- Department of Bioengineering, Marmara University, Istanbul, Turkey
Marine biotechnology can be considered as the use of marine bioresources as the target or source of biotechnological applications. Despite many successes have been achieved in marine biotechnology, still many gaps remain to be filled in our basic knowledge on marine science before it could be fully exploited. Systems biology focuses on complex interactions within biological systems, using a holistic approach instead of the traditional reductionism. Marine ecosystems, with different levels of organization at different scales, are ideal benchmark for designing integrative and systems biology approaches. On the other hand, except a few pioneering applications to marine ecology, systems biology approach to marine science is still at its infancy, and marine biotechnology could not benefit from the blessings of systems biology yet. Here, with its inherent holistic philosophy and multidisciplinary nature, systems biology perspective was proposed as a new driver toward seeking solutions to the grand challenges of marine biotechnology.
Marine, which is the world's largest environment covering more than 70% of the earth's surface, features several types of extreme ecosystems and hosts the greatest diversity of life (Sala and Knowlton, 2006). Marine prokaryotic and eukaryotic organisms are very diverse and this biodiversity is encountered in many different habitats and it shapes various abiotic and biotic interactions which influence the operation of the individual components of ecosystems and the ecosystems as a whole. Marine has been considered as a promising resource of biotechnology for many years (Ngo et al., 2012; Vinothkumar and Parameswaran, 2013), but still harbor an unexpended potential for biotechnological applications. For instance, the development of efficient cell factories stands out as one of the major bottlenecks for further advancement of industrial biotechnology (Knuf and Nielsen, 2012). Marine, containing a wealth of unexplored habitats and organisms, may provide opportunities for design of efficient cell factories, and therefore, on its own, harbor a significant potential for overcoming the bottlenecks in biotechnological applications.
Marine biotechnology can be thought of as the use of marine bioresources as the target or source of biotechnological applications, and includes marine organisms used as feedstock (i.e., for production of food, fuel, materials, or bioactive compounds), products extracted from marine organisms or developed in laboratories using the knowledge of the natural processes or properties of marine organisms (including products created from marine DNA via genetic engineering or synthetic biology), processes catalyzed by marine organisms (or derivatives thereof), ecosystem services (such as biosensors and bioremediation), the application of biotechnology knowledge to fish health and welfare, and understanding and mapping of ecosystems based on generic biotechnological tools and knowledge (Pulz and Gross, 2004; Leary et al., 2009).
Many biotechnological advancements have already resulted in successes in the field of food security (in the form of new vaccines and molecular-based diagnostics), human health (biomedical, pharmaceutical and nutraceutical applications with bioactive compounds), fisheries (the development of molecular markers that help to avoid overfishing, etc.), environmental recovery or restoration (i.e., marine organism based bioremediation) and energy (by the use of marine organisms to produce sustainable and renewable energy). Marine biotechnology has already helped to increase production, to reduce the use of antibiotics and improve fish welfare (Sommerset et al., 2005). Biomedical, pharmaceutical and nutraceutical applications of marine biotechnology made significant contributions to human health. Some marine organisms contain, or produce, bioactive or structural compounds that can be used to manage pain or reduce inflammation, to treat cancer or other diseases, and also as new materials for dressing wounds, or to regenerate tissue. Over 20,000 novel marine natural products from marine organisms have been discovered (Hu et al., 2011). The marine ecosystems are still untapped reservoir of biologically active compounds, which have considerable potential to supply food ingredients toward development of new functional foods (Kim, 2013). Marine organisms (algae, invertebrates, vertebrates, and microorganisms) produce a number of metabolites and active compounds that can be incorporated in a range of nutraceuticals containing active ingredients such as antioxidants, essential oils and vitamins that support good health. Many biotechnology advances were achieved in production of functional ingredients, including enzymes, for the food industry (Freitas et al., 2012). Examples of applications in energy field include the use of micro- and macroalgae for the production of biofuel (Rajkumar et al., 2014) and the development of small bioactive molecules to increase the sustainability of marine renewable energy sources (Trincone, 2013).
