- 1Department of Earth, Ocean and Atmospheric Science, Florida State University, Tallahassee, FL, USA
- 2Chesapeake Biological Laboratory, University of Maryland, Solomons, MD, USA
In contrast to other oligotrophic water bodies the Gulf of Mexico (GOM) hosts an abundance of hydrocarbon seeps, which likely influences the microbial assemblages it hosts particularly regarding the availability of labile carbon in the aphotic GOM. The aphotic zone receives direct injection of seep methane (CH4), but CH4 from an unknown source has been reported at supersaturated concentrations relative to the atmosphere in the GOM photic zone. Here we used iTag sequencing of 16S rRNA genes to characterize GOM microbial communities and to relate changes in microbial community structure to the properties inherent to their oceanic province-seafloor to the photic zone, seep and non-seep. Along this trajectory water column communities were distinct in the euphotic zone compared to the mesopelagic and deep-sea. In the euphotic zone the relative abundance of a cyanobacterial species (Prochlorococcus) was significantly correlated with both CH4 and chlorophyll a concentrations and was abundant in some deep-chlorophyll maximum (DCM) samples. The relative abundance of microorganisms related to known hydrocarbon degraders were also significantly correlated with CH4 in the euphotic zone, but no canonical methanotrophs were observed. In the mesopelagic to the seafloor canonical methanotrophs were identified, but only a Marine Group II Euryarchaeota was significantly correlated with CH4. Overall, depth and the associated environmental conditions were the primary drivers in structuring microbial communities over the GOM water column. Further, CH4 concentrations and relative microbial abundances covaried significantly from the seafloor to the photic zone in the GOM. The lack of a significant relationship between canonical methanotrophs and CH4 in the aphotic zone, even when sampling at seep sites, may suggest methane-oxidation by unknown microorganisms. Similarly their absence in the CH4 maximum and DCM suggested that CH4 is either oxidized by unrecognized methanotrophs or escapes the CH4 biofilter and fluxes to the atmosphere.
Introduction
Analyses of spatial patterns of marine microbial plankton revealed that in several marine provinces dominant microbial players are stratified in the vertical water column (Giovannoni and Stingl, 2005 and references therein). This observed stratified vertical distribution was extended to resolve global scale microbial distributions and revealed disparate communities from surface to deep-water, with different environmental drivers influencing the structure of these communities (Zinger et al., 2011; Sunagawa et al., 2015). One of the primary organizing principles determining microbial plankton structure is the availability of sunlight throughout the water column, particularly for photosynthetic Cyanobacteria (Giovannoni and Stingl, 2005; Delong et al., 2006; Zinger et al., 2011). It would seem that microbial plankton in the oligotrophic Gulf of Mexico (GOM) are bounded by these very same organizing principles. However, the GOM has numerous natural hydrocarbon seeps that provide a carbon source, and in particular CH4, for heterotrophic microbes near seep sites. Additionally, well over 30 years ago CH4 at supersaturated concentrations relative to the atmosphere were reported in the near-surface water column in the GOM (Swinnerton and Lamontagne, 1974; Brooks et al., 1981) and more recently by Finke et al. (2011). The source of this CH4 remains elusive. Thus, both the deep-sea and the near-surface photic zone have labile fixed carbon in the form of CH4 that likely influences microbial structure.
There are few studies that have described microbial community stucuture and the environmental factors that influence this structure in the GOM. Recently, Gillies et al. (2015) found that dissolved oxygen concentrations played a significant role in influencing microbial abundances in the northern GOM coastal hypoxic zone. King et al. (2013) also analyzed bacterioplankton in the coastal northern GOM, but their study extended to the mesopelagic. They reported that depth, temperature and, similar to Gillies et al. (2015), oxygen were the primary drivers influencing microbial abundances in the GOM. Together these studies suggest that similar organizing principles as those discussed above shape microbal communities in the GOM, but hydrocarbon concentrations were not considered in these studies. Yet, several reports revealed the profound influence that oil and gas input during the Deepwater Horizon oil spill can have on microbial abundances from the photic zone to the deep-sea in the GOM (Hazen et al., 2010; Valentine et al., 2010; Mason et al., 2012; Redmond and Valentine, 2012; Dubinsky et al., 2013). In particular, CH4 concentrations reported in the deep-sea plume were 10–1000 times higher than what is typically observed at GOM hydrocarbon seeps (Joye et al., 2011) which ranges from 4 nM to 20 μM (Solomon et al., 2009; Wankel et al., 2010). During the oil spill CH4 oxidation rates were some of the highest reported in the pelagic ocean (Crespo-Medina et al., 2014). Active CH4 oxidation was occurring soon after the spill began (Mason et al., 2012; Rivers et al., 2013), but canonical methanotrophs were not abundant in 16S rRNA gene data (Mason et al., 2012). Collectively, these studies illustrated how hydrocarbons, and in particular, CH4 can shape microbial community structure in the GOM.
Given that methane is a potent greenhouse gas absorbing infrared radiation 25 times more effectively than CO2 on a molecule per molecule basis (Lelieveld et al., 1998; Petit et al., 1999) there is an impetus to understand both the sources and sinks for this gas. In fact, the most significant perturbations to climate forcing, or radiative forcing, are changes related to greenhouse gases such as CH4 (Forster et al., 2007). Methane has made a significant contribution to global warming over the past few 100 years. The ocean is a CH4 source contributing 1–4% of annual emissions to the atmosphere (Karl et al., 2008). Yet, to our knowledge there are no studies that present a characterization of the bacterioplankton along with CH4 data in the water column at natural seeps in the GOM. Further, what microorganisms are associated with the methane maxima in the GOM is not currently known. To fill this knowledge gap we sought to examine the correlations between CH4 concentrations and other environmental variables and relative microbial abundances over the entire GOM water column, from the photic zone to the deep-sea, at hydrocarbon seep and non-seep sites. We hypothesized that this carbon source may play an important role in structuring pelagic microbial communities. Further, we sought to evaluate the relative abundances of canonical methanotrophs where CH4 concentrations were highest—above seeps and in the chlorophyll/CH4 maxima in the photic zone. Specifically, our research aim was to better understand the relationship between microbial distributions as they covary with CH4 concentrations. Here we used iTag sequencing of bacterial and archaeal 16 rRNA genes in 58 samples using Illumina's MiSeq platform. We determined CH4, oxygen, nitrate, nitrite, ammonium and phosphorus concentrations. We then evaluated the relationships between these environmental variables and relative microbial abundances.
