- 1Key Laboratory of Tropical Marine Bio-Resources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2South China Sea Bio-Resource Exploitation and Utilization Collaborative Innovation Center, Guangzhou, China
- 3Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences, Guangzhou, China
- 4Guangdong Key Laboratory of Animal Conservation and Resource Utilization, Guangdong Public Laboratory of Wild Animal Conservation and Utilization, Institute of Zoology, Guangdong Academy of Sciences, Guangzhou, China
The noble scallop, Chlamys nobilis, is an important bivalve mollusk with high commercial value and is usually farmed in the waters of southern China. To date, very little is known about the genetic diversity and population structure of C. nobilis. In this study, 10 microsatellite loci of four farmed C. nobilis populations were compared with one another and compared wild population in southern China. A total of 83 alleles were found. Surprisingly, the level of genetic diversity of the farmed C. nobilis populations was higher than that of the wild population. Although the population genetic of wild population was completely in the Hardy–Weinberg equilibrium, due to heterozygote deficiency, significant deviations from the Hardy–Weinberg equilibrium were found in all farmed populations, suggesting a genetic admixture caused by the mixing of seeds from various hatcheries. The Fst and AMOVA values showed significant genetic differences between wild and farmed populations. The Bayesian assignment also confirmed that genetic admixture was significant and widespread in artificial breeding of C. nobilis. Furthermore, the UPGMA tree topology and PCA demonstrated that the genetic diversity of wild population can be clearly distinguished from farmed populations. In a nutshell, the findings of this study not only fill the knowledge gaps in genetic diversity of wild and farmed C. nobilis populations, but also serve as a guide for maintaining the genetic diversity of C. nobilis in both farmed and wild populations.
Introduction
Scallops are important animal protein source for humans (Liu et al., 2007; Tan et al., 2019). The global scallop landings have increased remarkably in the past decades (Food and Agriculture Organisation (FAO), 2020). The main reason for the dramatic increase in production is the development of scallop farming operations in Japan and China (Bourne, 2000; Tan et al., 2020b). The noble scallop, Chlamys nobilis, is an economically important bivalve species that naturally inhabits the coastal waters of southern China and Japan. In China, C. nobilis was initiated farmed in the Fujian province in the 1970s and later introduced to the Guangdong and Hainan provinces (Tan et al., 2020a).
With the expansion of the scale of C. nobilis aquaculture, the mass mortality rate of farmed C. nobilis has become more prominent since 2006. The production of C. nobilis seed in the hatchery is currently poorly managed due to the lack of necessary information about the origin of broodstock and seeds. In hatcheries, broodstock are usually selected from farmed stocks (Yuan et al., 2009). It is generally believed that the reproduction of animals in small or isolated populations will result in reduced heterozygosity and inbreeding depression, as well as reduced survival rates (Reed and Frankham, 2003). To avoid those potential problems, scallop farmers usually purchase scallop seeds from various sources or use seeds produced by mixing broodstocks obtained from various sources. As a result, there are significant variations in aquaculture traits among scallops in different farms, such as growth, survival rate, and disease resistance. It is worth noting that mixing population seeds from different sources may lead to a higher levels of genetic diversity and fewer heterozygotes in the populations (Ferguson, 1995; Thai et al., 2007), which might explain the high mortalities within scallop populations (Nagashima et al., 2005).
Molecular markers are an important tool for studying population genetic and can provide useful information about genetic diversity of populations (Williams and Benzie, 1998; Azuma et al., 2008; Janson et al., 2015; Phinchongsakuldit et al., 2015), as well as understand the connectivity among populations and its adaptive potential to adapt to different environments (Petersen et al., 2010). To date, genetics studies on bivalves has been very limited and mainly focusing on bivalve species with important commercial value (Benzie and Williams, 1997; Nagashima et al., 2005; Sato et al., 2005; Song et al., 2006; Gurney-Smith et al., 2017). For C. nobilis, only information on few microsatellites (Ma and Yu, 2009a, b) and mitochondrial DNA variation in farmed and wild stocks of C. nobilis in southern China (Yuan et al., 2009) are available. It is found that the genetic diversity of wild C. nobilis population is lower than that of farmed population from the same region in southern China, and the genetic composition of farmed C. nobilis is highly affected by hatchery operations (Yuan et al., 2009).
In order to verify the discovery of Yuan et al. (2009), in this study, we use 10 microsatellite markers to examine the genetic diversity of wild and farmed C. nobilis populations in coastal water of southern China. Microsatellites were chosen in this study because of their neutral, clear scoring of alleles, hypervariability, codominance, and abundance properties, making them the best choice for studying genetic variation of individuals and populations (Zhan et al., 2009). The aim of this study was to use microsatellites to explore the genetic variation of C. nobilis among hatcheries, and between hatchery and wild populations. Understanding the genetic diversity and differentiation among C. nobilis populations is essential for establishing appropriate guidelines for establishment and maintenance of farmed C. nobilis populations.
