- 1Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao, China
- 2Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 3Department of Biological Sciences, Boise State University, Boise, ID, United States
In the present study, based on both morphologic and phylogenetic analyses, a new genus, Monourostylopsis n. gen., and new species, Metaurostylopsis alrasheidi n. sp. as well as a new combination, Monourostylopsis antarctica (Jung et al., 2011) n. comb. (original combination: Metaurostylopsis antarctica Jung et al., 2011), are suggested. The new genus is diagnosed mainly by having three or more frontoterminal cirri, a midventral complex with midventral pairs and a single midventral row, one right marginal row and two or more left marginal rows. The new genus can be easily separated from the morphologically similar genera mainly by having single right marginal row (vs. two or more right marginal rows). Based on live observation and protargol staining, the morphology and morphogenesis of a new species, M. alrasheidi n. sp. isolated from China, were investigated. The new species can be characterized by: two types of cortical granules; about 22 adoral membranelles; three or four frontoterminal, four or five transverse cirri; about eight midventral pairs and a midventral row of three or four unpaired midventral cirri; three or four left and right marginal rows. The main morphogenetic features of Metaurostylopsis alrasheidi n. sp. can be summarized as: (1) the entire parental ciliature, including the oral apparatus, is renewed; (2) the oral primordium of the proter probably originates within a pouch; (3) the oral primordium of the opisthe forms de novo on the cell surface; (4) the anlagen of marginal rows and dorsal kineties are formed intrakinetally, and (5) the fusion of macronuclear nodules results in an irregular branched mass prior to karyokinesis. In the phylogenetic trees, all the available Metaurostylopsis sequences cluster together in a clade with full support (ML/BI: 100/1.00) revealing that the genus is monophyletic within the large group of core urostylids.
Introduction
Hypotrichs have been widely regarded as the most complex and highly differentiated ciliate group (Foissner, 1982, 2016; Tuffrau and Fleury, 1994; Berger, 1999, 2006, 2008, 2011; Song et al., 2009; Li et al., 2018). Recent faunistic studies have revealed numerous new hypotrichous taxa, suggesting that the diversity of this group is still underestimated (Kaur et al., 2019; Luo et al., 2019; Dong et al., 2020; Lu et al., 2020; Ma et al., 2020; Wang et al., 2020a). Furthermore, the extensive studies on morphogenesis and molecular phylogeny of hypotrichs have led to a better understanding of their systematics and evolutionary relationships (Song and Shao, 2017; Lyu et al., 2018; Kim and Min, 2019; Shao et al., 2019; Wang et al., 2020b; Zhang et al., 2020).
Urostylids, with a diversity of more than 200 species, are one of the largest groups in hypotrichs, and are commonly found in marine, soil and freshwater habitats. Morphologically, they are characterized by a midventral complex composed of zigzag-patterned midventral cirral pairs (Berger, 2006; Kumar et al., 2010; Chen et al., 2011; Pan et al., 2016; Kim et al., 2017; Hu et al., 2019; Jung and Berger, 2019). Based mainly on the presence of frontoterminal cirri, the clearly differentiated frontal cirri and several morphogenetic features, Song et al. (2001) transferred Urostyla marina Kahl, 1932 to a newly established urostylid genus, Metaurostylopsis, with M. marina (Kahl, 1932) Song et al., 2001 as the type species. In the following decades, more species of this genus have been identified and described (Song and Wilbert, 2002; Lei et al., 2005; Shao et al., 2008a, b; Chen et al., 2011, 2013; Jung et al., 2011; Lu et al., 2016). Nevertheless, detailed ontogenetic and molecular information has consistently questioned the assignments of some Metaurostylopsis species and further revisions of the genus have been made. Based on comprehensive analyses, Song et al. (2011) recognized five species in the genus Metaurostylopsis (M. marina, M. rubra Song and Wilbert, 2002, M. salina Lei et al., 2005, M. struederkypkeae Shao et al., 2008, and M. cheni Chen et al., 2011) and reassigned M. sinica Shao et al., 2008 into a new genus Apourostylopsis. While M. flavicans Wang et al., 2011 and M. songi Lei et al., 2005 were treated as incertae sedis. In the same year, a new Metaurostylopsis species, M. antarctica Jung et al., 2011 was reported. Subsequently, Chen et al. (2013) transferred the two incertae sedis, M. flavicans and M. songi, into a new genus Neourostylopsis and provided improved diagnoses for both the genus Metaurostylopsis and Apourostylopsis. Most recently, Lu et al. (2016) added a new species, Metaurostylopsis parastruederkypkeae to the genus. As of this writing, seven species are included in Metaurostylopsis.
In the present work, morphological, ontogenetic, and molecular data for a new urostylid species, Metaurostylopsis alrasheidi n. sp., are provided. In addition, morphological, morphogenetic, and phylogenetic analyses comparing Metaurostylopsis antarctica and other Metaurostylopsis species are carried out, leading to the establishment of a new genus for Metaurostylopsis antarctica.
Materials and Methods
Sampling, Observation, and Identification
Metaurostylopsis alrasheidi n. sp. was discovered on June 3, 2019 in a freshwater sample from Lake Weishan (34°46′14″N, 117°12′56″E) (Figures 1A–D), China. A raw culture was established in Petri dishes with rice grains added to facilitate the growth of bacteria as food source for the ciliates. A single cell was transferred with a micropipette to a mini Petri dish containing mineral water with squeezed rice grains to establish a clonal culture which was maintained for about 1 month at room temperature about 25°C, and from which DNA extraction was done.
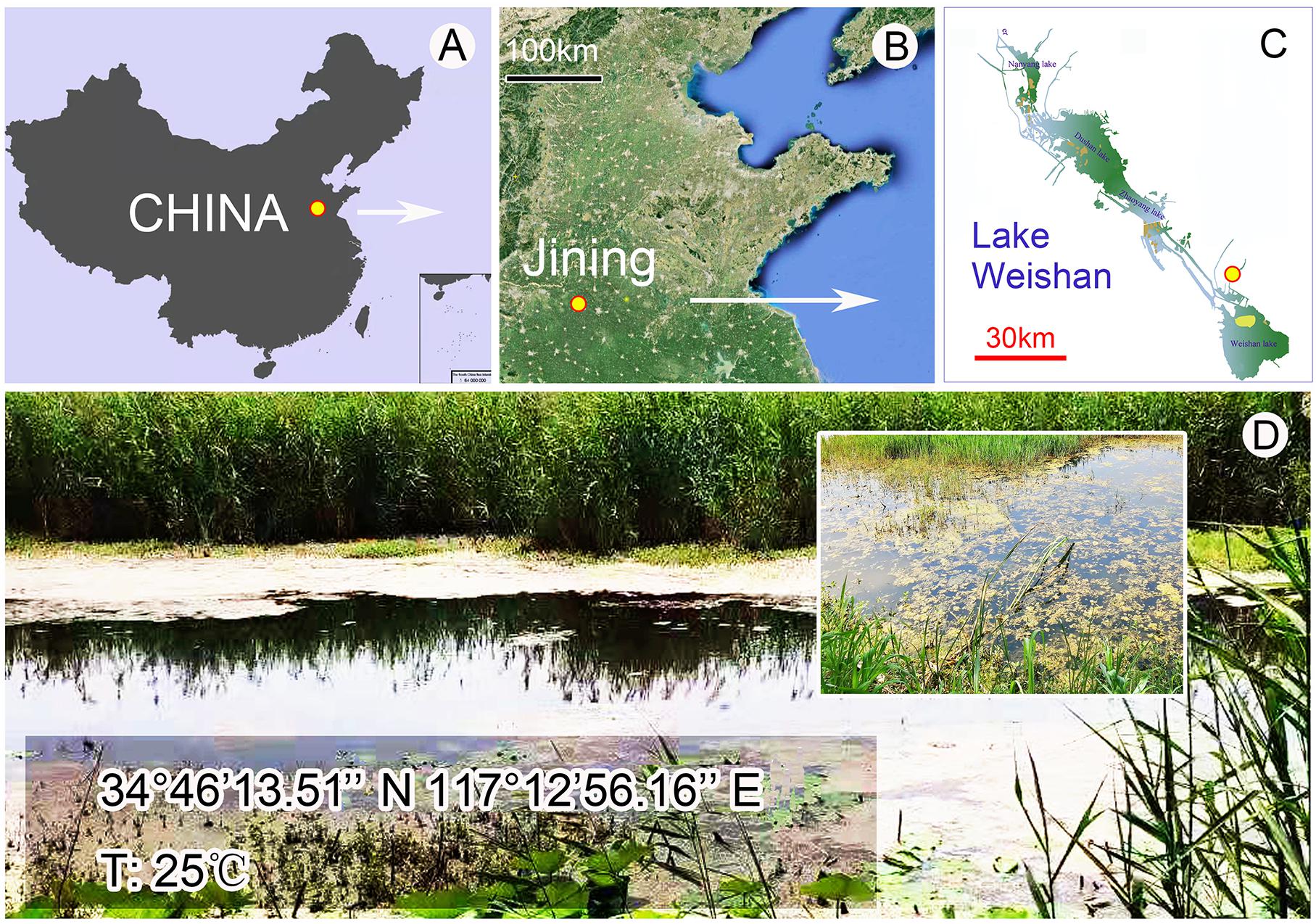
Figure 1. Location of the sampling site. (A–C) Map showing the location of the Lake Weishan, China. (D) Photographs of the Lake Weishan where Metaurostylopsis alrasheidi n. sp. was collected.
Live observation, protargol preparation, and drawings of protargol-impregnated specimens, live cells, and dividers are according to Shao et al. (2019). Classification is mainly according to Jankowski (2007) and terminology is according to Berger (2006).
DNA Extraction, PCR, and Sequencing
A total of five clonal cells were isolated, washed several times with sterilized freshwater. Subsequently, the five cells were picked into three 1.5 ml microfuge tubes with as little water as possible, two tubes with one cell each and three cells in the third tube. Genomic DNA of cells was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD) following the manufacturer’s instructions. The PCR primers for 18S rRNA gene amplification were 82SF (5′-GAAACTGCGAATGGCTC-3′) (Jerome et al., 1996) and 5.8SR (5′-TACTGATATGCTTAAGTTCAGCGG-3′) (Gao et al., 2012). Q5®HotStart High-Fidelity 2 × Master Mix DNA Polymerase (NEB, Ipswich, MA) (Wang et al., 2017a) was selected for PCR, with parameters according to Lian et al. (2019). The PCR target product was sent to Tsingke, Biological Technology Company (Qingdao, China) to sequence bidirectionally using primers 82SF, 5.8SR, Pro + B (5′-GGTTAAAAAGCTCGTAGT-3′), 900F (5′-CGATCAGATACCGTCCTAGT-3′), and 900R (5′-ACTAGGACGGTATCTGATCG-3′) (Wang et al., 2017b). Seqman 5.0 (DNAStar) was used to assemble contigs, which included the partial 18S rRNA gene and ITS1-5.8S rRNA-ITS2 sequence. The sequence was aligned using 18SR (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) (Medlin et al., 1988) to remove ITS1-5.8S rRNA-ITS2. The 18S rRNA gene sequence with a length of 1,647 bp was used for phylogenetic analyses.
