Abstract
Background:
Alexandrium pacificum is a dinoflagellate species notorious for its rapid growth resulting in large-scale blooms. This study aimed to investigate the molecular mechanisms of A. pacificum under laboratory-simulated rapid growth conditions from the perspective of H3K4me3 modification regulation.
Methods and results:
Western blot was used to detect the modification abundance of H3K4me3 in A. pacificum cultured under different conditions, including high light (HL), high nitrogen (HN), and f/2 medium (control, CT), in the rapid growth exponential phase. The results showed that the modification abundance of H3K4me3 under HL or HN was greater than that under CT. Chromatin immunoprecipitation-sequencing was used to explore the acting genes of H3K4me3 under different conditions for the first time. Nitrogen metabolism and endocytosis were significantly associated with H3K4me3 regulation under HL. Furthermore, H3K4me3 was also significantly associated with the vitamin metabolism pathway under HN.
Conclusions:
These findings demonstrate that H3K4me3 plays a potentially important role in the regulation of the rapid growth of A. pacificum. Such knowledge of a histone modification regulatory network in this dinoflagellate, lays a necessary foundation for future research in related fields.
Highlights
The regulatory mechanism of H3K4me3 was studied for the first time in Alexandrium pacificum under different growth conditions.
ChIP-seq analysis of species without genomic data was performed using a close species.
Nitrogen metabolism and endocytosis were related to H3K4me3 regulation in A. pacificum under high light conditions.
Vitamin metabolism was related to H3K4me3 regulation under high nitrogen conditions.
Introduction
Alexandrium pacificum is one of the main blooming microalgae species in many coastal areas. Paralytic shellfish toxin produced by A. pacificum can significantly harm or kill marine organisms, and ultimately human, through food chain transfer via bivalve mollusks (Grattan et al., 2016). Variations in nutrient concentrations, especially nitrogen, are closely related to the occurrence and termination of blooms, and the accumulation of nitrogencan induce the rapid growth of algal cells (Yu et al., 2020; Baek et al., 2021). The biomass of Alexandrium tamarense under high nitrogen concentrations (2,646 µmol L-1) was greater than that observed under low nitrogen concentrations when f/2 medium was used for culturing (Zhang et al., 2006). Irradiance is another factor that plays a vital role in stimulating growth (Laabir et al., 2011). It was found that the increase of illumination from 10 to 90 μmol photons m-2 s-1 was directly proportional to the growth rate of A. pacificum (Cao et al., 2011; Karabashev and Evdoshenko, 2016). However, knowledge of the intercellular mechanisms underlying the rapid growth of A. pacificum is limited. Previous studies have focused on the gene expression A. pacificum under different conditions through transcriptome technology (Zhang et al., 2014; Liu et al., 2021). Proteins related to environmental stress (nutrients, light, and temperature) at different growth stages of A. pacificum have also been identified, including, S-adenosylmethionine synthetase (AdoMetS), ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), and heat shock proteins (HSPs)which are involved in macromolecular biosynthesis, photosynthesis, stress response, and cell cycle regulation (Uribe et al., 2008; Wang et al., 2012). However, the research into the growth mechanisms of A. pacificum, including the epigenetic regulation of molecular mechanisms, remains limited.
The genomic structure of dinoflagellates is unique; the genome is immense, and the DNA display a status of liquid crystal, and the chromosomes are permanently condensed (Hackett et al., 2004; Chow et al., 2010). Moreover, there appears to be a high degree of DNA redundancy in the dinoflagellate genome. Noncoding repetitive sequences comprise up to 60% of dinoflagellate genomes (Jaeckisch et al., 2011). In genome structure, non-coding sequences generally comprise promoters, enhancers, cis-acting elements, and trans-acting factors that regulate downstream genes to respond to biological needs. These non-coding regulatory regions are usually combined with transcription factors and epigenetic modifiers (histone modifications, DNA methylation, RNA methylation, and chromatin remodeling) to respond to environmental signals through the regulation of downstream gene expression (Koenecke et al., 2016). In dinoflagellates, these non-coding sequences comprise 60% of the genome, indicating that the existence of a prodigious and complex regulatory network. Therefore, the study of this regulatory role in dinoflagellates is very important for researching physiological activity and response mechanisms to the environment.
In dinoflagellates, histones exist with a very low protein/DNA ratio (1:10) (Hamkalo and Rattner, 1977; Hou and Lin, 2009; Lin, 2011), which is significantly lower than the ratio close to 1:1 in other eukaryotes. However, unlike in other eukaryotes, histones in dinoflagellates are not involved in nucleosome assembly (Figueroa et al., 2014). This leads to questions regarding the role that histones play in dinoflagellates. A previous study reported that several genes are responsible for histone modification in A. pacificum, including histone acetyltransferases, deacetylases, methylases, and demethylases (Riaz and Sui, 2018). The presence of histone modifying enzymes indicates that histone modifications may play an important role in A. pacificum. In higher plants, histone modification plays an important role in regulating the expression of genes at the epigenetic level, in response to the environment and stress, as well as being involved in a variety of biological processes such as growth and development (Yamamoto et al., 2020; Moreno-Perez et al., 2021). Therefore, it is reasonable to expect that histone modifications also play an essential role in A. pacificum.
Histone modifications, as a form of epigenetic regulation, play an important role in plant growth, development, and stress response (Liu et al., 2010). However, their effects on algae remain poorly understood, especially in dinoflagellates. In microalgae, studies on histone modifications have focused on marine diatoms and green algae. For example, in Nannochloropsis spp., H3K27me3, H3K27ac, and H3K9me3 showed different modification levels under different carbon dioxide conditions (Wei and Xu, 2018). In the macroalgae Ectocarpus, Bourdareau et al. (2021) identified 47 histone modifications and a new histone modification, H2AZR38me1; however, H3K27me3 modification, which is common in higher plants, and a corresponding multi-comb complex, were absent. This indicated that there are differences in histone modification and regulation mechanisms between algae and higher plants (Bourdareau et al., 2021).
Previous studies have reported various modification types such as histone methylation, acetylation, phosphorylation, and ubiquitination (Riaz et al., 2019; Zhu et al., 2022). Among the histone modifications studied previously, H3K4me3 modification was the most conservative site (Marinov and Lynch, 2015), which provided a great advantage for using commercial H3K4me3 antibodies in subsequent studies. H3K4me3 is an activator of gene expressions in multiple biological processes, as well as a flexible modification in response to the environment in plants (Lauberth et al., 2013; Yan et al., 2019). Genes occupied by H3K4me3 in plants, especially in the absence of H3K4me1 and H3K4me2, generally display low tissue specificity but high levels of constitutive expression in Arabidopsis (Ha et al., 2011). However, H3K4me3 distribution broadened considerably along with differentially expressed genes during drought stress in Arabidopsis (Dijk et al., 2010), and showed differential trimethylation for a proportion of genes differentially expressed during drought stress in rice (Zong et al., 2013). This suggests that H3K4me3 may also be associated with tightly regulated pathways. Ngan et al. (2015) explored the relationship between the lipid production capacity of Chlamydomonas reinhardtii under nitrogen and sulfur starvation, and the regulation of five histone modifications including H3K4me3, and found that these histone modifications showed differential modification levels under nitrogen and sulfur restriction. H3K4me3 modification in soybean was found to promote carbon and nitrogen metabolism in rhizobia, supporting effective nitrogen fixation and promote material exchange between the and rhizobia (Wang et al., 2020). This suggests that H3K4me3 modification plays an important role in the growth and development of plants. Therefore, it is speculated that H3K4me3 may also play an important role in the growth of A. pacificum. In this study, epigenomicprofiles of H3K4me3 were employed to investigate its involvement in the responses of A. pacificum rapid growth to high light and high nitrogen. This study aimed to generate the first global H3K4me3 profile in A. pacificum through chromatin-immunoprecipitation (ChIP) followed by ChIP-sequencing (ChIP-seq) to disclose the regulation mechanism of H3K4me3 sustaining bloom progression and to decipher the regulation networks behind the utilization of light and nutrients in rapid growth.
Materials and methods
Algal culture and treatment conditions
Alexandrium pacificum was obtained from the Key Laboratory of Marine Genetics and Breeding, Ministry of Education, Ocean University of China. The algal cells were grown at 20 ± 1°C in a f/2 medium (Guillard and Ryther, 1962; Guillard, 1975) under a 12 hr light: 12 hr dark photo cycle using cool white fluorescent lights at a set intensity of 30 μmol photon m–2 s–l (Guillard, 1975).
Alexandrium pacificum was initially inoculated at 4,000 cells/mL into 3 L Erlenmeyer flasks containing 2 L of modified f/2 medium for processing. The three treatment conditions were high light (HL), high nitrogen (HN), and control (CT), as shown in Table 1. Cell growth was then followed for 25 days. Samples (1 mL) for cell counting were fixed in 2% Lugol’s iodine solution every 2 days, and then a micro-slide under a microscope (Nikon YS-100, Tokyo, Japan) was used to enumerate the cell concentration. The growth curve of A. pacificum was established to observe its growth state and help to determine sampling time. For counting, the samples were collected at a fixed time (between 11:00 and 12:00 pm) every day. Before each sampling, the cells were evenly distributed by gently shaking the flask. For each treatment condition, there were three biological replicates and three technical replicates.
