- 1State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
- 2Shenzhen Institute of Guangdong Ocean University, Shenzhen, China
Corals live in a symbiotic relationship with various bacteria that are fundamental to host fitness, health, and survival. Though the diversity of symbiotic bacteria has been revealed in the early life stages of some corals, the dynamic bacterial community profiles of one coral are still poorly characterized, particularly the stage-specific bacteria. In this study, the bacterial communities in the patent, eggs, and 4-day-old planula larvae of a hermaphrodite coral Dipsastraea favus were investigated by high-throughput sequencing of 16S ribosomal RNA gene. As a result, dynamic profiles of bacterial community in the parent, eggs and larvae of D. favus were suggested. The bacterial diversity in the planula larvae was a bit higher than that in the D. favus parent, and distinct stage-specific symbiotic bacteria were detected, e.g., Oceanospirillaceae, Kordia, and Legionellaceae in D. favus larvae, and Kiloniellales and Prosthecochloris in adult D. favus. The dynamic change of bacterial community in coral adults and larvae may expand our understanding of the complex relationship between coral host and its symbiotic microbiota.
Introduction
Coral holobionts harbor a complex consortium of microorganisms, including zooxanthellae, bacteria, archaea, fungi, viruses, and protists (Knowlton and Rohwer, 2003). Coral microbial flora has been attracting attention for their potentials to protect corals from environmental stresses and their relevance with coral hosts, such as defense against pathogens, promotion of coral health, development, nutrition-based symbioses, and environmental adaption (Krediet et al., 2013; Soffer et al., 2015; Lema et al., 2016; Yang et al., 2016; Zhou et al., 2017; Bernasconi et al., 2019; Babbin et al., 2021; Gavish et al., 2021; Kitamura et al., 2021). For example, the confirmation of anaerobic nitrogen metabolism in coral microorganisms sheds new light on coral and reef productivity (Babbin et al., 2021). Pollock et al. (2018) found evidence of coral–microbe phylosymbiosis, in which coral microbiome composition and richness reflect coral phylogeny.
The majority of coral microbiology studies focus on coral adults (Rohwer et al., 2002; Ceh et al., 2011). To date, the coral–bacteria symbiosis particularly the relationship between the coral host and its intracellular bacteria is just beginning to be acknowledged (Kitamura et al., 2021; Maire et al., 2021). Bernasconi et al. (2019) indicated that the uptake and structure of bacterial communities were developmental, supporting the idea that microbial communities are likely to play specific roles within the distinct life history stages of the coral host. The establishment of coral microbial community in the early developmental stages of corals is fundamental to coral fitness, health, and survival (Apprill et al., 2009; Lema et al., 2014; Lema et al., 2016). However, though the diversity of symbiotic bacteria has been revealed in early life stages of some corals (Sharp et al., 2010; Sharp et al., 2012; Lema et al., 2014; Zhou et al., 2017; Epstein et al., 2019), we still rarely know about the dynamic profiles of bacteria at the different development stages of one coral because it is difficult to get enough male and female gametes and larvae at the early development stage of corals. Thus, investigation on the dynamic community profiles of symbiotic bacteria at the different life stages of one coral is with great importance for our understanding of the possible roles of bacterial symbionts, particularly stage-specific bacteria, in the host’s development and coral–bacteria symbioses (Sharp et al., 2010; Lema et al., 2014).
Dipsastraea favus is a hermaphrodite, broadcast-spawning species coral belonging to the family Merulinidae (Shlesinger and Woesik, 2021), which is able to release gamete bundles with both sperms and eggs during its spawning period. In this study, with the seawater as control, the bacterial diversity in the coral D. favus parent, eggs and larvae was investigated, respectively, using high-throughput sequencing of 16S ribosomal RNA gene, with the aim to reveal the coral’s dynamic profiles of symbiotic bacteria and consequently find stage-specific bacteria for the future understanding of D. favus–microbe symbioses and utilization of D. favus symbiotic microbes.
