMADID: 25335). 5 micrograms of Cy3 and Cy5 labeled samples were hybridized. Hybridizations were done using the CGH Hybridization kit of Agilent (Part Number: 5190-0404). Hybridization was performed in Agilent’s Surehyb Chambers at 65° C for 40 h. The hybridized slides were washed using Agilent aCGH wash buffers (Part No: 5188-5221/22) and scanned using an Agilent Microarray Scanner G2505C at 3-micron resolution. The microarray data discussed in this manuscript has been deposited in the NCBI Gene Expression Omnibus (GEO) under the GEO series accession number GSE288219.
2.4 Array CGH data analysis
Image analysis and data normalization were performed by applying the linear dye normalization method using Agilent Feature Extraction Software. Data were further normalized via the centralization algorithm to ensure that log-ratio distributions were centered and comparable across probes. CNVs were then identified using the widely validated Aberration Detection Method II (ADM-2), which integrates probe error information in addition to log-ratio values, providing robust detection even in the presence of noisy probes. ADM-2 is the standard algorithm for Agilent array-CGH analysis and is widely used for high-confidence CNV calling (Delahaye et al., 2012; Chehbani et al., 2022; Bui et al., 2024; Mademont-Soler et al., 2024). To ensure stringent and reliable CNV identification, we applied a minimum requirement of three consecutive probes with an average absolute log2 ratio ≥0.3 (default threshold 6.0, minProbes = 3, minAvgAbsLogRatio = 0.25), effectively removing single-probe artifacts arising from mismatches between probe and target sequences. Additionally, the Fuzzy Zero correction was enabled to reduce spurious long, low-level aberrations caused by correlated probe errors, such as those resulting from sequence divergence. Only genomic regions meeting all these criteria were considered significant. These thresholds provide both statistical support and effect-size filtering, ensuring high specificity in the detection of true copy number changes.
3 Results
This study utilized a custom-designed tiling microarray to explore the overall distinctions between clinical isolates associated with complicated/cerebral malaria and uncomplicated malaria caused by P. vivax. Probes were designed based on the reference genome P. vivax Sal-1. The tiling array facilitated the comprehensive detection of copy number variations (CNVs) throughout the genome. CNVs were identified in the field isolates showing complicated malaria by comparing it with the field isolates showing uncomplicated malaria. The widely accepted ADM-2 algorithm, employing filtering criteria of at least 3 probes and an average log fold change of 0.3, was employed for CNV analysis. The terms “amplification” and “deletion” denote the copy number differences between the test and the control. Deletions indicate regions where probe signal intensities in the test strain are close to background levels, indicating the absence of hybridization.
The size of each chromosome with the number of genes reported here are as per PlasmoDB v68.0. A circos plot of whole genome CNV data of the test has been generated to show the regions in chromosomes exhibiting CNV (Figure 1).
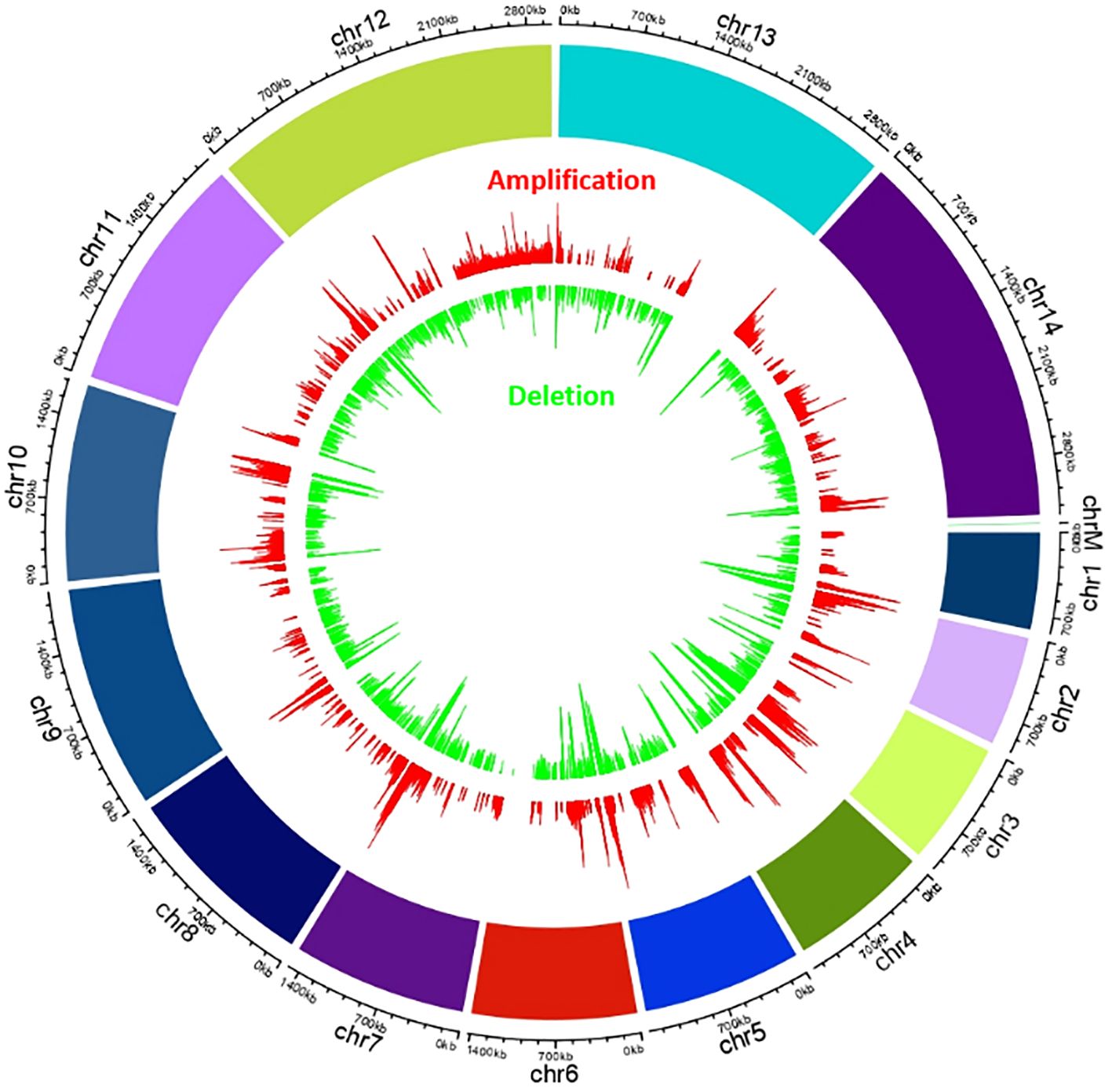
Figure 1. Circos plot of whole genome variation in P.vivax isolates showing cerebral malaria manifestation, representing probe-based chromosome-wise copy number variations. The outer red bars are showing amplifications and green inner bars are showing deletions with fold (log base2).
3.1 Genome-wide variability in gene copy numbers
Amplification/deletion intervals varied from approximately 100bp to 1429 kb. Amplification and deletion were present in all the 14 chromosomes (Supplementary Table 2). Since maintaining a continuous P. vivax culture has been a challenge, therefore many proteins remain uncharacterized. Many of the regions with CNVs have genes encoding these hypothetical proteins.