Table 1
| Sample | Treatments |
|---|---|
| High nitrogen (HN) | Nitrate nitrogen concentration: 2.646 mmol L-1, 30 μmol photos m-1 s-1 |
| High light (HL) | Nitrate nitrogen concentration: 882 µmol L-1, 60 μmol photos m-1 s-1 |
| Control (CT) | Nitrate nitrogen concentration: 882 µmol L-1, 30 μmol photos m-1 s-1 |
Treatment conditions of A. pacificum.
Detection of H3K4me3 modification abundance
The abundance of H3K4me3 in A. pacificum at the exponential phase (12 days) under the different treatment conditions (HN, HL, and CT) was detected by Western blot with three biological replicates (Liu et al., 2022). The histone was extracted following the nucleoprotein extraction method, and quantified by the Bradford protein quantification kit (P0006, Beyotime Biotechnology, Haimen, China). Given that A. pacificum histone H3 sequences are very similar to human H3 sequences and show conservation, including the H3K4 locus (Riaz et al., 2019), a H3K4me3 commercial antibody raised from human H3 (PTM-613, Jingjie Biotechnology Co., Ltd. Hangzhou, China) was used in the western blot assay to determine H3K4me3 abundance under the different treatment conditions, with H3 as the internal reference (Chen et al., 2013).
ChIP
Alexandrium pacificum was grown in 2 L of f/2 medium under the three treatment conditions. Samples at the exponential phase (12 days, ~ 4 × 107cells) under the three treatment conditions were collected in a 50 mL centrifuge tube. Formaldehyde (final concentration: 1%) was added for cross-linking reaction at 25°Cand maintained for 30 min. Glycine (final concentration of 0.125 M) was added to quench the cross-linking reaction for 10 min at 25°C. The collected cells were washed with the sterilized water three times and centrifugated at 3,000 rpm for 5 min at 4°C to remove the formaldehyde. The cross-linked A. pacificum cells were frozen with liquid nitrogen and stored at -80°C. Chromatin extraction was carried out (Gendrel et al., 2005), and then the chromatin was sheared into 200–500 bp fragments using a chromatin shearing non-contact ultrasound instrument (S220 focused-ultrasonicator, Covaris, USA). Reverse cross-linking and gel electrophoresis (1.5% agarose gel) was performed to examine the effect of sonication. The ChIP kit of Abcam (ab117137, ABCAM, Cambridge, UK) and H3K4me3 antibody (PTM-613, PTM, Hangzhou, China) were used for immunoprecipitation. Reverse cross-linking and DNA purification was carried out following the kit instructions. Qubit2.0 (Invitrogen, Waltham, USA) was employed to quantify DNA concentration. Library construction and sequencing were performed by Wuhan IGENEBOOK Biotechnology Co., Ltd.
Sequencing and data analysis
The DNA library was sequenced with the HiSeq 2000 system for 50 nt single-end sequencing. Respectively, low-quality reads were filtered out with Trimmomatic (v. 0.4, Bolger et al., 2014). The clean reads were mapped to the symbiosis dinoflagellate (Fugacium kawagutii) genome (PRJNA630740) by BWA (v.: 0.7.15-r1140). Potential polymerase chain reaction (PCR) duplicates were removed with the software Samtools (v.1.3.1, Danecek et al., 2021). The software MACS2 (v. 2.1.1.20160309, Zhang et al., 2008) was used to call peaks using default parameters (model fold, 5, 50; bandwidth, 300 bp; q value, 0.05) and then the peak result was obtained for the subsequent analysis.
To further explore the binding site characteristics of H3K4me3 and to investigate the mechanism of H3K4me3 regulation on gene expression, the gene corresponding to the nearest transcription start site was found from the peak summit position (if there was no summit site, the midpoint was taken) for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation (Ashburner et al., 2000; Kanehisa and Goto, 2000). A diagram of the number of genes associated with H3K4me3 under the different conditions was then generated using a webtool (https://www.omicshare.com/tools). To determine the interval of up-regulation or down-regulation difference modification between samples, the number of reads in the peak region was counted using the DiffBind package (v.1.16.3, Wang et al., 2008), and the difference analysis was performed on two different groups of samples (Stark and Brown, 2011). The standard condition for screening the difference peak is a false discovery rate < 0.05 and a fold change > 0. From the summit position of the significant difference peak (if there is no summit site, the midpoint was taken), the nearest gene was selected for GO and KEGG annotation.
To study the unique mechanism of H3K4me3 regulation on the rapid growth of A. pacificum under the conditions of HL and HN, the genes associated with H3K4me3 only in HL or HN, but not in CT, were further analyzed using KEGG enrichment (Kanehisa and Goto, 2000). The genes enriched in the top 10 KEEG items were used to draw KEEG bubble diagrams, (https://www.omicshare.com/tools). The metabolic pathways and related genes with a number of genes > 3 and p-value < 0.05 in the enriched metabolic pathways were then further analyzed through a network of pathways and genes.
ChIP-qPCR
To validate the ChIP-Seq results, nine genes associated with H3K4me3 were selected from the ChIP-Seq data for ChIP-quantitative qPCR. The peak sequences (Supplementary 1) of the selected genes were used for primers design. The PCR primers of the genes were designed using Premier 5.0 software (https://www.PremierBiosoft.com) and presented in Table 2. The ChIP-qPCR was performed in 20 μl reactions using Universal Blue qPCR SYBR Green Master Mix (Ye Sen Biotech Co., Ltd., Shanghai, China). The reactions consisted of 10 μl of Universal Blue qPCR SYBR Green Master Mix, 0.4 μl of forward and reverse primer (10 μmol L-1), 1 μl of immunoprecipitated DNA or input DNA, and 8.2 μl of H2O. ChIP-qPCR was performed with three biological replicates, and the results were calculated as the percentage of input DNA (Haring et al., 2007; Solomon et al., 2021).
Table 2
| Gene name | Forward | Reverse |
|---|---|---|
| RibBA | TCCGCTCCAGTTCTGAGATGGTTGT | AAGGATTGGCTTTGAGGGTTGGGTA |
| Euk | ACAACACCAAACGAAACCACACCA | TTGTGATGTGTTGAGTTGTCGCGTG |
| Mok12 | CGGTCATCCTCAACAGAAATCCAAC | ACAAGAAATAACAATACAGGGCATCC |
| L14 | TCTTGGCAAATGCTTTCGCAGTAGT | ATGATTGATAGGGATAGTTGGGGGC |
| E1 | GTAAGCAGAACTGGCGATGAGGGAT | CGTCCTGCTGTCAAAATGAACCAAC |
| RNA gene | GTCATCCTCAACAGAAATCCAACTACG | GACAAGAAATAACAATACAGGGCATCC |
| cytochrome b gene | TTTGGCACAGCTTTTAACATAGCATATTG | ATTCATTTCGGTTTCTCTTATTTCTCACA |
| ATP synthase CF1 | AGGAGGAATTGTTTGGAATCTTTGGT | TGCTTTTTGCTTGACGGGCAGTATG |
| 18S rRNA | GGCGATGAGGGATGAACCTACCG | GACCACCGTCCTGCTGTCAAAAT |
ChIP-qPCR primers.
Results
Physiological responses of A. pacificum under different conditions
To observe the growth status and determine the sampling time, counting and growth curves were carried out for A. pacificum under three treatment conditions. It was found that cells of A. pacificum exhibited a low growth rate from days 1 to 3, then grew rapidly from 5 to 15 days (Figure 1). After 15 days the algae growth tends to stabilize. The results showed that high irradiation and nitrogen conditions will promote the growth of Alexandrium pacificum. The 12th day of the rapid growth period was selected as the sampling time for subsequent experiments.