Materials and methods
Coral spawning and larval cultivation
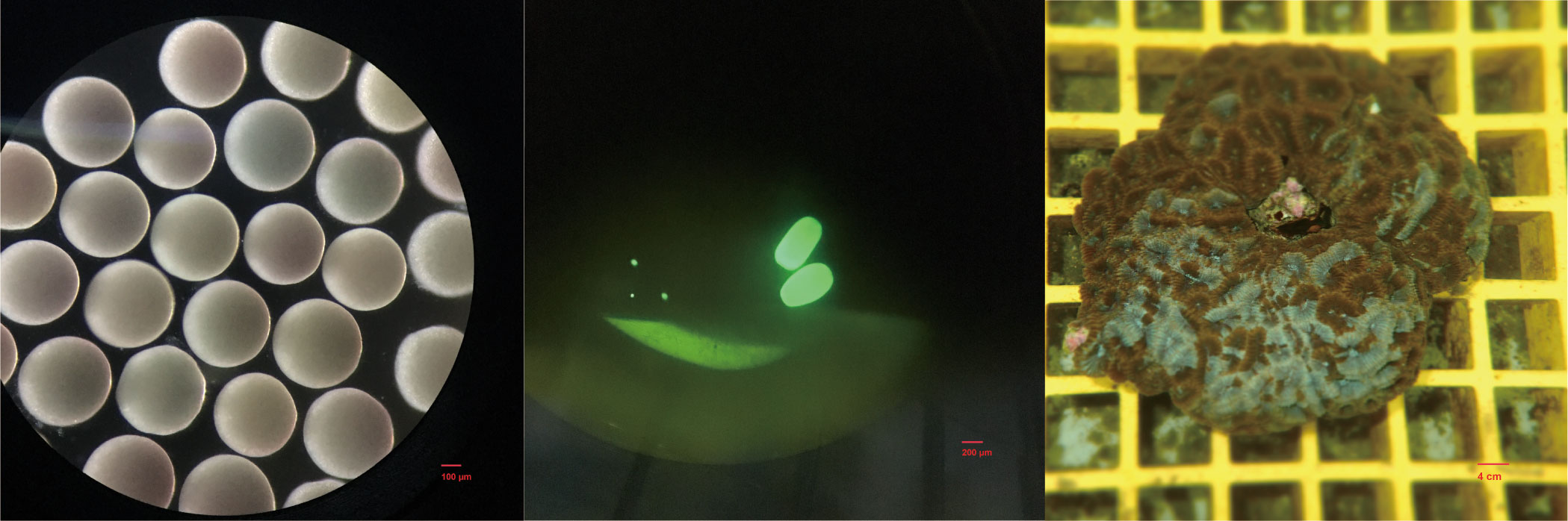
Eight colonies of hermaphrodite D. favus (ca. 30 × 40 × 5 cm3), 10 m away from each other, were collected at the Egong bay (114°29’E, 22°29’N), at a depth of 5 m, Shenzhen, China, approximately 1 month before spawning, and cultivated in pools with circulating natural seawater, and the temperature was kept at 26°C. The spawning occurred on 9 June 2017. The gamete bundles from each of the eight coral parents were captured and transferred to separate experimental tanks. After floating to the surface, the gamete bundles ruptured and released eggs and sperm. The eggs were collected and fixed immediately after being washed by sterile seawater. The seawater in tanks was not changed after spawning to avoid loss of sperm and kept at 26°C. Approximately 4 days after fertilization, most larvae tended to swim close to the substrate and searched for settlement locations, indicating that they were ready to settle and metamorphose soon; thus, we collected the 4-day-old planula larvae before they became settled juveniles. Because of the number limitation of eggs, no juveniles were collected in this study. The coral eggs, 4-day-old larvae, and parent fragments (Figure 1) were washed by sterile seawater several times to remove the surface microbes and fixed in RNALater. In order to provide enough biomass for successful PCR amplification, at least 50 eggs and 20 individuals of 4-day-old larvae were collected as one replicate for each coral parent. Approximately 2,000 ml of seawater in tanks was filtered by a 0.22-μm filter as one replicate. All samples with three duplicates were frozen in −80°C for further processing.
DNA extraction and high-throughput sequencing
For coral adult DNA extraction, fragments of approximately 1 cm2 including mucus, tissue, and skeleton were ground in liquid nitrogen by mortar and pestle. Proteinase (20 ml) was added to egg using Qiagen DNeasy Plant Mini Kit (cat. no. 69104), following the manufacturer’s instructions. The extracted DNA was preserved at −20°C. DNA samples, including eggs, larvae, adult clones, and tank seawater (3 replications for each), were PCR-amplified using the following primers: forward primer (CCTACGGRRBGCASCAGKVRVGAAT) and reverse primer (GGACTACNVGGGTWTCTAATCC), which were designed by GENEWIZ (Suzhou, China), targeting the hypervariable regions V3–V4 of 16S ribosomal RNA gene for bacteria. The PCR solution contained 2.5 μl of TranStart Buffer, 2 μl of dNTPs, 0.5 μl of TransStart Taq DNA polymerase, 20 ng of total DNA, 1 μl of each primer, and ddH2O in a final volume of 25 μl. The PCR amplification reaction was performed with the following procedure: initial denaturing at 94°C for 3 min; 35 cycles at 94°C for 5 s, 57°C for 1.5 min, and 72°C for 10 s; final extension at 72°C for 5 min. The library quality of PCR products was validated using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The library concentration was quantified by Qubit2.0 Fluorometer (Invitrogen, Carlsbad, CA). All the amplicon products were sequenced on an Illumina Miseq platform according to the manufacturer’s instructions, using 2X300 bp mode at GENEWIZ (Suzhou, China).
Sequence analysis
Analysis of the 16S rDNA sequences was conducted using the bioinformatics package QIIME 1.6 (Caporaso et al., 2010). A total of 74,9631 raw reads were generated from the nine samples (three replications for coral parent, eggs, and larvae, respectively). The sequences containing the adapter were discarded by Trimmomatic (Bolger et al., 2014). The paired-end sequence was merged using Pandaseq (Masella et al., 2012) with the parameter (minimum sequence length of 430 bp and maximum length of 470 bp). The chimera sequences detected by Usearch 61 were abandoned. The clean reads were clustered using UCLUST. Operational taxonomic units (OTUs) were selected at 97% identity by pick_open_reference_otus.py with the greengenes database (DeSantis et al., 2006). The OTUs annotated as chloroplast, mitochondria, and unassigned were discarded. After quality control, an average of 27,839 sequences were selected from each sample, ranging from 11,299 to 46,463. For a fair comparative analysis, samples were subsampled to an equal depth of 11,229 and were conducted by core_diversity_analyses.py. Summarized taxonomic abundance at the phylum, class, order, family, and genus levels was analyzed by QIIME.
