- 1Department of Biological Sciences, University of Southern California, Los Angeles, CA, United States
- 2School of Molecular Sciences, Arizona State University, Tempe, AZ, United States
- 3Department of Earth Sciences – Geochemistry, Utrecht University, Utrecht, Netherlands
Nutritional modes of unicellular eukaryotes range from pure photoautotrophy of some phytoplankton to pure heterotrophy of species typically called protozoa. Between these two extremes lies a functional continuum of nutrient and energy acquisition modes termed mixotrophy. Prymnesium parvum is an ecologically important mixotrophic haptophyte alga that can produce toxins and form ecosystem disruptive blooms that result in fish kills and changes in planktonic food web structure. We investigated carbon and nitrogen acquisition strategies of single cells of P. parvum using a combined experimental-imaging approach employing labeling of live cells with stable isotope tracers (13C and 15N) followed by measurement of cellular isotopic ratios using nanometer-scale secondary ion mass spectrometry (NanoSIMS). With this method, we were able to quantify the relative contributions of photosynthesis and heterotrophy to the nutrition of the alga. Our results suggest that P. parvum relies on predation primarily for nitrogen, while most carbon for cellular building blocks is obtained from inorganic sources. Our analysis further revealed that nitrogen assimilation can vary up to an order of magnitude among individual cells, a finding that would be difficult to determine using other methods. These results help to improve our understanding of mixotrophy across the enormous diversity of eukaryotes, one cell and one species at a time.
Introduction
It is often assumed that the vast diversity of the eukaryotic domain can be neatly divided into plant-like (exclusively photoautotrophic) or animal-like (exclusively heterotrophic) categories, with the few taxa that combine both capabilities (i.e., mixotrophy) viewed as curiosities. However, accumulating research and recent reviews on protists offer a strikingly different view of our domain (Raven et al., 2009; Mitra et al., 2016; Caron, 2017; Stoecker et al., 2017). Mixotrophy is very common across the eukaryotic tree (Raven et al., 2009), is ubiquitous in aquatic ecosystems, and is of great importance in aquatic biogeochemical cycles where it influences carbon and energy flow in food webs (e.g., Sanders, 2011; Mitra et al., 2014; Ward and Follows, 2016). In recent attempts to include mixotrophy in marine global biogeochemical models, mixotrophy has been shown to increase primary productivity where inorganic nutrients are limiting, to result in larger average cell size of plankton (diameter), and to increase the proportion of carbon exported into the deep ocean (Mitra et al., 2014; Ward and Follows, 2016). Further, mixotrophy is a key strategy influencing the distributions and abundances of planktonic protistan taxa (Jones, 1994, 2000; Hartmann et al., 2013; Leles et al., 2017), including many harmful algal bloom (HAB) species (Tillmann, 2003; Burkholder et al., 2008).
The term mixotrophy has been broadly applied to protists that combine photoautotrophic with heterotrophic nutrition (primarily phagotrophy) (Flynn et al., 2013; Mitra et al., 2016). Far from a curiosity, many phytoplankton taxa have phagotrophic members, including numerous dinoflagellates, chrysophytes, cryptophytes, prasinophytes, and haptophytes (Sanders and Porter, 1988; Raven et al., 2009; Flynn et al., 2013). The great abundances of mixotrophs in most aquatic systems suggest this nutritional mode should be considered the rule rather than the exception (Matantseva and Skarlato, 2013).
The range of energy and nutrient acquisition strategies exhibited by three mixotrophic algae was recently illustrated by a comparative analysis of gene expression under different light regimes and prey availabilities for cultures of the haptophyte, Prymnesium parvum, and two chrysophyte algae (Dinobryon sp. and Ochromonas sp., strain CCMP1393). P. parvum and Dinobryon sp. showed differential expression of thousands of genes between light and dark regimes, while Ochromonas sp. showed differential expression of only ~50 genes between the two regimes (Liu et al., 2016). These observations led to the conclusion that P. parvum and Dinobryon sp. are both predominantly photosynthetic, but the study also revealed subtle differences in nutritional strategy between those two algae (Liu et al., 2016). Ochromonas strain 1393, on the other hand, is more heterotrophic as shown by a less dramatic response of gene expression to the light regime. A study of another Ochromonas species (strain BG-1) also demonstrated the predominantly heterotrophic nature of the alga by showing that the availability of bacterial prey led to the differential expression of >7X more genes than the availability of light (Lie et al., 2017).
The mixotrophic alga P. parvum (Carter, 1937) is a member of the Haptophyta (prymnesiophytes), a large and extremely abundant group of photosynthetic plankton including coccolithophorids (e.g., Emiliania huxleyi), whose chloroplasts arose via endosymbiosis with a red alga (Yoon et al., 2002; Burki et al., 2016). P. parvum is a small (8–16 μm), ellipsoid cell with phagotrophic ability assisted by a tube-like feeding appendage (the haptonema) emerging between two anterior flagella. It occurs in freshwater, brackish, and coastal marine waters worldwide (Green et al., 1982; Moestrup, 1994; Barkoh and Fries, 2010), and is classified as an Ecosystem Disruptive Algal Bloom (EDAB) species (Sunda et al., 2006), due to its production of a variety of potent toxins (e.g., prymnesins) (Rasmussen et al., 2016). Some of these blooms result in massive fish kills and deaths of other gill-breathing organisms as well as other algae, protists, and bacteria (Tillmann, 2003; Hambright et al., 2015; Roelke et al., 2016). Such devastating harmful blooms have undergone a dramatic range expansion in recent decades, especially in western North America freshwater systems (Sallenave, 2010; Roelke et al., 2016).
P. parvum appears to rely on phagotrophy to supplement a predominantly photoautotrophic nutritional mode (Carvalho and Granéli, 2010; Granéli et al., 2012; Liu et al., 2016). The alga is capable of photosynthetic growth in axenic culture but cannot maintain growth or survival in continuous darkness, even with an abundance of prey (Brutemark and Granéli, 2011). Experimental and observational studies have indicated that mixotrophy and toxin production in this species may be particularly important under inorganic nutrient limitation (Granéli et al., 2012; Liu et al., 2015b; Roelke et al., 2016). Tillmann (2003) provided evidence that P. parvum toxins are crucial to its mixotrophic strategy, and are used to immobilize and kill prey prior to ingestion. Recent transcriptomic evidence has supported the contention that inorganic nutrient limitation stimulates toxin production in P. parvum (Manning and La Claire, 2010). Based on comparison of gene expression under different growth conditions, such work has also intimated that P. parvum may take up organic carbon from both ciliate and bacterial prey for energy and carbon skeletons (fatty acids), and obtain nitrogen (amino acids) from ciliate prey and iron from bacteria (Liu et al., 2015a). Transcriptomic evidence also suggests that P. parvum may supplement energy from photosynthesis with organic carbon from prey capture, especially when grown under light limitation (Liu et al., 2016).