Figure 1
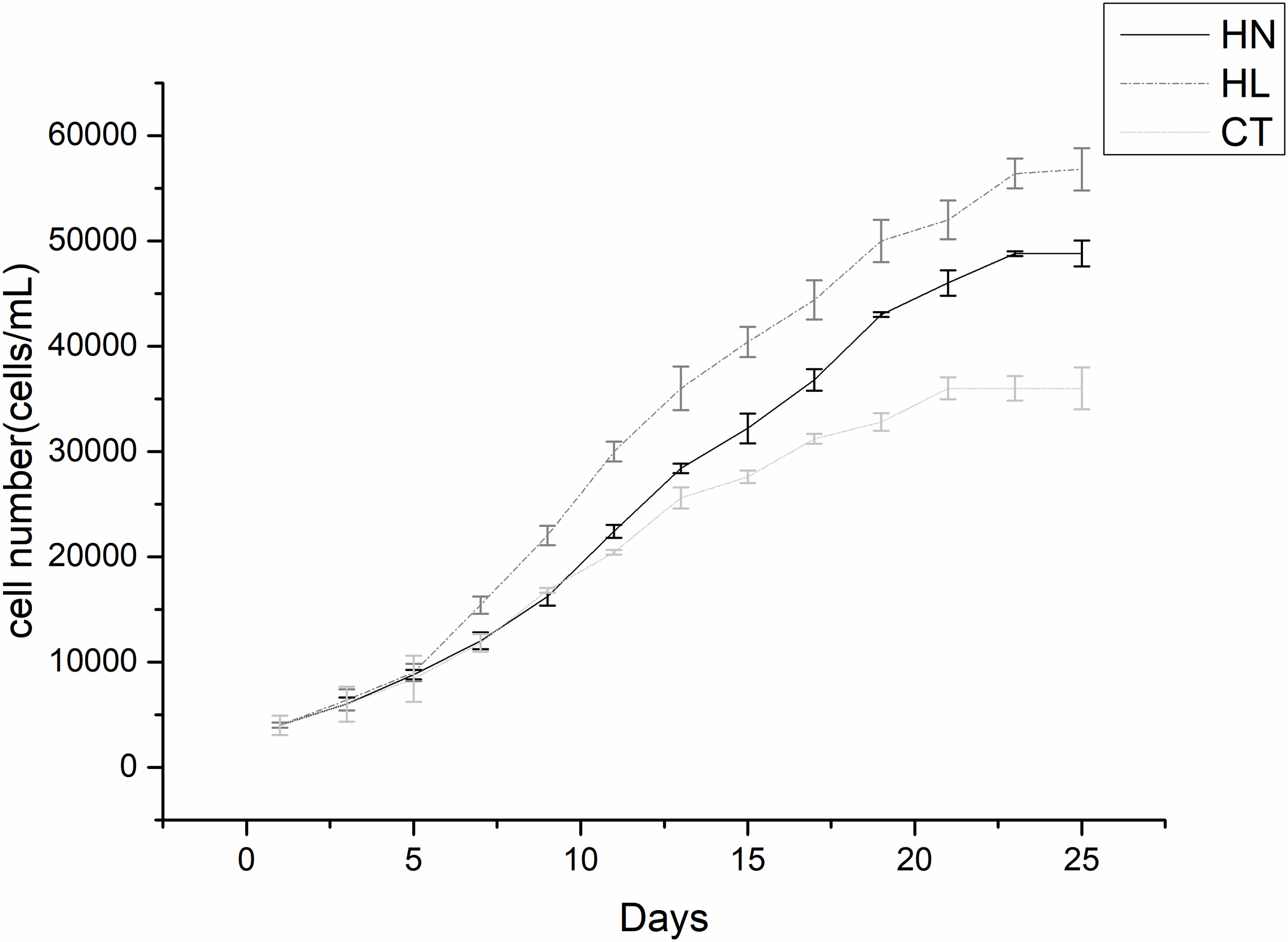
Growth curves of Alexandrium pacificum under different conditions. High light (HL), high nitrogen (HN), and f/2 normal medium condition (control [CT]). Each point is the mean of three independent experiments.
H3K4me3 abundance of A. pacificum under different conditions
The modification abundance of H3K4me3 of A. pacificum under the different treatment conditions is shown in Figure 2. The abundance of H3K4me3 under HN and HL was greater than that under CT during the exponential rapid growth period of A. pacificum.
Figure 2
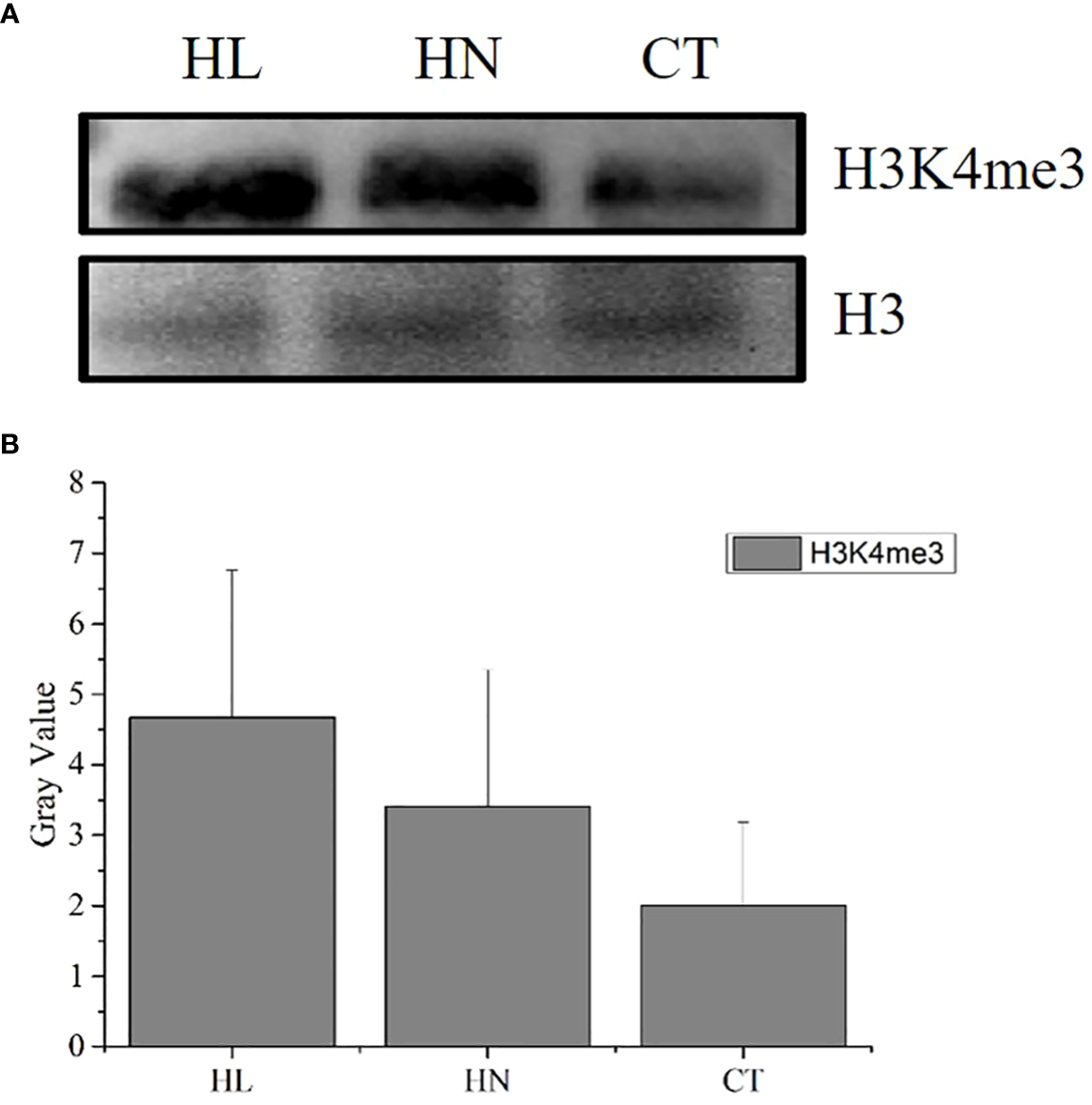
H3K4me3 abundance detection of Alexandrium pacificum under different conditions. (A) Western blot results of H3K4me3 modification in A pacificum under HN, HL, and CT; (B) Bar graph of the blot-spot density. Each column is the mean of three independent experiments, statistically significant differences were assessed using SPSS Statistics (v. 17.0; IBM Corp., Armonk, NY, USA) based on Tukey’s test at p < 0.05.
H3K4me3 associated genes of A. pacificum under different conditions
Over 30 million uniquely mapped reads were obtained per library. More than 35% of the ChIP-Seq reads could be aligned to the Fugacium kawagutii genome. In total, around 2,878–7,822 peaks were called under each condition; and 156 genes from HL, 254 genes from HN, and 803 from CT were found to be specifically associated with H3K4me3 (Figure 3). A total of 36 genes were associated with H3K4me3 modification under HN and HL, but not under CT.
Figure 3
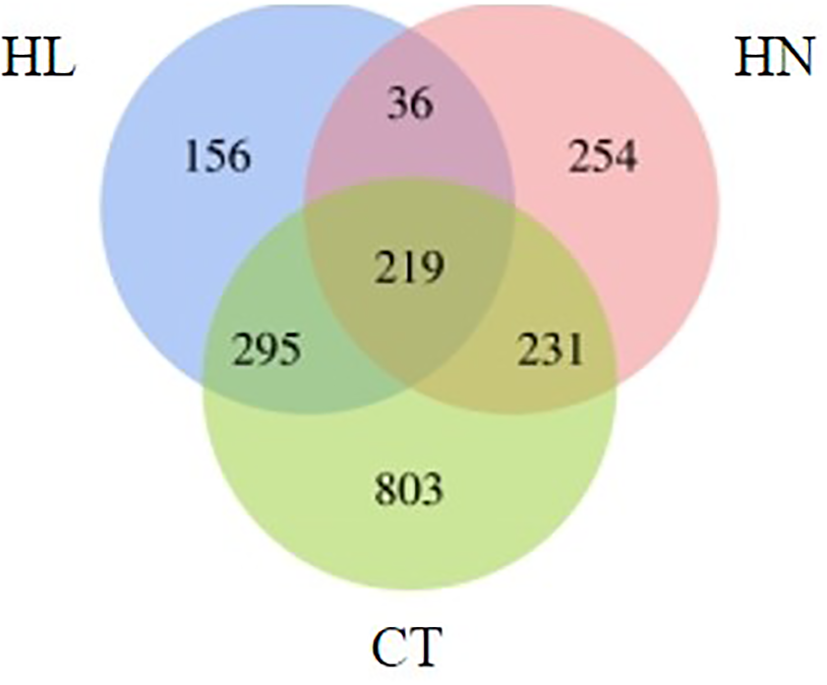
Venn diagram of the number of genes associated with H3K4me3 under different conditions: high light (HL), high nitrogen (HN), and normal f/2 medium condition (CT).