Materials and Methods
Sample and DNA Extraction
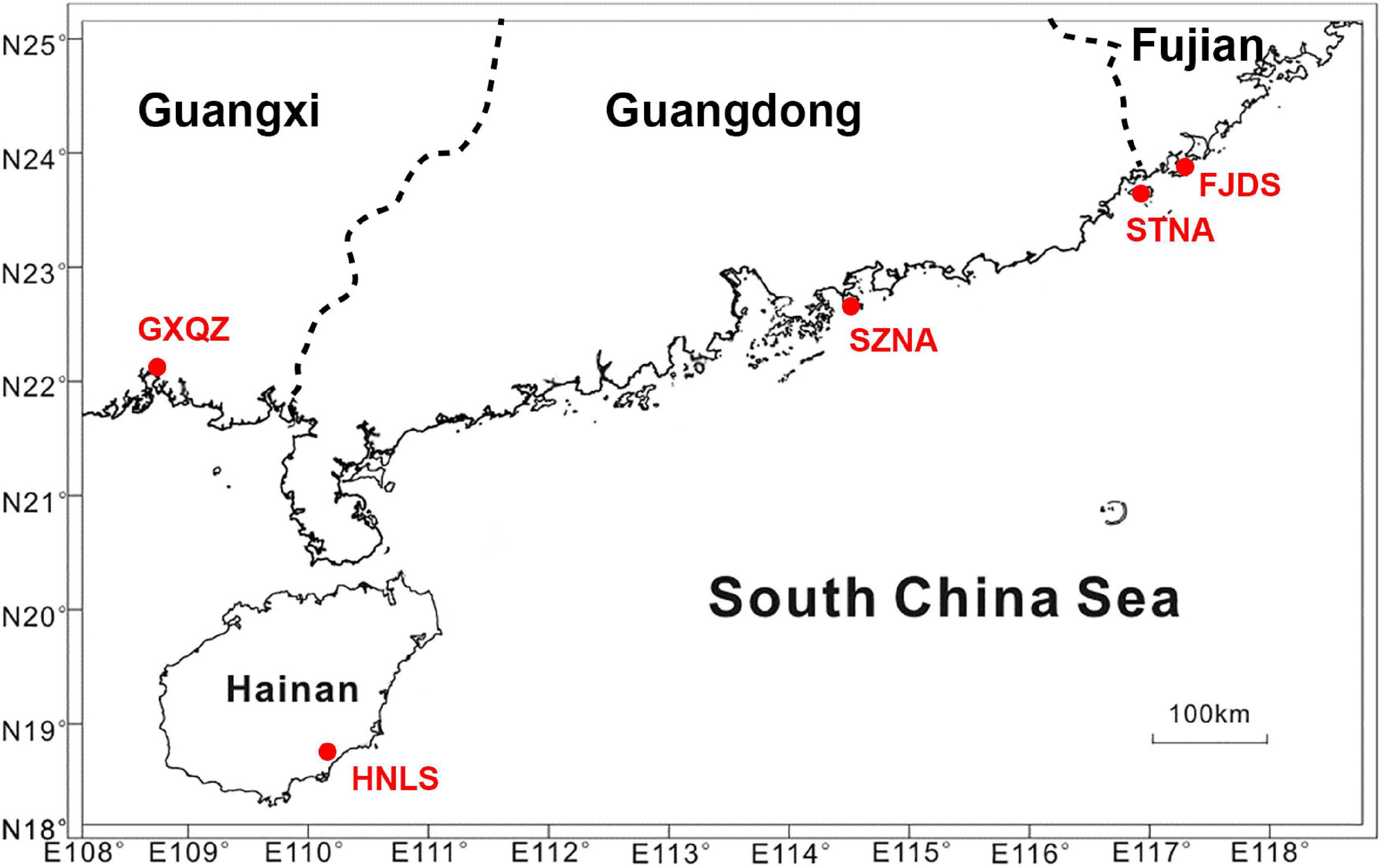
A total of 152 adult C. nobilis were randomly collected from four C. nobilis farms [Dongshan (Fujian province, FJDS) (30 individuals), Nan’ao (Shenzhen city of Guangdong province, SZNA) (30 individuals), Nan’ao island (Shantou city of Guangdong province, STNA) (32 individuals), and Qinzhou (Guangxi province, GXQZ) (30 individuals)] and one wild population (Lingshui, Hainan province, HNLS) (30 individuals) along the southern coast of China (Figure 1).
The adductor muscle was extracted and stored in 70% ethanol. The genomic DNA was extracted individually using phenol-chloroform (Sambrook and Russell, 2000). Subsequently, 0.8% agarose gel electrophoresis was used to screen for the integrity of the extracted genomic DNA, and then ND-1000 UV-Vis spectrophotometer (NanoDrop, Wilmington, DE, United States) was used to determine the quality and quantity of extracted genomic DNA.
Data Collection
In order to avoid scoring errors, nine previously reported microsatellite loci (Ma and Yu, 2009a, b) and a newly developed microsatellite locus (Cn224) with a unique band size of were selected. The sequences of primers and microsatellite core, as well as the optimal annealing conditions are summarized in Table 1. Amplifications were conducted in 20-μL reactions (0.25–0.5 U Taq polymerase (Tiangen, Beijing, China), 80 ng of template DNA, 0.2 mM dNTPs, 1 × PCR buffer, 0.2–1 μM primers (each), and 1.0–2.0 mM MgCl2). For PCR, the initial denaturation was set at 95°C for 5 min, and then at 94°C for 30 s for 30 cycles, followed by primer-specific annealing at 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products were then separated by electrophoresis on an 8% non-denaturing polyacrylamide gel, and then observed using silver staining. For allele size determination, each gel contains a control DNA sample and a 50-bp DNA ladder (Takara).
Data Analysis
In order to identify the scoring errors, invalid alleles, and large-allele drop out in microsatellite products, MICRO-CHECHER was used according to the manufacturing instructions (Oosterhout et al., 2004). For the analysis of microsatellite loci variation in the five populations, POPGENE 1.31 software was use to calculate the number of alleles, allele frequency, effective number of alleles, as well as expected and observed heterozygosities (Yeh et al., 1999). Locus polymorphism [polymorphism information content (PIC)] and allelic richness (R) were estimated according to the formula described by Botstein et al. (1980) and was calculated (EIMousadik and Petit, 1996), respectively. An independent T-test was performed, followed by a Turkey multiple comparison test (Turkey HSD) to test for significant differences in genetic diversity between C. nobilis from hatchery and wild populations.
GENEPOP4.0 was used to evaluate the statistical significance of Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium test (LD) (Raymond and Rousset, 1995). Chi-square tests were used to test for significant differences, while sequential Bonferroni correction was used to adjust the significance of p values for multiple tests (Rice, 1989).
Genetic variation among populations were evaluated using pairwise Fst values, and bootstrap analysis (1000 permutations) was used to test for their significant difference using FSTAT2.9.3. Analysis of molecular variance (AMOVA) was used to compare hierarchical genetic structure between and within populations using ARLEQUIN 3.0 (Excoffier et al., 2005).
In order to evaluate individual migrant events across ocean currents, GENECLASS 2.0 was used to estimate the probability of each individual being assigned to a particular population based on multilocus genotype data (Piry et al., 2004).
Principal component analysis (PCA) between populations was performed using NTSYS software (Rohlf, 2005). To evaluate the genetic relationships among populations, POPULATION was used to calculate A matrix of pairwise DA distance (Nei, 1972), and then a UPGMA dendrogram was constructed using PHYLIP3.5c (Felsenstein, 1993).
