- 1Zoological Institute, Christian-Albrechts University Kiel, Kiel, Germany
- 2Evolution, Ecology and Genetics, Research School of Biology, Australian National University, Canberra, ACT, Australia
- 3Department of Biological Sciences, The University of North Carolina at Charlotte, Charlotte, NC, USA
The phenotype of an animal cannot be explained entirely by its genes. It is now clear that factors other than the genome contribute to the ecology and evolution of animals. Two fundamentally important factors are the associated microbiota and epigenetic regulations. Unlike the genes and regulatory regions of the genome, epigenetics and microbial composition can be rapidly modified, and may thus represent mechanisms for rapid acclimation to a changing environment. At present, the individual functions of epigenetics, microbiomes, and genomic mutations are largely studied in isolation, particularly for species in marine ecosystems. This single variable approach leaves significant questions open for how these mechanisms intersect in the acclimation and adaptation of organisms in different environments. Here, we propose that the starlet sea anemone, Nematostella vectensis, is a model of choice to investigate the complex interplay between adaptation as well as physiological and molecular plasticity in coastal ecosystems. N. vectensis' geographic range spans four distinct coastlines, including a wide thermocline along the Atlantic coast of North America. N. vectensis is a particularly powerful invertebrate model for studying genome-environment interactions due to (1) the availability of a well-annotated genome, including preexisting data on genome methylation, histone modifications and miRNAs, (2) an extensive molecular toolkit including well-developed protocols for gene suppression and transgenesis, and (3) the simplicity of culture and experimentation in the laboratory. Taken together, N. vectensis has the tractability to connect the functional relationships between a host animal, microbes, and genome modifications to determine mechanisms underlying phenotypic plasticity and local adaptation.
Introduction
Evidence is growing that climate change has profound effects on marine ecosystems (Kroeker et al., 2011; Beaugrand et al., 2012). However, our empirical understanding and ability to make predictions about the response of species in these ecosystems is very limited. One of the major limitations is the general lack of understanding of the mechanisms, genetic and non-genetic, that are involved in acclimation and adaptation. Epigenetic modifications and animal-microbe interactions play significant roles in core biological functions, but little is known about the importance of these non-genetic processes for responses to shifting environments.
Traditional theory and research since the Modern Synthesis have focused on the balance of mutation and selection as the central explanation for the adaptation of populations to their environment and as the generator for phenotypic novelty. These approaches have made tremendous inroads to determine how populations adapt to different environments. However, some organisms also have a remarkable ability to acclimate to environmental change during their lifetime. The mechanisms for acclimation are generally assumed to be due to shifts in the regulation of gene expression. A focus on gene regulation alone is surely incomplete because the phenotype of an animal cannot be explained entirely by its genes. As hypothesized by Waddington (2012) almost a century ago, epigenetic mechanisms that result from modifications of the genome without changes in the underlying nucleotide sequence can also have an important impact on the phenotype. In addition, in 1927, the microbiologist Ivan E. Wallin hypothesized in his book, Symbionticism and the Origin of Species, that the acquisition of bacterial endosymbionts favors the origin of new species (Wallin, 1927).
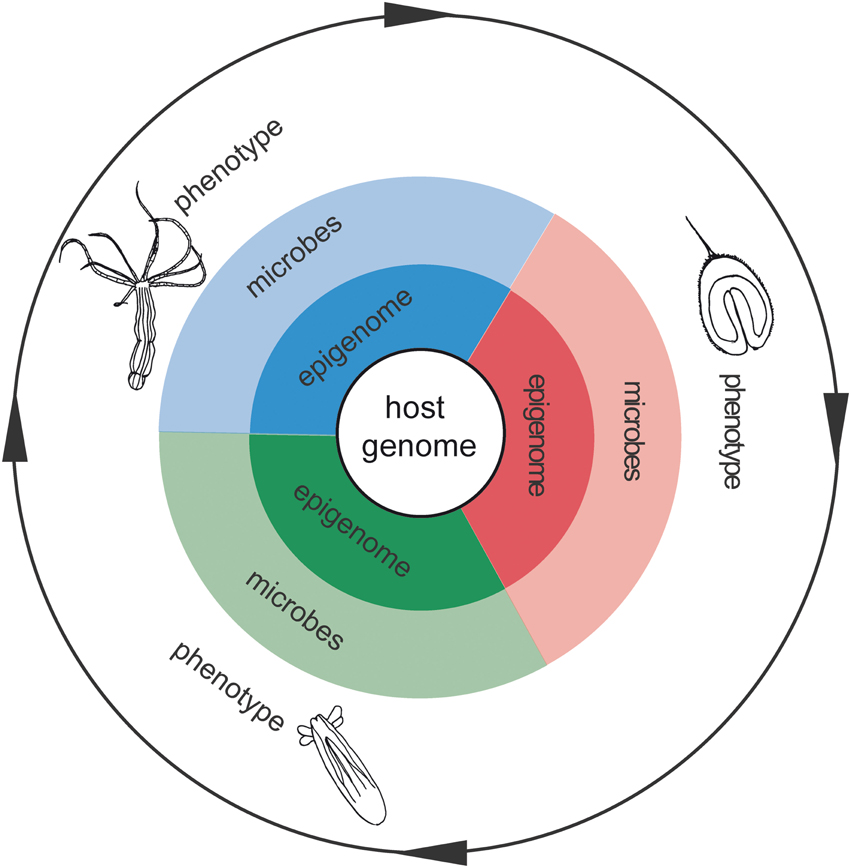
In addition to the genome sequence, epigenetic regulations and microbial communities are important factors contributing to the development and dynamic homeostasis of animals. Moreover, since microbes and epigenetic modifications can potentially be inherited, these environment-induced changes could have transgenerational impacts for adaptation. However, the functions of epigenetic regulations, microbiomes, and gene expression are generally studied separately and little is known about their interactions. As a marine organism, Nematostella vectensis offers a rare combination of physiological and molecular tools that make it a very useful model to study the complex interplay and synergies of microbes and epigenetic regulations in acclimation and adaptation to a changing environment (Figure 1).