Despite many successes have been achieved in marine biotechnology, still many gaps remain to be filled in our basic knowledge on marine science before it could be fully exploited. Still there remains a variety of problems that needs to be overcome in marine science, including understanding the role of biodiversity and the impact of global change in maintaining the functionality of ecosystems; understanding relationships between disturbances owing to human behavior and ecosystems; assessing marine ecosystems' health; recovering ecosystem structure and functioning through restoration; conserving, protecting and managing the seas using the ecosystem approach and spatial planning, and modeling ecosystems for better management (Borja, 2014). According to Antonio Trincone, the grand challenge of marine biotechnology in the coming era is to foster knowledge, i.e., to continue to build our basic knowledge of marine environment (Trincone, 2014). Only through this understanding, we can fully understand and make use of what marine organisms have to offer to society in terms of new technology and applications, and we owe much of the commercial success of biotechnological developments and applications to the basic knowledge and academic research findings garnered across a spectrum of scientific disciplines. The specific challenges of marine biotechnology include improvement of cultivation techniques for certain marine organisms for efficient and profitable production schemes; advanced researches on organisms' genetics; bridging the findings of new strains and their commercial applications; understanding symbiotic relationships, the biology and chemistry of defense mechanisms, chemo-ecology of marine invasions; and strategies found in prokaryotes to adapt to extreme environments (Trincone, 2014).
Historically, researchers have investigated ecosystems focusing on individual or few components of biodiversity and trying to understand individual component's roles. However, it became apparent that understanding the entire ecosystem requires the study of all biodiversity components, from the genetic structure of populations, to species, habitats and ecosystem integrity, including food-webs and complex biophysical interrelationships within the system. So, integration of data obtained from different levels (molecular, cellular, tissue/organ, organism/individual, population, community, ecosystem) via principles and tools of various scientific disciplines (biology, ecology, mathematics, physics, computer sciences, etc.) is crucial to fully understand the nature of an ecosystem and to accelerate the biotechnological achievements. From this perspective, with its inherent holistic philosophy and multidisciplinary nature, systems biology not only offers novel solutions to the grand challenges of marine biotechnology, but also helps us to protect the ocean and the organisms that inhabit it through deep understanding of the complexity of the marine ecosystems.
Within the last decade of systems biology, successful experimental and computational methods for reconstruction and analysis of advanced biological network models were developed, as well as the implementation of various design strategies for the production of fuels and chemicals. As a consequence of various applications of these systems biology approaches, significant leap was observed in industrial and medical biotechnologies in the last decade. Even though marine ecosystems, with different levels of organization at different scales, are ideal benchmark for designing integrative and systems biology approaches, however, except pioneering applications of the system biology to marine ecology (Karsenti et al., 2011; Mock and Kirkham, 2012), the holistic approach of systems biology to marine science is still at its infancy, and marine biotechnology could not benefit from these blessings of systems biology yet.
The Genome Sequencing Projects of Marine Organisms Should be Encouraged
The availability of genome sequences for several organisms has allowed a transition from molecular biology to systems biology. Although marine accounts for over 90% of the biosphere and constitutes a major contribution to biodiversity, the number of genome sequencing projects on marine organisms is very limited, i.e., among the completed sequencing projects, only 2.3% of them belonged to marine organisms (Figure 1). In addition, the great majority (97.7%) of these marine-based genome sequencing projects were microbial. These marine projects include genomes of marine haloarchaea and halobacteria, microorganisms from marine oxygen minimum zones, bacterioplankton clades, dark ocean microorganisms, marine red alga, marine protista, carbon monoxide oxidizing thermophiles, marine microbial communities from multiple species of wood-boring bivalves, hydrocarbon-degrading bacteria (including glucose-oxidizing and sulfate-reducing bacteria), ammonia-oxidizing bacteria, bacterial symbionts of gutless marine worms, actinomycetes, flavobacteria, and other marine bacterial strains. The recent years have clearly shown that these genome studies gave important clues on the marine life in terms of symbiosis (Müller et al., 2004), defense mechanisms (Thakur et al., 2005) and biopolymer production (Sogutcu et al., 2012), and expected to accelerate marine biotechnology especially via better understanding of the organisms' genetics and metabolism, improvement of cultivation techniques for these marine organisms, discovery of novel pathways for energy and carbon use, development of efficient and profitable production schemes by use of different hydrocarbons for energy sources, and discovery of novel commercial applications of marine organisms, in near future.