The longest region showing CNV of approximately 1450 kb is on chromosome 12 (1575586-3004875) covering 316 genes that shows continuous amplification (Figure 2), followed by chromosome 14 (1221441–1677668) with 100 genes and chromosome 10 (496-429938) with 96 genes, both having amplifications.
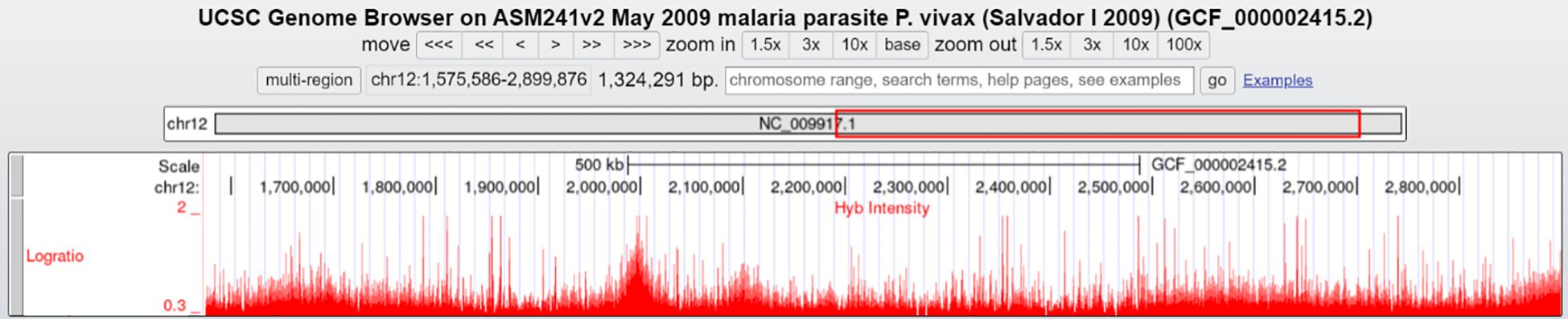
Figure 2. Continuous amplification of 1450 kb in chromosome 12 (1575586-3004875) covering 316 genes (UCSC Genome Browser). Visualization of a 1,450 kb amplified region on chromosome 12 (coordinates 1,575,586–3,004,875) covering 316 genes, as depicted in the UCSC Genome Browser. The log ratio (red peaks) represents hybridization intensity, indicating continuous copy number amplification across the region.
3.2 Copy number variation in regions having drug-resistance genes
Regions having genes encoding ABC transporters from different families showed CNVs (Supplementary Table 3). Orthologs of Plasmodium ABC transporters, known to drive drug resistance in microorganisms and human tumor cells, may similarly contribute to drug resistance in Plasmodium (Koenderink et al., 2010). There are three genes encoding ABC transporters (putative) that are in regions showing amplification while two genes were in regions showing deletion. Three of the genes (PVX_097025, PVX_080100, PVX_118100) encoding multidrug resistance proteins that belong to ABC transporter families are within regions of amplifications. Figure 3 shows the continuous amplification in the region containing PVX_097025, highlighting the CNV at this locus. Amplification in an ortholog of the gene PVX_080100 in P. falciparum (PF3D7_0523000) has been reported to be associate with mefloquine resistance (Cheeseman et al., 2009; Menard and Dondorp, 2017) (Rovira-Vallbona et al., 2023). Increased copy number of multidrug resistance gene-1 (mdr1) has been reported to be associated with increased drug resistance in P. falciparum isolates as well as in P. vivax (Imwong et al., 2008; Suwanarusk et al., 2008; Anderson et al., 2009; Rovira-Vallbona et al., 2023). Suwanarusk et al. (2008) showed that pvmdr1 amplification in Thai P. vivax isolates was linked to higher IC50 values for mefloquine and artesunate, while isolates lacking amplification were more susceptible, supporting a functional role of CNVs in drug resistance.
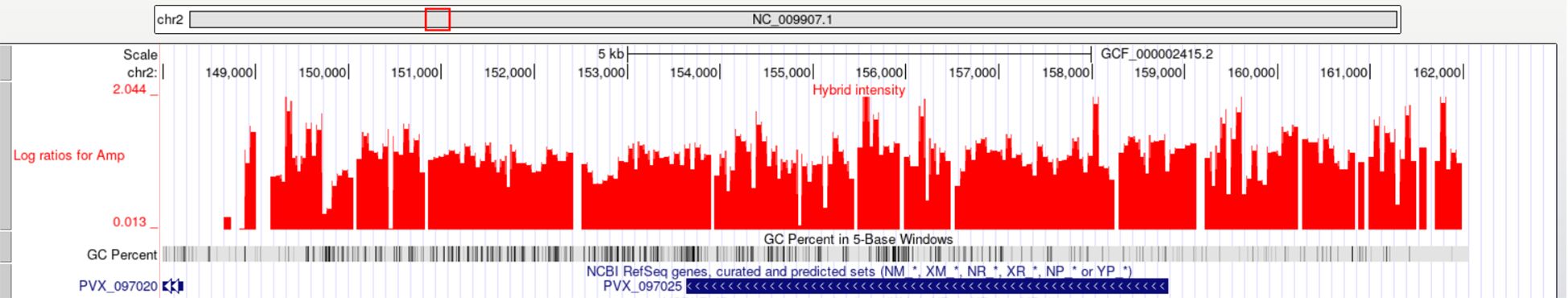
Figure 3. Amplification of P. vivax gene PVX_097025 (multidrug resistance-associated protein 1, putative) on chromosome 2. The UCSC Genome Browser plot shows log2 ratio intensities (red peaks) indicating copy number gain in the region spanning PVX_097025.
In our data, we have also seen CNVs in regions having chloroquine resistance genes, a putative chloroquine resistance marker protein (PVX_118062 in chr 12) having amplification and putative chloroquine resistance transporters (PVX_087980 in chr 1) having deletion.
There is amplification observed in the gene responsible for producing the enzyme hydroxymethylpterin pyrophosphokinase-dihydropteroate synthetase (PPPK-DHPS). PPPK-DHPS plays a crucial role in the folate pathway of P. falciparum and is a specific target of the drug sulfadoxine (Kasekarn et al., 2004). Sulfadoxine resistance has been reported in both P. falciparum and P. vivax (Marfurt et al., 2008). Amplification in this gene might be playing a role in drug resistance against sulfadoxine.
3.3 Copy number variation in regions having surface antigens
Regions with genes for surface antigens such as 6-cysteine proteins, tryptophan-rich antigens (TRAgs), serine-repeat antigen (SERA), apical membrane antigen (AMA), have shown CNVs. There are 24 TRAgs showing CNVs and all of them have amplifications. In P. vivax species, TRAgs often occur in clusters along chromosomes, suggesting their proliferation via gene duplication and diversification. Certain members of the P. vivax TRAg family have demonstrated binding properties to red blood cells, and in certain instances, interactions with partner molecules have been documented (Kundu et al., 2023). One of the TRAgs encoded by PVX_088850 which lies within region of amplification in our data, has been reported to be interacting with PvMSP7, suggesting possible establishment or stabilization of protein complex at P.vivax merozoite surface (Tyagi et al., 2016).