ChIP-qPCR
ChIP-qPCR was performed to confirm the results of ChIP-Seq, as shown in Figure 4. A total of nine genes (from 156 + 36+254+219 in Figure 3) involved in various biological processes were selected for ChIP-qPCR, including riboflavin biosynthesis protein RibBA (RibBA), eukaryotic translation initiation factor 2 (Euk), cell wall alpha-1,3-glucan synthase mok12 (Mok12), ribosomal protein L14 (L14), ubiquitin-activating enzyme E1 (E1), small subunit ribosomal RNA gene (RNA gene), cytochrome b gene (CB), ATP synthase CF1 subunit alpha (ATP), and 18S rRNA gene (18S). Sample immunoglobulin G was used as the negative control in the ChIP-qPCR assay. Most selected genes displayed significant enrichment under HL and HN compared to under CT. The relative enrichment of immunoprecipitated DNA and immunoglobulin G (negative control) of each gene was more than 10-fold.
Figure 4
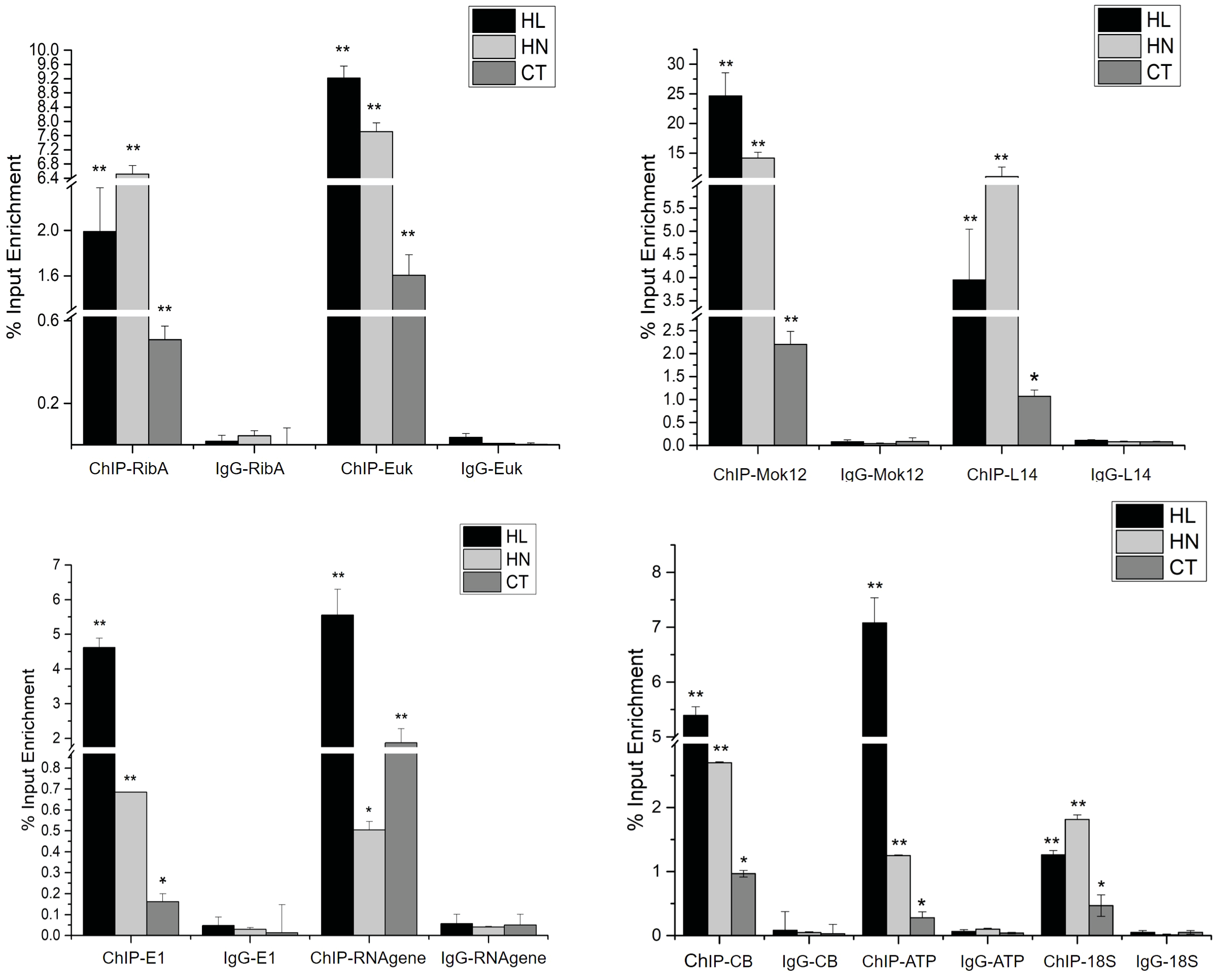
The result of ChIP-qPCR under high light (HL), high nitrogen (HN), and normal (CT) conditions. IP DNA was the experimental group and IgG DNA was the negative control group. Each column is the mean of three independent experiments. Significant differences are indicated by asterisks (**p < 0.01; *p < 0.05).
Analysis of H3K4me3-associated genes unique to the HL condition in A. pacificum
KEGG enrichment analysis showed that pathways related to growth and nutrient uptake, such as nitrogen metabolism, endocytosis, glyoxylate, and dicarboxylate metabolism, folate biosynthesis, phenylalanine metabolism, and protein processing in the endoplasmic reticulum, were all significantly enriched (Figure 5). In these enriched pathways, H3K4me3-associated genes unique to the HL condition, such as nitrate reductase and glutamine synthetase, were involved in nitrogen metabolism. In addition, several H3K4me3-related genes such as protein 26, E3 ubiquitin-protein ligase, and heat shock protein 70 (HSP70) were involved in the endocytosis pathway (Supplementary 2).
Figure 5
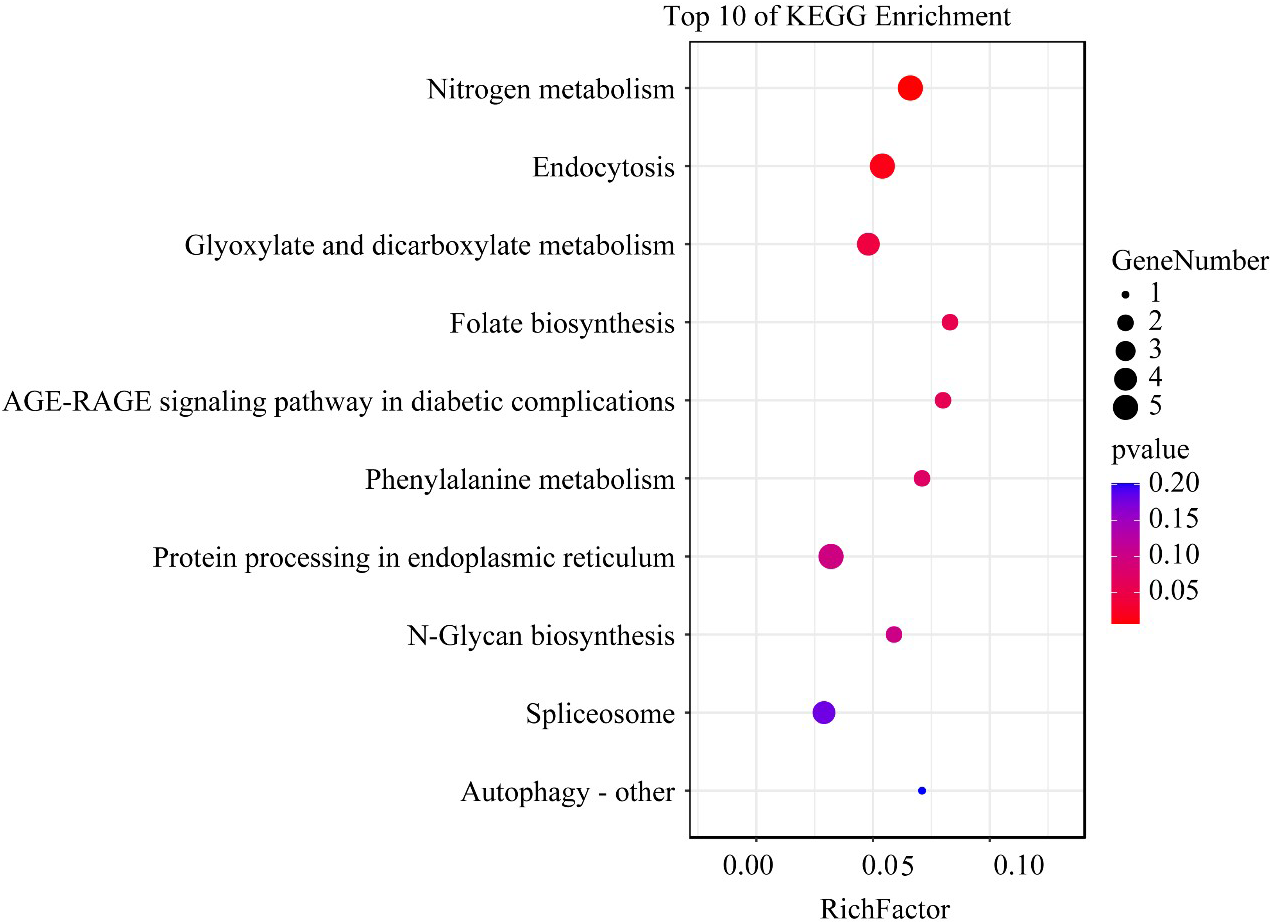
The top 10 enriched KEGG pathway terms of H3K4me3 associated genes unique to the HL condition in Alexandrium pacificum. RichFactor represents the ratio of the number of genes located in the KEGG term to the total number of genes located in the KEGG term in all annotated genes. The size of the circle represents the number of genes enriched in the term. The gradient of color from blue to red represents the size of the p-value changes.