Materials and Methods
Sample Collection
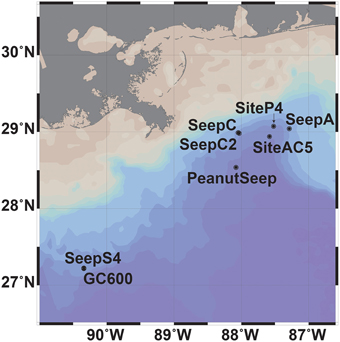
Eight sites in the GOM were sampled during expeditions in September and October 2012 (R/V Weatherbird II) and in May and June 2013 (R/V Pelican) (Figure 1). Sites were located in the Northeastern GOM continental shelf and varied from gas seep sites (SeepC and SeepC2), a gas and asphalt volcanism site (PeanutSeep), a prospective seep site with observed intermittent oil sheens (Oscar Garcia-Pineda, personal communication) (SeepA), an active oil seep with visible oil at the surface (GC600 and SeepS4) and non-seep sites (SiteAC5 and SiteP4). Collection depths spanned the near-surface to the seafloor at these eight sites. In the euphotic zone the deep-chlorophyll maximum was targeted for collection (Supplemental Table 1).
Oxygen, Chlorophyll a and nutrients
Oxygen concentrations were determined using the Winkler method (Environmental Protection Agency's Standard Operating Procedure of Dissolved Oxygen Micro Method, Winkler Titration, LG501). Chlorophyll a samples were concentrated on 25 mm Whatman GF/F filters from 1 to 2 L of seawater and stored at −20°C. Chlorophyll a was extracted using the methods described in the Environmental Protection Agency Method 445.0, “In Vivo Determination of Chlorophyll a in Marine and Freshwater Algae by Fluorescence”; however, no mechanical tissue grinder or HCl were used. Chlorophyll a concentrations were determined using a fluorometer with a chlorophyll a standard (Anacystis nidulans chlorophyll a). For nutrients, 60 ml of seawater was filtered through a Whatman GF/D filter (EMD Millipore, Billerica, MA) into two 30 ml Nalgene bottles using a swinnex filter holder and syringe and stored at −20°C. Nutrient concentrations were determined by the marine chemistry lab at the University of Washington following the WOCE Hydrographic Program using a Technicon AAII system (http://www.ocean.washington.edu/story/Marine+Chemistry+Laboratory).
Methane
Methane samples were collected from niskin bottles as soon as the CTD rosette was shipboard. 150 ml volume glass vials were filled with sample water until a reverse meniscus was present. To preserve samples vials were then injected with 0.5 ml of a 8 M KOH solution, using a technique to prevent injection of air into the sample. Finally, 10 ml of helium gas was injected into the vial as headspace. Vials were stored inverted until shorebased analysis via gas chromatography. A complete description of our analytical techniques can be found in Magen et al. (2014).
Microbial Sample Collection and DNA Extractions
From each station, up to 10 L of seawater were collected and filtered with a peristaltic pump. A 2.7 μM Whatman GF/D pre-filter was used and samples were concentrated on 0.22 μM Sterivex filters (EMD Millipore). Sterivex filters were sparged and filled with RNAlater. DNA was extracted directly off of the filter by placing half of the Sterivex filter in a Lysing matrix E (LME) glass/zirconia/silica beads Tube (MP Biomedicals, Santa Ana, CA) using the protocol described in Gillies et al. (2015) which combines phenol:chloroform:isoamyalcohol (25:24:1) and bead beating. Genomic DNA was stored at −80°C until purified. DNA was purified using a QIAGEN (Valencia, CA) AllPrep DNA/RNA Kit. DNA quantity was determined using a Qubit 2.0 Fluorometer (Life Technologies, Grand Island, NY).
16S rRNA Gene Sequencing and Analysis
16S rRNA genes were amplified from 10 ng of purified DNA in duplicate using primers 515F and 806R that amplify both bacteria and archaea, targeting the V4 region of E. coli in accordance with the protocol described by Caporaso et al. (2011, 2012) and used by the Earth Microbiome Project (http://www.earthmicrobiome.org/emp-standard-protocols/16s/), with a slight modification, specifically, the annealing temperature was modified to 60°C. PCR amplicons were purified using Agencourt AMPure XP PCR Purification beads (Beckman Coulter, Indianapolis, IN). Sequencing was carried out using the MiSeq (Illumina, San Diego, CA) platform. Sequences were analyzed using the QIIME version 1.7.0 (Caporaso et al., 2010a) pipeline. Raw sequences were demultiplexed and then quality filtered using the default parameters in QIIME. These sequences are available at: http://mason.eoas.fsu.edu and in the NCBI's sequence read archive (PRJNA294680). Sequences were then clustered into operational taxonomic units (OTU)s, which was defined as ≥97% 16S rRNA gene sequence similarity, using UCLUST (Edgar, 2010) using the open reference clustering protocol (http://qiime.org/tutorials/open_reference_illumina_processing.html). The resulting representative sequences were aligned using PyNAST (Caporaso et al., 2010b) and given a taxonomic classification using RDP (Wang et al., 2007), retrained with the Greengenes version 13.5 (McDonald et al., 2012). The resulting OTU table was filtered to keep only OTUs that had at least 10 observations, and then converted to relative abundance, by summing the total sequence count in a sample and then dividing each sample OTU count by this total. Those species that were significantly correlated with CH4 concentrations (see below) were further classified using RDP (Wang et al., 2007), Silva (Pruesse et al., 2007) and Greengenes (McDonald et al., 2012).
Statistics
Nutrient, CH4 and oxygen data were interpolated using Ocean Data View (Schlitzer, 2013). The differences in relativized OTU abundances in communities in samples that were categorized as either below or above 4 nM CH4 (CH4 concentrations in equilibrium with the atmosphere are >4 nM, Joye et al., 2011) were analyzed using nonparametric statistics (Mann-Whitney) to test for statistically significant differences using METAGENassist (Arndt et al., 2012) and the application of a False Discovery Rate (FDR) to account for multiple comparisons. Further, statistical analyses were carried out in R, including determination of Spearman correlation coefficients among all variables. The QIIME generated, normalized (by conversion to relative abundance), OTU abundances in the 58 different samples were then analyzed using non-metric multidimensional (NMDS) scaling in R using metaMDS in the Vegan package. P-values were derived from 999 permutations of the data.