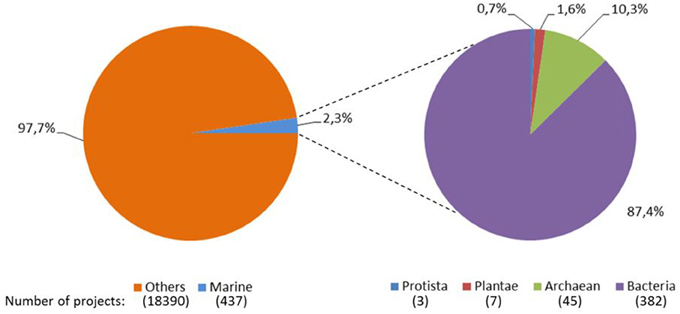
Figure 1. Pie distribution of the completely sequenced genomes. Data obtained from Joint Genome Institute Genomes Online Database (Pagani et al., 2012) as April 6th, 2014.
Genome-Scale Metabolic Models of Marine Organisms will Enable Development of Novel Rational Metabolic Engineering Strategies
Despite all the significant achievements in microbial improvement in the past few decades, the traditional genetic engineering strategies cannot meet today's needs. Rational metabolic engineering strategies based on the genome information are required. Genome scale metabolic models have proven to be valuable for predicting organism phenotypes from genotypes (Herrgård et al., 2008; Henry et al., 2010; Ateş et al., 2011, 2013). Being a valuable guide for identification and filling of knowledge gaps, once a metabolic network is reconstructed, mathematical methods such as convex analysis and linear programming can be applied to simulate the cellular behavior under different genetic and physiological conditions, and to develop metabolic engineering strategies for the construction of strains with desired and improved properties. The achievements of genome scale metabolic models belong to different industrial application fields, including food and nutrients, biopharmaceuticals, biopolymers, microbial biofuel, and bioremediation (Xu et al., 2013). Metabolic reconstructions of marine organisms and their corresponding models will allow the formulation of hypotheses about the presence of certain enzymatic activities, the production of metabolites, and purposeful modification of metabolic pathways and other cellular networks to achieve desired cellular phenotype and performance.
The Complex Community Composition and its Effect on Functionality Should be Investigated Through Metagenomics Projects
The molecular and ecological details as well as influencing factors of marine processes are still poorly understood due to complexity of the communities and lack of cultivability of most of its members. On the other hand, metagenomics technologies, which have the potential to unravel the underlying complex interspecies interactions within a community, currently offer new and promising strategies for marine biodiscovery and biotechnology. Although marine contains a wealth of unexplored habitats and communities, 120 marine-based metagenomics projects constitute only one quarter of the published metagenomics studies (a total of 497 projects), and are mostly dominated by studies on bacterial communities (Figure 2). The metagenomics projects of complex marine communities should be encouraged to develop our limited knowledge of the variation, functioning and ecology of these communities.
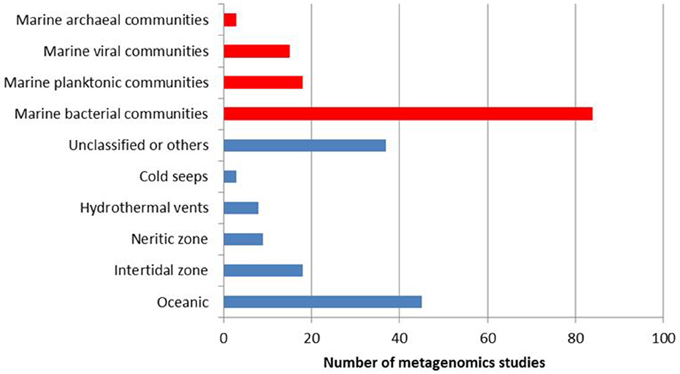
Figure 2. Distribution of completed metagenomics projects on marine ecosystems according to biological domain (in red) and ecosystem subtype (in blue). Data obtained from Joint Genome Institute Genomes Online Database (Pagani et al., 2012) as April 6th, 2014.
