- Department of Human Ecology, Food Science and Biotechnology, College of Agricultural and Related Sciences, Delaware State University, Dover, DE, United States
Fish-borne illness is associated with pathogenic bacteria, Vibrio spp., transmitted from contaminated aquaculture water to fish. But little is known about the correlation between water quality in aquaculture and subsequent bacterial contamination in fish products. The degree of bacterial transmission from Vibrio spp. contaminated aquaculture water to fish was investigated. V. anguillarum, V. parahaemolyticus, and V. vulnificus were administered to aquaculture water and the amounts of Vibrio spp. and total bacteria in the aquaculture water and fish filets were assessed using a multiplex real-time PCR assay. Following the challenge, the counts of total bacteria and Vibrio spp. simultaneously increased in both the water and fish filets with levels of bacteria in the fish directly proportional to that in the water. The total bacterial population in the water decreased to pre-challenge levels over time. However, Vibrio spp. counts in fish filets were nearly unchanged during the same time period. There is a strong correlation between the amount of bacteria that are present in aquaculture waters and those that can be observed in the subsequent fish muscle tissue. In addition, the numbers of Vibrio spp. in the random sampled aquaculture fish filets were highly associated with the levels in the aquaculture water. Consequently, populations of total bacteria and Vibrio spp. in aquaculture water is highly correlated with bacterial number of aquacultured fish product, therefore the surveillance of Vibrio populations in the aquaculture water could be an indicator of the microbial contaminations of fish filets.
Introduction
Owing to the exponential consumption of fishery products in the past 50 years, a rapid increase in seafood production, including finfish and shellfish, has been demanded. A great deal of this demand was met by wild-caught fish, but as world fisheries reached over-exploitation and many marine species have been depleted, aquaculture systems have grown at an unprecedented rate (Sapkota et al., 2008). While annual aquaculture production has increased from 55.7 million tons in 2009 to 73.8 million tons in 2014, total captured fishery production has been stationary around 90~93 million tons from 2009 to 2014 (FAO, 2016).
Microbiological water quality of aquaculture systems can ultimately determine the success or failure of an aquaculture enterprise. Microorganisms in aquaculture systems play pivotal roles in economic losses due to factors including low productivity, nutrient cycling, disease control and environmental impacts (Moriaty, 1997). Aquacultured seafood products associated with microbial contamination may not only cause a significant economic loss, but are also potential seafood safety and human health concerns. Fish can acquire pathogenic bacteria from contaminated aquaculture water and highly infectious bacteria or toxin-producing microbes in fish may lead to consumption of unsafe fish (Meals, 2004). Bacteria have been implicated in food-borne illness including infection causing Listeria monocytogenes, Salmonella spp., Escherichia coli, Vibrio spp., Shigella spp., and toxin-producing Staphylococcus aureus and Clostridium botulinum (FAO, 2005).
The incidence of human infections caused by Vibrio spp. increased 141% between 1996 to 1998 and 54% between 2006 to 2008, and 7% between 2011 to 2013 (CDC, 2014). Vibrio may infect those who consume raw or undercooked seafood, as well as fish farm employees who improperly handle or process contaminated fish products (Jahncke, 2007; Iwamoto et al., 2010). V. parahaemolyticus is the most commonly identified Vibrio species, occupying 48% of total cases in the USA (CDC, 2014). V. vulnificus is also a common Vibrio spp. (10% in 2014) associated with seafood-borne illness in the USA (CDC, 2014). They are highly invasive and may cause acute gastroenteritis, septicemia, and can lead to death (Su and Liu, 2007; Jones and Oliver, 2009; Scallan et al., 2011).
Vibrio anguillarum causes hemorrhagic septicemia or vibriosis, which is a severe disease affecting a multitude of fish species, leads economic losses to the worldwide aquaculture industry (Actis et al., 2011). It has been reported that V. anguillarum may be transmitted to fish by contaminated water through the mouth or anus for systemic colonization within the gut (Kanno et al., 1989; Olsson et al., 1996; O'Toole et al., 2004). There have also been reports that skin is an alternative route for the invasion of V. anguillarum into fish (O'Toole et al., 2004).
Many researchers have shown possible types of microbial contaminants in aquaculture systems and the route of infection into the fish, however, subsequent amounts of bacterial residues in fish products have not been fully characterized. Here, the degree of bacterial transmission from Vibrio spp. contaminated aquaculture water to fish was investigated. We challenged fish with a practical dose of three Vibrio spp. (V. anguillarum, V. parahaemolyticus, and V. vulnificus) in an aquaculture system and investigated the transmission of these Vibrio spp. to the fish. The exact amounts of bacterial residues in the fish filets were measured. Using a multiplex real-time PCR, the Vibrio spp. and total bacteria population relationships between aquaculture water and fish products were clarified.
Materials and Methods
Fish and Rearing Condition
Hybrid striped bass (Morone saxatilis × Morone chrysops) were purchased from Delmarva Aquatics (Smyrna, DE, USA). They were held and reared in 17,740 L of fresh water in a closed recirculating aquaculture system (RAS) with an installed sump tank, 1.5 hp water pump, PBF-10 prop washed bead filter, and inline UV sterilizer. The water temperature and the room humidity were maintained at 26 ± 1°C and 60–70%, respectively. Aeration was provided by a central regenerative air blower and individual tank air diffusers and light/dark cycles were set at 12 h. The fish were fed twice daily based on total biomass in the system. Feed-levels began at 0.03% total biomass when the fish were smaller, and reduced to 0.01% as they grew. Diet was Cargill Hybrid Striped Bass Food (Cargill Inc., Minneapolis, MN, USA) including 45% crude protein and 12% crude fat.
Preparation of Vibrio spp.