These studies have provided insight into the general trophic activities of P. parvum, but a complementary line of evidence quantifying the behavior of individual cells is needed to test and further refine our understanding of the specific contributions of mixotrophy for this species and to serve as a source of new hypotheses generated at the single-cell level. The experimental-imaging approach combining exposure of live cells to substrates enriched in stable isotope tracers followed by nanometer-scale secondary ion mass spectrometry (NanoSIMS) imaging of the cells is now well-established as a powerful tool in microbial ecology, offering the unique capability of imaging and quantifying isotopic ratios at very high spatial resolution (up to 50 nm lateral resolution). This technique has been successfully applied in numerous studies to investigate uptake, metabolism, and transfer of various isotopically labeled substances in individual cells, symbiotic consortia, and complex natural microbial communities (Orphan et al., 2001; Lechene et al., 2007; Carpenter et al., 2013; Kopp et al., 2013; Bonnet et al., 2016; Tai et al., 2016). This approach was recently applied for the first time to the study of a chrysophyte mixotrophic alga (Ochromonas sp. strain BG-1) to demonstrate that this species relies heavily on carbon and nitrogen uptake from prey for energy and nutrients, while photosynthesis represents a minor contribution to its overall nutrition (Terrado et al., 2017).
We investigated the mixotrophic potential of P. parvum at the single cell level by comparing carbon and nitrogen uptake from prey and inorganic nutrients using stable isotope tracers and imaging of cells with NanoSIMS. The primary goal of this study was to quantify and compare the amounts of carbon and nitrogen that P. parvum obtains from inorganic sources (using 13C-bicarbonate and 15N-nitrate) vs. nutrients obtained from live prey (using a 13C- and 15N-labeled ciliate protist, Uronema marina, ranging from 18 to 25 μm in length). Variability among P. parvum cells with respect to C and N uptake within each experimental treatment was also examined to gain insight on potential variability in natural populations. Whole P. parvum cells were imaged using NanoSIMS at time zero and 48 h, shortly after all prey were killed by the alga. We also imaged cells in an unlabeled control treatment to assess the effects of cell topography on the measured isotopic fractionation. This study provides important insights into how mixotrophy contributes to energy and nutrient budgets in P. parvum, an organism of considerable ecological interest.
Materials and Methods
Organisms and Cultures
Prymnesium parvum strain UOBS-LP0109 (Texoma1) was isolated from Lake Texoma, Oklahoma, USA, and rendered axenic by repetitively micropipetting single cells through rinses of sterile medium. The alga was grown in sterile L1 medium without silica following the recipe of the Provasoli-Guillard National Center for Marine Algae and Microbiota protocol (see https://ncma.bigelow.org/algal-recipes) at 18‰ (1:1 0.2 μm-filtered, aged natural seawater: ultrapure water (Barnstead GenPure xCAD Plus, Thermo Fisher Scientific, Waltham, USA). P. parvum is a euryhaline alga, but has been demonstrated to be most toxic at low-to-intermediate salinities (Roelke et al., 2016). For that reason we chose to conduct all experimental work at 18‰. The axenicity of the P. parvum culture used for the experiment was confirmed by the lack of bacterial or fungal growth after inoculating 3 ml of the culture into 7 ml of 0.5% yeast extract broth at 18‰ and observing for 2 weeks.
The bacterivorous ciliate U. marina was originally isolated from Buzzards Bay, Massachusetts, USA, and was used as prey for P. parvum. It is also a relatively euryhaline species, and was chosen as prey because it grows readily at 18‰ and has been previously shown to be suitable prey item for P. parvum (Liu et al., 2015a). The ciliate was maintained on its attendant bacterial flora by periodic subculturing into the same medium used to culture P. parvum, with addition of yeast extract (~0.02% final concentration) and a sterilized rice grain to promote bacterial growth.
Ciliate prey for the NanoSIMS experiments were grown by subculturing ciliates in a medium designed to promote the growth of their attendant bacteria (final concentrations: 362 μM NaH2PO4·H2O; 501 μM NH4Cl; 0.04% glucose; 18‰). Labeled ciliates (13C and 15N) were grown with labeled bacteria in medium made with 15NH4Cl (98 atom % 15N; Sigma-Aldrich, St. Louis, USA) and D-glucose-1-13C (99 atom % 13C, Sigma-Aldrich). Both labeled and unlabeled ciliates were cleaned of remaining attendant bacteria, and concentrated prior to use as prey in the NanoSIMS experiment through 3 rounds of centrifugation (1,500 g for 5 min at 4°C; Sorvall RC5C plus, Thermo Fisher) and rinsed using sterilized 18‰ water (Liu et al., 2015a). Ciliates remained alive and highly motile following this procedure.
Stable Isotope Labeling Experiments
Experiments were designed to determine the degree to which P. parvum acquired carbon and nitrogen directly from the ciliate prey, relative to these elements from the dissolved inorganic pools. This was done by comparing reciprocal labeling of the carbon and nitrogen pools (inorganic pools of carbon and nitrogen, or these elements contained in ciliate prey). Predator (P. parvum) and prey (U. marina) abundances were chosen based on preliminary studies of the ability of the smaller alga (cell volume ~200 μm3) to attack and subdue the larger ciliate prey (cell volume ~800–1,000 μm3). Our observations in the laboratory revealed that several P. parvum are required to immobilize and kill a ciliate, therefore we conducted simple bioassay experiments with a set number of algae (~2 × 105 ml−1) but varying abundances of ciliates in order to determine the optimal ratio of P. parvum to U. marina that led to the complete elimination of ciliates within 48–72 h (unpublished data). We chose this time frame to ensure that all prey would be killed by the time of stable isotope analysis. Based on that work, we chose starting concentrations of P. parvum and ciliates of ~2 × 105 ml−1 and ~1 × 104 ml−1, respectively. Furthermore, to our knowledge, cannibalism is not known in P. parvum. We have grown it to very high abundances (>106/ml) and we have not witnessed it, even after extensive behavioral monitoring with microscopy.
Batch (50 ml) cultures of P. parvum were grown in 125 ml Erylenmeyer flasks at 18°C and 200 μE m−2 s−1 light intensity (12:12 h light:dark cycle) on an orbital shaker (60 rpm), and was used in all experiments while still in exponential growth. Cultures were grown in the same medium for maintaining P. parvum, with an addition of 95 μM NaHCO3. No attempt was made to limit inorganic nutrients in the experiments for three reasons. First, in order to directly compare the outcomes of the treatments with labeled inorganic carbon and nitrogen to treatments with labeled ciliate carbon and nitrogen, it was necessary to provide the same amounts of inorganic nutrient or ciliates in all treatments (labeled or unlabeled). Second, release of inorganic carbon or nitrogen when isotope-labeled ciliate prey are killed could be confounded by subsequent uptake (i.e., erroneously attributed to direct uptake from prey). Maintaining high concentrations of unlabeled inorganic carbon and nitrogen in the medium at all times minimized this potential artifact. Finally, our preliminary work (noted above) demonstrated that our strain of P. parvum remained toxic in the presence of inorganic nutrients and therefore there was no reason to limit nutrients to stimulate predation.