The Functional Genomics Datasets are Crucial to Understand the Dynamic Properties of an Organism at Cellular and/or Organismal Levels
The application of high throughput genome-wide profiling techniques to marine organisms allows the acquisition of functional genomics datasets, which focuses on the dynamic aspects such as gene transcription (transcriptome), translation (proteome), protein-protein interactions (interactome), metabolism (metabolome and fluxome), as opposed to the static aspects of the genomic information. The promise of functional genomics is to expand and synthesize genomic, proteomic and metabolic knowledge into an understanding of the dynamic properties of an organism at cellular and/or organismal levels. The application of functional genomics approaches to marine organisms would provide a more complete picture of how biological function arises from the information encoded in an organism's genome, allow reconstruction of functional interaction networks, and pave the way for modeling and predicting the evolution of marine processes under changing environmental conditions, and also for inspiring and providing some solutions for grand challenges in marine biotechnology. For instance, metabolome datasets, representing profiles of extracellular and intracellular metabolites, would give important clues on cell-cell communication leading to understanding symbiotic relationships, and the biology and chemistry of defense mechanisms in marine organisms. Differentially expressed genes and proteins, and metabolic/signaling pathways utilized under extreme conditions could be identified via transcriptome and proteome datasets in order to understand adaptation strategies found in marine organisms. In addition, integration of proteomics and metabolomics data with genome-scale mathematical models will accelerate the performance of the models in rational design of microbial cell factories.
Community Approaches for Reconstruction of Functional Interaction Networks and Development of Mathematical Models Should be Encouraged
Although we possess general knowledge about the major processes within a marine community, we are presently unable to decipher what role individual (micro)organisms have and how their individual actions influence others in the community. Community systems biology approaches (Zengler and Palsson, 2012) will enable us to decode these complex relationships and will therefore improve our understanding of individual members of the community and the modes of interactions in which they engage. Investigation of marine systems requires the use of computer models for a better understanding of the processes, functioning and interrelationships among system components. In addition, computer models are important design tools and preliminary testing methods for consortial interactions which can save time and money. Traditional ecology modeling approaches have used differential equations, game theory and stochastic methods to gain systems-based insight. Stoichiometric modeling methods are attractive in recent years due to their applicability to the growing genomics and functional genomics datasets and because they do not require extensive condition-dependent kinetic parameter sets. These models require only stoichiometric knowledge of system relevant metabolic reactions and the assumption of a pseudo steady-state. The two most widely applied stoichiometric modeling approaches for microbial communities include flux balance analysis (FBA) (Stolyar et al., 2007) and community elementary flux mode analysis (cEFMA) (Taffs et al., 2009). Each approach had its theoretical and computational advantages and disadvantages. Consortium approach through annotation jamborees, which provide a forum for bringing researchers together to build an organism-specific knowledge base, has been proven to be successful in reconstruction of genome-scale stoichiometric metabolic models for various model organisms (Herrgård et al., 2008; Thiele and Palsson, 2010; Thiele et al., 2013). This approach should be upgraded to ecosystem/metagenomic-level and be initiated for marine systems.
Marine is an Untapped Valuable Resource for Microbial Consortia Engineering
The methodology of microbial consortia engineering is based on assembling microbial consortia through enabling, encouraging or enforcing interactions between distinct cell populations and their environment. The common aim is to capitalize on both the capabilities of individual microbes and their interactions to create useful systems-level emergent properties like enhanced productivity, stability or metabolic functionality (Bernstein and Carlson, 2012). The adaptation of marine organisms to a wide range of environmental conditions (temperature, salinity, tides, pressure, radiation, light) render them an enormous reservoir for biotechnological improvements (Trincone, 2014) and marine should be thought of an untapped resource of microorganisms to develop artificial microbial consortia, which describe systems composed of two or more wild-type populations whose interactions do not typically occur naturally. Two specific examples are the co-culturing of marine fungus, Pestolotia sp., with gram negative bacterial strain CNJ-328 to produce an antibiotic (Cueto et al., 2001), and photoautotrophic microalgae Chlorella vulgaris with the filamentous fungi Aspergillus niger to produce biofuel (Zhang and Hu, 2012).
Fostering knowledge on marine ecosystem will accelerate development of multi-scale artificial microbial consortia since it requires a priori knowledge of each population's native eco-physiology.
Marine systems biology can be defined as the application of systems biology approaches to marine biotechnology. Interestingly, marine ecosystems, with different levels of organization at different scales, are ideal benchmark for designing systems biology approaches. Systems biology should be regarded as the new driver for marine biotechnology and the systems biology perspective may provide a breakthrough in marine biotechnology in near future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Financial support to Kazim Y. Arga by The Scientific and Technological Research Council of Turkey (TUBITAK) through project 110M613, and Marmara University Research Fund through projects FEN-A-130511-0163, FEN-B-090414-0089, FEN-C-YLP-101013-0403, FEN-C-YLP-101013-0404 is greatly acknowledged.