Phylogenetic Analyses
In this work, 18S rRNA gene sequences of Metaurostylopsis alrasheidi n. sp. and 65 other Hypotrichia relatives were selected to perform phylogenetic analyses. According to Chen et al. (2020), Holosticha diademata (KF306396), Holosticha cf. heterofoissneri (KP717081), Holosticha heterofoissneri (KP717082), Uncinata bradburyae (EF123706), and Uncinata gigantea (KP717083) were selected as outgroup species.
The 66 18S rRNA gene sequences were aligned using the MUSCLE algorithm on the webserver GUIDANCE 21 (Sela et al., 2015). Both primers were removed using BioEdit 7.0 (Hall, 1999). The final alignment of 1,709 positions was used to construct phylogenetic trees. Maximum likelihood (ML) analysis with 1,000 bootstrap replicates was performed using RAxML-HPC2 on XSEDE 8.2.12 on the online server CIPRES Science Gateway (Stamatakis, 2014), with the GTRGAMMA model as the optimal choice. Bayesian inference (BI) analysis was carried out using MrBayes on XSEDE 3.2.6 (Ronquist et al., 2012) on CIPRES Science Gateway with the GTR + I + G model selected under Akaike Information Criterion (AIC) by jModelTest 2 (Darriba et al., 2012). Four Markov chain Monte Carlo (MCMC) chains were run for 1,000,000 generations, with sampling every 100 generations, the first 2,500 trees as burn-in. SeaView 4.6.1 (Gouy et al., 2010) and MEGA X (Kumar et al., 2018) were used to adjust the tree topologies.
Results
ZooBank Registration
Present work: LSIDurn:lsid:zoobank.org:pub:0085C8A1-6EE5-4E85-A245-D30CF9E18FC2.
Monourostylopsis n. gen.: LSIDurn:lsid:zoobank.org:act:CF7 BB200-33C6-4BBE-8998-2299FA70A0E7.
Metaurostylopsis alrasheidi n. sp.: LSIDurn:lsid:zoobank. org:act:077DAC39-8F8B-4BCD-849A-D07BE1C0C790.
Taxonomy and Morphological Description of the New Species
Subclass Hypotrichia Stein, 1859.
Order Urostylida Jankowski, 1979.
Family Urostylidae Bütschli, 1889.
Genus Metaurostylopsis Song et al., 2001
Metaurostylopsis alrasheidi n. sp. (Figures 2–5 and Table 1).
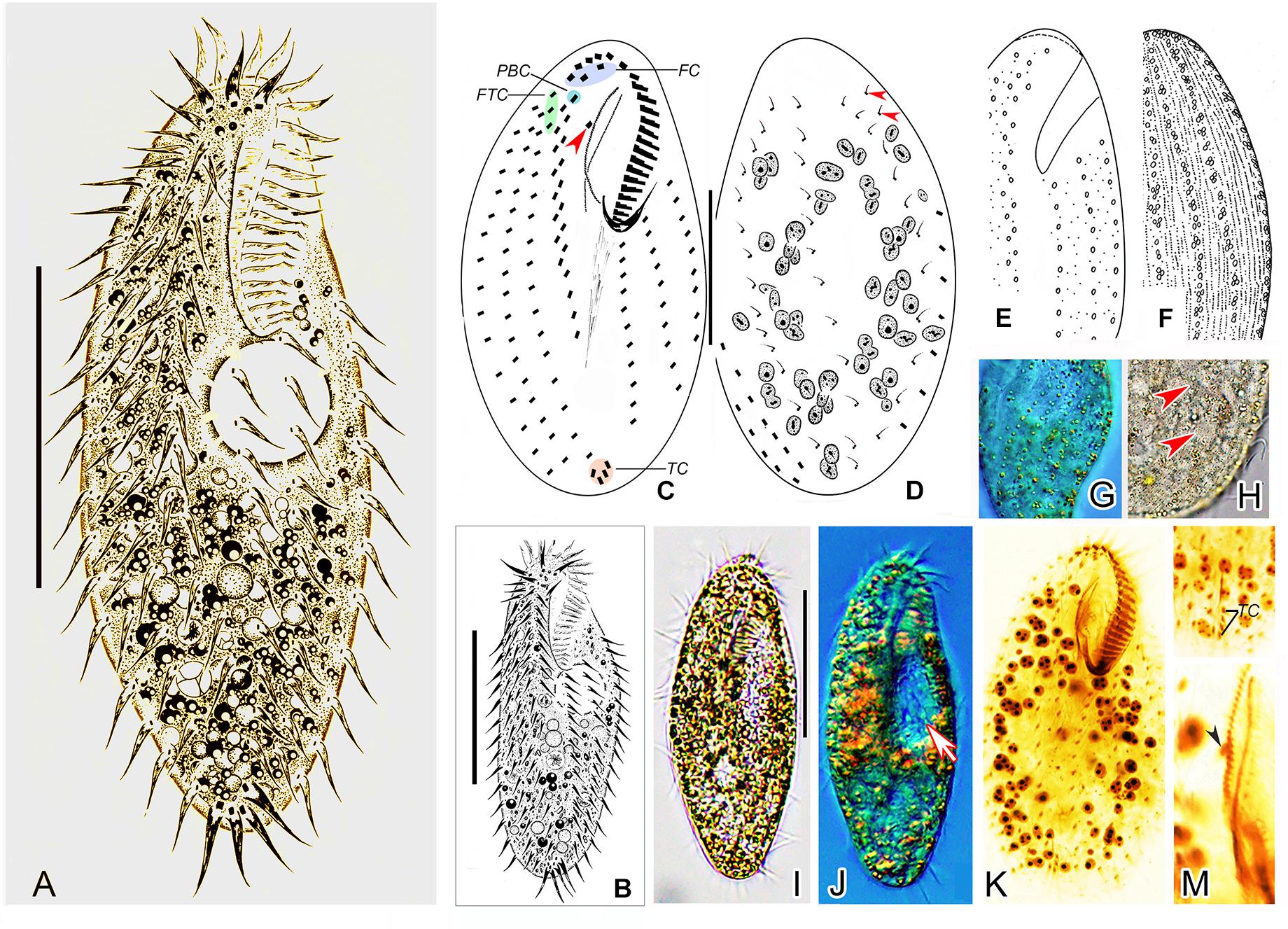
Figure 2. Morphology of Metaurostylopsis alrasheidi n. sp. from life (A,B,E,F,H,I, bright field; G,J, differential interference contrast) and after protargol preparation (C,D,K–M). (A,B) Ventral views showing representative individuals. (C,D) Ventral (C) and dorsal (D) views, arrowhead in (C) indicates the buccal cirrus, arrowheads in (D) mark the extra dorsal bristles. (E–H) Arrangement of cortical granules on ventral (E,G) and dorsal (F,H) side, arrowheads in (H) indicate small cortical granules. (I,J) Ventral views of different individuals, arrow in (J) shows the contractile vacuole. (K) Ventral view of the holotype specimen to show the ciliature and nuclear apparatus. (L) Showing the J-shaped transverse cirri. (M) Details of the undulating membranes, arrowhead indicates the buccal cirrus. FC, frontal cirri; FTC, frontoterminal cirri; PBC, parabuccal cirrus; TC, transverse cirri. Scale bars = 40 μm.
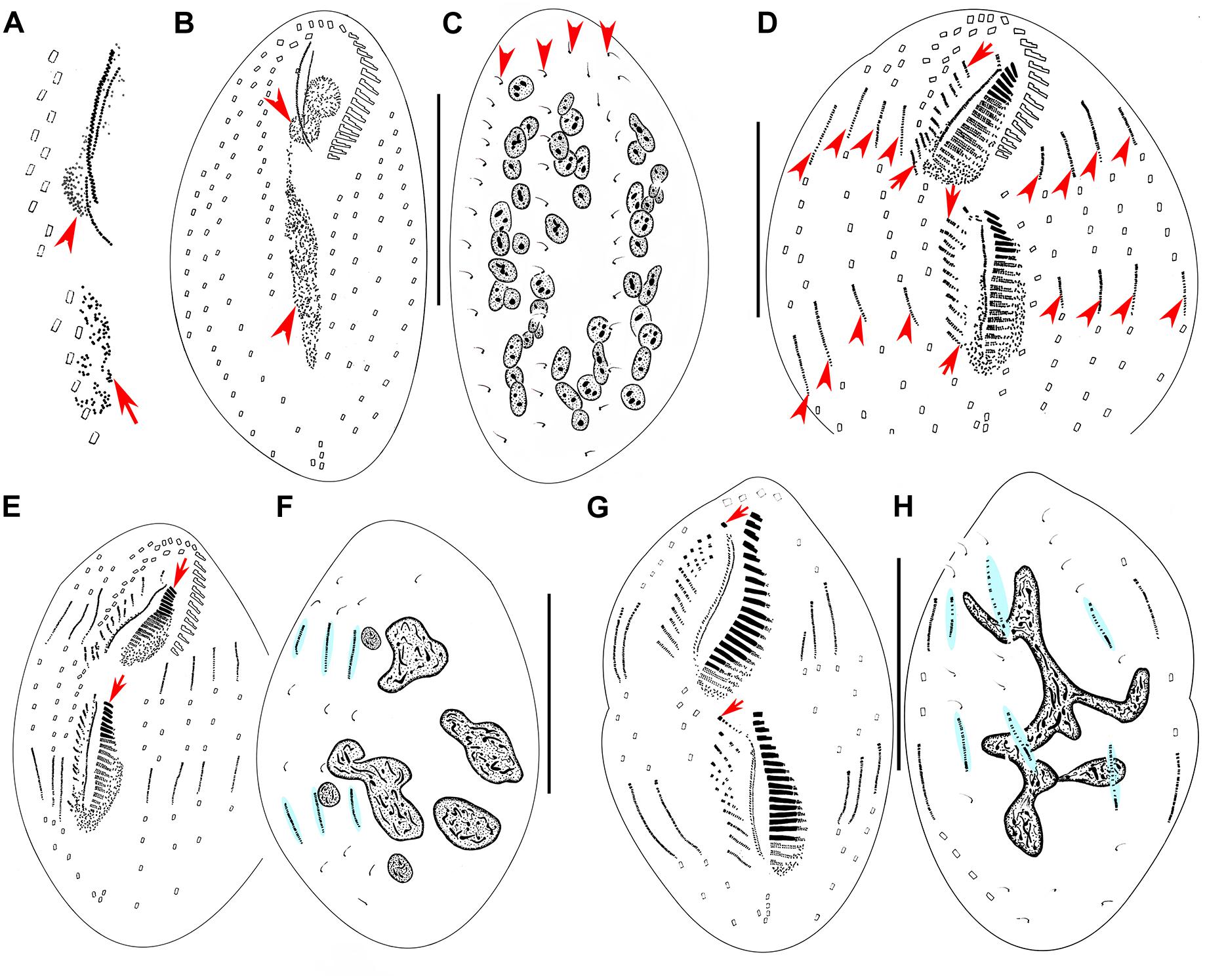
Figure 3. Morphogenesis of Metaurostylopsis alrasheidi n. sp. from early to middle stages after protargol impregnation. (A) Newly formed oral primordium in proter (arrowhead) and opisthe (arrow) at very early stage. (B,C) Ventral (B) and dorsal (C) views of an early divider to show the development of oral primordia for proter and opisthe (arrowheads), and the unchanged dorsal ciliature (arrowheads). (D) Ventral view of an early-middle divider, showing the streak-like frontoventral transverse cirral anlagen (arrows) and the intrakinetally formed marginal anlagen (arrowheads). (E,F) Ventral (E) and dorsal (F) views of the same early-middle divider, to show the newly formed adoral membranelles (arrows), and dorsal kinety anlagen (shaded in light-blue). (G,H) Ventral (G) and dorsal (H) views of a middle divider, showing the leftmost frontal cirrus (arrowheads) forming from the undulating membrane anlagen, dorsal kinety anlagen (shaded in light-blue) and macronuclear nodules fusing into a mass with few branches in (H). Scale bars = 50 μm.