Statistical analysis
Alpha diversity statistics including OTU, Chao1, and Shannon index were calculated in QIIME by alpha_diversity.py. The differences among groups were analyzed by one-way ANOVA with Tukey’s HSD test. OTU count data were square-root-transformed. Bray–Curtis distance matrix was built to examine the patterns of community structure and was visualized using non-metric multidimensional scaling (NMDS) analysis. Pearson correlation vectors were overlaid to demonstrate which taxon has strong correlation with group difference. Multidimensional statistical analysis was performed in PRIMER v7 with the PERMANOVA+ add-on (PRIMER-e, Quest Research Limited, Auckland, NZ).
Results and discussion
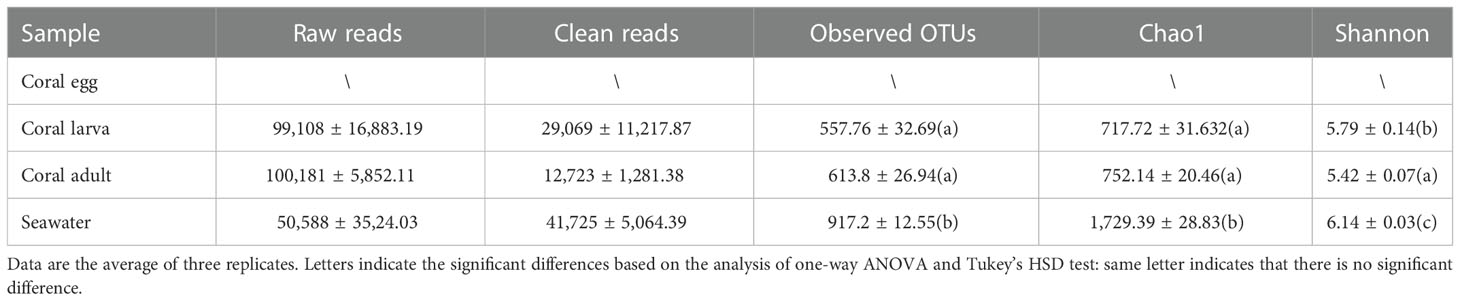
A total of 250,555 high-quality 16S rRNA gene sequences (clean reads in Table 1) were obtained from all of the nine larva and adult samples of D. favus and seawater sample (three replications each). The rarefaction curves showed that the diversity reached a plateau (Figure S1). It was a pity that no 16S rRNA gene sequences were retrieved from the egg samples by egg DNA sequencing. The number of 16S rRNA gene reads, number of OTUs, and alpha diversity (Shannon and Chao) estimate were calculated with OTUs at 3% dissimilarity (Table 1). The alpha diversity of the bacterial community of D. favus larvae is a bit higher than that of adult D. favus based on the Shannon index, and the bacterial diversity shows statistical difference among the coral parent, larvae, and seawater samples (Table 1). The main shared bacterial OTUs among the three samples belong to the class of Rhodobacteraceae and Alteromonadaceae (Table S1). The shared OTUs in the coral parent and its larvae mainly belong to Bacteriodaceae and Flavobacteriales, despite the fact that these bacteria were not detected in the seawater (Table S2). In the case of seawater control, its bacterial community is composed of Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes (Figure 2).
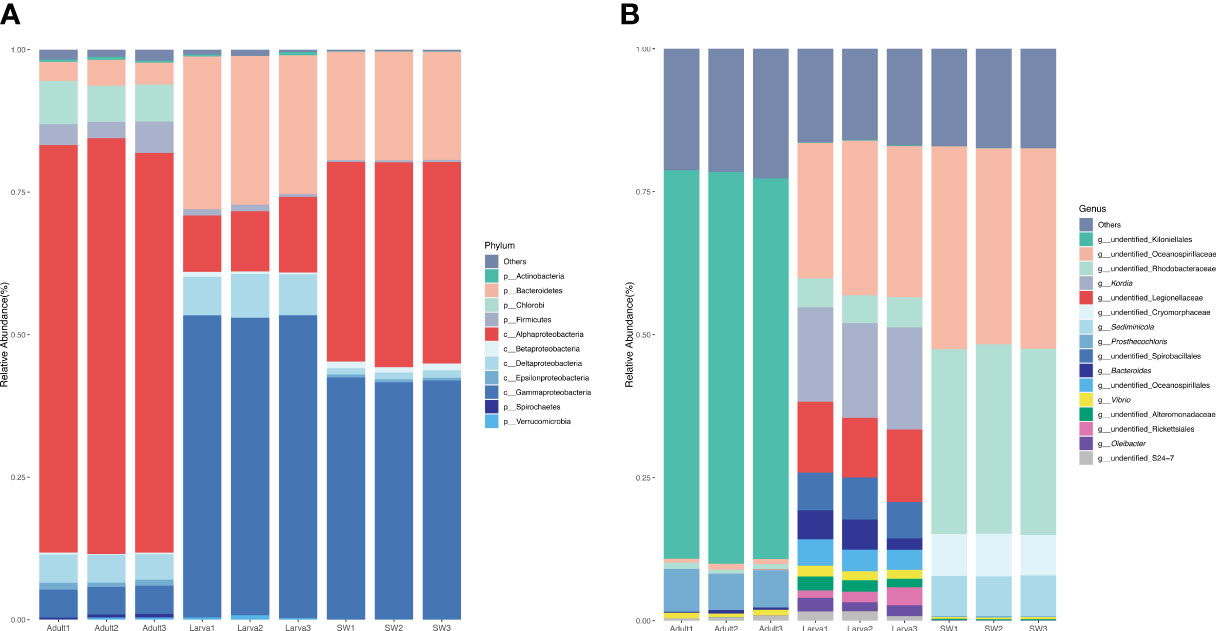
Figure 2 Percentage of the microbes in different samples at phylum (A) and genus (B) levels. A sequence number of less than 0.5% of the total sequence in the sample is classified as other.