Analysis of H3K4me3 associated genes unique to the HN condition in A. pacificum
H3K4me3-associated genes unique to the HN condition were analyzed by KEGG enrichment. Vitamin B6 metabolism was significantly enriched. Furthermore, other pathways related to vitamin metabolism including folate, thiamine, pantothenate, and coenzyme A biosynthesis, and nicotinate and nicotinamide metabolism, were also enriched. In addition, the citrate cycle, mitogen−activated protein kinase signaling, plant hormone signal transduction, and RNA polymerase inositol phosphate metabolism pathways were also enriched (Figure 6). KEGG analysis revealed that the most affected pathway in A. pacificum under the HN condition was the vitamin metabolic pathway. The three genes of phosphoserine aminotransferase, exosome complex exonuclease RRP6, and pyridoxine 4-dehydrogenase involved in vitamin B6 metabolism were significantly enriched (Supplementary 2).
Figure 6
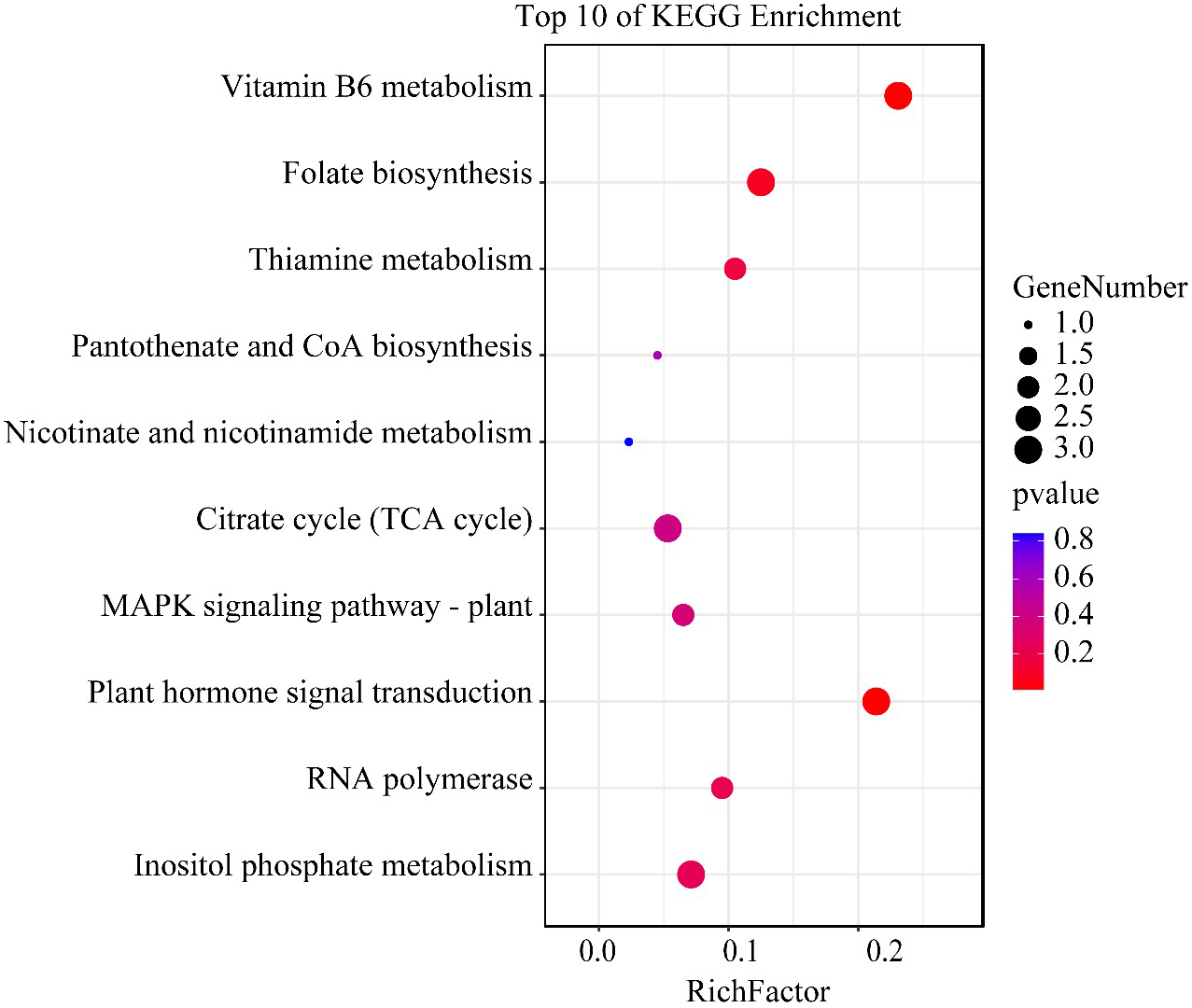
The top 10 enriched KEGG pathway terms of H3K4me3 associated genes unique to the HN condition in Alexandrium pacificum. RichFactor represents the ratio of the number of genes located in the KEGG term to the total number of genes located in the KEGG term in all annotated genes. The size of the circle represents the number of genes enriched in the term. The gradient of color from blue to red represents the size of the p value changes.
Discussion
H3K4me3 antibody selection and genome mapping protocol
For H3K4me3 modification, approximately 5.5% of the homologous sequences are covered by H3K4me3 peaks in the human genome (effective genome) and rhesus macaque genome, accounting for 132,294 homologous pairs. Additionally, there is a total of 79,865 human-mouse (more evolutionary distant) homologous region pairs with H3K4me3 signals (Lu et al., 2018), which indicate that H3K4me3 modification is highly conservative. The conservatism of H3K4me3 is important in the selection of commercial antibodies in ChIP. The antibody is the most critical factor in ChIP-seq, and directly affects the success of the ChIP experiment. The selection of the human H3K4me3 antibody is supported by several aspects. First, the N-terminal of the A. pacificum histone is conservative and divergent, and the two H3 variants are similar to human H3 sequences, both have conserved all the key residues (including the H3K4) of the histone code except H3K18 and H3S28 (Liu et al., 2019; Riaz et al., 2019). In addition, the human histone H3 antibody has been successfully used to detect histone H3 of A. pacificum (Liu et al., 2019). Moreover, mass spectrometry on the protein at the position of the H3 antibody was performed and the conserved modification site of H3K4 was found (Zhu et al., 2022). This led to the successful use of the H3K4me3 commercial antibody raised from human H3 for chromatin immunoprecipitation.
Due to the large size of the dinoflagellate genome, which contains highly redundant and repetitive sequences (Davidson et al., 2002; Hou and Lin, 2009), complete sequencing, assembly, and annotation of the dinoflagellate genome is nearly impossible within the constraints of current technology and expenditure. To date, less than 250 of the more than 8 million estimated eukaryotic species have been fully sequenced at the chromosome level (He et al., 2015). Nevertheless, information from several related species is often required to determine common processes and their evolutionary plasticity in order to understand the overarching principles of developmental biology. While detailed maps of developmental gene regulatory networks are well established for the sea urchin, Stronglyocentrotus purpuratus, a model organism, comparative studies using related species including sea stars and sea cucumbers, which have not been fully sequenced to date, are required to resolve longstanding questions related to factors involved in sea urchin development (Davidson et al., 2002). The genome of the dinoflagellate, F. kawagutii, was previously completed alongside the assembly, annotation, and uploading of the genome (Lin et al., 2015). In the present study, the genome of F. kawagutii was used to complete the genome-matching process. Finally, approximately 35% of the ChIP-seq reads were aligned to the genome of F. kawagutii. The results were verified by ChIP-qPCR, indicating that the results of the ChIP-seq had a certain credibility, and that the single gene in the results (including the target genes of interest in the subsequent analysis) had a high reliability. The analysis of the ChIP-seq results can help us acquire knowledge of H3K4me3-related genes under different conditions.
However, 65% of the reads were not aligned with the genome of F. kawagutii, leading to the omission of a large amount of relevant information from the results and making it impossible to judge the most important regulatory information of H3K4me3 overall. In addition, most of the peak sequences obtained by H3K4me3 were in gene regulatory regions. The gene information obtained by this alignment method cannot be correlated with the transcriptome data of A. pacificum, which introduces significant difficulties in the follow-up study of H3K4me3-related genes. Moreover, the method of the comparing genomes of other species may also have a large impact on the accuracy of the peak annotation and results analysis. For example, the annotation information of the peak sequence is the annotation information of F. kawagutii obtained by aligning the genome of F. kawagutii, and although the peak sequence is conservative, it does not exclude the possibility that the gene in which it is located is not conservative, which may also cause deviation in the subsequent gene analysis results. Therefore, it is necessary to continue the search for more suitable alignment genomes or more appropriate research methods to solve them.
H3K4me3 participates in nitrogen metabolism and endocytosis in A. pacificum under HL
The growth curves showed that A. pacificum grew more rapidly under HL than under the other conditions. H3K4me3 modification abundance was greater in the fast-growing log-phase under this condition than under the other conditions, indicating that H3K4me3 modification may be involved in the regulation of the rapid growth of A. pacificum. ChIP-seq results showed that among the H3K4me3-related genes specific to the HL condition, genes related to nitrogen metabolism, endocytosis, and mitogen-activated protein kinase signal transduction were significantly enriched. Therefore, these metabolic pathways and related genes may be involved in the regulation of the rapid growth of A.pacificum under HL in response to the change of H3K4me3.