For the challenge with Vibrio spp. to the aquaculture tanks, 3 Vibrio spp.: V. anguillarum HB155721 was purchased from Carolina Biological Supply Co. (Burlington, NC, USA) and V. parahaemolyticus O1:Kuk, and V. vulnificus MLT1009 were provided from U.S. Department of Agriculture–Agricultural Research Service, Dover, DE (USDA-ARS). V. parahaemolyticus O1:Kuk strain may potentially cause human illness, because this strain is closely related with strain O3:K6 which was associated with quite a number of Asian outbreaks (Depaola et al., 2003). V. vulnificus MLT1009 has VvhA (virulent factor of V. vulnificus) gene that induces autophagy-related cell death (Song et al., 2016). Each was cultivated overnight in Tryptic Soy Broth, (TSB, Carolina Biological Supply Co.) supplemented with 2% additional NaCl at 37°C, and washed twice with normal saline (0.85% NaCl) buffer. The washed Vibrio spp. were suspended in the saline buffer. To enumerate the bacteria, the suspended Vibrio spp. were serial decimal diluted with the saline buffer and plated on Tryptic Soy Agar (TSA, Carolina Biological Supply Co.) supplemented with 2% NaCl. After incubating for 24 h at 37°C, the colonies were counted. The Vibrio spp. were inactivated by UV radiation just prior to the challenge. V. anguillarum and V. vulnificus were irradiated by UV light for 8 h and V. parahaemolyticus was irradiated overnight. These inactivated Vibrio spp. were administered into the experimental aquaculture tanks.
Preparation of Aquarium and Experimental Design
Sixteen hybrid striped bass were transferred from a rearing tank to each experimental tank and adapted to the experimental tank environments with increasing salinity gradually for 2 weeks. The average weight of the fish was ~450 g (ranging from 340 to 567 g). They were held in separate but identical closed RASs each containing 1,005 L of seawater at 32 ppt salinity. Each system consisted of 1 culture tank, seawater pump, solids filter, and covered biofiltration sump tank filled with bio-ball media (Figure 1). The water temperature and room humidity were controlled at 26 ± 1°C and 60–70%, respectively. Aeration was provided by a diaphragm aerator with individual air diffusers placed in the biofilter sump and light/dark cycles were maintained as 12/12 h. The fish were fed a minimal maintenance ration of two pellets per fish every other day during the trials.

Figure 1. Picture of experimental aquaculture system for the Vibrio spp. challenging trial. Seawater stream circulated through the square water quality controller contained solids filter and covered biofiltration sump with bio-ball media, the circular fish culture tank, seawater pump, and again the controller. Four experimental groups (control, group2, group3, and group4) were allocated to four identical and separate experimental tanks.
Four experimental groups (control, group2, group3, and group4) were allocated to four experimental tanks. After the adaptation of the fish in the experimental tanks, 2 fish and 500 ml of water were collected from each experimental tank to determine the baseline data (day 0 sampling). On the day following collection of day 0 samples, heat-inactivated cultures of the three Vibrio spp. were inoculated into experimental tanks except for control group at a final concentration of 2 Log cells ml−1 in group 2, 3 Log cells ml−1 in group 3, and 4 Log cells ml−1 in group 4. After 1 h circulation of the inoculated bacteria, two fish and water samples were collected from each experimental tank as a day 1 sampling. The sampling was repeated exactly at 2, 4, and 6 days. Immediately after removing fish, they were euthanized by a manually applied blunt force trauma (cranial concussion) followed by pithing and were fileted for bacterial analyses. The fish fileting was hygienically conducted at bio safety level 2 clean laboratory. This trial was duplicated by exactly the same procedures. The overall experimental design, including fish sampling, was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Delaware State University.
Random Sampling and Processing from Different Aquaculture Systems
To clarify the correlation of bacterial populations between fish and the quality of their surrounding water, randomized aquaculture fish and water samples were collected from the Aquaculture Research Facility at Delaware State University in Dover, DE, USA and Horn Point Research Facility at University of Maryland in Cambridge, MD, USA. The samples were collected from eight independent aquaculture systems, including five water tanks and three ponds. The fish were mummychog (Fundulus heteroclitus), tilapia (Oreochromis niloticus), hybrid striped bass (Morone Chrysops × Morone saxatilis), blue catfish (Ictalurus furcatus), and striped bass (Morone saxatilus) cultured inside the facility. as Additionally, baitfish, large size channel catfish (Ictalurus punctatus), and small size channel catfish (Ictalurus punctatus) were cultured in outdoor ponds. The size and age of the collected fish were random and widely variable. Mummychog, the smallest fish, was less than 10 cm and blue catfish, the largest one, was about 50 cm. The age of each fish ranged anywhere from 1 month to 2 years old. Two fish and water samples were collected from each tank. The samples were processed similarly to the aquarium experiments described previously and skinless filets were used for quantifying Vibrio spp. and total bacteria.
Quantification of Vibrio spp. and Total Bacteria in Water and Fish Samples
Detection of the three Vibrio spp. and total bacteria in fish filets and aquaculture water samples was carried out as described in a previous study (Kim and Lee, 2014). Briefly, the filets were cut into small pieces and 25 g of fish tissue was homogenized with 75 ml of 0.85% saline buffer by a Bag Mixer (Interscience, St. Nom, France). The homogenized fish filets were centrifuged at 150 × g for 5 min to remove large fish tissue and the supernatants were re-centrifuged at 10,000 × g for 10 min. The pellets were re-suspended and plated on Thiosulfate Citrate Bile Salts Sucrose agar (TCBS; Difco, Sparks, MD, USA) and TSA using Eddy Jet 2 spiral plater (Neutec Group Inc., Farmingdale, NY, USA). Colonies were enumerated on TCBS and TSA at 1 and 3 days incubation, respectively. The pellets were also used for the extraction of bacterial genomic DNA. The DNA was extracted by a boiling method with TZ buffer (20 mg ml−1 Triton X-100 and 2.5 mg ml−1 sodium azide in 0.1 M Tris-HCl, pH 8.0) and 6% chelex solution (Bio-rad Laboratories, Inc., Hercules, CA, USA) from the pellets and precipitated using a Quick-Precip Plus Solution (Edgebio, Gaithersburg, MD, USA) and absolute ethanol (Fisher Scientific, Fair Lawn, NJ, USA). The precipitated DNA was dissolved in nuclease-free water.
The collected aquaculture water samples were also analyzed by culture-dependent enumeration of the bacterial counts using the Eddy Jet 2 spiral plater. In order to retrieve bacteria, the water samples were vacuum filtered through polyethersulfone membrane filters (0.22 μm pore size, 25 mm diameter; Whatman, Buckinghamshire, UK). The filters were soaked in TZ buffer and 6% Chelex solution and then vortexed for 2 min. The bacterial DNA from the filters was extracted by boiling and precipitation methods as described above.