The dominant 16S rRNA sequences from 4-day-old coral larvae are Gammaproteobacteria and Bacteroidetes (Figure 2). In contrast, Alphaproteobacteria dominates the 16S rRNA gene sequences recovered from adult F. favus, representing ~71% of the total sequences (Figure 2). The most abundant OTUs in the larvae are unidentified Oceanospirillaceae (OTU2) and Kordia sp. (OTU5), while the unidentified Kiloniellales (OTU1) and Prosthecochloris sp. (OTU10) are abundant in the adults (Figure 3). Based on non-metric multi-dimensional scaling analysis and permutational multivariate analysis of variance (PERMANOVA), we found obvious variations in the bacterial community composition among the D. favus adults, larvae, and seawater (PERMANOVA: p = 0.03) (Figure 4), e.g., Oceanospirillaceae, Kordia, and Legionellaceae in D. favus larvae, Kiloniellales and Prosthecochloris in adult D. favus, while Oceanospirillaceae, Rhodobacteraceae, and Sediminicole in seawater. Clearly, Figure 4 shows that the D. favus adults and larvae host different predominant bacteria. Similarly, significant stage-specific changes were detected in bacterial profiles during the development of scleractinian coral Acropora gemmifera (Zhou et al., 2017). Lema et al. (2014) suggested dynamic overall bacterial communities associated with the coral Acropora millepora at early life stages (larva, 2-week-old juveniles, and 3-, 6-, 12-month-old juveniles). Damjanovic et al. (2020) also suggested that Pocillopora acuta recruits harbored dynamic and diverse bacterial assemblages.
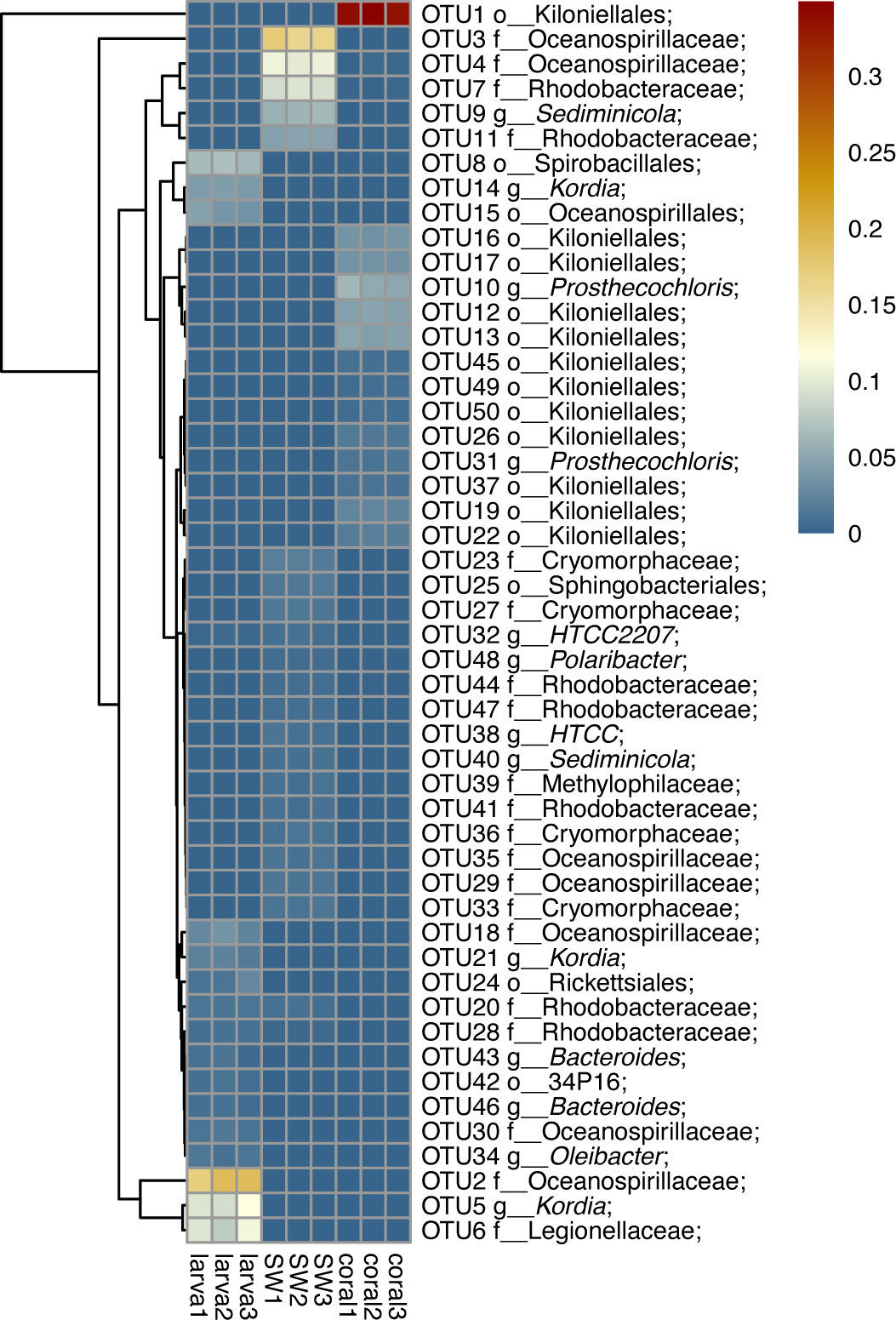
Figure 3 Heat map reflecting the relative abundance and distributions of the top 20 OTUs. The scales 0.01 and 0.1 show the abundance at 5% and 10%, respectively.