The Ecology of Nematostella
N. vectensis is native to the Atlantic coast of the United States and Canada and was likely introduced to the Pacific coast of the United States and the southeastern coasts of England (Hand and Uhlinger, 1994; Reitzel et al., 2008). In all of these locations, this species occurs in brackish habitats, particularly in tidally restricted pools in the high marsh (Figure 2A). N. vectensis primarily burrows into soft sediments and extends its tentacles at the surface to capture prey, which include snails, copepods, and insect larvae (Frank and Bleakney, 1978). Research experimentally characterizing the food web architecture in coastal estuaries of the Atlantic seaboard has shown that N. vectensis occupies a central node in the biological community as an infaunal predator (Kneib, 1985, 1991; Posey and Hines, 1991).
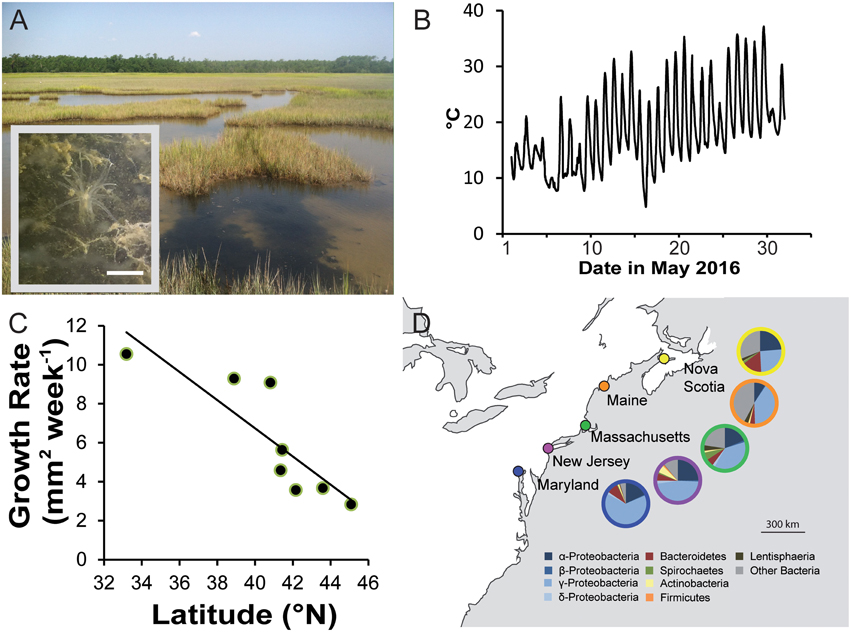
Figure 2. (A) Natural habitat of N. vectensis along the Nonesuch River, Maine, USA; small insert: single polyp of N. vectensis burrowed in the sediment. (B) Temperature data (May 2016) collected by a temperature logger positioned in a pool occupied by N. vectensis in Sippewissett Marsh, Massachusetts, USA. (C) Relationship of growth rate and latitude of origin for individuals cultured at 29°C (data from Reitzel et al., 2013a). (D) Contribution of host biogeography to the overall pattern of bacterial diversity between individual Nematostella polyps sampled from the US east coast. Map showing the location of the sampled populations: Pie charts representing the mean relative abundances of bacterial classes at the five different populations(data from Mortzfeld et al., 2015).
Similar to many estuarine organisms, N. vectensis is tolerant of a wide range of temperature and other environmental variables (e.g., salinity, oxygen concentration, pH). Because it is a predominately sessile species, individuals must have wide physiological plasticity and ability to acclimate to thrive in these habitats. Natural populations have been documented in tide pools with temperatures ranging from −1.5 to 41°C (Reitzel et al., 2013a) and salinities from 9 to 51‰; (reviewed by Hand and Uhlinger, 1994). Within these tidally restricted pools, the environment also has extreme temperature-oscillations with >20°C fluctuations daily (Figure 2B), exerting cellular stress evidenced by upregulation of heat shock genes (Reitzel, unpublished data). N. vectensis' native geographic range spans a pronounced thermocline (~10°C mean temperature over 10° latitude) (Hand and Uhlinger, 1994; Reitzel et al., 2008), which appears to have resulted in different thermal optima for growth and tolerance for high temperatures (Reitzel et al., 2013a, Figure 2C).