Results
Genetic Variation Within C. nobilis Population
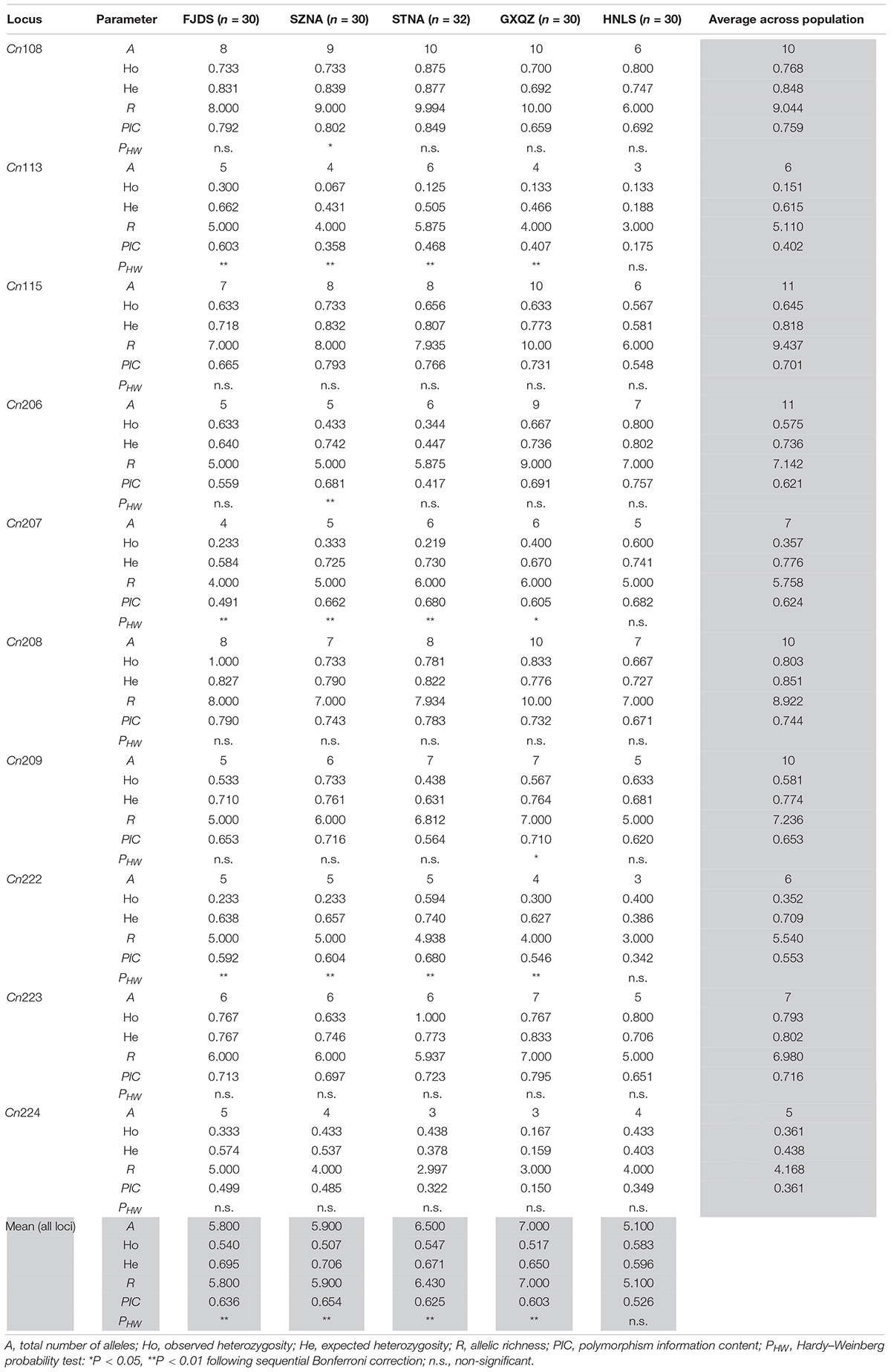
All 10 microsatellite loci were polymorphic in all five C. nobilis populations. 83 different alleles were observed in these 10 loci, ranging in size from 66 to 464 bp. Microsatellites Cn206 and Cn115 have the highest polymorphisms (11 alleles), whereas locus Cn224 has the least polymorphism (five alleles). Moderate-to-high polymorphism information content (PIC) was detected at most microsatellite loci in each population (Table 2).
The intra-population allele frequencies of all 10 microsatellite loci revealed differences among the five populations (Figure 2). The mean number of alleles in all loci of the wild C. nobilis population (HNLS) (5.1) was significantly lower than that of the farmed C. nobilis populations (5.8–7.0). Since the number of alleles is sensitive to the sample size, allele richness was used for comparison. Consistent with the mean number of alleles, the allele richness of the wild C. nobilis population was also the lowest (5.1). Moreover, the average expected heterozygosity of the wild C. nobilis population (0.596) was significantly lower than that of the farmed C. nobilis populations (0.650–0.706). The polymorphism information content (PIC) also showed similar trends, in which the PIC of wild C. nobilis population (0.526) was significantly lower than that of farmed C. nobilis populations (0.603–0.654). However, observed heterozygosity showed the opposite trend. The observed heterozygosity of the wild C. nobilis population (0.583) was significantly higher than that of the farmed C. nobilis populations (0.507–0.547) (Table 2). Nevertheless, there was no statistical difference in genetic diversity between wild and farmed C. nobilis populations (P > 0.05).