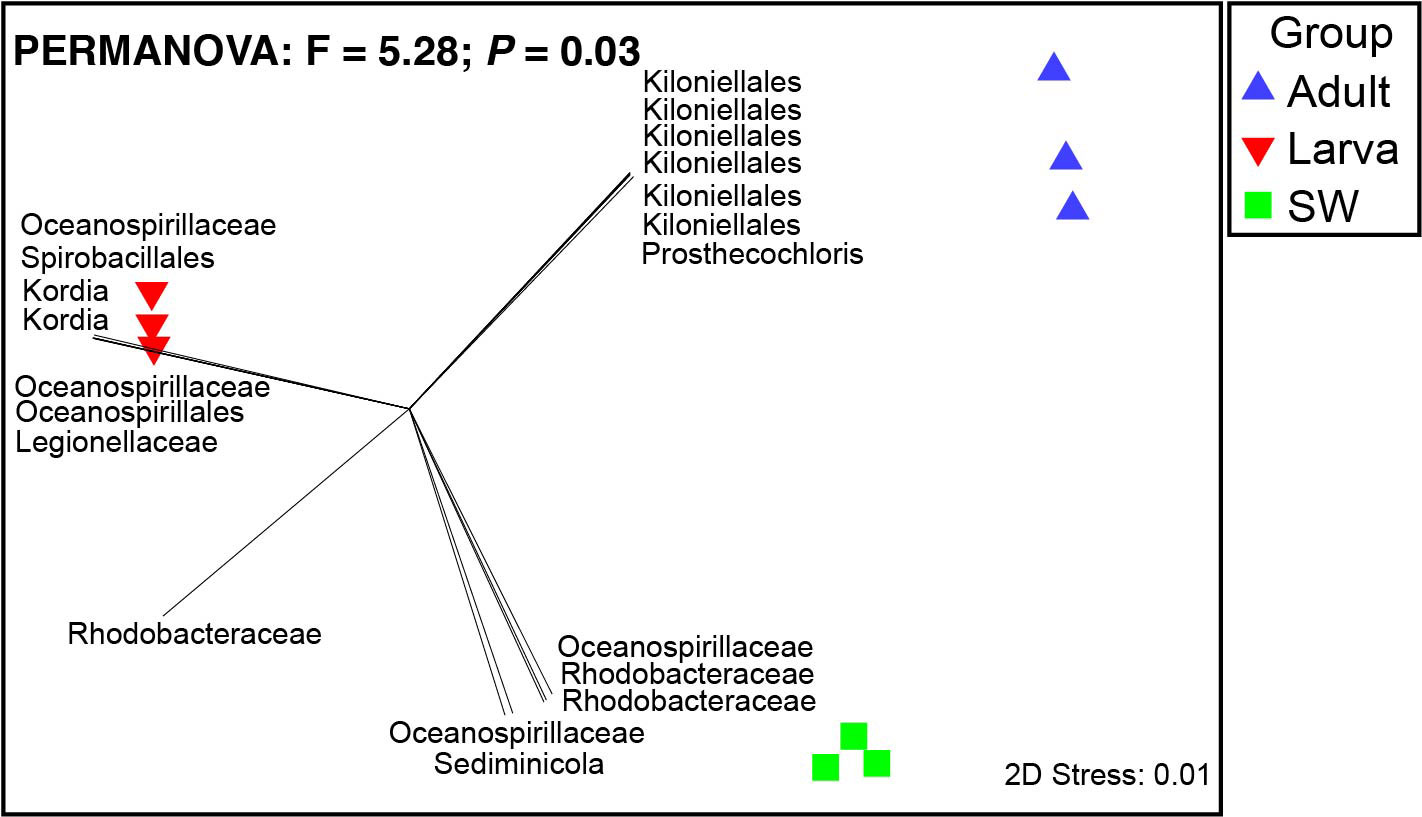
Figure 4 Non-metric multi-dimensional scaling analysis of microbial communities in seawater, coral larvae, and adults. Pearson correlation vectors represent the most resolved taxonomy for bacterial OTUs.
We detected abundant Oceanospirillaceae in the larvae of D. favus rather than in adult D. favus (Figures 3, 4). Oceanospirillaceae has been identified from some stony corals (Morrow et al., 2012; Rodriguez-Lanetty et al., 2013). Oceanospirillaceae was only detected in 1-week old juveniles of A. millepora and disappeared in the later development juveniles (Lema et al., 2014). The same pattern occurred in the Caribbean coral Porites astreoides, which harbors high abundant Oceanospirillaceae in 4-day-old larvae while less Oceanospirillaceae is presented in 11-day-old larvae (Sharp et al., 2012). Previous studies demonstrated that the Oceanospirillales, belonging to Oceanospirillaceae, are able to metabolize DMSP in the coral A. millepora, which indicates that Oceanospirillaceae may play an important role in the coral–microbiome interactions because DMSP is a carbon and sulfur source for bacteria and produced by symbiotic dinoflagellates (Bourne et al., 2013). Moreover, the genus Kordia, which was abundant in the D. favus larvae (Figures 3, 4), was previously detected in the sea urchin and the sea anemone (Becker et al., 2009; Herrera et al., 2017). The genus Kordia is one of many genera affiliated with the family Flavobacteriaceae of the phylum Bacteroidetes, well known for its degradation of high-molecular-weight organic matters. The genome of Kordia antarctica IMCC3317T encodes many carbohydrate-active enzymes, some of which are clustered into approximately seven polysaccharide utilization loci, thereby demonstrating the potential for polysaccharide utilization (Lim et al., 2020).