Distribution in high marsh estuarine environments typically results in reduced genetic connectivity between locations and can tend to provide the foundation for local adaptation (Bilton et al., 2002; Virgilio et al., 2006). Population genetics research has revealed N. vectensis populations have significant genetic differentiation among locations (Pearson et al., 2002; Darling et al., 2004, 2009; Reitzel et al., 2008) and strong phylogeographic structure (Reitzel et al., 2013b). Comparisons of alleles and allele frequencies also support a hypothesis that some of this genetic variation is a result of natural selection shown by SNPs from expressed sequence tags and whole-genome sequencing (Reitzel et al., 2008, 2010, 2013b; Sullivan et al., 2009). Although there is significant genetic and phenotypic variation between populations of N. vectensis, individuals from geographically distant populations are interfertile (Hand and Uhlinger, 1994; Reitzel et al., 2008) confirming them as all belonging to the same species.
N. vectensis, like many cnidarians, can reproduce both sexually and asexually (Williams, 1975; Hand and Uhlinger, 1992; Reitzel et al., 2007). For sexual reproduction, eggs are released from females in a matrix of gelatinous material and fertilized by sperm free-spawned from males (Hand and Uhlinger, 1992). The ontogeny of N. vectensis is well characterized in laboratory conditions and includes two principal developmental transitions. The first event occurs approximately 1-week post fertilization when the planula larvae metamorphose first into a primary polyp with four tentacles and later into a juvenile polyp with several tentacles. These juvenile polyps need up to 6 more months to reach the second major event, the transition from a juvenile to a sexually mature polyp. Asexual reproduction occurs through two distinct types of binary fission (Reitzel et al., 2007). Although natural populations, particularly in the native range, are composed of a mix of genetically unique (indicative of sexual reproduction) and clonal genotypes (indicative of asexual reproduction), populations in the introduced range are primarily composed of clonal genotypes that extend over large geographic distances, likely due to anthropogenic dispersal (Darling et al., 2009). N. vectensis is readily collected from diverse field locations throughout its range, thus future studies of cycles in reproduction in natural conditions would be attainable.
Nematostella—a Marine Model System for Functional Genomics
Cnidaria belong to an early-branching group of metazoans and have preserved much of the genetic complexity of the common metazoan ancestor. Interestingly, most of the signaling pathways that regulate development (Technau et al., 2005) and innate immunity (Miller et al., 2007) important in bilaterian animals, including receptors, ligands, antagonists, and signaling components, have been identified in cnidarians (Putnam et al., 2007b; Chapman et al., 2010; Shinzato et al., 2011). Thus, fundamental principles that are general to all metazoan life—including humans—can be investigated in this animal group.
N. vectensis is a particularly powerful cnidarian model due to its the availability of a well assembled and annotated genome (Putnam et al., 2007a), and an extensive molecular toolkit including a set of mature protocols, and the ability to reliably procure all developmental stages on a weekly basis. Spawning is induced by a shift in temperature and exposure to light (Fritzenwanker and Technau, 2002). It is easily cultured in laboratory in high numbers (Hand and Uhlinger, 1992, 1995) and clonally propagated to eliminate genetic confounding effects. In addition, N. vectensis has the unique advantage amongst marine invertebrates of having extensive sequencing of transcriptomes (Helm et al., 2013; Tulin et al., 2013), preexisting data on genome methylation (Zemach et al., 2010), histone modifications (Schwaiger et al., 2014), and miRNAs (Moran et al., 2014). These existing data result in an exceptionally well-annotated genome, an essential but still rare tool for marine species.
Among marine organisms, tools for functional studies are limited to a few cnidarian species, where transgenesis has been established (Künzel et al., 2010; Renfer et al., 2010). In addition, morpholino-mediated knock-down experiments were established in N. vectensis (Rentzsch et al., 2008) and the hydrozoan Clytia hemisphaerica (Houliston et al., 2010). More recently, the CRISPR/Cas9 mediated genome-editing system has become an approach of choice to study gene function in-vivo and was recently established in N. vectensis (Ikmi et al., 2014), opening new opportunities to study gene-environment interactions at a functional level.
These characteristics, together with an exceptionally high acclimation potential to varying abiotic factors, make N. vectensis a very promising model to understand how environmental factors affect the composition and function of microbiota, the consequences of host-microbe interactions during rapid acclimation of a holobiont to changing environmental conditions, and adaptation in separate geographic regions.
Acclimations by Epigenetic Modifications in Cnidaria
Long-lived sessile organisms are likely to experience a broad range of environmental conditions within their lifetime. It would therefore be advantageous for such species to be able to acclimate to certain environmental stressors and to “learn” how to better respond to subsequent exposures to this stress. Some recent studies provide evidence for this type of acclimation process in reef building corals. It has been shown that corals transplanted into warmer waters are able to better respond to acute heat stress than colonies that remain in cooler waters (Palumbi et al., 2014). Indeed, pre-exposure to a mild thermal stress is likely to initiate an acclimation mechanism protecting corals from bleaching in several recent heat stress events (Ainsworth et al., 2016). Despite this ecological and physiological evidence for acclimation, little is known about the molecular mechanisms underlying this type of response. However, it is clear that the genome of the hosts did not change during these short periods of acclimation.