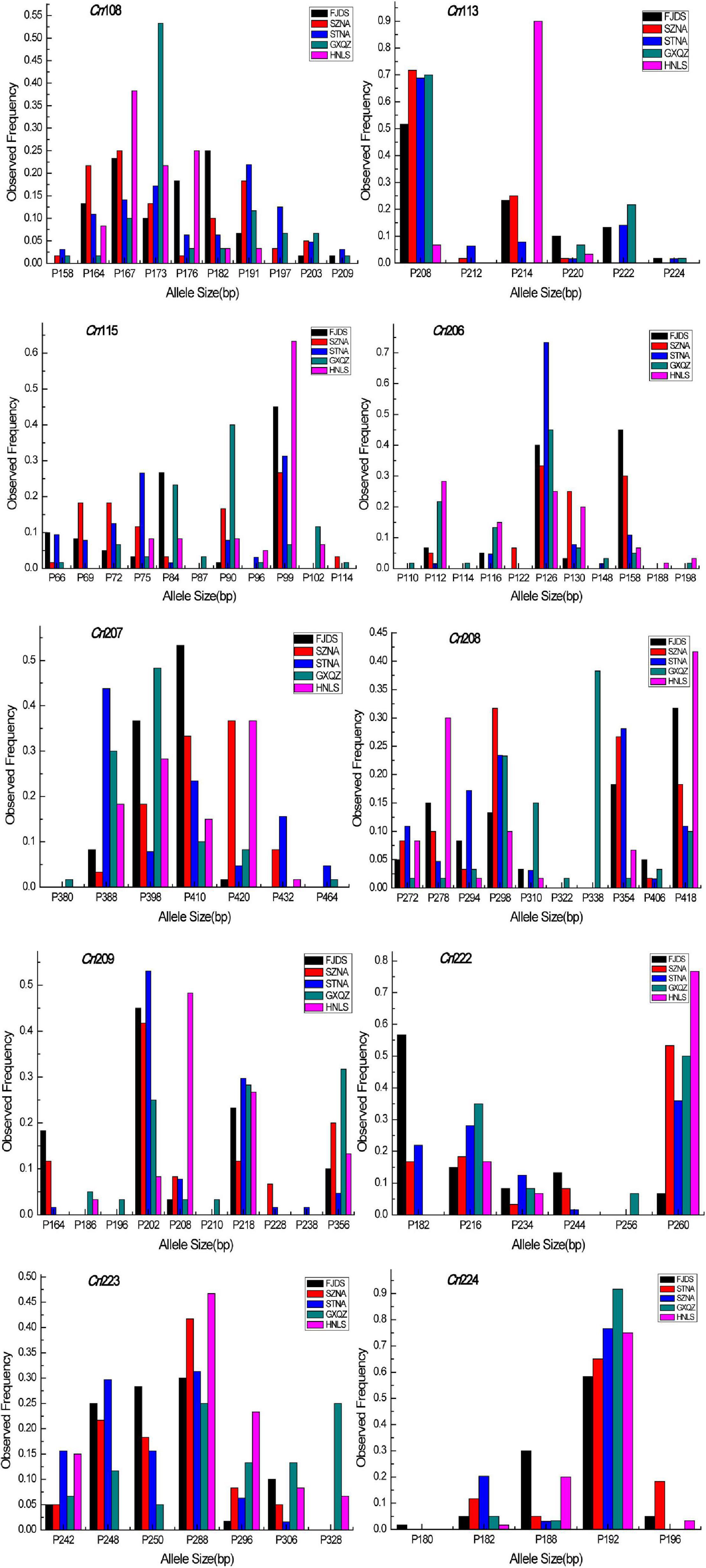
Figure 2. Allele size frequency distributions of the 10 microsatellite loci in five C. nobilis populations in this study.
The results of MICRO-CHECKER revealed that there were no major allele dropout, scoring errors and null alleles in the wild C. nobilis population. However, there were null alleles at few microsatellite loci in every farmed C. nobilis populations. In farmed C. nobilis populations, significant deviations from Hardy–Weinberg equilibrium were observed in 10 of the 50 tests at the 5% level after sequential Bonferroni correction for multiple tests, indicating heterozygote deficiencies. However, for the wild C. nobilis population, all microsatellite loci were at the H-W equilibrium value. No linkage disequilibrium was detected among any loci in this study, indicating that these loci were genetically independent.
Genetic Differentiation and Relationships Among Populations
Pairwise Fst analyses revealed that the genetic differentiation between wild and farmed C. nobilis populations (0.129–0.186) was significantly higher than that among farmed C. nobilis populations (0.046–0.14) (Table 3). Similar results have been recorded from the Analysis of molecular variances (AMOVA) of microsatellites, in which the variations within individuals, among individuals within populations, and among populations were 71.40% (P = 0.000), 16.81% (P = 0.000), and 11.79% (P = 0.000), respectively, and there were significant genetic differentiation among the five populations (Table 4).
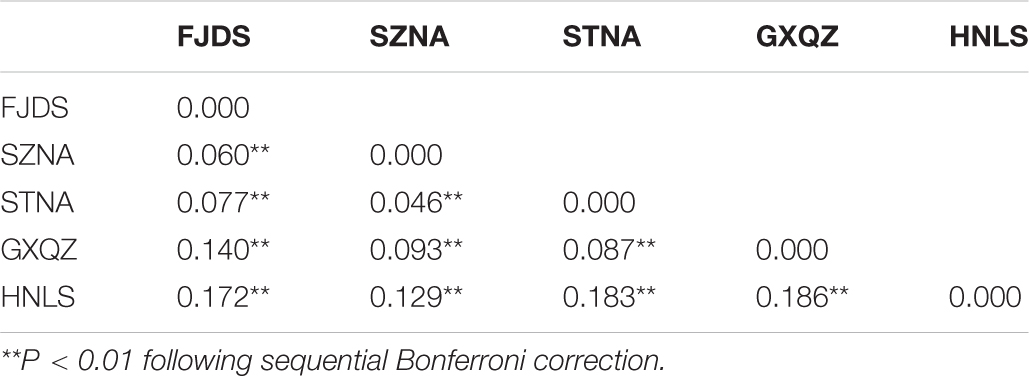
Table 3. Pairwise Fst values between five noble scallop populations based on 10 microsatellite loci.
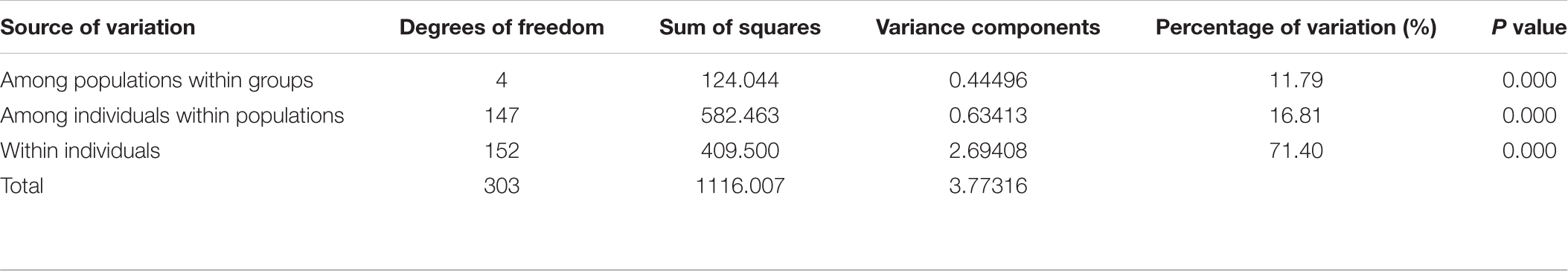
Table 4. Analysis of molecular variance (AMOVA) of 10 microsatellite loci in five C. nobilis populations.