Results and Discussion
Water Column Properties and Chemistry
Maximum CH4 concentrations (24 and 45 nM) were observed in two samples collected at or near gas seep site GC600 (Figures 1, 2, Supplementary Table 1). Of the remaining sites and depths, with one exception, sample SeepC2_025_1109 m, the highest CH4 concentrations were observed in the photic zone (Figure 2, Supplementary Table 1), despite obtaining several additional samples directly above seeps. Specifically, CH4 values ranged from 11 to 14 nM with an average depth of 84 m (Figure 2, Supplementary Table 1). These values were well above CH4 concentrations in equilibrium with the atmosphere at < 4 nM (Joye et al., 2011). Typical GOM seawater nutrient and oxygen profiles were observed (see for example Shiller and Joung, 2012; Figure 1) for nitrate, nitrite, ammonium and phosphate (Figure 2, Supplementary Table 1), although we acknowledge the dearth of profiles for the GOM. Chlorophyll a ranged from 0.92 μg/L to undetectable below the euphotic zone (Figure 2, Supplementary Table 1). Further, a deep-chlorophyll maximum (DCM) was observed at an average depth of 83 m (Figure 2). Spearman correlation analysis revealed that CH4 concentrations were significantly correlated with salinity, nitrite, and chlorophyll a in the euphotic zone (Figure 3). A co-located CH4/DCM in the GOM has been previously reported (for example, Finke et al., 2011). Below the euphotic zone CH4 concentrations were inversely correlated with phosphate and nitrate, compared to ammonium concentrations, which were positively correlated with CH4 (Figure 3).
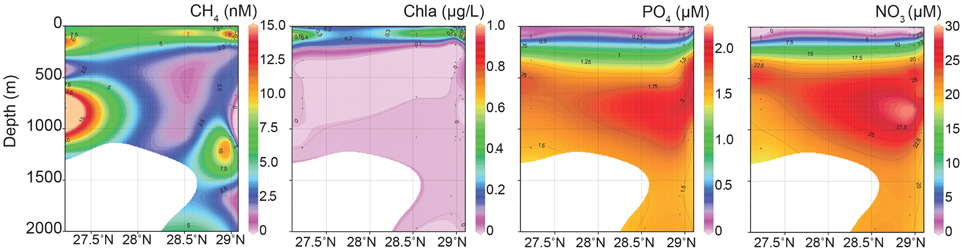
Figure 2. Interpolation plots of methane, chlorophyll a, phosphate and nitrate data from eight stations sampled.
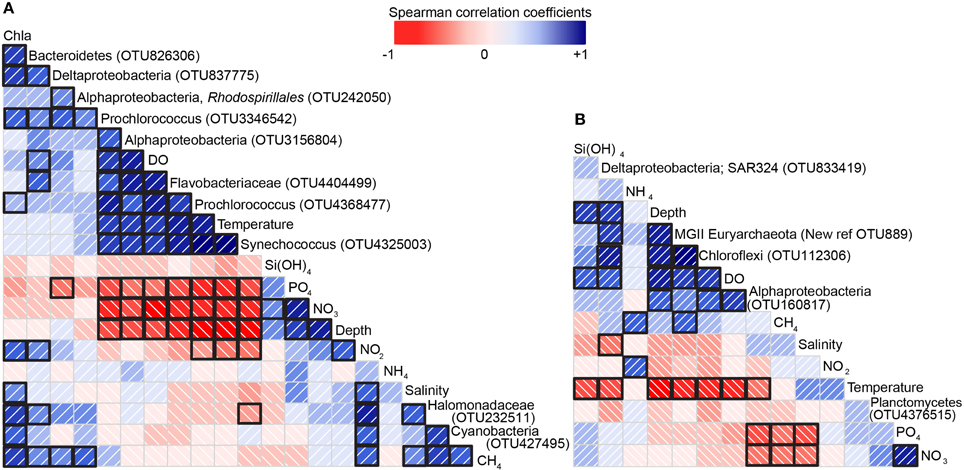
Figure 3. Correlation matrix showing the sign and magnitude of the correlation (Spearman) coefficients between biological, chemical and physical data. (A) Correlation matrix for 0–200 m and (B) correlation matrix for 200–2000 m. Blue indicates a positive value and red a negative value (see scale bar). The diagonal lines also indicate the direction of the correlation. The intensity of each of these colors increases as the correlation value moves away from zero (see scale bar). Correlations that were statistically significant (p ≤ 0.05) are shown by a black square around the borders (e.g.,  , has a black border indicating a significant correlation vs.
, has a black border indicating a significant correlation vs.  , indicating a correlation that was not significant).
, indicating a correlation that was not significant).
Surface to Seafloor Microbial Community Structure
Proteobacteria was the most dominant phyla over the entire water column, with an average relative abundance of 35% in the photic zone/43% below the photic zone (Figure 4A). Of the proteobacteria, gammaproteobacteria comprised 40.6% photic/42.8% aphotic, alphaproteobacteria were 40.0% photic/12.8% aphotic and deltaproteobacteria were 19.1% photic/42.0% aphotic (Figure 4B). King et al. (2013) analyzed the microbial communities in the Northern GOM, from the nearshore to offshore and from surface to deep with the same 16S rRNA gene primers we used here. They found a similar distribution of alpha and gammaproteobacteria, in that alphaproteobacteria abundances were higher in the photic zone and decreased with depth, while gammaproteobacteria showed an inverse trend. Cyanobacteria were also abundant in the photic zone (avg. 28%) compared to avg. 0.4% in the aphotic zone (Figure 4). Overall, these patterns were consistent with the global scale synthesis of pelagic verses benthic communities (Zinger et al., 2011). Thaumarchaeota increased with depth (avg. 5.6% photic/21.1% aphotic) (Figure 4). The increase in Thaumarchaeota (Crenarchaeota) abundances with depth was first reported more than a decade ago (Karner et al., 2001). In the GOM Tolar et al. (2013) reported that Thaumarchaeota abundance was strongly correlated with depth, consistent with our findings. Euryarchaeota showed a similar distribution over the water column (avg. 11.0% photic/11.6% aphotic) (Figure 4). This homogenous distribution with depth was also reported by Karner et al. (2001) in the North Pacific subtropical gyre. In contrast, Tolar et al. (2013) reported that Euryarchaeota were generally less abundant and decreased with depth in their GOM samples. Marine Group A (MGA; formerly referred to as SAR406) increased from 5.5% in the photic zone to 8.2% in the aphotic zone (Figure 4). A similar distribution, with an increase in relative abundance with depth of MGA, was recently shown by fluorescent in situ hybridization in the Northeast subarctic Pacific Ocean (Allers et al., 2013). The relative abundances of Actinobacteria, Bacteriodetes, Chloroflexi, Planctomycetes, and Acidobacteria changed with increasing depth in the water column but were never more than 3% of the overall microbial community.