While the roles for DNA mutation and natural selection have been at the center of research to understand the relationship between genotype and phenotype, recent studies have uncovered that epigenetic mechanisms can profoundly influence how a genome is interpreted depending on the context (Suzuki and Bird, 2008). These epigenetic mechanisms do not change the nucleotide sequence but can have large effects on the physical structure of the DNA molecule and thus its accessibility to the transcriptional machinery (Weber et al., 2007). Epigenetic marks include the packaging of chromatin by nucleosomes made of different histone variants, modifications of these histones, chemical modifications of individual nucleotides, non-coding RNAs, and RNA editing (Liebers et al., 2014). These mechanisms can be triggered by factors such as developmental stage, environmental cues or bacterial infections (Pereira et al., 2016), and enable condition-specific expression of the genome. An in-depth understanding of the effects of epigenetic processes in organismal biology has the potential to radically alter how we view the genotype-phenotype map and to improve our mechanistic understanding for the interactions between the individual and its environment in the wild.
DNA methylation is probably the best studied epigenetic mechanism, where the addition of a methyl group to cytosine residues is mediated by DNA methyltransferases (Colot and Rossignol, 1999; Suzuki and Bird, 2008). Depending on the context, DNA methylation results in enhanced or repressed activity by making the DNA more or less available to other molecules, thereby determining expression of specific transcripts in particular cells or as a result of particular environmental shifts. These chemical modifications explain why identical twins, despite sharing exactly the same DNA, show differences in appearance, how in utero exposure can affect offspring, and how maternal behavior influences offspring behavior (Flintoft, 2005; Suzuki and Bird, 2008; Hou et al., 2012). Despite these fundamental insights, the role of epigenetics in the biology of animals, particularly marine species, remains largely unexplored (Gavery and Roberts, 2010).
In animals, studies on DNA methylation have traditionally focused on vertebrates due, in part, to the absence of this chemical modification in the invertebrate model species Drosophila melanogaster (Raddatz et al., 2013) and Caenorhabditis elegans (Lyko and Maleszka, 2011). It is now clear that both of these species have secondarily lost their methylation machinery that is otherwise broadly conserved in the animal kingdom (Yi, 2012). As a consequence, researchers have been exploring alternative models for epigenetic research. Notably, these include social insects, where DNA methylation (Kucharski et al., 2008), histone modifications (Simola et al., 2013), and micro-RNAs (Guo et al., 2013) have been shown to play important roles in the division of labor. Among marine invertebrates, relatively little is known about the mechanisms of epigenetic regulation. However, N. vectensis is the only marine invertebrate where existing data is available on genome methylation (Zemach et al., 2010), histone modifications (Schwaiger et al., 2014), and miRNAs (Moran et al., 2013). Thus, we suggest that N. vectensis is uniquely positioned as an organism of choice to study how these epigenetic alterations of the genome link molecular changes with acclimation observed at the organismal and physiological level.
The Extended Phenotype of Nematostella—Bacteria and their Role in Acclimation
The diversity of microbes colonizing a multicellular organism has been proposed to be a result of the coevolution between the eukaryotic species and the associated microbial community, influenced by both the environment and the host (Ley et al., 2006). Often the bacterial community is specific for a certain host species (Ley et al., 2008; Franzenburg et al., 2013), which indicates regulation of its microbiome. The microbiota can have strong effects on host fitness, influencing a broad range of changes in host physiology, including facilitation of nutrient supply, immune system maturation, development, and resistance against pathogens (Fraune and Bosch, 2010; Sommer and Bäckhed, 2013). Although invertebrates do not possess an adaptive immune system, their innate immune system provides them with means to interact with their associated microbes (Franzenburg et al., 2012, 2013) and has to fulfil the challenging tasks of attracting beneficial bacteria, repelling pathogens and preventing commensals from becoming harmful.
Recently, animal–microbe interactions have been recognized as important drivers of animal evolution and diversification (Brucker and Bordenstein, 2012; McFall-Ngai et al., 2013). Mutualistic associations between animals and microbes can evolve by distinct selective forces. Because association with a beneficial microbiota increases the host's fitness, selective pressures should act on regulatory mechanisms in the host, e.g., immune system, ensuring specific bacterial colonizers. Additionally, bacteria can be selected if they are beneficial to the host, since the host's fitness ensures the future availability of the host as habitat (Ley et al., 2006; Bordenstein and Theis, 2015). These interlinked dependencies between the host and its associated microbes led to the “hologenome theory of evolution,” considering the holobiont as a unit undergoing natural selection (Rosenberg et al., 2007). Therefore, microbial symbionts represent a specific form of genetic inheritance (Gilbert et al., 2010), being either vertically acquired through the egg or horizontally transmitted through the environment. Changes in the bacterial community can produce phenotypic variation of the metaorganism and may impact on the ecological tolerance and distribution of the metaorganism, providing immense potential for adaptation to changing environmental conditions (Reshef et al., 2006). These changes can arise through changes in the relative abundance of associated microorganisms, the introduction of new members, genetic variation in the microorganisms or horizontal gene transfer. Plasticity in bacterial colonization can contribute to thermotolerance of the holobiont (Dunbar et al., 2007), prevent growth of external pathogens (Woodhams et al., 2007; Fraune et al., 2014) or determine host body color (Tsuchida et al., 2010). Therefore, it can be hypothesized that molecular communications between host and microbe select for a core microbiota in a given host species which contributes to adaptation when the environment changes.