Bayesian assignment placed all wild individuals in their own population with a score of 100. However, farmed populations suffered a relatively high proportion of misclassifications (Table 5).
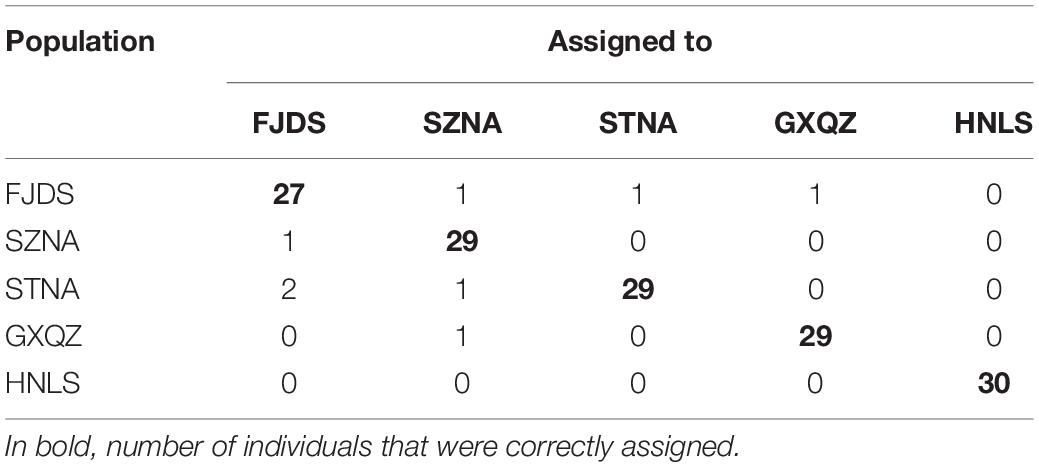
Table 5. Results of assignment tests based on the Bayesian method for 10 microsatellite loci of C. nobilis individuals.
Genetic relationships between populations were graphically illustrated by a dendrogram (Figure 3) and PCA (Figure 4). The UPGMA dendrogram showed that four farmed C. nobilis populations were clustered together and the wild population (HNLS) formed another cluster. Similar findings were observed in the PCA scatter plots where two major groups can be seen (i.e., FJDS-GXQZ and HNLS).
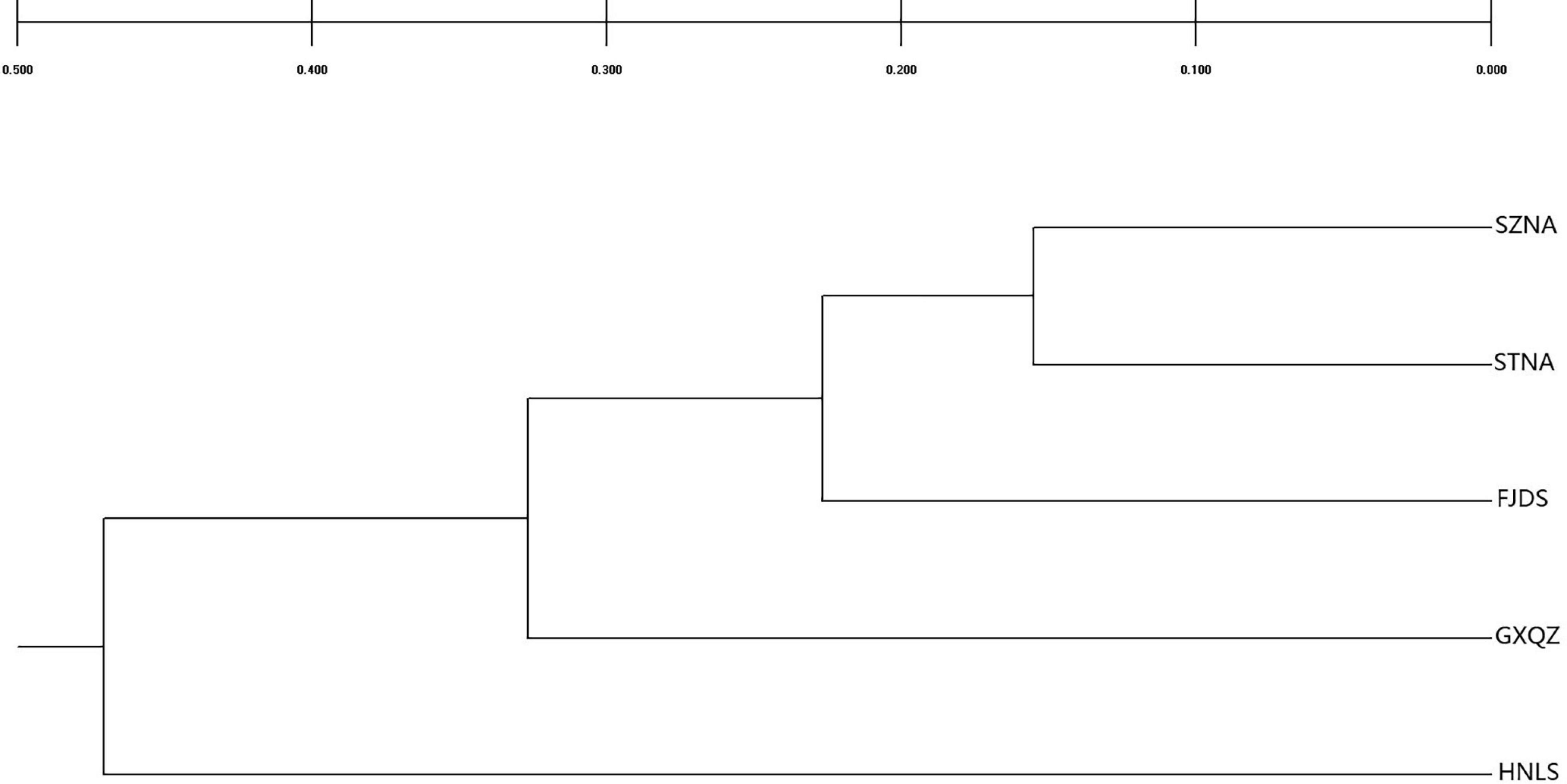
Figure 3. UPGMA dendrogram showing the phylogenetic relationships among five samples of C. nobilis, based on a matrix of genetic distance (Nei, 1972). Bootstrap values are given for nodes at least 62% or more support.

