- 1Joint Laboratory for Deep Blue Fishery Engineering of Pilot National Laboratory for Marine Science and Technology (Qingdao), Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 2Key Laboratory of South China Sea Fishery Resources Exploitation and Utilization, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
Protein metabolism can provide energy for various biological activities of organisms. However, there have been few studies on the succession of gastrointestinal microbiota structure during the metabolism of protein by the host. In this study, physiological and biochemical, qRT-PCR, and 16S rDNA high throughput sequencing methods were used to detect the activity characteristics of proteases and the related gene expression levels in each gastrointestinal tissue, as well as the distribution characteristics and succession of the gastrointestinal microbiota structure of pond-cultured Japanese flounder (Paralichthys olivaceus). The results showed that the activity of pepsin, trypsin, and chymotrypsin in the pyloric caecum and gut, and the relative expression levels of the related genes, increased first and then decreased, but the aspartate aminotransferase (ASP) and alanine aminotransferase (ALA) activity showed the opposite tendency. These reflected that the nutrient composition of gastrointestinal tract contents was dynamic, which caused obvious changes to the alpha diversity and structure of gastrointestinal microbiota. Lactobacillus, Acinetobacter, Bacteroides, Escherichia-Shigella, Prevotella, the genera represented by the Lachnospiraceae_NK4A136_group and MND1, Sphingomonas, Alistipes, Alloprevotella, Enterobacter, Helicobacter, Myroides, Ruminiclostridium, and Romboutsia were the main gastrointestinal microbiota based on the dominance, commonality, and colonization. The relative abundance of Lactobacillus and Acinetobacter in the three gastrointestinal tissues increased first and then decreased. According to the number of shared and dominant genera, the consistency of succession of some genera, and the increase in the similarity of microbiota structure among the three tissues, we inferred that the microbiota was affected by the gastrointestinal tract nutrient composition. Gastrointestinal microbiota genes were mainly enriched in the metabolism pathway according to the KEGG pathway, and the ratio of Firmicutes to Bacteroidetes, the representative of energy accumulation, followed an at-first increasing and then decreasing trend in each tissue. This shows that the main gastrointestinal microbiota played a synergistic role in the energy accumulation of the host. This study will provide a reference for the development of high-efficiency compound feed formulae and improve feed efficiency by regulating the gastrointestinal tract microecology for pond-cultured Japanese flounder.
1 Introduction
Protein is one of the most important nutrients for organisms because it participates in essential physiological processes, such as the maintenance and repair of damaged tissues and the production of enzymes, hormones, and antibodies, through the anabolism and catabolism of protein during growth and reproduction (Tu et al., 2015; Santos et al., 2020a; Santos et al., 2020b). Proteases are enzymes that specifically break peptide bonds between amino acids during protein degradation. Pepsin, trypsin, and chymotrypsin are major proteases derived from pepsinogen, trypsinogen, and chymotrypsinogen, respectively. Pepsinogen, a precursor of pepsin, belonging to the aspartic proteinase family, is activated in the acidic environment of the stomach and converted to the biologically active pepsin (Foltmann, 1981; Korbova and Kohout, 1981; Chi et al., 2013; Nazemroaya et al., 2020). Trypsinogen and chymotrypsinogen, the pancreatic serine proteases of teleosts, are activated to perform their biologically active function in the gut (Berger and Schechter, 1970; Psochiou et al., 2007; Santos et al., 2020a; Santos et al., 2020b). The synthesis of pepsinogen, trypsinogen, and chymotrypsinogen is regulated by the expression of the respective genes (pepsinogen, trypsinogen, and chymotrypsinogen). The gastrointestinal tract is the center of nutrient metabolism in a multicellular organism, and food is the main source of protein. After ingestion, protein nutrients in food enter the gastrointestinal tract. On receipt of this stimulation, the organism up-regulates the expression of these genes to promote the synthesis of proteases, which will degrade protein nutrients into free amino acids. These amino acids are then transported through the blood circulation by related transporter proteins and used in protein anabolism and catabolism (Bröer, 2008; Santigosa et al., 2008; Verri et al., 2017; Wei et al., 2020).
Gastrointestinal tissues with digestive function in cultured fish mainly include the stomach, pyloric caecum, and gut. In recent years, the gut microbiota has become a hot research field that has progressed through the development of molecular biology, omics, and bioinformatic technologies. Studies have reported trillions of microorganisms colonizing the gut, and their composition and activity are closely related to characteristics of the host such as the species, feeding habits, and physiological stage, and environmental factors such as diet nutrition, environmental bacteria, and stress (Li et al., 2012; Nicholson et al., 2012; Banerjee and Ray, 2017; Dwivedi et al., 2017; Jiang et al., 2019; Walburn et al., 2019; Horlick et al., 2020; Jiang et al., 2020). Likewise, microbiota colonizing the gut can, in turn, participate in and influence many of the host’s physiological activities, including nutrition, energy absorption, immunity, and especially adaption to extreme environments (Shabat et al., 2016; Zhang et al., 2016; Legrand et al., 2018). There have been 130 glycoside hydrolases, 22 polysaccharide lyases, and 16 carbohydrate esterases described in all living organisms, most of which are encoded by microbial genomes (Cantarel et al., 2009; Flint et al., 2012). Therefore, microbiota colonizing the gastrointestinal tract can perform metabolic functions that alter the contents, such as secreting vitamins, organic acids, amino acids, growth-promoting factors, digestive enzymes, and other biologically active metabolites, and participate in host physiological activities. However, the nutrient composition of gastrointestinal tract contents is constantly changing during nutrient metabolism, and different gastrointestinal tissues also contain different nutrient contents that lead to changes in the structure of gastrointestinal microbiota. In other words, the host interacts with the gastrointestinal microbiota by changing the nutrient levels in the gastrointestinal environment.
Japanese flounder (Paralichthys olivaceus) is the economic species in China. However, there have been few studies published on changes to the gastrointestinal microbiota structure during nutrient metabolism in this species. In this study, the composition and distribution of gastrointestinal microbiota and trends in protease activities and related gene expression patterns were investigated, and the effect of the gastrointestinal microbiota during nutrient metabolism of pond-cultured Japanese flounder were analyzed. These will provide a reference for the development of high-efficiency compound feed formulae and the screening of native probiotics for pond-cultured Japanese flounder.
2 Materials and methods
2.1 Source and aquaculture management of Japanese flounder
The Japanese flounder used in this experiment were pond-cultured on a farm in Qingdao, Shandong Province, China. One Japanese flounder culture pond (35.63°N, 119.89°E) on this farm was randomly selected as the experimental pond. The size of this pond was approximately 6660 m2, and the average water depth was 3 m. The average body length of the pond-cultured Japanese flounder was 47.97 ± 0.54 cm, and the average weight was 1043.44 ± 38.06 g.
The pond-cultured Japanese flounders were usually fed frozen Ammodytes personatus twice a day (6: 00 and 16: 00), the amount of which was 3%–5% of the fish’s body weight. The frozen A. personatus were purchased in large quantities and stored in the refrigerated warehouse. Before feeding, A. personatus were taken out of the warehouse and naturally thawed to a certain extent, and then washed several times with fresh water. During this experiment, the water conditions were as follows: the temperature was 20–22°C, the dissolved oxygen was higher than 5 mg/L, and the salinity was 28–31. After starvation for 24 h, the experiment was started. Since the experiment only involved one complete feeding cycle, the pond-cultured Japanese flounders were only fed once (6: 00) during the experiment.
2.2 Collection of samples
On the basis of our previous study, we set the sampling times as 0 h (before feeding), 6 h after ingestion, and 12 h after ingestion. At each sampling time, nine individuals were randomly selected from the Japanese flounder culture pond as the sample fish. These individuals were completely narcotized with MS-222 (Sigma-Aldrich, St. Louis, MO, USA) to minimize suffering, and the collection of all fish samples was strictly conducted according to the regulations from the local government and the Institutional Animal Care and Use Committee of the Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences. The weight and length of each were measured. After cleaning with 75% alcohol, the blood samples (2 mL) were quickly collected from the tail vein of each fish with a disposable syringe and placed in the refrigerator at 4°C for 4 h.
Then, these individuals were dissected and the gastrointestinal tracts were taken out with sterile scissors and forceps under sterile conditions. After removing the fatty tissue, blood vessels, and other tissues from the surface of gastrointestinal tracts and cleaning with 75% alcohol, the samples were squeezed gently to remove any of the remaining contents in the gastrointestinal tracts. Then, the gastrointestinal tracts were rinsed with pre-cooled sterile 1.5% saline three to five times. Each gastrointestinal tract, including the stomach, pyloric caecum, and gut, was differentiated according to its structural characteristics, separated with sterilized scissors, packed in sterile and RNAase-free centrifuge tubes, and stored in liquid nitrogen.
2.3 Enzymes activity assay and bacterial DNA extraction
The blood samples were taken out of the refrigerator and centrifuged for 10 min at 4°C and 4000 r/min. Then the supernatant (serum) was aspirated. Serum samples from three individuals were mixed into one sample. The mixed serum samples were used to determine the aspartate aminotransferase (ASP) and alanine aminotransferase (ALA) activities with ELISA kits (Meilian, Shanghai, China) and RT-6100 (Rayto, Shenzhen, China) following the manufacturer’s instructions.
Gastrointestinal tissues from three individuals were randomly selected for the detection of protease activities. Each gastrointestinal tissue sample was taken out of liquid nitrogen and then thawed naturally on the ice. A certain amount of tissue was weighed, after adding the pre-cooled tissue homogenate according to the ratio of 1: 9 (w/v), the T10 Basic S25 tissue homogenizer (IKA, Staufen, Germany) was used to homogenize the sample, which was centrifuged at 5000 r/min for 30 min at 4°C. The supernatant was then collected and used to determine the pepsin, trypsin, and chymotrypsin activities with ELISA kits (Meilian, Shanghai, China) and RT-6100, following the manufacturer’s instructions.