Figure 4. Morphogenesis of Metaurostylopsis alrasheidi n. sp. from middle-late to late stages after protargol impregnation, hatched lines show cirri originated from the same cirral anlage. (A,B) Ventral (A) and dorsal (B) views of the same middle-late divider, arrows indicate the leftmost frontal cirrus. (C) Ventral view of a divider, arrows show the migration of newly formed frontoterminal cirri. (D,E) Ventral (D) and dorsal (E) views of the same divider, arrows mark the frontoterminal cirri migrating toward their final positions. Scale bars = 50 μm.
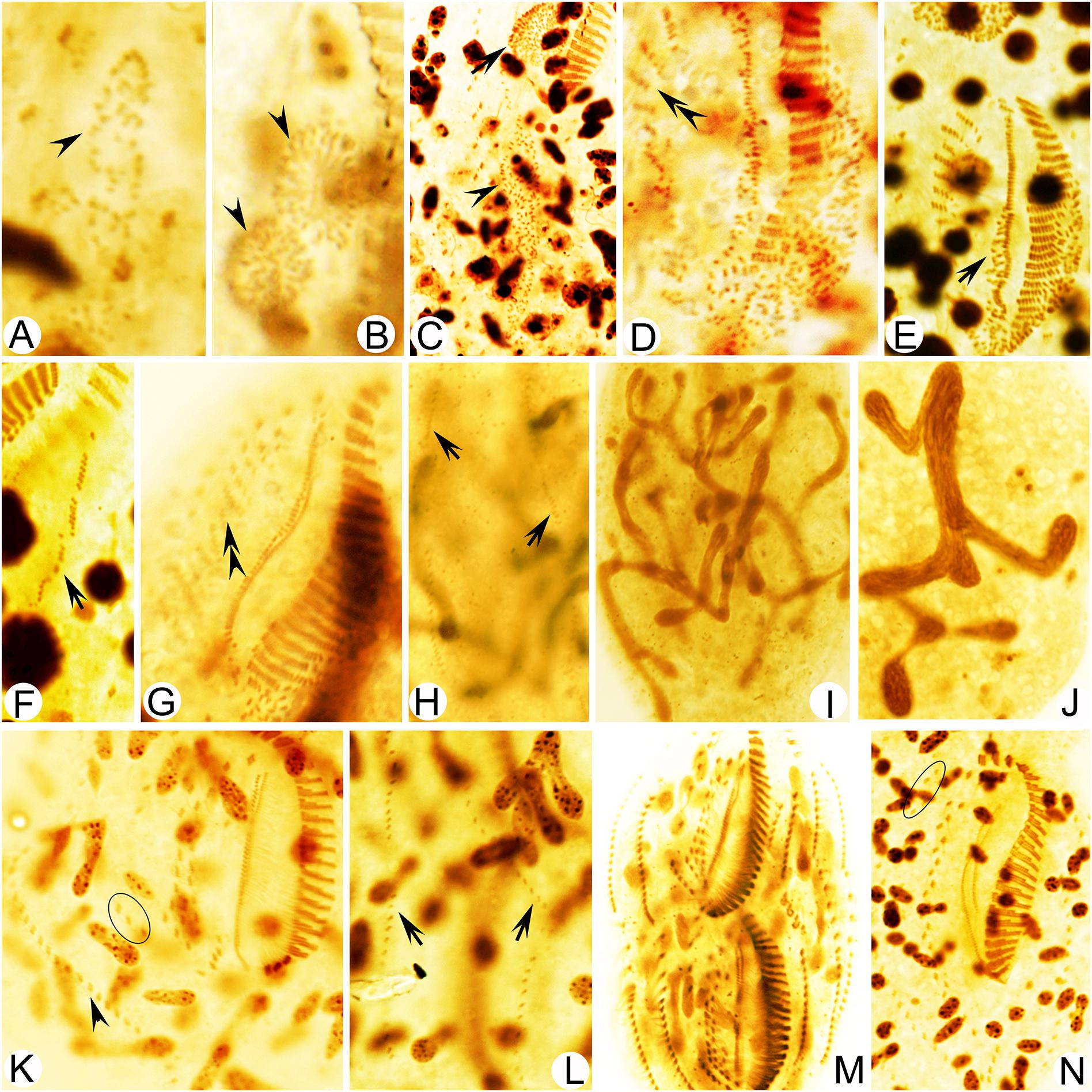
Figure 5. Photomicrographs of Metaurostylopsis alrasheidi n. sp. during morphogenesis after protargol impregnation. (A,B) Ventral view of a very early divider, arrowheads point to the oral primordium of the opisthe (A) and the proter (B). (C) The oral primordium continues to grow by further proliferation for proter (arrow) and opisthe (arrowhead) at early stage. (D,E) Ventral views, to show the formation of streak-like frontoventral transverse cirral anlagen (double-arrowheads) and newly formed undulating membrane anlagen (arrow) at early-middle stage. (F,G) Ventral views of middle dividers, to show the intrakinetally formed left marginal anlagen (arrow) and the newly developed cirri (double-arrowhead) formed from the frontoventral transverse cirral anlagen. (H) Dorsal view to show the dorsal kinety anlagen (arrows). (I,J) Macronuclear nodules fusing into a branching mass. (K) Ventral view of a late divider to show the newly formed right marginal cirri (arrowhead) and frontoterminal cirri (circle). (L) Dorsal view to show the new dorsal kineties, no caudal cirri are formed. (M,N) Ventral views of very late dividers, to show the immigration of frontoterminal cirri (circle). Scale bars = 50 μm.
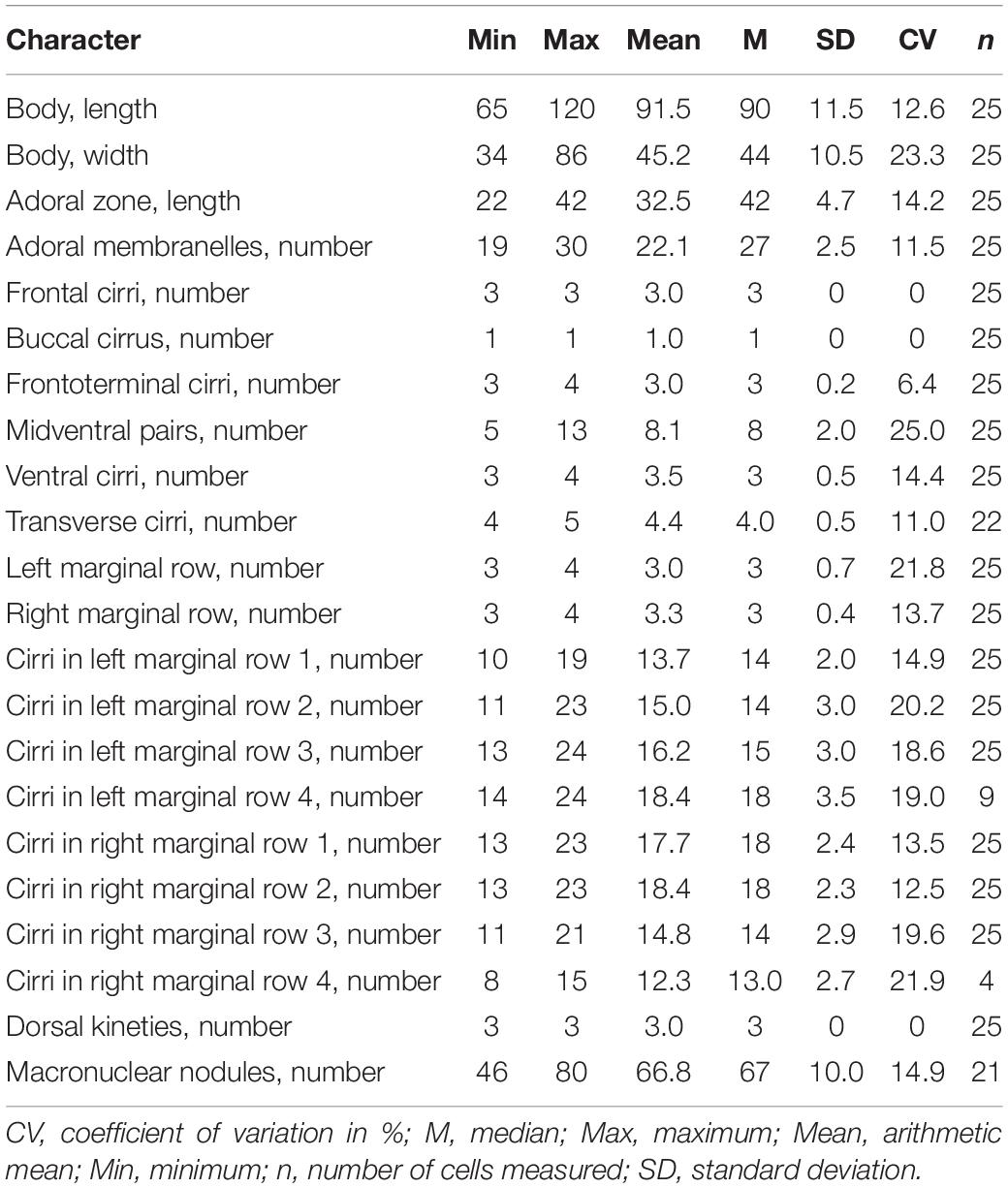
Table 1. Morphometric characterization of Metaurostylopsis alrasheidi n. sp. based on protargol-stained specimens (measurements in μm).