Using the genomic DNA extracted from fish filets and water samples, quantification of Vibrio spp. and total bacteria in the samples were determined using the multiplex real-time PCR assay reported by Kim and Lee (2014). Species specific target genes, tlh for V. parahaemolyticus, toxR for V. anguillarum, vvhA for V. vulnificus were used for detection of each Vibrio spp. and 16S rDNA was used for detection of total bacteria. The 25 μL reaction mixture consisted of 12.5 μL of 2X master mix, 800 nM each of the toxR forward and reverse primers and probe; 200 nM each of the 16S rDNA forward and reverse primers and probe; and 50 nM each of the tlh and vvhA forward and reverse primers and probe. The cycling parameters consisted of a 95°C initial denaturation hold for 2 min followed by 35 cycles of amplification, with each cycle consisting of denaturation at 95°C for 15 s and a combined annealing/extension step at 60°C for 50 s. The detection limits of the multiplex assay were 1 cell ml−1 in seawater, 10 cells g−1 in fish for Vibrio spp. and 4 cells ml−1 in seawater and 40 cells g−1 in fish for total bacteria.
Statistical Analyses
Analysis of variation (ANOVA) was used to evaluate a significant difference between bacterial populations through experimental groups and periods. Tukey post hoc test was applied to determine which groups and days were statistically different. Also, Pearson's correlation was used to assess whether there is a correlation between inoculated bacterial counts in water and the final counts in fish filets. All bacterial concentrations were transformed to log10 scale, and the IBM SPSS Statistics for Windows, Version 22.0 (IBM Corp., Armonk, NY, USA) was used to explore the statistical significance. A confidence interval at the 95% level (P < 0.05) was considered in all cases.
Results
Culture-Dependent Enumeration of Vibrio spp. and Total Bacterial Populations in Fish and Water Samples
To detect culturable bacteria in the fish and water inoculated with UV-irradiated Vibrio spp. in the aquaculture system, the samples were plated on TSA and TCBS agar. The general medium TSA was used to detect total bacteria and selective medium TCBS was for detection of Vibrionaceae bacteria. Through all experimental periods including before and after Vibrio challenging, total bacteria were counted on TSA from 2.3 log to 3.5 log cells ml−1 in the water samples and from 2.2 log to 4.0 log cells g−1 in the fish samples. In the case of Vibrionaceae, between 1.2 and 2.6 log cells ml−1 in the water samples and below 1.7 log cells g−1 in the fish samples were counted on TCBS agar. Some TCBS agar plated fish samples did not detect any colonies. There were no significant differences between control and Vibrio challenged groups determined by culture-dependent method. Also, there was no change of bacterial number in fish and water samples during all experimental periods. Only minor differences among individual water and fish samples were found.
Multiplex Real-Time PCR Detection of Vibrio spp. and Total Bacteria in Fish and Water Samples Challenged Vibrio spp.
The inoculated Vibrio spp. and total bacteria counts in the aquaculture water and fish were detected by multiplex real-time PCR assay. In the water samples, total bacteria counts in control tank had no significant (p = 0.717) change through the experimental periods around 3.2 log cells ml−1 (Figure 2A), whereas, the counts of total bacteria in Vibrio challenged tanks peaked on day 1 and then decreased (Figures 2B–D). The bacterial numbers in the water at day 1 were directly proportional to the amount of challenged Vibrio spp. (Control, 3.13 ± 0.39; Group 2, 3.48 ± 0.67; Group 3, 3.98 ± 0.65, Group 4, 4.81 ± 0.56 log cells ml−1). In the fish filet samples, the number of total bacteria in the control group and challenge group 2 were relatively constant from days 0 through 6 of the experiment remaining around 3.7 log cells g−1 (Figures 2A,B). However, the total bacteria numbers in challenge groups 3 and 4 eventually accumulated from 3.87 ± 0.08 log cells g−1 to 4.76 ± 0.26 log cells g−1 in group 3 and from 3.95 ± 0.12 log cells ml−1 to 5.28 ± 0.20 log cells g−1 in group 4 (Figures 2C,D) over the experiment.
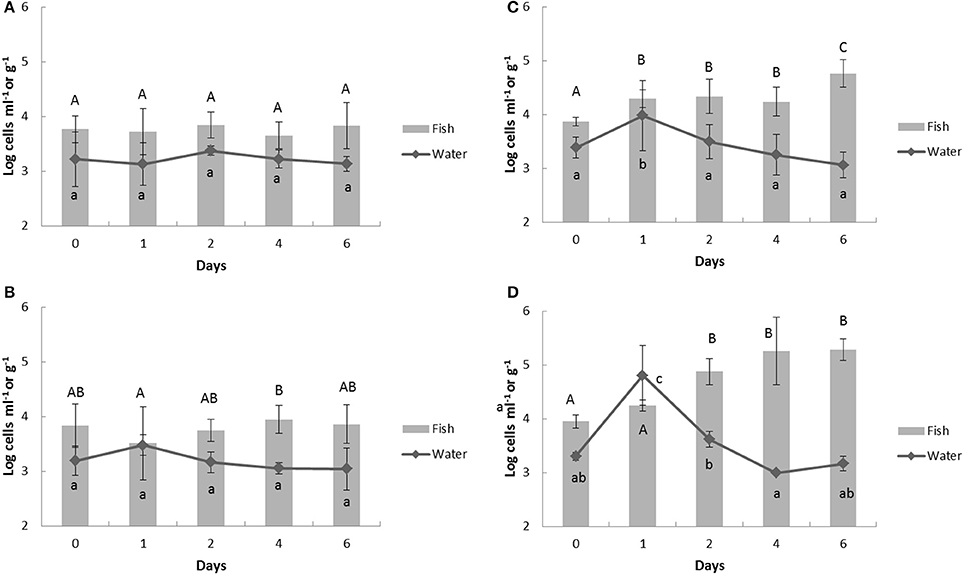
Figure 2. Change of total bacteria counts in aquaculture water and fish detected by multiplex real-time PCR assay in response to heat- inactivated Vibrio spp. challenge. Bold (A) control group; (B) group 2; (C) group 3; (D) group 4. The Vibrio spp. were challenged in the aquaculture water at day 1 and the samples were collected at 24 h before the challenge (day 0), 1 h after challenge (day 1), and further 2, 4, 6 days. Upper case represents significantly different grouping among fish samples and lower case shows the significant grouping between water samples by Tukey test.