The gastrointestinal tissues of another three individuals were taken out of liquid nitrogen and then ground up with liquid nitrogen, and the total genomic DNA of the microbiota was extracted using a DNA Extraction Kit (DNeasy PowerSoil Kit, Qiagen, Germany) following the manufacturer’s instructions, respectively. The concentration of DNA was examined with NanoDrop and agarose gel. The genomic DNA was used as a template for PCR amplification with the barcoded primers and Tks Gflex DNA Polymerase (Takara, Japan). For the microbiota diversity analysis, the V3-V4 variable regions of 16S rDNA were amplified using PCR with universal primers 343 F (5′-TACGGRAGGCAGCAG-3’) and 798 R (5′-AGGGTATCTAATCCT-3’). The amplicon quality was visualized using gel electrophoresis, purified with AMPure XP beads (Agencourt, Beverly, Massachusetts, USA), and amplified for another round of PCR. After being purified with the AMPure XP beads again, the final amplicon was quantified. Equal amounts of purified amplicon were pooled for subsequent sequencing. The paired-end reads (2 × 250 bp) were generated through sequencing.
2.4 Gene expression detection
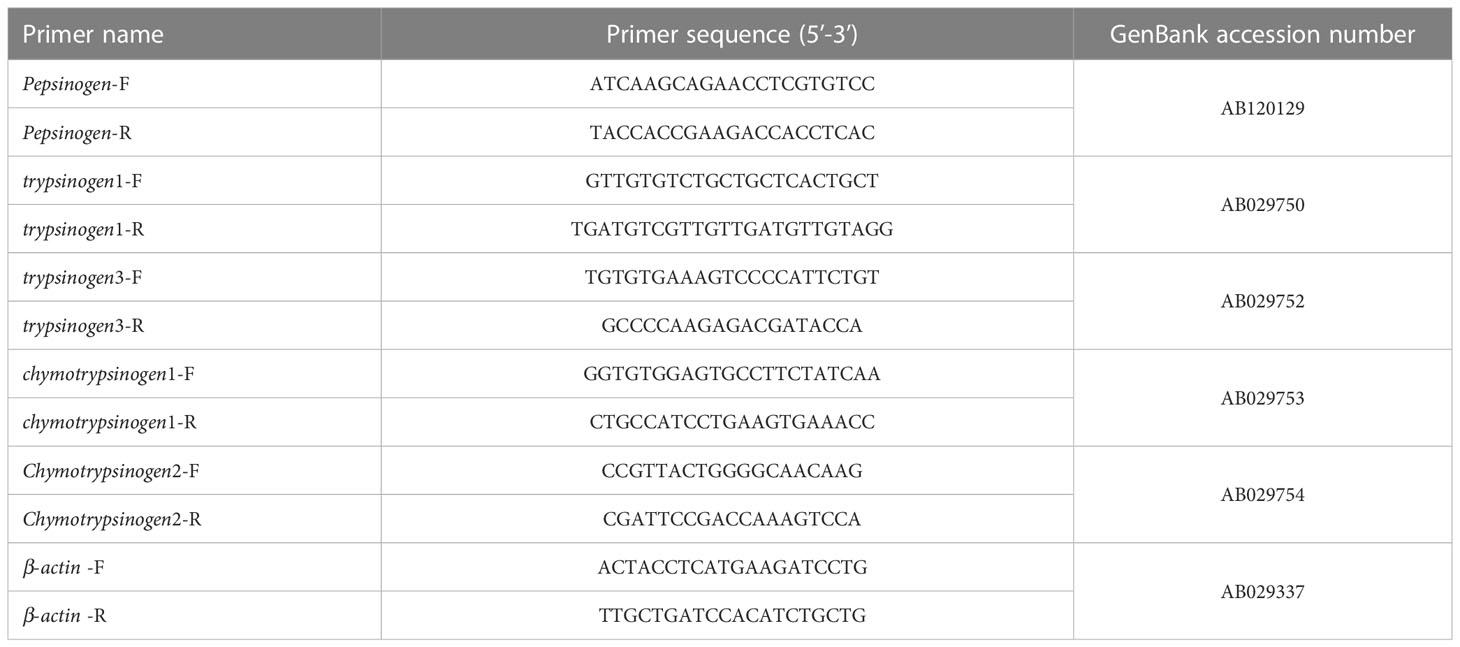
The total RNA of the gastrointestinal tissues of the final three individuals was extracted using the RNAiso Plus reagent (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The purity and concentration of the RNA were assessed by the NanoDrop, and the integrity of the RNA was checked by 1% agarose gel electrophoresis. The first strand of cDNA was synthesized using the PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time) (Takara, Dalian, China) according to the manufacturer’s instructions. The real-time quantitative polymerase chain reaction (qRT-PCR) method was used to determine the gene expression levels of pond-cultured Japanese flounder gastrointestinal tracts during nutrient metabolism. Primers of the pepsinogen, trypsinogen1, trypsinogen3, chymotrypsinogen1, chymotrypsinogen2, and the housekeeping gene (β-actin) (Kim et al., 2004) for the qRT-PCR assays are listed in Table 1. The qRT-PCR was performed with SYBR Green Real-time PCR Master Mix (TaKaRa, Dalian, China) using the Mastercycler® ep realplex Real-time PCR System (Eppendorf, Hamburg, Germany). The melting curve was also plotted to confirm the presence of a single PCR product.
2.5 Statistical analysis
The protease and the metabolic enzyme activities were calculated according to the standard curve method. The target gene expression level was normalized to the level of β-actin and quantified using the 2-ΔΔCT method.
The paired-end reads were then preprocessed using Trimmomatic software (Bolger et al., 2014) to detect and cut off ambiguous bases (N). We also filtered out low-quality sequences with an average quality score below 20 using the sliding window trimming approach. After trimming, the paired-end reads were assembled using FLASH software (Reyon et al., 2012). The parameters of the assembly were: 10 bp of minimal overlapping, 200 bp of maximum overlapping, and a 20% maximum mismatch rate. The sequences underwent further denoising as follows: Reads with ambiguous, homologous sequences or those below 200 bp in length were abandoned, reads with 75% of the bases above Q20 were retained, and reads with chimera were detected and removed. These steps were achieved using QIIME software (Caporaso et al., 2010) (version 1.8.0). After these treatments, effective tags were acquired from all samples. Several effective tags were clustered into operational taxonomic units (OTUs) when their shared sequencing identity was higher than 97% using Vsearch (v 2.4.2) (Rognes et al., 2016). Representative sequences of each OTU were selected and annotated with species using the RDP Classifier (v 2.2) (Edgar, 2013) and Silva database (v 123) (Quast et al., 2013), where the threshold was higher than 0.7. To control for differences in the sampling effect, we rarefied each sample to 22,951 sequences (the minimum sequence amount among all samples) per sample before calculating the diversity indices. The relative abundance and alpha and beta diversity analyses were performed based on this homogenization process. A Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted using Tax4fun (0.3.1).
The K-S test was used to analyze the normal distribution characteristic of the data. The one-way analysis of variance (ANOVA) was used to analyze the data with SPSS 17.0, and Duncan’s test was used to make multiple comparisons of the differences among the different sampling times in the gene expression and the changes in the serum indicators during the whole experiment. The Kruskal–Wallis H test was used to detect the differences in the values of gastrointestinal microbiota among the three sampling times. The significance level was set at p < 0.05, and the results are expressed as the mean ± SD.
3 Results
3.1 Characteristics of protease and serum metabolic enzyme activity
The pepsin activity of pond-cultured Japanese flounder increased first and then decreased during the whole experiment (Table 2). However, there was no significant difference in the stomach among the three sampling times (P > 0.05). Trends of trypsin activities in the pyloric caecum and gut were all first increasing and then decreasing. The trypsin activity in the pyloric caecum was significantly higher at 6 h than 12 h after ingestion (P < 0.05); however, there was no significant difference in activity in the gut among the three sampling times (P > 0.05). Chymotrypsin activity in the pyloric caecum and gut increased first and then decreased. Meantime, the chymotrypsin activity in the gut was significantly higher at 6 h after ingestion than at the other two sampling times (P < 0.05), however, there was no significant difference among the three sampling times in the pyloric caecum (P > 0.05).