Colonizing bacteria are also a vital component of cnidarian holobionts (Lesser et al., 2004; Fiore et al., 2010; Fraune et al., 2014). In N. vectensis bacterial colonization is characterized by a stable associated bacterial community, which is dynamic but highly conserved in response to host development. Nevertheless, environmental changes induce a robust tuning of bacterial colonization (Mortzfeld et al., 2015), e.g., by the increase of rare bacterial species. In addition, analysis of bacterial communities of Nematostella polyps from five different populations revealed a strong correlation between host biogeography and bacterial diversity (Figure 2D). This correlation is caused by the sum of host genetics and environmental factors the animals were confronted with, such as temperature, salinity, food, and substrate. Future studies will elucidate the contribution of each factor, host genetic, and environment, to overall bacterial diversity.
Perspectives
The “systems view” in biology posits that the properties of a complex system cannot be found in its isolated components but rather emerge from their interactions. As a consequence, environmental adaptation can only be fully understood—functionally and evolutionary—by integrating all potential levels. Therefore, our main hypotheses are (i) Epigenetic modifications contribute to environmental acclimation and can be inherited (ii) Changes in the bacterial colonization and diversity contribute to the thermal acclimation response and (iii) Synergistic interactions between host transcription, epigenetic regulations, and bacterial colonization determine the potential for adaptation.
Knocking down or inhibiting DNA methyltransferases is likely to have broad indiscriminate effects on the genome and subsequent detrimental effect on fitness. Therefore it is unlikely that it would be a viable approach to test the first hypothesis. Instead, function of epigenetics in organismal acclimation could be tested by mimicking the specific effects of methylation without interfering with the methylation machinery. For instance if the silencing of a gene by methylation is suspected to have a role in acclimation to high temperatures, this could be tested by silencing this gene by other means. The role of bacteria could be tested by creating germ-free animals and re-infecting these animals with specific strains or combination of stains to determine functional relationships between the anemone and its microbial community (Fraune et al., 2014). Hypotheses pertaining to the interactions between these two types of mechanisms could then be tested by combining these two types of experimental manipulations (e.g., addition of one bacterial species plus repression on one methylated gene in germ-free anemone cultured at high temperature) to discern combinatorial effects that may be additive, synergistic, or antagonistic.
To date only few marine species offer the molecular tools required for a detailed mechanistic understanding of these acclimation mechanisms. Researchers are now in a position to leverage the unique characteristics of the sea anemone N. vectensis as a model for field ecology, genomics, and animal-microbial interactions to connect the roles of the host, its associated microbes and epigenetic regulations to responses to shifts in the environment.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are supported by the Human Frontier Science Program (Young Investigators' Grant) RGY0079/2106. SFr is also supported by the CRC 1182 (Project B1) Deutsche Forschungsgemeinschaft (DFG). AR was also supported by National Science Foundation Grants 1545539 and 1536530. AR would like to thank Whitney Leach for field assistance.
References
Ainsworth, T. D., Heron, S. F., Ortiz, J. C., Mumby, P. J., Grech, A., Ogawa, D., et al. (2016). Climate change disables coral bleaching protection on the Great Barrier Reef. Science 352, 338–342. doi: 10.1126/science.aac7125
Beaugrand, G., McQuatters-Gollop, A., Edwards, M., and Goberville, E. (2012). Long-term responses of North Atlantic calcifying plankton to climate change. Nat. Clim. Change 3, 263–267. doi: 10.1038/nclimate1753
Bilton, D., Paula, J., and Bishop, J. (2002). Dispersal, genetic differentiation and speciation in estuarine organisms. Estuar. Coast. Shelf Sci. 55, 937–952. doi: 10.1006/ecss.2002.1037
Bordenstein, S. R., and Theis, K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13:e1002226. doi: 10.1371/journal.pbio.1002226
Brucker, R. M., and Bordenstein, S. R. (2012). Speciation by symbiosis. Trends Ecol. Evol. 27, 443–451. doi: 10.1016/j.tree.2012.03.011
Chapman, J. A., Kirkness, E. F., Simakov, O., Hampson, S. E., Mitros, T., Weinmaier, T., et al. (2010). The dynamic genome of Hydra. Nature 464, 592–596. doi: 10.1038/nature08830
Colot, V., and Rossignol, J. L. (1999). Eukaryotic DNA methylation as an evolutionary device. Bioessays 21, 402–411.