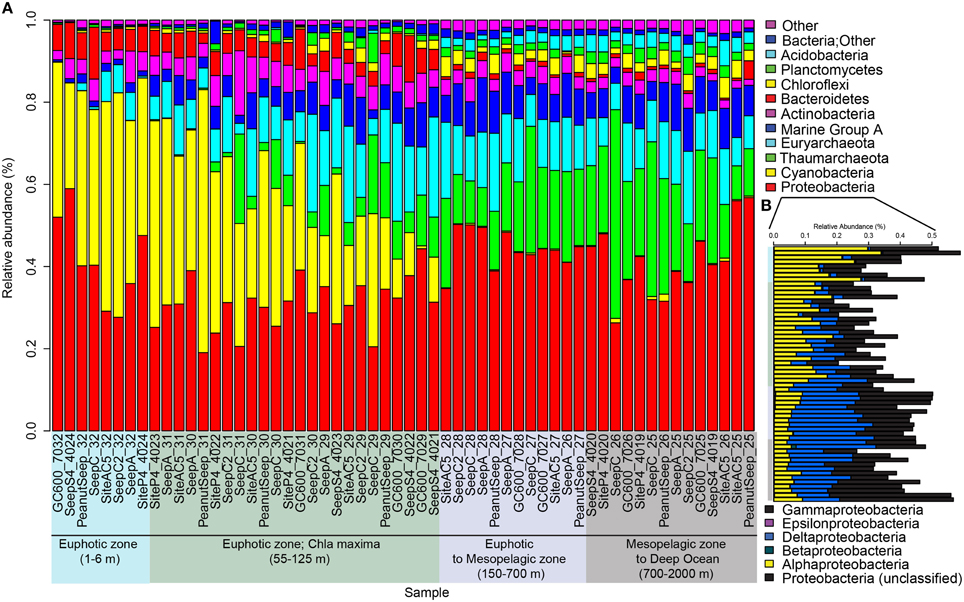
Figure 4. Bar graphs of relativized 16S rRNA gene iTag sequence data. Graph (A) shows phyla level microbial community composition. Only the more abundant bacterial and archaeal groups are shown. Less abundant groups were summed under “Other.” Graph (B) shows the relative abundances of sub-phyla in the Proteobacteria. Samples are sorted by depth on the x-axis in both (A,B). Graph (B) shows the sample depths using the same color scheme shown in (A).
Beta Diversity
Beta diversity analysis (nonmetric multidimensional scaling) of 10,912 OTUs revealed distinct microbial communities that were structured by depth and the associated environmental conditions with these depths (Figure 5). The differences in microbial community structure with depth was previously reported by Zinger et al. (2011). No clustering by season was observed in the NMDS ordination. Given the disparate communities in the euphotic zone compared to the mesopelagic and deep-sea we elected to focus on these two environments independently in the following analyses.
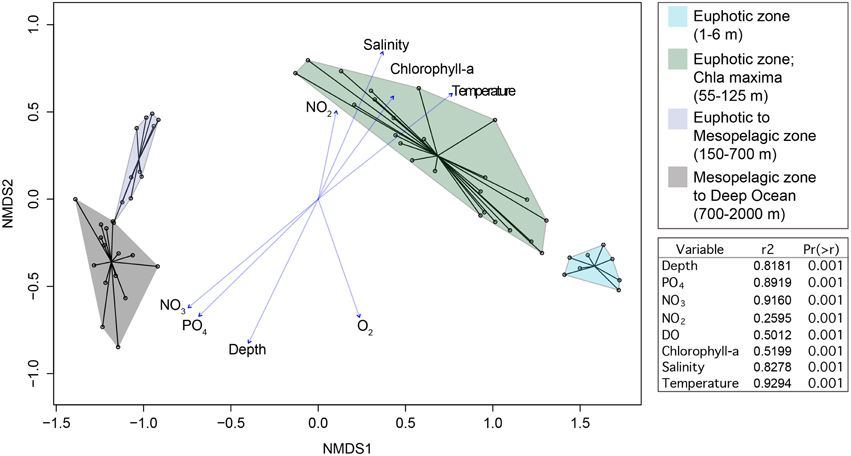
Figure 5. NMDS ordination of relativized 16S rRNA gene iTag sequence data. Samples grouped by depth as shown in the ordination plot. Vectors are shown for environmental variables that were significantly correlated with NMDS ordination axes. Goodness of fit statistics (r2) and p-values are shown.
Euphotic Zone Bacterioplankton Structure and Diversity
Nonparametric statistics were used to determine which species were significantly different in samples with above or below background CH4 concentrations (< 4 nM, Joye et al., 2011) in photic zone samples (0–200 m). Specifically, samples were categorized as above or below background CH4 concentrations and the relative abundances of 8156 OTUs were tested for significance using the Mann-Whitney test. The relative abundances of 10 microbial species were significantly different in samples that were above or below background. These species were members of the Cyanobacteria, Bacteroidetes, and Proteobacteria phyla (Figure 3A).
We next directly correlated relative abundances of these 10 species with CH4 concentrations. Only those species that were significantly correlated with CH4 at ≤ 0.05 p-value are discussed here. A Cyanobacteria (OTU427495) classified as a Family II, GPIIa [Ribosomal Database Project (RDP) (Cole et al., 2013)] was positively correlated with CH4 (Spearman correlation coefficient = 0.53, p-value = 0.00) and chlorophyll a (Figure 3A). In fact, this species was highly abundant in the DCM, comprising up to 15% relative abundance in some samples. This species was 100% similar to bacterioplankton from the tropical Atlantic (Lekunberri et al., 2013), from 125 m North Pacific subtropical gyre seawater (Eiler et al., 2011) and from the Northern South China Sea (AC FJ598533).
Members of the cyanobacterial clade have been shown to evolve CH4 when microcosms in which orthophosphate concentrations are low are supplemented with methylphosphonate (MPn) (Karl et al., 2008). These authors demonstrated that Trichodesmium from the North Pacific gyre evolves CH4 when utilizing MPn. The mechanism suggested was MPn hydrolysis of phosphonate compounds by a carbon-phosphorus pathway. This pathway was actively expressed by the Cyanobacteria, Trichodesmium erythreum ISM101, when faced with phosphate-deficient conditions (Dyhrman et al., 2006). When comparing OTU427495 to cultured representatives it is most similar (99%) to Prochlorococcus marinus SS120, isolated from 120 m in the Sargasso Sea (Moore et al., 1995). P. marinus SS120 encodes an ABC-type transporter for potentially using phosphonates (Dufresne et al., 2003). To our knowledge however it is unknown whether this Cyanobacteria utilizes MPn and evolves CH4 when orthophosphate is depleted.