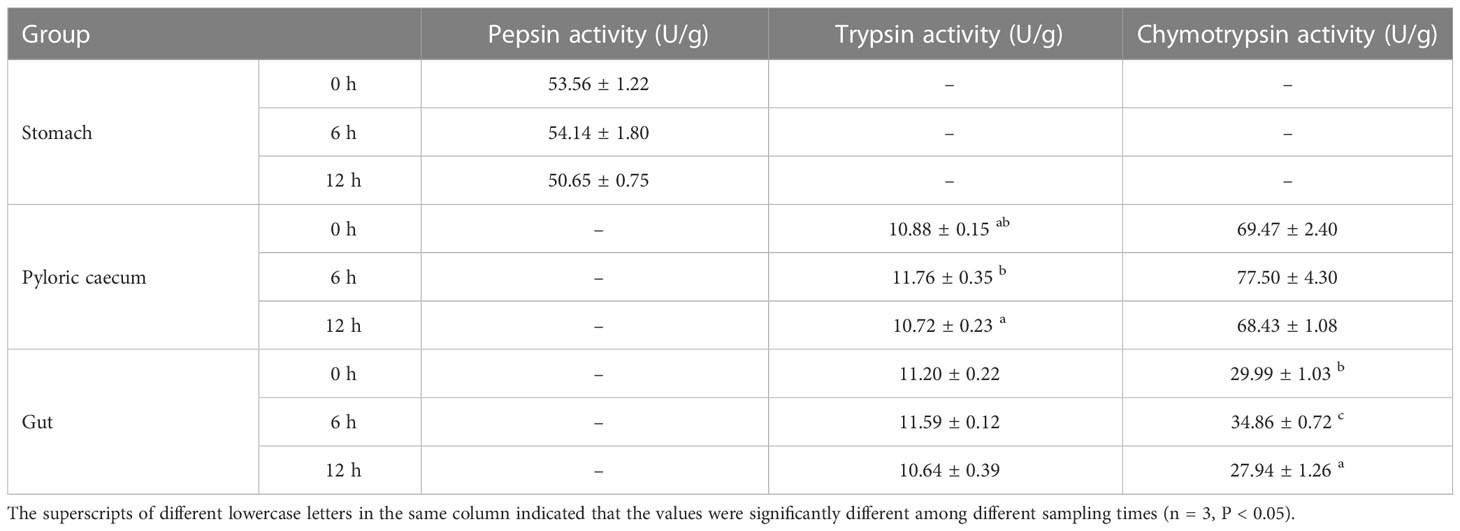
Table 2 Protease activities of each gastrointestinal tissue of pond-cultured Japanese flounder (Fresh weight).
The characteristics of serum ASP and ALA activity are shown in Table 3 during nutrient metabolism of pond-cultured Japanese flounder. The activity of these two metabolic enzymes decreased first and then increased with digestion and absorption, and the higher values were obtained at 0 h (95.00 U/L) and 12 h (16.23 U/L), respectively. Serum ASP and ALA activity were significantly lower at 6 h than at the other two sampling times (P < 0.05).
3.2 Protease gene expression pattern in gastrointestinal tissue
The expression characteristics of protease genes, including pepsinogen, trypsinogen1, trypsinogen3, chymotrypsinogen1, and chymotrypsinogen2, during nutrient metabolism in gastrointestinal tissue of each pond-cultured Japanese flounder, are presented in Figures 1A–I. The relative expression levels of pepsinogen in the stomach, and trypsinogen1, trypsinogen3, chymotrypsinogen1, and chymotrypsinogen2 in the pyloric caecum and gut increased first and then decreased, and the values were significantly higher at 6 h than at the other two sampling times (P < 0.05).
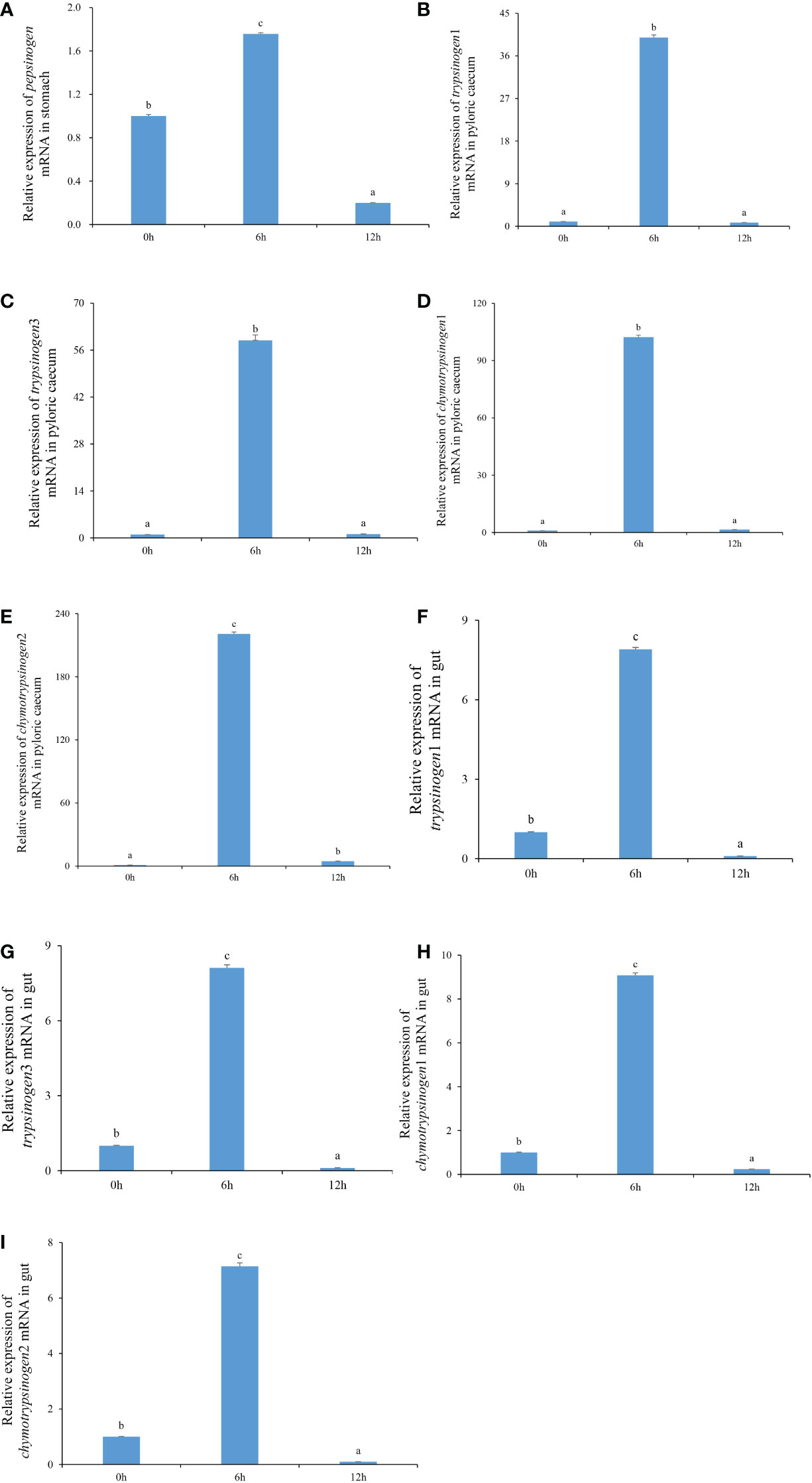
Figure 1 The expression levels of pepsinogen, trypsinogen1, trypsinogen3, chymotrypsinogen1, and chymotrypsinogen2 in the gastrointestinal tissues of pond-cultured Japanese flounder. (A) pepsinogen in the stomach; (B) trypsinogen1 in the pyloric caecum; (C) trypsinogen3 in the pyloric caecum; (D) chymotrypsinogen1 in the pyloric caecum; (E) chymotrypsinogen2 in the pyloric caecum; (F) trypsinogen1 in the gut; (G) trypsinogen3 in the gut; (H) chymotrypsinogen1 in the gut; (I) chymotrypsinogen2 in the gut. Different lowercase letters represented that values were significantly different among different sampling times for each gene (n = 3, P < 0.05).
3.3 Changes to alpha diversity of gastrointestinal microbiota
During nutrient metabolism, the alpha diversity of microbiota in each gastrointestinal tissue of pond-cultured Japanese flounder is presented in Figure 2. The Chao 1 and Shannon indices of microbiota decreased first and then increased in the stomach, but the difference among the three sampling times was not significant (P > 0.05) (Figure 2A). The microbiota in the pyloric caecum showed gradually increasing trends in the Chao1 and Shannon indices during nutrient metabolism, and the Chao1 index was significantly higher at 12 h than at 0 h (P < 0.05) (Figure 2B). The changes in the Chao1 and Shannon indices of microbiota in the gut were the same as those in the pyloric caecum, however, there was no significant difference among the three sampling times (P > 0.05) (Figure 2C).
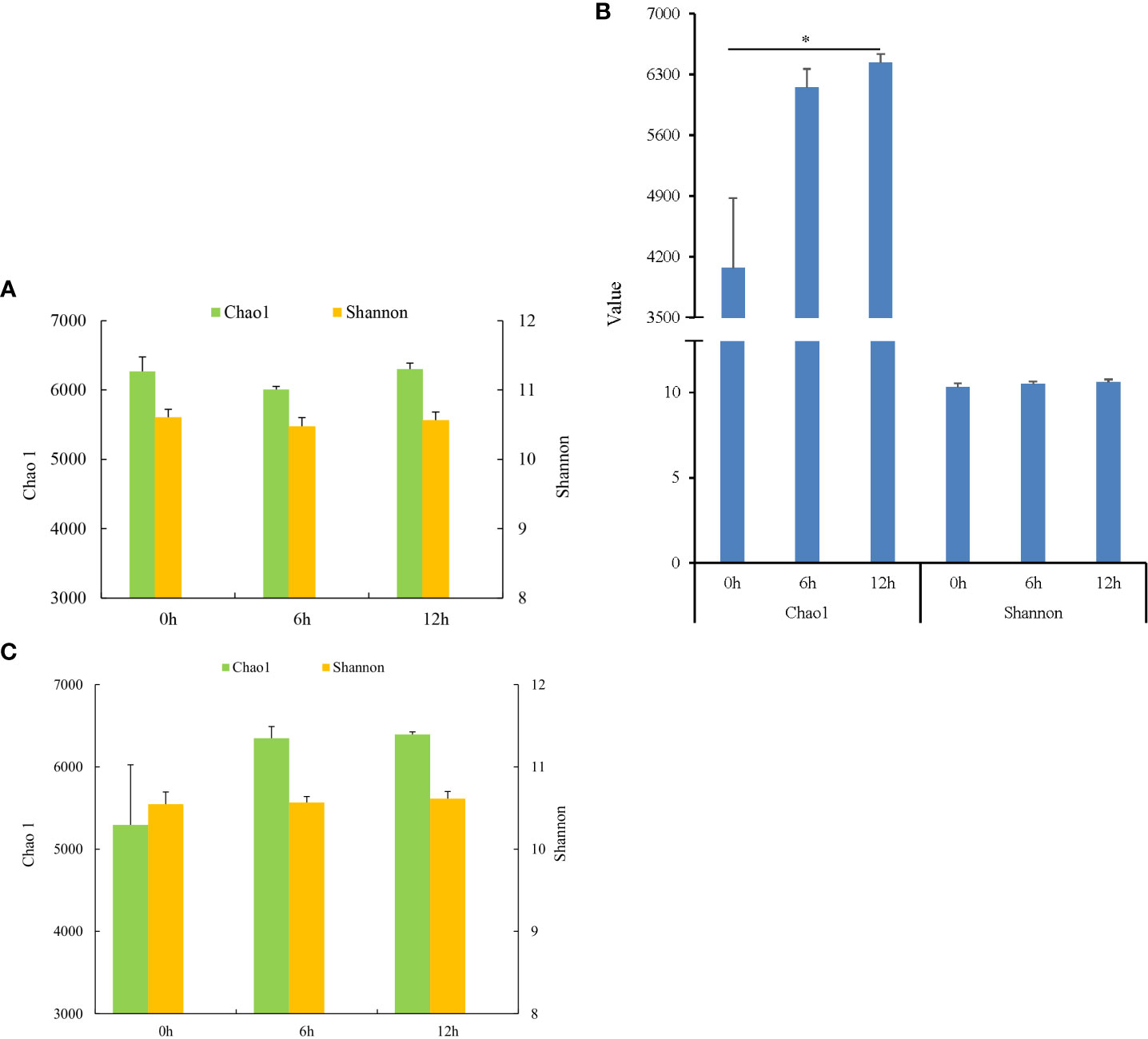
Figure 2 Alpha diversities of gastrointestinal microbiota of Japanese flounder. (A) stomach; (B) pyloric caecum; (C) gut. * represented that values were significantly different among different sampling times (n = 3, P < 0.05).