Darling, J. A., Reitzel, A. M., and Finnerty, J. R. (2004). Regional population structure of a widely introduced estuarine invertebrate: Nematostella vectensis Stephenson in New England. Mol. Ecol. 13, 2969–2981. doi: 10.1111/j.1365-294X.2004.02313.x
Darling, J., Kuenzi, A., and Reitzel, A. (2009). Human-mediated transport determines the non-native distribution of the anemone Nematostella vectensis, a dispersal-limited estuarine invertebrate. Mar. Ecol. Prog. Ser. 380, 137–146. doi: 10.3354/meps07924
Dunbar, H. E., Wilson, A. C. C., Ferguson, N. R., and Moran, N. A. (2007). Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 5:e96. doi: 10.1371/journal.pbio.0050096
Fiore, C. L., Jarett, J. K., Olson, N. D., and Lesser, M. P. (2010). Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 18, 455–463. doi: 10.1016/j.tim.2010.07.001
Flintoft, L. (2005). Epigenetics: identical twins: epigenetics makes the difference. Nat. Rev. Genet. 6, 667. doi: 10.1038/nrg1693
Frank, P. G., and Bleakney, J. S. (1978). Asexual reproduction, diet, and anomalies of the anemone Nematostella vectensis in Nova Scotia. Can. Field Nat. 92, 259–263.
Franzenburg, S., Fraune, S., Künzel, S., Baines, J. F., Domazet-Loso, T., and Bosch, T. C. G. (2012). MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc. Natl. Acad. Sci. U.S.A. 109, 19374–19379. doi: 10.1073/pnas.1213110109
Franzenburg, S., Walter, J., Künzel, S., Wang, J., Baines, J. F., Bosch, T. C. G., et al. (2013). Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl. Acad. Sci. U.S.A. 110, E3730–E3738. doi: 10.1073/pnas.1304960110
Fraune, S., Anton-Erxleben, F., Augustin, R., Franzenburg, S., Knop, M., Schröder, K., et al. (2014). Bacteria-bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance. ISME J. 9, 1543–1556. doi: 10.1038/ismej.2014.239
Fraune, S., and Bosch, T. C. (2010). Why bacteria matter in animal development and evolution. Bioessays 32, 571–580. doi: 10.1002/bies.200900192
Fritzenwanker, J. H., and Technau, U. (2002). Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa). Dev. Genes Evol. 212, 99–103. doi: 10.1007/s00427-002-0214-7
Gavery, M. R., and Roberts, S. B. (2010). DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (Crassostrea gigas). BMC Genomics 11:483. doi: 10.1186/1471-2164-11-483
Gilbert, S. F., McDonald, E., Boyle, N., Buttino, N., Gyi, L., Mai, M., et al. (2010). Symbiosis as a source of selectable epigenetic variation: taking the heat for the big guy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 671–678. doi: 10.1098/rstb.2009.0245
Guo, X., Su, S., Skogerboe, G., Dai, S., Li, W., Li, Z., et al. (2013). Recipe for a busy bee: microRNAs in Honey Bee caste determination. PLoS ONE 8:e81661. doi: 10.1371/journal.pone.0081661
Hand, C., and Uhlinger, K. (1995). Asexual reproduction by transverse fission and some anomalies in the sea anemone Nematostella vectensis. Invertebr. Biol. 114, 9–18. doi: 10.2307/3226948
Hand, C., and Uhlinger, K. R. (1992). The Culture, sexual and asexual reproduction, and growth of the sea anemone Nematostella vectensis. Biol. Bull. 182, 169–176. doi: 10.2307/1542110
Hand, C., and Uhlinger, K. R. (1994). The unique, widely distributed, estuarine sea anemone, Nematostella vectensis Stephenson: a review, new facts, and questions. Estuaries 17, 501. doi: 10.2307/1352679
Helm, R. R., Siebert, S., Tulin, S., Smith, J., and Dunn, C. W. (2013). Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC Genomics 14:266. doi: 10.1186/1471-2164-14-266
Hou, L., Zhang, X., Wang, D., and Baccarelli, A. (2012). Environmental chemical exposures and human epigenetics. Int. J. Epidemiol. 41, 79–105. doi: 10.1093/ije/dyr154
Houliston, E., Momose, T., and Manuel, M. (2010). Clytia hemisphaerica: a jellyfish cousin joins the laboratory. Trends Genet. 26, 159–167. doi: 10.1016/j.tig.2010.01.008
Ikmi, A., McKinney, S. A., Delventhal, K. M., and Gibson, M. C. (2014). TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nat. Commun. 5, 5486. doi: 10.1038/ncomms6486
Kneib, R. (1985). Predation and disturbance by grass shrimp, Palaemonetespugio Holthuis, in soft-substratum benthic invertebrate assemblages. J. Exp. Mar. Bio. Ecol. 93, 91–102. doi: 10.1016/0022-0981(85)90151-0
Kneib, R. (1991). Indirect effects in experimental studies of marine soft-sediment communities. Am. Zool. 31, 874–885. doi: 10.1093/icb/31.6.874
Kroeker, K. J., Micheli, F., Gambi, M. C., and Martz, T. R. (2011). Divergent ecosystem responses within a benthic marine community to ocean acidification. Proc. Natl. Acad. Sci. U.S.A. 108, 14515–14520. doi: 10.1073/pnas.1107789108