Diagnosis
Body 60–115 × 20–60 μm in vivo; two types of cortical granules: larger yellow-greenish ones and smaller colorless ones; about 22 adoral membranelles; three frontal, three or four frontoterminal, one buccal, four or five transverse cirri; 5–13 midventral pairs and a midventral row of three or four unpaired ventral cirri; three or four left and right marginal rows respectively; invariably three dorsal kineties; over 40 macronuclear nodules; freshwater habitat.
Type Locality
Lake Weishan (34°46′14″N, 117°12′56″E), Shandong Province, China, a freshwater lake crowded with water plants, algae and decaying plant matter.
Material Deposited
The protargol slide (accession no. SWY2019060301-1) containing the holotype and several paratype slides (accession no. SWY2019060301-2–9) are deposited in the Laboratory of Protozoology, Ocean University of China.
Dedication
We dedicate this new species to our eminent colleague, Prof. Khaled A.S. Al-Rasheid, King Saud University, Saudi Arabia, in recognition of his contributions to ciliatology.
Description
Cell size about 60–115 × 20–60 μm in vivo, with ratio of length to width about 2–3:1, body outline broad to elongated elliptical (Figures 2A,B,I,J). Cortex rather flexible, but not contractile. Two types of cortical granules on both ventral and dorsal sides (Figures 2E–H): larger ones about 1.0 μm in size, yellow-greenish, wheat-grain shaped along the cirral rows, dorsal kineties and between dorsal dikinetids, giving cells a yellow-greenish appearance; smaller ones colorless, about 0.4–0.5 μm in diameter, relatively densely arranged. Cytoplasm colorless, usually with many tiny lipid droplets (about 0.6–1.0 μm across) and food vacuoles containing algae. Contractile vacuole about 15–18 μm across, located at two-fifth of body length, behind buccal vertex, near left body margin, collecting canals not observed (Figures 2A,B,J). On average 67 (46–80) spherical to elliptical macronuclear nodules (Figure 2K). Micronuclei not easily distinguished from macronuclear nodules in stained individuals. Locomotion unremarkable, by moderately fast crawling on bottom of Petri dishes and debris.
Adoral zone extending about 38% of body length, with cilia up to 10 μm long in vivo, consisting of 19–30 (on average 22) membranelles (Table 1), distal end of adoral zone of membranelles (AZM) bending only slightly to the right of the midline in protargol preparations (Figures 2C,K). Paroral and endoral of nearly equal length, slightly curved and optically intersect at about posterior quarter of the paroral (Figures 2C,K,M). Three clearly differentiated frontal cirri (Figures 2C,K and Table 1). Single buccal cirrus located at level of anterior one-third of paroral (Figures 2C,K,M and Table 1). Three or four frontoterminal cirri behind the distal end of adoral zone (Figures 2C,K and Table 1). Midventral complex extending to about half of cell length, composed of five to thirteen pairs of cirri arranged in typical zigzag pattern and a midventral row of three or four unpaired ventral cirri posteriorly (Figures 2A–C,K and Table 1). Four or five transverse cirri in J-shaped row (Figures 2A–C,K,L and Table 1). Both right and left marginal cirri composed of three to four rows (Figures 2A–C,K and Table 1). Three complete dorsal kineties and two or three dorsal bristles to right of dorsal kinety 3 anteriorly (Figure 2D, arrowheads), with cilia 2–3 μm long.
Divisional Ontogenesis
Stomatogenesis and Development of Frontal-Ventral-Transverse Cirral Anlagen
Stomatogenesis commences with the proliferation of densely arranged basal bodies forming a slender anarchic field, that is, the opisthe’s oral primordium, which occurs de novo to the left of posterior midventral pairs (Figures 3A, 5A). Simultaneously, a small field of basal bodies, the proter’s oral primordium, appears de novo in a subsurface pouch beneath the buccal field and near the parental undulating membranes (UM). Subsequently, the opisthe’s oral primordium widens and lengthens between the midventral complex and the inner left marginal row during which time the parental structures remain unchanged. At the same time, the proter’s oral primordium continues to develop within the pouch, which is clearly outlined (Figures 3B, 5B,C). The oral primordium then differentiates into new adoral membranelles, and the UM anlage appears to the right of the oral primordium in both proter and opisthe (Figures 3D,E, 5D–G). During this time, the parental oral structures remain unchanged, while the frontal-ventral-transverse cirral anlagen (FVTA) appear as several streak-like collections of basal bodies to the right of the UM anlage for both proter and opisthe (Figures 3D,E, 5D,E,G). In the next stage, the differentiation of adoral membranelles is almost complete, forming new structures for both proter and opisthe. The UM anlagen split and give rise to the paroral and endoral membranes and the leftmost frontal cirrus is generated from the anterior end of the UM anlage (=FVTA I) (Figures 3G, 5G). Simultaneously the FVTA begin to break apart and differentiate into new cirri. In the following stages, the new cirri formed from the FVTA and migrate to their final positions as follows: FVTA II produces the middle frontal cirrus and the buccal cirrus; FVTA III provides the rightmost frontal cirrus and the parabuccal cirrus (III/2); FVTA IV to n-2 each develops a midventral pair, with the last two or three streaks forming one transverse cirrus; FVTA n-1 provides a short midventral row composed of three or four cirri and one transverse cirrus; FVTA n, the last anlage, develops three or four frontoterminal cirri and probably the rightmost transverse cirrus (Figures 4A,C,D, 5K,M,N).
Development of Marginal Rows and Dorsal Kineties
At the early-to-middle stages, the marginal anlagen are formed intrakinetally both in proter and opisthe. Subsequently, these anlagen elongate toward both ends and generate new cirri and gradually replace the parental structures (Figures 3D,E,G, 4A,C,D, 5F,K,M,N). In the same way, the dorsal kineties develop by intrakinetal basal body proliferation. During cell division, no caudal cirri are formed. As some key stages were not observed, the origins of the extra dorsal bristles (usually only two) are not clear (Figures 3C,F,H, 4B,E, 5H,L).
Division of Nuclear Apparatus
Although macronuclear replication bands were not definitely identified in M. alrasheidi n. sp., they have been identified (but seen with some difficulty) in M. rubra and M. marina which, otherwise, have the same macronuclear morphology as M. alrasheidi n. sp. during division and replication bands were very possibly overlooked in the new species (Song et al., 2001; Song and Wilbert, 2002). During the middle stage, all macronuclear nodules fuse into a single somewhat branched complex which then divides into numerous nodules (Figures 3C,F,H, 4B,E, 5I,J,N). Division of the micronuclei was not observed.
Phylogeny Analyses Based on 18S rRNA Gene Sequence
The partial 18S rRNA gene sequence of Metaurostylopsis alrasheidi n. sp. has been submitted into GenBank database (acc. no. MT911525), with 1,643 bp long and G + C content of 44.92%.
According to sequence comparisons of the 18S rRNA gene, the sequence of Metaurostylopsis alrasheidi n. sp. differs by 8 bp from an unidentified Metaurostylopsis sequence (FJ870098) (from a taxonomic research lab), 13–14 bp from five populations of M. cheni, 16 bp from M. salina, 34–44 bp from four M. struederkypkeae sequences, and 73 bp from Monourostylopsis antarctica (Jung et al., 2011) n. comb (Figure 6).
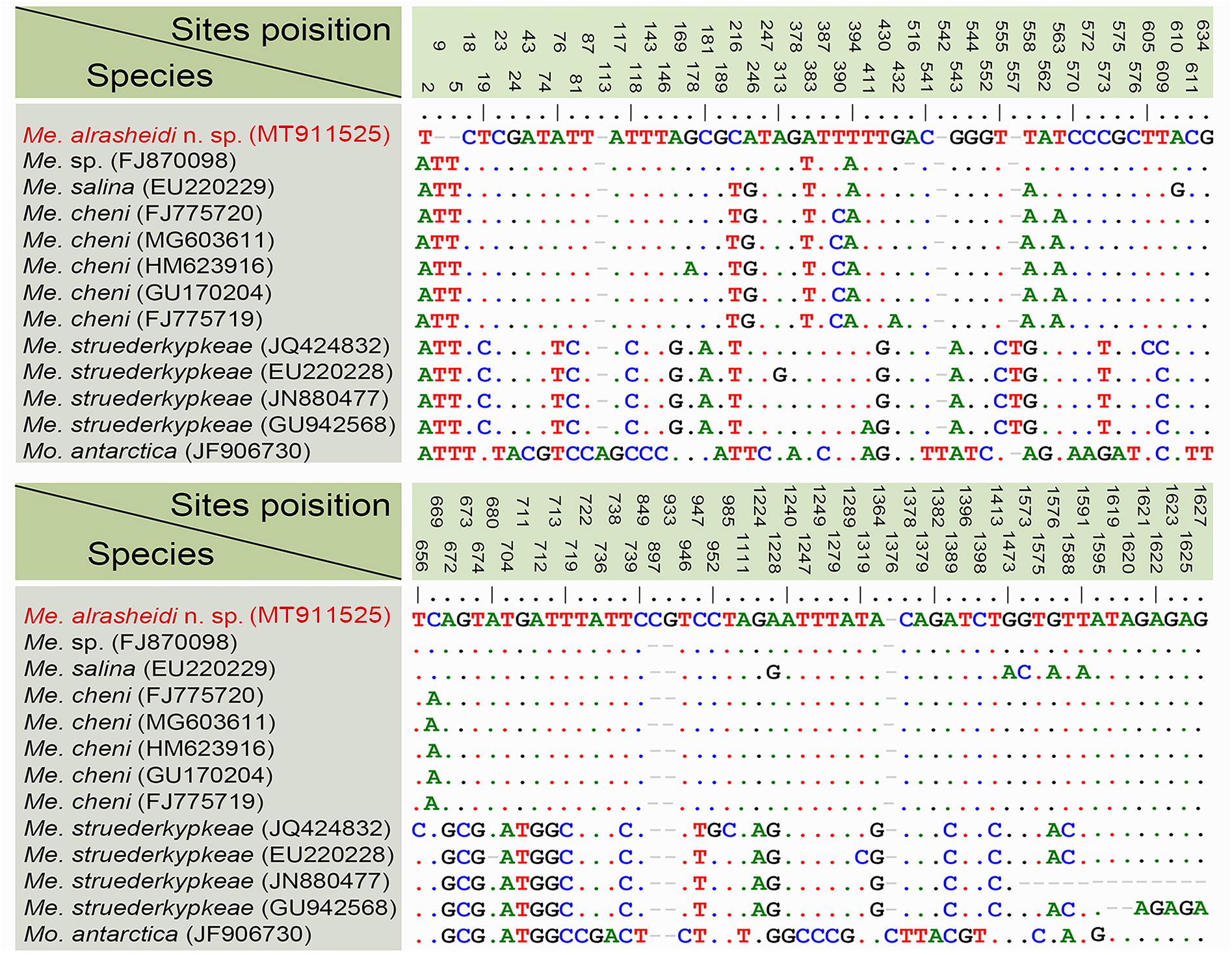
Figure 6. Nucleotide differences among populations of Metaurostylopsis species and Monourostylopsis antarctica (Jung et al., 2011) n. comb., based on 18S rRNA gene sequences. The vertical numbers indicate the unmatched site positions. The new sequence is in red. Short lines (–) suggest the insertions and deletions. Solid dots represent matched sites.