3.4 Succession of gastrointestinal microbiota structure
The top 10 phyla (according to the average relative abundances in the three parallel samples of each gastrointestinal tissue in each treatment) in each sample are presented in Figure 3A, which includes Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Gemmatimonadetes. The relative abundances of Proteobacteria, Firmicutes, and Bacteroidetes were all higher than 20%. Meantime, the ratio of Firmicutes to Bacteroidetes increased first and then decreased at each sampling time along the gastrointestinal tract. The highest ratio was shown in the pyloric caecum (Figure 3B). In the same gastrointestinal tissue, the ratio of Firmicutes to Bacteroidetes increased first and then decreased with the progress of nutrient metabolism.
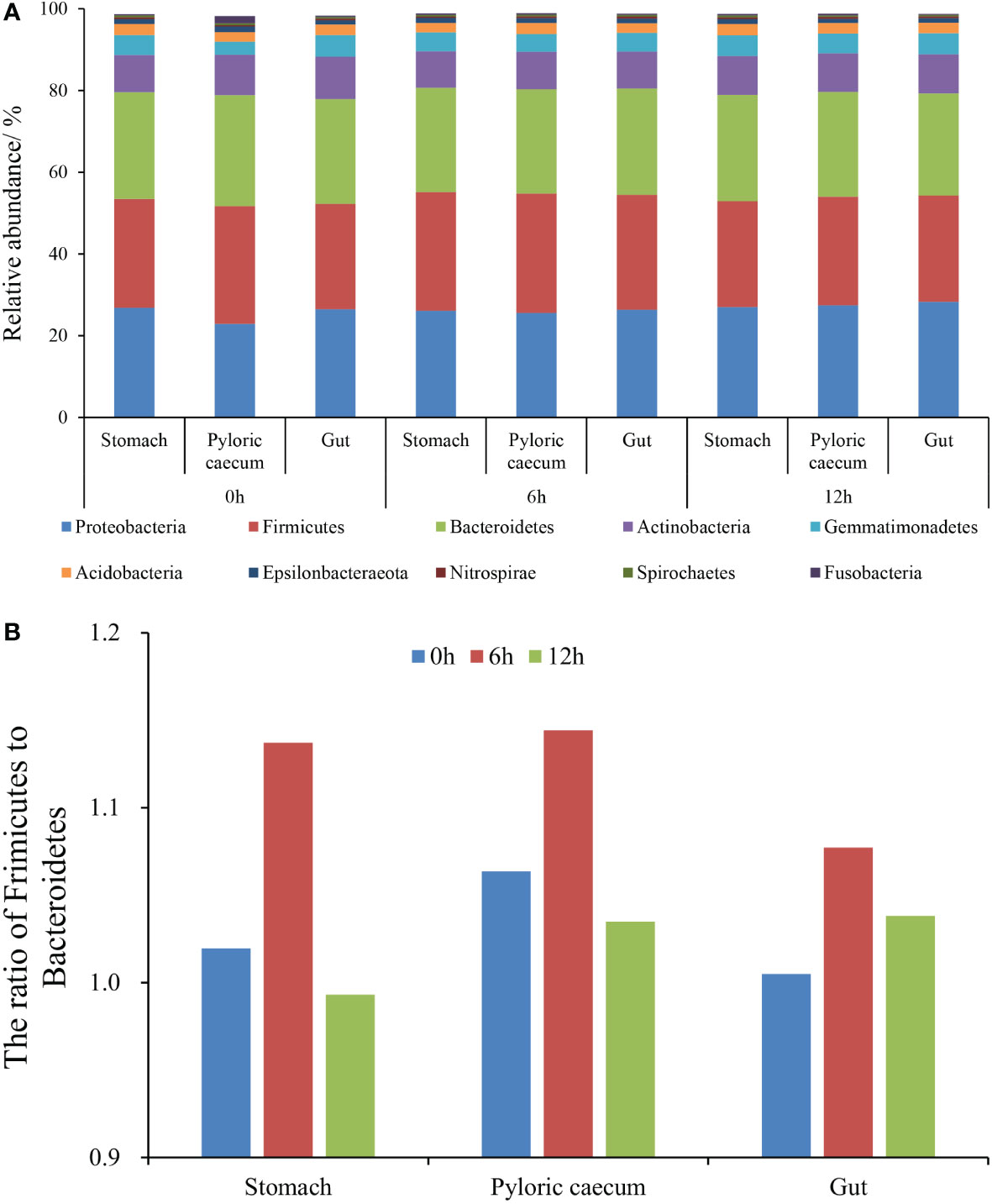
Figure 3 The structural characteristics of gastrointestinal microbiota at the phylum level. (A) The top-ten phyla with relative abundance; (B) The ratio of Firmicutes to Bacteroidetes.
The genus-level compositions of the top 15 microbiota, according to the average relative abundance of each sample, are shown in Figure 4. Lactobacillus, the genera represented by Lachnospiraceae_NK4A136_group, Acinetobacter, Bacteroides, Escherichia-Shigella, Sphingomonas, and Prevotella were the dominant and shared genera in the three gastrointestinal tissues during nutrient metabolism of the pond-cultured Japanese flounder. These microbiota were also the colonizing microbiota in the gastrointestinal tract of pond-cultured Japanese flounder. In the stomach, the relative abundances of Lactobacillus, the genera represented by Lachnospiraceae_NK4A136_group, Acinetobacter, Romboutsia, Helicobacter, and Escherichia-Shigella increased first and then decreased (Figure 4A). The relative abundances of Bacteroides, Alistipes, Sphingomonas, Myroides, Pseudomonas, and Haliangium presented a gradually decreasing trend, while the relative abundance of the other dominant genera gradually increased in the stomach. In the pyloric caecum, relative abundances of Lactobacillus, the genera represented by Lachnospiraceae_NK4A136_group, Prevotella, Acinetobacter, Ruminiclostridium, Enterobacter, and Myroides increased first and then decreased (Figure 4B). The relative abundances of Bacteroides, Glycomyces, Fusobacterium, Alloprevotella, Escherichia-Shigella, and Helicobacter gradually decreased, while the relative abundances of the other dominant genera showed a gradually decreasing trend in the pyloric caecum. In the gut, the relative abundances of Bacteroides, Lactobacillus, the genera represented by Lachnospiraceae_NK4A136_group, Acinetobacter, Alistipes, Romboutsia, Sphingomonas, Ruminiclostridium, Helicobacter, Escherichia-Shigella, Pseudomonas, Enterobacter, and Myroides increased first and then decreased, while the relative abundances of Prevotella, Glycomyces, and Alloprevotella gradually decreased (Figure 4C). Apart from Glycomyces in the pyloric caecum, the relative abundances of dominant genera in each gastrointestinal tissue did not significantly differ among the three sampling times (P > 0.05).