Nitrogen is essential for the growth of marine phytoplankton (Li et al., 2021). The rapid growth of algae is closely related to an increase in nitrogen metabolic rate under HL. A previous study showed that the influence of light is very important on the activity of nitrate reductase; nitrate reductase activity increased with increasing light in the range of 10–90 μmol m-2 s-1 (Li et al., 2013). Therefore, under the condition of HL, nitrate reductase should play a major role in nitrogen metabolism. In our study, the genes related to H3K4me3 unique to HL, encoding metabolic enzymes within the nitrogen metabolism, especially those governing the rate-limiting steps, including nitrate reductase and glutamine synthetase gene, suggesting that H3K4me3 may contribute to the rapid growth of A. pacificum cells under HL conditions by nitrogen metabolism pathway.
Autotrophic, heterotrophic, and mixotrophic nutrition are the three main ways for dinoflagellates to uptake nutrients (Legrand and Carlsson, 1998). Studies have shown that dinoflagellate can ingest carbohydrates, some heterotrophic bacteria, and cyanobacteria through endocytosis (Feinstein et al., 2002; Zhang et al., 2011). Endocytosis plays an important role in the uptake of extracellular nutrients, the regulation of cell surface receptor expression, plasma membrane homeostasis, and antigen presentation (Couto and Zipfel, 2016; Liao et al., 2017). HSP70 plays an important role in transmembrane transport, and E3 ubiquitin ligase-mediated ubiquitination regulates plasma membrane-resident receptor-like kinase endocytosis and intracellular degradation (Zhou et al., 2018). E3 ubiquitin ligase-mediated endocytosis of the plasma membrane receptor BR INSENSITIVE1 is mainly needed for signaling attenuation because the blocking of its internalization resulted in the enhancement of brassinosteroid responses in Arabidopsis thaliana (Irani et al., 2012). Regarding the present ChIP-seq results, the genes related to endocytosis affected by H3K4me3, including vacuolar sorting-related protein 26, HSP70, and E3 ubiquitin ligase, were significantly enriched. Previously, it has been reported that Prorocentrum shikokuense, when forming blooms, can ingest Synechococcus, Isochrysis galbana, and Skeletonema to acquire nutrients (Jeong et al., 2005). This indicates that endocytosis may be an important approach for the uptake of external nutrients and intracellular nutrient signal transduction during the rapid growth of A. pacificum, and may be conducive to maintaining a sufficient nutrient supply for the rapidly growing algal cells.
H3K4me3 participates in vitamin metabolism in A. pacificum under HN
The growth curves showed that A.pacificum grew faster under HN than under CT (normal f/2 condition). The H3K4me3 modification abundance was greater in the fast-growing algae under this condition than under CT, indicating that H3K4me3 may also be involved in the regulation of the rapid growth of A. pacificum under HN. The results of the differential analysis of H3K4me3-related genes between HN and CT showed that genes involved in vitamin metabolism were significantly enriched. The vitamin metabolism pathyway is closely related to dinoflagellate growth (Lin et al., 2019).
Vitamin B6 is an important coenzyme in over 100 different cellular reactions and processes including those of amino acid metabolism, heme and chlorophyll biosynthesis, ethylene biosynthesis, fatty acid metabolism, transcriptional regulation, and response to oxidative stress (Rueschhoff, 2009; Colinas et al., 2016). The growth of dinoflagellates requires vitamin B, for example, Prorocentrum donghaiense and Lingulodinium polyedrum need vitamin B12 during tbloomings (Lin et al., 2014). Furthermore, microalgae growth can require different combinations of three types of vitamins B: vitamin B12 (cobalamin), B1 (thiamine), and B7 (biotin) (Fischer and Bacher, 2011). Thiamine plays an important role in carbon metabolism, and is a coenzyme factor of various carbohydrate-related enzymes in primary metabolism and amino acid-related enzymes (Tang et al., 2010). The active form of thiamine is thiamine pyrophosphate, which is essential for all organisms. The cofactor is associated with several enzymes involved in primary carbohydrate and branched-chain amino acid metabolism, including pyruvate dehydrogenase, transketolase, α-keto-acid decarboxylase, and α-ketoacid oxidase (Suzuki et al., 2010). Pantothenate (vitamin B5) is essential for fatty acid and anabolic steroids. It is also involved in steroid violet, melatonin, and heme synthesis, the citric acid cycle, choline acetylation, antibodies, and other necessary intermediate metabolites (Webb and Smith, 2011). However, there are no reports of vitamin B6 utilization in dinoflagellates, and the enrichment of the vitamin B6 metabolic pathway in A. pacificum suggested that this may be a unique form of vitamin utilization and synthesis, which may contribute to the formation and succession of its red tide.
Conclusion
In this study, H3K4me3-associated genes of A.pacificum under the conditions of HL and HN were investigated. H3K4me3 was involved in the rapid growth of A. pacificum under HL and HN conditions, including acting on genes related to nitrogen metabolism, endocytosis, and vitamin metabolism (Figure 7). To the best of the authors’ knowledge, this study revealed the epigenetic regulatory network of a red tide microalgae which induced red tide outbreak for the first time, providing important information for further understanding the mechanisms of harmful algal blooms. However, the detailed mechanisms behind the involvement of these metabolic pathways in algal growth need to be further investigated and explored.
Figure 7
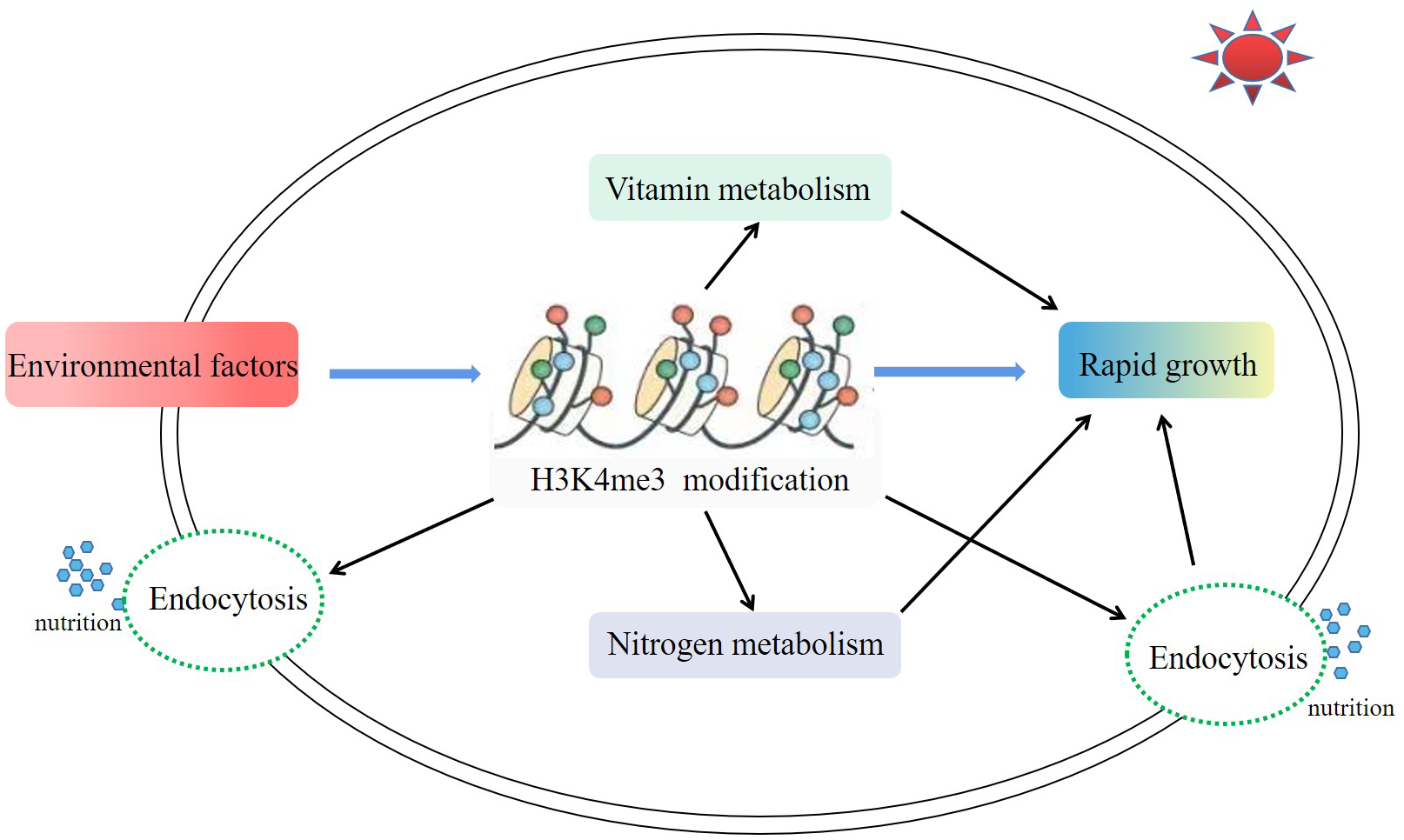
Major metabolic pathways associated with H3K4me3 in Alexandrium pacificum under high light and high nitrogen conditions. Environmental factors (high nitrate and high light intensity) enhanced the modification enrichment of H3K4me3. A higher enrichment level of H3K4me3 further promoted the vitamin metabolism, nitrogen metabolism, and endocytosis pathways, and finally promoted the rapid growth of A. pacificum.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 41676091 and 41176098).