The topologies of the ML and BI trees are mostly congruent. As a result, only the ML tree with nodal supports inferred from both algorithms is shown (Figure 7). According to our 18S rRNA gene tree, M. alrasheidi n. sp. clusters with Metaurostylopsis sp. (FJ870098), M. salina (EU220229) and five M. cheni populations (GU170204, FJ775719, HM623916, MG603611, FJ775720) with full support, forming a sister clade of four M. struederkypkeae populations (JQ424832, GU942568, EU220228, JN880477). Monourostylopsis antarctica (Jung et al., 2011) n. comb. (JF906730) is sister to all Metaurostylopsis species with strong to full support (ML/BI: 97/1.00).
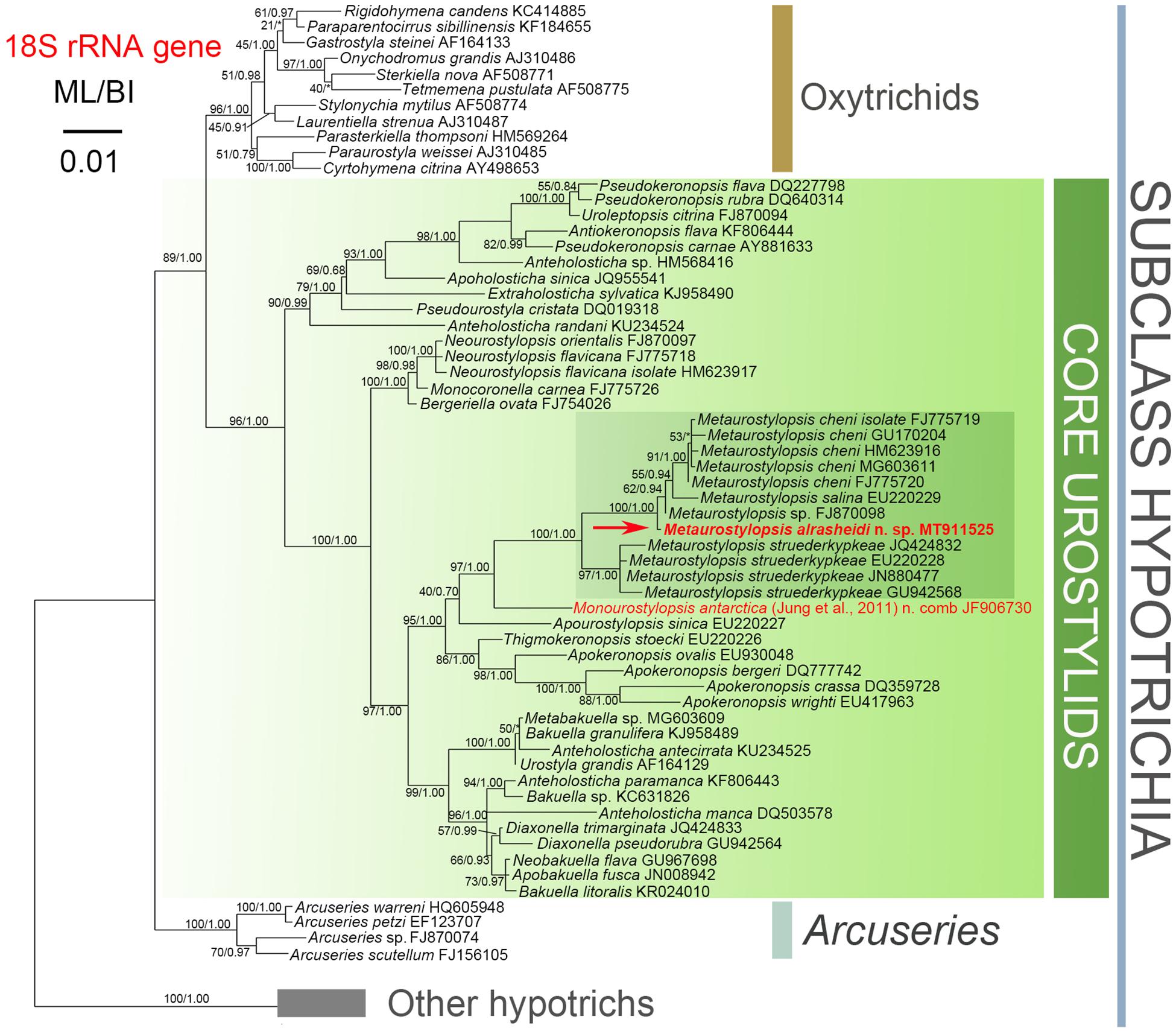
Figure 7. Maximum likelihood (ML) phylogenetic tree with 18S rRNA gene as gene marker. The newly characterized sequence is in red (arrow). Numbers near nodes represent bootstrap values of ML and posterior probability values of Bayesian inference (BI), respectively. Asterisks (*) indicate the disagreement between ML and BI. The scale bar represents 1 substitution for every 100 nucleotides.
Geographic Distribution
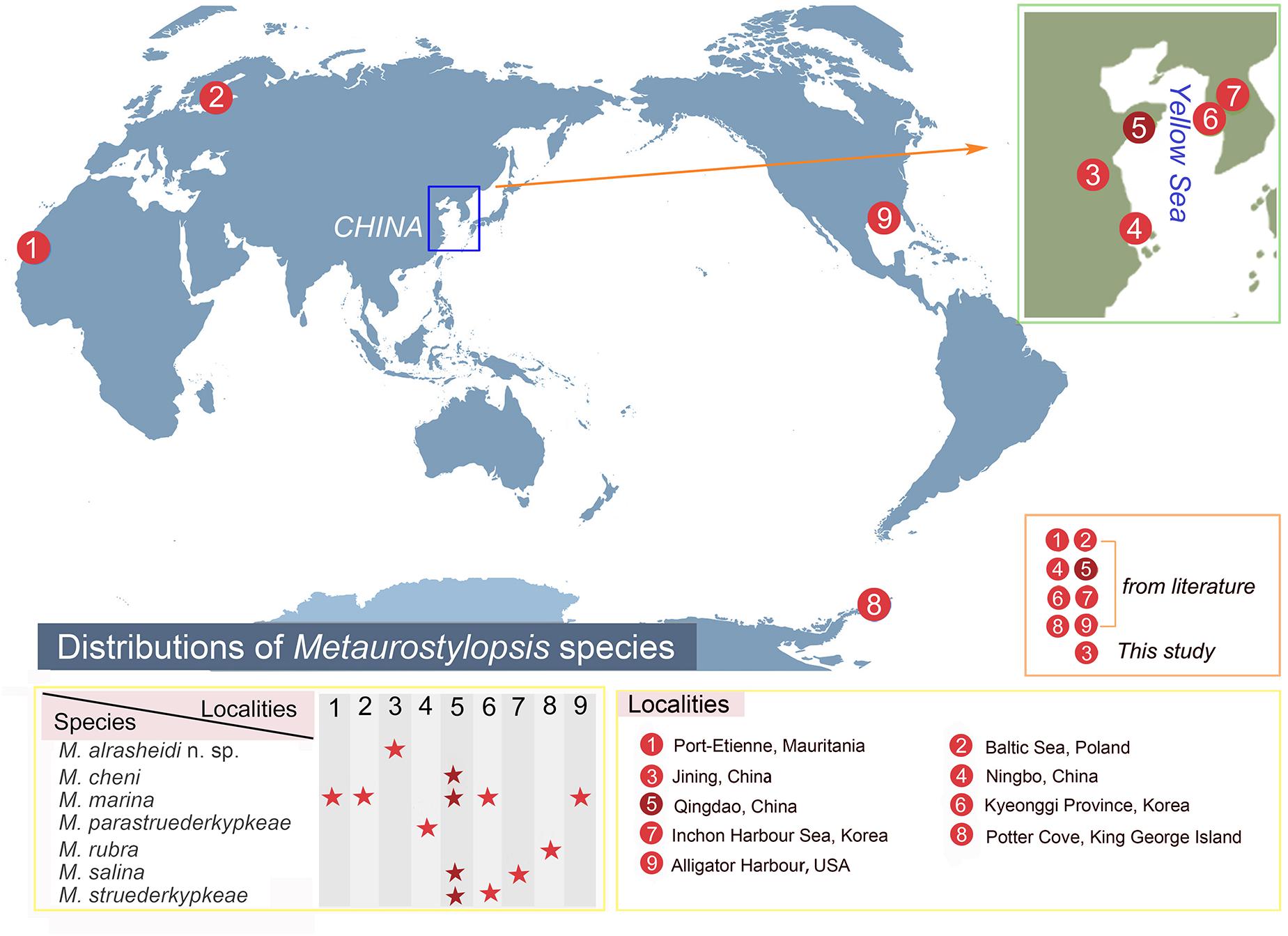
Metaurostylopsis alrasheidi n. sp. was isolated from freshwater habitat. All the other Metaurostylopsis species have been reported in marine or brackish water (Song et al., 2001; Lei et al., 2005; Berger, 2006; Shao et al., 2008a, b; Chen et al., 2011; Lu et al., 2016). These studies have shown that Metaurostylopsis species can be found in a variety of habitats (Figure 8). Metaurostylopsis marina, has been reported in Mauritania, Poland, China, Korea, and United States, and is probably a cosmopolitan species. While most of the other Metaurostylopsis species have been found in one place only, this likely reflects undersampling.