Interestingly, in our samples CH4 and chlorophyll a concentrations were significantly correlated; in contrast CH4 and phosphate concentrations were not (Figure 3A). It has been previously reported that in the GOM the near surface CH4 maximum is coincident with the deep-chlorophyll maximum (DCM) (Finke et al., 2011). These authors also reported that in samples from the GOM DCM addition of MPn (or other previously identified CH4 precursors) did not influence CH4 production, but that light exposure did. They suggest a hitherto unrecognized photosynthetic process in the DCM as responsible for the observed CH4 maxima in the GOM. The correlations we reported here provided some evidence that indeed photosynthetic processes may be involved in CH4 formation in the GOM DCM, but that the mechanism is unresolved. Although we acknowledge that 1) while statistically significant, the correlation between the Cyanobacteria (OTU427495) species discussed above and CH4 concentrations at 0.53 is moderate and 2) the CH4 source was not explicitly tested herein and that the relative abundance of a Cyanobacteria closely related to a Prochlorococcus sp. in the GOM DCM (and CH4 maximum) may not be a CH4 source.
Except for Bacteroidetes OTU826306, close relatives of the remaining species that were significantly correlated with CH4 concentrations in the euphotic zone have been shown to degrade hydrocarbons. Specifically, in the Oceanospirillales a Halomonadaceae species (OTU232511) was positively correlated with CH4 (0.70, p = 0.00) (Figure 3A). This species was 100% similar to bacteria from 300 m in the Challenger Deep, Mariana Trench (Acc. AB703798), 99% similar to a diffuse hydrothermal vent clone (Acc. HG819048), a clone from the eastern tropical South Pacific oxygen minimum zone (Stevens and Ulloa, 2008) and an oil sheen from the Deepwater Horizon wreckage in the GOM (Acc. KF786504). In the deep-sea plume that resulted from Deepwater Horizon oil spill Oceanospirillales was highly abundant (Hazen et al., 2010; Redmond and Valentine, 2012). A single cell genome sequence of this Oceanospirillales revealed a capacity for aliphatic hydrocarbon degradation (Mason et al., 2012). Further, the Oceanospirillales Alcanivorax borkumensis is a ubiquitous n-alkane degrading marine bacterium (Schneiker et al., 2006). Further, several Halomonadaceae are capable of growth on hydrocarbons (for example Gasperotti et al., 2015). However, whether the Halomonadaceae discussed here is capable of oxidizing CH4 is unknown.
Alphaproteobacteria OTU242050 was also significantly correlated with CH4 (0.46, p = 0.01). This microorganism was classified as a Rhodospirillales with 100% similarity to bacteria associated with a marine sponge (AC JQ240926), the South China Sea (Zhang et al., 2011) and a shallow CH4 seep (Wasmund et al., 2009). Redmond and Valentine (2012) reported that surface samples with a visible oil sheen collected during the Deepwater Horizon oil spill, were dominated by Rhodospirillales, other Alphaproteobacteria and Cyanobacteria. Members of the Rhodospirillales clade have been shown to degrade hydrocarbons (Kodama et al., 2008; Zhao et al., 2008), but not, to our knowledge, short chain n-alkanes. Thus, the relationship between CH4 and Rhodospirillales requires further investigation.
Deltaproteobacteria OTU837775 was significantly correlated with CH4 (0.39, p = 0.02) (Figure 3A). This species was 100% similar to bacterioplankton SAR324 from an oxygen minimum zone of the Arabian Sea (AC KJ589823), an oil sheen from the Deepwater Horizon wreckage in the GOM (AC KF786619) and the Sargasso Sea (Sjöstedt et al., 2014). Further, deltaproteobacteria OTU837775 16S rRNA gene sequence was 100% similar to the uncultured SAR324 clone HF130_05G09 presented by Pham et al. (2008) and also discussed by Chitsaz et al. (2011). SAR324 have been reported in numerous marine habitats, for example in the North Pacific Subtropical Gyre (Delong et al., 2006) and in oxygen minimum zones (Wright et al., 2012). Swan et al. (2011) sequenced two SAR324 single cell genomes (SAGs), of which SAG AAA240-J09 was 93% similar to Deltaproteobacteria OTU837775. In this report they suggested that SAR324 have the potential to oxidize CH4 and other C1 compounds. Recently, Sheik et al. (2014) reported that deep-sea SAR324 possess an actively transcribed particulate hydrocarbon monooxygenase (pHMO). Whether this enzyme acts on CH4 or C2-C4 gases only is currently unresolved. The significant relationship between CH4 and the relative abundance of Deltaproteobacteria OTU837775 suggested that CH4 may indeed be a substrate utilized by SAR324.
Finally, in the Bacteroidetes, Cytophagales OTU826306 (correlation coefficient with CH4 = 0.33, p = 0.05), was most similar to microorganisms from a variety of marine environments including surface seawater (Li et al., 2014), seawater particles (D'Ambrosio et al., 2014) and corals (Sunagawa et al., 2010). As discussed above, the Cytophagales species described here is not closely related to a known hydrocarbon degrader, thus why its abundance is significantly correlated with CH4 concentrations is unknown.
Mesopelagic to Deep-sea Bacterioplankton Structure and Diversity
Nonparametric statistics were used to determine which species in the mesopelagic to the seafloor were significantly different in samples with CH4 above or below background concentrations. Specifically, samples from below 200 m were categorized as above or below and the relative abundances of 6928 OTUs were then tested for significance using the Mann-Whitney test. The relative abundances of five species were significantly different in samples that were either above or below background CH4 concentrations. Of these five species, however, only one was significantly correlated with CH4 (Figure 3B). This species was an unclassified Euryarchaeota (new reference OTU 889), that was 100% similar to archaea in the deep-sea hydrocarbon plume that resulted from the Deepwater Horizon oil spill (Redmond and Valentine, 2012) and to Euryarchaeota from 2940 m in the Japan Trench (Kawagucci et al., 2012). It was also 99% similar to a Marine Group II Euryarchaeota (MGII) from a Guaymas Basin hydrothermal plume (Dick and Tebo, 2010). This MGII species (new reference OTU 889) was also positively correlated with depth and dissolved oxygen and inversely correlated with temperature. The role of MGII in biogeochemical cycling is largely uncharacterized given that there are currently no cultured representatives described in the literature. To date, one MGII has been assembled from metagenomic data revealing a photo-heterotrophic metabolism (Iverson et al., 2012). The MGII species discussed above is not closely related (91% similar, 16S rRNA gene) to the MGII presented by Iverson et al. (2012). Given the photo-heterotrophic lifestyle of a surface ocean MGII, and the distant phylogenetic relationships between these MGII species there are few clues provided as to the ecological role of MGII in the deep-sea, particularly as it relates to CH4. Thus, why the relative abundance of the MGII species described herein was significantly correlated with CH4 is unknown.