V. anguillarum was not detected in our aquaculture systems before challenges (Figure 3). After the challenge, the V. anguillarum showed a sharp peak of bacterial counts at day 1 and the levels in the water were directly proportional with the challenged amounts (Group 2, 1.97 ± 0.12; Group 3, 2.85 ± 0.16; Group 4, 3.85 ± 0.01 log cells ml−1). After day 1, the counts of V. anguillarum quickly decreased in the water and fish (Figure 3). At day 6, V. anguillarum was not detected in the 2 log cells ml−1 challenged fish filets, whereas, 0.78 ± 0.43 log cells g−1 and 1.90 ± 0.21 log cells g−1 of V. anguillarum remained in each in the 3 log and 4 log cells ml−1challenged fish (Figures 3C,D).
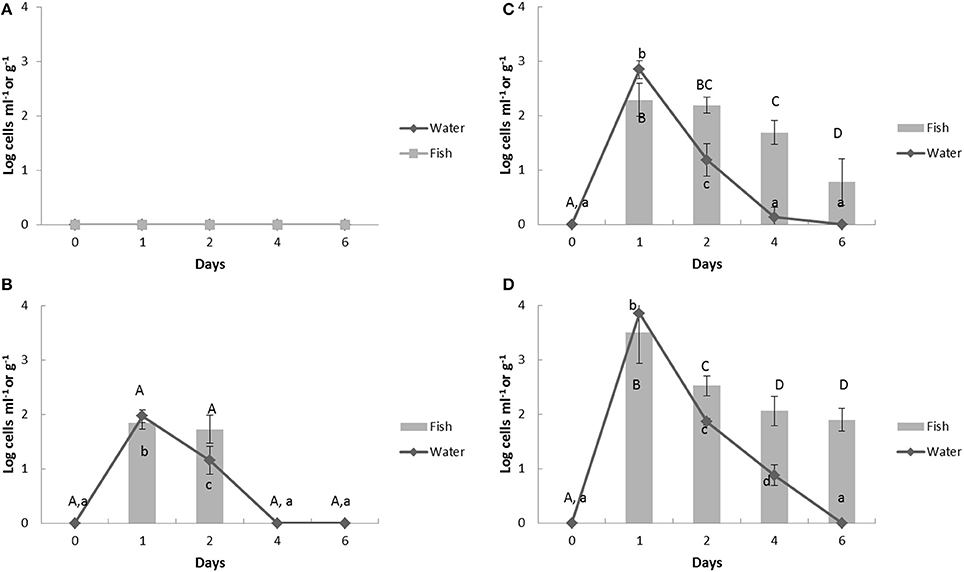
Figure 3. Change of Vibrio anguillarum counts in aquaculture water and fish by multiplex real-time PCR assay in response to heat- inactivated Vibrio spp. challenge. Bold (A) control group; (B) group 2; (C) group 3; (D) group 4. The Vibrio spp. were challenged in the aquaculture water at day 1 and the samples were collected at 24 h before the challenge (day 0), 1 h after challenge (day 1), and further 2, 4, 6 days. The upper case represents significantly different grouping among fish samples and lower case shows the significant grouping between water samples by Tukey test.
Low levels (under 1 log cells ml−1) of V. parahaemolyticus already inhabited in the control aquaculture water, but were not detected in fish (Figure 4A). After the challenge, V. parahaemolyticus counts in the water showed similar patterns with V. anguillarum (Figures 4B–D), which were the highest number at day 1 and then decreasing. However, the V. parahaemolyticus counts in the fish did not decrease but maintained through day 1 to day 6, (Figures 4B–D). On day 6, the residues of V. parahaemolyticus were 2.19 ± 0.23, 2.36 ± 0.53, and 3.21 ± 0.34 log cells g−1 in Group 2, 3, and 4 fish filets, respectively.
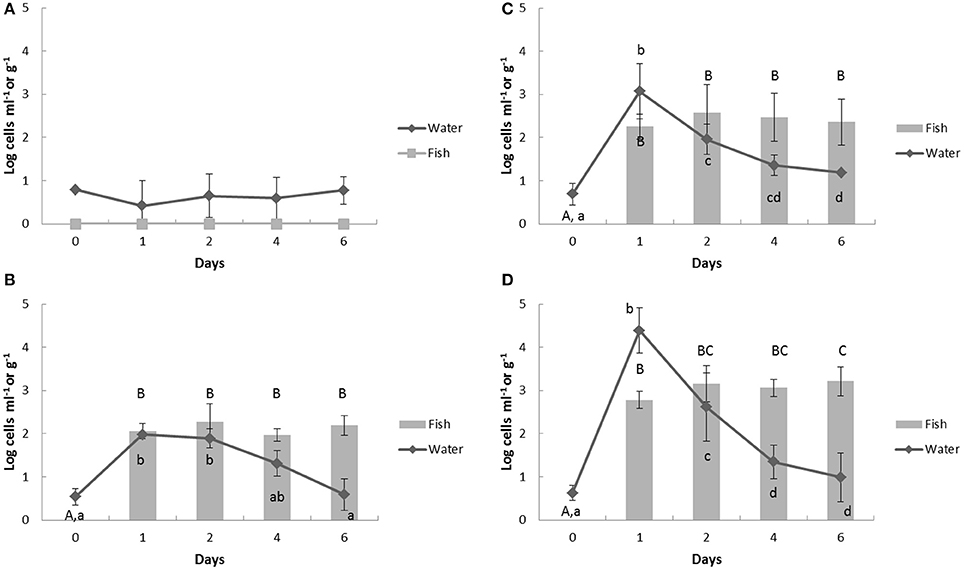
Figure 4. Change of Vibrio parahaemolyticus counts in aquaculture water and fish by multiplex real-time PCR assay in response to heat- inactivated Vibrio spp. challenge. Bold (A) control group; (B) group 2; (C) group 3; (D) group 4. The Vibrio spp. were challenged in the aquaculture water at day 1 and the samples were collected at 24 h before the challenge (day 0), 1 h after challenge (day 1), and further 2, 4, 6 days. The upper case represents significantly different grouping among fish samples and lower case shows the significant grouping between water samples by Tukey test.