Kucharski, R., Maleszka, J., Foret, S., and Maleszka, R. (2008). Nutritional control of reproductive status in honeybees via DNA methylation. Science 319, 1827–1830. doi: 10.1126/science.1153069
Künzel, T., Heiermann, R., Frank, U., and Müller, W. (2010). Migration and differentiation potential of stem cells in the cnidarian Hydractinia analysed in eGFP-transgenic animals and chimeras. Dev. Biol. 348, 120–129. doi: 10.1016/j.ydbio.2010.08.017
Lesser, M. P., Mazel, C. H., Gorbunov, M. Y., and Falkowski, P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000. doi: 10.1126/science.1099128
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Ley, R. E., Peterson, D. A., and Gordon, J. I. (2006). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848. doi: 10.1016/j.cell.2006.02.017
Liebers, R., Rassoulzadegan, M., and Lyko, F. (2014). Epigenetic regulation by heritable RNA. PLoS Genet. 10:e1004296. doi: 10.1371/journal.pgen.1004296
Lyko, F., and Maleszka, R. (2011). Insects as innovative models for functional studies of DNA methylation. Trends Genet. 27, 127–131. doi: 10.1016/j.tig.2011.01.003
McFall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Miller, D. J., Hemmrich, G., Ball, E. E., Hayward, D. C., Khalturin, K., Funayama, N., et al. (2007). The innate immune repertoire in cnidaria–ancestral complexity and stochastic gene loss. Genome Biol. 8:R59. doi: 10.1186/gb-2007-8-4-r59
Moran, Y., Fredman, D., Praher, D., Li, X. Z., Wee, L. M., Rentzsch, F., et al. (2014). Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 24, 651–663. doi: 10.1101/gr.162503.113
Moran, Y., Praher, D., Fredman, D., and Technau, U. (2013). The evolution of microRNA pathway protein components in Cnidaria. Mol. Biol. Evol. 30, 2541–2552. doi: 10.1093/molbev/mst159
Mortzfeld, B. M., Urbanski, S., Reitzel, A. M., Künzel, S., Technau, U., and Fraune, S. (2015). Response of bacterial colonization in Nematostella vectensis to development, environment and biogeography. Environ. Microbiol. 18, 1764–1781. doi: 10.1111/1462-2920.12926
Palumbi, S. R., Barshis, D. J., Traylor-Knowles, N., and Bay, R. A. (2014). Mechanisms of reef coral resistance to future climate change. Science 344, 895–898. doi: 10.1126/science.1251336
Pearson, C. V., Rogers, A. D., and Sheader, M. (2002). The genetic structure of the rare lagoonal sea anemone, Nematostella vectensis Stephenson (Cnidaria; Anthozoa) in the United Kingdom based on RAPD analysis. Mol. Ecol. 11, 2285–2293. doi: 10.1046/j.1365-294x.2002.01621.x
Pereira, J. M., Hamon, M. A., and Cossart, P. (2016). A lasting impression: epigenetic memory of bacterial infections? Cell Host Microbe 19, 579–582. doi: 10.1016/j.chom.2016.04.012
Posey, M. H., and Hines, A. H. (1991). Complex predator-prey interactions within an estuarine benthic community. Ecology 72, 2155–2169. doi: 10.2307/1941567
Putnam, N. H., Srivastava, M., Hellsten, U., Dirks, B., Chapman, J., Salamov, A., et al. (2007a). Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94. doi: 10.1126/science.1139158
Putnam, N. H., Srivastava, M., Hellsten, U., Dirks, B., Chapman, J., Salamov, A., et al. (2007b). Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94. doi: 10.1126/science.1139158
Raddatz, G., Guzzardo, P. M., Olova, N., Fantappié, M. R., Rampp, M., Schaefer, M., et al. (2013). Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc. Natl. Acad. Sci. U.S.A. 110, 8627–8631. doi: 10.1073/pnas.1306723110
Reitzel, A. M., Chu, T., Edquist, S., and Genovese, C. (2013a). Physiological and developmental responses to temperature by the sea anemone Nematostella vectensis. Mar. Ecol. Prog. Ser. 484, 115–130, doi: 10.3354/meps10281
Reitzel, A. M., Darling, J., Sullivan, J., and Finnerty, J. (2008). Global population genetic structure of the starlet anemone Nematostella vectensis: multiple introductions and implications for conservation policy. Biol. Invasions. 10, 1197–1213. doi: 10.1007/s10530-007-9196-8
Reitzel, A. M., Burton, P. M., Krone, C., and Finnerty, J. R. (2007). Comparison of developmental trajectories in the starlet sea anemone Nematostella vectensis: embryogenesis, regeneration, and two forms of asexual fission. Invertebr. Biol. 126, 99–112. doi: 10.1111/j.1744-7410.2007.00081.x
Reitzel, A. M., Herrera, S., Layden, M. J., Martindale, M. Q., and Shank, T. M. (2013b). Going where traditional markers have not gone before: utility of and promise for RAD sequencing in marine invertebrate phylogeography and population genomics. Mol. Ecol. 22, 2953–2970. doi: 10.1111/mec.12228
Reitzel, A. M., Sullivan, J., and Finnerty, J. (2010). Discovering SNPs in protein coding regions with StellaSNP: illustrating the characterization and geographic distribution of polymorphisms in the estuarine anemone. Estuar. coasts 33, 930–943. doi: 10.1007/s12237-009-9231-3