It should be noted that a Methylococcales (OTU724366) was observed in several deep-sea samples, but was never more than 1% in relative abundance (Supplemental Figure 1). Methylococcales require CH4 and other C1 compounds for both carbon and energy (Bowman, 2005). Surprisingly, its relative abundance was not significantly correlated with CH4. It is discussed here because it was the only canonical methanotroph observed in any of our GOM samples. This species was 100% similar to a microorganism from a remnant hydrocarbon plume sampled 2 months after the Deepwater Horizon oil spill ceased (Yang et al., 2014) and to microorganisms from hydrothermal vent fluids (Dick and Tebo, 2010; Yoshida-Takashima et al., 2012). There are several interpretations as to why canonical methanotrophs were low in relative abundance, one being that the primers used herein were biased against them. It should be noted, however, that in Mason et al. (2012) a different 16S rRNA gene primer set was used for pyrotag sequencing and 16S rRNA genes were annotated in metagenomes and metatranscriptomes from Deepwater Horizon oil spill deep-sea plumes and uncontaminated seawater from plume depth revealing relative abundances of canonical methanotrophs < 1%, yet methane-oxidation was an active and abundant process. Together, the data suggests that unrecognized marine methanotrophs mediate methane-oxidation, rather than primer bias against canonical methanotrophs as an explanation for why their relative abundances were low in our study.
Conclusion
Unlike other large oligotrophic water bodies the GOM is rich in hydrocarbon seeps. These hydrocarbon seeps inject fixed carbon into the deep-sea water column. As these hydrocarbons migrate toward the surface they likely influence the microbial community structure. Given the paucity of information regarding the microbial community structure in the GOM water column outside of the Deepwater Horizon oil spill we characterized the microbial communities at seep and non-seep sites from the seafloor to the near surface in the water column. We found that depth and the inherent properties of those depths from the seafloor to the near-surface photic zone were the dominant organizing principles structuring water column microbial communities. In the photic zone we observed a coincident CH4 and chlorophyll a maxima. At this maxima a Cyanobacteria putatively identified as Prochlorococcus was abundant and significantly correlated with both CH4 and chlorophyll a. Whether this microorganism acts as a CH4 source by an unidentified photosynthetic process is unknown, but warrants future research. No canonical methanotrophs were identified in the photic zone CH4 maximum, yet several relatives of hydrocarbon degraders were present. In the mesopelagic to the deep-sea a MGII Euryarchaeota was significantly correlated with CH4, but whether it can oxidize CH4 aerobically is unknown. Canonical methanotrophs were identified in deep-sea samples but their relative abundances did not co-vary significantly with CH4. Collectively our data revealed that similar to other oligotrophic water masses, disparate microbial communities comprised the euphotic zone compared to the mesopelagic and deep-sea in the GOM. Unlike other water bodies, however, our findings suggested that CH4 input from the many natural GOM seeps to the deep-sea and from an unknown source in the euphotic zone did appear to significantly influence the relative abundance of some of the key microbial players. Future research efforts will be directed toward linking CH4 production and consumption rates with specific microbial species.
Author Contributions
CVR participated in the research cruises to obtain samples, prepped samples and carried out sequencing, data analyses and helped write the manuscript. CM and SB determined methane concentrations in samples. SB, KLR, and LEG participated in cruises and collected samples. JPC provided methane data. OUM conceived of the experimental design, prepped samples, helped with sequencing, carried out data analyses and helped write the manuscript.
Funding
The participation of JPC, SB, and CM was funded by grants from BP/The Gulf of Mexico Research Initiative (GoMRI) to the Deep-C research consortium administered by Florida State University and to the consortium Ecosystem Impacts of Oil and Gas Inputs to the Gulf-2, (ECOGIG-2) administered by the University of Georgia. This is ECOGIG contribution # 305. CVR was funded for one summer by the GoMRI Deep-C internship program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Deep-C and ECOGIG consortiums for the opportunity to participate in GoMRI funded cruises to obtain the samples that we describe herein. Data is available at GRIIDCC UDI R4.x268.185:0001. Finally we thank the Captains and crews of the R/V Pelican and Weatherbird II.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmars.2015.00069
References
Allers, E., Wright, J. J., Konwar, K. M., Howes, C. G., Beneze, E., Hallam, S. J., et al. (2013). Diversity and population structure of Marine Group A bacteria in the Northeast subarctic Pacific Ocean. ISME J. 7, 56–68. doi: 10.1038/ismej.2012.108
Arndt, D., Xia, J., Liu, Y., Zhou, Y., Guo, A. C., Cruz, J. A., et al. (2012). METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res. 40:W88–W95. doi: 10.1093/nar/gks497
Bowman, J. P. (2005). “Methylococcales ord. nov,” in Bergey's Manual® of Systematic Bacteriology, eds D. J. Brenner, N. R. Krieg, J. T. Staley, G. M. Garrity, D. R. Boone, P. De Vos, M. Goodfellow, F. A. Rainey, and K.-H. Schleifer (New York, NY: Springer), 248–270.
Brooks, J. M., Reid, D. F., and Bernard, B. B. (1981). Methane in the upper water of the Northwestern Gulf of Mexico. J. Geophys. Res. 8, 1029–11040. doi: 10.1029/jc086ic11p11029
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2, 66–67. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F., Costello, E. K., et al. (2010a). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 35–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 (Suppl.), 4516–4522. doi: 10.1073/pnas.1000080107
Chitsaz, H., Yee-Greenbaum, J. L., Tesler, G., Lombardo, M.-J., Dupont, C. L., Badger, J. H., et al. (2011). Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat. Biotechnol. 29, 915–921. doi: 10.1038/nbt.1966
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2013). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Crespo-Medina, M., Meile, C. D., Hunter, K. S., Diercks, A., Asper, V. L., Orphan, V. J., et al. (2014). The rise and fall of methanotrophy following a deepwater oil-well blowout. Nat. Geosci. 7, 423–427. doi: 10.1038/ngeo2156
D'Ambrosio, L., Ziervogel, K., MacGregor, B., Teske, A., and Arnosti, C. (2014). Composition and enzymatic function of particle-associated and free-living bacteria: a coastal/offshore comparison. ISME J. 11, 2167–2179. doi: 10.1038/ismej.2014.67
Delong, E. F., Preston, C. M., Mincer, T., Rich, V., Hallam, S. J., Frigaard, N., et al. (2006). Community genomics among stratified microbial assemblages in the Ocean's Interior. Science 311, 496–503. doi: 10.1126/science.1120250
Dick, G. J., and Tebo, B. M. (2010). Microbial diversity and biogeochemistry of the Guaymas Basin deep-sea hydrothermal plume. Environ. Microbiol. 12, 1334–1347. doi: 10.1111/j.1462-2920.2010.02177.x
Dubinsky, E. A., Conrad, M. E., Chakraborty, R., Bill, M., Borglin, S. E., Hollibaugh, J. T., et al. (2013). Succession of hydrocarbon-degrading bacteria in the aftermath of the deepwater horizon oil spill in the gulf of Mexico. Environ. Sci. Technol. 47, 10860–10867. doi: 10.1021/es401676y
Dufresne, A., Salanoubat, M., Partensky, F., Artiguenave, F., Axmann, I. M., Barbe, V., et al. (2003). Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. U.S.A. 100, 10020–10025. doi: 10.1073/pnas.1733211100
Dyhrman, S. T., Chappell, P. D., Haley, S. T., Moffett, J. W., Orchard, E. D., Waterbury, J. B., et al. (2006). Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439, 68–71. doi: 10.1038/nature04203
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eiler, A., Hayakawa, D. H., and Rapp,é, M. S. (2011). Non-random assembly of bacterioplankton communities in the subtropical north pacific ocean. Front. Microbiol. 2:140. doi: 10.3389/fmicb.2011.00140
Finke, N., Crespo-Medina, M, Schweers, J., and Joye, S. B. (2011). “Aerobic methane production in surface waters of the Gulf of Mexico,” in American Geophysical Union, Fall Meeting 2011, Abstract #A24A-05.