In contrast to D. favus larvae, we detected abundant bacteria Kiloniellales (Figures 3, 4) in the adult D. favus, which were previously detected in sponges and coral larvae (Sharp et al., 2012; Cleary et al., 2013). In the coral Montastraea annularis, Kiloniellales is significantly more abundant in healthy corals than the diseased corals and able to utilize nitrates through denitrification, indicating its positive function on coral’s health (Soffer et al., 2015). The future research on the metabolic function of Kiloniellales associated with corals may explain its high abundance in D. favus. Another abundant bacterium in adult D. favus is Prosthecochloris (Figures 3, 4). Prosthecochloris sp. is a green sulfur bacterium, frequently detected in corals Platygyra carnosa and Porites lutea (Li et al., 2014; Cai et al., 2017). Prosthecochloris sp. may provide organic and nitrogenous nutrients to coral host (Yang et al., 2016).
In this study, the most abundant bacteria shared by the larvae and adults are the unidentified Rhodobacteraceae (Table S1), which has been detected at the early development stages of corals (Sharp et al., 2012; Zhou et al., 2017). Despite the fact that Rhodobacteraceae is generally associated with diseased corals (Roder et al., 2014), the OTUs belonging to Rhodobacteraceae are also observed in healthy corals, such as the Oculina patagonica and Isopora palifera (Chen et al., 2011; Rubio-Portillo et al., 2016). The genus Roseobacter belonging to the family Rhodobacteraceae is reported to have an important function, that is, supplying fixed organic nitrogen to Symbiodiniaceae or defending against pathogenic bacteria (Nissimov et al., 2009). The Alteromans (OTU836759) and HTCC2207 (OTU805719) belonging to the Alteromonadaceae were also detected in both D. favus larvae and adults in this study (Table S1). A similar bacterial community was detected in the larvae of D. favus like other corals, indicating that these bacteria may be important at the early stages of corals.
Based on this study, some core bacteria were shared among the coral larvae, adults, and seawater despite their small proportion, suggesting that the coral D. favus is able to obtain bacteria from environmental seawater. The fact that abundant bacterial population in the coral larvae, adults, and the seawater is different (Figure 4) indicates that the selection of bacterial partners by the coral host is not random. Similarly, some studies also suggest the possible horizontal transmission of symbiotic bacteria from the environmental seawater to corals during their development because of the dynamic microbial community change. For instance, Lema et al. (2014); Lema et al. (2016) demonstrated horizontal (environmental) uptake of coral bacteria, e.g., nitrogen-fixing bacteria. Apprill et al. (2009) revealed that the composition of bacteria differed between life stages, i.e., prespawned oocyte bundles, spawned eggs, and week-old planulae, and concluded that the onset of association between microorganisms and the coral P. meandrina appeared to occur through horizontal uptake by planulae older than 79 h. Epstein et al. (2019) found that only a small number of bacterial strains were shared between offspring and their respective parents, revealing bacterial establishment as largely environmentally driven at the early life stages and suggested that recruits harbored more variable microbiomes compared to their parents, indicating that winnowing occurs as corals mature (Epstein et al., 2019).
On the contrary, Sharp et al. (2010) suggested a bacterial vertical transmission strategy based on the finding of bacteria in the ectoderm layer in newly released planula larvae of P. astreoides, but it was not known whether there were bacteria in the gametes of P. astreoides. Leite et al. (2017) found bacteria from the gamete bundles to the planula larvae and parental colony of M. hispida based on sequencing, and suggested that spawning coral M. hispida could vertically transfer microbes to the offspring based on the similar bacterial community in different stages of M. hispida, but the presence of bacterial cells in association with the (female and male) gametes of M. hispida through microscopy was not verified. Zhou et al. (2017) found that the microbial communities in the eggs of A. gemmifera were highly similar to those in parental colonies, indicating a transmission from coral parents to eggs. However, the possibility that some bacteria are eventually transmitted inside the gametes or larvae from the environments cannot be excluded. Most likely, the coral offspring can be partly colonized postspawning through the uptake of microbial associates released by the parental colonies into the surrounding seawater. In the case of coral D. favus in this study, no bacteria were detected by the 16S rRNA gene amplification from the egg samples, but we could not exclude the presence of bacteria in D. favus eggs because the failure in detecting the bacterial sequences in eggs may result from the very low abundance of bacteria or the primers and PCR procedure used. Meanwhile, in this study, some of the bacteria shared between the coral parents and larvae, and stage-specific bacteria were not detected in the seawater, suggesting the possible vertically transfer. Whether there is a vertical transmission of bacteria by coral D. favus needs future in-depth study, e.g., the confirmation of bacteria in the eggs or gametes of D. favus using electron microscopy and fluorescence in situ hybridization (FISH). Meanwhile, the characteristics of the symbiotic community profiles of D. favus in different regions need to be wholly investigated using abundant samples from all life stages, e.g., gametes, larvae, and the early juvenile.