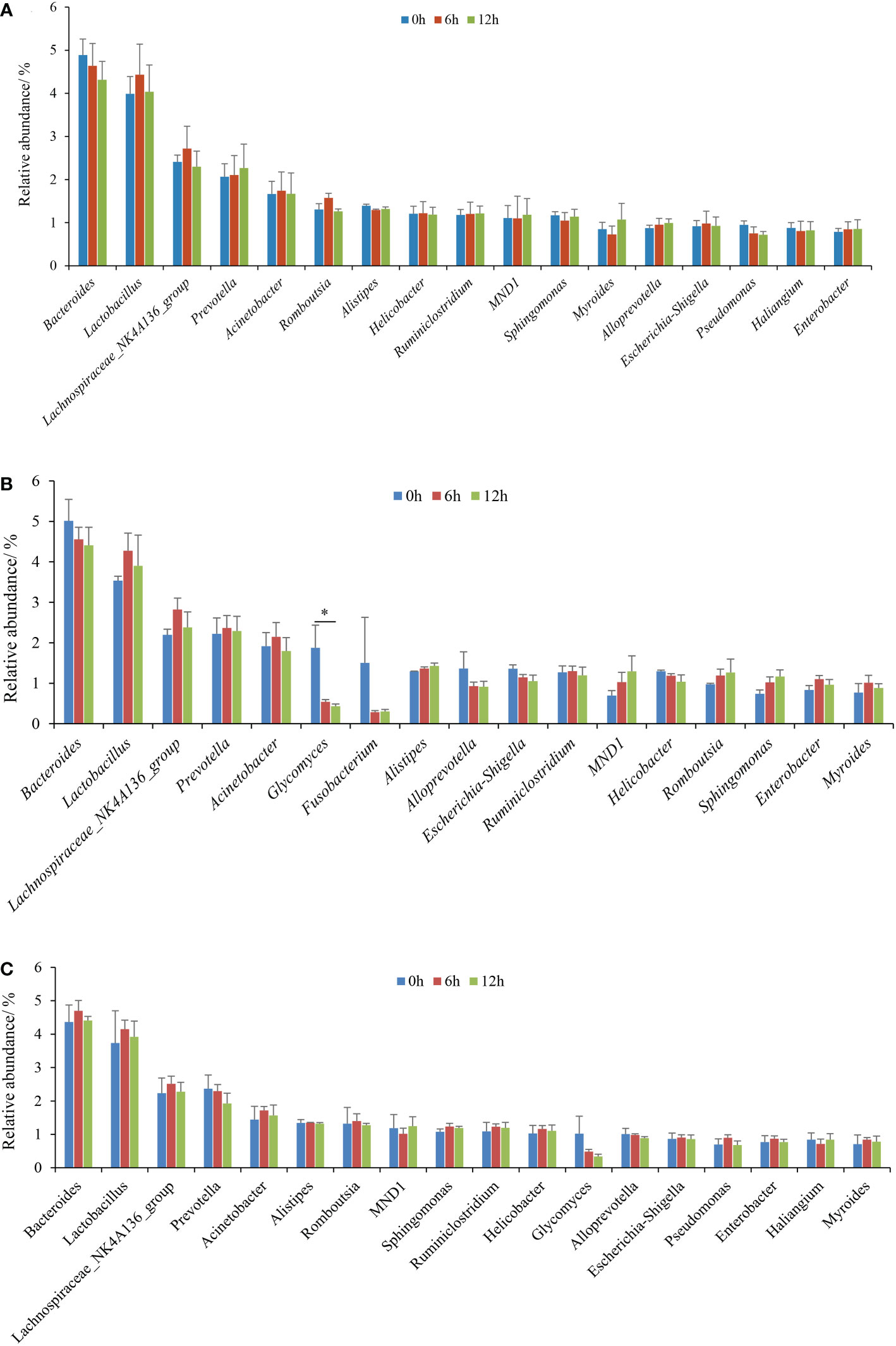
Figure 4 The dominant (top 15, according to the average relative abundances of the three parallel samples for each gastrointestinal tissue in each treatment) genera of gastrointestinal microbiota of pond-cultured Japanese flounder. (A) stomach; (B) pyloric caecum; (C) gut. * represented significant differences for the same genus among the different sampling times (p < 0.05).
3.5 Beta diversity analysis
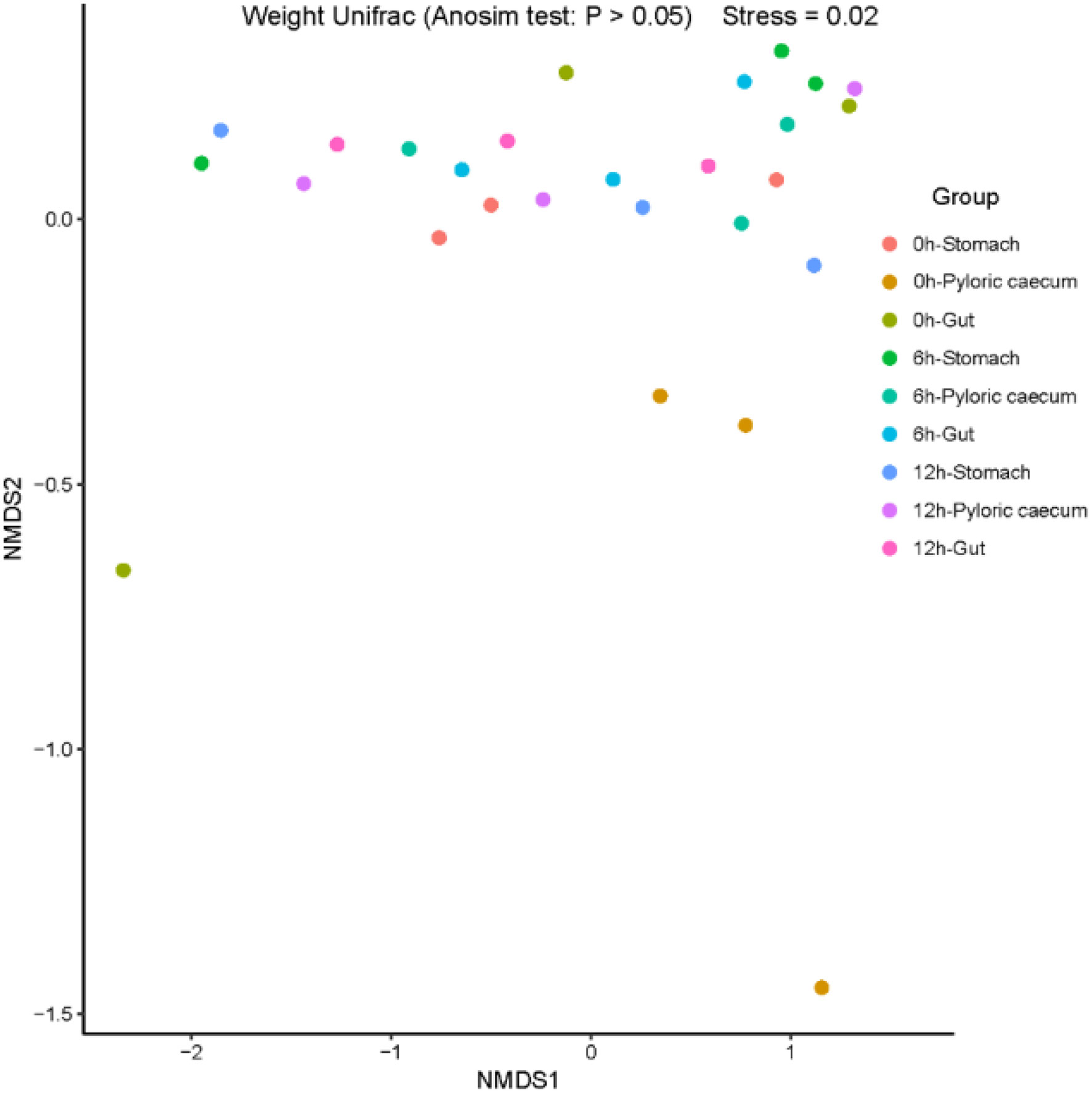
Nonmetric multidimensional scaling (NMDS) is a data analysis method that simplifies research objects (samples or variables) in a multi-dimensional space and low-dimensional space for positioning, analysis, and classification while retaining the original relationship between objects. ANOSIM analysis is a method for analyzing the overall differences among different samples. According to the ANOSIM analyses performed in the study, there were no significant differences in the microbiota structure among the samples (p > 0.05) (Figure 5), which indicated that the diversity of the gastrointestinal microbiota was not significantly correlated with digestion.
The number of genera in each sample was obtained on a common basis among the three replicates for each sample (Figure 6). During nutrient metabolism, the number of genera in each gastrointestinal tissue showed an increasing trend. After ingestion, the number of unique genera in the pyloric caecum obviously decreased, and the number of unique genera in the other samples tended to show a gradual increasing trend.