Before Vibrio challenge, V. vulnificus existed in the experimental aquaculture systems at relatively high levels, around 2.1 log cells ml−1, and fish, about 2.0 log cells g−1 (Figure 5A). Similar to the other challenges, the levels of V. vulnificus in the water increased on day 1 and then dramatically decreased in groups 3 and 4 (Figures 5C,D). Uniquely, in the group 2 water, the level of V. vulnificus was highest at day 2 and then decreased (Figure 5B). The V. vulnificus counts finally settled back to original levels (day 0, 2.29 ± 0.09; day 1, 4.13 ± 0.77; day 6, 2.42 ± 0.18 log cells ml−1 in group 4) in the water. However, the residues of V. vulnificus in the fish were significantly higher on day 6 than those on day 0 in all challenge groups (group 2, day 0, 2.47 ± 0.16, day 6, 3.08 ± 0.34; group 3, day 0, 2.43 ± 0.25, day 6, 3.39 ± 0.20; group 4, day 0, 2.56 ± 0.09, day 6, 4.14 ± 0.26 log cells g−1; p < 0.001; Figures 5B–D).
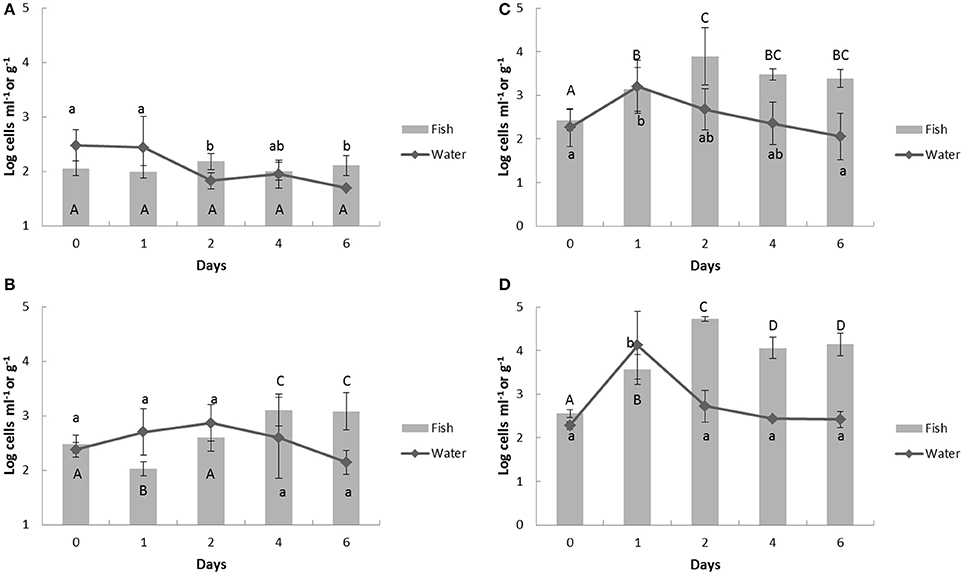
Figure 5. Change of Vibrio vulnificus counts in aquaculture water and fish by multiplex real-time PCR assay in response to heat- inactivated Vibrio spp. challenge. Bold (A) control group; (B) group 2; (C) group 3; (D) group 4. The Vibrio spp. were challenged in the aquaculture water at day and the samples were collected at 24 h before the challenge (day 0), 1 h after challenge (day 1), and further 2, 4, 6 days. The upper case represents significantly different grouping among fish samples and lower case shows the significant grouping between water samples by Tukey test.
Relationship between Challenged Vibrio spp. Counts in Water and the Residual Population in Fish Fillets
On the final day 6, strong correlations were observed between challenged Vibrio amounts in water and their residual population in fish filets (total bacteria, r = 0.794, p < 0.001; V. anguillarum, r = 0.754, p < 0.001; V. parahaemolyticus, r = 0.943, p < 0.001; V. vulnificus, r = 0.942, p < 0.001; Figure 6).
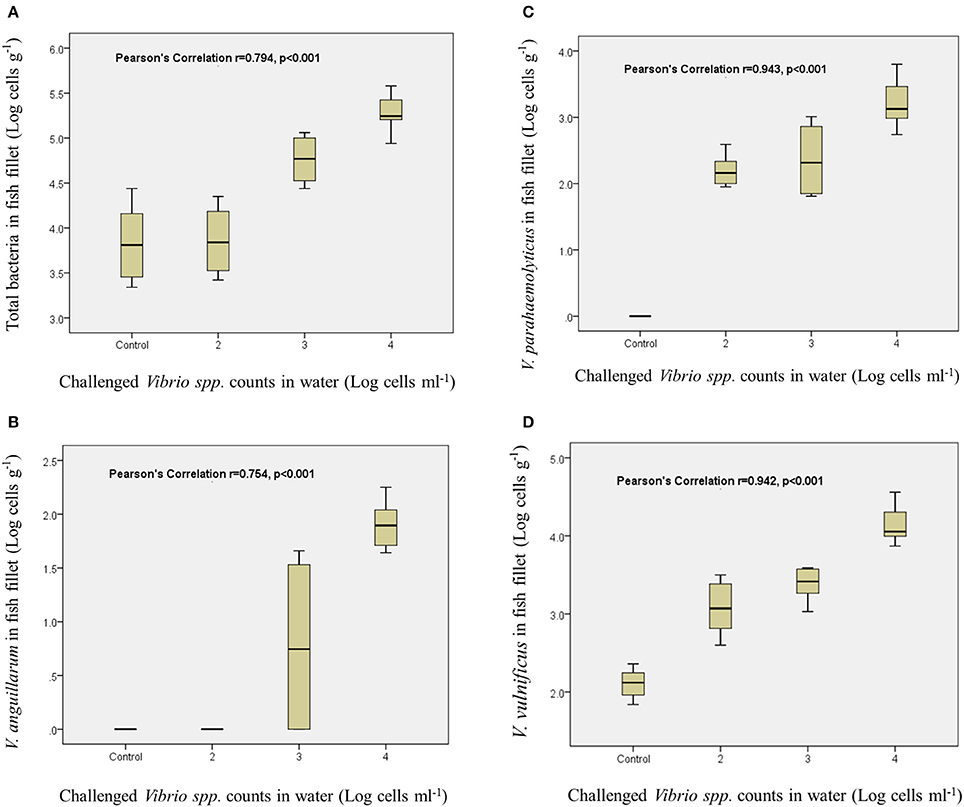
Figure 6. Each figure represents the correlation between challenged Vibrio spp. counts in water and total bacteria counts in fish fillet (A), V. anguillarum counts in fish fillet (B), V. parahaemolyticus counts in fish fillet (C), and V. vulnificus counts in fish fillet (D).