According to the results from this study, the larvae of D. favus harbor a bit higher number of diverse bacteria in comparison to adult D. favus. This may be explained by the different selective ability of corals at different life stages. It has been proposed that morphology change at different life stages may provide different complex niches, which may lead to a significant shift in the microbial community structure (Zhou et al., 2017). An initial higher diversity of bacterial community at the early life stage of corals, followed by low species diversity associated with the adults, has been observed (Franzenburg et al., 2013; Lema et al., 2014). The same situation occurs in the coral Acropora gemmifera, which has higher bacterial diversity at the larvae and 6-day-old juvenile stages than in adults (Zhou et al., 2017). The higher diversity of bacteria at the early life stage (e.g., larvae period) of corals provides a higher possibility for nutrient cycling due to the presence of special bacteria, such as Alteromonas and Roseobacter, which have been suggested to help coral larvae acquire additional nutrients in the oligotrophic environment and consequently increase the survival rate and fitness of corals at the early life stage (Lema et al., 2016). Moreover, the higher diversity of bacteria in the larvae may increase the possibility for more production of antibiotics, which can protect coral larvae from pathogens (Sharp et al., 2012; Lema et al., 2014).
In summary, we investigated the dynamic profiles of bacteria in the coral D. favus adult and its eggs and larvae by the high-throughput sequencing method, with the seawater as control. The D. favus parent and its larvae host variant bacterial community with larvae have a higher diversity. The shared bacteria among D. favus adult and its larvae, and seawater suggest that D. favus is able to acquire specific bacteria from the environment. The observed stage-specific bacteria, e.g., Oceanospirillaceae, Kordia, and Legionellaceae in D. favus larvae, and Kiloniellales and Prosthecochloris in adult D. favus, could be an important clue to explain the biological or ecological functions of bacteria in D. favus holobionts.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI with SRR number SRR12687352-12687360 and SRR12687344-12687349.
Author contributions
JX, ZX, and XY finished the experiment. JX, GC, and YX analyzed the data. BL and BX participated in the experiment design. JX and ZL designed the study plan and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (42176146 and 41776138), the Major National Scientific Research Project, China (2013CB956100), and the Shenzhen Science and Technology R&D Fund (KJYY20180213182720347).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1055848/full#supplementary-material
References
Apprill A., Marlow H. Q., Martindale M. Q., Rappe M. S. (2009). The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 3, 685–699. doi: 10.1038/ismej.2009.3
Babbin A. R., Tamasi T., Dumit D., Weber L., Rodríguez M. V. I., Schwartz S. L., et al. (2021). Discovery and quantification of anaerobic nitrogen metabolisms among oxygenated tropical Cuban stony corals. ISME J. 15, 1222–1235. doi: 10.1038/s41396-020-00845-2
Becker P. T., Gillan D. C., Eeckhaut I. (2009). Characterization of the bacterial community associated with body wall lesions of Tripneustes gratilla (Echinoidea) using culture-independent methods. J. Invertebr. Pathol. 100, 127–130. doi: 10.1016/j.jip.2008.11.002
Bernasconi R., Stat M., Koenders A., Paparini A., Bunce M., Huggett M. J. (2019). Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01529
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bourne D. G., Dennis P. G., Uthicke S., Soo R. M., Tyson G. W., Webster N. (2013). Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J. 7, 1452–1458. doi: 10.1038/ismej.2012.172
Cai L., Zhou G., Tian R. M., Tong H., Zhang W., Sun J., et al. (2017). Metagenomic analysis reveals a green sulfur bacterium as a potential coral symbiont. Sci. Rep. 7, 9320. doi: 10.1038/s41598-017-09032-4
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Meth. 7, 335–336. doi: 10.1038/nmeth.f.303
Ceh J., Van Keulen M., Bourne D. G. (2011). Coral-associated bacterial communities on ningaloo reef, Western Australia. FEMS Microbiol. Ecol. 75, 134–144. doi: 10.1111/j.1574-6941.2010.00986.x
Chen C. P., Tseng C. H., Chen C. A., Tang S. L. (2011). The dynamics of microbial partnerships in the coral Isopora palifera. ISME J. 5, 728–740. doi: 10.1038/ismej.2010.151
Cleary D. F., Becking L. E., de Voogd N. J., Pires A. C., Polonia A. R., Egas C., et al. (2013). Habitat- and host-related variation in sponge bacterial symbiont communities in Indonesian waters. FEMS Microbiol. Ecol. 85, 465–482. doi: 10.1111/1574-6941.12135
Damjanovic K., Blackall L. L., Menéndez P., van Oppen M. J. H. (2020). Bacterial and algal symbiont dynamics in early recruits exposed to two adult coral species. Coral Reefs 39, 189–202. doi: 10.1007/s00338-019-01871-z
DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Epstein H. E., Torda G., Munday P. L. (2019). Parental and early life stage environments drive establishment of bacterial and dinoflagellate communities in a common coral. ISME J. 13, 1635–1638. doi: 10.1038/s41396-019-0358-3
Franzenburg S., Fraune S., Altrock P. M., Kunzel S., Baines J. F., Traulsen A., et al. (2013). Bacterial colonization of hydra hatchlings follows a robust temporal pattern. ISME J. 7, 781–790. doi: 10.1038/ismej.2012.156