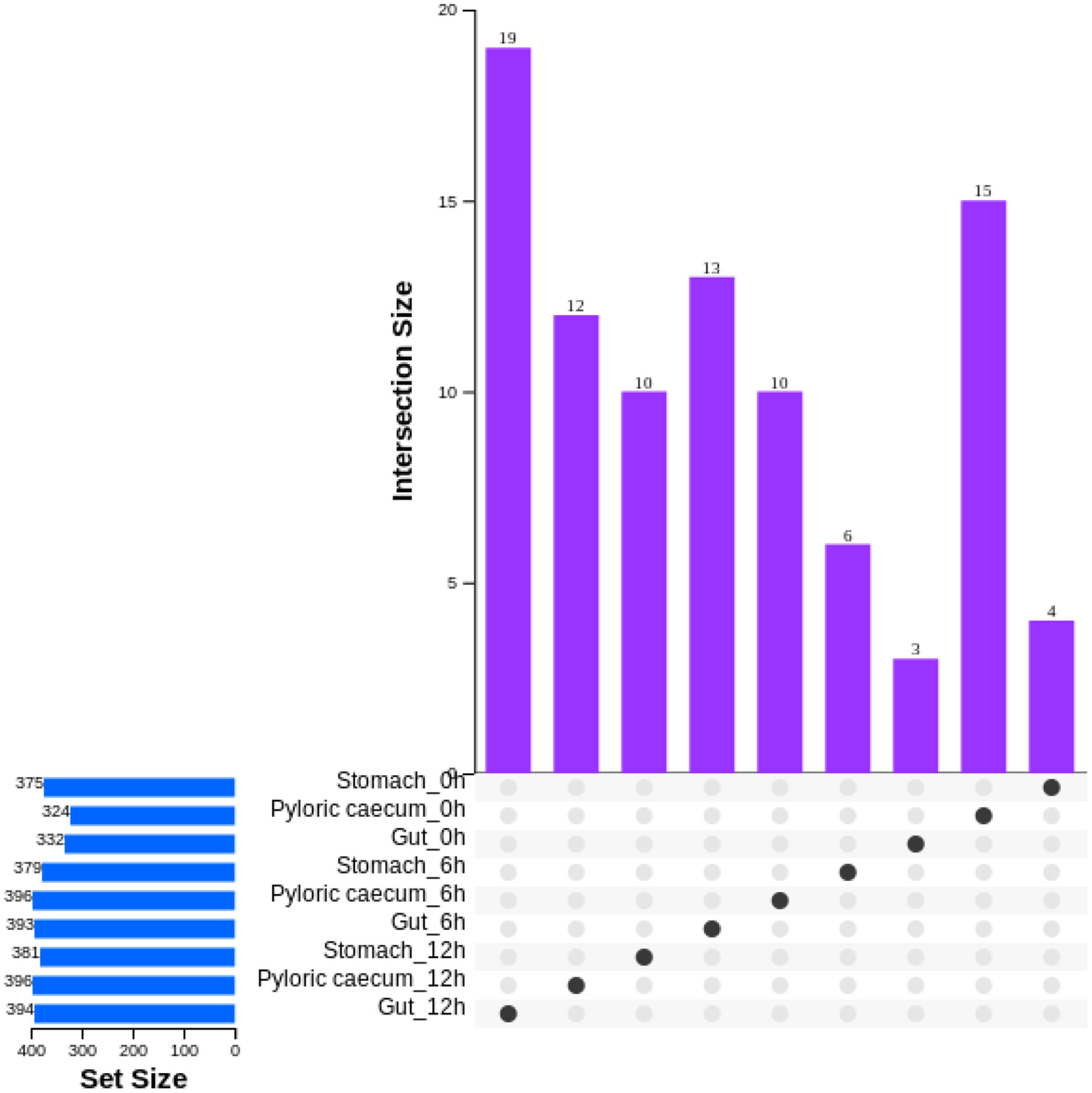
Figure 6 The characteristic of genera numbers in each gastrointestinal tissue of pond-cultured Japanese flounder.
3.6 Functional characteristics of gastrointestinal microbiota
According to the relative abundance of dominant KEGG pathways (an average relative abundance higher than 1%) in each gastrointestinal tissue at level 1, the metabolism pathway, genetic information processing, environmental information processing, and cellular processes were ranked according to the relative abundance (Figure 7). The relative abundance of cellular processes in the stomach and environmental information processing and genetic information processing in the pyloric caecum gradually decreased during nutrient metabolism. The relative abundance of environmental information processing and genetic information processing in the stomach and gut increased first and then decreased during nutrient metabolism. However, there was no significant difference among the different sampling times in each gastrointestinal tissue for each dominant KEEG pathway (P > 0.05).
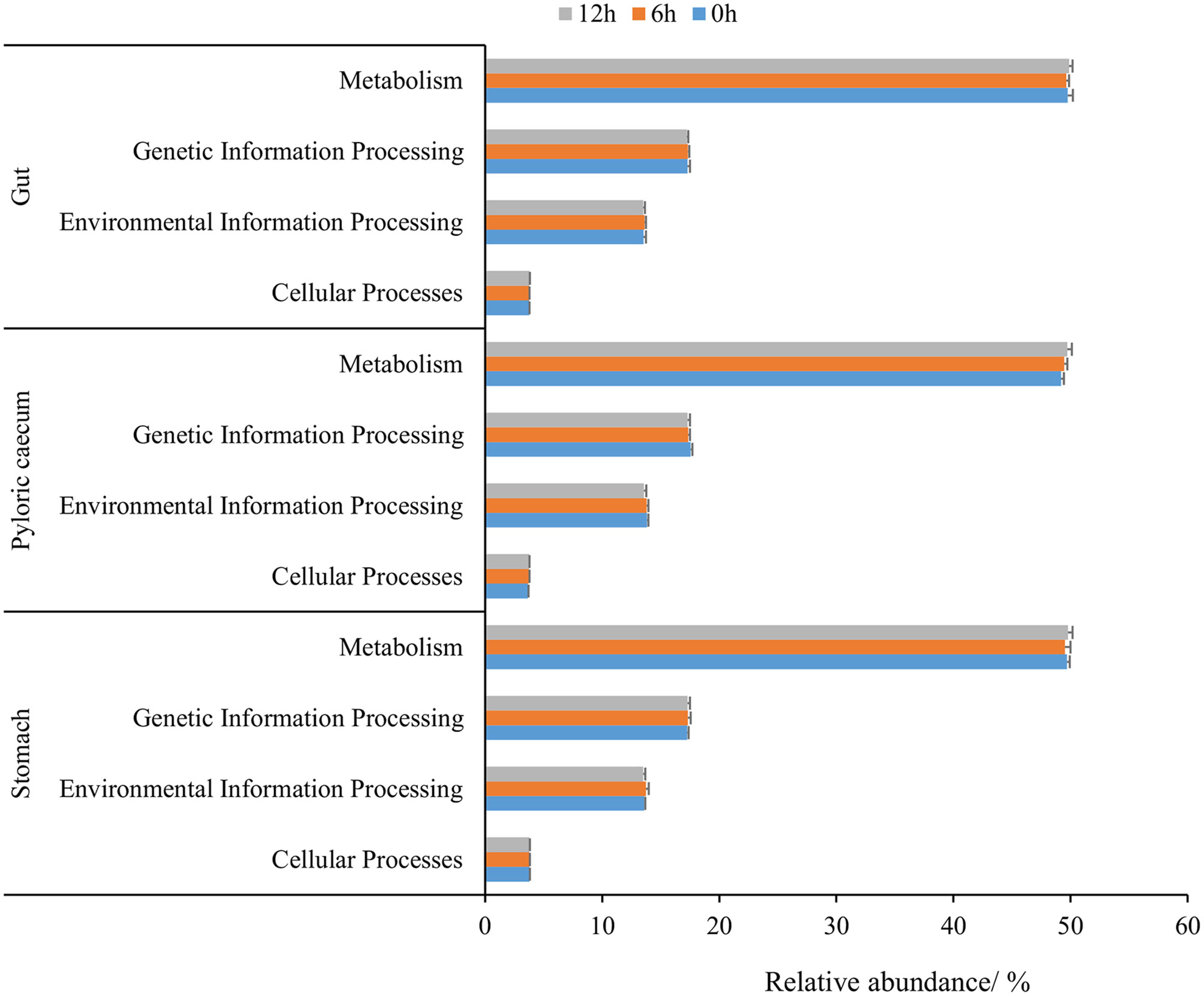
Figure 7 The dominant KEGG pathway of gastrointestinal microbiota of pond-cultured Japanese flounder.
4 Discussion
4.1 Digestion and metabolism of protein
Proteases, including pepsin, trypsin, and chymotrypsin, are key enzymes for the digestion of protein nutrients in animals. The synthesis of these proteases is mainly regulated by related genes. In this study, all protease activities increased first and then decreased during the nutrient metabolism of pond-cultured Japanese flounder. However, there were no significant differences in pepsin activity, trypsin activity in the gut, or chymotrypsin activity in the pyloric caecum among the different sampling times. Moreover, the expression levels of pepsinogen, trypsinogen1, trypsinogen3, chymotrypsinogen1, and chymotrypsinogen2 all significantly changed during nutrient metabolism. Although these genes can regulate the synthesis of pepsinogen, trypsinogen, and chymotrypsinogen, the activations are also related to the specific environment in the gastrointestinal tract. Some studies have reported that protease activity is affected by both intrinsic and extrinsic factors, such as the physiological stage and health status of the organism, nutrient level of the feed, temperature, and exogenous protease (Liu et al., 2019; Choi et al., 2020; Hassani et al., 2020; Santos et al., 2020a; Santos et al., 2020b).. The feed, living environment, species, and physiological stage among the Japanese flounder used in this study have no differences. Interestingly, after being ingested, the organism was stimulated to up-regulate the expression of protease-related genes, promoting the synthesis and secretion of protease, and leading to an increase in protease activity. With the gradual digestion of protein, the protein nutrient level of the gastrointestinal tract contents was gradually reduced, and the synthesis and secretion of protease were reduced, leading to the gradual decline of protease activity. The constant nutrient level fluctuations in the gastrointestinal tract meant that the responses of the organism also changed dynamically; therefore, there were dynamic changes in protease-related gene expression and protease activity. This might have been the key reason for the dynamic changes in protease activities and related gene expression of pond-culture Japanese flounder.
ASP and ALA are important amino acid transferases and play important roles in protein metabolism (de Sousa and Sodek, 2003; de la Torre et al., 2014). Under normal circumstances, ASP and ALA activities in serum are low, but when animals are subjected to malnutrition or stress, serum ASP and ALA activities in serum increase (Lemaire et al., 1991; Jia et al., 2021). In this study, the lowest ASP and ALA activities were recorded at 6 h after ingestion, which might be related to the abundant nutrition in the gastrointestinal tract, and ASP and ALA were mostly involved in protein metabolism. Karlsson et al. (2006) reported that free amino acids concentrations in the plasma of Oncorhynchus mykiss had the highest values at 6 h after ingestion, which also indicated that there is a stage of vigorous metabolism during the digestion of feed. This also revealed the importance of sample collection time in studies related to physiological characteristics. The study on the activity characteristics of digestive enzymes and metabolic enzymes will provide a reference for the development of a high-efficiency formula feed for large-size Japanese flounder.