Forster, P., Ramaswamy, V., Artaxo, P., Berntsen, T., Betts, R., Fahey, D. W., et al. (2007). “Changes in atmospheric constituents and in radiative forcing,” in Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, eds S. Solomon, D. Qin, M. Manning, Z. Chen, M. Marquis, K. B. Averyt, M. Tignor and H. L. Miller (Cambridge; New York, NY: Cambridge University Press).
Gasperotti, A. F., Studdert, C. A., Revale, S., and Seitz, M. K. H. (2015). Draft genome sequence of halomonas sp. KHS3, a polyaromatic hydrocarbon-chemotactic strain. Genome Announc. 3, 8–9. doi: 10.1128/genomeA.00020-15
Gillies, L. E., Thrash, J. C., deRada, S., Rabalais, N. N., and Mason, O. U. (2015). Archaeal enrichment in the hypoxic zone in the northern Gulf of Mexico. Environ. Microbiol. doi: 10.1111/1462-2920.12853. [Epub ahead of print].
Giovannoni, S. J., and Stingl, U. (2005). Molecular diversity and ecology of microbial plankton. Nature 437, 343–348. doi: 10.1038/nature04158
Hazen, T. C., Dubinsky, E. A., DeSantis, T. Z., Andersen, G. L., Piceno, Y. M., Singh, N., et al. (2010). Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330, 204–208. doi: 10.1126/science.1195979
Iverson, V., Morris, R. M., Frazar, C. D., Berthiaume, C. T., Morales, R. L., and Armbrust, E. V. (2012). Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335, 587–590. doi: 10.1126/science.1212665
Joye, S. B., MacDonald, I. R., Leifer, I., and Asper, V. (2011). Magnitude and oxidation potential of hydrocarbon gases released from the BP oil well blowout. Nat. Geosci. 4, 160–164. doi: 10.1038/ngeo1067
Karl, D. M., Beversdorf, L., Björkman, K. M., Church, M. J., Martinez, A., and Delong, E. F. (2008). Aerobic production of methane in the sea. Nat. Geosci. 1, 473–478. doi: 10.1038/ngeo234
Karner, M. B., DeLong, E. F., and Karl, D. M. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510. doi: 10.1038/35054051
Kawagucci, S., Yoshida, Y. T., Noguchi, T., Honda, M. C., Uchida, H., Ishibashi, H., et al. (2012). Disturbance of deep-sea environments induced by the M9.0 Tohoku Earthquake. Sci. Rep. 2:270. doi: 10.1038/srep00270
King, G. M., Smith, C. B., Tolar, B., and Hollibaugh, J. T. (2013). Analysis of composition and structure of coastal to mesopelagic bacterioplankton communities in the northern gulf of Mexico. Front. Microbiol. 3:438. doi: 10.3389/fmicb.2012.00438
Kodama, Y., Stiknowati, L. I., Ueki, A., Ueki, K., and Watanabe, K. (2008). Thalassospira tepidiphila sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 58, 711–715. doi: 10.1099/ijs.0.65476-0
Lekunberri, I., Sintes, E., de Corte, D., Yokokawa, T., and Herndl, G. J. (2013). Spatial patterns of bacterial and archaeal communities along the Romanche Fracture Zone (tropical Atlantic). FEMS Microbiol. Ecol. 85, 537–552. doi: 10.1111/1574-6941.12142
Lelieveld, J., Crutzen, P. J., and Dentener, F. J. (1998). Changing concentration, lifetime and climate forcing of atmospheric methane. Tellus 50B, 128–150. doi: 10.1034/j.1600-0889.1998.t01-1-00002.x
Li, J., Li, N., Li, F., Zou, T., Yu, S., Wang, Y., et al. (2014). Spatial diversity of bacterioplankton communities in surface water of Northern South China Sea. PLoS ONE 9:e113014. doi: 10.1371/journal.pone.0113014
Magen, C., Lapham, L. L., Pohlman, J. W., Marshall, K., Bosman, S., Casso, M., et al. (2014). A simple headspace equilibration method for measuring dissolved methane. Limnol. Oceanogr. Methods 12, 637–650. doi: 10.4319/lom.2014.12.637
Mason, O. U., Hazen, T. C., Borglin, S., Chain, P. S. G., Dubinsky, E. A., Fortney, J. L., et al. (2012). Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 6, 1715–1727. doi: 10.1038/ismej.2012.59
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Moore, L. R., Rocap, G., and Chisholm, S. W. (1995). Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Ecol. Prog. Ser. 116, 259–275. doi: 10.3354/meps116259
Petit, J. R., Jouzel, J., Raynaud, D., Barkov, N. I., Barnola, J. M., Basile, I., et al. (1999). Climate and atmospheric history of the past 420,000 years from the Vostok ice core, Antarctica. Nature 399, 429–436. doi: 10.1038/20859
Pham, V. D., Konstantinidis, K. T., Palden, T., and DeLong, E. F. (2008). Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environ. Microbiol. 10, 2313–2330. doi: 10.1111/j.1462-2920.2008.01657.x
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Redmond, M. C., and Valentine, D. L. (2012). Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc. Natl. Acad. Sci. U.S.A. 109, 20292–20297. doi: 10.1073/pnas.1108756108
Rivers, A. R., Sharma, S., Tringe, S. G., Martin, J., Joye, S. B., and Moran, M. A. (2013). Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 7, 2315–2329. doi: 10.1038/ismej.2013.129
Schlitzer, R. (2013). Ocean Data View. Available online at: http://odv.awi.de.