Discussion
Comparison of Metaurostylopsis alrasheidi n. sp. With Closely Related Species (Table 2)
With respect to the ciliature, that is, frontal and transverse cirri clearly differentiated, buccal cirri present, more than two frontoterminal cirri, midventral complex with midventral pairs and a single short midventral row, more than two marginal rows on each body side; pretransverse cirri and caudal cirri absent, the new isolate corresponds well with the diagnosed characteristics of the genus Metaurostylopsis (Chen et al., 2013).
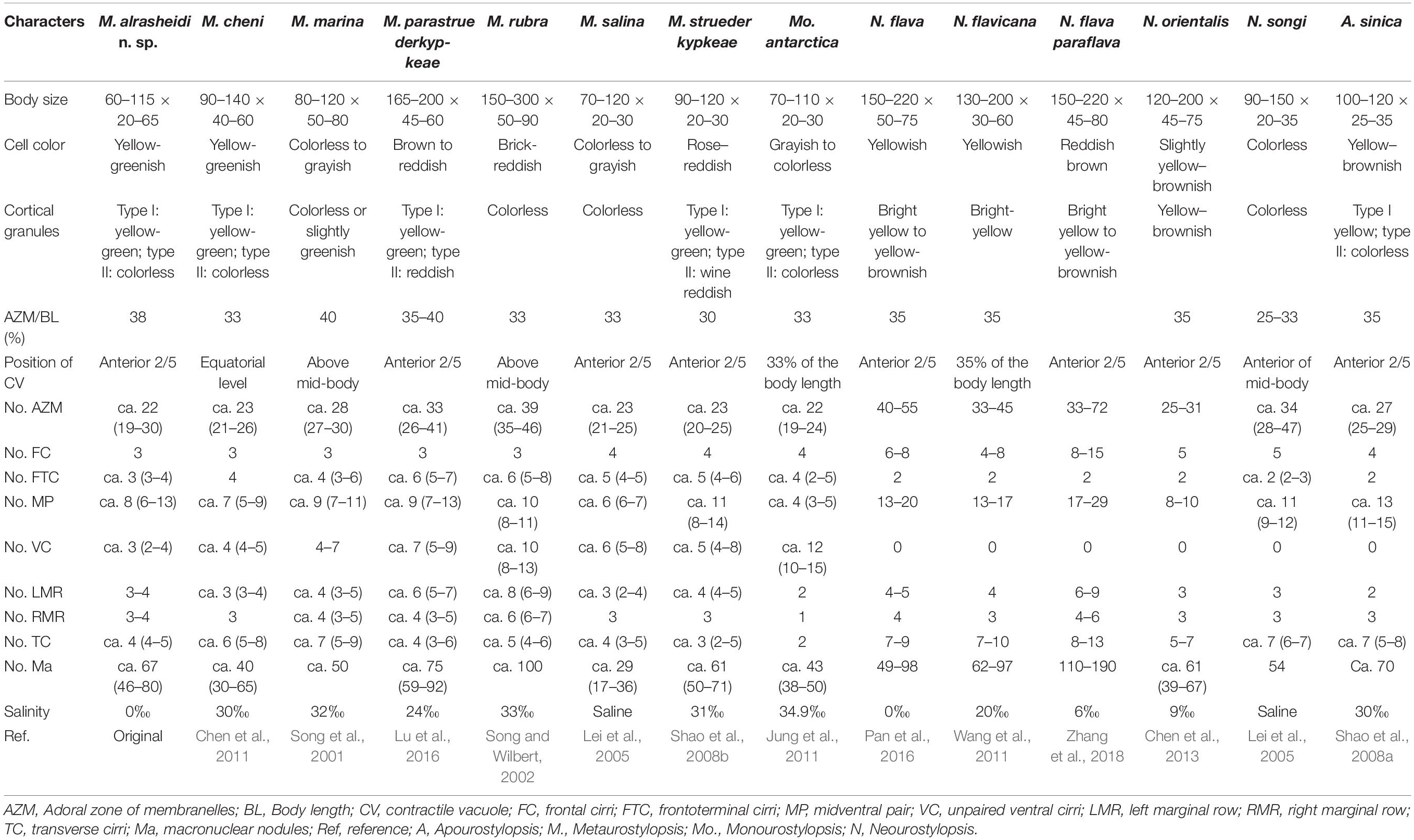
Table 2. Morphometric comparison of Metaurostylopsis alrasheidi n. sp. with closely related species.
Metaurostylopsis alrasheidi n. sp. is most similar to M. cheni, especially in body shape, yellow-greenish cell color, two types of cortical granules, and most aspects of the ciliature, but differs from the latter by having a relatively smaller body size in vivo 60–115 × 20–60 μm vs. 90–140 × 40–60 μm, a dense distribution pattern of the smaller type of cortical granules on dorsal side (vs. sparse), contractile vacuole located anteriorly (2/5 of body length vs. equatorial region), relatively more midventral pairs (5–13 vs. 5–9) and less transverse cirri (4–5 vs. 5–8) (Chen et al., 2011). The 18S rRNA gene sequence of the newly isolated organism differs from that of M. cheni (GU170204) by 13 nucleotides (Figure 6), which also reveals these two species are not conspecific.
Metaurostylopsis alrasheidi n. sp. differs from M. marina in body shape (broad to elongated elliptical vs. oval), two (vs. one) types of cortical granules, relatively fewer frontoterminal cirri (3–4 vs. 3–6), frontal cirri (3 vs. 4), transverse cirri (4–5 vs. 5–9), and different habitat (freshwater vs. marine) (Song et al., 2001).
Metaurostylopsis parastruederkypkeae is distinguished from M. alrasheidi n. sp. by having reddish body (vs. yellow-greenish), reddish cortical granules (vs. yellow-greenish), more frontoterminal cirri (5–7 vs. 3–4), more adoral membranelles (28–47 vs. 19–30), and more cirri in midventral row (5–9 vs. 3–4) (Lu et al., 2016).
Metaurostylopsis rubra can be easily distinguished from M. alrasheidi n. sp., by its distinctly larger body size (150–300 × 50–90 vs. 60–115 × 20–60 μm), brick-reddish body color (vs. yellow-greenish), one (vs. two) type of cortical granules, and higher numbers of membranelles (35–46 vs. 19–30) (Song and Wilbert, 2002).
Metaurostylopsis salina differs from M. alrasheidi n. sp. mainly in terms of having only one (vs. two) type of cortical granules and relatively more cirri in the midventral row 5–8 (vs. 3–4) (Shao et al., 2008a).
Metaurostylopsis struederkypkeae can be easily separated from M. alrasheidi n. sp. by its cell coloration (rose-reddish vs. yellow-greenish), the number of frontal cirri (4 vs. 3), frontoterminal cirri (4–6 vs. 3–4), and the distribution of the smaller cortical granules (in irregular rows vs. evenly distributed) (Shao et al., 2008b).
Metaurostylopsis antarctica resembles M. alrasheidi n. sp. closely in terms of body size, number of membranelles and types of cortical granules, but can be separated from the new species by having fewer midventral pairs (2–5 vs. 5–13), and fewer left and right marginal rows (2 vs. 3–4, 1 vs. 3–4 respectively) and transverse cirri (2 vs. 4–5) (Jung et al., 2011).
The character of cortical granules, that is, two types of cortical granules, of the new species is distinguishable from some of the congeners with only one type of cortical granules present (Table 2). However, it is possible that earlier researchers overlooked the tiny type of cortical granules. Considering the habitat, Metaurostylopsis alrasheidi n. sp. can be separated from all the other congeners (freshwater vs. marine or brackish, Table 2).
All the Neourostylopsis species can be separated from M. alrasheidi n. sp. by having fewer frontoterminal cirri (2 vs. 3–4), lack of a midventral row (vs. midventral row present) (Lei et al., 2005; Wang et al., 2011; Chen et al., 2013; Pan et al., 2016; Zhang et al., 2018).
Apourostylopsis sinica (Shao et al., 2008) Song et al., 2011 differs from our new species in its more transverse cirri (5–8 vs. 4–5), pretransverse cirri present (vs. absent), and midventral row absent (vs. present) (Shao et al., 2008a).
Emended Diagnosis of Genus Metaurostylopsis and Reassignment of the Species, M. antarctica Jung et al., 2011
In the most recent revision of the genus Metaurostylopsis, Chen et al. (2013) provided an improved diagnosis for the genus Metaurostylopsis, which were defined as marine urostylids with characteristics mentioned in the above section. However, the new species in the present study, Metaurostylopsis alrasheidi n. sp., which was isolated from a freshwater lake, fits the morphological characteristics of the genus Metaurostylopsis very well. Moreover, our new species clusters in a clade together with all the other Metaurostylopsis sequences, indicating the close relationship among them and supporting the classification of the new species in the genus Metaurostylopsis. Thus, an emended diagnosis including marine, brackish and freshwater habitats is supplied.
As discussed above, the genus Metaurostylopsis is characterized by having more than one marginal row on each body side. However, M. antarctica was characterized by having only one right marginal row, which does not fit the diagnosis of Metaurostylopsis. Also, in the phylogenetic trees, M. antarctica is placed as sister to the clade including all the other Metaurostylopsis sequences with strong to full support. Therefore, we propose that M. antarctica should be excluded from the genus Metaurostylopsis. As M. antarctica also can be separated from other morphologically similar genera, i.e., Neourostylopsis and Apourostylopsis, by having only one right marginal row (vs. two or more right marginal rows) (Table 2), this suggests that M. antarctica represents a new genus. Hence, an emended diagnosis of the genus Metaurostylopsis and diagnosis of a new genus, Monourostylopsis n. gen., are supplied here.
Metaurostylopsis Song et al., 2001
Improved Diagnosis
Marine, brackish or freshwater urostylids with frontal and transverse cirri clearly differentiated; three or more frontoterminal cirri; midventral complex with midventral pairs extending to the level of the buccal vertex and a single midventral row; buccal and transverse cirri present; pretransverse ventral and caudal cirri absent; more than two marginal rows on each body side.
Type Species
Urostyla marina Kahl, 1932.