Figure 4. Principal component analysis plotting the relationships of C. nobilis populations, based on a matrix of genetic distance (Nei, 1972).
Discussion
Maintaining genetic diversity is important for both wild and farmed populations as it offers genotypes that respond adaptively to changing environments (Fisher, 1958); in particular, heterozygous individuals are often superior to homozygous individuals in many aquaculture traits, including growth, fertility, and disease resistance (Thai et al., 2007). In aquaculture, as a result of limited broodstock and inbreeding activities, many shellfish species have reduced genetic variability compared to their wild stocks (Hedgecock and Sly, 1990; Evans et al., 2004; Li et al., 2007). In this study, we found that a high level of genetic diversity was maintained in the wild C. nobilis population (HNLS). However, the most genetic parameters (except Ho) of farmed C. nobilis populations were significantly higher than wild C. nobilis population. In agreement with the findings of Yuan et al. (2009) that based on mitochondrial 16S rRNA and COI genes, the farmed population of C. nobilis has a higher genetic diversity index than the wild population. In addition, similar observations have also been documented in other marine organisms, such as Salvelinus alpinus (Primmer et al., 1999) and Salmo salar L. (Elliott and Reilly, 2003).
In farmed stocks, it is generally believe that genetic variation is positively associated with the number of broodstock used. However, we found that all hatchery populations showed significant heterozygosity deficiency. In hatchery populations, heterozygote deficiency is typically considered to result from inbreeding. Although there was significant heterozygosity deficiency in all hatchery populations, the diversity levels of hatchery populations were still higher than the wild population. Therefore, the possibility explanation for this phenomenon is that the possibility of inbreeding was very small, which maybe due to the mixing of broodstock during selection and propagation process (Pucher et al., 2013). The results of Bayesian assignment test confirmed that the genetic admixture of germplasms in the farmed C. nobilis populations is significant. Therefore, the effect of reduced genetic diversity caused by heterozygote deficiency was offset by the effect of increasing genetic diversity of broodstock from multiple wild populations. It is not unclear whether this high polymorphism caused by genetic admixture is beneficial to the survival of this species. In Vietnam, Thai et al. (2007) found that the levels of genetic diversity of carp in most hatchery was similar to that of wild carp populations, and the high genetic diversity of farmed carp was mainly contributed by hybridization with imported stocks.
The highest mean heterozygosity observed across all loci was in the wild population (HNLS) compared to all hatchery populations and was nearly identical to its mean expected heterozygosity because the wild population was in HWE. However, all examined farmed populations showed significant deviations from Hardy–Weinberg equilibrium. These deviations were mainly caused by heterozygote deficiency. In farmed C. nobilis populations, heterozygote deficiency is typically considered to be caused by a limited number broodstock and/or, inbreeding (Kohlmann et al., 2005; An et al., 2011). In our case, the significant deviations from HWE in hatchery populations were caused by genetic admixture. Accordingly, others have also observed high proportions of deviation from HWE in previous studies of C. nobilis (Hui et al., 2006).
The genetic differentiation measured by the Fst values is generally described as negligible (0–0.05), moderate (0.05–0.15), great (0.15–0.25), and very great (above 0.25) (Wright, 1978; Hartl and Clark, 1997). In our study, negligible and moderate genetic differentiation was identified in four farmed C. nobilis populations. Great genetic differentiation was found between wild and hatchery C. nobilis populations, except for between wild and SZNA. The AMOVA further indicated that there was significant genetic differentiation among the five populations. The significant genetic differences among Chinese farmed populations and the population of Hainan were clearly differentiated from other populations, which Yuan et al. (2009) also found. However, they did not analyze the cause of this significant differentiation. The results from the Bayesian assignment test indicate that it may be caused by founder effects and admixture.
Bayesian assignment test showed that the genetic admixture was significant and widespread in artificial breeding of C. nobilis. In this study, individuals from the wild C. nobilis population form a single genetic group, which suggests that the wild population in Lingshui, Hainan province did not mixed with other populations. Therefore, more attention should be paid to its protection.
The UPGMA tree topology and PCA showed there is a clear division between wild and farmed C. nobilis populations. This genetic pattern showed that STNA and SZNA farmed populations are closely related, and that STNA and SZNA could have derived from similar broodstock.
Recently, the reduction of wild scallop resources has led to an increase in the transfer of seed and stock for aquaculture. Contradict with previous reports (Yuan et al., 2009), our data revealed that due to genetic admixture, the genetic diversity of farmed C. nobilis populations is higher than that of the wild population. Although the four farmed C. nobilis populations still have considerable genetic variation, heterozygote deficiency and significant genetic differentiation populations were detected. Therefore, in order to achieve sustainable development of C. nobilis aquaculture and hatchery seed production, it is important that hatcheries to maintain an effective population sizes and avoid the breeding of closely related broodstock.
Conclusion
In conclusion, both wild C. nobilis population (HNLS), and farmed C. nobilis populations recorded high levels of genetic diversity. Interestingly, the genetic diversity of farmed C. nobilis populations was significantly higher than that of wild C. nobilis population, but the farmed C. nobilis populations showed significant heterozygosity deficiency. Since genetic admixture occur in the artificial breeding of C. nobilis, the effect of reducing genetic diversity due to heterozygote deficiency was offset by the effect of increased genetic diversity in broodstock from multiple wild populations. These findings of this study not only fill the knowledge gaps in the genetic diversity of wild and farmed C. nobilis, but also serve as guidelines for maintaining the genetic diversity of C. nobilis in both farmed and wild populations.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
YHZ and ZY conceived the study. HM and DY carried out the field and laboratory work, participated in the data analysis, and drafted the manuscript. SX and YQ collected the noble scallop populations. YZ and JL contributed to the microsatellite analysis. All authors approved the manuscript for publication.