Acknowledgments
Thanks to Professor Senjie Lin from Xiamen University for generously providing the genomic information of Fugacium kawagutii.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, accession number PRJNA881615.
Author contributions
JQ and ZS mainly designed the experimental protocol and participated in the wrote of the paper. JQ was responsible for the experiment and date analysis. ZZ participated in the experiment and revised the writing. YL participated in the design of the experimental scheme. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1011663/full#supplementary-material
References
1
AshburnerM.BallC. A.BlakeJ. A.BotsteinD.ButlerH.CherryJ. M.et al. (2000). Gene ontology: Tool for the unification of biology. the gene ontology consortium. Nat. Genet.25 (1), 25–29. doi: 10.1038/75556
2
BaekS. S.KwonY. S.PyoJ.ChoiJ.KimY. O.ChoK. H. (2021). Identification of influencing factors of A. catenella bloom using machine learning and numerical simulation. Harmful Algae103, 102007. doi: 10.1016/j.hal.2021.102007
3
BolgerA.LohseM.UsadelB. (2014). Trimmomatic: A flexible trimmer for illumine sequence data. Bioinformatics30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
4
BourdareauS.TirichineL.LombardB.LoewD.ScornetD.WuY.et al. (2021). Histone modifications during the life cycle of the brown alga Ectocarpus. Genome Biol.22 (1), 12. doi: 10.1186/s13059-020-02216-8
5
CaoC.ZhengB.ChenZ.HuangM.ZhangJ. (2011). Eutrophication and algal blooms in channel type reservoirs: A novel enclosure experiment by changing light intensity. J. Environ. Sci.23 (10), 1660–1670. doi: 10.1016/s1001-0742(10)60587-6
6
ChenQ.ChenX.WangQ.ZhangF.LouZ.ZhangQ.et al. (2013). Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice. PloS Genet.9 (1), e1003239. doi: 10.1371/journal.pgen.1003239
7
ChowM. H.YanK. T.BennettM. J.WongJ. T. (2010). Birefringence and DNA condensation of liquid crystalline chromosomes. Eukaryot Cell9 (10), 1577–1587. doi: 10.1128/EC.00026-10
8
ColinasM.EisenhutM.TohgeT.PesqueraM.FernieA. R.WeberA. P. M.et al. (2016). Balancing of B6 vitamers is essential for plant development and metabolism in arabidopsis. Plant Cell28, 439–453. doi: 10.1105/tpc.15.01033
9
CoutoD.ZipfelC. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol.16 (9), 537–552. doi: 10.1038/nri.2016.77
10
DanecekP.BonfieldJ. K.LiddleJ.MarshallJ.OhanV.PollardM. O.et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience10 (2), 1–4. doi: 10.1093/gigascience/giab008
11
DavidsonE. H.RastJ. P.OliveriP.RansickA.CalestaniC.YuhC. H.et al. (2002). A genomic regulatory network for development. Science295 (5560), 1669–1678. doi: 10.1126/science.1069883
12
DijkK.DingY.MalkaramS.RiethovenJ.LiuR.YangJ.et al. (2010). Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol.10, 238. doi: 10.1186/1471-2229-10-238
13
FeinsteinT. N.TraslavinaR.SunM. Y.LinS. J. (2002). Effects of light on photosynthesis, grazing, and population dynamics of the heterotrophic dinoflagellate Pfiesteria piscicida (Dinophyceae). J. Phycol.38 (4), 659–669. doi: 10.1046/j.1529-8817.2002.01175.x
14
FigueroaR. I.CuadradoA.StükenA.RodríguezF.FragaS. (2014). Ribosomal DNA organization patterns within the dinoflagellate genus Alexandrium as revealed by FISH: Life cycle and evolutionary implications. Protist165 (3), 343–363. doi: 10.1016/j.protis.2014.04.001
15
FischerM.BacherA. (2011). Biosynthesis of vitamin B2 and flavocoenzymes in plants. Adv. Bot. Res.58, 93–152. doi: 10.1016/B978-0-12-386479-6.00003-2
16
GendrelA.LippmanZ.MartienssenR.VincentC. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods2 (3), 213–218. doi: 10.1038/nmeth0305-213
17
GrattanL. M.HolobaughS.MorrisJ. G. (2016). Harmful algal blooms and public health. Harmful Algae57, 2–8. doi: 10.1016/j.hal.2016.05.003
18
GuillardR. R. L. (1975). “Culture of phytoplankton for feeding marine invertebrates,” in Culture of marine invertebrate animals. Eds. SmithW. L.ChanleyM. H. (New York: Plenum Press), 26–60.
19
GuillardR. R. L.RytherJ. H. (1962). Studies of marine planktonic diatoms. i. cyclotella nana hustedt and detonula confervacea cleve. Can. J. Microbiol.8, 229–239. doi: 10.1139/m62-029
20
HackettJ. D.AndersonD. M.ErdnerD. L.BhattacharyaD. (2004). Dinoflagellates: a remarkable evolutionary experiment. Am. J. Bot.91 (10), 1523–1534. doi: 10.3732/ajb.91.10.1523
21
HamkaloB. A.RattnerJ. (1977). The structure of a mesokaryote chromosome. Chromosoma60 (1), 39–47. doi: 10.1007/BF00330409
22
HaM.NgD. W. K.LiW. H.ChenZ. J. (2011). Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res.21 (4), 590–598. doi: 10.1101/gr.116467.110
23
HaringM.OffermannS.DankerT.HorstI.PeterhanselC.StamM. (2007). Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods3 (1), 11. doi: 10.1186/1746-4811-3-11
24
HeX.CicekA. E.WangY.SchulzM. H.LeH. S.Bar-JosephZ. (2015). De novo ChIP-seq analysis. Genome Biol.16, 205. doi: 10.1186/s13059-015-0756-4
25
HouY.LinS. (2009). Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: gene content estimation for dinoflagellate genomes. PloS One4 (9), e6978. doi: 10.1371/journal.pone.0006978
26
IraniN. G.RubboS. D.MylleE.BeginJ.Schneider-PizońHnilikováJ.et al. (2012). Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol.8 (6), 583–589. doi: 10.1038/nchembio.958
27
JaeckischN.YangI.WohlrabS.GlocknerG.KroymannJ.VogelH.et al. (2011). Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii. PloS One6 (12), e28012. doi: 10.1371/journal.pone.0028012
28
JeongH. J.ParkJ. Y.NhoJ. H.ParkM. O.HaJ. H.SeongK. A.et al. (2005). Feeding by red-tide dinoflagellates on the cyanobacterium Synechococcus. Aquat. Microb. Ecol.41, 131–143. doi: 10.3354/ame041131
29
KanehisaM.GotoS. (2000). KEGG: kyoto encyclopedia of genes and genomes. nucleic acids research. Nucleic Acids Res.28 (1), 27–30. doi: 10.1093/nar/28.1.27
30
KarabashevG. S.EvdoshenkoM. A. (2016). Narrowband shortwave minima in spectra of backscattered light from the sea obtained from ocean color scanners as a remote indication of algal blooms. Oceanologia58 (4), 279–291. doi: 10.1016/j.oceano.2016.05.001
31
KoeneckeN.JohnstonJ.GaertnerB.NatarajanM.ZeitlingerJ. (2016). Genome-wide identification of drosophila dorso-ventral enhancers by differential histone acetylation analysis. Genome Biol.17 (1), 196. doi: 10.1186/s13059-016-1057-2
32
LaabirM.JauzeinC.GenovesiB.MasseretE.GrzebykD.CecchiP.et al. (2011). Influence of temperature, salinity and irradiance on the growth and cell yield of the harmful red tide dinoflagellate Alexandrium catenella colonizing Mediterranean waters. J. Plankton Res.33 (10), 1550–1563. doi: 10.1093/plankt/fbr050
33
LauberthS. M.NakayamaT.WuX.FerrisA. L.TangZ.HughesS. H.et al. (2013). H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell152 (5), 1021–1036. doi: 10.1016/j.cell.2013.01.052
34
LegrandC.CarlssonP. (1998). Uptake of high molecular weight dextran by the dinoflagellate Alexandrium catenella. Aquat. Microbial. Ecol.16, 81–86. doi: 10.3354/ame016081
35
LiaoD.CaoY.SunX.EspinozaC.NguyenC. T.LiangY.et al. (2017). Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor lysin motif receptor Kinase5 (LYK5) protein abundance. New Phytol.214 (4), 1646–1656. doi: 10.1111/nph.14472
36
LinS. (2011). Genomic understanding of dinoflagellates. Res. Microbiol.162 (6), 551–569. doi: 10.1016/j.resmic.2011.04.006
37
LinS.ChengF.SongB.ZhongX.LinX.LiW.et al. (2015). The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science350 (6261), 691–694. doi: 10.1126/science.aad0408
38
LinS. J.JiN. J.LuoH. (2019). Recent progress in marine harmful algal bloom research. Oceanol. Et Limnol. Sin.50 (03), 495–510. doi: 10.11693/hyhz20180800191
39