Four experimental treatments were carried out. These included two unlabeled treatments and two combining 13C and 15N labeled inorganics or 13C and 15N labeled prey. Treatments 1–3 were performed in triplicates:
• Treatment 1 used labeled ciliates (as described above) + unlabeled inorganics.
• Treatment 2 used unlabeled ciliates + labeled inorganics [NaH13CO3 (98 atom % 13C, Sigma-Aldrich) was added as 100% of the total bicarbonate (95 μM) while Na15NO3 (98 atom % 15N, Sigma-Aldrich) was added as 50% of the total nitrate (441 μM Na15NO3 and 441 μM NaNO3)].
• Treatment 3 used unlabeled ciliates + unlabeled inorganics. Material from this treatment was not imaged in NanoSIMS but was included in a comparison of growth rates across the first three treatments.
• Treatment 4 used unlabeled ciliates + unlabeled inorganics. This treatment was run for a duration of 30 min, and was included to ensure sufficient material to test sample preparation procedures for field emission scanning electron microscope (FESEM) and NanoSIMS, and to quantify the effects of topography on isotopic fractionation. Based on evidence that P. parvum attacks prey very quickly (Tillmann, 2003), we confirmed beforehand that this was a suitable duration to observe predation, but not so long that ciliates would be completely consumed.
Treatments 1–3 were sampled every 24 h for the determination of P. parvum and ciliate densities. Aliquots for cell counts (3 ml) were preserved with Lugol's solution (final concentration 2.5%) and enumerated using a Palmer-Maloney counting chamber at 200 x on a compound light microscope (BX51; Olympus, Waltham, USA). The growth rate of P. parvum was calculated as the slope of natural log algal abundance against time (72 h).
NanoSIMS samples (5 ml aliquots) were taken from Treatment 1 and Treatment 2 immediately after the addition of labeled inorganics or labeled ciliates to P. parvum cultures (T0) and 48 h later (T48). The samples were preserved with glutaraldehyde at a final concentration of 1% and stored at 4°C.
Hence, five samples were prepared for analysis using NanoSIMS: Treatments 1 and 2, each at time zero (T0) and 48 h (T48) time points, and Treatment 4 (unlabeled control):
1) T0 labeled ciliates
2) T0 labeled inorganics
3) T48 labeled ciliates
4) T48 labeled inorganics
5) Treatment 4 (unlabeled control)
Specimen Fixation and Preparation for NanoSIMS
Fixed subsamples were pipetted onto 13 mm diam., 1.2 μm pore size Millipore polycarbonate filters (Billerica, MA, USA) held in Millipore Swinnex cartridges between two Teflon gaskets. Three of these filter-cartridge assemblies were affixed to 10 ml plastic syringes to process material from each sample. Approximately 500 μl of material was pipetted onto each filter to ensure deposition of enough P. parvum and U. marina cells without clogging the filters. Material on filters was then rinsed three times in PBS buffer to remove excess glutaraldehyde, and dehydrated in a series of 50, 70, 90, and 3X 100% ethanol. Rinses and dehydration were carried out by removing the cartridge from the syringe at the end of each step, quickly drawing the next solution into the syringe, reattaching it to the cartridge, and pushing the liquid through the filter using the syringe plunger (i.e., not by gravity filtration). Filters were then removed from the cartridges while immersed in 100% ethanol and dried in a Tousimis Autosamdri 815 carbon dioxide critical point dryer (Rockville, MD, USA). Dried filters were affixed to 12.5 mm aluminum pin mount stubs with conductive carbon double-stick tape and coated with ~5–10 nm of platinum in a Cressington 108 sputter coater (Watford, UK). Prepared material on stubs was examined with a JEOL 7001 FESEM (Tokyo, Japan) to ensure adequate cellular preservation and abundance prior to NanoSIMS measurements. This sample preparation method has previously been shown to be suitable for preserving cell ultrastructure and retaining 13C and 15N signal within fixed cells (Carpenter et al., 2013; Tai et al., 2016).
NanoSIMS Measurements and Image Analysis
Measurements of carbon and nitrogen isotopes in P. parvum and U. marina were carried out using a Cameca 50L NanoSIMS high resolution imaging mass spectrometer (Gennevilliers, France) at Arizona State University. The instrument was operated in Cesium mode (Cs+ primary ion beam), which maximizes yield of negatively charged secondary ions from the sample surface. Electron multiplier detectors 1–5 were set to collect the following species: 12C−, 13C−, 12C14N−, 12C15N−, and 31P−. For each session, prior to collection of data from samples, tuning, and measurements were made on a cyanoacrylate standard. Target cells for NanoSIMS imaging were selected by using the CCD camera to navigate to areas on the specimen mount surface with abundant cells and a minimal filter topography. This preliminary observation was followed by higher magnification observation and final selection using the secondary electron image and/or the 12C14N− secondary ion image. For imaging of the two T0 samples, care was taken to image P. parvum and U. marina in separate fields of view (FOV), to ensure that isotopically enriched material from labeled ciliates did not become redeposited upon nearby P. parvum cells, thus affecting measurements of those cells.
Data from samples were collected using a 75 nm diameter primary beam with 2–4 pA current measured at the sample (FC0 Faraday cup). Images ranged from 5–40 μm rasters, with dwell times of 10,000 to 15,000 μs/pixel and 2–40 planes. Upon selecting each FOV, secondary ion beam centering and focusing of secondary and primary ion beams with EOS and EOP (immersion lenses) were carried out. Mass peaks on each of the five electron multiplier detectors were checked in High Mass Resolution (HMR) scans. Prior to data collection, presputtering was conducted on an area slightly larger than the selected FOV using high L1 lens setting, and D1-0 aperture for a beam current of a few nA.
Images were analyzed using Look@NanoSIMS (Polerecky et al., 2012) and Cameca WinImage (Gennevilliers, France). Corrections for dead time and quasi-simultaneous arrival (QSA) were applied and drift-corrected planes were accumulated (combined into a single image) for further analysis. Regions of interest (ROIs) were hand-drawn around individual P. parvum cells, or portions of U. marina cells in the accumulated images, and 13C and 15N abundances in individual cells were quantified using the secondary ion counts accumulated over all pixels in the respective ROI as 13C/(12C+13C) and 12C15N/(12C14N+12C15N), respectively. Statistical tests comparing isotopic ratios in various pairs of samples [e.g., 13C/(12C+13C)] in T48 labeled inorganic vs. T0 labeled inorganic) were conducted in Look@NanoSIMS. Data were first analyzed with Levene's test to check for equal variances (homoscedasticity). Those with equal variances were analyzed with ANOVA, while the others were analyzed with the Kruskal-Wallis test.
Results
Growth of P. parvum and Ciliate Abundances
The growth rates of P. parvum over the 72 h period were similar among the different treatments (p > 0.05 One-way ANOVA), yielding averages of 0.48 ± 0.02 d−1, 0.57 ± 0.14 d−1, and 0.54 ± 0.05 d−1 for Treatment 1, 2, and 3, respectively. The predation of ciliates occurred rapidly. The abundances of ciliates were reduced by 70% after 24 h, and no ciliates were observed after 48 h.