Schneiker, S., Martins dos Santos, V. A. P., Bartels, D., Bekel, T., Brecht, M., Buhrmester, J., et al. (2006). Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat. Biotechnol. 24, 997–1004. doi: 10.1038/nbt1232
Sheik, C. S., Jain, S., and Dick, G. J. (2014). Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ. Microbiol. 16, 304–317. doi: 10.1111/1462-2920.12165
Shiller, A. M., and Joung, D. (2012). Nutrient depletion as a proxy for microbial growth in Deepwater Horizon subsurface oil/gas plumes. Environ. Res. Lett. 7:045301. doi: 10.1088/1748-9326/7/4/045301
Sjöstedt, J., Martiny, J. B. H., Munk, P., and Riemann, L. (2014). Abundance of broad bacterial taxa in the sargasso sea explained by environmental conditions but not water mass. Appl. Environ. Microbiol. 80, 2786–2795. doi: 10.1128/AEM.00099-14
Solomon, E. A., Kastner, M., MacDonald, I. R., and Leifer, I. (2009). Considerable methane fluxes to the atmosphere from hydrocarbon seeps in the Gulf of Mexico. Nat. Geosci. 2, 561–565. doi: 10.1038/ngeo574
Stevens, H., and Ulloa, O. (2008). Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ. Microbiol. 10, 1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., et al. (2015). Structure and function of the global ocean microbiome. Science 348, 1–9. doi: 10.1126/science.1261359
Sunagawa, S., Woodley, C. M., and Medina, M. (2010). Threatened corals provide underexplored microbial habitats. PLoS ONE 5:e9554. doi: 10.1371/journal.pone.0009554
Swan, B. K., Martinez-Garcia, M., Preston, C. M., Sczyrba, A., Woyke, T., Lamy, D., et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333, 1296–1299. doi: 10.1126/science.1203690
Swinnerton, J. W., and Lamontagne, R. A. (1974). Oceanic distribution of low-molecular-weight hydrocarbons baseline measurements. Environ. Sci. Technol. 8, 657–663. doi: 10.1021/es60092a006
Tolar, B. B., King, G. M., and Hollibaugh, J. T. (2013). An analysis of thaumarchaeota populations from the northern gulf of Mexico. Front. Microbiol. 4:72. doi: 10.3389/fmicb.2013.00072
Valentine, D. L., Kessler, J. D., Redmond, M. C., Mendes, S. D., Heintz, M. B., Farwell, C., et al. (2010). Propane respiration jump-starts microbial response to a deep oil spill. Science. 330, 208–211. doi: 10.1126/science.1196830
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wankel, S. D., Joye, S. B., Samarkin, V. A., Shah, S. R., Friederich, G., Melas-Kyriazi, J., et al. (2010). New constraints on methane fluxes and rates of anaerobic methane oxidation in a Gulf of Mexico brine pool via in situ mass spectrometry. Deep Sea Res. II Top. Stud. Oceanogr. 57, 2022–2029. doi: 10.1016/j.dsr2.2010.05.009
Wasmund, K., Kurtböke, D. I., Burns, K. A., and Bourne, D. G. (2009). Microbial diversity in sediments associated with a shallow methane seep in the tropical Timor Sea of Australia reveals a novel aerobic methanotroph diversity. FEMS Microbiol. Ecol. 68, 142–151. doi: 10.1111/j.1574-6941.2009.00667.x
Wright, J. J., Konwar, K. M., and Hallam, S. J. (2012). Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10, 381–394. doi: 10.1038/nrmicro2778
Yang, T., Nigro, L. M., Gutierrez, T., D′Ambrosio, L., Joye, S. B., Highsmith, R., et al. (2014). Pulsed blooms and persistent oil-degrading bacterial populations in the water column during and after the Deepwater Horizon blowout. Deep Sea Res. II Top. Stud. Oceanogr. doi: 10.1016/j.dsr2.2014.01.014. [Epub ahead of print].
Yoshida-Takashima, Y., Nunoura, T., Kazama, H., Noguchi, T., Inoue, K., Akashi, H., et al. (2012). Spatial distribution of viruses associated with planktonic and attached microbial communities in hydrothermal environments. Appl. Environ. Microbiol. 78, 1311–1320. doi: 10.1128/AEM.06491-11
Zhang, Y., Jiao, N., Sun, Z., Hu, A., and Zheng, Q. (2011). Phylogenetic diversity of bacterial communities in South China Sea mesoscale cyclonic eddy perturbations. Res. Microbiol. 162, 320–329. doi: 10.1016/j.resmic.2010.12.006
Zhao, H. P., Wang, L., Ren, J. R., Li, Z., Li, M., and Gao, H. W. (2008). Isolation and characterization of phenanthrene-degrading strains Sphingomonas sp. ZP1 and Tistrella sp. ZP5. J. Hazard. Mater. 152, 1293–1300. doi: 10.1016/j.jhazmat.2007.08.008
Keywords: ITag, microbial community structure, Gulf of Mexico, methane seep, methanotroph, methane photic zone
Citation: Rakowski CV, Magen C, Bosman S, Rogers KL, Gillies LE, Chanton JP and Mason OU (2015) Methane and microbial dynamics in the Gulf of Mexico water column. Front. Mar. Sci. 2:69. doi: 10.3389/fmars.2015.00069
Received: 24 June 2015; Accepted: 31 August 2015;
Published: 16 September 2015.
Edited by:
Hongyue Dang, Xiamen University, ChinaReviewed by:
J. Michael Beman, University of California, Merced, USABarbara J. Campbell, Clemson University, USA
Copyright © 2015 Rakowski, Magen, Bosman, Rogers, Gillies, Chanton and Mason. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olivia U. Mason, Earth, Ocean and Atmospheric Science, Florida State University, Rm 307 OSB, 117 North Woodward Avenue, Tallahassee, FL 32306-4320, USA,b21hc29uQGZzdS5lZHU=
 Chrisoulla V. Rakowski
Chrisoulla V. Rakowski Cedric Magen
Cedric Magen Samantha Bosman
Samantha Bosman Kelsey L. Rogers
Kelsey L. Rogers Lauren E. Gillies
Lauren E. Gillies Jeffrey P. Chanton1
Jeffrey P. Chanton1 Olivia U. Mason
Olivia U. Mason