References
Ateş, Ö., Arga, K. Y., and Toksoy Öner, E. (2013). The stimulatory effect of mannitol on levan biosynthesis: lessons from metabolic systems analysis of Halomonas smyrnensis AAD6. Biotechnol. Prog. 29, 1386–1397. doi: 10.1002/btpr.1823
Ateş, Ö., Toksoy Öner, E., and Arga, K. Y. (2011). Genome-scale reconstruction of metabolic network for a halophilic extremophile, Chromohalobacter salexigens DSM 3043. BMC Syst. Biol. 5:12. doi: 10.1186/1752-0509-5-12
Bernstein, H. C., and Carlson, R. P. (2012). Microbial consortia engineering for cellular factories: in vitro to in silico systems. Comput. Struct. Biotechnol. J. 3:e201210017. doi: 10.5936/csbj.201210017
Borja, A. (2014). Grand challenges in marine ecosystems ecology. Front. Mar. Sci. 1:1. doi: 10.3389/fmars.2014.00001
Cueto, M., Jensen, P. R., Kauffman, C., Fenical, W., Lobkovsky, E., and Clardy, J. (2001). Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 64, 1444–1446. doi: 10.1021/np0102713
Freitas, A. C., Rodrigues, D., Rocha-Santos, T. A., Gomes, A. M., and Duarte, A. C. (2012). Marine biotechnology advances towards applications in new functional foods. Biotechnol. Adv. 30, 1506–1515. doi: 10.1016/j.biotechadv.2012.03.006
Henry, C. S., DeJongh, M., Best, A. A., Frybarger, P. M., Linsay, B., and Stevens, R. L. (2010). High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977–982. doi: 10.1038/nbt.1672
Herrgård, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., et al. (2008). A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 26, 1155–1160. doi: 10.1038/nbt1492
Hu, G. P., Yuan, J., Sun, L., She, Z. G., Wu, J. H., Lan, X. J., et al. (2011). Statistical research on marine natural products based on data obtained between 1985 and 2008. Mar. Drugs 9, 514–525. doi: 10.3390/md9040514
Karsenti, E., Acinas, S. G., Bork, P., Bowler, C., Vargas, C. D., Raes, J., et al. (2011). A holistic approach to marine eco-systems biology. PLoS Biol. 9:e1001177. doi: 10.1371/journal.pbio.1001177
Knuf, C., and Nielsen, J. (2012). Aspergilli: systems biology and industrial applications. Biotechnol. J. 7, 1147–1155. doi: 10.1002/biot.201200169
Leary, D., Vierros, M., Hamon, G., Arico, S., and Monagle, C. (2009). Marine genetic resources: a review of scientific and commercial interest. Mar. Policy 33, 183–194. doi: 10.1016/j.marpol.2008.05.010
Mock, T., and Kirkham, A. (2012). What can we learn from genomics approaches in marine ecology? From sequences to eco-systems biology. Mar. Ecol. 33, 131–148. doi: 10.1111/j.1439-0485.2011.00479.x
Müller, W. E., Grebejuk, V. A., Thakur, N. L., Thakur, A. N., Batel, R., Krasko, A., et al. (2004). Oxygen-controlled bacterial growth in the sponge Suberitesdomuncula: toward a molecular understanding of the symbiotic relationship between sponge and bacteria. Appl. Environ. Microbiol. 70, 2332–2341. doi: 10.1128/AEM.70.4.2332-2341.2004
Ngo, D. H., Vo, T. S., Ngo, D. N., Wijesekara, I., and Kim, S. K. (2012). Biological activities and potential health benefits of bioactive peptides derived from marine organisms. Int. J. Biol. Macromol. 51, 378–383. doi: 10.1016/j.ijbiomac.2012.06.001
Pagani, I., Liolios, K., Jansson, J., Chen, I. M. A., Smirnova, T., Nosrat, B., et al. (2012). The Genomes OnLine Database (GOLD) v. 4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 40, D571–D579. doi: 10.1093/nar/gkr1100
Pulz, O., and Gross, W. (2004). Valuable products from biotechnology of microalgae. Appl. Microbiol. Biotechnol. 65, 635–648. doi: 10.1007/s00253-004-1647-x
Rajkumar, R., Yaakob, Z., and Takriff, M. S. (2014). Potential of the micro and macro algae for biofuel production: a brief review. Bioresources 9, 1606–1633.