Field Emission Scanning Electron Microscope (FESEM) Imaging
Chemically fixed, critical point dried, and sputter coated P. parvum cells, and those of its prey, U. marina, from each of the four isotopically labeled samples, plus an unlabeled control (Treatment 4; see Methods), were observed with FESEM prior to NanoSIMS imaging. Cellular preservation of both species was observed to be generally good, with most cells remaining intact, and retaining good membrane integrity and good overall shape, including cilia or flagella (Figure 1). Both species were abundant in the two T0 samples, and in the unlabeled control. However, U. marina was not observed in either of the two T48 samples with FESEM, nor in NanoSIMS secondary electron or ion images, due to predation by P. parvum.
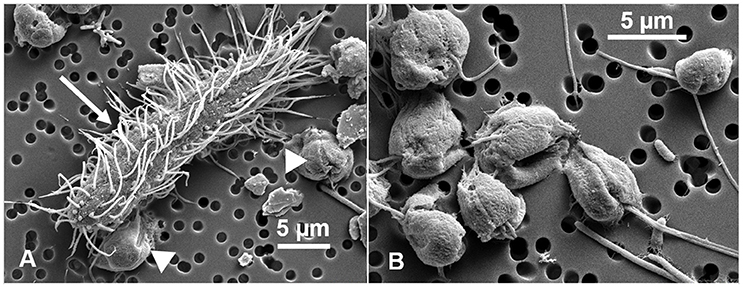
Figure 1. Scanning electron micrographs of the organisms used in these experimental treatments. Cells shown here were sampled from the unlabeled control treatment (see Methods). (A) Uronema marina (arrow), used as prey for Prymnesium parvum (arrow heads). (B) P. parvum cells.
NanoSIMS Imaging—T0 and Unlabeled Control Samples
Because the P. parvum cells are relatively large, 20 whole P. parvum cells from the unlabeled control (Treatment 4) were imaged using NanoSIMS to assess the effect of topography on the measured 13C/C and 15N/N ratios. The effect was relatively minor for 13C, with the 13C/C ratio varying by about 5% (mean = 0.0112, SD = 0.0005), whereas it was substantial for 15N, with the 15N/N ratio varying by about 55% (mean = 0.00367, SD = 0.00200) (Figure 2). This variation was taken into account when comparing enrichments in samples incubated with the label.
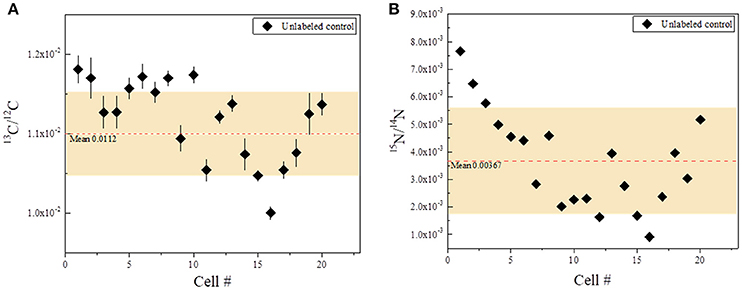
Figure 2. Carbon and nitrogen isotopic ratios from the unlabeled control (Treatment 4), showing effects of topography on isotopic fractionation. Each data point represents a single, whole P. parvum cell (20 cells total). Red dashed lines indicate the mean value for each ratio. The shaded area indicates one standard deviation for all data points. Bars represent standard errors. Where bars are not visible, it indicates that the error is smaller than the data point. (A) 13C/12C ratios. Mean ratio (red dashed line) = 0.0112. (B). 15N/14N ratios. Mean ratio (red dashed line) = 0.00367.
Four to five whole P. parvum cells were imaged in each of the two T0 samples (T0 labeled inorganics and T0 labeled ciliates). All of these cells fell within the ranges for both isotopes obtained from the unlabeled control sample with the exception of a single cell from the T0 labeled ciliate sample, which was slightly above this range for both ratios (1.21 × 10−2 for 13C/C and 8.49 × 10−3 for 15N/N) (Figure 3). However, statistical analysis showed no significant difference in 13C/C in P. parvum between the T0 inorganic labeled and unlabeled samples, or between T0 ciliate labeled and unlabeled samples (Table 1). However, the 15N/N ratios in P. parvum cells from the T0 ciliate labeled treatment were significantly, although only slightly, larger than in the unlabeled control (Table 1).
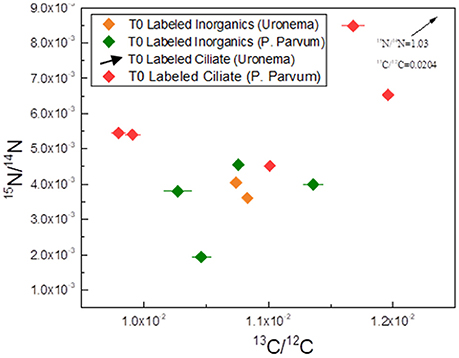
Figure 3. Carbon and nitrogen isotopic ratios measured from treatments 2 and 3—Labeled Inorganics and Labeled Ciliates, respectively—at Time point zero (T0). Each data point represents a single, whole P. parvum cell, or an approximate half of a U. marina cell. The arrow indicates a single U. marina cell from T0 Labeled Ciliate sample with a 15N/14N ratio exceeding the range of the graph. This ratio is indicated, and the position of the point along the X-axis (13C/12C) is indicated by the arrow. Note that the range for each ratio is the same as in Figure 2 to illustrate that the T0 samples are similar to the unlabeled controls in their isotopic composition (except for the U. marina cell indicated by the arrow).
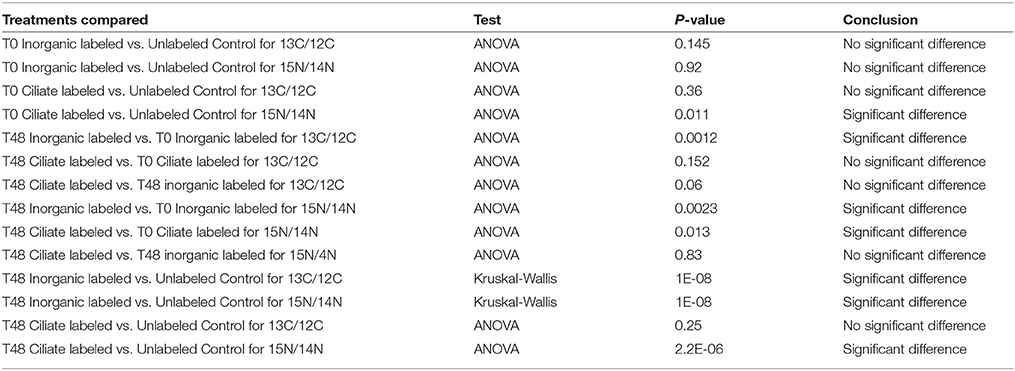
Table 1. Results of statistical tests (ANOVA or Kruskal-Wallis) for comparisons of different experimental treatments.