Quantification of Vibrio spp. and Total Bacteria in Random Fish and Water Samples Obtained from Different Aquaculture System
To further clarify the correlation of bacterial populations between fish and its surrounding aquaculture water, eight separate aquaculture systems were assessed. In randomly sampled aquaculture systems, total bacteria were detected from about 3.1 log to 4.1 log cells ml−1 in the water and 2.6 log to 5.1 log cells g−1 in the fish (Table 1). While V. parahaemolyticus and V. vulnificus were detected in water and fish held in aquaculture tanks 1 and 6 by multiplex real-time PCR, Vibrionaceae bacteria in tank 6 was not detected on TCBS agar. In the aquaculture tank 3, only V. vulnificus was detected. Also, in the tank 4 and 5, V. anguillarum and V. vulnificus were detected from aquaculture water and fish by culture independent multiplex PCR but Vibrionaceae was detected from only water by culture dependent assay. There was no detection of Vibrio spp. in aquaculture tanks 2, 7, and 8 (Table 1).
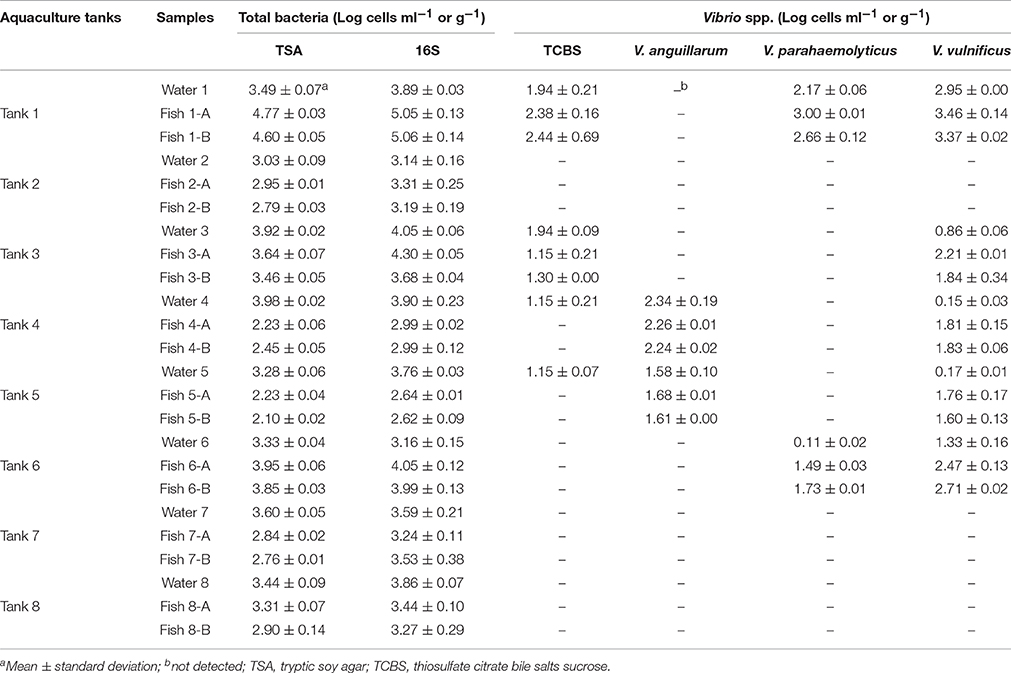
Table 1. Quantification of total bacteria and Vibrio spp. in randomly collected aquaculture fish and their surrounding aquaculture water from different aquaculture systems by culture methods and multiplex real-time polymerase chain reaction (PCR) assay.
Discussion
Wild caught fish production has been slowing down and aquaculture production is continuously increasing (FAO, 2016). Vibrio spp. are present in marine and estuarine environment worldwide (Austin and Austin, 2001) and it is important to public health to monitor Vibrio infection in fish from as a result of contaminated aquaculture water. This study describes the correlation between bacterial counts in aquaculture water and fish in relationship to the levels of Vibrio spp. that are transferred from aquaculture water to cultured fish.
The Vibrio parahaemolyticus and vulnificus strains used for this trial are highly invasive and lethal to humans, while the V. anguillarum strain used in this study threatens only the health of fish not humans. Since the IACUC was seriously concerned about the researchers' safety and animal welfare, we used inactivated Vibrio strains at their insistence. In order to inactivate the Vibrio spp., we chose UV radiation because UV sterilizers are commonly used in general aquaculture farms including our aquaculture facility to maintain water quality. We found that the proper UV radiation time for Vibrio inactivations was 8 h for V. anguillarum and V. vulnificus and overnight for V. parahaemolyticus. At those exposures of radiation, they were not culturable on TCBS and TSA but maintained their intact cell wall/membranes. Also, by means of inactivation of the Vibrio spp., the counts of total and Vibrionaceae bacteria in aquaculture water and fish detected by culture-dependent methods were not significantly changed even though Vibrio spp. were challenged into the aquaculture system.
Unlike the culture-dependent approach, real-time PCR can detect non-culturable or damaged/injured bacteria. Coinciding with the challenge of Vibrio spp. in the aquaculture water, the bacterial counts in the water resulted from the PCR assay increased. The greatest bacterial counts commonly have been shown to peak on the challenge day (day 1) and then the bacterial counts decrease in the aquaculture water (Figures 2–5); it was considered because the challenged Vibrios having UV treatment were gradually removed by the biofilter installed the aquaculture system (Stabili et al., 2016). The secretions and feces produced by the fish as well as natural microbes inhabited in the water might also be other factors to decline populations of the challenged Vibrios.
We have conducted two identical, but independent, trials to confirm our data. Between the two trials, the patterns of bacterial counts in aquaculture water and fish following Vibrio challenges were closely similar. Two of the bacteria, V. parahaemolyticus and V. vulnificus, were already present in our aquaculture system; 1 log cells ml−1 of V. parahaemolyticus and more than 2 log cells ml−1 of V. vulnificus. Only V. vulnificus was detected in the fish filet prior to challenge (Figure 5). These results suggest that aquacultured fish filet might not be contaminated with some Vibrio strains that are present in the aquaculture water at low levels (<1 log cells ml−1), while Vibrio spp. in aquaculture water could transfer to fish when the bacterial counts are over 2 log cells ml−1. In this context, our challenged Vibrio spp., 2 log and more cells ml−1, transmitted to the fish.