Sala, E., and Knowlton, N. (2006). Global marine biodiversity trends. Annu. Rev. Environ. Resour. 31, 93–122. doi: 10.1146/annurev.energy.31.020105.100235
Sogutcu, E., Emrence, Z., Arikan, M., Cakiris, A., Abaci, N., Toksoy Öner, E., et al. (2012). Draft genome sequence of Halomonas smyrnensis AAD6. J. Bacteriol. 194, 5690. doi: 10.1128/JB.00559-12
Sommerset, I., Krossøy, B., Biering, E., and Frost, P. (2005). Vaccines for fish in aquaculture. Expert Rev. 4, 89–101. doi: 10.1586/14760584.4.1.89
Stolyar, S., Van Dien, S., Hillesland, K. L., Pinel, N., Lie, T. J., Leigh, J. A., et al. (2007). Metabolic modeling of a mutualistic microbial community. Mol. Syst. Biol. 3:92. doi: 10.1038/msb4100131
Taffs, R., Aston, J. E., Brileya, K., Jay, Z., Klatt, C. G., McGlynn, S., et al. (2009). In silico approaches to study mass and energy flows in microbial consortia: a syntrophic case study. BMC Syst. Biol. 3:114. doi: 10.1186/1752-0509-3-114
Thakur, N., Peroviæ-Ottstadt, S., Batel, R., Korzhev, M., Diehl-Seifert, B., Müller, I., et al. (2005). Innate immune defense of the sponge Suberites domuncula against gram-positive bacteria: induction of lysozyme and AdaPTin. Mar. Biol. 146, 271–282. doi: 10.1007/s00227-004-1438-z
Thiele, I., and Palsson, B. Ø. (2010). Reconstruction annotation jamborees: a community approach to systems biology. Mol. Syst. Biol. 6:361. doi: 10.1038/msb.2010.15
Thiele, I., Swainston, N., Fleming, R. M., Hoppe, A., Sahoo, S., Aurich, M. K., et al. (2013). A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 31, 419–425. doi: 10.1038/nbt.2488
Trincone, A. (2013). Marine enzymes for Biocatalysis: Sources, Biocatalytic Characteristics and Bioprocesses of Marine Enzymes. Cambridge: Elsevier.
Trincone, A. (2014). Increasing knowledge: the grand challenge in marine biotechnology. Front. Mar. Sci. 1:2. doi: 10.3389/fmars.2014.00002
Vinothkumar, S., and Parameswaran, P. S. (2013). Recent advances in marine drug research. Biotechnol. Adv. 31, 1826–1845. doi: 10.1016/j.biotechadv.2013.02.006
Xu, C., Liu, L., Zhang, Z., Jin, D., Qiu, J., and Chen, M. (2013). Genome-scale metabolic model in guiding metabolic engineering of microbial improvement. Appl. Microbiol. Biotechnol. 97, 519–539. doi: 10.1007/s00253-012-4543-9
Zengler, K., and Palsson, B. O. (2012). A road map for the development of community systems (CoSy) biology. Nat. Rev. Microbiol. 10, 366–372. doi: 10.1038/nrmicro2763
Keywords: marine biotechnology, systems biology, genomics, metabolic engineering, metagenomics, functional genomics, microbial consortia engineering
Citation: Gov E and Arga KY (2014) Systems biology solutions to challenges in marine biotechnology. Front. Mar. Sci. 1:14. doi: 10.3389/fmars.2014.00014
Received: 28 April 2014; Accepted: 03 June 2014;
Published online: 20 June 2014.
Edited by:
Donatella De Pascale, National Research Council- CNR, Naples, ItalyReviewed by:
Laura Ghigliotti, University of Genoa, ItalyLuigi Mandrich, Institute of Protein Biochemistry - National Research Council, Italy
Copyright © 2014 Gov and Arga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazim Y. Arga, Faculty of Engineering, Marmara University Goztepe Campus, Building D, 34722 Göztepe, Kadikoy, Istanbul, Turkey e-mail:a2F6aW0uYXJnYUBtYXJtYXJhLmVkdS50cg==
 Esra Gov
Esra Gov Kazim Y. Arga
Kazim Y. Arga