Funding
This work was supported by the National Natural Science Foundation of China (31702340, 31872566, and 32002387), National Marine Genetic Resource Center, Guangdong Academy of Sciences (GDAS) Special Project of Science and Technology Development (2018GDASCX-0107), the Chinese Ministry of Science and Technology through the National Key Research and Development Program of China (2018YFD0901400 and 2020YFD0901100), Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0404), the Network Service Local Plan STS of the Chinese Academy of Sciences (KFJ-STS-QYZD-158), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA13020202), the Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences (ISEE2018PY01 and ISEE2018ZD02), the Open Foundation of State Key Laboratory of Loess and Quaternary Geology (SKLLQG1813 and SKLLQG1918), the China Agriculture Research System of MOF and MARA, and the Science and Technology Planning Project of Guangdong Province, China (2017B030314052).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, H. S., Kim, E. M., Lee, J. H., Noh, J. K., An, C. M., Yoon, S. J., et al. (2011). Population genetic structure of wild and hatchery black rockfish Sebastes inermis in Korea, assessed using cross-species microsatellite markers. Genet. Mol. Res. 10, 2492–2504. doi: 10.4238/2011.october.13.6
Azuma, N., Kunihiro, Y., Sasaki, J., Mihara, E., Mihara, Y., Yasunaga, T., et al. (2008). Genetic variation and population structure of Hair Crab (Erimacrusisenbeckii) in Japan inferred from Mitochondrial DNA sequence analysis. Mar. Biotechnol. 10, 39–48. doi: 10.1007/s10126-007-9033-1
Benzie, J. A. H., and Williams, S. T. (1997). Genetic structure of Giant Clam (Tridacna maxima) populations in the West Pacific is not consistent with dispersal by present-day ocean currents. Evolution 51, 768–783. doi: 10.2307/2411153
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314–331.
EIMousadik, A., and Petit, R. J. (1996). High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.)Skeels] endemic to Morocco. Theor. Appl. Genet. 92, 832–839. doi: 10.1007/s001220050200
Elliott, N. G., and Reilly, A. (2003). Likelihood of a bottleneck even inthe history of the Australian population of Atlantic salmon (Salmosalar L.). Aquaculture 215, 31–44. doi: 10.1016/s0044-8486(02)00055-8
Evans, B., Bartlett, J., Sweijd, N., Cook, P., and Elliott, N. G. (2004). Loss of genetic variation at microsatellite loci in hatchery produced abalone in Australia (Haliotis rubra) and South Africa (Haliotis midae). Aquaculture 233, 109–127. doi: 10.1016/j.aquaculture.2003.09.037
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50.
Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package) version 3.50. Seattle: University of Washington.
Ferguson, M. M. (1995). “The role of molecular genetic markers in the management of cultured fishes,” in Molecular Genetics in Fisheries, eds G. R. Carvalho and T. J. Pitcher (London: Chapman & Hall), 81–104. doi: 10.1007/978-94-011-1218-5_4
Fisher, R. A. (1958). The Genetical Theory Of Natural Selection, 2nd Revised Edn. New York: Dover Publication.
Food and Agriculture Organisation (FAO) (2020). Fishery and aquaculture statistics. Global production by production source 1950-2018 (FishstatJ). Rome: FAO Fisheries and Aquaculture Department.
Gurney-Smith, H. J., Wade, A. J., and Abbott, C. L. (2017). Species composition and genetic diversity of farmed mussels in British Columbia. Can. Aquac. 466, 33–40. doi: 10.1016/j.aquaculture.2016.08.038
Hartl, D. L., and Clark, A. G. (1997). Principles of population genetics, 3rd edn. Sunderland: Sinauer Associates.
Hedgecock, D., and Sly, F. (1990). Genetic drift and effective population sizes of hatchery-propagated stocks of the Pacific oyster Crassostrea gigas. Aquaculture 88, 21–38. doi: 10.1016/0044-8486(90)90316-f
Hui, M., Bao, Z. M., Zhan, A. B., Hu, X., Lu, W., Chang, D., et al. (2006). Ten polymorphic dinucleotide microsatellite markers of the noble scallop Chlamys nobilis. Mol. Ecol. Notes 6, 1033–1035. doi: 10.1111/j.1471-8286.2006.01420.x
Janson, S., Wouters, J., Bonow, M., Svanberg, I., and Olsén, K. H. (2015). Population genetic structure of crucian carp (Carassius carassius) in man-made ponds and wild populations in Sweden. Aquac. Int. 23, 359–368. doi: 10.1007/s10499-014-9820-4
Kohlmann, K., Kersten, P., and Flajshans, M. (2005). Microsatellite-based genetic variability and differentiation of domesticated, wild and feral common carp (Cyprinus carpio L) populations. Aquaculture 247, 253–266. doi: 10.1016/j.aquaculture.2005.02.024
Li, Q., Xu, K. F., and Yu, R. H. (2007). Genetic variation in Chinese hatchery populations of the Japanese scallop (Patinopecten yessoensis) inferred from microsatellite data. Aquaculture 269, 211–219. doi: 10.1016/j.aquaculture.2007.04.017
Liu, B. Z., Dong, B., Xiang, J. H., and Zaizhao, W. (2007). The phylogeny of native and exotic scallops cultured in China based on 16S rDNA sequences. Chin. J. Oceanol. Limnol. 25, 85–90. doi: 10.1007/s00343-007-0085-x
Ma, H. T., and Yu, Z. N. (2009a). Development of twenty-two polymorphic microsatellite loci in the noble scallop, Chlamys nobilis. Conserv. Genet. 10, 1587–1590. doi: 10.1007/s10592-008-9800-1
Ma, H. T., and Yu, Z. N. (2009b). Isolation and characterization of twenty-three microsatellite loci in the noble scallop, Chlamys nobilis. Conserv. Genet. Resour. 1, 131–134. doi: 10.1007/s12686-009-9032-9
Nagashima, K., Sato, M., Kawamata, K., Nakamura, A., and Ohta, T. (2005). Genetic structure of Japanese scallop population in Hokkaido, analyzed by mitochondrial haplotype distribution. Mar. Biotechnol. 7, 1–10. doi: 10.1007/s10126-004-3046-9