Gavish A. R., Shapiro O. H., Kramarsky-Winter E., Vardi A. (2021). Microscale tracking of coral-vibrio interactions. ISME Commun. 1, 1–18. doi: 10.1038/s43705-021-00016-0
Herrera M., Ziegler M., Voolstra C. R., Aranda M. (2017). Laboratory-cultured strains of the sea anemone Exaiptasia reveal distinct bacterial communities. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00115
Kitamura R., Miura N., Ito M., Takagi T., Yamashiro H., Nishikawa Y.. (2021). Specific detection of coral-associated Ruegeria, a potential probiotic bacterium, in corals and subtropical seawater. Mar Biotechnol. 23, 576–589. doi: 10.1007/s10126-021-10047-2
Knowlton N., Rohwer F. (2003). Multispecies microbial mutualisms on coral reefs: The host as a habitat. Am. Nat. 162, S51–S62. doi: 10.1086/378684
Krediet C. J., Ritchie K. B., Paul V. J., Teplitski M. (2013). Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc. Biol. Sci. 280, 20122328. doi: 10.1098/rspb.2012.2328
Leite D. C., Leao P., Garrido A. G., Lins U., Santos H. F., Pires D. O., et al. (2017). Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00176
Lema K. A., Bourne D. G., Willis B. L. (2014). Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol. Ecol. 23, 4682–4695. doi: 10.1111/mec.12899
Lema K. A., Clode P. L., Kilburn M. R., Thornton R., Willis B. L., Bourne D. G. (2016). Imaging the uptake of nitrogen-fixing bacteria into larvae of the coral Acropora millepora. ISME J. 10, 1804–1808. doi: 10.1038/ismej.2015.229
Li J., Chen Q., Long L. J., Dong J. D., Yang J., Zhang S. (2014). Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci. Rep. 4, 7320. doi: 10.1038/srep07320
Lim Y., Kang I., Cho J.-C. (2020). Genome characteristics of Kordia antarctica IMCC3317T and comparative genome analysis of the genus kordia. Sci. Rep. 10, 14715. doi: 10.1038/s41598-020-71328-9
Maire J., Girvan S. K., Barkla S. E., Perez-Gonzalez A., Suggett D. J., Blackall L. L.. (2021) Intracellular bacteria are common and taxonomically diverse in cultured and in hospite algal endosymbionts of coral reefs. ISME J. 15, 2028–2042. doi: 10.1038/s41396-021-00902-4
Masella A. P., Bartram A. K., Truszkowski J. M., Brown D. G., Neufeld J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinf. 13, 31. doi: 10.1186/1471-2105-13-31
Morrow K. M., Moss A. G., Chadwick N. E., Liles M. R. (2012). Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 78, 6438–6449. doi: 10.1128/AEM.01162-12
Nissimov J., Rosenberg E., Munn C. B. (2009). Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica. FEMS Microbiol. Lett. 292, 210–215. doi: 10.1111/j.1574-6968.2009.01490.x
Pollock F. J., McMinds R., Smith S., Bourne D. G., Willis B. L., Medina M.. (2018). Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat Commun. 9, 4921. doi: 10.1038/s41467-018-07275-x
Roder C., Arif C., Bayer T., Aranda M., Daniels C., Shibl A., et al. (2014). Bacterial profiling of white plague disease in a comparative coral species framework. ISME J. 8, 31–39. doi: 10.1038/ismej.2013.127
Rodriguez-Lanetty M., Granados-Cifuentes C., Barberan A., Bellantuono A. J., Bastidas C. (2013). Ecological inferences from a deep screening of the complex bacterial consortia associated with the coral, Porites astreoides. Mol. Ecol. 22, 4349–4362. doi: 10.1111/mec.12392
Rohwer F., Seguritan V., Azam F., Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Rubio-Portillo E., Santos F., Martinez-Garcia M., de Los Rios A., Ascaso C., Souza-Egipsy V., et al. (2016). Structure and temporal dynamics of the bacterial communities associated to microhabitats of the coral Oculina patagonica. Environ. Microbiol. 18, 4564–4578. doi: 10.1111/1462-2920.13548
Sharp K. H., Distel D., Paul V. J. (2012). Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 6, 790–801. doi: 10.1038/ismej.2011.144
Sharp K. H., Ritchie K. B., Schupp P. J., Ritson-Williams R., Paul V. J. (2010). Bacterial acquisition in juveniles of several broadcast spawning coral species. PloS One 5, e10898. doi: 10.1371/journal.pone.0010898
Shlesinger T., Woesik R. (2021). Different population trajectories of two reef-building corals with similar life-history traits. J. Anim. Ecol. 90, 1379–1389. doi: 10.1111/1365-2656.13463
Soffer N., Zaneveld J., Vega Thurber R. (2015). Phage-bacteria network analysis and its implication for the understanding of coral disease. Environ. Microbiol. 17, 1203–1218. doi: 10.1111/1462-2920.12553
Yang S. H., Lee S. T. M., Huang C. R., Tseng C. H., Chiang P. W., Chen C. P., et al. (2016). Prevalence of potential nitrogen-fixing, green sulfur bacteria in the skeleton of reef-building coral Isopora palifera. Limnol. Oceanogr. 61, 1078–1086. doi: 10.1002/lno.10277
Keywords: Dipsastraea favus, life stage, bacterial community, high-throughput sequencing, horizontal transmission
Citation: Xu J, Chai G, Xiao Y, Xie Z, Yang X, Liao B, Xiao B and Li Z (2023) The community profiles of symbiotic bacteria at the different life stages of coral Dipsastraea favus. Front. Mar. Sci. 10:1055848. doi: 10.3389/fmars.2023.1055848
Received: 28 September 2022; Accepted: 04 January 2023;
Published: 19 January 2023.
Edited by:
Senjie Lin, University of Connecticut, United StatesCopyright © 2023 Xu, Chai, Xiao, Xie, Yang, Liao, Xiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyong Li, enlsaUBzanR1LmVkdS5jbg==
 Jianjun Xu1
Jianjun Xu1 Guangjun Chai
Guangjun Chai Yilin Xiao
Yilin Xiao Baohua Xiao
Baohua Xiao Zhiyong Li
Zhiyong Li