4.2 Structural characteristics of gastrointestinal microbiota
Numerous studies have reported that feed composition, nutrient level, and additives could obviously change the structure and composition of the gut microbiota of cultured animals (Zhao et al., 2018; Bi et al., 2019; Hassani et al., 2020; Abou-Kassem et al., 2021). In this study, the pond-cultured Japanese flounder were fed twice a day according to 3-5% fish body weight and were basically satiated. Although the cultured fish could catch wild plankton and small fish, the proportion of these food sources was very small. Therefore, we did not consider their effect on the structure of the gastrointestinal microbiota. The feed of the pond-cultured Japanese flounder, the frozen A. personatus, had been washed several times with fresh water before being supplied to the fish to minimize the effects of microbiota carried by the frozen A. personatus on the gastrointestinal microbiota of the Japanese flounder. The protein nutrient digestion and metabolism data indicated that the changes in nutrient substrates in the gastrointestinal tissue led to changes in the structure and alpha diversity of the microbiota in this study. Although the differences in relative abundances of dominant genera were not significant, changes in relative abundances of part genera were more obvious, such as Bacteroides, Lactobacillus, the genera represented by Lachnospiraceae_NK4A136_group, Acinetobacter, Prevotella, and Romboutsia. It was reasoned that the changes in gastrointestinal microbiota were dynamic and complex and susceptible to change by the nutrient level in gastrointestinal tract contents. Based on the principles of dominance, commonality, and colonization, we classified Bacteroides, Lactobacillus, the genera represented by Lachnospiraceae_NK4A136_group and MND1, Acinetobacter, Escherichia-Shigella, Sphingomonas, Prevotella, Alistipes, Alloprevotella, Enterobacter, Helicobacter, Myroides, Ruminiclostridium, and Romboutsia as the main microbiota in the gastrointestinal tracts of pond-cultured Japanese flounder in this experiment.
Lactobacillus is generally the potential probiotic in aquaculture, and it is often provided in combination with Bifidobacterium in feed to promote the growth performance of the host (Hassani et al., 2020). Some strains belonging to Bacteroides could synthesize and secrete polysaccharide A, which has anti-inflammatory properties (Hiippala et al., 2020). Shao et al. (2020) provided evidence that Sphingomonas sp. PDD-57b-25 and Acinetobacter towneri isolated from wastewater had higher ammonia nitrogen, nitrite nitrogen, and total phosphorus assimilation capacities. The increased relative abundance of Prevotella in the human gut may improve glucose metabolism induced by dietary fiber (Kovatcheva-Datchary et al., 2015). Escherichia-Shigella may be detrimental to the response of patients with a heart valve replacement to anticoagulant therapy (Wang et al., 2020). However, no studies have been performed on the pathogenicity of Escherichia-Shigella in fish farming. Some strains belonging to Ruminiclostridium play an important role in cellulose degradation (Rettenmaier et al., 2021). Some strains belonging to Enterobacter, Alistipes, Alloprevotella, and Helicobacter might be associated with human clinical infection. However, the relative abundances of these main genera were not elevated, and there are no relative reports on these genera causing disease in aquaculture to date. Therefore, these aquaculture products can be safely and normally eaten.
A study reported that the microbes isolated from field-caught Drosophila melanogaster promote amino acid harvest to increase the lifespan of undernourished flies (Yamada et al., 2015). The gut microbiota can assist the host with the absorption of energy to maintain normal physiological activities, especially under extreme environmental conditions (Shabat et al., 2016; Zhang et al., 2016). The changes in the ratio of Firmicutes to Bacteroidetes, the representative of energy accumulation (Ley et al., 2006; Cox et al., 2015), was higher at 6 h after ingestion than the other sampling times in this study, indicating that the nutrient metabolism was strong and energy accumulation was fast at this time. This change was consistent with the change in the protease activity and the related gene expression, indicating that, with the change in nutrient protein level, there was a rapid change in the gastrointestinal microbiota structure that allowed the host to obtain as much energy as possible for physiological activities. These also reflected that the gastrointestinal microbiota cooperated with the host for protein nutrition metabolism. Moreover, most gastrointestinal microbiota mainly participated in the metabolism pathway to supply the energy for pond-cultured Japanese flounder, which might be related to the rapid growth and development of the fish. The relative abundances of dominant genera colonizing the gastrointestinal tract remained high during the protein metabolism of pond-cultured Japanese flounder, which indicated that the main colonizing genera were the same genera participating in the host’s physiological activities. It is possible that the same kind of fish ingesting the same feed for a certain time in the same ocean has led to a high similarity in gastrointestinal microbiota structures among fish. A study reported that short-term dietary changes can obviously affect the composition of gut microbiota (David et al., 2014). These all indicate that feed has an important influence on the structural characteristics of the gastrointestinal microbiota. The relevant study on the gastrointestinal microbiota will help in the screening of the native and functional probiotics for large-size Japanese flounder.
5 Conclusions
In summary, the changes in protease activities, related gene expression, and serum metabolic enzyme activities indicated that the protein metabolism of pond-cultured Japanese flounder was normal. The nutritional composition of the gastrointestinal tract contents underwent dynamic changes during this process, which led to the succession in the gastrointestinal microbiota structure. According to the principles of dominance, commonality, and colonization, Bacteroides, Lactobacillus, the genera represented by Lachnospiraceae_NK4A136_group and MND1, Acinetobacter, Escherichia-Shigella, Sphingomonas, Prevotella, Alistipes, Alloprevotella, Enterobacter, Helicobacter, Myroides, Ruminiclostridium, and Romboutsia were classified as the main microbiota. Based on the changes in the ratio of Firmicutes to Bacteroidetes and the KEGG pathway analysis of gastrointestinal microbiota, we conclude that the main colonizing microbiota seems to have major synergistic effects on the protein metabolism of pond-cultured Japanese flounder.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, accession number: PRJNA876070 https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA876070.
Ethics statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences.
Author contributions
YX: conceptualization, methodology, validation, writing—review and editing, funding acquisition; YJ: conceptualization, methodology, validation, formal analysis, investigation, writing—original draft preparation; JW: validation, investigation; LL: formal analysis, data curation; AC, BW, and KW: data curation; XL: supervision. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Natural Science Foundation of Shandong Province (ZR2020QC207), Fund of Key Laboratory of South China Sea Fishery Resources Exploitation & Utilization, Ministry of Agriculture and Rural Affairs, P. R. China (FREU2020-06), Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO. 20603022021011, 2020TD47), and China Agriculture Research System of MOF and MARA (CARS-47).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZM declared a shared affiliation with the authors JW and LL to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abou-Kassem D. E., Mahrose K. M., El-Samahy R. A., Shafi M. E., El-Saadony M. T., Abd El-Hack M. E., et al. (2021). Influences of dietary herbal blend and feed restriction on growth, carcass characteristics and gut microbiota of growing rabbits. Ital. J. Anim. Sci. 20, 896–910. doi: 10.1080/1828051X.2021.1926348
Banerjee G., Ray A. K. (2017). Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis 72, 1–11. doi: 10.1007/s13199-016-0441-8
Berger A., Schechter I. (1970). Mapping the active site of papain with the aid of peptide substrates and inhibitors. Philosophical transactions of the royal society of london. Ser. B Biol. Sci. 257, 249–264. doi: 10.1098/rstb.1970.0024
Bi Y., Cox M. S., Zhang F., Suen G., Zhang N., Tu Y., et al. (2019). Feeding modes shape the acquisition and structure of the initial gut microbiota in newborn lambs. Environ. Microbiol. 21, 2333–2346. doi: 10.1111/1462-2920.14614
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bröer S. (2008). Amino acid transport across mammalian intestinal and renal epithelia. Physiol. Rev. 88, 249–286. doi: 10.1152/physrev.00018.2006
Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009). The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, 233–238. doi: 10.1093/nar/gkn663
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chi L., Xu S., Xiao Z., Lin F., Ma D., Zhao C., et al. (2013). Pepsinogen a and c genes in turbot (Scophthalmus maximus): Characterization and expression in early development. Comp. Biochem. Phys. B 165, 58–65. doi: 10.1016/j.cbpb.2013.03.002
Choi D. G., He M., Fang H., Wang X. L., Li X. Q., Leng X. J. (2020). Replacement of fish meal with two fermented soybean meals in diets for rainbow trout (Oncorhynchus mykiss). Aquacult. Nutr. 26, 37–46. doi: 10.1111/anu.12965
Cox A. J., West N. P., Cripps A. W. (2015). Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endo. 3, 207–215. doi: 10.1016/S2213-8587(14)70134-2
David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
de la Torre F., Cañas R. A., Pascual M. B., Avila C., Cánovas F. M. (2014). Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J. Exp. Bot. 65, 5527–5534. doi: 10.1093/jxb/eru240
de Sousa C. A. F., Sodek L. (2003). Alanine metabolism and alanine aminotransferase activity in soybean (Glycine max) during hypoxia of the root system and subsequent return to normoxia. Environ. Exp. Bot. 50, 1–8. doi: 10.1016/S0098-8472(02)00108-9
Dwivedi M., Ansarullah, Radichev I., Kemp E. H. (2017). Alteration of immune-mechanisms by human microbiota and development and prevention of human diseases. J. Immunol. Res. 2017, 6985256. doi: 10.1155/2017/6985256
Edgar R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Flint H. J., Scott K. P., Duncan S. H., Louis P., Forano E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306. doi: 10.4161/gmic.19897
Foltmann B. (1981). Gastric proteinases-structure, function, evolution and mechanism of action. Essays Biochem. 17, 52–84.