Amplification is present in regions with cytoadherence-linked asexual protein (CLAG) genes located on chromosomes 7 and 14. In P. falciparum, CLAG proteins has been shown to be involved in the binding of infected erythrocytes to host endothelial cells, a process termed cytoadherence (Holt et al., 1999; Newbold et al., 1999; Gupta et al., 2015) and involved in nutrient uptake and solute transport (Nguitragool et al., 2011; Gupta et al., 2018a). CNVs are found in regions containing genes associated with merozoite surface proteins (MSP-1, MSP-3, and MSP-7), with eight occurring in amplified regions and six in deleted regions. Using the UCSC Genome Browser, we observed that region having several MSP paralogs lying in close proximity on the chromosome was amplified (Figure 4). Additionally, CNVs are present in 23 genes encoding SERA proteins, all located on chromosome 4 of the P. vivax genome, including four in amplified regions and 19 in deleted regions. They form a multigene family and are conserved among Plasmodium species. Recent studies implicate this gene family in a number of aspects in parasite biology and induction of protective immune response (Arisue et al., 2020).
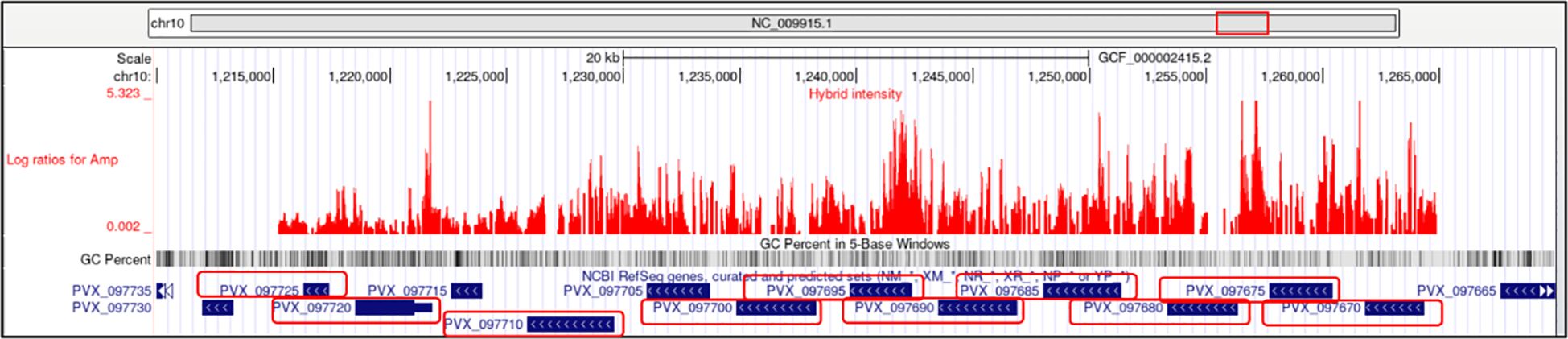
Figure 4. Amplification of merozoite surface protein (MSP) paralogs on chromosome 10 in P. vivax. The UCSC Genome Browser plot shows log2 ratio intensities (red peaks) indicating amplified regions in the chromosome 10 containing MSP genes. Gene IDs corresponding to MSP paralogs (PVX_097700, PVX_097710, PVX_097720, PVX_097725, PVX_097690, PVX_097695, PVX_097675, PVX_097680, PVX_097670) are highlighted with red boxes.
Amplifications in regions with genes for reticulocyte-binding proteins or PvRBP (PVX_090325 and PVX_090330 in chr 5, PVX_098582 and PVX_098585 in chr 7, PVX_094255 in chr 8, and PVX_121920 in chr 14) are present. A study by Kosaisavee et al. also reported variations in the copy number in the reticulocyte-binding proteins Pvrbp2a and Pvrbp2b by a quantitative real-time SYBR Green PCR assay (Kosaisavee et al., 2012). Additionally, a recent study suggests that copy number variations in PvRBP2 of Plasmodium vivax may contribute to its ability to infect Duffy-negative individuals (Pestana et al., 2024).
Rhoptry proteins, such as rhoptry-associated protein and rhoptry neck protein, have amplifications in their genes, specifically PVX_097590 in chr 10, PVX_117880 in chr 12, and PVX_101485 in chr 14.
Some of the regions with genes for ETRAMPS or early transcribed membrane proteins (PVX_096070 in chr 3, PVX_003565 in chr 4, PVX_088870 and PVX_090230 in chr 5, PVX_086915 in chr 7, PVX_118680 and PVX_121950 in chr 12) were shown to have amplifications. ETRAMPs are expressed during Plasmodium intracellular phases and inserted at the parasite parasitophorous vacuole membrane (PVM) (Brandsma et al., 2022).
There are gains in regions having two genes encoding Pvstp1, which is phylogenetically closely related to P. falciparum SURFIN4.2’, a protein exposed on the parasite-infected erythrocyte (iE) surface, and is thus considered to be exposed on P. vivax-iE (Winter et al., 2005).
Our data showed CNVs in 117 unique vir genes present in the sub-telomeric regions of chromosomes. There are 84 genes that are within amplified regions and 10 in regions with deletions. There are 23 vir genes that have both deletion and amplifications (Supplementary Table 3). The vir (variant interspersed repeat) genes in P. vivax have been proposed to play a crucial role in cytoadherence, facilitating the adherence of infected red blood cells to host endothelial cells. This phenomenon might be contributing to the pathogenesis and severity of malaria, as cytoadherence can lead to microvascular obstruction and tissue damage, exacerbating the clinical manifestations of P. vivax infection (Carvalho et al., 2010; Chotivanich et al., 2012; De las Salas et al., 2013; Marín-Menéndez et al., 2013; Totino and Lopes, 2017). It has also been hypothesized that vir genes play a role in malaria pathogenesis and that P. vivax may exploit the extensive sequence diversity within these genes to enhance virulence (Merino et al., 2006; Gupta et al., 2014; Na et al., 2021).
In our analysis, we have also seen CNVs in genes encoding Plasmodium exported proteins of unknown functions. There are 73 gains and 13 losses in the regions harboring these genes. These proteins are secreted by the parasite into the host erythrocyte, where they modulate various cellular processes. In P. falciparum, some exported proteins have been reported to be involved in cytoadherence, allowing infected red blood cells to sequester in different tissues and evade clearance by the immune system while others are implicated in modifying the host cell membrane, leading to the formation of structures such as Maurer’s clefts, which serve as trafficking hubs for exported proteins. Some exported proteins help permeabilize the RBC to allow nutrients and wastes to be exchanged with the blood plasma to facilitate rapid growth and parasite proliferation (Elsworth et al., 2014).