Renfer, E., Amon-Hassenzahl, A., Steinmetz, P. R. H., and Technau, U. (2010). A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc. Natl. Acad. Sci. U.S.A. 107, 104–108. doi: 10.1073/pnas.0909148107
Rentzsch, F., Fritzenwanker, J. H., Scholz, C. B., and Technau, U. (2008). FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development 135, 1761–1769. doi: 10.1242/dev.020784
Reshef, L., Koren, O., Loya, Y., Zilber-Rosenberg, I., and Rosenberg, E. (2006). The coral probiotic hypothesis. Environ. Microbiol. 8, 2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Schwaiger, M., Schönauer, A., Rendeiro, A. F., Pribitzer, C., Schauer, A., Gilles, A. F., et al. (2014). Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res. 24, 639–650. doi: 10.1101/gr.162529.113
Shinzato, C., Shoguchi, E., Kawashima, T., Hamada, M., Hisata, K., Tanaka, M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. doi: 10.1038/nature10249
Simola, D. F., Ye, C., Mutti, N. S., Dolezal, K., Bonasio, R., Liebig, J., et al. (2013). A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Res. 23, 486–496. doi: 10.1101/gr.148361.112
Sommer, F., and Bäckhed, F. (2013). The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
Sullivan, J. C., Wolenski, F. S., Reitzel, A. M., French, C. E., Traylor-Knowles, N., Gilmore, T. D., et al. (2009). Two alleles of NF-kappaB in the sea anemone Nematostella vectensis are widely dispersed in nature and encode proteins with distinct activities. PLoS ONE 4:e7311. doi: 10.1371/journal.pone.0007311
Suzuki, M. M., and Bird, A. (2008). DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476. doi: 10.1038/nrg2341
Technau, U., Rudd, S., Maxwell, P., Gordon, P. M. K., Saina, M., Grasso, L. C., et al. (2005). Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 21, 633–639. doi: 10.1016/j.tig.2005.09.007
Tsuchida, T., Koga, R., Horikawa, M., Tsunoda, T., Maoka, T., Matsumoto, S., et al. (2010). Symbiotic bacterium modifies aphid body color. Science 330, 1102–1104. doi: 10.1126/science.1195463
Tulin, S., Aguiar, D., Istrail, S., and Smith, J. (2013). A quantitative reference transcriptome for Nematostella vectensis early embryonic development: a pipeline for de novo assembly in emerging model systems. Evodevo 4:16. doi: 10.1186/2041-9139-4-16
Virgilio, M., Backeljau, T., and Abbiati, M. (2006). Mitochondrial DNA and allozyme patterns of Hediste diversicolor (Polychaeta: Nereididae): the importance of small scale genetic structuring. Mar. Ecol. Prog. Ser. 326, 157–165. doi: 10.3354/meps326157
Waddington, C. H. (2012). The epigenotype. 1942. Int. J. Epidemiol. 41, 10–13. doi: 10.1093/ije/dyr184
Wallin, I. E. (1927). Symbionticism and the Origin of Species/by Ivan E. Wallin. Baltimore, MD : Williams & Wilkins Company.
Weber, M., Hellmann, I., Stadler, M. B., Ramos, L., Pääbo, S., Rebhan, M., et al. (2007). Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 39, 457–466. doi: 10.1038/ng1990
Williams, R. (1975). A redescription of the brackish-water sea anemone Nematostella vectensis Stephenson, with an appraisal of congeneric species. J. Nat. Hist. 9, 51–64. doi: 10.1080/00222937500770051
Woodhams, D. C., Vredenburg, V. T., Simon, M.-A., Billheimer, D., Shakhtour, B., Shyr, Y., et al. (2007). Symbiotic bacteria contribute to innate immune defenses of the threatened mountain yellow-legged frog, Rana muscosa. Biol. Conserv. 138, 390–398. doi: 10.1016/j.biocon.2007.05.004
Yi, S. (2012). Birds do it, bees do it, worms and ciliates do it too: DNA methylation from unexpected corners of the tree of life. Genome Biol. 13:174. doi: 10.1186/gb-2012-13-10-174
Keywords: Nematostella vectensis, micobiota, epigenomics, acclimation, adaptation, biological
Citation: Fraune S, Forêt S and Reitzel AM (2016) Using Nematostella vectensis to Study the Interactions between Genome, Epigenome, and Bacteria in a Changing Environment. Front. Mar. Sci. 3:148. doi: 10.3389/fmars.2016.00148
Received: 29 June 2016; Accepted: 02 August 2016;
Published: 17 August 2016.
Edited by:
Robert Brucker, Rowland Institute at Harvard, USAReviewed by:
Sandra Breum Andersen, University of Oxford, UKGrigory Genikhovich, University of Vienna, Austria
Copyright © 2016 Fraune, Forêt and Reitzel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian Fraune, c2ZyYXVuZUB6b29sb2dpZS51bmkta2llbC5kZQ==
Sylvain Forêt, c3lsdmFpbi5mb3JldEBhbnUuZWR1LmF1
Adam M. Reitzel, YXJlaXR6ZTJAdW5jYy5lZHU=
 Sebastian Fraune
Sebastian Fraune Sylvain Forêt
Sylvain Forêt Adam M. Reitzel
Adam M. Reitzel