Hassani M. H. S., Jourdehi A. Y., Zelti A. H., Masouleh A. S., Lakani F. B. (2020). Effects of commercial superzist probiotic on growth performance and hematological and immune indices in fingerlings. Acipenser baerii. Aquacult. Int. 28, 377–387. doi: 10.1007/s10499-019-00468-1
Hiippala K., Kainulainen V., Suutarinen M., Heini T., Bowers J. R., Jasso-Selles D., et al. (2020). Isolation of anti-inflammatory and epithelium reinforcing Bacteroides and Parabacteroides spp. from a healthy fecal donor. Nutrients 12, 935. doi: 10.3390/nu12040935
Horlick J., Booth M. A., Tetu S. G. (2020). Alternative dietary protein and water temperature influence the skin and gut microbial communities of yellowtail kingfish (Seriola lalandi). PeerJ 8, e8705. doi: 10.7717/peerj.8705
Jia Y., Gao Y., Gao Y., Li W., Guan C. (2021). Growth performance, hematological and biochemical parameters, and hepatic antioxidant status of spotted knifejaw Oplegnathus punctatus in an offshore aquaculture net pen. Aquaculture 541, 736761. doi: 10.1016/j.aquaculture.2021.736761
Jiang Y., Liu X., Xu Y., Shi B., Wang B. (2020). Microbiota characteristics in Sebastes schlegelii intestine in early life stages. J. Oceanol. Limnol. 38, 275–287. doi: 10.1007/s00343-019-9011-2
Jiang Y., Wang Y., Zhang Z., Liao M., Li B., Rong X., et al. (2019). Responses of microbial community structure in turbot (Scophthalmus maximus) larval intestine to the regulation of probiotic introduced through live feed. PloS One 14, e0216590. doi: 10.1371/journal.pone.0216590
Karlsson A., Eliason E. J., Mydland L. T., Farrell A. P., Kiessling A. (2006). Postprandial changes in plasma free amino acid levels obtained simultaneously from the hepatic portal vein and the dorsal aorta in rainbow trout (Oncorhynchus mykiss). J. Exp. Boil. 209, 4885–4894. doi: 10.1242/jeb.02597
Kim D. H., Han H. J., Lee D. C., Park S. L. (2004). Bacterial enteritis and the development of the larval digestive tract in olive flounder, Paralichthys olivaceus (Temminck & schlegel). J. Fish Dis. 27, 497–505. doi: 10.1111/j.1365-2761.2004.00553.x
Korbova L., Kohout J. (1981). Gastric acid proteinases and their zymogens. Acta Univ. Carol. Med. Monogr. 101, 1–144.
Kovatcheva-Datchary P., Nilsson N., Akrami R., Lee Y. S., De Vadder F., Arora T., et al. (2015). Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 22, 971–982. doi: 10.1016/j.cmet.2015.10.001
Legrand T. P. R. A., Catalano S. R., Wos-Oxley M. L., Stephens F., Landos M., Bansemer M. S., et al. (2018). The inner workings of the outer surface: Skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02664
Lemaire P., Drai P., Mathieu A., Lemaire S., Carrière S., Giudicelli J., et al. (1991). Changes with different diets in plasma enzymes (GOT, GPT, LDH, ALP) and plasma lipids (cholesterol, triglycerides) of sea-bass (Dicentrarchus labrax). Aquaculture 93, 63–75. doi: 10.1016/0044-8486(91)90205-L
Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li X., Yu Y., Feng W., Yan Q., Gong Y. (2012). Host species as a strong determinant of the intestinal microbiota of fish larvae. J. Microbiol. 50, 29–37. doi: 10.1007/s12275-012-1340-1
Liu H. P., Wen B., Chen Z. Z., Gao J. Z., Liu Y., Zhang Y. C., et al. (2019). Effects of dietary vitamin c and vitamin e on the growth, antioxidant defence and digestive enzyme activities of juvenile discus fish (Symphysodon haraldi). Aquacult. Nutr. 25, 176–183. doi: 10.1111/anu.12841
Nazemroaya S., Nematollahi M. A., Yazdanparast R., Farahmand H., Rezaie A., Najafabadi M. Z. (2020). Pepsinogen expression during larval development of a Persian gulf sparid, sobaity. Aquaculture 523, 735131. doi: 10.1016/j.aquaculture.2020.735131
Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.12238
Psochiou E., Sarropoulou E., Mamuris Z., Moutou K. A. (2007). Sequence analysis and tissue expression pattern of Sparus aurata chymotrypsinogens and trypsinogen. Comp. Biochem. Phys. B 147, 367–377. doi: 10.1016/j.cbpb.2007.01.020
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rettenmaier R., Kowollik M. L., Klingl A., Liebl W., Zverlov V. (2021). Ruminiclostridium herbifermentans sp. nov., a mesophilic and moderately thermophilic cellulolytic and xylanolytic bacterium isolated from a lab-scale biogas fermenter fed with maize silage. Int. J. Syst. Evol. Micr. 71, 4692. doi: 10.1099/ijsem.0.004692
Reyon D., Tsai S. Q., Khayter C., Foden J. A., Sander J. D., Joung J. K. (2012). FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465. doi: 10.1038/nbt.2170
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Santigosa E., Sánchez J., Médale F., Kaushik S., Pérez-Sánchez J., Gallardo M. A. (2008). Modifications of digestive enzymes in trout (Oncorhynchus mykiss) and sea bream (Sparus aurata) in response to dietary fish meal replacement by plant protein sources. Aquaculture 282, 68–74. doi: 10.1016/j.aquaculture.2008.06.007
Santos W. M., Costa L. S., López-Olmeda J. F., Costa N. C. S., Santos F. A. C., Gamarano P. G., et al. (2020a). Effects of dietary protein levels on activities of protease and expression of ingestion and protein digestion-related genes in Nile tilapia juveniles. Aquac. Res. 51, 2973–2984. doi: 10.1111/are.14635
Santos W. M., Costa L. S., López-Olmeda J. F., Costa N. C. S., Santos F. A. C., Oliveira C. G., et al. (2020b). Dietary protein modulates digestive enzyme activities and gene expression in red tilapia juveniles. Animal 14, 1802–1810. doi: 10.1017/S1751731120000543
Shabat S. K. B., Sasson G., Doron-faigenboim A., Durman T., Yaacoby S., Miller M. E. B., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
Shao Y., Zhong H., Mao X., Zhang H. (2020). Biochar-immobilized Sphingomonas sp. and Acinetobacter sp. isolates to enhance nutrient removal: potential application in crab aquaculture. Aquacult. Env. Interac. 12, 251–262. doi: 10.3354/aei00364
Tu Y., Xie S., Han D., Yang Y., Jin J., Liu H., et al. (2015). Growth performance, digestive enzyme, transaminase and GH-IGF-I axis gene responsiveness to different dietary protein levels in broodstock allogenogynetic gibel carp (Carassius auratus gibelio) CAS III. Aquaculture 446, 290–297. doi: 10.1016/j.aquaculture.2015.05.003
Verri T., Barca A., Pisani P., Piccinni B., Storelli C., Romano A. (2017). Di-and tripeptide transport in vertebrates: The contribution of teleost fish models. J. Comp. Physiol. B 187, 395–462. doi: 10.1007/s00360-016-1044-7
Walburn J. W., Wemheuer B., Thomas T., Copeland E., O’Connor W., Booth M., et al. (2019). Diet and diet-associated bacteria shape early microbiome development in yellowtail kingfish (Seriola lalandi). Microb. Biotechnol. 12, 275–288. doi: 10.1111/1751-7915.13323
Wang L., Liu L., Liu X., Xiang M., Zhou L., Huang C., et al. (2020). The gut microbes, Enterococcus and Escherichia-Shigella, affect the responses of heart valve replacement patients to the anticoagulant warfarin. Pharmacol. Res. 159, 104979. doi: 10.1016/j.phrs.2020.104979
Wei Y., Liang M., Xu H. (2020). Fish protein hydrolysate affected amino acid absorption and related gene expressions of IGF-1/AKT pathways in turbot (Scophthalmus maximus). Aquacult. Nutr. 26, 145–155. doi: 10.1111/anu.12976
Yamada R., Deshpande S. A., Bruce K. D., Mak E. M., Ja W. W. (2015). Microbes promote amino acid harvest to rescue undernutrition in drosophila. Cell Rep. 10, 865–872. doi: 10.1016/j.celrep.2015.01.018
Zhang Z., Xu D., Wang L., Hao J., Wang J., Zhou X., et al. (2016). Convergent evolution of rumen microbiomes in high-altitude mammals. Curr. Biol. 26, 1873–1879. doi: 10.1016/j.cub.2016.05.012
Keywords: Paralichthys olivaceus, protein metabolism, protease activity, gene expression, gastrointestinal microbiota
Citation: Jiang Y, Wang J, Lin L, Xu Y, Cui A, Wang K, Liu X and Wang B (2023) Responses of the gastrointestinal microbiota to the protein metabolism of pond-cultured Japanese flounder (Paralichthys olivaceus). Front. Mar. Sci. 10:1033094. doi: 10.3389/fmars.2023.1033094
Received: 05 September 2022; Accepted: 10 February 2023;
Published: 01 March 2023.
Edited by:
Yun Li, Ocean University of China, ChinaReviewed by:
Omid Safari, Ferdowsi University of Mashhad, IranZhenhua Ma, South China Sea Fisheries Research Institute (CAFS), China
Quanqi Zhang, Ocean University of China, China
Copyright © 2023 Jiang, Wang, Lin, Xu, Cui, Wang, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongjiang Xu, eHV5akB5c2ZyaS5hYy5jbg==
†These authors have contributed equally to this work
 Yan Jiang
Yan Jiang Jun Wang
Jun Wang Lin Lin
Lin Lin Yongjiang Xu
Yongjiang Xu Aijun Cui1
Aijun Cui1 Bin Wang
Bin Wang