3.4 Copy number variations in important genes
Some genes encoding for proteins belonging to the ubiquitin system machinery and proteasomal degradation pathway were seen to be in the amplified or deleted regions. 20 genes encoding for proteins of the ubiquitin system machinery and 9 genes encoding for proteasomal degradation pathway have shown CNVs. When the parasite is exposed to host molecule there is an increase in transcription levels of genes encoding for proteins related to the Ubiquitin Proteasome (UPS) System (Pereira et al., 2018). The proteasome is the main engine of Plasmodium protein degradation. Protein ubiquitylation plays key roles in cell biology, for example in tagging proteins for proteasomal degradation, or targeting proteins to distinct subcellular locations. The transition from intracellular schizont to extracellular merozoite stages in the asexual blood stage cycle is associated with a general increase in ubiquitylation levels (Green et al., 2020). Ubiquitin modification is associated with altered parasite susceptibility to multiple antimalarials (Ng et al., 2017).
Amplification in 12 genes encoding Phist proteins are present. Studies in P. falciparum have shown that these proteins are involved in different processes including surface display of PfEMP1, gametocytogenesis, changes in cell rigidity. Some phist genes have been shown to be differentially expressed in cerebral malaria and pregnancy-associated malaria, indicating a significant role of Phist in these complications (Warncke et al., 2016).
Approximately 80 genes responsible for encoding proteins forming ribosomal complexes have exhibited copy number variations. This observation aligns with expectations, given the parasite’s multifaceted life cycle necessitating a robust protein synthesis machinery.
Another important amplification is seen in the gene encoding for putative plasmepsin IV (PVX_086040). Plasmepsin IV (Plm IV), an aspartic protease within the food vacuole of the malaria parasite Plasmodium falciparum, participates in the degradation of host hemoglobin by the parasite (Gutiérrez-de-Terán et al., 2006).
3.5 Copy number variation in intergenic region
CNVs have been seen in the intergenic region as well (Supplementary Table 3). CNVs in intergenic regions can also affect gene expression, although indirectly. While intergenic regions do not contain genes themselves, they often harbor important regions such as promoter and other regulatory elements such as enhancers and insulators, which play crucial roles in regulating the expression of nearby genes (Nelson et al., 2004). When CNVs occur in intergenic regions, they can disrupt the regulatory landscape by altering the number or arrangement of regulatory elements. This disruption can affect the expression of neighboring genes by modifying the accessibility of their regulatory elements to transcription factors and other regulatory proteins.
Additionally, CNVs in intergenic regions can sometimes influence gene expression through long-range interactions between regulatory elements and target genes. These interactions can occur through chromatin looping and other mechanisms, allowing distal regulatory elements to exert control over gene expression. Disruption of these long-range interactions by CNVs can consequently impact gene expression levels. In our study, several CNVs were located within intergenic regions upstream of protein-coding genes. While the precise functional impact of these CNVs remains to be determined, their proximity to genes encoding surface antigens, drug resistance proteins, and stress-related proteins suggests that they could potentially influence gene regulation. However, experimental studies are required to clarify whether these intergenic CNVs play a regulatory role in P. vivax.
Although direct functional validation of intergenic CNVs in Plasmodium vivax has not yet been performed, evidence from malaria research and studies in other human diseases suggests that intergenic regions can have important regulatory roles. In P. falciparum, intergenic regions can give rise to the transcription of regulatory lncRNAs, which may influence gene expression, sexual development, and parasite transmission (Batugedara et al., 2023). Similarly, in humans, CNVs in non-coding regions have been shown to alter gene regulation and impact phenotypic outcomes (Zhang and Lupski, 2015). By analogy, intergenic CNVs in P. vivax may also influence nearby gene expression and contribute to parasite adaptability, highlighting a potential mechanism by which non-coding genomic variation could affect parasite biology. Future studies are required to experimentally validate these effects.
4 Discussion
In this study, we report CNVs observed in P. vivax complicated malaria (cerebral malaria) field isolates compared to uncomplicated cases, although we cannot conclude whether these CNVs contribute to disease severity. All patient samples in this study were collected from a single location, Bikaner, North-west India, over the time period 2007-2008, which may limit the generalizability of the CNV patterns observed. However, obtaining cerebral malaria (CM) samples for larger cohort studies is challenging due to the comparative infrequency of such cases. In this context, the three CM samples analyzed here provide a sufficient basis for a preliminary investigation. While limited in scale, they offer unique insights into potential CNV patterns associated with severe disease and establish a foundation for future studies. To our knowledge, no previous whole-genome CNV analysis has been performed in P. vivax cerebral malaria cases. Future studies across multiple geographic regions and larger sample sizes will be needed to confirm and extend these findings. vivaxSignificant gains and losses can be seen in regions harboring important genes such as surface antigens, drug resistance, Plasmodium exported proteins etc. Copy number variations (CNVs) observed in surface antigen and drug resistance genes may contribute to P. vivax adaptability, immune evasion, and treatment outcomes. Gene families such as msp7 and vir, which exhibited CNVs in our dataset, are known to undergo duplication and diversification, a strategy that enhances immune evasion and may influence parasite pathogenicity. Amplifications within vir genes, in particular, could facilitate antigenic variation and cytoadherence, processes implicated in disease severity (Merino et al., 2006; Carvalho et al., 2010; Bernabeu et al., 2012; Gupta et al., 2014; Garzón-Ospina et al., 2016; Castillo et al., 2017; Na et al., 2021) Additionally, CNVs detected in drug resistance-related loci, including pvmdr1, have been associated in previous studies with altered drug sensitivity, suggesting that structural variation in these genes may also influence treatment response in P. vivax (Suwanarusk et al., 2008). At this stage of the investigation, it’s not possible to speculate whether the variances observed in the test (cerebral malaria field isolates) contribute directly to the severity of the disease. Rather, we are reporting that these CNVs were observed in cerebral malaria patient isolates when compared with uncomplicated malaria cases. These findings highlight differences in parasite CNV patterns between patient groups showing the same disease manifestation, but further functional studies would be required to establish any causal relationship. These variances extend beyond coding regions to intergenic locations, suggesting possible implications for gene regulation. Although functional validation is lacking, intergenic CNVs in P. vivax may, based on parallels with P. falciparum and human studies, influence gene regulation and parasite adaptability, underscoring the need for experimental confirmation (Batugedara et al., 2023; Zhang and Lupski, 2015). While our study did not directly examine host genetic factors or immune responses, these remain plausible influences on the CNV patterns observed in the P. vivax cerebral malaria isolates. For instance, amplification of the gene encoding pvmdr1 in our dataset, previously linked to multidrug resistance in P. vivax, has been reported that host-imposed selection pressures, such as antimalarial drug exposure or immune challenges may shape genomic variation (Suwanarusk et al., 2008). Supporting this, studies in P. falciparum have shown that CNVs frequently arise under selective pressures such as drug treatment, where gene amplifications confer resistance (Qidwai, 2020; Luth et al., 2024). This study is an initial effort to employ aCGH tiling array to identify any variations or disparities in genomic segments between parasite isolates causing cerebral malaria and uncomplicated malaria. Understanding these genomic variations is essential for elucidating the mechanisms of drug resistance, immune evasion and survivability of P. vivax, ultimately guiding the development of effective malaria control strategies.
Data availability statement
The microarray data discussed in this manuscript has been deposited in the NCBI Gene Expression Omnibus (GEO) under the GEO series accession number GSE288219.