Two U. marina cells were imaged in the T0 labeled inorganic sample, and their 13C/C and 15N/N ratios fell within the range of values obtained for the unlabeled control (Figure 3). In contrast, two U. marina cells imaged for the T0 labeled ciliate treatment showed a relatively small but significant enrichment in 13C (mean 13C/C of 0.0185) and a substantial enrichment in 15N (mean 15N/N of 0.386).
NanoSIMS Imaging—T48 Samples
Twenty-five whole P. parvum cells were imaged with NanoSIMS for the T48 labeled inorganic sample, while 22 whole P. parvum cells were imaged for the T48 labeled ciliate sample (no ciliates remained in either T48 sample). Carbon and nitrogen isotopic ratios from the two T48 samples and the unlabeled control are plotted in Figure 4, while representative NanoSIMS images are shown in Figure 5.
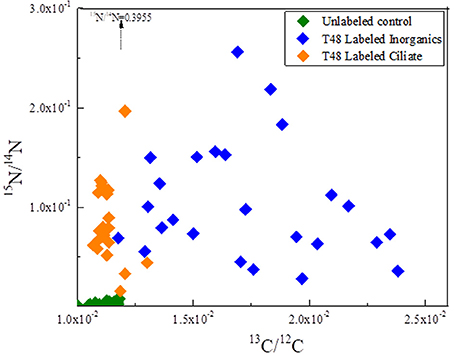
Figure 4. Carbon and nitrogen isotopic ratios from the two T48 samples and the unlabeled control. Each data point represents a single, whole P. parvum cell (Ciliates were not observed in T48 samples.) The arrow indicates a single P. parvum cell from T48 Labeled Ciliate sample with a 15N/14N ratio exceeding the range of the graph. This ratio is indicated, and the position of the point along the X-axis (13C/12C) is indicated by the arrow. The X-axis intersects the Y-axis at the mean 15N/14N ratio, while the Y-axis intersects the X-axis at the mean 13C/12C ratio. Some points from the unlabeled control are thus not visible and lie below the Y-axis boundary.
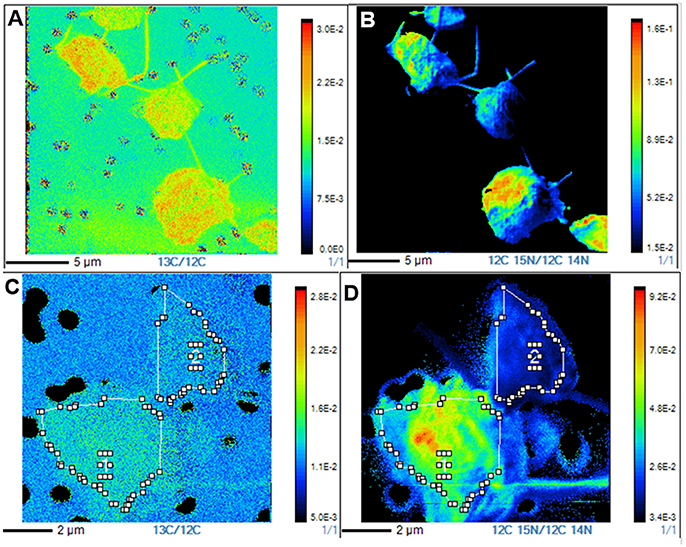
Figure 5. NanoSIMS 13C/12C and 15N/14N isotope ratio images of representative P. parvum cells from the two T48 experiments. (A) 13C/12C isotope ratio image from a T48 labeled inorganic nutrients sample. (B) 15N/14N ratio image from a T48 labeled inorganic nutrients sample. (C) 13C/12C isotope ratio image from a T48 labeled ciliate sample. (D) 15N/14N isotope ratio image from a T48 labeled ciliate sample.
The 13C enrichment in the P. parvum cells from the T48 labeled inorganic sample was relatively low and variable (mean 13C/C ratio of 0.0172, SD = 0.003; Figure 4). Nevertheless, it was significant when compared to the T0 labeled inorganic sample as well as to the unlabeled control (Table 1). In contrast, most of the P. parvum cells from the T48 labeled ciliate sample had 13C/C ratios within the range defined by the T0 labeled ciliate sample or the unlabeled control, with only 3 out of 22 exhibiting significant enrichment (13C/C ratios in the range 0.0119–0.0127). Overall, P. parvum cells from the T48 labeled ciliate sample had no significant 13C enrichment (Table 1).
Irrespective of the N source, the 15N enrichment in the P. parvum cells from the T48 samples was relatively high and variable (Figure 4). For the T48 labeled inorganic sample, the average 15N/N ratio was 0.093 (SD = 0.051), whereas it was 0.089 (SD = 0.054) for the T48 labeled ciliate sample. Both of these values were significantly larger than the values in the corresponding T0 samples as well as in the unlabeled control samples, but not significantly different from each other (Table 1).
To estimate the importance of the different C and N sources in the nutrition of P. parvum, we calculated the relative amounts of C and N assimilated by the P. parvum cells during the incubation interval of Δt = 2 days as
This calculation takes into account both the measured enrichment in the P. parvum cells (subscripts T48 and T0) as well as the enrichment of the corresponding assumed C and N source (subscript SRC).
For the labeled inorganic incubation (Treatment 2), we assumed that the DIC and nitrate were the only labeled C and N sources, and hence used (13C/C)SRC = 0.99 and (15N/N)SRC = 0.49 (see Methods). Using the average isotopic ratios measured in the P. parvum cells, we obtained rC = 0.0033 mol C (mol C)−1 d−1 and rN = 0.092 mol N (mol N)−1 d−1; i.e., a roughly 30-fold lower value of rC relative to rN.
For the labeled ciliate incubation (Treatment 1), we assumed that the ciliates were the only labeled source of C and N, and hence used the average values measured by NanoSIMS (13C/C)SRC = 0.0185 and (15N/N)SRC = 0.386 (see Results above). For the P. parvum cells whose values of (13C/C)T48 and (13C/C)T0 were not significantly different, we obtained rC = 0 and rN = 0.11 mol N (mol N)−1 d−1, whereas for the three P. parvum cells that showed significant 13C enrichment we obtained rC = 0.094 mol C (mol C)−1 d−1 and rN = 0.20 mol N (mol N)−1 d−1. This suggests that while most of the P. parvum cells preyed on the ciliates to gain nitrogen, a few of them likely used them as a significant source of both C and N. Additionally, because the rN values estimated for the inorganic and ciliate labeled treatments were similar P. parvum appears to exhibit no preference for nitrogen assimilation from inorganic nutrients (nitrate) vs. predation on U. marina.