Species Assignable
Metaurostylopsis marina (Kahl, 1932) Song et al., 2001 (type species); Metaurostylopsis rubra Song and Wilbert, 2002; Metaurostylopsis salina Lei et al., 2005; Metaurostylopsis struederkypkeae Shao et al., 2008; Metaurostylopsis cheni Chen et al., 2011; Metaurostylopsis parastruederkypkeae Lu et al., 2016; Metaurostylopsis alrasheidi n. sp.
Monourostylopsis n. gen.
Diagnosis
Urostylids with frontal and transverse cirri clearly differentiated; three or more frontoterminal cirri; midventral complex with midventral pairs extending to the level of the buccal vertex and a single midventral row; buccal and transverse cirri present; pretransverse ventral and caudal cirri absent; one right marginal row and two or more left marginal rows.
Type Species
Monourostylopsis antarctica (Jung et al., 2011) n. comb. (original combination: Metaurostylopsis antarctica Jung et al., 2011).
Species Assignable
Monourostylopsis antarctica (Jung et al., 2011) n. comb. (type species).
Etymology
The generic name is a composite of the Greek mono- (single), referring to the single right marginal row, and the posterior part (-urostylopsis) of the genus-group name Metaurostylopsis.
Morphogenetic Comparison
As of this writing, the morphogenetic process of four Metaurostylopsis species, i.e., M. alrasheidi n. sp., M. cheni, M. marina, and M. rubra has been investigated (Wilbert and Song, 2005; Chen et al., 2011; Song et al., 2011). The morphogenetic pattern of the genus Metaurostylopsis shows high stability, and shares similar morphogenetic features with Apourostylopsis, Monourostylopsis, and Neourostylopsis as follows: (1) in the proter, the oral primordium (OP) is formed de novo in a subcortical pouch beneath (i.e., dorsal to) the buccal field, the new undulating membranes (UM) anlage is generated near the OP, and the parental oral structures are completely replaced; (2) in the opisthe, both the OP and the UM anlage develop apokinetally on the cell surface; (3) the FVT cirral anlagen are formed apokinetally, and none of the old ciliature seems to be involved in the development of the new primordia; (4) marginal anlagen and dorsal anlagen develop intrakinetally; (5) all macronuclear nodules fuse to a single branching mass during the middle divisional stage (Wilbert and Song, 2005; Shao et al., 2008a; Chen et al., 2011; Jung et al., 2011; Song et al., 2011; Zhang et al., 2018).
Ontogenetic features of Metaurostylopsis show striking differences when compared with genera Apourostylopsis and Neourostylopsis, namely: (1) FVTA n-1 forms a row of unpaired ventral cirri which migrates along with the midventral pairs, whereas in Apourostylopsis and Neourostylopsis, no unpaired ventral cirri are generated; (2) FVTA n formed a transverse cirrus and more than two frontoterminal cirri in Metaurostylopsis (i.e., typical Metaurostylopsis mode), whereas the FVTA n forms two frontoterminal cirri, a pretransverse and a transverse cirrus in Apourostylopsis (i.e., typical urostylid mode), and whether FVTA n forms pretransverse cirrus in Neourostylopsis is variable (Wilbert and Song, 2005; Shao et al., 2008a; Chen et al., 2011; Jung et al., 2011; Song et al., 2011; Song and Shao, 2017; Zhang et al., 2018).
Phylogenetic Analyses
Consistent with previous studies (Shao et al., 2008a; Chen et al., 2011; Song et al., 2011; Lu et al., 2016), we confirm the monophyly of genus Metaurostylopsis based on the phylogenetic analyses in present work.
In the 18S rRNA gene tree, our new taxon, Metaurostylopsis alrasheidi n. sp., clusters with Metaurostylopsis species and falls into the monophyletic clade of Metaurostylopsis, which can also be supported by the shared morphological features, such as, similar body shape and size, as well as semblable ciliature. Additionally, 8–44 different nucleotides of 18S rRNA gene are discovered between M. alrasheidi n. sp. and its congeners, further indicating the validity of our isolate as a distinct species. It is worth noting that the genus Metaurostylopsis forms a sister clade to Monourostylopsis n. gen., with which it shares some similar characteristics: (1) three or more frontoterminal cirri; (2) midventral complex with midventral pairs and a single midventral row; (3) buccal and transverse cirri present; (4) pretransverse ventral and caudal cirri absent. However, in addition to its molecular phylogenetic position, Metaurostylopsis can be clearly separated from Monourostylopsis n. gen. morphologically by having more than two marginal rows on each body side (vs. one left marginal row and two right marginal rows) (Jung et al., 2011).
Data Availability Statement
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GenBank (accession: MT911525).
Author Contributions
XL and TZ conceived the study. WS carried out the live observation, protargol staining, DNA extraction, and data analyses. All authors contributed to the manuscript and approved the final version.
Funding
This work was financially supported by the National Natural Science Foundation of China (Nos. 32030015 and 31900319) and the China Postdoctoral Science Foundation Grants (Nos. BX20180348 and 2018M642955).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to express our gratitude to Dr. Xumiao Chen, Institute of Oceanology, Chinese Academy of Sciences, for her kind help and advice on the identification of this species.
Footnotes
References
Berger, H. (1999). Monograph of the Oxytrichidae (Ciliophora, Hypotrichia). Vol. 78, Berlin: Springer, 1–1080. Monographiae Biologicae.
Berger, H. (2006). Monograph of the Urostyloidea (Ciliophora, Hypotricha). Vol. 85, Berlin: Springer, 1–1304. Monographiae Biologicae.
Berger, H. (2008). Monograph of the Amphisiellidae and Trachelostylidae (Ciliophora, Hypotricha). Vol. 88, Dordrecht: Springer, 1–737. Monographiae Biologicae.
Berger, H. (2011). Monograph of the Gonostomatidae and Kahliellidae (Ciliophora, Hypotricha). Vol. 90, Dordrecht: Springer, 1–740. Monographiae Biologicae.
Chen, L., Dong, J., Wu, W., Xin, Y., Warren, A., Ning, Y., et al. (2020). Morphology and molecular phylogeny of a new hypotrich ciliate, Anteholosticha songi nov. spec., and an American population of Holosticha pullaster (Müller, 1773) Foissner et al., 1991 (Ciliophora, Hypotrichia). Eur. J. Protistol. 72:125646. doi: 10.1016/j.ejop.2019.125646
Chen, X., Huang, J., and Song, W. (2011). Ontogeny and phylogeny of Metaurostylopsis cheni sp. n. (Protozoa, Ciliophora), with estimating the systematic position of Metaurostylopsis. Zool. Scr. 40, 99–111. doi: 10.1111/j.1463-6409.2010.00451.x
Chen, X., Shao, C., Liu, X., Huang, J., and Al-Rasheid, K. A. S. (2013). Morphology and phylogenies of two hypotrichous brackish-water ciliates from China, Neourostylopsis orientalis n. sp. and Protogastrostyla sterkii (Wallengren, 1900) n. comb., with establishment of a new genus Neourostylopsis n. gen. (Protista, Ciliophora, Hypotrichia). Int. J. Syst. Evol. Microbiol. 63, 1197–1209. doi: 10.1099/ijs.0.049403-0
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Dong, J., Li, L., Fan, X., Ma, H., and Warren, A. (2020). Two Urosoma species (Ciliophora, Hypotrichia): a multidisciplinary approach provides new insights into their ultrastructure and systematics. Eur. J. Protistol. 72:125661. doi: 10.1016/j.ejop.2019.125661
Foissner, W. (1982). Ökologie und taxonomie der hypotrichida (Protozoa: Ciliophora) einiger österreichischer Böden). Arch. Protistenk. 126, 19–143. doi: 10.1016/S0003-9365(82)80065-1
Foissner, W. (2016). Terrestrial and semiterrestrial ciliates (Protozoa, Ciliophora) from venezuela and Galápagos. Denisia 35:912.
Gao, F., Katz, L. A., and Song, W. (2012). Insights into the phylogenetic and taxonomy of philasterid ciliates (Protozoa, Ciliophora, Scuticociliatia) based on analyses of multiple molecular markers. Mol. Phylogenet. Evol. 64, 308–317. doi: 10.1016/j.ympev.2012.04.008
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analyses program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hu, X., Lin, X., and Song, W. (2019). Ciliate Atlas: Species found in the South China Sea. Beijing: Science Press.
Jankowski, A. V. (2007). “Phylum ciliophora doflein, 1901. Review of taxa,” in Protista: Handbook on Zoology, Part 2, ed. A. F. Alimov (St. Petersburg: Nauka), 415–993.