Ethics statement
The studies involving humans were approved by Institute Ethics and Research Board, S.P. Medical College, Bikaner, Rajasthan. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Sample collection was previously approved by the Ethics Committee of SP Medical College Hospital (Approval No. F. (Acad)SPMC/2003/2395). Authorization to utilize these samples for further studies was granted through IERB approval No. F29(Acad)SPMC/2020/3151, dated 05.09.2020.
Author contributions
ST: Investigation, Formal Analysis, Writing – original draft. PB: Methodology, Writing – review & editing. SK: Writing – review & editing, Methodology, Investigation. DK: Methodology, Investigation, Writing – review & editing. MA: Formal Analysis, Writing – review & editing, Data curation, Methodology. RM: Writing – review & editing, Conceptualization. SR: Conceptualization, Writing – review & editing. AD: Supervision, Methodology, Conceptualization, Writing – review & editing, Visualization, Resources.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Acknowledgments
We extend our gratitude to all patients for their voluntary consent and participation. We are grateful to Birla Institute of Technology and Science (BITS), Pilani (Pilani campus), and SP Medical College, Bikaner, for providing the essential facilities needed for this research. We also extend our thanks to Genotypic Private Limited, Bengaluru, for their collaboration in microarray design, hybridization, and data generation.
Conflict of interest
Authors MA, RM, and SR were employed by the company Genotypic Technology Pvt. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmala.2025.1667330/full#supplementary-material
References
Alexandre M. A., Ferreira C. O., Siqueira A. M., Magalhães B. L., Mourão M. P. G., Lacerda M. V., et al. (2010). Severe plasmodium vivax malaria, Brazilian amazon. Emerg. Infect. Dis. 16, 1611–1614. doi: 10.3201/eid1610.100685
Anderson T. J. C., Patel J., and Ferdig M. T. (2009). Gene copy number and malaria biology. Trends Parasitol. 25, 336–343. doi: 10.1016/j.pt.2009.04.005
Angstadt A. Y., Berg A., Zhu J., Miller P., Hartman T. J., Lesko S. M., et al. (2013). The effect of copy number variation in the phase II detoxification genes UGT2B17 and UGT2B28 on colorectal cancer risk. Cancer 119, 2477–2485. doi: 10.1002/cncr.28009
Arisue N., Palacpac N. M. Q., Tougan T., and Horii T. (2020). Characteristic features of the SERA multigene family in the malaria parasite. Parasitol. Vectors 13, 1–10. doi: 10.1186/s13071-020-04044-y
Batugedara G., Lu X. M., Hristov B., Abel S., Chahine Z., Hollin T., et al. (2023). Novel insights into the role of long non-coding RNA in the human malaria parasite, Plasmodium falciparum. Nat. Commun. 14, 5086. doi: 10.1038/s41467-023-40883-w
Bernabeu M., Lopez F. J., Ferrer M., Martin-Jaular L., Razaname A., Corradin G., et al. (2012). Functional analysis of Plasmodium vivax VIR proteins reveals different subcellular localizations and cytoadherence to the ICAM-1 endothelial receptor. Cell. Microbiol. 14, 386–400. doi: 10.1111/j.1462-5822.2011.01726.x
Brandsma A. M., Hilmer C., Rauch M., Matuschewski K., and Montagna G. N. (2022). Plasmodium early transcribed membrane proteins appear tailored to the host range of malaria parasites. Int. J. Parasitol. 52, 135–143. doi: 10.1016/j.ijpara.2021.08.005
Bui H. T. P., Huy Do D., Ly H. T. T., Tran K. T., Le H. T. T., Nguyen K. T., et al. (2024). De novo copy number variations in candidate genomic regions in patients of severe autism spectrum disorder in Vietnam. PloS One 19, e0290936. doi: 10.1371/journal.pone.0290936
Buyon L. E., Elsworth B., and Duraisingh M. T. (2021). The molecular basis of antimalarial drug resistance in Plasmodium vivax. Int. J. Parasitol. Drugs Drug Resist. 16, 23–37. doi: 10.1016/j.ijpddr.2021.04.002
Carvalho B. O., Lopes S. C. P., Nogueira P. A., Orlandi P. P., Bargieri D. Y., Blanco Y. C., et al. (2010). On the cytoadhesion of plasmodium vivax–infected erythrocytes. J. Infect. Dis. 202, 638–647. doi: 10.1086/654815
Castillo A. I., Pacheco M. A., and Escalante A. A. (2017). Evolution of the merozoite surface protein 7 (MSP7) family in Plasmodium vivax and P. falciparum: a comparative approach. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 50, 7–19. doi: 10.1016/j.meegid.2017.01.024
Chaignat E., Yahya-Graison E. A., Henrichsen C. N., Chrast J., Schütz F., Pradervand S., et al. (2011). Copy number variation modifies expression time courses. Genome Res. 21, 106–113. doi: 10.1101/gr.112748.110
Chang H.-H., Moss E. L., Park D. J., Ndiaye D., Mboup S., Volkman S. K., et al. (2013). Malaria life cycle intensifies both natural selection and random genetic drift. Proc. Natl. Acad. Sci. U. S. A. 110, 20129–20134. doi: 10.1073/pnas.1319857110
Cheeseman I. H., Gomez-Escobar N., Carret C. K., Ivens A., Stewart L. B., Tetteh K. K., et al. (2009). Gene copy number variation throughout the Plasmodium falciparum genome. BMC Genomics 10, 353. doi: 10.1186/1471-2164-10-353
Chehbani F., Tomaiuolo P., Picinelli C., Baccarin M., Castronovo P., Scattoni M. L., et al. (2022). Yield of array-CGH analysis in Tunisian children with autism spectrum disorder. Mol. Genet. Genomic Med. 10, e1939. doi: 10.1002/mgg3.1939
Chomczynski P. (1993). A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 15, 532–534.