Discussion
Prior to this study of P. parvum, the only previous application of stable isotope tracers and NanoSIMS to investigate protistan mixotrophy was completed by Terrado et al. (2017) on the chrysophyte alga Ochromonas sp. strain BG-1. That study showed that strain BG-1 relied heavily on heterotrophy (predation on bacteria) for acquisition of both carbon and nitrogen (84–99 and 88–95% of total uptake respectively). Uptake of inorganic forms of these elements from photoautotrophy occurred only after prey were depleted, and was insufficient to support population growth (Terrado et al., 2017). The inferred strategy of P. parvum determined in the present study stands in marked contrast to those findings. Our results suggest that P. parvum relied mostly on photoautotrophy for carbon assimilation into cellular biomass, while predation served as a strategy to supplement energy and nitrogen, but much less for carbon integrated into cellular building blocks. However, no preference was demonstrated for nitrogen uptake from prey vs. inorganic sources (nitrate) when both were available. Hence, in the spectrum of strategies exhibited by mixotrophic protists, P. parvum lies close to the pure phototrophy end of the spectrum (also supported by data from Carvalho and Granéli, 2010; Brutemark and Granéli, 2011), while Ochromonas strain BG-1 lies much closer to the pure heterotrophy end of the mixotrophic spectrum.
Previous NanoSIMS imaging of protists has demonstrated that their large cell topography can have a significant influence on isotopic fractionation, and that this effect must be quantified through measurement of unlabeled cells (Carpenter et al., 2013; Kopp et al., 2015). Hence, we imaged 20 whole P. parvum cells from the unlabeled control sample to assess the effects of topography on fractionation of both carbon and nitrogen isotopes (13C/C and 15N/N). This was done to ensure that we could unequivocally detect enrichments in our labeled treatments (i.e., T48 labeled ciliate and T48 labeled inorganic samples)—i.e., whether these putative enrichments fall outside the range of variability caused by topography. Although the values for both isotopic ratios as measured from the unlabeled control sample show a fairly wide range (Figure 2), our statistical tests revealed significant differences in 15N/N in both T48 samples compared to the unlabeled control, and in 13C/C in the labeled T48 inorganic sample compared to the unlabeled control (Table 1; Figure 4). This analysis confirmed that the level of precision achieved here was sufficient to meet the goals of the study.
As expected, the 13C/C and 15N/N ratios measured from whole P. parvum cells of our two T0 samples (T0 inorganic labeled and T0 ciliate labeled) showed ranges that were not significantly different from the unlabeled control (Table 1; Figure 3), with the exception of 15N in the T0 ciliate labeled sample. We suggest this signal is attributable to some assimilation of the 15N label after immediate feeding while we were sampling T0 samples. For the prey ciliate cells (U. marina), those from the T0 labeled inorganics sample fell in the center of the ranges for P. parvum cells from the unlabeled control for both isotopes (13C/C and 15N/N), but the T0 labeled ciliates were significantly enriched in both isotopes as expected (Figure 3).
We found no significant difference in 13C/C between the T48 labeled ciliate sample and the unlabeled control, or between the former and the T0 labeled ciliate sample. We conclude that, in our experimental design, P. parvum cells overall obtained no detectable carbon (i.e., for anabolic integration into cellular biomass) from ingestion of labeled ciliates after 48 h. However, three of the 22 imaged P. parvum cells did exhibit small but significant 13C enrichment (Figure 4 and Results). Hence, it appears that a small amount of carbon assimilation for integration into cellular biomass does occur through predation for some individual P. parvum, although this source appears to be a less important carbon source overall than photoautotrophy. That no significant difference in 13C enrichment between T48 labeled inorganic and T48 labeled ciliate samples was observed is due to the few data points that overlap between the two data sets (Figure 4), while the p-value of 0.06 is close to significance (Table 1). No significant difference in 15N enrichment between the two T48 samples was observed (Table 1; Figure 4), indicating that P. parvum had no preference for assimilation of nitrogen from inorganic nutrients (nitrate) or live prey when both are available.
Our results after 48 h (T48 labeled ciliate and T48 labeled inorganics treatments) demonstrated a much greater assimilation of prey nitrogen than prey carbon (Figure 4). A roughly 30-fold greater assimilation of 15N than 13C was observed in the treatment with labeled ciliates, although there was considerable variability among individual cells, as would be expected (see next paragraph). This was surprising in that we expected nitrogen and carbon assimilation would be more closely in balance. Specifically, the assimilation of nitrogen from prey by the alga was consistent with our expectation (i.e., prey can be a significant source of nitrogen for the mixotroph), but the low level of carbon assimilation was unexpected. A previous transcriptomic study of P. parvum (Liu et al., 2015a) indicated that the availability of ciliate prey had a clear effect on gene expression in the alga, resulting in increased expression of genes involved in fatty acid oxidation compared to treatments lacking prey (Liu et al., 2015a). These prior results seemed to imply that carbon from prey was readily utilized (and presumably assimilated) by P. parvum. However, given that fatty acid oxidation can result in either complete breakdown to CO2 via the TCA cycle (i.e., for energy production), or anabolic processes (conversion to succinate through the glyoxylate cycle), our NanoSIMS data help to clarify which of the two pathways are favored for prey-derived fatty acids. Specifically, our data suggest that P. parvum utilizes fatty acids derived from ciliate prey mostly for energy generation and to a lesser extent, in some individuals, for anabolic processes. Thus, our data further refine the picture of P. parvum as a predominantly photoautotrophic species that can rely on predation to supplement its nitrogen requirements. In a similar NanoSIMS-tracer study of the cellulolytic protist Oxymonas sp.—a gut symbiont of the termite Paraneotermes —Carpenter et al. (2013) found a roughly two order of magnitude enrichment in 13C, but no detectable enrichment in 15N after a 6-week labeling experiment, thus providing another example of unbalanced uptake of C and N.
The high levels of variability in isotopic enrichment—up to an order of magnitude for 15N, but also wide for 13C—among P. parvum cells in each of the two T48 treatments (Figure 4) appears to indicate that individual cells of P. parvum within a natural population encompass a correspondingly wide range of carbon and nitrogen uptake from various sources. This is possibly a function of cell size, point in the cell cycle, or—in the case of the T48 ciliate labeled sample—proximity to freshly killed prey. (This latter possibility makes sense because the ratio of predator to prey was 20:1.) We believe that assessment of such cell-to-cell variability, which is not possible to infer from molecular or other standard approaches, is an important outcome of this study. With such data in hand, it becomes possible to estimate averages for populations with much greater accuracy. It also becomes possible to then ask why such variability exists, and what effect the variation may have for populations, predator-prey relationships, production of toxins/harmful blooms, ecosystems, and biogeochemical cycles. In any case, it illustrates that in addition to the great diversity of mixotrophic strategies across higher taxa, evaluating the diversity across cells of a given species and/or population can refine our understanding of mixotrophy.
Our stable isotope labeling and NanoSIMS imaging analysis of single cells yields new insights on the cell physiology and ecology of P. parvum. It offers evidence that P. parvum relies on photoautotrophy for the production of cellular carbon, but uses predation mostly as a supplemental source of nitrogen and perhaps an energy source from carbon. It also shows P. parvum displays no preference for uptake of the nitrogen from prey or inorganic nutrients (nitrate) when both sources are available. One might conclude, however, that nitrogen derived from prey could be important when inorganic nitrogen sources are limiting. In addition, our study reveals a wide range of cell physiologies with respect to uptake of carbon and nitrogen across this population, perhaps reflecting the opportunistic nature of individual cells encountering (and utilizing) prey biomass, thus highlighting the capability to make far more accurate estimates of population level processes and the activity of an average cell within it. Future studies using methods such as the one presented here will continue to yield important insight into the vast and largely unexplored range of nutrient and energy acquisition strategies that fall under the very broad umbrella of mixotrophy—a strategy that is only very recently gaining recognition as one of enormous importance in biogeochemical cycles.