Jerome, C. A., Simon, E. M., and Lynn, D. H. (1996). Description of Tetrahymena empidokyrea n. sp., a new species in the Tetrahymena pyriformis sibling species complex (Ciliophora, Oligohymenophorea), and an assessment of its phylogenetic position using small-subunit rRNA sequences. Can. J. Zool. 74, 1898–1906. doi: 10.1139/z96-214
Jung, J. H., Baek, Y. S., Kim, S., Choi, H. G., and Min, G. S. (2011). A new marine ciliate, Metaurostylopsis antarctica nov. spec. (Ciliophora, Urostylida) from the Antarctic Ocean. Acta Protozool. 50, 289–300. doi: 10.4467/16890027AP.11.026.0063
Jung, J. H., and Berger, H. (2019). Monographic treatment of Paraholosticha muscicola (Ciliophora, Keronopsidae), including morphological and molecular biological characterization of a brackish water population from Korea. Eur. J. Protistol. 68, 48–67. doi: 10.1016/j.ejop.2018.12.004
Kaur, H., Shashi, L., Negi, R. K., and Kamra, K. (2019). Morphological and molecular characterization of Neogastrostyla aqua nov. gen., nov. spec. (Ciliophora, Hypotrichia) from River Yamuna, Delhi; comparison with Gastrostyla-like genera. Eur. J. Protistol. 68, 68–79. doi: 10.1016/j.ejop.2019.01.002
Kim, K. S., Jung, J. H., and Min, G. S. (2017). Morphology and molecular phylogeny of two new ciliates, Holostichides heterotypicus n. sp. and Holosticha muuiensis n. sp. (Ciliophora: Urostylida). J. Eukaryot. Microbiol. 64, 873–884. doi: 10.1111/jeu.12421
Kim, K. S., and Min, G. S. (2019). Morphology and molecular phylogeny of Oxytricha seokmoensis sp. nov. (Hypotrichia: Oxytrichidae), with notes on its morphogenesis. Eur. J. Protistol. 71:125641. doi: 10.1016/j.ejop.2019.125641
Kumar, S., Kamra, K., and Sapra, G. R. (2010). Ciliates of the silent valley National Park, India: Urostyloid hypotrichs of the region with a note on the habitat. Acta Protozool. 49, 339–364.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lei, Y., Choi, J. K., Xu, K., and Petz, W. (2005). Morphology and infraciliature of three species of Metaurostylopsis (Ciliophora, Stichotrichia): M. songi n. sp., M. salina n. sp., and M. marina (Kahl 1932) from sediments, saline ponds, and coastal waters. J. Eukaryot. Microbiol. 52, 1–10. doi: 10.1111/j.1550-7408.2005.3294rr.x
Li, F., Li, Y., Luo, D., Miao, M., and Shao, C. (2018). Morphology, morphogenesis, and molecular phylogeny of a new soil ciliate, Sterkiella multicirrata sp. nov. (Ciliophora, Hypotrichia) from China. J. Eukaryot. Microbiol. 65, 627–636. doi: 10.1111/jeu.12508
Lian, C., Zhang, T., Al-Rasheid, K. A. S., Yu, Y., Jiang, J., and Huang, J. A. (2019). Morphology and SSU rDNA-based phylogeny of two Euplotes species from China: E. wuhanensis sp. n. and E. muscicola Kahl, 1932 (Ciliophora, Euplotida). Eur. J. Protistol. 67, 1–14. doi: 10.1016/j.ejop.2018.10.001
Lu, B., Wang, C., Huang, J., Shi, Y., and Chen, X. (2016). Morphology and SSU rDNA sequence analysis of two hypotrichous ciliates (Protozoa, Ciliophora, Hypotrichia) including the new species Metaurostylopsis parastruederkypkeae n. sp. J. Ocean Univ. China 15, 866–878. doi: 10.1007/s11802-016-3148-9
Lu, X., Wang, Y., Al-Farraj, S. A., El-Serehy, H., Huang, J., and Shao, C. (2020). The insights into the systematic relationship of Gastrostyla-affinitive genera, with report on a new saline soil ciliate genus and new species (Protozoa, Ciliophora). BMC Evol. Biol. 20:92. doi: 10.1186/s12862-020-01659-8
Luo, X., Huang, J. A., Li, L., Song, W., and Bourland, W. A. (2019). Phylogeny of the ciliate family Psilotrichidae (Protista, Ciliophora), a curious and poorly-known taxon, with notes on two algae-bearing psilotrichids from Guam, USA. BMC Evol. Biol. 19:125. doi: 10.1186/s12862-019-1450-z
Lyu, Z., Wang, J., Huang, J., Warren, A., and Shao, C. (2018). Multigene-based phylogeny of Urostylida (Ciliophora, Hypotrichia), with establishment of a novel family. Zool. Scr. 47, 243–254. doi: 10.1111/zsc.12267
Ma, J., Zhao, Y., Zhang, T., Shao, C., Al-Rasheid, K. A. S., and Song, W. (2020). Cell-division pattern and phylogenetic analyses of a new ciliate genus Parasincirra n. g. (Protista, Ciliophora, Hypotrichia), with a report of a new soil species, P. sinica n. sp. from northwest China. BMC Evol. Biol.
Medlin, L., Elwood, H. J., Stickel, S., and Sogin, M. L. (1988). The characterization of enzymatically amplified eukaryotes 16S-like ribosomal RNA coding regions. Gene 71, 491–500. doi: 10.1016/0378-1119(88)90066-2
Pan, X., Fan, Y., Gao, F., Qiu, Z., Al-Farraj, S. A., Warren, A., et al. (2016). Morphology and systematics of two freshwater urostylid ciliates, with description of a new species (Protista, Ciliophora, Hypotrichia). Eur. J. Protistol. 52, 73–84. doi: 10.1016/j.ejop.2015.11.003
Ronquist, F., Teslenko, M., Van Der, M., Daniel, La, Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large Model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sela, I., Ashkenazy, H., Katoh, K., and Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. doi: 10.1093/nar/gkv318
Shao, C., Hu, C., Fan, Y., Warren, A., and Lin, X. (2019). Morphology, morphogenesis and molecular phylogeny of a freshwater ciliate, Monomicrocaryon euglenivorum euglenivorum (Ciliophora, Oxytrichidae). Eur. J. Protistol. 68, 25–36. doi: 10.1016/j.ejop.2019.01.001
Shao, C., Miao, M., Song, W., Warren, A., Al-Rasheid, K. A. S., Al-Quraishy, S. A., et al. (2008a). Studies on two marine Metaurostylopsis spp. from China with notes on morphogenesis in M. sinica nov. spec. (Ciliophora, Urostylida). Acta Protozool. 47, 95–112.
Shao, C., Song, W., Al-Rasheid, K. A. S., Yi, Z., Chen, X., Al-Farraj, S. A., et al. (2008b). Morphology and infraciliature of two new marine urostylid ciliates: Metaurostylopsis struederkypkeae n. sp. and Thigmokeronopsis stoecki n. sp. (Ciliophora, Hypotrichida) from China. J. Eukaryot. Microbiol. 55, 289–296. doi: 10.1111/j.1550-7408.2008.00327.x
Song, W., Petz, W., and Warren, A. (2001). Morphology and morphogenesis of the poorly-known marine urostylid ciliate, Metaurostylopsis marina (Kahl, 1932) nov. gen., nov. comb. (Protozoa, Ciliophora, Hypotrichida). Eur. J. Protistol. 37, 63–76. doi: 10.1078/0932-4739-00802
Song, W., and Shao, C. (2017). Ontogenetic Patterns of Hypotrich Ciliates (in Chinese). Beijing: Science Press.
Song, W., Warren, A., and Hu, X. (2009). Free-Living Ciliates in the Bohai and Yellow Seas, China. Beijing: Science Press.
Song, W., and Wilbert, N. (2002). Faunistic studies on marine ciliates from the Antarctic benthic area, including descriptions of one epizoic form, 6 new species and, 2 new genera (Protozoa: Ciliophora). Acta Protozool. 41, 23–61.
Song, W., Wilbert, N., Li, L., and Zhang, Q. (2011). Re-evaluation on the diversity of the polyphyletic genus Metaurostylopsis (Ciliophora, Hypotricha): ontogenetic, morphologic, and molecular data suggest the establishment of a new genus Apourostylopsis n. g. J. Eukaryot. Microbiol. 58, 11–21. doi: 10.1111/j.1550-7408.2010.00518.x
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Wang, C., Zhang, T., Wang, Y., Katz, L. A., Gao, F., and Song, W. (2017a). Disentangling sources of variation in SSU rDNA sequences from single cell analyses of ciliates: impacts of copy number variation and experimental errors. P. Roy. Soc. B-Biol. Sci. 284:20170425. doi: 10.1098/rspb.2017.0425
Wang, J., Li, J., and Shao, C. (2020a). Morphology, morphogenesis, and molecular phylogeny of a novel saline soil ciliate, Heterourosomoida sinica n. sp. (Ciliophora, Hypotrichia). Eur. J. Protistol. 73:125666. doi: 10.1016/j.ejop.2019.125666
Wang, J., Li, L., Warren, A., and Shao, C. (2017b). Morphogenesis and molecular phylogeny of the soil ciliate Rigidohymena quadrinucleata (Dragesco and Njine, 1971) Berger, 2011 (Ciliophora, Hypotricha, Oxytrichidae). Eur. J. Protistol. 60, 1–12. doi: 10.1016/j.ejop.2017.04.006
Wang, J., Zhao, Y., Lu, X., Lyu, Z., Warren, A., and Shao, C. (2020b). Does the Gonostomum-patterned oral apparatus in Hypotrichia carry a phylogenetic signal? Evidence from morphological and molecular data based on extended taxon sampling using three nuclear genes (Ciliophora, Spirotrichea). Sci. China Life Sci. 63. doi: 10.1007/s11427-020-1667-3
Wang, Y., Hu, X., Huang, J., Al-Rasheid, K. A. S., and Warren, A. (2011). Characterization of two urostylid ciliates, Metaurostylopsis flavicana spec. nov. and Tunicothrix wilberti (Lin & Song, 2004) Xu et al., 2006 (Ciliophora, Stichotrichia), from a mangrove nature protection area in China. Int. J. Syst. Evol. Microbiol. 61, 1740–1750. doi: 10.1099/ijs.0.024935-0
Wilbert, N., and Song, W. (2005). New contributions to the marine benthic ciliates from the Antarctic area, including description of seven new species (Protozoa, Ciliophora). J. Nat. Hist. 39, 935–973. doi: 10.1080/00222930400001509
Zhang, T., Dong, J., Cheng, T., Duan, L., and Shao, C. (2020). Reconsideration of the taxonomy of the marine ciliate Neobakuella aenigmatica Moon et al., 2019 (Protozoa, Ciliophora, Hypotrichia). Mar. Life Sci. Technol. 2, 97–108. doi: 10.1007/s42995-020-00032-4
Zhang, T., Qi, H., Zhang, T., Sheng, Y., Warren, A., and Shao, C. (2018). Morphology, morphogenesis and molecular phylogeny of a new brackish water subspecies, Neourostylopsis flava paraflava nov. subsp. (Ciliophora, Hypotrichia, Urostylidae), with redefinition of the genus Neourostylopsis. Eur. J. Protistol. 66, 48–62. doi: 10.1016/j.ejop.2018.07.004
Keywords: Metaurostylopsis, Monourostylopsis n. gen., ontogenesis, 18S rRNA gene, systematics, taxonomy
Citation: Song W, Qiao Y, Dong J, Bourland WA, Zhang T and Luo X (2020) Ontogeny and Phylogeny of a New Hypotrichous Ciliate (Protista, Ciliophora), Metaurostylopsis alrasheidi n. sp., With Establishment of a New Genus Monourostylopsis n. gen.. Front. Mar. Sci. 7:602317. doi: 10.3389/fmars.2020.602317
Received: 03 September 2020; Accepted: 27 October 2020;
Published: 26 November 2020.
Edited by:
Hongbo Pan, Shanghai Ocean University, ChinaReviewed by:
Lingyun Chen, Northwest Normal University, ChinaXiangrui Chen, Ningbo University, China
Copyright © 2020 Song, Qiao, Dong, Bourland, Zhang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tengteng Zhang, dGVuZ3R6aGFuZ0Bmb3htYWlsLmNvbQ==; Xiaotian Luo, bHVveGlhb3RpYW5AaWhiLmFjLmNu
 Wenya Song
Wenya Song Yu Qiao1
Yu Qiao1 Jingyi Dong
Jingyi Dong William A. Bourland
William A. Bourland Xiaotian Luo
Xiaotian Luo