Chotivanich K., Udomsangpetch R., Suwanarusk R., Pukrittayakamee S., Wilairatana P., Beeson J. G., et al. (2012). Plasmodium vivax adherence to placental glycosaminoglycans. PloS One 7, e34509. doi: 10.1371/journal.pone.0034509
Costa G. L., Amaral L. C., Fontes C. J. F., Carvalho L. H., de Brito C. F. A., and de Sousa T. N. (2017). Assessment of copy number variation in genes related to drug resistance in Plasmodium vivax and Plasmodium falciparum isolates from the Brazilian Amazon and a systematic review of the literature. Malar. J. 16, 152. doi: 10.1186/s12936-017-1806-z
Delahaye A., Bitoun P., Drunat S., Gérard-Blanluet M., Chassaing N., Toutain A., et al. (2012). Genomic imbalances detected by array-CGH in patients with syndromal ocular developmental anomalies. Eur. J. Hum. Genet. EJHG 20, 527–533. doi: 10.1038/ejhg.2011.233
De las Salas B., Segura C., Pabón A., Lopes S. C. P., Costa F. T. M., and Blair S. (2013). Adherence to human lung microvascular endothelial cells (HMVEC-L) of Plasmodium vivax isolates from Colombia. Malar. J. 12, 347. doi: 10.1186/1475-2875-12-347
Douglas N. M., Anstey N. M., Buffet P. A., Poespoprodjo J. R., Yeo T. W., White N. J., et al. (2012). The anaemia of Plasmodium vivax malaria. Malar. J. 11, 135. doi: 10.1186/1475-2875-11-135
Elsworth B., Crabb B. S., and Gilson P. R. (2014). Protein export in malaria parasites: an update. Cell. Microbiol. 16, 355–363. doi: 10.1111/cmi.12261
Faway E., Musset L., Pelleau S., Volney B., Casteras J., Caro V., et al. (2016). Plasmodium vivax multidrug resistance-1 gene polymorphism in French Guiana. Malar. J. 15, 540. doi: 10.1186/s12936-016-1595-9
Feng X., Carlton J. M., Joy D. A., Mu J., Furuya T., Suh B. B., et al. (2003). Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc. Natl. Acad. Sci. 100, 8502–8507. doi: 10.1073/pnas.1232502100
Ford A., Kepple D., Abagero B. R., Connors J., Pearson R., Auburn S., et al. (2020). Whole genome sequencing of Plasmodium vivax isolates reveals frequent sequence and structural polymorphisms in erythrocyte binding genes. PloS Negl. Trop. Dis. 14, e0008234. doi: 10.1371/journal.pntd.0008234
Gadalla N. B., Adam I., Elzaki S.-E., Bashir S., Mukhtar I., Oguike M., et al. (2011). Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicrob. Agents Chemother. 55, 5408–5411. doi: 10.1128/AAC.05102-11
Garzón-Ospina D., Forero-Rodríguez J., and Patarroyo M. A. (2016). Evidence of functional divergence in MSP7 paralogous proteins: a molecular-evolutionary and phylogenetic analysis. BMC Evol. Biol. 16, 256. doi: 10.1186/s12862-016-0830-x
Gil J. P. and Fançony C. (2021). Plasmodium falciparum multidrug resistance proteins (pfMRPs). Front. Pharmacol. 12. doi: 10.3389/fphar.2021.759422
Green J. L., Wu Y., Encheva V., Lasonder E., Prommaban A., Kunzelmann S., et al. (2020). Ubiquitin activation is essential for schizont maturation in Plasmodium falciparum blood-stage development. PloS Pathog. 16, e1008640. doi: 10.1371/journal.ppat.1008640
Gupta A., Balabaskaran-Nina P., Nguitragool W., Saggu G. S., Schureck M. A., and Desai S. A. (2018a). CLAG3 self-associates in malaria parasites and quantitatively determines nutrient uptake channels at the host membrane. mBio 9, e02293–e02217. doi: 10.1128/mBio.02293-17
Gupta H., Macete E., Bulo H., Salvador C., Warsame M., Carvalho E., et al. (2018b). Drug-resistant polymorphisms and copy numbers in plasmodium falciparum, Mozambique. Emerg. Infect. Dis. 24, 40–48. doi: 10.3201/eid2401.170864
Gupta P., Pande V., Das A., and Singh V. (2014). Genetic Polymorphisms in VIR Genes among Indian Plasmodium vivax Populations. Korean J. Parasitol. 52, 557–564. doi: 10.3347/kjp.2014.52.5.557
Gupta A., Thiruvengadam G., and Desai S. A. (2015). The conserved clag multigene family of malaria parasites: essential roles in host-pathogen interaction. Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 18, 47–54. doi: 10.1016/j.drup.2014.10.004
Gutiérrez-de-Terán H., Nervall M., Ersmark K., Liu P., Janka L. K., Dunn B., et al. (2006). Inhibitor binding to the plasmepsin IV aspartic protease from plasmodium falciparum. Biochemistry 45, 10529–10541. doi: 10.1021/bi0609669
Hastings P., Lupski J. R., Rosenberg S. M., and Ira G. (2009). Mechanisms of change in gene copy number. Nat. Rev. Genet. 10, 551–564. doi: 10.1038/nrg2593
Heinberg A., Siu E., Stern C., Lawrence E. A., Ferdig M. T., Deitsch K. W., et al. (2013). Direct evidence for the adaptive role of copy number variation on antifolate susceptibility in Plasmodium falciparum. Mol. Microbiol. 88, 702–712. doi: 10.1111/mmi.12162
Henrichsen C. N., Chaignat E., and Reymond A. (2009). Copy number variants, diseases and gene expression. Hum. Mol. Genet. 18, R1–R8. doi: 10.1093/hmg/ddp011
Holt D. C., Gardiner D. L., Thomas E. A., Mayo M., Bourke P. F., Sutherland C. J., et al. (1999). The cytoadherence linked asexual gene family of Plasmodium falciparum: are there roles other than cytoadherence? Int. J. Parasitol. 29, 939–944. doi: 10.1016/S0020-7519(99)00046-6
Imwong M., Pukrittayakamee S., Pongtavornpinyo W., Nakeesathit S., Nair S., Newton P., et al. (2008). Gene amplification of the multidrug resistance 1 gene of plasmodium vivax isolates from Thailand, Laos, and Myanmar. Antimicrob. Agents Chemother. 52, 2657–2659. doi: 10.1128/aac.01459-07
Kasekarn W., Sirawaraporn R., Chahomchuen T., Cowman A. F., and Sirawaraporn W. (2004). Molecular characterization of bifunctional hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase from Plasmodium falciparum. Mol. Biochem. Parasitol. 137, 43–53. doi: 10.1016/j.molbiopara.2004.04.012
Kennedy D. A. and Dwyer G. (2018). Effects of multiple sources of genetic drift on pathogen variation within hosts. PloS Biol. 16, e2004444. doi: 10.1371/journal.pbio.2004444
Kirov G., Rees E., Walters J. T. R., Escott-Price V., Georgieva L., Richards A. L., et al. (2014). The penetrance of copy number variations for schizophrenia and developmental delay. Biol. Psychiatry 75, 378–385. doi: 10.1016/j.biopsych.2013.07.022
Kochar D. K., Das A., Kochar S. K., Saxena V., Sirohi P., Garg S., et al. (2009). Severe Plasmodium vivax malaria: a report on serial cases from Bikaner in northwestern India. Am. J. Trop. Med. Hyg. 80, 194–198. doi: 10.4269/ajtmh.2009.80.194