Oosterhout, C. V., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Petersen, J. L., Ibarra, A. M., and May, B. (2010). Nuclear and mtDNA lineage diversity in wild and cultured Pacific lion-paw scallop, Nodipectensubnodosus (Baja California Peninsula, Mexico). Mar. Biotechnol. 157, 2751–2767. doi: 10.1007/s00227-010-1534-1
Phinchongsakuldit, J., Chaipakdee, P., Collins, J. F., Jaroensutasinee, M., and Brookfield, J. F. Y. (2015). Population genetics of cobia (Rachycentron canadum) in the Gulf of Thailand and Andaman Sea: fisheries management implications. Aquac. Int. 21, 197–217. doi: 10.1007/s10499-012-9545-1
Piry, S., Alapetite, A., Cornuet, M. J., Paetkau, D., Baudouin, L., and Estoup, A. (2004). GENECLASS2: a software for genetic assignment and first-generation migrant detection. J. Hered. 95, 536–539. doi: 10.1093/jhered/esh074
Primmer, C. R., Aho, T., Piironen, J., Estoup, A., Cornuet, J. M., and Ranta, E. (1999). Microsatellite analysis of hatchery stocks and naturalpopulations of Arctic charr, Salvelinus alpinus, from the Nordicregion: implications for conservation. Hereditas 130, 277–289. doi: 10.1111/j.1601-5223.1999.00277.x
Pucher, J., Steinbronn, S., Mayrhofer, R., Schad, I., El-Matbouli, M., and Focken, U. (2013). “Improved sustainable aquaculture systems for small-scale farmers in northern Vietnam,” in Sustainable land use and rural development in southeast Asia: Innovations and policies for mountainous areas, eds H. L. Fröhlich, P. Schreinemachers, K. Stahr, and G. Clemens (Berlin: Springer), 281–317. doi: 10.1007/978-3-642-33377-4_8
Raymond, M., and Rousset, F. (1995). GENEPOP Version 4.0: population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249. doi: 10.1093/oxfordjournals.jhered.a111573
Reed, D. H., and Frankham, R. (2003). Correlation between Fitness and Genetic Diversity. Conserv. Biol. 17, 230–237. doi: 10.1046/j.1523-1739.2003.01236.x
Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution 43, 223–225. doi: 10.2307/2409177
Rohlf, F. J. (2005). NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.2. Setauket: Exeter Software.
Sambrook, J. D., and Russell, W. (2000). Molecular cloning: a laboratory manual, 2nd edn. New York: Cold Spring Harbor Laboratory Press.
Sato, M., Kawamata, K., Zaslavskaya, N., Nakamura, A., Ohta, T., Nishikiori, T., et al. (2005). Development of microsatellite markers for Japanese scallop (Mizuhopecten yessoensis) and their application to a population genetic study. Mar. Biotechnol. 7, 713–728. doi: 10.1007/s10126-004-0127-8
Song, L., Xu, W., Li, C., Li, H., Wu, L., Xiang, J., et al. (2006). Development of expressed sequence tags from the bay scallop, Argopecten irradians irradians. Mar. Biotechnol. 8, 161–169. doi: 10.1007/s10126-005-0126-4
Tan, K. S., Leng, X., Zhao, Y., Hongxing, L., Cheng, D., Ma, H., et al. (2019). Amino acid variations in polymorphic noble scallops, Chlamys nobilis. J. Food Process. Preserv. 43:e14262.
Tan, K. S., Zhang, H. K., and Zheng, H. P. (2020b). Selective breeding of edible bivalves and its implication of global climate change. Rev. Aquac. 4, 2559–2572. doi: 10.1111/raq.12458
Tan, K. S., Zhang, H. K., Lim, L. S., and Zheng, H. (2020a). Selection breeding program of Nan’ao Golden Scallop Chlamys nobilis with higher nutritional values and less susceptible to stress. Aquaculture 517:734769. doi: 10.1016/j.aquaculture.2019.734769
Thai, B. T., Burridge, C. P., and Austin, C. M. (2007). Genetic diversity of common carp (Cyprinus carpio L.) in Vietnam using four microsatellite loci. Aquaculture 269, 174–186. doi: 10.1016/j.aquaculture.2007.05.017
Williams, S. T., and Benzie, J. A. H. (1998). Evidence of a biogeographic break between populations of a high dispersal starfish: congruent regions within the Indo-West pacific defined by color morphs, mtDNA, and Allozyme Data. Evolution 52, 87–99. doi: 10.2307/2410923
Wright, S. (1978). Evoulution And The Genetics Of Populations, Vol Iv: Variability Within And Among Natural Populations. Chicago: University of Chicago Press.
Yeh, F. C., Yang, R., and Boyle, T. (1999). A Microsoft Window Based Freeware for Population Genetic Analysis. Version 1.31. Canada: University of Alberta.
Yuan, T., He, M. X., and Huang, L. M. (2009). Intraspecific genetic variation in mitochondrial 16S rRNA and COI genes in domestic and wild populations of Huaguizhikong scallop Chlamys nobilis Reeve. Aquaculture 289, 19–25. doi: 10.1016/j.aquaculture.2009.01.004
Keywords: Chlamys nobilis, cultivated population, genetic admixture, microsatellite, population structure, wild population
Citation: Ma H, Yu D, Xiao S, Qin Y, Zhang Y, Li J, Zhang Y and Yu Z (2021) A Significant Genetic Admixture in Farmed Populations of the Noble Scallop Chlamys nobilis Revealed by Microsatellite DNA Analysis in Southern China. Front. Mar. Sci. 8:721292. doi: 10.3389/fmars.2021.721292
Received: 06 June 2021; Accepted: 28 July 2021;
Published: 17 August 2021.
Edited by:
JInghui Fang, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (CAFS), ChinaCopyright © 2021 Ma, Yu, Xiao, Qin, Zhang, Li, Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haitao Ma, aHRtYUBzY3Npby5hYy5jbg==; Yuehuan Zhang, eWh6aGFuZ0BzY3Npby5hYy5jbg==; Ziniu Yu, Y2FybHp5dUBzY3Npby5hYy5jbg==
†These authors have contributed equally to this work
 Haitao Ma
Haitao Ma Dongmei Yu
Dongmei Yu Shu Xiao
Shu Xiao Yanping Qin
Yanping Qin Yang Zhang
Yang Zhang Jun Li
Jun Li Yuehuan Zhang
Yuehuan Zhang Ziniu Yu
Ziniu Yu