Author Contributions
KC designed the experiments, fixed and prepared the samples, imaged the samples with FESEM and NanoSIMS, analyzed and interpreted the data, and led the development of the manuscript; MB imaged the samples with NanoSIMS and analyzed the data; LP analyzed and interpreted the data and contributed to the development of the manuscript; AL designed and carried out stable isotope tracer experimental treatments and measured P. parvum growth rates; KH and DC designed the experiments, interpreted the data and contributed to the development of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank the Moore Foundation (Grant GBMF3299 to DC and KH) for funding and Peter Williams (Arizona State University) for helpful discussions on NanoSIMS analysis. Electron micrographs presented in this article were acquired at the Center for Electron Microscopy and Microanalysis at the University of Southern California with technical assistance from John Curulli and Casey Barr.
References
Barkoh, A., and Fries, L. T. (2010). Aspects of the origins, ecology, and control of Golden Alga Prymnesium parvum: introduction to the featured collection. J. Am. Water Resour. Assoc. 46, 1–5. doi: 10.1111/j.17521688.2009.00394.x
Bonnet, S., Berthelot, H., Turk-Kubo, K., Cornet-Bartaux, V., Fawcett, S. E., Berman-Frank, I., et al. (2016). Diazotroph derived nitrogen supports diatoms growth in the South West Pacific: a quantitative study using nanoSIMS. Limnol. Oceanogr. 61, 1549–1562. doi: 10.1002/lno.10300
Brutemark, A., and Granéli, E. (2011). Role of mixotrophy and light for growth and survival of the toxic haptophyte Prymnesium parvum. Harmful Algae 10, 388–394. doi: 10.1016/j.hal.2011.01.005
Burkholder, J. M., Glibert, P. M., and Skelton, H. M. (2008). Mixotrophy, a major mode of nutrition for harmful algal species in eutrophic waters. Harmful Algae 8, 77–93. doi: 10.1016/j.hal.2008.08.010
Burki, F., Kaplan, M., Tikhonenkov, D. V., Zlatogursky, V., Minh, B. Q., Radaykina, L. V., et al. (2016). Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proc. R. Soc. B 283:20152802. doi: 10.1098/rspb.2015.2802
Caron, D. (2017). Acknowledging and incorporating mixed nutrition into aquatic protistan ecology, finally. Environ. Microbiol. Rep. 9, 41–43. doi: 10.1111/1758-2229.12514
Carpenter, K. J., Weber, P. K., Davisson, M. L., Pett-Ridge, J., Haverty, M. I., and Keeling, P. J. (2013). Correlated SEM, FIB-SEM, TEM, and nanoSIMS imaging of microbes from the hindgut of a lower termite: methods for in situ functional and ecological studies of uncultivable microbes. Microsc. Microanal. 19, 1490–1501. doi: 10.1017/S1431927613013482
Carvalho, W. F., and Granéli, E. (2010). Contribution of phagotrophy versus autotrophy to Prymnesium parvum growth under nitrogen and phosphorus sufficiency and deficiency. Harmful Algae 9, 105–115. doi: 10.1016/j.hal.2009.08.007
Flynn, K. J., Stoecker, D. K., Mitra, A., Raven, J. A., Glibert, P. M., Hansen, P. J., et al. (2013). Misuse of the phytoplankton-zooplankton dichotomy: the need to assign organisms as mixotrophs within plankton functional types. J. Plankton Res. 35, 3–11. doi: 10.1093/plankt/fbs062
Granéli, E., Edvardsen, B., Roelke, D. L., and Hagström, J. A. (2012). The ecophysiology and bloom dynamics of Prymnesium spp. Harmful Algae 14, 260–270. doi: 10.1016/j.hal.2011.10.024
Green, J. C., Hibberd, D. J., and Pienaar, R. N. (1982). The taxonomy of Prymnesium (Prymnesiophyceae) including a description of a new cosmopolitan species, P. patellifera sp. nov., and further observations on P. parvum N. Carter. Br. Phycol. J. 17, 363–382.
Hambright, D. K., Beyer, J. E., Easton, J. D., Zamor, R. M., Easton, A. C., and Hallidayschult, T. C. (2015). The niche of an invasive marine microbe in a subtropical freshwater impoundment. ISME J. 9, 256–264. doi: 10.1038/ismej.2014.103
Hartmann, M., Zubkov, M. V., Scanian, D. J., and Lepère, C. (2013). In situ interactions between photosynthetic picoeukaryotes and bacterioplankton in the Atlantic Ocean: evidence for mixotrophy. Environ. Microbiol. Rep. 5, 835–840. doi: 10.1111/1758-2229.12084
Jones, R. I. (1994). Mixotrophy in planktonic protists as a spectrum of nutritional strategies. Mar. Microb. Food Webs 8, 87–96.