Kochar D. K., Pakalapati D., Kochar S. K., Sirohi P., Khatri M. P., Kochar A., et al. (2007). An unexpected cause of fever and seizures. Lancet 370, 908. doi: 10.1016/S0140-6736(07)61417-2
Kochar D. K., Saxena V., Singh N., Kochar S. K., Kumar S. V., and Das A. (2005). Plasmodium vivax malaria. Emerg. Infect. Dis. 11, 132–134. doi: 10.3201/eid1101.040519
Koenderink J. B., Kavishe R. A., Rijpma S. R., and Russel F. G. M. (2010). The ABCs of multidrug resistance in malaria. Trends Parasitol. 26, 440–446. doi: 10.1016/j.pt.2010.05.002
Kosaisavee V., Lek-Uthai U., Suwanarusk R., Grüner A. C., Russell B., Nosten F., et al. (2012). Genetic diversity in new members of the reticulocyte binding protein family in thai plasmodium vivax isolates. PloS One 7, e32105. doi: 10.1371/journal.pone.0032105
Kundu P., Naskar D., McKie S. J., Dass S., Kanjee U., Introini V., et al. (2023). The structure of a Plasmodium vivax Tryptophan Rich Antigen domain suggests a lipid binding function for a pan-Plasmodium multi-gene family. Nat. Commun. 14, 5703. doi: 10.1038/s41467-023-40885-8
Kute V. B., Trivedi H. L., Vanikar A. V., Shah P. R., Gumber M. R., Patel H. V., et al. (2012). Plasmodium vivax malaria–associated acute kidney injury, India 2010–2011. Emerg. Infect. Dis. 18, 842–845. doi: 10.3201/eid1805.111442
Liu M., Fang L., Liu S., Pan M. G., Seroussi E., Cole J. B., et al. (2019). Array CGH-based detection of CNV regions and their potential association with reproduction and other economic traits in Holsteins. BMC Genomics 20, 181. doi: 10.1186/s12864-019-5552-1
Lo E., Hostetler J. B., Yewhalaw D., Pearson R. D., Hamid M. M. A., Gunalan K., et al. (2019). Frequent expansion of Plasmodium vivax Duffy Binding Protein in Ethiopia and its epidemiological significance. PloS Negl. Trop. Dis. 13, e0007222. doi: 10.1371/journal.pntd.0007222
Luth M. R., Godinez-Macias K. P., Chen D., Okombo J., Thathy V., Cheng X., et al. (2024). Systematic in vitro evolution in Plasmodium falciparum reveals key determinants of drug resistance. Science 386, eadk9893. doi: 10.1126/science.adk9893
Mackinnon M. and Marsh K. (2010). The selection landscape of malaria parasites. Science 328, 866–871. doi: 10.1126/science.1185410
Mademont-Soler I., Esteba-Castillo S., Jiménez-Xifra A., Alemany B., Ribas-Vidal N., Cutillas M., et al. (2024). Unexpected complexity in the molecular diagnosis of spastic paraplegia 11. Mol. Genet. Genomic Med. 12, e2475. doi: 10.1002/mgg3.2475
Marfurt J., de Monbrison F., Brega S., Barbollat L., Müller I., Sie A., et al. (2008). Molecular Markers of In Vivo Plasmodium vivax Resistance to Amodiaquine Plus Sulfadoxine-Pyrimethamine: Mutations in pvdhfr and pvmdr1. J. Infect. Dis. 198, 409–417. doi: 10.1086/589882
Marín-Menéndez A., Bardají A., Martínez-Espinosa F. E., Bôtto-Menezes C., Lacerda M. V., Ortiz J., et al. (2013). Rosetting in plasmodium vivax: A cytoadhesion phenotype associated with anaemia. PloS Negl. Trop. Dis. 7, e2155. doi: 10.1371/journal.pntd.0002155
Melo G. C., Monteiro W. M., Siqueira A. M., Silva S. R., Magalhães B. M. L., Alencar A. C. C., et al. (2014). Expression Levels of pvcrt-o and pvmdr-1 Are Associated with Chloroquine Resistance and Severe Plasmodium vivax Malaria in Patients of the Brazilian Amazon. PloS One 9, e105922. doi: 10.1371/journal.pone.0105922
Menard D. and Dondorp A. (2017). Antimalarial drug resistance: A threat to malaria elimination. Cold Spring Harb. Perspect. Med. 7, a025619. doi: 10.1101/cshperspect.a025619
Merino E. F., Fernandez-Becerra C., Durham A. M., Ferreira J. E., Tumilasci V. F., d’Arc-Neves J., et al. (2006). Multi-character population study of the vir subtelomeric multigene superfamily of Plasmodium vivax, a major human malaria parasite. Mol. Biochem. Parasitol. 149, 10–16. doi: 10.1016/j.molbiopara.2006.04.002
Na B.-K., Kim T.-S., Lin K., Baek M.-C., Chung D.-I., Hong Y., et al. (2021). Genetic polymorphism of vir genes of Plasmodium vivax in Myanmar. Parasitol. Int. 80, 102233. doi: 10.1016/j.parint.2020.102233
Naing C., Whittaker M. A., Nyunt Wai V., and Mak J. W. (2014). Is Plasmodium vivax malaria a severe malaria?: a systematic review and meta-analysis. PloS Negl. Trop. Dis. 8, e3071. doi: 10.1371/journal.pntd.0003071
Nautiyal A., Singh S., Parmeswaran G., and DiSalle M. (2005). Hepatic dysfunction in a patient with Plasmodium vivax infection. MedGenMed Medscape Gen. Med. 7, 8.
Nelson C. E., Hersh B. M., and Carroll S. B. (2004). The regulatory content of intergenic DNA shapes genome architecture. Genome Biol. 5, R25. doi: 10.1186/gb-2004-5-4-r25
Newbold C., Craig A., Kyes S., Rowe A., Fernandez-Reyes D., and Fagan T. (1999). Cytoadherence, pathogenesis and the infected red cell surface in Plasmodium falciparum. Int. J. Parasitol. 29, 927–937. doi: 10.1016/S0020-7519(99)00049-1
Ng C. L., Fidock D. A., and Bogyo M. (2017). Protein degradation systems as antimalarial therapeutic targets. Trends Parasitol. 33, 731–743. doi: 10.1016/j.pt.2017.05.009
Nguitragool W., Bokhari A. A. B., Pillai A. D., Rayavara K., Sharma P., Turpin B., et al. (2011). Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell 145, 665–677. doi: 10.1016/j.cell.2011.05.002
Pakalapati D., Garg S., Middha S., Kochar A., Subudhi A. K., Arunachalam B. P., et al. (2013). Comparative evaluation of microscopy, OptiMAL® and 18S rRNA gene based multiplex PCR for detection of Plasmodium falciparum & Plasmodium vivax from field isolates of Bikaner, India. Asian Pac. J. Trop. Med. 6, 346–351. doi: 10.1016/S1995-7645(13)60037-1
Pereira P. H. S., Curra C., and Garcia C. R. S. (2018). Ubiquitin proteasome system as a potential drug target for malaria. Curr. Top. Med. Chem. 18, 315–320. doi: 10.2174/1568026618666180427145308
Pestana K., Ford A., Rama R., Abagero B., Kepple D., Tomida J., et al. (2024). Copy number variations of plasmodium vivax DBP1, EBP/DBP2, and RBP2b in Ethiopians who are duffy positive and duffy negative. J. Infect. Dis. 230, 1004–1012. doi: 10.1093/infdis/jiae388