Within 1 h of Vibrio challenges to aquaculture water, the Vibrios were quickly transmitted into the fish filets. In other words, it believes that contaminations of Vibrio spp. in the fish are due to the increase of Vibrio counts in the aquaculture water. Once the fish were infected with Vibrios, the Vibrio residues in the fish filet did not decrease as the same levels in the aquaculture water. In case of group 4, V. anguillarum in the water was not detected on day 6, but 1.90 ± 0.21 log cells g−1 of V. anguillarum remained in the fish filet (Figure 3). The level of V. vulnificus in the fish filet was higher on day 2, 4, and 6 compared with day 1 and even total bacteria in the fish filet showed a tendency of bacterial accumulation steadily. Even though this study used UV-inactivated Vibrio spp., V. parahaemolyticus and V. vulnificus especially accumulated or at least persisted in the fish tissues. Therefore, if the Vibrios were active, they might be more invasively infected into the fish tissues. It is well known that fish have immune systems to defend bacterial diseases including humoral factors such as transferring, lysozyme, C-reactive protein, complement, and antibodies, as well as cellular factors, e.g., macrophages, and T lymphocytes (Ellis, 1999). In a previous study, the injection of V. anguillarum in the fish leads to infiltration of acidophilic granulocytes, which have phagocytic reactions (Chaves-Pozo et al., 2005). Also, Vibrio strains may induce antibody production followed by the activation of the classical complement pathway and these reactions result in the bacterial clearing (Ellis, 1999). However, pathogenic Vibrios are known to be resistant to serum bactericidal activity (Trust et al., 1981). Virulent V. anguillarum has shown resistance to complement-mediated bactericidal activity, though production of specific antibodies was induced (Boesen et al., 1999). These resistance properties against antibodies and/or inoculation of inactivated Vibrio strains may cause the maintenance or accumulation of Vibrio counts in the fish filets even though the Vibrios were no longer present at increased levels in the water.
Total bacteria counts in random fish and water samples detected by multiplex real-time PCR assay were similar with the counts by culture-dependent methods and agreed with a previous study (Kim and Lee, 2014). On the contrary, Vibrio counts between the PCR and culture method showed inconsistency. It is likely due to the detection limitations of TCBS medium containing highly selective components, which cannot cultivate some Vibrio spp. such as V. hoolisae and V. anguillarum (Amy et al., 1983; Oliver et al., 1991; Hardy diagnostics, 2013; Kim and Lee, 2014). All randomly assessed aquaculture systems showed similar microbial water quality which contained total bacterial counts between 3.03 and 3.98 log cells ml−1, whereas, the counts of total bacteria in random sampling fish filets were quite variable. All aquaculture water was managed by filtration and irradiation but the aquaculture fish could not be directly controlled. Also, each fish in the randomly selected aquaculture tanks were different species, sizes, and ages. It is supposed that different species or age of fish may result in different levels of infiltration and colonization of bacteria from the surrounding water. For this reason, we could not find any consistency in total bacterial number between fish and their surrounding aquaculture water in randomly collected samples. Nevertheless, Vibrio counts in fish were obviously associated with and directly proportional to Vibrio amounts in their surrounding aquaculture water. For example, as the count of V. anguillarum in tank 4 water was higher than that in tank 5 water, the counts of V. anguillarum in tank 4 fish were higher than in tank 5 fish (Table 1).
Taken together, Vibrio challenge trials showed that Vibrio spp. were transmitted from the aquaculture water to fish at almost the same time with the increase of Vibrio levels in the water. The minimum dose of Vibrio spp. which could contaminate fish filets was about 2 log or more cells ml−1 in aquaculture water. Once the three Vibrio spp. had been transmitted to the fish from the surrounding aquaculture water, the transmitted Vibrio spp. remain as residents in the fish filet for an extended period after which the levels of residues in fish were not directly associated with the amounts of Vibrio in the aquaculture water. The close correlation between the inoculated Vibrio spp. levels in the water and the final total bacteria or Vibrio counts in fish filets was also noteworthy. This result suggested that the more contamination of Vibrio spp. in the aquaculture water, the more total and Vibrio counts would harbor in fish filets.
Consequently, the surveillance of V. anguillarum, V. parahaemolyticus, V. vulnificus, and total bacterial populations in the aquaculture water could be used as an indicator of the microbial contaminations of fish filets The multiplex real-time PCR assay would be a fast and useful tool to detect the Vibrio spp. and total bacteria from the aquaculture water. However, based on data from our study, if the fish were infected with the Vibrio spp. (except V. anguillarum contamination less than 2 log cells mL−1), the Vibrio would remain in the fish filet for an extended period even after the aquaculture water has been purified through a filtration system. Our results suggest that the contamination-prevention of these Vibrio spp. in aquaculture systems should be a priority.
Ethics Statement
All procedures were followed in accordance with the ethical standards of the responsible committee (Delaware State University IACUC, Dover, USA) on the animal experimentation.
Author Contributions
JK participated in all parts of experiments at the aquarium and laboratory including data collections and analysis, and preparation of the manuscript. JL participated in most parts of the works including study design, data analyses and interpretation, and preparation of the manuscript.
Funding
This project was supported by 1890 Capacity Building Grants Program (Award no. 2011-38821-30923 and 2013-38821-21456) from the USDA National Institute of Food and Agriculture.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Actis, L. A., Tolmasky, M. E., and Crosa, J. H. (2011). “Vibriosis,” in Fish Diseases and Disorders, Viral, Bacterial, and Fungal Infections, 2nd Edn., vol. 3, eds P. T. K Woo and D. W. Bruno (Oxfordshire: CABI International), 570–605. doi: 10.1079/9781845935542.0570
Amy, P. S., Pauling, C., and Morita, R. Y. (1983). Starvation-survival of a marine Vibrio. Appl. Environ. Microbiol. 45, 1041–1048.
Austin, B., and Austin, D. A. (2001). “Bacterial fish pathogens,” in Diseases in Farmed and Wild Fish, 4th edn (Chichester: Springer; Praxis Publishing), 24–163.