Jones, R. I. (2000). Mixotrophy in planktonic protists: an overview. Freshw. Biol. 45, 219–226. doi: 10.1046/j.1365-2427.2000.00672.x
Kopp, C., Pernice, M., Domart-Coulon, I., Djediat, C., Spangenberg, J., Alexander, D. T. L., et al. (2013). Highly dynamic cellular-level response of symbiotic coral to a sudden increase in environmental nitrogen. mBio 4, e00052–e00013. doi: 10.1128/mBio.00052-13
Kopp, C., Wisztorski, M., Revel, J., Mehiri, M., Dani, V., Capron, L., et al. (2015). MALDI-MS and NanoSIMS imaging techniques to study cnidarian–dinoflagellate symbioses. Zoology 118, 125–131. doi: 10.1016/j.zool.2014.06.006
Lechene, C. P., Luyten, Y., McMahon, G., and Distel, D. L. (2007). Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science 317, 1563–1566. doi: 10.1126/science.1145557
Leles, S., Mitra, A., Flynn, K. J., Stoecker, D. K., Hansen, P. J., Calbet, A., et al. (2017). Oceanic protists with different forms of acquired phototrophy display diverse biogeographies and abundance. Proc. Biol. Sci. 284:20170664. doi: 10.1098/rspb.2017.0664
Lie, A. A. Y., Liu, Z., Terrado, R., Tatters, A. O., Heidelberg, K. B., and Caron, D. C. (2017). Effect of light and prey availability on gene expression of the mixotrophic chrysophyte, Ochromonas sp. BMC Genomics 18:163. doi: 10.1186/s12864-017-3549-1
Liu, Z., Jones, A. C., Campbell, V., Hambright, K. D., Heidelberg, K., and Caron, D. A. (2015a). Gene expression in the mixotrophic prymnesiophyte, Prymnesium parvum, responds to prey availability. Front. Microbiol. 6:319. doi: 10.3389/fmicb.2015.00319
Liu, Z., Jones, A. C., Campbell, V., Heidelberg, K. B., and Caron, D. A. (2016). Gene expression characterizes different nutritional strategies among three mixotrophic protists. FEMS Microbiol. Ecol. 92, 1–11. doi: 10.1093/femsec/fiw106
Liu, Z., Koid, A. E., Terrado, R., Campbell, V., Caron, D. A., and Heidelberg, K. B. (2015b). Changes in gene expression of Prymnesium parvum induced by nitrogen and phosphorus limitation. Front. Microbiol. 6:631. doi: 10.3389/fmicb.2015.00631
Manning, S. R., and La Claire, J. W. (2010). Prymnesins: toxic metabolites of the golden alga, prymnesium parvum carter (Haptophyta). Mar. Drugs 8, 678–704. doi: 10.3390/md8030678
Matantseva, O. V., and Skarlato, S. O. (2013). Mixotrophy in microorganisms: ecological and cytophysiological aspects. J. Evol. Biochem. Phys. 49, 377–388. doi: 10.1134/S0022093013040014
Mitra, A., Flynn, K. J., Burkholder, J. M., Berge, T., Calbet, A., Raven, J. A., et al. (2014). The role of mixotrophic protists in the biological carbon pump. Biogeosciences 11, 995–1005. doi: 10.5194/bg-11-995-2014
Mitra, A., Flynn, K. J., Tillmann, U., Raven, J. A., Caron, D., Stoecker, D. K., et al. (2016). Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition: incorporation of diverse mixotrophic strategies. Protist 167, 106–120. doi: 10.1016/j.protis.2016.01.003
Moestrup, O. (1994). “Economic aspect of ‘blooms’, nuisance species, and toxins,” in The Haptophyte Algae, eds J. C. Green and B. S. C. Leadbeater (New York, NY: Oxford University Press), 265–285.
Orphan, V. J., House, C. H., Hinrichs, K.-U., McKeegan, K. D., and DeLong, E. F. (2001). Methane-consuming Archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293, 484–487. doi: 10.1126/science.1061338
Polerecky, L., Adam, B., Milucka, J., Musat, N., Vagner, T., and Kuypers, M. M. (2012). Look@NanoSIMS - a tool for the analysis of nanoSIMS data in environmental microbiology. Environ. Microbiol. 14, 1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x
Rasmussen, S. A., Meier, S., Andersen, N. G., Blossom, H. E., Duus, J. Ø., Nielsen, K. F., et al. (2016). Chemodiversity of ladder-frame prymnesin polyethers in Prymnesium parvum. J. Nat. Prod. 79, 2250–2256. doi: 10.1021/acs.jnatprod.6b00345
Raven, J. A., Beardall, J., Flynn, K. J., and Maberly, S. C. (2009). Phagotrophy in the origins of photosynthesis in eukaryotes and as a complementary mode of nutrition in phototrophs:relation to Darwin's insectivorous plants. J. Exp. Bot. 60, 3975–3987. doi: 10.1093/jxb/erp282
Roelke, D. L., Barkoh, A., Brooks, B. W., Grover, J. P., Hambright, D., LaClaire, J. W. II., et al. (2016). A chronicle of a killer alga in the west: ecology, assessment, and management of Prymnesium parvum blooms. Hydrobiologia 764, 29–50. doi: 10.1007/s10750-015-2273-6
Sallenave, R. (2010). Toxic Golden Algae (Prymnesium parvum), Circular 647. Cooperative extension service, New Mexico State University, NM.
Sanders, R. W. (2011). Alternative nutritional strategies in protists: symposium introduction and a review of freshwater protists that combine photosynthesis and heterotrophy. J. Euk. Microbiol. 58, 181–184. doi: 10.1111/j.1550-7408.2011.00543.x
Sanders, R. W., and Porter, K. G. (1988). Phagotrophic phytoflagellates. Adv. Microb. Ecol. 10, 167–192. doi: 10.1007/978-1-4684-5409-3_5
Stoecker, D. K., Hansen, P. J., Caron, D. A., and Mitra, A. (2017). Mixotrophy in the marine plankton. Ann. Rev. Mar. Sci. 9, 331–335. doi: 10.1146/annurev-marine-010816-060617
Sunda, W. G., Granelli, E., and Gobler, C. J. (2006). Positive feedback and the development and persistence of ecosystem disruptive algal blooms. J. Phycol. 42, 963–974. doi: 10.1111/j.1529-8817.2006.00261.x
Tai, V., Carpenter, K. J., Weber, P. K., Nalepa, C. A., Perlman, S. J., and Keeling, P. J. (2016). Genome evolution and nitrogen fixation in bacterial ectosymbionts of a protist inhabiting wood-feeding cockroaches. Appl. Environ. Microbiol. 82, 4682–4695. doi: 10.1128/AEM.00611-16
Terrado, R., Pasulka, A. L., Lie, A. A., Orphan, V. J., Heidelberg, K. B., and Caron, D. A. (2017). Autotrophic and heterotrophic acquisition of carbon and nitrogen by a mixotrophic chrysophyte. ISME J. 11, 2022–2034. doi: 10.1038/ismej.2017.68
Tillmann, U. (2003). Kill and eat your predator: a winning strategy of the planktonic flagellate Prymnesium parvum. Aquat. Microgiol. J. 32, 73–84. doi: 10.3354/ame032073
Ward, B. A., and Follows, M. J. (2016). Marine mixotrophy increases trophic transfer efficiency, mean organism size, and vertical carbon flux. Proc. Nat. Acad. Sci. U.S.A. 113, 2958–2963. doi: 10.1073/pnas.1517118113
Keywords: algae, haptophyte, mixotrophy, NanoSIMS, protist, predation, Prymnesium parvum, stable isotope tracers
Citation: Carpenter KJ, Bose M, Polerecky L, Lie AAY, Heidelberg KB and Caron DA (2018) Single-Cell View of Carbon and Nitrogen Acquisition in the Mixotrophic Alga Prymnesium parvum (Haptophyta) Inferred From Stable Isotope Tracers and NanoSIMS. Front. Mar. Sci. 5:157. doi: 10.3389/fmars.2018.00157
Received: 13 January 2018; Accepted: 19 April 2018;
Published: 11 May 2018.
Edited by:
Matthew D. Johnson, Woods Hole Oceanographic Institution, United StatesReviewed by:
Robert Fischer, Wasser Cluster Lunz, AustriaAkkur Vasudevan Raman, Andhra University, India
Copyright © 2018 Carpenter, Bose, Polerecky, Lie, Heidelberg and Caron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kevin J. Carpenter, a2V2aW5Aa2V2aW5qY2FycGVudGVyLmNvbQ==
 Kevin J. Carpenter
Kevin J. Carpenter Maitrayee Bose2
Maitrayee Bose2 Lubos Polerecky
Lubos Polerecky Karla B. Heidelberg
Karla B. Heidelberg David A. Caron
David A. Caron