Phyo A. P., Dahal P., Mayxay M., and Ashley E. A. (2022). Clinical impact of vivax malaria: A collection review. PloS Med. 19, e1003890. doi: 10.1371/journal.pmed.1003890
Popovici J., Roesch C., Carias L. L., Khim N., Kim S., Vantaux A., et al. (2020). Amplification of Duffy binding protein-encoding gene allows Plasmodium vivax to evade host anti-DBP humoral immunity. Nat. Commun. 11, 953. doi: 10.1038/s41467-020-14574-9
Pös O., Radvanszky J., Buglyó G., Pös Z., Rusnakova D., Nagy B., et al. (2021). DNA copy number variation: Main characteristics, evolutionary significance, and pathological aspects. Biomed. J. 44, 548–559. doi: 10.1016/j.bj.2021.02.003
Qidwai T. (2020). Exploration of copy number variation in genes related to anti-malarial drug resistance in Plasmodium falciparum. Gene 736, 144414. doi: 10.1016/j.gene.2020.144414
Rovira-Vallbona E., Kattenberg J. H., Hong N. V., Guetens P., Imamura H., Monsieurs P., et al. (2023). Molecular surveillance of Plasmodium falciparum drug-resistance markers in Vietnam using multiplex amplicon sequencing, (2000–2016). Sci. Rep. 13, 13948. doi: 10.1038/s41598-023-40935-7
Sarkar S. and Bhattacharya P. (2008). Cerebral malaria caused by Plasmodium vivax in adult subjects. Indian J. Crit. Care Med. Peer-Rev. Off. Publ. Indian Soc Crit. Care Med. 12, 204–205. doi: 10.4103/0972-5229.45084
Sarkar S., Saha K., and Das C. S. (2010). Three cases of ARDS: An emerging complication of: Plasmodium vivax: malaria. Lung India 27, 154. doi: 10.4103/0970-2113.68323
Silva S. R., Almeida A. C. G., da Silva G. A. V., Ramasawmy R., Lopes S. C. P., Siqueira A. M., et al. (2018). Chloroquine resistance is associated to multi-copy pvcrt-o gene in Plasmodium vivax malaria in the Brazilian Amazon. Malar. J. 17, 267. doi: 10.1186/s12936-018-2411-5
Simam J., Rono M., Ngoi J., Nyonda M., Mok S., Marsh K., et al. (2018). Gene copy number variation in natural populations of Plasmodium falciparum in Eastern Africa. BMC Genomics 19, 372. doi: 10.1186/s12864-018-4689-7
Singh A. K., Olsen M. F., Lavik L. A. S., Vold T., Drabløs F., and Sjursen W. (2021). Detecting copy number variation in next generation sequencing data from diagnostic gene panels. BMC Med. Genomics 14, 214. doi: 10.1186/s12920-021-01059-x
Srisutham S., Madmanee W., Kouhathong J., Sutawong K., Tripura R., Peto T. J., et al. (2022). Ten-year persistence and evolution of Plasmodium falciparum antifolate and anti-sulfonamide resistance markers pfdhfr and pfdhps in three Asian countries. PloS One 17, e0278928. doi: 10.1371/journal.pone.0278928
Suwanarusk R., Chavchich M., Russell B., Jaidee A., Chalfein F., Barends M., et al. (2008). Amplification of pvmdr1 Associated with Multidrug-Resistant Plasmodium vivax. J. Infect. Dis. 198, 1558–1564. doi: 10.1086/592451
Totino P. R. and Lopes S. C. (2017). Insights into the Cytoadherence Phenomenon of Plasmodium vivax: The Putative Role of Phosphatidylserine. Front. Immunol. 8. doi: 10.3389/fimmu.2017.01148
Tyagi K., Hossain M. E., Thakur V., Aggarwal P., Malhotra P., Mohmmed A., et al. (2016). Plasmodium vivax Tryptophan Rich Antigen PvTRAg36.6 Interacts with PvETRAMP and PvTRAg56.6 Interacts with PvMSP7 during Erythrocytic Stages of the Parasite. PloS One 11, e0151065. doi: 10.1371/journal.pone.0151065
Vargas-Rodríguez R., del C. M., da Silva Bastos M., Menezes M. J., Orjuela-Sánchez P., and Ferreira M. U. (2012). Single-nucleotide polymorphism and copy number variation of the multidrug resistance-1 locus of Plasmodium vivax: local and global patterns. Am. J. Trop. Med. Hyg. 87, 813–821. doi: 10.4269/ajtmh.2012.12-0094
Ward K. E., Fidock D. A., and Bridgford J. L. (2022). Plasmodium falciparum resistance to artemisinin-based combination therapies. Curr. Opin. Microbiol. 69, 102193. doi: 10.1016/j.mib.2022.102193
Warncke J. D., Vakonakis I., and Beck H.-P. (2016). Plasmodium helical interspersed subtelomeric (PHIST) proteins, at the center of host cell remodeling. Microbiol. Mol. Biol. Rev. 80, 905–927. doi: 10.1128/mmbr.00014-16
Wellcome Trust Case Control Consortium, Craddock N., Hurles M. E., Cardin N., Pearson R. D., Plagnol V., et al. (2010). Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464, 713–720. doi: 10.1038/nature08979
Wicht K. J., Mok S., and Fidock D. A. (2020). Molecular mechanisms of drug resistance in plasmodium falciparum malaria. Annu. Rev. Microbiol. 74, 431–454. doi: 10.1146/annurev-micro-020518-115546
Winter G., Kawai S., Haeggström M., Kaneko O., von Euler A., Kawazu S., et al. (2005). SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J. Exp. Med. 201, 1853–1863. doi: 10.1084/jem.20041392
World Health Organization. (2010). World malaria report 2010. Geneva: World Health Organization. Available online at: https://www.who.int/publications/i/item/9789241564106
Keywords: copy number variations, aCGH, cerebral malaria, clinical isolates, Plasmodium vivax
Citation: Tahbildar S, Boopathi PA, Kochar SK, Kochar DK, Aiyaz M, Mugasimangalam RC, Rao SN and Das A (2025) Genomic dynamics of clinical Plasmodium vivax: comparative genomic hybridization in severe malaria cases. Front. Malar. 3:1667330. doi: 10.3389/fmala.2025.1667330
Received: 16 July 2025; Accepted: 22 October 2025;
Published: 11 November 2025.
Edited by:
Quique Bassat, Instituto Salud Global Barcelona (ISGlobal), SpainReviewed by:
Shrikant Nema, International Centre for Genetic Engineering and Biotechnology (India), IndiaFathia Ben-Rached, King Abdullah University of Science and Technology, Saudi Arabia
Copyright © 2025 Tahbildar, Boopathi, Kochar, Kochar, Aiyaz, Mugasimangalam, Rao and Das. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashis Das, YWRhc0BwaWxhbmkuYml0cy1waWxhbmkuYWMuaW4=; YXNoaXNkMjhAZ21haWwuY29t
†Present address: Pon Arunachalam Boopathi, Design and Development, Premier Medical Corporation Private Limited, Sarigam, Valsad, Gujarat, India
 Sampreeti Tahbildar
Sampreeti Tahbildar Pon Arunachalam Boopathi
Pon Arunachalam Boopathi Sanjay Kumar Kochar
Sanjay Kumar Kochar Dhanpat Kumar Kochar
Dhanpat Kumar Kochar Mohamed Aiyaz
Mohamed Aiyaz Raja C. Mugasimangalam
Raja C. Mugasimangalam Sudha N. Rao
Sudha N. Rao Ashis Das
Ashis Das