Boesen, H. T., Pedersen, K., Larsen, J. L., Koch, C., and Ellis, A. E. (1999). Vibrio anguillarum Resistance to Rainbow Trout (Oncorhynchus mykiss) Serum: Role of O-Antigen Structure of Lipopolysaccharide. Infect. Immun. 67, 294–301.
CDC (2014). Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet Surveillance Report for 2014 (Final Report). Atlanta, GA: U.S. Department of Health and Human Services, CDC.
Chaves-Pozo, E., Muñoz, P., López-Muñoz, A., Pelegrín, P., Ayala, A. G., Mulero, V., et al. (2005). Early innate immune response and redistribution of inflammatory cells in the bony fish gilthead seabream experimentally infected with Vibrio anguillarum. Cell Tissue Res. 320, 61–68. doi: 10.1007/s00441-004-1063-7
Depaola, A., Ulaszek, J., Kaysner, C. A., Tenge, B. J., Nordstrom, J. L., Wells, J., et al. (2003). Molecular, serological and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Envron. Microbiol. 69, 3999–4005. doi: 10.1128/AEM.69.7.3999-4005.2003
Ellis, A. E. (1999). Immunity to bacteria in fish. Fish Shellfish Immune 9, 291–308. doi: 10.1006/fsim.1998.0192
FAO (2016). The State of World Fisheries and Aquaculture 2016. Contributing to Food Security and Nutrition for All. Rome: FAO.
FAO (Fisheries and Aquaculture Department) (2005). “Fisheries and Aquaculture topics. Safety of fish and fish products,” in Topics Fact Sheets, ed L. Ababouch (Rome). Available online at: http://www.fao.org/fishery/topic/1522/en
Hardy diagnostics (2013). TCBS (Thiosulfate Citrate bile Salts Sucrose) Agar, Hardy User Group Observer ver. 7.0. Santa Maria, CA.
Iwamoto, M., Ayers, T., Mahon, B. E., and Swerdlow, D. L. (2010). Epidemiology of seafood-associated infections in the United States. Clin. Microbiol. Rev. 23, 399–411. doi: 10.1128/CMR.00059-09
Jahncke, M. (2007). JIFSAN Good Aquacultural Practices Manual, Section 2–Aquaculture Product Safety and Consumer Health, University of Maryland FDA. College Park, MD: JIFRAN University of Maryland.
Jones, M. K., and Oliver, J. D. (2009). Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77, 1723–1733. doi: 10.1128/IAI.01046-08
Kanno, T., Nakai, T., and Muroga, K. (1989). Mode of transmission of vibriosis among Ayu, Plecoglossus altivelis. J. Aquat. Anim. Health 1, 2–6.
Kim, J. Y., and Lee, J.-L. (2014). Multipurpose assessment for the quantification of Vibrio spp. and total bacteria in fish and seawater using multiplex real-time polymerase chain reaction. J. Sci. Food Agric. 94, 2807–2817. doi: 10.1002/jsfa.6699
Meals, D. W. (2004). The Farmer's Guide to Agriculture and Water Quality Issues: Pathogens and Water Quality, 2004, USDA-CSREES, the U.S. EPA's National Agriculture Compliance Assistance Center and North Carolina State University Cooperative Extension. Raleigh, NC: US EPA and NC State University.
Moriaty, D. H. W. (1997). The role of microorganisms in aquaculture ponds. Aquaculture 151, 333–349. doi: 10.1016/S0044-8486(96)01487-1
Oliver, J. D., Nilsson, L., and Kjelleberg, S. (1991). Formation of nonculturable Vibrio vulnificus cells and its relationship to the starvation state. Appl. Environ. Microbiol. 57, 2640–2644.
Olsson, J. C., Jöborn, A., Westerdahl, A., Blomberg, L., Kjelleberg, S., and Conway, P. L. (1996). Is the turbot, Scophthalmus maximus (L.), intestine a portal of entry for the fish pathogen Vibrio anguillarum. J. Fish Dis. 19, 225–234. doi: 10.1111/j.1365-2761.1996.tb00129.x
O'Toole, R., von Hofstena, J., Rosqvista, R., Olssonc, P.-E., and Wolf-Watz, H. (2004). Visualisation of Zebrafish infection by GFP-labelled Vibrio anguillarum. Microb. Pathogenesis 37, 41–46. doi: 10.1016/j.micpath.2004.03.001
Sapkota, A., Sapkota, A. R., Kucharski, M., Burke, J., McKenzie, S., Walker, P., et al. (2008). Aquaculture practices and potential human health risks: Current knowledge and future priorities. Environ. Int. 34, 1215–1226. doi: 10.1016/j.envint.2008.04.009
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Song, E. J., Lee, S.-J., Lim, H. S., Kim, J. S., Jang, K. K., Choi, S. H., et al. (2016). Vibrio vulnificus VvhA induceds autophagy-related cell death through the lipid raft-dependent c-Src/NOX signaling pathway. Sci. Rep. 6:27080. doi: 10.1038/srep27080
Stabili, L., Licciano, M., Gravina, M. F., and Giangrande, A. (2016). Filtering activity on a pure culture of Vibrio alginolyticus by the solitary ascidian Stylea plicata and the colonial ascidian Polyandrocarpa zorritensis: a potential service to imporuve microbiological seawater quality economically. Sci. Total Environ. 573, 11–18. doi: 10.1016/j.scitotenv.2016.07.216
Su, Y. C., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Keywords: total bacteria, Vibrio spp., aquaculture, fish, water quality, real-time PCR
Citation: Kim JY and Lee J-L (2017) Correlation of Total Bacterial and Vibrio spp. Populations between Fish and Water in the Aquaculture System. Front. Mar. Sci. 4:147. doi: 10.3389/fmars.2017.00147
Received: 07 February 2017; Accepted: 02 May 2017;
Published: 19 May 2017.
Edited by:
Pedro Fernandes, Universidade de Lisboa, PortugalReviewed by:
Clélia Neves Afonso, Escola Superior de Turismo e Tecnologia do Mar do Instituto Politécnico de Leiria, PortugalKonstantinos Ar. Kormas, University of Thessaly, Greece
Sucharit Basu Neogi, International Centre for Diarrhoeal Disease Research, Bangladesh
Copyright © 2017 Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jung-Lim Lee, amxlZUBkZXN1LmVkdQ==
 Ji Yeun Kim
Ji Yeun Kim Jung-Lim Lee
Jung-Lim Lee