LinJ. N.YanT.ZhangQ. C.WangY. F.LiuQ.ZhouM. J. (2014). In situ detrimental impacts of Prorocentrum donghaiense blooms on zooplankton in the East China Sea. Mar. pollut. Bull.88 (1-2), 302–310. doi: 10.1016/j.marpolbul.2014.08.026
40
LiH. M.ShiX. Y.DingY. Y.TangH. J. (2013). Illumination’s effect on the growth and nitrate reductase activity of typical red-tide algae in the East China Sea. Huan Jing Ke Xue34 (9), 3391–3397. doi: 10.13227/j.hjkx
41
LiuC.LuF.CuiX.CaoX. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol.61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
42
LiuY.ZhuZ.QiJ.SuiZ.ShangE.ZhangS.et al. (2021). Comparative transcriptome profiling reveals insights into the mechanisms related to explosive growth of Alexandrium pacificum. Front. Mar. Sci.8. doi: 10.3389/fmars.2021.751851
43
LiuY.ZhuZ.SuiZ.LiuH.RiazS. (2022). Calmodulin and its interactive proteins participate in regulating the explosive growth of Alexandrium pacificum (dinoflagellate). Int. J. Mol. Sci.23, 145. doi: 10.3390/ijms23010145
44
LiuH.RiazS.LiuY.KhurshidA.ZhangW.SuiZ.et al (2019). Cloning and expression analysis of histone H3 in Alexandrium catenella during the growth process. Marine Sci.43 (11), 19–27. doi: 10.11759/hykx20190401001
45
LiX. Y.YuR. C.GengH. X.LiY. F. (2021). Increasing dominance of dinoflagellate red tides in the coastal waters of yellow Sea. China Mar. pollut. Bull.168, 112439. doi: 10.1016/j.marpolbul.2021.112439
46
LuJ.CaoX.ZhongS. (2018). A likelihood approach to testing hypotheses on the co-evolution of epigenome and genome. PloS Comput. Biol.14 (12), e1006673. doi: 10.1371/journal.pcbi.1006673
47
MarinovG. K.LynchM. (2015). Diversity and divergence of dinoflagellate histone proteins. G3 (Bethesda)6 (2), 397–422. doi: 10.1534/g3.115.023275
48
Moreno-PerezA. J.Santos-PereiraJ. M.Martins-NoguerolR.DeAndres-GilC.Troncoso-PonceM. A.Venegas-CaleronM.et al. (2021). Genome-wide mapping of histone H3 lysine 4 trimethylation (H3K4me3) and its involvement in fatty acid biosynthesis in sunflower developing seeds. Plants (Basel)10 (4), 706. doi: 10.3390/plants10040706
49
NganC. Y.WongC. H.ChoiC.YoshinagaY.LouieK.JiaJ.et al. (2015). Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nat. Plants1 (8), 15107. doi: 10.1038/nplants.2015.107
50
RiazS.NiazZ.KhanS.LiuY.SuiZ. (2019). Detection, characterization and expression dynamics of histone proteins in the dinoflagellate Alexandrium pacificum during growth regulation. Harmful Algae87, 101630. doi: 10.1016/j.hal.2019.101630
51
RiazS.SuiZ. (2018). Molecular cloning, transcriptome profiling, and characterization of histone genes in the dinoflagellate Alexandrium pacificum. J. Microbiol. Biotechnol.28 (7), 1185–1198. doi: 10.4014/jmb.1802.01075
52
RueschhoffE. E. (2009). Vitamin B6 metabolism in arabidopsis thaliana. dissertations and theses - gradworks. (Raleigh, North Carolina: North Carolina State University).
53
SolomonE.CaldwellK.AllanA. (2021). A novel method for the normalization of ChIP-qPCR data. MethodX8, 101504. doi: 10.1016/j.taap.2020.114920
54
StarkR.BrownG. (2011). DiffBind: differential binding analysis of ChIP-seq peak data. r package version (Cambridge: University of Cambridge, Cancer Research UK- Cambridge Institute.), Vol. 100. 4–3.
55
SuzukiR.KatayamaT.KimB. J.WakagiT.ShounH.AshidaH.et al. (2010). Crystal structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism. J. Biol. Chem.285 (44), 34279–34287. doi: 10.1074/jbc.M110.156281
56
TangY. Z.KochF.GoblerC. J. (2010). Most harmful algal bloom species are vitamin B1 and B12 auxotrophs. Proc. Natl. Acad. Sci. United States America107 (48), 20756–20761. doi: 10.1073/pnas.1009566107
57
UribeP.FuentesD.ValdésJ.ShmaryahuA.ZúñigaA.HolmesD.et al. (2008). Preparation and analysis of an expressed sequence tag library from the toxic dinoflagellate Alexandrium catenella. Mar. Biotechnol.10 (6), 692–700. doi: 10.1007/s10126-008-9107-8
58
WangH.HernandezJ. M.Van MieghemP. (2008). Betweenness centrality in a weighted network. Phys. Rev. E Stat. Nonlin. Soft Matter Phys.77, 46105. doi: 10.1103/PhysRevE.77.046105
59
WangD.LinL.WangM.LiC.HongH. (2012). Proteomic analysis of a toxic dinoflagellate Alexandrium catenella under different growth phases and conditions. Chin. Sci. Bull.57 (25), 3328–3341. doi: 10.1007/s11434-012-5160-9
60
WangQ.YungW. S.WangZ.LamH. M. (2020). The histone modification H3K4me3 marks functional genes in soybean nodules. Genomics112 (6), 5282–5294. doi: 10.1016/j.ygeno.2020.09.052
61
WebbM. E.SmithA. G. (2011). Pantothenate biosynthesis in higher plants. Adv. Bot. Res.58, 203–255. doi: 10.1016/B978-0-12-386479-6.00001-9
62
WeiL.XuJ. (2018). Optimized methods of chromatin immunoprecipitation for profiling histone modifications in industrial microalgae nannochloropsis spp. J. Phycol.54, 358–367. doi: 10.1111/jpy.12623
63
YamamotoJ.HanQ.InubushiS.SugisawaN.HamadaK.NishinoH.et al. (2020). Histone methylation status of H3K4me3 and H3K9me3 under methionine restriction is unstable in methionine-addicted cancer cells, but stable in normal cells. Biochem. Biophys. Res. Commun.533 (4), 1034–1038. doi: 10.1016/j.bbrc.2020.09.108
64
YanH.LiuY.ZhangK.SongJ.XuW.SuZ. (2019). Chromatin state-based analysis of epigenetic H3K4me3 marks of Arabidopsis in response to dark stress. Front. Genet.10. doi: 10.3389/fgene.2019.00306
65
YuL.ZhangY.LiM.WangC.LinX.LiL.et al. (2020). Comparative metatranscriptomic profiling and microRNA sequencing to reveal active metabolic pathways associated with a dinoflagellate bloom. Sci. Total Environ.699, 134323. doi: 10.1016/j.scitotenv.2019.134323
66
ZhangY. J.CaoY.WangZ.HanB.YangY. (2006). Effects of limitation of nitrogen and phosphorus on the growth of Alexandrium tamarense. J. Trop. Subtrop. Bot.14 (006), 482–486. doi: 10.1186/gb-2008-9-9-r137
67
ZhangY.LiuT.MeyerC. A.EeckhouteJ.JohnsonD. S.BernsteinB. E.et al. (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol.9, R137. doi: 10.1186/gb-2008-9-9-r137
68
ZhangS.OUL.LuS. (2011). Effects of light and nutrients on the phagotrophic behaviors of three harmful dinoflagellates. Jinan Univ. (MA thesis)35 (4), 94–99. doi: 10.1088/1674-1137/35/2/019
69
ZhangS.SuiZ.ChangL.KangK.MaJ.KongF.et al. (2014). Transcriptome de novo assembly sequencing and analysis of the toxic dinoflagellate Alexandrium catenella using the illumina platform. Gene537 (2), 285–293. doi: 10.1016/j.gene.2013.12.041
70
ZhouJ.LiuD.WangP.MaX.LinW.ChenS.et al. (2018). Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. U.S.A.115 (8), E1906–E1915. doi: 10.1073/pnas.1712251115
71
ZhuZ.LiuY.QiJ.SuiZ. (2022). Identification of epigenetic histone modifications and analysis of histone lysine methyltransferases in Alexandrium pacificum. Harmful Algae119, 102323. doi: 10.1016/j.hal.2022.102323
72
ZongW.ZhongX.YouJ.XiongL. (2013). Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol. Biol.81 (1-2), 175–188. doi: 10.1007/s11103-012-9990-2
Summary
Keywords
Alexandrium pacificum, H3K4me3, histone, ChIP-seq, epigenetics, HABs
Citation
Qi J, Zhu Z, Liu Y and Sui Z (2022) First insight into H3K4me3 modification in the rapid growth of Alexandrium pacificum (dinoflagellates). Front. Mar. Sci. 9:1011663. doi: 10.3389/fmars.2022.1011663
Received
04 August 2022
Accepted
14 November 2022
Published
30 November 2022
Volume
9 - 2022
Edited by
Carolina Madeira, NOVA University Lisbon, Portugal
Reviewed by
Mickael Le Gac, Institut Français de Recherche pour l’Exploitation de la Mer, France; Kieng Soon Hii, University of Malaya, Malaysia
Updates
Copyright
© 2022 Qi, Zhu, Liu and Sui.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenghong Sui, suizhengh@ouc.edu.cn
†These authors have contributed equally to this work
This article was submitted to Marine Molecular Biology and Ecology, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.